
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 June 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01308
This article is part of the Research Topic Combating Antimicrobial Resistance - A One Health Approach View all 26 articles
 Konduri Ranjith1,2
Konduri Ranjith1,2 Jahnabi Ramchiary3
Jahnabi Ramchiary3 Jogadhenu S. S. Prakash3
Jogadhenu S. S. Prakash3 Kotakonda Arunasri1
Kotakonda Arunasri1 Savitri Sharma1
Savitri Sharma1 Sisinthy Shivaji1*
Sisinthy Shivaji1*The present work is an attempt to establish the functionality of genes involved in biofilm formation and antibiotic resistance in an ocular strain of Escherichia coli (L-1216/2010) which was isolated and characterized from the Vitreous fluid of a patient with Endophthalmitis. For this purpose, seven separate gene-specific knockout mutants were generated by homologous recombination in ocular E. coli. The genes that were mutated included three transmembrane genes ytfR (ABC transporter ATP-binding protein), mdtO (multidrug efflux system) and tolA (inner membrane protein), ryfA coding for non-coding RNA and three metabolic genes mhpA (3-3-hydroxyphenylpropionate 1,2-dioxygenase), mhpB (2,3-di hydroxyphenylpropionate 1,2-dioxygenase), and bdcR (regulatory gene of bdcA). Mutants were validated by sequencing and Reverse transcription-PCR and monitored for biofilm formation by XTT method and confocal microscopy. The antibiotic susceptibility of the mutants was also ascertained. The results indicated that biofilm formation was inhibited in five mutants (ΔbdcR, ΔmhpA, ΔmhpB, ΔryfA, and ΔtolA) and the thickness of biofilm reduced from 17.2 μm in the wildtype to 1.5 to 4.8 μm in the mutants. Mutants ΔytfR and ΔmdtO retained the potential to form biofilm. Complementation of the mutants with the wild type gene restored biofilm formation potential in all mutants except in ΔmhpB. The 5 mutants which lost their ability to form biofilm (ΔbdcR, ΔmhpA, ΔmhpB, ΔtolA, and ΔryfA) did not exhibit any change in their susceptibility to Ceftazidime, Cefuroxime, Ciprofloxacin, Gentamicin, Cefotaxime, Sulfamethoxazole, Imipenem, Erythromycin, and Streptomycin in the planktonic phase compared to wild type ocular E. coli. But ΔmdtO was the only mutant with altered MIC to Sulfamethoxazole, Imipenem, Erythromycin, and Streptomycin both in the planktonic and biofilm phase. This is the first report demonstrating the involvement of the metabolic genes mhpA and mhpB and bdcR (regulatory gene of bdcA) in biofilm formation in ocular E. coli. In addition we provide evidence that tolA and ryfA are required for biofilm formation while ytfR and mdtO are not required. Mitigation of biofilm formation to overcome antibiotic resistance could be achieved by targeting the genes bdcR, mhpA, mhpB, ryfA, and tolA.
Antimicrobial resistance (AMR) refers to the ability of microorganisms (bacteria, fungi, viruses, and parasites) to resist the inhibitory effect of an antimicrobial agent at the minimum inhibitory concentration (MIC) of the antimicrobial agent. AMR is a global phenomenon and bacteria, fungi and viruses are known to exhibit AMR. Various factors intrinsic to the microbe are also known to facilitate AMR such as their ability to inactivate the antimicrobial agent, to prevent its entry, to activate its efflux, to modify the antibiotic or the target, acquire resistance genes, and the ability to form a biofilm. Over the years with the development of the crystal violet method, XTT method, confocal microscopy and scanning electron microscopy it has been possible to monitor the dynamics of biofilm formation, and evaluate simultaneously the extent of AMR. These studies have clearly established that biofilm confers AMR to microorganisms (Hoyle et al., 1992; Elder et al., 1995; Ranjith et al., 2017). Several ocular pathogens like Pseudomonas aeruginosa, Staphylococcus aureus, S. epidermidis, Micrococcus luteus, Serratia marcescens, Neisseria spp., Moraxella spp., Bacillus spp., E. coli, Proteus mirabilis, Enterobacter agglomerans, and Klebsiella spp., also exhibited the potential to form biofilms and were resistant to antibiotics (Katiyar et al., 2012). Recently we demonstrated that ocular E. coli from patients with conjunctivitis, keratitis or endopthalmitis were resistant to one or more of the antibiotics tested and majority of the isolates were positive for biofilm formation. E. coli L-1216/2010, was the best biofilm forming ocular isolate, and in the biofilm phase it was 100 times more resistant to the 8 antibiotics tested compared to planktonic phase (Ranjith et al., 2017). In the same study it was also demonstrated by DNA microarray analysis that ocular E. coli L-1216/2010 in the biofilm phase over-expressed several genes (Ranjith et al., 2017). But, comparison of the expression pattern of specific genes in ocular E. coli L-1216/2010 and in pathogenic E. coli ABU strain 83972 (Hancock and Klemm, 2007) and in E. coli UPEC strain CFT073 (Schembri et al., 2003) were distinctly different though both are pathogens. For instance, fim genes coding for type 1 fimbriae, genes coding for the LPS lipid A moiety (waaB, waaP, waaJ, and waaR), yhjN coding for cellulose synthase regulator protein, were up regulated in ocular E. coli L-1216/2010 but not in E. coli ABU strain 83972 (Hancock and Klemm, 2007) and in E. coli UPEC strain CFT073 (Schembri et al., 2003). Further the 22 genes (c2418 to c2440) that constitute the pathogenicity island and rfaH a virulence regulator, which regulates expression of several virulent genes in E. coli were up regulated during biofilm formation in several pathogenic strains but not in ocular E. coli L-1216/2010 (Schembri et al., 2003; Ranjith et al., 2017). It was also observed that ocular E. coli in biofilm phase did not exhibit up regulation of the stress response encoding genes (such as cspG, cspH, pphA, ibpA, ibp, soxS, hha, and yfiD) as observed in ABU strains of E. coli. These results indicate differences between the ocular and the non-ocular pathogenic E. coli in expression of genes in the biofilm phase.
In the present study a few genes viz., mhpA [coding for 3-(3-hydroxyphenyl) propanoate hydroxylase], mhpB (coding for 2,3-dihydroxyphenylpropionate 1,2-), ryfA (coding for non-coding RNA), tolA (coding for the inner membrane protein -cell envelope integrity), bdcR (coding for the regulatory gene of bdcA), ytfR (coding for ABC transporter ATP-binding protein), and mdtO (coding for multidrug efflux system component) known to be up regulated in expression during biofilm formation in ocular E. coli L-1216/2010 (Ranjith et al., 2017) were mutated by homologous recombination based on the procedure of Datsenko and Wanner (2000) to validate the function of the above genes in biofilm formation in ocular pathogenic E. coli. The choice of the seven genes for gene specific mutation was based on the following criteria:
(a) genes known to be required for biofilm formation like ryfA (Bak et al., 2015) in E. coli and tolA in P. aeruginosa (Whiteley et al., 2001) and E. coli (Vianney et al., 2005) and (b) genes up regulated in expression during biofilm phase like bdcR, mdtO, mhpA, mhpB, and ytfR (Ranjith et al., 2017) but whose specific role in biofilm formation has not yet been demonstrated.
Such studies would help to identify genes involved in biofilm formation in ocular E. coli directly, establish whether genes known to be involved in biofilm formation in other organisms have a similar function in ocular E. coli and may also establish if there is a relationship between biofilm formation and resistance to antimicrobial agents and vice a versa.
Ocular E. coli L-1216/2010 which was isolated and characterized from the Vitreous fluid of a patient with Endophthalmitis and resistant to ciprofloxacin and positive for biofilm formation was used in the present study (Ranjith et al., 2017). The bacterium was cultured in LB medium at 37°C. Several mutant strains of this bacterium (Table 1) were generated by homologous recombination (see below). Growth was monitored spectrophotometrically at 600 nm for 24 h.
Culture (200 μl of 1:10 dilution of 0.5 McF culture) was added to a 96 well plate and incubated for 72 h at 37°C. XTT [2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide] (50 μl of 1 mg/ml) (Sigma, United States) and 4.2 μl (0.5 mM) Menadione and 145.8 μl of 1X PBS was added to each well and incubated in the dark for 3 h and quantified using a spectrophotometer (SpectroMax M3, Molecular Devices, CA, United States) at 490 nm. Wells without culture served as the control and the control OD at 490 nm was <0.12.
Escherichia coli L-1216/2010 and the mutants were cultured on μ- chamber slide 8 well (ibidi, Gmbh, Germany) for 72 h as above and the biofilms were rinsed twice gently with autoclaved water and fixed with 4% formaldehyde (Himedia-Secunderabad, India) for 45 min. Fixed biofilms were then washed twice as above and stained with 200 μl of 1.67 μM Syto®9 a nuclear fluorescent dye (Invitrogen, United States) for 30 min. Stained biofilms were washed again with autoclaved water. Confocal images were taken using Zeiss confocal laser scanning microscope (Carl Zeiss LSM 880, Jena, Germany). Argon Laser was excited at 450–490 nm and a 40× objective was used set at Zoom 2.
After staining of the cells in the biofilm with Syto 9, the biofilms were stained in the dark with 0.025% Calcofluor white M2R (Sigma Chemical Co., St. Louis, MO, United States) for 30 min. This dye binds to β-linked polysaccharides and fluoresces under long-wave UV light (Wood, 1980) and biofilm could be visualized (blue) using confocal microscopy. Calcofluor white has been used to study exopolysaccharides (EPSs) involved in biofilm formation in a variety of organisms (Zogaj et al., 2001; Ledeboer and Jones, 2005). Calcofluor white was excited at 363-nm using a 455/30 band-pass filter (Cowan et al., 2000).
Curli and cellulose nanofibers production in E. coli L-1216/2010 and the mutants was monitored by the Congo red-binding assay method (Arita-Morioka et al., 2015) using Congo red-containing YESCA agar plates. E. coli L-1216/2010 and the mutants were cultured overnight and then 10 μl of the culture were spotted on YESCA media plates containing 10 g/l casamino acids, 10 g/ml Coomassie brilliant blue G-250, 1 g/l yeast extract, and 20 g/L agar. The plates were incubated at 30°C for 3 days and the colonies were photographed. Colonies positive for curli and cellulose nanofibers production were red and colonies positive only for cellulose nanofibers were pink in color and could be easily differentiated.
