
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 April 2019
Sec. Aquatic Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00650
The cyanobacterial iron-stress-inducible isiA gene encodes a chlorophyll-binding protein that provides flexibility in photosynthetic strategy enabling cells to acclimate to low iron availability. Here, we report on the diversity and abundance of isiA genes from 14 oceanic stations encompassing large natural gradients in iron availability. Synechococcus CRD1 and CRD2-like isiA genes were ubiquitously identified from tropical and subtropical waters of the Pacific, Atlantic, and Indian Oceans. The relative abundance of isiA-containing Synechococcus cells ranged from less than 10% of the total Synechococcus population in regions where iron is replete such as the North Atlantic subtropical gyre, to over 80% in low-iron but high-nitrate regions of the eastern equatorial Pacific. Interestingly, Synechococcus populations in regions with both low iron and low nitrate concentrations such as the subtropical gyres in the North Pacific and South Atlantic had a low relative abundance of the isiA gene. Indeed, fitting our data into a multiple regression model showed that ∼80% of the variation in isiA relative abundances can be explained by nitrate and iron concentrations, whereas no other environmental variables (temperature, salinity, Chl a) had a significant effect. Hence, isiA has a predictable biogeographical distribution, consistent with the perceived biological role of IsiA as an adaptation to low-iron conditions. Understanding such photosynthetic strategies is critical to our ability to accurately estimate primary production and map nutrient limitation on global scales.
Picocyanobacteria belonging to the Synechococcus genus are among the most abundant photosynthetic organisms in open ocean ecosystems (Partensky et al., 1999; Scanlan and West, 2002) and are among the key contributors of primary production (Liu et al., 1998; Flombaum et al., 2013) and carbon cycling (Guidi et al., 2016). Their biogeography is associated with different habitats driven by several major abiotic factors including light (Grébert et al., 2018), temperature (Zwirglmaier et al., 2008), macronutrients and iron (Sohm et al., 2015; Farrant et al., 2016). The versatility of light-harvesting systems in the Synechococcus genus has substantially contributed to their ability to adapt to different underwater light environments (Stomp et al., 2004; Six et al., 2007) and exploit different ecological niches (Ting et al., 2002; Scanlan et al., 2009; Grébert et al., 2018). Their ability to change photosynthetic strategy in response to variation in iron availability (Bibby et al., 2001) is of particular interest as iron limits marine primary production in large oceanographic regions (Martin and Fitzwater, 1988; Behrenfeld et al., 1996; Boyd et al., 2000).
The chlorophyll-binding IsiA protein complex has been demonstrated to play a central role in the light-harvesting strategy of many cyanobacteria (Boekema et al., 2001; Yeremenko et al., 2004; Snow et al., 2015). Expressed by the iron-stress-inducible isiA gene of cyanobacteria (Bibby et al., 2001; Geiss et al., 2001a), the IsiA proteins form six-transmembrane helices, bind chlorophyll and form photosynthetic ‘supercomplexes’ with the reaction centers of photosystem I (PSI) (Bibby et al., 2001). Most knowledge of the IsiA protein has been acquired by the study of model cyanobacteria like the freshwater species Synechococcus PCC 7942 (Ghassemian and Straus, 1996; Boekema et al., 2001) and Synechocystis PCC 6803 (Yeremenko et al., 2004; Havaux et al., 2005). However, the gene has also been identified and characterized widely in marine cyanobacterial species (including Synechococcus, Prochlorococcus, and other marine Cyanobacterium spp.), through experiments conducted under iron limitation (Leonhardt and Straus, 1992; Bibby et al., 2003; Hewson et al., 2009; Richier et al., 2012), genomic (Palenik et al., 2006; Dufresne et al., 2008; Rivers et al., 2009) and metagenomic analyses (Bibby et al., 2009). Although isiA may be expressed under other growth conditions, including high salt (Vinnemeier et al., 1998), high heat (Vinnemeier et al., 1998; Kojima et al., 2006) and high light (Havaux et al., 2005), acclimation to iron stress appears to be the primary functional role for this chlorophyll binding complex (Ghassemian and Straus, 1996; Bibby et al., 2001; Ryan-Keogh et al., 2012).
As a single copy gene, not all oceanic strains of Synechococcus possess an isiA gene (Rivers et al., 2009; Scanlan et al., 2009). The non-isiA-harboring Synechococcus str. WH8102 and isiA-harboring str. WH8020 were isolated from the south Sargasso Sea (high dust iron input) and coastal New England shelf (highly seasonal iron dynamics), respectively, which suggests the presence of isiA in a genome might be an adaptation response to consistent low or variable iron conditions. Indeed, comparative genomics has identified Synechococcus str. WH8020 tend to have more iron-responsive genes in general compared to str. WH8102 (Mackey et al., 2015).
The IsiA-PSI supercomplex increases the functional absorption cross-section of PSI under iron stress conditions (Bibby et al., 2001; Boekema et al., 2001; Ryan-Keogh et al., 2012). However, not all of the chlorophyll associated with IsiA is photosynthetically active and overexpression of IsiA uncoupled to photosynthetic reaction centers is commonly observed when iron is depleted and macronutrients are replete (Schrader et al., 2011; Ryan-Keogh et al., 2012). There is evidence that both prokaryotic and eukaryotic phytoplankton species employ similar photosynthetic strategies that result in accumulation of energetically uncoupled chlorophyll under iron stress, which can comprise as much as 40% of total chlorophyll (Macey et al., 2014). These disconnected pools of chlorophyll elevate fluorescence yield, making it possible to map regions of iron limitation remotely (Behrenfeld et al., 2006, 2009; Lin et al., 2016). Knowledge of the biogeographical distribution of these chlorophyll-binding antenna systems, especially under high-nutrient low-chlorophyll (HNLC) conditions, is central to our ability to accurately interpret satellite chlorophyll data, to estimate ocean primary production, and to identify oceanic regions with potential iron limitation (Behrenfeld et al., 2006, 2009; Behrenfeld and Milligan, 2013; Lin et al., 2016).
Targeted studies on the diversity and biogeographical distribution of isiA genes in the global ocean, and their association with iron limitation, are scarce. In this study, we designed isiA gene primers specific for marine Synechococcus spp., and investigated the biogeographical distribution, diversity and abundance of isiA genes from 14 oceanic stations that differ widely in iron availability using clone libraries and qPCR. In particular, we aimed to derive a relationship between the abundance of the isiA gene and iron availability, which would enable the use of isiA as a genetic indicator of iron limitation in marine cyanobacteria.
Water samples were collected between 2008 and 2011, onboard the R/V Da Yang I, R/V Ke Xue I, and R/V Dong Fang Hong II (Table 1). In total, we sampled 14 stations (Figure 1), including one station in the South China Sea (SCS), three stations in the equatorial Pacific Ocean (two in the typical HNLC eastern equatorial Pacific Ocean, EEP1 and EEP2; and one in the western Pacific warm pool, WPWP), two stations in the western part of the North Pacific subtropical gyre (N18 and N7), two stations in the Indian Ocean (NIO and SIO), four in the Atlantic Ocean (At7, At17, At27, and At37), and two in the Bering Sea (BS23 and BS24). At all stations, samples were taken from just below the water surface (5 m depth) using a rosette sampler with 20-L Niskin bottles. In addition, at four stations (SCS, EEP1, SIO, and NIO) we investigated a depth profile with samples from 5, 50, 100, and 200 m depth. From each sample, 2-L of water was filtered through 0.22-μm pore size 47-mm diameter polycarbonate filters (Millipore, Bedford, MA, United States), and immediately frozen and stored at -80°C until DNA extraction. An extra 2-mL of seawater was sampled in triplicate, fixed with glutaraldehyde (final concentration 0.1%, Marie et al., 1997), quick-frozen in liquid nitrogen and stored in a freezer at -20°C for later flow cytometry analysis.
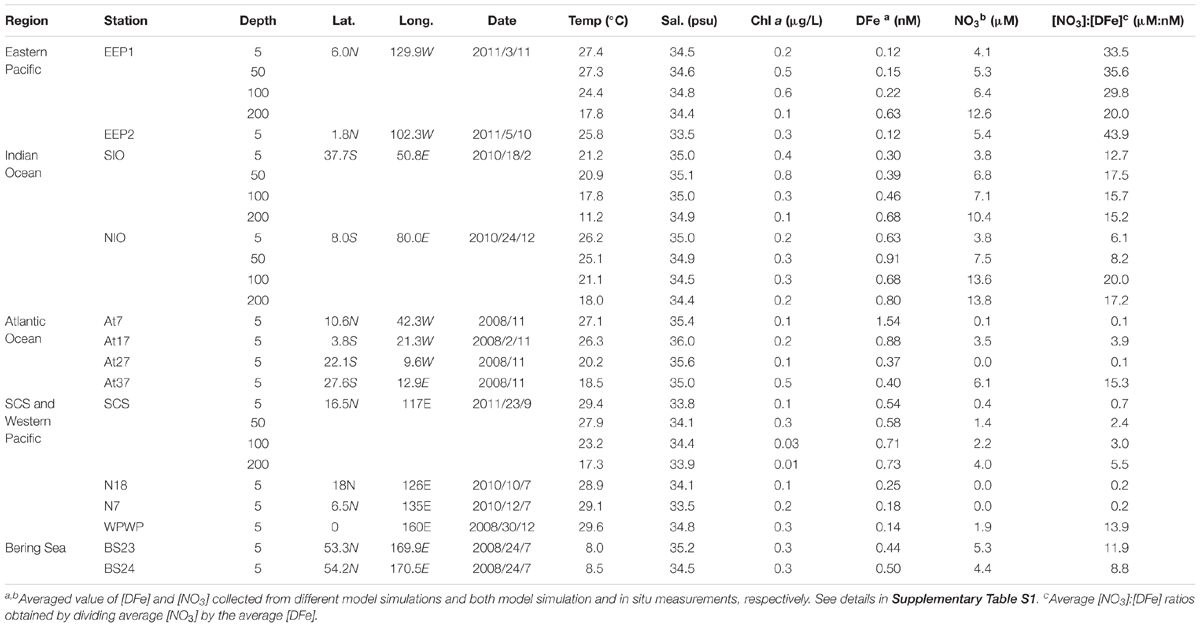
Table 1. Sampling information and relevant environmental parameters in the surface water of each station.
