- 1Department of Civil and Environmental Engineering, Korea Advanced Institute of Science and Technology, Daejeon, South Korea
- 2Department of Biological Sciences, Virginia Institute of Marine Science, College of William and Mary, Gloucester Point, VA, United States
Salinity and pH have direct and indirect impacts on the growth and metabolic activities of microorganisms. In this study, the effects of salt and alkaline stresses on the kinetic balance between nitrous oxide (N2O) production and consumption in the denitrification pathway of Dechloromonas aromatica strain RCB were examined. N2O accumulated transiently only in insignificant amounts at low salinity (≤0.5% NaCl) and circumneutral pH (7.0 and 7.5). As compared to these control conditions, incubation at 0.7% salinity resulted in substantially longer lag phase and slower growth rate, along with the increase in the amounts of transiently accumulated N2O (15.8 ± 2.8 μmoles N2O-N/vessel). Incubation at pH 8.0 severely inhibited growth and resulted in permanent accumulation of 29.9 ± 1.3 μmoles N2O-N/vessel from reduction of 151 ± 20 μmoles NO3−/vessel. Monitoring of temporal changes in nirS1, nirS2, and nosZ transcription suggested that the nosZ/(nirS1+nirS2) ratios were indicative of whether N2O was produced or consumed at the time points where measurements were taken. The salt and alkaline stresses altered the N2O consumption kinetics of the resting D. aromatica cells with expressed nitrous oxide reductases. The N2O consumption rates of the cells subjected to the salt and alkaline stress conditions were significantly reduced from 0.84 ± 0.007 μmoles min−1 mg protein−1 of the control to 0.27 ± 0.02 μmoles min−1 mg protein−1 and 0.31 ± 0.03 μmoles min−1 mg protein−1, respectively, when the initial dissolved N2O concentration was 0.1 mM. As the rates of N2O production from NO2− reduction was not significantly affected by the stresses (0.45–0.55 μmoles min−1 mg protein−1), the N2O consumption rate was lower than the N2O production rate at the stress conditions, but not at the control condition. These results clearly indicate that the altered kinetics of expressed nitrous oxide reductase and the resultant disruption of kinetic balance between N2O production and consumption was another cause of enhanced N2O emission observed under the salt and alkaline stress conditions. These findings suggest that canonical denitrifiers may become a significant N2O source when faced with abrupt environmental changes.
Introduction
Nitrous oxide (N2O), currently constituting 320 ppbv of the atmosphere, is a potent greenhouse gas with approximately 300 times higher global warming potential than carbon dioxide and the most influential ozone depletion agent (Ravishankara et al., 2009; Montzka et al., 2011). Due to the ever increasing global demand for food production, which inevitably accompanies increasing demands for nitrogen fertilizers, a steady increase in anthropogenic N2O emission is anticipated in the foreseeable future (Godfray et al., 2010). The occurrences of summer heat waves in the Arctic regions that have become increasingly regular in the recent years may also contribute to substantial increase in N2O emissions from thawing permafrost tundra soils, providing positive feedback to global warming (Voigt et al., 2017). Therefore, continued research efforts are warranted for identification of yet unknown N2O sources and development of environmental management strategies to minimize N2O emissions from its sources.
Unlike carbon dioxide, N2O emitted to the atmosphere is predominantly of biological origin with nitrification and denitrification as the two major contributing pathways (Ciais et al., 2013). In denitrification, the stepwise reduction of NO3− and NO2− to dinitrogen (N2) via NO and N2O, N2O may accumulate as a transient intermediate or the final product (Betlach and Tiedje, 1981; Philippot et al., 2011; Thomson et al., 2012; Lycus et al., 2017). Bacterial and fungal denitrifiers lacking N2O reductase gene, thus releasing N2O as the final product of denitrification, have been often pointed out as the major N2O producers (Philippot et al., 2011; Shoun et al., 2012). Even in the canonical denitrifiers capable of reducing NO3−/NO2− to N2, trace amounts of N2O is often transiently detected during the course of denitrification, although the magnitudes of accumulation vary even within closely related taxa (Betlach and Tiedje, 1981; Liu et al., 2013; Chang et al., 2018). Certain environmental conditions, e.g., acidic pH and low copper bioavailability, may amplify N2O production via several mechanisms of action including inhibition of Nos-type N2O reductase (NosZ) activity, reduced/delayed transcription of nosZ, and/or disrupted synthesis of functional NosZ (Bergaust et al., 2010; Liu et al., 2010; Felgate et al., 2012; Chang et al., 2018).
Several of the key enzymes catalyzing the different steps of denitrification, including NO-forming nitrite reductases (NirS and NirK) and NosZ, are intracellularly located in the periplasm, which undergoes substantial physiological alterations in response to the changes to the extracellular environment (e.g., shifts in pH or salinity) (Zumft, 1997; Sleator and Hill, 2002; Wilks and Slonczewski, 2007). Despite the possibility, apart from the stress exerted by acidic pH, the effects of environmental stressors on N2O release from denitrification of a canonical denitrifier have not yet been thoroughly investigated (Zhao et al., 2013). Periodic fluctuations or abrupt changes of pH or salinity are not unexpected in natural and built environments where denitrification occur. In activated-sludge WWTPs, fluctuations in pH within circumneutral range (6.0–8.0) are often recorded (Fang et al., 2010). Abrupt increases in soil salinity due to intrusions of seawater into fertilized coastal agricultural areas have recently become increasingly frequent as consequences of climate change (Nicholls and Cazenave, 2010). Although impacts of such rapid temporal shifts in the environmental conditions on the microbial compositions have been observed to be substantial in various ecological settings, little has been investigated regarding the physiological responses of isolates representing key functional groups of biogeochemical cycling to such rapid temporal shifts in the environmental conditions (Weber et al., 2008; Baho et al., 2012; Han et al., 2017; Ho et al., 2018).
Dechloromonas spp. and closely related taxa affiliated to the Betaproteobacteria class are often among the dominant denitrifying population in activated sludge tanks of wastewater treatment plants and are also found in abundance in soils and freshwater sediments (Zhang et al., 2011; Chourey et al., 2013; McIlroy et al., 2016). The isolates affiliated to this organismal group, when monitored for progression of denitrification at their optimal growth conditions (i.e., neutral pH, optimal temperature, and low salinity) in batch experiments, exhibited minimal transient N2O accumulation, presumably owing to their high affinity to N2O (Horn et al., 2005; Yoon et al., 2016; Chang et al., 2018; Suenaga et al., 2018). Perhaps, to maximize the energy yield from limited amounts of NO3−/NO2−, these denitrifiers may have evolved to maintain kinetic balance between production and consumption of N2O, such that loss of this energy-yielding terminal electron acceptor is minimized (Sanford et al., 2012). In this study, we have investigated N2O production from the denitrification pathway in Dechloromonas aromatica strain RCB, a model organism representing the Dechloromonas-like denitrifiers, upon incubation under salt and alkaline stresses. Whether and how these growth-limiting stresses may affect the balance between the N2O-producing and consuming steps of the denitrification pathway were examined, as this stress-induced N2O emissions may potentially account for substantial portions of N2O emissions from the environments with sudden fluctuations in pH and/or salinity.
Materials and Methods
Medium and Culturing Conditions
A phosphate-buffered minimal salt medium containing, per liter, 0.5 g NaCl, 1.64 g Na2PO4, 1.15 g KH2PO4 and 1 mL 1000X trace metal solution was used for maintenance of the D. aromatica strain RCB culture and preparation of precultures for the experiments performed in this study (Myers and Nealson, 1990; Yoon et al., 2016). The medium was boiled with N2 gas (>99.999%) sparging to remove dissolved O2. Anoxic culture bottles were prepared by dispensing 96.5-mL aliquots of the cooled medium into 160-mL serum bottles using the Hungate technique (Löffler et al., 2005). The serum bottles were then sealed with butyl rubber stoppers (Geo-Microbial Technologies, Inc., Ochelata, OK, United States) and autoclaved. Sterilized anoxic stock solutions of sodium acetate, KNO3, and NH4Cl were prepared separately and were added aseptically to the culture bottles to the target concentrations of 10, 5.0, and 0.5 mM, respectively. The aerobic controls were prepared by replacing 10% of the N2 headspace of the prepared serum bottles with O2 (>99.995%, Deokyang Co., Ulsan, South Korea), instead of adding KNO3. Filter-sterilized 200X vitamin stock solution was also added to the culture bottles (Wolin et al., 1964). The media used for the high-pH incubation experiments (at pH 7.5 and 8.0) were prepared with different Na2HPO4:KH2PO4 ratios, although the total phosphate buffer concentration was maintained constant at 20 mM. The media with NaCl concentrations higher than 0.05% (w/v) were prepared by adding appropriate volumes of 26% (w/v) NaCl solution. For each batch experiment, 0.5 mL of D. aromatica strain RCB preculture (pH 7.0, 0.05% NaCl) at the late exponential phase (OD600nm ∼ 0.12) was inoculated to the prepared media bottles. All incubations and experiments were performed in dark at 25°C with shaking at 160 rpm. The final pH was always within 0.2 of the initial value at any examined experimental condition.