Adhesion of cells to the substratum was assessed using the procedure of Schmidt-Emrich et al. (2016). Overnight culture was adjusted to 0.5 McFarland units, diluted 100-fold with the medium and 100 μl of the diluted inoculum was dispensed into a single well of a 96 well plate (Thermo Fisher Scientific, NunclonTM, Denmark) containing 100 μl of fresh medium. The plate was incubated at 37 °C for 4 h. The broth was then discarded by inverting the plate and gently tapping it after which it was washed thrice with 200 μl of phosphate buffered saline (PBS, Sigma-Aldrich Corporation, St. Louis, MO, United States) and allowed to dry for 30 min. The bacteria in the biofilm adhering to the plate were then stained with 200 μl of 2.5 μM SYTO9 in dark and incubated for 15 min at room temperature on a shaker and quantified using a spectrophotometer (SpectroMax M3, Molecular Devices, CA, United States) at 490 nm. Wells without culture served as the control.
Swimming motility is an individual and random-directional movement in liquid medium, and essential for initial attachment to develop biofilm. Swarming motility is a coordinated bacterial social movement across the top of a solid surface, accompanied by hyper-flagellation critical for surface colonization after initial attachment. Swimming and swarming motilities were investigated (Bak et al., 2015) on specific agar plates as indicated below. For this purpose 1 μl of 0.5 mcF (∼5 × 104 cells) of each mutant was inoculated with a pipette tip onto swim agar (0.3% Bacto Agar, 1% tryptone and 0.5% NaCl) or swarm agar (0.6% Eiken Agar, 0.5% glucose, 1% tryptone, 0.5% yeast extract and 0.5% NaCl). The swimming assay was performed at 30 °C for 12 h, and swarming assay at 37 °C for 16 h, respectively. The diameter of the swimming circle or the diameter of swarming area was measured and normalized against that of a wild type strain (Bak et al., 2015).
The minimum inhibitory concentration (MIC) of the antibiotic was determined by the micro-dilution method as described by the European Committee on Antimicrobial susceptibility Testing (CLSI, 2012). Antibiotics were obtained from commercial sources and each concentration was tested thrice. The susceptibility of the strains was also determined after the formation of the biofilm. For this purpose, the cultures were incubated as above in the 96 well plate at 37°C for 48 h for biofilm formation. Planktonic cells were discarded, bound cells washed with milliQ water and then the antibiotic dissolved in BHI medium (HiMedia, Mumbai, India) was added to the biofilm and incubated for 16 h. Biofilm formation was then monitored by the crystal violet method. The concentration of the antibiotic that inhibited the formation of the biofilm was determined. E. coli isolate L-1216/2010 in the planktonic phase (where in antibiotics were added after 24 h of growth) was used as controls for this experiment.
Plasmids pKD3 and pKD46 were maintained in DH5α in LB medium with Ampicillin (100 μg/ml) (HiMedia, Mumbai, India) and the plasmids were isolated from an overnight 5 ml culture using Qiagen plasmid mini kit (Qiagen, Heilden, Germany). pKD46 plasmid containing the lambda red recombinase (plasmid gifted by CDFD, Hyderabad, India) gene was transformed into ocular E. coli by CaCl2 heat shock method (42°C for 45 s) and was plated on to LB agar plates supplemented with Ampicillin (100 μg/ml) and incubated at 30°C overnight. Colonies were then selected and grown in the same medium for future use.
Electro-competent cells of E. coli pKD46 were prepared using a standard protocol. An overnight culture diluted 1:100 in LB medium was inoculated into fresh LB medium and incubated at 30°C until the O.D600 reached 0.1.L-arabinose (10 mM, final concentration) was added to the culture and incubated further to OD600 of 0.6 to induce the expression of recombinase gene. The cells were then pelleted by centrifugation at 4000 rpm, washed thrice with autoclaved chilled MilliQ water, and finally resuspended in MilliQ water. The prepared electro-competent cells were stored at -80°C until use.
Mutant strains of E. coli were generated by homologous recombination based on the procedure of Datsenko and Wanner (2000). E. coli L-1216/2010 was first transformed by the CaCl2 heat shock method with pKD46 which expresses the Recombinase gene (E. coli pKD46) to facilitate homologous recombination). Subsequently, electro-competent cells of E. coli pKD46 were electroporated with a linear DNA fragment of the target gene using the Gene-Pulser-Xcellelectroporator (BioRad, Osaka, Japan), at 2.5 kV, 200 Ohm, and 25 μF. For this purpose the electro-competent ocular E. coli pKD46 cells were transferred to pre-chilled electroporation cuvettes (in ice for 10 min) and 5 μg of the PCR amplified linear DNA fragment was added and electroporated. LB medium (1 ml) (0.5% Yeast extract, 1% Peptone, 1% sodium chloride, and 1.5% Agar) was added immediately to the cells after applying the pulse and the electroporated cells were incubated at 37°C for 2 h and plated on an LB plate containing chloramphenicol (25 μg/ml) (HiMedia). Transformants were then selected on chloramphenicol plates and mutants of the target gene were validated by sequencing and Reverse transcription-PCR. Prior to electroporation the linear fragment was obtained by using hybrid primers (FH and RH). FH consisted of 36 nucleotides of the 5′ end of the gene of interest along with the forward primer of the cassette and RH contained 36 nucleotides of the 3′ end of the gene of interest along with the reverse primer of the cassette. When these primers were used they amplified a linear fragment from pKD3 which would contain 36 nucleotides of the target gene from either end plus the FRT (Flippase Recognition Target) region, the chloramphenicol resistance gene cassette and the FRT minimal region (1100 bp). This linear fragment was amplified (Table 1) for 35 cycles (each cycle: 94°C for 5 min, 94°C for 1 min, 63°C for 90 s, 72°C for 2 min, and finally 72°C for 15 min) using pfu polymerase enzyme (Thermo Fisher Scientific, MA, United States).
For complementation studies the genes bdcR, mhpA, mhpB, ryfA, and tolA were amplified along with 200 bp upstream region using pfu polymerase (Table 1). Primers were designed based on the whole genome sequence of E. coli MG1655. Amplification was done for 35 PCR cycles as follows: 94°C for 5 min, 35× (94°C for 1 min, 56°C for 60 s, 72°C for 1.30 min), 72°C for 15 min. The PCR amplified products were then purified using Nucleospin Gel and PCR cleanup kit (Nucleospin, Duren, Germany) and then digested using Hind III/NdeI and XhoI/SacI restriction enzymes for 30 min at 37°C. These digested products would be ligated to pET28a(+) vector (as below).
Plasmid pET28a(+) (gift from J S SPrakash, University of Hyderabad, India) was isolated using plasmid isolation kit (Qiagen, Hilden, Germany) and 1 μg of pET28a(+) was digested with 1 μl of HindIII/NdeI and 1 μl of XhoI/SacI (NEB, Massachusetts, United Kingdom) for 30 min at 37°C. Later the digested vector was purified using Nucleospin Gel and PCR cleanup kit (Duren, Germany) and then ligated to NdeI and SacI digested target gene using T4DNA ligase (Thermo Fisher Scientific, MA, United States). Ligation was performed using final concentration of 0.06 pmol of the digested PCR amplified product and 0.02 pmol of the digested vector at 22°C for 1 h. The ligation reaction mixture was inactivated at 65°C for 10 min and used for transforming DH5α to increase the copy number of the plasmid. Subsequently it was used to transform the respective competent mutant ocular E. coli cells by heat shock method.
Reverse transcription-PCR was performed to validate the expression of genes in the mutants and complemented strains using primers as in Table 1. Amplification was done for 40 cycles as follows: 94°C for 5 min, 40× (94°C for 30 s, 60°C for 30 s).
The predicted functional interactions among genes bdcR, mdtO, mhpA, mhpB, tolA, and ytfR was ascertained using STRING1, Search Tool for the Retrieval of Interacting Genes/Proteins) analysis.
In an earlier study we had demonstrated that E. coli L-1216/2010 isolated from the vitreous of an endophthalmitis patient exhibited very good ability to form biofilm and in the biofilm phase it was 100 fold more resistant to several antibiotics (Ranjith et al., 2017). We had also identified several genes that were up-regulated in expression during biofilm phase (Ranjith et al., 2017). In the present study, a few of the genes up-regulated in expression during biofilm phase viz., bdcR, mdtO, mhpA, mhpB, ryfA, tolA, and ytfR (Ranjith et al., 2017) were mutated by homologous recombination (Datsenko and Wanner, 2000) to validate the function of the above genes in biofilm formation in ocular pathogenic E. coli. Following homologous recombination the mutants were validated by PCR and Reverse transcription-PCR. PCR confirmed the insertion of the antibiotic cassette in the target gene such that the PCR amplicon on sequencing revealed that the antibiotic cassette was intact and 36 bp of the target was present upstream and downstream of the antibiotic cassette. Further Reverse transcription-PCR also indicated that the gene is not expressed in the mutant (Supplementary Figure 1).
Five (ΔbdcR, ΔmhpA, ΔmhpB, ΔryfA, and ΔtolA) of the seven mutants showed significant decrease in biofilm formation (p < 0.05) as monitored by XTT method. In all the five mutants biofilm formation by the XTT method appeared to be as in E. coli ATCC 25922 which served as the negative control. In contrast, the mutants ΔmdtO and ΔytfR retained their ability to form biofilm, similar to wild type E. coli L-1216/2010 (p > 0.05) (Figure 1A). The inability to form the biofilm in the above five mutants was not dependent on its growth which was not affected compared to the control (Figure 1B). Even the growth curves of the mutants compared to E. coli L-1216/2010 was monitored and no significant difference was observed (Figure 1C).
