- 1Food Microbiology Laboratory, Laboratory Sciences and Services Division, International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B), Bangladesh, India
- 2School of Chemical, Biological and Environmental Engineering, Oregon State University, Corvallis, OR, United States
- 3Division of Foodborne, Waterborne, and Environmental Diseases, Centers for Disease Control and Prevention, Atlanta, GA, United States
- 4Department of Environmental Health, Rollins School of Public Health, Emory University, Atlanta, GA, United States
- 5School of Biological and Population Health Sciences, Oregon State University, Corvallis, OR, United States
- 6Eawag, Swiss Federal Institute of Aquatic Science and Technology, Dübendorf, Switzerland
- 7Swiss Tropical and Public Health Institute, Basel, Switzerland
- 8University of Basel, Basel, Switzerland
Third generation cephalosporins (3GC) are one of the main choices for treatment of infections caused by multidrug-resistant (MDR) Gram-negative bacteria. Due to their overuse, an increasing trend of resistance to 3GC has been observed in developing countries. Here, we describe fecal colonization of 3GC-resistant (3GCr) Escherichia coli in healthy infants (1–12 months old) living in rural areas of Bangladesh. We found that stool samples of 82% of infants (n = 100) were positive for 3GCr E. coli with a mean ± standard deviation of 6.21 ± 1.32 log10 CFU/g wet weight of stool. 3GCr E. coli encompasses an average one third (33%) of the total E. coli of stool. Almost 77% (n = 63) of these 3GCr E. coli were MDR (or resistant to ≥3 classes of antibiotics). Around 90% (n = 74) of 3GCr E. coli were extended spectrum beta-lactamase (ESBL)-producing in which blaCTX–M–group–1 was the predominant (96%, n = 71) ESBL-gene followed by blaTEM (41%, n = 30) and blaOXA–1 (11%, n = 8). A significant proportion (26.5%, n = 22) of 3GCr E. coli was pathogenic, comprising two types, enteroaggregative (EAEC, n = 19) and enteropathogenic (EPEC, n = 3). Colonization of 3GCr E. coli in infant guts was not associated with demographic characteristics such as age, sex, mode of delivery, maternal and infant antibiotic use, disease morbidity, and feeding practices. The high rate of colonization of 3GCr E. coli in infants’ guts is a serious public health concern which needs immediate attention and warrants further studies to explore the cause.
Introduction
The rapid rise of multidrug-resistant (MDR) bacterial infections is a major public health concern and a growing threat to the global health security. Unregulated use of broad spectrum antibiotics and widespread reservoirs of these pathogens are main contributors to this problem (Hilty et al., 2012). Broad spectrum antibiotics, in particular third generation cephalosporins (3GC), are among the most frequently prescribed drugs for the treatment of infections caused by Enterobacteriaceae (Pereira et al., 2004; World Health Organization [WHO], 2017). Failure of treatment with these antibiotics has increasingly been reported due to the emergence of extended spectrum beta-lactamases (ESBLs) during the last two decades (Pitout and Laupland, 2008). Several studies have suggested that children are more likely to be exposed to antibiotics both directly (Alexander et al., 2011; Saari et al., 2015) and indirectly, through exposure to antibiotics taken by their mothers (Verani et al., 2010; Macones et al., 2012; Ledger and Blaser, 2013; de Tejada, 2014). This direct and/or indirect consumption of antibiotics might thus affect infants’ intestinal microflora, including Escherichia coli, which is one of the first bacterial species that colonizes the infant gut (Hewitt and Rigby, 1976).
Antimicrobial resistance in commensal bacteria is worrisome due to its ability to spread to pathogens (Munk et al., 2018). Recent studies have showed that school children and children up to 2 years of age were colonized by E. coli resistant to broad spectrum antibiotics and ciprofloxacin, respectively (Gurnee et al., 2015; Ferjani et al., 2017). However, there is no information on the carriage rate and abundance of this antibiotic-resistant E. coli in relation to the total number of E. coli present in the gut. In addition, there is no data available on the carriage rate of MDR E. coli, including ESBL-producing E. coli, among infants under 1 year old. Therefore, this study evaluated the prevalence and rate of colonization of this organism during the early life of infants. We determined the prevalence, abundance, and carriage rate of 3GC-resistant (3GCr) E. coli, including pathogenic E. coli, by analyzing culturable E. coli in infant’s stool samples.
Materials and Methods
Ethics Statement
The research and the ethical review committees of icddr,b approved and monitored the progress of the study. Informed written consent was obtained from mothers of all infants either by signature or, for those who were not literate, by thumbprint after verbal communication. Samples were identified with codes to preserve anonymity. A witness also signed each informed-consent form. All authors vouch for the completeness and accuracy of the data and analyses presented.
Study Design, Site, and Enrollment of Participants
We conducted a cross-sectional study of children <1 year of age living in five rural villages of Matlab and Hajiganj, sub-districts of Chandpur district of Bangladesh, between March and October 2017. Hajigonj has a total area of 189 km2 with 327,367 people living in 64,257 households at 11 unions, whereas Matlab Uttar has a total area of 279 km2 with 382,195 people in 62,418 households at 15 unions (DGHS Health Bulletin, 2014). According to the The World Bank (2016) the crude birth rate for Bangladesh is 18.95 per 1000 people as of 2016, so an approximation for the number of infants in Hajigonj and Matlab Uttar is 6,200 and 7,250, respectively, or less than 13,500 total infants (2016). For study sites we included one union from each sub-district. A total of 100 households (50 from each union) containing one infant (age ≤1 year) in each household were enrolled in the study after obtaining written informed consent from the mothers of enrolled infants. A pre-tested survey questionnaire was used to collect information on age, sex, mode of delivery, maternal and child antibiotic consumption, disease morbidity, and feeding practices.
Sample Collection
A total of 100 stool samples were collected from 100 infants located in the selected households using sterile stool containers provided earlier to all the mothers on the date of the interview. Assuming an infant population in Hajigong and Matlab Uttar of 13,500, the sample size of 100 stool samples would imply a margin of error of approximately 10% with 95% confidence interval for prevalence rates of 3GCr E. coli (Dhand and Khatkar, 2014). The field staff collected the samples on the following day and transported it to icddr,b laboratory (Dhaka, Bangladesh) on ice for microbiological analyses within 4 h.
Enumeration and Isolation of E. coli
Both total and 3GCr E. coli were enumerated using the drop plate method as described previously (Herigstad et al., 2001). In brief, MacConkey agar plates (Becton Dickson, MD) with and without fixed concentrations of cefotaxime (1.0 μg/mL) were used to enumerate 3GCr E. coli and total E. coli, respectively. A total of four 10-fold serial dilutions (10−1 to 10−4) of each stool sample were made and 50 μl from each dilution was inoculated onto MacConkey agar plates with and without antibiotic added to the culture media. All plates were incubated at 37°C for 18 h and the number of colony forming units (CFUs) per gram wet weight of stool sample were counted from the dilution of readable range. Proportion of 3GC-sensitive (3GCs) E. coli CFUs per gram feces (CFU/g) count was calculated by subtracting 3GCr CFU/g count from corresponding total E. coli CFU/g. Further, proportion of 3GCr E. coli count was measured in respect to the total E. coli count obtained on MacConkey agar plate. At least three colonies from each sample were confirmed as E. coli by API-20E (bioMerieux, France) and stored at −80°C for further characterization.
Antibiotics Susceptibility Test
Antibiotic susceptibility of E. coli (one isolate per sample) was determined by standard disk diffusion technique following the Clinical and Laboratory Standards Institute (CLSI) guidelines (Patel, 2017). The antibiotics used in this study were ampicillin (10 μg), gentamycin (10 μg), tetracycline (30 μg), meropenem (10 μg), imipenem (10 μg), ceftriaxone (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefepime (30 μg), colistin (10 μg), ciprofloxacin (5 μg), nalidixic acid (30 μg), azithromycin (15 μg), trimethoprim/sulfamethoxazole (25 μg), nitrofurantoin (30 μg), and chloramphenicol (30 μg) (Oxoid, United Kingdom). The zone of inhibition was measured and the isolates were classified as resistant, intermediate, or sensitive according to the interpretation guideline provided by the CLSI (Patel, 2017). An isolate was considered MDR if resistant to three or more classes of antibiotics.
