
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 March 2019
Sec. Infectious Agents and Disease
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00632
This article is part of the Research Topic Unraveling the Biology, Genetics, and Host/Environmental Interactions of Acinetobacter View all 17 articles
The dif/Xer system of site-specific recombination allows resolution of chromosomal dimers during bacterial DNA replication. Recently, it was also shown to be involved in horizontal transfer of a few known Xer-dependent mobile elements. Here, we show that plasmids of various Acinetobacter species, including clinically important strains, often contain multiple pdif sites that are mainly located within their accessory regions. Chromosomes of Acinetobacter strains may also contain additional dif sites, and their similarity with plasmid pdif sites is higher than with the main chromosomal site dif1. We further identify putative mobile genetic elements containing pdif sites on both flanks of adaptive genes and analyze their distribution in Acinetobacter species. In total, we describe seven mobile elements containing genes with various adaptive functions from permafrost strains of A. lwoffii group. All of them are also spread in modern plasmids of different Acinetobacter species including A. baumannii. We could not detect pdif sites and corresponding mobile elements in closely related bacterial genera, including Psychrobacter and Moraxella. Thus, the widespread distribution of dif modules is a characteristic feature of Acinetobacter species and may contribute to their high adaptability both in the environment and in the clinic.
Acinetobacter strains are common everywhere due to their ability to adapt to various environments (Touchon et al., 2014). Many of them – first of all, Acinetobacter baumannii, but also Acinetobacter nosocomialis, Acinetobacter pittii, Acinetobacter lwoffii, Acinetobacter ursingii, Acinetobacter haemolyticus, Acinetobacter parvus, Acinetobacter junii – are among the most important pathogens in the clinic (Turton et al., 2010; Peleg et al., 2012; Evans and Amyes, 2014; Touchon et al., 2014). The rise of antimicrobial resistance in these strains is a burning medical problem, with a large number of mobile genetic elements involved in the spread of the resistance determinants (Martins et al., 2015; Da Silva and Domingues, 2016; Pagano et al., 2016).
Recently a novel class of mobile genetic elements whose transposition likely depends on the action of the dif/Xer recombination system was discovered in Acinetobacter species. Initially, a discrete DNA module that contained the gene encoding OXA-24 carbapenemase, flanked by conserved inverted repeats homologous to the XerC/XerD binding sites (dif sites), was revealed in an A. baumannii plasmid pABVA01a. It was shown that this element (later designated dif module) is also present in a different context in another Acinetobacter plasmid, suggesting its horizontal transfer (D’Andrea et al., 2009). Similar observations were soon made by other laboratories when studying plasmids containing the same gene OXA-24 (Merino et al., 2010; Grosso et al., 2012) and related carbapenemase genes OXA-72 (a variant of OXA-24) or OXA-58 (Povilonis et al., 2013; Blackwell and Hall, 2017). Subsequent studies demonstrated that other antibiotic resistance genes can also be included in dif modules. In particular, Blackwell and Hall (2017) described two novel dif modules containing genes encoding resistance to tetracycline and erythromycin.
The dif sites found in Acinetobacter plasmids (pdif sites) are similar to the chromosomal dif1 site recognized by the dif/Xer system of site-specific recombination, which plays an essential role in the resolution of chromosomal and plasmid dimers (Carnoy and Roten, 2009; Castillo et al., 2017). Orphan pdif sites could also be revealed in a number of plasmids (Carnoy and Roten, 2009; Blackwell and Hall, 2017; Cameranesi et al., 2018), and the reaction of site-specific recombination could be observed between the two sites present in different plasmids, suggesting that this system is involved in the horizontal transfer of dif modules (Cameranesi et al., 2018).
Further studies have shown that the spread of dif sites and dif modules is not limited to plasmids of clinical Acinetobacter strains and that they can include a variety of genes, not only determinants of antibiotic resistance. In particular, we recently described a novel dif module containing chromium resistance genes (chrAB dif). Initially discovered in plasmids from ancient (permafrost) strains of A. lwoffii it was also found in plasmids of modern Acinetobacter strains (Mindlin et al., 2018). We also identified two other potential dif modules, kup dif and sulP dif, but did not characterize them in detail (Mindlin et al., 2018). Recently, putative new dif modules containing genes with potential adaptive functions were found in plasmids of the clinical isolate of A. baumannii D36 (Hamidian and Hall, 2018). This suggested that formation of mobile elements flanked by pdif sites and carrying various adaptive genes may be a characteristic feature of plasmids of the whole Acinetobacter genus.
In the present work, we investigated the distribution of dif sites in more than 180 Acinetobacter plasmids belonging to different species. The main emphasis was made on the identification and analysis of dif modules from ancient A. lwoffii plasmids and studies of their distribution in modern strains. As a result, we identified four novel adaptive dif modules, in addition to chrAB dif, kup dif, and sulP dif, and revealed their broad distribution in plasmids in environmental and clinical strains of the Acinetobacter genus. We also showed that most Acinetobacter chromosomes contain additional dif sites besides the main site dif1, which may contribute to the distribution of dif modules in this genus.
Acinetobacter strains ED23-35, ED45-23, ED9-5a, EK30A, and VS15 used in this study were previously isolated from 15 thousand to 3 million years old permafrost sediments collected from different regions of Kolyma Lowland. We here in referred to these strains as “ancient” strains, which never come into contact with clinical isolates. Initially they were identified as A. lwoffii based on metabolic testing and from 16S rRNA gene sequence analysis (Petrova et al., 2002; Kurakov et al., 2016; Mindlin et al., 2016). In this work, based on data from a recent article by Nemec et al. (2018), revising the taxonomy of the Acinetobacter lwoffii group we showed that strain ED9-5a actually belongs to the Acinetobacter pseudolwoffii species. The remaining permafrost strains do belong to A. lwoffii. In addition, we analyzed two sets of Acinetobacter strains obtained from publicly available databases (“modern” strains). These were (i) 38 strains of A. baumannii described by Shintani et al. (2015), and (ii) 30 strains belonging to other Acinetobacter species obtained from public databases before January 2018. Bacteria were grown in lysogeny broth (LB) medium or solidified LB medium (LA) at 30°C.
Species identification was confirmed by comparison of partial rpoB gene sequences with the reference Acinetobacter genomes1. The rpoB gene nucleotide identity level of 98–100% was regarded as evidence that the compared genomes belong to the same species.
The genomes were sequenced with a Roche GS FLX genome sequencer (Roche, Switzerland) using the Titanium protocol to prepare shotgun genomic libraries. The GS FLX reads were assembled into contigs using the GS De Novo Assembler (Roche); protein coding genes were identified and annotated using the RAST web server. To identify potential plasmid contigs we used read coverage depth and annotation information, such as existence of genes for mobilization and/or replication of plasmids. Using 454 assembly graph file (output of the GS De Novo Assembler) we extended and merge our potential plasmid contigs with each other. Some additional merging of contigs was done using PCR. All manually assembled regions were checked using PCR. A plasmid was considered as finished if it was circular according to the 454 assembly graph and PCR check. The 454 reads were mapped back to the finished sequences using 454 GS Reference Mapper, and read mapping was visualized in IGV browser and manually inspected for potential missassemblies.
