
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 30 April 2019
Sec. Systems Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00876
This article is part of the Research TopicOmics and Systems Approaches to Study the Biology and Applications of Lactic Acid Bacteria View all 20 articles
Lactic Acid Bacteria (LAB) are extensively employed in the production of various fermented foods, due to their safe status, ability to affect texture and flavor and finally due to the beneficial effect they have on shelf-life. More recently, LAB have also gained interest as production hosts for various useful compounds, particularly compounds with sensitive applications, such as food ingredients and therapeutics. As for all industrial microorganisms, it is important to have a good understanding of the physiology and metabolism of LAB in order to fully exploit their potential, and for this purpose, many systems biology approaches are available. Systems metabolic engineering, an approach that combines optimization of metabolic enzymes/pathways at the systems level, synthetic biology as well as in silico model simulation, has been used to build microbial cell factories for production of biofuels, food ingredients and biochemicals. When developing LAB for use in foods, genetic engineering is in general not an accepted approach. An alternative is to screen mutant libraries for candidates with desirable traits using high-throughput screening technologies or to use adaptive laboratory evolution to select for mutants with special properties. In both cases, by using omics data and data-driven technologies to scrutinize these, it is possible to find the underlying cause for the desired attributes of such mutants. This review aims to describe how systems biology tools can be used for obtaining both engineered as well as non-engineered LAB with novel and desired properties.
Lactic acid bacteria (LAB) are traditionally used as starters in fermented food production. LAB produce lactic acid, the presence of which reduces growth of pathogens and other undesirable microbes in the products, while contributing to an appealing acidic taste. LAB are also capable of producing various aromatic compounds, the formation of which are initiated by lipolysis and proteolysis, and these also contribute to the pleasant organoleptic characteristics of different fermented products such as yogurt, cheese, and butter (Song et al., 2017). LAB are thus highly industrially relevant microorganisms and represent a multi-billion dollars business worldwide (Johansen, 2017).
Several important phenotypical characteristics of LAB such as acidification rate (Martinussen et al., 2013), robustness to environmental stresses and phage resistance (Garneau and Moineau, 2011; Papadimitriou et al., 2016), contribution to flavor and texture formation (Broadbent et al., 2005; Chen J. et al., 2017), bio-protection activity (Siedler et al., 2019) and probiotic function (Ljungh and Wadström, 2006), are important for real-life applications and are continuously being investigated by both the academic and industrial community. Traditional non-GMO (genetically modified organism) strain modification approaches such as random mutagenesis and screening, selection using toxic analogs and adaptive evolution, have a long history of successful use for improving properties of LAB. The use of mutagenesis for improving LAB is only constrained by the genetic repertoire of the bacteria, but a large screening input is usually required to identify desired mutants. The latter two methods are specific and very selective, but their implementation is largely condition-dependent. For analog selection, the target pathway should be tightly associated with anabolism, and an effective metabolite analog should also be easily accessible. For adaptive evolution, the selection is dependent on fitness improvements under stressful conditions. Rational strain modification methods, such as metabolic engineering and synthetic biology, have demonstrated their effectiveness to endow LAB with new/improved characteristics useful for food applications beyond what traditional approaches are capable of delivering. However, since LAB are often present in the final product destined for human consumption, there are still hurdles that have to be overcome before recombinant DNA technologies can be widely implemented, e.g., regulatory issues and skepticism of consumers.
The understanding of LAB physiology has fundamentally changed since the emergence of high-throughput genome sequencing technologies. By using whole-genome sequencing to compare the genome sequences of model LAB strains and their derivatives generated using traditional strain improvement procedures, researchers have gained great insight into how phenotypes are affected by genetic variations. Modern sequencing technologies allow for entire genome landscapes of multiple bacteria to be generated simultaneously within less than 24 h. These genome sequence data can be used as references, when doing physiological characterization or when using various systems biology approaches to address different fundamental and practical questions.
Due to their Generally Recognized as Safe (GRAS) status, simple metabolism, and a very high glycolytic flux (Koebmann et al., 2002a), LAB recently have emerged as promising cell factories for production of high-value biochemicals, including food ingredients and pharmaceutical precursors. This has been aided by genome-scale metabolic models, where researchers have used in silico simulations to find the best way to reroute LAB metabolic networks to optimize production of various compounds. In this review we will discuss four main approaches used for improving LAB: (1) Adaptive Laboratory evolution (ALE) in combination with meta-omics analysis for characterization of mutants; (2) Systems biology tools for elucidating microbial interactions and metabolic capacities; (3) a detailed analysis on metabolic flux regulation for LAB model strain – Lactococcus lactis; (4) metabolic engineering of L. lactis as a novel microbial cell factory.
In contrast to the comparative genomics approach for studying the inter/intraspecies relationship of LAB, ALE studies focus on genomic adaptation, including single nucleotide variations (SNV), deletions and insertions, to specific environmental stresses or metabolic perturbations, and its correlation to phenotypical changes, where the latter are typically studied using a combination of omics analysis, e.g., transcriptomics, proteomics, metabolomics, and physiological characterization. In an ALE setup, the number of accumulated mutations is normally controlled by limiting the number of generations of propagation in a subculturing or chemostat system. In contrast to natural evolution, the type and dose of selection stress, which serve as an input variable, can easily be defined in a laboratory environment. This approach puts a limit on the complexity of the subsequent comparative systems biology study, by limiting the number of output variations. The eventual multi-omics analysis and data integration with systems biology modeling are expected to provide the foundation for a better understanding of the evolutionary driving force of LAB, and to guide the rational design of ALE conditions to improve the performance of industrial LAB (Bachmann et al., 2017).
Adaptive laboratory evolution has been extensively used in stress physiological studies of LAB. As LAB are industrially important workhorses, LAB often undergo a variety of environmental stresses e.g., heat, salt and acid stress in different manufacturing conditions. Heat is one of the common stresses that LAB need to handle well in industrial processes. For instance, the mesophilic L. lactis, the main constituent in starters used for making semi-hard cheeses, such as cheddar and Gouda, is exposed to high temperatures during the syneresis step where the moisture content in the cheese grains is reduced. Such suboptimal temperatures are not lethal to L. lactis, but the physiological properties of L. lactis are significantly altered to manage the harsh conditions. LAB normally display a multi-level cellular response to environmental stresses, where multi-omics serves as desirable analysis tools for understanding the mechanism. To study the high-temperature physiology and its molecular basis, Chen et al. (2015a) applied ALE on the model L. lactis strain MG1363 with gradually increased incubation temperature. After an 800-generation session of experimental adaptation at high temperatures using a serial-transfer regime, a thermal-tolerant variant was isolated. The authors characterized the mutant with comprehensive systems biology analysis. On the metabolic level, the mutant had expanded its glycolysis capacity at high temperatures, where the overall glycolytic flux in the mutant was 13% higher compared to that of the wild-type strain at 38°C. On the transcriptomic level, a variety of solute transport activities had been enhanced in the mutant including ones for carbohydrates, amino acids and ions. Alterations in cell-membrane fatty acid composition were also observed for the mutant, which had an increased amount of saturated fatty acids (C14:0 and C16:0) in the cell membrane of the thermally tolerant mutants. Increased amounts of saturated fatty acids help maintain the fluidity of cell membranes at high temperatures and thereby stabilizing the cross-membrane transport activities (Los and Murata, 2000). On the genomic level, the authors identified 13 mutations through whole genome resequencing, and subsequently used reverse engineering and physiological characterization to determine the contribution of individual mutations to the overall phenotype. It was found that SNVs in groESL (encoding chaperon proteins) and rpoC (encoding RNA polymerase subunit) contributed significantly to thermal tolerance of the mutant. The SNVs in groESL led to overexpression of molecular chaperones, and the SNVs in rpoC caused the alteration in global gene expression, and as a consequence of this, the saturated fatty acid synthesis pathway was overexpressed. The integrated multifaceted analysis indicated that a stable cell membrane structure in combination with other events, such as overexpression of molecular chaperones, is important for the cells to support a high-energy turnover rate under heat stress conditions. The study demonstrates how multi-omics analysis can help understand the stress physiology of LAB on the molecular level.
Besides the environmental stresses, LAB such as L. lactis often encounter nutrient starvation conditions during cheese ripening. In the early stage of ripening, acidification continues with limited lactose (3–5% lactose of milk depending on moisture) in the curd after whey drainage (Ardö and Nielsen, 2007). After the carbohydrate is completely exhausted, the cells stop growing or eventually lyse (Ganesan et al., 2007). The global catabolite regulator CcpA plays an important role in the regulation of the metabolic shift under these carbohydrate limitation/starvation conditions, but how LAB reshape the cellular metabolism has still not been elucidated. In contrast to traditional ALE with the serial-passage regime, the growth rate can be controlled by limiting specific nutrients in a chemostat ALE setup, which is used to mimic the slow growth and metabolic activity of LAB during cheese ripening (Van Mastrigt et al., 2018). Price et al. (2017) performed a glucose-limited ALE for L. lactis in a chemostat setup to study the response to carbohydrate starvation. After a few generations of adaption, fast-growing L. lactis mutants emerged and finally dominated the population. Whole genome resequencing of the isolates obtained from parallel evolution lineages indicated that the SNVs causing a site-specific change in the amino acid sequence in CcpA (Met-19) was responsible for the adaptation. To study the global regulatory role of the mutated CcpA, they performed a transcriptomics analysis, and found that glucose transport in the mutant had changed. The mutation in CcpA led to an overexpression of PTSMan but a downregulation of PTSCel. Both PTSMan and PTSCel participate in the glucose transport in L. lactis, but PTSMan has a higher substrate affinity for glucose compared to PTSCel (Castro et al., 2009). The CcpA-mediated transcriptional change caused a 3-fold increased rate in glucose uptake in the mutant, which was further corroborated by the flux analysis with C14-glucose. These findings suggest that there is an evolutionary advantage associated with altering global regulators when compared to altering expression of individual genes, at least in response to carbohydrate starvation (Goel et al., 2015).
Since the first LAB genome sequence (L. lactis subsp. lactis IL1403) was announced in 2001 (Bolotin et al., 2001), whole genome sequencing has significantly accelerated systems biology research on LAB. Based on the genome annotation, Genome-Scale Metabolic models (GEMs) with Constraint-Based Reconstruction and Analysis (COBRA) have been used to predict the nutrient requirements for growth, and metabolic patterns for LAB under different conditions (Wu et al., 2017). This information is paramount for both starter culture producers as well as for the food industries relying on LAB. The accuracy of metabolic models depends on correct genome annotations, and subsequent manual curation with biochemical, genetic and cell physiological data (Kitano, 2002a). Several studies have suggested that the NADH/NAD+ ratio, which depends on the sugar uptake rate, has a role in the shift between the homolactic and mixed-acid fermentation in LAB (Garrigues et al., 1997), and that LAB with a low glycolytic flux need more energy to produce biomass. During mixed-acid fermentation, one extra ATP is generated when acetate is formed from acetyl-CoA. The experimental observation that biomass yield is maximized under starvation conditions has also been predicted using GEMs (Oliveira et al., 2005). In another case, GEMs and COBRA were used to predict the outcome of an ALE, where Lactobacillus plantarum was grown on the non-conventional substrate glycerol. The question addressed was how the metabolic network could be reshaped to support fast growth. L. plantarum grows slowly on glycerol, but only under aerobic conditions. Teusink et al. (2009) adapted L. plantarum on glycerol using a complex medium. After approximately 800 generations, fast-growing mutants could be isolated. Metabolic flux analysis showed that lactate rather than acetate was the major product from glycerol dissimilation. It was assumed that a mixed-acid fermentation mode was needed to support a high-energy yield on slowly fermentable sugars. In this case, the specific growth rate of the mutant (0.26 h-1) on glycerol was still substantially lower than the growth rate on other fast-fermentable sugars. To explain the observations, the authors applied flux balance analysis (FBA) using the uptake rates of glycerol, citrate (already present in the medium) and oxygen as constraints. As the flux model was set to maximize the biomass yield under aerobic conditions, the oxygen consumption played an important role in FBA. Under aerobic conditions, the model predicted that more ATP could be generated when lactate was formed, and that lactate production would support faster growth. The good correlation between the model prediction and experimental outcome demonstrates that COBRA is a valuable tool for predicting how the metabolic network of LAB adjusts over the course of an evolution experiment.
Adaptive laboratory evolution in combination with genomic analysis can also help disclose fitness-associated gene functions in LAB. LAB such as Lactobacillus and Lactococcus species, are facultative anaerobes. Albeit the lack of respiration, the presence of oxygen usually does not affect normal growth (Higuchi et al., 2000). The toxicity of oxygen toward LAB is believed to be due to generation of reactive oxygen species (ROSs) during oxygen metabolism (Chen et al., 2013). Due to their catalase negative nature, LAB are more vulnerable to the attack from H2O2-derived ROSs. Oxygen not only affects the cell fitness, but also tends to alter the acidification profile of LAB cultures (Larsen et al., 2015). Therefore, the study of oxygen related gene functions in LAB has high industrial relevance. The thioredoxin-thioredoxin reductase is the major system for maintaining the cellular redox homeostasis of LAB when oxidative stress is imposed (Vido et al., 2005; Serrano et al., 2007; Serata et al., 2012). Two thioredoxin reductase genes are annotated on the chromosome of L. lactis namely trxB1 and trxB2. The importance of trxB1 for regulating redox homeostasis has been demonstrated by single gene knockout experiments in L. lactis MG1363. The loss of functional TrxB1, however, is not lethal for L. lactis in the presence of oxygen (Vido et al., 2005). Chen et al. (2015b) introduced a deletion in trxB2, and noticed that aerobic growth of the mutant was substantially retarded. TrxB2 shares a high amino acid sequence similarity with TrxB1, but it lacks two conserved cysteine residues, which are necessary to function as a thioredoxin reductase. To elucidate the biological function of TrxB2 under aerobic conditions, the authors designed an ALE setup, where the mutant was subcultured under aerobic conditions for a prolonged period of time. Genome resequencing of adapted mutants revealed the occurrence of suppressor mutations in the ribonucleotide reduction pathways in mutants isolated from independent lineages. The research suggested an important role of TrxB2 as a flavodoxin reductase in the aerobic ribonucleotide reduction, which was supported by subsequent physiological characterization (Chen et al., 2015b). Often, the physiology studies of LAB under oxidative stress conditions focus on ROS, as ROS has detrimental effects on cell fitness (Guchte et al., 2002). However, oxygen-dependent anabolism should not be underestimated in terms of its importance for aerobic growth of LAB.
