
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 June 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01334
This article is part of the Research Topic Bacteria Present in Plant Nodules: Diversity, Biogeography and Plant Growth Promotion View all 14 articles
Whole genome analysis of the Bradyrhizobium genus using average nucleotide identity (ANI) and phylogenomics showed the genus to be essentially monophyletic with seven robust groups within this taxon that includes nitrogen-fixing nodule forming bacteria as well as free living strains. Despite the wide genetic diversity of these bacteria no indication was found to suggest that the Bradyrhizobium genus have to split in different taxa. Bradyrhizobia have larger genomes than other genera of the Bradyrhizobiaceae family, probably reflecting their metabolic diversity and different lifestyles. Few plasmids in the sequenced strains were revealed from rep gene analysis and a relatively low proportion of the genome is devoted to mobile genetic elements. Sequence diversity of recA and glnII gene metadata was used to theoretically estimate the number of existing species and to predict how many would exist. There may be many more species than those presently described with predictions of around 800 species in nature. Different arguments are presented suggesting that nodulation might have arose in the ancestral genus Bradyrhizobium.
Bradyrhizobium is one of the several genera of nitrogen fixing bacteria capable of forming symbiotic nodules in legumes. Bradyrhizobium strains were previously designated the slow growing Rhizobium and recognized as an independent genus in 1982 (Jordan, 1982). The number of species in Bradyrhizobium has increased largely in recent years1. Notably, there are photosynthetic bacteria among bradyrhizobia (Ladha and So, 1994) which need no Nod factors to induce nodules (Giraud et al., 2007) and this opened a new research area on plant nodulation. Non-symbiotic bradyrhizobia are dominant in forest soils (VanInsberghe et al., 2015).
Different Bradyrhizobium species are the main nodule symbionts of important crop legumes such as soybean (Xu et al., 1995; Delamuta et al., 2013), Lima bean (Durán et al., 2014) or peanuts (Steenkamp et al., 2008) and have been isolated from nodules of many tropical (Ramírez-Bahena et al., 2009; López-López et al., 2013; Delamuta et al., 2015) and temperate legumes (Vinuesa et al., 2005a; Stepkowski et al., 2007). The ancestral symbionts in Phaseolus could have been bradyrhizobia (Servín-Garcidueñas et al., 2014) with a later symbiont shift to Rhizobium in nodules of some temperate Phaseolus species.
A large diversity of bradyrhizobia has been revealed with the sequence of few genes used as molecular markers. Within bradyrhizobia, 16S rRNA genes are known not to provide adequate sequence diversity to clearly recognize distinct species (Willems et al., 2001). For this, markers such as recA, glnII, atpD, dnaK, gyrB, and rpoB, have been frequently used to characterize bradyrhizobial species and strains (Vinuesa et al., 2005b; Rivas et al., 2009; Delamuta et al., 2012). Novel markers such as ftsA gene provide congruent phylogenies to those derived from recA and glnII gene sequences (Kalita and Malek, 2019). Bradyrhizobium diversity knowledge has expanded recently with studies from native legumes in Africa (Gronemeyer et al., 2017; Jaiswal and Dakora, 2019; Puozaa et al., 2019), from Brazilian and Indian Chamaecrista (Santos et al., 2017; Rathi et al., 2018), from threatened native species in Brazil (Fonseca et al., 2012), from Genisteae plants in Poland (Kalita and Malek, 2017), indigenous trees in China (Yao et al., 2015) as examples. In addition, new bradyrhizobial species have been reported as well (Yao et al., 2015; Araujo et al., 2017; Gronemeyer et al., 2017; Helene et al., 2017; Ahnia et al., 2018; Bunger et al., 2018).
Additionally, some bradyrhizobial strains are capable of fixing nitrogen as endophytes of some plants (Piromyou et al., 2015). Growth-independent approaches have shown a broad distribution of bradyrhizobia associated with roots of many non-legume plants such as rice, maize and pines (Chaintreuil et al., 2000; Tan et al., 2001) and bradyrhizobia have been found in plant tumors (Rivas et al., 2004; Islam et al., 2008) and also in earthworm (Thakuria et al., 2010) and insect guts (Degli Esposti and Martinez Romero, 2017).
Nowadays there are publicly available genomes of 187 bradyrhizobial strains. Bradyrhizobia have characteristic large genomes with few plasmids and no symbiosis plasmids with one exception (Okazaki et al., 2015). In Bradyrhizobium chromosomes there are symbiosis islands which carry nod and nif genes that are responsible for nodulation and nitrogen fixation, respectively. Symbiosis markers that are commonly used toward symbiosis phylogenetic reconstructions are nifH and nod genes. In regard to nodulation genes, there is a large diversity of nodA or nodC genes (Stepkowski et al., 2007; Martínez-Romero et al., 2010) and the phylogenies from these genes support a vertical and also a horizontal transfer of these genes among bradyrhizobia (Moulin et al., 2004; Menna and Hungria, 2011). Symbiovars in relation to host specificity have been identified for some bradyrhizobial species (Rogel et al., 2011) and novel symbiovars have been described (Bejarano et al., 2014; Cobo-Diaz et al., 2014; Ramirez-Bahena et al., 2016; Delamuta et al., 2017; Salmi et al., 2018; Martins da Costa et al., 2019).
Novel metrics to recognize species are based on genome analysis and average nucleotide identity, ANI (Richter and Rossello-Mora, 2009) and phylogenomics (Wu and Scott, 2012) are proving to be very useful toward this goal. A phylogenomic study of Bradyrhizobium strains showed that the presence and type of flagellum are phylogenetically determined (Garrido-Sanz et al., 2019). Other phylogenomic-based studies have been reported for the beta-rhizobia (Beukes et al., 2017; Estrada-de Los Santos et al., 2018) highlighting the existence of novel genera. It is the aim of this work to use a similar genomic-based approach and present an up-dated global genomic-based analysis of the Bradyrhizobium genus to further support its taxonomic status. Previously, such a study was performed for Rhizobium, Sinorhizobium and Agrobacterium, shedding light on these genera taxonomical designations (Ormeño-Orrillo et al., 2015).
All Bradyrhizobium genome sequences available in GenBank as of December 2018 were retrieved. An all-versus-all average nucleotide identity (ANI) matrix was constructed using OrthoANI (Lee et al., 2016). Non-metric multidimensional analysis on the ANI matrix was performed with PAST to identify outlier genome sequences. Species-level clusters were defined at a 95% ANI cutoff value and representative genomes from each cluster were selected for further analysis. For the phylogenomic reconstruction, Prodigal was used for de novo prediction of protein-coding genes for each selected genome. Amino acid sequences of 31 conserved phylogenetic markers were retrieved from each genome using AMPHORA2 (Wu and Scott, 2012) and aligned with muscle (Edgar, 2004). Each alignment was processed with trimAl to identify and remove poorly aligned regions (Capella-Gutierrez et al., 2009). All alignments were concatenated and a maximum likelihood phylogeny was constructed with PhyML (Guindon et al., 2010). Tree node support was evaluated with bootstrap analysis of 1000 replicates.
The presence of plasmid replication systems in the sequenced genomes was evaluated by looking for homologs to the replication protein B (RepB) using BLASTP searches against a database of all the Bradyrhizobium proteins obtained in the previous section. The number and size of genomic islands in complete and almost complete genomes were determined with the IslandViewer 4 server (Bertelli et al., 2017).
Bradyrhizobium nucleotide sequences from the recombinase A protein (recA) and glutamine synthetase type II (glnII) genes were retrieved from the GenBank database and aligned using MAFFT (Katoh and Standley, 2013). The multiple sequence alignments were trimmed with Bioedit (Hall, 1999) recovering 336 or 454 bp fragments common to most recA or glnII sequences, respectively, shorter sequences were discarded. An all-versus-all identity distance matrix was constructed from the alignments with the dist.seqs command of mothur (Schloss et al., 2009). Sequences were classified into species-level OTUs with the cluster command of mothur. OTU abundances were used to calculate species richness estimators with SpadesR2 and EstimateS3, and to construct rarefaction curves with PAST (Hammer et al., 2001).
One hundred and eighty-seven genomes of strains named as Bradyrhizobium were available in GenBank at the time of writing this manuscript in December 2018 (Supplementary Table 1). A non-metric multidimensional scaling graph of ANI values shared by these genomes allowed the identification of a core set of 180 related genomes plus 7 outliers (Figure 1). Upon examination of selected phylogenetic markers (rrs, recA, and/or glnII), only the 180 related genomes corresponded to bona fide bradyrhizobia. The outliers were misnamed strains belonging to other genera (see Supplementary Table 1 for details).
