Response: Commentary: Manifold Routes to a Nucleus
- 1Department of Microbiology, Radboud University Nijmegen, Nijmegen, Netherlands
- 2Centro Andaluz de Biología del Desarrollo (CABD)-CSIC, Pablo de Olavide University, Seville, Spain
A Commentary on
Manifold Routes to a Nucleus
by Hendrickson, H. L., and Poole, A. M. (2018). Front. Microbiol. 9:2604. doi: 10.3389/fmicb.2018.02604
In the manuscript “Manifold Routes to a Nucleus”, Hendrickson and Poole question the assumption that the evolution of nuclear compartmentalization has been a singular event. They provide interesting observations on jumbophages and newly described archaea. However, their claim that bacteria from the Planctomycetes phylum display “nucleus-like compartmentation” is based on an outdated interpretation of the planctomycetal cell plan (Fuerst and Sagulenko, 2011). Various comprehensive reviews on the paradigm shift in the field of planctomycetal research were recently published (Devos, 2014a,b; Rivas-Marin and Devos, 2018; Wiegand et al., 2018) and are summarized in Figure 1.
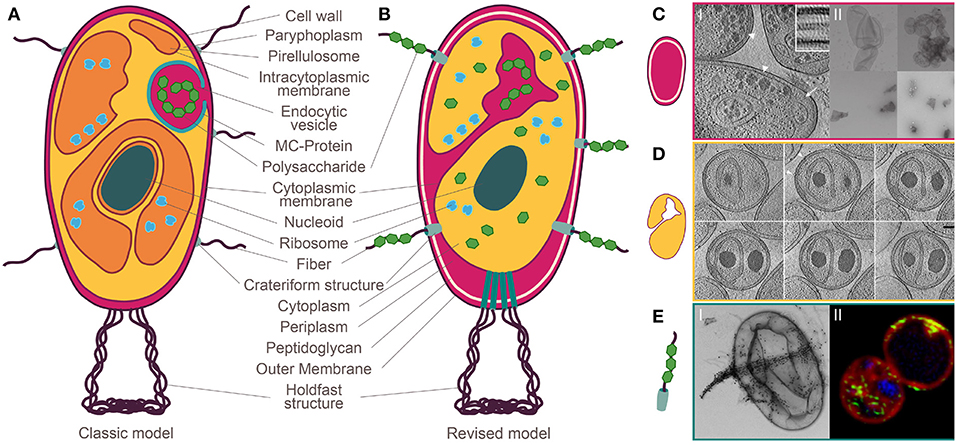
Figure 1. Planctomycetal cell plan before and after the paradigm shift. (A) Classic model: compartmentalization into paryphoplasm and pirellulosome with an additional nuclear compartment in Gemmata obscuriglobus. A peptidoglycan layer is missing in favor of a proteinaceous cell wall. Complex macromolecules are ingested by endocytosis. (B) Revised model: Gram-negative cell plan including an outer membrane, a cell wall formed of peptidoglycan and a cytoplasmic membrane. (C) Jeske et al. (2015) I: Cryo-electron tomography (CET) of the planctomycetal cell envelope shows three layers, II: preparation of peptidoglycan saccule (top) that have been destroyed by lysozyme (bottom). (D) Boedeker et al. (2017) Invaginations into the cytoplasm by extended periplasmic space. (E) Boedeker et al. (2017) I: Crateriform structures give rise to fibers that bind macromolecules such as dextran, II: Green-labeled dextran enters the cell through crateriform structures and is stored in the enlarged periplasmic space. Modified after Wiegand and Jogler (2018).
The concept of a nuclear structure outside the eukaryotes was introduced in 1991, when Fuerst and Webb postulated that the planctomycetal species Gemmata obscuriglobus harbors a bacterial “nucleus” (Fuerst and Webb, 1991). At this time, Planctomycetes were thought to share similarities with bacteria and eukaryotes alike [Figure 1, for review see (Fuerst and Sagulenko, 2011)]. Besides the suggested nucleoid (Fuerst and Webb, 1991), the cytoplasm of Planctomycetes was considered to be compartmentalized by an intracytoplasmic membrane (Lindsay et al., 2001). In contrast to all other Gram-negative bacteria, Planctomycetes were believed to lack a peptidoglycan sacculus and their outermost membrane was suggested to be the cytoplasmic one and to be surrounded by a proteinaceous cell wall instead (König et al., 1984; Liesack et al., 1986; Stackebrandt et al., 1986). Given this model of a planctomycetal cell (see Figure 1A), it was not surprising that other typical eukaryotic hallmark-traits such as endocytosis (Fuerst and Sagulenko, 2010; Lonhienne et al., 2010) and the separation of translation and transcription were suggested (Gottshall et al., 2014). Various interpretations were proposed, such as Planctomycetes are “beyond the bacterium” (Fuerst and Sagulenko, 2011), or that they represent intermediate steps between pro- and eukaryotes (Devos and Reynaud, 2010).