Cultivation of D. aromatica Strain RCB Under Salt and Alkaline Stressed Conditions
To examine whether elevated salinity or pH has any effect on N2O accumulation during the growth of D. aromatica strain RCB on denitrification, the strain RCB cells were incubated in the media with varying NaCl concentrations (0.5, 1.0, 3.0, 5.0, 7.0, 9.0, and 12 g L−1) and pH (7.0, 7.5, 8.0, and 8.5 buffered with 20 mM phosphate). The pH of the medium was fixed to 7.0 for the salinity experiments and the NaCl concentration was fixed to 0.5 g L−1 for the pH experiments. The cell density (OD600nm) and the amounts of N2O in the vessels were monitored until the end of the reaction, denoted by decrease in the cell density. All incubation experiments were performed in triplicates. To ensure that the adverse effects of elevated NaCl concentrations and pH were not specific to the denitrification pathway, cell growths were also monitored with suboxic D. aromatica cultures with 10% O2 in the headspace at the same stress conditions.
Based on the observations in these preliminary experiments, the salt stress condition with 7.0 g L−1 NaCl and the alkaline stress condition at pH 8.0 were selected for closer examination, as the highest N2O accumulation was observed at these incubation conditions. The strain RCB cultures were incubated in the medium adjusted to the target NaCl concentration or pH. Upon each measurement time point, the headspace N2O was measured and 1 mL of the aqueous phase was withdrawn with a sterile, N2-flushed plastic syringe. After measuring the OD600nm, the aqueous sample was filtered using 0.2 μm-pore-size syringe filters (Advantec, Inc., Tokyo, Japan) and the filtrate was stored at −20°C until further analyses. At the time points representative of different growth phases, samples were withdrawn for reverse transcription quantitative polymerase chain reaction (RT-qPCR) analyses. The extracted 400-μL cell suspensions were treated with 800 μL RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany), and after centrifugation at 5,000 × g for 10 min and removal of the supernatants, the pellets were stored at −80°C until further treatment. As sample volume loss was an issue in these batch experiments, overly frequent sampling was avoided and the time intervals between sampling were maintained at approximately 48 h. Monitoring of the culture vessels was continued until the end of the reaction, denoted by at least three consecutive measurements yielding statistically indistinguishable concentrations of the nitrogen species.
In order to confirm that NO3− reduction and N2O accumulation were biologically catalyzed, killed-cell controls were prepared. For these negative controls, the precultures were autoclaved for 40 min before inoculation. The concentrations of NO3−, NO2−, and N2O− were measured before and after 400 h of incubation to confirm that no abiotic reactions occurred at the examined experimental conditions. An additional control experiment was performed for the alkaline stress condition (pH 8.0) with 10% v/v C2H2 in the headspace to inhibit N2O reduction to N2, as the comparison between the amounts of N2O accumulated in samples with and without C2H2 would provide an additional evidence of N2O reductase activity in the unamended samples (Yoon et al., 2015a).
Kinetic Experiments With Non-growing Cells to Measure N2O Production and Consumption Upon Induced Salt and Alkalinity Stress Conditions
The kinetics of N2O production from NO2− reduction and N2O consumption were examined at the salt stress (7.0 g L−1 NaCl) and alkaline stress (pH 8.0) conditions with the non-growing D. aromatica strain RCB cells cultivated at the control conditions. All kinetics experiments were performed in triplicate with 100-mL D. aromatica strain RCB cultures in 160-mL serum bottles with N2 headspace. The strain RCB cultures were initially incubated at the control condition with 10 mM acetate and 5 mM NO3− as the electron donor and electron acceptor, respectively. Immediately after depletion of the initially added NO3− and the intermediates (NO2− and N2O), chloramphenicol (Sigma Aldrich, St. Louis, MO, United States) was added to the concentration of 35 μg mL−1 to inhibit de novo protein synthesis (Maalej et al., 2004). The absence of growth and linear N2O consumption in chloramphenicol-treated D. aromatica cells verified the effectiveness of chloramphenicol at the concentration tested (data not shown). The salt stress samples were prepared by adjusting the dissolved NaCl concentrations to 7.0 g L−1 with degassed 260 g L−1 NaCl brine, and the pH stress samples by adjusting the pH to 8.0 with degassed 0.5 M NaOH solution. An 1.0-mL aliquot was withdrawn from each vessel and stored at −20°C for determination of the total protein concentration.
Four sets of experiments were performed with these chloramphenicol-treated cells to determine the N2O production rates from NO2− reduction and N2O consumption rates at two different initial dissolved concentrations. For determination of the N2O production rates, NaNO2 was added to the chloramphenicol-treated cells to the concentration of 0.1 mM or 1.0 mM, and 6.0 mL of the N2 headspace was replaced with C2H2 gas (>99.9%, Taekyung Eco Co., Ansan, South Korea) to inhibit N2O reduction (Yoon et al., 2015a). The culture vessels for measurement of N2O consumption rates were prepared by adding 2.4 mL of autoclaved 99.999% N2O and 10% v/v N2O in N2 to the headspace to yield the target aqueous N2O concentrations of 1.0 and 0.1 mM, respectively. The increase or decrease in N2O concentrations in these vessels were monitored until the changes in the concentrations were no longer linear, and the reaction rates were determined from linear regression of the N2O production or consumption curves. These N2O production and consumption rates were normalized with total protein concentration determined with the Bradford protein assay using Bio-Rad Protein Assay Kit II (Hercules, CA, United States) according to the instruction provided by the manufacturer.
Analytical Procedures
Aqueous concentrations of NO3-N and NO2−-N were determined with the spectrophotometric detection method using the Griess reagents as previously described (Miranda et al., 2001). Vanadium chloride (VCl3) was used as the reducing agent to reduce NO3− to NO2−. The absorbance at 540 nm was measured using a Sunrise absorbance microplate reader (Tecan, Männedorf, Switzerland). The headspace N2O concentrations were monitored using a HP6890 series gas chromatograph equipped with an HP-PLOT/Q column and electron capture detector (Agilent, Palo Alto, CA, United States). For each measurement, 200 μL of the headspace gas was withdrawn and manually injected into the gas chromatograph with a 1700 series gas-tight syringe (Hamilton Company, Reno, NV, United States). The total amount of N2O in a culture vessel was calculated from the headspace concentration with the Henry’s constant adjusted for the salinity of the medium, as previously described (Schumpe et al., 1982; Schumpe, 1993; Sander, 2015; Yoon et al., 2015b). The cell density was determined by measuring the absorbance at 600 nm with a spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
Reverse Transcription Quantitative PCR (RT-qPCR) Analyses
The transcription profiles of the genes encoding the enzymes involved in denitrification at different incubation conditions were analyzed using RT-qPCR analyses performed as previously described (Supplementary Table S1; Yoon et al., 2015a). The RNAprotect-treated samples stored at −80°C were thawed on ice and 1-μL solution with 1010 copies of luciferase control mRNA (Promega, Madison, WI, United States) was added to each sample tube. Total RNA was extracted according to the protocol provided with the RNeasy Mini Kit (Qiagen). The total RNA collected in the column was eluted twice with 30 μL RNase-free water, and the resulting eluent was treated with RNase-free DNAse Set (Qiagen) and subsequently purified with RNeasy MinElute Kit (Qiagen). Of the 20 μL eluent, 10 μL was subjected to reverse transcription using SuperscriptTM III Reverse Transcriptase (Invitrogen, Carlsbad, CA, United States) as previously described (Yoon et al., 2015a). The rest of the eluent was stored at −20°C and later used for confirmation of absence of DNA in the purified RNA samples. After reverse transcription, 1 μL of RNase H (Invitrogen) was added to each reaction mixture, which was then incubated at 37°C for 20 min. The cDNA solution was diluted 5-fold with nuclease-free water and stored at −20°C.