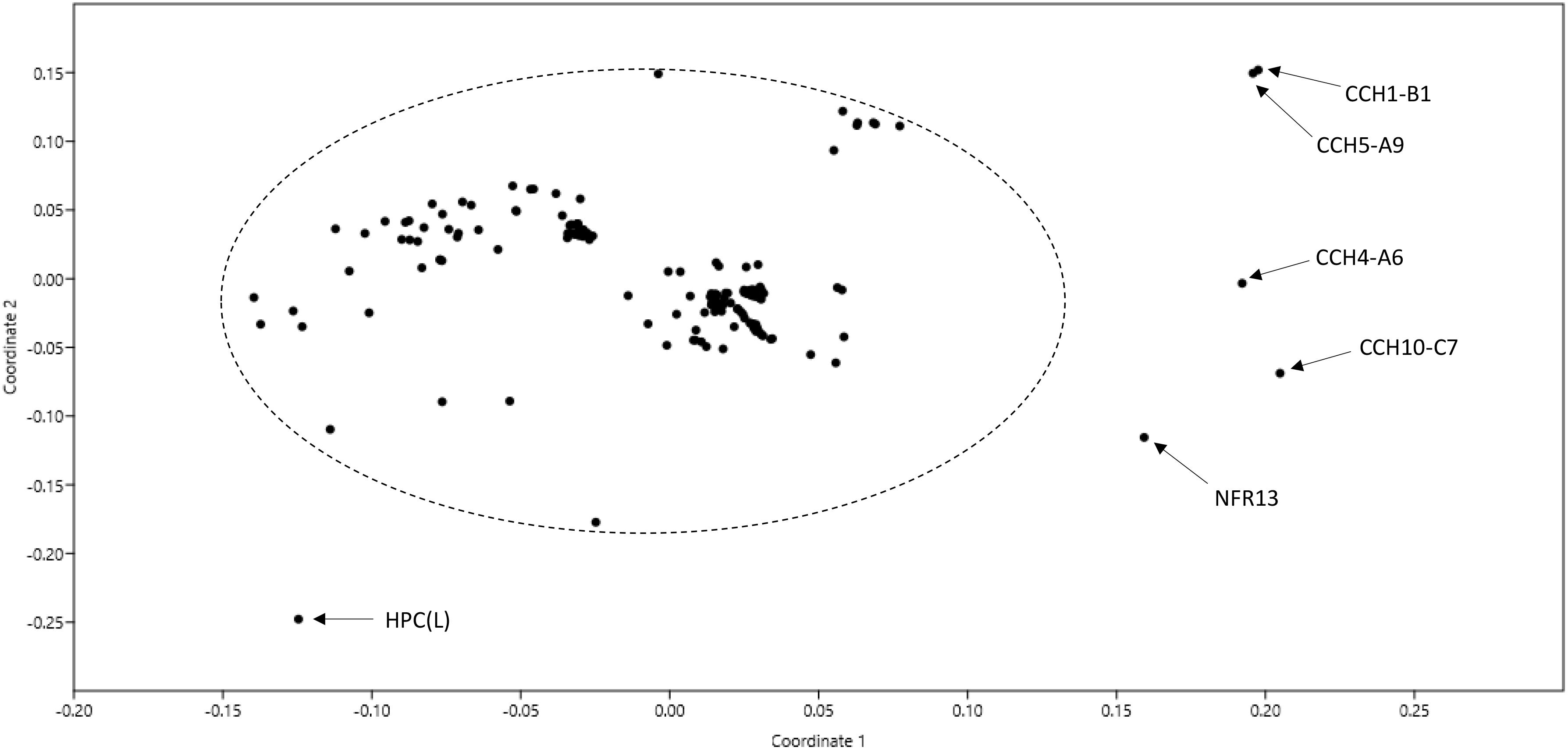
Figure 1. Non-metric multidimensional scaling (nMDS) of pair-wise ANI values of 187 genomes named as Bradyrhizobium in GenBank as of December 2018. The ellipse includes genomes of bona fide bradyrhizobia. Names of non-bradyrhizobial outlier strains are indicated.
Based on a 95% cutoff ANI value, the 180 bona fide bradyrhizobial genomes in GenBank were grouped into 93 species-level clusters (referred here as OTUs), the majority of which (72%) were represented by only one sequenced strain (Table 1). Twenty nine type strains were present among the 180 sequenced bradyrhizobia. The most represented species were Bradyrhizobium japonicum and Bradyrhizobium canariense, each with 15 strains; followed by B. diazoefficiens and B elkanii with 9 and 8 strains, respectively. The abundance of sequences from species able to nodulate soybean indicates the bias toward genomic studies directed at symbionts from this agronomical important legume.

Table 1. Classification of 180 Bradyrhizobium genome-sequenced strains into superclades, OTUs and species.
Besides Bradyrhizobium, other genera of the Bradyrhizobiaceae family with sequenced genomes are Bosea, Afipia, Rhodopseudomonas, Nitrobacter, Tardiphaga, Oligotropha, and Variibacter with 30, 24, 18, 6, 4, 3, and 1 sequences, respectively. A phylogenomic analysis of the family showed that most genera segregated as expected with the sole exception of Oligotropha and Afipia which intermingle (see Figure 2 for a condensed tree and Supplementary Figure 1 for a full tree). Bosea was the most distantly related genus in the family and its clustering with the other genera was not significantly supported by bootstrap analysis.
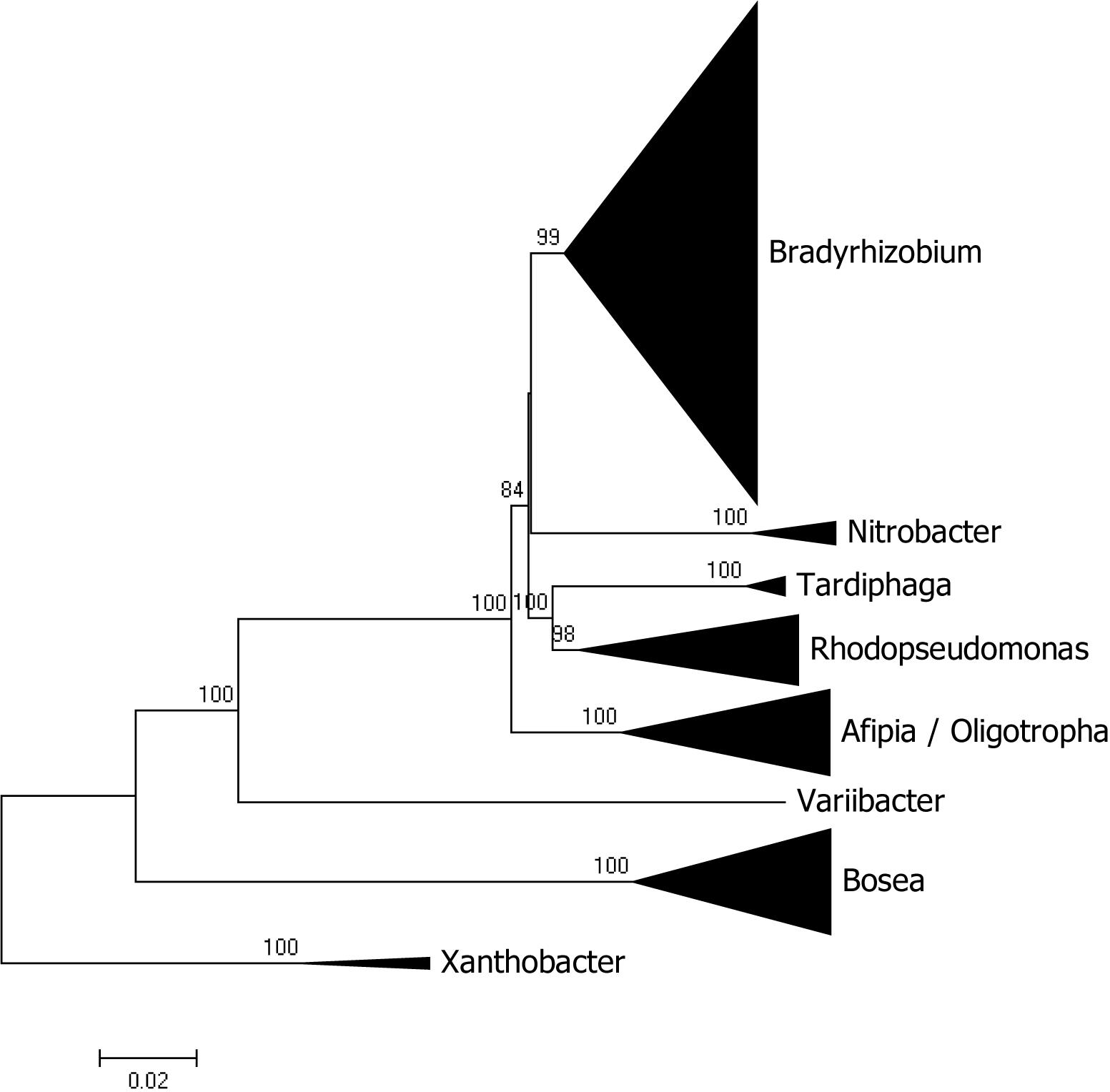
Figure 2. Phylogenomic tree of Bradyrhizobiaceae rooted with genomes of Xanthobacter. The tree was constructed with maximum likelihood using a concatenated alignment of 31 conserved proteins identified with AMPHORA2. Bootstrap node support values lower than 70% are not shown.
Two Bradyrhizobium superclades are recognized based on analysis of the 16S rDNA gene (Willems et al., 2001; Ormeño-Orrillo et al., 2006). Superclade I includes B. japonicum and related species while superclade II contains B. elkanii and allied species. The new phylogenomic analysis supports the existence of those clades but revealed a more complex structure within Bradyrhizobium with additional groups (see Figure 3 for a condensed tree and Supplementary Figure 2 for a full tree).
