- 1Department of Geography and Environment, Mount Allison University, Sackville, NB, Canada
- 2Department of Mathematics and Computer Science, Mount Allison University, Sackville, NB, Canada
- 3Department of Earth, Atmospheric and Planetary Science, Massachusetts Institute of Technology, Cambridge, MA, United States
Biogeochemical cycles in the ocean are strongly affected by the elemental stoichiometry (C:N:P) of phytoplankton, which largely reflects their macromolecular content. A greater understanding of how this macromolecular content varies among phytoplankton taxa and with resource limitation may strengthen physiological and biogeochemical modeling efforts. We determined the macromolecular basis (protein, carbohydrate, lipid, nucleic acids, pigments) of C:N:P in diatoms and prasinophytes, two globally important phytoplankton taxa, in response to N starvation. Despite their differing cell sizes and evolutionary histories, the relative decline in protein during N starvation was similar in all four species studied and largely determined variations in N content. The accumulation of carbohydrate and lipid dominated the increase in C content and C:N in all species during N starvation, but these processes differed greatly between diatoms and prasinophytes. Diatoms displayed far greater accumulation of carbohydrate with N starvation, possibly due to their greater cell size and storage capacity, resulting in larger increases in C content and C:N. In contrast, the prasinophytes had smaller increases in C and C:N that were largely driven by lipid accumulation. Variation in C:P and N:P was species-specific and mainly determined by residual P pools, which likely represent intracellular storage of inorganic P and accounted for the majority of cellular P in all species throughout N starvation. Our findings indicate that carbohydrate and lipid accumulation may play a key role in determining the environmental and taxonomic variability in phytoplankton C:N. This quantitative assessment of macromolecular and elemental content spanning several marine phytoplankton species can be used to develop physiological models for ecological and biogeochemical applications.
Introduction
The elemental stoichiometry (C:N:P) of phytoplankton and particulate organic matter (POM) in the surface ocean is often assumed to conform to the Redfield ratio of 106:16:1 (Redfield, 1958). However, the C:N:P of phytoplankton and of surface POM, largely derived from phytoplankton and their detritus, can greatly deviate from Redfield proportions (Quigg et al., 2003; Martiny et al., 2013a,b; Garcia et al., 2018). This variability in phytoplankton C:N:P may influence key biogeochemical cycles (Finkel et al., 2010) by affecting how efficiently phytoplankton biomass is remineralized by bacteria (Del Giorgio and Cole, 1998), exported to the deep ocean (Weber and Deutsch, 2010), or utilized by consumers (Sterner and Elser, 2002; Hessen et al., 2004). The C:N:P of phytoplankton reflects their macromolecular content (e.g., protein, carbohydrate, lipid, and nucleic acids; summarized in Figure 1; Elser et al., 2000; Geider and LaRoche, 2002; Finkel et al., 2016), which varies among major phytoplankton groups (Finkel et al., 2016) and within a species in response to environmental conditions (Geider and LaRoche, 2002). This phylogenetic and physiological variability in phytoplankton macromolecules may provide a mechanistic basis for modeling phytoplankton and ocean particulate C:N:P (Geider and LaRoche, 2002; Follows and Dutkiewicz, 2011).
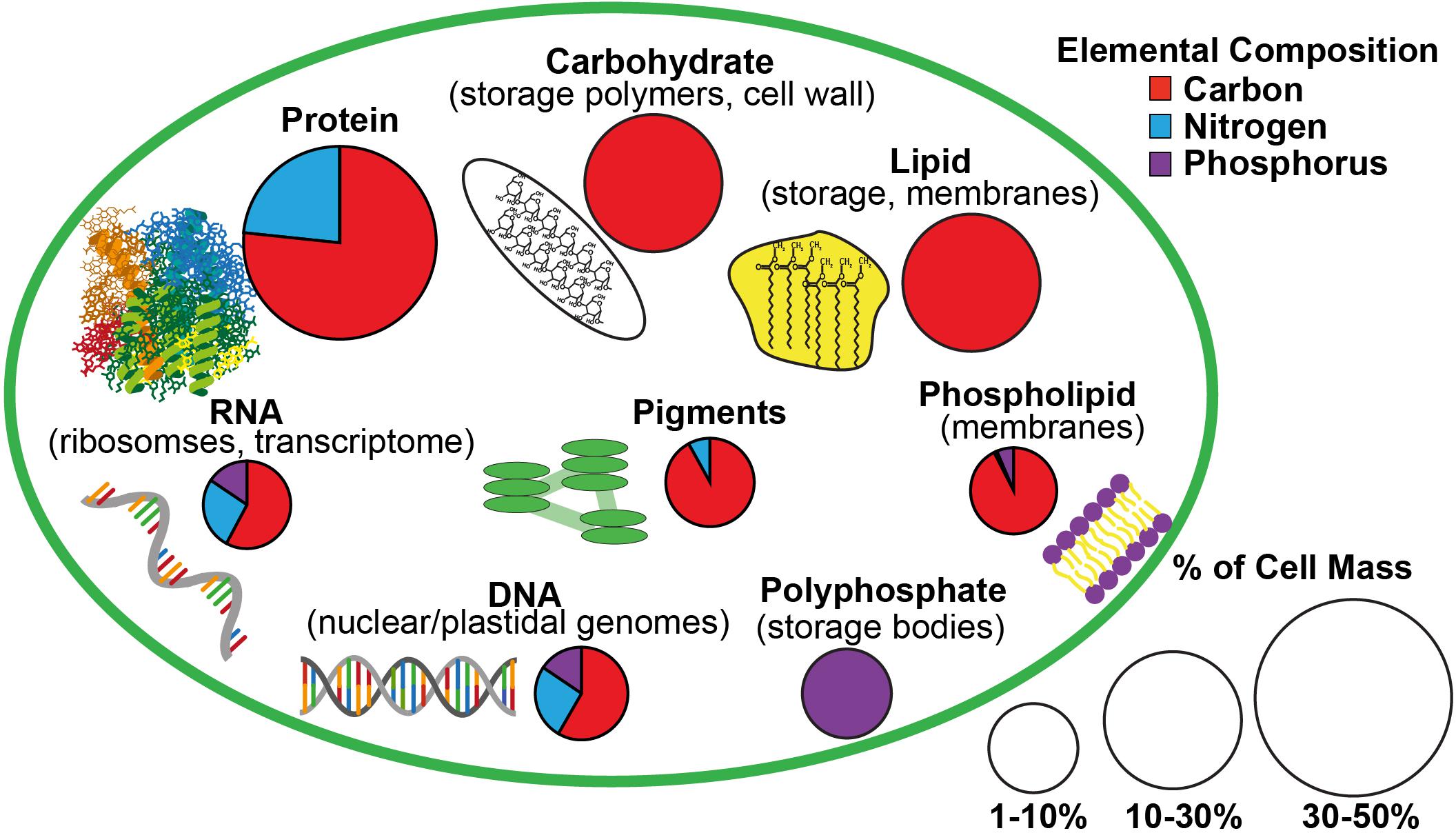
Figure 1. A schematic of the estimated macromolecular content of a microalgal cell and the allocation of carbon, nitrogen, and phosphorus to each macromolecular pool based on Geider and LaRoche (2002) and Dyhrman (2016). The size of the pie charts indicate the estimated contribution of each macromolecular pool to total cell mass while the portions of the pie charts indicate the relative content of carbon, nitrogen, and phosphorus in each type of macromolecule.
Fixed nitrogen is the limiting factor for phytoplankton growth over much of the global ocean (Moore et al., 2013) and its availability affects the C:N:P of phytoplankton cells (Geider and LaRoche, 2002; Hillebrand et al., 2013). When N limits phytoplankton growth, C:N tends to increase and N:P tends to decline (Rhee, 1978; Goldman et al., 1979; Elrifi and Turpin, 1985; Leonardos and Geider, 2004; Garcia et al., 2016). The macromolecular basis of this response is a decline in cell protein content and a reallocation of remaining protein content from photosynthetic and biosynthetic (e.g., ribosomes) components to N metabolism and acquisition (Young and Beardall, 2003; Hockin et al., 2012; Simionato et al., 2013). Nitrogen stress also tends to result in the accumulation of C and energy as carbohydrate or lipid (Piorreck et al., 1984; Breuer et al., 2012; Zhu et al., 2014). Many culture studies of phytoplankton C:N:P have examined responses to balanced, steady-state N limitation, but nutrient stress also occurs as unbalanced nutrient starvation and arrested growth due to rapid nutrient fluctuations in dynamic ocean environments (Goldman, 1988; Legendre and Rassoulzadegan, 1995). Unbalanced N starvation may also have a distinct effect on phytoplankton C:N:P as it has been shown to cause larger accumulations of C-rich lipid (Richardson et al., 1969; Lacour et al., 2012b) and larger decreases in N-rich proteins (Geider et al., 1993; Berges et al., 1996; Geider et al., 1998) compared to steady-state N limitation (Cullen et al., 1992; Halsey et al., 2013).
Storage macromolecules may play a particularly large role in the variability of phytoplankton C:N:P as they can represent large fractions of these elements and their utilization differs among phytoplankton taxa (Allen, 1984; Geider and LaRoche, 2002; Gillooly et al., 2005). The accumulation of C-rich carbohydrate and lipid storage during N stress varies among phytoplankton taxa, both in extent and in the relative distribution between these two pools (Piorreck et al., 1984; Gatenby et al., 2003; Breuer et al., 2012). A preference for carbohydrate or lipid storage may affect a species’ growth strategy as these storage products differ in their energetic efficiency (Sorguven and Ozilgen, 2013), utilization of intracellular space (Subramanian et al., 2013), and effect on cell buoyancy (Raven, 1984; Richardson and Cullen, 1995). Additionally, the few comprehensive studies of phytoplankton P allocation show the majority of total P is not accounted for by P-rich functional macromolecules (RNA, DNA, and phospholipids), particularly under N-limited conditions (Rhee, 1978; Leonardos and Geider, 2004; Mouginot et al., 2015; Garcia et al., 2016). The large amount of residual P not detected in these functional macromolecules is typically assumed to be in storage pools like polyphosphate bodies and orthophosphate in vacuoles that are difficult to quantify (Miyata et al., 1986; Diaz and Ingall, 2010; Martin et al., 2014; Dyhrman, 2016). The effect of C and P storage on phytoplankton C:N:P may vary with cell size as storage capacity may be greater in larger cells (Grover, 1991; Tozzi et al., 2004; Gillooly et al., 2005) and a greater benefit to larger cells in dynamic light and nutrient environments (Talmy et al., 2014).
Using macromolecular variation as a mechanistic basis to explain ocean C:N:P variability requires a comprehensive understanding of how elemental allocation to macromolecules differs among phytoplankton taxa and ocean conditions. Here we examine the effect of N starvation on phytoplankton elemental and macromolecular content accounting for all major C- and N-containing pools (protein, carbohydrate, lipid, pigments). We measured the major functional P-containing pools (RNA, DNA, phospholipids) and total P and describe the P not attributed to measured functional pools as residual P, which may be intracellular storage. Cell size and evolutionary history likely affect macromolecular storage capacity and adaptation to different nutrient regimes may affect the overall plasticity of macromolecular content across species. Hence we examined two phytoplankton classes, diatoms and prasinophytes, which represent distinct size classes and adaptation to different environmental niches. Diatoms are typically opportunists that exploit dynamic nutrient conditions (Sarthou et al., 2005) while the smaller prasinophytes appear to be adapted to more stable nutrient conditions (Cardol et al., 2008; Six et al., 2009). Additionally, C:N appears to be lower in prasinophytes than diatoms during nutrient replete conditions (Quigg et al., 2003; Garcia et al., 2018), possibly due to a relatively higher protein content in prasinophytes (Finkel et al., 2016). Portions of the macromolecular response to steady-state N limitation in diatoms and prasinophytes have been well characterized (Leonardos and Geider, 2004; Halsey et al., 2014; Halsey and Jones, 2015), yet it is unclear how the macromolecular response of these groups to non-steady state N starvation may differ. We anticipated that diatoms would display greater C:N:P variability as their larger cell size may allow more storage accumulation of carbohydrates and lipids and their adaptation to dynamic nutrient regimes may allow more variation in their N-rich functional macromolecules (protein, RNA, and pigments). We also hypothesized that protein, the largest fraction of phytoplankton N, would be the most variable macromolecular pool during N starvation and dominate variation in C:N and N:P. We find dramatic differences between diatoms and prasinophytes with regard to the impact of N starvation on C:N:P ratios, largely due to varying utilization of C-rich storage macromolecules and species-specific differences in P pools likely associated with storage.
Materials and Methods
Study Species and Growth Conditions
The diatoms Thalassiosira pseudonana (strain CCMP 1335) and Thalassiosira weissflogii (strain CCMP 1010) as well as the Arctic prasinophyte Micromonas sp. (strain CCMP 2099) were obtained from the National Center for Marine Algae and Microbiota (NCMA). The prasinophyte Ostreococcus tauri (strain OTH95, RCC745) was obtained from the Roscoff Culture Collection (RCC). The strains of O. tauri (mean cell volume of 1.8 ± 0.3 μm3) and T. pseudonana (mean cell volume of 158 ± 23 μm3) used are both coastal isolates while the strains of Micromonas sp. (mean cell volume of 1.8 ± 0.3 μm3) and T. weissflogii (mean cell volume of 1630 ± 215 μm3) were isolated from shelf waters of the Arctic Ocean and the Gulf Stream of the North Atlantic, respectively. Cell volumes for each strain were based on measurements of a minimum of 50 live, unstained nutrient-replete cells observed by light microscopy and assuming a cylindrical form for diatoms and a spherical form for both prasinophytes (Hillebrand et al., 1999).
All cultures were grown under a irradiance of 85 μmol photons m-2 s-1 provided by cool white fluorescent bulbs on a 12:12 light:dark cycle and a temperature of 18°C with the exception of Micromonas sp., which was grown at 6°C. These conditions have been shown to be at subsaturating growth irradiance and near optimum growth temperatures for each study species (Thompson, 1999; Strzepek and Price, 2000; Lovejoy et al., 2007; Six et al., 2008). Growth media was prepared from natural seawater (Cape Tormentine, Canada) with a salinity of ∼32 ppt and amended with half the f/2 concentrations (Guillard and Ryther, 1962; Guillard, 1975) of sodium phosphate, sodium silicate (not used for prasinophytes), trace metals, and vitamins. Media was further amended with 2 mM sodium bicarbonate and 60 or 120 μM sodium nitrate, respectively, for diatoms and prasinophytes. The lower nitrogen (N) concentration was necessary for diatom cultures to ensure carbon-replete conditions and a maximum pH of less than 9 at all growth phases. Media was adjusted to a pH of 7.95–8.00 with HCl and filter sterilized (Pall Acropak 0.8/0.2 μm capsule filter) before use. Cultures were maintained in 5 L glass bottles (Pyrex) and mixed by stirring with PTFE stir bars at ∼60 RPM and continuous bubbling with filter-sterilized (VWR, 0.2 μm PES syringe filter) air. To assess cell composition during nutrient-replete balanced exponential growth, all cultures were maintained as optically thin, semi-continuous batch cultures and were considered to be fully acclimated to these conditions after a minimum of 10 generations with less than 15% variation in growth rate. During this acclimation period cultures were maintained within ∼15–120% of the cell density at the time of sampling for cell composition to ensure similar optical conditions and growth rates throughout this period. After each culture was sampled at fully acclimated, nutrient-replete exponential growth, N starvation was imposed by diluting these cultures once with N-free media (the same media as described above without sodium nitrate added). This single dilution with N-free media was such that cultures reached a similar optical density and pH in stationary phase as observed during the nutrient-replete exponential phase.