A collection of plasmids isolated from different Acinetobacter species (Supplementary Table S1) was screened for the presence of pdif sites by BLAST software. The XerC/XerD (3916–3943 bp) and XerD/XerC (6899–6926) pdif sites of plasmid pM131-6 [NC_025120] were used as a reference. Only sites that were at least 75–80% identical to the reference were taken into account. The same algorithm was used in the search for the main and additional dif sites in Acinetobacter chromosomes.
For the assembly and analysis of plasmid genomes from ancient strains, the program UGENE2 was used. Similarity searches were performed using BLAST (Altschul et al., 1997) and REBASE (Roberts et al., 2009). Conserved domains and motifs were identified using the NCBI Conserved Domain Database (CDD) (Marchler-Bauer et al., 2011) and the Pfam database (Finn et al., 2009). Dif sites were clustered using blastclust from blast package with 95% identity threshold. Fasta files with dif sites were sorted according to blastclust results using custom perl script (dif sites from the same cluster follow each other, and clusters are sorted by size). The sorted fasta files were visualized in AliView as color blocks. The fasta files with dif sites were used to create sequence logos using WebLogo3 (Crooks et al., 2004).
Identification of dif modules in permafrost plasmids and determination of their mobility was carried out manually in several stages. Initially, plasmids containing at least two different pdif-sites were selected. Then, the regions between the pairs of pdif-sites were analyzed, and, based on the presence of a potentially adaptive gene (s) in this region candidates for mobile modules were revealed. At the final stage, using the Blast software, the distribution of the identified modules among the ancient (isolated from permafrost) and modern strains of Acinetobacter’species was investigated. The presence of the same module with an identity level of 98–99% in different regions of different plasmids (contigs) isolated from Acinetobacter strains belonging to different species was regarded as evidence of its mobility.
The level of resistance was determined by the agar dilution method (Mindlin et al., 2016). Overnight cultures of bacterial strains were diluted 10-fold with fresh LB and grown at 30°C with shaking for 3–4 h; the cultures were then induced for 1 h by the addition of tert-butyl hydroperoxide (tBHP) at 0.1 mM. Five μl of the bacterial suspension (about 5 × 106 cells per ml) were plated onto LA supplemented with Na2TeO3 (Tel) or tBHP. The concentrations tested were as follows: Tel – 0.01; 0.02; 0.04 and 0.1 mg/ml and tBHP – 0.05; 0.1; 0.2; 0.3 and 0.4 mM. The plates were incubated at 30°C for 24 h and visually inspected. In each case, three independent experiments were performed.
The accession numbers for nucleotide sequences of the plasmids deposited in this work in the GenBank database are as follows: CP032112.1 (pALWED1.2); CP032113-CP032116.1 (pALWED1.4-1.7); CP032117.1-CP032124.1 (pALWED2.2-2.9); CP032287.1-CP032289.1 (pALWED3.2-3.4); CP032290.1 (pALWED3.6); CP032102.1 (pALWEK1.1); CP032105.1-CP032107.1 (pALWEK1.2-1.4); CP032108.1- CP032111.1 (pALWEK1.6-1.9); CP032103.1 (pALWEK1.10); CP032104.1 (pALWEK1.11).
For analysis of the occurrence of pdif sites in Acinetobacter plasmids, we studied plasmids found in strains of various Acinetobacter species, divided into three groups (see Materials and Methods): (1) plasmids of permafrost strains of A. lwoffii and A. pseudolwoffii isolated and studied in our group (35 plasmids, see below) together with plasmids from A. lwoffii ZS207 (10 plasmids, see Supplementary Table S1); (2) plasmids from A. baumannii strains, described in Shintani et al. (2015) (65 plasmids, Supplementary Table S1); (3) plasmids isolated from strains belonging to others Acinetobacter species available from public databases before January 2018 (73 plasmids, Supplementary Table S1).
The presence of pdif sites and the number of their copies in each plasmid were determined by the BLAST tool. The results obtained for each set of plasmids are presented in Figure 1, Table 1, and Supplementary Table S1. Plasmid pdif sites were found in more than 50% of the Acinetobacter plasmids in all three groups (Table 1). In most cases, the number of sites was 1–4, while some plasmids contained 5–8 copies of pdif or more (Table 1). Most pdif sites were found in small to middle size plasmids (6–30 kb), while very small plasmids (<6 kb) as a rule did not contain them.
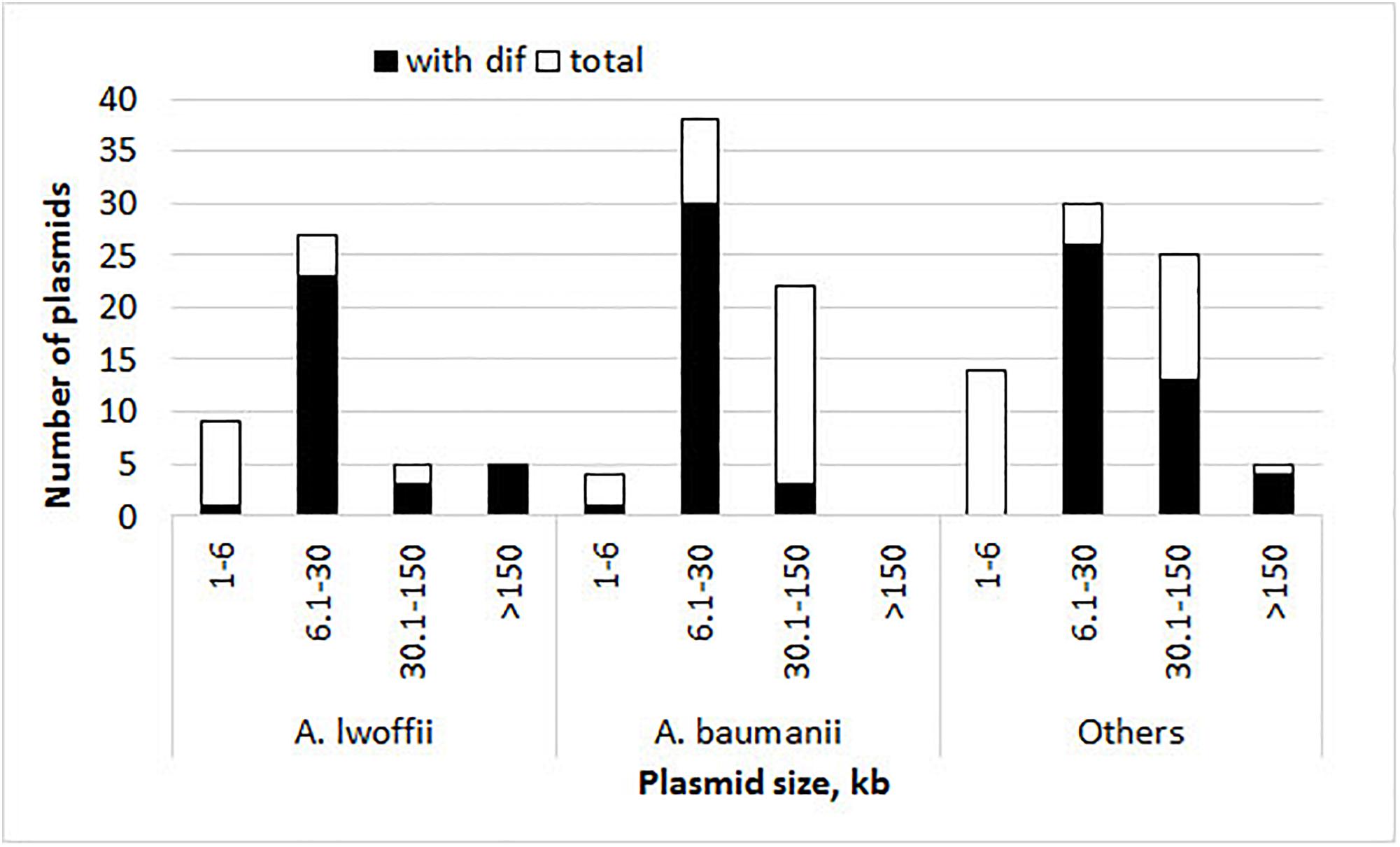
Figure 1. Histograms of plasmid size and their numbers with and without pdif sites. The height of the column indicates the total number of plasmids, and its black part shows the number of plasmids containing pdif sites.
