- 1División de Ciencias Naturales y Exactas, Departamento de Biología, Universidad de Guanajuato, Guanajuato, Mexico
- 2Laboratorio de Biohidrometalurgia, Departamento de Medio Ambiente y Energía, Centro de Investigación en Materiales Avanzados, Chihuahua, Mexico
- 3Laboratorio de Genómica Funcional y Comparativa, División de Biología Molecular, Instituto Potosino de Investigación Científica y Tecnológica, San Luis Potosí, Mexico
Plants are capable of perceiving microorganisms by coordinating processes to establish different forms of plant–microbe relationships. Plant colonization is governed in fungal and bacterial systems by secreted effector molecules, suppressing plant defense responses and modulating plant physiology to promote either virulence or compatibility. Proteins, secondary metabolites, and small RNAs have been described as effector molecules that use different mechanisms to establish the interaction. Effector molecules have been studied in more detail due to their involvement in harmful interactions, leading to a negative impact on agriculture. Recently, research groups have started to study the effectors in symbiotic interactions. Interestingly, most symbiotic effectors are members of the same families present in phytopathogens. Nevertheless, the quantity and ratio of secreted effectors depends on the microorganism and the host, suggesting a complex mechanism of recognition between the plant and their associated microorganisms. Fungi belonging to Trichoderma genus interact with plants by inducing their defense system and promoting plant growth. Research suggests that some of these effects are associated with effector molecules that Trichoderma delivers during the association with the plant. In this review, we will focus on the main findings concerning the effector molecules reported in Trichoderma spp. and their role during the interaction with plants, mainly in the molecular dialogue that takes place between them.
Introduction
One of the main challenges that agriculture production encounters today is to supply the demands of quality and quantity for the producer and consumer without affecting the environment. Various pathogenic agents attack the crops, among which filamentous fungi are the most destructive, causing important economic losses (Singh, 2014). To implement strategies to control plant diseases, it is necessary to understand the pathogenic process, determining how these fungi are established in plants and how they generate tissue damage and bypass plant defenses. In this sense, one of the main events currently under scrutiny corresponds to the early stages of the pathogen–plant interaction.
Now it is known that successful pathogens deliver a wide range of molecules to the plant, which allows them to overcome the obstacles presented by the plant, regarding perception, signaling, or active defense response (Di et al., 2016; Shen et al., 2018). Also, there is ambiguity about the classification of the molecules released by the pathogens, depending on the degree of response generated by the plant immune system and their impact on the pathogenic process (Thomma et al., 2011). Initially, the gene-for-gene model proposed that the interaction of an avirulent protein (Avr) from the pathogen with a plant counterpart, a resistant plant protein (R), triggers the plant defense response, avoiding disease progression. However, it has been determined that this interaction could also lead to the pathogenic processes, so it was redefined that avirulence factors can also be virulence factors (Flor, 1942; Bent and Mackey, 2007; Boller and Felix, 2009). For these pathogen-derived molecules, the term “effector” was proposed, defined as any given molecule that can alter the physiology, structure or function of another organism, facilitating the infection and/or triggering defense responses (Kamoun, 2006). This definition does not imply a positive or negative impact on the outcome of the host–pathogen interaction and therefore, can be applied to non-pathogenic interactions as well (Thomma et al., 2011).
In the plant–pathogen interaction, the participation of elicitors has also been reported, and those are described as a diverse group of molecules that induce plant defense in a weak and non-specific fashion and independent of races or cultivars (Alba et al., 2011). Additionally, it has been proposed that the participation of highly conserved pathogen-derived molecules, with essential functions in the pathogens, for example chitin and flagellin, are needed for the establishment of the interaction with the host. Overall, these molecules are termed Pathogen-Associated Molecular Patterns (PAMPs) and, since they have been described in non-pathogenic microorganisms, the more general concept of Microorganism-Associated Molecular Patterns (MAMPs) is also used (Jones and Dangl, 2006). Because of the wide distribution showed by elicitors, as well as their functional features, it was recently proposed to refer to them as PAMPs/MAMPs. In some cases, the distinction between PAMPs/MAMPs and effectors is not sufficiently defined, which is mainly related to the fact that PAMPs/MAMPs are highly conserved among genera, while effectors tend to be related to a single or few related species. However, new studies have uncovered that certain molecules classified as effectors are also widely distributed among several species (Thomma et al., 2011; Pazzagli et al., 2014). The debate about the functional classification of the molecules released by microorganisms remains open, and new evidence is necessary to clearly classify them and assign distinctive features for them. In order to avoid conceptual confusion, in this review, we will refer to effectors as those non-structural molecules derived from microorganisms that have a function modulating the plant defense pathways and/or participating in the establishment of associations with plants.
Plant defense response is based on the perception of PAMPs/MAMPs inducing primary PAMP- or MAMP-immunity (PTI/MTI), to counteract invading microorganisms (Jones and Dangl, 2006). The ‘effector-triggered immunity’ (ETI) is a second layer of defense, providing systemic resistance by sensing effectors from microorganisms to activate the induced systemic resistance (ISR), mediated by jasmonic acid (JA) and ethylene (ET). In this layer of defense, PDF1.2 (Plant defensin 1.2), Thi2.1 (Thionin) or Chib (Chitinase B) are commonly used as marker genes (Van Loon et al., 2006). Also, the systemic acquired resistance (SAR), regulated by salicylic acid (SA), leads to the expression of Pathogenesis-Related genes (PR) (Bari and Jones, 2009). Pathogens suppress the PTI producing a wide number of effectors with different functions, such as the prevention of the plant recognition or in the formation of infective structures (Kulkarni et al., 2005; de Jonge et al., 2010).
Traditionally, agriculture techniques to counteract phytopathogens involve the use of chemical formulations that have secondary effects such as toxicity and soil pollution. Safer agricultural strategies imply the use of biocontrol agents, displacing and eliminating phytopathogens. Among these agents, fungi classified in the Trichoderma genus are predominant. The antagonistic capacity of Trichoderma has been widely studied, and the mechanisms associated with it include competition for space and nutrients against its adversaries, antibiosis and mycoparasitism (Mukherjee et al., 2013).
Some Trichoderma species can effectively colonize plant roots and shoots and establish a molecular dialogue, having a positive effect in plants (Macías-Rodríguez et al., 2018; Manganiello et al., 2018). Mostafa and Gayed (1952) made the first observations in this regard, reporting that Trichoderma improves fresh and dry weight in cotton plants. More than 20 years later, Catska et al. (1975) reported that exudates from lettuce have a beneficial effect in conidia germination of Trichoderma viride, indicating that the fungus and plants obtain mutual benefits.
Moreover, an increasing amount of reports demonstrate that Trichoderma is an important plant endophyte that can interact with plants such as maize, cucumber, cotton, tomato, and Arabidopsis thaliana (Contreras-Cornejo et al., 2009; Vishnevetsky et al., 2010; Lopes et al., 2012; Mastouri et al., 2012; Reithner et al., 2014; Zhang et al., 2014; Lamdan et al., 2015). Trichoderma penetrates the first or second layers of cells of the epidermis in the root tissue or even colonizes intracellular spaces and grows between the plasma membrane and the plant cell wall (Yedidia et al., 1999; Nogueira-Lopez et al., 2018). Colonization by Trichoderma promotes plant growth, biomass gaining, higher seed germination, increased plant height, root development, shoot dry mass and leaf number, increased crop yield and improved plant vigor (Harman et al., 2004; Salas-Marina et al., 2015; Chagas et al., 2017). One of the most evident morphologic changes in plants triggered by Trichoderma is the increase of lateral roots, thus modifying root architecture. In this process, previous observations have demonstrated the participation of auxins (Contreras-Cornejo et al., 2009) as well as a cross-talk between ET and auxins through the signaling pathways mediated by MAP-kinases (Contreras-Cornejo et al., 2015). Also, the presence of Trichoderma not only modulates the levels of the hormones produced by the plant but Trichoderma itself can contribute with its own hormones or could provide intermediates for the synthesis of some phytohormones, as a part of the benefits reported in the Trichoderma–plant interaction (Guzmán-Guzmán et al., 2019).
Recent research has confirmed that these fungi activate plant defense pathways. However, how Trichoderma modulates the plant immune response to establish a beneficial interaction is one of the main challenges to be addressed. In the establishment of the beneficial association between Trichoderma and plants, the effectors may play key roles, as demonstrated in mycorrhizal systems such as Laccaria bicolor and Glomus intraradices (Kloppholz et al., 2011; Plett et al., 2011).
Here, we will focus on the filamentous fungi Trichoderma spp. and the efforts of the scientific community has done to identify their effector molecules, as well as their role in the establishment of a beneficial relationship with plants that promote growth and immune response activation.
Plant Defense Response and Its Activation by Trichoderma
One of the first observations related with the Trichoderma-induced ISR in plants was the induction of the hypersensitive response (HR) and synthesis of phytoalexins in grapevine cell cultures after the application of a cellulase from T. viride (Calderón et al., 1994). Trichoderma harzianum also increases the resistance of Phaseolus vulgaris against Botrytis cinerea and Colletotrichum lindemuthianum (Bigirimana et al., 1997). Arabidopsis mutants, impaired in JA biosynthesis, showed a similar level of root colonization to wild-type plants (Martínez-Medina et al., 2017). The opposite effect was observed when impaired plants in the SA synthesis were analyzed. These plants were unable to restrict the colonization by the fungus, indicating that SA-mediated response is implicated in the regulation of the Trichoderma–plant colonization, thus preventing vascular system invasion (Alonso-Ramírez et al., 2014). These two results suggested that ISR does not play a relevant role during Trichoderma plant colonization, while SAR pathway modulates the root colonization extend.
Moreover, other studies indicate that ISR induced by Trichoderma asperellum in cucumber was associated with an increase in chitinase and peroxidase activity, as well as the modulation of genes that are implicated in the JA/ET signaling pathways (Yedidia et al., 2000; Yedidia et al., 2003; Shoresh et al., 2005).
Trichoderma can also induce SAR defense even when ISR defense is activated and they can improve the plant resistance against phytopathogens, such as Sclerotinia sclerotiorum, where the effect of Trichoderma strains correlates with the production of cell-wall degrading enzymes by the plant (Contreras-Cornejo et al., 2011; Salas-Marina et al., 2011; Lopes et al., 2012). In pepper, Trichoderma stilbohypoxyli, Trichoderma Caribbaeum, and Trichoderma theobromicola altered the expression of the genes involved in the hypersensitive response and sesquiterpene phytoalexins biosynthesis (Bae et al., 2010). In a tripartite system, involving pathogenic nematodes, T. harzianum T-79 and tomato plants, the presence of T-79 differentially primes the responses related to SA and JA in the plant. T-79 initially primes the SA pathway leading to a faster defense response, protecting the plant from the invasion. Later, the nematode suppresses the JA-mediated defense pathway, but T-79 initiates its priming activity on this pathway, antagonizing the pathogen and reducing its development and reproduction. Finally, when the infection by nematodes has already been established, T-79 intensely activates the SA pathway, increasing the defense against subsequent attacks by juvenile nematodes. These results show that the effects of T-79 over the plants are a dynamic phenomenon, which can be adapted to signals from other organisms in the near environment (Martínez-Medina et al., 2016).