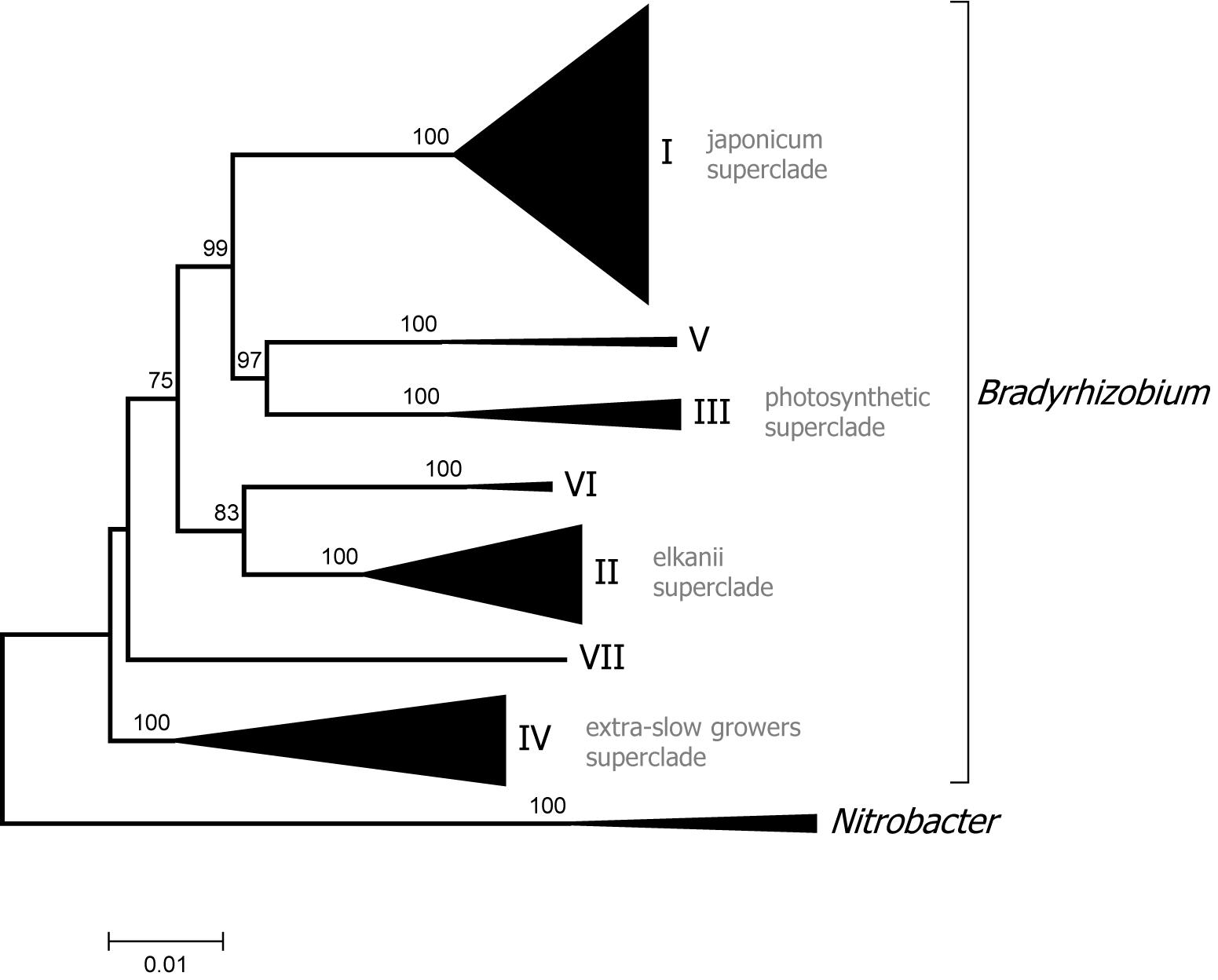
Figure 3. Phylogenomic tree of Bradyrhizobium. Superclades are indicated with Roman numerals. The tree was constructed with maximum likelihood using a concatenated alignment of 31 conserved proteins identified with AMPHORA2 (7672 aa alignment length). Bootstrap node support values lower than 70% are not shown.
Superclade III encompassed several photosynthetic strains isolated from Aeschynomene indica, as well as Bradyrhizobium oligotrophicum which also forms nodules in that legume (Okubo et al., 2013). Bradyrhizobium denitrificans, which comprise photosynthetic bacteria able to nodulate A. indica (Van Berkum et al., 2006), also belong to superclade III as shown by MLSA analysis (Ramírez-Bahena et al., 2013). Members of superclade III were the first to be recognized as having the capacity to induce nodulation in the absence of the canonical nod genes (Giraud et al., 2007).
Superclade IV includes strains isolated from several cultivated and wild legumes such as Phaseolus lunatus and Lupinus maria-josephi. Bacteria from this superclade closely group with B. elkanii in 16S rRNA phylogenies but its distinctiveness was first recognized by dnaK sequence analysis (Ormeño-Orrillo et al., 2006) and MLSA (Sanchez-Cañizares et al., 2011) and is now confirmed by phylogenomics. These bradyrhizobia have the characteristics of extra slow growth forming punctate colonies on YEM medium and usually strong alkali production.
Superclade V was formed by only two genomes one of them from strain Tv2a.2 from Tachigali versicolor, that was previously shown to occupy instable positions in the Bradyrhizobium phylogeny depending on the marker used (Parker, 2000). Superclade VI also grouped only two genomes including that of strain Ai1a.2 a representative of neotropical bradyrhizobia which possess a characteristic insertion in their 23S ribosomal gene sequence (Qian et al., 2003). Finally, superclade VII included a single strain, GAS401, which was isolated from a forest soil in the United States.
The wide genotypic and phenotypic diversity within Bradyrhizobium may suggest that it includes several genera. As early as 1990, strains now classified in superclade IV, were proposed to constitute the separate genus Photorhizobium because of their photosynthetic abilities and induction of stem nodulation (Eaglesham et al., 1990; Ladha and So, 1994). The most up to date, although not officially recognized, identity threshold for genus circumscription based on the 16S rRNA gene is 96.4% (Yarza et al., 2014). When applied to bradyrhizobia, superclade II strains are different enough to be considered a separate genus, however, the same threshold indicates that Bradyrhizobium, Rhodopseudomonas, Nitrobacter, and Afipia may constitute a single genus (Willems et al., 2001). Superclade II strains possess recombinant segments in their 16S ribosomal gene that explains their sequence divergence from other bradyrhizobia (Van Berkum et al., 2003) but this case of localized recombination with other bacteria do not justify their separation from the genus.
Recently, Qin et al. (2014) used the percentage of conserved proteins (POCP) as a genome metric for genus circumscription. According to their proposal, two bacteria may belong to the same genus if they share 50% or more of their proteins. In the case of Bradyrhizobium, all compared strains share >50% of their protein complements supporting a single genus. Different genera of the Bradyrhizobiaceae family had POCPs values between 40 and 50% except for most comparisons between bradyrhizobia and Rhodopseudomonas that can share more than half of their proteins. Thus, in general the POCP metric supports Bradyrhizobium as a single genus but indicate a close relationship with Rhodopseudomonas. Conservation of gene content may indicate not only phylogenetic relationship but also phenotypic similarity (Martín et al., 2003). Both Bradyrhizobium and Rhodopseudomonas include nitrogen fixing and photosynthetic strains, and, recently, a study suggest that Rhodopseudomonas can be mutualistic symbionts of some fungi (Le Roux et al., 2016). It will be worth to further explore the common features of both genera based on their genomic sequences.
Bradyrhizobia are considered bacteria with large genomes (Kundig et al., 1993), however the range of genome sizes in the genus is still unknown. A graphic showing the size distribution of the 180 genome assemblies of Bradyrhizobium strains is shown in Figure 4. Since only 15 of the genome sequences correspond to strains with closed replicon(s), we determined if the remaining sequences represent partial or whole genomes. A completeness analysis with BUSCO revealed that the four smaller assemblies, all <2 Mbp, represented partially sequenced genomes. Information available from those genomes revealed that they are derived from single cells or metagenomes likely explaining their incompleteness.
The smallest sequence representing a complete genome, was 6.1 Mbp in size and corresponded to strain GAS165 isolated from a forest soil. This size is similar to that found in Rhizobium and Sinorhizobium genomes. The largest complete assembly (11.7 Mbp) was also from an isolate obtained from soil (strain GAS478). Interestingly, both GAS 165 and GAS478 strains which belong to superclade IV, lacked symbiosis genes which may indicate that genome size in Bradyrhizobium is not related to its ability to engage in symbiosis with legumes. It is worth noting that the largest bradyrhizobial genome is close in size to that of Sorangium cellulosum (13 Mbp) the bacteria with the largest known genome (Schneiker et al., 2007).
Most Bradyrhizobium strains had genome sizes between 7 and 10 Mbp with a mean size of 8.6 Mbp (Figure 4). When compared to other genera in its family, bradyrhizobia have the largest genomes (Figure 5) followed by Tardiphaga and Bosea, while the smallest genomes were those from Nitrobacter. The three genera with the biggest genomes can interact with plants (De Meyer et al., 2012; De Meyer and Willems, 2012). On the other hand the smallest genomes are found in metabolically limited bacteria like Nitrobacter, Oligotropha or Variibacter or in intracellular pathogens like Afipia. Thus, genome size in the Bradyrhizobiaceae seems to be related to lifestyle.