Test for Extended Spectrum Beta-Lactamase (ESBL)
Extended spectrum beta-lactamase was tested by combination disk test (CDT) as described by CLSI (Patel, 2017). Disks containing a 3GC, including cefotaxime, CTX (30 μg) or ceftazidime, CAZ (30 μg) alone and in combination with clavulanic acid (CLA, 10 μg) were applied (Oxoid, United Kingdom). The zone of inhibition around the CTX or CAZ disk combined with CLA was compared with the inhibition zone around CTX or CAZ disks alone. The test was considered positive for ESBL-production if the inhibition zone diameter was ≥5 mm larger with CLA than without (Patel, 2017).
PCR for ESBL Genes and E. coli Pathotypes
All 3GCr E. coli were tested for ESBL genes considered priorities due to their clinical relevance, specifically: blaCTX–M–group–1, blaSHV, blaTEM, and blaOXA–1. These genes were tested by PCR using primer sequences and PCR conditions as described previously (Islam et al., 2017). The pathotypes of E. coli (EPEC, ETEC, EAEC, EIEC, and STEC) were identified by multiplex PCR of different pathogenic genes according to the procedure described earlier (Talukdar et al., 2013).
Statistical Data Analysis
Data were entered in SPSS 20.0 (IBM Inc., Chicago, IL, United States). Data cleaning, statistical analysis and graphical presentation were done in Stata 13.0 (College Station, TX, United States) and R-3.4.2 (R Core Team, 2014). E. coli concentrations were log10 transformed in order to assess the association between demographic variables with 3GCr E. coli carriage using chi-square test and non-parametric Mann–Whitney U-test. Population counts of the susceptible and resistant isolates from the same infant were compared using Wilcoxon Rank-Sum test on the paired data. Statistical significance was determined using alpha = 0.05 for all tests except for the Wilcoxon Rank-Sum test which stratified analyses by age (1–3, 4–6, 7–9, and 10–12 months). For this, the conservative Bonferroni correction was applied to adjust alpha to 0.0125 (0.5/4) to correct for multiple comparisons.
Results
Carriage of 3GCr E. coli in Infant’s Fecal Sample
Of the 100 stool samples from infants, 82% were positive for 3GCr E. coli. Mean count ± standard deviation for total E. coli was 6.86 ± 1.56 log10 E. coli CFU/g of stool while the mean count of 3GCr E. coli was 6.21 ± 1.32 log10 CFU/g. On average, 3GCr E. coli encompasses approximately one third of (33%) of the total E. coli present/g wet weight of stool.
Antibiotic Susceptibility of 3GCr E. coli
All the 3GCr (n = 82) E. coli isolates were tested for susceptibility against a panel of 13 antibiotics. Resistance to multiple antibiotics other than 3GCr was very common, with 77% (n = 63) of isolates classified as MDR. None of the isolates were resistant to colistin or carbapenem (Figure 1).
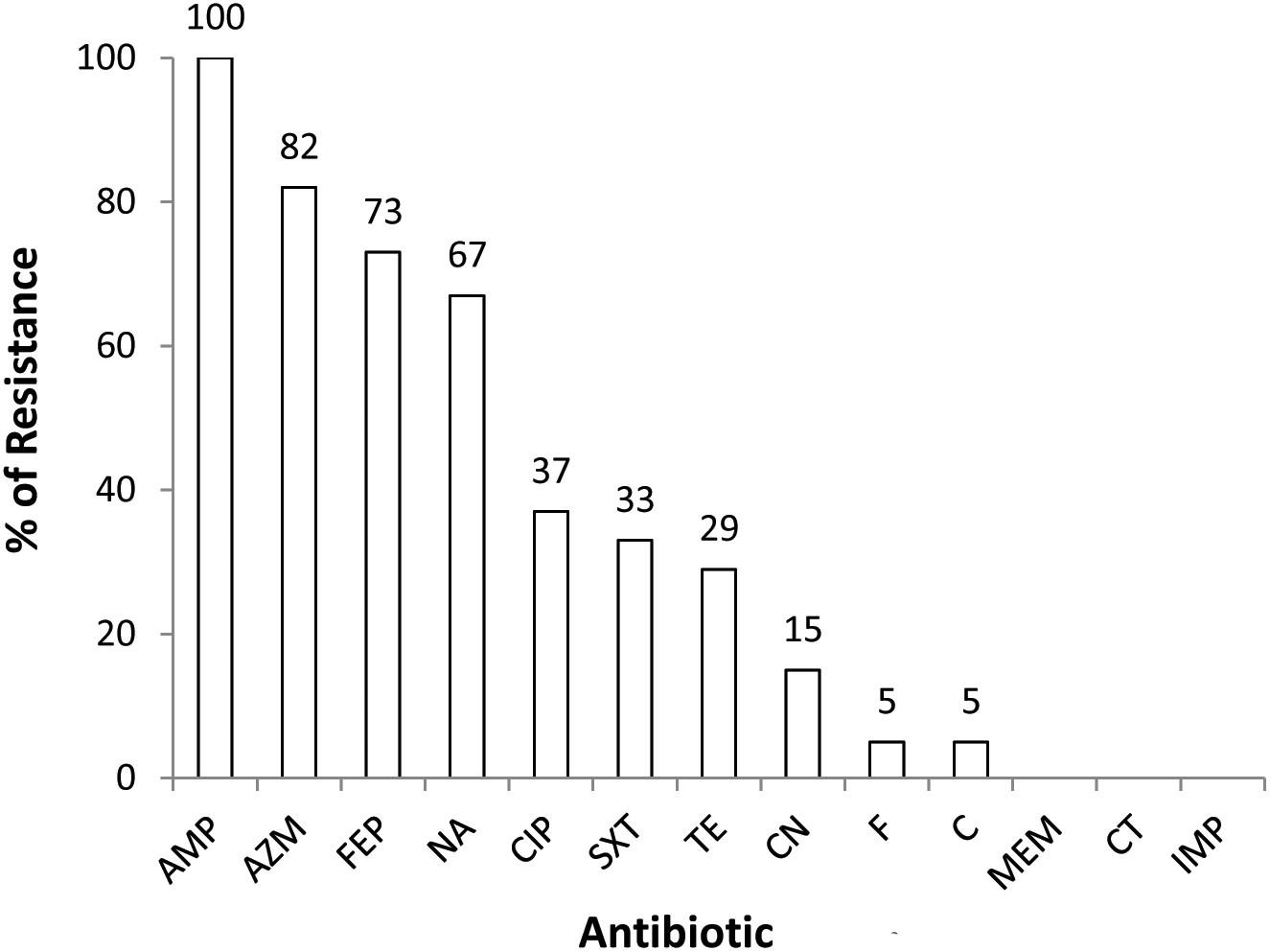
Figure 1. Prevalence of 3GCr E. coli resistant to different classes of antibiotics in infants aged from 1 to 12 months (n = 82). AMP, ampicillin; AZM, azithromycin; FEP, cefepime; NA, nalidixic acid; CIP, ciprofloxacin; SXT, sulfamethoxazole; TE, tetracycline; CN, gentamycin; F, nitrofurantoin; C, chloramphenicol; MEM, meropenem; CT, colistin; IMP, imipenem; 3GCr, third generation cephalosporins resistant.
