
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Microbiol., 22 March 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00549
This article is a correction to:
RfpA, RfpB, and RfpC are the Master Control Elements of Far-Red Light Photoacclimation (FaRLiP)
A Corrigendum on
RfpA, RfpB, and RfpC are the Master Control Elements of Far-Red Light Photoacclimation (FaRLiP)
Zhao, C., Gan, F., Shen, G., and Bryant, D. A. (2015). Front. Microbiol. 6:1303. doi: 10.3389/fmicb.2015.01303
In the original article, there was a mistake in Figure 1 as published. In Panel F, the lane labeled “ΔrfpB” should have been labeled “WT”, while the lane labeled “WT” should have been labeled “ΔrfpB”. The corrected Figure 1 appears below.
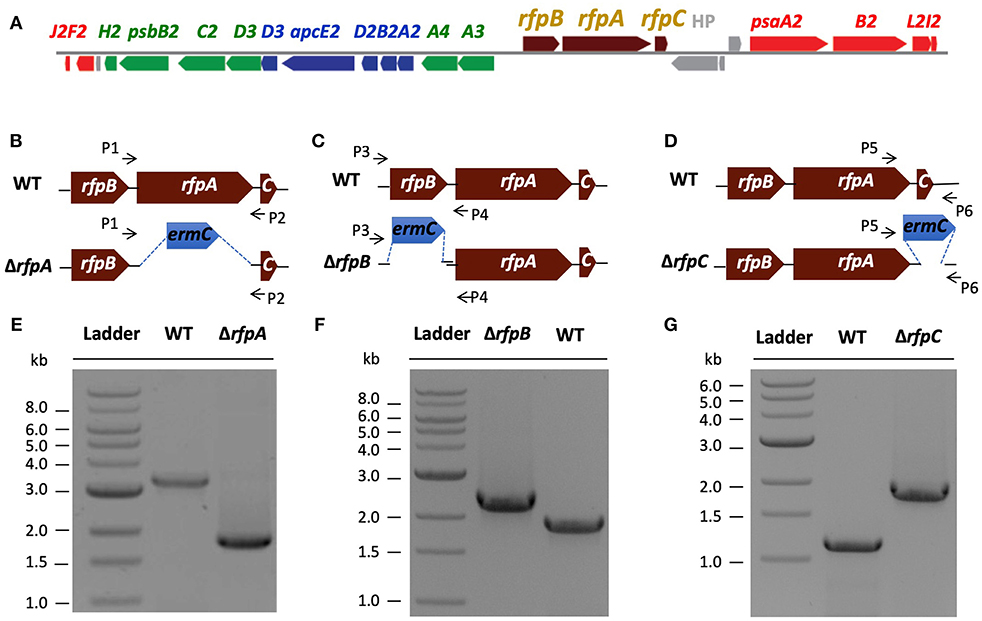
Figure 1. The organization of the FaRLiP gene cluster in Chl. fritschii PCC 9212 and construction and validation of rfpA, rfpB, and rfpC deletion mutants. (A) Gene organization of the FaRLiP gene cluster in Chl. fritschii PCC 9212. Red boxes represent genes encoding core subunits of PS I; green boxes represent genes encoding core subunits of PS II; blue boxes represent genes encoding core components of the phycobilisome; brown boxes represent regulatory rfp genes; and gray boxes represent genes that are not found in other FaRLiP clusters. (B) Schematic showing deletion of rfpA. The small arrows (P1 and P2) indicate the positions of the primers used for PCR verification of deletion. (C) Schematic showing deletion of rfpB. The small arrows (P3 and P4) indicate the positions of primers used for PCR verification of the deletion. (D) Schematic showing deletion of rfpC. The small arrows (P5 and P6) indicate the positions of primers used for PCR verification of the deletion. (E) Agarose gel electrophoresis of amplicons showing complete segregation of wild-type and mutant rfpA alleles. (F) Agarose gel electrophoresis of amplicons showing complete segregation of wild-type and mutant rfpB alleles. (G) Agarose gel electrophoresis of amplicons showing complete segregation of wild-type and mutant rfpC alleles.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Keywords: photosynthesis, photosystem I, photosystem II, phycobilisome, chlorophyll f, chlorophyll d, Chlorogloeopsis fritschii PCC 9212, Chroococcidiopsis thermalis PCC 7203
Citation: Zhao C, Gan F, Shen G and Bryant DA (2019) Corrigendum: RfpA, RfpB, and RfpC are the Master Control Elements of Far-Red Light Photoacclimation (FaRLiP). Front. Microbiol. 10:549. doi: 10.3389/fmicb.2019.00549
Received: 11 January 2019; Accepted: 04 March 2019;
Published: 22 March 2019.
Edited and reviewed by: Martin G. Klotz, Washington State University, United States
Copyright © 2019 Zhao, Gan, Shen and Bryant. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donald A. Bryant, ZGFiMTRAcHN1LmVkdQ==
†Present Address: Fei Gan, The Scripps Research Institute, La Jolla, CA, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.