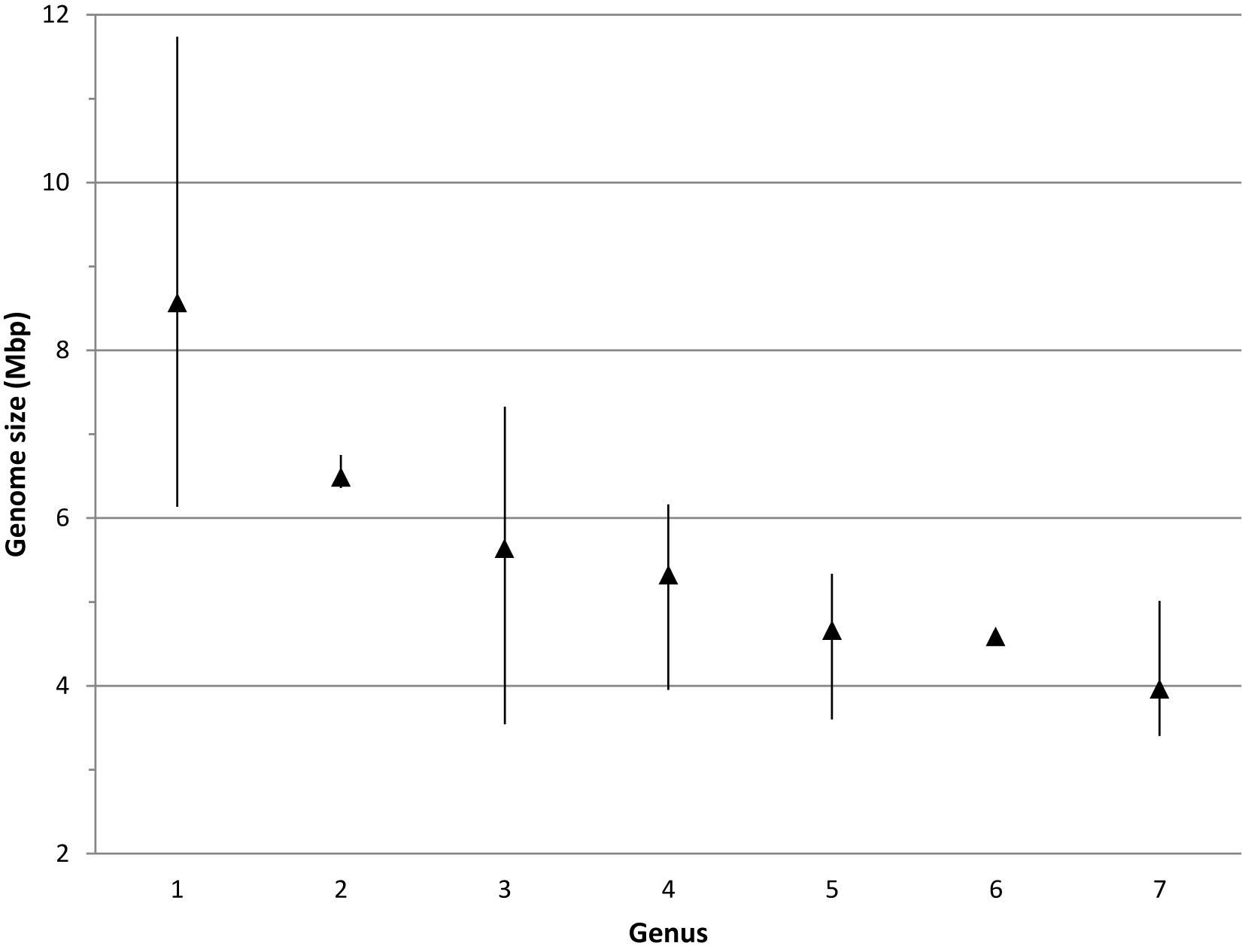
Figure 5. Genome size range in the different genera of the Bradyrhizobiaceae family. 1, Bradyrhizobium; 2, Tardiphaga; 3, Bosea; 4, Rhodopseudomonas; 5, Afipia/Oligotropha; 6, Variibacter; 7, Nitrobacter. Triangles indicate the mean genome size.
Given the large genome size in bradyrhizobia we wondered which the proportion of plasmids and genomic islands (GIs) was. Bradyrhizobia are typically regarded as unireplicon bacteria, however, strains with plasmids have been reported (Cytryn et al., 2008) including a single case of a symbiosis plasmid (Okazaki et al., 2015). The presence of extra-chromosomal replicons was evaluated in the genome-sequenced strains by searching for genes coding for homologs of the plasmid partitioning protein RepB. We found repB genes in 35 genomes (Supplementary Table 2) with almost half of the strains (n = 17) possessing a single homolog which suggested the presence of a single plasmid. The remaining strains may harbor up to 6 plasmids, although it should be noted that a single plasmid could harbor two repABC operons. The few completely-sequenced plasmids ranged in size from 136 to 285 kbp which is a size range similar to that found by pulse-field gel electrophoresis analysis of plasmids in a diverse population of Bradyrhizobium (Cytryn et al., 2008). Plasmids can represent from 2.7 to 6.6% of the genome in a single strain. The sequence of the single Bradyrhizobium symbiosis plasmid known to date was reported as a scaffold that is larger than other plasmids in the genus (736 kbp) and which represents 9.4% of the corresponding genome (Okazaki et al., 2015).
Mobile elements in unireplicon bacteria are typically present as clusters of genes known as genomic islands (GIs) (Juhas et al., 2009). As mutualistic symbionts of legumes, bradyrhizobia typically possess nodulation and nitrogen fixation genes grouped in a symbiosis island (SI) (Kaneko et al., 2002). The percentage of a Bradyrhizobium chromosome devoted to GIs was calculated using the Island Viewer 4 server applied on completely sequenced genomes (Table 2). It was found that from 5.2% and up to 17.8% of the chromosome can be regarded as the GI mobilome of individual strains.
Another aspect related to the wide diversity found among bradyrhizobia is related to their species richness. Up to December 2018, forty one Bradyrhizobium species have been described. As previously mentioned, there would be 93 species among the genome-sequenced Bradyrhizobium strains, i.e., more than twice the number of presently described species. A quick survey of some of the studies published on Bradyrhizobium diversity in different regions like North America (Ormeño-Orrillo et al., 2012), South America (Delamuta et al., 2012), Africa (Aserse et al., 2012), Asia (Vinuesa et al., 2008), and Australia (Stepkowski et al., 2012) suggest that the number of bradyrhizobial species must be larger than a hundred but the total number is yet unknown.
To estimate the number of potential Bradyrhizobium species we decided to treat the 5678 recA and 3575 glnII sequences available in the GenBank database as meta-samples of the worldwide population of bradyrhizobia. Although other phylogenetic markers have also been used to characterize bradyrhizobia, numbers of their available sequences were much lower in comparison to recA and glnII (2411, 1771, 1758, 1465 for dnaK, rpoB, atpD, gyrB, respectively). We refrain to use the 16S rDNA gene despite having a large number of sequences in the databases because it is already known that sequences of this gene are too conserved in bradyrhizobia to discriminate between species and in some cases are even unable to discriminate between Bradyrhizobium and closely related genera (Willems et al., 2001).
Pair wise comparisons between sequences from Bradyrhizobium type strains revealed that nucleotide identities of 98.2% for recA and 98.8% for glnII can be used as cutoff values to discriminate between currently described bradyrhizobial species. Using those cutoff levels, the worldwide sampled bradyrhizobia with sequenced recA genes can be clustered into 648 species-level OTUs while glnII data revealed 431 potential species. Rarefaction curves of both meta-samples did not leveled off (Figure 6) suggesting that there are still species to be discovered. Non-parametric richness estimation (Chao, 2005) applied to both meta-samples indicated that there could be from 750 to 880 species of Bradyrhizobium in nature (Figure 7). It is worth noting that both meta-samples are composed primarily of root nodule isolates. Non-symbiotic bradyrhizobia inhabiting niches such as soil or rhizosphere, or as endophytes of non-legume plants may encompass additional species.
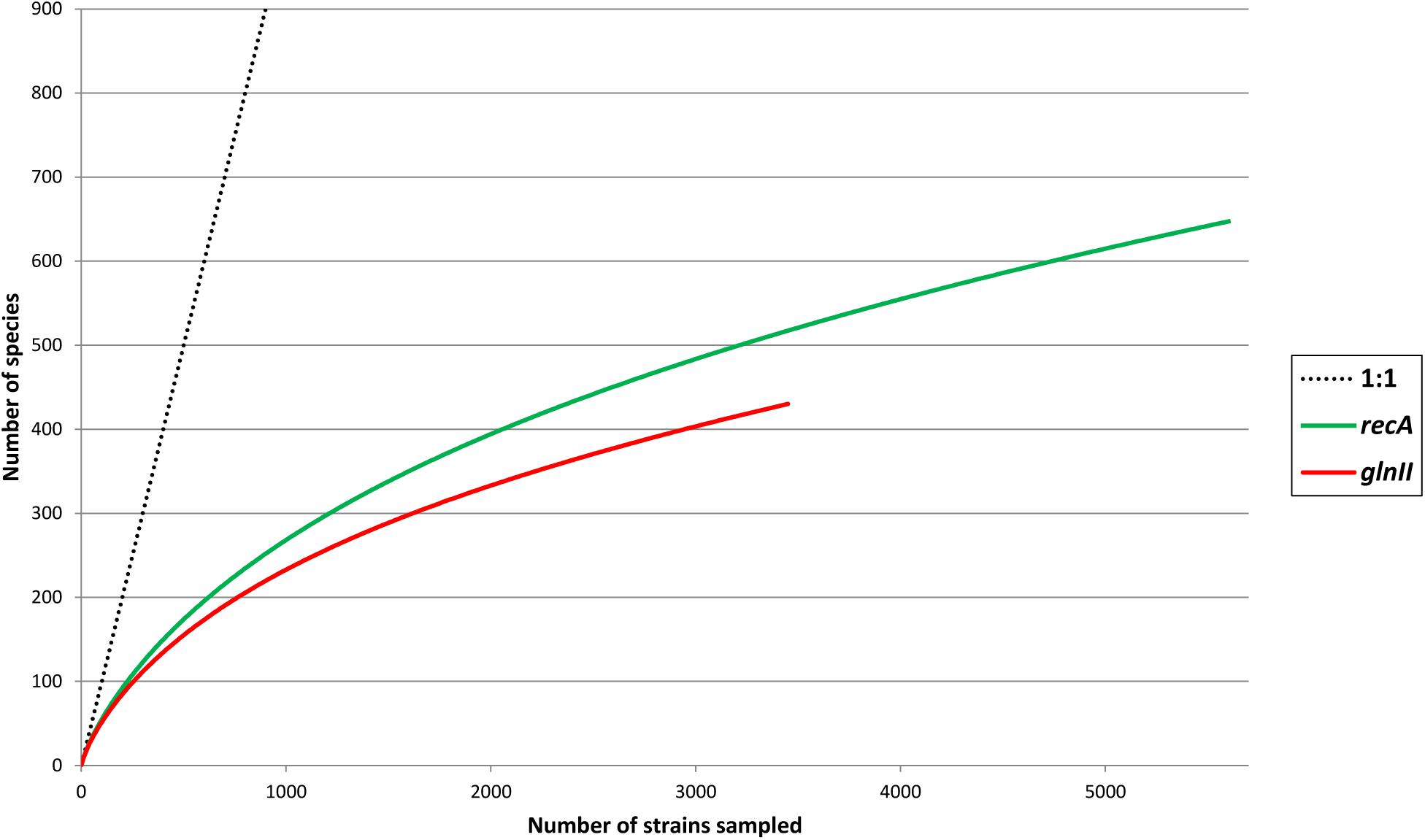
Figure 6. Rarefaction curves of species found in meta-samples of bradyrhizobia with sequenced recA or glnII genes. A hypothetical 1:1 line (each strain equal a new species) was drawn for reference to better represent the leveling of each meta-sample.