Prevalence of ESBLs Among 3GCr E. coli Isolates
CDT of all 3GCr E. coli (n = 82) isolates revealed that more than 90% (n = 74) were ESBL-producing. Among ESBL-producing isolates, 96% (n = 71) were positive for blaCTX–M–group–1, 41% (n = 30) for blaTEM, and 11% (n = 8) for blaOXA–1. Of the 8 CDT negative isolates, 4 were positive for blaCTX–M–group–1 indicating that these isolates might have co-produced ESBL and AmpC enzymes. Given the high rates of positivity for detection of blaCTX–M–group–1, further molecular characterization for resistance genes was not conducted; carriage of multiple mechanisms of resistance is possible but was not explored.
Prevalence of E. coli Pathotypes Among 3GCr E. coli Isolates
Analysis of virulence genes among 3GCr isolates revealed that 23% (n = 19) of the isolates were positive for genes specific for EAEC (aatA, aaiC) and 3.6% (n = 3) of the isolates were positive for genes specific for EPEC (bfp, eae). No other virulence genes were detected (lt, st, ipaH, and ial).
Determinants of 3GCr E. coli Carriage Among Infants
Statistical analysis of results did not reveal any significant association between the presence of 3GCr E. coli in infant stool and characteristics of the infant, including gender, religion, age, mode of delivery, feeding practice of child, diarrheal history, and maternal and child antibiotic consumption (Table 1). Moreover, association between the rate of colonization of 3GCr E. coli in the child’s gut and demographic characteristics of infant (age, p = 0.5; delivery mode, p = 0.8; infant and mother antibiotic consumption, p = 0.2, p = 0.4; and infant diarrhea, p = 0.2) were not statistically significant in non-parametric Mann–Whitney U-test (Table 2). Therefore, the presence or abundance of 3GCr E. coli in infant stools in this study could not be explained by some of the most common risk factors, including prior exposures to antibiotics.
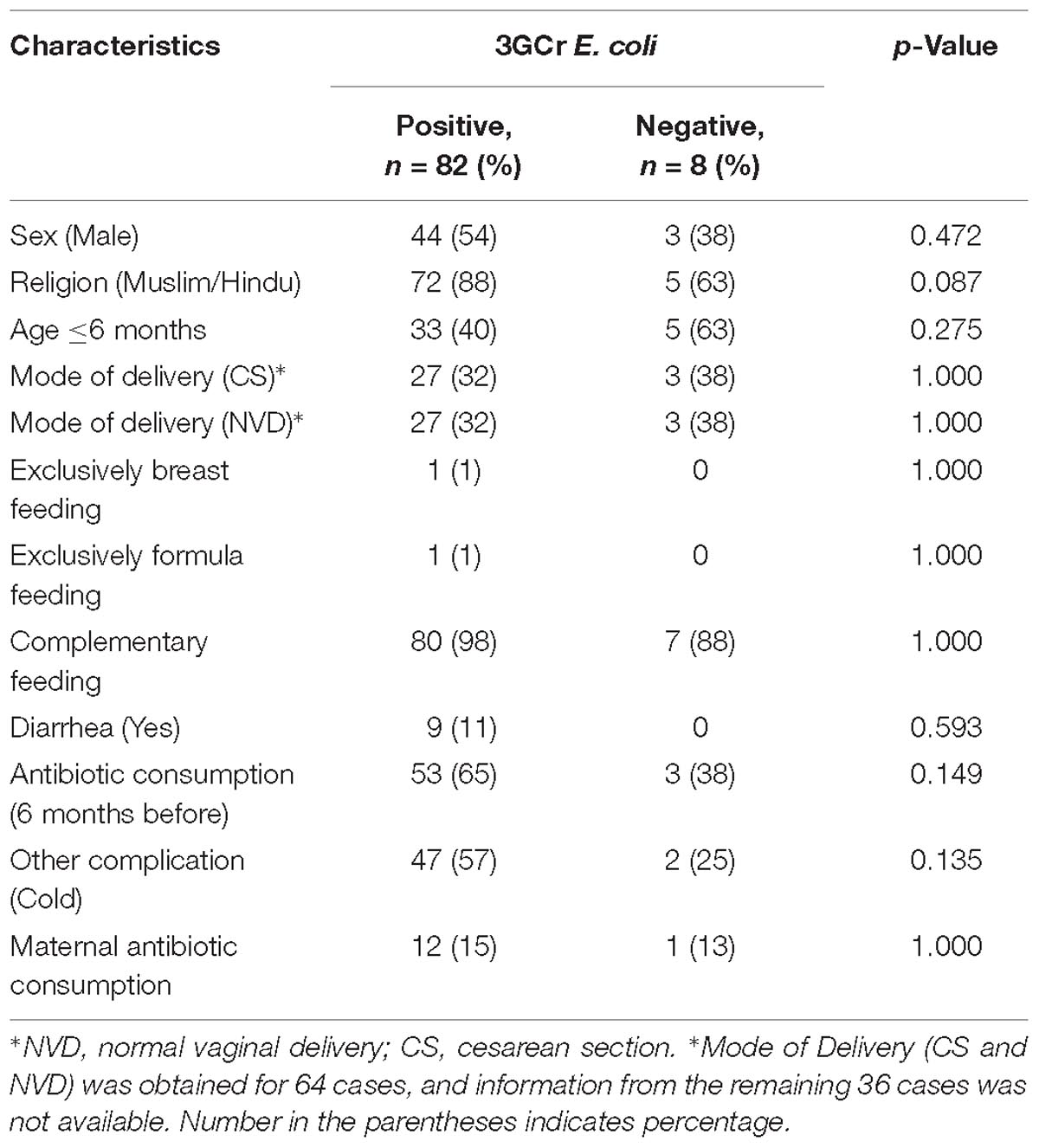
Table 1. Demographic characteristics of infants with and without fecal carriage of third generation cephalosporins resistant (3GCr) E. coli.
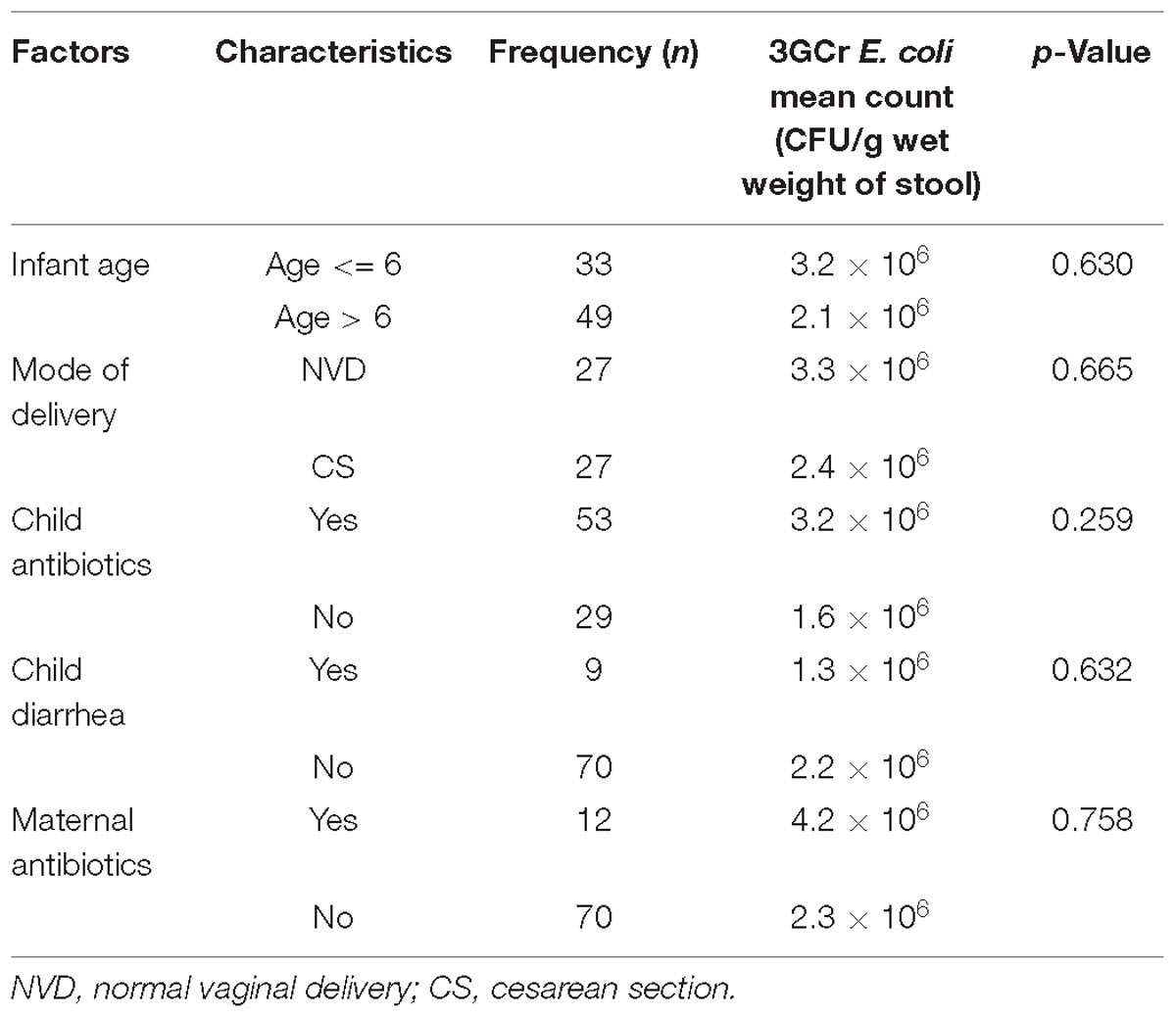
Table 2. Association between demographic variables and third generation cephalosporins resistant (3GCr) E. coli colonization in infants’ gut.
