- Department of Biochemistry and Molecular Biology, Faculty of Veterinary Medicine, Wrocław University of Environmental and Life Sciences, Wrocław, Poland
Initial attachment to host intestinal mucosa after oral infection is one of the most important stages during bacterial pathogenesis. Adhesive structures, widely present on the bacterial surface, are mainly responsible for the first contact with host cells and of host-pathogen interactions. Among dozens of different bacterial adhesins, type 1 fimbriae (T1F) are one of the most common adhesive organelles in the members of the Enterobacteriaceae family, including Salmonella spp., and are important virulence factors. Those long, thin structures, composed mainly of FimA proteins, are responsible for recognizing and binding high-mannose oligosaccharides, which are carried by various glycoproteins and expressed at the host cell surface, via FimH adhesin, which is presented at the top of T1F. In this review, we discuss investigations into the functions of T1F, from the earliest work published in 1958 to operon organization, organelle structure, T1F biogenesis, and the various functions of T1F in Salmonella-host interactions. We give special attention to regulation of T1F expression and their role in binding of Salmonella to cells, cell lines, organ explants, and other surfaces with emphasis on biofilm formation and discuss T1F role as virulence factors based on work using animal models. We also discuss the importance of allelic variation in fimH to Salmonella pathogenesis, as well as role of FimH in Salmonella host specificity.
Introduction
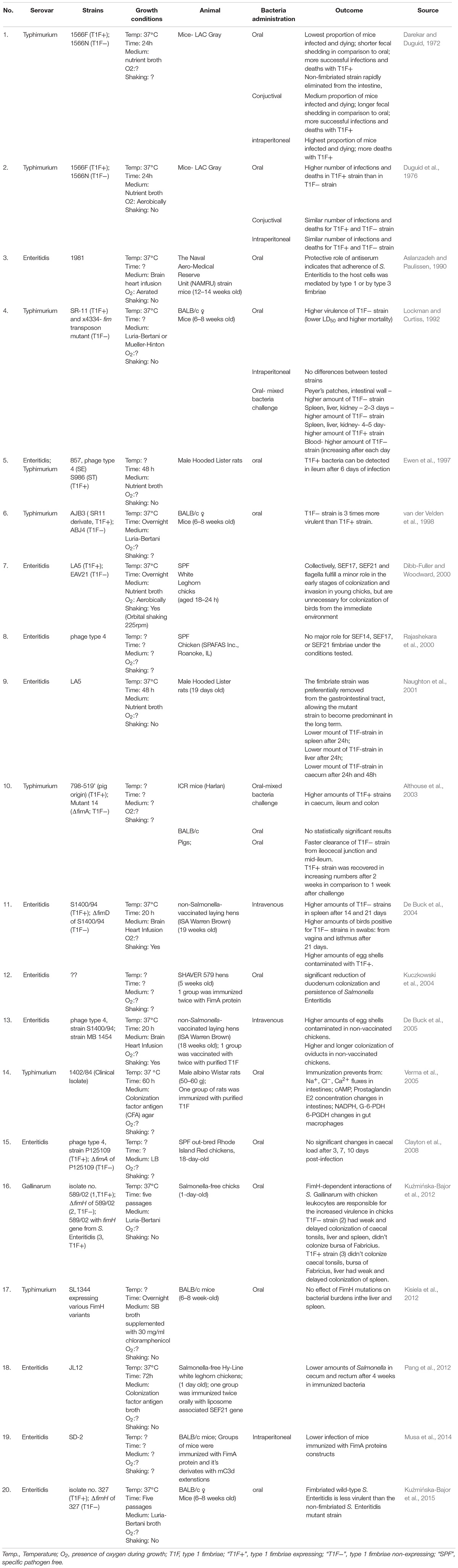
Adhesion to host tissues is thought to be one of the crucial events during Salmonella pathogenesis. Among dozens of different bacterial adhesins, type 1 fimbriae (T1F) are one of the most common adhesive organelles in the members of the Enterobacteriaceae family, including Salmonella spp., and are important virulence factors. The fim fimbrial cluster is one of seven most abundant fimbrial clusters (including fim, bcf, stb, sth, std, saf, and sti) in the genome of Salmonella spp. (Yue et al., 2012). The importance of T1F in Salmonella biology is confirmed by the fact that it is expressed in more than 80% of 1453 clinical isolates, representing 149 serovars (Duguid et al., 1966). The Salmonella fim cluster comprises 10 genes (fimA, fimI, fimC, fimD, fimH, fimF, fimZ, fimY, fimW, and stm0551) and an tRNA-Arg (Purcell et al., 1987; Boyd and Hartl, 1999). Of these, fimA, fimI, fimC, fimD, fimH, and fimF, comprise a single operon, under the control of fimA promoter region (PfimA). The six genes in this operon encode proteins involved in biogenesis and structure of T1F (Figure 1A). FimW, FimY, FimZ proteins and the STM0551 open reading frame are involved in transcriptional regulation of T1F and tRNA-Arg additionally controls expression of T1F on translational level. T1F are rod shaped structures composed of primarily 500 to 3000 FimA monomers (Hahn et al., 2002), with a single lectin-like protein, FimH, which is directly involved in the binding of high-mannose oligosaccharides carried by surface glycoproteins of eukaryotic cells and is placed on the top of the fimbrial shaft by FimF (Figure 1B).
Figure 1. Schematic representation of Salmonella type 1 fimbriae (T1F). (A) fim gene cluster organization, (B) structure of fimbrium, and (C) biogenesis by the chaperon-usher pathway.
Type 1 fimbriae are assembled by the chaperone-usher pathway (for a detailed review see Waksman and Hultgren, 2009; Werneburg and Thanassi, 2018; Figure 1C). All proteins needed for the assembly of T1F contain signal peptides. FimC acts as a chaperone for FimA, FimF and FimH, preventing premature polymerization in the periplasm, and takes part in folding and assembly of the fimbriae. FimA, FimF, and FimH contain hydrophobic N- and C-terminal extensions that are bound by a complementary hydrophobic groove in FimC. FimD is an usher outer-membrane protein that exports fimbrial proteins through the outer membrane and facilitates fimbriae subunit assembly. All of the proteins that constitute T1F are linked together by N- and C-terminal extensions, a process referred to as donor strand exchange (Remaut et al., 2006). T1F assembly starts with the binding of the FimC-FimH complex to the FimD usher protein. The FimC–FimF complex is next transferred into the FimD pocket, and the FimC bound to the C-terminal extension of FimH is exchanged for the N-terminal extension of FimF, resulting in the formation of the FimH–FimF complex. In the next step, donor strand exchange is repeated with FimA and further elongation of the fimbrial shaft is continued with FimA. Deletion of any one of fimA, fimF or fimH results in no fimbriae production (Zeiner et al., 2012), ascribing a shared role for all of these genes in pilus biogenesis. Though it has been speculated that fimI is required for regulation of fimbriae length, and therefore adhesion, the mechanism of this process remains unknown (Rossolini et al., 1993).
Since the first publication in 1958 describing T1F in Salmonella, 150 studies concerning this topic have been published consistently, showing constant interest of scientific community in this virulence factor (Supplementary Figure 1). In this review, we summarize current knowledge on the regulation of Salmonella T1F expression, the roles of different T1F encoding genes in virulence, and discuss perspectives of future work in this field.
Early Studies on T1F
The occurrence of T1F in Salmonella spp. was first described by Duguid and Gillies (Duguid and Gillies, 1958). This initial study focused mainly on the ability of different Salmonella serovars and isolates to produce fimbriae and on conditions that induced or inhibited T1F expression. Moreover, the authors analyzed agglutination of red blood cells (RBCs) isolated from different animal species caused by T1F-positive (T1F+) Salmonella, and indicated, for the first time, that Salmonella Gallinarum produces T1F that do not agglutinate RBCs from all species tested in this study (Supplementary Table 1). Follow-up experiments conducted by Duguid provided information on the fimbrial status of 149 serovars and 1442 isolates, and showed for the first time the mannose-dependent agglutination of RBCs (Duguid et al., 1966). Both studies revealed that induction of T1F in static liquid culture led to pellicle formation, and multiple passages of bacteria in these conditions usually led to an increase in the fraction of T1F+ bacteria. On the other hand, growth on solid agar resulted in nearly no T1F+ Salmonella. In another study from Duguid’s lab, it was observed that T1F mediates adhesion of Salmonella to RBCs, leukocytes and epithelial cells. That study also found that induction of T1F expression in Salmonella was associated with growth in static aerobic conditions for 24–48 h, with multiple passages leading to an increase in the fraction of T1F+ bacteria (Old and Duguid, 1970).
Regulation of T1F Expression
The aforementioned work revealed the impact of Salmonella growth conditions on T1F expression. It was later shown that, depending on environmental conditions, T1F expression undergoes phase variation (Silverman et al., 1979), and is either in the “off” phase, wherein the whole operon is not transcribed, or in the “on” phase, which results in the expression of fim operon mRNA. Expression state is heritable but also reversible, with the frequency of switching from “on” to “off” much higher (approx. 10-2 per generation) than the switch in the reverse direction (approx. 10-4 per generation; Isaacson and Kinsel, 1992; Isaacson et al., 1999; Patterson et al., 2012). The ability to switch fimbriae expression from the “on” to “off” state and vice-versa plays an important role in Salmonella pathogenesis, since presence or absence of these structures on the surface of bacteria can affect various stages of bacterial infection (see below). The mechanisms of phase variation among different bacterial species are diverse, and in the family Enterobacteriaceae, such mechanisms are clearly described only in E. coli. Moreover, despite morphological and functional similarities of T1F in E. coli and Salmonella, they are serologically (Clegg et al., 1985) and evolutionarily (Kisiela et al., 2013) unrelated. It is therefore not surprising that the regulatory mechanisms of fim operon phase variation in Salmonella are quite different from those in the E. coli. Based on phase variation experiments with Salmonella Typhimurium strain 798, Patterson et al. (2012) hypothesized, that phase variation controls not only expression of T1F but also regulates expression of determinants responsible for invasion, intracellular survival, O-antigen chain length, and sensitivity to complement.
