
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 February 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00325
This article is part of the Research Topic Industrial and Health Applications of Lactic Acid Bacteria and Their Metabolites View all 57 articles
Water kefir is a slightly alcoholic and traditionally fermented beverage, which is prepared from sucrose, water, kefir grains, and dried or fresh fruits (e.g., figs). Lactobacillus (L.) nagelii, L. hordei, and Saccharomyces (S.) cerevisiae are predominant and stable lactic acid bacteria and yeasts, respectively, isolated from water kefir consortia. The growth of L. nagelii and L. hordei are improved in the presence of S. cerevisiae. In this work we demonstrate that quantitative comparative proteomics enables the investigation of interactions between LAB and yeast to predict real-time metabolic exchange in water kefir. It revealed 73 differentially expressed (DE) in L. nagelii TMW 1.1827 in the presence of S. cerevisiae. The presence of the yeast induced changes in the changes in the carbohydrate metabolism of L. nagelii and affected reactions involved in NAD+/NADH homeostasis. Furthermore, the DE enzymes involved in amino acid biosynthesis or catabolism predict that S. cerevisiae releases glutamine, histidine, methionine, and arginine, which are subsequently used by L. nagelii to ensure its survival in the water kefir consortium. In co-culture with S. cerevisiae, L. nagelii profits from riboflavin, most likely secreted by the yeast. The reaction of L. nagelii to the presence of S. cerevisiae differs from that one of the previously studied L. hordei, which displays 233 differentially expressed proteins, changes in citrate metabolism and an antidromic strategy for NAD+/NADH homeostasis. So far, aggregation promotion factors, i.e., formation of a specific glucan and bifunctional enzymes were only detected in L. hordei.
Water kefir is a slightly alcoholic, traditionally fermented beverage, which is prepared from sucrose, water, kefir grains, and dried or fresh fruits (e.g., figs). Water kefirs, originating from definitely different sources, exhibit different species diversities. Still, the basic consortium, which mainly consists of lactic acid bacteria (LAB), acetic acid bacteria (AAB) and yeasts (Ward, 1891; Neve and Heller, 2002; Gulitz et al., 2011; Marsh et al., 2013; Laureys and De Vuyst, 2014) appears to be stable. L. hordei, L. nagelii, and S. cerevisiae are dominant LAB and yeast species, respectively, isolated from water kefir grains (Gulitz et al., 2011; Stadie et al., 2013; Laureys and De Vuyst, 2014). Among L. hordei and L. nagelii isolates from these water kefirs L. hordei TMW 1.1822 and L. nagelii 1.1827 were the most abundant isolates, which also produced dextrans and showed synergisms with concomitant yeasts (Stadie et al., 2013; Xu et al., 2018, 2019a).
In contrast to milk kefir, there is only very limited research on water kefir. Most of the available studies focused on its species diversity (Ward, 1891; Pidoux, 1989; Neve and Heller, 2002; Gulitz et al., 2011; Marsh et al., 2013; Laureys and De Vuyst, 2014; Martínez-Torres et al., 2017), or on the chemical and structural composition of the water kefir grains (Horisberger, 1969; Pidoux et al., 1988; Pidoux et al., 1990; Waldherr et al., 2010; Fels et al., 2018; Xu et al., 2018). To date, several attempts have been made to understand the interactions of the microorganisms in water kefir. For instance, Stadie et al. (2013) studied the metabolic interaction between LAB (L. hordei and L. nagelii) and yeasts (S. cerevisiae and Zygotorulaspora florentina) isolated from water kefir and inferred, that the growth of L. hordei TMW 1.1822 should be improved by nutrients produced by both yeasts, such as several amino acids (isoleucine, leucine, methionine, phenylalanine, tryptophan, tyrosine, and valine) and vitamin B6.
Another study explored the metabolite dynamics in a water kefir fermentation. The major metabolites produced were ethanol and lactic acid during 192 h of fermentation. Glycerol, acetic acid, and mannitol were produced in low concentrations. The prevailing volatile aroma compounds were ethyl acetate, isoamyl acetate, ethyl hexanoate, ethyl octanoate, and ethyl decanoate after 72 h (Laureys and De Vuyst, 2014). Further, the water kefirs were supplied with dried figs, apricots and raisins, respectively, as different nutrient sources delivering various concentrations. Also, the influence of oxygen has been investigated. It was concluded, that raisins led to low nutrient concentrations in the water kefir formulation, which favored the growth of L. hilgardii and Dekkera bruxellensis. In contrast, figs supplied the water kefir with high nutrient concentrations, which favored the growth of L. nagelii and S. cerevisiae. The presence of oxygen allowed the proliferation of AAB, resulting in high concentrations of acetic acid (Laureys et al., 2018). In addition, three main metabolic products were evaluated from the carbon flux from sucrose during 192 h of fermentation (Martínez-Torres et al., 2017). After 24 h, lactic and acetic acid have been postulated to be initially produced by L. hilgardii and subsequently produced by Acetobacter spp., mainly A. tropicalis. Ethanol was almost entirely oxidized to acetic acid, which could be further dissimilated by Acetobacter species.
However, these studies only determined total metabolite concentrations produced by the microorganisms during fermentation, but they did not reveal, how LAB, AAB and yeasts benefit from or affect each other through dynamic metabolite exchanges. Recently, we have shown that L. hordei TMW 1.1822 is highly adapted to the water kefir environment (Xu et al., 2019a) and its sucrose rich but amino- and fatty acids poor conditions. In the presence of abundant sucrose, it produces a dextran, which specifically induces the aggregation of S. cerevisiae as to ensure spatial proximity of the yeast cells in an initial step of granule formation (Xu et al., 2018). In a quantitative proteomic analysis we could quantify 233 differentially expressed proteins of L. hordei as its response to the co-culture with S. cerevisiae (Xu et al., 2019b). These were predicted to be involved in citrate and amino acids metabolism as well as maintenance of NAD+/NADH homeostasis. It appears that L. hordei benefits from S. cerevisiae by enhanced availability of amino acids, while it alleviates acid stress of the yeast via metabolism of arginine provided by the yeast.
In order to probe whether the response of L. hordei to S. cerevisiae and its role for the water kefir system are typical or unique as compared to other water kefir lactobacilli, we investigated L. nagelii and compared its response to co-culture with S. cerevisiae with that one of L. hordei.
L. nagelii TMW 1.1827 isolated from water kefir by Gulitz et al. (2011) was single-cultured anaerobically at 30°C in modified MRS (mMRS) medium (Stolz et al., 1995). Genomic DNA was isolated, as described previously (Xu et al., 2019a), and sent to GATC Biotech (Konstanz, Germany) for PacBio SingleMolecule RealTime sequencing. The whole genome sequences were annotated by the NCBI Prokaryotic Genome Annotation Pipeline and RAST, which is a SEED-based prokaryotic genome annotation service using default settings (Aziz et al., 2008; Overbeek et al., 2013), as described previously (Xu et al., 2019a), and their key features were summarized in Supplementary Table S1.
S. cerevisiae TMW 3.221 was pre-cultured in YPG medium (Xu et al., 2019b). Single-cultivated S. cerevisiae, L. nagelii and co-cultivated L. nagelii TMW 1.1827 and S. cerevisiae TMW 3.221 were prepared in water kefir medium (WKM) (Stadie et al., 2013). Cell counts were assessed by plating serial dilutions of co-cultivated L. nagelii and S. cerevisiae on mMRS agar plates, supplemented with cycloheximide and YPG agar plates, supplemented with chloromycetin, respectively. In the same way, single-cultivated L. nagelii was plated on mMRS agar plates and single-cultivated S. cerevisiae on YPG agar plates, as described previously by Xu et al. (2019b).
1% pre-cultured L. nagelii TMW 1.1827 and S. cerevisiae TMW 3.221 were separately inoculated into chemically defined medium (CDM) in triplicate, as described previously (Xu et al., 2019a). After 24 h of cultivation at 30°C, 1 ml of each culture and 1 ml of CDM as a control were mixed with 50 μl of 70% (v/v) perchloric acid (Sigma-Aldrich, St. Louis, MO, United States) and subsequently incubated overnight at 4°C for protein precipitation. After centrifugation (12,000 rpm, 10 min), the supernatant was collected and filtered by 0.2 μm PhenexTM Regenerated Cellulose Membrane (Phenomenex, Aschaffenburg, Germany) for the detection of amino acids and organic acids as below. Amino acids were analyzed on a Dionex Ultimate 3000 HPLC system (Dionex, Idstein, Germany) using a Gemini C18 column (Phenomenex, Aschaffenburg, Germany) with UV detection at 338 and 269 nm. Quantification was executed employing calibration adjustment by external HPLC grade standards and the Chromeleon software version 6.80 (Dionex, Idstein, Germany).