Proteins as Effectors in Trichoderma–Plant Interactions
Proteins were the first molecules proposed as effectors and have been widely studied in pathogenic systems. There is not much information about Trichoderma effector proteins, less than 20 effectors have been experimentally analyzed during the interaction with plant systems (Table 1), among them the following:
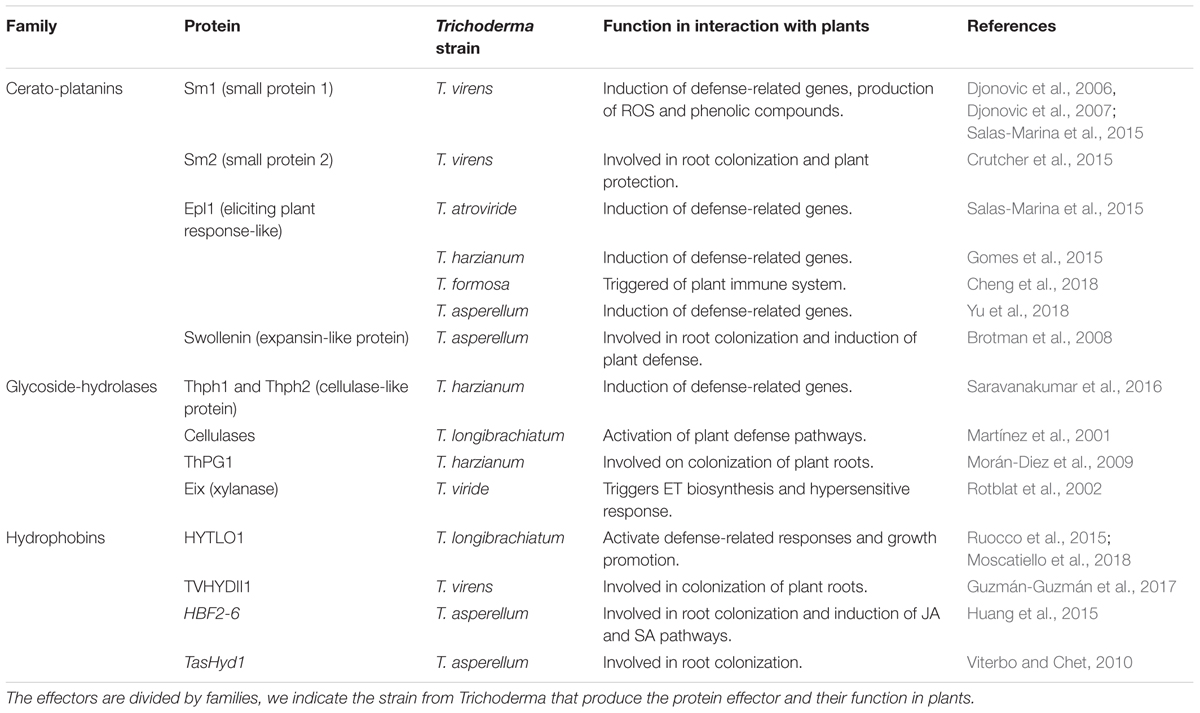
Table 1. Protein effectors from Trichoderma functionally validated during the interaction with plants.
Cerato-Platanins
These are non-catalytic secreted proteins, which contribute to virulence in pathogens, acting as expansin-like proteins weakening cellulose aggregates from the cell wall (Baccelli et al., 2013). The Small Proteins (Sm) Sm1/2/3 from Trichoderma virens and their orthologs in Trichoderma atroviride Eliciting plant-response (Epl) Epl1/2/3 are produced in the association with plants, playing different roles. Transcripts and proteins corresponding to Sm1/Epl1 are detected as the most abundant in cultures supplemented with glucose or during the interaction with tomato or maize plants (Djonovic et al., 2006; Seidl et al., 2006; Gaderer et al., 2015). In maize, only the monomeric form of Sm1 or Epl1 proteins can induce the plant defense response and the dimeric form blocks the activation of ISR. When T. virens and T. atroviride strains were cultured in the presence of maize seedlings, Sm1 and Epl1 were produced as a glycosylated monomer and as a non-glycosylated dimer, respectively. These results suggest that the signals released by the plant influence the state of glycosylation and multimerization of Sm1/Epl1 proteins that may control the Trichoderma–plant molecular dialogue (Vargas et al., 2008). However, Sm2/Epl2 proteins are quantitatively fewer present that Sm1/Epl1 during the interaction with maize, there is evidence showing that Sm2 and Epl2 are more relevant in the activation of defense and root colonization in maize. The mechanism employed by Sm2/Epl2 to induce plant defense is unknown, but a strong reduction of the protection level of maize seedlings against Cochliobolus heterostrophus was observed when plants were treated with the sm2/epl2 knockout strains in contrast with sm1/epl1 knockout strains (Crutcher et al., 2015; Gaderer et al., 2015).
Hydrophobins
These are small surface-active proteins, which are only found in fungi. The genomes from T. virens and T. atroviride contain 17 sequences encoding for hydrophobins (Kubicek et al., 2011), and the upregulation of some of them has been reported during Trichoderma–plant interactions. The expression of the hydrophobin-encoding gene HFB2-6 was down-regulated in the presence of 1% poplar leaves powder, whilst the gene upregulated under 1% poplar root powder, suggesting that HFB2-6 has a function in root colonization. Moreover, the recombinant hydrophobin purified from E. coli activated the JA and SA signal transduction pathways when used on poplar seedling. Also, the upregulation of two genes related to auxin signaling was observed. Therefore, this protein seems to be involved in the promotion of growth and defense of poplar plants (Huang et al., 2015). TVHYDII1, from T. virens, participates in the colonization of tomato roots; this was demonstrated by using null and overexpressing tvhydii1 strains (Guzmán-Guzmán et al., 2017). Likewise, the TasHyd1 from T. asperellum participates in the colonization of cucumber plants (Viterbo and Chet, 2010). The purified hydrophobin HYTLO1 from Trichoderma longibrachiatum activates the defense response and promotes plant growth. Moreover, knockout strains significantly decreased their antagonistic activity and their capability to promote plant growth (Ruocco et al., 2015). Therefore, HYTLO1 has a dual role in the process of interaction with plants.
Glycoside-Hydrolases
This is a wider group of proteins with enzymatic activity that are secreted by Trichoderma; some of them have been characterized and implicated in the Trichoderma–plant interaction. The xylanase Eix (ethylene induced xylanase) from T. viride triggers ET biosynthesis and the hypersensitive response in tobacco plants, highlighting that the elicitation of ET biosynthesis is not related with Eix enzymatic activity (Sharon et al., 1993; Rotblat et al., 2002). The cellulases, Thph1 and Thph2 from T. harzianum, were applied on maize leaves and this treatment led to the transient elevation of free cytosolic calcium and the production of reactive oxygen species (ROS). The ΔThph1- or ΔThph2 null mutants were not able to upregulate the expression of genes related to the jasmonate/ET signaling pathway in maize. By using the yeast two-hybrid system, Thph1 and Thph2 can bind to the autophagocytosis associated protein (ZmATG3) and germin-like protein (ZmGL) of the plant, respectively. The identification of this molecular interaction opens the possibility to analyze the cellular localization of these targets as well as their participation in the ISR (Saravanakumar et al., 2016, 2018).
CFEM and Small Secreted Cysteine-Rich Proteins (SSCPs)
In Trichoderma spp., one of the most abundant groups of secreted proteins corresponds to small proteins containing four or more cysteine residues. The SSCPs could be grouped into protein families with functions such as hydrophobins, some glycosyl hydrolases, cerato-platanins, CFEM proteins (Common in several Fungal Extracellular Membrane) as well as proteins with unknown function (Druzhinina et al., 2012).
Lamdan et al. (2015) reported a secretome analysis from the interaction between T. virens and maize, detecting 13 SSCPs negatively regulated by the presence of the roots. Plant inoculation with independent knockouts strains, for four of these genes encoding the SSCPs (ID 92810, ID 71692, ID 111486, and ID 77560), showed an improvement in ISR activity compared with the wild-type strain, without affecting root colonization. SSCPs could act as negative effectors reducing the defense levels in the plant and may be important for the fine-tuning of ISR by Trichoderma. Bioinformatic predictions indicated the abundant presence of SSCPs in T. atroviride and T. virens genomes, some of them containing CFEM domains, which are present in cell surface proteins with important roles in the interaction with other organisms (Pérez et al., 2011; Druzhinina et al., 2012). Possibly, but not in all cases, CFEM proteins participate as negative regulators of plant defense. In the bioinformatic analysis reported by Guzmán-Guzmán et al. (2017), there are 32 sequences grouped as CFEM; among them the tacfem1 gene, which was upregulated when the fungus was co-cultivated with A. thaliana, suggesting a possible role during the establishment of the interaction. Also, in T. virens, the SSCP gene ID 19757 was upregulated in the interaction with both maize and tomato, while the ID 17705 gene was upregulated only by the presence of maize plants, rendering it a promising candidate for further analysis during the T. virens-maize interaction (Morán-Diez et al., 2015).
Hunting for New Trichoderma Protein Effectors
One of the main strategies to identify effector proteins is to analyze their upregulation in the presence of plants (Djonovic et al., 2006; Plett et al., 2011; Guzmán-Guzmán et al., 2017). Trichoderma transcriptome and secretome analyses under different culture conditions led to describe that cellulases, small proteins, and cytochrome p450, among others, are highly represented (Lamdan et al., 2015; Morán-Diez et al., 2015; Rocha et al., 2016). Recently, a catalog of 233 putative effector proteins was reported from T. virens, T. atroviride, and Trichoderma reesei, identified and grouped in 18 families. The expression pattern of some of these genes was analyzed during the Trichoderma–Arabidopsis interaction, observing the upregulation of genes grouped in LysM proteins, Serine-proteases, Thioredoxins, Hydrophobins, CFEM, and Cerato-platanin families. Down-regulation was also observed, as in the case of the Tvmp1 gene, encoding for a Metalloprotease (Guzmán-Guzmán et al., 2017), and for the Tvcyt2 gene, which encodes a p450-cytochrome (Ramírez-Valdespino et al., 2018). However, more than 200 sequences from this in silico analysis were not studied further and their possible function during Trichoderma–plants interaction remains unknown.
The LysM effectors play relevant roles in the establishment of pathogenic interactions: they protect the fungal mycelium by either covering the surface of the hypha, thereby interfering with the enzymatic activity of the plant delivered chitinases or hijacking fungal cell wall derived chitin fragments, thereby avoiding the stimulation of the immune response (de Jonge et al., 2010; Kombrink et al., 2017). LysM encoding genes are present in Trichoderma genomes, but they are not characterized as well (Mendoza-Mendoza et al., 2018). In the catalog of putative effector proteins, there are 15 LysM encoding genes, six belonging to T. atroviride and nine to T. virens. Additionally, the tvlysm1 gene was upregulated in co-culture of T. virens with Arabidopsis, suggesting a role during Trichoderma interaction with the plant (Guzmán-Guzmán et al., 2017). Additionally, the Tal6 protein from T. atroviride, was reported as an inhibitor of conidia germination (Seidl et al., 2013); however, their direct participation with plants has not been determined.