Direct Regulation
In Salmonella, there are three major regulatory proteins, FimZ, FimY, and FimW (each expressed under its own promoter), that control fim operon expression primarily through regulation of the fimA promotor (PfimA; Yeh et al., 1995, 2002b; Tinker and Clegg, 2000, 2001). FimZ and FimY are both necessary for positive regulation of T1F expression (Yeh et al., 1995; Tinker and Clegg, 2000), with FimZ as a dominant activator (Saini et al., 2009), whereas FimW was found to be a negative regulator (Tinker et al., 2001; Figure 2). Additionally, a putative phosphodiesterase encoded by an open reading frame, stm0551, that is located between fimY and fimW, was found to down-regulate T1F expression in S. Typhimurium (Wang et al., 2012). At the end of fim cluster, a tRNAArg encoded by fimU is present.
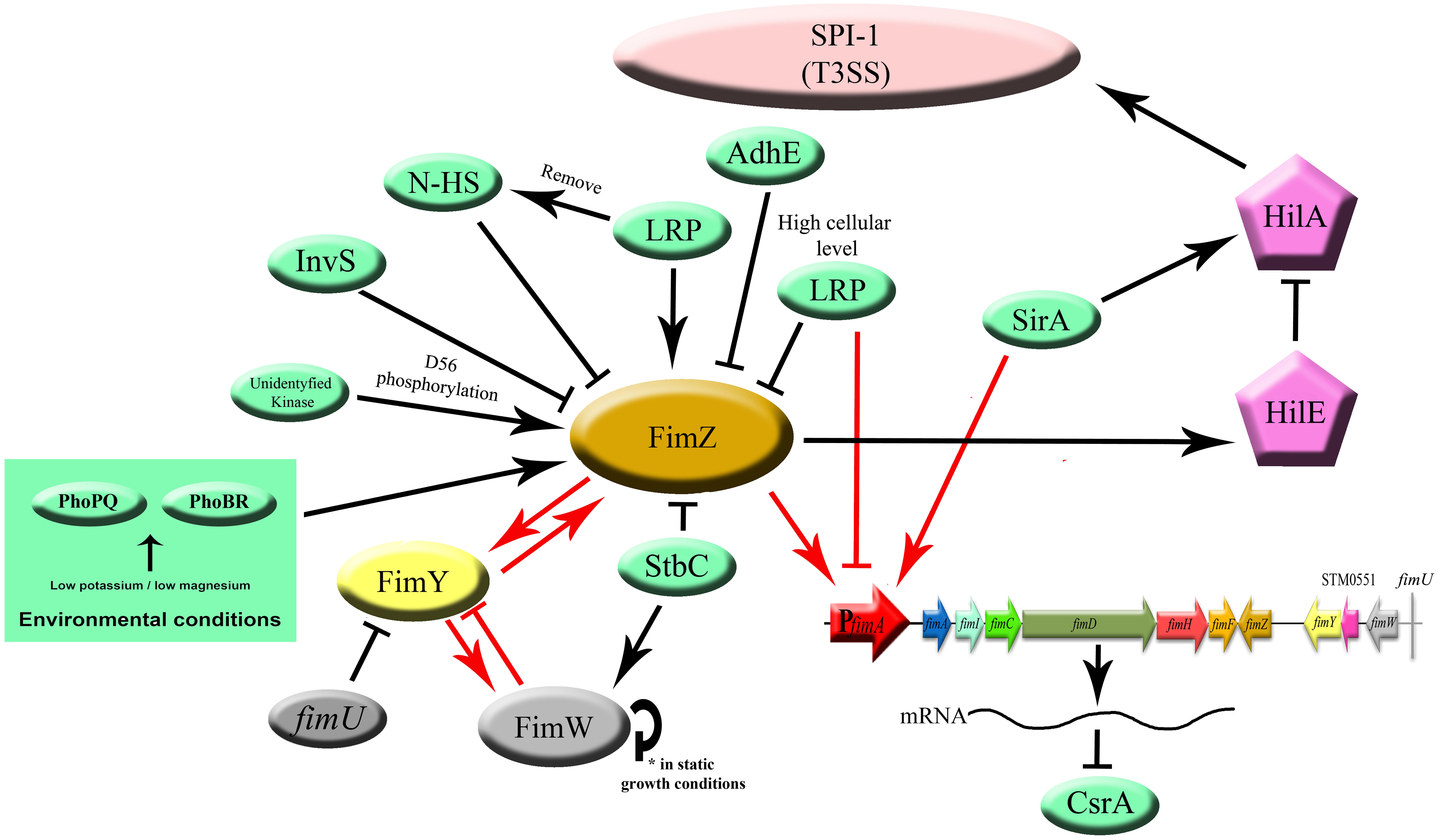
Figure 2. Regulation of T1F expression in Salmonella. Proteins directly involved in T1F expression are represented as ovals. Proteins involved in activation of T3SS (HilE, HilA) are represented as pentagons. All proteins encoded by the fim operon, including three major regulatory proteins, FimZ, FimY, and FimW are colored according to the scheme in the operon figure (Figure 1). Red arrows indicate direct promoter activation. Black arrows indicate another type of activation (not specified).
FimZ, which is thought to be a dominant activator of T1F expression, has high homology to a family of DNA binding proteins associated with response regulators of two-component regulatory systems and is able to bind the region upstream (from –47 to –98 nucleotides) of the fimA transcription initiation site, similar to other classical activators (Yeh et al., 2002b). It has been proposed that initial activation of FimZ could be driven by phosphorylation of aspartate 56 (D56; Zeiner et al., 2013) mediated by a yet unidentified kinase (Saini et al., 2009). Constitutive phosphorylation stemming from D56E mutation resulted in fimbriated bacteria even under non-inducing conditions. On the other hand, a null phenotype (D56A) blocked fimbriation completely. The authors suggested 31 putative sensory kinases that can phosphorylate FimZ at D56, however, this phosphorylation has not been directly linked to any one of these kinases. FimZ is able to regulate its own expression (Yeh et al., 2002a; Saini et al., 2009), however, no binding of FimZ to its own regulatory region was detected, and therefore the mechanism of such regulation is still uncertain. Moreover, FimZ is considered to be an important control protein of other Salmonella regulatory systems and coordinates different aspects of infection.
Activation of the fimA promoter by FimZ requires the presence of a second regulator – FimY (Yeh et al., 1995), which acts upstream of FimZ in the regulatory pathway, but does not interact directly with either FimZ or FimW (Zeiner et al., 2013). FimY and FimZ were found to cross activate each other’s promotor regions (PfimY and PfimZ), and then, through accumulation of both proteins, strongly stimulate T1F expression (Saini et al., 2009). Interestingly, fimZ cloned into a multicopy plasmid can overcome the lack of FimY (Zeiner, 2012). On the other hand, FimY is able to induce expression of fimW, which results in repression of PfimY. Wang et al. (2014) provided evidences that the FimY of S. Typhimurium is a DNA-binding protein and binds to a region within the fimZ promoter (Wang et al., 2014).
A third major regulatory protein, FimW, acts as a repressor of T1F expression and was found to be highly expressed in conditions favoring poor or no Salmonella fimbriation (i.e., growth on solid medium). Moreover, FimW was found to auto-regulate its own expression, but the exact mechanism is not known (Tinker et al., 2001). As with FimY, there has been no demonstrated direct interaction between FimW and the fimA promoter region, despite the presence of a putative DNA binding site (Clegg et al., 2011). However, direct interactions with FimZ have been observed (Zeiner et al., 2013). The authors suggested that FimW may have a modulatory role in FimZ-induced activation of the T1F fimA promoter by preventing FimZ binding to PfimA. According to Saini et al. (2009), expression of two T1F activators, FimZ and FimY, is negatively controlled by FimW, since FimY activates PfimW and, therefore, initiates a negative feedback loop.
In 1986, Feutrier et al. (1986) reported that T1F expression can be regulated by a Tn10 element, later recognized as the fimU coding tRNAArg, which has an impact on translation of fim regulatory genes (Clouthier et al., 1998). This tRNA is indispensable for efficient translation of fimY mRNA, which contains a number of rare AGA and AGG arginine codons that are recognized by the tRNAArg (Swenson et al., 1994; Clouthier et al., 1998). There are five rare arginine AGA codons in T1F – three in fimY and two in fimW – however, fimU deficiency only affects expression of FimY (Tinker and Clegg, 2001).
Global Regulation
Salmonella T1F expression can be also regulated by transcription factors involved in global regulation of genes engaged in metabolism, stress response or production of virulence factors. So far, the protein products of lrp, sirA, iprA, stbC, yqiC, invS, arcZ/hfq, and adhE have been implicated in T1F regulation (Figure 2).
Leucine responsive regulatory protein (Lrp) is an 18.8 kDa DNA binding protein known to regulate many fimbrial genes and to induce the E. coli fim operon (Blomfield et al., 1993). McFarland et al. (2008) reported that Lrp-negative mutants of S. Typhimurium do not express T1F. Lrp directly affects fimZ expression, probably by displacement of histone-like nucleoid-structuring (N-HS) protein, a global repressor of gram-negative bacteria (reviewed in Dorman, 2004), which binds to fimZ promoter via AT-rich sequences (Navarre et al., 2006). More recent studies revealed that direct binding of Lrp to the fimA promoter is indispensable for activation as well as repression of T1F expression (Baek et al., 2011). This activation or repression depends on cellular Lrp levels, as excessive or insufficient levels of Lrp results in inhibition of T1F expression. The same study also showed that Lrp expression is directly related to the nutritional conditions of the environment, and that T1F expression can be linked to cellular nutritional status via Lrp levels. sirA involved in Salmonella biofilm formation also takes part in positive regulation of T1F expression (Teplitski et al., 2006). Since SirA can directly bind to PfimA, it was hypothesized that this protein is necessary for initiation of fim operon transcription and increases the expression of T1F. iprA in S. Typhimurium, another gene that increases the expression of T1F, is highly conserved among Enterobacteriaceae. A deletion mutant of iprA showed decreased expression of the fim operon and fimbrial stbB, stcB, stdA, and stfE, and increased expression of stbA (Herman et al., 2016).