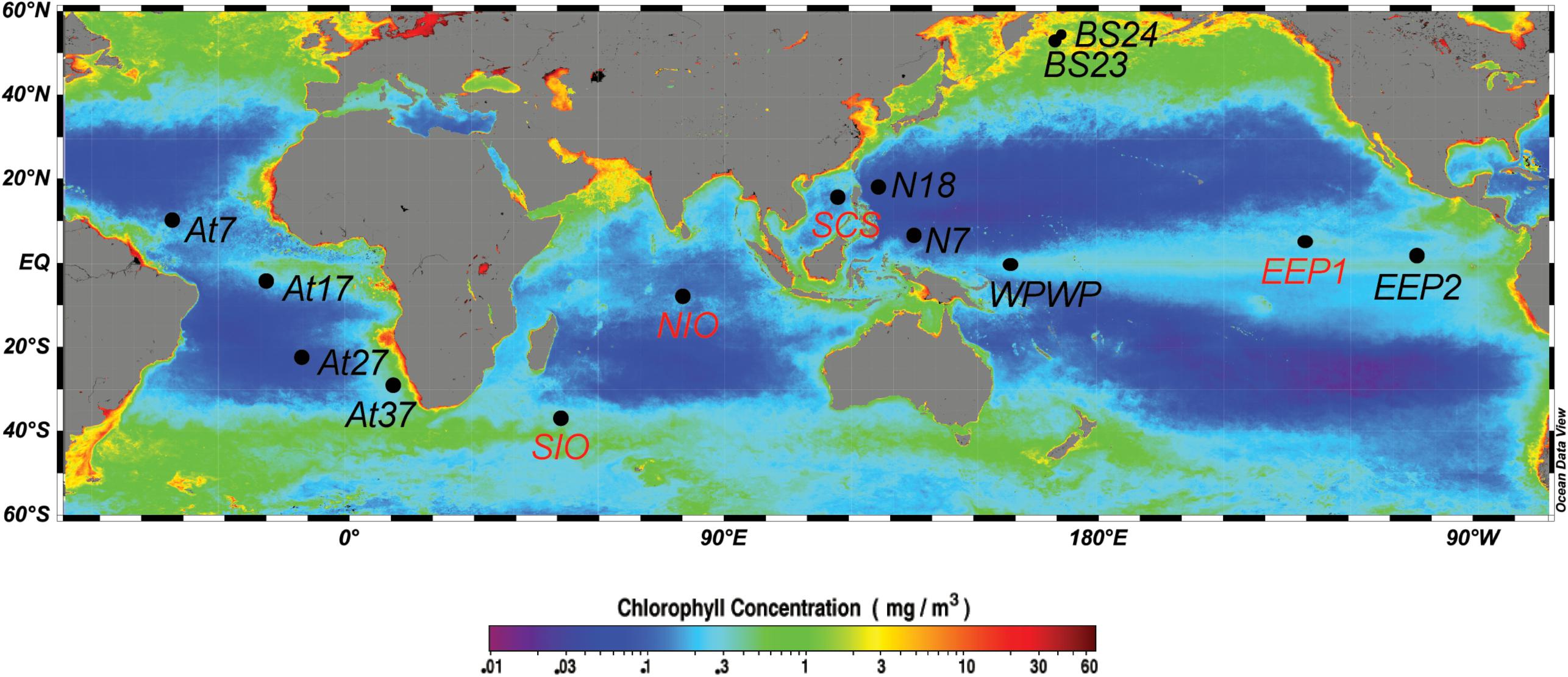
Figure 1. Map showing the 14 sampling stations, including eight surface stations (black labels: BS23, BS24, N18, N7, At7, At17, At27, At37) and four vertically resolved stations (red labels: SCS, EEP1, SIO, NIO). Sampling sites were mapped using Ocean Data View (Schlitzer, 2012), with the base map of annually averaged (2008) surface chlorophyll a concentration extracted from the NASA website (http://oceancolor.gsfc.nasa.gov/).
Profiles of temperature, salinity, and total chlorophyll a at each station were measured with a SeaBird CTD (SBE 9/11 plus, SeaBird Inc., United States) equipped with auxiliary sensors for chlorophyll a fluorescence. NOx (nitrate+nitrite) concentrations were measured after reduction of nitrate to nitrite with a Technicon AA3 Auto Analyzer (Braun-Lube, GmbH) using standard protocols (Grasshoff et al., 1999) (Table 1). Nitrite concentrations were measured similarly but without the reduction step and subtracted from NOx to calculate the nitrate concentration [NO3]. The detection limits for NOx were 0.03 μM.
Dissolved iron (DFe) concentrations were retrieved from two different models on different time scales (Table 1 and Supplementary Table S1). (1) NASA’s Ocean Biogeochemical Model (NOBM1) according to Gregg et al. (2003), from which we retrieved daily (at the sampling day, based on daily simulation), monthly (at the sampling month, based on monthly simulation) and annually averaged (at the sampling year, based on monthly simulation) DFe concentrations at a special resolution of 1.25 × 0.67 degree (longitude-latitude). (2) The Ecco2-Darwin model from which we retrieved the monthly (at the sampling month, based on monthly simulation) and annually averaged (at the sampling year, based on monthly simulation) DFe at 1 × 1 degree (longitude-latitude) (data file available to download2) (Behrenfeld et al., 2009; personal communication with Oliver Jahn and Stephanie Dutkiewicz). DFe concentrations at the four stations for which we collected depth profiles were only retrieved from the Ecco2-Darwin model (at both monthly and annually time scale). A caveat of this approach is that the simulated DFe concentrations may not represent the exact DFe concentration in situ, and thus may introduce additional variance in the correlation between isiA and iron. However, using the mean of two independent models and different time scales could help mitigate some of this variation, and possibly retrieve the closest-to-real iron and nitrogen conditions the ambient Synechococcus had been experiencing. To assess the reliability of this modeling approach, we also retrieved NO3 concentrations from the models, which matched well with the NO3 concentrations measured in our study (Supplementary Table S1). Therefore, the averaged DFe and NO3 concentrations based on two models at two or three simulation time scales and the NO3 in situ measurements were used for our statistical analysis.
The molar ratio of [NO3]:[DFe] is widely used in biogeochemistry models (Fung et al., 2000; Tagliabue et al., 2016; Browning et al., 2017) to indicate iron and/or nitrate limitation. It is thus applied for this study to characterize relative iron availability as it also describes conditions where nitrate and iron are co-limiting (Moore et al., 2013; Browning et al., 2017). Available observational data from earlier studies near our sampling stations show that this ratio provides a good indication of nutrient limitation patterns in the oceans (Supplementary Table S2). More specifically, [NO3]:[DFe] ratios <1 μM/nM indicate that nitrate is the primary limiting nutrient, whereas ratios >10 (Browning et al., 2017) or >5 μM/nM (Biller and Bruland, 2014) suggest iron is primarily limiting.
Abundances of Synechococcus were determined using 2-mL seawater aliquots on an Epics Altra II flow cytometer (Beckman Coulter, Brea, CA, United States) with a 306C-5 argon laser (Coherent, Santa Clara, CA, United States), according to the method of Jiao et al. (2002). They were identified and distinguished from other autotrophs based on their positions in plots of side scatter versus red fluorescence and orange fluorescence versus red fluorescence. Event rates were set to 50–200 events/second (0.1–1 mL h-1) in order to enhance the particle capturing sensitivity. Fluorescent microspheres of 1 μm diameter (Polysciences Inc., Warrington, PA, United States) were added to all samples as an internal standard to calibrate flow rate and cell size. All samples were run in triplicate. The data were analyzed with EXPOTM32 MultiCOMP software (Beckman Coulter, United States).
The genomic DNA of the bacterial community was extracted using a commercial fast DNA kit (Qiagen, United States). Software package Primer 5.0 and Geneious R10 (Biomatters Ltd., United States) was used to generate new isiA primers targeting Synechococcus spp. based on the conserved protein sequences. Detailed information on the primer design is given in the “Results” section, Supplementary Figures S1, S2 and a collection of all isiA and pcb genes used as references is summarized in Supplementary Table S3.