The differences in the experimental design in each case, where Trichoderma–Plant transcriptome/secretome are analyzed, makes it difficult to compare among all data sets to determine the functional relevance of these genes. In the case of T. virens, two secretomes obtained from hydroponic cultures, were analyzed looking for proteins differentially expressed during their interaction with maize seedlings. One of them analyzed the soluble proteins, while the other focused only on the proteins found in the plant apoplast. The main differences between both studies are related to the culture medium used and the time length of interaction. Lamdan et al. (2015) obtained the secretome from hydroponic cultures in sucrose-supplemented MS media after 96 h of interaction, a total of 280 soluble proteins were detected, of which 86% (241 proteins) contained a predicted signal peptide, identifying members of glycoside hydrolases (GH), LysM proteins, CFEM, lipases, and SSCPs, including Sm1 as the most abundant protein. From all secreted proteins, 66 were identified as differentially expressed, 32 were increased, while the remaining 34 showed a decrease when maize seedlings were present. Nogueira-Lopez et al. (2018) reported 43 secreted proteins, obtained from apoplastic space of maize roots cultured in Hoagland’s solution after 60 h of interaction with T. virens, and using a filtered-pipeline, nine of them were classified as putative effector proteins. Both secretomes share 13 putative candidates, being the GH group; the most representative proteins with five protein family members (Supplementary Table S1). The low number of common sequences could be due to the filtered-pipeline used in each study or due to the differences in growth conditions and sample collection times; without leaving aside the possibility that the roots could retain some proteins limiting their detection in the media (Lamdan et al., 2015).
The comparison between each secretome with the proposed T. virens effectors from the in silico analysis showed differences related to the number of effector candidates, which is expected given that the predicted secretome includes all sequences with the potential of being effectors.
Considering the full list of the apoplastic secretome (43 proteins), the proteins found with differential expression in the complete soluble secretome (66 proteins) and the effector candidates from the in silico prediction (84 proteins), we only found the CFEM member (ID 92810) and the GH member (ID 42143) in the three research works, while 22 common sequences were found between the soluble secretome (Lamdan et al., 2015) with the predicted effectors (Guzmán-Guzmán et al., 2017). Whereas, five sequences from the in silico analysis are present among the apoplastic secretome proteins, but they were not considered as effectors.
The differences observed among the experimental strategies to identify new protein effectors in the association of Trichoderma with plants suggests that there is still no consensus to consider products with the potential to participate as effectors, which represents a relevant topic to analyze further. The in silico analysis can be less precise when identifying effector molecules, but they provide a wide range of candidates to determine biological functions and the best way to know the function of a putative effector is to perform a wide and specialized characterization. Understanding and identifying the effector proteins will provide tools to deepen our understanding on how plant-beneficial fungi interaction is established and which could be the differences between a beneficial and a pathogenic system.
Secondary Metabolites as Effectors in Trichoderma–Plant Interactions
One of the main characteristics of fungi is the production of a huge diversity of secondary metabolites (SMs); compounds with potential application in food, pharmaceutical and agricultural industries (Brakhage, 2013). It is generally assumed that SMs are not essential for growth and development in fungi but play important functions in the sensing, signaling and counteracting processes for the organisms present in their environment (Macheleidt et al., 2016). SMs comprise compounds of low molecular weight, diffusible in the culture medium or volatile, which are synthesized through a great variety of pathways (Brakhage, 2013). The synthesis of SMs is usually different between strains, and is influenced by growth conditions (Yu and Keller, 2005). At the molecular level, this environmental influence is related to the availability of regulatory elements. In T. reesei, the SOR cluster contains two genes encoding for transcription factors involved in biosynthesis of sorbecillinoids, a group of yellow pigments with antimicrobial activities. YPR2, one of those transcription factors, carries out its major function in constant darkness and in the presence of cellulose as a carbon source. The function of YPR is positively related to the levels of alamethicin and to the production of orsellinic acid in darkness (Hitzenhammer et al., 2019). In T. atroviride and T. virens, it has been shown that heterotrimeric G proteins, mitogen-activated protein kinases (MAPK) and transcription factors are involved in the signaling pathway leading to SMs synthesis, which also respond to environmental conditions as a type of nutrients, pH, light or temperature (Reithner et al., 2005; Mukherjee and Kenerley, 2010). Antibiotic activity has been reported for SMs produced by Trichoderma species against various yeast, filamentous fungi and bacteria, causing growth inhibition or cell death. These SMs, acting synergistically with hydrolytic enzymes, are more likely to be implicated in the effectiveness of the strain producing them as a biological control agent (Reino et al., 2008).
Although the metabolites produced by plants could also affect the association with microorganisms, either by favoring it or by restricting it, in this section we will focus on those SMs produced by Trichoderma species that have shown impact in their capacity as plant symbionts.
Lactones
These are generally very pleasant, potent, flavor materials, which are widely distributed in nature (Kapfer et al., 1989). The best-known SM for Trichoderma species is 6-pentyl-2H-pyran-2-one (6-PP), derived from linoleic acid and the pathway of its formation has been elucidated by metabolization of [U-14C] and of [1-14C] linoleic acid in T. harzianum (Serrano-Carreon et al., 1993). In plants, 6-PP interferes with the signaling pathway involving auxins and ET, promotes plant growth, and regulates root architecture by inhibiting the growth of the primary root and inducing the formation of lateral shoots, by modulating the expression of genes encoding for auxin transporters. The modification related to the lateral shoots is mediated by the TIR1, AFB2, and AFB3 receptors and the ARF7 and ARF19 transcription factors, while the sensitization in the main root is mediated by EIN2 (Garnica-Vergara et al., 2016). In field, the application of 6PP (1 μM) or a spore suspension of T. harzianum T22 (108 spore/liter) in Vitis vinifera, increased polyphenol content, antioxidant activity and weight in fruits (Pascale et al., 2017). Cremenolide is a 10-member lactone isolated from Trichoderma cremeum, that improved the root length and fresh weight in tomato seedlings, but changes in plant height were not observed (Vinale et al., 2016).
Peptaibols
These are small linear peptides of non-ribosomal synthesis, which usually have a high content of 2-amino-isobutyric acid (Aib) bound to unconventional amino acids, such as ethyl-valine, isovaline, and hydroxyproline. There are at least 190 compounds of this type synthesized by Trichoderma species1.
In T. virens, 18-mer peptaibols are produced through the activity of the Non-Ribosomal Peptide Synthase (NRPS) encoded by the gene Tex1. This kind of peptaibols activates the plant defense in Cucumis sativus against Pseudomonas syringae. Cucumber plants growing in contact with T. virens increase the expression of three genes involved in the synthesis of phytoalexins, hpl, pal1, and prx, which encode for a hydroxy peroxide lyase, phenylalanine ammonia lyase, and peroxidase, respectively. The tex1 null mutants lose their ability to produce 18-mer peptaibols, leading to a lower expression of hpl, pal1, and prx genes in plants. The use of two synthetic peptaibols TvBI and TvBII on cucumber seedlings activated systemic protection against bacteria and induced the expression of hpl, pal1, and prx (Viterbo et al., 2007). Null mutants in ppt1 gene, which codes for a 4-phosphopantetheinyl transferase in T. virens, are affected in the synthesis of 11, 14, and 18-mer peptaibols, although their capability to colonize roots is not affected. Furthermore, antibiosis on phytopathogens, as well as the ability to induce the synthesis of SA and of camalexin in Arabidopsis plants, is compromised in these strains (Velázquez-Robledo et al., 2011).
Alamethicin is one of the most studied peptaibols due to its ability to induce defense responses in plants such as callose deposition, expression of genes related to plant defense, production of SMs, and accumulation of phenolic compounds, that together can improve the vigor of plants and their response to stressing conditions (Rippa et al., 2010). In A. thaliana, the presence of alamethicin interferes with the synthesis of methy farnesoate (MeFA), an SM related to herbivory, by altering the presence of the miRNA163, which targets genes encoding methyltransferases relevant in the synthetic pathway of the MeFA (Ng et al., 2011). It is proposed that alamethicin increases ion permeability in the cell membrane (Duclohier and Wróblewski, 2001). Trichokonin VI (TK VI) is produced by T. longibrachiatum SMF2, and it has been shown that this peptaibol interferes with the GORK channel, a rectifying K+ channel gated outwardly, which alters the root structure by inhibiting cell division and elongation in the main root. TK VI increases the auxin content and interrupts its gradient at the tips of the roots, interfering with the local synthesis and its polar transport (Shi et al., 2016).
Polyketides
These are one of the most abundant groups of SMs in nature, which includes macrolides, polyenes, and polyphenols. They have been studied in detail because the group includes compounds with an impact on human health such as sterigmatocystin, aflatoxin B1 and lovastatin. They are produced by polyketide synthases (PKSs), multi-domain proteins similar to fatty acid synthases, which condense acetyl coenzyme A or malonyl coenzyme A units, to form carbon chains of variable length (Chiang et al., 2010). In Trichoderma arundinaceum, the production of 4 aspinolides has been reported, in particular, aspinolide C participates in the induction of genes PR1b1, PR-P2 involved in the signaling pathway mediated by SA. During the interaction of T. virens with maize plants, the expression of the defense-related genes pal1 and aos (allene oxide synthase) is increased. This upregulation is related to SMs produced by T. virens through the activity of the PKS/NPRS encoded by the Tex 13 gene. Strains affected in Tex13 retained their ability to increase the expression of the gene aos in plant, but they could not upregulate the pal1 gene (Mukherjee et al., 2012).
Terpenes
These are a highly diverse family of SMs at the structural and stereochemical level. These molecules are derived from long polyisoprenoid diphosphates that can be cyclized to generate single or multiple ring products. The cyclization reactions are carried out by high-affinity terpene cyclase enzymes, which generate a single product or by promiscuous enzymes that can generate up to 52 different products (Christianson, 2008). Cytochrome p450 enzymes are involved in the reactions of synthesis and/or modification of terpenes. Recently 477 cytochrome P450s have been identified from seven Trichoderma species (Chadha et al., 2018). Cytochrome p450 activity is needed for the synthesis of SMs, which are related to the mycoparasitic capacity and/or its association with plants. The enzyme encoded by the G3 gene in Trichoderma hamatum is activated in response to Sclerotinia and Sclerotium species (Carpenter et al., 2008). Through the generation of mutant and overexpressing strains of the Tvcyt2 gene of T. virens, five terpene-like compounds were identified as involved in the antagonistic activity against Rhizoctonia solani, in the activation of genes related to the JA pathway in Arabidopsis plants as well as in the promotion of growth in Arabidopsis and tomato seedlings (Ramírez-Valdespino et al., 2018).
