
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 March 2019
Sec. Virology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00432
The influenza virus protein PA-X modulates the host immune responses and viral pathogenicity through suppression of host protein expression. The endonuclease active site in the N-terminal region, the basic amino acid cluster in the C-terminal PA-X-specific region, and N-terminal acetylation of PA-X by NatB are important for the shutoff activity of PA-X. Here, we focused on the shutoff activity of PA-X derived from the A/California/04/2009 and A/WSN/33 viruses because these two PA-X proteins differ in their shutoff activity. Mutagenesis analysis revealed that proline and serine at positions 28 and 65, respectively, play a central role in this difference. Furthermore, we found that P28 and S65 also affect the shutoff activity of PA-X derived from other influenza virus subtypes. These data demonstrate that P28 and S65 contribute to enhanced shutoff activity of PA-X.
Influenza A virus protein PA-X is encoded in the PA segment and expressed via a ribosomal frameshift (Jagger et al., 2012). It comprises the N-terminal amino acids of PA and the C-terminal PA-X-specific amino acids that result from the frameshift (Jagger et al., 2012). PA-X plays a role in virus-induced host shutoff by cleaving mRNA using its endonuclease activity. The endonuclease active site, which is in the N-terminal region, as well as several amino acids around the endonuclease active site and a cluster of basic amino acids in the C-terminal region of PA-X are important for PA-X shutoff activity (Desmet et al., 2013; Oishi et al., 2015, 2018a; Hayashi et al., 2016). Previously, we identified NatB as a host protein involved in the shutoff activity of PA-X by using a single-gene deletion yeast library (Oishi et al., 2018b). During the course of infection, PA-X inhibits host antiviral responses, such as the induction of IFN-β production and anti-viral antibody production in vivo (Hayashi et al., 2015; Hu et al., 2018; Nogales et al., 2018). PA-X also disables antiviral stress-induced translation arrest in infected cells by inhibiting stress granule formation (Khaperskyy et al., 2014). Although several studies have attempted to elucidate the role of PA-X in viral pathogenicity by using different subtypes of PA-X-deficient mutant viruses, its role in this process remains controversial (Jagger et al., 2012; Gao et al., 2015; Hayashi et al., 2015; Lee et al., 2017; Hussain et al., 2019).
A previous study showed that the shutoff activity of PA-X derived from A/California/04/2009 (H1N1pdm09) was higher than that of PA-X derived from A/WSN/33 (H1N1) (Hayashi et al., 2016). However, the amino acid(s) responsible for the enhanced activity of the PA-X protein of A/California/04/2009 (H1N1pdm09) relative to that of A/WSN/33 (H1N1) have not been identified. Here, we attempted to identify residues that contribute to enhanced PA-X shutoff activity.
To pinpoint the PA-X region that contributes to the different shutoff activity between CA04 PA-X and WSN PA-X, we constructed a series of chimeric PA-X proteins between CA04 PA-X and WSN PA-X (Figure 1A). To examine the shutoff activity of each chimeric PA-X protein, we performed the shutoff assay. Since PA-X suppresses its own expression via its shutoff activity (Oishi et al., 2015), the expression of each wild-type or chimeric PA-X was also assessed by western blotting. The suppression of firefly luciferase activity by CA04 PA-X was stronger than that by WSN PA-X (Figure 1B), suggesting that the shutoff activity of CA04 PA-X is higher than that of WSN PA-X, as reported previously (Hayashi et al., 2016). WSN70-CA04 and WSN140-CA04, which possessed the N-terminal 70 or 140 amino acids of WSN PA-X, respectively, and the C-terminal region of CA04 PA-X, showed low shutoff activity like WSN PA-X. CA0470-WSN and CA04140-WSN, which possessed the N-terminal 70 and 140 amino acids of CA04 PA-X, respectively, and the C-terminal region of WSN PA-X, showed high shutoff activity like CA04 PA-X. WSN70-CA04 showed statistically higher shutoff activity than WSN140-CA04, whereas CA0470-WSN showed comparable shutoff activity to CA04140-WSN (Figure 1B). The protein expression levels of CA04 PA-X and WSN PA-X were comparable, even though CA04 PA-X showed higher shutoff activity than WSN PA-X (Figure 1B). The protein expression level of WSN140-CA04 was the highest among the chimeras tested, whereas the expression levels of the other chimeric PA-X proteins were comparable (Figure 1B). These results demonstrate that the N-terminal 70 amino acids of CA04 PA-X contribute to high PA-X shutoff activity. They also suggest that the amino acid residues between positions 70 and 140 may play roles for the shutoff activity of PA-X.