Adaptive laboratory evolution and multiomics analysis have been demonstrated to be useful tools for addressing fundamental questions in the natural evolution of LAB. The comparative genomics study of different L. lactis isolates suggests that the dairy-associated L. lactis diverged from the plant isolates through a long history of natural evolution (Siezen et al., 2008). The phenotypic properties are quite different between the non-dairy (plant, meat and bovine rumen) isolates and dairy-associated isolates in terms of nutrient requirements and stress tolerance in different environments. Some unique traits e.g., excellent flavor formation capacity from amino acid catabolism of the plant-associated L. lactis strains has also been used for enhancing the flavor formation in dairy fermentation (Tanous et al., 2005). However, the important question of gene gain and loss during the natural evolution of the plant isolates adapting the dairy niche has not been fully addressed. Bachmann et al. (2012) adapted the plant-derived L. lactis strain KF147 in milk for nearly 1,000 generations. Isolates obtained from three independent evolutionary lineages had adapted to growing well in milk. The integrated comparative genomics and transcriptomics analysis revealed that the adapted strains had variations that resulted in improved utilization of milk proteins and loss or down-regulation of pathways important for using plant materials. Such gene gain and loss are a typical feature found in the natural evolution of LAB accommodated by a dairy niche (Makarova and Koonin, 2007; Hanemaaijer et al., 2015).
It is a common practice that mixed starter cultures (the combination of different strains) are used for food fermentations, e.g., for production of cheese and yogurt. One consideration is that the process with a single strain is more vulnerable to bacteriophage predation, where strain-specific phages can result in fermentation failure. This problem is alleviated when mixed strains are used, as strains unaffected by these phages will ensure that the fermentation does not fail (Smid et al., 2014).
The use of mixed strains also confers the culture a broader metabolic capacity, which is important to achieve a desirable flavor and organoleptic property of the final product. In particular for cheeses made using mesophilic starters cultures this is relevant. The mesophilic starter cultures are typically composed of L. lactis subsp. cremoris, L. lactis subsp. lactis and its citrate-positive variant (L. lactis subsp. biovar diacetylactis). During milk fermentation, both L. lactis subsp. cremoris and L. lactis subsp. lactis contribute to acidification. During the subsequent cheese ripening stage, the roles of different strains for flavor formation become distinct (Ardö and Nielsen, 2007).
Mixed strains can be rationally designed by blending two or more well-characterized industrial strains on the condition that their growth dependency and product profiles meet the quality requirement such as the mesophilic/thermophilic cultures used in cheese/yogurt manufacturing (Sieuwerts, 2016). On the other hand, mesophilic undefined starter cultures are commonly used for the manufacture of the European continental semi-hard cheeses such as Cheddar, Gouda, and Danbo. Such cultures derive from back-slopping cultures, which were often collected from the batches yielding good quality cheese from artisanal cheese operations, and saved at –80°C to minimize changes in LAB composition during storage (Smid et al., 2014). Such undefined starters have a long history of successful use for manufacturing cheeses with a rich flavor, and are highly phage resistant, most likely due to the coevolution of the strains and exchange of phage resistance mechanisms.
To study the undefined starter culture, it is necessary first to characterize starter composition, i.e., determine the strains it contains and the corresponding amounts, before characterizing single-strain physiology and genetic background (Temmerman et al., 2004). Culture dependent approaches are commonly used, however, are tedious and the empirical choice of medium for enumeration does not ensure the recovery of the entire community of strains (Erkus et al., 2013; Frantzen et al., 2016). Culture-independent approaches for characterization scarcely provide detailed information about community dynamics and interaction in practice. It is desirable to transfer systems biology strategies used for characterizing single strains to mixed strains, but an understanding of community-level genomics is necessary before systems biology approaches can be applied to study microbial consortia.
Metagenomics analysis provides insights into the species composition and its dynamics in a culture-independent manner. The holistic decoding of the genomic material in an undefined starter community via metagenomics studies definitely benefits the research of starter LAB e.g., high-resolution surveillance of the microbial community dynamics or construction of community-based GEMs. Such knowledge will facilitate the general understanding of growth, metabolism and physiology during mixed culture preparation, cheese manufacturing and ripening (Jonnala et al., 2018). Use of 16S amplicon sequencing and shotgun metagenomics are typical in metagenomics studies of microbial consortia. Sequencing amplicons of conserved regions in 16S rRNA using Next-Generation Sequencing (NGS) technologies provide information about species-level community composition on how composition changes in response to different abiotic factors (De Filippis et al., 2017). There are two limitations, which should be considered when using 16S amplicon sequencing to determine composition. First, the variation in 16S rRNA allele numbers in LAB prevents a precise prediction of abundance, if the copy number is not known for individual strains. Second, 16S amplicon sequencing only enables the species-level differentiation. Especially for undefined mesophilic starter cultures, the resolution of 16S amplicon sequencing is low, as it is mainly composed of the L. lactis species with a low 16S rRNA sequence variation. For example, Porcellato and Skeie (2016) accessed the impact of elevated cooking temperature on cheese microbiota composition. The authors used 16S amplicon sequencing and found that a high temperature led to a reduced number of live L. lactis cells during ripening. L. lactis subsp. lactis is generally more stress tolerant compared to L. lactis subsp. cremoris, but the 16S amplicon sequencing could not deliver the subspecies information of the inactivated L. lactis in this particular study. Furthermore, 16S amplicon sequencing only provides information about rRNA sequences, but hardly provides deep insight into the metagenome.
With improved quality and decreased price of second-generation shotgun sequencing, it is now possible to do deep-sequencing metagenomics and determine the entire genomic content (core/pan-genome) of a microbial ecosystem, including the low-abundant strains. Using this approach, genome decay (plasmid-loss) and the species-level dynamic shift of the community due to the biotic/abiotic factors can be revealed while comparing the reads to marker databases for abundance calculation (De Filippis et al., 2017). Nevertheless, there are only few published studies where metagenomics analysis has been used to study starter culture composition at the strain-level. The high coverage sequencing eases the identification of metabolic genes in the strain community, but the challenges remains as to which strains the genes belong. One technical shortcoming of second-generation sequencing platforms is that only short reads (<800 bp) are generated, which prevents the strain-level differentiation with the de novo-based assembly based metagenomics analysis. Especially in the mesophilic-undefined starters, the dominating L. lactis species exhibits a high intra-subspecies genome similarity (Figure 1). Therefore, the typical contig binning steps by GC content, coverage or metabolic networks are difficult to apply on the strain-level assembly (Albertsen et al., 2013; Biggs and Papin, 2015).
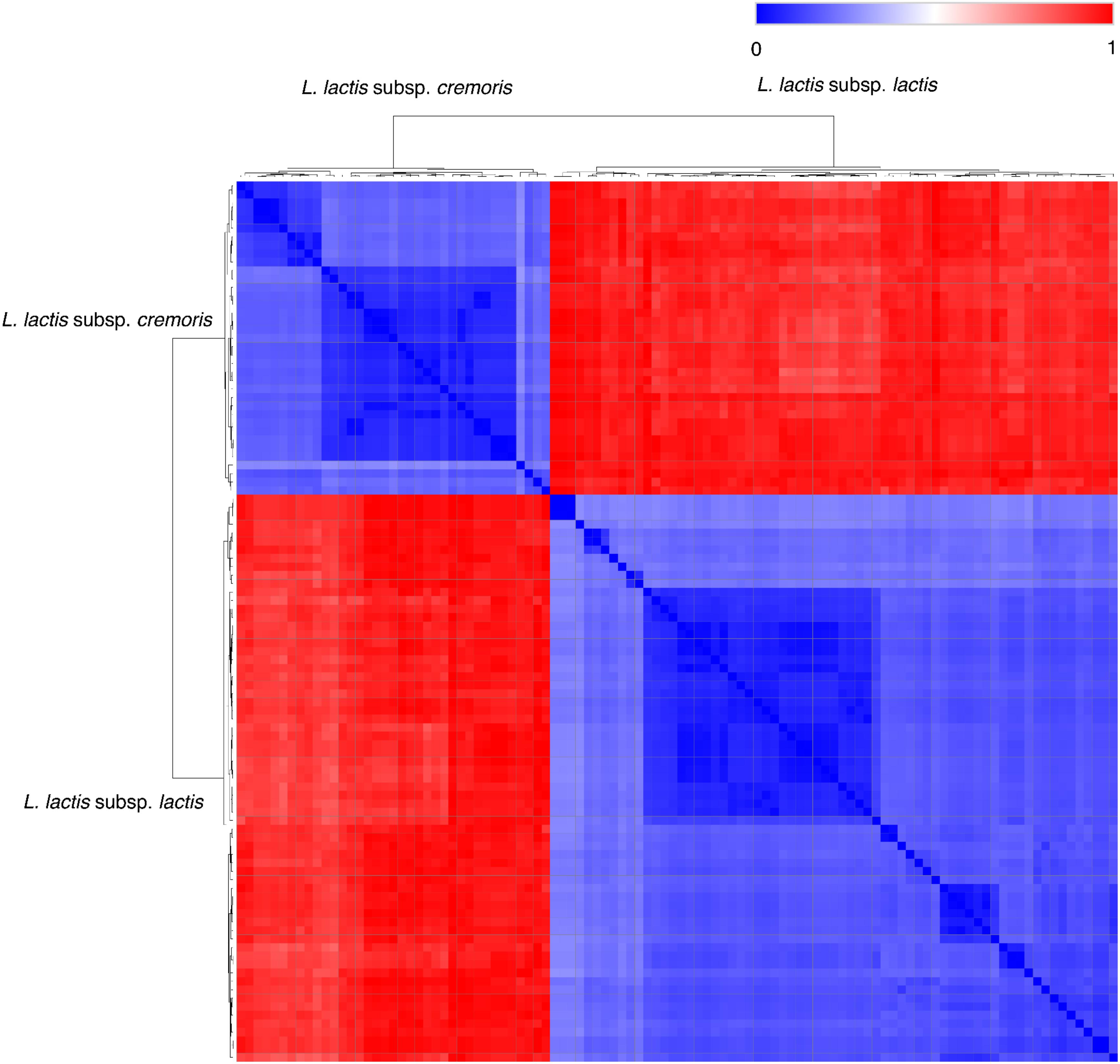
Figure 1. Hierarchical clustering of 103 published lactococcal genomes. The genome sequencing data were obtained from NCBI. The distance of phylogeny was calculated using MASH. The distance data generated by MASH (Ondov et al., 2016) was used to performing the hierarchical clustering on Morpheus available from the Broad Institute (https://software.broadinstitute.org/morpheus/).
Recently, several new data analysis tools based on reference-mapping approaches have been developed and validated which can be used for strain-level metagenomics data analysis (Scholz et al., 2016; Albanese and Donati, 2017; Truong et al., 2017; Zolfo et al., 2017). The emergence of these new tools has attracted the attention from the LAB scientific community as it now is possible to achieve a more profound understanding of the starter culture community (structure and function) and its contribution to the quality of fermented products (Ercolini, 2017). The development of the third-generation long-read sequencing technology is booming. Researchers have demonstrated single-read sequencing up to 0.9 Mbp length using the MinION system (Jain et al., 2018). Considering the comparable small genome size of LAB (1.2–4.9 Mbp) (Douillard and de Vos, 2014), in the future, with further improvements of throughput and accuracy, the third-generation genome sequencing platforms with long-read sequencing ability will definitely facilitate de novo assembly of single-strain genomes in undefined starter cultures, and eventually facilitate system-level understanding of the community (Turaev and Rattei, 2016). Another promising approach for looking at single-cell genomics in microbial communities is to use single-cell sequencing technologies. The concept is to first sort and compartmentalize single cells in water-in-oil droplets, where the DNA is isolated and barcoded in an isolated environment prior to whole genome amplification of single chromosomes followed by sequencing (Gawad et al., 2016). The main challenge for this technology is the resolution. If the sequencing accuracy can be improved in the future, it can largely facilitate the strain-level genomics study of microbial consortia.
Genomics data are the basis of systems biology (Kitano, 2002b). Postgenomic analysis tools such as transcriptomics, proteomics and metabolomics have also been widely used to understand the physiology of LAB at a systems level. A large number of these kinds of analyses have been applied to single strains and they provide great insights into how cells respond to different abiotic/biotic perturbations. To thoroughly understand industrially relevant properties of LAB, useful omics data should be generated from the LAB consortia in action, i.e., in dairy fermentations that are similar to those taking place in dairy plants, and there are only few such studies. The yogurt culture is a well-described mutualistic system, in which defined Streptococcus thermophilus (S. thermophilus) and Lactobacillus delbrueckii subsp. bulgaricus (L. bulgaricus) strains are mixed for co-culture fermentation of bovine milk. In the mutualistic life of the yogurt culture, the growth profiles of S. thermophilus and L. bulgaricus show a clear protocooperation relationship (Sieuwerts, 2016). As the LAB blended in the yogurt starter culture are normally well-documented, it serves as a simplified model system for the implementation of system biology study for microbial community. The balanced growth of these two bacteria is important for fast acidification and desirable texture and flavor formation of yogurt (Chen C. et al., 2017; Yamauchi et al., 2019). In yogurt fermentations, both the growth and acidification are stimulated when cocultures of the two LAB are used. It appears that the protocooperation occurs in the manner of nutrient exchange, where production of formic acid and carbon dioxide from S. thermopilus stimulates the growth of L. bulgaricus, and the proteolytic activity of L. bulgaricus provides essential peptides and amino acids to S. thermophilus. The integrated transcriptomics and proteomics analysis of S. thermophilus in coculture with L. bulgaricus confirmed the mutualistic effects, where upregulation of anabolic pathways in both S. thermophilus and L. bulgaricus compared to monoculture cultivation was observed (Herve-Jimenez et al., 2009; Sieuwerts et al., 2010). Meta-transcriptomics and meta-proteomics analyses revealed a dynamic response to both biotic and abiotic factors at different stages of the mutualistic growth, results that largely confirm the previously observed phenotypic behavior between the cooperation of these two bacteria. Interestingly, ion homeostasis was affected in S. thermophilus in the coculture with L. bulgaricus (Herve-Jimenez et al., 2009; Sieuwerts et al., 2010). Reduced ion transport and increased ion-chelating activity in S. thermophilus was shown to be correlated with H2O2 production by L. bulgaricus. The major oxidative stress reponse system, however, was not induced in S. thermophilus. One reason for this was that L. bulgaricus produces minor amounts of H2O2 during growth in milk. The adaptation of the catalase-negative S. thermophilus to H2O2 by control of the intracellular ion homeostasis gives an indirect way to circumvent the oxidative stress.