Interestingly, multiple copies of pdif sites were found both in very large and in middle-size plasmids (22–400 kbp) (Figure 1 and Table 2).
To determine the localization of pdif sites relative to the backbone and accessory plasmid regions, we analyzed their distribution in four plasmids, two middle size (pALWED2.3 and pOXA58-AP_882) and two large (pALWED2.1 and pXBB1-9), each containing more than 10 sites (Table 2). For all four plasmids, pdif sites were distributed unevenly across the plasmid sequence; notably, they were absent from the backbone regions and formed clusters in certain plasmid regions (Figure 2).
To our knowledge, there is currently no data on the distribution of typical pdif sites in the plasmids of bacteria from other systematic groups. To get insight into their abundance in closely related genera to Acinetobacter, we analyzed a total of 31 plasmids from 16 strains of Psychrobacter (2117–44793 bp) and 8 plasmids from 6 strains of Moraxella (1313–44215 bp) described by Shintani et al. (2015) and Moghadam et al. (2016). No pdif sites were detected in any of these plasmids. Therefore, the presence of a large number of pdif sites may be a unique feature of Acinetobacter plasmids.
The primary search for dif modules was performed on the set of plasmids from five permafrost strains from our collection (ED23-35, ED45-23, ED9-5a, EK30A and VS15; Supplementary Table S1) (Petrova et al., 2002; Mindlin et al., 2016). In five fully sequenced genomes, a total of 35 plasmids of various sizes were found. In addition to previously described 8 plasmids (Mindlin et al., 2016, 2018), we have now assembled 27 new plasmids from the same strains (see details in Materials and Methods).
In 10 of the plasmids, sequences homologous to pdif sites were absent or only a single pdif site was detected (Supplementary Table S1). The remaining 25 plasmids with two or more pdif sites were analyzed for the presence of dif modules containing adaptive genes surrounded by two pdif sites (see Materials and Methods). In total, putative dif modules were detected in 12 plasmids; 6 of them contained one module; 4 contained two modules, and 2 contained three different modules (Supplementary Table S2).
In addition to the three modules identified previously (chrAB dif, kup dif, and sulP dif) (Mindlin et al., 2018), we revealed four new putative modules with adaptive genes: terC dif, add dif, ohr dif, and sulP-uspA dif (Figure 3).
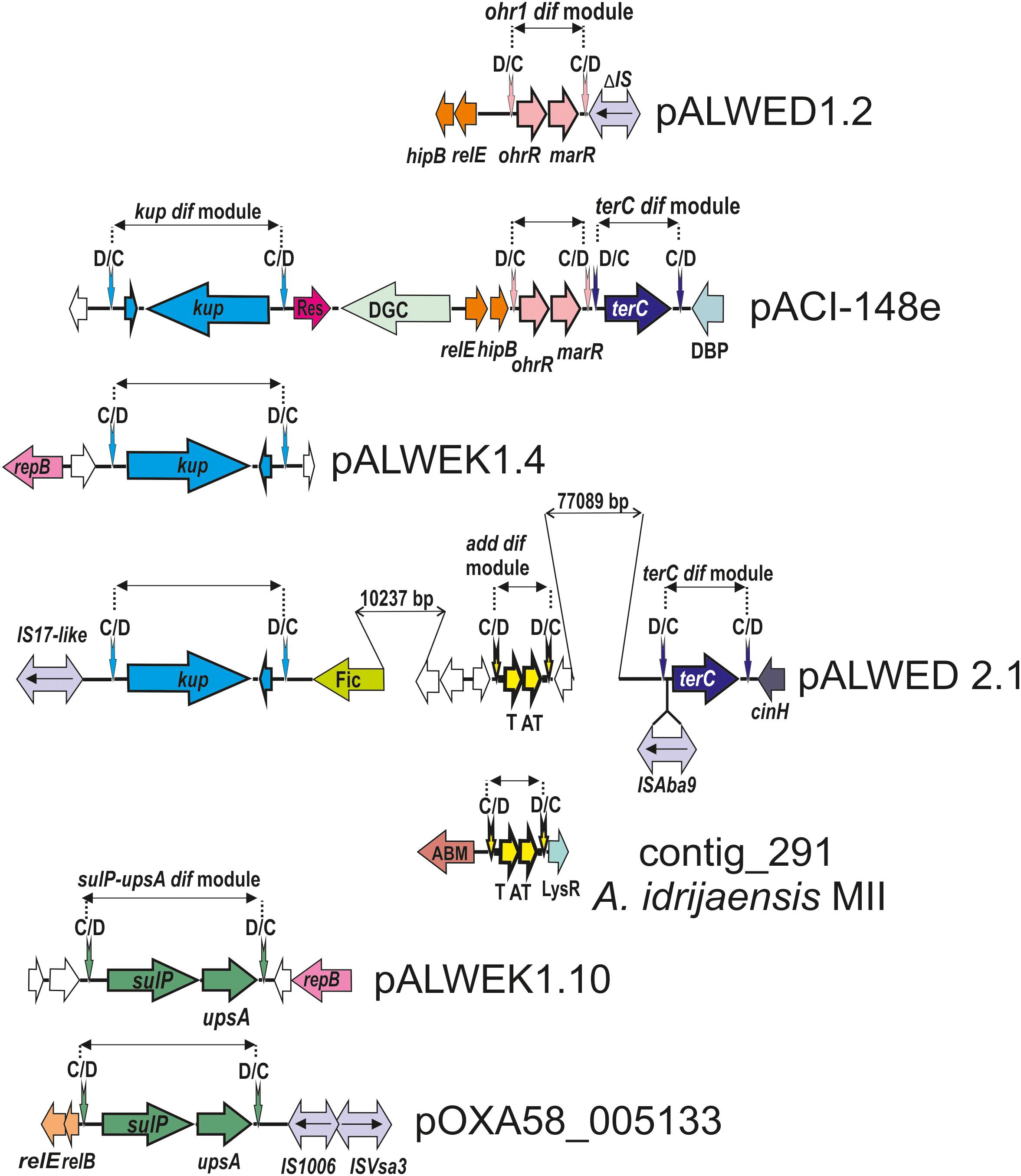
Figure 3. Comparative genetic structures of plasmid regions containing dif modules. The locations and directions of genes and ORFs are shown by arrows. The components of the dif modules are represented by arrows with different colors. Vertical arrows indicate pdif sites with the orientation of the subsites shown above. Numbers (bp) show the size of the sequences located between different genes. Designations of gene products are as follows: DBP, DNA-binding protein; DGC, diguanylate cyclase; Res, resolvase; T, toxin; AT, antitoxin; Fic, Fic/DOC family protein; ABM, antibiotic biosynthesis monooxigenase; LysR, LysR family transcriptional regulator; unnamed ORFs, hypothetical proteins. Designations of IS-s: ISVsa3 -100% with ISVsa3; IS1006 – 1 mismatch with IS1006; ISAba9 – 2 mismatches with ISAba9; IS17-like -96% identity with IS17; ΔIS – truncated copy of an IS5 family element (IS427 group). The picture is drawn to scale.