The primer sets used for the RT-qPCR analyses were designed de novo from the genome sequence of D. aromatica strain RCB available at the NCBI genome database (accession number: CP000089.1) using Primer3 software (Rozen and Skaletsky, 2000). The primers targeting nirS1 (Daro_3274), nirS2 (Daro_3323), norB (Daro_3191; the gene encoding nitric oxide reductase), nosZ (Daro_1575), and recA (Daro_4152; the single-copy housekeeping gene encoding recombinase A) were designed and the calibration curves were constructed with the dilution series of PCR2.1 plasmids (Invitrogen) with the amplicons inserted (Supplementary Table S1). Amplification was performed with QuantStudioTM 3 Real-Time PCR System (Thermo Fisher Scientific) using SYBR Green detection chemistry. The remaining eluent stored after the DNase treatment step was analyzed along with the cDNA samples, to confirm that DNA was completely removed by the DNase treatment. The copy numbers of luc genes in the cDNA samples were quantified to ensure that the RNA recovery rates were acceptable (>10%). The transcript copy numbers of nirS1, nirS2, norB, and nosZ were normalized with the transcript copy number of recA (Park et al., 2017). The samples withdrawn from triplicate cultures were independently treated through the RNA extraction and reverse transcription steps, serving as the biological replicates of the RT-qPCR analyses. Each qPCR reaction was performed in duplicate and the average CT value of the technical replicates was taken.
Statistical Analyses
The statistical analyses in this study was performed with the R statistical software package (version 3.5.0). The statistical comparisons were performed with Kruskal–Wallis tests and subsequent pairwise statistical comparisons of subsets (n = 2) were performed with Tukey’s HSD post hoc tests. Unless otherwise mentioned, triplicate experiments were performed and the average values were presented with the standard deviations of the triplicate samples as errors.
Results
Effects of Salt or Alkaline Stress on N2O Evolution From Denitrifying D. aromatica Strain RCB
The effects of salt and alkaline stresses on growth and N2O production of D. aromatica strain RCB were monitored in anaerobic batch cultures with 10 mM acetate and 5.0 mM NO3− provided as the sole electron donor and acceptor, respectively (Figure 1, Table 1, and Supplementary Figure S1). No significant accumulation of N2O was observed in the cultures incubated with salt (NaCl) concentrations up to 5.0 g L−1 despite the lower observed growth rates. At the NaCl concentration of 7.0 g L−1, a significant increase (p < 0.05) in N2O accumulation associated with severe growth inhibition (the lag phase prolonged to ∼146 h and the growth rate diminished to 0.032 h−1) was observed. The accumulated N2O was eventually completely transformed, presumably to N2; however, the maximum amount of accumulated N2O reached 15.8 ± 2.8 μmoles N2O-N/vessel at the peak (t = 222 h). No growth was observed at 9.0 g L−1 NaCl, although 1.97 ± 0.38 μmoles N2O-N/vessel (0.40% of added NO3−) was produced and remained in the medium until the end of incubation. Although neither NO3− nor NO2− concentration was monitored in these preliminary experiments, the absence of cell growth suggested that only insignificant portion of initially added NO3− was consumed. The elevated pH also had unexpectedly pronounced impacts on N2O production and growth of D. aromatica strain RCB. At pH 8.0, the cell density reached only up to an OD600nm value of 0.018 (from the initial value of 0.001) and exponential growth was not observed. The amounts of the accumulated N2O-N in the vessels reached a maximum of 31.7 ± 2.06 μmoles and the produced N2O permanently remained in the vessels until the ends of the experiments. In the cultures adjusted to 12.0 g L−1 NaCl or pH 8.5, neither cell growth nor N2O accumulation was observed, suggesting that higher NaCl concentrations or pH are lethal to these cells. In the killed-cell controls, neither N2O accumulation nor change in NO3− concentration was observed for 400 h, confirming against the possibility of abiotic N2O production at any of the incubation conditions examined (data not shown). Growth inhibition was observed in the strain RCB cultures incubated with 10% (v/v) O2 as the electron acceptor at the elevated NaCl concentrations and pH, confirming that the salt and alkaline stresses were not specific to denitrifying cells (Table 1 and Supplementary Figure S2).
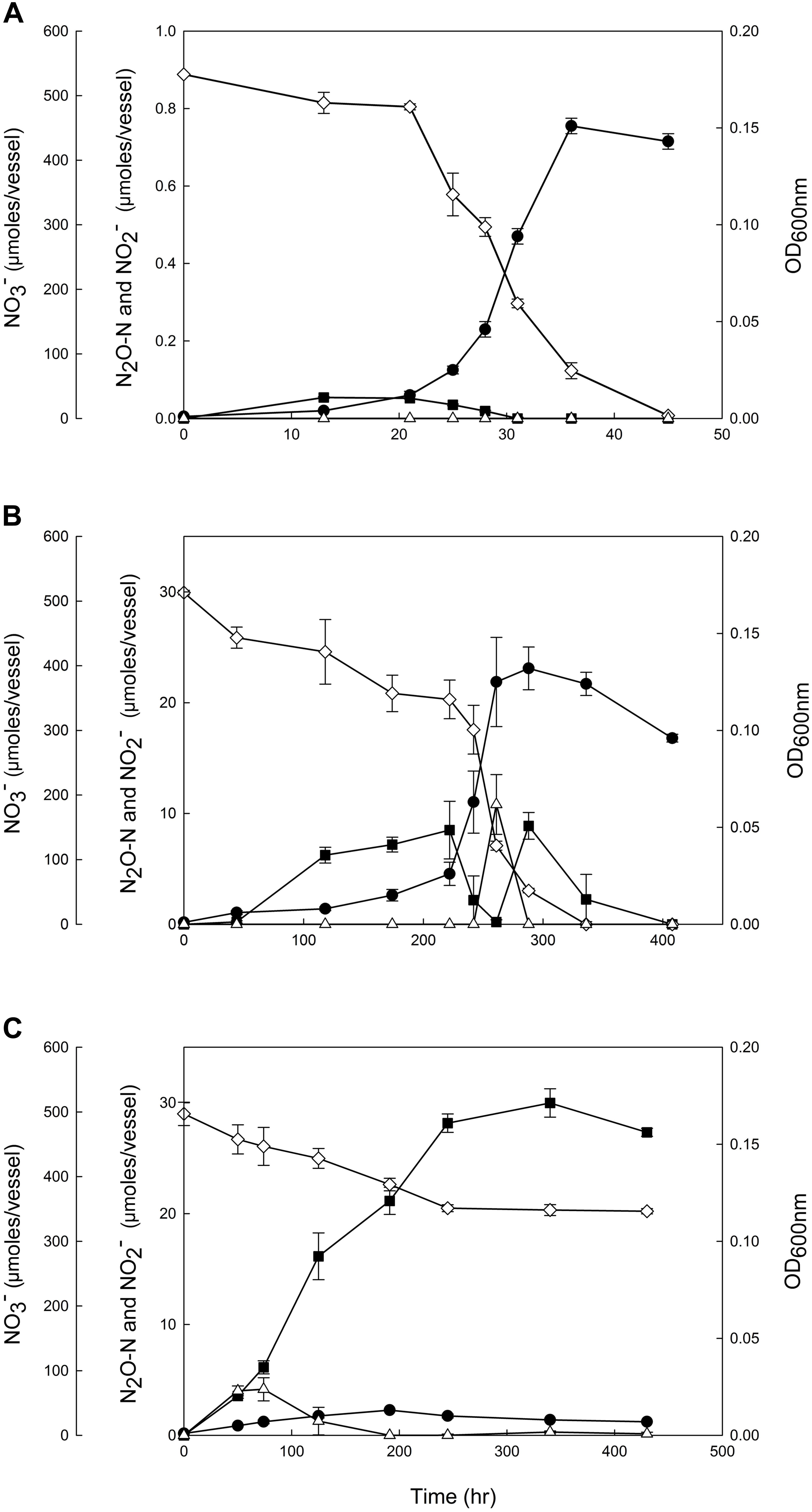
Figure 1. Reduction of 5 mM NO3− by the denitrifying Dechloromonas aromatica RCB cultures incubated at (A) the control condition (0.05% w/v NaCl, pH 7.0), (B) the salt stress condition (0.70% w/v NaCl, pH 7.0) and (C) the alkaline stress condition (0.05% w/v NaCl, pH 8.0). The amounts of NO3− ( ), NO2− (Δ), and N2O-N (
), NO2− (Δ), and N2O-N ( ) and the absorbances at 600 nm (
) and the absorbances at 600 nm ( ) were monitored until the nitrogen oxides were depleted or upon confirmation of the termination of denitrification reaction. Each reaction vessel is a 160 mL serum bottle with the initial aqueous phase volume of 100 mL. The data points represent the averages of triplicate cultures and the error bars the standard deviations of the triplicate measurements.