Consumption and production of sugars and organic acids of L. nagelii and S. cerevisiae grown in CDM for 24 h were quantified by a Dionex UltiMate 3000 HPLC system (Dionex, Idstein, Germany) with Rezex ROA-Organic Acid H+ column (Phenomenex, Aschaffenburg, Germany) and RI-101 detector (Shodex, München, Germany), as described previously (Xu et al., 2019a). For sugar analysis, 500 μl of each sample were mixed with 250 μl of a 10% (w/v) ZnSO4∗7H2O solution and afterward added with 250 μl 0.5 M NaOH. After incubation for 20 min at 25°C, the supernatant was obtained by centrifugation and filtered as described above. Analytes were separated at a constant flow rate of 0.7 ml/min with a column temperature of 85°C for 30 min. Sulfuric acid (Rotipuran, Roth, Karlsruhe, Germany) solution with a concentration of 5 mM served as mobile phase.
Co-cultivated L. nagelii and S. cerevisiae, as well as single-cultured L. nagelii and S. cerevisiae were incubated anaerobically in WKM at 30°C for 10 h in triplicate and prepared for proteomic analysis, as previously described (Xu et al., 2019a). First of all, these samples were treated with trichloroacetic acid (TCA, 6.25% w/v), centrifuged (5,000 rpm, 5 min) at 4°C, washed with acetone and reconstituted in lysis buffer [8 M urea, 5 mM EDTA di-sodium salt, 100 mM (NH)4HCO3, 1 mM dithiothreitol (DDT)]. Subsequently, the cells were mechanically disrupted with acid-washed glass beads (G8772, 425–600 μm, Sigma, Germany). Proteins were reduced with 10 mM DTT at 30°C for 30 min, and subsequently carbamidomethylated with 55 mM chloroacetamide in the dark for 60 min. Finally, proteins were digested by trypsin and desalted by C18 solid phase extraction using Sep-Pak columns (Waters, WAT054960). Purified peptide samples were dried in a SpeedVac concentrator (Acid-Resistant CentriVap Vacuum Concentrator, Labconco) and resuspended in an aqueous solution containing 1.9% acetonitrile and 0.1% formic acid to a final concentration of 0.25 μg/μl.
Generated peptides were analyzed on a Dionex Ultimate 3000 nano LC system, coupled to a Q-Exactive HF mass spectrometer (Thermo Scientific, Bremen, Germany), as described previously (Xu et al., 2019b). Peptides were delivered to a trap column (75 μm × 2 cm, self-packed with Reprosil-Pur C18 ODS-3 5 μm resin, Dr. Maisch, Ammerbuch, Germany) at a flow rate of 5 μl/min in solvent A0 (0.1% formic acid in water). Peptides were separated on an analytical column (75 μm × 40 cm, self-packed with Reprosil-Gold C18, 3 μm resin, Dr. Maisch, Ammerbuch, Germany), using a 120 min linear gradient from 4 to 32% solvent B (0.1% formic acid, 5% DMSO in acetonitrile) and solvent A1 (0.1% formic acid, 5% DMSO in water) at a flow rate of 300 nl/min. The mass spectrometer was operated in data dependent mode, automatically switching between MS1 and MS2 spectra. MS1 spectra were acquired over a mass-to-charge (m/z) range of 360–1,300 m/z at a resolution of 60,000 (at m/z 200) using a maximum injection time of 50 ms and an AGC target value of 3e6. Up to 20 peptide precursors were isolated (isolation window 1.7 m/z, maximum injection time 25 ms, AGC value 1e5), fragmented by higher-energy collisional dissociation (HCD), using 25% normalized collision energy (Letort et al., 2002) and analyzed at a resolution of 15,000 with a scan range from 200 to 2,000 m/z.
To enable differentiation of L. nagelii and S. cerevisiae proteins and their identification, peptide and protein identification plus quantification were performed with MaxQuant (version 1.5.7.4) by searching the MS2 data against all protein sequences obtained from UniProt – reference proteome S. cerevisiae S288c (6,724 entries, downloaded 13.03.2017) and all protein sequences from L. nagelii TMW 1.1827 (cf. section “Comparative Genomic Features and Growth Characteristics of L. nagelii in the Presence of S. cerevisiae,” GenBank CP018180 – CP018183), using the embedded search engine Andromeda (Cox et al., 2011), as previously described (Xu et al., 2019a). Carbamidomethylated cysteine was a fixed modification. Oxidation of methionine, and N-terminal protein acetylation were variable modifications. Precursor and fragment ion tolerances were 10 ppm and 20 ppm, respectively. Label-free quantification and data matching between consecutive analyses were enabled within MaxQuant. Search results were filtered for a minimum peptide length of seven amino acids, 1% peptide and protein false discovery rate (FDR) plus common contaminants and reverse identifications. MaxQuant output files were further analyzed using Perseus (version 1.5.6.0) (Tyanova et al., 2016). iBAQ intensities were log2-transformed for further statistical analysis. NCBI annotation, PSORTb subcellular localization, SEED category (subcategory and subsystem) as previously annotated (cf. section “Strain Culture, Whole-Genome Sequencing, and Cell Counts”) were added to the matrix through identifier matching. For the comparison between two groups, t-tests were performed. Log2 fold change ≥ 2 or ≤-2 and -Log10 P-value ≥ 2 (p-value ≤ 0.05) were considered to be significantly differentially expressed proteins of L. nagelii TMW 1.1827 in the presence of S. cerevisiae TMW 3.221.
A genomic atlas of L. nagelii TMW 1.1827 was generated using Artemis and DNA plotter1 (Carver et al., 2008) as described previously (Xu et al., 2019a). Subcellular localization of proteins was predicted, using the tool PSORTb (Version 3.0.22) (Gardy et al., 2004; Yu et al., 2010). All the annotated EC numbers from RAST were imported into iPath 3.03 (Yamada et al., 2011) for generating an overview of complete metabolic pathways and biosynthesis of other secondary metabolites.
The sucrose metabolism, pyruvate metabolism, and amino acid biosynthesis pathways of L. nagelii TMW 1.1827 were constructed based on the self-constructed overview on the key reactions involved in sucrose metabolism, pyruvate metabolism, and amino acid biosynthesis pathways of L. hordei TMW 1.1822 as described previously (Xu et al., 2019a). Enzymes involved in each reaction step were manually checked, whether they were present in translated open reading frames (ORFs) annotated from both, NCBI and RAST. The figure of the biosynthesis pathways of amino acids and riboflavin was generated using the KEGG PATHWAY mapping tool4 by importing EC numbers only involved in amino acid biosynthesis and riboflavin metabolism.
Genomic differences between L. nagelii TMW 1.1827 and L. hordei TMW 1.1822 were identified using Blast Diagnostic Gene findEr (BADGE) (Behr et al., 2016) under modified settings. The “min_DMG_occurance” was set to 0.00000000000001. The “megablast_perc_identity_cut” value was set to 90, while both, the “megablast_within_groub_qscov” and the “megablast_between_group_qscov” value was set to 0.90. The dc_mode was enabled. Additionally, BADGE was run on protein level using default protein-level options. The BADGE output was divided in pan and core genome. The genome comparison was graphically visualized by the BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011) using the annotated and translated ORFs of the pan genome as reference. Furthermore, the genomic differences between L. nagelii TMW 1.1827 and L. nagelii DSM 13675 were identified by BADGE using default settings.
The whole-genome sequence of L. nagelii TMW 1.1827 was submitted to GenBank designated as BioSample SAMN06052354, referred to as accession numbers CP018180 to CP018183. An additional file containing all metadata of L. nagelii TMW 1.1827 from NCBI and RAST annotation is deposited as Supplementary Material. The mass spectrometry proteomics data have been deposited to the ProteomeXchange via the PRIDE partner repository with the dataset identifier PXD0125135.
The genomic size of L. nagelii TMW 1.1827 is 2.41 Mbp and exhibits a GC content of 36.68% (shown in Table 1). L. nagelii TMW 1.1827 exhibits a total number of 2,391 coding sequences (CDS), including all three plasmids (shown in comparison with L. hordei in Table 1 and visualized in Supplementary Figure S1). So far, the only published whole genome sequences of L. nagelii strains result from a comparative genomics project together with 211 other LAB strains (Sun et al., 2015). L. nagelii DSM 13675 isolated from wine, was associated to a different environment than water kefir and therefore faces different conditions. Those differences in the adaptation to distinct environmental conditions were also displayed in the genomes. For the two L. nagelii strains from wine and water kefir the annotated differences could be referred to genes related to carbohydrate metabolism, namely enzymes of citrate and concomitant acetolactate metabolism, which were only found in the water kefir isolate L. nagelii TMW 1.1827. Also, the water kefir isolate differed from the wine isolate by galactose PTS and metabolism including the tagatose pathway. As citrate and galactose are present or absent, respectively, in both environments, a specific adaptation to the respective source of isolation cannot be deduced from this. The genomic reflection of environmental adaptation observed in strains of L. hordei isolated from widely different environments of malted barley (DSM 19519; Sun et al., 2015) or water kefir TMW 1.1822; Xu et al., 2019a), respectively, was more decisive and markedly resides in sucrose metabolism.