Age-Wise Distribution of Fecal Carriage of 3GCr E. coli in Respect to 3GCs E. coli
We calculated the mean count of 3GCr E. coli among infants of the same age groups and plotted the distribution with 1 month intervals in order to determine if the CFU count of resistant microflora changes with infant age. Analysis of mean count of 3GCr up to 12 months showed that log10 CFU mean count of 3GCr even at 3 months of age was high (6.43 log10) and consistent in subsequent months (Figure 2), indicating early appearance of 3GCr E. coli in infants. However, the percentage of infants infected with resistant E. coli population (3GCr and MDR) was progressively increased as infant age grown by months (Figure 3).
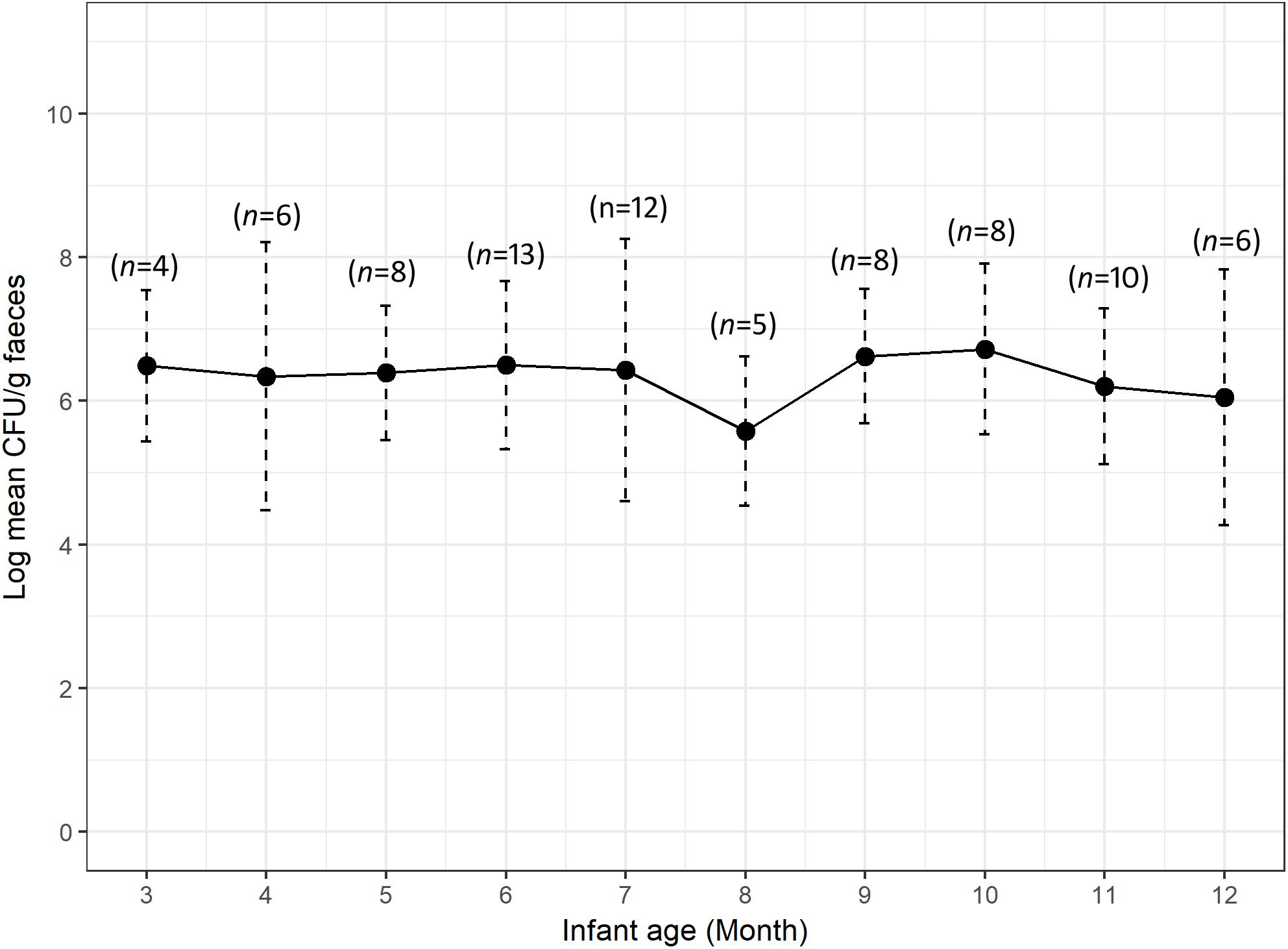
Figure 2. Variations in carriage rates of 3GCr E. coli in infants at different ages from 3 to 12 months. The log10 mean count of 3GCr CFU from infant at 1 and 2 month of age was excluded because only one infant of each month was obtained for these time periods. Error bars are the standard errors of the results for each age group. 3GCr, third generation cephalosporins resistant.