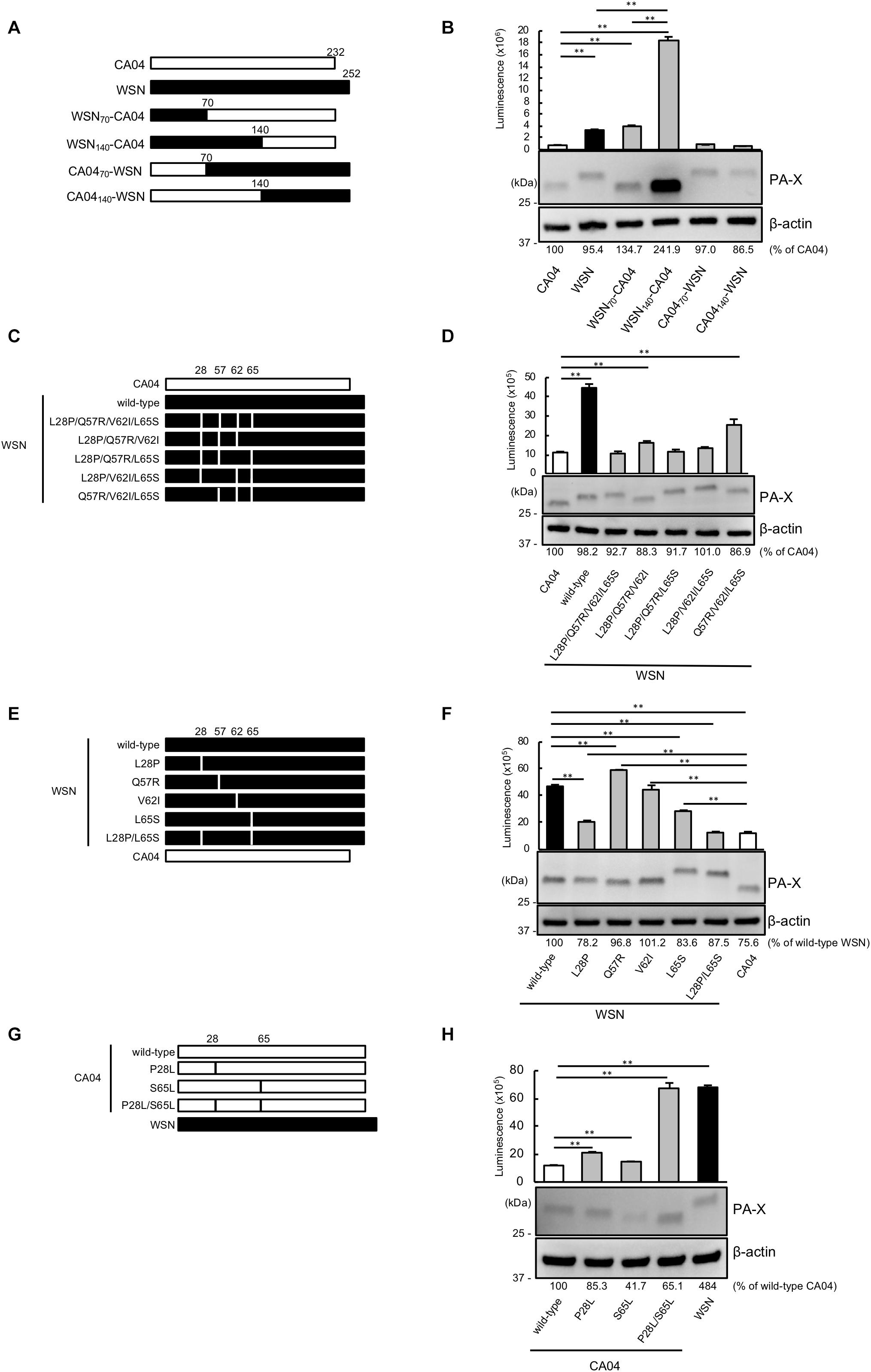
Figure 1. Proline at position 28 and serine at position 65 are determinants of the higher shutoff activity of CA04 PA-X compared with WSN PA-X. (A,C,E,G) Schematic diagrams of chimeric WSN/CA04 PA-X proteins (B,D,F,H) Shutoff activities of PA-X. Human 293 cells were transfected with a plasmid encoding firefly luciferase together with an empty plasmid or a plasmid encoding the indicated PA-X. At 24 h post-transfection, luciferase activities were assessed by using a luciferase assay. Expression of wild-type and mutant PA-X was also evaluated by western blotting using an anti-FLAG antibody. (B,F) ∗∗ indicates P < 0.01, according to a one-way ANOVA followed by Tukey’s test. (D,H) ∗∗ indicates P < 0.01, according to a one-way ANOVA followed by Dunnett’s test.
To locate the amino acid residues that are important for the high shutoff activity of PA-X, we compared the N-terminal 70 amino acid sequences of WSN PA-X and CA04 PA-X and found four differences, at positions 28, 57, 62, and 65 (Table 1). To confirm that these four positions affected the shutoff activity of PA-X, we prepared WSN PA-X mutants possessing four amino acid substitutions (L28P/Q57R/V62I/L65S) or three amino acid substitutions in different combinations (L28P/Q57R/V62I, L28P/Q57R/L65S, L28P/V62I/L65S, and Q57R/V62I/L65S) (Figure 1C). The shutoff activity of WSN PA-X L28P/Q57R/V62I/L65S, L28P/Q57R/L65S, and L28P/V62I/L65S was comparable to that of CA04 PA-X (Figure 1D). In contrast, the shutoff activity of WSN PA-X L28P/Q57R/V62I and Q57R/V62I/L65S was lower than that of CA04 PA-X (Figure 1D). All tested PA-Xs were expressed at similar levels (Figure 1D). The L65S mutation affected the mobility of PA-X mutants for reasons that are currently unknown (Figure 1D). These results suggest that both L28P and L65S might contribute to the enhanced shutoff activity of PA-X.
To further explore these results, we constructed and assessed the shutoff activity of WSN PA-X mutants that each possessed a single mutation at position 28, 57, 62, or 65 (Figure 1E). The shutoff activity of WSN PA-X possessing the V62I mutation was comparable to that of wild-type WSN PA-X. The shutoff activity of WSN PA-X possessing the Q57R mutation was lower than that of wild-type WSN PA-X, whereas the shutoff activity of WSN PA-X possessing the L28P or L65S mutation was significantly higher than that of wild-type WSN PA-X. The shutoff activity of the L28P or L65S mutant was significantly lower than that of CA04 PA-X (Figure 1F). Therefore, we examined the shutoff activity of a WSN PA-X mutant that possessed both the L28P and L65S mutations (Figure 1E) and found that the double mutant showed similar shutoff activity to CA04 PA-X (Figure 1F).