For comparative insights the whole genome sequences of L. hordei TMW 1.1822 and L. nagelii TMW 1.1827 were compared to each other using BADGE. As visualized in Figure 1, the core genome of both microorganisms included 1,380 CDS, which displays 56.0% of the whole genome of L. hordei TMW 1.1822 and 57.7% of the whole genome of L. nagelii TMW 1.1827. The main components of the core genome were found in the SEED categories of protein, carbohydrate and amino acid metabolism. The accessory genome of L. hordei TMW 1.1822 as compared to that one of L. nagelii TMW 1.1827 was dominated by additional genes for carbohydrate and amino acid metabolism, and cell wall biosynthesis. Corresponding results were found for L. nagelii TMW 1.1827, except for the SEED category of cell wall formation, which was substituted by CDS involved in DNA metabolism (shown in Figure 2). Since both microorganisms are associated to water kefir, representing an environment rich in sugar, it was not surprising, that L. nagelii TMW 1.1827 and L. hordei TMW 1.1822 mainly adapted to it by additional genes coding for carbohydrate metabolism.
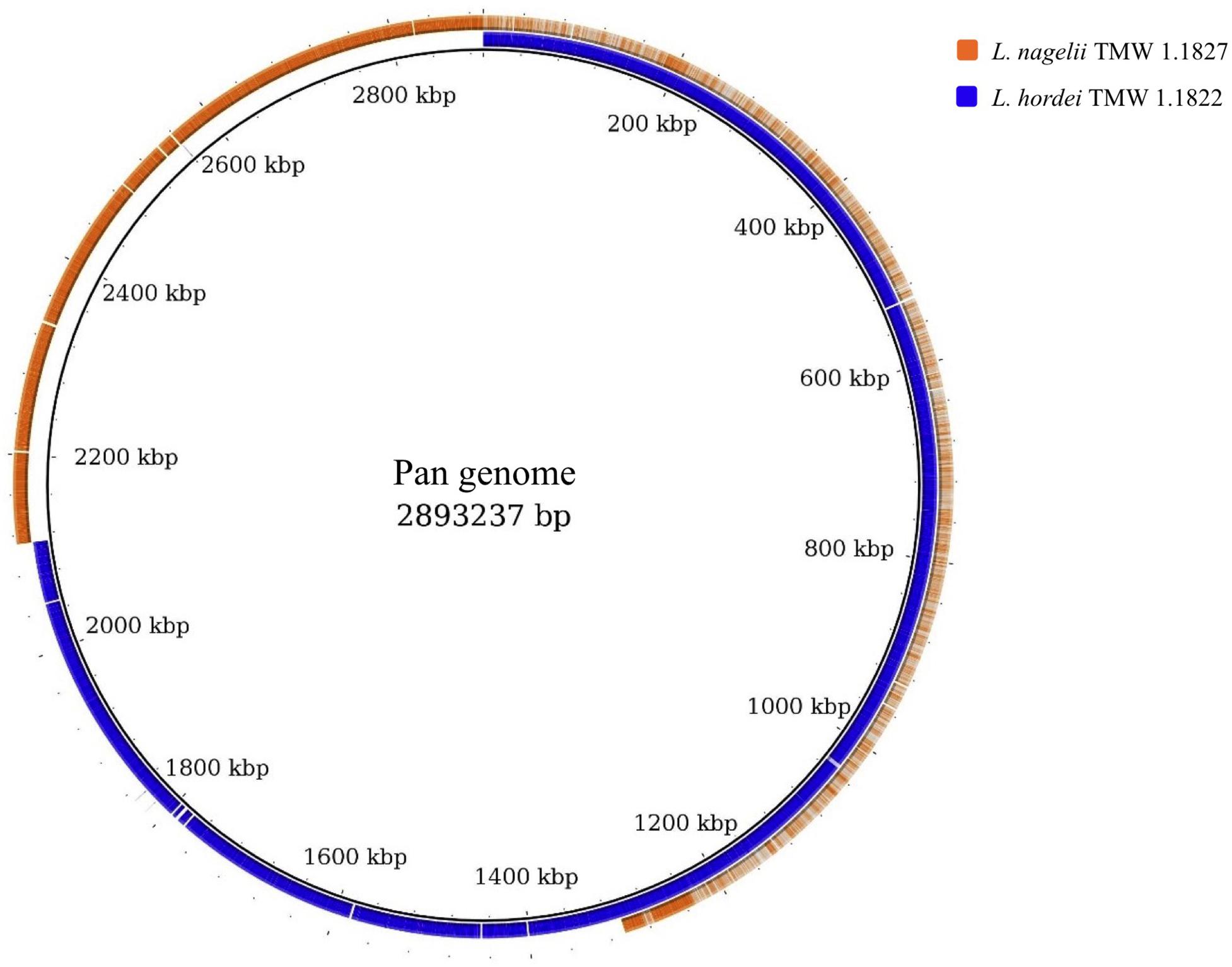
Figure 1. Whole genome comparison as visualized by BRIG (Alikhan et al., 2011). CDS of the pan genome was used as reference and the genomes of both microorganisms were aligned to this reference. As a result, the structures of the genomes and the pan genome did not reflect the physical structure of the chromosomes or plasmids. The core genome was approximately half of the pan genome and was detected from the beginning until about 1,300 kbp. Strain specific genes were displayed in the range of approximately 1,300 kbp until the end.
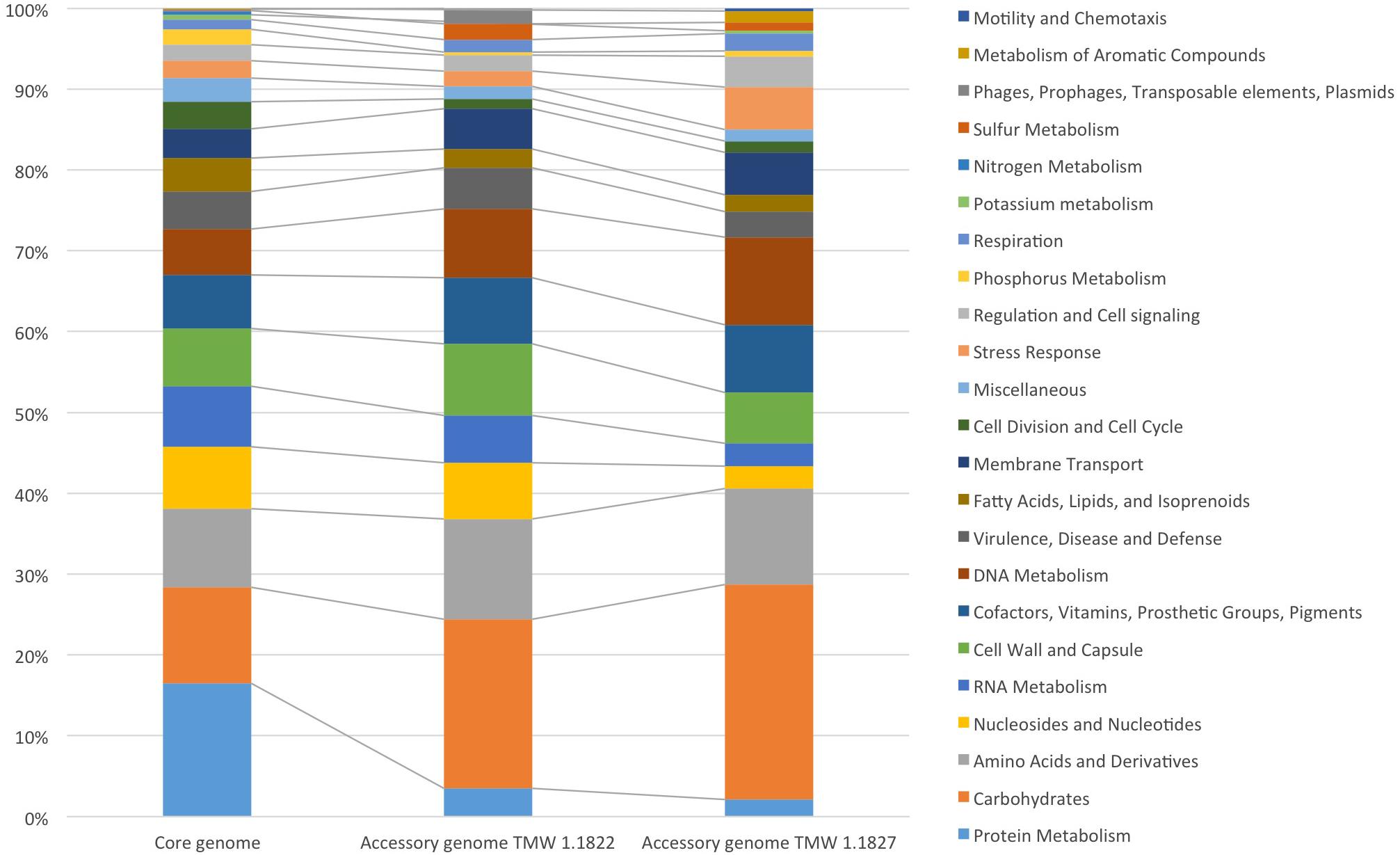
Figure 2. Annotated SEED categories of proteins, divided in the core and accessory genomes of L. hordei TMW 1.1822 and L. nagelii TMW 1.1827, which was done by BADGE analysis. The proportion of proteins assigned to each SEED category with respect to the total number of proteins is shown in the bar chart.