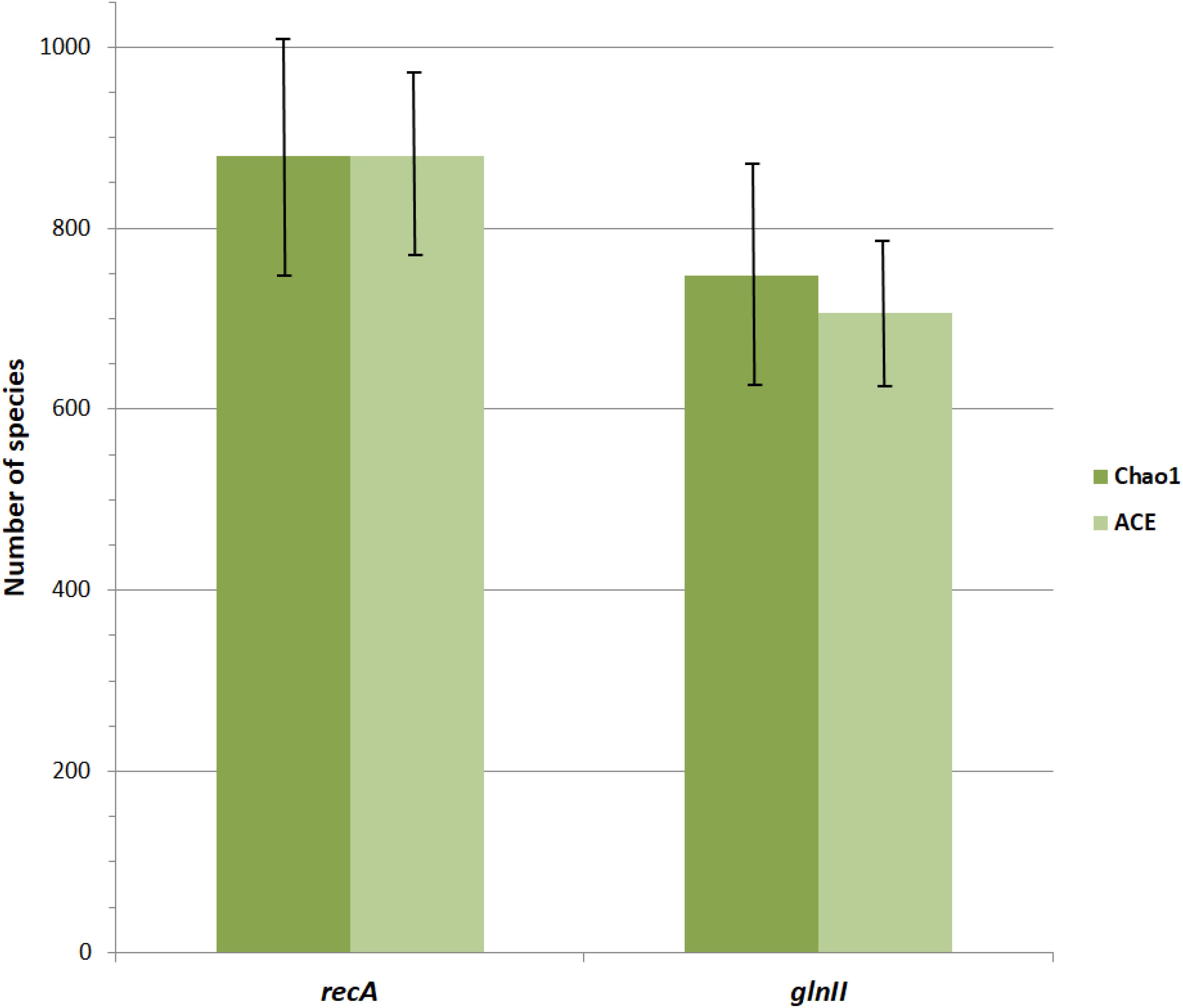
Figure 7. Chao1 and ACE non-parametric estimates of Bradyrhizobium species richness based on meta-samples of strains with sequenced recA or glnII genes. The recA metasample was rarefied to the number of glnII sequences (n = 3464). Vertical lines indicate standard errors.
The large genomic and phenomic diversity, as well as the high number of predicted species may indicate that the Bradyrhizobium genus is old. Interestingly, up till now this is the only nodule bacteria genus that contains photosynthetic bacteria and rhizobia that do not need Nod factors for nodulation. It is notable that it contains some very efficient strains for nitrogen fixation but also many generalists that form nodules in tropical legumes which in general are considered to precede temperate legumes. Several authors have noted that more “primitive” legume plants form nodules predominantly with Bradyrhizobium, thus lending support to the hypothesis that these rhizobia are their ancestral symbionts (Fonseca et al., 2012; Yao et al., 2014, 2015; Santos et al., 2017; Rathi et al., 2018). Similarly, in Phaseolus the ancestral symbionts were bradyrhizobia with a later symbiont shift to Rhizobium in nodules of some temperate species (Servín-Garcidueñas et al., 2014). The large diversity and number of different nod genes in bradyrhizobia (Martínez-Romero et al., 2010) may be suggestive that nodulation arose in bradyrhizobia. This has been a subject of discussion (Martínez-Romero, 1994; Parker, 2015; Sprent et al., 2017) with alternative views placing the origin of nod genes in beta-proteobacteria (Aoki et al., 2013) based on Nod factor exporter gene phylogenies. Since these exporters are not strictly required for nodulation (Cárdenas et al., 1996), we can suppose that accessory nod genes such as those for transporting Nod factors may be later additions after the emergence of common nodulation genes in Bradyrhizobium, then we may conciliate other possible origins for accessory nod genes.
During this article reviewing process, two papers that we must mention were published. The first one by Tindall (2019) stated that Bradyrhizobiaceae is an illegitimate name that needs to be replaced by Nitrobacteraceae because the latter contains Nitrobacter and was proposed earlier than Bradyrhizobiaceae and therefore takes precedence. Since this nomenclatural change did not affect our conclusions and because Bradyrhizobiaceae is how the family is still known, we choose to retain the name throughout this paper. However, we recognize that future studies will probably have to use Nitrobacteraceae as the proper family name. The second paper reported a phylogenomic analysis of bradyrhizobia and related taxa (Avontuur et al., 2019). Similar to our findings, Avontuur et al. (2019) confirmed the distinctiveness of the japonicum, elkanii and photosynthetic superclades and described additional infrageneric groups, albeit with some differences probably due to the use of different sets of genes for the phylogenomic reconstructions. In their analysis, our superclade IV was scattered in three groups named jicamae, soil 1 and soil 2; and our superclade VI was included in the elkanii group. In the first case we recognized that the larger number of genes used by Avontuur et al. (2019) in comparison to us, may have better resolved strains from our superclade IV, however, in the latter case we consider that superclade VI can be rightly segregated from the elkanii group by the presence of an insertion in their 23S ribosomal genes. Unlike us, Avontuur et al. (2019) tried to relate symbiotic and photosynthetic lifestyles with the infrageneric structure found within Bradyrhizobium but no clear relationship was found probably because those lifestyles are coded by accessory genes that can be gained and loss. On the other hand, our study includes aspects that were not covered by Avontuur et al. (2019) like species richness in Bradyrhizobium and the mobilome and putative plasmid content. We also provide a more thoroughly consideration of genome size in the Bradyrhizobiaceae and whether or not bradyrhizobia may be split into different genera. Thus, our analysis and those of Avontuur et al. (2019) provide complementary views on the highly diverse Bradyrhizobium genus.
EO-O and EM-R contributed conception and design of the study. EO-O performed the bioinformatics analysis. Both authors analyzed the results and wrote manuscript, and contributed to manuscript revision, read and approved the submitted version.