All abovementioned genes positively regulate T1F expression, but several other genes expressed by Salmonella suppress T1F expression. It was shown that deletion of yqiC (Wang et al., 2016) or arcZ (Monteiro et al., 2012) activates T1F expression in S. Typhimurium . arcZ and stbC are examples of cross-talk between different fimbrial systems (Monteiro et al., 2012; Wu et al., 2012). ArcZ, a protein involved in biofilm formation, plays a role in T1F expression by mediating the switch between T1F and curli fimbriae expression, as upregulation of T1F expression was observed in an arcZ deletion mutant (Papenfort et al., 2009; Monteiro et al., 2012). The stbC gene product, an Stb fimbriae usher, also plays a role in the T1F regulatory network. An stbC mutant had higher fimZ expression and lower fimW expression, and therefore stimulated T1F production. Depending on environmental conditions, T1F expression can also be affected by other factors, e.g., by proteins like AdhE (fermentative alcohol dehydrogenase) that are involved in carbohydrate metabolism (Chuang et al., 2008). Overexpression of AdhE is known to inhibit T1F expression, whereas the absence of AdhE stimulates T1F expression even in Salmonella growing on solid agar.
Crosstalk Between T1F and Other Virulence Factors
Expression of T1F can have an impact on expression of other virulence factors. It was shown that the activated fim operon prevents expression of plasmid-encoded fimbriae (Pef) by regulating CsrA activity, demonstrating a hierarchy in expression of different types of fimbriae (Sterzenbach et al., 2013). Binding of the polycistronic fimAICDHF mRNA to CsrA, a global post-transcriptional regulator, decreases CsrA activity, and thereby inhibits its positive effect on Pef expression (Figure 2).
FimZ, involved in direct regulation of fim operon, is also involved in regulation of motility. FimZ overexpression correlates with decreased Salmonella motility and invasion (Clegg and Hughes, 2002), as the Salmonella invasion process requires precisely regulated, hierarchical expression of different proteins. The invasion phenotype is dependent on the activation of Salmonella Pathogenicity Island 1 Type 3 Secretion System (SPI-1 T3SS) by a variety of positive and negative regulatory genes, with hilA and invF as two key activators (Lee et al., 1992; Bajaj et al., 1995). It was shown that hilA expression can be regulated by the level of hilE expression, and hilE is regulated by fimY and fimZ (Baxter and Jones, 2005). Induction of hilE, stimulated by fimYZ, leads to repression of hilA and subsequent down-regulation of invasion genes encoded by SPI-1 (Figure 2).
Recent work has shown that two-component regulatory systems, such as PhoPQ and PhoBR, are able to activate fimZ, and therefore to increase T1F expression, as well as upregulate expression of hilE. As activation of PhoPQ and PhoBR is induced by low magnesium and low phosphate concentrations, respectively, it directly links regulation of the fim operon with environmental conditions (Baxter and Jones, 2015). PhoPQ, together with several other regulatory systems (SsrA/B, OmpR/EnvZ), positively regulates InvS expression (Colgan et al., 2016), a Salmonella small RNA essential for invasion (Wang et al., 2017). InvS reduces expression of fimZ and increases expression of flhD, an important regulator of flagella expression, but the exact mechanism is still unclear. Saini et al. (2010) proposed that the three systems involved in Salmonella infection are subject to dynamic regulation. First, Salmonella use flagella to swim to site of invasion, then SPI-1 T3SS is expressed and finally T1F are expressed. The authors suggested that the most significant regulator of these processes is the flagellar regulator FliZ, as it regulates both SPI-1 and T1F expression; however, no direct effect of FliZ on fim cluster expression was observed.
Taking together, it seems that specific environmental signals can promote Salmonella fimbrial phase variations, and that this process is specifically related to virulence. What is more, a main regulator of T1F expression, FimZ, plays an important role not only in fimbrial phase regulation, but also acts as a one of the global regulons of a wide range of phenotypes in various stages of Salmonella infection.
T1F and Adhesion of Salmonella to Cells, Organ Explants, Cell Lines and Their Role in Biofilm Formation
The role of T1F in adhesion of Salmonella to eukaryotic cells has been extensively studied. Work on T1F-dependent binding in Salmonella was conducted primarily using yeast agglutination and guinea pig hemagglutination as controls for T1F expression (Supplementary Table 1). Other often used models are primary animal cell cultures or organ explants (Supplementary Table 1). For example, S. Typhimurium adhesion to and infection of isolated rat small intestine enterocytes occurred in a mannose-sensitive (MS) manner, suggesting the involvement of T1F (Lindquist et al., 1987). Similar MS adhesion by T1F+ S. Typhimurium was also reported in isolated intestines of 1-day old chicks as well as isolated rat enterocytes (Oyofo et al., 1989). Adhesion of Salmonella Enteritidis to human buccal cells and mouse small intestine epithelial cells was mediated by T1F and blocked by preincubation of bacteria with D-mannose (Aslanzadeh and Paulissen, 1990). However, when the S. Enteritidis wild-type (WT) strain and a fimD deletion mutant were analyzed for adhesion to chicken duodenal explants, no significant difference between these two strains was found (Allen-Vercoe and Woodward, 1999). Dendritic cells (DC) can send dendrites between enterocytes, which enables direct contact of DC with Salmonella. Using T1F+ S. Typhimurium, it was found that bacteria can bind to murine bone marrow-derived DCs in a MS manner (Guo et al., 2007).
There are 20 articles on Salmonella T1F-dependent binding to established in vitro cell lines (Table 1). It should be mentioned that the majority of these studies were performed with human non-intestinal cell lines, such as epithelioid cervix carcinoma Hela cells and HeLa derivative HEp-2 cells. Studies on S. Typhimurium and Salmonella Braenderup have shown that T1F+ strains adhered to and invaded HeLa cells in higher numbers than non-fimbriated strains (Horiuchi et al., 1992; Bäumler et al., 1996). Many studies with HEp-2 cells have shown that Salmonella also adhere to these cells in a MS manner, and therefore such binding is mediated by T1F (Tavendale et al., 1983; Old et al., 1986; Ernst et al., 1990). In a study by Hancox et al. (1997), WT S. Typhimurium bound better to the HEp-2 and HeLa cell lines than isogenic fimH mutants. However, according to early investigations conducted by Jones and Richardson (1981), S. Typhimurium adhesion and invasion to HeLa cells occurred in a mannose-resistant (MR) manner, and therefore was probably not mediated by T1F. Furthermore, results presented by Bäumler et al. (1996) showed that T1F did not contribute to adhesion and invasion of S. Typhimurium to HEp-2 cells.
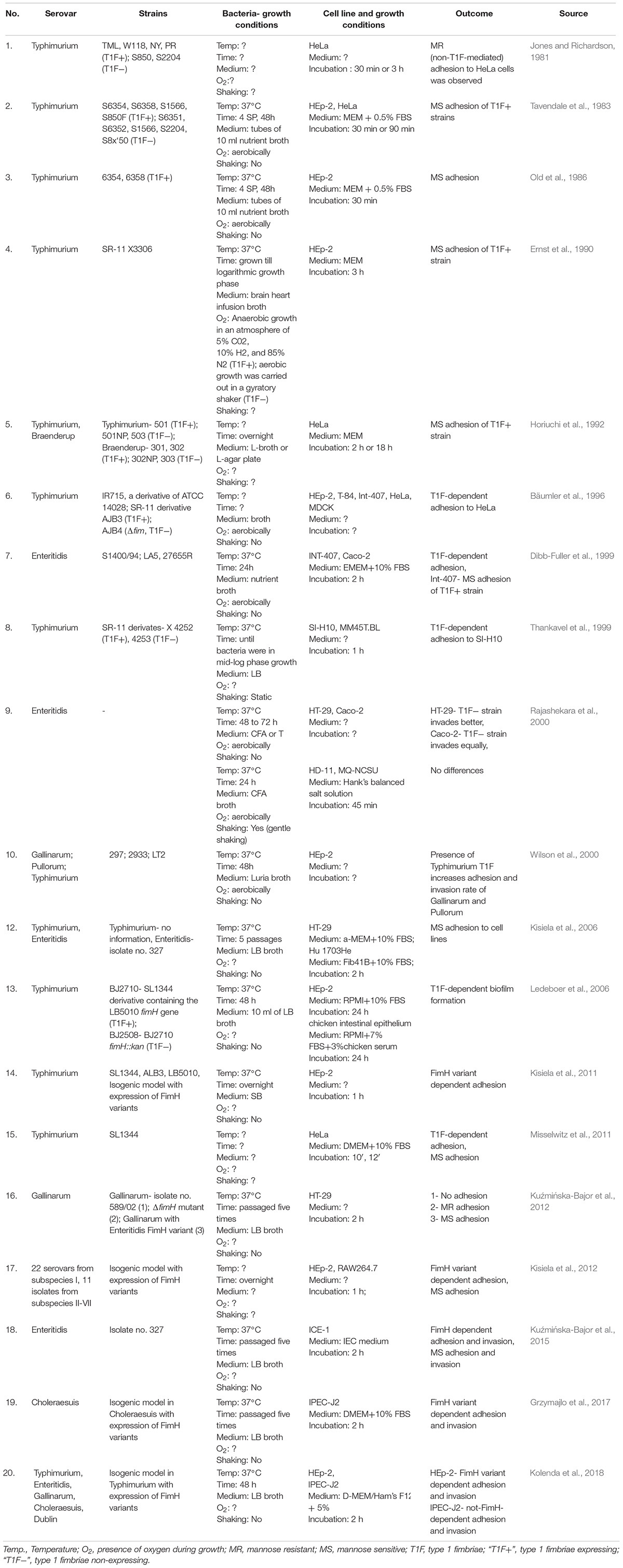
Table 1. Summary of assays performed to study the role of T1F in Salmonella adhesion and/or invasion of cell lines.
The majority of studies using cell lines originating from intestinal epithelial cells, e.g., IPEC-J2, ICE-1, or HT-29, showed T1F-dependent and/or MS binding of Salmonella to cells (Kisiela et al., 2006; Kuźmińska-Bajor et al., 2012, 2015; Grzymajlo et al., 2017). There are also reports which showed no contribution of T1F to adhesion to these type of cells (Rajashekara et al., 2000; Kolenda et al., 2018). These conflicting results obtained with cell lines may stem from various experimental procedures during cultivation of bacteria, differences in adhesion assays or use of different Salmonella and non-fimbriate strains (Table 1).