While the cell yield of single cultivated L. nagelii TMW 1.1827 was only slightly increased upon co-cultivation with S. cerevisiae after 8 and 12 h of fermentation (Figure 3A), it declined significantly slower in co-cultivated L. nagelii as compared to single-cultivated L. nagelii until 24 h. I appears that the co-culture with S. cerevisiae preconditions L. nagelii toward an increased tolerance to the (e.g., increasingly acidic) environmental conditions. On the other hand, the cfu of S. cerevisiae were reduced upon co-cultivation with L. nagelii (Figure 3B). This indicates that L. nagelii TMW 1.1827 affects the growth of S. cerevisiae much more than L. hordei TMW 1.1822 (Xu et al., 2019b). To get insights into the reasons of these differences, prediction of dynamic metabolite exchanges were explored by proteomics in this study for L. nagelii and compared with those previously determined for L. hordei (Xu et al., 2019b).
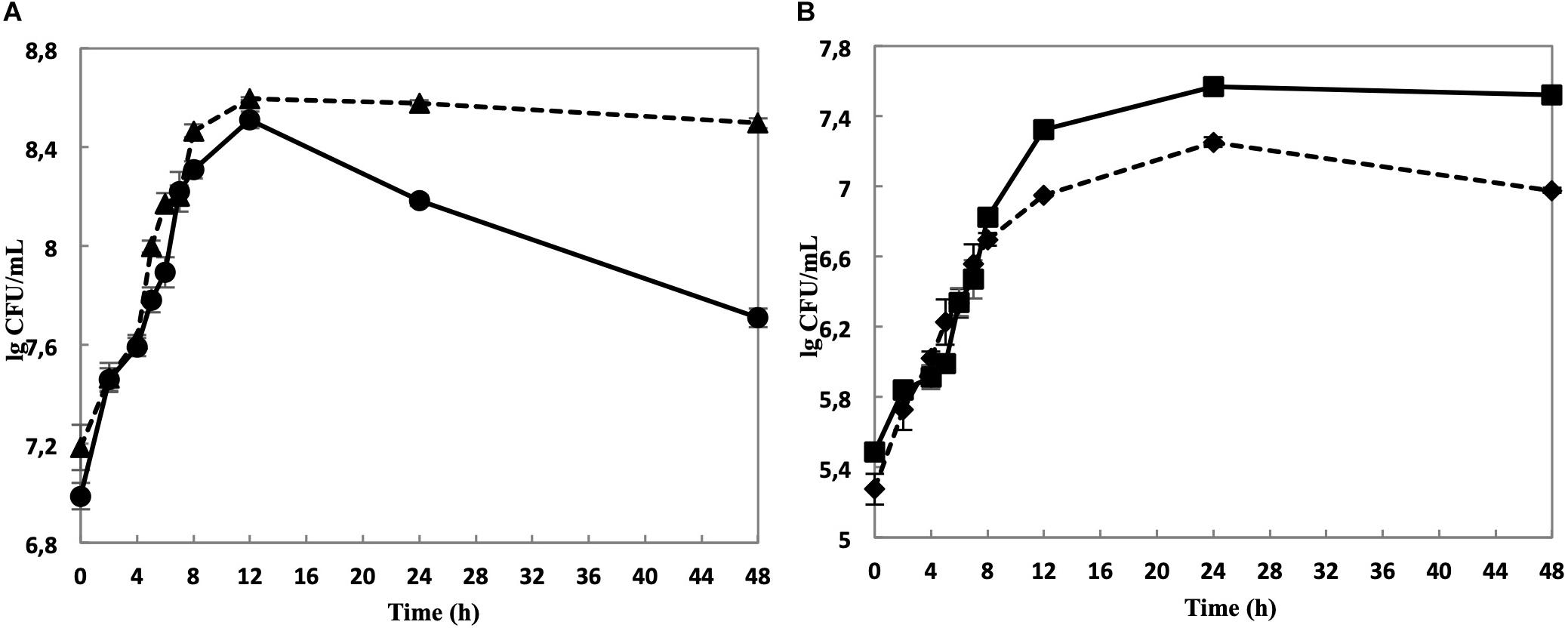
Figure 3. Cell counts of L. nagelii TMW 1.1827 in single culture ( ) and co-cultivation with S. cerevisiae TMW 3.221 (
) and co-cultivation with S. cerevisiae TMW 3.221 ( ) (A). Cell counts of S. cerevisiae TMW 3.221 in single culture (
) (A). Cell counts of S. cerevisiae TMW 3.221 in single culture ( ) and in co-cultivation with L. nagelii TMW 1.1827 (
) and in co-cultivation with L. nagelii TMW 1.1827 ( ) (B).
) (B).
As shown in Figure 4, 1,243 proteins of L. nagelii TMW 1.1827 were identified and quantified by proteomic analysis, comprising about 52% of the genes annotated by whole genome analysis. A comprehensive overview of the complete metabolic pathways and significantly differentially expressed (DE) proteins of L. nagelii in the presence of S. cerevisiae is provided in Supplementary Figure S2. As shown in Figure 5, there were 73 DE proteins in L. nagelii regulated in the presence of S. cerevisiae. Those up/down-regulated proteins of L. nagelii were most abundant in the SEED categories “amino acids and derivatives” (9 out of 69), “carbohydrates” (5 out of 93) “nucleosides and nucleotides” (4 out of 61) and “cofactors, vitamins” (7 out of 54) (shown in Figure 5).
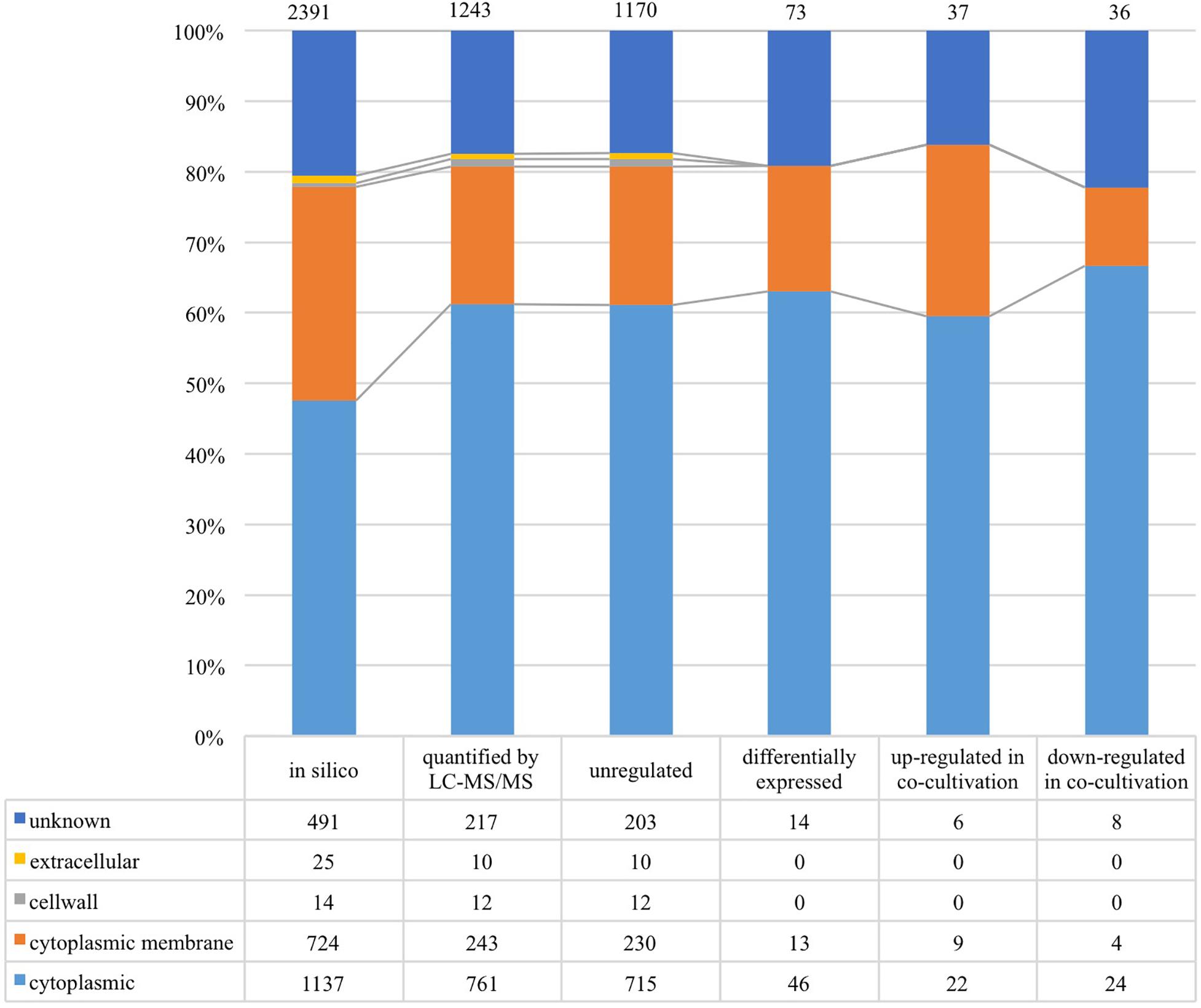
Figure 4. Subcellular localization of L. nagelii proteins (in silico, quantified by LC-MS/MS, unregulated, differentially expressed, up-regulated in co-cultivation, down-regulated in co-cultivation), which were predicted by PSORTb. The proportion of L. nagelii proteins assigned to each respective subcellular compartment and the group “unknown” with respect to the total number of L. nagelii proteins is shown by the bar chart. The table below shows the respective absolute numbers.