However, the concept of the planctomycetal cell plane evolved (Figure 1B). Others and ourselves have found that Planctomycetes possess a peptidoglycan cell wall (Figure 1C; Jeske et al., 2015; Van Teeseling et al., 2015) and the three-dimensional reconstruction of a G. obscuriglobus cell revealed interconnections of all previously suggested compartments, including the nucleus-like structure (Santarella-Mellwig et al., 2010) and an interconnected tubulovesicular network (Acehan et al., 2013). These findings of a continuous cytoplasm with invaginations of the cytoplasmic membrane called for a reinterpretation of the classic cell plan (Devos, 2014a,b; Wiegand et al., 2018). We also investigated members of different phylogenetic branches of Planctomycetes in a comprehensive study involving mutagenesis and state-of-the-art microscopic techniques and demonstrated that canonical Planctomycetes are a variation of, but not an exception to, the Gram-negative cell plan (Devos, 2014a,b; Boedeker et al., 2017). Most importantly, all investigated planctomycetal species lack compartmentalization by additional membranes (Figure 1D; Boedeker et al., 2017). Consequently, there is no separate nucleus-like compartment in Planctomycetes. However, unusual large pores known as crateriform structures are found in the planctomycetal outer membrane (Figure 1E; König et al., 1984; Schlesner and Hirsch, 1984; Fuerst et al., 1991; Ward et al., 2006; Boedeker et al., 2017). Such structures are unique among bacteria and can be found on the surface of all Planctomycetes (Wiegand et al., 2018). We believe that these crateriform structures have mistakenly been interpreted as “nuclear pores” in a recent study (Sagulenko et al., 2017). Despite these findings, alternative interpretations are still presented (Sagulenko et al., 2014;Feijoo-Siota et al., 2017).
However, we do acknowledge that one aspect of the potential planctomycetal “compartmentalization” requires further investigation: one branch of the phylum, the one of the anammox bacteria, was shown to contain a separated subcellular compartment, the anammoxosome (Neumann et al., 2014). To obtain energy, ammonium is anaerobically oxidized to dinitrogen gas in this compartment whilst forming toxic hydrazine as intermediate metabolite [for review see (Peeters and van Niftrik, 2018)]. Therefore, it was called the “bacterial mitochondrion” (Jogler, 2014). Specific ladderane lipids were thought to protect the anammox membrane against leakage of hydrazine into the cytosol where it would harm the DNA (Sinninghe Damste et al., 2002). In their study, Hendrickson and Pool argue that this is an example for chemical protection of the DNA by an organelle. However, recently it was shown that ladderane membranes are as permeable to hydrazine as straight-chain lipid bilayers (Moss et al., 2018), representing a problem for the authors' hypothesis.
Regarding the main aspect of their article “Manifold Routes to a Nucleus,” Hendrickson and Poole unfortunately do not mention most of the above discussed articles (Speth et al., 2012; Devos, 2014a,b; Rivas-Marin and Devos, 2018; Wiegand et al., 2018). By acknowledging that “the exact nature of this compartmentation is a matter of ongoing debate” they neglect the fact that the described findings do not question the nature of the compartments but their very existence. This disregard leaves the reader under the impression that there is a bacterial nucleus. However, many of their suggestions suffer incoherence when confronted with the most recent data.