In most cases, plasmids containing the same modules originated from different permafrost strains (Supplementary Table S2) suggesting their horizontal transfer. To test this hypothesis, we analyzed the distribution of all modules among plasmids (contigs) of modern Acinetobacter strains belonging to different Acinetobacter species (Supplementary Table S2). Below, we briefly describe characteristics and distribution of each of these modules (Table 3).
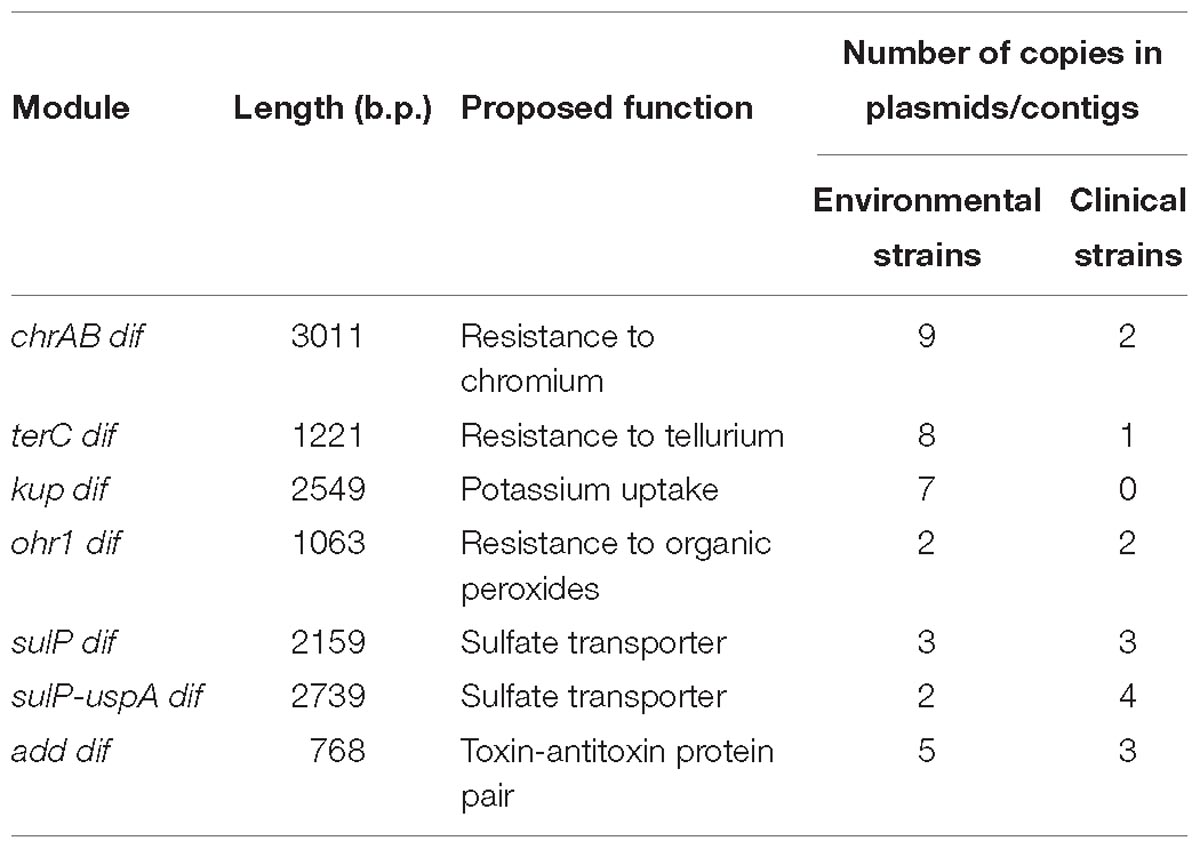
Table 3. The dif modules found in permafrost A. lwoffii strains and their distribution in modern strains.
We revealed the terC dif module (1221 bp) in four permafrost plasmids, pALWED1.4 from A. lwoffii ED23-35, pALWED2.1 from A. lwoffii ED45-23, pALWED3.6 from A. pseudolwoffii ED9-5a and pALWEK1.1 from A. lwoffii EK30A (Supplementary Table S2). It contained a single orf, identified as terC (978 bp), encoding resistance to tellurium. We also found the terC dif module in two modern plasmids and several contigs from modern strains belonging to different species of Acinetobacter (Supplementary Table S2 and Figure 3).
Up to date, genetic determinants of resistance to tellurium were found in the genomes of different bacteria both in chromosomes and plasmids (Chasteen et al., 2009). We also detected the terC gene in the sequenced chromosomes of Acinetobacter strains, belonging to different species including Acinetobacter baylyi ADP1 [NC_005966.1] widely used in genetic studies.
However, the plasmidic and chromosomal terC genes were related only distantly (42–60% identity at the protein sequence level), and the chromosomal terC gene was not flanked by dif sites.
To determine the functional activity of the terC dif module, we compared the level of tellurium resistance of the permafrost strains of A. lwoffii and A. pseudolwoffii containing the terC dif module with the strain A. lwoffii VS15 that lacked this module in its plasmids. The results of three independent experiments showed that strains ED23-35, ED9-5a and EK30A have a significantly higher resistance level (MIC from 0.04 to 0.1 mg/ml) compared to strain VS15 (MIC < 0.01 mg/ml); whereas strain ED45-23, characterized by very slow growth, does not differ from VS15. Thus, additional studies are needed to confirm the contribution of the terC dif module into the tellurium resistance of corresponding host strains.
Most prokaryotic chromosomes as well as many plasmids contain toxin-antitoxin (TA) systems consisting of a pair of genes that encode 2 components, a stable toxin and its cognate unstable antitoxin. TA systems are also known as addiction modules (Yamaguchi et al., 2011; Schuster and Bertram, 2013). In particular, many type II toxins are mRNA-specific endonucleases that arrest cell growth through RNA cleavage, thus preventing the process of translation (Bertram and Schuster, 2014). We revealed a number of type II TA modules in permafrost plasmids. Amongst them we found two related modules surrounded by pdif sites (both 768 bp long). Both modules contained two genes, one (297 bp) encoding a killer protein and another (291 bp) – an antidote protein. The first module was located in large plasmid pALWED2.1; the second was detected in medium-sized (22771 bp) plasmid pALWED2.3 and large plasmid pALWEK1.1. The difference in nucleotide sequences of the modules was 4%.
