Perchlorate-Coupled Carbon Monoxide (CO) Oxidation: Evidence for a Plausible Microbe-Mediated Reaction in Martian Brines
by Myers, M. R., and King, G. M. (2017). Front. Microbiol. 8:2571. doi: 10.3389/fmicb.2017.02571
Myers and King (2017) described perchlorate-coupled carbon monoxide oxidation by two haloarchaeal isolates that were presumed to be monocultures, Halobaculum sp. WSA2 and Haloarcula sp. PCN7. Although both isolates had been subjected to a battery of standard microbiological analyses, results from genome sequence analyses indicated that the former isolate was axenic while the latter was mixed. Subsequent purification and characterization efforts yielded an axenic isolate, Halobacterium sp. PCN9 and a purified Haloarcula sp. PCN7. Halobacterium sp. PCN9 oxidized CO and reduced perchlorate, and was shown to account for the results originally reported by Myers and King (2017).
Halobaculum sp. WSA2 was identified as a nitrate-respiring (denitratating, reducing nitrate to nitrite) CO oxidizer on the basis of a single 16S rRNA gene sequence deposited with NCBI as MF767889; it was most closely related to Halobaculum roseum D90T (98.19% identity). Haloarcula sp. PCN7, was identified as a denitrifying (respiring nitrate to dinitrogen) CO oxidizer that yielded two distinct 16S rRNA gene sequences during PCR amplification as described by McDuff et al. (2016). These two sequences (deposited with NCBI as MK490920 and MK490921) were most closely related to the 16S rRNA gene sequence of Haloarcula taiwanensis and Haloarcula japonica (99.37% and 99.45% identity, respectively) based on BLAST analyses. Since halobacterial genomes often contain multiple 16S rRNA genes (e.g., Cui et al., 2009), the presence of these two sequences was not considered unusual.
However, genomic analyses of paired-end DNA sequences generated on an Illumina Miseq using 2 × 250 bp chemistry at Michigan State University subsequent to the work reported by Myers and King (2017) revealed that the outcome of enriching Haloarcula sp. PCN7 was a mixed culture, while Halobaculum sp. WSA2 was confirmed to be a pure culture (IMG Metagenome/Genome IDs 3300032142 and 2756170195, respectively). For the mixed culture, two sets of contigs were evident after read assembly and annotation. One set included the two 16S rRNA genes along with numerous other genes that were most closely related to Haloarcula. Notably, these contigs did not contain genes essential for denitrification (e.g., nitric oxide and nitrous oxide reductase genes), or CO oxidation (e.g., form I CO dehydrogenase genes).
A second set of contigs was comprised of genes most closely related to Halobacterium, including a 16S rRNA gene that was 98.66% identical to Halobacterium noricense (available as Ga0334598_104419 from IMG metagenome ID 3300032142). In addition, the Halobacterium contigs contained genes essential for CO oxidation (e.g., coxL available as Ga0334598_11585 from IMG metagenome ID 3300032142).
Additional transfers of the original mixed culture ultimately yielded two distinct isolates. Haloarcula sp. PCN7 was obtained as a monoculture that was morphologically indistinguishable from the original mixed culture, but neither oxidized CO nor denitrified, a finding consistent with results from genome sequence annotation. Absence of Halobacterium from this derivative of the original mixed culture was supported by results from a PCR analysis (Figure 1) using primers designed for the Halobacterium 16S rRNA gene sequence identified by genome analysis (HalobacF: 5′-ATGCTAGTTGTGCGGGTTCA−3′ and reverse primer HalobacR:. 5′-CGAGCAGATTTCCCAACGGA−3′). PCR with these primers was positive for the original mixed culture, positive for a Halobacterium isolate derived from it, and negative for the Haloarcula sp. PCN7 monoculture (Figure 1). Haloarcula sp. PCN7 has been deposited as an axenic culture with the Deutsche Sammlung von Mikroorganismen und Zellculturen (DSM 103645).