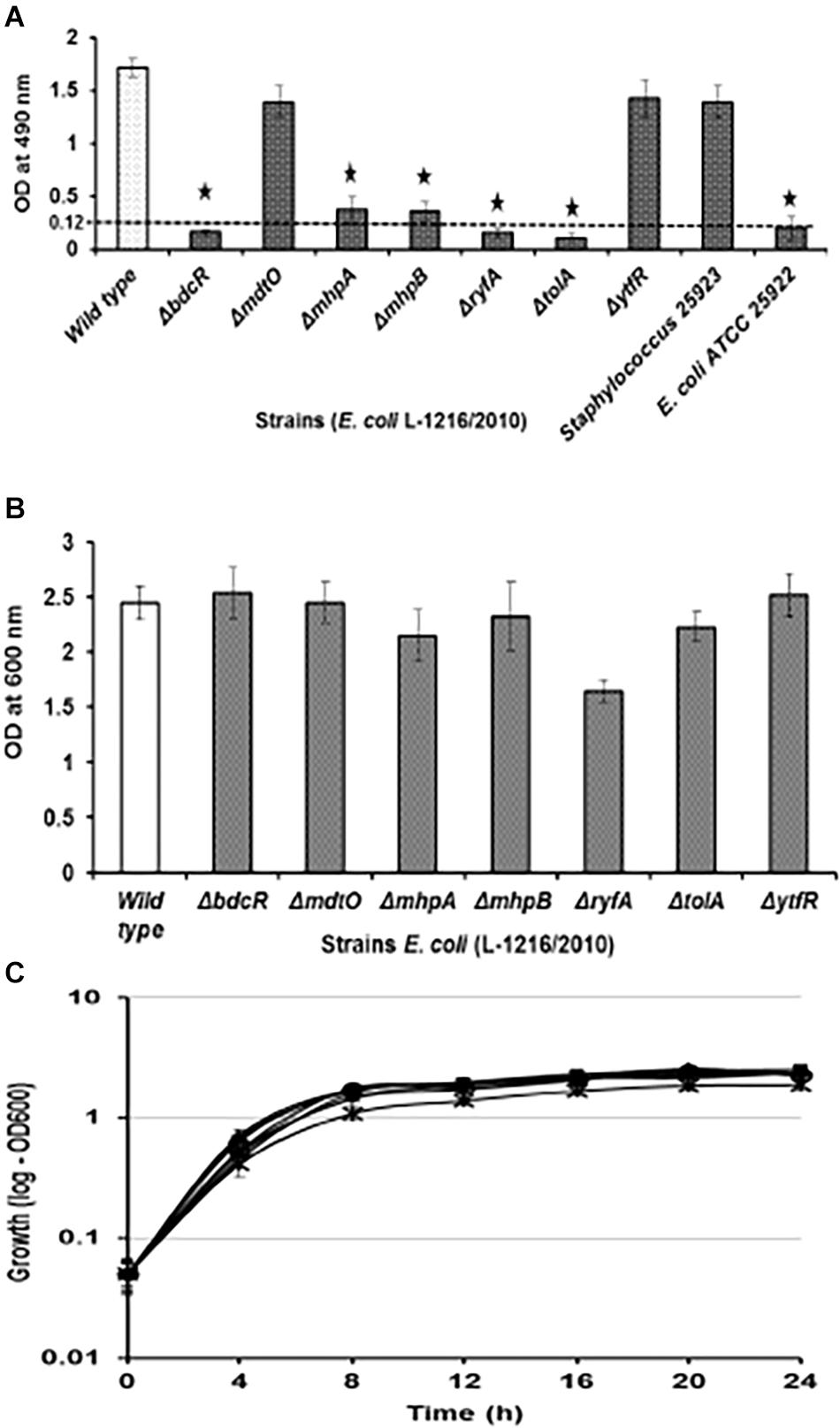
Figure 1. (A) Biofilm formation in ocular Escherichia coli L-1216/2010 (control) and the 7 mutants (ΔbdcR, ΔmdtO,ΔmhpA, ΔmhpB, ΔryfA,ΔtolA, and ΔytfR) by XTT assay. Staphylococcus 25923 and E. coli ATCC 25922 served as the positive and negative controls, respectively. ∗ indicates significant decrease from the control (ocular E. coli L-1216/2010) and similar to E. coli ATCC 25922 in the negative control. (B) Growth of ocular E. coli L-1216/2010 (control) and the 7 mutants (ΔbdcR, ΔmdtO,ΔmhpA, ΔmhpB, ΔryfA, ΔtolA, and ΔytfR) monitored at 600 nm at 24 h. Growth in all the mutants was similar to the wild type (p > 0.05). (C) Growth curve of E. coli L-1216/2010 (wild type) and the 7 mutants  ΔbdcR,
ΔbdcR,  ΔmdtO,
ΔmdtO,  ΔmhpA,
ΔmhpA,  ΔmhpB,
ΔmhpB,  ΔryfA,
ΔryfA,  ΔtolA,
ΔtolA,  ΔytfR, and
ΔytfR, and  control monitored spectrophotometrically at 600 nm.
control monitored spectrophotometrically at 600 nm.
Confocal laser scanning microscopy also confirmed that the luxuriant biofilm that formed after 72 h in E. coli L-1216/2010 was significantly decreased in ΔbdcR, ΔmhpA, ΔmhpB, ΔryfA, and ΔtolA and was unaffected in ΔytfR and ΔmdtO (Figure 2). The thickness of the biofilm reduced significantly from 16.8 μm in the wild type to as low as 1.5 μm in ΔtolA.
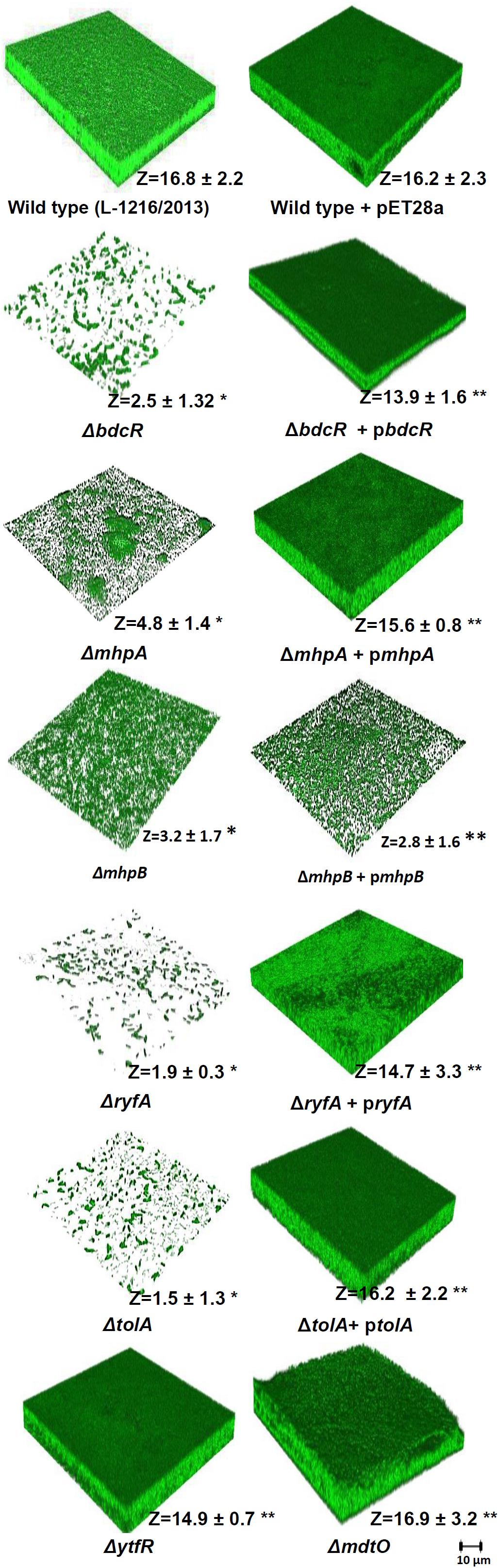
Figure 2. Biofilm formation in ocular E. coli L-1216/2010 (wild type) and the 7 mutants (ΔbdcR,ΔmhpA,ΔmhpB,ΔryfA,ΔtolA,ΔytfR, andΔmdtO) and the mutant complemented strains of ΔbdcR + pbdcR, ΔmhpA + pmhpA, ΔmhpB+ pmhpB, ΔryfA+ pryfA, and ΔtolA +ptolA. The image of E. coli L-1216/2010 + pET28A served as control for all the complemented strains.∗Indicates significant decrease in thickness compared to E. coli L-1216/2010 (control) (p ≤ 0.05). ∗∗ indicates no significant decrease in thickness compared to E. coli L-1216/2010 (control) (p ≥ 0.05). Z values are expressed in μm. All images were recorded at the same magnification. The scale bar is shown in the figure.
Confocal laser scanning microscopy indicated that in four of the five mutants following complementation with the respective gene (ΔbdcR + pbdcR, ΔmhpA + pmhpA, ΔryfA + pryfA, and ΔtolA + ptolA) biofilm formation was restored as in the control (Figure 2). Complementation also increased the thickness of the biofilm (range 13.9–16.3 μm) as observed in wild type ocular E. coli (16.8 + 2.2 μm) (p > 0.05). But, complementation did not restore the biofilm in ΔmhpB+ pmhpB (Figure 2). Quantitation of the biofilm by the XTT method also confirmed that except for the mutant ΔmhpB complementation was efficient in restoring biofilm potential in the remaining four mutants (Figure 3).