Bacteria form biofilms to survive and persist within the host and environment (Flemming et al., 2016). Biofilm formation starts with adhesion to biotic and abiotic surfaces. It was shown that T1F are up-regulated during biofilm formation on cholesterol gallstones, but hyper fimbriation had a negative impact on biofilm formation. (Crawford et al., 2010; Gonzalez-Escobedo and Gunn, 2013). Moreover, T1F were found to contribute to biofilm formation on HEp-2 cells, murine and chicken intestinal epithelium, and plastic surfaces (Boddicker et al., 2002; Ledeboer et al., 2006).
T1F and Animal Models
The role of T1F in Salmonella pathogenesis has been investigated using various animal models (20 studies was found, summarized in Table 2). The first such study on S. Typhimurium showed that a T1F+ strain was more infectious and virulent in mouse models than a non-fimbriated strain (Darekar and Duguid, 1972). In these studies, mice infected with T1F+ bacteria also excreted S. Typhimurium in their feces for a longer period of time. Similar results were obtained with follow-up experiments conducted by Duguid et al. (1976). A drawback of the aforementioned studies is that T1F- Salmonella strains used for testing were “natural” derivatives of T1F+ strains “induced” or “not-induced” for T1F production. Different results were obtained depending on the method used for generation of isogenic T1F- mutants of S. Typhimurium or S. Enteritidis, i.e., when transposon mutagenesis or the Datsenko-Wanner method were used. The work of Lockman and Curtiss (1992) revealed that a T1F-S. Typhimurium mutant was more virulent than the parental T1F+ strain. The authors hypothesized that the lower virulence of the T1F+ strain was due to sequestration of T1F+ bacteria in the liver, spleen and kidneys. The higher virulence of the T1F-S. Typhimurium strain in mouse infection models compared to the parental isogenic strain was further confirmed by van der Velden et al. (1998), who proposed that this phenomenon is caused by the expression of other virulence genes in the absence of T1F (van der Velden et al., 1998). Similar results were obtained with a S. Enteritidis ΔfimH mutant (Kuźmińska-Bajor et al., 2015). Using a mouse model, it was shown that the higher virulence of the T1F-S. Enteritidis compared to T1F+ bacteria could be attributed to higher systemic spread within the host. The authors suggested that T1F, which are responsible for adhesion to the intestinal mucosa, are a limiting factor in spreading bacteria outside of the intestinal tract. The direct involvement of T1F in the intestinal phase of infection was analyzed in a rat infection model, where expression of T1F in S. Enteritidis and S. Typhimurium adhering to enterocytes was demonstrated by immunohistochemistry (Ewen et al., 1997). However, long-term infection studies of rats challenged with the mixture of WT S. Enteritidis and a ΔfimD mutant revealed that the presence of T1F gives bacteria an advantage in the early stages of intestine infection, though higher counts of the T1F- strain were found in the intestine 6 days post infection (Naughton et al., 2001). In a 1-day old chicken model, the T1F-S. Enteritidis strain (which was also a mutant for the SEF14 and SEF17 fimbriae) had a lower ability than the S. Enteritidis WT strain to colonize the spleen, liver and caeca after 24 h of infection (Dibb-Fuller and Woodward, 2000). Similar results were obtained for S. Gallinarum in a study by Kuźmińska-Bajor et al. (2012), where a fimH deletion mutant had weak and delayed colonization of caecal tonsils, liver and spleen in comparison to its WT parent strain. In a study using laying hens, the T1F- strain of S. Enteritidis (ΔfimD deletion) was present in blood, caeca and oviducts for a longer time than the WT strain, though the WT strain was more frequently isolated from eggs (De Buck et al., 2004). On the other hand, T1F-S. Enteritidis (fimA single mutant) did not show any significant differences with the WT strain in colonization of the spleen and liver, and shedding of bacteria in feces of 5-day-old chickens (Rajashekara et al., 2000). The only difference found in the T1F- strain was a higher number of bacteria in the caecum 14 days post infection.
Receptors for T1F
The discovery of T1F-dependent, MS agglutination of yeast and guinea pig RBCs led to the hypothesis that oligosaccharide chains containing mannose residues are receptors for T1F. Therefore, the carbohydrate specificity of S. Typhimurium T1F was analyzed by inhibition of agglutination with linear and branched mannose-containing oligosaccharides or glycosides of D-mannose. It was found that binding of T1F+ S. Typhimurium to yeast cells and guinea pig RBCs was inhibited most efficiently by high mannose oligosaccharides (Firon et al., 1983, 1984).
One of the intriguing issues concerning Salmonella T1F is the innate T1F receptors expressed at the surface of host cells. Leusch et al. (1991) analyzed binding of various Salmonella serovars to glycoproteins expressed in the intestine, egg white, blood, spleen and bile. It was found that most of the Salmonella Typhi, Paratyphi A and B, and Java isolates bound to carcinoembryonic antigen (CEA). It was also revealed, that S. Typhi bound with the highest affinity to CEAs and an unknown glycoprotein, NCA-55. However, MS binding was only tested and shown for adhesion of S. Typhi to CEA. In another study, an extracellular matrix protein, laminin, was found as a receptor for S. Enteritidis and S. Typhimurium T1F, and the glycan part of laminin was bound by T1F in a MS manner (Kukkonen et al., 1993). Another possible receptor for S. Typhimurium T1F is a 60-kDa glycoprotein that was isolated from the brush border of a rat’s small intestine, but the protein was not further characterized (Ghosh et al., 1996). This glycoprotein interacted with isolated T1F in MS manner. S. Enteritidis T1F receptor, responsible for infection of chicken eggs during egg production, was found in the isthmus of the chicken reproductive tract. Binding of T1F+ S. Enteritidis to isthmus sections and secretions were blocked by mannose and mediated by mannosylated glycoproteins, which were detected in isthmus using lectins (De Buck et al., 2003). The best characterized Salmonella T1F receptor to date is pancreatic secretory granule membrane major glycoprotein GP2, first identified as a transcytotic receptor of M cells for T1F+ S. Typhimurium in human and mice. Translocation of FimH-positive S. Typhimurium through M cells leads to increased numbers of bacteria in the mesenteric lymph nodes and the immune response to antigens expressed by these bacteria (Hase et al., 2009). The adhesion of S. Typhimurium expressing FimH variants from serovars Typhimurium, Enteritidis, Dublin and Choleraesuis to porcine GP2 was shown recently (Kolenda et al., 2018).
A study on binding of S. Enteritidis, Salmonella Choleraesuis, Salmonella Dublin and S. Abortus-ovis FimH proteins to cell lysates from intestinal cell lines originating from various potential hosts (pig, sheep and cow) revealed that FimH binds in an MS manner to different glycoproteins, depending on serovar host range. FimH from generalist S. Enteritidis bound to surface membrane proteins of about 130 kDa, while FimH from host specialists bound to a protein of about 55 kDa (Grzymajło et al., 2013). A recent study identified a 55 kDa receptor as calreticulin (CRT; Grzymajlo et al., 2017). It was shown that CRT isolated from porcine IPEC-2 cells was bound specifically by S. Choleraesuis FimH and not by FimH from S. Enteritidis, suggesting that host-specificity of Salmonella serovars is dependent on both pathogen and host factors.
Glycosphingolipids are common components of the plasma membrane of cells. Studies by Li et al. (2003a,b) revealed that glucosylceramide (GlcCer) and monosialodihexosylganglioside (GM3) are possible receptors for S. Enteritidis T1F (Li et al., 2003a,b). The binding of GlcCer and GM3 isolated from intestinal mucosa and chicken oviductal tracts bound to T1F-expressing S. Enteritidis and this binding was blocked by anti-T1F antibodies. The only non-glycan-mediated binding of T1F was shown in S. Typhimurium, which bound to plasminogen (Kukkonen et al., 1998). This interaction was blocked by a lysine analog and not mannose.
Role of FimH Allelic Variation in Pathogenesis
FimH adhesin located at the top of T1F is directly involved in binding to different high-mannose oligosaccharides carried by surface glycoproteins of eukaryotic cells. Despite being very high, reaching 99% sequence homology, it became increasingly clear that significant micro-heterogeneity, associated with differences in the amino acid sequences of FimH adhesins, exists among type 1 fimbriae from different serovars and affects their binding to mannosylated oligosaccharides (Figure 3 and Supplementary Table 2).

Figure 3. Sequence logo for FimH variable sites identified in 29 Salmonella serovars. FimH variable sites were extracted from articles discussed in this review. Detailed information about serovars can be found in Supplementary Table 1.