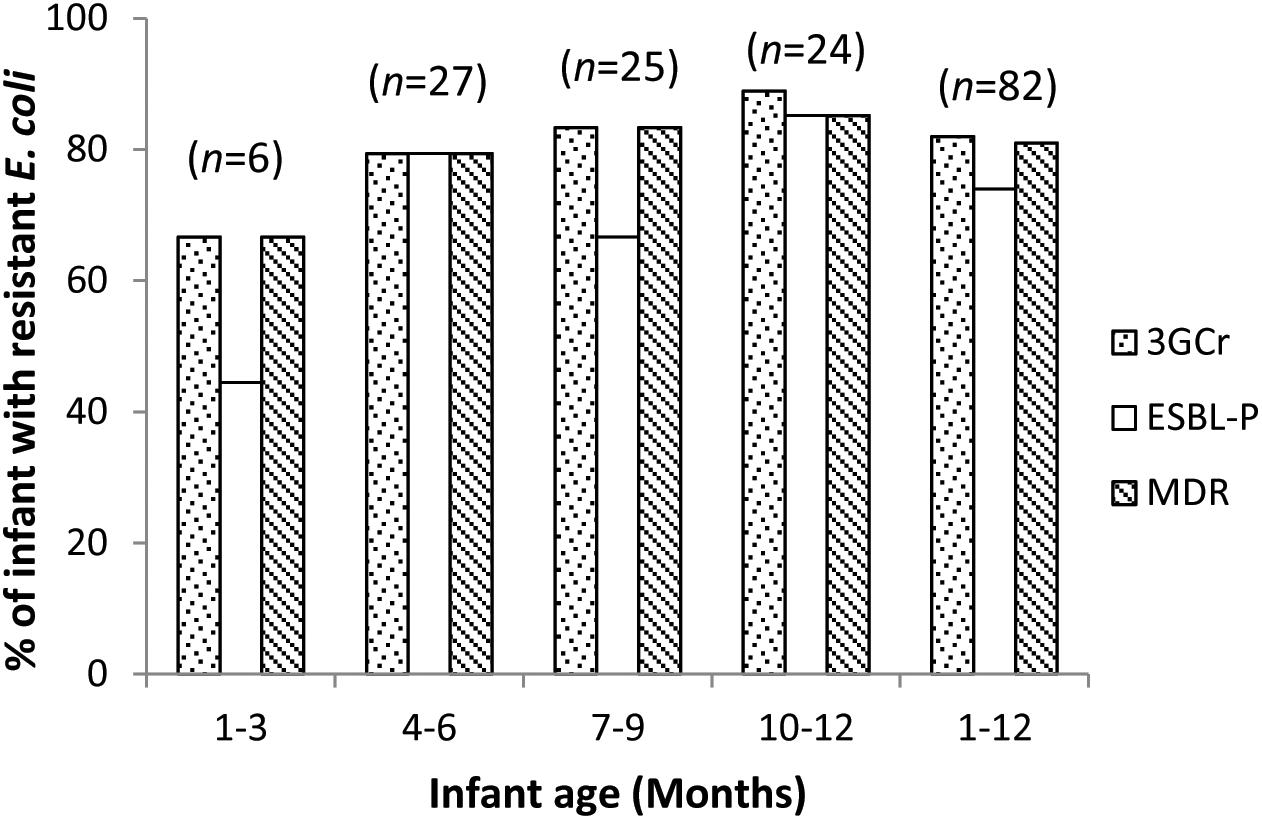
Figure 3. Age wise distribution of prevalence of 3GCr, MDR, and ESBL-P E. coli in infant stool samples. 3GCr, third generation cephalosporins resistant; ESBL-P, ESBL-producing; MDR, multidrug-resistant.
We compared the differences between 3GCr and 3GCs E. coli counts among infants of the same age groups at 1–3, 4–6, 7–9, and 10–12 months of age to examine whether 3GCr E. coli co-exist with the 3GCs favorably without selective pressure of antibiotics. There was no significant difference in the population of 3GCr and 3GCs E. coli using Wilcoxon Rank-Sum test at any age group except for the oldest one, 10–12 months (p = 0.011) (Figure 4), suggesting that 3GCr E. coli can stably persist like 3GCs E. coli from early months of life. Notably, statistical significance in the difference in 3GCr and 3GCs E. coli amongst infants 10–12 months old is borderline significant compared to the Bonferroni adjusted significance level of alpha = 0.0125 for the age-stratified Wilcoxon Rank-Sum test.
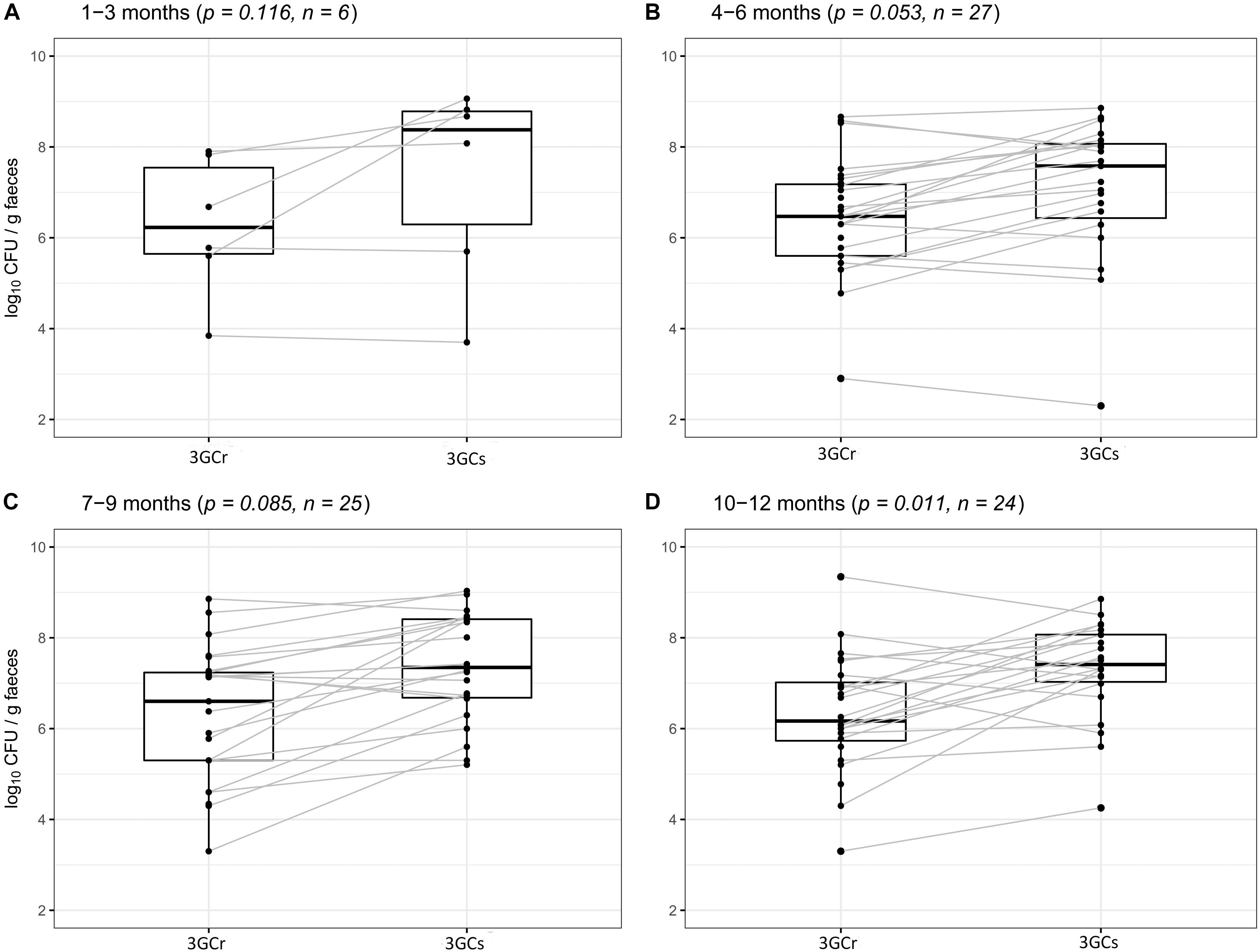
Figure 4. Comparative analysis of carriage rates (in CFU/g wet weight of stool) of 3GCr and 3GCs E. coli within similar age group of infants. (A) 1–3 months of age, (B) 4–6 months of age, (C) 7–9 months of age, and (D) 10–12 months of age. 3GCr, third generation cephalosporins resistant; 3GCs, third generation cephalosporins sensitive; CFU, colony forming unit.