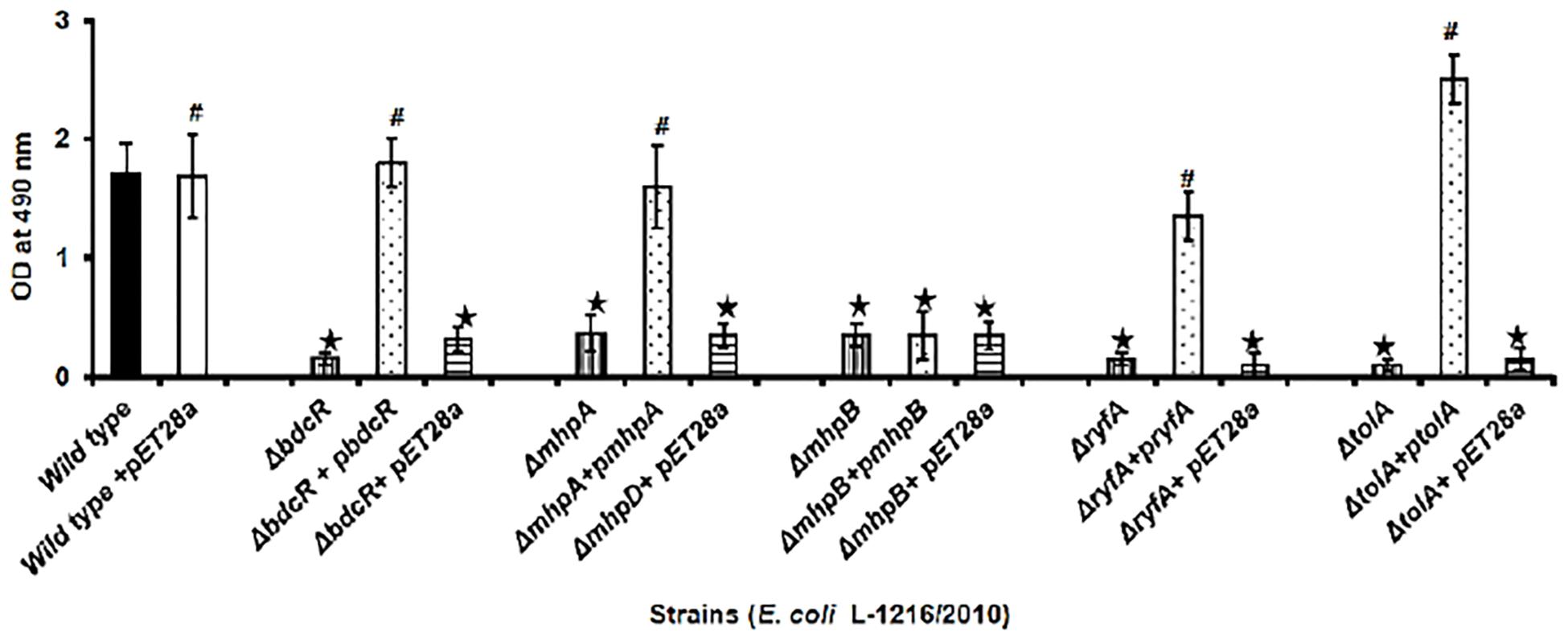
Figure 3. Quantification of biofilm formation by the XTT method in ocular E. coli L-1216/2010 (control), the 5 mutants (ΔbdcR, ΔmhpA, ΔmhpB, ΔryfA, and ΔtolA) and the 5 mutant complemented strains (ΔbdcR + pbdcR, ΔmhpA + pmhpA, ΔmhpB + pmhpB, ΔryfA+pryfA, and ΔtolA +ptolA). Control and mutants complemented with pET28A served as control for the complemented strains. ∗ indicates significant decrease in biofilm formation compared to E. coli L-1216/2010 (control) (p ≤ 0.05). # indicates no significant decrease in biofilm formation compared to E. coli L-1216/2010 (control) (p > 0.05).
Using a dual staining strategy with Syto 9 for cells (green) and Calcofluor white for EPS (blue) it was observed that in the wild type cells EPS thickness increased from 2.53 μm at 4 h to 32.80 μm by 96 h. Maximum thickness was observed at 72 h. In contrast all the 5 mutants showed a significant decrease in the thickness of EPS. In 3 mutants, namely ΔbdcR,ΔmhpA and ΔmhpB EPS thickness increased up to 24 h but remained unchanged subsequently except ΔbdcR which showed substantial decrease in thickness from 72 to 96 h. The other two mutants ΔtolA and ΔryfA did not show any significant increase in EPS between 4 to 96 h. We also observed that cells of ΔmhpA and ΔmhpB had reduced EPS production and appeared to aggregate in to clumps between 48–96 h (Figure 4A).
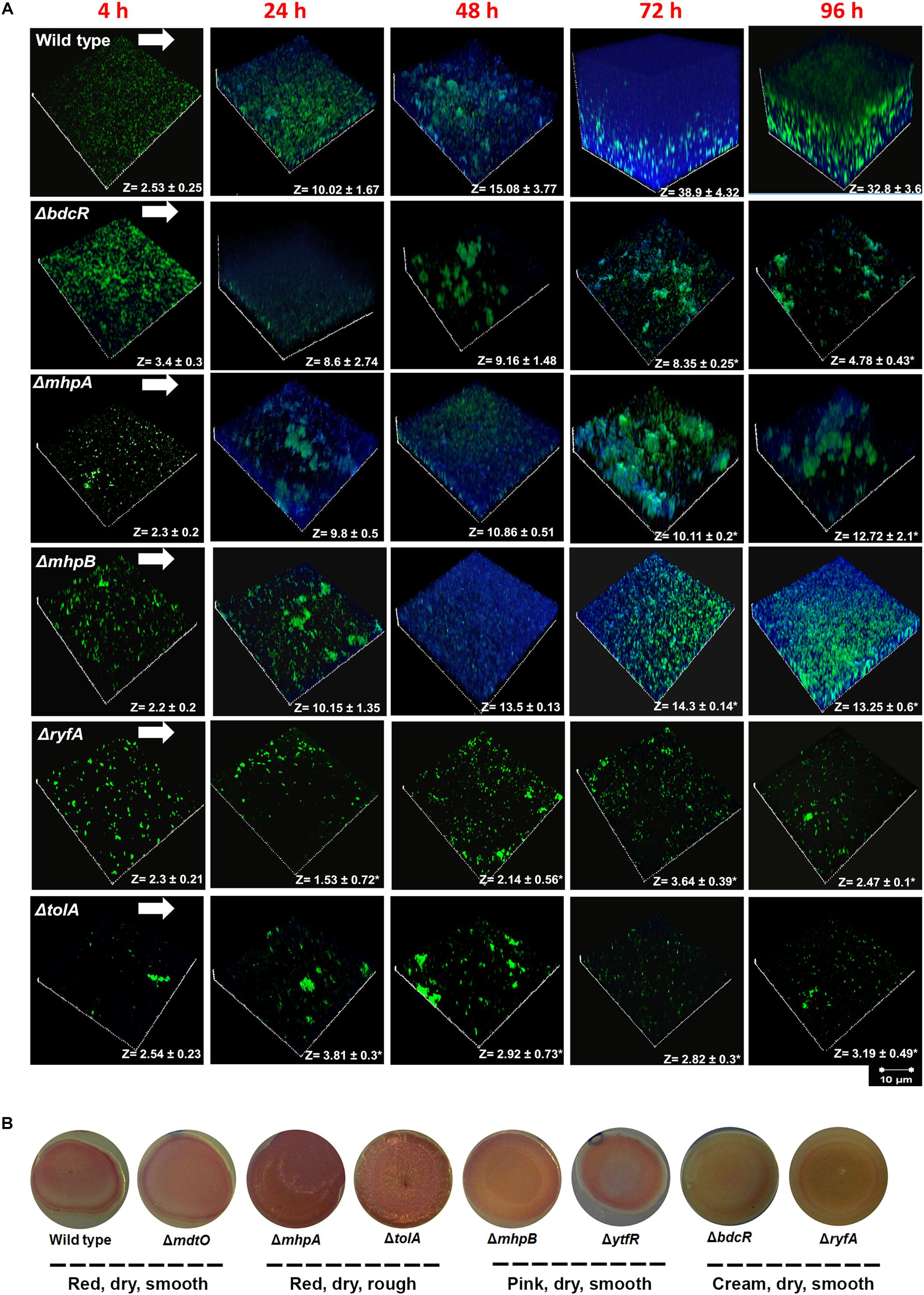
Figure 4. (A) EPS production in ocular E. coli L-1216/2010 (wild type l) and the 5 mutants (ΔbdcR, ΔmhpA, ΔmhpB, ΔryfA, and ΔtolA). Z values are expressed in μm. All images were recorded at the same magnification. The scale bar is shown in the figure. ∗ indicates significant decrease in thickness compared to E. coli L-1216/2010 (wild type) (p ≤ 0.05). (B) Production of curli and cellulose nanofibers in ocular E. coli L-1216/2010 (wild type) and the 7 mutants (ΔbdcR, ΔmdtO ΔmhpA, ΔmhpB, ΔryfA, ΔtolA, andΔytfR).
It was observed using Congo red-containing YESCA agar plates that out of 7 mutants, three were red in color, dry and either smooth (ΔmdtO) or rough (ΔmhpA and ΔtolA), two (ΔmhpB and ΔytfR) were pink, dry and smooth, and the remaining two (ΔbdcR and ΔryfA) were cream in color, dry and smooth. The wild type was also observed to be red, dry and smooth. These morphological features indicated that the wild type and 3 mutants that were dry and red are positive for curli fimbriae and cellulose nanofibers production. (Römling, 2005; Milanov et al., 2015), the 2 mutants that were pink and smooth were positive for cellulose nanofibers production (Zogaj et al., 2003), and the 2 mutants that were cream colored were negative for both curli fimbriae and cellulose nanofibers production.(Figure 4B).
Swimming and swarming of the wild type and mutant cells was monitored overnight and the results indicated that compared to E. coli L-1216/2010 swimming was significantly reduced in two mutants (ΔryfA and ΔtolA, p < 0.05) (Figure 5A). In contrast swarming was significantly reduced only in tolA (p < 0.05) (Figure 5B). Results of adhesion indicated that the number of cells attached to the substratum appeared to be similar in all except ΔtolA where less cells appeared to be attached (Figure 5C).