To confirm these results, we prepared and examined the shutoff activity of CA04 PA-X mutants possessing single or double mutations at P28L and/or S65L (Figure 1G). Both CA04 PA-X P28L and S65L showed lower shutoff activity than wild-type CA04 PA-X; the shutoff activity of CA04 PA-X P28L was lower than that of CA04 PA-X S65L (Figure 1H). Furthermore, the shutoff activity of CA04 PA-X P28L/S65L was similar to that of wild-type WSN PA-X (Figure 1H). The expression of wild-type or each CA04 PA-X mutant was comparable to that of WSN PA-X by western blotting. These results show that the amino acid residues P28 and S65 are responsible for the higher shutoff activity of CA04 PA-X relative to that of WSN PA-X.
Since PA has the same N-terminal sequence as PA-X, we next assessed the viral polymerase activity of PA possessing the mutation at position 28 or 65 or both positions by using a luciferase-based minigenome assay. The polymerase activity of WSN PA possessing the L28P or L65S mutation was higher than that of wild-type WSN PA. Mutant WSN PA that possessed both the L28P and L65S mutations showed the highest polymerase activity (Figure 2A). To further support these results, we evaluated the polymerase activity of CA04 PA mutants. Both CA04 PA P28L and CA04 PA S65L showed lower polymerase activities than wild-type CA04 PA. CA04 PA P28L/S65L showed the lowest polymerase activity (Figure 2B). The expression level of each PA mutant was comparable to that of each wild-type PA (Figures 2A,B). These data indicate that both proline and serine at positions 28 and 65, respectively, of PA are important for viral polymerase activity.
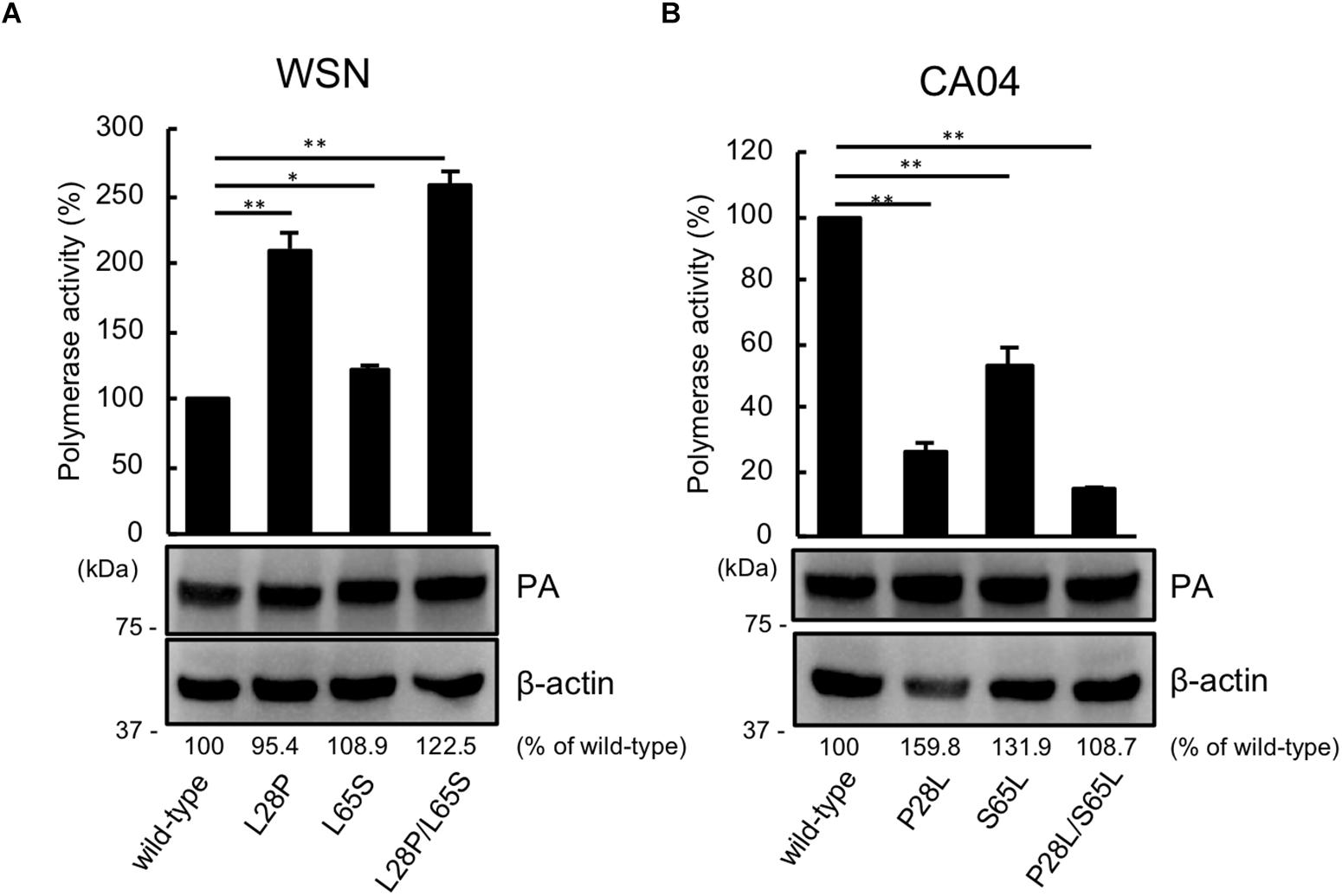
Figure 2. Proline at position 28 and serine at position 65 are important for virus polymerase activity. Viral polymerase activity of WSN (A) and CA04 (B) PA mutants. Cells were transfected with plasmids encoding PB2, PB1, NP, and wild-type or mutant PA, with a plasmid for the expression of viral RNA encoding firefly luciferase, and with a plasmid encoding Renilla luciferase as a transfection control. Polymerase activity was calculated by normalization of the firefly luciferase activity to the Renilla luciferase activity. The expression of each mutant PA was analyzed by western blotting with an anti-PA antibody. ∗∗ and ∗ indicate P < 0.01 and P < 0.05, respectively, according to a one-way ANOVA followed by Dunnett’s test.