The cheese environment is a more complex ecosystem than yogurt and also the cultures used are more complex. Typically, different kinds of LAB e.g., lactococci and streptococci are involved in acidification and cheese ripening. Cheese ripening is one of the most important processes in cheese manufacturing in terms of flavor formation. However, ripening is a slow process. Not only starter LAB but also other non-starter LAB (NSLAB) usually have an important contribution to flavor formation during cheese ripening. To accelerate cheese maturation, one option is to elevate the ripening temperature to increase the rate of the biochemical transformations taking place. Meta-transcriptomics (RNA-seq) data shows that expression of genes involved in proteolysis, lipolysis and amino acids/fatty acids catabolism are promoted in NSLAB at higher ripening temperatures, and in turn, an accelerated maturation has been noticed (De Filippis et al., 2016).
Besides meta-transcriptomics, meta-proteomics and metabolomics are also important tools for studying LAB. Currently only very few works have used these two technologies to study LAB communities. Reasons could be due to the challenges in sample preparation, proteins and metabolites identification compared to sequence-based metagenomics and meta-transcriptomics data in the community study (Blackburn and Martens, 2016; Smirnov et al., 2016).
The aforementioned ALE and metabolomics tools have been harnessed to obtain LAB that exhibit improved properties. We have also discussed the application of these systems biology tools to elucidate interactions taking place in LAB communities and to determine metabolic capacities. In the following part, we will focus on the model strain of LAB – L. lactis to elaborate its metabolic flux regulation in more detail and illustrate its metabolic engineering potential for valuable biochemicals production as a novel microbial cell factory.
The glycolysis of L. lactis comprises the typical EMP pathway with different carbohydrates entering at different points. Sugars enter cells of L. lactis by either the PEP-dependent PTSs or sugar-specific permeases. For glucose uptake, there are two distinct PTSs, PTSMan and PTSCel, and a proton-motive force dependent permease and the kinetic properties of these transport systems in MG1363 have been characterized (Castro et al., 2009). Glycolytic enzymes are in general highly expressed to sustain the large glycolytic flux in L. lactis, accounting for about 20% of the total soluble protein (Puri, 2014). Their genes are in general located close to the origin of replication for higher expression (Cocaign-Bousquet et al., 2002). Transcriptional regulation of the glycolytic genes in L. lactis primarily is through the carbon catabolite repression (CCR), which has been verified experimentally in L. lactis (Deutscher et al., 2006). The HPr protein, at high levels of fructose 1,6-bisphosphate (FBP) and ATP, is phosphorylated and then forms a complex with CcpA acting as a global regulator binding to the cre sites on chromosome. The well-conserved consensus in Gram-positive bacteria for the cre site is TGNNANCGNTNNCA, which is roughly palindromic with the central CG base always present. A study on the CcpA regulon in MG1363 proposed the consensus WGWAARCGYTWWMA, which is specific for L. lactis (Zomer et al., 2007). The binding of CcpA to cre sites can lead to either activation or repression, depending on the position relative to promoters also seen for Bacillus (Henkin, 1996). When a cre site is upstream of, inside, or downstream of the promoter of a gene, transcription is activated, repressed, or aborted accordingly (Cocaign-Bousquet et al., 2002). Results also have indicated that the interaction between CcpA and the transcription machinery may be dependent on the helix side of CcpA binding, because the strongest repression was observed for cre sites that were consecutively separated by around 10.5 bp, equal to a full helical turn of double-stranded DNA (Zomer et al., 2007). The expression of a number of glycolytic enzymes was found to be up-regulated by CcpA, including phosphofructokinase (PFK), pyruvate kinase (PYK) and lactate dehydrogenase (LDH) (the members of the las operon) (Luesink et al., 1998; Kok et al., 2017), phosphoglucose isomerase (PGI), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), enolase (Guédon et al., 2002).
In the intracellular environment, assuming constant environmental factors, such as pH, temperature, viscosity etc., the reaction rate of an enzyme depends on concentrations of the enzyme, substrates, products and other effectors, such as allosteric regulators, that can modulate the enzymatic activity. Reaction rates would in turn change metabolite concentrations, forming a dynamical system. The metabolic flux of a pathway is the overall conversion rate of metabolites by the pathway resulting from the dynamical interactions between involved enzymes and metabolites. The factors determining the flux can be divided into two levels. The first is the enzyme level, which accounts for changes in fluxes caused by changes in gene expression level. The second is the metabolic level referring to changes in fluxes which are not caused by altered gene expression but by changes in metabolite concentrations and the inherent kinetic properties of enzymes such as maximum velocity and substrate affinity. The statement that an enzyme has “control” on a flux should refer to the phenomenon that a change in the enzyme level leads to change in the flux but not the direct regulatory mechanism. “Regulation” should refer to the exact mechanism causing the change (Chan, 2014).
Control of fluxes by individual enzymes can be quantified by Flux Control Coefficients (FCCs) in the theory of Metabolic Control Analysis (MCA), which is defined by the rate of fractional change of the steady-state flux with respect to the fractional change of the enzyme activity. Finding the “rate-limiting” step or an enzyme with high FCC of the glycolytic flux in L. lactis can have direct industrial relevance, e.g., speeding up the production properly and increasing the productivity. An earlier study trying to inhibit the activity of GAPDH by the specific inhibitor iodoacetate indicated that GAPDH had a high FCC of about 0.9 on glycolytic flux in non-growing cells of L. lactis subsp. cremoris Wg2 (Poolman et al., 1987). Similar results were obtained in another strain NCDO2118 with GAPDH having a FCC equal to 0.7 (Even et al., 1999). Later, the control of glycolytic flux by glycolytic enzymes has been extensively studied by the Jensen group by experimental estimation of FCCs in the laboratory strains L. lactis IL1403 and MG1363. The results are summarized in Table 1.
Interestingly, usually each individual enzyme appears to have no control on growth rate and glycolytic flux at the wild-type enzyme level, including LDH (Andersen et al., 2001a), GAPDH (Solem et al., 2003), PFK, PYK (Koebmann et al., 2005) and triosephosphate isomerase (TPI) (Solem et al., 2008) for MG1363; TPI, enolase (Koebmann et al., 2006) and phosphoglycerate mutase (PGM) (Solem et al., 2010) for IL1403. Among these enzymes, some are present in the wild type in significant excess for attaining maximum glycolytic flux, such as LDH, TPI and GAPDH whereas some enzymes appear to be optimally expressed in the wild type for maximum glycolytic flux, such as PFK, PYK, PGM and enolase (ENO). For the latter set of enzymes, when the expression level is increased or decreased slightly, the growth rate and glycolytic flux decrease. This property of maximum growth rate and glycolytic flux in the wild type also leads to a zero FCC at the wild-type level. Figure 2 illustrates the two different scenarios leading to zero FCCs in the wild type encountered in the experimental studies of glycolytic flux control by glycolytic enzymes. In the literature, nonetheless, the possible consequences and interpretation of these observations have not been discussed thoroughly.
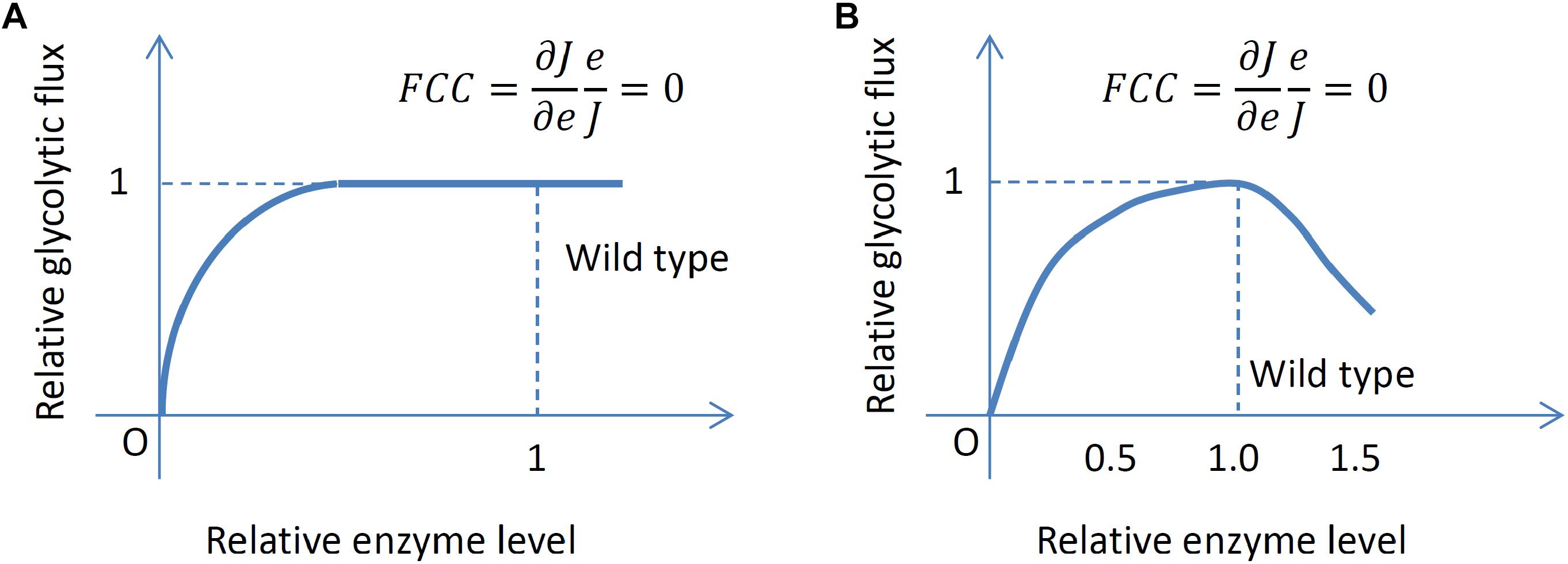
Figure 2. Two different scenarios leading to a zero FCC. (A) An enzyme is present in excess and perturbing its level slightly does not change the flux. (B) The level of an enzyme allows maximum flux and perturbing the level slightly always decreases the flux.
The reason for the zero flux control for these important enzymes also remains elusive. One possible explanation is that glycolysis is already running at its maximum possible rate or the control is distributed over many enzymes (Koebmann et al., 2002a). Another conjecture is that glycolysis is so optimized throughout evolution that the true FCCs cannot be measured due to optimal regulation of protein expressions which somehow counteracts the effect of modulating an enzyme by reallocating the protein expression profile (Teusink et al., 2011). If this is true, the calculated rate is not the defined partial derivative because the concentrations of other enzymes are also functions of the concentration of the perturbed enzyme. The conflicting results on the role of GAPDH from different studies also highlight the difficulty of studying flux control. In the earlier study, nearly full control of glycolytic flux by GAPDH in L. lactis Wg2 and NCDO2118 was found (Poolman et al., 1987; Even et al., 1999) but zero control was found in L. lactis MG1363 (Solem et al., 2003). One possible explanation is the intrinsic difference between the two strains as the GAPDH level was found to be two-fold higher in MG1363 compared to Wg2. Another possibility is the difference in experimental methods. GAPDH’s activity in Wg2 was only inhibited but not increased whereas both under- and over-expression of GAPDH were included in the study of MG1363. This can lead to contradictory estimation of FCCs.
Another approach used to distinguish between hierarchical regulation and metabolic regulation was proposed by Kuile and Westerhoff (2001) by estimating coefficients for the two types of regulation. This approach is to a certain extent working in a reverse sense to the aforementioned approach which estimates the FCC of an enzyme by growing strains with different activities of the enzyme under the same condition and then measuring the changes in activities and fluxes. In contrast, the same strain is cultured under different growth conditions, for example, in chemostat at different dilution rates, or different starvation conditions (Rossell et al., 2006). Then the relative change in enzyme level, measured by enzyme assay, is divided by the relative change in fluxes to define the “hierarchical regulation coefficient” (HRC). It is equal to one in the ideal case of pure hierarchical regulation. The “metabolic regulation coefficient” (MRC) can then be computed by 1 – HRC to account for the change in fluxes not accountable by hierarchical regulation.
In L. lactis, several studies have been conducted using this approach. For instance, MG1363 has been grown in chemostat at a dilution rate of 0.1 h-1 at different pH, from 4.7 to 6.6 (Even et al., 2003). It was found that when taking the inhibitory effect of pH on enzyme activities into account, metabolic regulation was the dominant force controlling glycolytic flux. Also among the hierarchical regulation, post-transcriptional regulation of gene expression was found to be more prominent than transcriptional regulation by comparing change in mRNA transcript level with change in enzyme activity. MG1363 was grown in chemostat at different dilution rates from 0.15 to 0.6 h-1 and transcriptomes, proteomes, enzyme activities were quantified simultaneously (Puri, 2014). Similar conclusions were reached. For dilution rates between 0.15 and 0.5 h-1, the changes in flux through most enzymes were predominantly caused by metabolic regulation instead of hierarchical regulation except for alcohol dehydrogenase (ADHE) and possibly pyruvate formate lyase (PFL) whose concentrations decreased as the dilution rate increased and the flux through mixed-acid fermentation pathway decreased. So these two enzymes probably controlled the switch between fermentation modes but not the glycolytic flux. Significant hierarchical regulation only occurred during transition from 0.5 to 0.6 h-1 in which the expression of several enzymes were found to have changed, probably due to the effect of CCR by the regulatory protein CcpA. Indeed, similar results of the lack of significant change in expression of glycolytic enzymes have also been observed in an accelerostat study on IL1403 in which the dilution rate increased very slowly from 0.1 to 0.6 h-1 to obtain different steady states (Lahtvee et al., 2011). Only PGM was found to show changes in expression.
Metabolic regulation is not easy to discover because it usually involves interactions between an enzyme and metabolites other than substrates and products of that enzyme. Extensive in vitro enzyme characterization is required to identify possible effector metabolites and experiments for confirmation of in vivo regulatory roles can even be more difficult to design. As mentioned above some studies indicated that metabolic regulation was the main driving force for flux regulation and meanwhile many pieces of knowledge on particular regulatory relationships are available, nonetheless, a clear and integrative picture of how different types of metabolic regulation work together to explain most of the known experimental results still remains elusive.