The discovery that fimH allelic variants found in two S. Typhimurium strains (LB5010 and SL1344), which differ in amino acid at positions 61 and 118, are responsible for different adhesion phenotypes, started a new period in T1F studies (Boddicker et al., 2002). The FimH variant from strain SL1344, with glycine and phenylalanine at positions 61 and 118, respectively, mediated low binding of Salmonella to HEp-2 cells and the FimH variant from strain LB5010, with alanine and serine at positions 61 and 118, respectively, mediated a high level of Salmonella binding to the same cells. When the amino acid sequence of Salmonella Enteritidis FimH was compared with the low-binding variant of S. Typhimurium FimH adhesin, it was found that S. Enteritidis FimH also represent the low-binding phenotype with glycine in position 61 and phenylalanine in position 118 (Kisiela et al., 2006). However, follow-up experiments revealed that only the substitution from phenylalanine to serine at position 118 contributes to a change from the low to high binding phenotype, at least in the case of S. Enteritidis (Grzymajlo et al., 2010). It has been previously mentioned that T1F mediate binding of Salmonella to HEp-2 cells. Substitution at position 158 from asparagine to tyrosine within the S. Typhimurium FimH increases adhesion of bacteria to HEp-2 cells and DCs (Guo et al., 2009). Screening of various Salmonella serovars for FimH variation and MS adherence to HEp-2 cells, and biofilm formation further confirmed the role of sequence variability in the binding properties of Salmonella FimHs (Dwyer et al., 2011). The magnum opus on the association with and the role of fimH allelic variants in the pathogenesis of salmonellosis caused by Salmonella serovars with different host ranges was published by Kisiela et al. (2012). The authors investigated the effects of sequence variation on binding of FimH adhesins to purified high-mannose type glycoproteins, epithelial and macrophage-like cell lines. Based on these results, FimH high- and non-binding variants were linked with host specialists (host-restricted and host-adapted serovars) and FimH low-binding variants were linked with host generalists (host-unrestricted serovars). However, in mouse infection models no significant differences in colonization of spleen and liver were observed for S. Typhimurium isogenic strains expressing FimH variants representing high-, low- or non-binding phenotypes, and no contribution of FimH variation to disease phenotype was found. In a more recent study, Yue et al. (2015) searched for the presence of FimH variants among S. Typhimurium strains isolated from human and bovine hosts and analyzed host-specific adhesion to human and bovine cell lines (Yue et al., 2015). Sequence analysis of fimH from 580 isolates revealed that the presence of valine residue at position 245 was found more often in human isolates and alanine at the same position was found more often in bovine isolates. An adhesion assay showed that substitution from valine to alanine at position 245 in FimH from S. Typhimurium increased binding to cells of bovine origin without affecting binding to cells of human origin. This study also showed that FimH variants from Salmonella specialists mediated binding to cell lines in a host-specific manner. For example, FimH from swine-associated S. Typhisuis and S. Choleraesuis serovars bound better to porcine IPEC-1 and IPEC-J2 cells, whereas FimH from human-associated S. Typhi and S. Newport isolates of human origin bound better to human RKO, HCT116 and Caco-2 cells. What is more, FimH from bovine-associated S. Dublin isolates bound better to bovine C8 and CMS cells, and FimH from poultry-associated S. Gallinarum isolates bound better to avian LMH cells than to all other aforementioned cells. In another study, Schifferli’s group analyzed the role of FimH allelic variants in adhesion of S. Newport strains isolated from bovine, porcine and human hosts to cell lines of the same host origin (De Masi et al., 2017). It was shown that strains of bovine and porcine origin, carrying a FimH variant with phenylalanine at position 15, alanine at position 32 and arginine at position 89 bound better to cell lines of bovine (CMS, J8) and porcine origin (IPEC-J2) than to human cell lines. On the other hand, Salmonella strains of human origin, carrying a FimH variant with leucine at position 15, valine at position 32 and glutamine at position 89 bound better to human cell lines (RKO, Caco-2) than to bovine or swine cell lines.
Studies employing random mutagenesis and 3D structure predictions of the FimH from S. Typhimurium showed that mutations in the predicted lectin domain, the interdomain and the pilin domain can lead to a change in FimH binding properties. It was proposed that binding of FimH to its receptor under shear force leads to activation of allosteric properties in FimH variants, which can alter the binding properties of these FimH variants compared to their binding under static conditions (Kisiela et al., 2011). For example, a FimH variant from S. Typhimurium SL1344 exhibited a higher degree of binding to the same glycoproteins when tested under shear conditions than in a static adhesion assay. On the other hand, FimH variants that had higher binding properties in static conditions had weaker binding under shear stress.
The GP2 of both human and mouse was shown to be the receptor for T1F of S. Typhimurium (Hase et al., 2009). The influence of fimH variation on binding to human and porcine GP2 isoforms expressed in SF9 cells was tested in a study by Kolenda et al. (2018). The authors used FimH variants from five Salmonella serovars and found that binding to different GP2 isoforms was FimH variant-dependent and not GP2-host origin-dependent or GP2-variant-dependent. Another example of an association of FimH variation with recognition of host proteins can be found in the previously described study by Grzymajło et al. (2013), which identified substitutions in positions L57P and N101S in FimH as altering receptor specificity and possibly contributing to changes in host range of Salmonella serovars.
Lee and Yeh observed that T1F production in S. Choleraesuis is dependent on amino acid variation at residue 63. They analyzed the expression of T1F in S. Choleraesuis, showing that only 4 out of 120 strains expressed T1F (Lee and Yeh, 2016). All strains expressing T1F had a valine residue at position 63 and strains without T1F expression had glycine at the same position.
Fimbriae of serovars not agglutinating RBCs and yeast cells (MR T1F), i.e., Salmonella Gallinarum and S. Paratyphi B, were initially called type 2 fimbriae, but genetic and microscopic analysis showed that these fimbriae were T1F (Crichton et al., 1989; Kisiela et al., 2005). In the case of S. Gallinarum T1F, the loss of MS binding was linked to a single amino acid substitution, from threonine to isoleucine, at position 78 of FimH (Kisiela et al., 2005). Whether the MR phenotype of S. Gallinarum FimH confers the inactive T1F phenotype because T1F is unable to bind to any receptors or whether the MR phenotype leads to changes in receptor specificity was investigated by Guo et al. (2009) who provided proof that the latter is, in fact, true by demonstrating that S. Gallinarum T1F mediates binding to chicken leukocytes. Another study investigating the role of FimH variation in the pathogenesis of S. Gallinarum showed that expression of MS S. Enteritidis FimH in S. Gallinarum leads to decreased or no colonization of liver, spleen and caecal tonsils, thus proving a significant role for FimH variation in bacterial host specificity (Kuźmińska-Bajor et al., 2012). Investigations by Guo et al. (2009) and Kuźmińska-Bajor et al. (2012) revealed that T1F variation can confer a significant advantage for Salmonella Gallinarum during pathogenesis in chickens, an observation that could explain such a drastic change like the switch from MS to MR phenotype.
Future Prospects
In this article, we have presented both early and recent studies that describe the importance of T1F in Salmonella infections. During the last 60 years, investigations have been carried out to establish the role of T1F in Salmonella pathogenesis. Although much has been revealed about the functions of T1F, many unanswered questions remain to be addressed in future studies.
T1F mediate binding to host tissues, in which FimH is directly involved, is one of the first steps of Salmonella pathogenesis. Designing or finding inhibitors that can block the binding of T1F to receptors could provide new options for prevention and treatment of Salmonella infections. Unfortunately, the crystal structure of FimH from Salmonella is yet to be resolved, which significantly hampers structure-based inhibitor design and the search for new inhibitors. Design of new T1F inhibitors could be also aided by more complete identification and characterization of receptors expressed on host tissues. While some studies have reported proteins binding with T1F, knowledge of the range of receptors present in Salmonella serovars other than S. Typhimurium and S. Choleraesuis is still limited. Moreover, already identified receptors, such as CRT and GP2, require further research to study interactions between FimH and glycosylation as well as to describe the accessibility and expression of these receptors in host organs.
Although binding of T1F+ Salmonella to intestines has been shown, there are no studies about spatial expression of T1F within the host during Salmonella infection. Such studies would allow researchers to more fully elucidate where and for how long T1F are expressed, which could improve the search for T1F receptors by allowing efforts to focus on host sites directly involved in T1F-mediated Salmonella adhesion. It was proposed that T1F might have different functions in the intestinal phase of Salmonella infection in host specialists and generalists. Taking into account the recent developments in microbiomics and the importance of inflammation during Salmonella gastroenteritis, it would be interesting to investigate how or if T1F contribute to the development of inflammatory diarrhea caused by Salmonella generalists and whether the host range associated SNPs in FimH can alter infection outcomes.
Advances in next generation sequencing (NGS) technologies have resulted in the availability of thousands of Salmonella genomes, providing a great opportunity to study the genetic organization of the fim gene clusters present in many Salmonella serovars, representing different host specialists and host generalists. NGS data can be used to assess variability in T1F coding regions and to explore how that variability translates into T1F regulation, expression, and host specific actions. NGS also brings new tools for analyzing regulation of T1F in Salmonella. As a large part of our review recapitulates regulation of T1F expression in Salmonella (mainly S. Typhimurium), it is clear that there is still a considerable amount work to be done in this area, in particular there is a need for a systematic assessment of T1F expression in vitro and in vivo. One approach to achieving this would be to prepare a mutant library of the fim gene cluster of one Salmonella host generalist and specialist (e.g., S. Typhimurium or S. Enteritidis and S. Typhi or S. Gallinarum) and compare the influence of particular mutations on expression of T1F and on crosstalk between T1F and other virulence factors. Use of NGS could also facilitate studies of spatial expression of T1F in vivo and assessment of immune responses during host infection.
The role of fimH allelic variation on expression of T1F and other virulence factors is another interesting topic that requires further research. A study by Lee and Yeh (2016) showed that a single SNP (V63G) in fimH is responsible for a lack of T1F expression in S. Choleraesuis. However, it should be noted that this result does not accord with our observations of expression of the same FimH variant in a WT S. Choleraesuis isolate, in isogenic (V63G) model generated in another S. Choleraesuis isolate (Grzymajlo et al., 2017). A deleterious effect of the V63G mutation was observed on FimH expression in S. Typhimurium SL1344, but the same variant was expressed in one S. Choleraesuis isolate in a study by Kisiela et al. (2012). Two isogenic models generated for expression of different FimH variants in S. Typhimurium (Kisiela et al., 2012; Kolenda, 2018) showed differences in T1F expression, although it must be noted that only one antibody was used to assess T1F expression. These data indicate that there is a phenotypic variation in T1F expression in Salmonella that is associated with SNPs in FimH sequence as well as other unknown factors. It is possible that lack of T1F expression results in increased expression of other virulence factors that are beneficial for successful host infection.
Author Contributions
RK and KG conceptualized, wrote original draft, visualized, and prepared the figures and tables. MU wrote the review and edited the manuscript. All authors discussed the results and contributed to the final manuscript.
Funding
RK work was supported by the Deutsche Forschungsgemeinschaft (DFG SCHI 1147/3-1). KG work was funded by the Polish National Science Centre by decision number DEC2013/09/D/NZ6/02413. The project is financed under the program of the Minister of Science and Higher Education “Strategy of Excellence – University of Research” in 2018–2019 project number 0019/SDU/2018/18 in the amount of PLN 700,000.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Part of the results published in this manuscript was taken from Ph.D. thesis of RK, which is cited in the text and accessible online at: https://refubium.fu-berlin.de/handle/fub188/9629.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01017/full#supplementary-material
References
Allen-Vercoe, E., and Woodward, M. J. (1999). The role of flagella, but not fimbriae, in the adherence of Salmonella enterica serotype enteritidis to chick gut explant. J. Med. Microbiol. 48, 771–780. doi: 10.1099/00222615-48-8-771
Althouse, C., Patterson, S., Fedorka-Cray, P., and Isaacson, R. E. (2003). Type 1 Fimbriae of Salmonella enterica serovar typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 71, 6446–6452. doi: 10.1128/IAI.71.11.6446-6452.2003
Aslanzadeh, J., and Paulissen, L. J. (1990). Adherence and pathogenesis of Salmonella enteritidis in mice. Microbiol. Immunol. 34, 885–893.