Trichothecenes
Trichoderma brevicompactum produces trichodermin through a pathway involving the activity of p450 enzyme, encoded by Tri5 gene. The overexpressing strains of this gene had a negative effect in tomato plants, decreasing the root length and plant size (Malmierca et al., 2015). Harzianum A (HA), isolated from T. arundinaceum, did not show any effect over growth of tomato seedlings. However, mutant strains that do not produce HA were impaired to upregulate the expression of genes involved in plant defense at the same level. HA could be sensitizing the plant cell to induce those genes faster and at higher levels (Malmierca et al., 2012).
Volatile Organic Compounds (VOCs)
The VOCs group includes several small compounds with different chemical natures, playing relevant roles as essential signals in interactions among plant roots, microbes, and insects (Schenkel et al., 2015). The effect by the VOCs produced by 25 different Trichoderma strains on the plants were analyzed using A. thaliana as a host, in two independent research works. The general results showed that one of the strains produced VOCs with a negative impact on plant growth, 10 of the strains did not have any obvious effect on the plants, while fourteen of them had a positive effect on total biomass and on chlorophyll content. The analysis of the VOCs produced by each strain determined the presence of great diversity of compounds, suggesting the participation of several mechanisms to generate the final effect on plants (Lee et al., 2016; Nieto-Jacobo et al., 2017). This blend of compounds present in VOCs makes it difficult to determine which of the metabolites is responsible for the effects observed on the plants. Some strategies that could help to define their biological role are based on genetic manipulation in order to generate strains affected in key elements of the VOCs biosynthesis. Trichodiene (TD) is a VOC used as a substrate by the sesquiterpene synthase Tri5 to produce the compound Harzianum A (HA) in T. arundinaceum. The heterologous expression of the gene Tri5 in T. harzianum led to the production of TD. VOCs released by this Tri5-transformant and TD itself induced the expression of tomato defense genes related to SA (Malmierca et al., 2015). In T. atroviride, the mutation of genes encoding membrane-bound NADPH oxidases (Nox), leads to the alteration of VOCs profiles. Loss of function of Nox1 or the regulator NoxR, in the presence of a functional Nox2 enzyme leads to the production of VOCs with inhibitory effects on plant growth (Cruz-Magalhães et al., 2019).
Phytohormones
Phytohormones are important growth regulators with relevant roles on metabolism and plant defense responses. Several root-associated microbes are able to produce phytohormones that have an effect in plants (Egamberdieva et al., 2017). T. virens produces Indole-3-acetic acid (IAA) and indole-3-acetaldehyde (IAAId), which promotes plant growth and development in A. thaliana (Contreras-Cornejo et al., 2009).
Cytokinins (CKs) are essential molecules that regulate plant growth and development (Osugi and Sakakibara, 2015). ET is a volatile hormone that regulates a range of processes, from seed germination, organ senescence, among others (Bleecker and Kende, 2000). CKs promote hyphal branching and help in oxidative stress tolerance in Magnaporthe oryzae (Chanclud et al., 2016), whilst in mycorrhizal fungi, ET affects spore germination and growth (Barker and Tagu, 2000). There are no reports in Trichoderma about CKs or ET production, though in the genome of T. atroviride, the genes necessary to synthesize both compounds are present, suggesting that it could produce these phytohormones, with a potential impact in the association with plants (Guzmán-Guzmán et al., 2019). Abscisic acid (ABA) is involved in seed dormancy and development, abiotic stress response, among other roles in plants, and there is evidence of its production by several fungi on which ABA was proposed as a factor promoting plant colonization (Charpenter et al., 2014; Chanclud and Morel, 2016). There were six genes related to the ABA biosynthesis pathway identified in T. atroviride. However, no homolog to bcaba3/ataba3, a gene encoding a key enzyme in last steps of the biosynthesis of ABA was found, suggesting that T. virens is unlikely to produce this phytohormone and that it could only provide an ABA intermediary (Guzmán-Guzmán et al., 2019). Additionally, it was proposed that T. virens and T. atroviride modulate ABA-regulated responses, such as stomatal aperture and leaf transpiration in A. thaliana (Contreras-Cornejo et al., 2015).
Factors That Influence the Effect of SMs on Plants
Three relevant factors related to the positive effect generated on the plants by SMs are, the dose used of each SM, the physiological state, as well as the genetic background of the plants tested (Vinale et al., 2009; Li et al., 2019). The effect of alamethicin over plants has been tested at high doses such as 10 and 50 mM. When these are applied to Arabidopsis seedlings, they generate plant death and this toxicity is related to cleavage of ribosomal RNA and cellular lysis. The use of alamethicin at lower doses (5 μM) suggest that a threshold in the concentration of this SM is required to trigger only plant-cell permeabilization and induce programed cell death as a hypersensitive response, showing similarities with the response elicited by avirulent pathogens or by compounds that mimic a pathogen attack (Rippa et al., 2007; Rippa et al., 2010). Koninginins A, B, C, E, and G tested at 10-3 M, inhibited growth of etiolated wheat coleoptiles at different rates, varying between 54 and 65% and up to 100% (Reino et al., 2008). Treatments of 6-PP over A. thaliana seedlings in doses from 50 to 175 μM increased shoot, root and total plant biomass while at 200 μM, no increase in biomass was observed. 6-PP also raised both lateral root number and density in a dose- dependent manner, showing an inhibition of primary root growth from 125 μM onwards, without cell damage (Garnica-Vergara et al., 2016).
The plant genetic background is a determinant in the response generated by specific strains of Trichoderma. In maize, the use of T. harzianum T22, one of the most widely used commercial strains, induced strong positive growth over eight maize hybrids tested, it had little effect on growth over eight other hybrids and it even negatively affected growth of two other maize hybrids (Harman, 2006). While in tomato, independent symbiotic interactions between T. harzianum T22 or T. atroviride P1 with four Solanum lycopersicum lines or the wild Solanum habrochaites accession demonstrated that genetic variability is a determinant in the response shown by plants related to growth, weight, resistance against B. cinerea and the expression of genes involved in plant defense (Tucci et al., 2011).
We propose that many SMs can be classified as effectors and the different Trichoderma–plant systems developed by different work groups make evident the utility of using them as a biological model. Metabolomic analysis, involving the study of the concentrations, structures and interactions of thousands of SMs represents a useful tool to identify molecules with potential biotechnological application in the improvement of plant yield and vigor.
Trichoderma’s Small RNAs as Putative Effectors
Thus far, plant analysis has focused on the identification of proteins involved in the plant immune response. However, several lines of evidence have shown that plants also use non-coding RNAs against pathogens (Pumplin and Voinnet, 2013) and symbionts (Pieterse et al., 2014). Small RNAs (sRNAs) play important roles in plant immune responses against virus, bacteria, fungi, and oomycetes (Pumplin and Voinnet, 2013). sRNAs are 20–30 nucleotide long non-coding, sequence specific regulatory RNA molecules that mediate gene silencing to regulate physiological and developmental processes (Chang et al., 2012; Pumplin and Voinnet, 2013). In plants, sRNAs are processed from double-stranded or single-stranded RNA with hairpin structures by Dicer-like (DCL) proteins, which release RNA duplexes. After processing, sRNAs are loaded into RNA-induced silencing complexes (RISC), which contains one member of the ARGONAUTE (AGO) protein family, leading to transcriptional gene silencing by guiding heterochromatin formation, inhibiting mRNA translation or inducing mRNA degradation. Several members of the sRNA biogenesis machinery are involved in plant immunity, including DCL, AGO, DCL associated proteins and RNA dependent RNA polymerases (RDRs). Mutants in such genes show defects in sRNAs accumulation and are impaired in pathogen response (Seo et al., 2013).
During the last decade, cross-kingdom RNA interference (RNAi) between host and phytopathogens has demonstrated its role in the successful colonization of plant tissues by the phytopathogens or in their avoidance by the plant. For instance, host-induced gene silencing (HIGS), a technology developed to protect crops from fungal infections by expressing dsRNA in planta to silence virulence genes, enhances plant resistance to Fusarium verticillioides or Blumeria spp. upon infection of the host plant (Nowara et al., 2010; Tinoco et al., 2010). HIGS technology is also effective against Puccinia spp. (Yin et al., 2011, 2014), Fusarium spp. (Koch et al., 2013; Hu et al., 2015) and S. sclerotiorum (Andrade et al., 2016).
B. cinerea, produces complementary sRNAs against plant immunity related genes in Arabidopsis and Lycopersicum esculentum (Weiberg et al., 2013). Upon infection of Arabidopsis by B. cinerea, the fungal sRNAs associate with Arabidopsis AGO1 protein, interfering with plant target mRNAs, for example, those encoding MAPKs. These findings indicate that the fungus hijacks the host RNAi machinery to silence the host own genes (Weiberg et al., 2013; Wang M. et al., 2017). Moreover, Arabidopsis ago1-27 mutants are more resistant to B. cinerea infection, supporting that the fungus uses the plant RNAi machinery to silence host genes. Transferred fungal sRNAs into the plant cell are detected in B. cinerea dcl1 or dcl2 single mutants, but not in a dcl1/dcl2 double mutant, leading to a diminished fungal virulence, indicating that the biogenesis of sRNAs is required for pathogenesis (Weiberg et al., 2013). Transgenic plants expressing one of these sRNAs (Bc-siR37) silence Arabidopsis genes encoding a pectin lyase, a WRKY transcription factor, and a receptor-like kinase (Wang M. et al., 2017). The causal agent of wheat stripe rust disease, Puccinia striiformis, silences the mRNA that encodes for the PR-2 protein, through Pst-milR1, a miRNA-like sRNA (Wang B. et al., 2017). In Verticillium dahliae, it was shown that AGO2 does not play a role in the infection of Arabidopsis. This pathogen infected Arabidopsis ago2-1 normally, whereas in ago1-27 mutant, the rate of infection was reduced (Wang et al., 2016).
Successful establishment of the Trichoderma–plant interaction during early stages of root colonization implies the activation of cell detoxification and protection mechanisms in the fungi (Ruocco et al., 2009; Estrada-Rivera et al., 2019). Therefore, these fungi possess effective systems that efficiently scavenge harmful compounds from the cell. This has been partially shown with the suppression of phytoalexin and plant genes related to the defense mechanisms in Lotus japonicus and Arabidopsis during its interaction with Trichoderma koningii and T. atroviride. Application of these fungi to plant roots induced a rapid accumulation of host transcripts that encode key enzymes of SAR and ISR and those involved in phytoalexin synthesis. The expression of these genes is transient and decreased to levels of the control plants. Trichoderma resembles mycorrhizal fungi in the establishment of symbiotic associations rather than fungal pathogens (Masunaka et al., 2011; Estrada-Rivera et al., 2019). Production of sRNAs by filamentous fungi, including Trichoderma species, has been documented. Although analysis of mutants in the biogenesis components of sRNAs in fungi have shown their role on their biology (Chang et al., 2012; Carreras-Villaseñor et al., 2013), much is yet to be explored regarding the roles of sRNAs in this kingdom. There is no direct evidence for the role of beneficial microorganisms sRNAs on the suppression of plant immunity to establish a symbiotic relationship. In our group, we sequenced sRNAs libraries during Arabidopsis–T. atroviride interaction at different interaction times. Mapping the sRNAs over the T. atroviride genome revealed that 37 sRNAs of the fungus matched with genes of the host plant. Interestingly, target genes encode lytic enzymes, MAPKs putatively involved in plant immunity, proteins with a domain of unknown function, disease resistance protein, NBS-LRR class family proteins, and S-Adenosyl-L-Methionine-dependent methyltransferases superfamily proteins, among others. This indicates that T. atroviride could be using sRNAs as effector molecules, similarly to B. cinerea and V. dahliae, to establish a symbiotic relationship with Arabidopsis through interfering mRNAs involved in plant immunity, chromatin modifications and cell wall degrading enzymes, among others that remain to be determined. However, more research is needed to unravel the molecular mechanisms mediated by the fungal sRNAs that allows us to better understand the mechanisms that lead to the establishment of this beneficial association.
