
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 April 2019
Sec. Fungi and Their Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00864
This article is part of the Research TopicSensing and Signaling of the Environment by Fungi in Health and Disease View all 20 articles
Understanding stress responses and signaling pathways in fungi became a fundamental need for the discovery of new specific antifungal targets for fighting emerging life-threatening pathogens and drug resistance. Ionic liquids constitute a unique class of chemicals, which structural diversity and tunable physical and chemical properties can provide a great diversity of stimuli. In this study, we propose the use of ionic liquids as tools to unravel signaling of stress responses in the filamentous fungus Aspergillus nidulans. We assessed how three ionic liquids with distinct effects over the cell wall and plasma membrane affect the biosynthesis of sphingolipids and accumulation of free sphingoid bases in this fungus. The stress imposed by each ionic liquid triggered the sphingolipid biosynthetic pathway and led to distinct profiles of sphingoid bases accumulation. Dodecyltributylphosphonium chloride and 1-decyl-3-methylimidazolium chloride induced the accumulation of sphingosine and of a yet unknown sphingoid base, respectively, while cholinium decanoate did not seem to accumulate any of these intermediates. This study brings further light to the roles of sphingoid bases in A. nidulans. In particular, sphingosine as a possible response mediator to cell wall damage induced by dodecyltributylphosphonium chloride, and involvement of an unknown sphingoid base in the response to plasma membrane permeabilization caused by 1-decyl-3-methylimidazolium chloride. In addition, we completed the genetic assignment of the glucosylceramide pathway in A. nidulans through the identification of the sphingolipid Δ4-desaturase gene (AN4405). The knowledge established reinforces the idea of targeting sphingolipids biosynthesis in the search of improved antifungal compounds.
During the last few decades, fungi have been considered an increasing concern due to the rise of life-threating infections (Richardson and Warnock, 2012). Invasive fungal infections are associated with high mortality rates, killing about 1.5–2 M people every year, exceeding the deaths reported for malaria or tuberculosis (Brown et al., 2012; Denning and Bromley, 2015). The increasing number of diseases caused by fungi is mostly related to the increase of the population at risk, mainly immunocompromised individuals, such as HIV/AIDS, cancer, transplant and diabetes patients (Araujo et al., 2010; Slavin et al., 2015). Despite the growing need for efficient antifungal treatment, essentially four classes of drugs are clinically used to treat invasive fungal infections: polyenes, azoles, echinocandins and pyrimidine analogs (Vermes et al., 2000; Roemer and Krysan, 2014). The emergence of resistance to antifungal drugs is being reported more frequently, challenging the efficacy of the treatments in the near future (Kathiravan et al., 2012; Arendrup and Perlin, 2014; Perfect, 2017). Therefore, understanding of stress responses and signaling pathways in fungi became a fundamental need for discovery of new specific antifungal targets to fight antifungal drug resistance.
Ionic liquids emerge as a source of unusual and diverse tools to activate stress mechanisms in filamentous fungi. They constitute a unique class of chemicals, able to provide stimuli not often encountered in nature. Ionic liquids are generally defined as salts that are liquid below 100°C (Stark and Seddon, 2007). There are millions of possible formulations and several hundred are already well characterized (Dong et al., 2007; Stark and Seddon, 2007). Our group has for long been studying the effects of ionic liquids in filamentous fungi. Combining gene and protein expression with organic and analytical chemistry, previous studies have elucidated how particular ionic liquids affect the stress response of filamentous fungi (Petkovic et al., 2009, 2011, 2012; Petkovic and Silva Pereira, 2012; Hartmann and Silva Pereira, 2013; Martins et al., 2013; Hartmann et al., 2015, 2016; Alves et al., 2016). At sub-inhibitory concentrations, they can alter the fungal metabolic footprint (Petkovic et al., 2009), activate the biosynthesis of osmolytes (Martins et al., 2013) and uncommon secondary metabolites (Alves et al., 2016), and increase the expression of multidrug transporters (Martins et al., 2013). Recently, the effects of some 1-alkyl-3-methylimidazolium chlorides and cholinium alkanoates on the plasma membrane of Aspergillus nidulans have been assessed (Hartmann et al., 2015). While the first were shown to permeabilize the fungal plasma membrane, the biocompatible cholinium alkanoates did not cause this effect. Some ionic liquids have also been reported to cause damage to the cell wall of filamentous fungi (Petkovic et al., 2012; Hartmann and Silva Pereira, 2013). In yeast and filamentous fungi, damages to the cell wall can activate the cell wall integrity pathway, a response mechanism that overcomes the stress imposed over this cellular structure (Levin, 2005; Fujioka et al., 2007; Valiante et al., 2015). In Saccharomyces cerevisiae, cross-talk mechanisms between this pathway and other signaling pathways have been described, including the activation of the cell wall integrity pathway by sphingolipid intermediates (Levin, 2005). In A. nidulans, we have preliminarily observed a similar response upon exposure to ionic liquids that damage the cell wall (Hartmann and Silva Pereira, 2013, 2016; Hartmann et al., 2016).
Sphingolipids are essential components of all eukaryotic cell membranes and play an important role in a variety of biological processes, like heat stress response, signal transduction and apoptosis (Jenkins et al., 1997; Cheng et al., 2003; Hannun and Obeid, 2008). In fungi, sphingolipids such as inositol phosphorylceramides and glucosylceramides play a key role in growth and pathogenesis (Fernandes et al., 2018). Recent studies have shown that inhibition of the synthesis of these sphingolipids can attenuate the virulence of Candida and Aspergillus species (Zhong et al., 2000; Levery et al., 2002). The sphingolipid biosynthetic pathway has been characterized in greater detail in the yeast S. cerevisiae (Dickson and Lester, 2002) and in dimorphic fungi such as Candida albicans (Oura and Kajiwara, 2008) and Cryptococcus neoformans (Singh et al., 2012). However, knowledge on this pathway in filamentous fungi, such as A. nidulans, is still limited. After formation of sphingoid bases (such as dihydrosphingosine and phytosphingosine) and ceramides (Nagiec et al., 1994; Haak et al., 1997; Merrill, 2002; Singh and Del Poeta, 2016), two branches can follow up, yielding inositol phosphorylceramides or glucosylceramides. Studies have shown that the intermediates of glucosylceramide pathway can play important roles in fungal cells. In C. albicans disruption of sphingolipid Δ8-desaturase leads to accumulation of sphingosine and reduced hyphal growth (Oura and Kajiwara, 2008). In C. neoformans, mutation of the sphingolipid C9-methyltransferase gene alters the composition and rigidity of the membrane (Singh et al., 2012). Fusarium graminearum strains lacking the MT2 gene, encoding a sphingolipid C9-methyltransferase, exhibit severe growth defects and produce abnormal conidia (Ramamoorthy et al., 2009). Only very recently, some of the genes of the glucosylceramide pathway in A. nidulans were assigned and their deletion observed to promote resistance to cell wall damaging agents, such as congo red and calcofluor white (Fernandes et al., 2016).
In the present study, we propose using ionic liquids as tools to unravel poorly characterized stress responses in A. nidulans. Three ionic liquids were selected – dodecyltributylphosphonium chloride ([P4 4 4 12]Cl), 1-decyl-3-methylimidazolium chloride ([C10mim]Cl) and cholinium decanoate – each displaying a distinct mechanism of toxicity (Petkovic et al., 2012; Hartmann and Silva Pereira, 2013; Hartmann et al., 2015). Toxicity evaluation, gene expression analysis by quantitative real-time PCR (qRT-PCR) and sphingoid base accumulation profiling by high-performance liquid chromatography (HPLC) were the main used methodologies. Collectively, the obtained data are suggestive of participation of specific sphingoid bases in the stress responses imposed by ionic liquids in either the cell wall and/or membrane of A. nidulans.
The ionic liquids used in this study are depicted in Figure 1. Dodecyltributylphosphonium chloride, [P4 4 4 12]Cl, and 1-decyl-3-methylimidazolium chloride, [C10mim]Cl, were provided by IoLiTec Ionic Liquids Technologies, the first relying on a custom synthesis (Adamová et al., 2012). Cholinium decanoate was synthesized as previously described (Petkovic et al., 2010). Ionic liquids were characterized by 1H, 13C, and 31P NMR spectroscopy, mass spectrometry, CHN elemental analysis, and halide and water content analyses.
The salts and supplements used to prepare the culture media were in general purchased from Sigma-Aldrich; except Dichloran-Glycerol (DG18) agar, yeast extract and bacteriological peptone that were purchased from Oxoid; sucrose from Alfa Aesar; casein hydrolysate from Fluka; agar and NaCl from Panreac AppliChem; NaOH from J.M.G. Santos and glycerol from Fisher Bioreagents. Molecular biology reagents used for PCR reactions were purchased from NZYTech. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) was purchased from Sigma-Aldrich. The reagents used for sphingolipid extraction: pyridine, butanol, ammonium hydroxide and 40% methylamine in water were purchased from Acros Organics; ethanol, methanol and chloroform (and dimethyl sulfoxide used in MTT assays) were purchased from Fisher Chemical and diethylether from Panreac AppliChem. Sphingoid bases standards (phytosphingosine, sphingosine and dihydrosphingosine) were purchased from Avanti Polar Lipids. All solvents used in chromatographic analyses were of the highest analytical grade and water was obtained from a Milli-Q system (Millipore).
The Aspergillus nidulans strains used in this study are described in Supplementary Table S1. Strain A4 (used as the reference strain) and strain A1149 were obtained from the Fungal Genetics Stock Center (Kansas City, MO, United States). A1149 served as parental strain to generate ΔAN4405, ΔAN4592, ΔAN5688, ΔAN7375, ΔAN8806 deletion mutants and a pyrG+ prototroph strain (used as control strain for experiments comparing the deletion mutants). All strains were cultivated on DG18 agar plates with the appropriate supplements. Cultures were incubated in the dark, for 6–7 days, at 30°C. Asexual spores (conidia) were harvested using a saline solution (NaCl 8.5 g⋅l-1) and collected after passing through three layers of miracloth. The harvested spores were resuspended in a saline solution, to be used immediately, or in a cryoprotective saline solution containing 10% (v/v) glycerol, to be stored at -20°C or -80°C.
Liquid minimal medium was prepared with glucose (10 g⋅l-1), thiamine (0.01 g⋅l-1), 5% (v/v) nitrate salts solution [NaNO3 (120.0 g⋅l-1), KCl (10.4 g⋅l-1), MgSO4⋅7H2O (10.4 g⋅l-1) and KH2PO4 (30.4 g⋅l-1)], 0.1% (v/v) trace elements solution [ZnSO4⋅7H2O (22.0 g⋅l-1); H3BO3 (11.0 g⋅l-1); MnCl2⋅4H2O (5.0 g⋅l-1); FeSO4⋅7H2O (5.0 g⋅l-1); CoCl2⋅6H2O (1.7 g⋅l-1); CuSO4⋅5H2O (1.6 g⋅l-1); Na2MoO4⋅2H2O (1.5 g⋅l-1), and Na4EDTA (50.0 g⋅l-1)] and the pH was adjusted to 6.5 with NaOH. Whenever necessary, media were supplemented with uracil (1.12 g⋅l-1), uridine (1.2 g⋅l-1), and/or pyridoxin (0.05 mg⋅l-1). Fungal cultures (100 ml of liquid minimal medium) were inoculated with freshly harvested spores (106 conidia per ml) and incubated for 24 h at 30°C and 200 rpm. Ionic liquids were then added to the cultures, at the MIC, and incubated for 4 h. Longer incubation periods (8, 12, or 24 h) were used for cultures exposed to [P4 4 4 12]Cl. Negative controls without ionic liquids were also prepared. At the end of the treatments, mycelia were recovered by vacuum filtration and immediately frozen in liquid nitrogen. All the samples were stored at -80°C until lyophilized.