Despite the absence of a nucleus-like structure, the phylum Planctomycetes and the extended PVC superphylum is still a relevant model for endomembrane formation—maybe related to DNA protection—and in any case for cell organization and complexity.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
DPD is supported by a grant from the Spanish Mineco #BFU-2016-7826-P. CJ is supported by the Deutsche Forschungsgemeinschaft JO 893/4-1.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Acehan, D., Santarella-Mellwig, R., and Devos, D. P. (2013). A bacterial tubulovesicular network. J. Cell Sci. 127, 277–280. doi: 10.1242/jcs.137596
Boedeker, C., Schuler, M., Reintjes, G., Jeske, O., Van Teeseling, M. C., Jogler, M., et al. (2017). Determining the bacterial cell biology of Planctomycetes. Nat. Commun. 8:14853. doi: 10.1038/ncomms14853
Devos, D. P. (2014a). PVC bacteria: variation of, but not exception to, the Gram-negative cell plan. Trends Microbiol. 22, 14–20. doi: 10.1016/j.tim.2013.10.008
Devos, D. P. (2014b). Re-interpretation of the evidence for the PVC cell plan supports a Gram-negative origin. Antonie Van Leeuwenhoek 105, 271–274. doi: 10.1007/s10482-013-0087-y
Devos, D. P., and Reynaud, E. G. (2010). Evolution. Intermediate steps. Science 330, 1187–1188. doi: 10.1126/science.1196720
Feijoo-Siota, L., Rama, J. L. R., Sanchez-Perez, A., and Villa, T. G. (2017). Considerations on bacterial nucleoids. Appl. Microbiol. Biotechnol. 101, 5591–5602. doi: 10.1007/s00253-017-8381-7
Fuerst, J. A., and Sagulenko, E. (2010). Protein uptake by bacteria: an endocytosis-like process in the planctomycete Gemmata obscuriglobus. Commun. Integr. Biol. 3, 572–575. doi: 10.4161/cib.3.6.13061
Fuerst, J. A., and Sagulenko, E. (2011). Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9, 403–413. doi: 10.1038/nrmicro2578
Fuerst, J. A., Sambhi, S. K., Paynter, J. L., Hawkins, J. A., and Atherton, J. G. (1991). Isolation of a bacterium resembling Pirellula species from primary tissue culture of the giant tiger prawn (Penaeus monodon). Appl. Environ. Microbiol. 57, 3127–3134.
Fuerst, J. A., and Webb, R. I. (1991). Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U.S.A. 88, 8184–8188. doi: 10.1073/pnas.88.18.8184
Gottshall, E. Y., Seebart, C., Gatlin, J. C., and Ward, N. L. (2014). Spatially segregated transcription and translation in cells of the endomembrane-containing bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U.S.A. 111, 11067–11072. doi: 10.1073/pnas.1409187111
Jeske, O., Schüler, M., Schumann, P., Schneider, A., Boedeker, C., Jogler, M., et al. (2015). Planctomycetes do possess a peptidoglycan cell wall. Nat. Commun. 6:7116. doi: 10.1038/ncomms8116
Jogler, C. (2014). The bacterial 'mitochondrium'. Mol. Microbiol. 94, 751–755. doi: 10.1111/mmi.12814
König, E., Schlesner, H., and Hirsch, P. (1984). Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch. Microbiol. 138, 200–205. doi: 10.1007/BF00402120
Liesack, W., König, H., Schlesner, H., and Hirsch, P. (1986). Chemical-composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella Planctomyces Group. Arch. Microbiol. 145, 361–366. doi: 10.1007/BF00470872
Lindsay, M. R., Webb, R. I., Strous, M., Jetten, M. S., Butler, M. K., Forde, R. J., et al. (2001). Cell compartmentalisation in Planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175, 413–429. doi: 10.1007/s002030100280
Lonhienne, T. G., Sagulenko, E., Webb, R. I., Lee, K. C., Franke, J., Devos, D. P., et al. (2010). Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U.S.A. 107, 12883–12888. doi: 10.1073/pnas.1001085107
Moss, F. R. III., Shuken, S. R., Mercer, J. A. M., Cohen, C. M., Weiss, T. M., Boxer, S. G., et al. (2018). Ladderane phospholipids form a densely packed membrane with normal hydrazine and anomalously low proton/hydroxide permeability. Proc. Natl. Acad. Sci. U.S.A. 115, 9098–9103. doi: 10.1073/pnas.1810706115