Conclusion
The consequence in the field of the presence of Trichoderma on plants can be indirect, for example by exerting antagonistic activity on potential phytopathogens, by attacking them, and by colonizing the rhizosphere, so they can avoid the contact of the pathogen with plant tissue. Direct beneficial effects on plants by Trichoderma are related to root colonization, although in many cases it has been shown that direct contact may not be necessary. Effectors of Trichoderma may play a key role in the success of colonization of the plant, first by establishing the initial contact and subsequently maintaining the fungus–plant interaction (Figure 1). Many of the effectors that have been identified in Trichoderma, and in other fungal symbionts, have reported activities described in their pathogenic counterparts. Therefore, the success of the interaction must not only rely on molecules playing these functions. The physiological state of both participants, as well as their threshold of perception toward the molecular signals exchanged are also important factors. This regulation does not depend solely on the genetic background of the fungus, the signals generated by the plant are also important. The study of effectors in Trichoderma is a relatively recent topic; however, thanks to technological advances to detect, identify and quantify molecules (proteins, secondary metabolites, and RNAs), the scientific community already has an extensive catalog of effector candidates. In most cases, it is necessary to carry out biological validation and determine the spectrum of action of these effectors on different plants at the level of cultivars, species and even genera. Moreover, the analysis of the interaction, considering longer times, may indicate how Trichoderma can change the plant physiology, during its complete life cycle. Due to the versatility shown by Trichoderma to associate with a wide variety of plants, it would be possible to determine the relevance of those candidates in different plant systems. In this review, we have focused mainly on the participation of the effectors in the interaction with plants, but its relevance in fungus–fungus associations is still pending, highlighting the idea of a multidirectional molecular exchange in the rhizosphere.
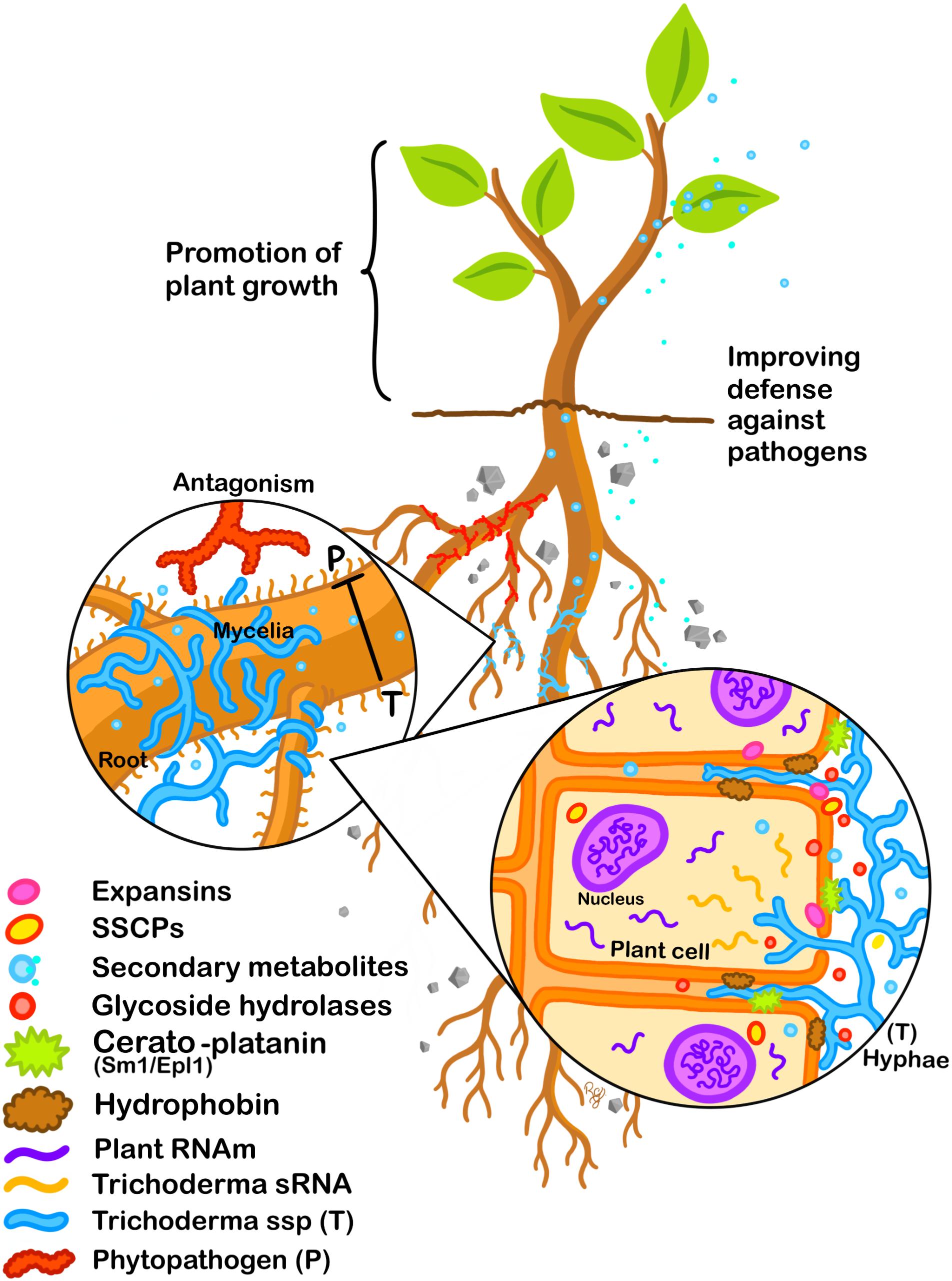
Figure 1. Effectors participate during Trichoderma–plant interaction. Antagonist activity of Trichoderma against phytopathogenic fungi. The release of molecules from Trichoderma with activity as effectors is highlighted. These molecules will modulate the plant hormonal balance as well as its defense response, allowing colonization. The beneficial association will result in the improvement of plant growth and in the resistance against phytopathogens.
Author Contributions
CR-V, SC-F, and VO-M contributed equally with ideas and discussion material. VO-M coordinated the work. CR-V wrote the review.
Funding
Research conducted by the groups, related to the topics of this review was supported by CONACyT grants CB-286709 and FC-2016-1538 conferred to VO-M and SC-F, respectively.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Bernardo Franco-Bárcenas Ph.D. for his useful suggestions and corrections and Roberto Gámez-Ramírez for his support to make the drawing in Figure 1.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01030/full#supplementary-material
Footnotes
References
Alba, J. M., Glas, J. J., Schimmel, B. C., and Kant, M. R. (2011). Avoidance and suppression of plant defenses by herbivores. J. Plant Interact. 6, 221–227. doi: 10.1080/17429145.2010.551670
Alonso-Ramírez, A., Poveda, J., Martín, I., Hermosa, R., Monte, E., and Nicolás, C. (2014). Salicylic acid prevents Trichoderma harzianum from entering the vascular system of roots. Mol. Plant Pathol. 15, 823–831. doi: 10.1111/mpp.12141
Andrade, C. M., Tinoco, M. L. P., Rieth, A. F., Maia, F. C. O., and Aragao, F. J. L. (2016). Host-induced gene silencing in the necrotrophic fungal pathogen Sclerotinia Sclerotiorum. Plant Pathol. 65, 626–632. doi: 10.1111/ppa.12447
Baccelli, I., Luti, S., Bernardi, R., Scala, A., and Pazzagli, L. (2013). Cerato-platanin shows expansin-like activity on cellulosic materials. Appl. Microbiol. Biotechnol. 98, 175–184. doi: 10.1007/s00253-013-4822-0
Bae, H., Roberts, D. P., Lim, H. S., Strem, M. D., Park, S. C., Ryu, C. M., et al. (2010). Endophytic Trichoderma isolates from tropical environments delay disease onset and induce resistance against Phytophthora capsica in hot pepper using multiple mechanisms. Mol. Plant Microbe Interact. 24, 336–351. doi: 10.1094/MPMI-09-10-0221
Bari, R., and Jones, J. D. (2009). Role of plant hormones in plant defense responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Barker, S. J., and Tagu, D. (2000). The roles of auxins and cytokinins in mycorrhizal symbionses. J. Plant Growth Regul. 19, 144–154.
Bent, A. F., and Mackey, D. (2007). Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. doi: 10.1146/annurev.phyto.45.062806.094427
Bigirimana, J., De Mejer, G., Poppe, J., Elad, Y., and Hofte, M. (1997). Induction of systemic resistance on bean (Phaseolus vulgaris) by Trichoderma harzianum. mededelingen van de faculteit landbouwkundige en toegepaste biologische wetenschappen, universiteit. Gent 62, 1001–1007.
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Brakhage, A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. doi: 10.1038/nrmicro2916
Brotman, Y., Briff, E., Viterbo, A., and Chet, I. (2008). Role of swollenin, an expansin-like protein from Trichoderma, in plant root colonization. Plant Physiol. 147, 779–789. doi: 10.1104/pp.108.116293
Calderón, A. A., Zapata, J. M., and Barceló, R. (1994). Peroxidase-mediated formation of resveratrol oxidation products during the hypersensitive-like reaction of grapevine cells to an elicitor from Trichoderma viride. Physiol. Mol. Plant Pathol. 44, 289–299. doi: 10.1016/s0885-5765(05)80031-1
Carpenter, M. A., Ridgway, H. J., Stringer, A. M., Hay, A. J., and Stewart, A. (2008). Characterisation of a Trichoderma hamatum monooxygenase gene involved in antagonistic activity against fungal plant pathogens. Curr. Genet. 53, 193–205. doi: 10.1007/s00294-007-0175-5
Carreras-Villaseñor, N., Esquivel-Naranjo, E. U., Villalobos-Escobedo, J. M., Abreu-Goodger, C., and Herrera-Estrella, A. (2013). The RNAi machinery regulates growth and development in the filamentous fungus T richoderma atroviride. Mol. Microbiol. 89, 96–112. doi: 10.1111/mmi.12261
Catska, V., Afifi, A. F., and Vancura, V. (1975). The effect of volatile and gaseous metabolites of swealling seeds on germination of fungal spores. Folia Microbiol. 20, 152–156. doi: 10.1007/bf02876772
Chadha, S., Mehetre, S. T., Bansal, R., Kuo, A., Aerts, A., Grigoriev, I. V., et al. (2018). Genome-wide analysis of the cytochrome p450 of Trichoderma spp.: annotation and evolutionary relationships. Fungal Biol. Biotechnol. 4:12. doi: 10.1186/s40694-018-0056-3
Chagas, L. F., Orozco, B. S., and Rodrigues, G. (2017). Rice growth influence by Trichoderma spp. with natural phosphate fertilization under greenhouse conditions. IJDR 7, 13147–13152.