Baek, C.-H., Kang, H.-Y., Roland, K. L., and Curtiss, R. (2011). Lrp acts as both a positive and negative regulator for type 1 fimbriae production in Salmonella enterica serovar Typhimurium. PLoS One 6:e26896. doi: 10.1371/journal.pone.0026896
Bajaj, V., Hwang, C., and Lee, C. A. (1995). hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18, 715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x
Bäumler, A. J., Tsolis, R. M., and Heffron, F. (1996). Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64, 1862–1865.
Baxter, M. A., and Jones, B. D. (2005). The fimYZ genes regulate Salmonella enterica serovar typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect. Immun. 73, 1377–1385. doi: 10.1128/IAI.73.3.1377-1385.2005
Baxter, M. A., and Jones, B. D. (2015). Two-component regulators control hilA expression by controlling fimZ and hilE expression within Salmonella enterica serovar Typhimurium. Infect. Immun. 83, 978–985. doi: 10.1128/IAI.02506-14
Blomfield, I. C., Calie, P. J., Eberhardt, K. J., McClain, M. S., and Eisenstein, B. I. (1993). Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 175, 27–36.
Boddicker, J. D., Ledeboer, N. A., Jagnow, J., Jones, B. D., and Clegg, S. (2002). Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 45, 1255–1265.
Boyd, E. F., and Hartl, D. L. (1999). Analysis of the type 1 pilin gene cluster fim in Salmonella: its distinct evolutionary histories in the 5′ and 3′ regions. J. Bacteriol. 181, 1301–1308.
Chuang, Y.-C., Wang, K.-C., Chen, Y.-T., Yang, C.-H., Men, S.-C., Fan, C.-C., et al. (2008). Identification of the genetic determinants of Salmonella enterica serotype Typhimurium that may regulate the expression of the type 1 fimbriae in response to solid agar and static broth culture conditions. BMC Microbiol. 8:126. doi: 10.1186/1471-2180-8-126
Clayton, D. J., Bowen, A. J., Hulme, S. D., Buckley, A. M., Deacon, V. L., et al. (2008). Analysis of the role of 13 major fimbrial subunits in colonisation of the chicken intestines by Salmonella enterica serovar Enteritidis reveals a role for a novel locus. BMC Microbiol. 8:228. doi: 10.1186/1471-2180-8-228
Clegg, S., and Hughes, K. T. (2002). FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184, 1209–1213.
Clegg, S., Hull, S., Hull, R., and Pruckler, J. (1985). Construction and comparison of recombinant plasmids encoding type 1 fimbriae of members of the family Enterobacteriaceae. Infect. Immun. 48, 275–279.
Clegg, S., Wilson, J., and Johnson, J. (2011). More than one way to control hair growth: regulatory mechanisms in enterobacteria that affect fimbriae assembled by the chaperone/usher pathway. J. Bacteriol. 193, 2081–2088. doi: 10.1128/JB.00071-11
Clouthier, S. C., Collinson, S. K., White, A. P., Banser, P. A., and Kay, W. W. (1998). tRNA(Arg) (fimU) and expression of SEF14 and SEF21 in Salmonella enteritidis. J. Bacteriol. 180, 840–845.
Colgan, A. M., Kröger, C., Diard, M., Hardt, W.-D., Puente, J. L., Sivasankaran, S. K., et al. (2016). The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 12:e1006258. doi: 10.1371/journal.pgen.1006258
Crawford, R. W., Reeve, K. E., and Gunn, J. S. (2010). Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J. Bacteriol. 192, 2981–2990. doi: 10.1128/JB.01620-09
Crichton, P. B., Yakubu, D. E., Old, D. C., and Clegg, S. (1989). Immunological and genetical relatedness of type-1 and type-2 fimbriae in salmonellas of serotypes Gallinarum, Pullorum and Typhimurium. J. Appl. Bacteriol. 67,283–291.
Darekar, M. R., and Duguid, J. P. (1972). The influence of fimbriation on the infectivity of Salmonella typhimurium. Proc. Indian Acad. Sci. Sect. B 75, 283–293. doi: 10.1007/BF03045722
De Buck, J., Van Immerseel, F., Haesebrouck, F., and Ducatelle, R. (2004). Effect of type 1 fimbriae of Salmonella enterica serotype enteritidis on bacteraemia and reproductive tract infection in laying hens. Avian Pathol. 33, 314–320. doi: 10.1080/0307945042000220561
De Buck, J., Van Immerseel, F., Haesebrouck, F., and Ducatelle, R. (2005). Protection of laying hens against Salmonella Enteritidis by immunization with type 1 fimbriae. Vet. Microbiol. 105, 93–101. doi: 10.1016/j.vetmic.2004.10.008
De Buck, J., Van Immerseel, F., Meulemans, G., Haesebrouck, F., and Ducatelle, R. (2003). Adhesion of Salmonella enterica serotype enteritidis isolates to chicken isthmal glandular secretions. Vet. Microbiol. 93, 223–233.
De Masi, L., Yue, M., Hu, C., Rakov, A. V., Rankin, S. C., and Schifferli, D. M. (2017). Cooperation of adhesin alleles in Salmonella-host tropism. mSphere 2:e00066-17. doi: 10.1128/mSphere.00066-17
Dibb-Fuller, M. P., Allen-Vercoe, E., Thorns, C. J., and Woodward, M. J. (1999). Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiol. 145, 1023–1031. doi: 10.1099/13500872-145-5-1023
Dibb-Fuller, M. P., and Woodward, M. J. (2000). Contribution of fimbriae and flagella of Salmonella enteritidis to colonization and invasion of chicks. Avian Pathol. 29, 295–304. doi: 10.1080/03079450050118412
Dorman, C. J. (2004). H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2, 391–400. doi: 10.1038/nrmicro883
Duguid, J. P., Anderson, E. S., and Campbell, I. (1966). Fimbriae and adhesive properties in Salmonellae. J. Pathol. Bacteriol. 92, 107–138. doi: 10.1002/path.1700920113
Duguid, J. P., Darekar, M. R., and Wheater, D. W. (1976). Fimbriae and infectivity in Salmonella typhimurium. J. Med. Microbiol. 9, 459–473. doi: 10.1099/00222615-9-4-459
Duguid, J. P., and Gillies, R. R. (1958). Fimbriae and haemagglutinating activity in Salmonella, Klebsiella, Proteus and Chromobacterium. J. Pathol. Bacteriol. 75, 517–523. doi: 10.1002/path.1700750239
Dwyer, B. E., Newton, K. L., Kisiela, D., Sokurenko, E. V., and Clegg, S. (2011). Single nucleotide polypmorphisms of fimH associated with adherence and biofilm formation by serovars of Salmonella enterica. Microbiol. Read. Engl. 157, 3162–3171. doi: 10.1099/mic.0.051425-0
Ernst, R. K., Dombroski, D. M., and Merrick, J. M. (1990). Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect. Immun. 58, 2014–2016.
Ewen, S. W., Naughton, P. J., Grant, G., Sojka, M., Allen-Vercoe, E., Bardocz, S., et al. (1997). Salmonella enterica var Typhimurium and Salmonella enterica var enteritidis express type 1 fimbriae in the rat in vivo. FEMS Immunol. Med. Microbiol. 18, 185–192.
Feutrier, J., Kay, W. W., and Trust, T. J. (1986). Purification and characterization of fimbriae from Salmonella enteritidis. J. Bacteriol. 168, 221–227.
Firon, N., Ofek, I., and Sharon, N. (1983). Carbohydrate specificity of the surface lectins of Escherichia coli, Klebsiella pneumoniae, and Salmonella typhimurium. Carbohydr. Res. 120, 235–249.
Firon, N., Ofek, I., and Sharon, N. (1984). Carbohydrate-binding sites of the mannose-specific fimbrial lectins of enterobacteria. Infect. Immun. 43, 1088–1090.
Flemming, H.-C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Ghosh, S., Mittal, A., Vohra, H., and Ganguly, N. K. (1996). Interaction of a rat intestinal brush border membrane glycoprotein with type-1 fimbriae of Salmonella typhimurium. Mol. Cell. Biochem. 158, 125–131.
Gonzalez-Escobedo, G., and Gunn, J. S. (2013). Identification of Salmonella enterica serovar Typhimurium genes regulated during biofilm formation on cholesterol gallstone surfaces. Infect. Immun. 81, 3770–3780. doi: 10.1128/IAI.00647-13
Grzymajlo, K., Kuzminska-Bajor, M., Jaworski, J., Dobryszycki, P., and Ugorski, M. (2010). The high-adhesive properties of the FimH adhesin of Salmonella enterica serovar Enteritidis are determined by a single F118S substitution. Microbiol. Read. Engl. 156, 1738–1748. doi: 10.1099/mic.0.039206-0
Grzymajło, K., Ugorski, M., Kolenda, R., Kęedzierska, A., Kuźmińska-Bajor, M., and Wieliczko, A. (2013). FimH adhesin from host unrestricted Salmonella Enteritidis binds to different glycoprotein ligands expressed by enterocytes from sheep, pig and cattle than FimH adhesins from host restricted Salmonella Abortus-ovis, Salmonella Choleraesuis and Salmonella Dublin. Vet. Microbiol. 166, 550–557. doi: 10.1016/j.vetmic.2013.07.004
Grzymajlo, K., Ugorski, M., Suchanski, J., Kedzierska, A. E., Kolenda, R., Jarzab, A., et al. (2017). The novel type 1 fimbriae fimh receptor calreticulin plays a role in Salmonella host specificity. Front. Cell. Infect. Microbiol. 7:326. doi: 10.3389/fcimb.2017.00326
Guo, A., Cao, S., Tu, L., Chen, P., Zhang, C., Jia, A., et al. (2009). FimH alleles direct preferential binding of Salmonella to distinct mammalian cells or to avian cells. Microbiol. Read. Engl. 155, 1623–1633. doi: 10.1099/mic.0.026286-0
Guo, A., Lasaro, M. A., Sirard, J.-C., Kraehenbühl, J.-P., and Schifferli, D. M. (2007). Adhesin-dependent binding and uptake of Salmonella enterica serovar Typhimurium by dendritic cells. Microbiol. Read. Engl. 153, 1059–1069. doi: 10.1099/mic.0.2006/000331-0
Hahn, E., Wild, P., Hermanns, U., Sebbel, P., Glockshuber, R., Häner, M., et al. (2002). Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 323, 845–857.