The new degenerate primer pair isiA33F (5′-ACY TAT GAC TGG TGG GC-3′) and isiA654R (5′-CCD CCC ATV ACR TCY TC-3′) (with variable nucleotides Y = C/T, D = A/G/T, V = A/G/C, and R = A/G) was then tested for specificity and cross-reactivity with marine Synechococcus str. CC9605 (as a positive control), str. WH8102 (as a negative control) and field samples from station SIO. A predicted ∼625 bp fragment was obtained by PCR performed with Ex Taq PCR Mix (TaKaRa, Dalian, China). The PCR program was run in a T3 thermocycler (Biometra, Germany) and consisted of an initial 94°C denaturation step for 5 min, 35 cycles of denaturation at 94°C for 45 s, annealing at 52°C for 45 s and extension at 72°C for 45 s. Amplification products were gel-purified, ligated into pMD18-T vectors (TaKaRa, Dalian, China), and transferred to Escherichia coli DH5α-competent cells (TaKaRa, Dalian, China). Positive clones were randomly chosen and screened with vector M13-F/M-13R using an ABI model 3730 automated DNA sequence analyzer with BigDye terminator chemistry (Applied Bio Systems, Perkin-Elmer).
DNA sequences were analyzed, aligned and translated into protein sequences (∼200 amino acids) using MEGA version 6 (Tamura et al., 2013) and Genieous R10 (Biomatters Ltd., United States). Multiple alignments of MAFFT v7.388 E-INS-i (Katoh and Standley, 2013) with BLOSUM62 scoring matrix was used for the sequence alignments. DNA and protein sequences were afterward blasted against the NCBI gene bank using the BLASTN and BLASTP tools to aid the selection of the closest reference sequences. Operational taxonomic units (OTUs) of sequences sharing identities of at least 90% for DNA and 97% for protein were identified and their Shannon diversity index was calculated using the DOTUR program (Schloss and Handelsman, 2005). These thresholds were tested to be most suitable for grouping closely related isiA/IsiA phylotypes with different references.
Maximum likelihood phylogenetic trees using 97% identity IsiA protein sequences were then constructed and compared both in MEGA using the Gamma LG model and RAxML3. Bootstrap analysis was used to estimate the reliability of phylogenetic reconstructions under 100 replicates. The final IsiA protein sequence phylogenetic tree was edited with iTOL4 (Letunic and Bork, 2016).
The abundance of Synechococcus isiA genes was determined using the quantitative PCR (qPCR) method and running with the CFX 96TM real-time system (Bio-Rad, Singapore). The specific primer pair for amplifying isiA genes (isiA33F, 5′-ACY TAT GAC TGG TGG GC-3′ and isiA348R, 5′-CCG DAG DGT RTG CCA GAT-3′; target isiA genes between length 33 bp and 348 bp) was designed using the same method as for the PCR primers. These qPCR primers were then checked for specificity and cross-reactivity with Synechococcus str. CC9605 (as a positive control) and str. WH8102 (as a negative control). Standard curves were constructed using a mixture of equal amounts of the plasmid DNA of the three most dominant isiA phylotype clones (374-WPWP-5m, 440-BS23-5m, and 367-SIO-50m). Using ten-fold increments, the standard concentrations were adjusted from 107 to 101 gene copies μL-1 of isiA. The reaction mixture (25 μL) contained 12.5 μL SYBR Premix Ex Taq II (TakaRa, Dalian, China), 0.5 μM of each primer and 1 μL template DNA, and qPCR thermal cycling conditions were the same as described for the PCR amplification. The amplification efficiencies of qPCR were always between 80 and 95% with R2 values > 0.99, and the sensitivity ranged from 102 to 108 isiA gene copies L-1 in the environmental samples (Supplementary Table S6). The specificity of the qPCR reactions was confirmed by melting curve analysis, agarose gel electrophoresis and sequencing analysis.
The environmental isiA sequences obtained in this study have been deposited in the NCBI GenBank database under accession numbers MF772983–MF773424.
Redundancy analysis (RDA) was applied to investigate correlations between isiA phylotypes and the measured environmental parameters. RDA was performed using forward selection with the Ordistep function in the R package vegan (Oksanen et al., 2015) using R version 3.3.3 (R Development Core Team, 2017). Significance was based on a permutation test with 999 permutations to select only those environmental parameters that contributed significantly (Padjusted < 0.05) to the RDA model (Zuur et al., 2009).
Multiple regression was used to investigate the relationship between the relative abundance of isiA-containing Synechococcus (isiA:cell, expressed as number of isiA gene copies L-1 divided by the number of Synechococcus cells L-1) and the measured environmental parameters. From a biological perspective, we are particularly interested in the hypothesis that iron limitation favors a high relative abundance of isiA-containing Synechococcus cells. That is, we would like to know whether the isiA:cell ratio is negatively correlated with iron availability, but positively correlated with other potentially limiting nutrients. The number of isiA gene copies L-1 and the number of Synechococcus cells L-1 were both approximately log-normally distributed, whereas [DFe] and [NO3] were normally distributed. We therefore investigated the following multiple regression model:
where we used log base 10. We note that, from a statistical perspective, the use of ratios of two variables can be problematic in regression analysis, because it can lead to spurious correlations (e.g., Kronmal, 1993). However, log(x:y) is equivalent to log(x) - log(y), and therefore the above regression model can also be written in a slightly more elaborate form as:
Regression analysis showed that the value a0 was very close to 1, which supports the use of log(isiA:cell) as the response variable in the regression model. A normal quantile–quantile (Q–Q) plot of the standardized residuals was used to assess the goodness-of-fit of the regression model.
IsiA gene diversity and abundance was retrieved from several oceanic environments, including samples from the equatorial Pacific Ocean (EEP1, EEP2, WPWP), equatorial Atlantic Ocean (At17), Northern Indian Ocean (NIO), Southern Indian Ocean (SIO), SCS, and Bering Sea (BS23, BS24) (Table 1 and Figure 1). In addition to surface samples, stations EEP1, SCS, SIO and NIO were sampled over depth profiles to 200 m. Abundance of isiA was also investigated in the western part of the North Pacific subtropical gyre (N7, N18), the North Atlantic subtropical gyre (At7) and southern Atlantic (At27, At37), although we did not investigate isiA gene diversity at these stations. The stations encompass a wide range of dissolved iron (DFe) concentrations, from <0.2 nM to >1.5 nM (Table 1). The [NO3]:[DFe] ratio ranged from <1 μM/nM at stations At7, At27, SCS, N18 and N7 to >30 μM/nM at stations EEP1 and EEP2 (Table 1 and Supplementary Tables S1, S5).
The previously published primer pairs isiAfw (WWAGNAR)/isiArev (PYFADT) (with a product length of ∼860 bp) and isiAfw (WWAGNAR)/isiArev (HLWHA) (with a product length of ∼1,000 bp), which were designed based on freshwater isiA genes (Geiss et al., 2001a,b), were first tested with our samples but both had poor amplification results. Therefore, we screened a total of 28 marine Synechococcus genomes for the isiA gene and designed new PCR primers. Out of those 28 genomes, 12 genomes harbored the isiA gene (Supplementary Table S3) and their translated IsiA protein sequences were aligned and screened manually for conserved regions. A completely conserved peptide sequence near the N-terminal (YDWWAG), modified from Geiss et al. (2001a,b), was chosen to design the degenerated forward primer isiA33F (Supplementary Figure S1). Several other conserved regions (213–218 bp, EDVMGG; 306–311 bp, PYFSDT; 346–351 bp, HFWHAL) were also found downstream the protein sequences (Supplementary Figure S1). The nucleotide sequences encoding these peptides were aligned, and reverse primers (e.g., isiA654R, isiA933R and isiA1053R) were designed after sequence comparisons. Testing on sample SIO-50m, the primer pair isiA33F (5′-ACY TAT GAC TGG TGG GC-3′) and isiA654R (5′-CCD CCC ATV ACR TCY TC-3′) generated the best amplification results in terms of diversity (Supplementary Figure S3) and thus was chosen for this study.
To assess the specificity of qPCR primers, an extra 28 homologous prochlorophyte-chlorophyll-binding pcb genes of 10 Prochlorococcus str. were recruited for screening together with the 12 isiA sequences (Supplementary Table S3 and Supplementary Figures S1, S2). A conserved region specific for isiA was manually found between 111 and 116 bp encoding the peptide sequence IWHTLR (Supplementary Figure S1), corresponding to their reverse gene primer isiA348R. This peptide sequence distinguishes IsiA from Pcb and the resulting qPCR product is short enough (∼300 bp) to yield high quantification efficiency. Applying the PCR and qPCR primers, ∼600 bp partial isiA/pcb fragments were amplified from all isiA-containing Synechococcus and some Prochlorococcus genomes, and ∼300 bp short isiA fragments were only retrieved from isiA-containing Synechococcus genomes, respectively (Supplementary Figure S2). Further sequencing of those fragments retrieved isiA genes covering all known isiA-harboring Synechococcus clades as well as unknown phylotypes (Figure 2 and Supplementary Table S4).