We then asked whether P28 and S65 of PA-X are conserved among human and avian influenza isolates (Table 2). We found that over 90% of human isolates of H1N1pdm09 viruses, H5N1 viruses, and H7N9 viruses had P28 and S65 (CA04 type). In contrast, almost all human isolates of H3N2 viruses had L28 and L65 in PA-X (WSN type). Many human isolates of H1N1pre09 virus had L28 and P65. To examine the effect of P65 on the shutoff activity of PA-X, we introduced the mutation L65P into wild-type WSN PA-X, resulting in a PA-X mutant that possessed L28 and P65, which are found in many human H1N1pre09 isolates. The shutoff activity of WSN PA-X L65P was higher than that of wild-type WSN PA-X, but still lower than that of WSN PA-X L28P/L65S (Figure 3A). Again, the protein expression level of each WSN PA-X mutant was comparable to that of wild-type WSN PA-X (Figure 3A). The mutation at position 65 affected the mobility of PA-X for unknown reasons (Figure 3A).
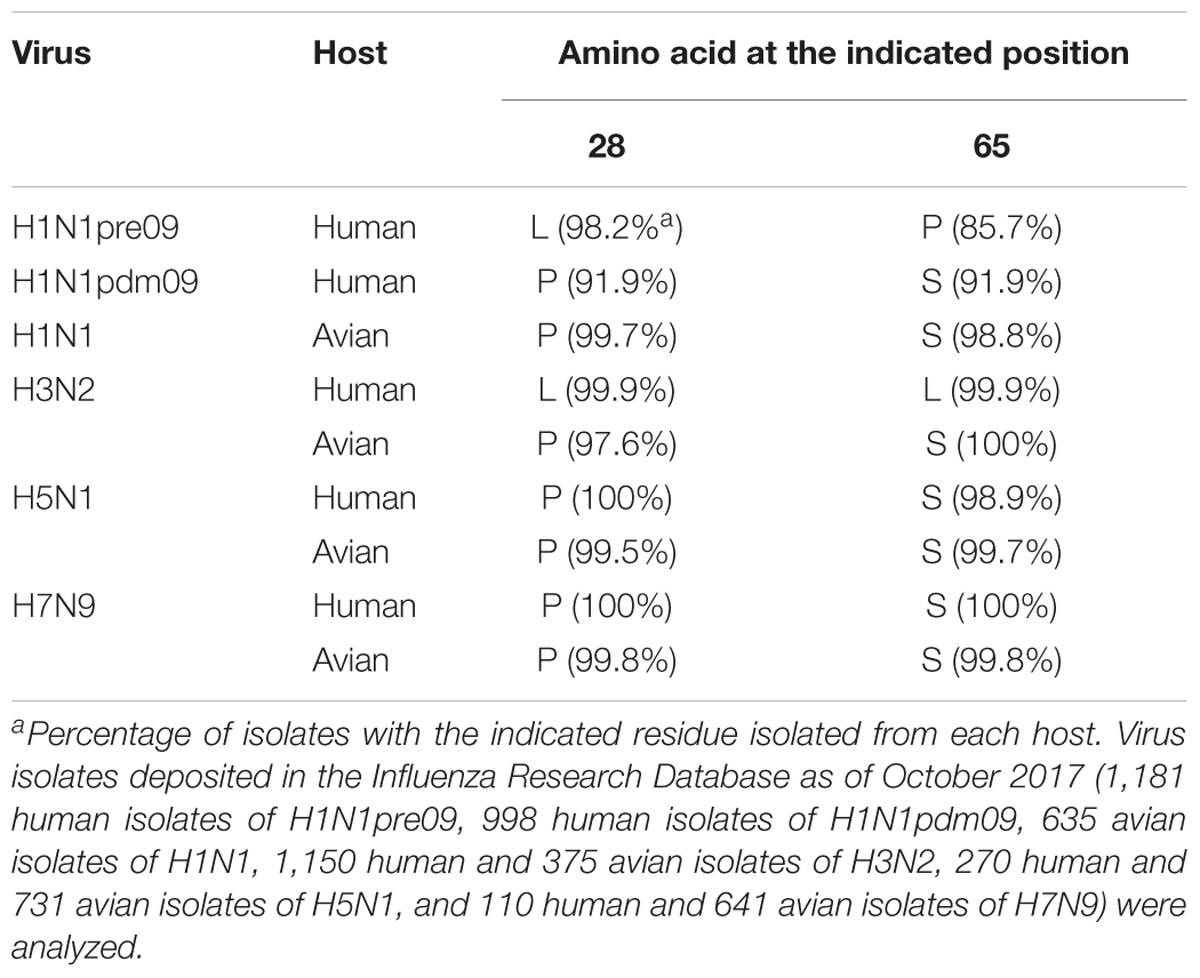
Table 2. Conservation of amino acid residues at positions 28 and 65 in PA-X among different virus subtypes.