Hancox, L. S., Yeh, K. S., and Clegg, S. (1997). Construction and characterization of type 1 non-fimbriate and non-adhesive mutants of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 19, 289–296.
Hase, K., Kawano, K., Nochi, T., Pontes, G. S., Fukuda, S., Ebisawa, M., et al. (2009). Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response. Nature 462, 226–230. doi: 10.1038/nature08529
Herman, A., Serfecz, J., Kinnally, A., Crosby, K., Youngman, M., Wykoff, D., et al. (2016). The bacterial iprA gene is conserved across Enterobacteriaceae, is involved in oxidative stress resistance, and influences gene expression in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 198, 2166–2179. doi: 10.1128/JB.00144-16
Horiuchi, S., Inagaki, Y., Okamura, N., Nakaya, R., and Yamamoto, N. (1992). Type 1 pili enhance the invasion of Salmonella braenderup and Salmonella typhimurium to HeLa cells. Microbiol. Immunol. 36, 593–602.
Isaacson, R. E., Argyilan, C., Kwan, L., Patterson, S., and Yoshinaga, K. (1999). Phase variable switching of in vivo and environmental phenotypes of Salmonella typhimurium. Adv. Exp. Med. Biol. 473, 281–289.
Isaacson, R. E., and Kinsel, M. (1992). Adhesion of Salmonella typhimurium to porcine intestinal epithelial surfaces: identification and characterization of two phenotypes. Infect. Immun. 60, 3193–3200.
Jones, G. W., and Richardson, L. A. (1981). The attachment to, and invasion of HeLa cells by Salmonella typhimurium: the contribution of mannose-sensitive and mannose-resistant haemagglutinating activities. J. Gen. Microbiol. 127, 361–370. doi: 10.1099/00221287-127-2-361
Kisiela, D., Laskowska, A., Sapeta, A., Kuczkowski, M., Wieliczko, A., and Ugorski, M. (2006). Functional characterization of the FimH adhesin from Salmonella enterica serovar enteritidis. Microbiol. Read. Engl. 152, 1337–1346. doi: 10.1099/mic.0.28588-0
Kisiela, D., Sapeta, A., Kuczkowski, M., Stefaniak, T., Wieliczko, A., and Ugorski, M. (2005). Characterization of FimH adhesins expressed by Salmonella enterica serovar gallinarum biovars gallinarum and pullorum: reconstitution of mannose-binding properties by single amino acid substitution. Infect. Immun. 73, 6187–6190. doi: 10.1128/IAI.73.9.6187-6190.2005
Kisiela, D. I., Chattopadhyay, S., Libby, S. J., Karlinsey, J. E., Fang, F. C., Tchesnokova, V., et al. (2012). Evolution of Salmonella enterica virulence via point mutations in the fimbrial adhesin. PLoS Pathog. 8:e1002733. doi: 10.1371/journal.ppat.1002733
Kisiela, D. I., Chattopadhyay, S., Tchesnokova, V., Paul, S., Weissman, S. J., Medenica, I., et al. (2013). Evolutionary analysis points to divergent physiological roles of type 1 fimbriae in Salmonella and Escherichia coli. mBio 4:e00625-12. doi: 10.1128/mBio.00625-12
Kisiela, D. I., Kramer, J. J., Tchesnokova, V., Aprikian, P., Yarov-Yarovoy, V., Clegg, S., et al. (2011). Allosteric catch bond properties of the FimH adhesin from Salmonella enterica serovar Typhimurium. J. Biol. Chem. 286, 38136–38147. doi: 10.1074/jbc.M111.237511
Kolenda, R. (2018). Expression and Characterization of Pancreatic Secretory Granule Membrane Major Glycoprotein GP2 of Animal Origin. Available at: https://refubium.fu-berlin.de/handle/fub188/9629 (accessed June 12, 2018).
Kolenda, R., Burdukiewicz, M., Schiebel, J., Rödiger, S., Sauer, L., Szabo, I., et al. (2018). Adhesion of Salmonella to pancreatic secretory granule membrane major glycoprotein GP2 of human and porcine origin depends on FimH sequence variation. Front. Microbiol. 9:1905. doi: 10.3389/fmicb.2018.01905
Kuczkowski, M., Wieliczko, A., Kisiela, D., Mazurkiewicz, M., and Ugorski, M. (2004). Cellular response and protective effect in hens immunised with Salmonella Enteritidis recombinant fimbrial SefA, FimA and AgfA proteins. Bull. Vet. Inst. Pulawy 48, 375–382.
Kukkonen, M., Raunio, T., Virkola, R., Lähteenmäki, K., Mäkelä, P. H., Klemm, P., et al. (1993). Basement membrane carbohydrate as a target for bacterial adhesion: binding of type I fimbriae of Salmonella enterica and Escherichia coli to laminin. Mol. Microbiol. 7, 229–237.
Kukkonen, M., Saarela, S., Lähteenmäki, K., Hynönen, U., Westerlund-Wikström, B., Rhen, M., et al. (1998). Identification of two laminin-binding fimbriae, the type 1 fimbria of Salmonella enterica serovar Typhimurium and the G Fimbria of Escherichia coli, as plasminogen receptors. Infect. Immun. 66,4965–4970.
Kuźmińska-Bajor, M., Grzymajło, K., and Ugorski, M. (2015). Type 1 fimbriae are important factors limiting the dissemination and colonization of mice by Salmonella Enteritidis and contribute to the induction of intestinal inflammation during Salmonella invasion. Front. Microbiol. 6:276. doi: 10.3389/fmicb.2015.00276
Kuźmińska-Bajor, M., Kuczkowski, M., Grzymajło, K., Wojciech,Ł, Sabat, M., Kisiela, D., et al. (2012). Decreased colonization of chicks by Salmonella enterica serovar gallinarum expressing mannose-sensitive FimH adhesin from Salmonella enterica serovar enteritidis. Vet. Microbiol. 158, 205–210. doi: 10.1016/j.vetmic.2012.01.029
Ledeboer, N. A., Frye, J. G., McClelland, M., and Jones, B. D. (2006). Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect. Immun. 74, 3156–3169. doi: 10.1128/IAI.01428-05
Lee, C. A., Jones, B. D., and Falkow, S. (1992). Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. U.S.A. 89, 1847–1851.
Lee, C.-A., and Yeh, K.-S. (2016). The non-fimbriate phenotype is predominant among Salmonella enterica serovar choleraesuis from swine and those non-fimbriate strains possess distinct amino acid variations in FimH. PLoS One 11:e0151126. doi: 10.1371/journal.pone.0151126
Leusch, H. G., Drzeniek, Z., Markos-Pusztai, Z., and Wagener, C. (1991). Binding of Escherichia coli and Salmonella strains to members of the carcinoembryonic antigen family: differential binding inhibition by aromatic alpha-glycosides of mannose. Infect. Immun. 59, 2051–2057.
Li, W., Watarai, S., and Kodama, H. (2003a). Identification of glycosphingolipid binding sites for SEF21-fimbriated Salmonella enterica serovar enteritidis in chicken oviductal mucosa. Vet. Microbiol. 93, 73–78.
Li, W., Watarai, S., and Kodama, H. (2003b). Identification of possible chicken intestinal mucosa receptors for SEF21-fimbriated Salmonella enterica serovar Enteritidis. Vet. Microbiol. 91, 215–229.
Lindquist, B. L., Lebenthal, E., Lee, P. C., Stinson, M. W., and Merrick, J. M. (1987). Adherence of Salmonella typhimurium to small-intestinal enterocytes of the rat. Infect. Immun. 55, 3044–3050.
Lockman, H. A., and Curtiss, R. (1992). Virulence of non-type 1-fimbriated and nonfimbriated nonflagellated Salmonella typhimurium mutants in murine typhoid fever. Infect. Immun. 60, 491–496.
McFarland, K. A., Lucchini, S., Hinton, J. C. D., and Dorman, C. J. (2008). The leucine-responsive regulatory protein, Lrp, activates transcription of the fim operon in Salmonella enterica serovar typhimurium via the fimZ regulatory gene. J. Bacteriol. 190, 602–612. doi: 10.1128/JB.01388-07
Misselwitz, B., Kreibich, S. K., Rout, S., Stecher, B., Periaswamy, B., and Hardt, W.-D. (2011). Salmonella enterica serovar Typhimurium binds to HeLa cells via Fim-mediated reversible adhesion and irreversible type three secretion system 1-mediated docking. Infect. Immun. 79, 330–341. doi: 10.1128/IAI.00581-10
Monteiro, C., Papenfort, K., Hentrich, K., Ahmad, I., Le Guyon, S., Reimann, R., et al. (2012). Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 9, 489–502. doi: 10.4161/rna.19682
Musa, H.-H., Zhang, W.-J., Lv, J., Duan, X.-L., Yang, Y., Zhu, C.-H., et al. (2014). The molecular adjuvant mC3d enhances the immunogenicity of FimA from type I fimbriae of Salmonella enterica serovar Enteritidis. J. Microbiol. Immunol. Infect. 47, 57–62. doi: 10.1016/j.jmii.2012.11.004
Naughton, P. J., Grant, G., Bardocz, S., Allen-Vercoe, E., Woodward, M. J., and Pusztai, A. (2001). Expression of type 1 fimbriae (SEF 21) of Salmonella enterica serotype enteritidis in the early colonisation of the rat intestine. J. Med. Microbiol. 50, 191–197. doi: 10.1099/0022-1317-50-2-191
Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238. doi: 10.1126/science.1128794
Old, D. C., and Duguid, J. P. (1970). Selective outgrowth of fimbriate bacteria in static liquid medium. J. Bacteriol. 103, 447–456.
Old, D. C., Roy, A. I., and Tavendale, A. (1986). Differences in adhesiveness among type 1 fimbriate strains of Enterobacteriaceae revealed by an in vitro HEp2 cell adhesion model. J. Appl. Bacteriol. 61, 563–568.