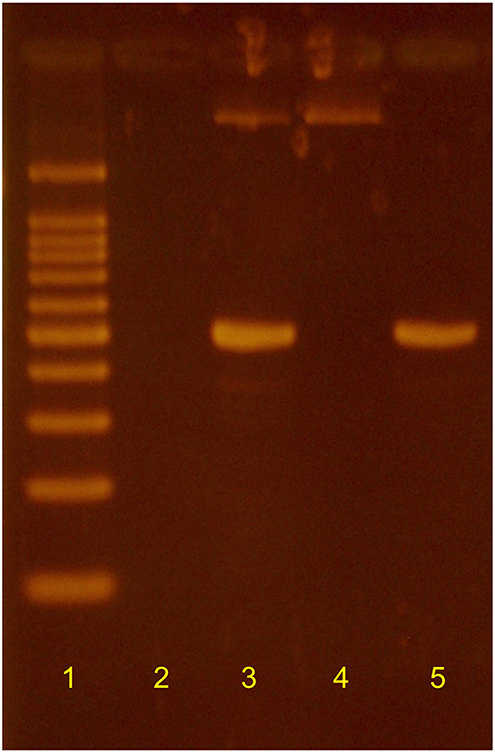
Figure 1. Results of PCR using Halobacterium-specific primers based on the 16S rRNA gene sequence of Halobacterium sp. PCN9. Lane 1: l00 bp ladder; 2: negative control; 3: Halobacterium sp. PCN9 monoculture; 4: Haloarcula sp. PCN7 monoculture; 5: Haloarcula sp. PCN7 mixed culture as used in original analyses of CO-coupled perchlorate reduction (Myers and King, 2017).
The second isolate in the original mixed culture was identified as a member of the genus Halobacterium, and designated as Halobacterium sp. PCN9. This isolate has been deposited with the American Type Culture Collection as TSD-126 and with the Belgian Coordinated Collection of Microorganisms as LMG 31022. The 16S rRNA sequence for Halobacterium sp. PCN9 was identical to the Halobacterium sequence found during genomic analysis of the original mixed Haloarcula sp. PCN7 culture. The genome of Halobacterium sp. PCN9 also harbored genes for CO oxidation as indicated by successful amplification of the coxL gene as described by McDuff et al. (2016); the sequence for this gene (Genbank accession MK262901) was identical to that observed in the mixed culture.
Halobacterium sp. PCN9 oxidized CO under oxic conditions as described by Myers and King (2017) in contrast to the Haloarcula sp. PCN7 monoculture (Figure 2A). CO uptake assays were also conducted with 10 mM perchlorate as an electron acceptor under anoxic conditions as described by Myers and King (2017). Halobacterium sp. PCN9 reduced perchlorate to chlorate (Figure 3) while simultaneously oxidizing CO (Figure 2B) as described in Myers and King (2017). Although the Haloarcula sp. PCN7 monoculture could not oxidize CO (Figure 2A), a 1:1 mixture (based on A600) of Halobacterium sp. PCN9 and the Haloarcula sp. PCN7 monoculture oxidized CO, reduced perchlorate, and produced chlorate as reported by McDuff et al. (2016) (Figures 2B, 3).
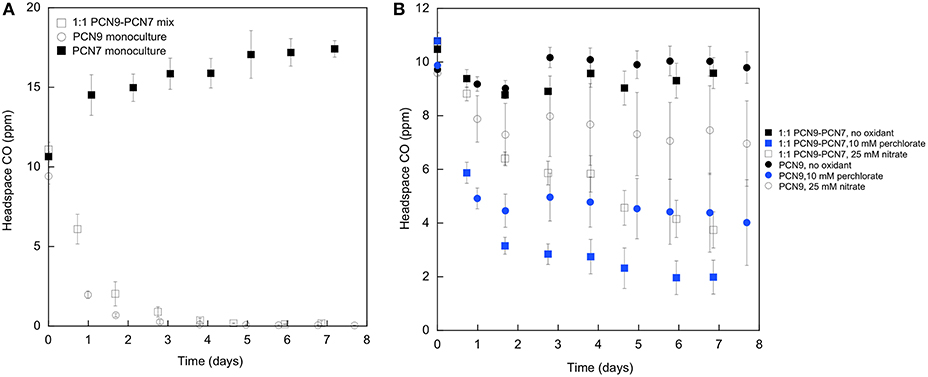
Figure 2. (A) CO uptake under oxic conditions by monocultures of Halobacterium sp. PCN9 and Haloarcula sp. PCN7, and a 1:1 mixed culture of both. (B) CO uptake under anoxic conditions by monocultures of Halobacterium sp. PCN9 and Haloarcula sp. PCN7, and a 1:1 mixed culture of both with and without perchlorate. Assays as described by Myers and King (2017).