Discussion
We found an extremely high prevalence of both 3GCr E. coli (82%) and ESBL-producing E. coli (74%) in stool samples of healthy infants living in rural areas of Bangladesh. Despite the relatively small sample size (n = 100) chosen based on logistic constraints, the high prevalence rates are likely representative of Bangladeshi infants under 1 year old in the study area within a margin of error of 10% (with 95% confidence interval). Even with the associated uncertainty, this is the highest prevalence of ESBL-producing E. coli in healthy human guts observed to date. For example, in a study in Tunisia, the prevalence of MDR E. coli was 6.6% in children aged between 6 and 12 years (Ferjani et al., 2017). The rates of 3GCr E. coli in healthy children at various ages was reported as 2.9% in Sweden, 2.7% in Portugal, and 10% in Senegal (Guimaraes et al., 2009; Kaarme et al., 2013). The fecal carriage rate of ESBL-producing Enterobacteriaceae in healthy children (0–59 months) was much higher (59%) in central Africa (Farra et al., 2016). None of these studies have reported the data on culturable 3GCr E. coli as a proportion of total culturable E. coli in stool samples. In our study we found that around one third of the total E. coli colonies obtained in culture were 3GCr, which is alarming.
The difference between 3GCr and 3GCs E. coli gives an indication of fitness costs for the maintenance of resistance within the gut microbial community. The proportion of 3GCr amongst the total E. coli (sum of 3GCr and 3GCs E. coli) was not significant in infants at different age groups, indicating that the competitiveness of resistant bacteria with normal residential flora within the gut is not influenced by age. There may be a low fitness cost associated with persistence and dissemination of resistance. Indeed, Cottell et al. (2012) suggest a low fitness cost associated with plasmid (pCT) carrying the resistance gene blaCTX–M–14 for E. coli, as the E. coli were able to persist and disseminate readily even in the absence of selective pressure from antibiotics (Cottell et al., 2012). Our findings also displayed congruence with previous reports demonstrating that tetracycline- and ampicillin-resistant isolates persist continuously without any selective pressure of antibiotics in the gut of different age groups of children (Karami et al., 2006, 2008).
The implication of this high load of 3GCr E. coli is substantial in the context of child health safety. Antibiotic resistant E. coli and other common commensals of the gut including Klebsiella spp. and Acinetobacter spp. are amongst the leading causes of community-acquired serious infection in Southeast Asia. In a cross sectional study at five sites across Southeast Asia, Saha et al. (2018) found that only 17% of possible serious bacterial infections (pSBI) identified in young children were resistant to first line antibiotics. Resistant infections, as compared to susceptible infections, are linked with worse outcomes. For example, in Tanzania, children with septicemia caused by bacteria producing extended-spectrum beta-lactamases were almost twice as likely to die compared to non-ESBL infections (71% mortality rate vs. 39%) (Blomberg et al., 2007). In the present study, the observed high carriage rate and high relative proportion of culturable 3GCr E. coli may harbinger higher rates of 3GCr E. coli as a proportion of pSBI infections. Further research linking fecal carriage of resistant bacteria to risks of resistant infections is warranted.
Apart from increasing the risk of resistant infections, high carriage of 3GCr Enterobacteriaceae in the gut results in shedding through infant stool and thus contributes to an elevated risk of exposure to nearby people and animals. There is a common belief among illiterate or less literate mothers in the community that infant stool is not a health hazard or harmful, especially in comparison to adult stool (Yeager et al., 1999; Gil et al., 2004). According to a national survey during the period from 2012 to 2013 in Bangladesh, feces of about 61% children of age 0–2 years were disposed unsafely where the percentage was much higher in rural areas (67%) compared to urban areas (40%) (Bangladesh Bureau of Statistics [BBS] and United Nations Children’s Fund [UNICEF], 2014). Thus household members including mothers or caregivers are exposed to fecal MDR bacteria through unsafe disposal of infant feces. Similarly, improper disposal of infant’s stool in front yards or nearby ditches may contribute to transmission of these resistant bacteria to domestic and wild birds and/or other animals (Hasan et al., 2012).
The 3GCr E. coli isolates in this study were predominantly resistant to azithromycin and ciprofloxacin, among other antibiotics (Figure 1). Azithromycin is the first line of choice for treatment of shigellosis in children (Centers for Disease Control and Prevention [CDC], 2006) and a second line of choice for treatment of shigellosis in adults (World Health Organization [WHO], 2005). Although no infants were reported to be suffering from shigellosis during the study period, shigellosis has been identified as a major contributor to moderate-to-severe diarrheal disease in neighboring Mirzapur (Kotloff et al., 2013). Management of this infection might be complicated due to the high prevalence of azithromycin resistance in the study community. Among other antibiotics, ciprofloxacin resistance was found among 37% of 3GCr isolates, which is even higher than a previous report that showed resistance in 19% of E. coli obtained from healthy children (Gurnee et al., 2015). Interestingly, only 29% of E. coli isolates in this study were resistant to tetracycline, a first generation antibiotic which is less commonly used for the treatment of E. coli infections in the community (Calva et al., 1996; Domínguez et al., 2002). Tetracycline is not prescribed in children due to its effect on the growth of bones and teeth (Sánchez et al., 2004). It suggests that by reducing the use of antibiotics in humans and animals, it is possible to reduce the burden of resistant microorganisms and this can eventually restore the efficacy of the existing antibiotics even in a setting like Bangladesh where overuse of antibiotics and burden of AMR, both are way too high.
Our study demonstrated that infant’s guts serve as a reservoir of E. coli resistant to multiple antibiotics including 3GCr and fluoroquinolones, which are critical for the treatment of many infectious diseases in humans (World Health Organization [WHO], 2017). High rates of ESBL-producers among 3GCr isolates in the present study is alarming because patients with community acquired infections as well as their household members carrying ESBL-producing Enterobacteriaceae may spread resistance to other people in their community (Valverde et al., 2008). This can be explained by the overall high prevalence of ESBL E. coli infections in the community. A recent study in Bangladesh has reported that 34% of all clinical isolates of E. coli from patients with extra-intestinal infections were ESBL-producing (Khan et al., 2018).
The probable cause of colonization with ESBL-producing Enterobacteriaceae among pre-school children in Laos was reported due to misuse of antibiotics (Stoesser et al., 2015). In our study, we did not observe this: there was no significant association between colonization and reported previous use of antibiotic treatment among infants. Previous studies have suggested that acquisition of antimicrobial resistant bacteria or antimicrobial resistant genes in the infant gut might occur during and/or after the delivery (Zhang et al., 2011). Specifically, mothers’ flora during normal vaginal delivery or environmental flora during caesarian (C-section) delivery colonize the infant gut (Zhang et al., 2011). Therefore, AMR bacteria from the mother or hospital environment may contribute to infant carriage. For example, a study carried out in Tunisia showed that 20% of patients acquired 3GCr E. coli in their gut due to nosocomial infection (Maamar et al., 2016). In our study, we did not find any significant differences in the level of colonization with 3GCr E. coli between infants with normal vaginal delivery and infants delivered through C-section (Tables 1, 2). In Bangladesh, a recent study showed that delivery by C-section increased from 3.5 to 23% between 2004 and 2014 (Khan et al., 2017) and it is a common practice that prophylactic antibiotics are used before and after the surgery. During post-operative care mothers start to breastfeed the newborn while still on antibiotic treatment. This leads to transfer of antibiotics in its active form to infants and thus their gut microbiota may shift to survive in an antibiotic selective environment (Mathew, 2004). Further in Dhaka, Bangladesh, a significant proportion of newborns (98%) receive antibiotics (sulfonamides, fluoroquinolones, metronidazole, penicillins, etc) before 6 months of age (Rogawski et al., 2017), which renders the selective environment for antibiotic-resistant bacteria. Even in a very low concentration of antibiotics, fitness cost for microorganisms to become resistant is lower than becoming antibiotic susceptible (Sandegren, 2014). Surprisingly, in our study neither history of antibiotic use or previous record of hospitalization was associated with 3GCr colonization nor were gender, religion and feeding practices. The lack of association may be due to the high rate of colonization combined with relatively small sample size (n = 100). The low proportion of infants without 3GCr limits statistical analyses. Therefore, further studies, particularly focused on larger sample sizes, are needed to identify the causes of the high rate of antibiotic resistance carriage among infants under 1 year old in this setting.