This study was funded by Grant 238-2015-FONDECYT-DE from Fondo Nacional de Ciencia y Tecnología (Peru).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Ministry of Education of Peru (MINEDU) is acknowledged for providing funds to acquire the high-performance computing server used in this study. We thank J. Martínez Romero for technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01334/full#supplementary-material
Ahnia, H., Bourebaba, Y., Duran, D., Boulila, F., Palacios, J. M., Rey, L., et al. (2018). Bradyrhizobium algeriense sp. nov., a novel species isolated from effective nodules of Retama sphaerocarpa from Northeastern Algeria. Syst. Appl. Microbiol. 41, 333–339. doi: 10.1016/j.syapm.2018.03.004
Aoki, S., Ito, M., and Iwasaki, W. (2013). From beta- to alpha-proteobacteria: the origin and evolution of rhizobial nodulation genes nodIJ. Mol. Biol. Evol. 30, 2494–2508. doi: 10.1093/molbev/mst153
Araujo, J., Flores-Felix, J. D., Igual, J. M., Peix, A., Gonzalez-Andres, F., Diaz-Alcantara, C. A., et al. (2017). Bradyrhizobium cajani sp. nov. isolated from nodules of Cajanus cajan. Int. J. Syst. Evol. Microbiol. 67, 2236–2241. doi: 10.1099/ijsem.0.001932
Aserse, A. A., Räsänen, L. A., Aseffa, F., Hailemariam, A., and Lindström, K. (2012). Phylogenetically diverse groups of Bradyrhizobium isolated from nodules of Crotalaria spp., Indigofera spp., Erythrina brucei and Glycine max growing in Ethiopia. Mol. Phylogenet. Evol. 65, 595–609. doi: 10.1016/j.ympev.2012.07.008
Avontuur, J. R., Palmer, M., Beukes, C. W., Chan, W. Y., Coetzee, M. P. A., Blom, J., et al. (2019). Genome-informed Bradyrhizobium taxonomy: where to from here? Syst. Appl. Microbiol. doi: 10.1016/j.syapm.2019.03.006 [Epub ahead of print].
Bejarano, A., Ramirez-Bahena, M. H., Velazquez, E., and Peix, A. (2014). Vigna unguiculata is nodulated in Spain by endosymbionts of Genisteae legumes and by a new symbiovar (vignae) of the genus Bradyrhizobium. Syst. Appl. Microbiol. 37, 533–540. doi: 10.1016/j.syapm.2014.04.003
Bertelli, C., Laird, M. R., Williams, K. P., Simon Fraser University Research Computing Group, Lau, B. Y., Hoad, G., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Beukes, C. W., Palmer, M., Manyaka, P., Chan, W. Y., Avontuur, J. R., Van Zyl, E., et al. (2017). Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 8:1154. doi: 10.3389/fmicb.2017.01154
Bunger, W., Gronemeyer, J. L., Sarkar, A., and Reinhold-Hurek, B. (2018). Bradyrhizobium ripae sp. nov., a nitrogen-fixing symbiont isolated from nodules of wild legumes in Namibia. Int. J. Syst. Evol. Microbiol. 68, 3688–3695. doi: 10.1099/ijsem.0.002955
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cárdenas, L., Dominguez, J., Santana, O., and Quinto, C. (1996). The role of the nodI and nodJ genes in the transport of Nod metabolites in Rhizobium etli. Gene 173, 183–187. doi: 10.1016/0378-1119(96)00166-7
Chaintreuil, C., Giraud, E., Prin, Y., Lorquin, J., Ba, A., Gillis, M., et al. (2000). Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl. Environ. Microbiol. 66, 5437–5447. doi: 10.1128/aem.66.12.5437-5447.2000
Chao, A. (2005). “Species estimation and applications,” in Encyclopedia of Statistical Sciences, eds N. Balakrishnan, C. B. Read, and B. Vidakovic (New York, NY: Wiley), 7907–7916.
Cobo-Diaz, J. F., Martinez-Hidalgo, P., Fernandez-Gonzalez, A. J., Martinez-Molina, E., Toro, N., Velazquez, E., et al. (2014). The endemic Genista versicolor from Sierra Nevada National Park in Spain is nodulated by putative new Bradyrhizobium species and a novel symbiovar (sierranevadense). Syst. Appl. Microbiol. 37, 177–185. doi: 10.1016/j.syapm.2013.09.008
Cytryn, E. J., Jitacksorn, S., Giraud, E., and Sadowsky, M. J. (2008). Insights learned from pBTAi1, a 229-kb accessory plasmid from Bradyrhizobium sp. strain BTAi1 and prevalence of accessory plasmids in other Bradyrhizobium sp. strains. ISME J. 2, 158–170. doi: 10.1038/ismej.2007.105
De Meyer, S. E., Coorevits, A., and Willems, A. (2012). Tardiphaga robiniae gen. nov., sp. nov., a new genus in the family Bradyrhizobiaceae isolated from Robinia pseudoacacia in Flanders (Belgium). Syst. Appl. Microbiol. 35, 205–214. doi: 10.1016/j.syapm.2012.02.002
De Meyer, S. E., and Willems, A. (2012). Multilocus sequence analysis of Bosea species and description of Bosea lupini sp. nov., Bosea lathyri sp. nov. and Bosea robiniae sp. nov., isolated from legumes. Int. J. Syst. Evol. Microbiol. 62, 2505–2510. doi: 10.1099/ijs.0.035477-0
Degli Esposti, M., and Martinez Romero, E. (2017). The functional microbiome of arthropods. PLoS One 12:e0176573. doi: 10.1371/journal.pone.0176573
Delamuta, J. R., Ribeiro, R. A., Menna, P., Bangel, E. V., and Hungria, M. (2012). Multilocus sequence analysis (MLSA) of Bradyrhizobium strains: revealing high diversity of tropical diazotrophic symbiotic bacteria. Braz. J. Microbiol. 43, 698–710. doi: 10.1590/S1517-83822012000200035
Delamuta, J. R., Ribeiro, R. A., Ormeño-Orrillo, E., Parma, M. M., Melo, I. S., Martínez-Romero, E., et al. (2015). Bradyrhizobium tropiciagri sp. nov. and Bradyrhizobium embrapense sp. nov., nitrogen-fixing symbionts of tropical forage legumes. Int. J. Syst. Evol. Microbiol. 65, 4424–4433. doi: 10.1099/ijsem.0.000592
Delamuta, J. R. M., Menna, P., Ribeiro, R. A., and Hungria, M. (2017). Phylogenies of symbiotic genes of Bradyrhizobium symbionts of legumes of economic and environmental importance in Brazil support the definition of the new symbiovars pachyrhizi and sojae. Syst. Appl. Microbiol. 40, 254–265. doi: 10.1016/j.syapm.2017.04.005
Delamuta, J. R. M., Ribeiro, R. A., Ormeño-Orrillo, E., Melo, I. S., Martínez-Romero, E., and Hungria, M. (2013). Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum Group IA strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 63(Pt 9), 3342–3351. doi: 10.1099/ijs.0.049130-0
Durán, D., Rey, L., Mayo, J., Zúñiga-Dávila, D., Imperial, J., Ruiz-Argüeso, T., et al. (2014). Bradyrhizobium paxllaeri sp. nov. and Bradyrhizobium icense sp. nov., nitrogen-fixing rhizobial symbionts of Lima bean (Phaseolus lunatus L.) in Peru. Int. J. Syst. Evol. Microbiol. 64, 2072–2078. doi: 10.1099/ijs.0.060426-0
Eaglesham, A. R. J., Ellis, J. M., Evans, W. R., Fleischman, D. E., Hungria, M., and Hardy, R. W. F. (1990). “The first photosynthetic N2-fixing Rhizobium: characteristics,” in Nitrogen Fixation: Achievements and Objectives, eds P. M. Gresshoff, L. E. Roth, G. Stacey, and W. E. Newton (Boston, MA: Springer), 805–811. doi: 10.1007/978-1-4684-6432-0_69
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Estrada-de Los Santos, P., Palmer, M., Chávez-Ramírez, B., Beukes, C., Steenkamp, E. T., Briscoe, L., et al. (2018). Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 9:E389. doi: 10.3390/genes9080389
Fonseca, M. B., Peix, A., De Faria, S. M., Mateos, P. F., Rivera, L. P., Simoes-Araujo, J. L., et al. (2012). Nodulation in Dimorphandra wilsonii Rizz. (Caesalpinioideae), a threatened species native to the Brazilian Cerrado. PLoS One 7:e49520. doi: 10.1371/journal.pone.0049520
Garrido-Sanz, D., Redondo-Nieto, M., Mongiardini, E., Blanco-Romero, E., Duran, D., Quelas, J. I., et al. (2019). Phylogenomic analyses of Bradyrhizobium reveal uneven distribution of the lateral and subpolar flagellar systems, which extends to Rhizobiales. Microorganisms 7:E50. doi: 10.3390/microorganisms7020050
Giraud, E., Moulin, L., Vallenet, D., Barbe, V., Cytryn, E., Avarre, J. C., et al. (2007). Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316, 1307–1312. doi: 10.1126/science.1139548
Gronemeyer, J. L., Bunger, W., and Reinhold-Hurek, B. (2017). Bradyrhizobium namibiense sp. nov., a symbiotic nitrogen-fixing bacterium from root nodules of Lablab purpureus, hyacinth bean, in Namibia. Int. J. Syst. Evol. Microbiol. 67, 4884–4891. doi: 10.1099/ijsem.0.002039
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9.