One example of the metabolic regulation of glycolysis is the regulation of the phosphorylation of HPr protein (HPr/HPr-Ser-P) by FBP, ATP and inorganic phosphate (Pi) mentioned previously. Since HPr helps sugar uptake through PTS but HPr-Ser-P does not, a high level of FBP due to a high rate of sugar uptake causes more HPr to be phosphorylated into HPr-Ser-P, which eventually slows down the sugar uptake thus forming a negative feedback loop. This loop may help to stabilize the glycolytic flux, especially against sudden changes in sugar availability (Teusink et al., 2011). The question of whether this negative feedback loop poses a bottleneck on maximum glycolytic flux, nevertheless, remains unanswered.
Besides the role in PTS, FBP has also been known to be an activator for PYK (Thompson, 1987). A kinetic study of glycolytic intermediates in glucose-pulse experiments using NMR found that the FBP level rose to a peak during glucose uptake and started to drop after glucose was exhausted and until a certain low FBP level, PEP started to accumulate and remained at a high level during glucose starvation (Voit et al., 2006). The authors proposed that the low FBP level reflecting low supply of glucose could serve as a way to preserve high PEP pool by inhibiting PYK which consumes PEP during sugar starvation for future rapid sugar uptake through PTS. Others, nonetheless, observed that such an activation relationship was also preserved in other organisms including those without PTS and remained conservative about the role of this FBP-PYK relation in glycolysis (Teusink et al., 2011). They suggested another possible role in which a high FBP level could serve as a signal for PYK to remove the phosphoglycerate compounds in favor of a high flux through GAPDH which operated close to thermodynamic equilibrium and was thus sensitive to mass action.
Another interesting example is how the glycolytic flux responds to cofactor levels, e.g., NADH/NAD+ and ATP/ADP. NADH/NAD+ ratio was first proposed by Garrigues et al. (2001) to be an important factor for regulating the glycolytic flux in L. lactis NCDO2118 based on findings in a study where the strain was exponentially growing on three sugars, glucose, galactose and lactose, with decreasing glycolytic fluxes. The following observations were made: (i) the in vitro activity of GAPDH was almost completely inhibited by a NADH/NAD+ ratio higher than 0.05; (ii) the NADH/NAD+ ratio positively correlated with the glycolytic flux and was as high as 0.08 on glucose (severe inhibition of GAPDH expected); (iii) high pools of metabolites upstream of GAPDH were found including FBP, GAP and dihydroxyacetone phosphate (DHAP) (suggesting insufficient GAPDH activity to metabolize GAP). A later study by the same group on MG1363 found the same correlation between NADH/NAD+ ratio and glycolytic flux, but the factors determining the ratio remained unknown (Garrigues et al., 2001).
Glycolytic kinetics in non-growing cells of L. lactis has been studied using in vivo NMR by Neves et al. (1999). The kinetic model built in the study fitted with experimental data predicted that conversion of PEP into pyruvate by PYK is inhibited by a high ATP surplus, i.e., high ATP/ADP ratio, which in turn inhibits NAD+ regeneration by LDH and in this way, restricts the glycolytic flux. This somehow provided an explanation for the positive correlation between NADH/NAD+ ratio and glycolytic flux observed in other studies (Garrigues et al., 1997). Later NMR study by the same group focusing on the role of NADH and NAD+ found that GAPDH was able to sustain a flux as high as in the wild-type MG1363 in a LDH-knockout strain in which the NADH concentration was 1.5 mM while the inhibitory constant of NADH for GAPDH was found to be 0.4 mM (Neves et al., 2002). The authors, to a certain extent, dismissed the control by GAPDH and NADH, and proposed ATP, ADP and Pi as important regulatory metabolites in glycolysis. When interpreting these results, however, one should bear in mind that that in vivo NMR studies often deal with non-growing L. lactis, so the results obtained can possibly be different from ones derived using exponentially growing cells.
To test the in vivo role of ATP/ADP ratio in growing L. lactis, Koebmann et al. (2002b) decreased the intracellular ATP/ADP ratios in MG1363 by expressing ATPase using a synthetic promoter library. Surprisingly, for these strains growing exponentially on glucose, the glycolytic flux showed no significant change over a large range of ATP/ADP ratio from around 9 in the wild type to around 5 in the strain with the highest expression of ATPase. When these strains were in a non-growing state (achieved by resuspending cells in media without amino acids and vitamins), however, the glycolytic flux increased with ATPase activities until a level close to that observed for growing wild type cells. These observations suggest the possibility that glycolysis already operates at its maximal rate in the wild type. Another possible situation suggested by the authors is that although lowering the ATP/ADP ratio might stimulate glycolysis (e.g., by increasing the activities of kinases in the pay-off phase), it might eventually reduce the activity of PFK, which becomes a bottleneck countering the effect, known as the risk of a “turbo design” in which part of the desirable products are invested in the first place as input (Teusink et al., 1998).
Other sources of metabolic regulation considered to be important for regulating glycolytic flux include the inhibition of PYK by Pi (Ramos et al., 2004), inhibition of PFK by PEP (Papagianni and Avramidis, 2012) and inhibition of GAPDH by NADH (Garrigues et al., 1997, 2001). Hoefnagel et al. (2002) integrated these three inhibitive relations and the activation of PYK by FBP in a single kinetic model with all rate equations and parameters adopted from literature without fitting. The model succeeded in simulating the observed kinetic behavior during glucose run-out experiments including the rapid increase in PEP and Pi, decrease in ATP and slow depletion of FBP. The authors reasoned the following sequence of kinetic responses upon glucose depletion: (1) Less Pi is retained in G6P, F6P and FBP; (2) Pi increases and inhibits PK; (3) PEP increases due to inhibited PK and no glucose for PTS and thus less pyruvate available; (4) Less substrate for LDH and thus NADH accumulates; (5) GAPDH is inhibited by NADH.
Due to its high glycolytic flux and its safe background, L. lactis has been engineered into a cell factory for production of a broad range of interesting biochemicals, from biofuels and food ingredients to vitamins and pharmaceutical precursors (Liu, 2017). Most of these biochemicals are derived from glycolytic precursors. Early efforts on metabolic engineering of L. lactis were primarily directed at the manipulation of the pyruvate node. The native metabolic pathways can drive the pyruvate flux to lactate via LDH, or to formate and acetyl-CoA by PFL. Then acetyl-CoA can either be converted into ethanol via the bi-functional ADHE or to acetate via phosphotransacetylase (PTA) and acetate kinase (ACK), subsequently (Figure 3).
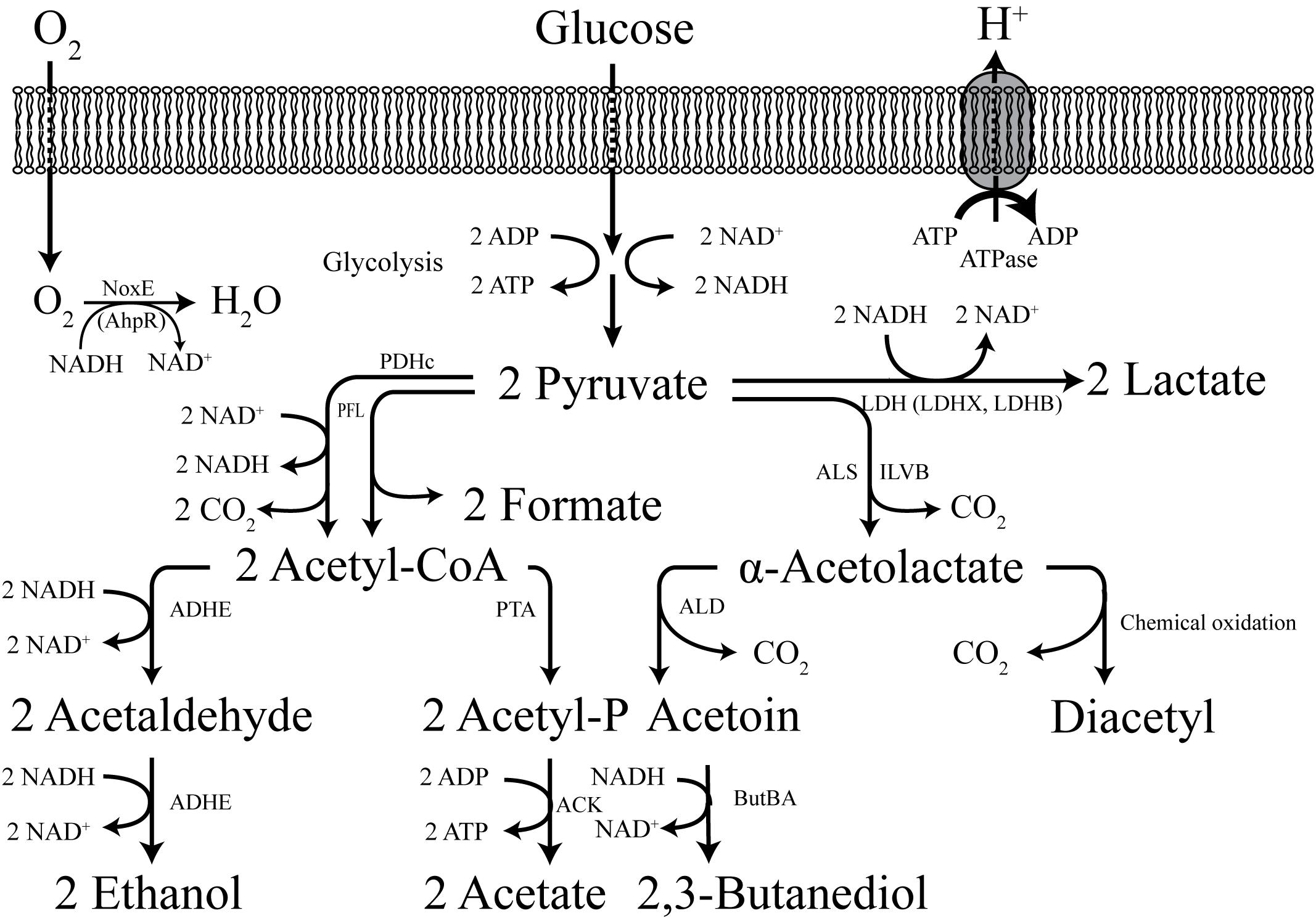
Figure 3. The central metabolism of L. lactis. LDH, lactate dehydrogenase; LDHX and LDHB, LDH homologs; ALS (ILVB), α-acetolactate synthase; ALD, a -acetolactate decarboxylase; ButBA, diacetyl reductase and butanediol dehydrogenase; PDHc, pyruvate dehydrogenase complex; PFL, pyruvate formate lyase; PTA, phosphotransacetylase; ADHE, alcohol dehydrogenase; ACK, acetate kinase; Nox, NADH oxidase; ATPase, (F1F0)-ATPase.
The natural main product of L. lactis is L-lactate. L. lactis ATCC 19435 has been reported to be able to produce around 100 g/L lactate when using a medium containing whole wheat flour hydrolysate, with a high productivity of 3.0 g/L/h (Hofvendahl and Hahn-Hägerdal, 1997). One of the limiting factors that have prevented the widespread use of L. lactis as a cell factory for lactate has been its fastidious nature and a relatively low pH tolerance when compared with various Lactobacillus species or yeast (van Hylckama Vlieg et al., 2006; Mazzoli et al., 2014).
Cheaper substrates, e.g., lignocellulose hydrolysates, could serve as a potential feedstock for some L. lactis strains that are able to metabolize both the pentoses and hexoses contained, and one research group has investigated the non-dairy strain L. lactis IO-1, which is capable of metabolizing xylose (Tanaka et al., 2002). It was demonstrated that xylose could be metabolized via both the phophoketolase pathway and the pentose phosphate pathway into lactate. The flux distribution between the two different pathways was affected by xylose concentration, and a more homolactic profile was attained at high xylose concentrations. Sirisansaneeyakul et al. (2007) used immobilized L. lactis IO-1 for producing lactate from glucose and achieved an extremely high volumetric productivity of 4.5 g/L/h using a packed bed of encapsulated cells where the broth was recycled. Another interesting work demonstrated that by expressing the Escherichia coli chaperone DnaK in L. lactis NZ9000, it was possible to improve multiple-stress tolerance (high temperature, salts, lactate and alcohol) and to increase lactate production (Abdullah-Al-Mahin et al., 2010).
The demand for liquid fuels is growing and microbial-based production of biofuels could be an attractive way to increase the supply and decrease the cost. There have been attempts to use L. lactis for producing ethanol, but most of them only had limited success. Possible reasons for this could be a too low expression level of heterologous pyruvate decarboxylase (PDC) and alcohol dehydrogenase (AdhB) from Zymomonas mobilis. Liu et al. (2005) introduced PDC from Zymobacter palmae in L. lactis and only observed the accumulation of acetaldehyde. Similarly, Nichols et al. (2003) expressed PDC and AdhB from Z. mobilis in L. lactis and still failed to drive the flux from lactate fermentation. However, our group several years ago successfully constructed a recombinant L. lactis, which was able to produce ethanol as the sole fermentation product (87% of the carbon flux was directed to ethanol production) (Solem et al., 2013). The homo-ethanol producer incorporated the knock-out of several genes encoding the three LDH homologs (ldh, ldhB, and ldhX), PTA and native ADHE with the introduction of codon-optimized PDC/AdhB from Z. mobilis under the synthetic promoter library. Further, this strain was modified by introducing a functional lactose-metabolism. By developing a cheap growth medium containing whey waste and processed corn steep liquor (Liu et al., 2016b), efficient and cheap production of ethanol was accomplished with a titer of 41 g/L ethanol. On the other hand, we also explored the possibility to use the non-dairy L. lactis strain KF147 to produce ethanol as the only dominant product from xylose, although the final titer was low. This work demonstrated the great potential of using the more metabolically diverse non-dairy L. lactis strains for bio-production from xylose containing feedstocks (Petersen et al., 2017).
Butanol has a higher energy density and lower hygroscopicity than ethanol (Peralta-Yahya et al., 2012; Generoso et al., 2015). There have been attempts at engineering L. lactis into producing butanol isomers, as they are excellent fuel additives. One advantage of using L. lactis as a butanol-producing host is its high butanol tolerance (L. lactis can tolerate more than 2% while E. coli can only tolerate 1%) (Hviid et al., 2017). Liu et al. (2010) introduced the Clostridium beijerinckii P260 thiolase, a key enzyme for re-directing acetyl-CoA to butanol, into L. lactis, and enabled production of 28 mg/L butanol, which demonstrates that it is feasible to use L. lactis as a production platform. L. lactis was also reported to be able to produce isobutanol via the valine degradation pathway (Priyadharshini et al., 2015). In both of these studies the performance of producing strains was clearly insufficient for commercial production, and one reason for the low titer could be a too low availability of acetyl-CoA, the precursor for butanol/isobutanol. In L. lactis, acetyl-CoA can be either formed by PFL or by the pyruvate dehydrogenase complex (PDHc) (Figure 3), where PFL is only active in the absence of oxygen and PDHc is quite sensitive to the NADH/NAD+ ratio (Snoep et al., 1993). It thus appears that one way to ensure an adequate supply of acetyl-CoA is to introduce a robust PDHc. One of the native enzymes in the isobutanol pathway of L. lactis is KDC (α-ketoisovalerate decarboxylase), and this enzyme has been widely used in different strain platforms for efficient isobutanol production (Atsumi et al., 2008).