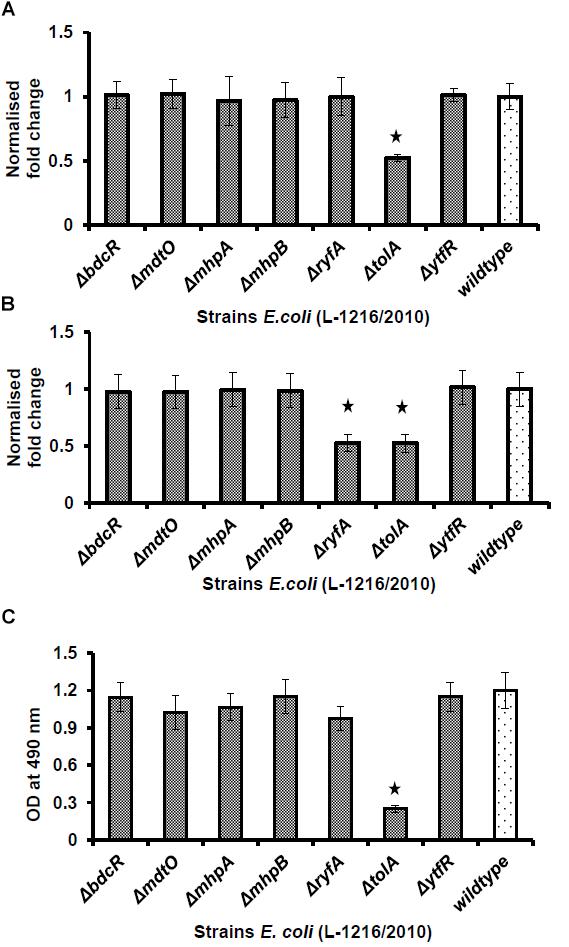
Figure 5. Swimming (A) and swarming (B) of ocular E. coli L-1216/2010 (wild type) and the 7 mutants (ΔbdcR, ΔmdtO, ΔmhpA, ΔmhpB, ΔryfA, ΔtolA, and ΔytfR). The diameter of the swimming and swarming zone in the wild type cells was 22 ± 3.75 mm- and 14.35 ± 2.45 mm, respectively. ∗ indicates significant decrease compared to E. coli L-1216/2010 (wild type) (p ≤ 0.05). (C) Quantification of adhesion by the syto9 staining method of ocular E. coli L-1216/2010 (wild type) and the 7 mutants (ΔbdcR, ΔmdtO, ΔmhpA, ΔmhpB, ΔryfA, ΔtolA, and ΔytfR). The adhesion of all mutants were similar to wild type cells except ΔtolA.
The five mutants which did not form the biofilm (ΔbdcR, ΔmhpA, ΔmhpB, ΔryfA, and ΔtolA) following exposure to 9 different antibiotics in the planktonic phase exhibited MICs similar to that observed for the wild type E. coli L-1216/2010 (Table 2). Mutant ΔytfR which retained its ability to form a biofilm also had a similar MIC compared to the wild type E. coli in the planktonic phase. But, mutant ΔmdtO which retained its ability to form a biofilm showed reduced MIC to 4 out of the 9 antibiotics tested (viz., Sulfamethoxazole, Imipenem, Streptomycin and Erythromycin) (Table 2) in the planktonic phase. We also checked the MIC of the mutants ΔytfR and ΔmdtO in response to the above four antibiotics in the their biofilm phase and observed that the MIC compared to the control was altered only in ΔmdtO in the biofilm phase implying that the mutation had not affected its ability to form biofilm but has altered its sensitivity to antibiotics (Table 3). Finally we also checked the antibiotic sensitivity of the complemented strains in the biofilm phase and the results indicated that the complemented strains ΔbdcR + pbdcR, ΔmhpA + pmhpA, and ΔmhpB+ pmhpB did not differ in their sensitivity to all the 9 tested antibiotics compared to the wild type cells whereas ΔtolA + ptolA appeared to be more resistant to 2 of the 9 antibiotics (namely Gentamycin and Erythromycin) (Table 4).
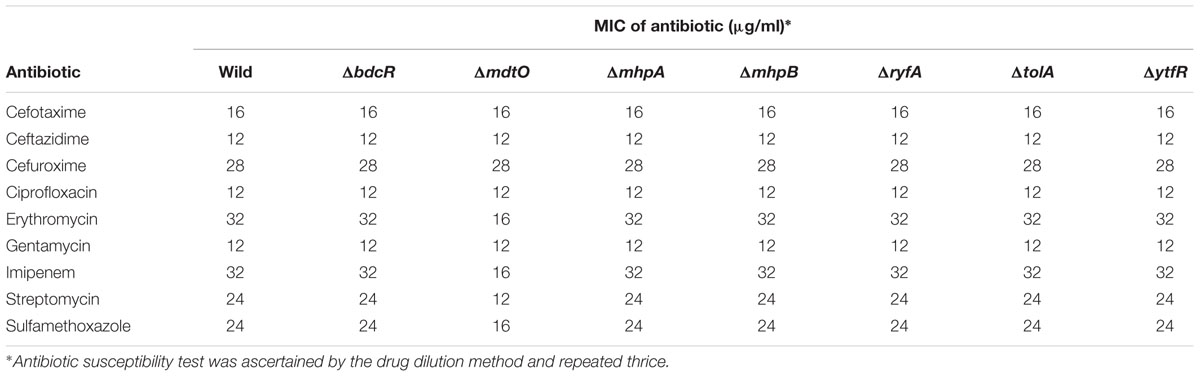
Table 2. Antibiotic sensitivity of the ocular E. coli L-1216/2010 and the 7 mutants to 9 different antibiotics.
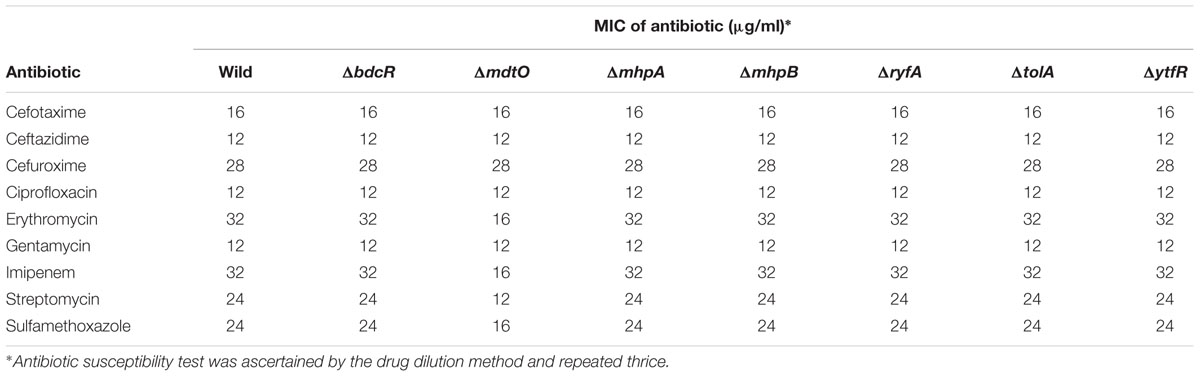
Table 3. Antibiotic susceptibility of ocular E. coli L-1216/2010 and mutants ΔytfR and ΔmdtO in the biofilm phase to 4 different antibiotics.
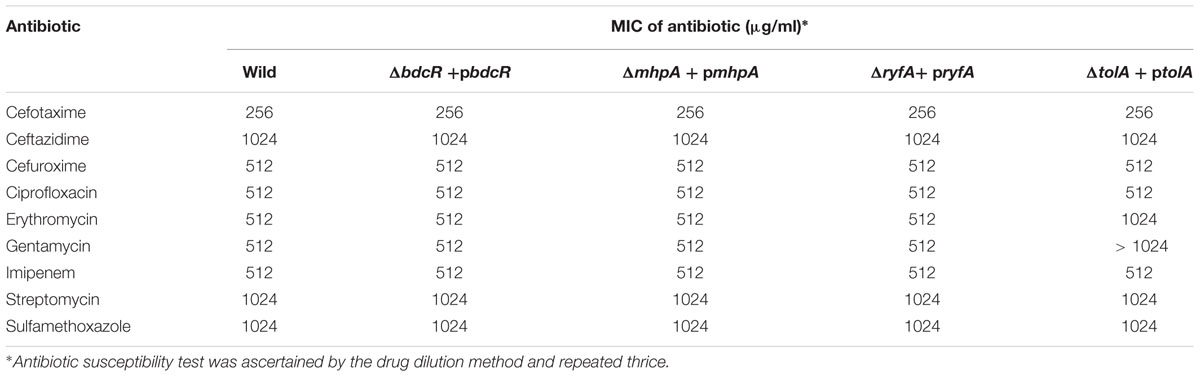
Table 4. Antibiotic susceptibility of ocular E. coli L-1216/2010 and mutants complemented with their respective genes in the biofilm phase to 9 different antibiotics.
STRING network analysis indicated that genes bdcR, mdtO, mhpA, mhpB, tolA, and ytfR were organized in to two functional arms namely microbial metabolism and transmembrane transporter activity. Genes bdcR, mhpA, mhpB, and tolA were part of the microbial metabolism arm and interacted with genes involved in pyruvate metabolism (genes mhpF, plfB, tdcE, and adhE), 3-(3-hydroxy) phenyl propionate catabolic process (genes mhpA and mhpB), acetaldehyde dehydrogenase activity (genes mhpF and adhE) and genes involved in biofilm such as c-di-GMP-binding biofilm dispersal mediator protein (bdcR). The remaining 2 genes mdtO and ytfR were part of the transmembrane transport activity and interacted with genes involved in transport such as protein transport genes (tolA, tolB, tolQ, and tolR) and transmembrane transport genes (ompF, tolC, acrD, acrA, JW0451, macB, emrA, macA, mdtP, mdtO, mdtN, hrsA, ytfT, ytfF, and ytfR) (Figure 6). The interaction between the genes was mediated by ompR encoding for DNA-binding transcriptional dual regulator, ompF encoding for outer membrane porin 1a, adhE and mhpF encoding for aldehyde dehydrogenase, tolC encoding for transport channel and tolA encoding for inner membrane protein member of Tol-Pal system. The network also included genes coding for the “two component signal transduction system” ompR and rcsA.