Sampling
Triplicate cultures were sampled daily for cell density by collecting and preserving a 1–3 ml aliquot of each culture replicate. T. pseudonana and Micromonas sp. were preserved in 0.5% glutaraldehyde and T. weissflogii and O. tauri were preserved in 2% Lugol’s solution. The preservation technique used for each study species was based on preliminary tests of various concentrations of Lugol’s solution and glutaraldehyde (0.5, 1, and 2%) to select the method that resulted in no significant change compared to the initial cell density of unpreserved cultures. Cell density in all samples was determined within 2 h of collection by light microscopy using a hemocytometer except for samples of T. weissflogii, which were counted using a Sedgwick-Rafter chamber. Cell density was used to track the growth rate of each culture with the onset of N starvation to determine sampling points for cell composition (Figure 2). Growth rate (μ) was calculated as the daily exponential rate of change in cell density: μ (d-1) = ln(Nf /Ni )/dt, where Nf is the cell density at a given sampling point, Ni is the cell density from the previous day measured at the same time of day and dt is the time interval in days between these two sampling points.
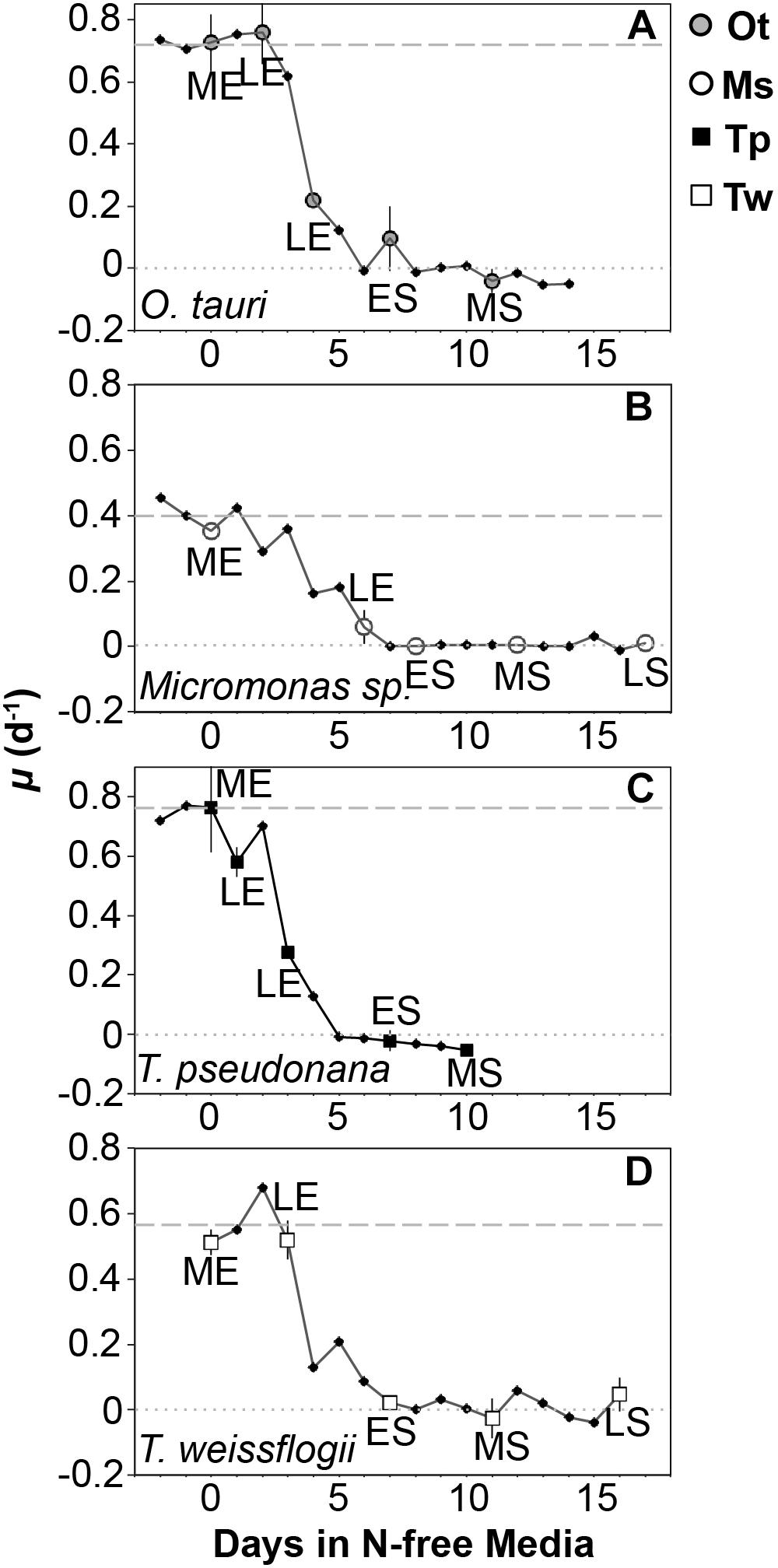
Figure 2. Growth rate (μ, d-1) from N-replete balanced growth to N starvation in (A) T. pseudonana, (B) T. weissflogii, (C) O. tauri, and (D) Micromonas sp. The larger symbols also shown in the legend indicate sampling points for macromolecular composition at mid-exponential (ME), late-exponential (LE), early-stationary (ES), mid-stationary (MS), and late-stationary (LS) growth phases. Criteria for defining these sampling points are provided in the text. The dashed line indicates the μmax for a species determined over a minimum of 10 generations during N-replete, balanced growth. The dotted line indicates a growth rate of 0. Error bars indicate one standard deviation among triplicate cultures.
Samples for elemental and macromolecular composition, dissolved nutrient concentrations, and bacterial contamination were collected during nutrient-replete, balanced exponential growth and at four additional points spanning late-exponential and stationary phase (Figure 1). Late-exponential sampling points characterized cell composition during the onset of N starvation 1–5 days after dilution to N-free media in each experiment. Stationary phase sampling was performed at early-stationary phase near N-starved cessation of growth (6–7 days after dilution to N-free media), at mid-stationary phase 6 days after the cessation of growth (10–12 days after dilution to N-free media) and at late-stationary phase either 10 days after cessation of growth (T. weissflogii and Micromonas sp.) or when cell densities consistently declined for 3 days after the mid-stationary phase (T. pseudonana and O. tauri) depending on whichever event occurred first. Element and macromolecular data from the late-stationary sampling of T. pseudonana and O. tauri are not presented or included in our data analyses as bacterial biomass was estimated to be greater than 10% of phytoplankton biomass at these sampling points (see Bacterial Enumeration section for details). Two additional samples were also collected for elemental composition only in late-exponential phase and stationary phase.
Samples for cell composition were collected by filtration under gentle vacuum pressure (<18 kPa or 5 in Hg) and low light. Samples for particulate carbon (C), nitrogen (N), phosphorus (P), carbohydrate, lipid, and pigments were collected on pre-combusted (4 h at 450°C) Whatman GF/F filters (effective pore size, 0.7 μm). Samples for protein, RNA, and DNA were collected on Whatman Nucleopore 25 mm polycarbonate membrane filters. Different filter types were used across these samples due to the varying compatibility of filter materials with a particular macromolecular assay. Polycarbonate filters with a pore size of 0.8 μm were used for both diatom species, while filters with a pore size of 0.4 and 0.6 μm were used for O. tauri and Micromonas sp., respectively, as these pore sizes were shown to be required to fully retain these smaller species. Filtrate from samples collected on pre-combusted GF/F filters was collected for dissolved nutrient analyses. All macromolecule samples were frozen immediately after collection in liquid nitrogen and stored at -80°C. Samples for particulate C, N, P and dissolved nutrient analyses were immediately placed in a -20°C freezer after collection. Carbohydrate, lipid, and pigment samples were freeze-dried prior to analyses to prevent dilution of the extracting solutions used in each analysis by variable amounts of seawater (∼100–200 μl) retained on GF/F filters. Abundance of contaminating bacteria was sampled by preserving 1 ml aliquots of each culture replicate in 0.1% glutaraldehyde (electron microscopy-rade, Sigma #G5882). Bacterial samples were allowed to fix at room temperature for 15 min, then frozen in liquid nitrogen and stored at -80°C.
Elemental and Nutrient Analyses
Filters collected for particulate C and N were dried at 60°C for 2 days, pelleted in pressed tin capsules and analyzed with a Costech CHN analyzer using acetanilide as a standard.
Samples for total particulate phosphorus were dried and extracted by hydrolysis with 0.1M HCl at 90°C (Solorzano and Sharp, 1980). Phosphorus was quantified by the ammonium molybdate method (Chen et al., 1956), modified for a microplate format, using a SpectraMax M3 microplate reader (Molecular Devices). Collected filtrate was thawed immediately before analyses and dissolved inorganic nitrogen (nitrate, nitrite, and ammonium), phosphate, and silicate were quantified by colorimetry using an autoanalyzer.
Macromolecular Analyses
Carbohydrate was analyzed colorimetrically using the TPTZ (2,4,6-Tris(2-pyridyl) –s triazine) method originally developed for dissolved carbohydrates (Myklestad et al., 1997), following a two-stage acid hydrolysis (Pakulski and Benner, 1992) and neutralization with sodium hydroxide.
Lipids were extracted and purified using modifications of the methods of Folch et al. (1957) to ensure full extraction (see Supplementary Text for detail) using only pre-combusted glass materials and HPLC-grade solvents. Dried lipid extracts containing 20–140 μg of total lipid were quantified by acid-dichromate colorimetry (Pande et al., 1963) using a spectrophotometer (Shimadzu UV-1800) with glyceryl tripalmitate (Sigma-Aldrich, Cat. #T5888) used as a reference standard. For analysis of the phosphorus contained in lipids, a portion of each total lipid extract described above was air-dried at room temperature to remove all chloroform and then extracted and analyzed as with total P samples.
Pigments were quantified by high performance liquid chromatography (HPLC) (Van Heukelem et al., 1994; Van Heukelem and Thomas, 2001) on an Agilent 1100 HPLC (Agilent Technologies, Santa Clara, CA, United States) using authenticated standards (DHI Lab, Horshølm, Denmark).
Protein was extracted from samples on polycarbonate filters by bead milling (Lysing Matrix D, MP Biomedicals) in 2% SDS (sodium dodecyl sulfate) buffer. Bead milling was performed four times for 1 min at 6.5 m s-1, with samples placed on ice for 2 min between each round of bead milling to prevent degradation by heating. Extracted protein was then quantified with the BioRad DC Assay, which is based on the Lowry method (Lowry et al., 1951), using a microplate reader (SpectraMax M3, Molecular Devices) and bovine gamma globulin (BioRad) as a standard. Protein quantification by these methods showed less than 5% variation compared to quantification after extraction with a more comprehensive protein solubilization buffer containing protease inhibitors (Brown et al., 2008) in preliminary tests with T. pseudonana and Micromonas sp., indicating that the simple SDS buffer used provides full protein extraction.
Nucleic acids were measured according to Berdalet et al. (2005) with modification of the sample extraction and scaled to a microplate format (see Supplementary Text for detail). RNA was quantified against an E. coli ribosomal RNA standard (Ambion #7940) and DNA was quantified against a type IX calf thymus DNA standard (Sigma # D4522). RNA was also quantified in samples from mid-exponential and mid-stationary growth phases after extraction with Trizol (Thermo Fisher Scientific), which is based on the phenol-chloroform extraction procedure (Chomczynski and Sacchi, 1987), to provide an additional verification of cellular RNA content. The extraction protocol provided by the Trizol manufacturers was used with modifications added to reduce the loss of RNA during various isolation and cleaning steps and account for consistent losses of RNA during solvent partitioning using a parallel RNA standard (Ambion #4940) spike-recovery test with each extraction (see Supplementary Information). Both RNA quantification methods provided similar results for all species at both growth conditions with the exception of N replete O. tauri and Micromonas sp., for which the Berdalet et al. (2005) method provided significantly higher (Student’s T-test, p < 0.01) RNA values (Supplementary Figure 1). Only results produced with the Berdalet et al. (2005) method are shown due to its greater consistency, simultaneous quantification of DNA, and greater apparent extraction efficiency (see Supplementary Information).
DNA quantification met or exceeded the expected genomic DNA content estimated from the known genome sizes of T. pseudonana (Armbrust et al., 2004), O. tauri (Derelle et al., 2002), and Micromonas pusilla (Worden et al., 2009), a species closely related to Micromonas sp., and assuming a mass of 10-3 pg for each mega-base pair of DNA (Doležel et al., 2003). A full genome sequence for T. weissflogii CCMP 1010 has not been published. T. pseudonana was assumed to be diplontic (Chepurnov et al., 2004) while O. tauri and Micromonas sp. were assumed to be haplontic (Graham et al., 2008). Measured DNA contents in N replete O. tauri and Micromonas sp. were 48.0 and 15.6% higher than genomic DNA estimates. Measured DNA content in T. pseudonana was 110% higher than its estimated genomic DNA content assuming diploidy. This higher DNA content in T. pseudonana may be the result of polyploidy, which has been observed in diatoms, particularly of the genus Thalassiosira (Von Dassow et al., 2008; Koester et al., 2010). The presence of multinucleate cells, which have been shown to occur in diatoms under nutrient or toxicity stress (Badour, 1968; Oey and Schnepf, 1970) or spontaneously (Von Dassow et al., 2006, 2008) may also explain T. pseudonana DNA content exceeding estimates of genomic DNA content.
The elemental content of macromolecular pools was calculated using a mean elemental stoichiometry for protein, carbohydrates, lipids, and nucleic acids (Geider and LaRoche, 2002) and the known elemental stoichiometry of phytoplankton pigments (Wright et al., 2005) as approximated in Figure 1. The cellular P not accounted for in RNA, DNA, and lipid P measurements was assumed to be residual P. This residual P likely represents intracellular storage of P as orthophosphate (PO4) or polyphosphate ([PO4]2[PO3]n) or the surface adsorption of orthophosphate (Sañudo-Wilhelmy et al., 2004; Dyhrman, 2016). As such, residual P was assumed to have the molecular formula of orthophosphate (PO4) in order to calculate its mass for comparison to the mass of other macromolecules.
Bacterial Enumeration
The abundance of contaminating bacteria at all sampling points was measured with a flow cytometer (BD Accuri C6) according to Marie et al. (2005) using DNA fluorescence (produced after staining with SYBR Green I dye) and a lack of chlorophyll fluorescence to discriminate heterotrophic bacteria from microalgal cells. Assuming contaminating bacteria are coccoid with a relatively large mean cell diameter for marine bacteria (0.7 μm), these bacteria would have a C content of 65 fg C cell-1 (Romanova and Sazhin, 2010). Using this estimate of C content, bacterial biomass was estimated to be 0.89–6.74% of particulate biomass (retained on a GF/F filter) in all samples. Additionally, the pore sizes of the filters (∼0.7 μm for glass fiber and 0.8 μm for polycarbonate filters) used to sample diatom cultures, which contained the highest bacterial abundances, are similar to or greater than the relatively large assumed bacterial cell size and thus unlikely to efficiently retain these bacteria. Given these measurements and assumptions, bacterial biomass was considered to be relatively low with no significant effect on the reported elemental or macromolecular measurements at each of these sampling points. Late-stationary sampling points for T. pseudonana and O. tauri were excluded from this study since bacterial biomass was deemed too high (10–13% of sampled biomass) to assume negligible effect on measurements of microalgal cell composition at these points.