) were monitored until the nitrogen oxides were depleted or upon confirmation of the termination of denitrification reaction. Each reaction vessel is a 160 mL serum bottle with the initial aqueous phase volume of 100 mL. The data points represent the averages of triplicate cultures and the error bars the standard deviations of the triplicate measurements.
Based on these results, a salt stress condition (7.0 g L−1) and an alkaline stress condition (pH 8.0) were selected for closer examination, whereby the cell growth and progression of denitrification reaction were monitored in strain RCB cultures (Figure 1). In the control cultures (0.5 g L−1 of NaCl, pH 7.0), NO3− and its intermediate, NO2− and N2O, were completely reduced within 45 h. Accumulation of N2O occurred during the early phase of growth (t < 31 h), reaching the maximum of 0.054 ± 0.002 μmoles N2O-N/vessel at t = 13 h; however, this amount was merely ∼0.1% of the total amount of NO3− reduced beyond NO2−. The concentration of NO2− remained below the detection limit throughout the incubation.
In the batch cultures of D. aromatica strain RCB incubated at the salt stress condition, the lag phase was remarkably prolonged and the exponential growth did not take off until >118 h after inoculation. The exponential growth rate, 0.032 ± 0.001 h−1, was also significantly lower than that of the control, 0.218 ± 0.014 h−1 (p < 0.05). Substantial accumulation of N2O (up to 8.5 ± 2.6 μmoles N2O-N/bottle) was observed during the lag phase and the early exponential phase (44–222 h), but accumulated N2O was almost completely consumed (to 0.19 ± 0.018 h−1) during the delayed exponential growth phase. Unexpected N2O accumulation was observed in the early stationary phase (up to 8.9 ± 2.6 μmoles N2O-N/bottle) before N2O was eventually consumed after depletion of NO3−. A modest transient accumulation of NO2− (up to 10.8 ± 2.7 μmoles /bottle) observed toward the end of the exponential growth phase (t = 261 h) suggested a temporal imbalance between NO3−-to-NO2− reduction and NO2− reduction.
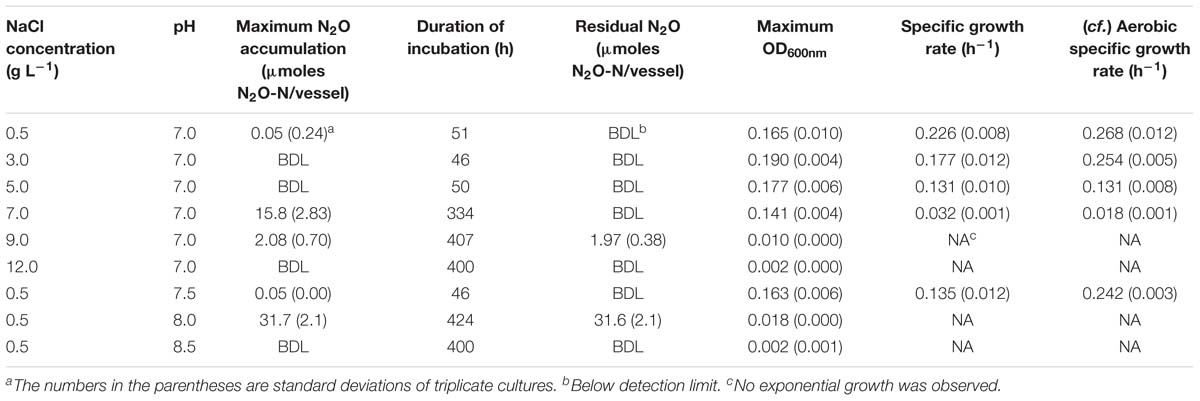
Table 1. N2O-N accumulation and cell growth in denitrifying D. aromatica strain RCB cultures grown on 10 mM acetate and 5 mM NO3− at varying NaCl concentrations and pH.
The growth of D. aromatica strain RCB was substantially inhibited at the alkaline stress condition (pH 8.0). Despite the absence of discernable exponential growth phase and the low cell density that was limited to a maximum of OD600nm = 0.013 ± 0.001, NO3− concentration decreased from 497.1 ± 1.0 to 346.6 ± 4.0 μmoles NO3−/bottle over 250 h of incubation. Accumulation of NO2− remained modest <4.16 ± 1.0 μmoles NO2−/bottle and was sustained only at the initial phase of incubation (<191 h). A significantly larger fraction of reduced NO3− (18.7 ± 2.4%) was recovered as N2O at the end of incubation than any other conditions examined; however, the reaction stoichiometry suggested against the possibility that the N2O reductase was completely inactivated. Indeed, significantly larger amounts of N2O (262.2 ± 5.0 μmoles N2O-N/vessel) accumulated in the strain RCB cultures amended with acetylene (C2H2), an inhibitor of N2O reductase, at the same incubation condition (Supplementary Figure S3). This discrepancy between the amounts of accumulated N2O in the samples with and without C2H2 amendment supported the presence of N2O reductase activity at pH 8.0. The near perfect nitrogen mass balance in the C2H2-amended cultures also indicated that NO accumulation was negligible. In contrast to the other incubation conditions, denitrification activity and cell growth were terminated after 245 h (336 h in the cultures amended with C2H2), as indicated by the stable NO3− and N2O concentrations and decreasing cell density, presumably due to the modest increase in the pH to ∼8.2 (data not shown).
RT-qPCR Analyses of nirS, norB, and nosZ Transcription
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analyses were performed to examine whether the increased transient or permanent accumulation of N2O at the stress conditions can be explained in terms of transcription of the genes encoding the enzymes of the denitrification cascade. The nirS1, nirS2, norB, nosZ, and recA (a single-copy housekeeping gene used for normalization of the RT-qPCR data) transcripts were quantified with the samples collected from the control, salt-stressed, and alkaline-stressed cultures at different stages of growth (Figure 2). The two copies of nirS genes, with 59% translated amino acid identity, exhibited distinct transcriptional patterns (Jones et al., 2008). Transcription of nirS1 appeared to be inducible by nitrogen oxides, whereas transcription of nirS2 was maintained relatively constant throughout the incubation at all three experimental conditions. In the samples taken during the exponential growth phases (t = 31 h for the control cultures and t = 288 h for the salt-stress cultures), the nirS1 transcription levels were at least 4.8-fold higher than those of nirS2. Both nirS1 and nirS2 were highly expressed at pH 8.0 throughout the incubation periods, as compared to other incubation conditions, despite severely inhibited cell growth (Figure 2).