Conclusion
The high rate of intestinal carriage with MDR microorganisms among infants in rural Bangladesh is a serious concern that can jeopardize the management of infectious diseases. In addition, shedding of high number of MDR microorganisms through infant feces increases the risk of widespread transmission of these microorganisms in the community and environment. This study raises important questions about how the acquisition of resistant microorganisms takes place in infants’ guts within the first 3 months of life, what are the major drivers of acquisition, and what are the implications on infant health and well-being. Future studies should explore the source of acquisition of resistance in infants, to understand whether such resistance is primarily acquired from the environment, vertically from the child’s mother, or through selective pressure from pediatric antibiotic use.
Author Contributions
MAI and TJ conceived the development. MAI, TJ, MK, KL, TN-D, and MM designed the study and developed the protocol. MA, SR, and MBA contributed to the experiments, collection, and assembly of the data. MRI, MBA, and TJ contributed to the data entry and statistical analysis. MAI and MBA performed the first draft of the manuscript. TJ, MK, KL, TN-D, MM, MA, and MAI revised the manuscript and prepared the final draft of the manuscript.
Funding
This study was supported by REACH catalyst grant, United Kingdom (icddr,b Grant No. GR-01507). The icddr,b acknowledges with gratitude the funding support of REACH program, United Kingdom. The icddr,b is also thankful to the following donors: the governments of Bangladesh, Canada, Sweden, and United Kingdom for providing core/unrestricted support. KL was supported by National Institute for Allergy and Infectious Diseases grant number K01AI103544. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alexander, V. N., Northrup, V., and Bizzarro, M. J. (2011). Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. J. Pediatr. 159, 392–397. doi: 10.1016/j.jpeds.2011.02.035
Bangladesh Bureau of Statistics [BBS], and United Nations Children’s Fund [UNICEF] (2014). Bangladesh Multiple Indicator Cluster Survey 2012-2013, ProgotirPathey: Final Report. Dhaka: Bangladesh Bureau of Statistics.
Blomberg, B., Manji, K. P., Urassa, W. K., Tamim, B. S., Mwakagile, D. S., Jureen, R., et al. (2007). Antimicrobial resistance predicts death in Tanzanian children with bloodstream infections: a prospective cohort study. BMC Infect. Dis. 7:43. doi: 10.1186/1471-2334-7-43
Calva, J. J., Sifuentes-Osornio, J., and Céron, C. (1996). Antimicrobial resistance in fecal flora: longitudinal community-based surveillance of children from urban Mexico. Antimicrob. Agents Chemother. 40, 1699–1702. doi: 10.1128/AAC.40.7.1699
Centers for Disease Control and Prevention [CDC] (2006). Outbreaks of multidrug-resistant Shigella sonnei gastroenteritis associated with day care centers–Kansas, Kentucky, and Missouri, 2005. MMWR Morb. Mortal. Wkly Rep. 55, 1068–1071.
Cottell, J. L., Webber, M. A., and Piddock, L. J. (2012). Persistence of transferable extended-spectrum-beta-lactamase resistance in the absence of antibiotic pressure. Antimicrob. Agents Chemother. 56, 4703–4706. doi: 10.1128/AAC.00848-12
de Tejada, B. M. (2014). Antibiotic use and misuse during pregnancy and delivery: benefits and risks. Int. J. Environ. Res. Public Health 11, 7993–8009. doi: 10.3390/ijerph110807993
DGHS Health Bulletin (2014). Matlab (Uttar) Upazila Health Complex. Dhaka: Ministry of Health & Family Welfare.
Dhand, N. K., and Khatkar, M. S. (2014). Statulator: An Online Statistical Calculator. Sample Size Calculator for Estimating a Single Proportion. Available at: http://statulator.com/SampleSize/ss1P.html (accessed January 28, 2019).
Domínguez, E., Zarazaga, M., Sáenz, Y., Briñas, L., and Torres, C. (2002). Mechanisms of antibiotic resistance in Escherichia coli isolates obtained from healthy children in Spain. Microbial. Drug Resist. 8, 321–327. doi: 10.1089/10766290260469589
Farra, A., Frank, T., Tondeur, L., Bata, P., Gody, J., Onambele, M., et al. (2016). High rate of faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae in healthy children in Bangui, Central African Republic. Clin. Microbiol. Infect. 22, 891.e1–891.e4. doi: 10.1016/j.cmi.2016.07.001
Ferjani, S., Saidani, M., Hamzaoui, Z., Alonso, C. A., Torres, C., Maamar, E., et al. (2017). Community fecal carriage of broad-spectrum cephalosporin-resistant Escherichia coli in Tunisian children. Diagn. Microbiol. Infect. Dis. 87, 188–192. doi: 10.1016/j.diagmicrobio.2016.03.008
Gil, A., Lanata, C., Kleinau, E., and Penny, M. (2004). Children’s Feces Disposal Practices in Developing Countries and Interventions to Prevent Diarrheal Diseases. McMurray, PA: Environmental Health Project.
Guimaraes, B., Barreto, A., Radhouani, H., Figueiredo, N., Gaspar, E., Rodrigues, J., et al. (2009). Genetic detection of extended-spectrum β-Lactamase–containing Escherichia coli isolates and vancomycin-resistant enterococci in fecal samples of healthy children. Microbial. Drug Resist. 15, 211–216. doi: 10.1089/mdr.2009.0910
Gurnee, E. A., Ndao, I. M., Johnson, J. R., Johnston, B. D., Gonzalez, M. D., Burnham, C.-A. D., et al. (2015). Gut colonization of healthy children and their mothers with pathogenic ciprofloxacin-resistant Escherichia coli. J. Infect. Dis. 212, 1862–1868. doi: 10.1093/infdis/jiv278
Hasan, B., Sandegren, L., Melhus, Å, Drobni, M., Hernandez, J., Waldenström, J., et al. (2012). Antimicrobial drug–resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg. Infect. Dis. 18, 2055–2058. doi: 10.3201/eid1812.120513
Herigstad, B., Hamilton, M., and Heersink, J. (2001). How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Methods 44, 121–129. doi: 10.1016/S0167-7012(00)00241-4
Hewitt, J., and Rigby, J. (1976). Effect of various milk feeds on numbers of Escherichia coli and Bifidobacterium in the stools of new-born infants. Epidemiol. Infect. 77, 129–139. doi: 10.1017/S0022172400055601
Hilty, M., Betsch, B. Y., Bögli-Stuber, K., Heiniger, N., Stadler, M., Küffer, M., et al. (2012). Transmission dynamics of extended-spectrum β-lactamase–producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin. Infect. Dis. 55, 967–975. doi: 10.1093/cid/cis581
Islam, M. A., Islam, M., Hasan, R., Hossain, M. I., Nabi, A., Rahman, M., et al. (2017). Environmental spread of New Delhi metallo-β-lactamase-1-producing multidrug-resistant bacteria in Dhaka, Bangladesh. Appl. Environ. Microbiol. 83, e793-17. doi: 10.1128/AEM.00793-17
Kaarme, J., Molin, Y., Olsen, B., and Melhus, Å (2013). Prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in healthy Swedish preschool children. Acta Paediatr. 102, 655–660. doi: 10.1111/apa.12206
Karami, N., Hannoun, C., Adlerberth, I., and Wold, A. E. (2008). Colonization dynamics of ampicillin-resistant Escherichia coli in the infantile colonic microbiota. J. Antimicrob. Chemother. 62, 703–708. doi: 10.1093/jac/dkn263
Karami, N., Nowrouzian, F., Adlerberth, I., and Wold, A. E. (2006). Tetracycline resistance in Escherichia coli and persistence in the infantile colonic microbiota. Antimicrob. Agents Chemother. 50, 156–161. doi: 10.1128/AAC.50.1.156-161.2006
Khan, E. R., Aung, M. S., Paul, S. K., Ahmed, S., Haque, N., Ahamed, F., et al. (2018). Prevalence and molecular epidemiology of clinical isolates of Escherichia coli and Klebsiella pneumoniae harboring extended-spectrum beta-lactamase and carbapenemase Genes in Bangladesh. Microb. Drug Resist. doi: 10.1089/mdr.2018.0063 [Epub ahead of print].