2,3-butanediol (2,3-BDO) is also considered to be an excellent biofuel as well as a good platform-chemical with many applications, e.g., for making plastics, perfumes and pharmaceuticals (Ji et al., 2011). Crow (1990) characterized the two native 2,3-butanediol dehydrogenases from L. lactis and identified that one generates meso-2,3-BDO from acetoin whereas the other forms an optical isomer generated from diacetyl. Gaspar et al. (2011) overexpressed the native α-acetolactate synthase (Als) and acetoin reductase (ButA) in an LDH-deficient strain, and demonstrated that 67% of the glucose flux could be redirected to 2,3-BDO concurrently with the production of formate (0.65 mol/mol glucose) and ethanol (0.59 mol/mol glucose) anaerobically. Recently we constructed several recombinant L. lactis strains for high-titer and high-yield production of 2,3-BDO isomers: meso-2,3-BDO, (R,R)-2,3-BDO and (S,S)-2,3-BDO (Kandasamy et al., 2016; Liu et al., 2016a). We introduced EcBDH (butanediol dehydrogenase from Enterobacter cloacae) into our platform strain L. lactis where deletions had been introduced into the genes encoding LDH, PTA, ADHE and ButBA, then further introduced the capacity to metabolize lactose, which enabled high-titer (51 g/L) and high-yield (0.47 g/g lactose) production of meso-2,3-BDO from whey permeate (dairy byproduct). Similarly, we demonstrated that the alcohol dehydrogenase SadB from Achromobacter xylosooxidants was able to produce (R,R)-2,3-BDO with a titer of 32 g/L and a yield of 0.40 g/g lactose. Since the production pathway of meso-2,3-BDO and (R,R)-2,3-BDO requires one NADH per mole glucose, the excess of NADH is consumed by controlling the Nox activities through limiting oxygen levels. It is not feasible or very difficult to control oxygen levels in large scales, and for this reason we developed a robust strategy to facilitate chemicals production by fine-tunning the respiration capacity (Liu et al., 2017). The efficient production of meso-2,3-BDO and (R,R)-2,3-BDO demonstrated the great potential of L. lactis to become an efficient cell factory for synthesis of biochemicals. Regarding the synthesis of (S,S)-2,3-BDO, which is another optical isomer of 2,3-BDO, we developed a special strategy, which is to combine enzymatic reactions (glycolysis and from diacetyl to (S,S)-2,3-BDO) and a non-enzymatic reaction (from α-acetolactate to diacetyl). By using a metabolically engineered L. lactis strain a titer of 6.7 g/L (S,S)-2,3-BDO was achieved from glucose (Liu et al., 2016a). To the best of our knowledge, this is the first time this chemical has been produced by direct microbial fermentation. The key to the success was to use a combination of a biocompatible catalyst, for speeding up the conversion of α-acetolactate into diacetyl, and a robust diacetyl reductase from E. cloacae.
The long record of safe use of L. lactis for food fermentations makes L. lactis an excellent choice as a host for producing food ingredients. There have been many attempts to use this organism for producing diacetyl, which is a potent flavor compound that contributes to the buttery aroma of many fermented foods, such as cheese, butter and butter milk. L. lactis subsp. lactis bv. diacetylactis is a native producer of diacetyl, and is widely used in the food industry. Normally the diacetyl-forming ability of this strain is associated with citrate metabolism, where citrate is converted into pyruvate which boosts the pyruvate pool enabling α-acetolactate formation via α-acetolactate synthase (Hugenholtz and Starrenburg, 1992). Since the availability of citrate is very low in the food raw materials, there have been efforts to change the native metabolism of L. lactis, either by random mutagenesis or by rational design. Monnet et al. (2000) selected mutants of L. lactis subsp. lactis bv. diacetylactis that were deficient in α-acetolactate decarboxylase (ALD) and had an overall low LDH activity, and demonstrated formation of 6 mM diacetyl, 30 mM acetoin and 12 mM α-acetolactate when the strains were grown in milk supplemented with catalase under aerobic conditions. Hugenholtz et al. (2000) combined NADH-oxidase (NoxE) overexpression and ALD inactivation, and achieved 1.6 mM of diacetyl using resting cells under aerobic conditions, which corresponded to a conversion efficiency of 16% (57% α-acetolactate, 21% acetate). Guo et al. (2012) constructed a promoter library for driving the expression of NoxE and could achieve a slightly higher diacetyl concentration of 4.16 mM. Recently we developed a novel strategy in L. lactis, relying on a combination of metabolic engineering, respiration technology and metal-ion catalysis, which turned out to be successful. The homolactic L. lactis was converted into a homo-diacetyl producer with a very high titer (95 mM) and a high yield (87% of the theoretical maximum) (Liu et al., 2016a).
Another important flavor compound, acetoin ((3R)-acetoin) can be produced through the native pathway in L. lactis from pyruvate by the subsequent Als and ALD. It was reported that L. lactis subsp. lactis bv. diacetylactis could produce 5.4 mM acetoin under fully aerated conditions (100% oxygen saturation) after the citrate had been completely consumed (Bassit et al., 1993). We recently achieved high-level production of acetoin (306 mM, 27 g/L) using metabolically engineered L. lactis from whey permeate (Kandasamy et al., 2016; Liu et al., 2016c). The high titer is mainly due to the completely rerouted metabolism.
We also managed to produce another isomer of acetoin, (3S)-acetoin. It can also serve as a flavor compound, but has other applications as well, e.g., for the synthesis of novel optically active α-hydroxyketone derivatives, pharmaceutical precursors and liquid crystal composites (Xiao and Lu, 2014). (3S)-acetoin can be produced from diacetyl with the aid of diacetyl reductase (DAR). By manipulating cofactor availability and using metal ion catalysis for speeding up non-enzymatic oxidative decarboxylation of α-acetolactate, (3S)-acetoin can be produced at 66 mM (71% of the theoretical maximum) (Liu et al., 2016d). To the best of our knowledge this is the first time this isomer has been made by microbial fermentation.
Hols et al. (1999) successfully rerouted the carbon flux of L. lactis toward the production of alanine, which is a natural sweetener used as a food ingredient and as a pharmaceutical precursor. The engineered strain was deficient in LDH activity and equipped with alanine dehydrogenase (AlaDH) from Bacillus sphaericus. Using resting cells as biocatalyst, alanine (around 200 mM) was the only product formed after optimizing pH and ammonium concentration. Another AlaDH from Bacillus subtilis (natto) was demonstrated to have a potential for improving alanine levels in L. lactis NZ9000 fermentation broth (Ye et al., 2010).
Acetaldehyde is considered as the most important aroma compound in yogurt. In order to achieve formation of more acetaldehyde in dairy products, Bongers et al. (2005) overexpressed PDC from Z. mobilis and NoxE in the wild type of L. lactis. They found it was possible to redirect 50% of the carbon flux in resting cells to acetaldehyde, which accumulated to 9.5 mM. From this work it was clear that PDC with its low Km for pyruvate (0.3 mM) could efficiently drain the pyruvate pool, whereas this is more difficult to achieve using Als that has a very high Km for pyruvate (50 mM) (Zullian et al., 2014).
Vitamins are vital nutrients needed for the normal functioning of living organisms. Vitamin deficiencies occur commonly, and are often associated with certain health problems, however, deficiencies can be overcome by supplementation, e.g., by fortifying foods with vitamins. Several studies have focused on engineering L. lactis into synthesizing B vitamins, such as riboflavin (vitamin B2) and folate (vitamin B11) (Thakur et al., 2016). Burgess et al. (2004) overexpressed the rib operon (ribGBAH) (Figure 4) and achieved high level production of riboflavin (24 mg/L). Furthermore, they developed a strategy to select and isolate spontaneous riboflavin-overproducing L. lactis, which was to use the toxic riboflavin analog roseoflavin. Several mutants exhibited a significant higher-level of riboflavin (around 900 mg/L) (Burgess et al., 2004). Recently our group selected a riboflavin overproducer through the combination of roseoflavin resistance, random mutagenesis and microfluidic screening. The mutant could increase the riboflavin content in milk to 2.81 mg/L whereas the wild-type reduced the riboflavin content of milk to 0.66 mg/L (Chen J. et al., 2017). These results indicate a great potential for L. lactis to become an efficient producer of riboflavin, either by using GMO or non-GMO approaches. LeBlanc et al. (2005) carried out an animal test and found that feeding rats with live L. lactis cells overproducing riboflavin could stimulate their growth.
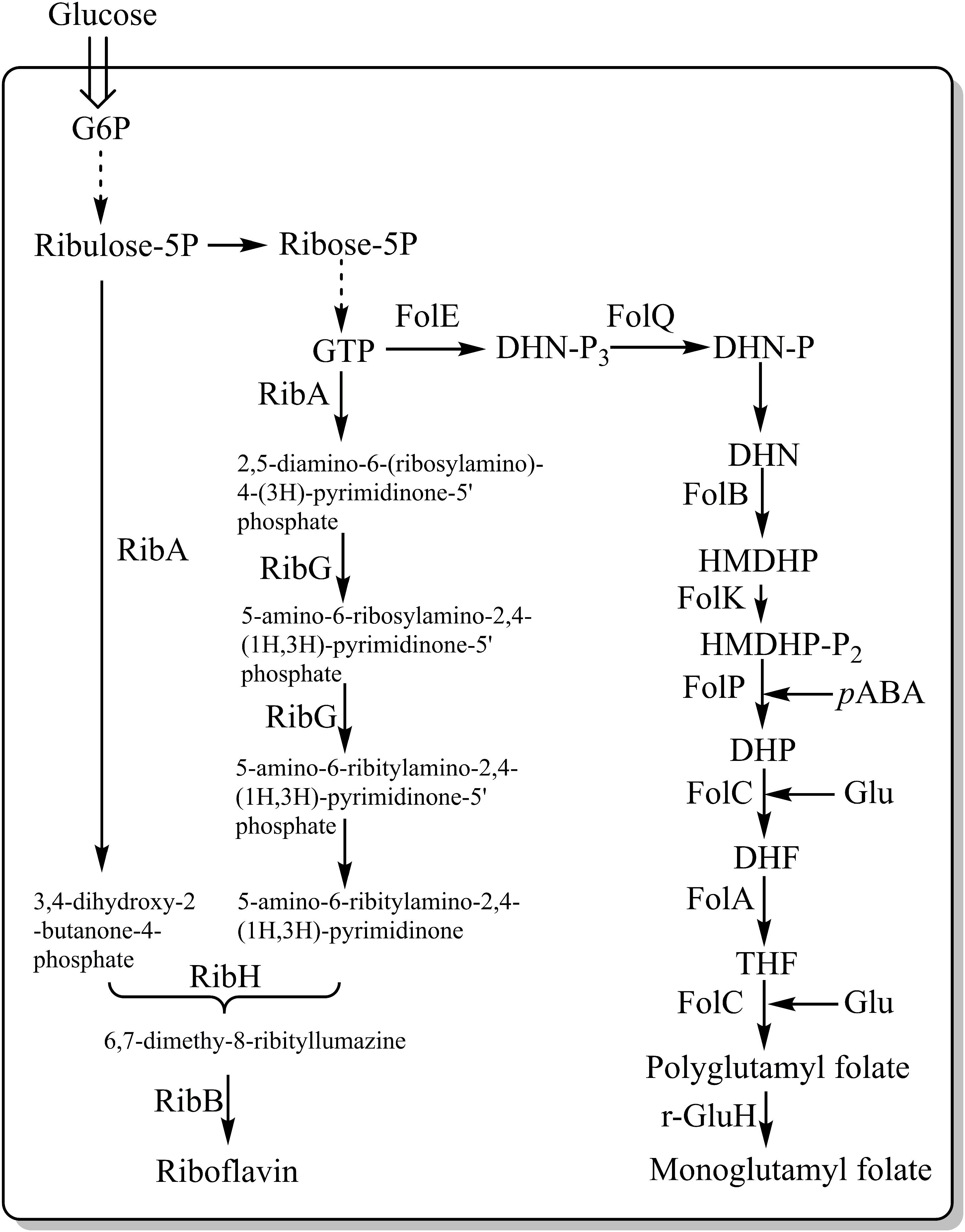
Figure 4. Metabolic pathways involved in the synthesis of riboflavin and folate in LAB. FolE, GTP cyclohydrolase I; FolQ, dihydroneopterin triphosphate pyrophosphohydrolase; FolB, dihydroneopterin aldolase; FolK, hydroxymethyldihydropterin pyrophosphokinase; FolP, dihydropteroate synthase; FolC, dihydrofolate synthase; FolA, dihydrofolate reductase; PabAB, chorismate synthetase component I and II; PabC, 4-amino-4-deoxychorismate lyase; Glu, glutamate; pABA, para-aminobenzoic acid; γ-GluH, gamma-glutamyl hydrolase; RibA, GTP cyclohydrolase II/3,4-dihydroxy-2-butanone-4-phosphate synthase; RibG, riboflavin-specific deaminase/reductase; RibH, riboflavin synthase (beta subunit); and RibB, riboflavin synthase (alpha subunit). Dotted arrows represent multiple consecutive steps in the pathways.
Lactococcus lactis is a good producer of folate, predominantly in the form of polyglutamyl. The Fol operon, which includes folA, folB, folKE, folP and folC, was found to be involved in folate biosynthesis (Figure 4). The overexpression of folKE in L. lactis resulted in an increase in extracellular folate production by 8-fold (from 10 to 80 ng/ml), while the total folate production increased by 2-fold (from 100 to 180 ng/ml) (Sybesma et al., 2003). Wegkamp et al. (2007) further documented that the overexpression of the folate operon and para-aminobenzoic acid (pABA) gene clusters, where pABA is the important precursor for folate synthesis, resulted in production of 2.7 mg/L folate per optical density unit at 600 nm, which is 80 times higher than what the wild type is capable of.