Data Analysis
Linear regressions as well as analysis of variance (ANOVA) and pairwise post hoc tests comparing proportional changes in elemental and macromolecular content among species were all performed using R statistical software (R Studio version 1.0.143). The error reported for all values indicates one standard deviation.
Results
Elemental Content and Stoichiometry
The elemental and macromolecular content of the species in this study were observed from N-replete steady-state growth to non-steady state N starvation. All species showed a decline in N quota (QN) to a consistent minimum (Figure 3B) as dissolved inorganic N fell below detection limits (Figure 3D) and replete levels of dissolved inorganic P (12.6 ± 4 μM) and silica (10.8 ± 2 μM) were maintained.
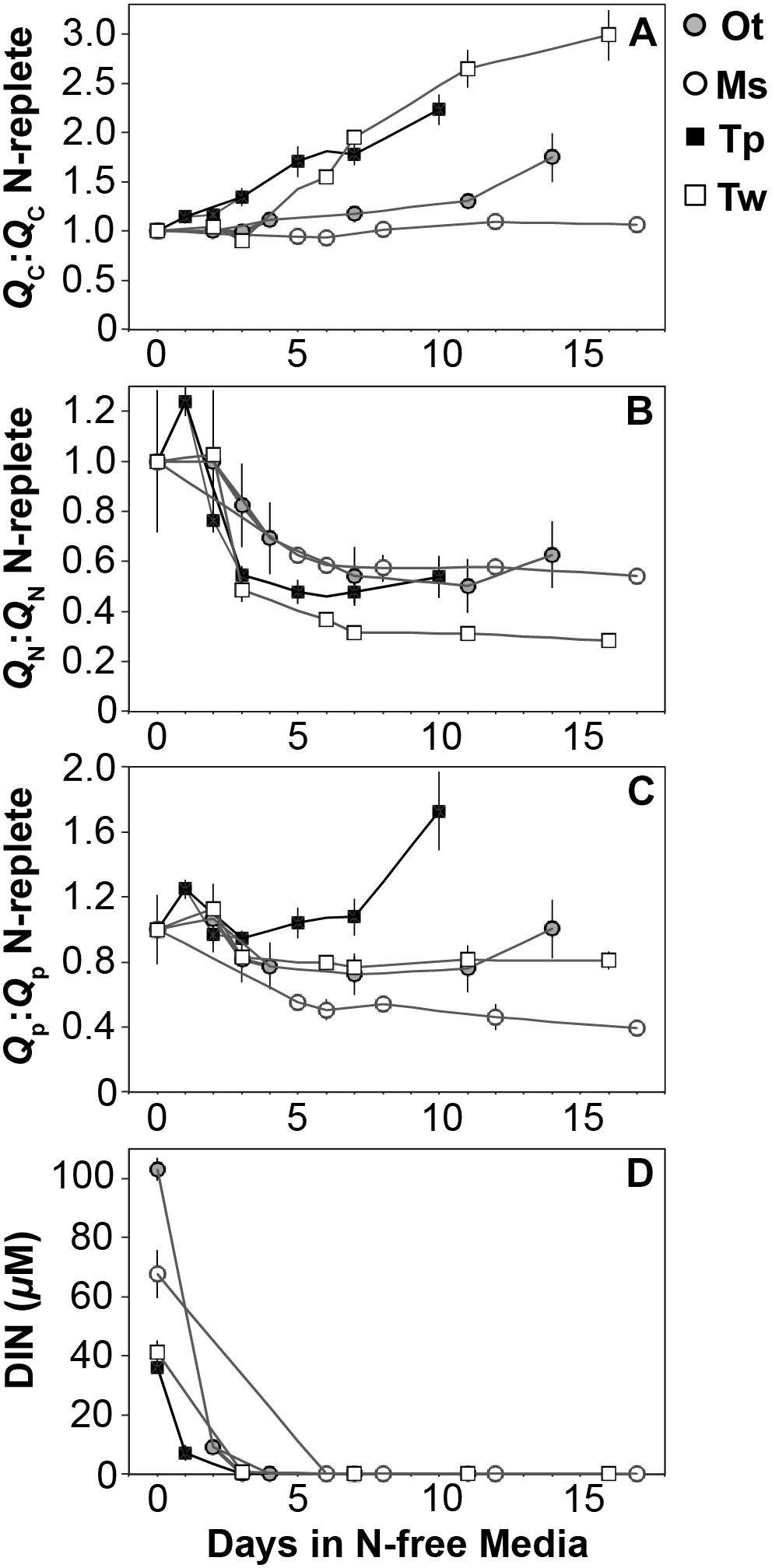
Figure 3. The ratio (mol:mol) of cellular (A) carbon, (B) nitrogen, and (C) phosphorus quota to N-replete quotas as well as (D) the available dissolved inorganic nitrogen from N-replete growth to N starvation. The decline in N quota to consistent minima in each species (B) and the removal of available DIN (D) indicate N starvation in all cultures. Error bars indicate one standard deviation among triplicate cultures.
All species showed an increase in cellular C quota (QC) with N starvation, but the extent of this increase varied considerably (Figure 3A and Table 1). Carbon accumulation with N starvation was greater in diatoms, particularly in the larger diatom species T. weissflogii (Table 2). The prasinophytes displayed far less change in QC than diatoms, particularly Micromonas sp., which had an increase in QC of only 8.95 ± 0.7% (Table 2). The decline in QN from N-replete growth to N starvation (Figure 3B and Table 1) was similar among T. pseudonana and both prasinophytes (-52.1 to -42.3%, Table 2), but significantly greater (p < 0.01) in T. weissflogii (-68.8 ± 5%). Though steady-state N-replete C:N was comparable across all species (6.32 ± 0.3, Figure 4A), under N starvation the greater C accumulation in diatoms resulted in a far greater increase in C:N compared to prasinophytes (Table 2 and Figure 4A).
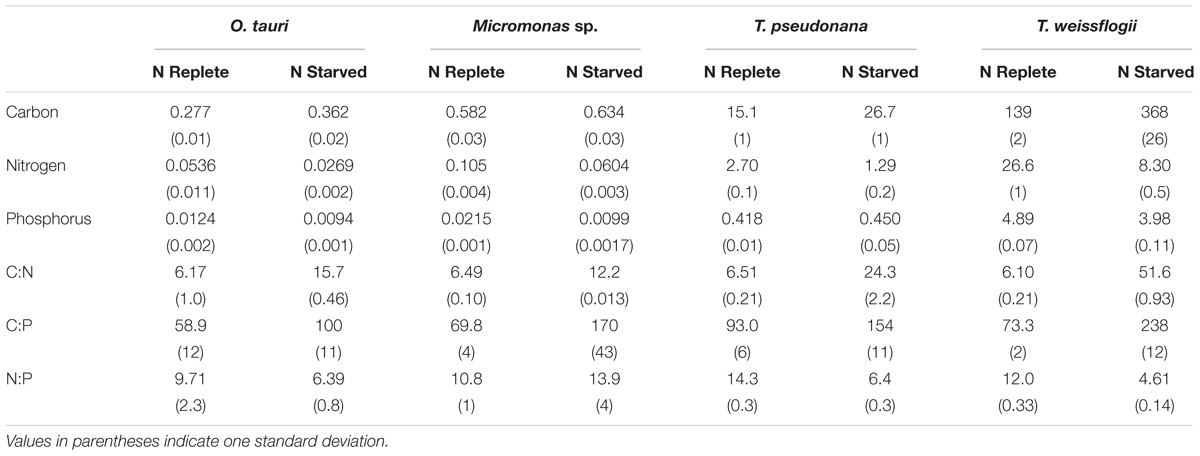
Table 1. The cellular elemental content (in pg cell-1) and molar elemental ratios of each species during N-replete, steady-state growth and during N-starved stationary phase (mid-stationary sampling point, 6 days after cessation of growth).
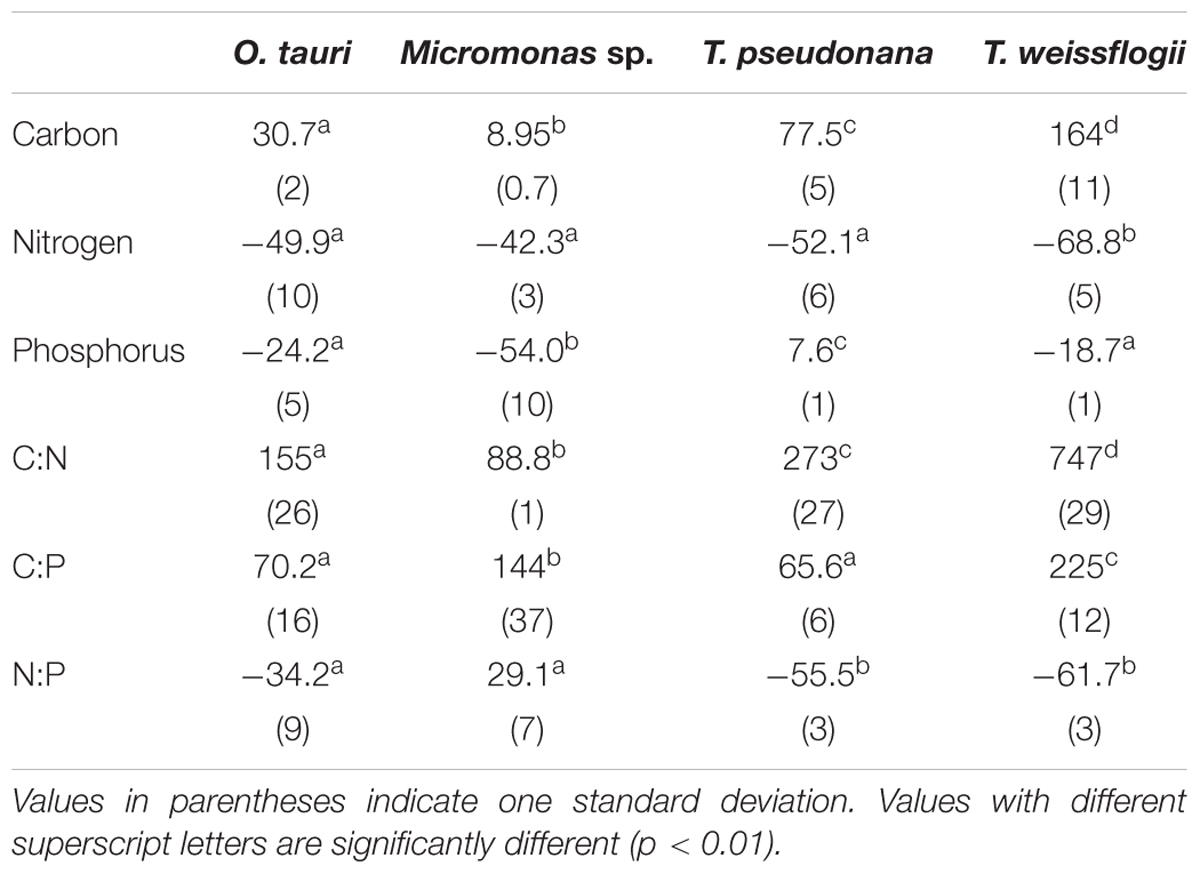
Table 2. Percent change in molar elemental content from N-replete mid-exponential growth (point ME in Figure 2) to N starved mid-stationary phase (point MS in Figure 2).
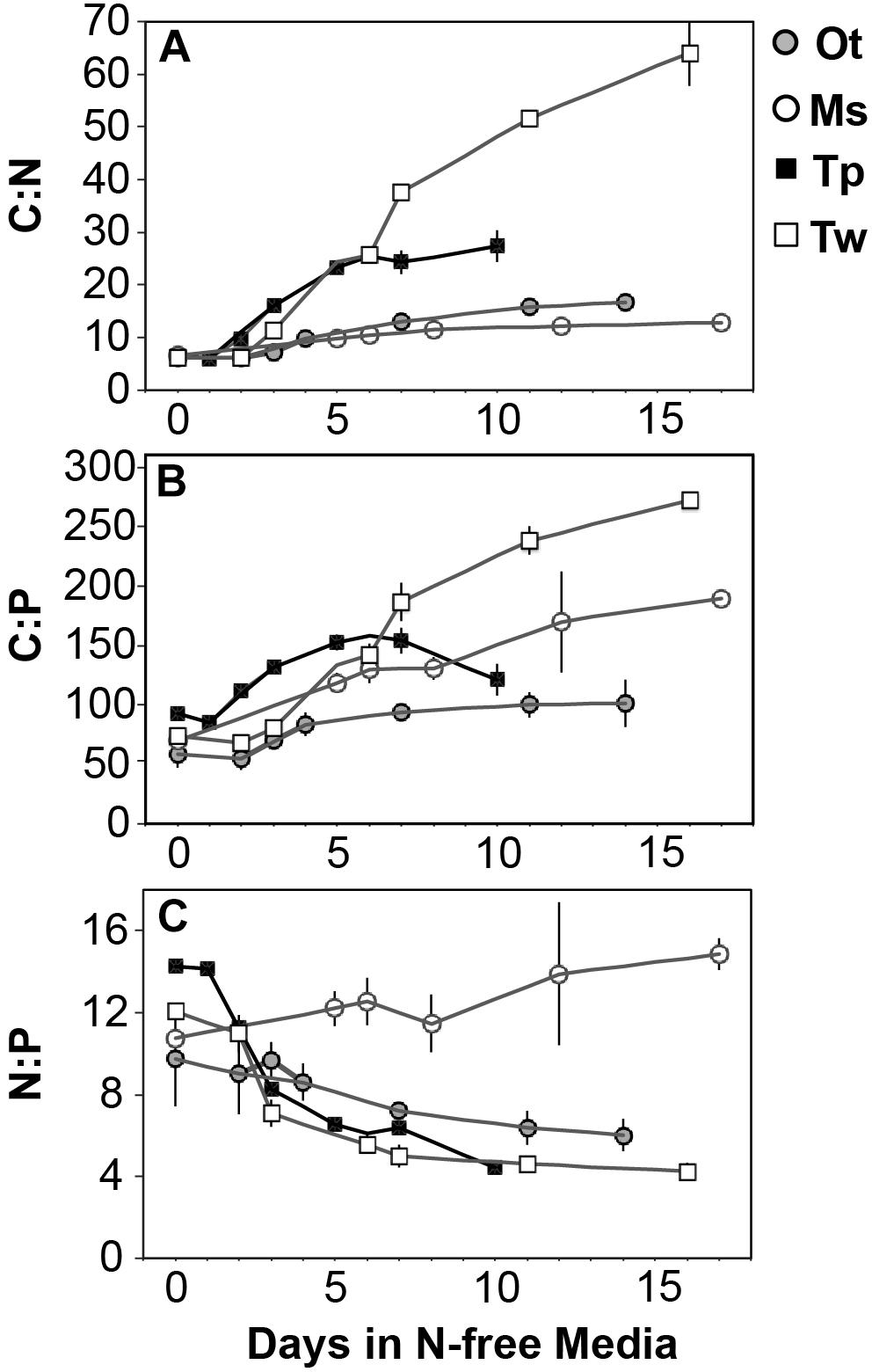
Figure 4. The change in molar ratios of (A) cellular carbon:nitrogen, (B) cellular carbon:phosphorus, (C) cellular nitrogen:phosphorus from N-replete growth to N starvation. Error bars indicate one standard deviation among triplicate cultures.