Approximately 100 mg of frozen mycelia was grounded with poly(vinylpolypirrolidone) (0.4 mg per mg of mycelia) in a Tissue Lyser LT (QIAGEN). The final powder was used in the extraction and purification of total RNA using the RNeasy Plant Mini Kit (QIAGEN), according to the manufacturer’s protocol. Genomic DNA digestion was done with the RNase-Free DNase Set (QIAGEN). Quality, integrity and quantity of the total RNA were analyzed in a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific) and by running 2 μg of RNA into 1% agarose gels. The complementary DNA (cDNA) was synthesized from 500 ng of the total RNA using an iScript cDNA Synthesis Kit (Bio-Rad) in a T100 Thermal Cycler (Bio-Rad). The reaction protocol consisted of 5 min at 25 °C, 30 min at 42°C and 5 min at 85°C. Based on the sequences of A. nidulans genes (Aspergillus Genome Database1), all primer pairs used in qRT-PCR experiments were designed using the GeneFisher2 web tool2, with the exception of those for chsB, fksA, agsB, which were previously designed in Fujioka et al. (2007). All primers were produced by Thermo Fisher Scientific (see Supplementary Table S2 for the list of the qRT-PCR primers used in this study). The qRT-PCR analysis was performed in a CFX96 Thermal Cycler (Bio-Rad), using the SsoFast EvaGreen Supermix (Bio-Rad), 250 nM of each oligonucleotide and the cDNA template equivalent to 5 ng of total RNA, at a final volume of 10 μl per well, in three technical replicates. The PCR conditions were: enzyme activation at 95°C for 30 s; 40 cycles of denaturation at 95°C for 10 s and annealing/extension at 59°C for 15 s; and melting curve obtained from 65°C to 95°C, consisting of 0.5°C increments for 5 s. Data analyses were performed using the CFX Manager software (Bio-Rad). The expression of each gene was taken as the relative expression compared to the time-zero (before incubation with the tested compounds). The expression of all target genes was normalized by the expression of γ-actin gene (internal control). Four biological replicates were performed. Statistical analysis of the qRT-PCR data was performed in the GraphPad Prism v8.0 software. Treatments with ionic liquids were compared with the control by Student’s t-test. Differences with a p-value below 0.05 were considered statistically significant.
Approximately 80 mg of dried mycelia of each condition were grounded in a Tissue Lyser LT (QIAGEN). Lipids were extracted in glass tubes with 5 ml of a solution containing 95% ethanol/water/diethylether/pyridine/4.2 M ammonium hydroxide (15:15:5:1:0.018; by volume), sonicated for 5 min and then incubated for 15 min at 60 °C. The extract was removed after centrifugation (1620 g for 5 min in a swing-bucket rotor) and the pellet extracted twice in the same manner. The lipid extracts were dried under a nitrogen flow. For methanolysis the dried extracts were resuspended in 5 ml of a solution containing methanol/ethanol/butanol/40% methylamine in water (4:3:1:4, by volume) and incubated at 52°C for 30 min to ensure the deacylation of lipids. The resulting extracts were dried under a nitrogen flow and resuspended in 3 ml of methanol/chloroform (2:1, by volume). One milliliter each of chloroform and alkaline solution (NH4OH in water, pH 9.0) were added to separate sphingoid bases to the organic layer. The mixture was centrifuged (1620 g for 5 min in a swing-bucket rotor), the aqueous phase was discarded and the organic phase was washed three times with 1 ml of alkaline solution, finally dried under a nitrogen flow. To perform alkaline hydrolysis the dried extracts were resuspended in 3 ml of 0.1 M KOH in methanol/chloroform (2:1, by volume) and incubated at 37°C for 1 h to break down cellular acylglycerolipids and phospholipids. After incubation, 1 ml each of chloroform and alkaline solution were added and the mixture centrifuged (1,620 g for 5 min in a swing bucket rotor). The aqueous phase was discarded and the organic phase was washed three times in the same way, then dried under nitrogen flow and finally resuspended in methanol (100 μl per each 20 mg of original dried mycelia) for further analysis of sphingoid bases by liquid chromatography. For the analysis by HPLC, 200 μl of extracts (or a 1:4 dilution for tests with glucosylceramide pathway mutants) underwent a 30 min pre-column derivatization with 25 μl o-phthalaldehyde solution, prepared by dissolving 5 mg o-phthalaldehyde and 5 μl β-mercaptoethanol in 100 μl of methanol, and adding 9.9 ml of 3% borate buffer (pH 10.5). The derivatized sphingoid bases were chromatographically separated using an Acquity UPLC System (Waters) with a fluorescence detector (acquisition with an energy gain of 1000), cooling auto sampler, and column oven. A Symmetry® C18 reverse phase column (250 × 4.6 mm), packed with end capped particles (5 μm, pore size 100 Å) was used at 40°C. Data were acquired using Empower 2 software, 2006 (Waters Corporation). Sample injections of 10 μl were made using a 50 μl loop operated in partial loop mode. The mobile phase, at a flow rate of 1 ml⋅min-1, consisted of 90% methanol. Each sample was run for 30 min and after each run the column was washed with water. Phytosphingosine, sphingosine and dihydrosphingosine were used as standards and their chromatographic profiles can be seen in Supplementary Figures S1–S3.
The genes AN4405, AN4592, AN5688, AN7375, or AN8806 were selected for gene replacement with A. fumigatus pyrG in A. nidulans A1149 strain, performed according to a previously established method (Szewczyk et al., 2006). Generation of the prototroph pyrG+ (gene AN6157) was performed by the same method. Deletion cassettes were constructed using a fusion PCR protocol. For each target gene, six primers (P1-P6) were designed, based on sequences from the Aspergillus Genome Database and analyzed using the NetPrimer web tool3. All primers used for generation of gene-replacement mutants are listed in Supplementary Table S3. PCRs reactions were performed in a T100 Thermal Cycler (Bio-Rad). A. fumigatus pyrG was amplified from plasmid pCDS60 (FGSC, Kansas City, MO, United States) using primers CDS164 and CDS165. Flanking fragment upstream and downstream of each target gene were amplified with primer pairs P1/P3 and P4/P6, respectively, using genomic DNA from A. nidulans A4 as template. The final cassette was produced by fusing the flanking regions with the A. fumigatus pyrG using nested primers P2 and P5 for each gene target. All PCR conditions are described in Supplementary Table S4. PCR products were cleaned with NZYGelPure kit (NZYTech). To produce transformable protoplasts, 108 freshly harvested conidia from A. nidulans A1149 were inoculated into 100 ml of liquid minimal medium with the appropriate nutritional supplements. The cultures were incubated overnight at 30°C, 90 rpm; then recovered by centrifugation and washed with 0.6 M MgSO4. The washed culture was resuspended in filter-sterilized enzymatic mix (300 mg lysing enzymes from Trichoderma harzianum, 150 μl β-glucuronidase from bovine liver, type B-1, and 150 mg Driselase from Basidiomycetes sp., all from Sigma-Aldrich) in osmotic medium (1.2 M MgSO4⋅7H2O and 10 mM sodium phosphate buffer pH 6.5; final pH adjusted to 5.8 with Na2HPO4) and incubated for 20 h at 30°C, 90 rpm, for digestion. Protoplasts were recovered by centrifugation and 10 ml of suspension were carefully overlaid with 5 ml of trapping buffer (0.6 M sorbitol and 100 mM Tris-HCl pH 7.0) and centrifuged at 1500 g, at 4°C, for 15 min in a swing-bucket rotor. The protoplasts phase was carefully removed and washed three times with 10 ml of ST10 buffer (1.2 M sorbitol and 10 mM Tris-HCl pH 7.5) and finally resuspended in 700 μl of STC buffer (1.2 M sorbitol, 10 mM Tris-HCl pH 7.5 and 10 mM CaCl2). For each transformation, 10 μl of cleaned fusion PCR product were added to 100 μl protoplast suspension, then vortexed shortly 6-8 times. 50 μl of freshly filtered polyethylene glycol (PEG) solution (25% (w/v) in STC buffer) were added to the mixture, vortexed 4–5 times and placed in an ice bath for 25 min. Another 1 ml of filtered PEG solution was then added, gently mixed using a micropipette and placed at room temperature for 25 min. Hundred microliter of the transformation mix were plated onto a selective medium containing glucose (5.0 g⋅l-1), yeast extract (5.0 g⋅l-1), sucrose (342.3 g⋅l-1), pyridoxin (0.05 mg⋅l-1), 0.1% (v/v) trace elements solution, 0.1% (v/v) vitamin solution [biotin (1.0 g⋅l-1), pyridoxin (1.0 g⋅l-1), thiamine (1.0 g⋅l-1), riboflavin (1.0 g⋅l-1), p-aminobenzoic acid (1.0 g⋅l-1) and nicotinic acid (1.0 g⋅l-1)] and 1.5% agar. The selective plates were incubated for 3–4 days at 30°C, and the isolated transformants were stroke onto complete medium plates [glucose (10.0 g⋅l-1), peptone (2.0 g⋅l-1), yeast extract (1.0 g⋅l-1), casein hydrolysate (1.0 g⋅l-1), 5% (v/v) nitrate salts solution, 0.1% (v/v) trace elements and 0.1% (v/v) vitamin solution and pyridoxine (0.05 mg⋅l-1), pH 6.5] and incubated at 30°C for 4 days. Three generations were obtained before DNA extraction with Quick-DNATM Fungal/Bacterial Microprep Kit (Zymo Research) and diagnostic PCR (identical protocol as fusion PCR) were done. The PCR products were digested using restriction enzymes ApaI, HpaI, BglII, or NcoI (NZYTech). Total amplification product size and differential digestion patterns allowed confirmation of correct gene replacement.
The minimal inhibitory concentrations (MIC) of the ionic liquids were determined as previously described (Petkovic et al., 2012). Ionic liquids were added to the minimal culture media at final concentrations ranging from 0.005 to 0.045 mM for [P4 4 4 12]Cl, 0.05 to 0.45 mM for [C10mim]Cl, and 2 to 6 mM for cholinium decanoate. For each condition, 1 ml was inoculated with 106 conidia and divided into four wells (0.2 ml each) of a 96-well microtiter plate. Cultures were incubated in the dark at 30°C, for 7 days, without agitation. Fungal growth or its absence was evaluated at the end of incubation, gauging by eye the formation of mycelium (turbidity). The lowest concentration that inhibited growth was taken as the MIC. The determined values of the MIC should not be taken as absolute ones but as an indication of the upper inhibitory concentration limits. In the same conditions, the MIC was also determined for strains A4 and ΔAN4405 in medium supplemented with exogenous sphingosine at final concentrations of 0.1, 0.5, 1.0, 2.5, 5.0, 7.5, and 10 μM. For analysis of growth in solid media, 1 μl of spore suspensions containing 108, 107, and 106 conidia per ml were used to inoculate plates containing minimal media (2% agar) supplemented with [P4 4 4 12]Cl (0.015, 0.02, and 0.025 mM), [C10mim]Cl (0.05, 0.075, and 0.1 mM) or cholinium decanoate (0.75, 1, and 1.25 mM). Plates were incubated in the dark for 48 h at 30°C. For each condition three biological replicates were prepared.