We also found variants of both add dif modules in plasmids and contigs of modern strains belonging to different Acinetobacter species (A. lwoffii, A. baumannii, Acinetobacter towneri, Acinetobacter johnsonii) (Supplementary Table S2). In most cases they were almost identical to the basic prototype modules (96–99% nucleotide identity level).
This genetic element contains two genes: a gene encoding organic hydroperoxide resistance, ohr (429 bp) and a second gene ohrR (432 bp) encoding transcription regulator from the MarR family of transcription factors (Sukchawalit et al., 2001; Atichartpongkul et al., 2010).
The ohr dif module was detected in plasmids from three permafrost strains A. lwoffii: pALWED1.2 from strain ED23-35; pALWVS1.1 from strain VS15 and pALWEK1.1 from strain EK30A (identity level 99%). However, the size and the boundaries of this module differed between the plasmids. We therefore analyzed its structure and distribution in more detail.
The ohr/ohrR genes with plasmid localization were first found by Dorsey et al. (2006) in the plasmid pMAC [AY541809.1] from a reference clinical strain A. baumannii ATCC 19606. The authors showed that A. baumannii ATCC 19606 is resistant to the organic peroxide-generating compounds cumene hydroperoxide (CHP) and tert-butyl hydroperoxide (t-BHP). We analyzed the genome of pMAC and detected typical pdif sites on both flanks of the ohr genes suggesting that pMAC contains an ohr dif module. The permafrost and clinical versions of the ohr dif module are overall closely related (96% nucleotide identity) but reveal some specific differences: (i) the ohr module from the modern strain is 5 bp longer than the ancient one (1068 bp vs.1063 bp); (ii) both modules contain identical ohrR genes but their ohrA genes differ by 3%; (iii) region adjacent to the XerD/XerC site contains multiple substitutions. We therefore designated the ancient variant ohr1 dif and the modern variant ohr2 dif and studied the distribution of each of them separately.
Although we did not find the ohr1 dif module in the plasmids of modern strains, it was revealed among unassembled genomic sequences of various Acinetobacter species, in most cases in small-size contigs (Supplementary Table S2). The ohr2 dif module was detected both in plasmids and in contigs from various Acinetobacter species (Supplementary Table S2).
Interestingly, two plasmids (plasmid unnamed 2 [CP027180] and plasmid unnamed 4 [CP027186] from the modern strains of A. baumannii AR_0070 and A. baumannii AR_0052, respectively) contained numerous copies of the ohr 2 module, 14 in the first plasmid and 8 in the second.
The location of both ohr dif modules in various sequence contexts in different plasmids and contigs (Figure 3) strongly suggests their ability to move. Both modules likely originated from a common ancestor and then distributed independently among Acinetobacter species.
To test the functional activity of the ohr1 dif module, we compared resistance of permafrost strains, containing (group 1: ED23-35, VS15 and EK30A) or lacking (group 2: ED45-23 and ED9-5a) this module, to tert-butyl hydroperoxide (t-BHP) (see Materials and Methods). The results of three independent experiments demonstrated a higher level of resistance of group 1 strains (MIC = 0.2–0.4 mM) in comparison with group 2 strains (MIC = 0.1 mM).
The gene sulP encodes sulfate permease (484 aa) that perform transport of inorganic anions into cells (Price et al., 2004). It is a member of the large and ubiquitous family of SulP proteins present in archaea, bacteria, fungi, plants and animals (Alper and Sharma, 2013).
We revealed two different mobile elements containing the sulP gene. The first module sulP dif (2159 bp), first found in our previous work (Mindlin et al., 2018), contained a small orf (198 bp) encoding hypothetical protein in addition to sulP; the second module sulP-uspA dif (2739 bp), detected in this work in the plasmid pALWEK1.10 from A. lwoffii EK30A, contained the uspA gene encoding a universal stress protein in addition to sulP. The sulP genes from the two modules are related (the identity level of 75%). The distribution of both dif modules in modern plasmids (contigs) is presented in Supplementary Table S2 and Figure 3 illustrates the location of the sulP-uspA dif module in different plasmids.
A putative 2549 bp dif module containing the kup gene (1878 bp) encoding a potassium uptake transporter (Zakharyan and Trchounian, 2001) and an orf of unknown function (210 bp) was first revealed in our previous work (Mindlin et al., 2018). In this work we detected almost identical kup dif module in plasmids of three other permafrost strains (Figure 3) and studied its distribution in modern strains. The kup dif modules with identity level of 99% were found in plasmids and contigs from Acinetobacter strains belonging to different species (Supplementary Table S2). Importantly, seven out of 10 modern plasmids/contigs containing this module were isolated from environmental strains. The origin of the host strains for the remaining three plasmids [CP026424.1; APOG01000001.1; JWHB01000051.1] could not be established. Therefore, this module, which likely controls the uptake of potassium, may increase the fitness of Acinetobacter strains living in the environment but not in the clinic.
To reveal whether the presence of multiple dif sites is restricted to Acinetobacter plasmids, we next analyzed the number and location of dif sites in the Acinetobacter chromosomes. Usually a single dif locus involved in the resolution of chromosomal concatenates is located near the chromosome terminus (Carnoy and Roten, 2009). Surprisingly, we found that the Acinetobacter chromosomes are organized in a different way. Besides the main dif site (dif1), we revealed additional dif sites in most completely sequenced circular chromosomes of different species of the Acinetobacter genus. In total, we analyzed 20 sequenced chromosomes of 13 Acinetobacter species, and only one of them did not contain additional dif sites.
As a rule, the number of discovered dif sites varied from one to five and they were located far from dif1 (e.g., A. lwoffii WJ10621 [CM001194.1]; A. lwoffii ZS207 [NZ_CP019143.1]; A. johnsonii XBB1 [CP010350.1]; A. nosocomialis SSA3 [CP020588.1]). Interestingly, we found two Acinetobacter strains that contained multiple additional dif sites in their chromosomes, Acinetobacter indicus SGAir0564 with 14 additional chromosomal dif sites [CP024620.1] and Acinetobacter sp. SWBY1 with 15 additional dif sites [CP026616.1].
It should be mentioned that strains containing multiple chromosomal dif sites were found occasionally when studying the distribution of different dif modules. In particular, the terC dif module, almost identical to that from plasmid pALWED1.4 (99% identity), was revealed in the chromosome of A. indicus SGAir0564 (CP024620.1). It was flanked by typical pdif sites (XerC/D and XerD/C) and was located at 2363397–2364617 bp, far from the main chromosomal dif1 site at position 1526563. Similarly, we found a variant of the add dif module in the chromosome of the Acinetobacter sp strain SWBY1 [CP026616.1].
To get insight into the origin and specific structure of the additional chromosomal dif sites, we compared their sequences with the main dif1 sites from Acinetobacter chromosomes and with pdif sites from A. baumannii and A. lwoffii (see Materials and Methods). In total 109 pdif sites from 23 plasmids of A. lwoffii, 115 pdif sites from 33 plasmids of A. baumannii, 20 main dif1 and 40 additional chromosomal Acinetobacter dif sites were analyzed.