Neumann, S., Wessels, H. J., Rijpstra, W. I., Sinninghe Damste, J. S., Kartal, B., Jetten, M. S., et al. (2014). Isolation and characterization of a prokaryotic cell organelle from the anammox bacterium Kuenenia stuttgartiensis. Mol. Microbiol. 94, 794–802. doi: 10.1111/mmi.12816
Peeters, S. H., and van Niftrik, L. (2018). Trending topics and open questions in anaerobic ammonium oxidation. Curr. Opin. Chem. Biol. 49, 45–52. doi: 10.1016/j.cbpa.2018.09.022
Rivas-Marin, E., and Devos, D. P. (2018). The Paradigms They Are a-Changin': past, present and future of PVC bacteria research. Antonie Van Leeuwenhoek 111, 785–799. doi: 10.1007/s10482-017-0962-z
Sagulenko, E., Morgan, G. P., Webb, R. I., Yee, B., Lee, K. C., and Fuerst, J. A. (2014). Structural studies of planctomycete Gemmata obscuriglobus support cell compartmentalisation in a bacterium. PLoS ONE 9:e91344. doi: 10.1371/journal.pone.0091344
Sagulenko, E., Nouwens, A., Webb, R. I., Green, K., Yee, B., Morgan, G., et al. (2017). Nuclear Pore-Like Structures in a Compartmentalized Bacterium. PLoS ONE 12:e0169432. doi: 10.1371/journal.pone.0169432
Santarella-Mellwig, R., Franke, J., Jaedicke, A., Gorjanacz, M., Bauer, U., Budd, A., et al. (2010). The compartmentalized bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 8:e1000281. doi: 10.1371/journal.pbio.1000281
Schlesner, H., and Hirsch, P. (1984). Assignment of ATCC 27377 to Pirella gen. nov. as Pirella staleyi comb. nov. Int. J. Syst. Bacteriol. 34, 492–495. doi: 10.1099/00207713-34-4-492
Sinninghe Damste, J. S., Strous, M., Rijpstra, W. I., Hopmans, E. C., Geenevasen, J. A., Van Duin, A. C., et al. (2002). Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nature 419, 708–712. doi: 10.1038/nature01128
Speth, D. R., Van Teeseling, M. C., and Jetten, M. S. (2012). Genomic analysis indicates the presence of an asymmetric bilayer outer membrane in planctomycetes and verrucomicrobia. Front. Microbiol. 3:304. doi: 10.3389/fmicb.2012.00304
Stackebrandt, E., Fischer, A., Hirsch, P., Roggentin, T., and Schlesner, H. (1986). The phylogeny of an ancient group of budding peptidoglycanless eubacteria – the genera Planctomyces and Pirella. Endocytobios. Cell Res. 3, 29–40.
Van Teeseling, M. C., Mesman, R. J., Kuru, E., Espaillat, A., Cava, F., Brun, Y. V., et al. (2015). Anammox Planctomycetes have a peptidoglycan cell wall. Nat. Commun. 6:6878. doi: 10.1038/ncomms7878
Ward, N., Staley, J. T., Fuerst, J. A., Giovannoni, S., Schlesner, H., and Stackebrandt, E. (2006). The order Planctomycetales, including the genera Planctomyces, Pirellula, Gemmata and Isosphaera and the Candidatus genera Brocadia, Kuenenia and Scalindua. Prokaryotes 2006, 757–793. doi: 10.1007/0-387-30747-8_31
Wiegand, S., and Jogler, C. (2018). Planctomyceten-außergewöhnlich, aber bakteriell: ein Paradigmenwechsel. BIOspektrum 24, 593–595. doi: 10.1007/s12268-018-0958-x
Keywords: nucleus, compartmentation, planctomycetes, nuclear pore, evolution
Citation: Jogler C, Wiegand S and Devos DP (2019) Commentary: Manifold Routes to a Nucleus. Front. Microbiol. 10:1198. doi: 10.3389/fmicb.2019.01198
Received: 09 April 2019; Accepted: 13 May 2019;
Published: 31 May 2019.
Edited by:
Ludmila Chistoserdova, University of Washington, United StatesReviewed by:
Svetlana N. Dedysh, Winogradsky Institute of Microbiology (RAS), RussiaCopyright © 2019 Jogler, Wiegand and Devos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Jogler, Y2hyaXN0aWFuQGpvZ2xlci5kZQ==; Damien P. Devos, ZGFtaWVucGRldm9zQGdtYWlsLmNvbQ==orcid.org/0000-0002-9453-4753
 Christian Jogler
Christian Jogler Sandra Wiegand
Sandra Wiegand Damien P. Devos2*
Damien P. Devos2*