Metabolic activity of the cultures after 4 h of exposure to each ionic liquid was verified by the reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT). Aspergillus nidulans (106 conidia per ml) was grown in minimal medium for 24 h, 30°C, without agitation, in 96-well plates, at a final volume of 200 μl per well (six wells per condition). The medium was carefully removed and 200 μl of fresh medium supplemented with either ionic liquid at the MIC concentration (only minimal medium for the control) was added to each well and incubated at 30°C for 4 h. The medium was then removed and the mycelia washed three times with saline. Hundred microliter of fresh minimal medium and 20 μl of a freshly prepared stock of MTT (5 mg⋅ml-1 in saline) were then added to each well and plates were incubated at 30°C, in the dark, for 4 h. The supernatant was then aspirated and 100 μl of dimethyl sulfoxide was added to each well to dissolve the resulting formazan crystals, overnight, in the dark. The plates were read in a Tecan Infinite M200 spectrophotometer, scoring the absorbance at 570 nm. The values of metabolic activity for each ionic liquid were obtained relatively to the control. Four biological replicates were performed.
Aspergillus nidulans (103 conidia per ml) was cultivated in 1 ml minimal medium in 12-well plates and grown for 24 h, at 30°C, without agitation. Prior to incubation plates were centrifuged at 600 g for 5 min to sediment conidia to the bottom of the wells. After growth, medium was carefully aspirated from each well and 1 ml of fresh medium supplemented with 100 mM of each ionic liquid was added. The plates were incubated for 4 h. Medium without supplementation was added for the control. After incubation, the medium from each well was carefully removed and the adhered mycelia were washed four times with a saline solution. Mycelia were stained with 1 ml calcofluor white, at the final concentration of 25 μM, and incubated at 30°C, for 30 min, in the dark. Residual dye was removed by carefully washing the mycelia with saline solution three times. The stained mycelia were visualized in a Nikon Eclipse Ts2-FL inverted fluorescence microscope with a 20× magnification and a C-LED 385 nm filter. Images were captured with a Nikon DS-Fi3 camera. This assay provided a qualitative analysis of the alterations provoked by exposure to the ionic liquids in the hyphal cell wall.
In previous reports, we have demonstrated that alkyltributylphosphonium chlorides, ionic liquids recalcitrant to degradation, damage the cell wall of A. nidulans and cause membrane permeabilization when a long alkyl substituent is present (Petkovic et al., 2012; Hartmann and Silva Pereira, 2013). In this study, we tested whether the stress imposed by [P4 4 4 12]Cl – an ionic liquid from this family that has an alkyl substituent composed of 12 carbon atoms (Figure 1) – could alter the biosynthesis of sphingolipids. After 4 h of exposure to [P4 4 4 12]Cl at the MIC (Table 1), A. nidulans cultures remained metabolically active (Supplementary Table S5) and the expression of genes involved in sphingolipid biosynthesis was further analyzed by qRT-PCR. A schematic representation of the sphingolipid biosynthetic pathway is depicted in Figure 2A (Levin, 2005; Li et al., 2006, 2007). Gene expression data suggested that the sphingolipid biosynthetic pathway was activated upon exposure to [P4 4 4 12]Cl. In detail, lcbA, the gene coding for a serine palmitoyltransferase that catalyzes the very first step in the biosynthesis of sphingoid bases (Cheng et al., 2001), underwent a significant 1.7-fold increase in its expression after exposure to the ionic liquid (Figure 2B and Supplementary Table S7). It is likely that, as in S. cerevisiae, this enzyme is comprised of two components (LCB1 and LCB2) (Nagiec et al., 1994). Aspergillus nidulans lcbA is homolog to the yeast LCB1. Although not described, AN1102 has very high similarity with LCB2, and putatively encodes another component of the serine palmitoyltransferase complex in A. nidulans (Supplementary Table S6). After exposure to [P4 4 4 12]Cl, AN1102 expression increased fivefold (Figure 2B and Supplementary Table S7). The up-regulation of both genes is consistent with the activation of sphingolipid biosynthesis under the ionic liquid stress.
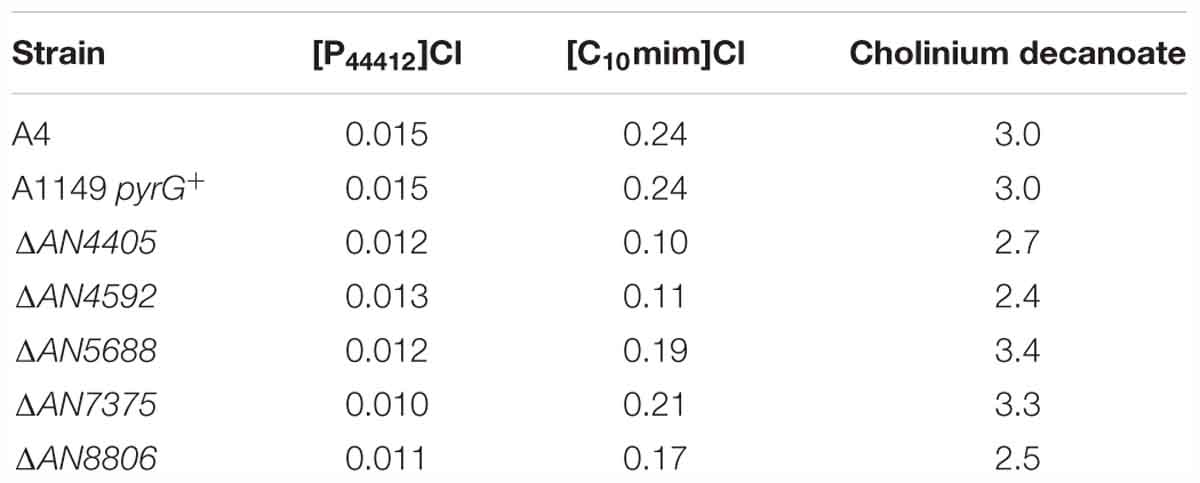
Table 1. Minimal inhibitory concentrations (in mM) of the tested ionic liquids against Aspergillus nidulans strains after growth in liquid minimal medium.
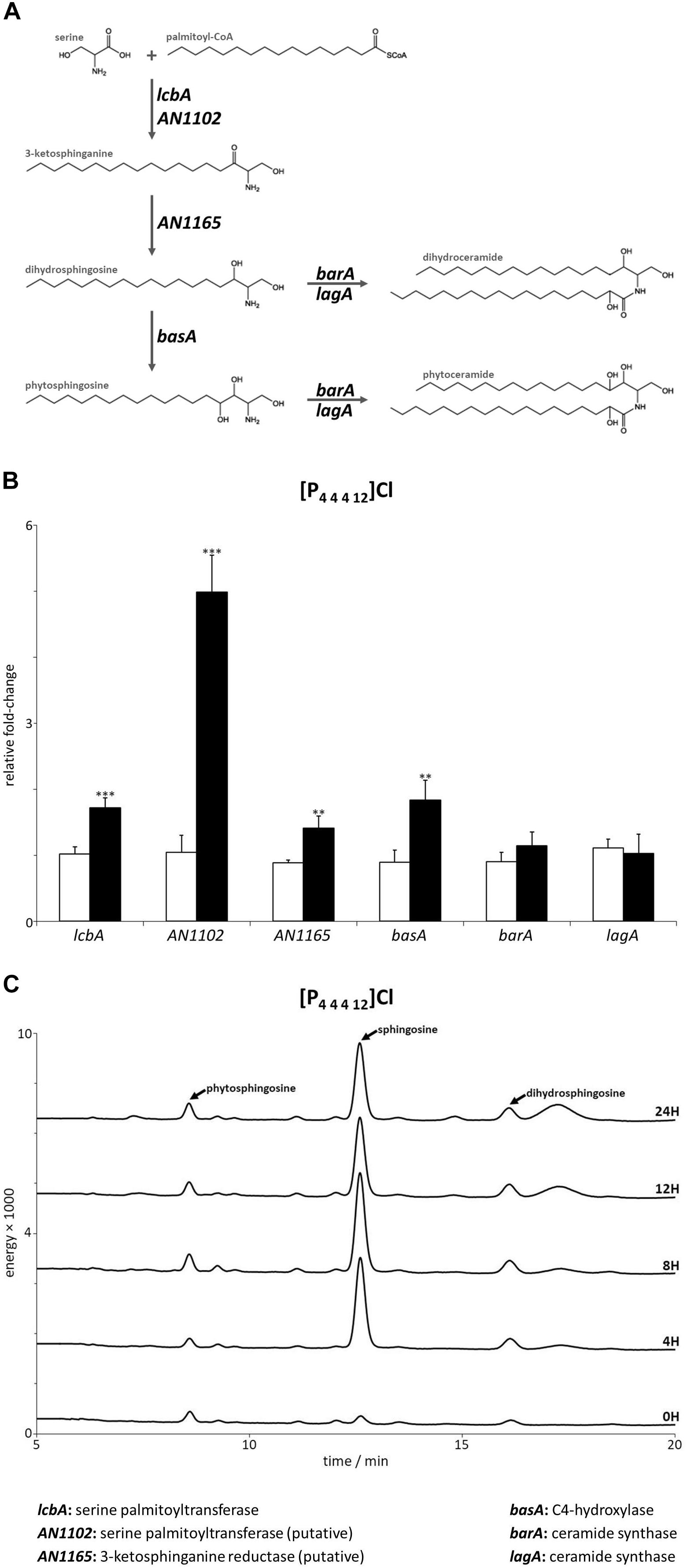
Figure 2. (A) Schematic representation of the first steps in the sphingolipid biosynthetic pathway. (B) Expression levels of the genes mediating the first steps of sphingolipid biosynthesis after 4 h of exposure to dodecyltributylphosphonium chloride ([P44412]Cl, black bars) compared to the control (white bars). Axis y represents the fold-change relative to time-zero (∗∗p < 0.01, ∗∗∗p < 0.001). (C) Sphingoid bases accumulation profile after 4, 8, 12, and 24 h of exposure to [P44412]Cl. The peaks assigned to phytosphingosine, sphingosine and dihydrosphingosine were confirmed by their standards.
The following steps of the sphingolipid biosynthetic pathway are involved in the formation of the sphingoid bases dihydrosphingosine and phytosphingosine (Figure 2A). These steps are catalyzed by enzymes encoded by AN1165 (putatively, Supplementary Table S6) (Beeler et al., 1998) and basA (Li et al., 2007), respectively. Both genes were significantly up-regulated 1.4 and 1.8-fold upon [P4 4 4 12]Cl stress compared to the control (Figure 2B and Supplementary Table S7). Interestingly, the synthesis of ceramides, the next step in the pathway, did not seem to be triggered, as inferred by the unaltered expression of the genes coding for ceramide synthases barA and lagA (Figure 2B and Supplementary Table S7) (Li et al., 2006). Collectively, these data expose that the stress imposed by [P4 4 4 12]Cl activated the genes involved in the synthesis of sphingoid bases but not ceramides, likely leading to accumulation of sphingoid bases.