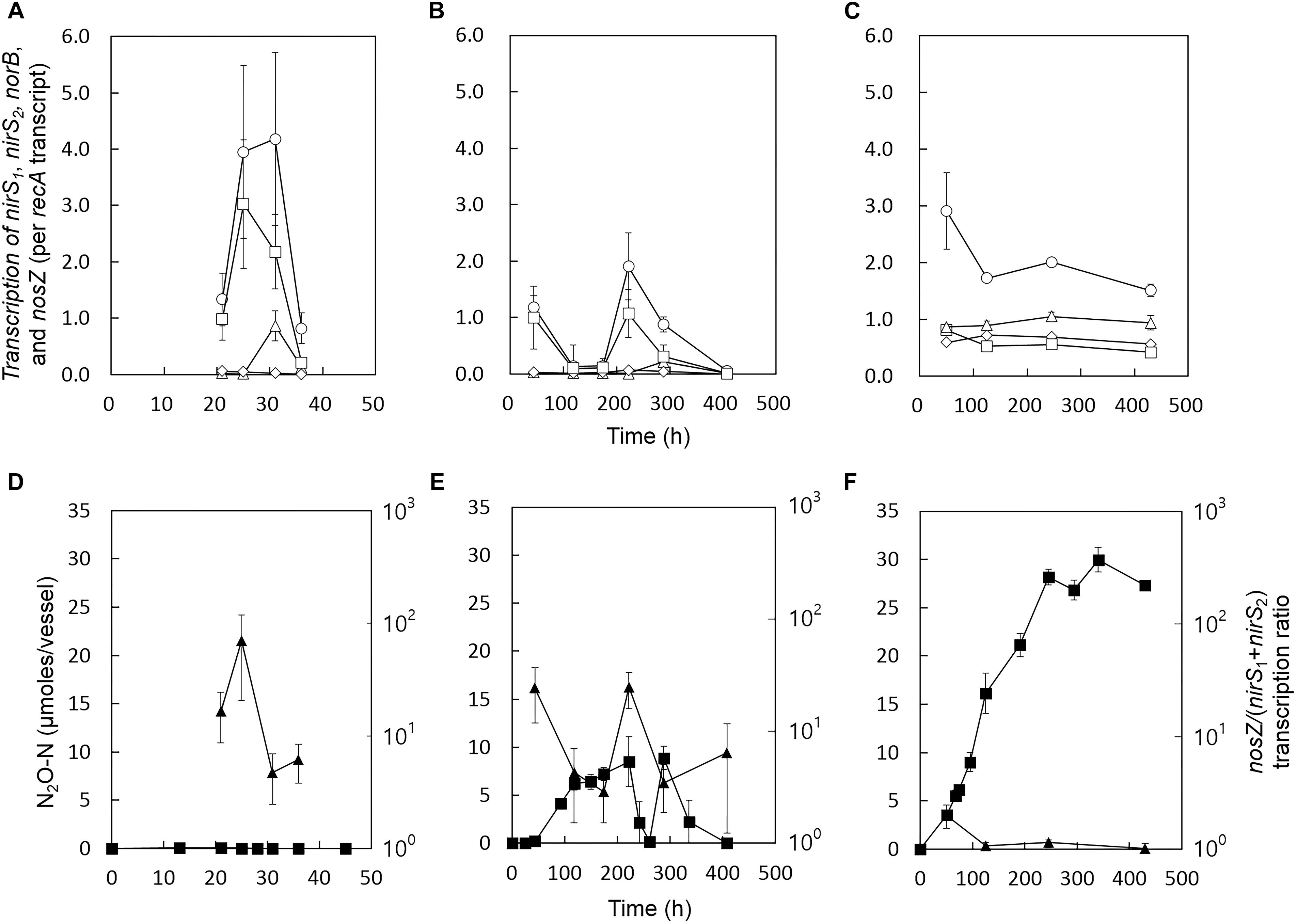
Figure 2. Transcription profiles of nirS1 (Δ), nirS2 ( ), norB (
), norB ( ), and nosZ (
), and nosZ ( ) in the D. aromatica strain RCB cultures incubated at (A) the control condition, (B) the salt stress condition, and (C) the alkaline stress condition. The samples for the RT-qPCR analyses were withdrawn from the same culture vessels used to construct Figure 1. The transcript copy numbers of the denitrification genes were normalized with the copy numbers of recA transcripts. The data points represent the averages of three biological replicates processed independently through RNA extraction and reverse transcription procedures. The error bars represent the standard deviations of the triplicate samples. The values of nosZ/(nirS1+nirS2) (
) in the D. aromatica strain RCB cultures incubated at (A) the control condition, (B) the salt stress condition, and (C) the alkaline stress condition. The samples for the RT-qPCR analyses were withdrawn from the same culture vessels used to construct Figure 1. The transcript copy numbers of the denitrification genes were normalized with the copy numbers of recA transcripts. The data points represent the averages of three biological replicates processed independently through RNA extraction and reverse transcription procedures. The error bars represent the standard deviations of the triplicate samples. The values of nosZ/(nirS1+nirS2) ( ) calculated with the nirS1, nirS2, and nosZ transcription data in panels (A–C) are presented in panels (D–F), respectively. The errors were calculated using the propagation of error. The amounts of N2O (
) calculated with the nirS1, nirS2, and nosZ transcription data in panels (A–C) are presented in panels (D–F), respectively. The errors were calculated using the propagation of error. The amounts of N2O ( ) are included for convenient comparison between the datasets.
) are included for convenient comparison between the datasets.
At all experimental conditions, nosZ was always the most highly transcribed of the denitrification genes. In the control and the salt-stressed samples, nosZ transcription fluctuated during the course of incubation, with increased transcription observed during the periods of active denitrification (characterized by NO3− reduction and/or N2O production) and down-regulated transcription upon depletion of the nitrogen oxides. In the salt stress samples, the period of down-regulated nosZ transcription (t = 118–174 h) coincided with the period of N2O accumulation. The peak nosZ/(nirS1+nirS2) value was observed at 222 h, consistent with the net N2O consumption observed between 222 and 261 h. In the alkaline stress samples, nosZ transcription was sustained relatively constant throughout the incubation period. The nosZ/(nirS1+nirS2) transcription ratios were consistently sustained relatively low (<2.0), agreeing with persistent N2O accumulation observed at this incubation condition. The transcription of norB generally followed the same trends as the transcription of nosZ, suggesting that the expressions of these two genes may be co-regulated in strain RCB. The nosZ/norB transcription ratios did not exhibit any evident relation with N2O production (Supplementary Figure S4). Although the periods with low nosZ/(nirS1+ nirS2) transcription ratios generally agreed with the periods of the net N2O production, the result from the transcription analysis can only serve as circumstantial evidence.
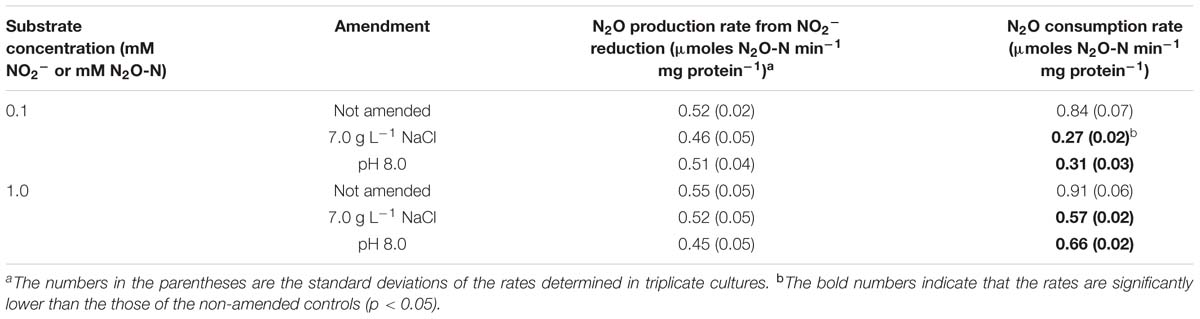
Table 2. N2O production and consumption rates of chloroamphenicol-treated D. aromatica strain RCB cells amended with NaCl (to 0.7% w/v) or NaOH (to pH 8.0).
NO2− and N2O Reduction Kinetics of Chloramphenicol-Treated D. aromatica Strain RCB Cells Under Imposed Salt and Alkaline Stress Conditions
A set of kinetic experiments were performed to examine the effects of high salt concentration and alkaline pH on the activities of N2O-producing and reducing reactions mediated by D. aromatica strain RCB (Table 2). The rates of N2O production from NO2− reduction were statistically indistinct across the chloroamphenicol-treated cells with or without salt or pH amendments (p > 0.05). The substrate concentration did not have significant effect on the N2O production rates, suggesting that the whole-cell half-saturation constant (Km,app) for NO2− reduction is substantially lower than 0.1 mM at all examined conditions. Contrastingly, the rates of N2O consumption were significantly lower at both stress conditions as compared to the control condition (p < 0.05). The potential N2O consumption rates measured with the control cultures were statistically indistinct at the two different dissolved N2O concentrations (1.0 and 0.1 mM) (p > 0.05). These rates were significantly higher than the measured N2O production rates (p < 0.05), supporting the near absence of transient N2O accumulation in the control cultures. In both salt- and pH-stressed cultures, the N2O consumption rates were significantly lower for the cultures amended with 0.1 mM N2O than those amended with 1.0 mM N2O (p < 0.05). While the N2O consumption rates were still higher than the N2O production rates at 1.0 mM, the consumption rates at 0.1 mM were significantly lower than the N2O production rates (p < 0.05). Although the Michaelis–Menten kinetics parameters were not determined, these observations suggest that the reaction kinetics of the N2O reductase, but not NO2− reductase, in D. aromatica strain RCB was substantially altered by both salt and alkaline stresses, such that the affinity of N2O reductase was substantially lowered in the stressed cells. As the concentration of N2O was sustained below 0.3 mM throughout the entire incubation periods at all three incubation conditions (Figure 1), the alteration of the N2O consumption kinetics in the salt- and pH-amended cultures provides additional, more succinct, mechanistic explanation for observed N2O accumulation.