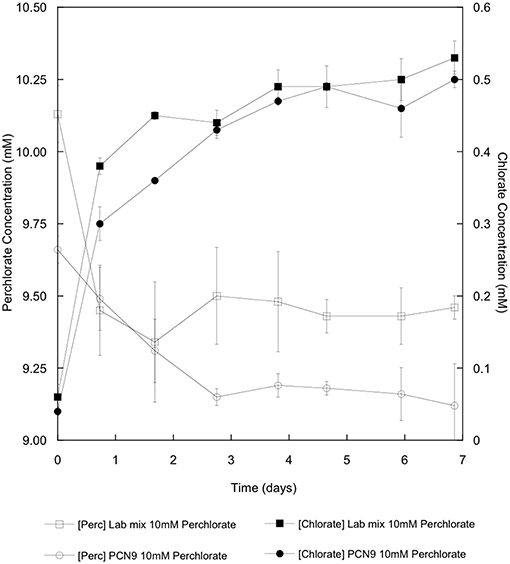
Figure 3. Perchlorate reduction (open symbols) and chlorate accumulation (closed symbols) for a monoculture of Halobacterium sp. PCN9 and a 1:1 mixed culture of Halobacterium sp. PCN9 and Haloarcula sp. PCN7 incubated anaerobically with conditions and assays as described by Myers and King (2017).
These results confirmed observations by Myers and King (2017) that extreme halophiles can couple CO oxidation to perchlorate reduction, but they revealed that a member of the genus Halobacterium (Halobacterium sp. PCN9) rather Haloarcula sp. PCN7 was responsible for the reported activity. In addition, analysis of the Halobaculum sp. WSA2 genome sequence confirmed that it was a monoculture, and thus responsible for CO-coupled perchlorate reduction as described by Myers and King (2017).
Funding
This work was supported in part by NSF EAR-1565499 and NASA awards NNX15AE82A and 15-EXO15_2-0147.
References
Cui, H.-L., Zhou, P.-J., Oren, A., and Liu, S.-Jd. (2009). Intraspecific polymorphism of 16S rRNA genes in two archaeal genera, Haloarcula and Halomicrobium. Extremophiles 13, 31–37. doi: 10.1007/s00792-008-0194-2
McDuff, S., King, G. M., Neupane, S., and Myers, M. R. (2016). Isolation and characterization of extremely halophilic CO-oxidizing Euryarchaeota from hypersaline cinder, sediments and soils and description of a novel CO oxidizer, Haloferax namakaokahaiae Mke2.3. FEMS Microbiol. Ecol. 92:28. doi: 10.1093/femsec/fiw028
Keywords: carbon monoxide, extreme halophile, perchlorate reduction, Mars, chlorate reduction
Citation: Myers MR and King GM (2019) Addendum: Perchlorate-Coupled Carbon Monoxide (CO) Oxidation: Evidence for a Plausible Microbe-Mediated Reaction in Martian Brines. Front. Microbiol. 10:1158. doi: 10.3389/fmicb.2019.01158
Received: 07 May 2019; Accepted: 07 May 2019;
Published: 29 May 2019.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2019 Myers and King. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gary M. King, Z2tpbmdtZSYjeDAwMDQwO2dtYWlsLmNvbQ==
 Marisa R. Myers
Marisa R. Myers Gary M. King
Gary M. King