Changes in cellular P quota (QP) and P stoichiometry in response to N starvation were complex and species specific (Figure 2C and Table 1). In N-replete steady-state growth, prasinophytes were enriched in phosphorus resulting in a lower C:P and N:P compared to diatoms (Figure 4 and Table 1). With the onset of N starvation, Micromonas sp. showed the largest reduction in QP while the larger diatom T. weissflogii and the smaller prasinophyte O. tauri displayed smaller declines. In contrast, QP increased slightly (7.6 ± 1%) in T. pseudonana with N starvation (Table 2). The smaller changes in diatom QP resulted in a larger decline in N:P in diatoms as compared to O. tauri, while N:P increased slightly in Micromonas sp. as its QN and QP both declined greatly with N starvation (Figure 3 and Figure 4C). Despite these differing responses in QP, C:P increased in all species with N starvation, although C:P declined at mid-stationary phase in T. pseudonana due to a large increase in QP at that sampling point (Figure 4B). As with C:N, C accumulation dominated the increase in C:P in all species except Micromonas sp., in which the increase in C:P was driven more by its relatively large decline in QP and little change in QC (Figure 4B and Figures 3A,C).
Macromolecular Content
The sum of the calculated C and N content of macromolecules closely matched direct measurements of QC and QN (Supplementary Figure 2) such that macromolecular measurements accounted for 98.3 ± 8% of measured cellular C (Supplementary Table 1). Measurements of protein, RNA, DNA, and pigments accounted for 94.4 ± 8% of N in all samples except at N-replete mid-exponential phase for the diatoms T. pseudonana (86.5 ± 3%) and T. weissflogii (70.3 ± 3%). The unmeasured residual N in N-replete diatoms is likely inorganic N stored in central vacuoles (Miyata et al., 1986; Grover, 1991), free amino acids, or other N-containing small metabolites that can represent a substantial pool of intracellular N (Dortch et al., 1984; Granum et al., 2002; Lourenço et al., 2004). Measurements of RNA, DNA, and phospholipids accounted for only 29.2 ± 3% of cellular P (Supplementary Table 1). We define the cellular P that is not accounted for by these macromolecules as residual P. Considering our additional verification of our RNA and DNA measurements (see Supplementary Information for more detail), the direct measurement of phosphorus in total lipid extracts, and the expectation that the contribution of other organic phosphorus pools (e.g., ATP, other free nucleotides) will be small (Geider and LaRoche, 2002; Dyhrman, 2016) we assume that the residual cellular P is in the form of either intracellular storage (e.g., polyphosphate bodies, orthophosphate in vacuoles) (Miyata et al., 1986; Dyhrman, 2016) or as surface-adsorbed phosphate (Sañudo-Wilhelmy et al., 2004; Fu et al., 2005).
The major macromolecular pools of protein, carbohydrate, and lipid represented 94.7 ± 9% of QC and their variation matches the observed changes in QC and QN with N starvation (Figure 5 and Supplementary Table 2). Diatoms displayed far greater increases in carbohydrate quota (370–829%) than prasinophytes (52.2–58.4%) during N starvation (Figure 5 and Table 3). The relative changes in lipid quotas were more similar across species (112–202%). The greater increase of QC and C:N observed in diatoms during N starvation (Figures 3A, 4A) was due primarily to the greater carbohydrate accumulation in diatoms while lipid accumulation dominated the more modest increases in QC and C:N in prasinophytes (Figure 5 and Table 3). Protein accounted for 84.7 ± 8% of QN and its similar relative decline in all species with N starvation (Figures 5A–D and Table 3) mirrored the changes in QN as both parameters were highly correlated in all species (r = 0.90–0.98, p < 0.001).
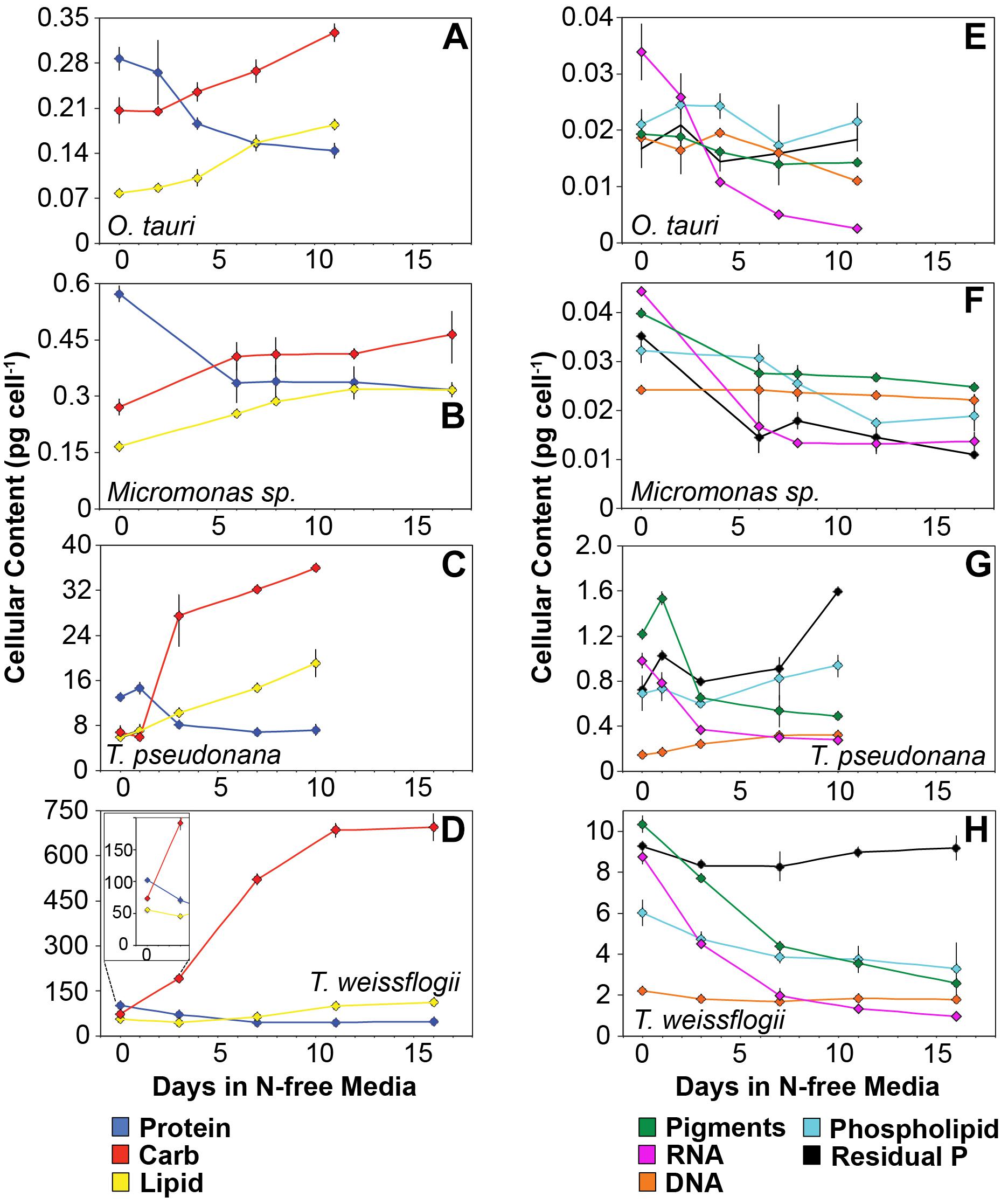
Figure 5. Changes in cellular content (pg cell-1) of (A–D) the major macromolecules protein (blue), carbohydrate (red), and lipid (yellow) as well as (E–H) the minor macromolecules pigments (green), RNA (purple), DNA (orange), phospholipid (cyan), and residual phosphorus (black) with N starvation. The inset in (B) highlights the macromolecular content of T. weissflogii at the beginning of the experiment as this is obscured by the large range of macromolecular content in this species. Error bars indicate one standard deviation among triplicate cultures. Mid-exponential and mid-stationary values for all macromolecular pools are also shown in Supplementary Table 2.
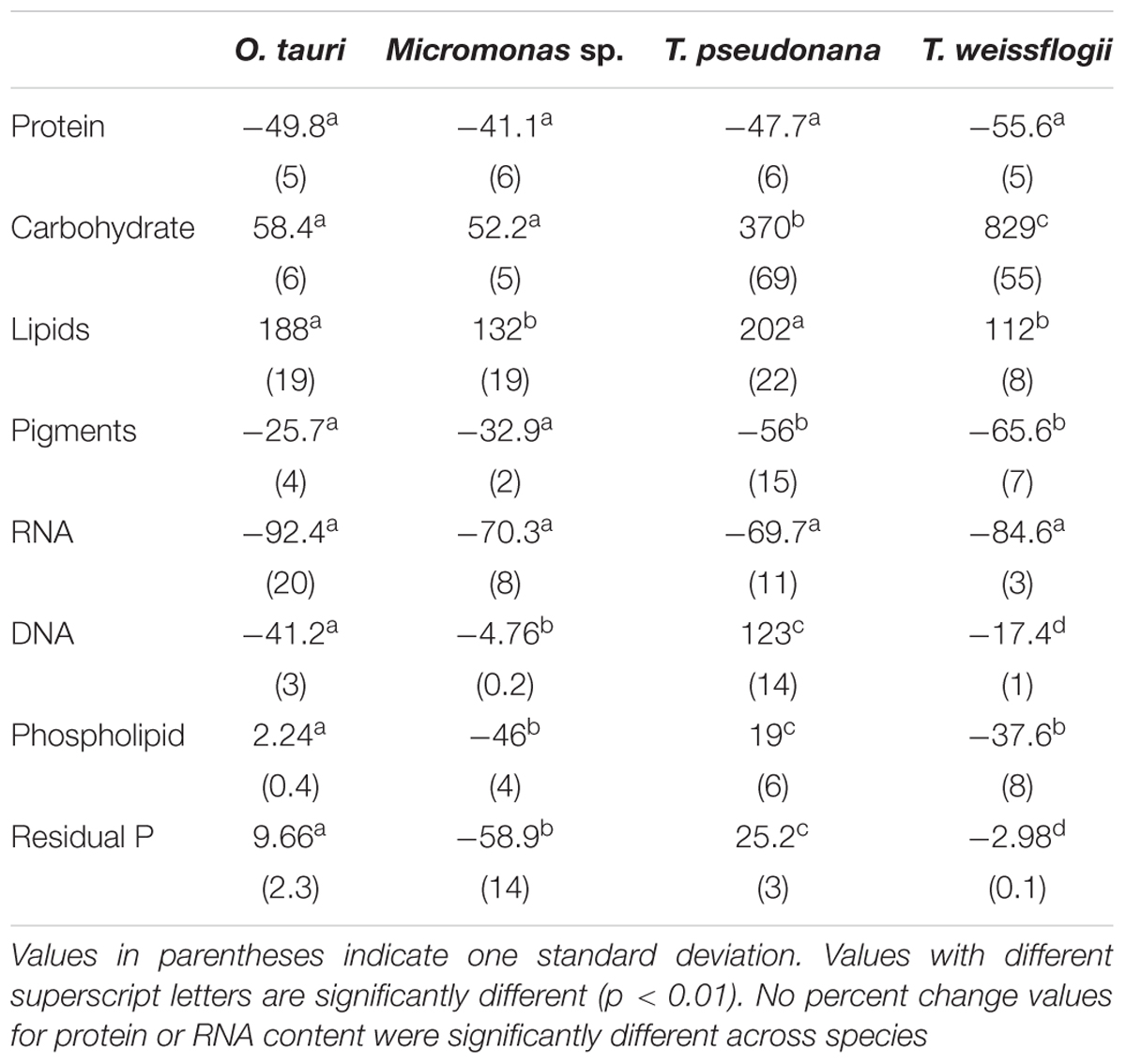
Table 3. Percent change in macromolecular content from N-replete mid-exponential growth (point ME in Figure 2) to N starved mid-stationary phase (point MS in Figure 2).
Minor pools of macromolecules (with respect to cell mass) made relatively small contributions to QC and QN, but dominated species-specific dynamics in QP. The small portion of QN not in proteins was mostly in nucleic acids and pigments. RNA was the most dynamic macromolecular pool in this study, showing a large and similar decline (-92.4 to -69.7%) in all species with N starvation. Pigments, particularly chlorophylls, also showed a large decline with N starvation (Figures 5E–H and Table 3) that was distinctly greater in diatoms (-65.6 to -56.0%) compared to prasinophytes (-32.9 to -25.7%). While these changes had a small impact on QN, RNA and residual P content had a large influence on cellular P stoichiometry. The greater decline in N:P with N starvation in diatoms reflects their maintenance of larger residual P pools relative to prasinophytes, but similar declines in protein and N content across species. Declines in both RNA and residual P accounted for a large decline in QP and slight increase in N:P in Micromonas sp., an elemental response that differed from that of all other species. This decrease in both RNA and residual P in Micromonas sp. also caused an increase in C:P despite little C accumulation. The comparatively small change in residual P in T. weissflogii and O. tauri correspond to their small declines in QP. The dynamics of residual P also dominated the response of QP in T. pseudonana, where a large increase in residual P during mid-stationary phase appeared to cause an increase in QP and a decline in C:P following its initial increase. Phospholipids and DNA represented the smallest portions of cellular P and were less variable than all other major macromolecules in response to N starvation (Figures 5E–H and Table 3) except in the case of T. pseudonana, in which the cellular DNA quota increased with N starvation while other species displayed only small declines.
Elemental Allocation to Macromolecules
All species in this study shared similar general patterns in C and N reallocation in response to N starvation. Protein represented the largest pool of cellular C and N in all species under N-replete steady-state growth. With the onset of N starvation, C was reallocated from protein to lipid and carbohydrate (Figure 6) while a consistently high proportion of N was maintained in protein and a smaller proportion on N was reallocated from RNA to DNA (Figures 7A–D). Within these shared general patterns, far more C was allocated to carbohydrate in T. weissflogii (Figure 6D) while in the other species the proportion of QC in both carbohydrate and lipid increased in a similar fashion (Figures 6A–C). The hyper-accumulation and higher C allocation of carbohydrate in T. weissflogii is also apparent in its large increase in carbohydrate:lipid with N starvation (Figure 8A). In contrast, there was only a slight increase in carbohydrate:lipid in T. pseudonana and a slight decline in carbohydrate:lipid for both prasinophytes (Figure 8A). The similar general patterns in N allocation pattern are also reflected in the protein:RNA ratio of each species, which was similar among diatoms and Micromonas sp. (12.70 ± 0.8) and lower in O. tauri (8.64 ± 1.7) during N-replete growth and increased in all species with N starvation as RNA declined more rapidly than protein (Figure 8B). The overall N allocation pattern is also clear in the sharp decline of RNA:DNA in each species from steady state N-replete values of 5.39 ± 2.0 in diatoms and 1.82 ± 0.1 in prasinophytes to, similarly, low levels in all species (0.60 ± 0.3) with N starvation (Figure 8C). One notable difference in N allocation among species was the slight increase in pigments as a proportion of QN in prasinophytes due to their small change in pigment quotas with N starvation (Figures 7A,B), whereas pigments decreased as a proportion of QN in diatoms (Figures 7C,D).