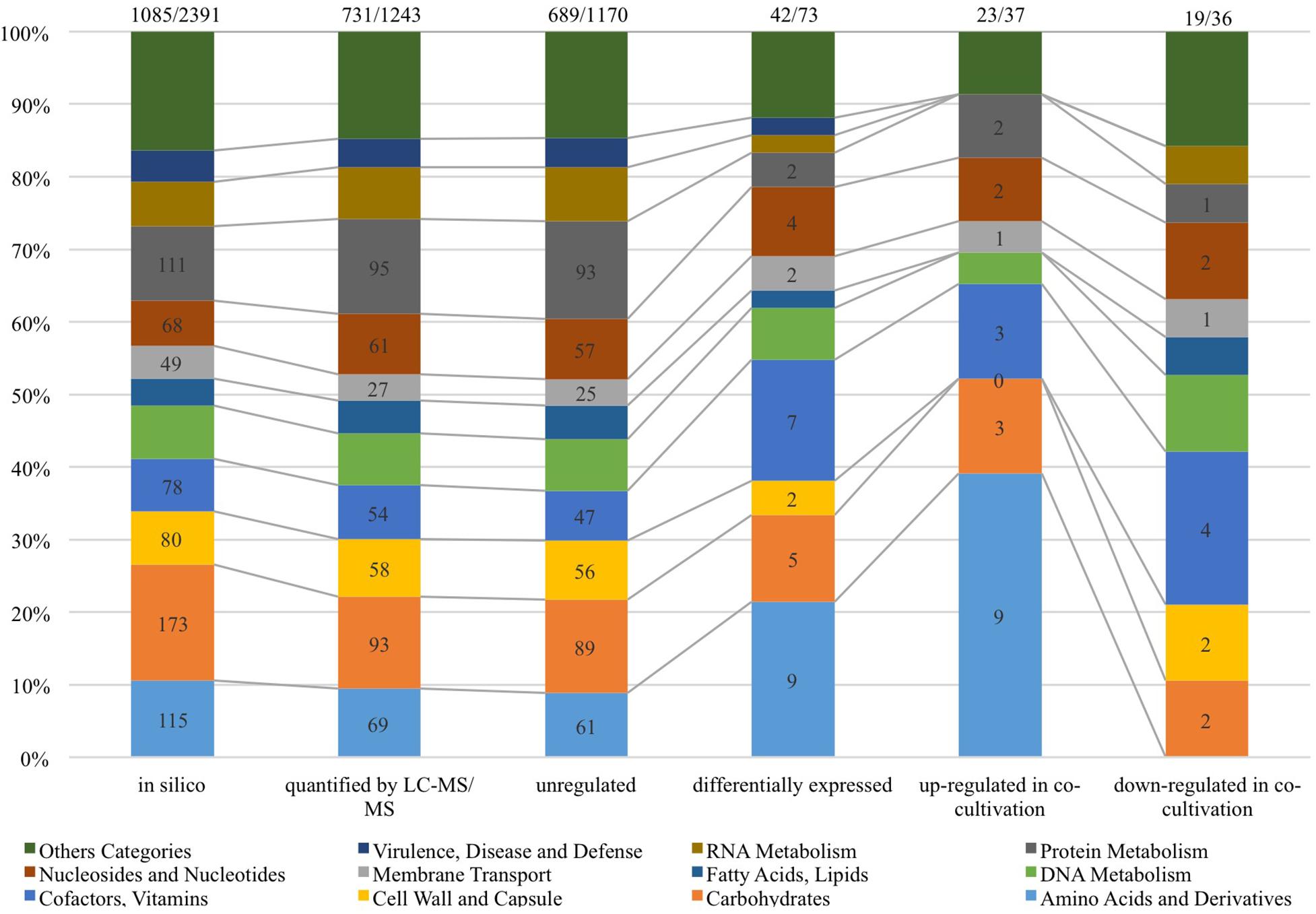
Figure 5. SEED categories of L. nagelii proteins (in silico, quantified by LC-MS/MS, unregulated, differentially expressed, up-regulated in co-cultivation, down-regulated in co-cultivation), which were predicted by SEED. The proportion of L. nagelii proteins assigned to each respective category of metabolism and the group “other categories” which is the sum of several small categories with respect to the total number of L. nagelii proteins is shown by the bar chart. The ratio on the top of each column is the number of predicted SEED categories accounts for the number of all coding DNA sequence (CDS).
The overview on the key reactions involved in sucrose metabolism of L. nagelii is provided in Figure 6. L. nagelii encoded and expressed an MFS-transporter specific for sucrose uptake. As previously demonstrated, L. nagelii also produces a glucan from sucrose by an extracellular glucansucrase (Xu et al., 2018). The residual fructose can then be transported into the cell by a fructose specific PTS and simultaneous phosphorylation. Once inside the cell, the phosphorylated fructose can directly enter the glycolytic pathway. All PTS, namely for sucrose, glucose, fructose, mannose, sorbose, and mannitol uptake, were identified by proteomic analysis as constitutively expressed upon co-culture. Whole genome sequence analysis of L. nagelii TMW 1.1827 confirmed the presence of the genes encoding all enzymes required for the EMP and PKP pathways (locus tags and IDs given in Supplementary Table S2). Thus, L. nagelii TMW 1.1827 should also be considered as facultatively heterofermentative, such as L. plantarum WCFS1 and Lactococcus lactis (Kleerebezem and Hugenholtz, 2003; Kleerebezem et al., 2003). This is contrary to the fact that L. hordei DSM 19519 and L. nagelii DSM 13675 were inferred as obligately homofermentative strains according to their phenotype (Sun et al., 2015). However, those strains have been isolated from different environments.
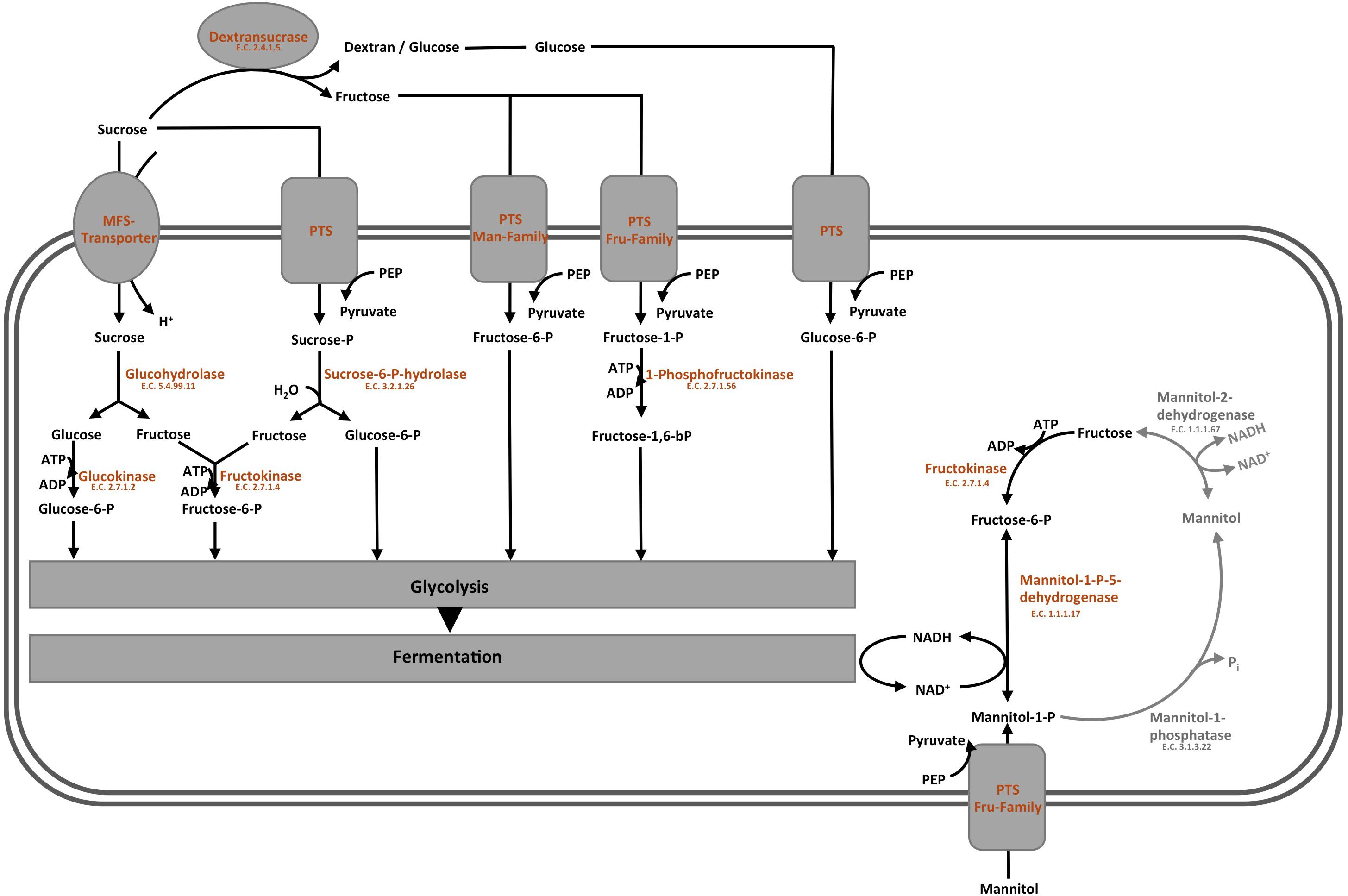
Figure 6. Modified overview on the key reactions involved in sucrose metabolism (Xu et al., 2019a) of L. nagelii TMW 1.1827 in the presence of S. cerevisiae TMW 3.221: the enzyme colored in blue was annotated by genomics but not quantified by proteomics, the enzymes colored in orange were both annotated by genomics and quantified by proteomics, the enzymes colored in gray were neither annotated by genomics nor quantified by proteomics.