Exopolysaccharides (EPSs) are long-chain polysaccharides that are secreted into their surroundings during bacterial growth. EPSs contribute to the texture of many dairy products (Laws et al., 2001). It has also been reported that they can function as prebiotics, cholesterol lowering nutraceuticals or immunomodulants (Nwodo et al., 2012). The biosynthesis of EPSs is complex and normally involves a large number of gene products. In L. lactis, the genes responsible for EPSs synthesis are located in a large operon containing 14 genes, epsRXABCDEFGHIJKL, which is found on a plasmid (van Kranenburg et al., 1997). van Kranenburg et al. (1999) demonstrated that by over-expressing edsD, encoding the priming glycosyltransferase, could increase the EPSs production from 113 to 133 mg/L. Further through increasing the expression level of the entire eps gene cluster, EPSs production was increased further to 343 mg/L (Boels et al., 2003a,b). However, an increase in the concentration of sugar-phosphates (Glu-6P, Glu-1P) or sugar nucleotides (UDP-glucose, UDP-galactose) did not affect production of EPSs significantly. Recently the EPSs biosynthetic pathways were re-engineered to enable production of hyaluronic acid (HA), which is another kind of polysaccharide with various applications in pharmaceuticals and foods (Shah et al., 2013). HA is synthesized from the polymerization of UDP-glucuronic acid and UDP-N-acetylglucosamine, two precursors of cell-wall components, through HA synthase. The recombinant expression of HA synthase from Streptococcus equi subsp. zooepidemicus in L. lactis resulted in 0.08 g/L HA, and the coexpression of HA synthase and uridine diphosphate-glucose dehydrogenase (UDP-GlcDH) significantly enhanced the HA production to 0.65 g/L (Chien and Lee, 2007). Prasad et al. (2010) coexpressed UDP-glucose pyrophosphorylase as well as HA synthase and UDP-GluDH and achieved 1.8 g/L HA in bioreactor experiment with controlled pH and aeration. These promising results demonstrate the great potential of L. lactis as a good platform for the production of functional polysaccharides.
In the last 10 years, L. lactis has been studied as a host for the expression of plant derived genes to produce plant metabolites. Song et al. (2012) expressed b-sesquiphellandrene synthase from Persicaria minor in L. lactis successfully and confirmed the production of b-sesquiphellandrene, which is a valuable sesquiterpene and has excellent antimicrobial and antioxidative properties. The co-expression of 3-hydroxy-3-methyglutaryl coenzyme A reductase, which is the limiting enzyme in the mevalonate pathway, increased the b-sesquiphellandrene level to 109 nM (Song et al., 2012). Hernández et al. (2007) cloned two genes from strawberry, encoding an alcohol acyltransferase and a linalool/nerolidol synthase and expressed them in L. lactis, which resulted in the production of octyl acetate (1.9 mM) and linalool (85 nM). The production of plant metabolites in L. lactis is just in the beginning phase and these previous results demonstrate a great potential.
In many single strain omics and systems biology studies, cells are scrutinized under steady-state conditions, which is indeed useful for the purpose of exploring and understanding the regulation of cellular metabolism of LAB, e.g., glycolysis. A large number of studies have also demonstrated how helpful this knowledge and understanding is for the design and engineering of robust LAB cell factories for producing different high-value chemicals. Systems biology methods including omics analysis also have allowed us to understand how LAB respond to environmental changes. In the future, other variables should be considered for the study of LAB communities e.g., temporal and spatial change of community composition due to biotic/abiotic factors in the production process, which significantly increase complexity of the research. These changes have showed great influences on the cellular states of the microbial community. So, in the future, the improved meta-omics analysis resolution, and use of properly simplified models are some of the keys for the systems biology study of the LAB community, which will benefit the understanding of these organisms in the real-life applications.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by Innovationsfonden (Grants 6150-00036B and 4150-00037B), Novo Nordisk Fonden (Grant NNF12OC0000818), and Statens Naturvidenskabelige Forskningsfond (FNU10-085115).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This review article includes some contents which first appeared in SC’s thesis (Chan, 2014) and JL’s thesis (Liu, 2017) for their doctoral degree (the only medium), individually, from Technical University of Denmark.
γ-GluH, gamma-glutamyl hydrolase; ACK, acetate kinase; ADHE, alcohol dehydrogenase; ALD, α-acetolactate decarboxylase; ALE, adaptive laboratory evolution; ALS (ILVB), α-acetolactate synthase; ATPase, (F1F0)-ATPase; ButBA, diacetyl reductase and butanediol dehydrogenase; FBA, flux balance analysis; FolA, dihydrofolate reductase; FolB, dihydroneopterin aldolase; FolC, dihydrofolate synthase; FolE, GTP cyclohydrolase I; FolK, hydroxymethyldihydropterin pyrophosphokinase; FolP, dihydropteroate synthase; FolQ, dihydroneopterin triphosphate pyrophosphohydrolase; Glu, glutamate; GMO, genetically modified organism; LAB, lactic acid bacteria; LDH, lactate dehydrogenase; Nox, NADH oxidase; pABA, para-aminobenzoic acid; PabAB, chorismate synthetase component I and II; PabC, 4-amino-4-deoxychorismate lyase; PDHc, pyruvate dehydrogenase complex; PFL, pyruvate formate lyase; PTA, phosphotransacetylase; PTS, phosphotransferase system; RibA, GTP cyclohydrolase II/3,4-dihydroxy-2-butanone-4-phosphate synthase; RibB, riboflavin synthase (a subunit); RibG, riboflavin-specific deaminase/reductase; RibH, riboflavin synthase (b subunit); SNV, single nucleotide variation.
Abdullah-Al-Mahin, Sugimoto, S., Higashi, C., Matsumoto, S., and Sonomoto, K. (2010). Improvement of multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 under conditions of thermal stress by heterologous expression of Escherichia coli dnaK. Appl. Environ. Microbiol. 76, 4277–4285. doi: 10.1128/AEM.02878-09
Albanese, D., and Donati, C. (2017). Strain profiling and epidemiology of bacterial species from metagenomic sequencing. Nat. Commun. 8, 1–13. doi: 10.1038/s41467-017-02209-5
Albertsen, M., Hugenholtz, P., Skarshewski, A., Nielsen, K. L., Tyson, G. W., and Nielsen, P. H. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538. doi: 10.1038/nbt.2579
Andersen, H. W., Pedersen, M. B., Hammer, K., and Jensen, P. R. (2001a). Lactate dehydrogenase has no control on lactate production but has a strong negative control on formate production in Lactococcus lactis. Eur. J. Biochem. 268, 6379–6389.
Andersen, H. W., Solem, C., Hammer, K., and Jensen, P. R. (2001b). Twofold reduction of phosphofructokinase activity in Lactococcus lactis results in strong decreases in growth rate and in glycolytic flux. J. Bacteriol. 183, 3458–3467.
Ardö, Y., and Nielsen, E. W. (2007). Biochemical, Chemical and Physical Processes in Cheese During Manufacture and Ripening. Copenhagen: University of Copenhagen.
Atsumi, S., Hanai, T., and Liao, J. C. (2008). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. doi: 10.1038/nature06450
Bachmann, H., Molenaar, D., Branco Dos Santos, F., and Teusink, B. (2017). Experimental evolution and the adjustment of metabolic strategies in lactic acid bacteria. FEMS Microbiol. Rev. 41, S201–S219. doi: 10.1093/femsre/fux024
Bachmann, H., Starrenburg, M. J. C., Molenaar, D., Kleerebezem, M., and Vlieg, J. (2012). Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 22, 115–124. doi: 10.1101/gr.121285.111
Bassit, N., Boquien, C. Y., Picque, D., and Corrieu, G. (1993). Effect of initial oxygen concentration on diacetyl and acetoin production by Lactococcus lactis subsp. lactis biovar diacetylactis. Appl. Environ. Microbiol. 59, 1893–1897.
Biggs, M. B., and Papin, J. A. (2015). Metabolic network-guided binning of metagenomic sequence fragments. Bioinformatics 32, 867–874. doi: 10.1093/bioinformatics/btv671
Blackburn, J. M., and Martens, L. (2016). The challenge of metaproteomic analysis in human samples. Exp. Rev. Proteom. 13, 135–138. doi: 10.1586/14789450.2016.1135058
Boels, I. C., Kleerebezem, M., and De Vos, W. M. (2003a). Engineering of carbon distribution between glycolysis and sugar nucleotide biosynthesis in Lactococcus lactis. Appl. Environ. Microbiol. 69, 1129–1135. doi: 10.1128/AEM.69.2.1129-1135.2003
Boels, I. C., Van Kranenburg, R., Kanning, M. W., Chong, B. F., De Vos, W. M., and Kleerebezem, M. (2003b). Increased exopolysaccharide production in Lactococcus lactis due to increased levels of expression of the NIZO B40 eps gene cluster. Appl. Environ. Microbiol. 69, 5029–5031. doi: 10.1128/AEM.69.8.5029-5031.2003
Bolotin, A., Wincker, P., Mauger, S., Jaillon, O., Malarme, K., Weissenbach, J., et al. (2001). The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11, 731–753. doi: 10.1101/gr.169701.There
Bongers, R. S., Hoefnagel, M. H. N., and Kleerebezem, M. (2005). High-level acetaldehyde production in Lactococcus lactis by metabolic engineering. Appl. Environ. Microbiol. 71, 1109–1113. doi: 10.1128/AEM.71.2.1109-1113.2005
Broadbent, J. R., Steele, J. L., and Fadiman, P. (2005). cheese flavor and the genomics of lactic acid bacteria. Asm News 71, 121–128.
Burgess, C., O’Connell-Motherway, M., Sybesma, W., Hugenholtz, J., and Van Sinderen, D. (2004). Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin-enriched foods. Appl. Environ. Microbiol. 70, 5769–5777.
Castro, R., Neves, A. R., Fonseca, L. L., Pool, W. A., Kok, J., Kuipers, O. P., et al. (2009). Characterization of the individual glucose uptake systems of Lactococcus lactis: mannose-PTS, cellobiose-PTS and the novel GlcU permease. Mol. Microbiol. 71, 795–806. doi: 10.1111/j.1365-2958.2008.06564.x
Chan, S. H. J. (2014). Elucidating Flux Regulation of the Fermentation Modes of Lactococcus lactis: A Mutlilevel Study. Ph.D. thesis, Technical University of Denmark, Kongens Lyngby, 167. doi: 10.11581/DTU:00000016
Chen, C., Zhao, S., Hao, G., Yu, H., Tian, H., and Zhao, G. (2017). Role of lactic acid bacteria on the yogurt flavour: a review. Int. J. Food Prop. 20, S316–S330. doi: 10.1080/10942912.2017.1295988
Chen, J., Shen, J., Ingvar Hellgren, L., Jensen, P. R., and Solem, C. (2015a). Adaptation of Lactococcus lactis to high growth temperature leads to a dramatic increase in acidification rate. Sci. Rep. 5:14199. doi: 10.1038/srep14199
Chen, J., Shen, J., Solem, C., and Jensen, P. R. (2015b). A new type of YumC-Like ferredoxin (flavodoxin) reductase is involved in ribonucleotide reduction. MBio 6, 1–8. doi: 10.1128/mBio.01132-15
Chen, J., Shen, J., Solem, C., and Jensen, P. R. (2013). Oxidative stress at high temperatures in Lactococcus lactis due to an insufficient supply of riboflavin. Appl. Environ. Microbiol. 79, 6140–6147. doi: 10.1128/AEM.01953-13
Chen, J., Vestergaard, M., Jensen, T. G., Shen, J., Dufva, M., Solem, C., et al. (2017). Finding the needle in the haystack - the use of microfluidic droplet technology to identify vitamin-secreting lactic acid bacteria. MBio 8, e00526-17. doi: 10.1128/mBio.00526-17
Chien, L.-J., and Lee, C.-K. (2007). Hyaluronic acid production by recombinant Lactococcus lactis. Appl. Microbiol. Biotechnol. 77, 339–346.
Cocaign-Bousquet, M., Even, S., Lindley, N. D., and Loubière, P. (2002). Anaerobic sugar catabolism in Lactococcus lactis: genetic regulation and enzyme control over pathway flux. Appl. Microbiol. Biotechnol. 60, 24–32. doi: 10.1007/s00253-002-1065-x
Crow, V. L. (1990). Properties of 2,3-butanediol dehydrogenases from Lactococcus lactis subsp. lactis in relation to citrate fermentation. Appl. Environ. Microbiol. 56, 1656–1665.
De Filippis, F., Genovese, A., Ferranti, P., Gilbert, J. A., and Ercolini, D. (2016). Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci. Rep. 6, 1–11. doi: 10.1038/srep21871
De Filippis, F., Parente, E., and Ercolini, D. (2017). Metagenomics insights into food fermentations. Microb. Biotechnol. 10, 91–102. doi: 10.1111/1751-7915.12421
Deutscher, J., Francke, C., and Postma, P. (2006). How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031.
Douillard, F. P., and de Vos, W. M. (2014). Functional genomics of lactic acid bacteria: from food to health. Microb. Cell Fact. 13(Suppl. 1):S8. doi: 10.1186/1475-2859-13-S1-S8
Ercolini, D. (2017). Crystal ball exciting strain-level resolution studies of the food microbiome. Microb. Biotechnol. Banner 10, 54–56. doi: 10.1111/1751-7915.12593
Erkus, O., De Jager, V. C. L., Spus, M., Van Alen-Boerrigter, I. J., Van Rijswijck, I. M. H., Hazelwood, L., et al. (2013). Multifactorial diversity sustains microbial community stability. ISME J. 7, 2126–2136. doi: 10.1038/ismej.2013.108
Even, S., Garrigues, C., Loubiere, P., Lindley, N. D., and Cocaign-Bousquet, M. (1999). Pyruvate metabolism in Lactococcus lactis is dependent upon glyceraldehyde-3-phosphate dehydrogenase activity. Metab. Eng. 1, 198–205.
Even, S., Lindley, N. D., and Cocaign-Bousquet, M. (2003). Transcriptional, translational and metabolic regulation of glycolysis in Lactococcus lactis subsp. cremoris MG 1363 grown in continuous acidic cultures. Microbiology 149, 1935–1944. doi: 10.1099/mic.0.26146-0
Frantzen, C., Kleppen, H. P., and Holo, H. (2016). Use of M17 and a milk-based medium enables isolation of two distinct and diverse populations of Lactococcus lactis strains from undefined mesophilic starter cultures. Int. Dairy J. 53, 45–50.