Khan, M. N., Islam, M. M., Shariff, A. A., Alam, M. M., and Rahman, M. M. (2017). Socio-demographic predictors and average annual rates of caesarean section in Bangladesh between 2004 and 2014. PLoS One 12:e0177579. doi: 10.1371/journal.pone.0177579
Kotloff, K. L., Nataro, J. P., Blackwelder, W. C., Nasrin, D., Farag, T. H., Panchalingam, S., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMS): a prospective, case-control study. Lancet 382, 209–222. doi: 10.1016/S0140-6736(13)60844-2
Ledger, W. J., and Blaser, M. J. (2013). Are we using too many antibiotics during pregnancy? BJOG 120, 1450–1452. doi: 10.1111/1471-0528.12371
Maamar, E., Ferjani, S., Jendoubi, A., Hammami, S., Hamzaoui, Z., Mayonnove-Coulange, L., et al. (2016). High prevalence of gut microbiota colonization with broad-spectrum cephalosporin resistant enterobacteriaceae in a tunisian intensive care unit. Front. Microbiol. 7:1859. doi: 10.3389/fmicb.2016.01859
Macones, G. A., Cleary, K. L., Parry, S., Stamilio, D. M., Cahill, A. G., Odibo, A. O., et al. (2012). The timing of antibiotics at cesarean: a randomized controlled trial. Am. J. Perinatol. 29, 273–276. doi: 10.1055/s-0031-1295657
Mathew, J. (2004). Effect of maternal antibiotics on breast feeding infants. Postgrad. Med. J. 80, 196–200. doi: 10.1136/pgmj.2003.011973
Munk, P., Knudsen, B. E., Lukjancenko, O., Duarte, A. S. R., Van Gompel, L., Luiken, R. E. C., et al. (2018). Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat. Microbiol. 3, 898–908. doi: 10.1038/s41564-018-0192-9
Patel, J. B. (2017). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute.
Pereira, L. M. P., Phillips, M., Ramlal, H., Teemul, K., and Prabhakar, P. (2004). Third generation cephalosporin use in a tertiary hospital in Port of Spain, Trinidad: need for an antibiotic policy. BMC Infect. Dis. 4:59. doi: 10.1186/1471-2334-4-59
Pitout, J. D., and Laupland, K. B. (2008). Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159–166. doi: 10.1016/S1473-3099(08)70041-0
R Core Team (2014). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rogawski, E. T., Platts-Mills, J. A., Seidman, J. C., John, S., Mahfuz, M., Ulak, M., et al. (2017). Use of antibiotics in children younger than two years in eight countries: a prospective cohort study. Bull. World Health Organ. 95:49. doi: 10.2471/BLT.16.176123
Saari, A., Virta, L. J., Sankilampi, U., Dunkel, L., and Saxen, H. (2015). Antibiotic exposure in infancy and risk of being overweight in the first 24 months of life. Pediatrics 135, 617–626. doi: 10.1542/peds.2014-3407
Saha, S. K., Schrag, S. J., El Arifeen, S., Mullany, L. C., Islam, M. S., Shang, N., et al. (2018). Causes and incidence of community-acquired serious infections among young children in south Asia (ANISA): an observational cohort study. Lancet 392, 145–159. doi: 10.1016/S0140-6736(18)31127-9
Sánchez, A. R., Rogers, R. S., and Sheridan, P. J. (2004). Tetracycline and other tetracycline-derivative staining of the teeth and oral cavity. Int. J. Dermatol. 43, 709–715. doi: 10.1111/j.1365-4632.2004.02108.x
Sandegren, L. (2014). Selection of antibiotic resistance at very low antibiotic concentrations. Upsala J. Med. Sci. 119, 103–107. doi: 10.3109/03009734.2014.904457
Stoesser, N., Xayaheuang, S., Vongsouvath, M., Phommasone, K., Elliott, I., Del Ojo Elias, C., et al. (2015). Colonization with Enterobacteriaceae producing ESBLs in children attending pre-school childcare facilities in the Lao People’s Democratic Republic. J. Antimicrob. Chemother. 70, 1893–1897. doi: 10.1093/jac/dkv021
Talukdar, P. K., Rahman, M., Rahman, M., Nabi, A., Islam, Z., Hoque, M. M., et al. (2013). Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One 8:e61090. doi: 10.1371/journal.pone.0061090
The World Bank (2016). Birth Rate, Crude (per 1,000 People) | Data. Available at: https://data.worldbank.org/indicator/SP.DYN.CBRT.IN?locations=BD (accessed January 28, 2019).
Valverde, A., Grill, F., Coque, T. M., Pintado, V., Baquero, F., Cantón, R., et al. (2008). High rate of intestinal colonization with extended-spectrum-β-lactamase-producing organisms in household contacts of infected community patients. J. Clin. Microbiol. 46, 2796–2799. doi: 10.1128/JCM.01008-08
Verani, J. R., Mcgee, L., Schrag, S. J., and Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases (2010). Prevention of perinatal group B streptococcal disease: revised guidelines from CDC, 2010. MMWR Recomm. Rep. 59, 1–36.
World Health Organization [WHO] (2005). Guidelines for the Control of Shigellosis, Including Epidemics Due to Shigella dysenteriae Type 1. Geneva: World Health Organization.
World Health Organization [WHO] (2017). Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-human Use. Geneva: World Health Organization.
Yeager, B. A., Huttly, S. R., Bartolini, R., Rojas, M., and Lanata, C. F. (1999). Defecation practices of young children in a Peruvian shanty town. Soc. Sci. Med. 49, 531–541. doi: 10.1016/S0277-9536(99)00119-7
Keywords: colonization, multidrug-resistant, E. coli, ESBL, third generation cephalosporins
Citation: Islam MA, Amin MB, Roy S, Asaduzzaman M, Islam MR, Navab-Daneshmand T, Mattioli MC, Kile ML, Levy K and Julian TR (2019) Fecal Colonization With Multidrug-Resistant E. coli Among Healthy Infants in Rural Bangladesh. Front. Microbiol. 10:640. doi: 10.3389/fmicb.2019.00640
Received: 14 August 2018; Accepted: 13 March 2019;
Published: 02 April 2019.
Edited by:
Patrícia Poeta, University of Trás-os-Montes and Alto Douro, PortugalReviewed by:
Peter Bergholz, North Dakota State University, United StatesXiaoqiang Liu, Northwest A&F University, China
Jian-Hua Liu, South China Agricultural University, China
Copyright © 2019 Islam, Amin, Roy, Asaduzzaman, Islam, Navab-Daneshmand, Mattioli, Kile, Levy and Julian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Aminul Islam, bWFpc2xhbUBpY2RkcmIub3Jn
†Present address: Mohammad Aminul Islam, Paul G. Allen School for Global Animal Health, Washington State University, Pullman, WA, United States
 Mohammad Aminul Islam
Mohammad Aminul Islam Mohammed Badrul Amin
Mohammed Badrul Amin Subarna Roy
Subarna Roy Muhammad Asaduzzaman
Muhammad Asaduzzaman Md. Rayhanul Islam
Md. Rayhanul Islam Tala Navab-Daneshmand
Tala Navab-Daneshmand Mia Catharine Mattioli3,4
Mia Catharine Mattioli3,4 Karen Levy
Karen Levy Timothy R. Julian
Timothy R. Julian