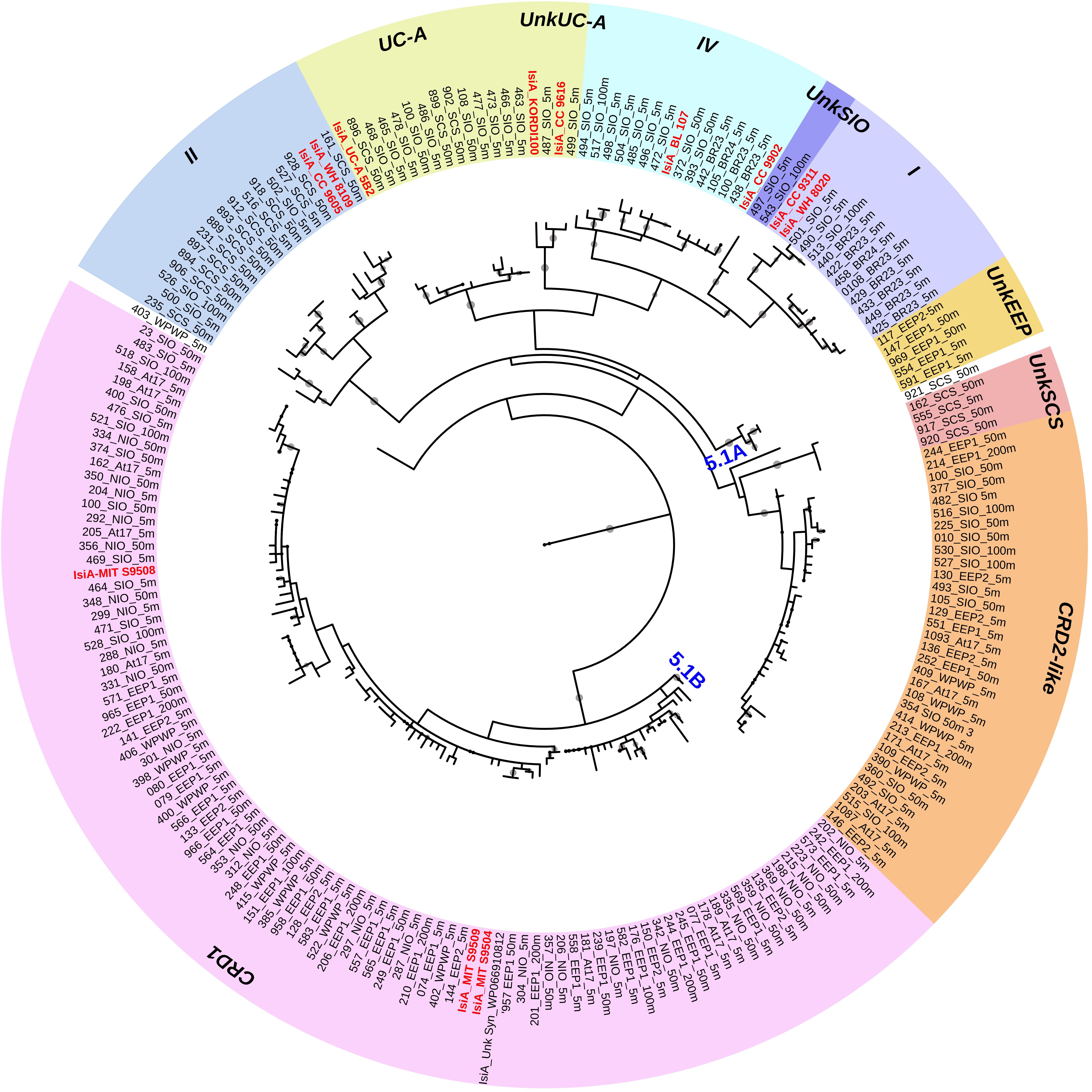
Figure 2. Circular maximum likelihood phylogenetic tree drawn based on 3% OTU (operational taxonomic units) cutoff of IsiA protein sequences. Each of the retrieved OTU is written in black in the form of clone number_station_depth, and known reference strains are indicated in red. Individual bootstrap values over 50% are indicated by circles of varying size on each branch. Gene phylotypes belonging to different Synechococcus clades are indicated by different colors. Further information on reference strains and the classification of each sequence can be found in Supplementary Tables S3, S4, respectively.
A total of 440 Synechococcus isiA sequences were amplified from all stations except for 100 m and 200 m of NIO and 200 m of SCS and SIO (probably due to low Synechococcus numbers) and were assigned to nine phylotypes belonging to different Synechococcus clades (Figures 2, 3A). Using the previously identified isiA genes (Supplementary Table S3) and classification of Synechococcus clades (Saito et al., 2005; Scanlan et al., 2009; Mazard et al., 2012; Ahlgren et al., 2014), the most abundant isiA sequences in our study were from clade CRD1 (accounting for ∼50% of the total sequences; Figure 3A), a clade that was originally recruited in the upwelling Costa Rica dome and later widely discovered in upwelling tropical/subtropical oceans (Saito et al., 2005; Sohm et al., 2015; Farrant et al., 2016), with three closest known reference strains MIT S9504, MIT S9508, MIT S9509 (Supplementary Table S3) (Cubillos-Ruiz et al., 2017). The second-most abundant clade was a group without known references (yet-to-be cultivated) but frequently co-existed with clade CRD1, and thus likely clade CRD2 according to Ahlgren et al. (2014) and Sohm et al. (2015) or EnvB according to Farrant et al. (2016) (labeled as IsiA-CRD2-like in our study) (Figures 2, 3). Other commonly amplified Synechococcus that harbor isiA belonged to clades I, IV (commonly found in cold waters with higher nutrients) (Sohm et al., 2015) and UC-A (inhabiting oligotrophic ocean with relatively low abundance of the population) (Choi et al., 2014). The remaining isiA sequences comprised another known clade II (commonly found in warm and oligotrophic waters) (Sohm et al., 2015) and three small unknown clades of unkSCS (exclusively retrieved from SCS), unkSIO (exclusively retrieved from SIO), and unkEEP (exclusively retrieved from EEP).
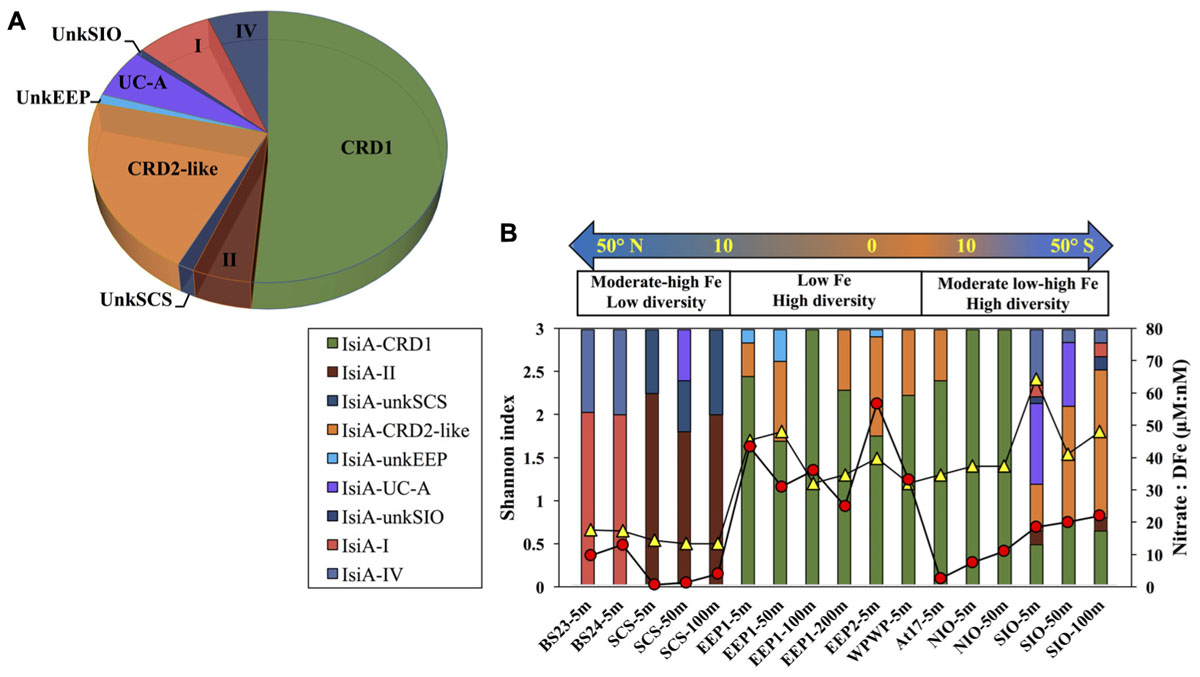
Figure 3. (A) Overall relative abundance of the nine isiA clades, across all stations. (B) Composition of isiA clades at each station arranged along a latitudinal gradient from 50° N to 50° S. The [NO3]:[DFe] ratio (red circles) and Shannon index of sequence diversity (yellow triangles) are also shown for each station.
Distinct variation in the diversity and composition of isiA genes was demonstrated along a latitudinal gradient, and among different iron habitats (as characterized by the [NO3]:[DFe] ratio) (Figure 3B). CRD1 and CRD2-like clades were most commonly retrieved together and dominant in 10°N–10°S warm waters, with high diversity of isiA genes (as represented by the Shannon diversity index). Although isiA genes of clades CRD1 and CRD2-like were obtained from a wide range of iron conditions, their distribution was mostly associated with high [NO3]:[DFe] ratios, as demonstrated by RDA (Figure 4). Clade II dominated at all three depths of station SCS, and all three depths were characterized by a low isiA diversity, and low [NO3]:[DFe] ratio (Figure 3B). The isiA genes of the high-latitude stations BS23 and BS24 with moderate-low [NO3]:[DFe] were co-dominated by clade I and clade IV, showed a low isiA diversity and in line with expectation that their isiA gene compositions at these stations was associated with low temperature and high latitude (Figures 3B, 4). By contrast, the three depths sampled at station SIO of moderate-high [NO3]:[DFe] ratios have also showed high isiA diversity, including all isiA-harboring clades (CRD1, CRD2, I, II, IV) recovered from other stations and also some clades unique to station SIO like UC-A and unkSIO. Yet, their isiA composition was also associated with low temperature and high latitude (Figures 3B, 4).