Oyofo, B. A., Droleskey, R. E., Norman, J. O., Mollenhauer, H. H., Ziprin, R. L., Corrier, D. E., et al. (1989). Inhibition by mannose of in vitro colonization of chicken small intestine by Salmonella typhimurium. Poult. Sci. 68,1351–1356.
Pang, Y., Wang, H., Li, Z., Piao, J., Piao, J., Chi, Y., et al. (2012). Immune response to liposome-associated recombinant SEF21 following oral immunization in chickens. Avian Dis. 56, 347–353. doi: 10.1637/9928-091011-Reg.1
Papenfort, K., Said, N., Welsink, T., Lucchini, S., Hinton, J. C. D., and Vogel, J. (2009). Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 74, 139–158. doi: 10.1111/j.1365-2958.2009.06857.x
Patterson, S. K., Borewicz, K., Johnson, T., Xu, W., and Isaacson, R. E. (2012). Characterization and differential gene expression between two phenotypic phase variants in Salmonella enterica serovar Typhimurium. PLoS One 7:e43592. doi: 10.1371/journal.pone.0043592
Purcell, B. K., Pruckler, J., and Clegg, S. (1987). Nucleotide sequences of the genes encoding type 1 fimbrial subunits of Klebsiella pneumoniae and Salmonella typhimurium. J. Bacteriol. 169, 5831–5834.
Rajashekara, G., Munir, S., Alexeyev, M. F., Halvorson, D. A., Wells, C. L., and Nagaraja, K. V. (2000). Pathogenic role of SEF14, SEF17, and SEF21 fimbriae in Salmonella enterica serovar enteritidis infection of chickens. Appl. Environ. Microbiol. 66, 1759–1763.
Remaut, H., Rose, R. J., Hannan, T. J., Hultgren, S. J., Radford, S. E., Ashcroft, A. E., et al. (2006). Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol. Cell 22, 831–842. doi: 10.1016/j.molcel.2006.05.033
Rossolini, G. M., Muscas, P., Chiesurin, A., and Satta, G. (1993). Analysis of the Salmonella fim gene cluster: identification of a new gene (fimI) encoding a fimbrin-like protein and located downstream from the fimA gene. FEMS Microbiol. Lett. 114, 259–265.
Saini, S., Pearl, J. A., and Rao, C. V. (2009). Role of FimW, FimY, and FimZ in regulating the expression of type I fimbriae in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191, 3003–3010. doi: 10.1128/JB.01694-08
Saini, S., Slauch, J. M., Aldridge, P. D., and Rao, C. V. (2010). Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes. J. Bacteriol. 192, 5767–5777. doi: 10.1128/JB.00624-10
Silverman, M., Zieg, J., Hilmen, M., and Simon, M. (1979). Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc. Natl. Acad. Sci. U.S.A. 76, 391–395.
Sterzenbach, T., Nguyen, K. T., Nuccio, S.-P., Winter, M. G., Vakulskas, C. A., Clegg, S., et al. (2013). A novel CsrA titration mechanism regulates fimbrial gene expression in Salmonella typhimurium. EMBO J. 32, 2872–2883. doi: 10.1038/emboj.2013.206
Swenson, D. L., Kim, K. J., Six, E. W., and Clegg, S. (1994). The gene fimU affects expression of Salmonella typhimurium type 1 fimbriae and is related to the Escherichia coli tRNA gene argU. Mol. Gen. Genet. 244, 216–218.
Tavendale, A., Jardine, C. K., Old, D. C., and Duguid, J. P. (1983). Haemagglutinins and adhesion of Salmonella typhimurium to HEp2 and HeLa cells. J. Med. Microbiol. 16, 371–380. doi: 10.1099/00222615-16-3-371
Teplitski, M., Al-Agely, A., and Ahmer, B. M. M. (2006). Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiol. Read. Engl. 152, 3411–3424. doi: 10.1099/mic.0.29118-0
Thankavel, K., Shah, A. H., Cohen, M. S., Ikeda, T., Lorenz, R. G., Curtiss, R., et al. (1999). Molecular basis for the enterocyte tropism exhibited by Salmonella typhimurium type 1 fimbriae. J. Biol. Chem. 274, 5797–5809. doi: 10.1074/jbc.274.9.5797
Tinker, J. K., and Clegg, S. (2000). Characterization of FimY as a coactivator of type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 68, 3305–3313.
Tinker, J. K., and Clegg, S. (2001). Control of FimY translation and type 1 fimbrial production by the arginine tRNA encoded by fimU in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 40, 757–768.
Tinker, J. K., Hancox, L. S., and Clegg, S. (2001). FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183, 435–442. doi: 10.1128/JB.183.2.435-442.2001
van der Velden, A. W. M., Bäumler, A. J., Tsolis, R. M., and Heffron, F. (1998). Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect. Immun. 66, 2803–2808.
Verma, A., Basu, S., Ghosh, S., Majumdar, S., and Ganguly, N. K. (2005). The effect of type-1 fimbrial immunization on gut pathophysiological response in rats infected with Salmonella enterica subsp. enterica serovar Typhimurium. Mol. Cell. Biochem. 273, 127–135. doi: 10.1007/s11010-005-8160-0
Waksman, G., and Hultgren, S. J. (2009). Structural biology of the chaperone–usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 7, 765–774. doi: 10.1038/nrmicro2220
Wang, K.-C., Hsu, Y.-H., Huang, Y.-N., Lin, J.-H., and Yeh, K.-S. (2014). FimY of Salmonella enterica serovar Typhimurium functions as a DNA-binding protein and binds the fimZ promoter. Microbiol. Res. 169, 496–503. doi: 10.1016/j.micres.2013.12.006
Wang, K.-C., Hsu, Y.-H., Huang, Y.-N., and Yeh, K.-S. (2012). A previously uncharacterized gene stm0551 plays a repressive role in the regulation of type 1 fimbriae in Salmonella enterica serotype Typhimurium. BMC Microbiol. 12:111. doi: 10.1186/1471-2180-12-111
Wang, K.-C., Huang, C.-H., Ding, S.-M., Chen, C.-K., Fang, H.-W., Huang, M.-T., et al. (2016). Role of yqiC in the pathogenicity of Salmonella and innate immune responses of human intestinal epithelium. Front. Microbiol. 7:1614. doi: 10.3389/fmicb.2016.01614
Wang, L., Cai, X., Wu, S., Bomjan, R., Nakayasu, E. S., Händler, K., et al. (2017). InvS coordinates expression of PrgH and FimZ and is required for invasion of epithelial cells by Salmonella enterica serovar Typhimurium. J. Bacteriol. 199:e00824-16. doi: 10.1128/JB.00824-16
Werneburg, G. T., and Thanassi, D. G. (2018). Pili assembled by the chaperone/usher pathway in Escherichia coli and Salmonella. EcoSal Plus 8, doi: 10.1128/ecosalplus.ESP-0007-2017
Wilson, R. L., Elthon, J., Clegg, S., and Jones, B. D. (2000). Salmonella enterica serovars gallinarum and pullorum expressing Salmonella enterica serovar typhimurium type 1 fimbriae exhibit increased invasiveness for mammalian cells. Infect. Immun. 68, 4782–4785. doi: 10.1128/IAI.68.8.4782-4785.2000
Wu, K.-H., Wang, K.-C., Lee, L.-W., Huang, Y.-N., and Yeh, K.-S. (2012). A constitutively mannose-sensitive agglutinating Salmonella enterica subsp. enterica serovar Typhimurium strain, carrying a transposon in the fimbrial usher gene stbC, exhibits multidrug resistance and flagellated phenotypes. Sci. World J. 2012:280264. doi: 10.1100/2012/280264
Yeh, K. S., Chen, T. H., Liao, C. W., Chang, C. S., and Lo, H. C. (2002a). PCR amplification of the Salmonella typhimurium fimY gene sequence to detect the Salmonella species. Int. J. Food Microbiol. 78, 227–234.
Yeh, K. S., Tinker, J. K., and Clegg, S. (2002b). FimZ binds the Salmonella typhimurium fimA promoter region and may regulate its own expression with FimY. Microbiol. Immunol. 46, 1–10.
Yeh, K. S., Hancox, L. S., and Clegg, S. (1995). Construction and characterization of a fimZ mutant of Salmonella typhimurium. J. Bacteriol. 177, 6861–6865.
Yue, M., Han, X., De Masi, L., Zhu, C., Ma, X., Zhang, J., et al. (2015). Allelic variation contributes to bacterial host specificity. Nat. Commun. 6:8754. doi: 10.1038/ncomms9754
Yue, M., Rankin, S. C., Blanchet, R. T., Nulton, J. D., Edwards, R. A., and Schifferli, D. M. (2012). Diversification of the Salmonella fimbriae: a model of macro- and microevolution. PLoS One 7:e38596. doi: 10.1371/journal.pone.0038596
Zeiner, S. (2012). Type 1 Fimbrial Structure and Regulation in Salmonella enterica Serovar Typhimurium. Ph.D. thesis, University of Iowa, Iowa City.
Zeiner, S. A., Dwyer, B. E., and Clegg, S. (2012). FimA, FimF, and FimH are necessary for assembly of type 1 fimbriae on Salmonella enterica serovar Typhimurium. Infect. Immun. 80, 3289–3296. doi: 10.1128/IAI.00331-12
Keywords: Salmonella, type 1 fimbriae, adhesion, invasion, regulation of expression
Citation: Kolenda R, Ugorski M and Grzymajlo K (2019) Everything You Always Wanted to Know About Salmonella Type 1 Fimbriae, but Were Afraid to Ask. Front. Microbiol. 10:1017. doi: 10.3389/fmicb.2019.01017
Received: 26 February 2019; Accepted: 24 April 2019;
Published: 14 May 2019.
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
Evgeni Sokurenko, University of Washington, United StatesKuang-Sheng Yeh, National Taiwan University, Taiwan
Ritwij Kulkarni, University of Louisiana at Lafayette, United States
Copyright © 2019 Kolenda, Ugorski and Grzymajlo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krzysztof Grzymajlo, a3J6eXN6dG9mLmdyenltYWpsb0B1cHdyLmVkdS5wbA==
 Rafal Kolenda
Rafal Kolenda Maciej Ugorski
Maciej Ugorski Krzysztof Grzymajlo
Krzysztof Grzymajlo