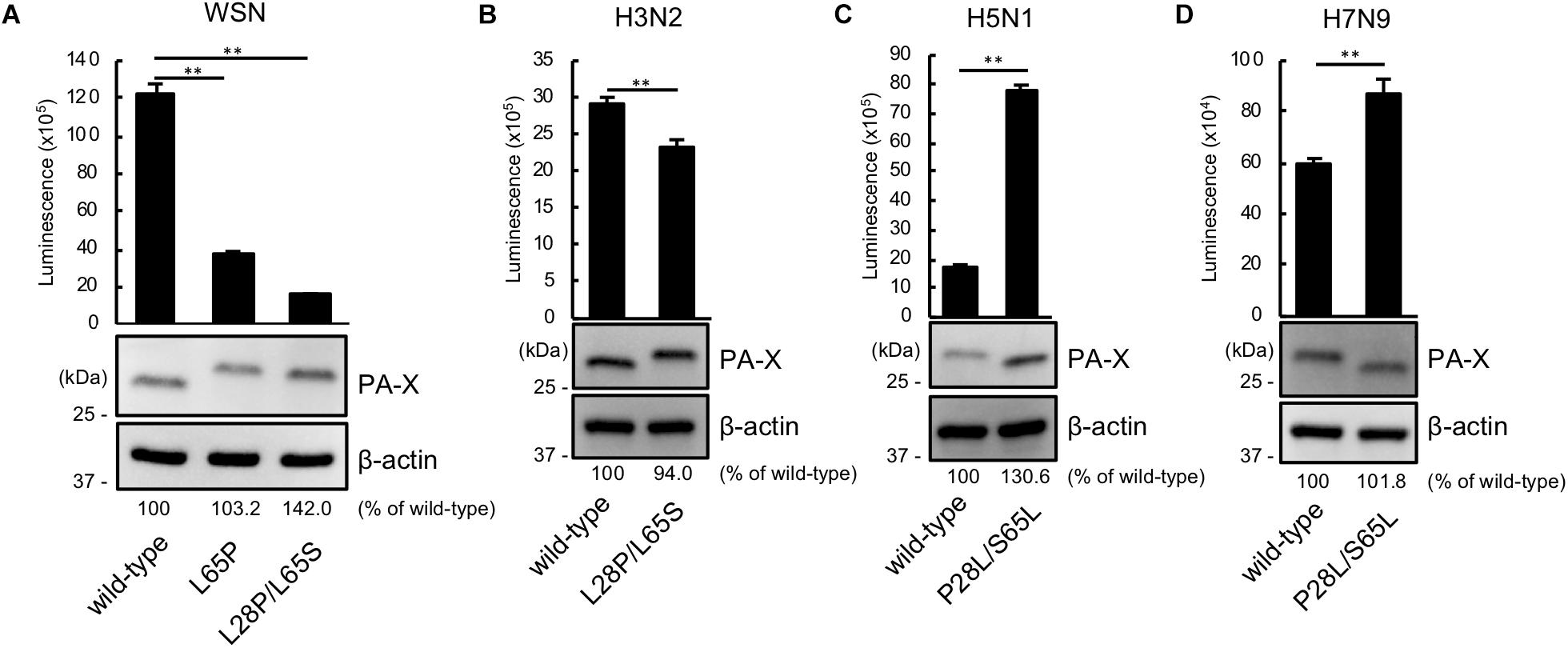
Figure 3. The importance of proline at position 28 and serine at position 65 in the shutoff activity of PA-X derived from other influenza A virus subtypes. (A,B) Shutoff activities and expression of wild-type or mutant PA-X derived from A/WSN/33 (H1N1) (A), A/Yokohama/UT2017/2003 (H3N2) (B), A/Vietnam/HN31604/2009 (H5N1) (C), and A/Anhui/1/2013 (H7N9) (D). Cells were transfected with a plasmid encoding firefly luciferase together with an empty plasmid or a plasmid encoding wild-type or mutant PA-X. At 24 h post-transfection, luciferase activities were assessed by using a luciferase assay. Luminescence data are presented as means ± SD (n = 3). Expression of wild-type and mutant PA-X was analyzed by western blotting using an anti-FLAG antibody. (A) ∗∗ indicates P < 0.01 according to a one-way ANOVA followed by Dunnett’s test. (B–D) ∗∗ indicates P < 0.01 according a two-tailed unpaired Student’s t-test.
Most avian isolates of H1N1, H3N2, H5N1, and H7N9 viruses possess P28 and S65 (CA04 type). This finding suggests that although the amino acids P28 and S65 are present in most avian isolates, these residues become L28 and L/P65 (low shutoff-type) upon adaptation to humans. We then examined the contribution of the amino acid residues at positions 28 and 65 to the activity of PA-X derived from H3N2 (WSN type), H5N1 (CA04 type), and H7N9 (CA04 type) viruses. To this end, we introduced the L28P and L65S mutations into PA-X derived from H3N2 virus, or the P28L and S65L mutations into PA-X derived from H5N1 and H7N9 viruses, and performed the shutoff assay. The H3N2 PA-X double mutant showed significantly higher shutoff activity than wild-type H3N2 PA-X (Figure 3B). Each H5N1 and H7N9 PA-X double mutant showed lower shutoff activity than its counterpart wild-type PA-X, although the reduction in the shutoff activity of the H7N9 PA-X mutant was limited (Figures 3C,D). Expression of the H5N1 PA-X mutant was higher than that of wild-type H5N1 PA-X, whereas expression of the H3N2 and H7N9 PA-X mutants was comparable to that of each wild-type PA-X (Figures 3B–D). These results demonstrate that the amino acid residues P28 and S65 of PA-X are required for the high shutoff activity of PA-X in multiple subtypes of influenza A virus.