To further confirm the accumulation of free sphingoid bases, they were extracted from mycelia of A. nidulans after exposure to [P4 4 4 12]Cl for 4, 8, 12, or 24 h and analyzed by liquid chromatography after derivatization with o-phthalaldehyde. The obtained chromatograms clearly reveal that the ionic liquid stress stimulated the accumulation of the sphingoid base sphingosine as early as 4 h, maintained up to 24 h of exposure to the ionic liquid (Figure 2C).
As known for other fungal species, sphingosine is synthesized through the glucosylceramide branch of sphingolipid biosynthesis (schematic representation in Figure 3A) (Del Poeta et al., 2014). In A. nidulans, part of the branch is known, namely Δ8-desaturase (AN4592), C9-methyltransferases (AN5688 and AN7375) and ceramide glucosyltransferase (AN8806) genes (Fernandes et al., 2016). However, the gene involved in the synthesis of sphingosine (i.e., coding for a sphingolipid Δ4-desaturase) remains unknown. To attempt its identification, the sequence of the previously verified dihydroceramide Δ4-desaturase from Schizosaccharomyces pombe (encoded by dsd1) (Garton et al., 2003) was used as query in protein-protein BLAST analysis (Supplementary Table S6). The best obtained hit was A. nidulans AN4405 gene (e = 6.0e-4, 93.0% query coverage, 54.0% identity), consistent with the predicted functions described in the Aspergillus Genome Database. The AN4405 gene model is located at chromosome III and comprises 1381 nucleotides with two predicted introns from 211 to 273 and 553 to 613; the open reading frame contains 1257 bp encoding 418 amino acids. A gene-replacement mutant (ΔAN4405) was generated and its sphingoid base accumulation profile after 24 h of growth was analyzed by liquid chromatography, further confirming its assignment as the gene coding for a sphingolipid Δ4-desaturase in A. nidulans (Supplementary Figure S4).
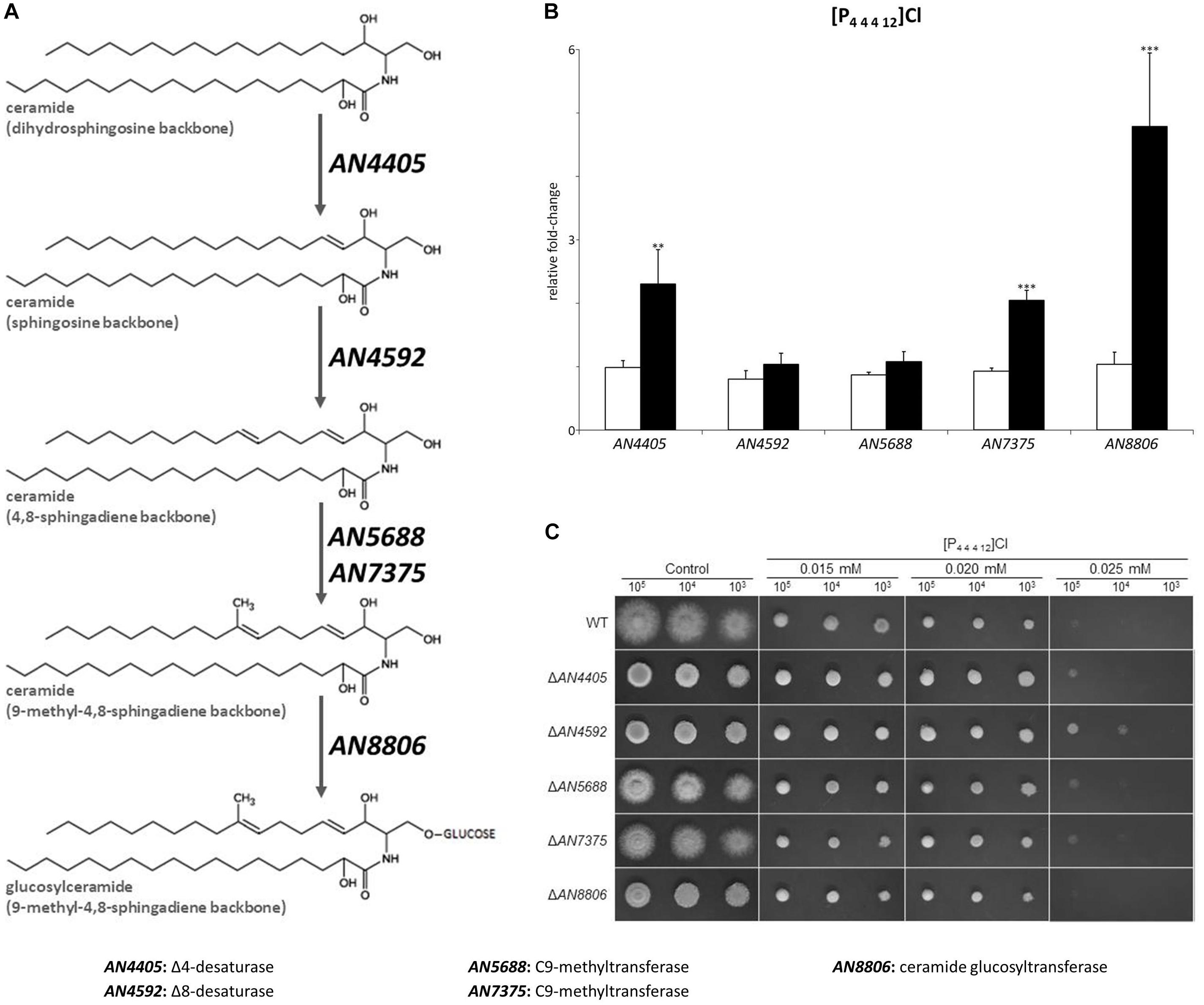
Figure 3. (A) Schematic representation of the glucosylceramide pathway. (B) Expression analysis of glucosylceramide pathway genes after 4 h of exposure to dodecyltributylphosphonium chloride ([P44412]Cl, black bars), compared to the control (white bars). Axis y represents the fold-change relative to time-zero (∗∗p < 0.01, ∗∗∗p < 0.001). (C) Growth assessment in solid media supplemented with 0.015, 0.020, or 0.025 mM of [P44412]Cl. 105, 104, and 103 conidia were inoculated in each plate.
The expression of genes involved in the glucosylceramide pathway after exposure to 4 h of [P4 4 4 12]Cl were also analyzed. Concordant to the observed accumulation of sphingosine, AN4405 underwent a significant increase of 2.3-fold compared to the control, while the gene responsible for the following step in the pathway, AN4592, did not alter its expression (Figure 3B and Supplementary Table S7). To additionally assess sphingosine involvement in the stress response imposed by [P4 4 4 12]Cl, the MIC values of wild-type and ΔAN4405 strains when exogenous sphingosine was added to the growth media were determined. Data show that increasing concentrations of exogenous sphingosine enabled ΔAN4405 to tolerate higher amounts of [P4 4 4 12]Cl, reaching values above the original wild-type MIC when 5 μM or more were present in the medium (Table 2). The wild-type strain was also able to increase its tolerance to the ionic liquid in medium with 2.5 μM or more of exogenous sphingosine.
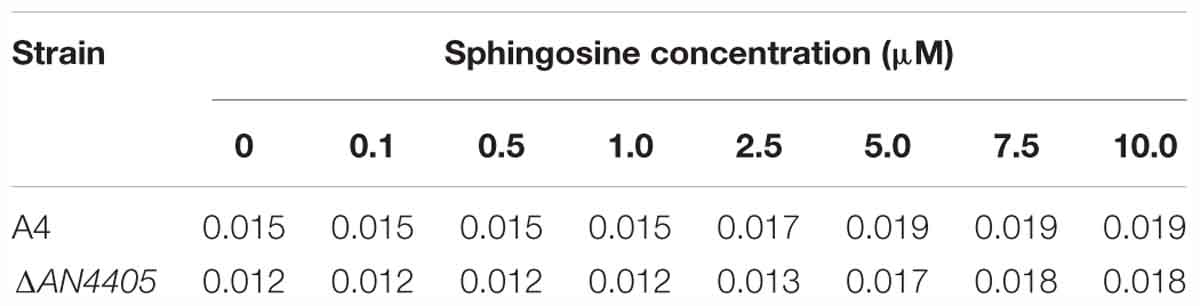
Table 2. Minimal inhibitory concentrations (in mM) for [P44412]Cl against Aspergillus nidulans strains A4 and ΔAN4405 after growth in liquid minimal medium supplemented with different concentrations of exogenous sphingosine (in μM).
To further evaluate the involvement of the glucosylceramide pathway in the stress response imposed by [P4 4 4 12]Cl, we generated gene-replacement mutants for the remaining genes of the glucosylceramide pathway and evaluated their susceptibility to the ionic liquid treatment. The determined MICs reveal higher susceptibility of all the mutants to [P4 4 4 12]Cl compared to the wild-type strain (Table 1), consistent with the involvement of the glucosylceramide pathway in the stress response. When looking at the growth on solid media, only the strain ΔAN4592 could grow well when 105 and 104 spores were inoculated in plates at the highest tested concentration of [P4 4 4 12]Cl (0.025 mM) (Figure 3C). Since this mutant is unable to utilize ceramides with a sphingosine backbone, it is likely that ΔAN4592 accumulates free sphingosine, reinforcing the potential role of this sphingoid base in signaling the stress induced by the ionic liquid.
As observed above, [P4 4 4 12]Cl, which is able to cause damage to both the cell wall and plasma membrane of A. nidulans (Petkovic et al., 2012; Hartmann and Silva Pereira, 2013), activated sphingolipid biosynthesis and induced the accumulation of sphingosine. We decided to test if ionic liquids with distinct effects would also affect the sphingolipid biosynthetic pathway in A. nidulans. Two ionic liquids, representative of families that we previously reported to have opposing effects over the plasma membrane (Hartmann et al., 2015), were selected: [C10mim]Cl (long alkyl substituent in the cation and recalcitrant to degradation) that permeabilizes the plasma membrane; and cholinium decanoate (long alkyl anion and biodegradable) that does not induce the same effect on the lipid bilayer. Similarly to [P4 4 4 12]Cl, after 4 h of exposure to either ionic liquid at the MIC, A. nidulans cultures were still metabolically active (Supplementary Table S5). We observed by microscopy that, unlike [P4 4 4 12]Cl, neither [C10mim]Cl or cholinium decanoate caused cell wall damage in A. nidulans hyphae after 4 h of exposure to 100 mM of ionic liquid (Figure 4A). This was further confirmed by qRT-PCR analysis of genes involved in the biosynthesis of chitin (chsA, chsB, chsC, chsD, chsF, chsG, csmA, and csmB), α-glucans (agsA, agsB) and β-glucans (fksA) after 4 h of exposure to each ionic liquid (Table 3). While the cell wall damaging [P4 4 4 12]Cl led to a significant increase in most tested cell wall biosynthetic genes, neither [C10mim]Cl or cholinium decanoate altered their expression (Table 3 and Supplementary Table S7). The exceptions were chsD and fksA, that presented an increase of 1.6- and 1.3-fold (chsD) and 1.6- and 1.7-fold (fksA) for [C10mim]Cl and cholinium decanoate, respectively. These values are nonetheless much lower than those obtained after exposure to [P4 4 4 12]Cl: 3.2-fold increase for chsD and 6.6-fold for fksA. Altogether, the data revealed, as expected, that each ionic liquid induce a distinct stress over A. nidulans, yet the activation of the sphingolipid biosynthetic pathway was observed for all (Figures 2B, 4B,C). Exposure to [C10mim]Cl significantly increased the expression of lcbA, AN1102, and basA 1.8-, 2.1-, and 1.3-fold, respectively (Figure 4B and Supplementary Table S7). Cholinium decanoate increased lcbA, AN1102, AN1165, and basA 2.0-, 1.5-, 1.9-, and 1.6-fold, respectively (Figure 4C and Supplementary Table S7). As observed for [P4 4 4 12]Cl, the expression of genes involved in ceramide synthesis (barA and lagA) remained unaltered (Figures 4B,C and Supplementary Table S7), suggesting that the stress imposed by [C10mim]Cl or cholinium decanoate also led to the accumulation of sphingoid bases.