To probe the principal fermentation type of L. nagelii TMW 1.1827 we determined fermentation metabolites upon its growth in CDM to find 40.1 mM lactate and 6.9 mM acetate after 24 h of fermentation. This way we could show for L. nagelii and also (previously) for L. hordei (Xu et al., 2019a) that the water kefir isolates of these species are indeed different from other ones even with respect to basic fermentation types. The data corroborate a homofermentative metabolism, in which energy generation via EMP and recycling of NAD+ by reducing pyruvate to lactate is favored. The small amount of acetate may reside from pyruvate by either generating formate via pyruvate formate lyase or by NADH and CO2 generation via the pyruvate dehydrogenase complex. Subsequently, the resulting acetyl-CoA may be metabolized to acetate. However, the latter option requires subsequent NAD+ recycling.
In the presence of S. cerevisiae, the 3-phosphoglycerate mutase of L. nagelii TMW 1.1827 was significantly down-regulated. As postulated previously, the expression of this enzyme is linked to the concentration of its substrate 3-phosphoglycerate (Smeianov et al., 2007). This indicates, that intermediates of early glycolytic steps may be used for other metabolic reactions or hexoses may rather enter PKP or PPP than EMP, resulting in less production of 3-phosphoglycerate. At the same time, the alcohol dehydrogenase (EC 1.1.1.1, 3.5 log2 fold change) of L. nagelii was significantly up-regulated in the presence of S. cerevisiae (shown in Figure 7). Yielding less ATP, but more reductive power, this metabolic switch to ethanol production may be important to keep the PKP running. L. nagelii TMW 1.1827 is capable of transporting and phosphorylating mannitol, possibly delivered by S. cerevisiae, inside the cell by a specific PTS and subsequent oxidation to fructose-6-P via mannitol-1-P-5-dehydrogenase. This provides evidence for the enhanced ethanol production as a recycling mechanism for the NADH, which is generated upon mannitol oxidation. Since fructose-6-P must not be phosphorylated prior to entering the EMP or PKP, there is less need for ATP generation upon acetate formation.
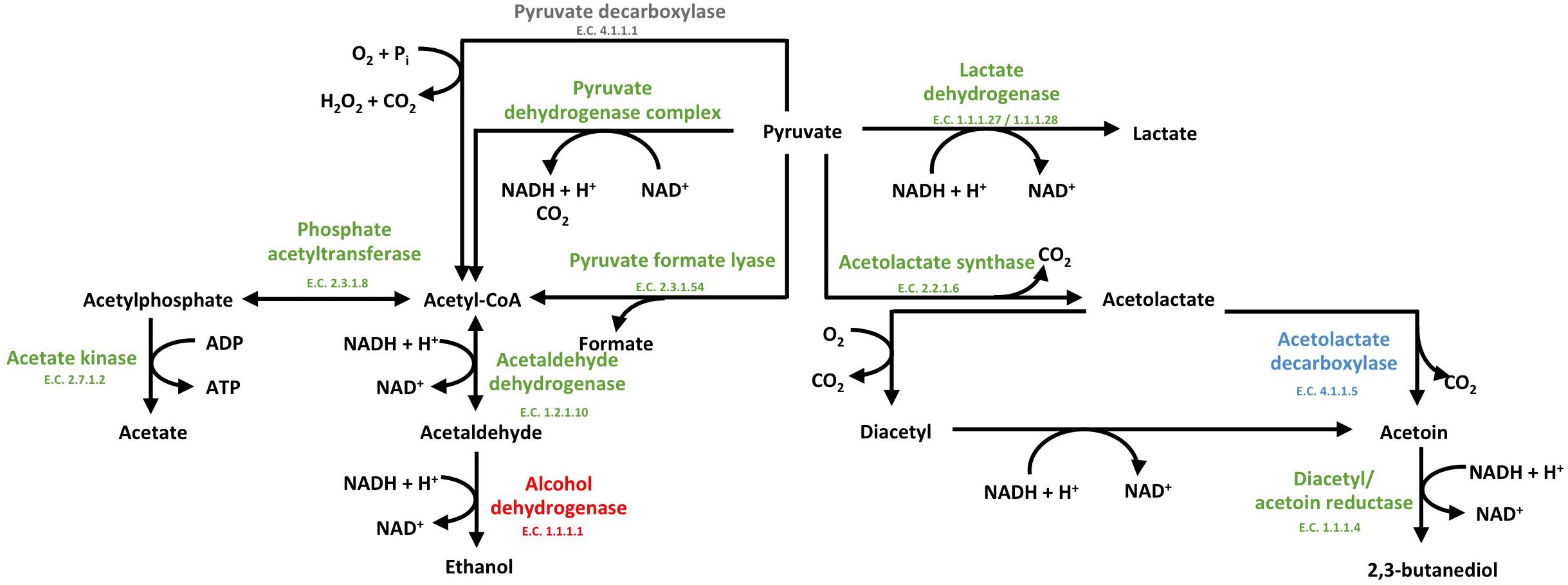
Figure 7. Modified predicted outline of pyruvate metabolism (Xu et al., 2019a) of L. nagelii TMW 1.1827 in the presence of S. cerevisiae TMW 3.221. The enzymes colored in red represented the up-regulated ones, while blue represented down-regulated proteins. Un-regulated proteins are indicated in green and proteins, neither annotated in the genome or proteome are colored in gray.
The expression of glucose-6-phosphate dehydrogenase, which is part of the PKP and PPP, was significantly up-regulated (Xu et al., 2019b). As discussed previously, this metabolic switch from EMP to PKP/PPP may also help L. nagelii to utilize gluconate, which appears to be a decisive trait in the water kefir environment (Xu et al., 2019a). Looking at the metabolic phenotype of other abundant LAB species in water kefir, these results are in line with the findings of Laureys et al. L. paracasei, which is the most dominant one during their water kefir grain growth (Laureys et al., 2018), is also facultatively heterofermentative lactobacilli as L. hordei TMW 1.1822 and L. nagelii TMW 1.1827.
Furthermore, α-acetolactate decarboxylase (-2.2 log2 fold change, shown in Figure 7) was significantly down-regulated in L. nagelii, blocking the direct decarboxylation of acetolactate to acetoin. Under aerobic conditions, acetolactate spontaneously decomposes into diacetyl enabling regeneration of two molecules of NAD+ upon reduction to 2,3-butanediol via diacetyl/acetoin reductase. Since oxygen is probably limited for L. nagelii due to the subsidence of the water kefir granules, this pathway for NAD+ regeneration may be completely disabled in the water kefir environment.
In contrast to L. nagelii, L. hordei possesses PTS specific for β-glucoside and cellobiose transport, and the expression of PTS belonging to the mannose–fructose–sorbose family were significantly up-regulated in L. hordei. This indicates a more restricted use of sugars by L. nagelii. The expression of EMP specific enzymes of L. hordei TMW 1.1822 was not influenced by S. cerevisiae. More decisively, L. hordei reacts to the presence of S. cerevisiae by down-regulation of alcohol dehydrogenase and up-regulation of diacetyl/ acetoin reductase and α-acetolactate decarboxylase, yielding 2,3-butanediol and NAD+ (Xu et al., 2019b). In conclusion, L. nagelii TMW 1.1827 displayed an antidromic strategy to maintain NAD+/NADH homeostasis after the metabolic switch induced by S. cerevisiae as compared to L. hordei TMW 1.1822.