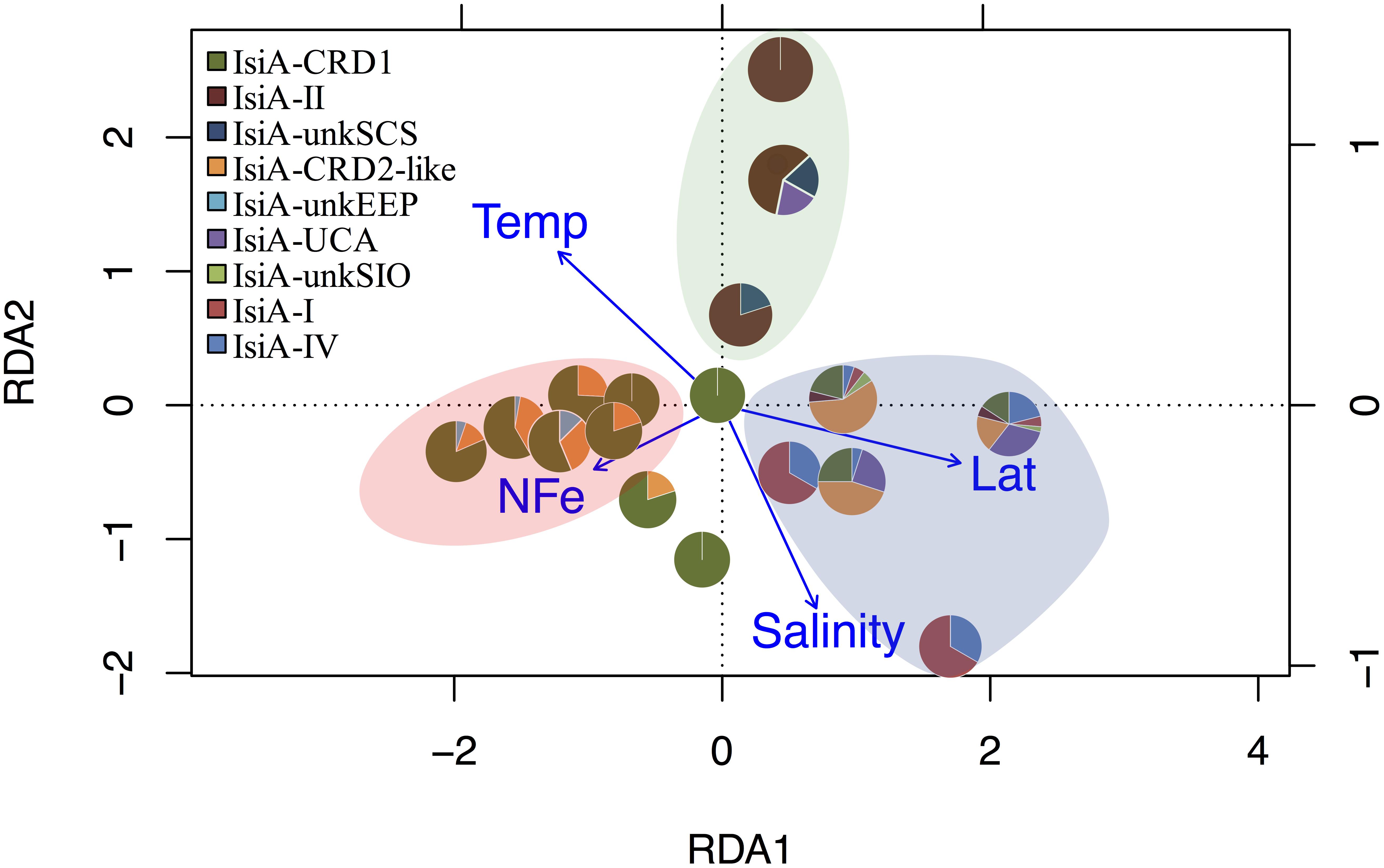
Figure 4. RDA tri-plot indicating the correlation between the isiA clades identified in this study and measured environmental parameters. Blue arrows denote the environmental variables that significantly correlated with sample ordination, and pie charts depict the relative abundances of the isiA clades at each station. Background shading in different colors points at three biogeographical clusters. The pink-beige cluster includes the low-Fe stations of EEP1, EEP2, and WPWP; the blue cluster includes the high-latitude cold stations of BS23, BS24, and SIO; the green cluster consists exclusively of the high-Fe and warm water at the three depths of station SCS.
In the surface waters (5 m depth) across all stations, isiA gene abundances were well above the detection limit of our qPCR assay at all stations, and ranged from 5.55 × 105 gene copies L-1 at station At7 in the north tropical Atlantic Ocean to 2.72 × 107 gene copies L-1 at station EEP2 in the eastern equatorial Pacific Ocean. At greater depths of 200 m at stations SCS, isiA gene abundance declined possibly below the detection limit of ∼800 gene copies L-1 (Supplementary Table S6). Synechococcus abundance ranged over ca. one order of magnitude in the surface, from 2.33 × 106 cells L-1 at station SCS to 3.81 × 107 cells L-1 at station NIO (Supplementary Table S6).
The relative abundance of isiA-containing Synechococcus, isiA:cell, varied from <10% to >80% (Figure 5 and Supplementary Tables S5, S6). High isiA:cell (>50%) were observed in potentially iron-limited waters (with high [NO3]:[DFe] ratio) of the eastern equatorial Pacific (5 m and 50 m at station EEP1, 5 m at EEP2), western equatorial Pacific (5 m at station WPWP) and the southern Indian Ocean (5 m, 50 m, and 200 m at station SIO). Deeper water depths at station EEP1 (100 m and 200 m), SIO (100 m) and NIO (100 m), 5 m at station NIO in the Northern Indian Ocean and 5 m at station At37 in the southern Atlantic Ocean also revealed relatively high values (40–50%) of isiA:cell, followed by moderate to low values (20–40%) for the Bering Sea (stations BR23, BR24), SCS (5–100 m at station SCS), 50 m at station NIO and equatorial Atlantic Ocean (station At17). Lowest isiA:cell values (<10%) were observed in the iron rich water (with low [NO3]:[DFe] ratio) of the North Atlantic subtropical gyre (station At7). Interestingly, water sampled from environments with both low-Fe and low-N concentrations in the western part of the North Pacific subtropical gyre (stations N7 and N18) and the South Atlantic subtropical gyre (station At27) also showed low isiA:cell (Figure 5 and Supplementary Tables S5, S6).
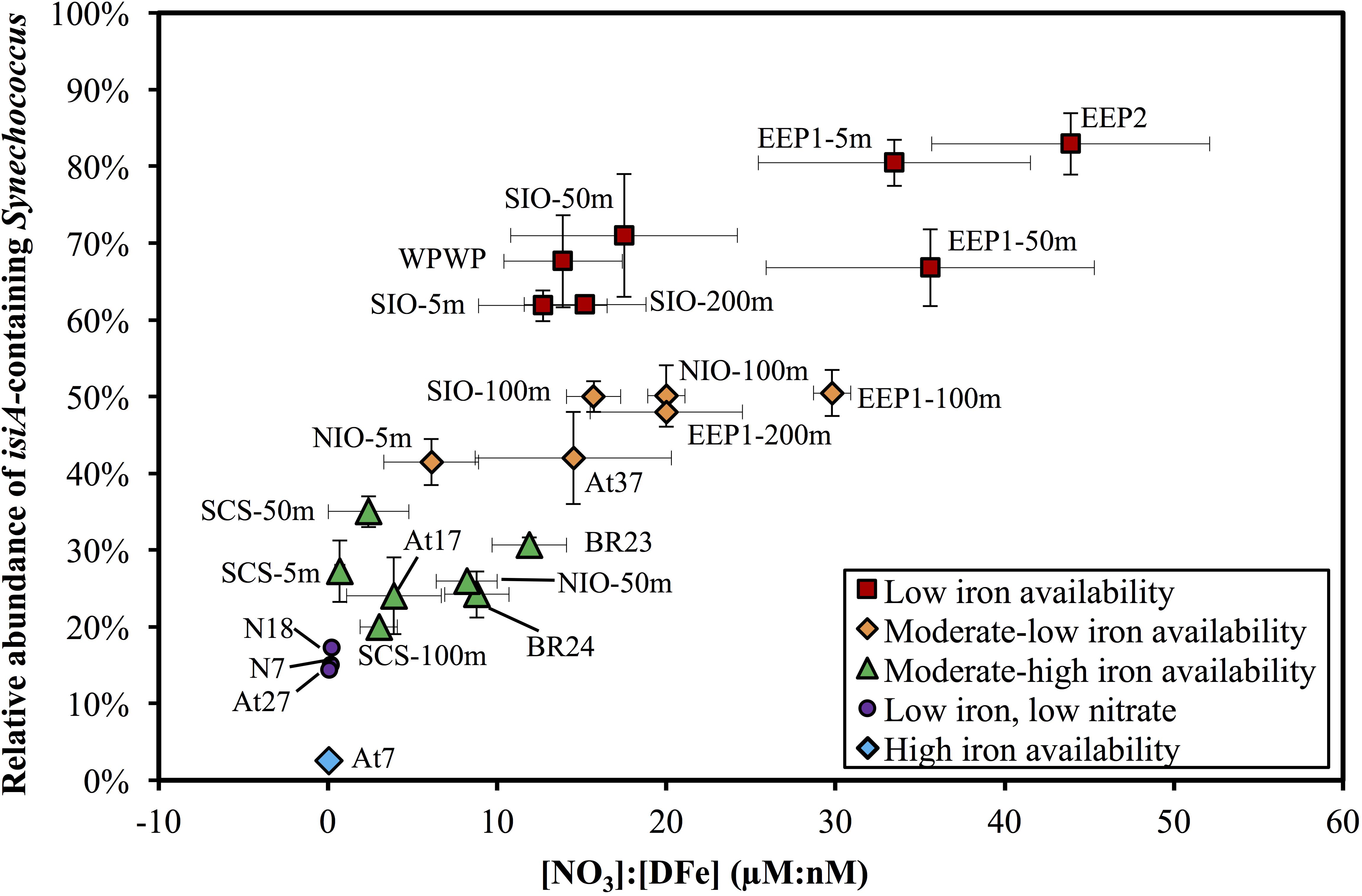
Figure 5. Scatter plot of the [NO3]:[DFe] ratio and the relative abundance of isiA-containing Synechococcus across the surface waters of all 14 stations and depth profile of four stations. Colored symbols represent five iron availability categories, see Supplementary Table S5 for the criteria used to distinguish these categories. Vertical and horizontal error bars on each dot represents the standard deviations of isiA:cell and [NO3]:[DFe], respectively.