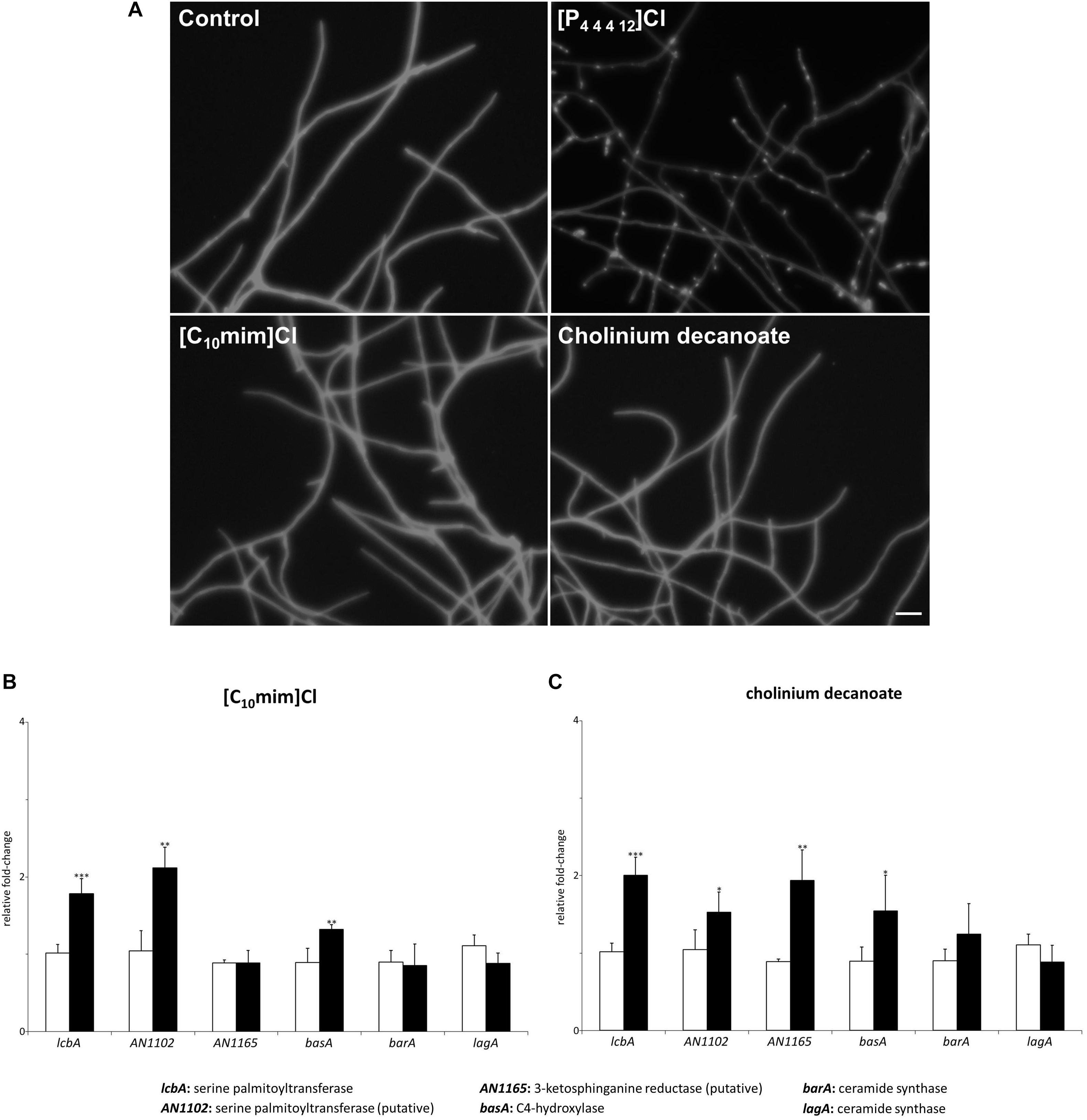
Figure 4. (A) Microscopic images of hyphae exposed to 100 mM of dodecyltributylphosphonium chloride ([P44412]Cl), 1-decyl-3-methylimidazolium chloride ([C10mim]Cl) or cholinium decanoate for 4 h and stained with calcofluor white (scale bar: 25 μM). (B,C) Expression levels of the genes mediating the first steps of sphingolipid biosynthesis after 4 h of exposure to [C10mim]Cl (B, black bars) or cholinium decanoate (C, black bars) compared to the control (white bars). Axis y represents the fold-change relative to time-zero (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
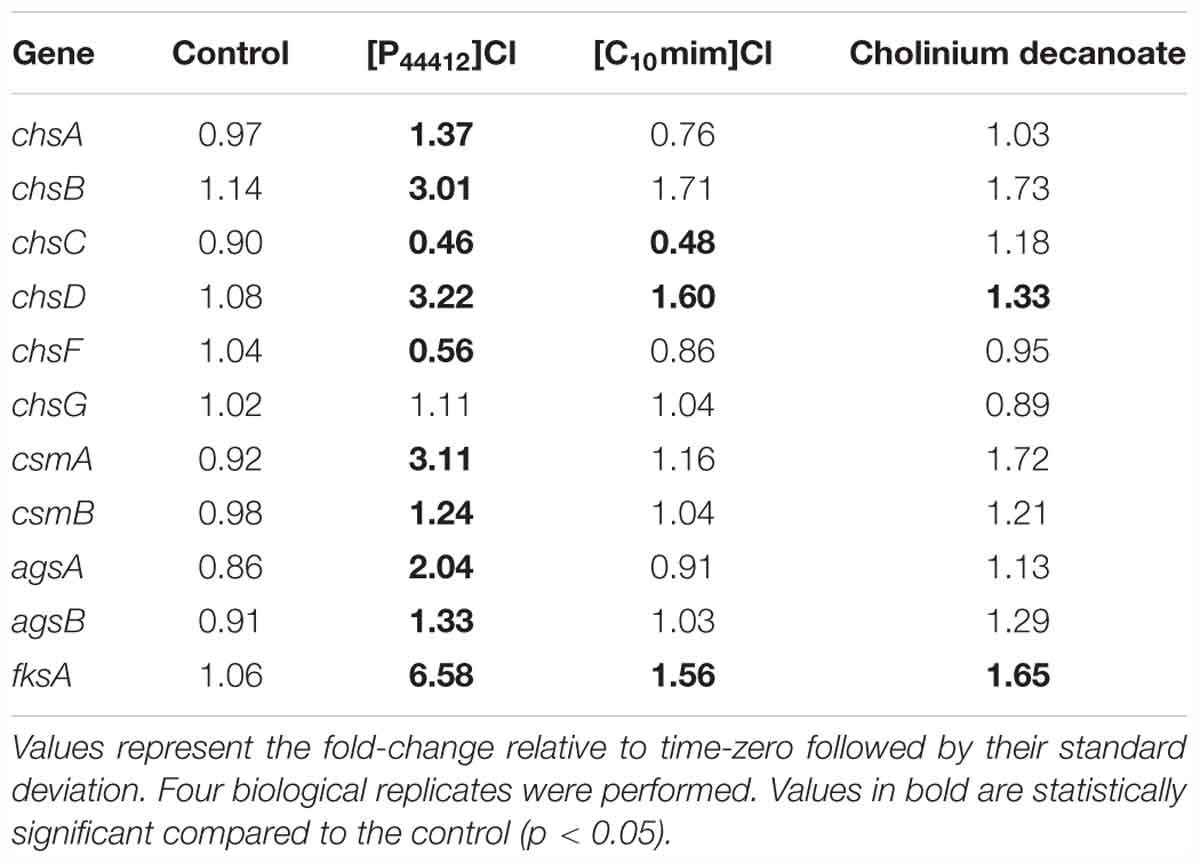
Table 3. Gene expression analysis (qRT-PCR) of genes involved in the synthesis of chitin, α-glucans and β-glucans after exposure to dodecyltributylphosphonium chloride ([P4 4 4 12]Cl), 1-decyl-3-methylimidazolium chloride ([C10mim]Cl) or cholinium decanoate for 4 h.
We additionally examined the alterations in expression of the glucosylceramide pathway genes after 4 h of exposure to the plasma membrane permeabilizing ionic liquid [C10mim]Cl. Only AN8806 had an increase (twofold) in its expression, while no increase was observed for AN4405, AN4592, AN5688, or AN7375 (Figure 5A and Supplementary Table S7). In fact, AN4592 and AN5688 significantly decreased their expression after exposure to the ionic liquid. Mutant strains of the glucosylceramide pathway displayed a higher susceptibility to the ionic liquid, as seen by lower MIC values compared to the wild-type strain (Table 1). However, when grown on solid media supplemented with [C10mim]Cl, all mutant strains showed similar susceptibility to the wild type, even at the highest concentration tested (Figure 5B), except ΔAN5688 – the only that grow when 103 conidia where inoculated in plates containing 0.1 mM of the ionic liquid. To further understand these differences, we assessed the sphingoid bases accumulation profile of the wild-type and mutant strains confronted with [C10mim]Cl for 4 h. The obtained chromatograms revealed that, unlike when A. nidulans was exposed to [P4 4 4 12]Cl (Figure 2C), no accumulation of sphingosine was noticed (Figure 5C). The lack of sphingosine accumulation is consistent with the observed unaltered expression of AN4405 after exposure to [C10mim]Cl (Figure 5A). Remarkably, wild-type and all mutant strains accumulated an unknown sphingoid base x (Figure 5C), which shows a distinct chromatographic eluting profile from phytosphingosine, sphingosine or dihydrosphingosine. The presence of sphingoid base x in all strains strongly suggests that its formation is independent of the glucosylceramide pathway. Nevertheless, the amounts accumulated of sphingoid base x were apparently correlated with the susceptibility of the mutant strains to the ionic liquid: ΔAN4405 and ΔAN4592 accumulated lower amounts and were more susceptible; and ΔAN5688, ΔAN7375 and ΔAN8806 accumulated higher amounts and were less susceptible. This suggests a potential role of sphingoid base x in signaling the membrane damage stress imposed by [C10mim]Cl.