Discussion
Viability and growth of microorganisms are largely dependent on environmental parameters such as pH and salinity (Sleator and Hill, 2002; Padan et al., 2005). The growth of non-halophilic neutrophilic denitrifier, D. aromatica strain RCB, was severely inhibited by NaCl concentrations and pH equal to or higher than 7.0 g L−1 and 8.0, respectively. Accumulation of N2O in substantial amounts (>1% of initially added NO3−) occurred only when strain RCB was grown at these salt and alkaline stress conditions. Transient or permanent N2O accumulation during denitrification under acid stress has been repeatedly observed with typical laboratory strains of canonical neutrophilic denitrifiers incubated at slightly acidic pH (∼6.0) (Bergaust et al., 2010; Kim et al., 2017). Both Paracoccus denitrificans and Shewanella loihica exhibited longer lag periods and slower metabolic turnover of NO3− at pH 6.0 than at their optimal pH-neutral condition and a significant increase in N2O accumulation was accompanied with the growth inhibition. The observations in this study clearly show that salt and alkaline stress may also lead to elevated N2O production in canonical denitrifiers.
The mechanisms underpinning the increased N2O production in stressed D. aromatica strain RCB cultures were (1) alteration in the expression of the genes encoding different steps of the denitrification pathway and (2) altered kinetic balance between N2O-producing and consuming reactions presumably due to the reduced affinity of the N2O reductase in stressed cells. In the previous experiments that examined denitrification at acidic pH, the transcription of nosZ and other denitrification genes were largely unaffected, as compared to the optimal growth conditions (Liu et al., 2014; Kim et al., 2017). Contrastingly, this study demonstrated that the nosZ/(nirS1+nirS2) transcription ratios were significantly lower at both alkaline and salt stress conditions at the time points where N2O accumulation was observed, suggesting that the altered transcription ratios may have been one of the causes for the disrupted balance between N2O production and consumption. In interpreting the presented data, the nosZ-to-nirS transcription ratio of 1:1 needs not be directly translated to 1:1 activity ratio for NO2− reduction and N2O reduction, as the reaction kinetics for the expressed enzymes may differ for NirS and NosZ (Wei et al., 2015). Nonetheless, the nosZ/(nirS1+nirS2) transcription ratios below 4.3 invariably overlapped with the incremental increases in the amounts of N2O. In the salt stress cultures, the recovery of nosZ/(nirS1+nirS2) transcription ratios in the mid-exponential growth phase to the values comparable to the level observed in exponential growth phase of the control experiment explained the temporal decline in the amounts of N2O in the vessels. The consistently low nosZ/(nirS1+nirS2) transcription ratios measured during the incubation of D. aromatica at pH 8.0 was also in agreement with the permanent N2O accumulation observed at this condition.
The results of the enzyme kinetics experiments with chloroamphenicol-treated cells indicated that the altered kinetic balance in the stressed D. aromatica cells lends another mechanistic explanation for elevated net N2O production in the pH- and salinity-stressed cells. The extent to which N2O accumulates in denitrification reaction is dependent on the difference between the rates of its production and consumption (Betlach and Tiedje, 1981). The recent investigations on the kinetics of N2O consumption by the nosZ-harboring organisms have revealed broadly varying kinetics among the different groups of N2O-reducing organisms (Yoon et al., 2016; Conthe et al., 2018; Suenaga et al., 2018). The minimal N2O accumulation observed in the previous observations of denitrifying isolates or enrichments suggest that the rates of N2O production are often surpassed by the rates of N2O consumption at non-acidic pH (Betlach and Tiedje, 1981; Liu et al., 2010; Chang et al., 2018). Consistent with these previous observations and the denitrification progression curve of the D. aromatica strain RCB control culture (Figure 1A), the rate of N2O consumption was significantly faster than the rate of N2O production in the chloramphenicol-treated cells at both high (1.0 mM NO2− or N2O) and low (0.1 mM NO2− or N2O) concentrations of the substrates. While the rates of N2O production were unaffected by the salt or alkaline stress, the rates of N2O consumption were severely affected. That the decreases in N2O-reducing rates were much more pronounced at the low substrate concentration suggested substantially reduced affinity of NosZ to N2O in the stressed cells. Such alterations in the whole-cell substrate utilization kinetics due to changes in the medium pH and salinity have been previously observed with ammonia oxidation in Nitrosomonas europaea and nitrite oxidation in Nitrobacter agilis (Hunik et al., 1992, 1993). The different responses of the NO2− and N2O reduction kinetics to the salt and alkaline stresses well explain the observed accumulation of N2O, as the rate of N2O production would exceed the rate of its consumption at the concentration ranges of NO2− and N2O relevant to the denitrifying strain RCB cultures.
Whether the increased net N2O production upon sudden exposure to the salt and alkaline stress is limited to the organisms closely related to D. aromatica or is widespread physiology among denitrifiers is yet unknown. The conflict with the previous reports regarding the effect of alkalic pH on N2O production from denitrification suggest that these stress-induced enhancement of N2O emissions may be limited to the neutrophilic, non-halophilic betaproteobacterial denitrifiers with nirS and clade II nosZ closely affiliated to those of D. aromatica (Šimek and Cooper, 2002; Liu et al., 2014; Kim et al., 2017). Even with a conservative extrapolation confining this physiological behavior to this specific organismal group, the findings in this study still have substantial implications to environmental N2O emissions. Dechloromonas spp. and closely affiliated organismal groups in the Rhodocyclaceae family, e.g., Azospira spp., have been often identified as one of the dominant denitrifier groups in soil and freshwater environments (Wei et al., 2015; Coyotzi et al., 2017; Bellini et al., 2018). Recent studies using high-throughput sequencing for microbial community profiling and metagenome analyses have revealed the abundance of the Rhodocyclaceae family of denitrifiers in estuarine environments susceptible to periodic seawater intrusions (Liu et al., 2015; Guo et al., 2017). As estuarine environments are often enriched in reactive nitrogen due to the run-off from neighboring agricultural soils, events of seawater intrusion may result in significant N2O emissions from denitrification by the Dechloromonas-like denitrifiers exposed to abrupt salt stress. Due to climate change that will undoubtedly bring about sea level rise, larger area of estuarine deltas will be affected by seawater intrusions, and thus, the N2O emission mechanism examined in this study may provide significant positive feedback to global warming (Nicholls and Cazenave, 2010).
Dechloromonas spp. and closely related organisms are also often the dominant denitrifiers in activated-sludge WWTPs, another important source of N2O (Zhang et al., 2011; McIlroy et al., 2016). Although pH of the sewers in the activated sludge tanks are relatively well maintained within the circumneutral range, temporal increases in pH to 8.0 due to, for example, shock loads of alkaline influent wastewater are not rarely observed (Fang et al., 2010). Caustic dosing, often used for mitigation of sewer biofilms, may also cause abrupt alkalization of activated sludge tanks (Gutierrez et al., 2014). Such alkaline stress suddenly exerted on the established population of Dechloromonas-like denitrifiers may cause an imbalance between production and consumption of N2O and result in substantially increased N2O emissions. This probable enhancement of N2O production upon alkaline shock may be one of the causes of highly fluctuating nature of N2O emissions from WWTPs (Kosonen et al., 2016; Thaler et al., 2017). In future research, a long-term investigation of the correlations between N2O emissions and fluctuating pH and expressions and activities of NirS/NirK and NosZ in an activated sludge tank may provide additional insights into N2O emission mechanism in activated sludge WWTPs.