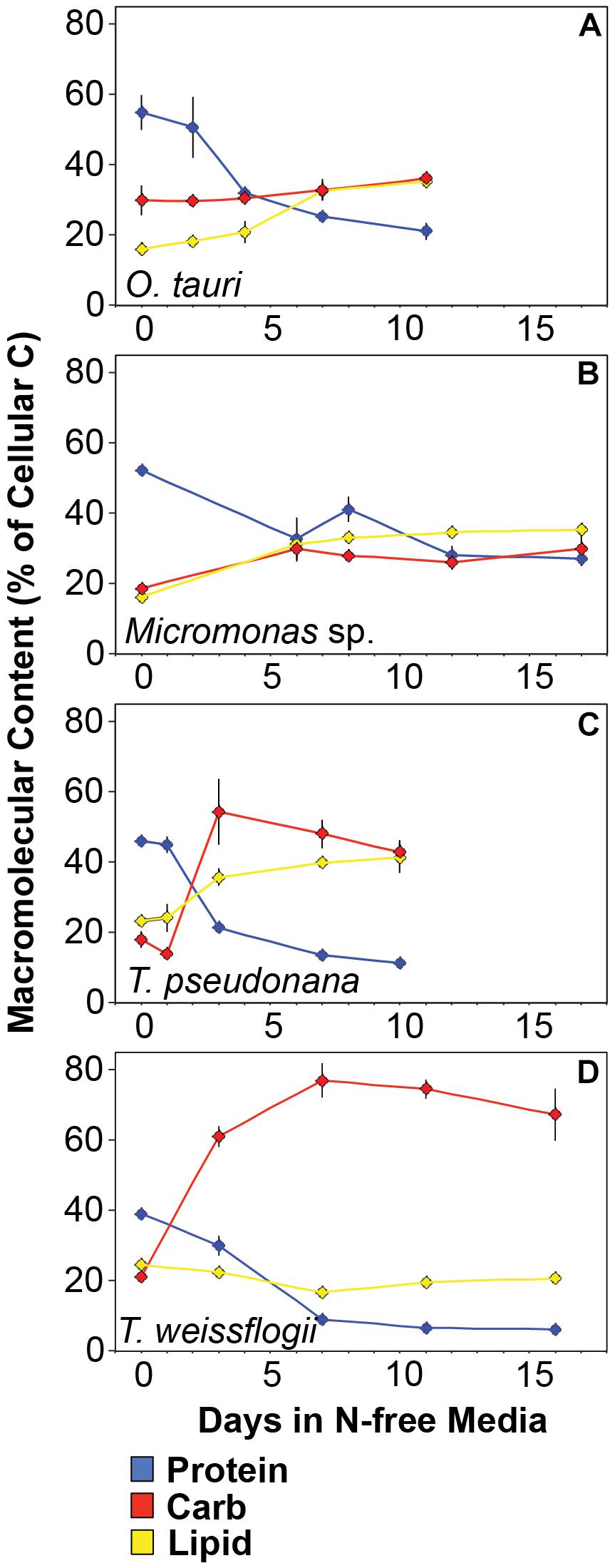
Figure 6. Changes in the allocation (%) of total cellular carbon among protein (blue), carbohydrates (red), and lipid (yellow) in (A) T. pseudonana, (B) T. weissflogii, (C) O. tauri, and (D) Micromonas sp. The estimated C content of the macromolecules shown accounted for 94.7 ± 9% of total cellular carbon. Error bars indicate one standard deviation among triplicate cultures.
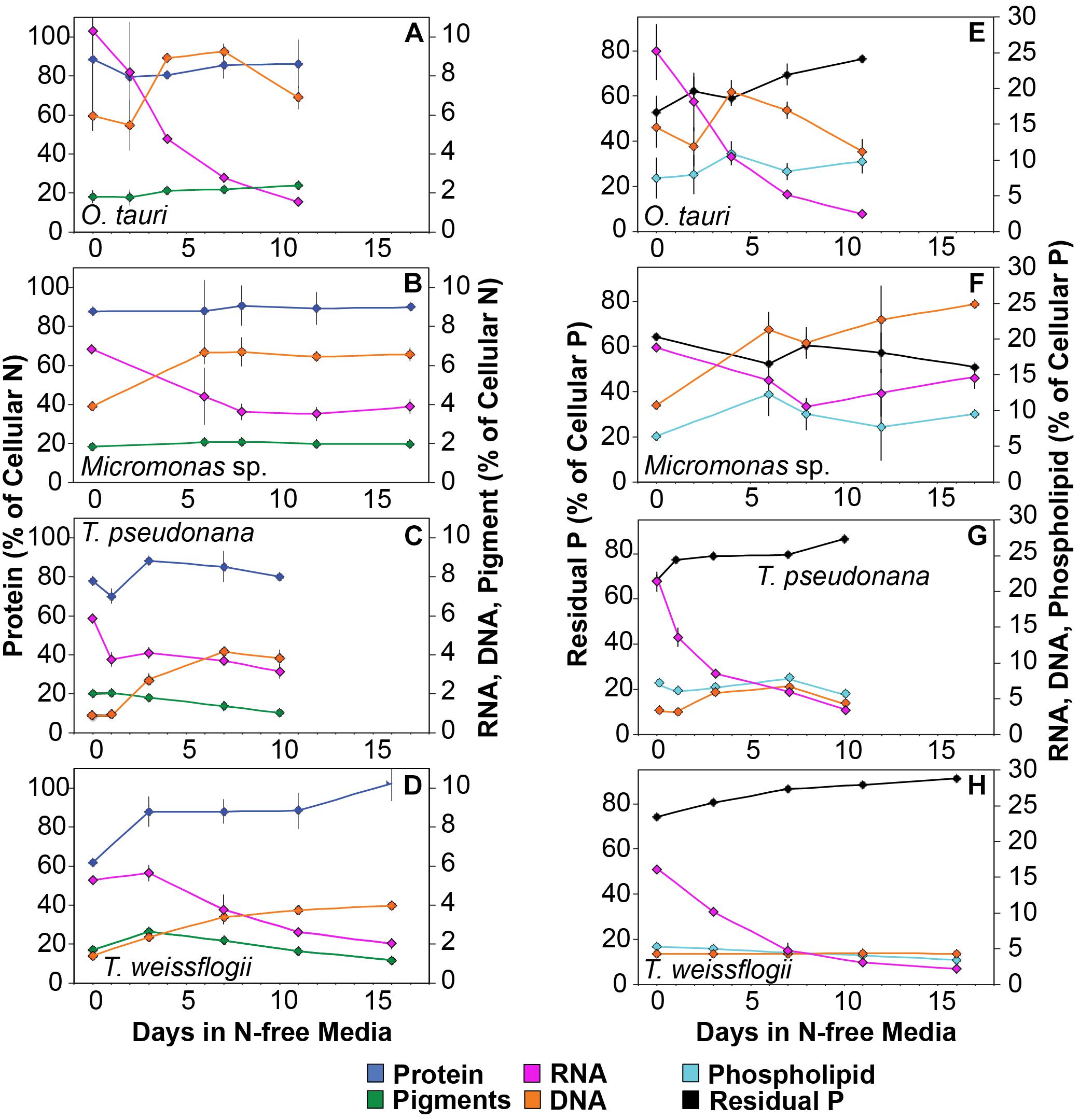
Figure 7. Changes in the allocation (%) of (A–D) total cellular nitrogen and (E–H) phosphorus among protein (blue), pigments (green), RNA (purple), DNA (orange), phospholipid (cyan), and residual phosphorus (black) with N starvation. Note that in the N allocation plots (A–D), protein (blue) is plotted on a different scale on the left axis while RNA (purple), DNA (orange), and pigments (green) are plotted on a smaller scale on the right axis. Note that in the P allocation plots (E–H) Residual phosphorus (black) is on the left axis with a different scale than RNA (purple), DNA (orange), and phospholipids (cyan), which are plotted on a smaller scale on the right axis. Error bars indicate one standard deviation among triplicate cultures.
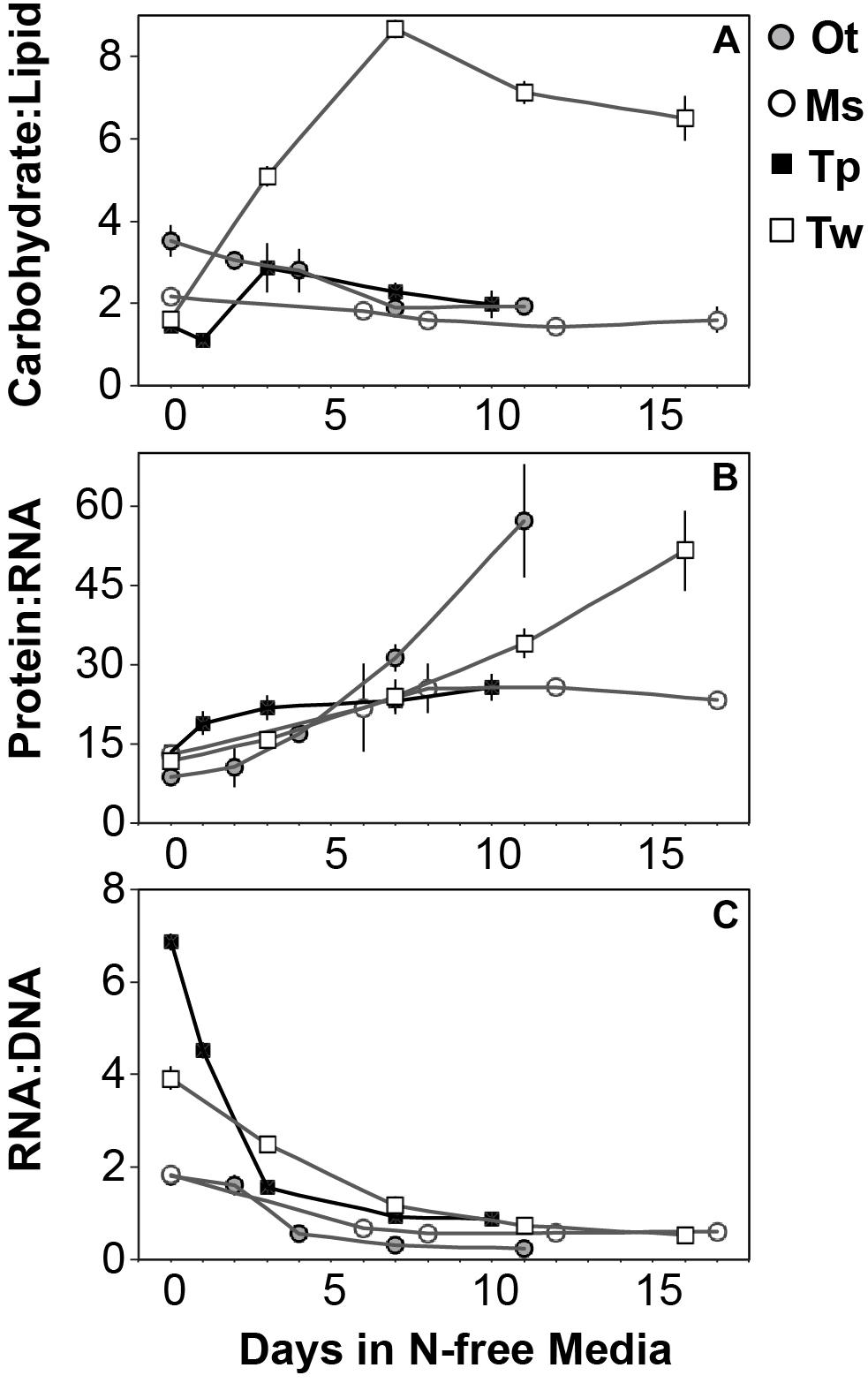
Figure 8. The mass ratios of (A) carbohydrate:lipid, (B) protein:RNA, and (C) RNA:DNA in each species with N starvation. Error bars indicate one standard deviation among triplicate cultures.
The majority of QP, in all species and at all growth phases, was allocated to residual P and this allocation was greater in diatoms (81.1 ± 7%) than in prasinophytes (60.5 ± 8%). The general pattern of changes in the allocation of QP during N starvation was similar among both diatoms and O. tauri as all showed a small increase in allocation to residual P, a declining allocation to RNA, and little to no change in the proportion of QP in DNA or phospholipids (Figures 7E–H). This general pattern was also reflected in the sharp decline in RNA:DNA observed in all species with N starvation (Figure 8C). Micromonas sp. showed a distinct pattern in P allocation during N starvation, with little change in allocation to RNA and residual P and an increase in the allocation to DNA (Figure 7F). The large declines in both residual P and RNA during N starvation in Micromonas sp. paralleled the decline in QP, thus Residual P and RNA accounted for similar allocations of declining QP throughout N starvation. Since DNA quotas did not change in Micromonas sp., DNA made up an increasing proportion of the declining QP with the onset of N starvation (Figure 6F).
Discussion
The diatoms and prasinophytes we examined represent distinct phyla, cell sizes and environmental niches, yet share some key similarities in the macromolecular responses to N starvation that dominate their elemental stoichiometry. The accumulation of C-rich carbohydrate and lipid with N starvation had the greatest impact on elemental stoichiometry, particularly C:N, in all species. However, diatoms displayed far greater increases in C content due mainly to carbohydrate accumulation while prasinophytes had more modest increases in C that were dominated by lipid accumulation. Unlike the distinct carbohydrate and lipid dynamics between diatoms and prasinophytes, the decline in protein and N quota overall was proportionally similar across species. There was also a similar relative decline in RNA in all species with N starvation. Notably, this similar relative decline in RNA exceeded the decline in protein in all species despite protein representing the vast majority of cellular N. The loss of RNA with N starvation did not dominate P dynamics as the majority of cellular P in all species was in residual pools, which are likely intracellular storage or surface adsorbed phosphate (Sañudo-Wilhelmy et al., 2004; Dyhrman, 2016). These residual P pools had strong, species-specific effects on cellular C:P and N:P, though the accumulation of C-rich macromolecules still dominated C:P, which increased in all species. Despite the general similarity of these biochemical responses across species, factors such as taxa and cell size clearly modify the extent of macromolecular responses and their influence on phytoplankton elemental stoichiometry.
Carbohydrate and Lipid Accumulation Dominate Variability in C:N
The greater increase in C:N (273–747%) in diatoms were driven by carbohydrate accumulation and the smaller increases in C:N (88.8–155%) in prasinophytes were driven by lipid accumulation. Increases in carbohydrate content in both diatoms and prasinophytes are generally attributed to the accumulation of storage polymers (Myklestad, 1974; Ral et al., 2004). The larger intracellular space of diatoms may allow more carbohydrate storage accumulation (Sicko-Goad et al., 1984). Such accumulation may be restricted in the comparatively small prasinophytes (Raven, 1984; Ral et al., 2004), with O. tauri being the smallest known free-living eukaryote (Chrétiennot-Dinet et al., 1995). Carbohydrate storage in diatoms occurs in a cytosolic vacuole that can greatly expand to fill intracellular space (Chiovitti et al., 2004) while prasinophytes store carbohydrates in a highly localized sheath around the pyrenoid of their single chloroplast (Deschamps et al., 2008), a structural constraint that may restrict their ability to accumulate carbohydrates. Compared to carbohydrate stores, lipid bodies represent C storage that is more densely packed (Subramanian et al., 2013) as well as more C and energy-rich (Sorguven and Ozilgen, 2013). These properties make lipid bodies a more space-efficient form of C storage (Subramanian et al., 2013), thus smaller cell size and structural constraints may also explain the greater increase in lipid content (132–188%, Figure 5 and Table 3) than carbohydrate (52.2–58.4%, Figure 5 and Table 3) in the N-starved prasinophytes. The greater carbohydrate accumulation in diatoms during N starvation may also be due to their high exudation of dissolved carbohydrates compared to other phytoplankton taxa (Myklestad et al., 1989; Granum et al., 2002), which is enhanced by nutrient starvation (Biddanda and Benner, 1997; Penna et al., 1999). These dissolved carbohydrates can form extracellular polymeric substances (EPS) that can aggregate and account for a large fraction of particulate C and carbohydrates in diatom cultures and field populations (Passow, 2000: Engel, 2004; Wetz and Wheeler, 2007; Taucher et al., 2015).