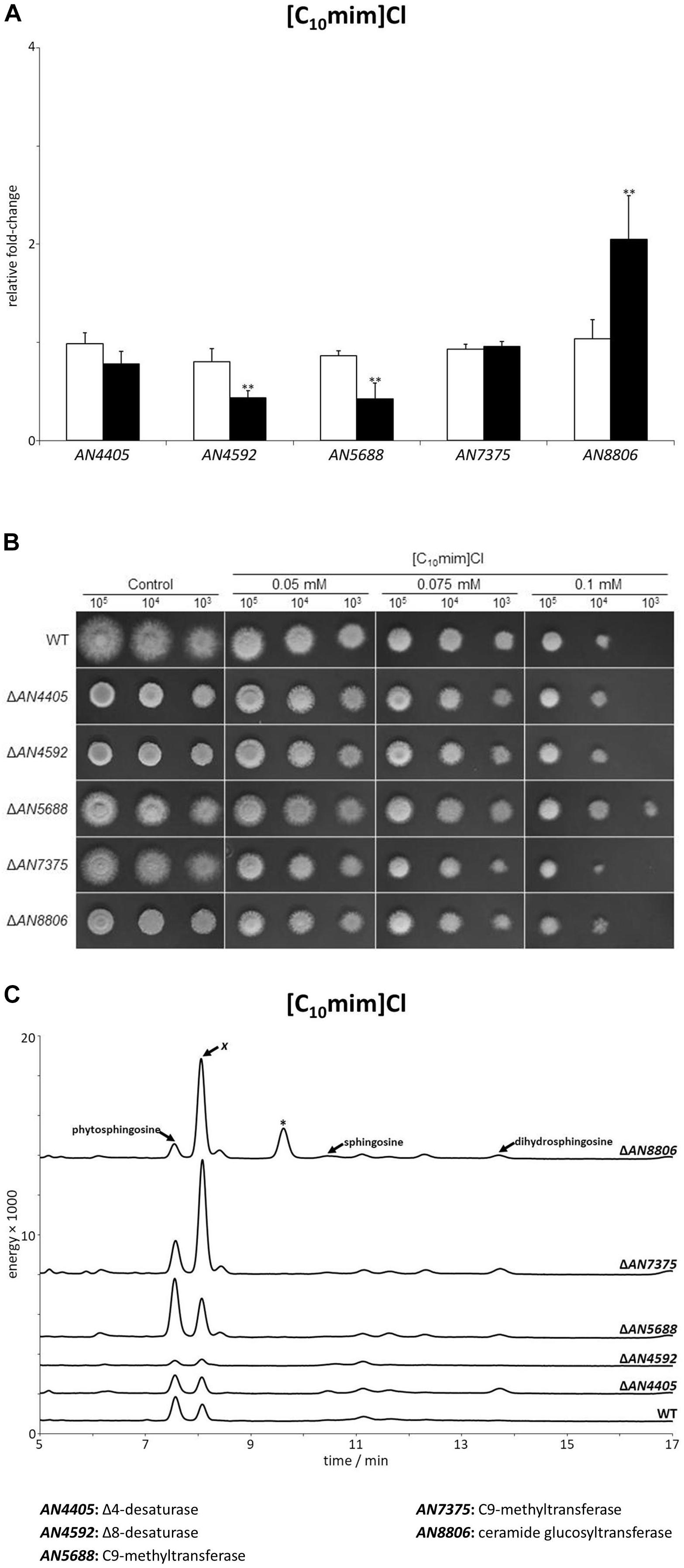
Figure 5. (A) Expression analysis of glucosylceramide pathway genes after 4 h of exposure to 1-decyl-3-methylimidazolium chloride ([C10mim]Cl, black bars), compared to the control (white bars). Axis y represents the fold-change relative to time-zero (∗∗p < 0.01). (B) Growth assessment in solid media supplemented with 0.05, 0.075, or 0.1 mM of [C10mim]Cl. 105, 104, and 103 conidia were inoculated in each plate. (C) Sphingoid bases accumulation profile of glucosylceramide pathway mutants and parental strain (wt) after 4 h of exposure to [C10mim]Cl. The peaks assigned to phytosphingosine, sphingosine and dihydrosphingosine were confirmed by their standards. The peak marked with an asterisk likely corresponds to 9-methyl-4,8-sphingadiene (more details in Supplementary Figure S5).
The last ionic liquid tested, cholinium decanoate, stands out from [P4 4 4 12]Cl and [C10mim]Cl due to its inability to cause plasma membrane permeabilization or cell wall damage (Figure 4A and Table 3), yet apparently also activating the sphingolipid biosynthetic pathway (Figure 4C). After 4 h of exposure to cholinium decanoate, only the AN8806 gene among those of the glucosylceramide pathway increased its expression, namely 2.6-fold (Figure 6A). Strains ΔAN4405, ΔAN4592, and ΔAN8806 were more susceptible to cholinium decanoate than the wild-type strain, whereas ΔAN5688 and ΔAN7375 presented a slightly higher tolerance (Table 3). A similar pattern was observed in solid media supplemented with the ionic liquid (Figure 6B). Strains ΔAN4592 and ΔAN8806 were unable to grow when 103 conidia were inoculated in plates containing 1 mM of cholinium decanoate (Figure 6B). On the other hand, we could observe some initial development of colonies for strains ΔAN5688 and ΔAN7375 when 103 conidia were inoculated on plates with the highest concentration of the ionic liquid (Figure 6B). When looking at the sphingoid base accumulation profile after 4 h of exposure to cholinium decanoate, we observe that strains with higher tolerance (ΔAN5688 and ΔAN7375) apparently presented high amounts of phytosphingosine.
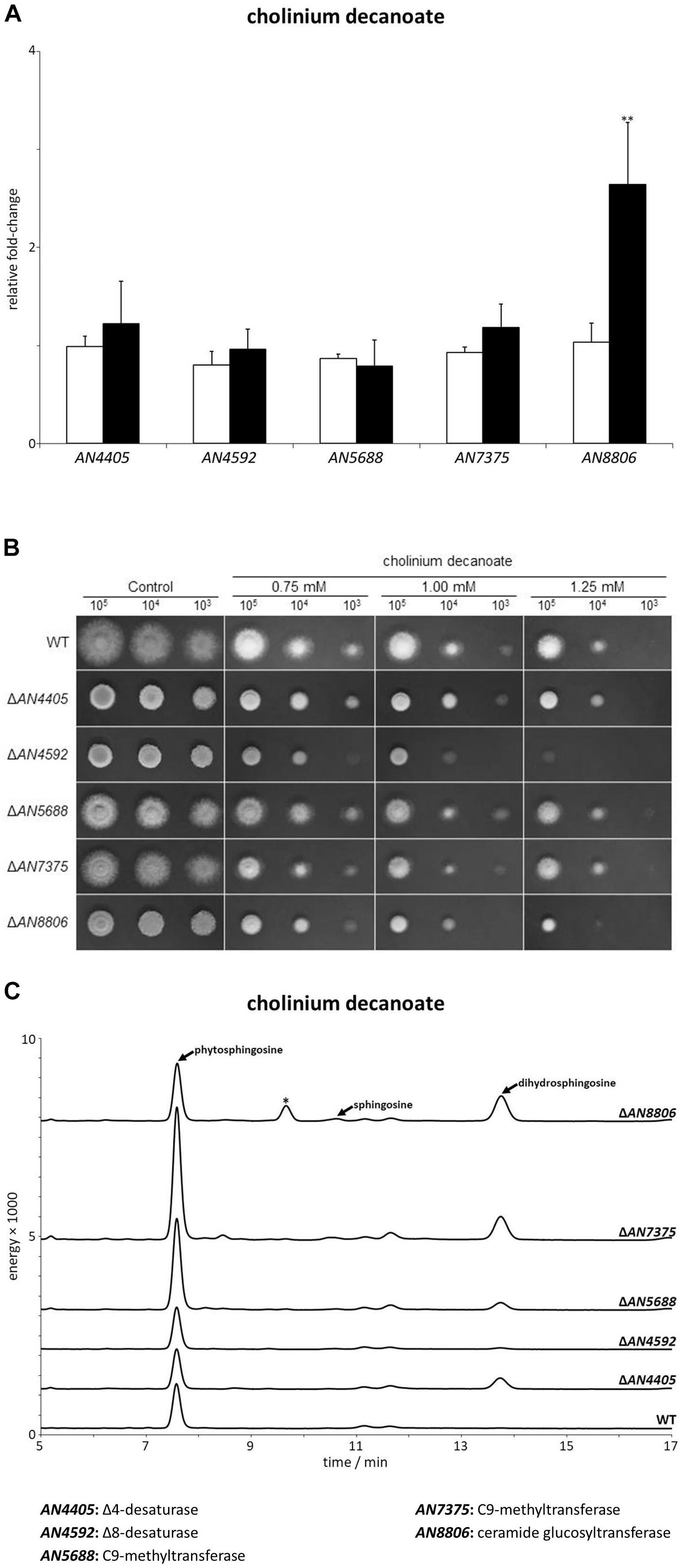
Figure 6. (A) Expression analysis of glucosylceramide pathway genes after 4 h of exposure to cholinium decanoate (black bars), compared to the control (white bars). Axis y represents the fold-change relative to time-zero (∗∗p < 0.01). (B) Growth assessment in solid media supplemented with 0.75, 1.0, or 1.25 mM of cholinium decanoate. 105, 104, and 103 conidia were inoculated in each plate. (C) Sphingoid bases accumulation profile of glucosylceramide pathway mutants and parental strain (wt) after 4 h of exposure to cholinium decanoate. The peaks assigned to phytosphingosine, sphingosine and dihydrosphingosine were confirmed by their standards. The peak marked with an asterisk likely corresponds to 9-methyl-4,8-sphingadiene (more details in Supplementary Figure S5).
In this work, we analyzed the response of A. nidulans to the stress induced by three ionic liquids, namely [P4 4 4 12]Cl, [C10mim]Cl and cholinium decanoate. Despite the distinct stresses imposed by each ionic liquid, we observed by gene expression analysis that all tested ionic liquids were able to activate the sphingolipid biosynthetic pathway, as seen by the increased expression of genes lcbA, AN1102, AN1165, and basA (Figures 2B, 4B,C). The accumulation of free sphingoid bases was also suggested due to the unaltered expression of the genes involved in synthesis of ceramides (barA and lagA). The gene expression data were validated by the chromatographic analysis of the ensuing sphingolipid extracts. Accumulation of free sphingoid bases upon the stress imposed by the ionic liquids could be a strong indicative of their role as signaling molecules. The lipid extraction protocol used in this study includes a mild alkaline hydrolysis that does not break down complex sphingolipids (Li et al., 2006). Consequently, only the free sphingoid bases are extracted but not those that are part of ceramides and of complex sphingolipids that make up cellular membranes. Both [P4 4 4 12]Cl and [C10mim]Cl led to a clear accumulation of specific free sphingoid bases: sphingosine (Figure 2C) and the unknown sphingoid base x (Figure 5C), respectively, none of which accumulated when cholinium decanoate was used instead (Figure 6C).
Upon exposure to [P4 4 4 12]Cl, A. nidulans accumulated sphingosine, suggesting involvement of this sphingoid base in the stress response to cell wall damage, the primary mechanism of toxicity of this family of ionic liquids (Petkovic et al., 2012; Hartmann and Silva Pereira, 2013). It has already been shown in S. cerevisiae that the cell wall integrity pathway can be regulated by the sphingolipid biosynthetic pathway through indirect activation of protein kinase Pkc1 by phytosphingosine (Levin, 2005). This sphingoid base has been demonstrated to be a lipid activator of protein kinase Pkh1 that in turn regulates downstream kinases including Ypk1/2 and Pkc1, which control cell wall integrity and growth, among other processes (Liu et al., 2005). Our data suggest that response to cell wall stress in A. nidulans involves the participation of sphingosine, instead of phytosphingosine. Supporting this, it has been observed that in mammals, PDK1, protein homologous to yeast Pkh1 can be activated by sphingosine (King et al., 2000). Indeed, we observed that addition of exogenous sphingosine to the growth medium increased the tolerance of the wild-type strain to [P4 4 4 12]Cl, reaching values above the original MIC (Table 2).