Helene, L. C. F., Delamuta, J. R. M., Ribeiro, R. A., and Hungria, M. (2017). Bradyrhizobium mercantei sp. nov., a nitrogen-fixing symbiont isolated from nodules of Deguelia costata (syn. Lonchocarpus costatus). Int. J. Syst. Evol. Microbiol. 67, 1827–1834. doi: 10.1099/ijsem.0.001870
Islam, M. S., Kawasaki, H., Muramatsu, Y., Nakagawa, Y., and Seki, T. (2008). Bradyrhizobium iriomotense sp. nov., isolated from a tumor-like root of the legume Entada koshunensis from Iriomote Island in Japan. Biosci. Biotechnol. Biochem. 72, 1416–1429. doi: 10.1271/bbb.70739
Jaiswal, S. K., and Dakora, F. D. (2019). Widespread distribution of highly adapted Bradyrhizobium species nodulating diverse legumes in Africa. Front. Microbiol. 10:310. doi: 10.3389/fmicb.2019.00310
Jordan, D. C. (1982). Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int. J. Syst. Bacteriol. 32, 136–139. doi: 10.1099/00207713-32-1-136
Juhas, M., Van Der Meer, J. R., Gaillard, M., Harding, R. M., Hood, D. W., and Crook, D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. doi: 10.1111/j.1574-6976.2008.00136.x
Kalita, M., and Malek, W. (2017). Molecular phylogeny of Bradyrhizobium bacteria isolated from root nodules of tribe Genisteae plants growing in southeast Poland. Syst. Appl. Microbiol. 40, 482–491. doi: 10.1016/j.syapm.2017.09.001
Kalita, M., and Malek, W. (2019). The ftsA gene as a molecular marker for phylogenetic studies in Bradyrhizobium and identification of Bradyrhizobium japonicum. J. Appl. Genet. 60, 123–126. doi: 10.1007/s13353-018-0479-9
Kaneko, T., Nakamura, Y., Sato, S., Minamisawa, K., Uchiumi, T., Sasamoto, S., et al. (2002). Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9, 189–197. doi: 10.1093/dnares/9.6.189
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kundig, C., Hennecke, H., and Gottfert, M. (1993). Correlated physical and genetic map of the Bradyrhizobium japonicum 110 genome. J. Bacteriol. 175, 613–622. doi: 10.1128/jb.175.3.613-622.1993
Ladha, J. K., and So, R. B. (1994). Numerical taxonomy of photosynthetic rhizobia nodulating Aeschynomene species. Int. J. Syst. Evol. Microbiol. 44, 62–73. doi: 10.1099/00207713-44-1-62
Le Roux, C., Tournier, E., Lies, A., Sanguin, H., Chevalier, G., Duponnois, R., et al. (2016). Bacteria of the genus Rhodopseudomonas (Bradyrhizobiaceae): obligate symbionts in mycelial cultures of the black truffles Tuber melanosporum and Tuber brumale. Springerplus 5:1085. doi: 10.1186/s40064-016-2756-6
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
López-López, A., Negrete-Yankelevich, S., Rogel, M. A., Ormeño-Orrillo, E., Martínez, J., and Martínez-Romero, E. (2013). Native bradyrhizobia from Los Tuxtlas in Mexico are symbionts of Phaseolus lunatus (Lima bean). Syst. Appl. Microbiol. 36, 33–38. doi: 10.1016/j.syapm.2012.10.006
Martín, M. J., Herrero, J., Mateos, A., and Dopazo, J. (2003). Comparing bacterial genomes through conservation profiles. Genome Res. 13, 991–998. doi: 10.1101/gr.678303
Martínez-Romero, E. (1994). Recent developments in Rhizobium taxonomy. Plant Soil 161, 11–20. doi: 10.1099/ijsem.0.003426
Martínez-Romero, J. C., Ormeño-Orrillo, E., Rogel, M. A., López-López, A., and Martínez-Romero, E. (2010). “Trends in rhizobial evolution and some taxonomic remarks,” in Evolutionary Biology: Concepts, Molecular and Morphological Evolution, ed. P. Pontarotti (Berlin: Springer), 301–315. doi: 10.1007/978-3-642-12340-5_18
Martins da Costa, E., Soares De Carvalho, T., Azarias Guimaraes, A., Ribas Leao, A. C., Magalhaes Cruz, L., De Baura, V. A., et al. (2019). Classification of the inoculant strain of cowpea UFLA03-84 and of other strains from soils of the Amazon region as Bradyrhizobium viridifuturi (symbiovar tropici). Braz. J. Microbiol. 50, 335–345. doi: 10.1007/s42770-019-00045-x
Menna, P., and Hungria, M. (2011). Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidence for the theory of monophyletic origin, and spread and maintenance by both horizontal and vertical transfer. Int. J. Syst. Evol. Microbiol. 61(Pt 12), 3052–3067. doi: 10.1099/ijs.0.028803-0
Moulin, L., Bena, G., Boivin-Masson, C., and Stepkowski, T. (2004). Phylogenetic analyses of symbiotic nodulation genes support vertical and lateral gene co-transfer within the Bradyrhizobium genus. Mol. Phylogenet. Evol. 30, 720–732. doi: 10.1016/s1055-7903(03)00255-0
Okazaki, S., Noisangiam, R., Okubo, T., Kaneko, T., Oshima, K., Hattori, M., et al. (2015). Genome analysis of a novel Bradyrhizobium sp. DOA9 carrying a symbiotic plasmid. PLoS One 10:e0117392. doi: 10.1371/journal.pone.0117392
Okubo, T., Fukushima, S., Itakura, M., Oshima, K., Longtonglang, A., Teaumroong, N., et al. (2013). Genome analysis suggests that the soil oligotrophic bacterium Agromonas oligotrophica (Bradyrhizobium oligotrophicum) is a nitrogen-fixing symbiont of Aeschynomene indica. Appl. Environ. Microbiol. 79, 2542–2551. doi: 10.1128/AEM.00009-13
Ormeño-Orrillo, E., Rogel-Hernández, M. A., Lloret, L., López-López, A., Martínez, J., Barois, I., et al. (2012). Change in land use alters the diversity and composition of Bradyrhizobium communities and led to the introduction of Rhizobium etli into the tropical rain forest of Los Tuxtlas (Mexico). Microb. Ecol. 63, 822–834. doi: 10.1007/s00248-011-9974-9
Ormeño-Orrillo, E., Servín-Garcidueñas, L. E., Rogel, M. A., González, V., Peralta, H., Mora, J., et al. (2015). Taxonomy of rhizobia and agrobacteria from the Rhizobiaceae family in light of genomics. Syst. Appl. Microbiol. 38, 287–291. doi: 10.1016/j.syapm.2014.12.002
Ormeño-Orrillo, E., Vinuesa, P., Zúñiga-Dávila, D., and Martínez-Romero, E. (2006). Molecular diversity of native bradyrhizobia isolated from Lima bean (Phaseolus lunatus L.) in Peru. Syst. Appl. Microbiol. 29, 253–262. doi: 10.1016/j.syapm.2005.09.002
Parker, M. A. (2000). Divergent Bradyrhizobium symbionts on Tachigali versicolor from Barro Colorado Island, Panama. Syst. Appl. Microbiol. 23, 585–590. doi: 10.1016/s0723-2020(00)80034-x
Parker, M. A. (2015). The spread of Bradyrhizobium lineages across host legume clades: from Abarema to Zygia. Microb. Ecol. 69, 630–640. doi: 10.1007/s00248-014-0503-5
Piromyou, P., Songwattana, P., Greetatorn, T., Okubo, T., Kakizaki, K. C., Prakamhang, J., et al. (2015). The type III secretion system (T3SS) is a determinant for rice-endophyte colonization by non-photosynthetic Bradyrhizobium. Microbes Environ. 30, 291–300. doi: 10.1264/jsme2.ME15080
Puozaa, D. K., Jaiswal, S. K., and Dakora, F. D. (2019). Phylogeny and distribution of Bradyrhizobium symbionts nodulating cowpea (Vigna unguiculata L. Walp) and their association with the physicochemical properties of acidic African soils. Syst. Appl. Microbiol. 42, 403–414. doi: 10.1016/j.syapm.2019.02.004
Qian, J., Kwon, S. W., and Parker, M. A. (2003). rRNA and nifD phylogeny of Bradyrhizobium from sites across the Pacific Basin. FEMS Microbiol. Lett. 219, 159–165. doi: 10.1016/s0378-1097(03)00043-0
Qin, Q.-L., Xie, B.-B., Zhang, X.-Y., Chen, X.-L., Zhou, B.-C., Zhou, J., et al. (2014). A proposed genus boundary for the Prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Ramírez-Bahena, M.-H., Chahboune, R., Peix, A., and Velázquez, E. (2013). Reclassification of Agromonas oligotrophica into the genus Bradyrhizobium as Bradyrhizobium oligotrophicum comb. nov. Int. J. Syst. Evol. Microbiol. 63, 1013–1016. doi: 10.1099/ijs.0.041897-0
Ramirez-Bahena, M. H., Flores-Felix, J. D., Chahboune, R., Toro, M., Velazquez, E., and Peix, A. (2016). Bradyrhizobium centrosemae (symbiovar centrosemae) sp. nov., Bradyrhizobium americanum (symbiovar phaseolarum) sp. nov. and a new symbiovar (tropici) of Bradyrhizobium viridifuturi establish symbiosis with Centrosema species native to America. Syst. Appl. Microbiol. 39, 378–383. doi: 10.1016/j.syapm.2016.06.001
Ramírez-Bahena, M. H., Peix, A., Rivas, R., Camacho, M., Rodríguez-Navarro, D. N., Mateos, P. F., et al. (2009). Bradyrhizobium pachyrhizi sp. nov. and Bradyrhizobium jicamae sp. nov., isolated from effective nodules of Pachyrhizus erosus. Int. J. Syst. Evol. Microbiol. 59, 1929–1934. doi: 10.1099/ijs.0.006320-0
Rathi, S., Tak, N., Bissa, G., Chouhan, B., Ojha, A., Adhikari, D., et al. (2018). Selection of Bradyrhizobium or Ensifer symbionts by the native Indian caesalpinioid legume Chamaecrista pumila depends on soil pH and other edaphic and climatic factors. FEMS Microbiol. Ecol. 94:fiy180.