The differing accumulation of C-rich macromolecules by diatoms and prasinophytes during N-starvation could also arise from distinct growth and energy utilization strategies. Carbohydrate storage may be preferred in the nutritionally dynamic niches where diatoms typically thrive (Sarthou et al., 2005) since carbohydrates are more rapidly accumulated and utilized than lipid stores (Siaut et al., 2011; Lacour et al., 2012a). Nitrogen starvation involves the down–regulation of photosynthetic components (e.g., proteins, pigments) to decrease cellular N demand and potentially limit harmful excess light absorption (Cullen et al., 1992). This decline in the photosynthetic apparatus during N starvation also involves the disassembly of lipids in photosynthetic thylakoid membranes and their reallocation to storage bodies prior to the de novo synthesis of storage lipids (Juergens et al., 2015). Micromonas sp. and O. tauri display far less decline in photosynthetic pigments and proteins (and thus presumably less decline in thylakoid membrane lipids) than diatoms during N starvation, instead relying on large inductions of non-photochemical quenching to dissipate excess light absorption (Liefer et al., 2018). The greater allocation of C to lipids in prasinophytes may be due to this strategy of maintaining photosynthetic components during N starvation. A smaller decline in thylakoid lipids prior to de novo synthesis of storage lipids may contribute to the greater allocation of C to lipids in prasinophytes during N starvation.
Similar Relative Declines in Protein and RNA Constrain Variability in Cellular N
The relative decline in cellular N (QN) and protein content was similar among all species in this study and thus had little effect on the differing responses in C:N between diatoms and prasinophytes. The similar percent decline in protein content across species (-41.1 to -55.6%) may indicate that the minimum protein quota for cell viability as a proportion of optimal protein quota is a shared physiological constraint on QN (Bonachela et al., 2013). We hypothesized that the decline in protein would exceed the decline in RNA during N starvation since protein represents the largest portion of QN and thus a metabolic response of minimizing QN seemed likely to disproportionately affect protein content. Instead, the similar decline in RNA across species (-69.7 to -92.4%) exceeded the decline in protein, which may be due to ribosome content (the largest cellular pool of RNA) being more closely tied to growth than any particular external resource (Elser et al., 2003).
The greater decline in RNA compared to protein observed here may also reflect a general resource allocation strategy in response to N stress. The redistribution and acquisition of N (protein-intensive processes) may be favored over maintaining biosynthesis and growth potential (ribosomes-intensive processes), as suggested by transcriptomic and proteomic studies of microalgae (Mock et al., 2008; Allen et al., 2011; Hockin et al., 2012). Biosynthetic and growth potential are directly related to ribosome/RNA content (Sterner and Elser, 2002), thus reducing RNA content during N starvation is an opportunity cost that may limit rapid recovery and growth when N is resupplied. This cost may be offset by N in RNA being reallocated to N metabolism and uptake during N starvation (Mock et al., 2008; Allen et al., 2011; Hockin et al., 2012), which may allow resistance and survival during N stress. Although the reduction in ribosome content was similar across species, the potential shared opportunity costs of this decline may be less severe for diatoms considering their adaptations for nutrient uptake. The success of diatoms in variable nutrient conditions has been attributed to their more rapid uptake (Lomas and Glibert, 2000) and greater storage capacity (Dortch et al., 1984; Lourenço et al., 2004) for N. These traits may lessen the opportunity cost of rebuilding N and P-rich ribosomes and explain the more rapid recovery of diatoms from N starvation as compared to prasinophytes (Liefer et al., 2018).
The small variation in N allocation across study species may be due to the effects of cell size and storage structures on intracellular N storage. During N-replete growth, N-rich macromolecules (protein, nucleic acids, chlorophylls) accounted for smaller proportions of QN in the diatoms T. pseudonana (86.5 ± 3%) and T. weissflogii (70.3 ± 3%) than in the prasinophytes (103 ± 11%) (Supplementary Table 1). We assume this greater residual N in diatoms represents intracellular storage of dissolved N, which appears to be greater in diatoms compared to other taxa (Dortch et al., 1984; Lourenço et al., 2004) due to their larger cell size (Grover, 1991; Tozzi et al., 2004) and use of a central storage vacuole (Miyata et al., 1986; Grover, 1991). The utilization of this greater intracellular N storage pool during N starvation may explain the significantly greater percent decline in QN in the larger diatom T. weissflogii (-68.8 ± 5%) compared to the other study species (-42.3 to -52.1%) (Figure 3B and Table 2). The size-dependent ability of diatoms to store dissolved N may allow greater declines in QN with N starvation as these N storage pools may represent a larger proportion of their N-replete QN (Supplementary Table 1).
Residual P Dominates the Variability in C:P and N:P During N Starvation
Nitrogen starvation caused responses in C:P and N:P that were species-specific and controlled by uncharacterized residual P pools. Here we show that this residual P can account for the majority of phytoplankton QP (70.7 ± 13%), rather than the macromolecular pools of RNA, DNA and phospholipids (Figures 7E–H). We assume this residual P represents the intracellular storage or surface adsorption of inorganic phosphorus since these are the largest known pools of cellular P other than nucleic acids and phospholipids (Sañudo-Wilhelmy et al., 2004; Dyhrman, 2016). Luxury uptake and storage of P by phytoplankton has long been recognized from culture studies (Rhee, 1978; Goldman et al., 1979; Elrifi and Turpin, 1985) and more recently supported by field studies (Orchard et al., 2010; Martin et al., 2014; Diaz et al., 2016), but methodological challenges have made quantification of internal P stores like polyphosphates (polyP) elusive (Dyhrman, 2016). Cell surface-adsorbed inorganic P is another potential source of residual P that has been shown to account for a majority of QP in some cases (Sañudo-Wilhelmy et al., 2004; Fu et al., 2005).
Residual P also had differing effects on elemental stoichiometry among species with implications for the variability in phytoplankton C:N:P observed across growth conditions, taxa, and ocean biomes. The decline in residual P with N starvation in Micromonas sp. caused a slight increase in N:P. This response differed from the decline in N:P that was observed in the other study species in which residual P was stable or increased (Figures 4C, 5E–H). Changes in residual P with N starvation also resulted in more species-specific variation in C:P as compared to the responses of C:N (Figures 4A,B). Similarly, large contributions from residual P pools such as intracellular storage may contribute to the large variations in C:P and N:P observed by others among phytoplankton species (Quigg et al., 2003; Garcia et al., 2018), and even among strains of the same taxa (Martiny et al., 2016). Studies of steady-state N and P limitation have shown large and variable amounts of residual P (Rhee, 1978) or surplus P accumulation (Goldman et al., 1979; Elrifi and Turpin, 1985) and linked these processes to large variations in C:P and N:P within species. Assuming a large and variable amount of phytoplankton P storage has also allowed recent biogeochemical models to reproduce global C:N:P patterns as a function of phytoplankton physiological responses (Daines et al., 2014; Moreno et al., 2018). Our quantifications of functional P pools and residual P can help constrain such models and provide insights as to how P allocation may vary among key phytoplankton taxa.
A greater role of residual P rather than RNA in determining QP has important implications for relating phytoplankton elemental content and growth dynamics. The Growth Rate Hypothesis (GRH) (Sterner and Elser, 2002) anticipates a positive relationship between QP and intrinsic growth rate since QP is expected to mostly reflect the ribosomal RNA content needed for cell biosynthesis. However, the luxury uptake and storage of P during non P-limiting conditions can cause P content to be decoupled from growth and RNA content (Elser et al., 2000; Gillooly et al., 2005; Daines et al., 2014). This decoupling has been observed in microalgae during N and light limitation when residual P is maintained while growth and RNA content decline (Rhee, 1978; Goldman et al., 1979; Mouginot et al., 2015). Similarly, we find that QP is decoupled from growth during the onset of N starvation as most of QP is allocated to residual P pools with varying dynamics among species. Considering this, the GRH prediction of a positive relationship between growth and QP may not apply to marine phytoplankton or rather this prediction is less applicable to N-limited communities than P-limited conditions for which QP, RNA content, and growth may be more tightly coupled. Better characterizations of P accumulation and storage allocation are needed to understand the true P growth requirements of phytoplankton (Gillooly et al., 2005; Cherif et al., 2017).
The Large Potential Impacts of C Accumulation and Residual P on Ocean Biogeochemistry
The differing resource allocation strategies we observed among diatoms and prasinophytes may greatly impact their respective roles in export production and other ocean biogeochemical processes. The greater carbohydrate accumulation in diatoms during N starvation could cause their exported biomass to have a higher C:N than that of prasinophytes. A higher C:N in sinking diatom biomass would result in lower C use efficiency for its consumers and a greater export of C to the deep ocean (Hessen et al., 2004; Finkel et al., 2010). The greater export production of diatoms compared to other phytoplankton taxa (Michaels and Silver, 1988; Buesseler, 1998) is often attributed to their larger mineralized cells and tendency to form aggregates, which result in faster sinking rates (Thornton, 2002; Sarthou et al., 2005). Greater carbohydrate production with N stress may be a physiological mechanism in diatoms that contributes to this rapid sinking. Intracellular accumulation of carbohydrates provides negative buoyancy (Raven, 1984; Richardson and Cullen, 1995) and carbohydrate exudation creates the EPS that allows fast-sinking aggregates to form (Passow, 2000). Recent work indicates that smaller taxa like prasinophytes make larger contributions to export production than previously thought due to their inclusion in such aggregates (Turner, 2015; Richardson, 2019). Unlike diatoms, the sinking and export of prasinophytes seems less likely to be enhanced by N stress given their preference for lipid rather carbohydrate accumulation observed here.
Our findings provide taxonomic and physiological constraints on phytoplankton C:N:P that can strengthen efforts to mechanistically quantify and predict ocean biogeochemical processes. The similar proportional decline in protein across species in this study may represent a useful constraint for predicting variable phytoplankton C:N and N:P in cellular trait-based models. In contrast, the presence of large residual P pools with species-specific variation presents a significant challenge for integrating phytoplankton biochemical traits into predictive models (Follows and Dutkiewicz, 2011). Global patterns in particulate C:P have been linked to increasing phytoplankton P requirements for biosynthesis (i.e., ribosomes) at low temperature (Toseland et al., 2013) and regional variation in P availability for phytoplankton growth (Galbraith and Martiny, 2015). Accounting for both the biochemical effects of temperature adaptation and variation in P storage with P availability has allowed recent modeling efforts to capture the full variability and much of the global patterns in phytoplankton and particulate C:P (Moreno et al., 2018). Our findings show that taxonomic variation in C accumulation and residual P pools like P storage can also greatly affect phytoplankton C:P and thus C export efficiency even in the absence of these proposed mechanisms. This highlights the impact that taxonomic variation in biochemical traits and the biogeography of phytoplankton communities may have on ocean C:N:P patterns (Weber and Deutsch, 2010; Martiny et al., 2013a). The presence of large, variable C and P storage pools provide additional mechanisms that may cause the observed flexibility in ocean particulate C:P (Galbraith and Martiny, 2015). Our work shows that these mechanisms may be affected by phytoplankton community structure and N availability in unexpected ways.
Author Contributions
JL, AI, MJF, and ZF contributed to the conceptualization and experimental design of the study. JL, AG, MHF, IB, and CB contributed to sample collection. JL, AG, and MHF performed all biochemical analyses. JL wrote the first manuscript draft. AI, MJF, AO, and ZF contributed to the writing and revisions. All authors read and approved the submitted version.
Funding
This work was supported by the Macromolecular Models of Marine Microbes grant from the Gordon and Betty Moore Foundation, ID#3378, as well as grants from the Simons Collaboration on Computational Biogeochemical Modeling of Marine Ecosystems/CBIOMES (Grant IDs: 549937 [ZF], 549935 [AI], 549931 [MJF]). Funding was also provided by the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canada Research Chairs program (ZF).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Hugh L. MacIntytre, Shannah Rastin, and Magda Waclawik of Dalhousie University for their assistance with CHN and pigment analyses and thoughtful discussions of this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00763/full#supplementary-material
References
Allen, A. E., Dupont, C. L., Oborník, M., Horák, A., Nunes-Nesi, A., McCrow, J. P., et al. (2011). Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473, 203–209. doi: 10.1038/nature10074
Allen, M. M. (1984). Cyanobacterial cell inclusions. Annu. Rev. Microbiol. 38, 1–25. doi: 10.1146/annurev.mi.38.100184.000245
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Badour, S. (1968). Experimental separation of cell division and silica shell formation in Cyclotella cryptica. Arch. Microbiol. 62, 17–33. doi: 10.1007/BF00407050
Berdalet, E., Roldán, C., Olivar, M. P., and Lysnes, K. (2005). Quantifying RNA and DNA in planktonic organisms with SYBR Green II and nucleases. Part A. Optimisation of the assay. Sci. Mar. 69, 1–16. doi: 10.3989/scimar.2005.69n11
Berges, J. A., Charlebois, D. O., Mauzerall, D. C., and Falkowski, P. G. (1996). Differential effects of nitrogen limitation on photosynthetic efficiency of photosystems I and II in microalgae. Plant Physiol. 110, 689–696. doi: 10.1104/pp.110.2.689
Biddanda, B., and Benner, R. (1997). Carbon, nitrogen, and carbohydrate fluxes during the production of particulate and dissolved organic matter by marine phytoplankton. Limnol. Oceanogr. 42, 506–518. doi: 10.4319/lo.1997.42.3.0506
Bonachela, J., Allison, S., Martiny, A., and Levin, S. (2013). A model for variable phytoplankton stoichiometry based on cell protein regulation. Biogeosciences 10, 4341–4356. doi: 10.5194/bg-10-4341-2013
Breuer, G., Lamers, P. P., Martens, D. E., Draaisma, R. B., and Wijffels, R. H. (2012). The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour. Technol. 124, 217–226. doi: 10.1016/j.biortech.2012.08.003
Brown, C. M., MacKinnon, J. D., Cockshutt, A. M., Villareal, T. A., and Campbell, D. A. (2008). Flux capacities and acclimation costs in Trichodesmium from the Gulf of Mexico. Mar. Biol. 154, 413–422. doi: 10.1007/s00227-008-0933-z
Buesseler, K. O. (1998). The decoupling of production and particulate export in the surface ocean. Glob. Biogeochem. Cycles 12, 297–310. doi: 10.1029/97GB03366
Cardol, P., Bailleul, B., Rappaport, F., Derelle, E., Béal, D., Breyton, C., et al. (2008). An original adaptation of photosynthesis in the marine green alga Ostreococcus. Proc. Natl. Acad. Sci. U.S.A. 105, 7881–7886. doi: 10.1073/pnas.0802762105
Chen, P., Toribara, T. T., and Warner, H. (1956). Microdetermination of phosphorus. Anal. Chem. 28, 1756–1758. doi: 10.1021/ac60119a033
Chepurnov, V. A., Mann, D. G., Sabbe, K., and Vyverman, W. (2004). “Experimental studies on sexual reproduction in diatoms,” in International Review of Cytology: A Survey of Cell Biology, ed. K. W. Jeon (London: Elsevier), 92–142.