In fungi, sphingosine is produced in the first step of the glucosylceramide pathway (Del Poeta et al., 2014), catalyzed by a sphingolipid Δ4-desaturase. Although the following steps of the pathway have been recently identified (Fernandes et al., 2016), up to now the gene responsible for the sphingolipid Δ4-desaturase function remained uncharacterized. We generated the deletion mutant for gene AN4405, homolog to S. pombe dsd1 (Garton et al., 2003). This strain was unable to produce sphingosine and accumulated dihydrosphingosine (the precursor sphingoid base), as observed by liquid chromatography (Supplementary Figure S4). This identification is further confirmed by the increased expression of gene AN4405 by the wild-type strain exposed to [P4 4 4 12]Cl, which accumulates sphingosine (Figures 2C, 3B). Supplementation of the growth medium with exogenous sphingosine restored the ability of the mutant strain ΔAN4405 to tolerate higher concentrations of the ionic liquid, at levels similar to the wild-type (Table 2). Accumulation of sphingosine appears to be indeed related to the cell wall stress response in A. nidulans.
Imidazolium-based ionic liquids have been frequently reported to be toxic toward many organisms, from bacteria to higher multicellular ones (Petkovic et al., 2011). This is likely due to their ability to interact with and permeabilize the plasma membrane, a cellular structure common to all these organisms. Molecular dynamic simulations proposed that the imidazolium cations interact and are inserted into the lipid bilayer, with the imidazolium ring placed along with the lipid heads and the alkyl chain deep into the hydrophobic portion (Yoo et al., 2014). This incorporation not only induces structural damage to the bilayer, but also impact on the activity of membrane proteins and ions flux (Ryu et al., 2015). These have also been investigated in model lipid membranes (Galluzzi et al., 2013) and diverse living organisms, such as fungi (Hartmann et al., 2015) and worms (Charan et al., 2015). When analyzing the effects of exposure to [C10mim]Cl no accumulation of sphingosine was observed. None of the strains, either wild-type or impaired in one of the steps of the glucosylceramide pathway, have shown accumulation of this sphingoid base. However, they all presented increase production of an unknown sphingoid base x (Figure 5C). The response to the stress imposed over the plasma membrane likely involves sphingoid base x, since higher its accumulation, lesser the strain susceptibility to [C10mim]Cl. We have attempted the identification of this unknown sphingoid base by mass spectrometry but a precise assessment was not possible since this compound, which is present in low levels, was masked by fatty acids (data not shown). The liquid chromatographic method used is blind to fatty acids since the sphingoid bases are only visible after the derivatization of the primary amines (Merrill et al., 2000).
The sphingoid base x elutes after phytosphingosine, hence we believe its formation involves modifications in the dihydrosphingosine or sphingosine backbone, e.g., addition of a double bond distinct from the Δ8-desaturation; this hypothesis will be further analyzed in the near future. The diversity of sphingoid bases already identified in plants includes compounds, e.g., formed through the addition of double bonds in the phytosphingosine backbone (Markham et al., 2006). Moreover, unusual modifications such as the presence of a Δ10-desaturation in a 4,8-sphingadiene backbone has been observed in the diatom Thalassiosira pseudonana (Michaelson et al., 2013) and, more recently, in F. graminearum (Tian et al., 2015). These recent reports reinforce the idea that uncommon structural modifications may exist in A. nidulans.
The last tested ionic liquid, cholinium decanoate, apparently did not provoke accumulation of any sphingoid base (Figure 6C). Although great amounts of phytosphingosine were present in the strains with higher tolerance to the ionic liquid (ΔAN5688 and ΔAN7375), higher amounts of this sphingoid base were also noticed in these mutant strains without any stress imposition (Supplementary Figure S5). Strain ΔAN4592 revealed to be the most susceptible to the ionic liquid stress, although no correlation could be made with the sphingoid base accumulation profile. It is possible that downstream effectors, not seen by the methodologies employed, could be involved in this increased susceptibility. Deeper understanding of the ionic liquid toxicity could help solving this question. Previous studies have shown that A. nidulans decreases cholinium decanoate concentrations in the growth medium along time (Hartmann et al., 2015) and that filamentous fungi can uptake the cholinium cation (Markham et al., 1993). A proteomic study by our group shed light on the mechanism of toxicity of cholinium-based ionic liquids (Martins et al., 2013). It was shown that major uptake of the cholinium cation led to the production of the toxic compound cyanide, which might be a cause of the observed growth inhibition provoked by this group of ionic liquids (Martins et al., 2013). Upon exposure to cholinium decanoate, neither sphingosine nor sphingoid base x were accumulated, further supporting the involvement and specificity of these sphingoid bases in signaling the stress imposed by cell wall damage or plasma membrane permeabilization, respectively.
Sphingoid bases – intermediates of the sphingoid biosynthetic pathway – have been demonstrated to play significant roles as signaling molecules in fungi, yet many aspects remain poorly understood. Our study sheds light to the roles of specific sphingoid bases in stress responses, namely sphingosine functioning in response to cell wall damage and of a yet unknown sphingoid base (possibly formed from a sphingosine or dihydrosphingosine backbone) in response to membrane damage. In addition, we also fill one important gap in the genetic assignment of the glucosylceramide pathway in A. nidulans through the identification of the sphingolipid Δ4-desaturase gene (AN4405). Our findings were made possible through the use of ionic liquids – compounds yet largely unexplored in biological sciences. Nonetheless, the structural diversity and tunable physical and chemical properties of these classes of compounds may provide a diversity of useful stimuli, such as cell wall damage, plasma membrane permeabilization and particular alterations in metabolism, to deepen our understanding of stress signaling responses in fungi. It comes without saying that the knowledge established reinforces the idea of targeting sphingolipids biosynthesis for the search of improved antifungal compounds.
DH contributed to planning, execution and analysis of all experiments. DP contributed to generation of mutants and respective experiments. CM contributed to experiments with dodecyltributylphosphonium chloride and wild-type strain. DH and CSP wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
We acknowledge funding from the European Research Council through grant ERC 2014-CoG-647928 and from Fundação para a Ciência e Tecnologia (FCT) through the grant UID/Multi/04551/2013 (Research unit GREEN-it “Bioresources for Sustainability”) and PTDC/AGR-TEC/1191/2014. DH is thankful to FCT for grant SFRH/BPD/121354/2016.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are extremely thankful to Maria C. Leitão (ITQB NOVA) for the support in the chromatographic analyses.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00864/full#supplementary-material
Adamová, G., Gardas, R. L., Nieuwenhuyzen, M., Puga, A. V., Rebelo, L. P. N., Robertson, A. J., et al. (2012). Alkyltributylphosphonium chloride ionic liquids: synthesis, physicochemical properties and crystal structure. Dalton Trans. 41, 8316–8332. doi: 10.1039/c1dt10466g
Alves, P. C., Hartmann, D. O., Núñez, O., Martins, I., Gomes, T. L., Garcia, H., et al. (2016). Transcriptomic and metabolomic profiling of ionic liquid stimuli unveils enhanced secondary metabolism in Aspergillus nidulans. BMC Genomics 17:284. doi: 10.1186/s12864-016-2577-6
Araujo, R., Pina-Vaz, C., and Rodrigues, A. G. (2010). “Mould infections: a global threat to immunocompromised patients,” in Combating Fungal Infections: Problems and Remedy, eds I. Ahmad, M. Owais, M. Shahid, and F. Aqil (Berlin: Springer), 1–21.
Arendrup, M. C., and Perlin, D. S. (2014). Echinocandin resistance: an emerging clinical problem? Curr. Opin. Infect. Dis. 27, 484–492. doi: 10.1097/qco.0000000000000111
Beeler, T., Bacikova, D., Gable, K., Hopkins, L., Johnson, C., Slife, H., et al. (1998). The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Delta mutant. J. Biol. Chem. 273, 30688–30694.
Brown, G. D., Denning, D. W., Gow, N. A. R., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv13. doi: 10.1126/scitranslmed.3004404
Charan, K. T. P., Ranjan, P., Manojkumar, K., Pothanagandhi, N., Jha, P. C., Khedkar, V. M., et al. (2015). Evaluation of imidazolium-based ionic liquids towards vermicidal activity: in vitro & in silico studies. RSC Adv. 5, 75415–75424. doi: 10.1039/c5ra13469b
Cheng, J., Park, T.-S., Chio, L.-C., Fischl, A. S., and Xiang, S. Y. (2003). Induction of apoptosis by sphingoid long-chain bases in Aspergillus nidulans. Mol. Cell. Biol. 23, 163–177. doi: 10.1128/MCB.23.1.163-177.2003
Cheng, J. J., Park, T. S., Fischl, A. S., and Ye, X. S. (2001). Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans. Mol. Cell. Biol. 21, 6198–6209. doi: 10.1128/mcb.21.18.6198-6209.2001
Del Poeta, M., Nimrichter, L., Rodrigues, M. L., and Luberto, C. (2014). Synthesis and biological properties of fungal glucosylceramide. PLoS Pathog. 10:e1003832. doi: 10.1371/journal.ppat.1003832
Denning, D. W., and Bromley, M. J. (2015). How to bolster the antifungal pipeline. Science 347, 1414–1416. doi: 10.1126/science.aaa6097
Dickson, R. C., and Lester, R. L. (2002). Sphingolipid functions in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1583, 13–25. doi: 10.1016/S1388-1981(02)00210-X
Dong, Q., Muzny, C. D., Kazakov, A., Diky, V., Magee, J. W., Widegren, J. A., et al. (2007). ILThermo: a free-access web database for thermodynamic properties of ionic liquids. J. Chem. Eng. Data 52, 1151–1159. doi: 10.1021/je700171f
Fernandes, C., de Castro, P., Singh, A., Fonseca, F., Pereira, M., Vila, T., et al. (2016). Functional characterization of the Aspergillus nidulans glucosylceramide pathway reveals that LCB Δ8-desaturation and C9-methylation are relevant to filamentous growth, lipid raft localization and Psd1 defensin activity. Mol. Microbiol. 102, 488–505. doi: 10.1111/mmi.13474
Fernandes, C. M., Goldman, G. H., and Del Poeta, M. (2018). Biological roles played by sphingolipids in dimorphic and filamentous fungi. mBio 9:e00642-18. doi: 10.1128/mBio.00642-18
Fujioka, T., Mizutani, O., Furukawa, K., Sato, N., Yoshimi, A., Yamagata, Y., et al. (2007). MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 6, 1497–1510. doi: 10.1128/ec.00281-06
Galluzzi, M., Zhang, S., Mohamadi, S., Vakurov, A., Podestà, A., and Nelson, A. (2013). Interaction of imidazolium-based room-temperature ionic liquids with DOPC phospholipid monolayers: electrochemical study. Langmuir 29, 6573–6581. doi: 10.1021/la400923d
Garton, S., Michaelson, L. V., Beaudoin, F., Beale, M. H., and Napier, J. A. (2003). The dihydroceramide desaturase is not essential for cell viability in Schizosaccharomyces pombe. FEBS Lett. 538, 192–196. doi: 10.1016/S0014-5793(03)00163-7
Haak, D., Gable, K., Beeler, T., and Dunn, T. (1997). Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272, 29704–29710. doi: 10.1074/jbc.272.47.29704
Hannun, Y. A., and Obeid, L. M. (2008). Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9, 139–150. doi: 10.1038/nrm2329
Hartmann, D. O., Petkovic, M., and Silva Pereira, C. (2016). Ionic liquids as unforeseen assets to fight life-threatening mycotic diseases. Front. Microbiol. 7:111. doi: 10.3389/fmicb.2016.00111
Hartmann, D. O., Shimizu, K., Siopa, F., Leitão, M. C., Afonso, C. A., Lopes, J. N. C., et al. (2015). Plasma membrane permeabilisation by ionic liquids: a matter of charge. Green Chem. 17, 4587–4598. doi: 10.1039/C5GC01472G
Hartmann, D. O., and Silva Pereira, C. (2013). A molecular analysis of the toxicity of alkyltributylphosphonium chlorides in Aspergillus nidulans. New J. Chem. 37, 1569–1577. doi: 10.1039/C3NJ00167A
Hartmann, D. O., and Silva Pereira, C. (2016). “Toxicity of ionic liquids: past, present and future,” in Ionic Liquids in Lipid Processing and Analysis: Opportunities and Challenges, eds X. Xu, Z. Guo, and L.-Z. Cheong (Urbana, IL: AOCS Press), 403–421.