Multiple regression analysis revealed a significant negative effect of iron (in nM) and a significant positive effect of nitrate (in μM) on the relative abundance of isiA-containing Synechococcus cells, according to the following regression model (R2 = 0.81, n = 24, p < 0.001):
The normal Q–Q plot of the standardized residuals indicates that the regression model fitted well to the data (Supplementary Figure S4). Interestingly, regressions with only iron (Log(isiA:cell) = -0.70[DFe] - 0.11) or only nitrate (Log(isiA:cell) = 0.05[NO3] - 0.68) were also significant (P < 0.001 and P < 0.01, respectively), but had a much lower explanatory power (R2 = 0.46 and R2 = 0.30, respectively; Supplementary Figures S5a,b). Hence, it is the combination of both iron and nitrate that determines the relative isiA-abundance. Other environmental variables (temperature, salinity, Chl a) were also fitted into the regression model, but none of these variables had a significant effect (P > 0.05; Supplementary Figures S5c–e). Altogether, variation in [DFe] and [NO3] explained more than 80% of the variation in the relative abundance of isiA-containing cells across stations and water depths.
We characterized distinct sets of isiA-harboring Synechococcus that have adapted to different ocean habitats defined by iron availability (Figures 3B, 4). The most ubiquitous isiA-containing clades, CRD1 and CRD2-like, were not only recovered and dominant in low-Fe habitats of the eastern equatorial Pacific (EEP1 and EEP2), but also at stations with higher Fe availability in both the Indian Ocean (e.g., NIO) and Atlantic Ocean (e.g., At17). This is interesting as previous studies have suggested that clades CRD1 and CRD2 were restricted to low-Fe environments (Sohm et al., 2015). Globally dominant Synechococcus clades, such as clades II (and possible III), were retrieved from some stations but not as commonly as CRD1 and CRD2-like. We therefore assume clades II and III have selectively lost or not gained the isiA gene during evolution. Indeed, all known genomes of clade III (e.g., WH8102, WH8103) and several clade II strains (e.g., KORDI-52) lack the isiA gene (Rivers et al., 2009; Scanlan et al., 2009) (Supplementary Table S3). This is in line with previous studies that have found no evidence for “low-Fe specialists” in those clades (Sohm et al., 2015; Farrant et al., 2016). One could assume that those species of clade II and III are likely to benefit from episodic iron enrichments whereas clade CRD1 and CRD-2like tend to proliferate in low-iron environments.
In addition to the more ubiquitous isiA-containing clades, several other isiA phylotypes were exclusively recovered from specific stations, such as unkEEP from the low-iron eastern equatorial Pacific (stations EEP1, EEP2), and thus seemed to have specifically adapted to that in situ ecological niche. At high-latitude stations (e.g., the Bering Sea), the known cold-water prevalent clades I and IV were the dominant isiA-containing Synechococcus. These results are generally in agreement with previous studies on cyanobacterial biogeographical distribution (Zwirglmaier et al., 2008; Huang et al., 2011; Mazard et al., 2012; Ahlgren et al., 2014; Sohm et al., 2015), especially the recently proposed close relationship between finer (than clade level) “ecologically significant taxonomic units” (ESTU) and specific ecological niches driven by temperature, iron, phosphate (Farrant et al., 2016) and light (Grébert et al., 2018) through the Tara ocean expedition. Interestingly, the high diversity of isiA-harboring Synechococcus revealed from station SIO in the present study, is consistent with the various low-iron “specialist” cyanobacteria observed earlier at an adjacent station east of Madagascar (TARA_052) (Farrant et al., 2016). This may suggest a special habitat off Madagascar supporting the prevalence of isiA-harboring and other genetically tuned low-iron adapted cyanobacteria.
Through our isiA gene quantification study, we have demonstrated a strong relationship between the relative abundance of isiA-containing Synechococcus cells and the iron and nitrate concentrations across a diverse range of ocean environments in the Atlantic, Pacific, and Indian Oceans. Specifically, the relative abundance of the isiA gene is negatively related with iron availability but positively related with nitrate availability. For instance, HNLC regions (e.g., stations EEP1 and EEP2) in which macronutrients such as nitrate are in ample supply but iron is limiting have a high relative abundance of the isiA gene. These results indicate that the presence of the isiA gene provides a clear selective advantage for Synechococcus assemblages living in iron-limited but nitrate-replete environments. In comparison, the isiA gene does not appear to provide a selective advantage in high-iron habitats (e.g., in the North Atlantic subtropical gyre), where the relative abundance of isiA-containing Synechococcus cells is low. Likewise, environments are limited by both nitrate and iron, e.g., stations N7 and N18 (Li et al., 2015), have a low relative abundance of the isiA gene as well, suggesting that the isiA gene does not offer a selective advantage in environments where nitrogen is limiting. These results indicate that both iron and nitrate exert a major selection pressure on the presence of the isiA gene in cyanobacterial populations.
Our results are consistent with previous studies pointing out that macronutrients such as nitrate must be considered when using isiA and other iron-responsive genes as biomarkers of iron limitation (Behrenfeld et al., 2006; Schrader et al., 2011; Mackey et al., 2015). Laboratory experiments have shown that accumulation of the IsiA protein is substantially less when experiencing synergetic limitation of nitrogen and iron rather than iron limitation alone (Schrader et al., 2011), and physiological signatures of iron limitation were only observed when nitrate is replete (Moore et al., 2013; Browning et al., 2017). The high relative abundance of the isiA gene that we found in HNLC regions is in agreement with the distinct fluorescence signatures reported from these HNLC regions, which have commonly been attributed to the accumulation of chlorophyll-binding proteins such as IsiA (Behrenfeld et al., 2006, 2009; Schrader et al., 2011; Behrenfeld and Milligan, 2013; Macey et al., 2014). Our results therefore strengthen the argument proposed by Behrenfeld et al. (2006, 2009) and Behrenfeld and Milligan (2013), that it is important to separate the functionally uncoupled chlorophyll molecules bound by IsiA from functional chlorophyll molecules for accurate primary production estimates, especially in HNLC areas.
Some laboratory studies indicated an increased expression of the isiA(BC) operon at high salt stress (Vinnemeier et al., 1998), heat stress (Vinnemeier et al., 1998; Kojima et al., 2006) and high light stress (Havaux et al., 2005). However, these studies were performed with freshwater Synechocystis and thus may not be relevant to marine Synechococcus species. Furthermore, these laboratory studies applied extremely high salinities, irradiance and temperatures that are not encountered in the oceans. In our field data, we found a significant relation between isiA and iron and nitrate availability, but the relative abundance of isiA did not show a significant relationship with salinity or temperature (Supplementary Figures S5c–e). Furthermore, the relative abundance of isiA was not positively associated with high light conditions either (most likely happens in the very surface water), as the relative abundance of isiA-containing Synechococcus cells tend to remain rather constant or even slightly increased with depth at several stations vertically sampled (SCS, SIO, NIO, EEP1; Supplementary Figure S6).
Our results suggest that the relative abundance of the isiA gene can be added to the ‘toolbox’ of molecular markers assessing the iron nutritional status of marine phytoplankton. Similar molecular approaches have previously been applied to the diatom Thalassiosira oceanica (Chappell et al., 2015) and marine cyanobacteria (Webb et al., 2001; Chappell et al., 2012), based on expression differences of known genes involved in acclimation to iron stress. Such biological responses at the gene level can be a powerful proxy for iron bioavailability, which is sometimes difficult to assess by chemical measurements alone because of the complex speciation of iron and its interactions with other molecules (Chappell et al., 2015; Browning et al., 2017).
In conclusion, this study provides novel information on the biogeographical distribution of the photophysiologically important isiA gene. Consistent with earlier results of a more limited geographical extent (e.g., Bibby et al., 2009), we found a close relationship with iron bioavailability in various ocean environments. More specifically, of all investigated oceanographic parameters, only iron and nitrate availability were strongly correlated with the relative abundance of isiA.
Therefore, we propose here that the relative abundance of isiA-containing Synechococcus cells (isiA:cell) is driven by adaptation to iron limitation and thus can be used as a potential biomarker for iron limitation under nitrate replete conditions. Obtaining estimates of the relative abundance of isiA-containing cells through qPCR could aid future iron limitation studies given the challenges of frequent contamination in trace-metal-clean techniques (Fitzwater et al., 1982) and the extremely complex speciation of iron ligands (Hutchins et al., 1999). Further laboratory work investigating iron-related physiology of IsiA proteins using model strains of the CRD1 and CRD2 clades will also provide exciting information on these special photosynthetic strategies, and how they enable these clades to dominate in low-iron environments. Lastly, an understanding of the environmental controls of the biogeography of isiA is useful for interpretation of the biophysical signatures of iron limitation that can be collected remotely and used to map the extent and dynamics of iron limitation in the ocean.