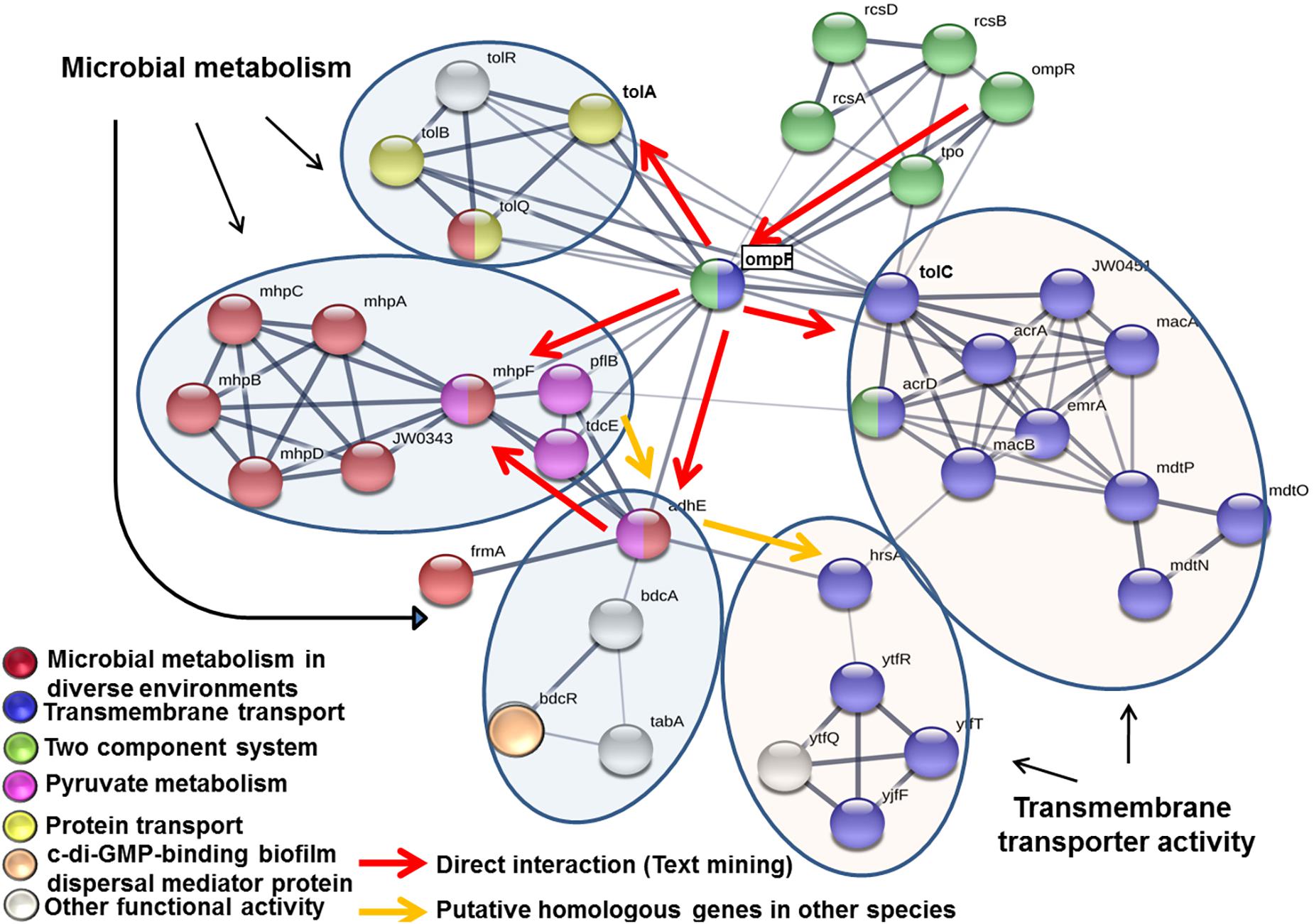
Figure 6. STRING network analysis of the genes bdcR, mdtO, mhpA, mhpB, ryfA, tolA, and ytfR that were mutated in ocular E. coli L-1216/2010. Each node in the network denotes a gene while edges between nodes indicate interactions between corresponding genes. The thickness of the edge color indicates the strength of interaction (0–1.0). In the present work, the edge confidence in the network ranged from 0.4 to 0.9. Nodes are colored based on the functional interactions. Genes involved in primary network interactions are circled.
Understanding the molecular basis of biofilm formation is very crucial in identifying targets to hack biofilms so that virulent and pathogenic bacteria could be controlled. Several studies have demonstrated that in E. coli in the biofilm phase several hundreds of genes are differentially expressed (Schembri et al., 2003; Beloin et al., 2004; Ren et al., 2004; Domka et al., 2006; Hancock and Klemm, 2007; Ranjith et al., 2017). In a recent study we demonstrated that in ocular E. coli L-1216/2010 also several genes were differentially expressed in the biofilm phase (Ranjith et al., 2017). Further, ocular E. coli could be differentiated from E. coli K12, a laboratory non-pathogenic strain of E. coli, with respect to expression of genes involved in cell cycle control, mitosis and meiosis, cell motility, intracellular trafficking, secretion, vesicular transport and defense mechanisms (Ranjith et al., 2017) implying that ocular E. coli is probably more like a pathogen. In fact this may indeed be so because the ocular E. coli L-1216/2010 and ABU E. coli (Ranjith et al., 2017) a pathogenic strain, were similar with respect to differential regulation of genes involved in intracellular trafficking, secretion and vesicular transport, defense mechanisms and replication, recombination, and repair which are normally associated with pathogenic bacteria. In the present study the question addressed is whether ocular pathogen are similar to other pathogens with respect to genes involved in biofilm formation and associated drug resistance.
Over the years it has been reasonably well established that genes involved in motility (Wood et al., 2006), that facilitate attachment to substratum (Prigent-Combaret et al., 2000; Ren et al., 2005), that code for production of extracellular polymers and adhesive factors (Danese et al., 2000; Schembri et al., 2004), that help in formation and maturation of micro-colonies and in cell signaling (quorum sensing) (Reisner et al., 2003) are associated with biofilm formation. A few of the genes such as tolA, ryfA (Whiteley et al., 2001), ymdB (Kim et al., 2013), csrA (Jackson et al., 2002), spoT and relA (de la Fuente-Nunez et al., 2014), and ydgG (Herzberg et al., 2006) have been shown to be required for biofilm formation by specific knock out of the gene (s) either in P. aeruginosa or in E. coli (Vianney et al., 2005). In the present study seven gene knock outs were generated in ocular E. coli L-1216/2010 to ascertain the role of these genes (bdcR, mdtO, mhpA, mhpB, tolA, ryfA, and ytfR) in biofilm formation. Earlier studies had indicated that knockout mutants of two genes ryfA and tolA had lost the potential to form biofilm (Whiteley et al., 2001; Vianney et al., 2005). In this study we confirm that in ocular E. coli, ryfA which codes for a small RNA (Bak et al., 2015) following deletion exhibits a decrease in biofilm development probably due to the observed decrease in EPS production, cellulose nanofibers and curli production. The mutant also exhibited significant decrease in swarming. Earlier studies had indicated that curli are important for biofilm formation by facilitating initial attachment to a surface and cell-to-cell cohesion (Pratt and Kolter, 1999). In an earlier study it was demonstrated that in P. aeruginosa, tolA gene was activated in biofilms (Whiteley et al., 2001) and mutation of the gene in E. coli MG1655 and two clinical strains of E. coli PHL881 and PHL885 led to a decrease in biofilm formation implying the importance of tolA in biofilm formation (Vianney et al., 2005). They also attributed the decrease in biofilm formation to the observation that in the tol mutants the adherence of cells to the substratum was lowered. The present study confirms both the observations that in ΔtolA mutants of ocular E. coli biofilm formation is decreased and it could be attributed to a decrease in adhesion of cells, swimming, swarming and production of EPS. Interestingly it was observed that in the mutant, both curli and cellulose nanofibers production was not affected implying that curli is not mandatory for biofilm formation. The reports on the requirement of curli for biofilm formation are contradictory with a few indicating that curli and cellulose nanofibers are not required for biofilm formation (Niba et al., 2007; Cegelski et al., 2009) unlike the report of Arita-Morioka et al. (2015). It also appears that since tolA interacts with other tol genes (tolB, tolQ, and tolR), with ompF involved in bacteriocin (tolA, tolB, tolQ, and tolR) and drug transport (acrD, acrA, JW0451, macB, emrA, mdtP, mdtO, and mdtN) and tolC which regulates genes involved in transmembrane transport activity (Figure 6) it may contribute to drug resistance. But mutants did not show any change in susceptibility to 9 antibiotics tested implying that tolA may not be directly involved in drug resistance.
We had earlier shown that many genes encoding dehydrogenases (mhpA, mhpB, mhpF, yiaY, yajO/ydbK, and yjjN) were up regulated during biofilm formation in ocular E. coli (Ranjith et al., 2017) implying their likely role in biofilm formation. But none of these genes were demonstrated to be required for biofilm formation. In this study we demonstrate that biofilm formation was decreased substantially in separate knockout mutants of mhpA and mhpB implying that these two genes are required for biofilm formation. mhpA and mhpB are specifically involved in 3-(3-hydroxy) phenyl propionate catabolic process and thus it is likely that these two genes may be associated with a metabolic function. In fact, String network analysis also indicated a close interaction between mhpA and mhpB with other microbial metabolic genes such as genes involved in pyruvate metabolism (genes mhpF, plfB, tdcE, and adhE) and genes coding for acetaldehyde dehydrogenases (genes mhpF and adhE) and could thus be involved in microbial metabolism, drug metabolism and support drug resistance a phenomenon associated with biofilm formation. But their role in drug resistance is unlikely since it was observed that the mutants susceptibility to the nine antibiotics tested remained unchanged. When we investigated the phenotypic characteristics it was observed that both the mutants (ΔmhpA and ΔmhpB) ability to swim, swarm and attachment remained unaffected. In addition it was observed that curli and nanofiber production was not affected in mphA but in contrast, ΔmhpB only curli production was affected.