Jenkins, G. M., Richards, A., Wahl, T., Mao, C., Obeid, L., and Hannun, Y. (1997). Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 272, 32566–32572. doi: 10.1074/jbc.272.51.32566
Kathiravan, M. K., Salake, A. B., Chothe, A. S., Dudhe, P. B., Watode, R. P., Mukta, M. S., et al. (2012). The biology and chemistry of antifungal agents: a review. Bioorg. Med. Chem. 20, 5678–5698. doi: 10.1016/j.bmc.2012.04.045
King, C. C., Zenke, F. T., Dawson, P. E., Dutil, E. M., Newton, A. C., Hemmings, B. A., et al. (2000). Sphingosine is a novel activator of 3-phosphoinositide-dependent kinase 1. J. Biol. Chem. 275, 18108–18113. doi: 10.1074/jbc.M909663199
Levery, S. B., Momany, M., Lindsey, R., Toledo, M. S., Shayman, J. A., Fuller, M., et al. (2002). Disruption of the glucosylceramide biosynthetic pathway in Aspergillus nidulans and Aspergillus fumigatus by inhibitors of UDP-Glc: ceramide glucosyltransferase strongly affects spore germination, cell cycle, and hyphal growth. FEBS Lett. 525, 59–64. doi: 10.1016/S0014-5793(02)03067-3
Levin, D. E. (2005). Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291. doi: 10.1128/mmbr.69.2.262-291.2005
Li, S. J., Bao, D. P., Yuen, G., Harris, S. D., and Calvo, A. M. (2007). basA regulates cell wall organization and asexual/sexual sporulation ratio in Aspergillus nidulans. Genetics 176, 243–253. doi: 10.1534/genetics.106.068239
Li, S. J., Du, L. C., Yuen, G., and Harris, S. D. (2006). Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 17, 1218–1227. doi: 10.1091/mbc.E05-06-0533
Liu, K., Zhang, X. P., Lester, R. L., and Dickson, R. C. (2005). The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 280, 22679–22687. doi: 10.1074/jbc.M502972200
Markham, J. E., Li, J., Cahoon, E. B., and Jaworski, J. G. (2006). Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281, 22684–22694. doi: 10.1074/jbc.m604050200
Markham, P., Robson, G. D., Bainbridge, B. W., and Trinci, A. P. (1993). Choline: its role in the growth of filamentous fungi and the regulation of mycelial morphology. FEMS Microbiol. Lett. 104, 287–300. doi: 10.1016/0378-1097(93)90601-w
Martins, I., Hartmann, D. O., Alves, P. C., Planchon, S., Renaut, J., Leitão, M. C., et al. (2013). Proteomic alterations induced by ionic liquids in Aspergillus nidulans and Neurospora crassa. J. Proteomics 94, 262–278. doi: 10.1016/j.jprot.2013.09.015
Merrill, A. H. (2002). De novo sphingolipid biosynthesis: a necessary, but dangerous, pathway. J. Biol. Chem. 277, 25843–25846. doi: 10.1074/jbc.r200009200
Merrill, A. H., Caligan, T. B., Wang, E., Peters, K., and Joyce, O. (2000). “Analysis of sphingoid bases and sphingoid base 1-Phosphates by high-performance liquid chromatography,” in Methods in Enzymology, eds A. H. Merrill and Y. A. Hannun (London: Academic Press), 3–9. doi: 10.1016/s0076-6879(00)12894-0
Michaelson, L. V., Markham, J. E., Zäeuner, S., Matsumoto, M., Chen, M., Cahoon, E. B., et al. (2013). Identification of a cytochrome b5-fusion desaturase responsible for the synthesis of triunsaturated sphingolipid long chain bases in the marine diatom Thalassiosira pseudonana. Phytochemistry 90, 50–55. doi: 10.1016/j.phytochem.2013.02.010
Nagiec, M. M., Baltisberger, J. A., Wells, G. B., Lester, R. L., and Dickson, R. C. (1994). The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc. Natl. Acad. Sci. U.S.A. 91, 7899–7902. doi: 10.1073/pnas.91.17.7899
Oura, T., and Kajiwara, S. (2008). Disruption of the sphingolipid Δ8-desaturase gene causes a delay in morphological changes in Candida albicans. Microbiology 154, 3795–3803. doi: 10.1099/mic.0.2008/018788-0
Perfect, J. R. (2017). The antifungal pipeline: a reality check. Nat. Rev. Drug Discov. 16, 603–616. doi: 10.1038/nrd.2017.46
Petkovic, M., Ferguson, J. L., Bohn, A., Trindade, J., Martins, I., Carvalho, M. B., et al. (2009). Exploring fungal activity in the presence of ionic liquids. Green Chem. 11, 889–894. doi: 10.1039/B823225C
Petkovic, M., Ferguson, J. L., Gunaratne, H. N., Ferreira, R., Leitao, M. C., Seddon, K. R., et al. (2010). Novel biocompatible cholinium-based ionic liquids - toxicity and biodegradability. Green Chem. 12, 643–649.
Petkovic, M., Hartmann, D. O., Adamová, G., Seddon, K. R., Rebelo, L. P. N., and Silva Pereira, C. (2012). Unravelling the mechanism of toxicity of alkyltributylphosphonium chlorides in Aspergillus nidulans conidia. New J. Chem. 36, 56–63. doi: 10.1039/c1nj20470j
Petkovic, M., Seddon, K. R., Rebelo, L. P. N., and Silva Pereira, C. (2011). Ionic liquids: a pathway to environmental acceptability. Chem. Soc. Rev. 40, 1383–1403. doi: 10.1039/c004968a
Petkovic, M., and Silva Pereira, C. (2012). “Pioneering biological processes in the presence of ionic liquids: the potential of filamentous fungi,” in Ionic Liquids UnCOILed: Critical Expert Overviews, eds N. V. Plechkova and K. R. Seddon (Hoboken, NJ: John Wiley & Sons, Inc.), 283–303. doi: 10.1002/9781118434987.ch9
Ramamoorthy, V., Cahoon, E. B., Thokala, M., Kaur, J., Li, J., and Shah, D. M. (2009). Sphingolipid C-9 methyltransferases are important for growth and virulence but not for sensitivity to antifungal plant defensins in Fusarium graminearum. Eukaryot. Cell 8, 217–229. doi: 10.1128/EC.00255-08
Richardson, M. D., and Warnock, D. W. (eds). (2012). “Fungi as human pathogens,” in Fungal Infection: Diagnosis and Management (Malden, MA: Wiley-Blackwell), 5–7.
Roemer, T., and Krysan, D. J. (2014). Antifungal drug development: challenges, unmet clinical needs, and new approaches. CSH Perspect. Med. 4:a019703. doi: 10.1101/cshperspect.a019703
Ryu, H., Lee, H., Iwata, S., Choi, S., Ki Kim, M., Kim, Y.-R., et al. (2015). Investigation of ion channel activities of gramicidin A in the presence of ionic liquids using model cell membranes. Sci. Rep. 5:11935. doi: 10.1038/srep11935
Singh, A., and Del Poeta, M. (2016). Sphingolipidomics: an important mechanistic tool for studying fungal pathogens. Front. Microbiol. 7:501. doi: 10.3389/fmicb.2016.00501
Singh, A., Wang, H., Silva, L. C., Na, C., Prieto, M., Futerman, A. H., et al. (2012). Methylation of glycosylated sphingolipid modulates membrane lipid topography and pathogenicity of Cryptococcus neoformans. Cell. Microbiol. 14, 500–516. doi: 10.1111/j.1462-5822.2011.01735.x
Slavin, M., van Hal, S., Sorrell, T. C., Lee, A., Marriott, D. J., Daveson, K., et al. (2015). Invasive infections due to filamentous fungi other than Aspergillus: epidemiology and determinants of mortality. Clin. Microbiol. Infect. 21, 490.e1–490.e10. doi: 10.1016/j.cmi.2014.12.021
Stark, A., and Seddon, K. R. (2007). “Ionic liquids,” in Kirk-Othmer Encyclopaedia of Chemical Technology, ed. A. Seidel (Hoboken, NJ: John Wiley & Sons, Inc.), 836–920.
Szewczyk, E., Nayak, T., Oakley, C. E., Edgerton, H., Xiong, Y., Taheri-Talesh, N., et al. (2006). Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111–3120. doi: 10.1038/nprot.2006.405
Tian, Y., Zhao, G. Y., Fang, W., Xu, Q., and Tan, R. X. (2015). Δ10(E)-sphingolipid desaturase involved in fusaruside mycosynthesis and stress adaptation in Fusarium graminearum. Sci. Rep. 5:10486. doi: 10.1038/srep10486
Valiante, V., Macheleidt, J., Föge, M., and Brakhage, A. A. (2015). The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front. Microbiol. 6:325. doi: 10.3389/fmicb.2015.00325
Vermes, A., Guchelaar, H.-J., and Dankert, J. (2000). Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 46, 171–179. doi: 10.1093/jac/46.2.171
Yoo, B., Shah, J. K., Zhu, Y., and Maginn, E. J. (2014). Amphiphilic interactions of ionic liquids with lipid biomembranes: a molecular simulation study. Soft Matter 10, 8641–8651. doi: 10.1039/c4sm01528b
Keywords: ionic liquids, cell wall, plasma membrane, sphingolipids, sphingoid base
Citation: Hartmann DO, Piontkivska D, Moreira CJS and Silva Pereira C (2019) Ionic Liquids Chemical Stress Triggers Sphingoid Base Accumulation in Aspergillus nidulans. Front. Microbiol. 10:864. doi: 10.3389/fmicb.2019.00864
Received: 09 January 2019; Accepted: 04 April 2019;
Published: 24 April 2019.
Edited by:
Gustavo Henrique Goldman, University of São Paulo, BrazilReviewed by:
Siddhartha Das, The University of Texas at El Paso, United StatesCopyright © 2019 Hartmann, Piontkivska, Moreira and Silva Pereira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina Silva Pereira, c3BlcmVpcmFAaXRxYi51bmwucHQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.