Environmental microbial communities are complex and dynamic systems consisting of actively interacting and competing microorganisms. The laboratory pure culture experiments with a single Dechloromonas strain may not be able to account for the N2O emissions associated with community dynamics, wherein halotolerant/halophilic or alkaliphilic/alkalitolerant denitrifiers may rapidly overtake the denitrifying community as a response to imposed salt or alkaline stress conditions. The overall N2O emissions from denitrification may be defined by such microbial community shifts, as well as the stress responses of the existing denitrifier communities. Therefore, investigation of the microbial community dynamics and associated changes to N2O emission patterns in complex environmental samples would be warranted for a more complete picture of the N2O dynamics in the environments affected by sudden changes to their physicochemical properties.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
HH performed the experiments and analyzed data. MS and HH carried out the statistical analyses. SY designed the research direction and experiments. SY, BS, and HH wrote the manuscript. All authors discussed the results.
Funding
This work was supported by “the R&D Center for reduction of Non-CO2 Greenhouse Gases (Grant No. 2017002420002)” funded by Korea Ministry of Environment (MOE) and National Research Foundation (Grant No. 2018H1D3A2063734).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01203/full#supplementary-material
References
Baho, D. L., Peter, H., and Tranvik, L. J. (2012). Resistance and resilience of microbial communities – temporal and spatial insurance against perturbations. Environ. Microbiol. 14, 2283–2292. doi: 10.1111/j.1462-2920.2012.02754.x
Bellini, M. I., Kumaresan, D., Tarlera, S., Murrell, J. C., and Fernández-Scavino, A. (2018). Identification of active denitrifiers by DNA-stable isotope probing and amplicon sequencing reveals Betaproteobacteria as responsible for attenuation of nitrate contamination in a low impacted aquifer. FEMS Microbiol. Ecol. 94:fix181. doi: 10.1093/femsec/fix181
Bergaust, L., Mao, Y., Bakken, L., and Frostegard, A. (2010). Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrous oxide reductase in Paracoccus denitrificans. Appl. Environ. Microbiol. 76, 6387–6396. doi: 10.1128/AEM.00608-10
Betlach, M. R., and Tiedje, J. M. (1981). Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Appl. Environ. Microbiol. 42, 1074–1084.
Chang, J., Gu, W., Park, D., Semrau, J. D., DiSpirito, A. A., and Yoon, S. (2018). Methanobactin from Methylosinus trichosporium OB3b inhibits N2O reduction in denitrifiers. ISME J. 12, 2086–2089. doi: 10.1038/s41396-017-0022-8
Chourey, K., Nissen, S., Vishnivetskaya, T., Shah, M., Pfiffner, S., Hettich, R. L., et al. (2013). Environmental proteomics reveals early microbial community responses to biostimulation at a uranium- and nitrate-contaminated site. Proteomics 13, 2921–2930. doi: 10.1002/pmic.201300155
Ciais, P., Sabine, C., Bala, G., Bopp, L., Brovkin, V., Canadell, J., et al. (2013). “Carbon and other biogeochemical cycles,” in Climate Change 2013: The Physical Science Basis Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds T. F. Stocker, D. Qin, G.-K. Plattner, M. Tignor, S. K. Allen, J. Boschung, et al. (Cambridge: Cambridge University Press), 465–570.
Conthe, M., Wittorf, L., Kuenen, J. G., Kleerebezem, R., van Loosdrecht, M. C. M., and Hallin, S. (2018). Life on N2O: deciphering the ecophysiology of N2O respiring bacterial communities in a continuous culture. ISME J. 12, 1142–1153. doi: 10.1038/s41396-018-0063-7
Coyotzi, S., Doxey, A. C., Clark, I. D., Lapen, D. R., van Cappellen, P., and Neufeld, J. D. (2017). Agricultural soil denitrifiers possess extensive nitrite reductase gene diversity. Environ. Microbiol. 19, 1189–1208. doi: 10.1111/1462-2920.13643
Fang, F., Ni, B.-J., Xie, W.-M., Sheng, G.-P., Liu, S.-G., Tong, Z.-H., et al. (2010). An integrated dynamic model for simulating a full-scale municipal wastewater treatment plant under fluctuating conditions. Chem. Eng. J. 160, 522–529. doi: 10.1016/j.cej.2010.03.063
Felgate, H., Giannopoulos, G., Sullivan, M. J., Gates, A. J., Clarke, T. A., Baggs, E., et al. (2012). The impact of copper, nitrate and carbon status on the emission of nitrous oxide by two species of bacteria with biochemically distinct denitrification pathways. Environ. Microbiol. 14, 1788–1800. doi: 10.1111/j.1462-2920.2012.02789.x
Godfray, H. C. J., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science 327, 812–818. doi: 10.1126/science.1185383
Guo, X.-P., Niu, Z.-S., Lu, D.-P., Feng, J.-N., Chen, Y.-R., Tou, F.-Y., et al. (2017). Bacterial community structure in the intertidal biofilm along the Yangtze Estuary. China. Mar. Pollut. Bull. 124, 314–320. doi: 10.1016/j.marpolbul.2017.07.051
Gutierrez, O., Sudarjanto, G., Ren, G., Ganigué, R., Jiang, G., and Yuan, Z. (2014). Assessment of pH shock as a method for controlling sulfide and methane formation in pressure main sewer systems. Water Res. 48, 569–578. doi: 10.1016/j.watres.2013.10.021
Han, D., Link, H., and Liesack, W. (2017). Response of Methylocystis sp. strain SC2 to salt stress: physiology, global transcriptome, and amino acid profiles. Appl. Environ. Microbiol. 83:e00866-17. doi: 10.1128/aem.00866-17
Ho, A., Mo, Y., Lee, H. J., Sauheitl, L., Jia, Z., and Horn, M. A. (2018). Effect of salt stress on aerobic methane oxidation and associated methanotrophs; a microcosm study of a natural community from a non-saline environment. Soil Biol. Biochem. 125, 210–214. doi: 10.1016/j.soilbio.2018.07.013
Horn, M. A., Ihssen, J., Matthies, C., Schramm, A., Acker, G., and Drake, H. L. (2005). Dechloromonas denitrificans sp. nov., Flavobacterium denitrificans sp. nov., Paenibacillus anaericanus sp. nov. and Paenibacillus terrae strain MH72, N2O-producing bacteria isolated from the gut of the earthworm Aporrectodea caliginosa. Int. J. Syst. Evol. Microbiol. 55, 1255–1265. doi: 10.1099/ijs.0.63484-0
Hunik, J., Meijer, H., and Tramper, J. (1992). Kinetics of Nitrosomonas europaea at extreme substrate, product and salt concentrations. Appl. Microbiol. Biotechnol. 37, 802–807. doi: 10.1007/BF00174849
Hunik, J., Meijer, H., and Tramper, J. (1993). Kinetics of Nitrobacter agilis at extreme substrate, product and salt concentrations. Appl. Microbiol. Biotechnol. 40, 442–448. doi: 10.1007/BF00170408
Jones, C. M., Stres, B., Rosenquist, M., and Hallin, S. (2008). Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25, 1955–1966. doi: 10.1093/molbev/msn146
Kim, H., Park, D., and Yoon, S. (2017). pH control enables simultaneous enhancement of nitrogen retention and N2O reduction in Shewanella loihica strain PV-4. Front. Microbiol. 8:1820. doi: 10.3389/fmicb.2017.01820
Kosonen, H., Heinonen, M., Mikola, A., Haimi, H., Mulas, M., Corona, F., et al. (2016). Nitrous oxide production at a fully covered wastewater treatment plant: results of a long-term online monitoring campaign. Environ. Sci. Technol. 50, 5547–5554. doi: 10.1021/acs.est.5b04466
Liu, B., Frostegård,Å, and Bakken, L. R. (2014). Impaired reduction of N2O to N2 in acid soils is due to a posttranscriptional interference with the expression of nosZ. mBio 5:e01383-14. doi: 10.1128/mBio.01383-14
Liu, B., Mao, Y., Bergaust, L., Bakken, L. R., and Frostegård, Å (2013). Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes. Environ. Microbiol. 15, 2816–2828. doi: 10.1111/1462-2920.12142
Liu, B., Mørkved, P. T., Frostegård, Å, and Bakken, L. R. (2010). Denitrification gene pools, transcription and kinetics of NO, N2O and N2 production as affected by soil pH. FEMS Microbiol. Ecol. 72, 407–417. doi: 10.1111/j.1574-6941.2010.00856.x