All main features of classical dif sites were revealed in all studied groups (Figure 4). All of them have a length of 28 bp and are composed of two 11 bp regions comprising the binding sites for XerD (more conserved) and XerC (less conserved), separated by a more variable 6 bp central region. The inner parts of the XerC and XerD binding motifs contain inverted repeats forming a palindrome and are highly conserved while their outer regions are more variable (Figure 4).
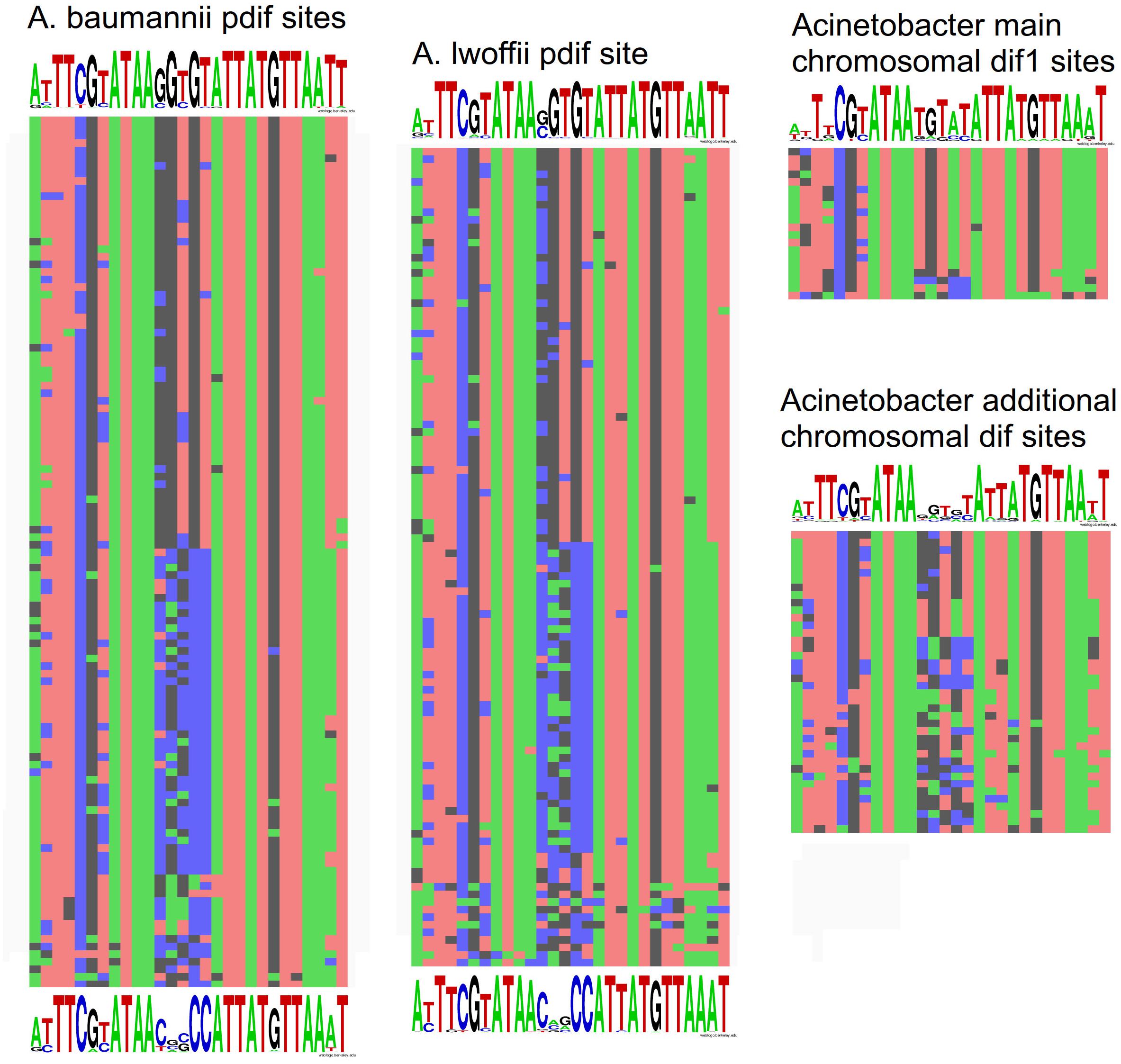
Figure 4. Nucleotide variability within dif sites in Acinetobacter plasmids and chromosomes. All dif sequences analyzed in this study are presented in Supplementary Table S1.
Comparison of dif sites from different groups revealed that (Figure 4): (i) pdif sites from plasmids of distant species A. baumannii and A. lwoffii (A. pseudolwoffii) are very close to each other in their consensus structure and form two clusters with different consensus sequences of five central nucleotides; (ii) additional chromosomal dif sites differ significantly in their structure from the main chromosomal dif1 site, especially in their central part, while (iii) they are more similar to the plasmid pdif sites from both clusters. It should be mentioned that the structure of the main chromosomal dif1 sites of Acinetobacter corresponds well to the consensus of dif sites from Proteobacteria chromosomes (Carnoy and Roten, 2009). Thus, it can be hypothesized that the plasmid pdif sites and additional chromosome dif sites have originated from a common genetic element, whose identity remains to be established.
The results of this and previous studies (D’Andrea et al., 2009; Merino et al., 2010; Grosso et al., 2012; Feng et al., 2016; Blackwell and Hall, 2017; Cameranesi et al., 2018; Hamidian and Hall, 2018) revealed the common presence of several (multiple) copies of dif sites and associated dif modules in plasmids and chromosomes of various Acinetobacter species including pathogens, commensals, and free-living bacteria belonging to different phylogenetic branches of the genus. Neither the additional copies of dif sites nor the mobile elements containing them are commonly found in other bacteria, even closely related to Acinetobacter, suggesting that their presence is a characteristic feature of the Acinetobacter genus. It should be noted that previous studies were mainly limited to the description of dif modules with antibiotic resistance genes (D’Andrea et al., 2009; Merino et al., 2010; Grosso et al., 2012; Povilonis et al., 2013; Blackwell and Hall, 2017). It was proposed that their distribution depends on the action of the site-specific recombination system dif/Xer. At the same time, the origin of the additional copies of dif sites, the mechanisms of their distribution and their role in the bacterial genome have not been studied in detail.
The results obtained in this study suggest that multiple dif sites present in Acinetobacter plasmids allow the formation of simple mobile elements containing various adaptive genes. In particular, analysis of the plasmid genomes from the ancient A. lwoffii (A. pseudolwoffii) strains revealed novel dif modules encoding different adaptive functions, including the resistance to chromium (chrAB dif, Mindlin et al., 2018), tellurium (terC dif) and organic peroxides (ohr1 dif), potassium (kup dif) and sulfate (sulP dif) transporters, toxin-antitoxin systems (add dif). All the detected genetic elements also have almost identical copies in the plasmids of modern Acinetobacter strains belonging to different species. It should be emphasized that the dif modules described in the present work were revealed in plasmids from only five studied permafrost strains of A. lwoffii group, and many more dif modules containing other adaptive genes are likely to be found in the future.