Chanclud, E., Kisiala, A., Emery, N. R., Chalvon, V., Ducasse, A., Romiti-Michel, C., et al. (2016). Cytokinin production by the rice blast fungus is a pivotal requirement for full virulence. PLoS Pathog. 12:e1005457. doi: 10.1371/journal.ppat.1005457
Chanclud, E., and Morel, J. B. (2016). Plant hormones: a fungal point of view. Mol. Plant Pathol. 17, 1289–1297. doi: 10.1111/mpp.12393
Chang, S., Zhang, Z., and Liu, Y. (2012). RNA interference pathway in fungi: mechanisms and functions. Annu. Rev. Microbiol. 66, 305–323. doi: 10.1146/annurev-micro-092611-150138
Charpenter, M., Sun, J., Wen, J., Mysore, K. S., and Oldroyd, G. E. (2014). Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the protein phosphatase 2A complex. Plant Physiol. 166, 2077–2090. doi: 10.1104/pp.114.246371
Cheng, C. H., Shen, B. N., Shang, Q. W., Liu, L. D., Peng, K. C., Chen, Y. H., et al. (2018). Gene-to-gene network análisis of the mediation of plant innate immunity by the eliciting plant response-like 1 (Epl1) elicitor of Thicoderma formosa. Mol. Plant Microbe Interact. 31, 683–691. doi: 10.1094/MPMI-01-18-0002-TA
Chiang, Y. M., Oakley, B. R., Keller, N. P., and Wang, C. C. (2010). Unraveling polyketide synthesis in members of the genus Aspergillus. Appl. Microbiol. Biothechnol. 86, 1719–1736. doi: 10.1007/s00253-010-2525-3
Christianson, D. W. (2008). Unearthing the roots of the terpenome. Curr. Opin. Chem. Biol. 12, 141–150. doi: 10.1016/j.cbpa.2007.12.008
Contreras-Cornejo, H. A., López-Bucio, J., Méndez-Bravo, A., Macías-Rodríguez, L., Ramos-Vega, M., Guevara-García, A. A., et al. (2015). Mitogen-Activated Protein Kinase 6 and ethylene and auxin signaling pathways are involved in Arabidopsis root-system architecture alteratios by Trichoderma atroviride. Mol. Plant Microbe Interact. 28, 701–710. doi: 10.1094/MPMI-01-15-0005-R
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Beltrán-Peña, E., Herrera-Estrella, A., and López-Bucio, J. (2011). Trichoderma-induced plant immunity likely involves both hormonal- and camalexin-dependent mechanisms in Arabidopsis thaliana and confers resistance against necrotrophic fungi Botrytis cinerea. Plant Signal. Behav. 6, 1554–1563. doi: 10.4161/psb.6.10.17443
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Cortés-Penagos, C., and López-Bucio, J. (2009). Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 149, 1579–1592. doi: 10.1104/pp.108.130369
Crutcher, F. K., Moran-Diez, M. E., Ding, S. L., Liu, J., Horwitz, B., Mukherjee, P. K., et al. (2015). A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens. Fungal Biol. 119, 476–486. doi: 10.1016/j.funbio.2015.01.004
Cruz-Magalhães, V., Nieto-Jacobo, M. F., van Zijll de Jong, E., Rostás, M., Padilla-Arizmendi, F., Kandula, D., et al. (2019). The NADPH oxidases Nox1 and Nox2 differentially regulate volatile organic compounds, fungistatic activity, plant growth promotion and nutrient assimilation in Trichoderma atroviride. Front. Microbiol. 23:3271. doi: 10.3389/fmicb.2018.03271
de Jonge, R., van Esse, H. P., Kombrink, A., Shinya, T., Desaki, Y., Bours, R., et al. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329, 953–955. doi: 10.1126/science.1190859
Di, X., Takken, F. L. W., and Tintor, N. (2016). How phytohormones shape interactions between plants and the soil-borne fungus Fusarium oxysporum. Front. Plant Sci. 7:170. doi: 10.3389/fpls.2016.00170
Djonovic, S., Pozo, M. J., Dangott, L. J., Howell, C. R., and Kenerley, C. M. (2006). Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant Microbe Interact. 19, 838–853. doi: 10.1094/mpmi-19-0838
Djonovic, S., Vargas, W. A., Kolomiets, M. V., Horndeski, M., Wiest, A., and Kenerley, C. M. (2007). A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–889. doi: 10.1104/pp.107.103689
Druzhinina, I. S., Shelest, E., and Kubicek, C. P. (2012). Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiol. Lett. 337, 1–9. doi: 10.1111/j.1574-6968.2012.02665.x
Duclohier, H., and Wróblewski, H. (2001). Voltage-dependent pore formation and antimicrobial activity by alamethicin and analogues. J. Membr. Biol. 184, 1–12. doi: 10.1007/s00232-001-0077-2
Egamberdieva, D., Wirth, S. J., Alqarawi, A. A., Abd Allah, E. F., and Hashem, A. (2017). Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 31:2104. doi: 10.3389/fmicb.2017.02104
Estrada-Rivera, M., Rebolledo-Prudencio, O. G., Pérez-Robles, D. A., Rocha-Medina, M. C., González-López, M. C., and Casas-Flores, S. (2019). Histone deacetylase HDA-2 is essential in Trichoderma to modulate multiple responses in Arabidopsis. Plant Physiol. 179, 1343–1361. doi: 10.1104/pp.18.01092
Gaderer, R., Lamdan, N., Frischmann, A., Sulyok, M., Krska, R., Horwitz, B. A., et al. (2015). Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize. BMC Microbiol. 15:2. doi: 10.1186/s12866-014-0333-0
Garnica-Vergara, A., Barrera-Ortiz, S., Muñoz-Parra, E., Raya-González, J., Méndez-Bravo, A., Macías-Rodríguez, L., et al. (2016). The volatile 6-pentyl-2H-pyran-2-one from Trichoderma atroviride regulates Arabidopsis thaliana root morphogenesis via auxin signaling and ETHYLENE INSENSITIVE 2 functioning. New Phytol. 209, 1496–1512. doi: 10.1111/nph.13725
Gomes, E. V., Costa, N., de Paula, R. G., de Azevedo, R. R., da Silva, F. L., Noronha, E. F., et al. (2015). The cerato-platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self wall protection. Sci. Rep. 5:17998. doi: 10.1038/srep17998
Guzmán-Guzmán, P., Alemán-Duarte, M. I., Delaye, L., Herrera-Estrella, A., and Olmedo-Monfil, V. (2017). Identification of effector-like proteins in Trichoderma spp. and role of hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 18:16. doi: 10.1186/s12863-017-0481-y
Guzmán-Guzmán, P., Porras-Troncoso, M. D., Olmedo-Monfil, V., and Herrera-Estrella, A. (2019). Trichoderma species: versatile plant symbionts. Phytopathology 109, 6–16. doi: 10.1094/PHYTO-07-18-0218-RVW
Harman, G. E. (2006). Overwiew of mechanisms and uses of Trichoderma spp. Phytopathology 96, 190–194. doi: 10.1094/PHYTO-96-0190
Harman, G. E., Petzoldt, R., Comis, A., and Chen, J. (2004). Interactions between Trichoderma harzianum strain T22 and maize inbred line Mo17 and effects of these interactions on diseases caused by Pythium ultimum and Colletotrichum graminicola. Phytopathology 94, 147–153. doi: 10.1094/PHYTO.2004.94.2.147
Hitzenhammer, E., Büschl, C., Sulyok, M., Schuhmacher, R., Kluger, B., Wischnitzki, E., et al. (2019). YPR2 is a regulator of light modulated carbon and secondary metabolism in Trichoderma reesei. BMC Genomics 20:211. doi: 10.1186/s12864-019-5574-8
Hu, Z., Parekh, U., Maruta, N., Trusov, Y., and Botella, J. R. (2015). Down-regulation of Fusarium oxysporum endogenous genes by host-delivered RNA interference enhances disease resistance. Front. Chem. 3:1. doi: 10.3389/fchem.2015.00001
Huang, Y., Mijiti, G., Wang, Z., Yu, W., Fan, H., Zhang, R., et al. (2015). Functional analysis of the class II hydrophobins gene HFB2-6 from the biocontrol agent Trichoderma asperellum ACCC30536. Microbiol. Res. 171, 8–20. doi: 10.1016/j.micres.2014.12.004
Kamoun, S. (2006). A catalogue of the effector secretome of plant pathogenic oomycets. Annu. Rev. Phytopathol. 44, 41–60. doi: 10.1146/annurev.phyto.44.070505.143436
Kapfer, G. F., Berger, R. G., and Drawert, F. (1989). Production of 4-decanolid by semi-continuous fermentation of Tyromyces sambuceus. Biotechnol. Lett. 11, 561–566. doi: 10.1007/bf01040036
Kloppholz, S., Kuhn, H., and Requena, N. (2011). A secreted fungal effector of Glomus intrarradices promotes symbiotic biotrophy. Curr. Biol. 21, 1204–1209. doi: 10.1016/j.cub.2011.06.044
Koch, A., Kumar, N., Weber, L., Keller, H., Imani, J., and Kogel, K. H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase–encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. U.S.A. 110, 19324–19329. doi: 10.1073/pnas.1306373110
Kombrink, A., Rovenich, H., Shi-Kunne, X., Rojas-Padilla, E., van den Berg, G. C., Domazakis, E., et al. (2017). Verticillium dahliae LysM effectors differentially contribute to virulence on plants hosts. Mol. Plant Pathol. 18, 596–608. doi: 10.1111/mpp.12520
Kubicek, C. P., Herrera, A., Seidl, V., Martinez, D., Druzhinina, I. S., Thon, M., et al. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12, R40. doi: 10.1186/gb-2011-12-4-r40
Kulkarni, R. D., Thon, M. R., Pan, H., and Dean, R. A. (2005). Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 6, R24.