Both, genomic and proteomic analyses revealed an incomplete TCA cycle in L. nagelii. In the case of L. nagelii, aconitate hydratase (EC 4.2.1.3), which catalyzes the stereo-specific isomerization of citrate to isocitrate via cis-aconitate, and isocitrate dehydrogenase (EC 1.1.1.42), which catalyzes the oxidative decarboxylation of isocitrate, producing 2-oxoglutarate and CO2, were significantly up-regulated (4.5 log2 fold change, 2.0 log2 fold change). Despite its incompleteness, the TCA cycle is an important supplier for compounds involved in other metabolic reactions and thus, isocitrate and 2-oxoglutarate may be useful for amino acid metabolism in L. nagelii.
Water kefir is a challenging environment for its habitants regarding low nutrient concentrations except for the excess sugar. Since lemon slices are added, it is not surprising that microorganisms in water kefir use citrate as a nutrient. L. nagelii is capable of direct citrate import using malate permease. Once inside the cell, citrate is converted by citrate lyase segregating one molecule of acetate. The resulting oxaloacetate may then be decarboxylated via oxaloacetate decarboxylase yielding pyruvate or is further used for amino acid biosynthesis. In contrast to L. nagelii, these enzymes are DE in L. hordei, which appears to be positively influenced in its metabolism of citrate as an additional carbon source upon co-culture with S. cerevisiae (Xu et al., 2019b). This may help to explain, why L. hordei is more abundant in the water kefir consortium than L. nagelii (Gulitz et al., 2011).
The concentration of amino acids in pure WKM is very low (<0.004 mmol/l, respectively) (Stadie et al., 2013). So respective metabolite quantification is way too low to obtain conclusive data on amino acids metabolism in WKM, namely on those metabolites, which are determinative for interaction of lactobacilli and yeasts. Indeed, this is a major reason to use quantitative proteomics for metabolic predictions. The in silico analysis of the genome and proteome of L. nagelii TMW 1.1827 did not reveal any known homologs of a cell wall proteinase (Prt). L. nagelii encodes the complete oligopeptide transport system OppABCDF (Tynkkynen et al., 1993; Detmers et al., 1998). Except from OppB, all genes of both annotated OppABCDF clusters were found to be present in the proteome of L. nagelii. Despite lacking an expressed OppB, the growth of L. nagelii was not impaired. This phenomenon was already described for other bacteria (Nepomuceno et al., 2007) indicating, that the function of OppB may be compensable by other trans-membrane proteins. In the presence of the yeast, the remaining proteins were widely un-regulated with the exception of OppF, which was significantly down-regulated in one cluster. Since OppF is responsible for coupling the energy of ATP hydrolysis with the import of oligopeptides, L. nagelii may reduce energy consumption caused by oligopeptide uptake. In contrast, L. hordei upregulated its OppABCDF system and a set of peptidases, suggesting that L. hordei benefits from peptides, which are more readily available in the presence of the yeast (Xu et al., 2019b). Since water kefir provides very limited resources of proteins and free amino acids, mainly originating from dried fruits and the yeast, these findings may also explain the fact, that the growth of L. hordei is stimulated in co-culture.
From genomic annotation, L. nagelii encodes several amino acid permeases and transporters. In the presence of S. cerevisiae, methionine aminopeptidase and amino acid permease were significantly up-regulated, suggesting that the yeast induces amino acid uptake in L. nagelii. However, it was not possible to specify from sequence comparison, which amino acids were ingested by L. nagelii. Still, this may be solved by a closer look at amino acid synthesis pathways and auxotrophies. The genomic analysis of L. nagelii TMW 1.1827 revealed the prototrophy for 13 amino acids and auxotrophy for 7 amino acids (Table 2). According to the quantitative proteomic analysis, nine enzymes of L. nagelii involved in histidine, methionine, glutamate, and arginine biosynthesis pathways were all significantly up-regulated in the presence of S. cerevisiae (shown in Figure 8 and Table 3). Since those biosynthesis pathways can also be used for amino acid catabolism, water kefir microorganisms may profit from amino acids secreted by the yeast, creating a symbiotic consortium. However, from in silico analysis, the direction of a respective metabolic pathway remains speculative. Still, together with physiological data on amino acid consumption and secretion of L. nagelii and S. cerevisiae, this can be solved for at least some of the predicted cases.
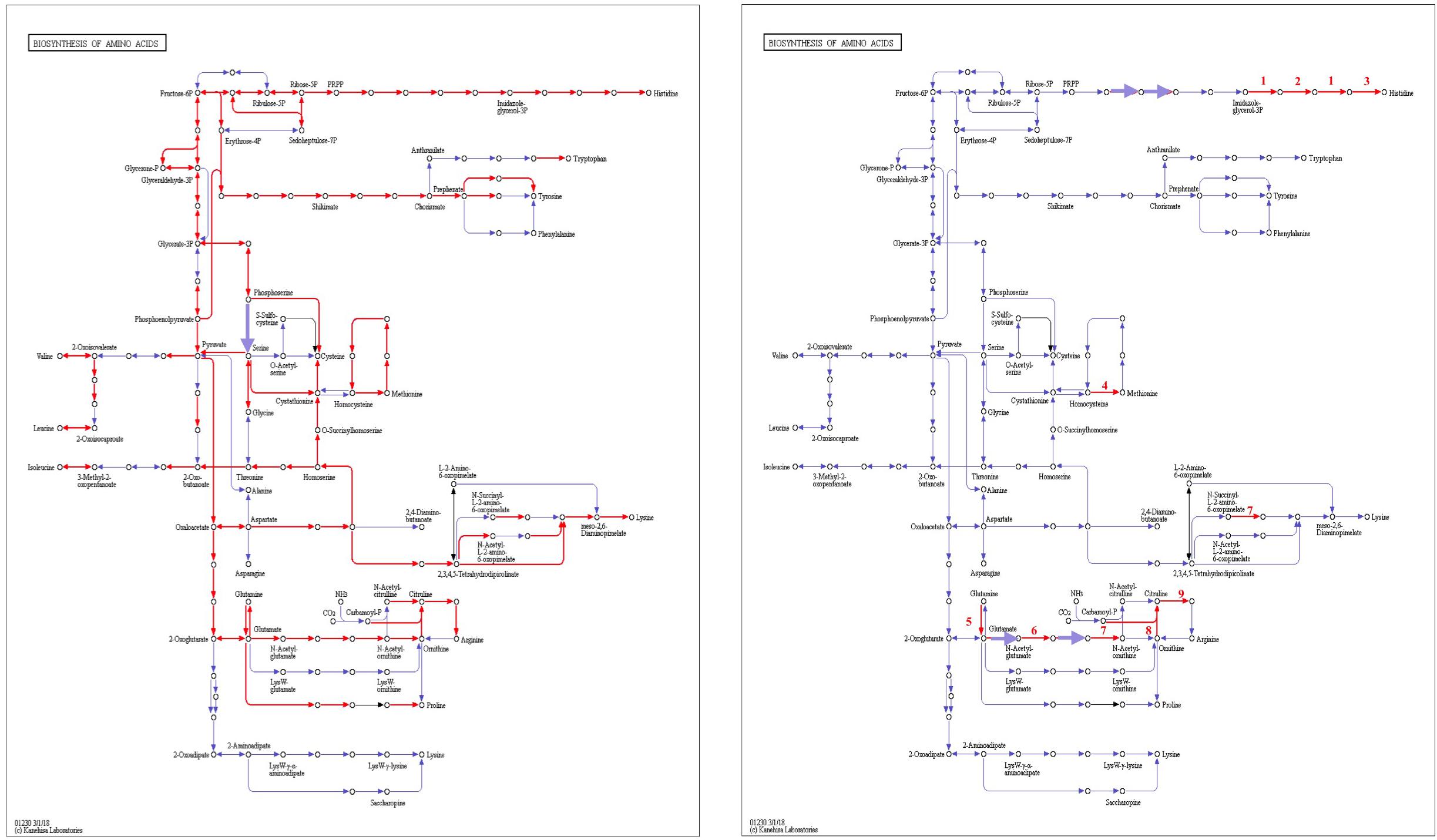
Figure 8. Biosynthesis of amino acids of L. nagelii TMW 1.1827 (A) and L. nagelii TMW 1.1827 in presence of S. cerevisiae TMW 3.221 (B). In (A), the red arrowed lines show the presence of enzymes annotated from genome. In (B), the red arrowed lines indicate up-regulated proteins.