QL and NJ designed the study and developed the experimental methods. QL conducted the experiments and drafted the manuscript. JH and TB helped with interpretation of the results and manuscript revision.
This work was supported by the National Key Research and Development Program of China (Grant No. 2016YFA0601400) and National Natural Science Foundation of China (Grant Nos. 91751207 and 91428308).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We warmly thank the crew of research vessels Da Yang I, Dong Fang Hong II, and Ke Xue I for assisting sample collection, Center for Open Ocean Research (Tianjin, China) for the opportunities to join the No. 21 and 22 open ocean cruises. Many thanks to Tingwei Luo (Xiamen University, China) and Dr. Yantao Liang (QIBEBT, China) for the FCM analysis. We thank Dr. Oliver Jahn and Dr. Stephanie Dutkiewicz (Massachusetts Institute of Technology, United States) for providing the simulated iron data from their Ecco2-Darwin model. We are most grateful to the reviewers for their helpful and constructive comments on the manuscript. We also thank deeply our late colleague Dr. Maayke Stomp (University of Amsterdam, Netherlands) for her help to retrieve the modeling iron data and supporting QL’s research in Amsterdam.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00650/full#supplementary-material
Ahlgren, N. A., Noble, A., Patton, A. P., Roache-Johnson, K., Jackson, L., Robinson, D., et al. (2014). The unique trace metal and mixed layer conditions of the Costa Rica upwelling dome support a distinct and dense community of Synechococcus. Limnol. Oceanogr. 59, 2166–2184. doi: 10.4319/lo.2014.59.6.2166
Behrenfeld, M. J., Bale, A. J., Kolber, Z. S., Aiken, J., and Falkowski, P. G. (1996). Confirmation of iron limitation of phytoplankton photosynthesis in the equatorial Pacific Ocean. Nature 383, 508–511. doi: 10.1038/383508a0
Behrenfeld, M. J., and Milligan, A. J. (2013). Photophysiological expressions of iron stress in phytoplankton. Annu. Rev. Mar. Sci. 5, 217–246. doi: 10.1146/annurev-marine-121211-172356
Behrenfeld, M. J., Westberry, T. K., Boss, E., O’Malley, R. T., Siegel, D. A., Wiggert, J. D., et al. (2009). Satellite-detected fluorescence reveals global physiology of ocean phytoplankton. Biogeosciences 6, 779–794. doi: 10.5194/bg-6-779-2009
Behrenfeld, M. J., Worthington, K., Sherrell, R. M., Chavez, F. P., Strutton, P., McPhaden, M., et al. (2006). Controls on tropical Pacific Ocean productivity revealed through nutrient stress diagnostics. Nature 442, 1025–1028. doi: 10.1038/nature05083
Bibby, T. S., Mary, I., Nield, J., Partensky, F., and Barber, J. (2003). Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424, 1051–1054. doi: 10.1038/nature01933
Bibby, T. S., Nield, J., and Barber, J. (2001). Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412, 743–745. doi: 10.1038/35089098
Bibby, T. S., Zhang, Y., and Chen, M. (2009). Biogeography of photosynthetic light-harvesting genes in marine phytoplankton. PLoS One 42:e4601. doi: 10.1371/journal.pone.0004601
Biller, D. V., and Bruland, K. W. (2014). The central California current transition zone: a broad region exhibiting evidence for iron limitation. Prog. Oceanogr. 120, 370–382. doi: 10.1016/j.pocean.2013.11.002
Boekema, E. J., Hifney, A., Yakushevska, A. E., and Piotrowski, M. A. (2001). A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412, 745–748. doi: 10.1038/35089104
Boyd, P. W., Watson, A. J., Law, C. S., Abraham, E. R., Trull, T., Murdoch, R., et al. (2000). A mesoscale phytoplankton bloom in the polar Southern Ocean stimulated by iron fertilization. Nature 407, 695–702. doi: 10.1038/35037500
Browning, T. J., Achterberg, E. P., Rapp, I., Engel, A., Bertrand, E. M., Tagliabue, A., et al. (2017). Nutrient co-limitation at the boundary of an oceanic gyre. Nature 551, 242–246. doi: 10.1038/nature24063
Chappell, P. D., Moffett, J. W., Hynes, A. M., and Webb, E. A. (2012). Molecular evidence of iron limitation and availability in the global diazotroph Trichodesmium. ISME J. 6, 1728–1739. doi: 10.1038/ismej.2012.13
Chappell, P. D., Whitney, L. P., Wallace, J. R., Darer, A. I., Jean-Charles, S., and Jenkins, B. D. (2015). Genetic indicators of iron limitation in wild populations of Thalassiosira oceanica from the northeast Pacific Ocean. ISME J. 9, 592–602. doi: 10.1038/ismej.2014.171
Choi, D. H., Noh, J. H., and Lee, J. H. (2014). Application of pyrosequencing method for investigating the diversity of Synechococcus subcluster 5.1 in open ocean. Microbes Environ. 29, 17–22. doi: 10.1264/jsme2.ME13063
Cubillos-Ruiz, A., Berta-Thompson, J. W., Becker, J. W., van der Donk, W. A., and Chisholm, S. W. (2017). Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 114, E5424–E5433. doi: 10.1073/pnas.1700990114
Dufresne, A., Ostrowski, M., Scanlan, D. J., Garczarek, L., Mazard, S., Palenik, B. P., et al. (2008). Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9:R90. doi: 10.1186/gb-2008-9-5-r90
Farrant, G. K., Doré, H., Cornejo-Castillo, F. M., Partensky, F., Ratin, M., Ostrowski, M., et al. (2016). Delineating ecologically significant taxonomic units from global patterns of marine picocyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 113, E3365–E3374. doi: 10.1073/pnas.1524865113
Fitzwater, S. E., Knauer, G. A., and Martin, J. H. (1982). Metal contamination and its effect on primary production measurements. Limnol. Oceanogr. 27, 544–551. doi: 10.1371/journal.pone.0084986
Flombaum, P., Gallegos, J. L., Gordillo, R. A., Rincón, J., Zabala, L. L., Jiao, N. Z., et al. (2013). Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829. doi: 10.1073/pnas.1307701110
Fung, I. Y., Meyn, S. K., Tegen, I., Doney, S. C., John, J. G., and Bishop, J. K. B. (2000). Iron supply and demand in the upper ocean. Global Biogeochem. Cycles 14, 281–291. doi: 10.1029/1999GB900059
Geiss, U., Vinnemeier, J., Kunert, A., Lindner, I., Gemmer, B., Lorenz, M., et al. (2001a). Detection of the isiA gene across cyanobacterial strains: potential for probing iron deficiency. Appl. Environ. Microbiol. 67, 5247–5253.
Geiss, U., Vinnemeier, J., Schoor, A., and Hagemann, M. (2001b). The iron-regulated isiA gene of Fischerella muscicola strain PCC 73103 is linked to a likewise regulated gene encoding a Pcb-like chlorophyll-binding protein. FEMS Microbiol. Lett. 197, 123–129.
Ghassemian, M., and Straus, N. A. (1996). Fur regulates the expression of iron-stress genes in the cyanobacterium Synechococcus sp. strain PCC 7942. Microbiol. 142, 1469–1476.
Grasshoff, K., Ehrhardt, M., and Kremling, K. (1999). Methods of Seawater Analysis, 3rd Edn. Weinheim: Wiley-VCH. doi: 10.1002/9783527613984
Grébert, T., Doré, H., Partensky, F., Farrant, G. K., Boss, E. S., Picheral, M., et al. (2018). Light color acclimation is a key process in the global ocean distribution of Synechococcus cyanobacteria. Proc. Natl. Acad. Sci. U.S.A 115, E2010–E2019. doi: 10.1073/pnas.1717069115
Gregg, W. W., Ginoux, P., Schopf, P. S., and Casey, N. W. (2003). Phytoplankton and iron: validation of a global three-dimensional ocean biogeochemical model. Deep Sea Res. Part II Top. Stud. Oceanogr. 5022, 3143–3169. doi: 10.1016/j.dsr2.2003.07.013
Guidi, L., Chaffron, S., Bittner, L., Eveillard, D., Larhlimi, A., Roux, S., et al. (2016). Plankton networks driving carbon export in the oligotrophic ocean. Nature 532, 465–470. doi: 10.1038/nature16942
Havaux, M., Guedeney, G., Hagemann, M., Yeremenko, N., Matthijs, H. C. P., and Jeanjean, R. (2005). The chlorophyll-binding protein IsiA is inducible by high light and protects the cyanobacterium Synechocystis PCC6803 from photooxidative stress. FEBS Lett. 579, 2289–2293. doi: 10.1016/j.febslet.2005.03.021
Hewson, I., Poretsky, R. S., Beinart, R. A., White, A. E., Shi, T., Bench, S. R., et al. (2009). In situ transcriptomic analysis of the globally important keystone N2-fixing taxon Crocosphaera watsonii. ISME J. 3, 618–631. doi: 10.1038/ismej.2009.8
Huang, S. J., Wilhelm, S., Harvey, H., Taylor, K., Jiao, N. Z., and Chen, F. (2011). Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J. 6, 285–297. doi: 10.1038/ismej.2011.106
Hutchins, D. A., Witter, A. E., Butler, A., and Luther, G. W. I. I. I. (1999). Competition among marine phytoplankton for different chelated iron species. Nature 400, 858–861. doi: 10.1038/23680
Jiao, N. Z., Yang, Y. H., Koshikawa, H., and Watanabe, M. (2002). Influence of hydrographie conditions on picoplankton distribution in the East China Sea. Aquat. Microb. Ecol. 30, 37–48. doi: 10.3354/ame030037
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kojima, K., Suzuki-Maenaka, T., Kikuchi, T., and Nakamoto, H. (2006). Roles of the cyanobacterial isiABC operon in protection from oxidative and heat stresses. Physiol. Plant 128, 507–519. doi: 10.1111/j.1399-3054.2006.00781.x
Kronmal, R. A. (1993). Spurious correlation and the fallacy of the ratio standard revisited. J. R. Stat. Soc. Ser. A 156, 379–392. doi: 10.2307/2983064
Leonhardt, K., and Straus, N. A. (1992). An iron stress operon involved in photosynthetic electron transport in the marine cyanobacterium Synechococcus sp. PCC 7002. Microbiology 138, 1613–1621.