Based on their distribution, three groups of known dif modules can be distinguished: (i) mobile elements that encode metabolic and regulatory functions (e.g., resistance to chromium and tellurium, potassium uptake) and ensure survival under natural conditions, and mainly found in plasmids of environmental strains of Acinetobacter (described in this study); (ii) mobile elements found in both clinical and natural strains (including dif modules encoding sulfate permease, organic hydroperoxide resistance and toxin/antitoxin protein pairs, described in this study); (iii) mobile elements containing genes of antibiotic resistance and found almost without exception in plasmids of clinical strains of Acinetobacter, mainly A. baumanni (D’Andrea et al., 2009; Merino et al., 2010; Grosso et al., 2012; Blackwell and Hall, 2017). It is also appropriate to place in this group an Acinetobacter module encoding a putative virulence factor (Sel1-repeat protein) described by Lean and Yeo (2017). According to our preliminary data, it is flanked by XerC/D and XerD/C sites and is widely distributed in A. baumannii plasmids, similarly to classical dif modules.
Despite their different distribution, comparative molecular analysis of the dif modules described in this work and those containing antibiotic resistance genes described previously (D’Andrea et al., 2009; Merino et al., 2010; Grosso et al., 2012; Povilonis et al., 2013; Blackwell and Hall, 2017) suggests that they belong to the same group of unique genetic elements characteristic to the genus Acinetobacter. The wide distribution of such modules among plasmids of various Acinetobacter species indicates their ability to transfer horizontally and suggests that they form a special group of mobile genetic elements.
We demonstrated that multiple dif sites and dif modules can also be found in most Acinetobacter chromosomes. The main dif1 site in the terminus region of the chromosome is required for the resolution of chromosome dimers by the XerC/XerD system and if it is transferred to other regions of the chromosome the process of dimer resolution is impaired, as shown for Escherichia coli (Cornet et al., 1996; Pérals et al., 2000). Thus, additional dif sites present in the Acinetobacter chromosomes are unlikely to function in the dimer resolution. Moreover, analysis of their sequences revealed that they are more closely related to pdif sites found in plasmids from various Acinetobacter species than to the chromosomal site dif1. Several examples of dif modules were also found in Acinetobacter chromosomes (this work). Thus, it can be proposed that the additional chromosomal dif sites may participate in the movement of dif modules encoding antibiotic resistance and various adaptive functions from plasmids to the chromosome and backward. Indeed, a dif module with the Acinetobacter-specific carbapenemase gene OXA-58 was recently found inserted into the XerC/XerD site of a prophage in the Proteus mirabilis chromosome (Girlich et al., 2017). As far as we know this is the first description of an XerC/XerD-dependent insertion of Acinetobacter antibiotic resistance genes within a bacterial chromosome.
It is natural to assume that the common role of all types of dif modules is to increase the fitness of their respective bacterial hosts in their habitats. Our results suggest that the dif modules possess the properties of mobile genetic elements capable to translocate between plasmids and chromosomes in Acinetobacter strains belonging to different species. Additional research is highly needed to assess the vast diversity of dif modules in this clinically important genus and reveal their contribution to horizontal gene transfer and plasmid variability.
AM, AB, and MP conducted the sequencing and assembly of plasmids. All authors performed the bioinformatic analysis. MP performed the experiments. SM, MP, and AM wrote the manuscript. All authors contributed to the discussion and provided comments on the manuscript.
This research was supported by the Grant of the Ministry of Science and Higher Education of Russian Federation 14.W03.31.0007.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to A. Kulbachinskiy and two reviewers for helpful comments and suggestions and for critical reading of the manuscript and to A. Petrenko for help in assembly of plasmids.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00632/full#supplementary-material
TABLE S1 | Distribution of dif sites in Acinetobacter plasmids and chromosomes.
TABLE S2 | Distribution of dif modules in ancient (permafrost) and modern Acinetobacter strains.
Alper, S. L., and Sharma, A. K. (2013). The SLC26 gene family of anion transporters and channels. Mol. Aspects Med. 34, 494–515. doi: 10.1016/j.mam.2012.07.009
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Atichartpongkul, S., Fuangthong, M., Vattanaviboon, P., and Mongkolsuk, S. (2010). Analyses of the regulatory mechanism and physiological roles of Pseudomonas aeruginosa OhrR, a transcription regulator and a sensor of organic hydroperoxides. J. Bacteriol. 192, 2093–2101. doi: 10.1128/JB.01510-09
Bertram, R., and Schuster, C. F. (2014). Post-transcriptional regulation of gene expression in bacterial pathogens by toxin-antitoxin systems. Front. Cell Infect. Microbiol. 4:6. doi: 10.3389/fcimb.2014.00006
Blackwell, G. A., and Hall, R. M. (2017). The tet39 determinant and the msrE-mphE genes in Acinetobacter plasmids are each part of discrete modules flanked by inversely oriented pdif (XerC-XerD) sites. Antimicrob. Agents Chemother. 61:e780-17. doi: 10.1128/AAC.00780-17
Cameranesi, M. M., Morán-Barrio, J., Limansky, A. S., Repizo, G. D., and Viale, A. M. (2018). Site-specific recombination at XerC/D sites mediates the formation and resolution of plasmid co-integrates carrying a blaOXA-58- and TnaphA6-resistance module in Acinetobacter baumannii. Front Microbiol. 9:66. doi: 10.3389/fmicb.2018.00066
Carnoy, C., and Roten, C. A. (2009). The dif/Xer recombination systems in Proteobacteria. PLoS One 4:e6531. doi: 10.1371/journal.pone.0006531
Castillo, F., Benmohamed, A., and Szatmari, G. (2017). Xer site specific recombination: double and single recombinase systems. Front Microbiol. 8:453. doi: 10.3389/fmicb.2017.00453
Chasteen, T. G., Fuentes, D. E., Tantaleán, J. C., and Vásquez, C. C. (2009). Tellurite: history, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 33, 820–832. doi: 10.1111/j.1574-6976.2009.00177.x
Cornet, F., Louarn, J., Patte, J., and Louarn, J. M. (1996). Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes Dev. 10, 1152–1161. doi: 10.1101/gad.10.9.1152
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Da Silva, G. J., and Domingues, S. (2016). Insights on the horizontal gene transfer of carbapenemase determinants in the opportunistic pathogen Acinetobacter baumannii. Microorganisms 4:E29. doi: 10.3390/microorganisms4030029
D’Andrea, M. M., Giani, T., D’Arezzo, S., Capone, A., Petrosillo, N., Visca, P., et al. (2009). Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 3528–3533. doi: 10.1128/AAC.00178-09