Here, we found that the amino acid residues at positions 28 and 65 of PA-X or PA affect the shutoff activity of PA-X or viral polymerase activity, respectively. Since PA-X shares its N-terminal 191 amino acids with PA (Jagger et al., 2012), we mapped the amino acids at positions 28 and 65 onto the N-terminal structure of PA. We used the crystal structure of PA derived from CA04 as a representative of CA04-type PA (Kowalinski et al., 2012; Figure 4A) and A/Victoria/3/75 (H3N2) as a representative of WSN-type PA (Dias et al., 2009; Figure 4B) because the crystal structure of PA derived from WSN is not available and the PA segment of A/Victoria/3/75, which possesses both L28 and L65, originates from H1N1 virus. Amino acid residues H41, E80, L106, P107, D108, E119, and K134 (shown in magenta), which form the endonuclease active site, are known to be important for both the endonuclease activity of PA (Dias et al., 2009) and the shutoff activity of PA-X (Jagger et al., 2012; Desmet et al., 2013; Oishi et al., 2018a). The amino acid residue at position 65 is located at the flexible loop (Figure 4, blue) that is formed by amino acid residues 53–73 (DuBois et al., 2012; Desmet et al., 2013). Since the L65P mutation together with Q57R and V62I radically change the conformation of the flexible loop (Dias et al., 2009; Kowalinski et al., 2012), a mutation at position 65 might affect the overall structure of the protein. The amino acid residue at position 28 is located in the linker region between two α-helices (Figure 4, green), and is far from the endonuclease active site. Other residues that are located far from the endonuclease active site, but still affect the shutoff activity of PA-X have been reported (Oishi et al., 2018a); the mechanistic role of these residues has not been determined.
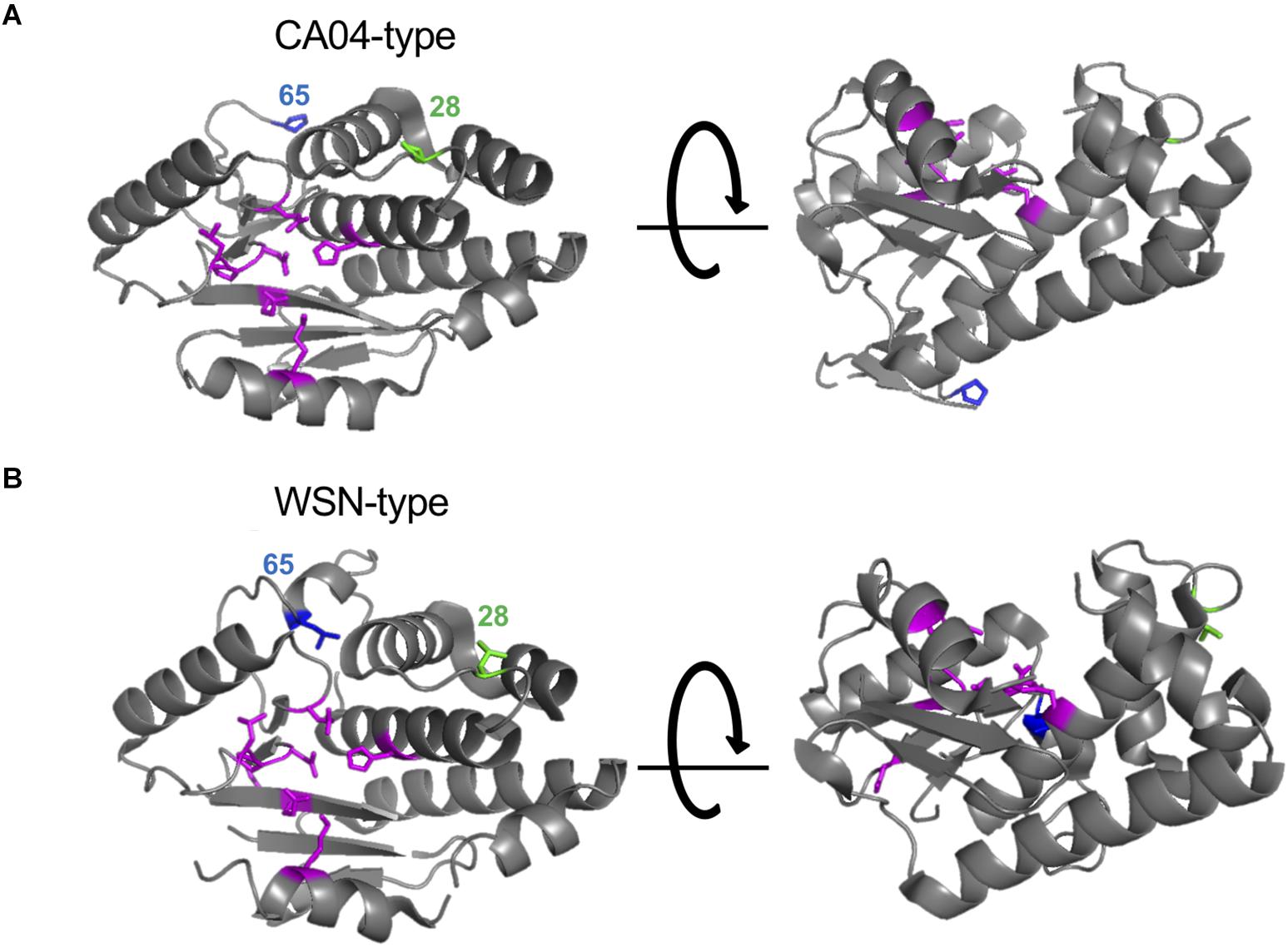
Figure 4. Structural positions of the amino acid residues at positions 28 and 65. The amino acid residues at positions 28 and 65 were mapped onto a ribbon diagram of the structure of the N-terminal region of PA derived from A/California/04/2009 (H1N1pdm09) (A) and A/Victoria/3/75 (H3N2) (B). Amino acid residues that form the endonuclease active site are shown in magenta. Amino acids at positions 28 or 65 are shown in green and blue, respectively.