Ganesan, B., Stuart, M. R., and Weimer, B. C. (2007). Carbohydrate starvation causes a metabolically active but nonculturable state in Lactococcus lactis. Appl. Environ. Microbiol. 73, 2498–2512. doi: 10.1128/AEM.01832-06
Garneau, J. E., and Moineau, S. (2011). Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Fact. 10:S20. doi: 10.1186/1475-2859-10-S1-S20
Garrigues, C., Loubiere, P., Lindley, N. D., and Cocaign-Bousquet, M. (1997). Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179, 5282–5287.
Garrigues, C., Mercade, M., Cocaign-Bousquet, M., Lindley, N. D., and Loubiere, P. (2001). Regulation of pyruvate metabolism in Lactococcus lactis depends on the imbalance between catabolism and anabolism. Biotechnol. Bioeng. 74, 108–115. doi: 10.1002/bit.1100
Gaspar, P., Neves, A. R., Gasson, M. J., Shearman, C. A., and Santos, H. (2011). High yields of 2,3-butanediol and mannitol in Lactococcus lactis through engineering of NAD+ cofactor recycling. Appl. Environ. Microbiol. 77, 6826–6835. doi: 10.1128/AEM.05544-11
Gawad, C., Koh, W., and Quake, S. R. (2016). Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188. doi: 10.1038/nrg.2015.16
Generoso, W. C., Schadeweg, V., Oreb, M., and Boles, E. (2015). Metabolic engineering of Saccharomyces cerevisiae for production of butanol isomers. Curr. Opin. Biotechnol. 33, 1–7. doi: 10.1016/j.copbio.2014.09.004
Goel, A., Eckhardt, T. H., Puri, P., de Jong, A., Branco dos Santos, F., Giera, M., et al. (2015). Protein costs do not explain evolution of metabolic strategies and regulation of ribosomal content: does protein investment explain an anaerobic bacterial Crabtree effect? Mol. Microbiol. 97, 77–92. doi: 10.1111/mmi.13012
Guchte, M., Serror, P., Chervaux, C., Smokvina, T., Ehrlich, S. D., and Maguin, E. (2002). Stress responses in lactic acid bacteria. Antonie van Leeuwenhoek 82, 187–216.
Guédon, E., Jamet, E., and Renault, P. (2002). Gene regulation in Lactococcus lactis: the gap between predicted and characterized regulators. Antonie van Leeuwenhoek 82, 93–112.
Guo, T., Kong, J., Zhang, L., Zhang, C., and Hu, S. (2012). Fine tuning of the lactate and diacetyl production through promoter engineering in Lactococcus lactis. PLoS One 7:e36296. doi: 10.1371/journal.pone.0036296
Hanemaaijer, M., Roling, W. F. M., Olivier, B. G., Khandelwal, R. A., Teusink, B., and Bruggeman, F. J. (2015). Systems modeling approaches for microbial community studies: from metagenomics to inference of the community structure. Front. Microbiol. 6:213. doi: 10.3389/fmicb.2015.00213
Henkin, T. M. (1996). The role of the CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135, 9–15.
Hernández, I., Molenaar, D., Beekwilder, J., Bouwmeester, H., and Vlieg, J. (2007). Expression of plant flavor genes in Lactococcus lactis. Appl. Environ. Microbiol. 73, 1544–1552. doi: 10.1128/AEM.01870-06
Herve-Jimenez, L., Guillouard, I., Guedon, E., Boudebbouze, S., Hols, P., Monnet, V., et al. (2009). Postgenomic analysis of Streptococcus thermophilus cocultivated in milk with Lactobacillus delbrueckii subsp. bulgaricus: involvement of nitrogen, purine, and iron metabolism. Appl. Environ. Microbiol. 75, 2062–2073. doi: 10.1128/AEM.01984-08
Higuchi, M., Yamamoto, Y., and Kamio, Y. (2000). Molecular biology of oxygen tolerance in lactic acid bacteria: functions of NADH oxidases and Dpr in oxidative stress. J. Biosci. Bioeng. 90, 484–493. doi: 10.1016/S1389-1723(01)80028-1
Hoefnagel, M. H. N., Van der Burgt, A., Martens, D. E., Hugenholtz, J., and Snoep, J. L. (2002). Time dependent responses of glycolytic intermediates in a detailed glycolytic model of Lactococcus lactis during glucose run-out experiments. Molecular Biology Reports 29, 157–161.
Hofvendahl, K., and Hahn-Hägerdal, B. (1997). L-lactic acid production from whole wheat flour hydrolysate using strains of Lactobacilli and Lactococci. Enzyme Microb. Technol. 20, 301–307. doi: 10.1016/S0141-0229(97)83489-8
Hols, P., Kleerebezem, M., Schanck, A. N., Ferain, T., Hugenholtz, J., Delcour, J., et al. (1999). Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 17, 588–592. doi: 10.1038/9902
Hugenholtz, J., Kleerebezem, M., Starrenburg, M., Delcour, J., De Vos, W., and Hols, P. (2000). Lactococcus lactis as a cell factory for high-level diacetyl production. Appl. Environ. Microbiol. 66, 4112–4114.
Hugenholtz, J., and Starrenburg, M. J. C. (1992). Diacetyl production by different strains of Lactococcus lactis subsp. lactis var. diacetylactis and Leuconostoc spp. Appl. Microbiol. Biotechnol. 38, 17–22. doi: 10.1007/BF00169412
Hviid, A. M. M., Ruhdal-Jensen, P., and Kilstrup, M. (2017). Butanol is cytotoxic to Lactococcus lactis while ethanol and hexanol are cytostatic. Microbiology 163, 453–461. doi: 10.1099/mic.0.000441
Jain, M., Koren, S., Miga, K. H., Quick, J., Rand, A. C., Sasani, T. A., et al. (2018). Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345. doi: 10.1038/nbt.4060
Ji, X.-J., Huang, H., and Ouyang, P.-K. (2011). Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol. Adv. 29, 351–364. doi: 10.1016/j.biotechadv.2011.01.007
Johansen, E. (2017). Future access and improvement of industrial lactic acid bacteria cultures. Microb. Cell Fact. 16, 230–235. doi: 10.1186/s12934-017-0851-1
Jonnala, B. R. Y., Mcsweeney, P. L. H., Sheehan, J. J., and Abram, F. (2018). Sequencing of the cheese microbiome and its relevance to industry. Front. Microbiol. 9:1020. doi: 10.3389/fmicb.2018.01020
Kandasamy, V., Liu, J., Dantoft, S. H., Solem, C., and Jensen, P. R. (2016). Synthesis of (3R)-acetoin and 2,3-butanediol isomers by metabolically engineered Lactococcus lactis. Sci. Rep. 6:36769. doi: 10.1038/srep36769
Koebmann, B., Solem, C., and Jensen, P. R. (2005). Control analysis as a tool to understand the formation of the las operon in Lactococcus lactis. FEBS J. 272, 2292–2303.
Koebmann, B., Solem, C., and Jensen, P. R. (2006). Control analysis of the importance of phosphoglycerate enolase for metabolic fluxes in Lactococcus lactis subsp. lactis IL1403. Syst. Biol. 153, 346–349. doi: 10.1049/ip-syb
Koebmann, B. J., Andersen, H. W., Solem, C., and Jensen, P. R. (2002a). Experimental determination of control of glycolysis in Lactococcus lactis. Antonie van Leeuwenhoek 82, 237–248.
Koebmann, B. J., Solem, C., Pedersen, M. B., Nilsson, D., and Jensen, P. R. (2002b). Expression of genes encoding F1-ATPase results in uncoupling of glycolysis from biomass production in Lactococcus lactis. Appl. Environ. Microbiol. 68, 4274–4282.
Kok, J., van Gijtenbeek, L. A., de Jong, A., van der Meulen, S. B., Solopova, A., and Kuipers, O. P. (2017). The evolution of gene regulation research in Lactococcus lactis. FEMS Microbiol. Rev. 41, S220–S243. doi: 10.1093/femsre/fux028
Kuile, B. H., and Westerhoff, H. V. (2001). Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett. 500, 169–171.
Lahtvee, P.-J., Adamberg, K., Arike, L., Nahku, R., Aller, K., and Vilu, R. (2011). Multi-omics approach to study the growth efficiency and amino acid metabolism in Lactococcus lactis at various specific growth rates. Microb. Cell Fact. 10:12. doi: 10.1186/1475-2859-10-12
Larsen, N., Werner, B. B., Vogensen, F. K., and Jespersen, L. (2015). Effect of dissolved oxygen on redox potential and milk acidification by lactic acid bacteria isolated from a DL-starter culture. J. Dairy Sci. 98, 1640–1651. doi: 10.3168/jds.2014-8971
Laws, A., Gu, Y., and Marshall, V. (2001). Biosynthesis, characterisation, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol. Adv. 19, 597–625.
LeBlanc, J. G., Burgess, C., Sesma, F., de Giori, G. S., and van Sinderen, D. (2005). Lactococcus lactis is capable of improving the riboflavin status in deficient rats. Br. J. Nutr. 94, 262–267. doi: 10.1079/BJN20051473
Liu, J. (2017). Harnessing the Metabolic Machinery of Lactococcus lactis for High-Yield Production of Biofuel and Biochemicals. Ph.D. thesis, Technical University of Denmark, Kongens Lyngby, 238.
Liu, J., Chan, S. H. J., Brock-Nannestad, T., Chen, J., Lee, S. Y., Solem, C., et al. (2016a). Combining metabolic engineering and biocompatible chemistry for high-yield production of homo-diacetyl and homo-(S,S)-2,3-butanediol. Metab. Eng. 36, 57–67. doi: 10.1016/j.ymben.2016.02.008
Liu, J., Dantoft, S. H., Würtz, A., Jensen, P. R., and Solem, C. (2016b). A novel cell factory for efficient production of ethanol from dairy waste. Biotechnol. Biofuels 9:33. doi: 10.1186/s13068-016-0448-7
Liu, J., Kandasamy, V., Würtz, A., Jensen, P. R., and Solem, C. (2016c). Stimulation of acetoin production in metabolically engineered Lactococcus lactis by increasing ATP demand. Appl. Microbiol. Biotechnol. 100, 9509–9517.
Liu, J., Solem, C., and Jensen, P. R. (2016d). Integrating biocompatible chemistry and manipulating cofactor partitioning in metabolically engineered Lactococcus lactis for fermentative production of (3S)-acetoin. Biotechnol. Bioeng. 113, 2744–2748. doi: 10.1002/bit.26038
Liu, J., Wang, Z., Kandasamy, V., Lee, S. Y., Solem, C., and Jensen, P. R. (2017). Harnessing the respiration machinery for high-yield production of chemicals in metabolically engineered Lactococcus lactis. Metab. Eng. 44, 22–29. doi: 10.1016/j.ymben.2017.09.001
Liu, S., Bischoff, K. M., Qureshi, N., Hughes, S. R., and Rich, J. O. (2010). Functional expression of the thiolase gene thl from Clostridium beijerinckii P260 in Lactococcus lactis and Lactobacillus buchneri. New Biotechnol. 27, 283–288. doi: 10.1016/j.nbt.2010.03.007
Liu, S., Dien, B. S., and Cotta, M. A. (2005). Functional expression of bacterial Zymobacter palmae pyruvate decarboxylase gene in Lactococcus lactis. Curr. Microbiol. 50, 324–328. doi: 10.1007/s00284-005-4485-x
Ljungh, A., and Wadström, T. (2006). Lactic acid bacteria as probiotics. Curr. Issues in Intest. Microbiol. 7, 73–89. doi: 10.1111/j.1574-6968.2010.02185.x
Los, D. A., and Murata, N. (2000). Regulation of enzymatic activity and gene expression by membrane fluidity. Sci. Signal. 62:e1. doi: 10.1126/stke.2000.62.pe1
Luesink, E. J., Van Herpen, R. E. M. A., Grossiord, B. P., Kuipers, O. P., and De Vos, W. M. (1998). Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30, 789–798. doi: 10.1046/j.1365-2958.1998.01111.x
Makarova, K. S., and Koonin, E. V. (2007). Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 189, 1199–1208. doi: 10.1128/JB.01351-06
Martinussen, J., Solem, C., Holm, A. K., and Jensen, P. R. (2013). Engineering strategies aimed at control of acidification rate of lactic acid bacteria. Curr. Opin. Biotechnol. 24, 124–129. doi: 10.1016/j.copbio.2012.11.009
Mazzoli, R., Bosco, F., Mizrahi, I., Bayer, E. A., and Pessione, E. (2014). Towards lactic acid bacteria-based biorefineries. Biotechnol. Adv. 32, 1216–1236. doi: 10.1016/j.biotechadv.2014.07.005
Monnet, C., Aymes, F., and Corrieu, G. (2000). Diacetyl and α-acetolactate overproduction by Lactococcus lactis subsp. lactis biovar diacetylactis mutants that are deficient in α-acetolactate decarboxylase and have a low lactate dehydrogenase activity. Appl. Environ. Microbiol. 66, 5518–5520. doi: 10.1128/AEM.66.12.5518-5520.2000
Neves, A. R., Ramos, A., Nunes, M. C., Kleerebezem, M., Hugenholtz, J., De Vos, W. M., et al. (1999). In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 64, 200–212.
Neves, A. R., Ventura, R., Mansour, N., Shearman, C., Gasson, M. J., Maycock, C., et al. (2002). Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? Kinetics of NAD+ and NADH pools determined in vivo by 13C NMR. J. Biol. Chem. 277, 28088–28098. doi: 10.1074/jbc.M202573200
Nichols, N. N., Dien, B. S., and Bothast, R. J. (2003). Engineering lactic acid bacteria with pyruvate decarboxylase and alcohol dehydrogenase genes for ethanol production from Zymomonas mobilis. J. Ind. Microbiol. Biotechnol. 30, 315–321.
Nwodo, U. U., Green, E., and Okoh, A. I. (2012). Bacterial exopolysaccharides: functionality and prospects. Int. J. Mol. Sci. 13, 14002–14015. doi: 10.3390/ijms131114002
Oliveira, A. P., Nielsen, J., and Förster, J. (2005). Modeling Lactococcus lactis using a genome-scale flux model. BMC Microbiol. 5:39. doi: 10.1186/1471-2180-5-39
Ondov, B. D., Treangen, T. J., Melsted, P., Mallonee, A. B., Bergman, N. H., Koren, S., et al. (2016). Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 1–14. doi: 10.1186/s13059-016-0997-x
Papadimitriou, K., Alegría,Á, Bron, P. A., De Angelis, M., Gobbetti, M., Kleerebezem, M., et al. (2016). Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 80, 837–890. doi: 10.1128/MMBR.00076-15.Address
Papagianni, M., and Avramidis, N. (2012). Engineering the central pathways in Lactococcus lactis: functional expression of the phosphofructokinase (pfk) and alternative oxidase (aox1) genes from Aspergillus niger in Lactococcus lactis facilitates improved carbon conversion rates under oxidizing conditons. Enzyme Microb. Technol. 51, 125–130.