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL). v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids. Res. 44, 242–245. doi: 10.1093/nar/gkw290
Li, Q., Legendre, L., and Jiao, N. J. (2015). Phytoplankton responses to nitrogen and iron limitation in the tropical and subtropical Pacific Ocean. J. Plankton Res. 37, 306–319. doi: 10.1093/plankt/fbv008
Lin, H. Z., Kuzminov, F. I., Park, J., Lee, S., Falkowski, P. G., and Gorbunov, M. Y. (2016). The fate of photons absorbed by phytoplankton in the global ocean. Science 351, 264–267. doi: 10.1126/science.aab2213
Liu, H., Campbell, L., Landry, M., Nolla, H., Brown, S., and Constantinou, J. (1998). Prochlorococcus and Synechococcus growth rates and contributions to production in the Arabian Sea during the 1995 Southwest and Northeast Monsoons. Deep Sea Res. Pt. II. Stud. Oceanogr. 45, 2327–2352. doi: 10.1126/science.aab2213
Macey, A. I., Ryan-Keogh, T., Richier, S., Moore, C. M., and Bibby, T. S. (2014). Photosynthetic protein stoichiometry and photophysiology in the high latitude North Atlantic. Limnol. Oceanogr. 59, 1853–1864. doi: 10.1016/S0967-0645(98)00073-3
Mackey, K. R., Post, A. F., McIlvin, M. R., Cutter, G. A., John, S. G., and Saito, M. A. (2015). Divergent responses of Atlantic coastal and oceanic Synechococcus to iron limitation. Proc. Natl. Acad. Sci. U.S.A. 112, 9944–9949. doi: 10.1073/pnas.1509448112
Marie, D., Partensky, F., Jacquet, S., and Vaulot, D. (1997). Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63, 186–193. doi: 10.1073/pnas.1509448112
Martin, J. H., and Fitzwater, S. E. (1988). Iron deficiency limits phytoplankton growth in the north-east Pacific subarctic. Nature 331, 341–343. doi: 10.1038/331341a0
Mazard, S., Ostrowski, M., Partensky, F., and Scanlan, D. J. (2012). Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environ. Microbiol. 14, 372–386. doi: 10.1111/j.1462-2920.2011.02514.x
Moore, C. M., Mills, M. M., Arrigo, K. R., Berman-Frank, I., Bopp, L., Boyd, P. W., et al. (2013). Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710. doi: 10.1038/ngeo1765
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2015). Vegan: Community Ecology Package. R package version 2.2–1. Available at: http://CRAN.Rproject.org/package=vegan (accessed December 12, 2018).
Palenik, B. Q., Ren, C. L., Dupont, G. S., Myers, J. F., Heidelberg, J. H., Badger, R., et al. (2006). Genome sequence of Synechococcus CC9311: insights into adaptation to a coastal environment. Proc. Natl. Acad. Sci. U.S.A. 103, 13555–13559. doi: 10.1073/pnas.0602963103
Partensky, F., Blanchot, J., and Vaulot, D. (1999). Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull. Inst. Oceanogr. Monaco 19, 457–476.
R Development Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Richier, S., Macey, A. I., Pratt, N. J., Honey, D. J., Moore, C. M., and Bibby, T. S. (2012). Abundances of iron-binding photosynthetic and nitrogen-fixing proteins of Trichodesmium both in culture and in situ from the North Atlantic. PLoS One 6:e35571. doi: 10.1371/journal.pone.0035571
Rivers, A. R., Jakuba, R. W., and Webb, E. A. (2009). Iron stress genes in marine Synechococcus and the development of a flow cytometric iron stress assay. Environ. Microbiol. 112, 382–396. doi: 10.1111/j.1462-2920.2008.01778.x
Ryan-Keogh, T. J., Macey, A. I., Cockshutt, A. M., Moore, C. M., and Bibby, T. S. (2012). The cyanobacterial chlorophyll-binding protein IsiA acts to increase the in vivo effective absorption cross-section of PSI under iron limitation. J. Phycol. 48, 145–154. doi: 10.1111/j.1529-8817.2011.01092.x
Saito, M. A., Rocap, G., and Moffett, J. W. (2005). Production of cobalt binding ligands in a Synechococcus feature at the Costa Rica upwelling dome. Limnol. Oceanogr. 50, 279–290. doi: 10.4319/lo.2005.50.1.0279
Scanlan, D. J., Ostrowski, M., Mazard, S., Dufresne, A., Garczarek, L., Hess, W. R., et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 732, 249–299. doi: 10.1128/MMBR.00035-08
Scanlan, D. J., and West, N. I. (2002). Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40, 1–12. doi: 10.1111/j.1574-6941.2002.tb00930.x
Schlitzer, R. (2012). Ocean Data View. Available at: http://odv.awi.de/ (accessed July 30, 2018).
Schloss, P. D., and Handelsman, J. (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005
Schrader, P. S., Milligan, A. J., and Behrenfeld, M. J. (2011). Surplus photosynthetic antennae complexes underlie diagnostics of iron limitation in a cyanobacterium. PLoS One 6:e18753. doi: 10.1371/journal.pone.0018753
Six, C., Thomas, J., Garczarek, L., Ostrowski, M., Dufresne, A., Blot, N., et al. (2007). Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study. Genome Biol. 8:R259. doi: 10.1186/gb-2007-8-12-r259
Snow, J. T., Polyviou, D., Skipp, P., Chrismas, N. A., Hitchcock, A., Geider, R., et al. (2015). Quantifying integrated proteomic responses to iron stress in the globally important marine diazotroph Trichodesmium. PLoS One 10:e0142626. doi: 10.1371/journal.pone.0142626
Sohm, J. A., Ahlgren, N. A., Thomson, Z. J., Williams, C., Moffett, J. W., Saito, M. A., et al. (2015). Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. ISME J. 10, 333–345. doi: 10.1038/ismej.2015.115
Stomp, M., Huisman, J., De Jongh, F., Veraart, A. J., Gerla, D., Rijkeboer, M., et al. (2004). Adaptive divergence in pigment composition promotes phytoplankton biodiversity. Nature 432, 104–107. doi: 10.1038/nature03044
Tagliabue, A., Aumont, O., DeAth, R., Dunne, J. P., Dutkiewicz, S., Galbraith, E., et al. (2016). How well do global ocean biogeochemistry models simulate dissolved iron distributions? Global Biogeochem. Cycles 30, 149–174. doi: 10.1002/2015GB005289
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Ting, C. S., Rocap, G., King, J., and Chisholm, S. W. (2002). Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10, 134–142. doi: 10.1016/S0966-842X(02)02319-3
Vinnemeier, J., Kunert, A., and Hagemann, M. (1998). Transcriptional analysis of the isiAB operon in salt-stressed cells of the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 169, 323–330. doi: 10.1111/j.1574-6968.1998.tb13336.x
Webb, E. A., Moffett, J. W., and Waterbury, J. B. (2001). Iron stress in open-ocean cyanobacteria (Synechococcus, Trichodesmium, and Crocosphaera spp.): identification of the IdiA Protein. Appl. Environ. Microbiol. 67, 5444–5452. doi: 10.1128/AEM.67.12.5444-5452.2001
Yeremenko, N., Kouril, R., Ihalainen, J. A., Haene, S., van Oosterwijk, N., Andrizhiyevskaya, E. G. et al. (2004). Supramolecular organization and dual function of the IsiA chlorophyll-binding protein in cyanobacteria. Biochemistry 43, 10308–10313. doi: 10.1021/bi048772l
Zuur, A., Ieno, E. N., Walker, N., Saveliev, A. A., and Smith, G. M. (2009). Mixed Effects Models and Extensions in Ecology with R. Berlin: Springer. doi: 10.1007/978-0-387-87458-6
Keywords: cyanobacteria, Synechococcus, iron limitation, antenna protein, isiA, chlorophyll
Citation: Li Q, Huisman J, Bibby TS and Jiao N (2019) Biogeography of Cyanobacterial isiA Genes and Their Link to Iron Availability in the Ocean. Front. Microbiol. 10:650. doi: 10.3389/fmicb.2019.00650
Received: 18 July 2018; Accepted: 14 March 2019;
Published: 04 April 2019.
Edited by:
George S. Bullerjahn, Bowling Green State University, United StatesReviewed by:
David J. Scanlan, University of Warwick, United KingdomCopyright © 2019 Li, Huisman, Bibby and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nianzhi Jiao, amlhb0B4bXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.