In ocular E. coli it was demonstrated that the genes bdcA and bdcR were upregulated more than 20 fold in the biofilm phase implying their need for biofilm formation (Ranjith et al., 2017). It has been demonstrated that bdcA deletion reduces biofilm dispersal by decreasing motility and thus does not affect biofilm formation (Ma et al., 2011). In accordance with the above observation in the present study it was observed that knockout mutants of bdcR, a negative regulator of bdcA, and showed reduced biofilm formation. But the mutants ability to swim, swarm and attach was not altered but EPS, curli and cellulose nanofiber production was significantly decreased. Thus it appears that bdcR is required for biofilm formation in ocular E. coli. It was also observed that unlike the wild type cells, knockout mutants of mhpA, mhpB, and bdcR showed clumping of cells at some time points during the biofilm phase.
Genes coding for ABC transporter ATP-binding protein and belonging to the ATP-binding cassette (ABC) transporter superfamily have been implicated in biofilm formation. But their involvement in biofilm formation appears to vary from organism to organism. For instance in Rhizobium leguminosarum mutation in the ATP-binding protein of an uncharacterized ABC transporter operon significantly reduced the number of viable cells and simultaneously inhibited biofilm formation suggesting that a functional transporter is essential for normal biofilm formation (Vanderlinde et al., 2010). In contrast, (Zhu et al., 2008; Ceruso et al., 2014) observed that in Listeria monocytogenes, inactivation of the putative ABC transporter or the permease component, caused enhanced biofilm-formation compared to the wild type, indicating that LMOf 2365_1875 negatively regulates biofilm formation. At the same time (Ceruso et al., 2014) also observed that LMOf 2365_1877 another mutant in the ABC transporter gene did not have any effect on biofilm formation. In the present study we observed that deletion of ytfR (a gene that codes for ABC transporter ATP-binding protein) in ocular E. coli did not alter its biofilm formation potential as observed in the above studies and also did not affect the motility, initial attachment and celluose nanofiber production but curli production was affected. It is possible that the function of ytfR is compensated by other genes of the ABC transporter superfamily. In fact, String analysis does indicate that ytfR closely interacts with other members of the ABC transporter superfamily like ytfT, yjfF, ytfQ, and hrsA which may functionally compensate for its deletion. Recently it was demonstrated by Pinweha et al. (2018) that in Burkholderia pseudomallei, ABC transporter mutant bpsl1039-1040 showed reduced biofilm formation as compared with the wild-type strain (P = 0.027) when cultured in LB medium supplemented with nitrate under anaerobic growth conditions. But this reduction in biofilm formation was not noticeable under aerobic conditions. Thus overall it would appear that ABC transporter gene function with respect to biofilm formation varies not only from organism to organism but also under the physiological condition it is tested.
Among the various genes that were upregulated in ocular E. coli we also noticed that genes involved in multidrug resistance like mdtO (synonym yjcQ) were also up regulated in their biofilm phase in ocular E. coli. This is of interest since biofilm formation is related to resistance to antibiotics (Kaplan, 2011) but the correlation appears to be inconsistent and the results are conflicting. For instance in P. aeruginosa and Acinetobacter baumannii biofilm production was significantly higher in multidrug resistant (MDR) isolates (Abidi et al., 2013; Gurung et al., 2013; Qi et al., 2016). But it was not so in MDR S. aureus (Eyoh et al., 2014). In this study we observed that in mdtO deletion mutant of ocular E. coli formation of biofilm was unaltered and all the attributes of a biofilm forming bacterium such as swimming, swarming, attachment and production of curli, and cellulose nanofibers were unaltered. But the susceptibility of the mdtO deletion mutant to Sulfamethoxazole, Imipenem, Erythromycin, and Streptomycin was increased as judged by the lower MICs. Thus the observations emphasize a role for the gene exclusively in drug resistance.
In the current study, out of the five deletion mutants (ΔbdcR, ΔmhpA, ΔmhpB, ΔtolA, and ΔryfA) complementation was successful in all the mutants except ΔmhpB in which the biofilm formation potential was not restored. Further the plasmid used for complementation had no effect on the growth. In two mutants of ABC transporter in Rhizobium leguminosarum attempts to restore wild-type phenotype of biofilm formation by complementation was also not successful (Vanderlinde et al., 2010) and was attributed to a dominant-negative effect as observed earlier for other ABC transporters (Bliss et al., 1996; Miyamoto et al., 2002; Eyoh et al., 2014). In the present case it needs to be established.
The STRING network analysis of the deleted genes in E. coli appeared to be involved in two functional arms namely “microbial metabolism” and “transmembrane transporter activity.” Genes mhpA, mhpB, bdcR, and tolA constituted the metabolism arm and deletion of these genes decreased biofilm formation but their susceptibility to antibiotics in the planktonic phase was unaltered. It appears that these genes involved in microbial metabolism are essential for biofilm formation and supports a previous study that acetate metabolism is related to the formation of biofilms in E. coli (Pruss et al., 2010). The four genes bdcR, mhpA, mhpB, and tolA of the metabolic arm also interacted with other metabolic genes of pyruvate metabolism, 3-(3-hydroxy) phenyl propionate catabolic process and acetaldehyde dehydrogenase activity regulated either directly or indirectly by their interaction with ompF (outer membrane porin). In contrast, E. coli cells mutated for genes ytfR and mdtO were implicated in “transmembrane transporter activity” and deletion of these genes did not alter their ability to produce biofilm as in wild type cells. But, one of the mutants ΔmdtO showed more sensitivity to Sulfamethoxazole, Imipenem, Streptomycin, and Erythromycin both in the planktonic and biofilm phase. Thus these genes appear to be more related to drug resistance. ompF also regulated this arm by indirectly interacting with mdtO through tolC and with ytfR through adhE. This arm also included genes involved in drug transport (ompF, acrD, acrA, JW0451, macB, emrA, mdtP, mdtO, and mdtN) and sugar transport genes (hrsA, ytfT, ytfF, and ytfR). It is also observed that genes coding for the “two component signal transduction system” ompR and rcsA (Pruss, 2017) were central to regulating the two arms through ompF. Thus it is possible that genes of the “microbial metabolism” arm are required for biofilm formation whereas genes of the “transmembrane transporter activity” are required for drug resistance and not essential for biofilm formation.
In the present study we mutated a total of seven genes in ocular E. coli L-1216/2010 out of which in five mutants (ΔbdcR, ΔmhpA, ΔmhpB, ΔryfA, and ΔtolA) biofilm formation was inhibited. In three of the mutants (ΔbdcR, ΔmhpA, and ΔmhpB) swimming, swarming and adhesion were not affected. In ΔtolA swimming, swarming and adhesion were affected. where as in ΔryfA only swarming was affected. In addition, all the five mutants exhibited decrease in EPS production and in two of the mutants (ΔbdcR and ΔryfA) curli and cellulose nanofibers production was also inhibited. Thus the only feature that appears to be shared by all the five mutants is their inability to produce EPS. In the remaining two mutants (ΔmdtO and ΔytfR) biofilm formation was unaltered. Out of these five genes which are demonstrated to be required for biofilm formation ryfA and tolA were earlier demonstrated to be required for biofilm formation (Vianney et al., 2005; Bak et al., 2015). Thus we report for the first time that genes bdcR, ΔmhpA, and ΔmhpB are required for biofilm formation in ocular E coli. We also report that gene mdtO, a transmembrane protein involved in the multi-drug transport of sulphanilamide drugs in planktonic phase, when mutated did not affect the biofilm formation in ocular E. coli L-1216/2010 but the mutant was more sensitive to Sulfamethoxazole, Imipenem, Erythromycin, and Streptomycin both in the planktonic and biofilm phase. Thus mitigation of biofilm formation to overcome antibiotic resistance could be achieved by targeting the genes bdcR, mhpA, mhpB, tolA, and ryfA.
KR performed the experiments and participated in the data analysis. JR and JP helped in generating knockout and complementation and provided the logistic support. KA helped in string analysis. SShi participated in manuscript writing and editing. SSha conceived the project, executed the project, and helped in manuscript writing and finalization. All authors read and approved the final manuscript.
KR thanks ICMR (2017-2836/CMB-BMS) for the SRF fellowship and HERF for funding the project.
The authors declare that the researchwas conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Indumathi Mariappan, for helping with the use of the confocal laser scanning microscope. We also thank Dr. Abhijit A. Sardesai from CDFD for pKD3 and pKD46 plasmids. Special thanks to SBN Chary for help with photography.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01308/full#supplementary-material
Abidi, S. H., Sherwani, S. K., Siddiqui, T. R., Bashir, A., and Kazmi, S. U. (2013). Drug resistance profile and biofilm forming potential of Pseudomonas aeruginosa isolated from contact lenses in Karachi-Pakistan. BMC Ophthalmol. 13:57. doi: 10.1186/1471-2415-13-57
Arita-Morioka, K., Yamanaka, K., Mizunoe, Y., Ogura, T., and Sugimoto, S. (2015). Novel strategy for biofilm inhibition by using small molecules targeting molecular chaperone DnaK. Antimicrob. Agents Chemother. 59, 633–641. doi: 10.1128/AAC.04465-14
Bak, G., Lee, J., Suk, S., Kim, D., Young Lee, J., Kim, K. S., et al. (2015). Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli. Sci. Rep. 5:15287. doi: 10.1038/srep15287
Beloin, C., Valle, J., Latour-Lambert, P., Faure, P., Kzreminski, M., Balestrino, D., et al. (2004). Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51, 659–674. doi: 10.1046/j.1365-2958.2003.03865.x
Bliss, J. M., Garon, C. F., and Silver, R. P. (1996). Polysialic acid export in Escherichia coli K1: the role of KpsT, the ATP-binding component of an ABC transporter, in chain translocation. Glycobiology 6, 445–452. doi: 10.1093/glycob/6.4.445
Cegelski, L., Pinkner, J. S., Hammer, N. D., Cusumano, C. K., Hung, C. S., Chorell, E., et al. (2009). Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 5, 913–919. doi: 10.1038/nchembio.242
Ceruso, M., Fratamico, P., Chirollo, C., Taglialatela, R., Cortesi, M. L., and Pepe, T. (2014). The capacity of Listeria Monocytogenes mutants with in-frame deletions in putative ATP-binding cassette transporters to form biofilms and comparison with the wild type. Ital. J. Food Saf. 3:1657.
CLSI (2012). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI/NCCLS Document M100-S22. Wayne, PA: Clinical and Laboratory Standards Institute.