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rivas, R., Martens, M., De Lajudie, P., and Willems, A. (2009). Multilocus sequence analysis of the genus Bradyrhizobium. Syst. Appl. Microbiol. 32, 101–110. doi: 10.1016/j.syapm.2008.12.005
Rivas, R., Willems, A., Palomo, J. L., Garcia-Benavides, P., Mateos, P. F., Martinez-Molina, E., et al. (2004). Bradyrhizobium betae sp. nov., isolated from roots of Beta vulgaris affected by tumour-like deformations. Int. J. Syst. Evol. Microbiol. 54, 1271–1275. doi: 10.1099/ijs.0.02971-0
Rogel, M. A., Ormeño-Orrillo, E., and Martínez Romero, E. (2011). Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst. Appl. Microbiol. 34, 96–104. doi: 10.1016/j.syapm.2010.11.015
Salmi, A., Boulila, F., Bourebaba, Y., Le Roux, C., Belhadi, D., and De Lajudie, P. (2018). Phylogenetic diversity of Bradyrhizobium strains nodulating Calicotome spinosa in the Northeast of Algeria. Syst. Appl. Microbiol. 41, 452–459. doi: 10.1016/j.syapm.2018.05.005
Sanchez-Cañizares, C., Rey, L., Duran, D., Temprano, F., Sanchez-Jimenez, P., Navarro, A., et al. (2011). Endosymbiotic bacteria nodulating a new endemic lupine Lupinus mariae-josephi from alkaline soils in Eastern Spain represent a new lineage within the Bradyrhizobium genus. Syst. Appl. Microbiol. 34, 207–215. doi: 10.1016/j.syapm.2010.11.020
Santos, J. M., Casaes Alves, P. A., Silva, V. C., Kruschewsky Rhem, M. F., James, E. K., and Gross, E. (2017). Diverse genotypes of Bradyrhizobium nodulate herbaceous Chamaecrista (Moench) (Fabaceae, Caesalpinioideae) species in Brazil. Syst. Appl. Microbiol. 40, 69–79. doi: 10.1016/j.syapm.2016.12.004
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schneiker, S., Perlova, O., Kaiser, O., Gerth, K., Alici, A., Altmeyer, M. O., et al. (2007). Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 25, 1281–1289.
Servín-Garcidueñas, L. E., Zayas-Del Moral, A., Ormeño-Orrillo, E., Rogel, M. A., Delgado-Salinas, A., Sánchez, F., et al. (2014). Symbiont shift towards Rhizobium nodulation in a group of phylogenetically related Phaseolus species. Mol. Phylogenet. Evol. 79, 1–11. doi: 10.1016/j.ympev.2014.06.006
Sprent, J. I., Ardley, J., and James, E. K. (2017). Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 215, 40–56. doi: 10.1111/nph.14474
Steenkamp, E. T., Stepkowski, T., Przymusiak, A., Botha, W. J., and Law, I. J. (2008). Cowpea and peanut in southern Africa are nodulated by diverse Bradyrhizobium strains harboring nodulation genes that belong to the large pantropical clade common in Africa. Mol. Phylogenet. Evol. 48, 1131–1144. doi: 10.1016/j.ympev.2008.04.032
Stepkowski, T., Hughes, C. E., Law, I. J., Markiewicz, L., Gurda, D., Chlebicka, A., et al. (2007). Diversification of lupine Bradyrhizobium strains: evidence from nodulation gene trees. Appl. Environ. Microbiol. 73, 3254–3264. doi: 10.1128/aem.02125-06
Stepkowski, T., Watkin, E., Mcinnes, A., Gurda, D., Gracz, J., and Steenkamp, E. T. (2012). Distinct Bradyrhizobium communities nodulate legumes native to temperate and tropical monsoon Australia. Mol. Phylogenet. Evol. 63, 265–277. doi: 10.1016/j.ympev.2011.12.020
Tan, Z., Hurek, T., Vinuesa, P., Muller, P., Ladha, J. K., and Reinhold-Hurek, B. (2001). Specific detection of Bradyrhizobium and Rhizobium strains colonizing rice (Oryza sativa) roots by 16S-23S ribosomal DNA intergenic spacer-targeted PCR. Appl. Environ. Microbiol. 67, 3655–3664. doi: 10.1128/aem.67.8.3655-3664.2001
Thakuria, D., Schmidt, O., Finan, D., Egan, D., and Doohan, F. M. (2010). Gut wall bacteria of earthworms: a natural selection process. ISME J. 4, 357–366. doi: 10.1038/ismej.2009.124
Tindall, B. J. (2019). The name Bradyrhizobiaceae Garrity et al. 2006 contains Nitrobacter Winogradsky 1892 (Approved Lists 1980), the nomenclatural type of the family Nitrobacteraceae Buchanan 1917 (Approved Lists 1980), is illegitimate and proposals to alter the wording of Rule 54 of the International Code of Nomenclature of Prokaryotes to clarify the fact that the family name Bradyrhizobiaceae Garrity et al. 2006 is replaced by the family name Nitrobacteraceae Buchanan 1917 (Approved Lists 1980) the only correct name. Int. J. Syst. Evol. Microbiol. 69, 998–1000. doi: 10.1099/ijsem.0.003253
Van Berkum, P., Leibold, J. M., and Eardly, B. D. (2006). Proposal for combining Bradyrhizobium spp. (Aeschynomene indica) with Blastobacter denitrificans and to transfer Blastobacter denitrificans (Hirsch and Muller, 1985) to the genus Bradyrhizobium as Bradyrhizobium denitrificans (comb. nov.). Syst. Appl. Microbiol. 29, 207–215. doi: 10.1016/j.syapm.2005.07.014
Van Berkum, P., Terefework, Z., Paulin, L., Suomalainen, S., Lindström, K., and Eardly, B. D. (2003). Discordant phylogenies within the rrn loci of rhizobia. J. Bacteriol. 185, 2988–2998. doi: 10.1128/jb.185.10.2988-2998.2003
VanInsberghe, D., Maas, K. R., Cardenas, E., Strachan, C. R., Hallam, S. J., and Mohn, W. W. (2015). Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J. 9, 2435–2441. doi: 10.1038/ismej.2015.54
Vinuesa, P., Leon-Barrios, M., Silva, C., Willems, A., Jarabo-Lorenzo, A., Perez-Galdona, R., et al. (2005a). Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int. J. Syst. Evol. Microbiol. 55, 569–575. doi: 10.1099/ijs.0.63292-0
Vinuesa, P., Silva, C., Lorite, M. J., Izaguirre-Mayoral, M. L., Bedmar, E. J., and Martínez-Romero, E. (2005b). Molecular systematics of rhizobia based on maximum likelihood and Bayesian phylogenies inferred from rrs, atpD, recA and nifH sequences, and their use in the classification of Sesbania microsymbionts from Venezuelan wetlands. Syst. Appl. Microbiol. 28, 702–716. doi: 10.1016/j.syapm.2005.05.007
Vinuesa, P., Rojas-Jiménez, K., Contreras-Moreira, B., Mahna, S. K., Prasad, B. N., Moe, H., et al. (2008). Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the Asiatic continent. Appl. Environ. Microbiol. 74, 6987–6996. doi: 10.1128/AEM.00875-08
Willems, A., Coopman, R., and Gillis, M. (2001). Phylogenetic and DNA-DNA hybridization analyses of Bradyrhizobium species. Int. J. Syst. Evol. Microbiol. 51, 111–117. doi: 10.1099/00207713-51-1-111
Wu, M., and Scott, A. J. (2012). Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28, 1033–1034. doi: 10.1093/bioinformatics/bts079
Xu, L. M., Ge, C., Cui, Z., Li, J., and Fan, H. (1995). Bradyrhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int. J. Syst. Bacteriol. 45, 706–711. doi: 10.1099/00207713-45-4-706
Yao, Y., Sui, X. H., Zhang, X. X., Wang, E. T., and Chen, W. X. (2015). Bradyrhizobium erythrophlei sp. nov. and Bradyrhizobium ferriligni sp. nov., isolated from effective nodules of Erythrophleum fordii. Int. J. Syst. Evol. Microbiol. 65, 1831–1837. doi: 10.1099/ijs.0.000183
Yao, Y., Wang, R., Lu, J. K., Sui, X. H., Wang, E. T., and Chen, W. X. (2014). Genetic diversity and evolution of Bradyrhizobium populations nodulating Erythrophleum fordii, an evergreen tree indigenous to the southern subtropical region of China. Appl. Environ. Microbiol. 80, 6184–6194. doi: 10.1128/AEM.01595-14
Keywords: nodulation, nitrogen-fixation, phylogenomics, legume symbionts, Rhodopseudomonas
Citation: Ormeño-Orrillo E and Martínez-Romero E (2019) A Genomotaxonomy View of the Bradyrhizobium Genus. Front. Microbiol. 10:1334. doi: 10.3389/fmicb.2019.01334
Received: 28 January 2019; Accepted: 28 May 2019;
Published: 13 June 2019.
Edited by:
Encarna Velázquez, University of Salamanca, SpainReviewed by:
Euan James, James Hutton Institute, United KingdomCopyright © 2019 Ormeño-Orrillo and Martínez-Romero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ernesto Ormeño-Orrillo, ZW9ybWVub0BsYW1vbGluYS5lZHUucGU=; Esperanza Martínez-Romero, ZW1hcnRpbmVAY2NnLnVuYW0ubXg=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.