We identified two amino acids at positions 28 and 65 that are important for high PA-X shutoff activity. The amino acids at positions 51–114 of CA04 PA were previously reported to be important for the higher shutoff activity of CA04 PA than WSN PA (Desmet et al., 2013). This region includes the amino acid at position 65 but not that at position 28. Since the authors of this report used a plasmid encoding the N-terminal 257 amino acids of PA to examine shutoff activity (Desmet et al., 2013), the shutoff activity of this truncated PA was extremely lower than that of PA-X (Desmet et al., 2013). Therefore, they may have failed to notice the subtle changes caused by the amino acid substitution at position 28.
Since PA-X suppresses its own expression via its shutoff activity, the expression of PA-X is largely affected by its own shutoff activity (Oishi et al., 2015). Expression of PA-X might also be downregulated by a reduction in the expression of host protein(s) involved in PA-X expression. In addition to these mechanisms, some amino acid residues of PA-X that are involved in protein stability (Oishi et al., 2018a) also decrease the expression of PA-X. This may explain why the expression of PA-X mutants may differ among mutants that show similar shutoff activity.
Here we reported that proline at position 28 and serine at position 65 contribute to enhanced shutoff activity of PA-X. Since these amino acid residues in PA also affect viral polymerase activity, they likely affect the mRNA cleavage step. The mechanism by which these amino acid residues contribute to enhanced shutoff activity of PA-X remains unknown. Further studies, such as determination of the crystal structure and cellular localization of PA-X, are necessary to elucidate this mechanism.
Human embryonic kidney 293 cells (ATCC) were maintained in DMEM (Sigma) containing 10% fetal calf serum (FCS) and penicillin-streptomycin.
Nucleotide sequences of PA-X derived from A/WSN/33 (H1N1), A/California/04/2009 (H1N1pdm09), A/Yokohama/ UT2017/2003 (H3N2), A/Vietnam/HN31604/2009 (H5N1), and A/Anhui/1/2013 (H7N9) were cloned into pCAGGS/MCS with a C-terminal FLAG tag (C-FLAG). Mutant PA-X with the C-FLAG was amplified with appropriate primers, and then cloned into the same plasmids. Primer sequences are available upon request. All constructs were sequenced to confirm the absence of unwanted mutations.
Two hundred and ninety three cells in four wells of a 24-well plate were transfected with a plasmid encoding firefly luciferase together with an empty plasmid or a plasmid encoding wild-type or mutant PA-X possessing a C-terminal FLAG tag. At 24 h post-transfection, the transfected cells from three of the wells were analyzed for firefly luciferase activity by using the Bright-Glo luciferase assay system (Promega). Data are shown as the average luminescence ± standard deviation (n = 3). To analyze the expression of PA-X, the transfected cells in the remaining well were lysed in SDS sample buffer at 24 h post-transfection. The cell lysates were sonicated, incubated for 10 min at 95°C, and then loaded onto an Any KD Mini-PROTEAN TGX Gel (Bio-Rad). Separated proteins were transferred to Immobilon-P PVDF membrane (Millipore) and detected by using anti-DYKDDDDK (FLAG) tag antibody clone 1E6 (Wako) or anti-β-actin antibody clone AC-74 (Sigma), followed by a sheep anti-mouse IgG-HRP secondary antibody (GE Healthcare).
A minigenome assay based on the dual-luciferase system was performed as previously described (Yamayoshi et al., 2014). Polymerase activity was calculated by standardization of the firefly luciferase activity to the Renilla luciferase activity. Data are shown as the average of the polymerase activity ± standard deviation (n = 3). To analyze the expression of PA, the transfected cells were lysed in SDS sample buffer and analyzed by western blotting using an anti-PA antibody (Hatta et al., 2000).
Amino acid positions were plotted on the crystal structure of the N-terminal region of PA derived from A/California/04/2009 (H1N1pdm09) (PDB accession: 4AVQ) and A/Victoria/3/75 (H3N2) (PDB accession: 2W69) using the PyMOL molecular graphics system.
One-way analysis of variance (ANOVA) followed by Dunnett’s test or Tukey’s test and the two-tailed unpaired Student’s t-test were performed using GraphPad Prism software. P < 0.01 were considered significantly different. No samples were excluded from the analysis.
All datasets generated for this study are included in the manuscript.
KO, SY, and YK designed the study, analyzed the data, and wrote the manuscript. KO performed the experiments. All authors reviewed and approved the manuscript.
This research was supported by Strategic Basic Research Programs from the Japan Science and Technology Agency (JST), by Leading Advanced Projects for Medical Innovation (LEAP) from the Japan Agency for Medical Research and Development (AMED) (JP18am001007), by Grants-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan (Nos. 16H06429, 16K21723, and 16H06434), by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from AMED (JP18fm0108006), by the NIAID-funded Center for Research on Influenza Pathogenesis (CRIP) (HHSN272201400008C), and by the NIH Functional Genomics award, “Characterization of novel genes encoded by RNA and DNA viruses” (U19 AI 107810) from the NIAID.