Cherif, M., Faithfull, C., Guo, J., Meunier, C. L., Sitters, J., Uszko, W., et al. (2017). An operational framework for the advancement of a molecule-to-biosphere stoichiometry theory. Front. Mar. Sci. 4:286. doi: 10.3389/fmars.2017.00286
Chiovitti, A., Molino, P., Crawford, S. A., Teng, R., Spurck, T., and Wetherbee, R. (2004). The glucans extracted with warm water from diatoms are mainly derived from intracellular chrysolaminaran and not extracellular polysaccharides. Eur. J. Phycol. 39, 117–128. doi: 10.1080/0967026042000201885
Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. doi: 10.1006/abio.1987.9999
Chrétiennot-Dinet, M., Courties, C., Vaquer, A., Neveux, J., Claustre, H., Lautier, J., et al. (1995). A new marine picoeucaryote: Ostreococcus tauri gen. et sp. nov. (Chlorophyta, Prasinophyceae). Phycologia 34, 285–292. doi: 10.2216/i0031-8884-34-4-285.1
Cullen, J. J., Yang, X., and MacIntyre, H. L. (1992). “Nutrient limitation of marine photosynthesis,” in Primary Productivity and Biogeochemical Cycles in the Sea, eds P. G. Falkowski and A. D. Woodhead (New York, NY: Plenum Publishing), 69–88. doi: 10.1007/978-1-4899-0762-2_5
Daines, S. J., Clark, J. R., and Lenton, T. M. (2014). Multiple environmental controls on phytoplankton growth strategies determine adaptive responses of the N: P ratio. Ecol. Lett. 17, 414–425. doi: 10.1111/ele.12239
Del Giorgio, P. A., and Cole, J. J. (1998). Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Syst. 29, 503–541. doi: 10.1146/annurev.ecolsys.29.1.503
Derelle, E., Ferraz, C., Lagoda, P., Eychenié, S., Cooke, R., Regad, F., et al. (2002). DNA libraries for sequencing the genome of Ostreococcus tauri (Chlorophyta, Prasinophyceae): the smallest free-living eukaryotic cell. J. Phycol. 38, 1150–1156. doi: 10.1046/j.1529-8817.2002.02021.x
Deschamps, P., Haferkamp, I., d’Hulst, C., Neuhaus, H. E., and Ball, S. G. (2008). The relocation of starch metabolism to chloroplasts: when, why and how. Trends Plant Sci. 13, 574–582. doi: 10.1016/j.tplants.2008.08.009
Diaz, J. M., Björkman, K. M., Haley, S. T., Ingall, E. D., Karl, D. M., Longo, A. F., et al. (2016). Polyphosphate dynamics at Station ALOHA, North Pacific subtropical gyre. Limnol. Oceanogr. 61, 227–239. doi: 10.1002/lno.10206
Diaz, J. M., and Ingall, E. D. (2010). Fluorometric quantification of natural inorganic polyphosphate. Environ. Sci. Technol. 44, 4665–4671. doi: 10.1021/es100191h
Doležel, J., Bartoš, J., Voglmayr, H., and Greilhuber, J. (2003). Nuclear DNA content and genome size of trout and human. Cytometry A 51, 127–128. doi: 10.1002/cyto.a.10013
Dortch, Q., Clayton, J. Jr., Thoresen, S., and Ahmed, S. (1984). Species differences in accumulation of nitrogen pools in phytoplankton. Mar. Biol. 81, 237–250. doi: 10.1007/BF00393218
Dyhrman, S. T. (2016). “Nutrients and their acquisition: phosphorus physiology in microalgae,” in The Physiology of Microalgae, eds M. A. Borowitzka, J. Beardall, and J. A. Raven (Heidelberg: Springer), 155–183.
Elrifi, I. R., and Turpin, D. H. (1985). Steady-state luxury consumption and the concept of optimum nutrient ratios: a study with phosphate and nitrate limited Selenastrum minutum (Chlorophyta). J. Phycol. 21, 592–602. doi: 10.1111/j.0022-3646.1985.00592.x
Elser, J., Acharya, K., Kyle, M., Cotner, J., Makino, W., Markow, T., et al. (2003). Growth rate–stoichiometry couplings in diverse biota. Ecol. Lett. 6, 936–943. doi: 10.1046/j.1461-0248.2003.00518.x
Elser, J. J., Fagan, W. F., Denno, R. F., Dobberfuhl, D. R., Folarin, A., Huberty, A., et al. (2000). Nutritional constraints in terrestrial and freshwater food webs. Nature 408, 578–580. doi: 10.1038/35046058
Engel, A. (2004). Distribution of transparent exopolymer particles (TEP) in the northeast Atlantic Ocean and their potential significance for aggregation processes. Deep Sea Res. Part I Oceanogr. Res. Pap. 51, 83–92. doi: 10.1016/j.dsr.2003.09.001
Finkel, Z. V., Beardall, J., Flynn, K. J., Quigg, A., Rees, T. A. V., and Raven, J. A. (2010). Phytoplankton in a changing world: cell size and elemental stoichiometry. J. Plankton Res. 32, 119–137. doi: 10.1093/plankt/fbp098
Finkel, Z. V., Follows, M. J., Liefer, J. D., Brown, C. M., Benner, I., and Irwin, A. J. (2016). Phylogenetic diversity in the macromolecular composition of microalgae. PLoS One 11:e0155977. doi: 10.1371/journal.pone.0155977
Folch, J., Lees, M., and Sloane Stanley, G. (1957). A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509.
Follows, M. J., and Dutkiewicz, S. (2011). Modeling diverse communities of marine microbes. Ann. Rev. Mar. Sci. 3, 427–451. doi: 10.1146/annurev-marine-120709-142848
Fu, F. X., Zhang, Y., Leblanc, K., Sañudo-Wilhelmy, S. A., and Hutchins, D. A. (2005). The biological and biogeochemical consequences of phosphate scavenging onto phytoplankton cell surfaces. Limnol. Oceanogr. 50, 1459–1472. doi: 10.4319/lo.2005.50.5.1459
Galbraith, E. D., and Martiny, A. C. (2015). A simple nutrient-dependence mechanism for predicting the stoichiometry of marine ecosystems. Proc. Natl. Acad. Sci. U.S.A. 112, 8199–8204. doi: 10.1073/pnas.1423917112
Garcia, N. S., Bonachela, J. A., and Martiny, A. C. (2016). Interactions between growth-dependent changes in cell size, nutrient supply and cellular elemental stoichiometry of marine Synechococcus. ISME J. 10, 2715–2724. doi: 10.1038/ismej.2016.50
Garcia, N. S., Sexton, J., Riggins, T., Brown, J., Lomas, M. W., and Martiny, A. C. (2018). High variability in cellular stoichiometry of carbon, nitrogen, and phosphorus within classes of marine eukaryotic phytoplankton under sufficient nutrient conditions. Front. Microbiol. 9:543. doi: 10.3389/fmicb.2018.00543
Gatenby, C. M., Orcutt, D. M., Kreeger, D. A., Parker, B. C., Jones, V. A., and Neves, R. J. (2003). Biochemical composition of three algal species proposed as food for captive freshwater mussels. J. Appl. Phycol. 15, 1–11. doi: 10.1023/A:1022929423011
Geider, R. J., and LaRoche, J. (2002). Redfield revisited: variability of C:N:P in marine microalgae and its biochemical basis. Eur. J. Phycol. 37, 1–17. doi: 10.1017/s0967026201003456
Geider, R. J., Laroche, J., Greene, R. M., and Olaizola, M. (1993). Response of the photosynthetic apparatus of Phaeodactylum tricornutum (Bacillariophyceae) to nitrate, phosphate, or iron starvation. J. Phycol. 29, 755–766. doi: 10.1111/j.0022-3646.1993.00755.x
Geider, R. J., Macintyre, H. L., Graziano, L. M., and McKay, R. M. L. (1998). Responses of the photosynthetic apparatus of Dunaliella tertiolecta (Chlorophyceae) to nitrogen and phosphorus limitation. Eur. J. Phycol. 33, 315–332. doi: 10.1080/09670269810001736813
Gillooly, J. F., Allen, A. P., Brown, J. H., Elser, J. J., Del Rio, C. M., Savage, V. M., et al. (2005). The metabolic basis of whole-organism RNA and phosphorus content. Proc. Natl. Acad. Sci. U.S.A. 102, 11923–11927. doi: 10.1073/pnas.0504756102
Goldman, J. C. (1988). “Spatial and temporal discontinuities of biological processes in pelagic surface waters,” in Toward a Theory on Biological-Physical Interactions in the World Ocean, ed. B. J. Rothschild (Dordrecht: Springer), 273–296. doi: 10.1007/978-94-009-3023-0_15
Goldman, J. C., McCarthy, J. J., and Peavey, D. G. (1979). Growth rate influence on the chemical composition of phytoplankton in oceanic waters. Nature 279, 210–215. doi: 10.1038/279210a0
Graham, J. E., Wilcox, L. W., and Graham, L. E. (2008). Algae. Upper Saddle River, NJ: Benjamin Cummings.
Granum, E., Kirkvold, S., and Myklestad, S. M. (2002). Cellular and extracellular production of carbohydrates and amino acids by the marine diatom Skeletonema costatum: diel variations and effects of N depletion. Mar. Ecol. Prog. Ser. 242, 83–94. doi: 10.3354/meps242083
Grover, J. P. (1991). Resource competition in a variable environment - Phytoplankton growing according to the variable-internal-stores model. Am. Nat. 138, 811–835. doi: 10.1086/285254
Guillard, R. R. (1975). “Culture of phytoplankton for feeding marine invertebrates,” in Culture of Marine Invertebrate Animals, eds W. L. Smith and M. H. Chanley (Boston: Springer), 29–60. doi: 10.1007/978-1-4615-8714-9_3
Guillard, R. R., and Ryther, J. H. (1962). Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 8, 229–239. doi: 10.1139/m62-029
Halsey, K. H., and Jones, B. M. (2015). Phytoplankton strategies for photosynthetic energy allocation. Ann. Rev. Mar. Sci. 7, 265–297. doi: 10.1146/annurev-marine-010814-015813
Halsey, K. H., Milligan, A. J., and Behrenfeld, M. J. (2014). Contrasting strategies of photosynthetic energy utilization drive lifestyle strategies in ecologically important picoeukaryotes. Metabolites 4, 260–280. doi: 10.3390/metabo4020260
Halsey, K. H., O’Malley, R. T., Graff, J. R., Milligan, A. J., and Behrenfeld, M. J. (2013). A common partitioning strategy for photosynthetic products in evolutionarily distinct phytoplankton species. New Phytol. 198, 1030–1038. doi: 10.1111/nph.12209
Hessen, D. O., Ågren, G. I., Anderson, T. R., Elser, J. J., and De Ruiter, P. C. (2004). Carbon sequestration in ecosystems: the role of stoichiometry. Ecology 85, 1179–1192. doi: 10.1890/02-0251
Hillebrand, H., Dürselen, C.-D., Kirschtel, D., Pollingher, U., and Zohary, T. (1999). Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 35, 403–424. doi: 10.1046/j.1529-8817.1999.3520403.x
Hillebrand, H., Steinert, G., Boersman, M., Malzahn, A., Meunier, C. L., Plum, C., et al. (2013). Goldman revisited: faster-growing phytoplankton has lower N : P and lower stoichiometric flexibility. Limnol. Oceanogr. 58, 2076–2088. doi: 10.4319/lo.2013.58.6.2076
Hockin, N. L., Mock, T., Mulholland, F., Kopriva, S., and Malin, G. (2012). The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol. 158, 299–312. doi: 10.1104/pp.111.184333
Juergens, M. T., Deshpande, R. R., Lucker, B. F., Park, J.-J., Wang, H., Gargouri, M., et al. (2015). The regulation of photosynthetic structure and function during nitrogen deprivation in Chlamydomonas reinhardtii. Plant Physiol. 167, 558–574. doi: 10.1104/pp.114.250530
Koester, J. A., Swalwell, J. E., Von Dassow, P., and Armbrust, E. V. (2010). Genome size differentiates co-occurring populations of the planktonic diatom Ditylum brightwellii (Bacillariophyta). BMC Evol. Biol. 10:1. doi: 10.1186/1471-2148-10-1
Lacour, T., Sciandra, A., Talec, A., Mayzaud, P., and Bernard, O. (2012a). Diel variations of carbohydrate and neutral lipids in nitrogen-sufficient and nitrogen-starved cyclostat cultures of Isochrysis sp. J. Phycol. 48, 966–975. doi: 10.1111/j.1529-8817.2012.01177.x
Lacour, T., Sciandra, A., Talec, A., Mayzaud, P., and Bernard, O. (2012b). Neutral lipid and carbohydrate productivities as a response to nitrogen status in Isochrysis sp. (T-ISO; Haptophyceae): starvation versus limitation. J. Phycol. 48, 647–656. doi: 10.1111/j.1529-8817.2012.01154.x
Legendre, L., and Rassoulzadegan, F. (1995). Plankton and nutrient dynamics in marine waters. Ophelia 41, 153–172. doi: 10.1080/00785236.1995.10422042
Leonardos, N., and Geider, R. J. (2004). Responses of elemental and biochemical composition of Chaetoceros muelleri to growth under varying light and nitrate: phosphate supply ratios and their influence on critical N : P. Limnol. Oceanogr. 49, 2105–2114. doi: 10.4319/lo.2004.49.6.2105
Liefer, J. D., Garg, A., Campbell, D. A., Irwin, A. J., and Finkel, Z. V. (2018). Nitrogen starvation induces distinct photosynthetic responses and recovery dynamics in diatoms and prasinophytes. PLoS One 13:e0195705. doi: 10.1371/journal.pone.0195705
Lomas, M. W., and Glibert, P. M. (2000). Comparisons of nitrate uptake, storage, and reduction in marine diatoms and flagellates. J. Phycol. 36, 903–913. doi: 10.1046/j.1529-8817.2000.99029.x
Lourenço, S. O., Barbarino, E., Lavín, P. L., Lanfer Marquez, U. M., and Aidar, E. (2004). Distribution of intracellular nitrogen in marine microalgae: calculation of new nitrogen-to-protein conversion factors. Eur. J. Phycol. 39, 17–32. doi: 10.1080/0967026032000157156
Lovejoy, C., Vincent, W. F., Bonilla, S., Roy, S., Martineau, M. J., Terrado, R., et al. (2007). Distribution, phylogeny, and growth of cold-adapted picoprasinophytes in Arctic Seas. J. Phycol. 43, 78–89. doi: 10.1111/j.1529-8817.2006.00310.x
Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275.