Dorsey, C. W., Tomaras, A. P., and Actis, L. A. (2006). Sequence and organization of pMAC, an A. baumannii plasmid harboring genes involved in organic peroxide resistance. Plasmid 56, 112–123. doi: 10.1016/j.plasmid.2006.01.004
Evans, B. A., and Amyes, S. G. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-13
Feng, Y., Yang, P., Wang, X., and Zong, Z. (2016). Characterization of Acinetobacter johnsonii isolate XBB1carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J. Antimicrob. Chemother. 71, 71–75. doi: 10.1093/jac/dkv324
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., et al. (2009). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. doi: 10.1093/nar/gkp985
Girlich, D., Bonnin, R. A., Bogaerts, P., De Laveleye, M., Huang, D. T., Dortet, L., et al. (2017). Chromosomal amplification of the blaOXA-58 carbapenemase gene in a Proteus mirabilis clinical isolate. Antimicrob. Agents Chemother. 61:e1697-16. doi: 10.1128/AAC.01697-16
Grosso, F., Quinteira, S., Poirel, L., Novais,Â, and Peixe, L. (2012). Role of common blaOXA-24/OXA-40-carrying platforms and plasmids in the spread of OXA-24/OXA-40 among Acinetobacter species clinical isolates. Antimicrob. Agents Chemother. 56, 3969–3972. doi: 10.1128/AAC.06255-11
Hamidian, M., and Hall, R. M. (2018). Genetic structure of four plasmids found in Acinetobacter baumannii isolate D36 belonging to lineage 2 of global clone 1. PLoS One 13:e0204357. doi: 10.1371/journal.pone.0204357
Kurakov, A., Mindlin, S., Beletsky, A., Shcherbatova, N., Rakitin, A., Ermakova, A., et al. (2016). The ancient small mobilizable plasmid pALWED1.8 harboring a new variant of the non-cassette streptomycin/spectinomycin resistance gene aadA27. Plasmid 8, 36–43. doi: 10.1016/j.plasmid.2016.02.005
Lean, S. S., and Yeo, C. C. (2017). Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: good, bad, who knows? Front. Microbiol. 8:1547. doi: 10.3389/fmicb.2017.01547
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39(Suppl. 1), D225–D229. doi: 10.1093/nar/gkq1189
Martins, N., Picao, R. C., Adams-Sapper, S., Riley, L. W., and Moreira, B. M. (2015). Association of class 1 and 2 integrons with multidrug-resistant Acinetobacter baumannii international clones and Acinetobacter nosocomialis isolates. Antimicrob. Agents Chemother. 59, 698–701. doi: 10.1128/AAC.02415-14
Merino, M., Acosta, J., Poza, M., Sanz, F., Beceiro, A., Chaves, F., et al. (2010). OXA-24 carbapenemase gene flanked by XerC/XerD-Like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54, 2724–2727. doi: 10.1128/AAC.01674-09
Mindlin, S., Petrenko, A., Kurakov, A., Beletsky, A., Mardanov, A., and Petrova, M. (2016). Resistance of ancient and modern Acinetobacter lwoffii strains to heavy metals and arsenic revealed by genome analysis. BioMed. Res. Int. 2016, 1–9. doi: 10.1155/2016/3970831
Mindlin, S., Petrenko, A., and Petrova, M. (2018). Chromium resistance genetic element flanked by XerC/XerD recombination sites and its distribution in environmental and clinical Acinetobacter strains. FEMS Microbiol. Lett. 365:fny047. doi: 10.1093/femsle/fny047
Moghadam, M. S., Albersmeier, A., Winkler, A., Cimmino, L., Rise, K., Hohmann-Marriott, M. F., et al. (2016). Isolation and genome sequencing of four Arctic marine Psychrobacter strains exhibiting multicopper oxidase activity. BMC Genomics 17:117. doi: 10.1186/s12864-016-2445-4
Nemec, A., Radolfová-Křížová, L., Maixnerová, M., Nemec, M., Clermont, D., Bzdil, J., et al. (2018). Revising the taxonomy of the Acinetobacter lwoffii group: the description of Acinetobacter pseudolwoffii sp. nov. and emended description of Acinetobacter lwoffii. Syst. Appl. Microbiol. 42, 159–167. doi: 10.1016/j.syapm.2018.10.004
Pagano, M., Martins, A. F., and Barth, A. L. (2016). Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz. J. Microbiol. 47, 785–792. doi: 10.1016/j.bjm.2016.06.005
Peleg, A. Y., de Breij, A., Adams, M. D., Cerqueira, G. M., Mocali, S., Galardini, M., et al. (2012). The success of acinetobacter species; genetic, metabolic and virulence attributes. PLoS One 7:e46984. doi: 10.1371/journal.pone.0046984
Pérals, K., Cornet, F., Merlet, Y., Delon, I., and Louarn, J. M. (2000). Functional polarization of the Escherichia coli chromosome terminus: the dif site acts in chromosome dimer resolution only when located between long stretches of opposite polarity. Mol. Microbiol. 36, 33–43. doi: 10.1046/j.1365-2958.2000.01847.x
Petrova, M. A., Mindlin, S. Z., Gorlenko, Zh. M, Kaliaeva, E. S., Soina, V. S., and Bogdanova, E. S. (2002). Mercury-resistant bacteria from permafrost sediments and prospects for their use in comparative studies of mercury resistance determinants. Genetika 38, 1569–1574.
Povilonis, J., Seputiene, V., Krasauskas, R., Juskaite, R., Miskinyte, M., Suziedelis, K., et al. (2013). Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 68, 1000–1006. doi: 10.1093/jac/dks499
Price, G. D., Woodger, F. J., Badger, M. R., Howitt, S. M., and Tucker, L. (2004). Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 101, 18228–18233. doi: 10.1073/pnas.0405211101
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2009). REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38, D234–D236. doi: 10.1093/nar/gkp874
Schuster, C. F., and Bertram, R. (2013). Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340, 73–85. doi: 10.1111/1574-6968.12074
Shintani, M., Sanchez, Z. K., and Kimbara, K. (2015). Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 6:242. doi: 10.3389/fmicb.2015.00242
Sukchawalit, R., Loprasert, S., Atichartpongkul, S., and Mongkolsuk, S. (2001). Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 183, 4405–4412. doi: 10.1128/JB.183.15.4405-4412.2001
Touchon, M., Cury, J., Yoon, E. J., Krizova, L., Cerqueira, G. C., Murphy, C., et al. (2014). The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol. Evol. 6, 2866–2882. doi: 10.1093/gbe/evu225
Turton, J. F., Shah, J., Ozongwu, C., and Pike, R. (2010). Incidence of Acinetobacter species other than A. baumannii among clinical isolates of Acinetobacter: evidence for emerging species. J. Clin. Microbiol. 48, 1445–1449. doi: 10.1128/JCM.02467-09
Yamaguchi, Y., Park, J., and Inouye, M. (2011). Toxin-antitoxin systems in bacteria and archaea. Ann. Rev. Genet. 45, 61–79. doi: 10.1146/annurev-genet-110410-132412
Keywords: plasmid pdif sites, additional chromosomal dif sites, terC dif, ohr1 dif, sulP dif, kup dif, add dif
Citation: Mindlin S, Beletsky A, Mardanov A and Petrova M (2019) Adaptive dif Modules in Permafrost Strains of Acinetobacter lwoffii and Their Distribution and Abundance Among Present Day Acinetobacter Strains. Front. Microbiol. 10:632. doi: 10.3389/fmicb.2019.00632
Received: 15 November 2018; Accepted: 13 March 2019;
Published: 29 March 2019.
Edited by:
Maria Alejandra Mussi, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Filipa Grosso, Universidade do Porto, PortugalCopyright © 2019 Mindlin, Beletsky, Mardanov and Petrova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mayya Petrova, cGV0cm92YUBpbWcucmFzLnJ1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.