Lamdan, N., Shalaby, S., Ziv, T., Kenerley, C. M., and Horwitz, B. A. (2015). Secretome of the biocontrol fungus Trichoderma virens co-cultured with maize roots: tole in induced systemic resistance. Mol. Cell Proteomics 14, 1054–1063. doi: 10.1074/mcp.m114.046607
Lee, S., Yap, M., Behringer, G., Hung, R., and Bennett, J. W. (2016). Volatile organic compounds emitted by Trichoderma species mediate plant growth. Fungal Biol. Biotechnol. 29:7. doi: 10.1186/s40694-016-0025-7
Li, M., Li, G., and Zhang, K. (2019). Non-volatile metabolites from Trichoderma spp. Metabolites 9:58. doi: 10.3390/metabo9030058
Lopes, F. A., Steindorff, A. S., Geraldine, A. M., Brandao, R. S., Monteiro, V. N., Lobo, M. Jr., et al. (2012). Biochemical and metabolic profiles of Trichoderma strains isolated from common bean crops in the Brazilian Cerrado, and potential antagonism against Sclerotinia sclerotiorum. Fungal Biol. 116, 815–824. doi: 10.1016/j.funbio.2012.04.015
Macheleidt, J., Mattern, D. J., Fischer, J., Netzker, T., Weber, J., Schroeckh, V., et al. (2016). Regulation and role of fungal secondary metabolites. Annu. Rev. Genet. 23, 371–392.
Macías-Rodríguez, L., Guzmán-Gómez, A., García-Juárez, P., and Contreras-Cornejo, H. A. (2018). Trichoderma atroviride promotes tomato development and alters the root exudation of carbohydrates, which stimulates fungal growth and the biocontrol of the phytopathogen Phytophtora cinnamomic in a tripartite interaction system. FEMS Microbiol. Ecol. 94
Malmierca, M. G., Cardoza, R. E., Alexander, N. J., McCormick, S. P., Hermosa, R., Monte, E., et al. (2012). Involvement of Trichoderma trichothecenes in the biocontrol activity and induction of plant defense-related genes. Appl. Environ. Microbiol. 78, 4856–4868. doi: 10.1128/AEM.00385-12
Malmierca, M. G., McCormick, S. P., Cardoza, R. E., Alexander, N. J., Monte, E., and Gutiérrez, S. (2015). Production of trichodiene by Trichoderma harzianum alters the perception of this biocontrol strain by plants and antagonized fungi. Environ. Microbiol. 17, 2628–2646. doi: 10.1111/1462-2920.12506
Manganiello, G., Sacco, A., Ercolano, M. R., Vinale, F., Lanzuise, S., Pascale, A., et al. (2018). Modulation of tomato response to Rhizoctonia solani by Trichoderma harzianum and its secondary metabolite harzianic acid. Front. Microbiol. 9:1966. doi: 10.3389/fmicb.2018.01966
Martínez, C., Blanc, F., Le Claire, E., Besnard, O., Nicole, M., and Baccou, J. C. (2001). Salicylic acid and ethylene pathways are differentially activated in melon cotyledons by active or heat-denatured cellulase from Trichoderma lingibrachiatum. Plant Physiol. 127, 334–344. doi: 10.1104/pp.127.1.334
Martínez-Medina, A., Appels, F. V. W., and van Wees, S. C. M. (2017). Impact of salycilic acid- and jasmonic acid-regulated defenses on root colonization by Trichoderma harzianum T-78. Plant Signal. Behav. 12, e1345404. doi: 10.1080/15592324.2017.1345404
Martínez-Medina, A., Fernández, I., Lok, G. B., Pozo, M. J., Pieterse, C. M., and Van Wees, S. C. (2016). Shifting from priming of salicylic acid- to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 13, 1363–1377. doi: 10.1111/nph.14251
Mastouri, F., Björkman, T., and Harman, G. E. (2012). Trichoderma harzianum enhances antioxidant defense of tomato seedlings and resistance to water deficit. Mol. Plant Microbe Interact. 25, 1264–1271. doi: 10.1094/MPMI-09-11-0240
Masunaka, A., Hyakumachi, M., and Takenaka, S. (2011). Plant growthpromoting fungus. Trichoderma koningi suppresses isoflavonoid phytoalexin vestitol production for colonization on/in the roots of Lotus japonicus. Microbes Environ. 26, 128–134. doi: 10.1264/jsme2.me10176
Mendoza-Mendoza, A., Zaid, R., Lawry, R., Hermosa, R., Monte, E., Horwitz, A., et al. (2018). Molecular dialogues between Trichoderma and roots: role of the fungal secretome. Fungal Biol. Rev. 32, 62–85. doi: 10.1016/j.fbr.2017.12.001
Morán-Diez, E., Hermosa, R., Ambrosino, P., Cardoza, R. E., Gutiérrez, S., Lorito, M., et al. (2009). The ThPG1 endopolygalacturonase is required for the Trichoderma harzianum-plant beneficial interaction. Mol. Plant Microbe Interact. 22, 1021–1031. doi: 10.1094/MPMI-22-8-1021
Morán-Diez, M. E., Trushina, N., Lamdan, N. L., Rosenfelder, L., Mukherjee, P. K., Kenerley, C. M., et al. (2015). Host-specific transcriptomic pattern of Trichoderma virens during interaction with maize or tomato roots. BMC Genomics 16:8. doi: 10.1186/s12864-014-1208-3
Moscatiello, R., Sello, S., Ruocco, M., Barbulova, A., Cortese, E., Nigris, S., et al. (2018). The hydrophobin HYTLO1 secreted by the biocontrol fungus Trichoderma longibrachiatum triggers a NAADP-mediated calcium signaling pathway in Lotus japonicus. Int. J. Mol. Sci. 19, E2596. doi: 10.3390/ijms19092596
Mostafa, M. A., and Gayed, S. K. (1952). Effect of Trichoderma metabolites on growth of cotton plants. Nature 169, 359–360. doi: 10.1038/169359a0
Mukherjee, P. K., Buensanteai, N., Morán-Diez, M. E., and Druzhinina, I. S. (2012). Functional analysis of non-ribosomal peptide synthetases (NRPSs) in Trichoderma virens reveals a polyketide synthase (PKS)/NRPS hybrid enzyme involved in the induced systemic resistance response in maize. Microbiology 158(Pt 1), 155–165. doi: 10.1099/mic.0.052159-0
Mukherjee, P. K., Horwitz, B., Herrera-Estrella, A., Schmoll, M., and Kenerley, C. M. (2013). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51, 105–129. doi: 10.1146/annurev-phyto-082712-102353
Mukherjee, P. K., and Kenerley, C. M. (2010). Regulation of morphogenesis and biocontrol properties in Trichoderma virens by a VELVET protein. Vel1. Appl. Environ. Microbiol. 76, 2345–2352. doi: 10.1128/AEM.02391-09
Ng, D. W., Zhang, C., Miller, M., Palmer, G., Whiteley, M., Tholl, D., et al. (2011). Cis- and trans-regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 23, 1729–1740. doi: 10.1105/tpc.111.083915
Nieto-Jacobo, M. F., Steyaert, J. M., Salazar-Badillo, F. B., Nguyen, D. V., Rostás, M., Braithwaite, M., et al. (2017). Environmental growth conditions of Trichoderma spp. affects indole acetic acid derivatives, volatile organic compounds, and plant growth promotion. Front. Plant Sci. 8:102. doi: 10.3389/fpls.2017.00102
Nogueira-Lopez, G., Greenwood, D. R., Middleditch, M., Winefield, C., Eaton, C., Steyaert, J. M., et al. (2018). The apoplastic secretome of Trichoderma virens during interaction with maize roots shows an inhibition of plant defense and scavenging oxidative stress secreted proteins. Front. Plant Sci. 5:409. doi: 10.3389/fpls.2018.00409
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Osugi, A., and Sakakibara, H. (2015). Q&A: how do plants respond to cytokinins and what is their importance? BMC Biol. 27:102. doi: 10.1186/s12915-015-0214-5
Pascale, A., Vinale, F., Manganiello, G., Nigro, M., Lanzuise, S., Ruocco, M., et al. (2017). Trichoderma and its secondary metabolites improve yield and quality of grapes. Crop Prot. 92, 176–181. doi: 10.1016/j.cropro.2016.11.010
Pazzagli, L., Seidl-Seiboth, V., Barsottini, M., Vargas, W. A., Scala, A., and Mukherjee, P. K. (2014). Cerato-platanins: elicitors and effectors. Plant Sci. 228, 79–87. doi: 10.1016/j.plantsci.2014.02.009
Pérez, A., Ramage, G., Blanes, R., Murgui, A., Casanova, M., and Martínez, J. P. (2011). Some biological features of Candida albicans mutants for genes coding fungal proteins containing the CFEM domain. FEMS Yest Res. 11, 273–284. doi: 10.1111/j.1567-1364.2010.00714.x
Pieterse, C. M., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C., and Bakker, P. A. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Plett, J. M. M., Kemppainen, M., Kale, S. D. D., Kohler, A., Legué, V., Brun, A., et al. (2011). A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr. Biol. 21, 1197–1203. doi: 10.1016/j.cub.2011.05.033
Pumplin, N., and Voinnet, O. (2013). RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 11:745. doi: 10.1038/nrmicro3120
Ramírez-Valdespino, C. A., Porras-Troncoso, M. D., Corrales-Escobosa, A. R., Wrobel, K., Martínez-Hernández, P., and Olmedo-Monfil, V. (2018). Functional characterization of TvCyt2, a member of the p450 monooxygenases from Trichoderma virens relevant during the association with plants and mycoparasitism. Mol. Plant Microbe Interact. 31, 289–298. doi: 10.1094/MPMI-01-17-0015-R
Reino, J. L., Guerrero, R. F., Hernández-Galan, R., and Collado, I. G. (2008). Secondary metabolites from species of the biocontrol agent Trichoderma. Phytochem. Rev. 7, 89–123. doi: 10.1007/s11101-006-9032-2
Reithner, B., Brunner, K., Schuhmacher, R., Peissl, I., Seidl, V., Krska, R., et al. (2005). The G protein alpha subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet. Biol. 42, 749–760. doi: 10.1016/j.fgb.2005.04.009
Reithner, B., Mach-Aigner, A. R., Herrera-Estrella, A., and Mach, R. L. (2014). Trichoderma atroviride transcriptional regulator Xyr1 supports the induction of systemic resistance in plants. Appl. Environ. Microbiol. 80, 5274–5281. doi: 10.1128/AEM.00930-14
Rippa, S., Adenier, H., Derbaly, M., and Béven, L. (2007). The Peptaibol Alamethicin Induces an rRNA-Cleavage-Associated Death in Arabidopsis thaliana. Chem. Biodivers. 4, 1360–1373. doi: 10.1002/cbdv.200790116
Rippa, S., Eid, M., Formaggio, F., Toníolo, C., and Béven, L. (2010). Hypersensitive-like response to the pore-former peptaibol alamethicin in Arabidopsis thaliana. ChemBioChem 11, 2042–2049. doi: 10.1002/cbic.201000262
Rocha, A. L., Maeda, N., Pereira, N., Kern, F., Elias, L., Simister, R., et al. (2016). Characterization of the cellulolytic secretome of Trichoderma harzianum during growth on sugarcane bagasse and analysis of the activity boosting effects of swwollenin. Biothechnol. Prog. 32, 327–336. doi: 10.1002/btpr.2217