Cowan, S. E., Gilbert, E., Liepmann, D., and Keasling, J. D. (2000). Commensal interactions in a dual-species biofilm exposed to mixed organic compounds. Appl. Environ. Microbiol. 66, 4481–4485. doi: 10.1128/aem.66.10.4481-4485.2000
Danese, P. N., Pratt, L. A., and Kolter, R. (2000). Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182, 3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
de la Fuente-Nunez, C., Reffuveille, F., Haney, E. F., Straus, S. K., and Hancock, R. E. (2014). Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10:e1004152. doi: 10.1371/journal.ppat.1004152
Domka, J., Lee, J., and Wood, T. K. (2006). YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72, 2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006
Elder, M. J., Stapleton, F., Evans, E., and Dart, J. K. (1995). Biofilm-related infections in ophthalmology. Eye 9(Pt 1), 102–109. doi: 10.1038/eye.1995.16
Eyoh, A. B., Toukam, M., Atashili, J., Fokunang, C., Gonsu, H., Lyonga, E. E., et al. (2014). Relationship between multiple drug resistance and biofilm formation in Staphylococcus aureus isolated from medical and non-medical personnel in Yaounde. Cameroon. Pan. Afr. Med. J. 17:186. doi: 10.11604/pamj.2014.17.186.2363
Gurung, J., Khyriem, A. B., Banik, A., Lyngdoh, W. V., Choudhury, B., and Bhattacharyya, P. (2013). Association of biofilm production with multidrug resistance among clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa from intensive care unit. Indian J. Crit. Care Med. 17, 214–218. doi: 10.4103/0972-5229.118416
Hancock, V., and Klemm, P. (2007). Global gene expression profiling of asymptomatic bacteriuria Escherichia coli during biofilm growth in human urine. Infect. Immun. 75, 966–976. doi: 10.1128/IAI.01748-06
Herzberg, M., Kaye, I. K., Peti, W., and Wood, T. K. (2006). YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188, 587–598. doi: 10.1128/JB.188.2.587-598.2006
Hoyle, B. D., Alcantara, J., and Costerton, J. W. (1992). Pseudomonas aeruginosa biofilm as a diffusion barrier to piperacillin. Antimicrob. Agents Chemother. 36, 2054–2056. doi: 10.1128/aac.36.9.2054
Jackson, D. W., Suzuki, K., Oakford, L., Simecka, J. W., Hart, M. E., and Romeo, T. (2002). Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184, 290–301. doi: 10.1128/jb.184.1.290-301.2002
Kaplan, J. B. (2011). Antibiotic-induced biofilm formation. Int. J. Artif. Organs 34, 737–751. doi: 10.5301/ijao.5000027
Katiyar, R., Vishwakarma, A., and Kaistha, S. (2012). Analysis of biofilm formation and antibiotic resistance of microbial isolates from intraocular lens following conventional extracapsular cataract surgery. Int. J. Res. Pure Appl. Microbiol. 2, 20–24.
Kim, T., Lee, J., and Kim, K. S. (2013). Escherichia coli YmdB regulates biofilm formation independently of its role as an RNase III modulator. BMC Microbiol. 13:266. doi: 10.1186/1471-2180-13-266
Ledeboer, N. A., and Jones, B. D. (2005). Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar typhimurium on HEp-2 cells and chicken intestinal epithelium. J. Bacteriol. 187, 3214–3226. doi: 10.1128/JB.187.9.3214-3226.2005
Ma, Q., Yang, Z., Pu, M., Peti, W., and Wood, T. K. (2011). Engineering a novel c-di-GMP-binding protein for biofilm dispersal. Environ. Microbiol. 13, 631–642. doi: 10.1111/j.1462-2920.2010.02368.x
Milanov, D. S., Prunic, B. Z., Velhner, M. J., Pajic, M. L., and Cabarkapa, I. S. (2015). Rdar morphotype- a resting stage of some Enterobacteriaceae. Food Feed. Res. 42, 43–50. doi: 10.5937/FFR1501043M
Miyamoto, A., Matsuyama, S., and Tokuda, H. (2002). Dominant negative mutant of a lipoprotein-specific molecular chaperone, LolA, tightly associates with LolCDE. FEBS Lett. 528, 193–196. doi: 10.1016/s0014-5793(02)03305-7
Niba, E. T., Naka, Y., Nagase, M., Mori, H., and Kitakawa, M. (2007). A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Res. 14, 237–246. doi: 10.1093/dnares/dsm024
Pinweha, P., Pumirat, P., Cuccui, J., Jitprasutwit, N., Muangsombut, V., Srinon, V., et al. (2018). Inactivation of bpsl1039-1040 ATP-binding cassette transporter reduces intracellular survival in macrophages, biofilm formation and virulence in the murine model of Burkholderia pseudomallei infection. PLoS One 13:e0196202. doi: 10.1371/journal.pone.0196202
Pratt, L. A., and Kolter, R. (1999). Genetic analyses of bacterial biofilm formation. Curr. Opin. Microbiol. 2, 598–603. doi: 10.1016/s1369-5274(99)00028-4
Prigent-Combaret, C., Prensier, G., Le Thi, T. T., Vidal, O., Lejeune, P., and Dorel, C. (2000). Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2, 450–464. doi: 10.1046/j.1462-2920.2000.00128.x
Pruss, B. M. (2017). Involvement of two-component signaling on bacterial motility and biofilm development. J. Bacteriol. 199, e00259-17. doi: 10.1128/JB.00259-17
Pruss, B. M., Verma, K., Samanta, P., Sule, P., Kumar, S., Wu, J., et al. (2010). Environmental and genetic factors that contribute to Escherichia coli K-12 biofilm formation. Arch. Microbiol. 192, 715–728. doi: 10.1007/s00203-010-0599-z
Qi, L., Li, H., Zhang, C., Liang, B., Li, J., Wang, L., et al. (2016). Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 7:483. doi: 10.3389/fmicb.2016.00483
Ranjith, K., Arunasri, K., Reddy, G. S., Adicherla, H., Sharma, S., and Shivaji, S. (2017). Global gene expression in Escherichia coli, isolated from the diseased ocular surface of the human eye with a potential to form biofilm. Gut Pathog 9:15. doi: 10.1186/s13099-017-0164-2
Reisner, A., Haagensen, J. A., Schembri, M. A., Zechner, E. L., and Molin, S. (2003). Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48, 933–946. doi: 10.1046/j.1365-2958.2003.03490.x
Ren, C. P., Beatson, S. A., Parkhill, J., and Pallen, M. J. (2005). The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J. Bacteriol. 187, 1430–1440. doi: 10.1128/JB.187.4.1430-1440.2005
Ren, D., Bedzyk, L. A., Thomas, S. M., Ye, R. W., and Wood, T. K. (2004). Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64, 515–524. doi: 10.1007/s00253-003-1517-y
Römling, U. (2005). Characterization of the rdar morphotype, a multicellular behavior in Enterobacteriaceae. Cell Mol. Life Sci. 62, 1234–1246. doi: 10.1007/s00018-005-4557-x
Schembri, M. A., Dalsgaard, D., and Klemm, P. (2004). Capsule shields the function of short bacterial adhesins. J. Bacteriol. 186, 1249–1257. doi: 10.1128/jb.186.5.1249-1257.2004
Schembri, M. A., Kjaergaard, K., and Klemm, P. (2003). Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48, 253–267. doi: 10.1046/j.1365-2958.2003.03432.x
Schmidt-Emrich, S., Stiefel, P., Rupper, P., Katzenmeier, H., Amberg, C., Maniura-Weber, K., et al. (2016). Rapid assay to assess bacterial adhesion on textiles. Materials 9:249. doi: 10.3390/ma9040249
Vanderlinde, E. M., Harrison, J. J., Muszynski, A., Carlson, R. W., Turner, R. J., and Yost, C. K. (2010). Identification of a novel ABC transporter required for desiccation tolerance, and biofilm formation in Rhizobium leguminosarum bv. viciae 3841. FEMS Microbiol. Ecol. 71, 327–340. doi: 10.1111/j.1574-6941.2009.00824.x
Vianney, A., Jubelin, G., Renault, S., Dorel, C., Lejeune, P., and Lazzaroni, J. C. (2005). Escherichia coli tol and rcs genes participate in the complex network affecting curli synthesis. Microbiology 151(Pt 7), 2487–2497. doi: 10.1099/mic.0.27913-0
Whiteley, M., Bangera, M. G., Bumgarner, R. E., Parsek, M. R., Teitzel, G. M., Lory, S., et al. (2001). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864. doi: 10.1038/35101627
Wood, P. J. (1980). Specificity in the interaction of direct dyes with polysaccharides. Carbohy. Res. 85, 271–287. doi: 10.1016/S0008-6215(00)84676-5
Wood, T. K., Gonzalez Barrios, A. F., Herzberg, M., and Lee, J. (2006). Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 72, 361–367. doi: 10.1007/s00253-005-0263-8
Zhu, X., Long, F., Chen, Y., Knochel, S., She, Q., and Shi, X. (2008). A putative ABC transporter is involved in negative regulation of biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 74, 7675–7683. doi: 10.1128/AEM.01229-08
Zogaj, X., Bokranz, W., Nimtz, M., and Römling, U. (2003). Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71, 4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003
Keywords: antimicrobial resistance, biofilm, ocular, E. coli, endophthalmitis, mutation
Citation: Ranjith K, Ramchiary J, Prakash JSS, Arunasri K, Sharma S and Shivaji S (2019) Gene Targets in Ocular Pathogenic Escherichia coli for Mitigation of Biofilm Formation to Overcome Antibiotic Resistance. Front. Microbiol. 10:1308. doi: 10.3389/fmicb.2019.01308
Received: 24 December 2018; Accepted: 27 May 2019;
Published: 21 June 2019.
Edited by:
Antoine Andremont, Paris Diderot University, FranceReviewed by:
Yosuke Tashiro, Shizuoka University, JapanCopyright © 2019 Ranjith, Ramchiary, Prakash, Arunasri, Sharma and Shivaji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sisinthy Shivaji, c2hpdmFzQGx2cGVpLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.