Peralta-Yahya, P. P., Zhang, F., Del Cardayre, S. B., and Keasling, J. D. (2012). Microbial engineering for the production of advanced biofuels. Nature 488, 320–328. doi: 10.1038/nature11478
Petersen, K. V., Liu, J., Chen, J., Martinussen, J., Jensen, P. R., and Solem, C. (2017). Metabolic characterization and transformation of the non-dairy Lactococcus lactis strain KF147, for production of ethanol from xylose. Biotechnol. J. 12:1700171. doi: 10.1002/biot.201700171
Poolman, B., Bosman, B., Kiers, J., and Konings, W. N. (1987). Control of glycolysis by glyceraldehyde-3-phosphate dehydrogenase in Streptococcus cremoris and Streptococcus lactis. J. Bacteriol. 169, 5887–5890. doi: 10.1128/jb.169.12.5887-5890.1987
Porcellato, D., and Skeie, S. B. (2016). Bacterial dynamics and functional analysis of microbial metagenomes during ripening of Dutch-type cheese. Int. Dairy J. 61, 182–188. doi: 10.1016/j.idairyj.2016.05.005
Prasad, S. B., Jayaraman, G., and Ramachandran, K. B. (2010). Hyaluronic acid production is enhanced by the additional co-expression of UDP-glucose pyrophosphorylase in Lactococcus lactis. Appl. Microbiol. Biotechnol. 86, 273–283. doi: 10.1007/s00253-009-2293-0
Price, C., Branco dos Santos, F., Hesseling, A., Uusitalo, J., Bachmann, H., Benavente, V., et al. (2017). Glucose limitation in Lactococcus shapes a single-peaked fitness landscape exposing membrane occupancy as a constraint. bioRxiv [Preprint]. doi: 10.1101/147926
Priyadharshini, S., Archana, M., Angeline Kiruba, D., Priyadharshini, M. N., and Malarvizhi, T. (2015). Production of isobutanol from Lactococcus lactis using valine catabolic degradation pathway. Int. J. Eng. Technol. Sci. Res. 2, 72–81.
Puri, P. (2014). On the Regulation of the Metabolic Shift and Protein Synthesis in Lactococcus Lactis. Groningen: University of Groningen.
Ramos, A., Neves, A., Ventura, R., Maycock, C., López, P., and Santos, H. (2004). Effect of pyruvate kinase overproduction on glucose metabolism of Lactococcus lactis. Microbiology 150, 1103–1111.
Rossell, S., van der Weijden, C. C., Lindenbergh, A., van Tuijl, A., Francke, C., Bakker, B. M., et al. (2006). Unraveling the complexity of flux regulation: a new method demonstrated for nutrient starvation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 103, 2166–2171. doi: 10.1073/pnas.0509831103
Scholz, M., Ward, D. V., Pasolli, E., Tolio, T., Zolfo, M., Asnicar, F., et al. (2016). Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat. Methods 13:435. doi: 10.1038/nmeth.3802
Serata, M., Iino, T., Yasuda, E., and Sako, T. (2012). Roles of thioredoxin and thioredoxin reductase in the resistance to oxidative stress in Lactobacillus casei. Microbiology 158, 953–962. doi: 10.1099/mic.0.053942-0
Serrano, L. M., Molenaar, D., Wels, M., Teusink, B., Bron, P. A., De Vos, W. M., et al. (2007). Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb. Cell Fact. 6:29. doi: 10.1186/1475-2859-6-29
Shah, M. V., Badle, S. S., and Ramachandran, K. B. (2013). Hyaluronic acid production and molecular weight improvement by redirection of carbon flux towards its biosynthesis pathway. Biochem. Eng. J. 80, 53–60. doi: 10.1016/j.bej.2013.09.013
Siedler, S., Balti, R., and Neves, A. R. (2019). Bioprotective mechanisms of lactic acid bacteria againtst fungal spoilage of food. Curr. Opin. Biotechnol. 56, 138–146.
Sieuwerts, S. (2016). Microbial interactions in the yoghurt consortium: current status and product implications. SOJ Microb. Infect. Dis. 4, 1–5.
Sieuwerts, S., Molenaar, D., Van Hijum, S. A. F. T., Beerthuyzen, M., Stevens, M. J. A., Janssen, P. W. M., et al. (2010). Mixed-culture transcriptome analysis reveals the molecular basis of mixed-culture growth in Streptococcus thermophilus and Lactobacillus bulgaricus. Appl. Environ. Microbiol. 76, 7775–7784. doi: 10.1128/AEM.01122-10
Siezen, R. J., Starrenburg, M. J. C., Boekhorst, J., Renckens, B., Molenaar, D., and Van Hylckama Vlieg, J. E. T. (2008). Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 74, 424–436. doi: 10.1128/AEM.01850-07
Sirisansaneeyakul, S., Luangpipat, T., Vanichsriratana, W., Srinophakun, T., Chen, H. H.-H., and Chisti, Y. (2007). Optimization of lactic acid production by immobilized Lactococcus lactis IO-1. J. Ind. Microbiol. Biotechnol. 34, 381–391.
Smid, E. J., Erkus, O., Spus, M., Wolkers-Rooijackers, J. C., Alexeeva, S., and Kleerebezem, M. (2014). Functional implications of the microbial community structure of undefined mesophilic starter cultures. Microb. Cell Fact. 13:S2. doi: 10.1186/1475-2859-13-S1-S2
Smirnov, K. S., Maier, T. V., Walker, A., Heinzmann, S. S., Forcisi, S., Martinez, I., et al. (2016). Challenges of metabolomics in human gut microbiota research. Int. J. Med. Microbiol. 306, 266–279. doi: 10.1016/j.ijmm.2016.03.006
Snoep, J. L., de Graef, M. R., Westphal, A. H., de Kok, A., Teixeira de Mattos, M. J., and Neijssel, O. M. (1993). Differences in sensitivity to NADH of purified pyruvate dehydrogenase complexes of Enterococcus faecalis, Lactococcus lactis, Azotobacter vinelandii and Escherichia coli: implications for their activity in vivo. FEMS Microbiol. Lett. 114, 279–283.
Solem, C., Dehli, T., and Jensen, P. R. (2013). Rewiring lactococcus lactis for ethanol production. Appl. Environ. Microbiol. 79, 2512–2518. doi: 10.1128/AEM.03623-12
Solem, C., Koebmann, B., and Jensen, P. R. (2008). Control analysis of the role of triosephosphate isomerase in glucose metabolism in Lactococcus lactis. IET Syst. Biol. 2, 64–72. doi: 10.1049/iet-syb
Solem, C., Koebmann, B. J., and Jensen, P. R. (2003). Glyceraldehyde-3-Phosphate dehydrogenase has no control over glycolytic flux in Lactococcus lactis MG1363. J. Bacteriol. 185, 1564–1571. doi: 10.1128/JB.185.5.1564-1571.2003
Solem, C., Petranovic, D., Koebmann, B., Mijakovic, I., and Jensen, P. R. (2010). Phosphoglycerate mutase is a highly efficient enzyme without flux control in Lactococcus lactis. J. Mol. Microbiol. Biotechnol. 18, 174–180. doi: 10.1159/000315458
Song, A. A. L., Abdullah, J. O., Abdullah, M. P., Shafee, N., Othman, R., Tan, E. F., et al. (2012). Overexpressing 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) in the Lactococcal mevalonate pathway for heterologous plant sesquiterpene production. PLoS One 7:e52444. doi: 10.1371/journal.pone.0052444
Song, A. A. L., In, L. L. A., Lim, S. H. E., and Rahim, R. A. (2017). A review on Lactococcus lactis: from food to factory. Microb. Cell Fact. 16:55.
Sybesma, W., Starrenburg, M., Kleerebezem, M., Mierau, I., De Vos, W. M., and Hugenholtz, J. (2003). Increased production of folate by metabolic engineering of Lactococcus lactis. Appl. Environ. Microbiol. 69, 3069–3076.
Tanaka, K., Komiyama, A., Sonomoto, K., Ishizaki, A., Hall, S., and Stanbury, P. (2002). Two different pathways for D-xylose metabolism and the effect of xylose concentration on the yield coefficient of L-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis IO-1. Appl. Microbiol. Biotechnol. 60, 160–167.
Tanous, C., Chambellon, E., Sepulchre, A., and Yvon, M. (2005). The gene encoding the glutamate dehydrogenase in Lactococcus lactis is part of a remnant Tn3 transposon carried by a large plasmid. J. Bacteriol. 187, 5019–5022. doi: 10.1128/JB.187.14.5019
Temmerman, R., Huys, G., and Swings, J. (2004). Identification of lactic acid bacteria: culture-dependent and culture-independent methods. Trends Food Sci. Technol. 15, 348–359. doi: 10.1016/j.tifs.2003.12.007
Teusink, B., Bachmann, H., and Molenaar, D. (2011). Systems biology of lactic acid bacteria: a critical review. Microb. Cell Fact. 10:S11. doi: 10.1186/1475-2859-10-S1-S11
Teusink, B., Walsh, M. C., Van Dam, K., and Westerhoff, H. V. (1998). The danger of metabolic pathways with turbo design. Trends Biochem. Sci. 23, 162–169.
Teusink, B., Wiersma, A., Jacobs, L., Notebaart, R. A., and Smid, E. J. (2009). Understanding the adaptive growth strategy of Lactobacillus plantarum by in silico optimisation. PLoS Comput. Biol. 5:e1000410. doi: 10.1371/journal.pcbi.1000410
Thakur, K., Tomar, S. K., and De, S. (2016). Lactic acid bacteria as a cell factory for riboflavin production. Microb. Biotechnol. 9, 441–451. doi: 10.1111/1751-7915.12335
Thompson, J. (1987). Regulation of sugar transport and metabolism in lactic acid bacteria. FEMS Microbiol. Lett. 46, 221–231.
Truong, D. T., Tett, A., Pasolli, E., Huttenhower, C., and Segata, N. (2017). Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 27, 626–638. doi: 10.1101/gr.216242.116
Turaev, D., and Rattei, T. (2016). High definition for systems biology of microbial communities: metagenomics gets genome-centric and strain-resolved. Curr. Opin. Biotechnol. 39, 174–181. doi: 10.1016/j.copbio.2016.04.011
van Hylckama Vlieg, J. E., Rademaker, J. L., Bachmann, H., Molenaar, D., Kelly, W. J., and Siezen, R. J. (2006). Natural diversity and adaptive responses of Lactococcus lactis. Curr. Opin. Biotechnol. 17, 183–190. doi: 10.1016/j.copbio.2006.02.007
van Kranenburg, R., Marugg, J. D., van Swam, I. I., Willem, N. J., and de Vos, W. M. (1997). Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24, 387–397.
van Kranenburg, R., Vos, H. R., Van Swam, I. I., Kleerebezem, M., and De Vos, W. M. (1999). Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: complementation, expression, and diversity. J. Bacteriol. 181, 6347–6353.
Van Mastrigt, O., Abee, T., Lillevang, S. K., and Smid, E. J. (2018). Quantitative physiology and aroma formation of a dairy Lactococcus lactis at near-zero growth rates. Food Microbiol. 73, 216–226. doi: 10.1016/j.fm.2018.01.027
Vido, K., Diemer, H., Dorsselaer, A., Van Leize, E., Juillard, V., Gruss, A., et al. (2005). Roles of thioredoxin reductase during the aerobic life of Lactococcus lactis. J. Bacteriol. 187, 601–610. doi: 10.1128/JB.187.2.601
Voit, E., Neves, A. R., and Santos, H. (2006). The intricate side of systems biology. Proc. Natl. Acad. Sci. 103, 9452–9457. doi: 10.1073/pnas.0603337103
Wegkamp, A., Van Oorschot, W., De Vos, W. M., and Smid, E. J. (2007). Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl. Environ. Microbiol. 73, 2673–2681. doi: 10.1128/AEM.02174-06
Wu, C., Huang, J., and Zhou, R. (2017). Genomics of lactic acid bacteria: current status and potential applications. Critic. Rev. Microbiol. 43, 393–404. doi: 10.1080/1040841X.2016.1179623
Xiao, Z., and Lu, J. R. (2014). Strategies for enhancing fermentative production of acetoin: a review. Biotechnol. Adv. 32, 492–503. doi: 10.1016/j.biotechadv.2014.01.002
Yamauchi, R., Maguin, E., Horiuchi, H., Hosokawa, M., and Sasaki, Y. (2019). The critical role of urease in yogurt fermentation with various combinations of Streptococcus thermophilus and Lactobacillus delbrueckii ssp. bulgaricus. J. Dairy Sci. 102, 1033–1043. doi: 10.3168/jds.2018-15192
Ye, W., Huo, G., Chen, J., Liu, F., Yin, J., Yang, L., et al. (2010). Heterologous expression of the Bacillus subtilis (natto) alanine dehydrogenase in Escherichia coli and Lactococcus lactis. Microbiol. Res. 165, 268–275. doi: 10.1016/j.micres.2009.05.008
Zolfo, M., Tett, A., Jousson, O., Donati, C., and Segata, N. (2017). MetaMLST: multi-locus strain-level bacterial typing from metagenomic samples. Nucleic Acids Res. 45, e7. doi: 10.1093/nar/gkw837
Zomer, A. L., Buist, G., Larsen, R., Kok, J., and Kuipers, O. P. (2007). Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189, 1366–1381. doi: 10.1128/JB.01013-06
Keywords: food fermentation, metabolic engineering, strain development, control analysis, screening and selection
Citation: Liu J, Chan SHJ, Chen J, Solem C and Jensen PR (2019) Systems Biology – A Guide for Understanding and Developing Improved Strains of Lactic Acid Bacteria. Front. Microbiol. 10:876. doi: 10.3389/fmicb.2019.00876
Received: 20 December 2018; Accepted: 04 April 2019;
Published: 30 April 2019.
Edited by:
Jan Kok, University of Groningen, NetherlandsReviewed by:
Fernanda Mozzi, CONICET Centro de Referencia para Lactobacilos (CERELA), ArgentinaCopyright © 2019 Liu, Chan, Chen, Solem and Jensen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Solem, chso@food.dtu.dk Peter Ruhdal Jensen, perj@food.dtu.dk
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.