Liu, J., Fu, B., Yang, H., Zhao, M., He, B., and Zhang, X.-H. (2015). Phylogenetic shifts of bacterioplankton community composition along the Pearl Estuary: the potential impact of hypoxia and nutrients. Front. Microbiol. 6:64. doi: 10.3389/fmicb.2015.00064
Löffler, F. E., Sanford, R. A., and Ritalahti, K. M. (2005). Enrichment, cultivation, and detection of reductively dechlorinating bacteria. Methods Enzymol. 397, 77–111. doi: 10.1016/S0076-6879(05)97005-5
Lycus, P., Lovise Bothun, K., Bergaust, L., Peele Shapleigh, J., Reier Bakken, L., and Frostegard, A. (2017). Phenotypic and genotypic richness of denitrifiers revealed by a novel isolation strategy. ISME J. 11, 2219–2232. doi: 10.1038/ismej.2017.82
Maalej, S., Denis, M., and Dukan, S. (2004). Temperature and growth-phase effects on Aeromonas hydrophila survival in natural seawater microcosms: role of protein synthesis and nucleic acid content on viable but temporarily nonculturable response. Microbiology 150, 181–187. doi: 10.1099/mic.0.26639-0
McIlroy, S. J., Starnawska, A., Starnawski, P., Saunders, A. M., Nierychlo, M., Nielsen, P. H., et al. (2016). Identification of active denitrifiers in full-scale nutrient removal wastewater treatment systems. Environ. Microbiol. 18, 50–64. doi: 10.1111/1462-2920.12614
Miranda, K. M., Espey, M. G., and Wink, D. A. (2001). A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5, 62–71. doi: 10.1006/niox.2000.0319
Montzka, S. A., Dlugokencky, E. J., and Butler, J. H. (2011). Non-CO2 greenhouse gases and climate change. Nature 476, 43–50. doi: 10.1038/nature10322
Myers, C. R., and Nealson, K. H. (1990). Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J. Bacteriol. 172, 6232–6238. doi: 10.1128/jb.172.11.6232-6238.1990
Nicholls, R. J., and Cazenave, A. (2010). Sea-level rise and its impact on coastal zones. Science 328:1517. doi: 10.1126/science.1185782
Padan, E., Bibi, E., Ito, M., and Krulwich, T. A. (2005). Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta 1717, 67–88. doi: 10.1016/j.bbamem.2005.09.010
Park, D., Kim, H., and Yoon, S. (2017). Nitrous oxide reduction by an obligate aerobic bacterium Gemmatimonas aurantiaca strain T-27. Appl. Environ. Microbiol. 83:e00502-17. doi: 10.1128/aem.00502-17
Philippot, L., Andert, J., Jones, C. M., Bru, D., and Hallin, S. (2011). Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Glob. Chang. Biol. 17, 1497–1504. doi: 10.1111/j.1365-2486.2010.02334.x
Ravishankara, A. R., Daniel, J. S., and Portmann, R. W. (2009). Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125. doi: 10.1126/science.1176985
Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Sander, R. (2015). Compilation of Henry’s law constants (version 4.0) for water as solvent. Atmos Chem. Phys. 15, 4399–4981. doi: 10.5194/acp-15-4399-2015
Sanford, R. A., Wagner, D. D., Wu, Q., Chee-Sanford, J. C., Thomas, S. H., Cruz-García, C., et al. (2012). Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl. Acad. Sci. U.S.A. 109, 19709–19714. doi: 10.1073/pnas.1211238109
Schumpe, A. (1993). The estimation of gas solubilities in salt solutions. Chem. Eng. Sci. 48, 153–158. doi: 10.1016/0009-2509(93)80291-w
Schumpe, A., Quicker, G., and Deckwer, W.-D. (1982). Gas solubilities in microbial culture media. Adv. Biochem. Eng. 24, 1–38. doi: 10.1007/3-540-11699-0_9
Shoun, H., Fushinobu, S., Jiang, L., Kim, S. W., and Wakagi, T. (2012). Fungal denitrification and nitric oxide reductase cytochrome P450nor. Philos. Trans. R. Soc. B Biol. Sci. 367, 1186–1194. doi: 10.1098/rstb.2011.0335
Šimek, M., and Cooper, J. E. (2002). The influence of soil pH on denitrification: progress towards the understanding of this interaction over the last 50 years. Eur. J. Soil Sci. 53, 345–354. doi: 10.1046/j.1365-2389.2002.00461.x
Sleator, R. D., and Hill, C. (2002). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26, 49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x
Suenaga, T., Riya, S., Hosomi, M., and Terada, A. (2018). Biokinetic characterization and activities of N2O-reducing bacteria in response to various oxygen Llevels. Front. Microbiol. 9:697. doi: 10.3389/fmicb.2018.00697
Thaler, K. M., Berger, C., Leix, C., Drewes, J., Niessner, R., and Haisch, C. (2017). Photoacoustic spectroscopy for the quantification of N2O in the off-gas of wastewater treatment plants. Anal. Chem. 89, 3795–3801. doi: 10.1021/acs.analchem.7b00491
Thomson, A. J., Giannopoulos, G., Pretty, J., Baggs, E. M., and Richardson, D. J. (2012). Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos. Trans. R. Soc. B Biol. Sci. 367, 1157–1168. doi: 10.1098/rstb.2011.0415
Voigt, C., Lamprecht, R. E., Marushchak, M. E., Lind, S. E., Novakovskiy, A., Aurela, M., et al. (2017). Warming of subarctic tundra increases emissions of all three important greenhouse gases – carbon dioxide, methane, and nitrous oxide. Glob. Change Biol. 23, 3121–3138. doi: 10.1111/gcb.13563
Weber, K. P., Gehder, M., and Legge, R. L. (2008). Assessment of changes in the microbial community of constructed wetland mesocosms in response to acid mine drainage exposure. Water Res. 42, 180–188. doi: 10.1016/j.watres.2007.06.055
Wei, W., Isobe, K., Nishizawa, T., Zhu, L., Shiratori, Y., Ohte, N., et al. (2015). Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. 9, 1954–1965. doi: 10.1038/ismej.2015.9
Wilks, J. C., and Slonczewski, J. L. (2007). pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 189, 5601–5607. doi: 10.1128/jb.00615-07
Wolin, E., Wolfe, R., and Wolin, M. (1964). Viologen dye inhibition of methane formation by Methanobacillus omelianskii. J. Bacteriol. 87,993–998.
Yoon, S., Cruz-Garcia, C., Sanford, R., Ritalahti, K. M., and Loffler, F. E. (2015a). Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3-/NO2- reduction pathways in Shewanella loihica strain PV-4. ISME J. 9, 1093–1104. doi: 10.1038/ismej.2014.201
Yoon, S., Sanford, R., and Löffler, F. E. (2015b). Nitrite control over dissimilatory nitrate/nitrite reduction pathways in Shewanella loihica strain PV-4. Appl. Environ. Microbiol. 81, 3510–3517. doi: 10.1128/aem.00688-15
Yoon, S., Nissen, S., Park, D., Sanford, R. A., and Löffler, F. E. (2016). Nitrous oxide reduction kinetics distinguish bacteria harboring clade I versus clade II NosZ. Appl. Environ. Microbiol. 82, 3793–3800. doi: 10.1128/aem.00409-16
Zhang, T., Shao, M.-F., and Ye, L. (2011). 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 6, 1137–1147. doi: 10.1038/ismej.2011.188
Zhao, W., Wang, Y., Liu, S., Pan, M., Yang, J., and Chen, S. (2013). Denitrification activities and N2O production under salt stress with varying COD/N ratios and terminal electron acceptors. Chem. Eng. J. 215, 252–260. doi: 10.1016/j.cej.2012.10.084
Keywords: denitrification, nitrous oxide, salt stress, alkaline stress, RT-qPCR, kinetics
Citation: Han H, Song B, Song MJ and Yoon S (2019) Enhanced Nitrous Oxide Production in Denitrifying Dechloromonas aromatica Strain RCB Under Salt or Alkaline Stress Conditions. Front. Microbiol. 10:1203. doi: 10.3389/fmicb.2019.01203
Received: 24 March 2019; Accepted: 13 May 2019;
Published: 05 June 2019.
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Koki Maeda, Japan International Research Center for Agricultural Sciences, JapanLars Reier Bakken, Norwegian University of Life Sciences, Norway
Bing-Jie Ni, The University of Queensland, Australia
Copyright © 2019 Han, Song, Song and Yoon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sukhwan Yoon, c3lvb244MEBrYWlzdC5hYy5rcg==
 Heejoo Han
Heejoo Han Bongkeun Song
Bongkeun Song Min Joon Song
Min Joon Song Sukhwan Yoon
Sukhwan Yoon