Marie, D., Simon, N., and Vaulot, D. (2005). “Phytoplankton cell counting by flow cytometry,” in Algal Culturing Techniques, ed. R. A. Andersen (London: Elsevier), 253–267. doi: 10.1016/B978-012088426-1/50018-4
Martin, P., Dyhrman, S. T., Lomas, M. W., Poulton, N. J., and Van Mooy, B. A. S. (2014). Accumulation and enhanced cycling of polyphosphate by Sargasso Sea plankton in response to low phosphorus. Proc. Natl. Acad. Sci. U.S.A. 111, 8089–8094. doi: 10.1073/pnas.1321719111
Martiny, A. C., Ma, L., Mouginot, C., Chandler, J. W., and Zinser, E. R. (2016). Interactions between thermal acclimation, growth rate, and phylogeny influence Prochlorococcus elemental stoichiometry. PLoS One 11:e0168291. doi: 10.1371/journal.pone.0168291
Martiny, A. C., Pham, C. T., Primeau, F. W., Vrugt, J. A., Moore, J. K., Levin, S. A., et al. (2013a). Strong latitudinal patterns in the elemental ratios of marine plankton and organic matter. Nat. Geosci. 6, 279–283. doi: 10.1038/ngeo1757
Martiny, A. C., Vrugt, J. A., Primeau, F. W., and Lomas, M. W. (2013b). Regional variation in the particulate organic carbon to nitrogen ratio in the surface ocean. Glob. Biogeochem. Cycles 27, 723–731. doi: 10.1002/gbc.20061
Michaels, A. F., and Silver, M. W. (1988). Primary production, sinking fluxes and the microbial food web. Deep Sea Res. Part A. Oceangr. Res. Pap. 35, 473–490. doi: 10.1016/0198-0149(88)90126-4
Miyata, K., Hattori, A., and Ohtsuki, A. (1986). Variation of cellular phosphorus composition of Skeletonema costatum and Heterosigma akashiwo grown in chemostats. Mar. Biol. 93, 291–297. doi: 10.1007/BF00508266
Mock, T., Samanta, M. P., Iverson, V., Berthiaume, C., Robison, M., Holtermann, K., et al. (2008). Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. Proc. Natl. Acad. Sci. U.S.A. 105, 1579–1584. doi: 10.1073/pnas.0707946105
Moore, C., Mills, M., Arrigo, K., Berman-Frank, I., Bopp, L., Boyd, P., et al. (2013). Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710. doi: 10.1038/ngeo1765
Moreno, A. R., Hagstrom, G. I., Primeau, F. W., Levin, S. A., and Martiny, A. C. (2018). Marine phytoplankton stoichiometry mediates nonlinear interactions between nutrient supply, temperature, and atmospheric CO 2. Biogeosciences 15, 2761–2779. doi: 10.5194/bg-15-2761-2018
Mouginot, C., Zimmerman, A. E., Bonachela, J. A., Fredricks, H., Allison, S. D., Van Mooy, B. A., et al. (2015). Resource allocation by the marine cyanobacterium Synechococcus WH8102 in response to different nutrient supply ratios. Limnol. Oceanogr. 60, 1634–1641. doi: 10.1002/lno.10123
Myklestad, S. (1974). Production of carbohydrates by marine planktonic diatoms. I. Comparison of nine different species in culture. J. Exp. Mar. Biol. Ecol. 15, 261–274. doi: 10.1016/0022-0981(74)90049-5
Myklestad, S., Holm-Hansen, O., Vårum, K. M., and Volcani, B. E. (1989). Rate of release of extracellular amino acids and carbohydrates from the marine diatom Chaetoceros affinis. J. Plankton Res. 11, 763–773. doi: 10.1093/plankt/11.4.763
Myklestad, S. M., Skånøy, E., and Hestmann, S. (1997). A sensitive and rapid method for analysis of dissolved mono-and polysaccharides in seawater. Mar. Chem. 56, 279–286. doi: 10.1016/S0304-4203(96)00074-6
Oey, J., and Schnepf, E. (1970). Über die auslösung der valvenbildung bei der diatomee Cyclotella cryptica. Arch. Microbiol. 71, 199–213. doi: 10.1007/BF00410153
Orchard, E. D., Benitez-Nelson, C. R., Pellechia, P. J., Lomas, M. W., and Dyhrman, S. T. (2010). Polyphosphate in Trichodesmium from the low-phosphorus Sargasso Sea. Limnol. Oceanogr. 55, 2161–2169. doi: 10.4319/lo.2010.55.5.2161
Pakulski, J. D., and Benner, R. (1992). An improved method for the hydrolysis and MBTH analysis of dissolved and particulate carbohydrates in seawater. Mar. Chem. 40, 143–160. doi: 10.1016/0304-4203(92)90020-B
Pande, S., Khan, R. P., and Venkitasubramanian, T. (1963). Microdetermination of lipids and serum total fatty acids. Anal. Biochem. 6, 415–423. doi: 10.1016/0003-2697(63)90094-0
Passow, U. (2000). Formation of transparent exopolymer particles, TEP, from dissolved precursor material. Mar. Ecol. Prog. Ser. 192, 1–11. doi: 10.3354/meps192001
Penna, A., Berluti, S., Penna, N., and Magnani, M. (1999). Influence of nutrient ratios on the in vitro extracellular polysaccharide production by marine diatoms from the Adriatic Sea. J. Plankton Res. 21, 1681–1699. doi: 10.1093/plankt/21.9.1681
Piorreck, M., Klaus-Hinnerk, B., and Pohl, P. (1984). Biomass production, total protein, chlorophylls, lipids and fatty acids of freshwater green and blue-green algae under different nitrogen regimes. Phytochemistry 23, 207–216. doi: 10.1016/S0031-9422(00)80304-0
Quigg, A., Finkel, Z. V., Irwin, A. J., Rosenthal, Y., Ho, T.-Y., Reinfelder, J. R., et al. (2003). The evolutionary inheritance of elemental stoichiometry in marine phytoplankton. Nature 425, 291–294. doi: 10.1038/nature01953
Ral, J.-P., Derelle, E., Ferraz, C., Wattebled, F., Farinas, B., Corellou, F., et al. (2004). Starch division and partitioning. A mechanism for granule propagation and maintenance in the picophytoplanktonic green alga Ostreococcus tauri. Plant Physiol. 136, 3333–3340. doi: 10.1104/pp.104.044131
Raven, J. A. (1984). A cost-benefit analysis of photon absorbtion by photosynthetic unicells. New Phytol. 98, 593–625. doi: 10.1111/j.1469-8137.1984.tb04152.x
Redfield, A. C. (1958). The biological control of chemical factors in the environment. Am. Sci. 46, 205–221.
Rhee, G.-Y. (1978). Effects of N: P atomic ratios and nitrate limitation on algal growth, cell composition, and nitrate uptake. Limnol. Oceanogr. 23, 10–25. doi: 10.4319/lo.1978.23.1.0010
Richardson, B., Orcutt, D. M., Schwertner, H. A., Martinez, C. L., and Wickline, H. E. (1969). Effects of nitrogen limitation on the growth and composition of unicellular algae in continuous culture. Appl. Microbiol. 18, 245–250. PMid: 16349857.
Richardson, T. L. (2019). Mechanisms and pathways of small-phytoplankton export from the surface ocean. Annu. Rev. Mar. Sci. 11, 57–74. doi: 10.1146/annurev-marine-121916-063627
Richardson, T. L., and Cullen, J. J. (1995). Changes in buoyancy and chemical composition during growth of a coastal marine diatom: ecological and biogeochemical consequences. Mar. Ecol. Prog. Ser. 128, 77–90. doi: 10.3354/meps128077
Romanova, N., and Sazhin, A. (2010). Relationships between the cell volume and the carbon content of bacteria. Oceanology 50, 522–530. doi: 10.1134/S0001437010040089
Sañudo-Wilhelmy, S. A., Tovar-Sanchez, A., Fu, F.-X., Capone, D. G., Carpenter, E. J., and Hutchins, D. A. (2004). The impact of surface-adsorbed phosphorus on phytoplankton Redfield stoichiometry. Nature 432, 897–901. doi: 10.1038/nature03125
Sarthou, G., Timmermans, K. R., Blain, S., and Treguer, P. (2005). Growth physiology and fate of diatoms in the ocean: a review. J. Sea Res. 53, 25–42. doi: 10.1016/j.seares.2004.01.007
Siaut, M., Cuine, S., Cagnon, C., Fessler, B., Nguyen, M., Carrier, P., et al. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11:7. doi: 10.1186/1472-6750-11-7
Sicko-Goad, L. M., Schelske, C. L., and Stoermer, E. F. (1984). Estimation of intracellular carbon and silica content of diatoms from natural assemblages using morphometric techniques. Limnol. Oceanogr. 29, 1170–1178. doi: 10.4319/lo.1984.29.6.1170
Simionato, D., Block, M. A., La Rocca, N., Jouhet, J., Maréchal, E., Finazzi, G., et al. (2013). The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot. Cell 12, 665–676. doi: 10.1128/EC.00363-12
Six, C., Finkel, Z. V., Rodriguez, F., Marie, D., Partensky, F., and Campbell, D. A. (2008). Contrasting photoacclimation costs in ecotypes of the marine eukaryotic picoplankter Ostreococcus. Limnol. Oceanogr. 53, 255–265. doi: 10.4319/lo.2008.53.1.0255
Six, C., Sherrard, R., Lionard, M., Roy, S., and Campbell, D. A. (2009). Photosystem II and pigment dynamics among ecotypes of the green alga Ostreococcus. Plant Physiol. 151, 379–390. doi: 10.1104/pp.109.140566
Solorzano, L., and Sharp, J. H. (1980). Determination of total dissolved phosphorus and particulate phosphorus in natural waters. Limnol. Oceanogr. 25, 754–758. doi: 10.4319/lo.1980.25.4.0754
Sorguven, E., and Ozilgen, M. (2013). Thermodynamic efficiency of synthesis, storage and breakdown of the high-energy metabolites by photosynthetic microalgae. Energy 58, 679–687. doi: 10.1016/j.energy.2013.06.003
Sterner, R. W., and Elser, J. J. (2002). Ecological Stoichiometry: the Biology of Elements from Molecules to the Biosphere. Princeton, NJ: Princeton University Press.
Strzepek, R., and Price, N. (2000). Influence of irradiance and temperature on the iron content of the marine diatom Thalassiosira weissflogii (Bacillariophyceae). Mar. Ecol. Prog. Ser. 206, 107–117. doi: 10.3354/meps206107
Subramanian, S., Barry, A. N., Pieris, S., and Sayre, R. T. (2013). Comparative energetics and kinetics of autotrophic lipid and starch metabolism in chlorophytic microalgae: implications for biomass and biofuel production. Biotechnol. Biofuels 6:150. doi: 10.1186/1754-6834-6-150
Talmy, D., Blackford, J., Hardman-Mountford, N. J., Polimene, L., Follows, M. J., and Geider, R. J. (2014). Flexible C : N ratio enhances metabolism of large phytoplankton when resource supply is intermittent. Biogeosciences 11, 4881–4895. doi: 10.5194/bg-11-4881-2014
Taucher, J., Jones, J., James, A., Brzezinski, M. A., Carlson, C. A., Riebesell, U., et al. (2015). Combined effects of CO2 and temperature on carbon uptake and partitioning by the marine diatoms Thalassiosira weissflogii and Dactyliosolen fragilissimus. Limnol. Oceanogr. 60, 901–919. doi: 10.1002/lno.10063
Thompson, P. (1999). Response of growth and biochemical composition to variations in daylength, temperature, and irradiance in the marine diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol. 35, 1215–1223. doi: 10.1046/j.1529-8817.1999.3561215.x
Thornton, D. (2002). Diatom aggregation in the sea: mechanisms and ecological implications. Eur. J. Phycol. 37, 149–161. doi: 10.1017/S0967026202003657
Toseland, A., Daines, S. J., Clark, J. R., Kirkham, A., Strauss, J., Uhlig, C., et al. (2013). The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Chang. 3, 979–984. doi: 10.1038/nclimate1989
Tozzi, S., Schofield, O., and Falkowski, P. (2004). Historical climate change and ocean turbulence as selective agents for two key phytoplankton functional groups. Mar. Ecol. Prog. Ser. 274, 123–132. doi: 10.3354/meps274123
Turner, J. T. (2015). Zooplankton fecal pellets, marine snow, phytodetritus and the ocean’s biological pump. Prog. Oceanogr. 130, 205–248. doi: 10.1016/j.pocean.2014.08.005
Van Heukelem, L., Lewitus, J., Kana, T. M., and Craft, N. E. (1994). Improved separations of phytoplankton pigments using temperature-controlled high-performance liquid-chromatography. Mar. Ecol. Prog. Ser. 114, 303–313. doi: 10.3354/meps114303
Van Heukelem, L., and Thomas, C. S. (2001). Computer-assisted high-performance liquid chromatography method development with applications to the isolation and analysis of phytoplankton pigments. J. Chromatogr. 910, 31–49. doi: 10.1016/S0378-4347(00)00603-4
Von Dassow, P., Chepurnov, V. A., and Armbrust, E. V. (2006). Relationships between growth rate, cell size, and induction of spermatogenesis in the centric diatom Thalassiosira weissflogii (Bacillariophyta). J. Phycol. 42, 887–899. doi: 10.1111/j.1529-8817.2006.00250.x
Von Dassow, P., Petersen, T. W., Chepurnov, V. A., and Virginia Armbrust, E. (2008). Inter- and intraspecific relationships between nuclear DNA content and cell size in selected members of the centric diatom genus Thalassiosira (Bacillariophyceae). J. Phycol. 44, 335–349. doi: 10.1111/j.1529-8817.2008.00476.x
Weber, T. S., and Deutsch, C. (2010). Ocean nutrient ratios governed by plankton biogeography. Nature 467, 550–554. doi: 10.1038/nature09403
Wetz, M. S., and Wheeler, P. A. (2007). Release of dissolved organic matter by coastal diatoms. Limnol. Oceanogr. 52, 798–807. doi: 10.4319/lo.2007.52.2.0798
Worden, A. Z., Lee, J.-H., Mock, T., Rouzé, P., Simmons, M. P., Aerts, A. L., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268–272. doi: 10.1126/science.1167222
Wright, S., Jeffrey, S., and Mantoura, R. (2005). Phytoplankton Pigments in Oceanography: Guidelines to Modern Methods. Paris: Unesco Publishing.
Young, E. B., and Beardall, J. (2003). Photosynthetic function in Dunaliella tertiolecta (Chlorophyta) during a nitrogen starvation and recovery cycle. J. Phycol. 39, 897–905. doi: 10.1046/j.1529-8817.2003.03042.x
Keywords: phytoplankton, diatoms, prasinophytes, stoichiometry, nitrogen, macromolecules
Citation: Liefer JD, Garg A, Fyfe MH, Irwin AJ, Benner I, Brown CM, Follows MJ, Omta AW and Finkel ZV (2019) The Macromolecular Basis of Phytoplankton C:N:P Under Nitrogen Starvation. Front. Microbiol. 10:763. doi: 10.3389/fmicb.2019.00763
Received: 08 October 2018; Accepted: 26 March 2019;
Published: 17 April 2019.
Edited by:
James Cotner, University of Minnesota Twin Cities, United StatesReviewed by:
Michael William Lomas, Bigelow Laboratory for Ocean Sciences, United StatesDedmer B. Van de Waal, Netherlands Institute of Ecology (NIOO-KNAW), Netherlands
Copyright © 2019 Liefer, Garg, Fyfe, Irwin, Benner, Brown, Follows, Omta and Finkel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Justin D. Liefer, amxpZWZlckBtdGEuY2E=
 Justin D. Liefer
Justin D. Liefer Aneri Garg
Aneri Garg Matthew H. Fyfe
Matthew H. Fyfe Andrew J. Irwin
Andrew J. Irwin Ina Benner1
Ina Benner1 Anne Willem Omta
Anne Willem Omta