YK has received speaker’s honoraria from Toyama Chemical, has received grant support from Chugai Pharmaceuticals, Daiichi Sankyo Pharmaceutical, Toyama Chemical, Tauns Laboratories, Inc., Tsumura & Co, and Denka Seiken Co., Ltd., and is a co-founder of FluGen.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Susan Watson for editing the manuscript.
Desmet, E. A., Bussey, K. A., Stone, R., and Takimoto, T. (2013). Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J. Virol. 87, 3108–3118. doi: 10.1128/JVI.02826-12
Dias, A., Bouvier, D., Crepin, T., McCarthy, A. A., Hart, D. J., Baudin, F., et al. (2009). The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458, 914–918. doi: 10.1038/nature07745
DuBois, R. M., Slavish, P. J., Baughman, B. M., Yun, M. K., Bao, J., Webby, R. J., et al. (2012). Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog. 8:e1002830. doi: 10.1371/journal.ppat.1002830
Gao, H., Sun, Y., Hu, J., Qi, L., Wang, J., Xiong, X., et al. (2015). The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci. Rep. 5:8262. doi: 10.1038/srep08262
Hatta, M., Asano, Y., Masunaga, K., Ito, T., Okazaki, K., Toyoda, T., et al. (2000). Epitope mapping of the influenza A virus RNA polymerase PA using monoclonal antibodies. Arch. Virol. 145, 895–903. doi: 10.1007/s007050050682
Hayashi, T., Chaimayo, C., McGuinness, J., and Takimoto, T. (2016). Critical role of the PA-X C-terminal domain of influenza a virus in its subcellular localization and shutoff activity. J. Virol. 90, 7131–7141. doi: 10.1128/JVI.00954-16
Hayashi, T., MacDonald, L. A., and Takimoto, T. (2015). Influenza a virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J. Virol. 89, 6442–6452. doi: 10.1128/JVI.00319-15
Hu, J., Ma, C., and Liu, X. (2018). PA-X: a key regulator of influenza a virus pathogenicity and host immune responses. Med. Microbiol. Immunol. 207, 255–269. doi: 10.1007/s00430-018-0548-z
Hussain, S., Turnbull, M. L., Wise, H. M., Jagger, B. W., Beard, P. M., Kovacikova, K., et al. (2019). Mutation of influenza a virus Pa-X decreases pathogenicity in chicken embryos and can increase the yield of reassortant candidate vaccine viruses. J. Virol. 93:e1551–18. doi: 10.1128/JVI.01551-18
Jagger, B. W., Wise, H. M., Kash, J. C., Walters, K. A., Wills, N. M., Xiao, Y. L., et al. (2012). An overlapping protein-coding region in influenza a virus segment 3 modulates the host response. Science 337, 199–204. doi: 10.1126/science.1222213
Khaperskyy, D. A., Emara, M. M., Johnston, B. P., Anderson, P., Hatchette, T. F., and McCormick, C. (2014). Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog. 10:e1004217. doi: 10.1371/journal.ppat.1004217
Kowalinski, E., Zubieta, C., Wolkerstorfer, A., Szolar, O. H., Ruigrok, R. W., and Cusack, S. (2012). Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 8:e1002831. doi: 10.1371/journal.ppat.1002831
Lee, J., Yu, H., Li, Y., Ma, J., Lang, Y., Duff, M., et al. (2017). Impacts of different expressions of PA-X protein on 2009 pandemic H1N1 virus replication, pathogenicity and host immune responses. Virology 504, 25–35. doi: 10.1016/j.virol.2017.01.015
Nogales, A., Martinez-Sobrido, L., Topham, D. J., and DeDiego, M. L. (2018). Modulation of innate immune responses by the influenza A NS1 and PA-X proteins. Viruses 10:E708. doi: 10.3390/v10120708
Oishi, K., Yamayoshi, S., and Kawaoka, Y. (2015). Mapping of a region of the PA-X protein of influenza a virus that is important for its shutoff activity. J. Virol. 89, 8661–8665. doi: 10.1128/JVI.01132-15
Oishi, K., Yamayoshi, S., and Kawaoka, Y. (2018a). Identification of novel amino acid residues of influenza virus PA-X that are important for PA-X shutoff activity by using yeast. Virology 516, 71–75. doi: 10.1016/j.virol.2018.01.004
Oishi, K., Yamayoshi, S., Kozuka-Hata, H., Oyama, M., and Kawaoka, Y. (2018b). N-Terminal acetylation by natb is required for the shutoff activity of influenza a virus PA-X. Cell Rep. 24, 851–860. doi: 10.1016/j.celrep.2018.06.078
Keywords: influenza A virus, PA-X, shutoff activity, mutagenesis analysis, mRNA degradation
Citation: Oishi K, Yamayoshi S and Kawaoka Y (2019) Identification of Amino Acid Residues in Influenza A Virus PA-X That Contribute to Enhanced Shutoff Activity. Front. Microbiol. 10:432. doi: 10.3389/fmicb.2019.00432
Received: 05 December 2018; Accepted: 19 February 2019;
Published: 06 March 2019.
Edited by:
Daniel Roberto Perez, University of Georgia, United StatesReviewed by:
Luis Martinez-Sobrido, University of Rochester, United StatesCopyright © 2019 Oishi, Yamayoshi and Kawaoka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seiya Yamayoshi, eWFtYXlvQGltcy51LXRva3lvLmFjLmpw Yoshihiro Kawaoka, a2F3YW9rYXlAc3ZtLnZldG1lZC53aXNjLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.