Rotblat, B., Enshell-Seijffers, D., Gershoni, J. M., Schuster, S., and Avni, A. (2002). Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J. 32, 1049–1055. doi: 10.1046/j.1365-313x.2002.01490.x
Ruocco, M., Lanzuise, S., Vinale, F., Marra, R., Turrà, D., Woo, S. L., et al. (2009). Identification of a new biocontrol gene in Trichoderma atroviride: the role of an ABC transporter membrane pump in the interaction with different plant-pathogenic fungi. Mol. Plant Microbe Interact. 22, 291–301. doi: 10.1094/MPMI-22-3-0291
Ruocco, M., Lanzuise, S., Woo, S. L., Vinale, F., Marra, R., Varlese, R., et al. (2015). Multiple roles and effects of a novel Trichoderma hydrophobin. Mol. Plant Microbe Interact. 28, 167–179. doi: 10.1094/MPMI-07-14-0194-R
Salas-Marina, M. A., Isordia-Jasso, M. I., Islas-Osuna, M. A., Delgado-Sánchez, P., Jiménez-Bremont, J. F., Rodríguez-Kessler, M., et al. (2015). The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum. Front. Plant Sci. 23:77. doi: 10.3389/fpls.2015.00077
Salas-Marina, M. A., Silva-Flores, M. A., Uresti-Rivera, E. E., Castro-Longoria, E., Herrera-Estrella, A., and Casas-Flores, S. (2011). Colonization of Arabidopsis roots by Trichoderma atroviride promotes growth and enhances systemic disease resistance through jasmonic acid/ethylene and salicylic acid pathways. Eur. J. Plant Pathol. 131, 15–26. doi: 10.1007/s10658-011-9782-6
Saravanakumar, K., Fan, L., Fu, K., Yu, C., Wang, M., Xia, H., et al. (2016). Cellulase from Trichoderma harzianum interacts with roots and triggers induced systemic resistence to foliar disease in maize. Sci. Rep. 10:35543. doi: 10.1038/srep35543
Saravanakumar, K., Wang, S., Dou, K., Lu, Z., and Chen, J. (2018). Yest two-hybrid and label-free proteomics-based screening of maize root receptor to cellulase of Trichoderma harzianum. Physiol. Mol. Plant Pathol. 104, 86–94. doi: 10.1016/j.pmpp.2018.10.002
Schenkel, D., Lemfack, M. C., Piechulla, B., and Splivallo, R. (2015). A meta-analysis approach for assessing the diversity and specificity of belowground root and microbial volatiles. Front. Plant Sci. 14:707. doi: 10.3389/fpls.2015.00707
Seidl, V., Marchetti, M., Schandl, R., Kubicek, C. P., Allmaier, G., and Kubicek, C. P. (2006). Epl1, the major secreted protein of Hypocrea atroviridis on glucose, is a member of a strongly conserved protein family comprising plant defense response elicitors. FEBS J. 273, 4346–4359. doi: 10.1111/j.1742-4658.2006.05435.x
Seidl, V., Zach, S., Frischmann, A., Spadiut, O., Dietzsch, C., Herwig, C., et al. (2013). Spore germination of Trichoderma atroviride is inhibited by its LysM protein TAL6. FEBS J. 280, 1226–1236. doi: 10.1111/febs.12113
Seo, J., Wu, J., Lii, Y., Li, Y., and Jin, H. (2013). Contribution of Small RNA Pathway Components in Plant Immunity. Mol. Plant Microbe Interact. 26, 617–625. doi: 10.1094/MPMI-10-12-0255-IA
Serrano-Carreon, L., Hathout, Y., Bensoussan, M., and Belin, J. M. (1993). Metabolism of linoleic acid or mevalonate and 6-pentyl-alpha-pyrone biosynthesis by Trichoderma species. Appl. Environ. Microbiol. 59, 2945–2950.
Sharon, A., Fuchs, Y., and Anderson, J. D. (1993). The elicitation of ethylene biosynthesis by a Trichoderma xylanase is not related to the cell wall degradation activity of the enzyme. Plant Physiol. 102, 1325–1329. doi: 10.1104/pp.102.4.1325
Shen, Q., Liu, L., Wang, L., and Wang, Q. (2018). Indole primes plant defense against necrotrophic fungal pathogen infection. PLoS One 13:e0207607. doi: 10.1371/journal.pone.0207607
Shi, W. L., Chen, X. L., Wang, L. X., Gong, Z. T., Li, S., Li, C. L., et al. (2016). Cellular and molecular insight into the inhibition of primary root growth of Arabidopsis induced by peptaibols, a class of linear peptide antibiotics mainly produced by Trichoderma spp. J. Exp. Bot. 67, 2191–2205. doi: 10.1093/jxb/erw023
Shoresh, M., Yedida, I., and Chet, I. (2005). Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 96, 76–84. doi: 10.1094/PHYTO-95-0076
Singh, H. B. (2014). Management of plant pathogens with microorganisms. Nat. Sci. Acad. 80, 443–454.
Thomma, B. P., Nürnberger, T., and Joosten, M. H. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15. doi: 10.1105/tpc.110.082602
Tinoco, M. L., Dias, B. B., Dall’Astta, R. C., Pamphile, J. A., and Aragao, F. J. (2010). In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol. 31:27. doi: 10.1186/1741-7007-8-27
Tucci, M., Roucco, M., De Masi, L., De Palma, M., and Lorito, M. (2011). The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 12, 341–354. doi: 10.1111/j.1364-3703.2010.00674.x
Van Loon, L. C., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Vargas, W. A., Djonovic, S., Sukno, S. A., and Kenerley, C. M. (2008). Dimerization controls the activity of fungal elicitors that trigger systemic resistance in plants. J. Biol. Chem. 283, 19804–19815. doi: 10.1074/jbc.M802724200
Velázquez-Robledo, R., Contreras-Cornejo, H. A., Macías-Rodrígez, L., Hernández-Morales, A., Aguirre, J., Casas-Flores, S., et al. (2011). Role of the 4-phosphopantetheinyl transferase of Trichoderma virens in secondary metabolism and induction of plant defense response. Mol. Plant Microbe Interact. 24, 1459–1471. doi: 10.1094/mpmi-02-11-0045
Vinale, F., Flematti, G., Sivasithamparam, K., Lorito, M., Marra, R., Skelton, B. W., et al. (2009). Harzianic acid, an antifungal and plant growth promoting metabolite from Trichoderma harzianum. J. Nat. Prod. 72, 2032–2035. doi: 10.1021/np900548p
Vinale, F., Strakowska, J., Mazzei, P., Piccolo, A., Marra, R., Lombardi, N., et al. (2016). Cremenolide, a new antifungal, 10-member lactone from Trichoderma cremeum with plant growth promotion activity. Nat. Prod. Res. 30, 2575–2581. doi: 10.1080/14786419.2015.1131985
Vishnevetsky, J., White, T. L. Jr., Palmateer, A. J., Flaishman, M., Cohen, Y., Elad, Y., et al. (2010). Improved tolerance toward fungal diseases in transgenic Cavendish banana (Musa spp AAA group) cv. grand nain. Transgenic Res. 20, 61–72. doi: 10.1007/s11248-010-9392-7
Viterbo, A., and Chet, I. (2010). TasHyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol Plant Pathol. 7:4. doi: 10.1111/j.1364-3703.2006.00335.x
Viterbo, A., Wiest, A., Brotman, Y., Chet, I., and Kenerley, C. (2007). The 18mer peptaibols from Trichoderma virens elicit plant defense responses. Mol. Plant Pathol. 8, 737–746. doi: 10.1111/j.1364-3703.2007.00430.x
Wang, M., Weiberg, A., Dellota, E. Jr., Yamane, D., and Jin, H. (2017). Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 14, 421–428. doi: 10.1080/15476286.2017.1291112
Wang, B., Sun, Y., Song, N., Zhao, M., Liu, R., Feng, H., et al. (2017). Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 215, 338–350. doi: 10.1111/nph.14577
Wang, M., Weiberg, A., Lin, F. M., Thomma, B. P., Huang, H. D., and Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants. 2, 16151. doi: 10.1038/nplants.2016.151
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhao, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Yedidia, I., Benhamou, N., and Chet, I. (1999). Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 65, 1061–1070.
Yedidia, I., Benhamou, N., Kapulnik, Y., and Chet, I. (2000). Induction and accumulation of PR proteins activity during early stages of root colonization by the mycoparasite Trichoderma harzianum strain T-203. Plant Physiol. Biochem. 38, 863–873. doi: 10.1016/s0981-9428(00)01198-0
Yedidia, I., Shoresh, M., Kerem, Z., Benhamou, N., Kapulnik, Y., and Chet, I. (2003). Concomitant induction of systemic resistance to Pseudomonas syringae pv. Lachrymans in cucumber by Trichoderma asperellum (T-203) and acumulation of phytoalexins. Am. Soc. Microbiol. 69, 7343–7353. doi: 10.1128/aem.69.12.7343-7353.2003
Yin, C., Jurgenson, J. E., and Hulbert, S. H. (2011). Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant-Microbe Interact. 24, 554–561. doi: 10.1094/MPMI-10-10-0229
Yin, C., Park, J. J., Gang, D. R., and Hulbert, S. H. (2014). Characterization of a tryptophan 2-monooxygenase gene from Puccinia graminis f. sp. tritici involved in auxin biosynthesis and rust pathogenicity. Mol. Plant-Microbe Interact. 27, 227–235. doi: 10.1094/MPMI-09-13-0289-FI
Yu, J. H., and Keller, N. (2005). Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43, 437–458. doi: 10.1146/annurev.phyto.43.040204.140214
Yu, W., Mijiti, G., Huang, Y., Fan, H., Wang, Y., and Liu, Z. (2018). Functional analysis of eliciting plant response protein Epl1-Tas from Trichoderma asperellum ACCC30536. Sci. Rep. 8:7974. doi: 10.1038/s41598-018-26328-1
Keywords: Trichoderma, effector molecules, plant–microbe interactions, secondary metabolites, effector proteins, small RNA
Citation: Ramírez-Valdespino CA, Casas-Flores S and Olmedo-Monfil V (2019) Trichoderma as a Model to Study Effector-Like Molecules. Front. Microbiol. 10:1030. doi: 10.3389/fmicb.2019.01030
Received: 30 November 2018; Accepted: 24 April 2019;
Published: 15 May 2019.
Edited by:
Sabrina Sarrocco, University of Pisa, ItalyReviewed by:
Paul Dean, Teesside University, United KingdomFrancesco Vinale, Istituto per la Protezione Sostenibile delle Piante (CNR), Italy
Copyright © 2019 Ramírez-Valdespino, Casas-Flores and Olmedo-Monfil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vianey Olmedo-Monfil, dmcub2xtZWRvQHVndG8ubXg=
 Claudia A. Ramírez-Valdespino
Claudia A. Ramírez-Valdespino Sergio Casas-Flores
Sergio Casas-Flores Vianey Olmedo-Monfil
Vianey Olmedo-Monfil