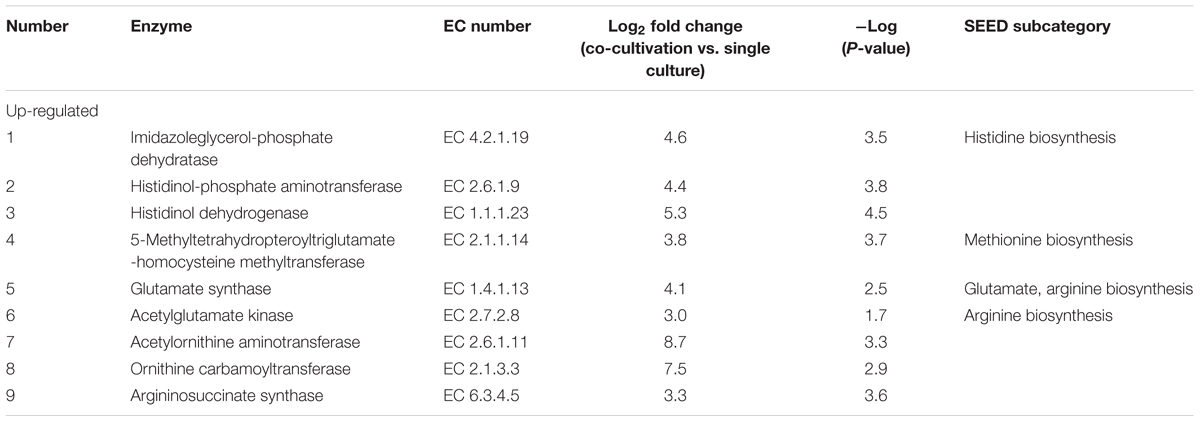
Table 3. Significantly differentially expressed proteins in L. nagelii TMW 1.1827 in response to S. cerevisiae TMW 3.221 involved in amino acid biosynthesis.
As shown most prominently in Figure 9, S. cerevisiae secreted glutamine in high amounts, whereas L. nagelii consumed the amino acid at high levels via an up-regulated amino acid permease involved in glutamine uptake. This suggests, that L. nagelii, even though it is capable of producing glutamine by itself, profits from the glutamine provided by the yeast via the up-regulated glutamate synthase. Since glutamine plays an important role in anaplerotic sequences of transamination reactions in the biosynthesis of other amino acids, and also as a nitrogen carrier for the production of amino sugars and nucleotides, the uptake of this amino acid may be crucial to persist in the water kefir environment. L. nagelii was predicted to produce glutamate by itself via the up-regulated glutamate synthase using glutamine and 2-oxoglutarate, which probably results from the incomplete TCA cycle. This was consistent with an un-regulated glutamine synthetase in the presence of S. cerevisiae. As already described for Lactobacillus crispatus ST1, this enzyme might exhibit additional functions, if displayed on the bacterial surface, which enable physical coherence of the water kefir consortium under stressful conditions (Kainulainen et al., 2012). As a result, the yeast aggregation promotion of L. hordei by its functional dextran (Xu et al., 2018) may be even enhanced by over expression of this enzyme. Furthermore, among all amino acids, the production of glutamate is of primary importance in the assimilation of nitrogen, representing a donor for amino groups in the synthesis of other amino acids (Bernard and Habash, 2009; Dincturk et al., 2011).
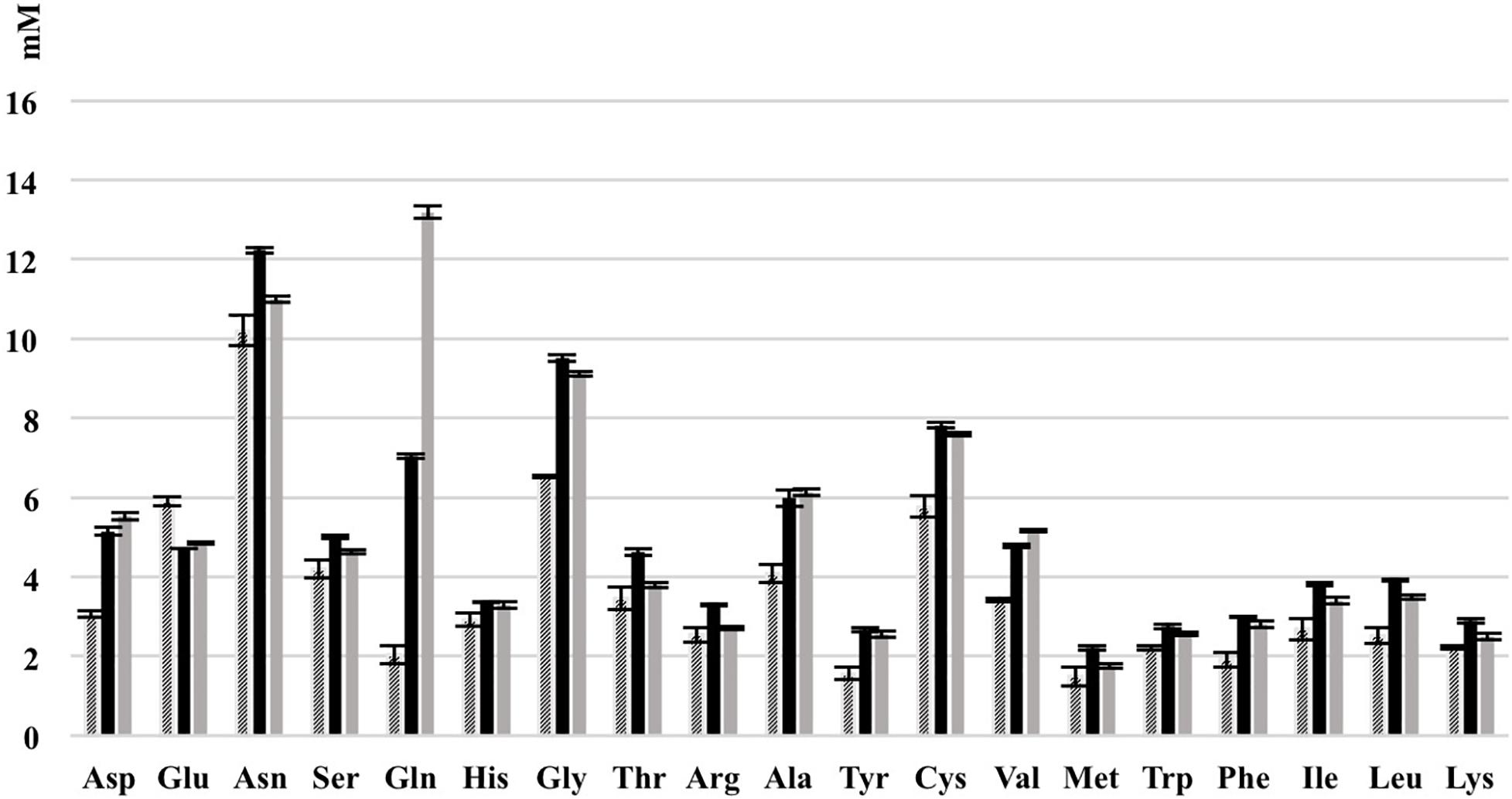
Figure 9. Consumption of amino acids of L. nagelii TMW 1.1827 and S. cerevisiae TMW 3.221 isolated from water kefir grown in CDM after 24 h. Black bar represents CDM, slash bar represents L. nagelii, gray bar represents S. cerevisiae.
Other amino acids were not produced, but partly consumed by the yeast after 24 h of fermentation in CDM. Therefore, it was not possible to determine real-time metabolic exchange (release/uptake) between L. nagelii and S. cerevisiae based on physiological data. Still, the label-free quantitative proteomic analysis enabled the investigation of the dynamic metabolic exchanges between microbial communities in water kefir. The DE enzymes involved in amino acid biosynthesis or catabolism predict that S. cerevisiae releases glutamine, histidine, methionine, and arginine, which are subsequently used by L. nagelii to ensure its survival in the water kefir consortium.
Functionally, the ADI pathway enables enhanced acid tolerance and energy provision in a variety of LAB genera such as Lactobacillus, Lactococcus, Leuconostoc, and Weissella (Tonon and Lonvaud-Funel, 2002; Fernández and Zúñiga, 2006; Rimaux et al., 2011). The system involves the three enzymes arginine deiminase (ADI), ornithine transcarbamylase (OTC), carbamate kinase (CK) and a transmembrane arginine/ornithine antiporter, which exchanges extracellular arginine against intracellular ornithine. While ADI and OTC were present in both, the genome and proteome of L. nagelii TMW 1.1827, CK and the arginine/ornithine antiporter were only detectable in the genome. Thus, it should be unable to convert carbamoyl-P to generate additional ATP in co-culture. In the energy rich environment of water kefir, this does not appear to be a disadvantage. Therefore, the fate of carbamoyl-P remains unclear. However, only OTC was significantly up-regulated in L. nagelii in co-culture with S. cerevisiae. This reaction may occur in both directions yielding citrulline or ornithine and carbamoyl-phosphate. L. nagelii did not encode any complete alternative acid tolerance systems, e.g., the agmatine deiminase (AGDI) system or the glutamate decarboxylase (GAD) system. Except for neutralization upon ammonia formation via the ADI system, acidification appears limited by the switch from lactic and acetic acid production to ethanol formation, when L. nagelii and S. cerevisiae were co-cultivated.
In contrast, all respective enzymes of L. hordei involved in ADI pathway were up-regulated in co-culture with the yeast. Although the fate of carbamoyl-phosphate and other incidental compounds remains unclear, L. hordei likely produces ammonia upon arginine hydrolysis to protect itself from pH stress by alkalization of its cytoplasm and proximal environment. Consequently, only L. hordei should reduce the acid stress for the yeast (Xu et al., 2019a).
Another limit in the water kefir environment is the limited availability of fatty acids. L. nagelii TMW 1.1827 appears to be deficient in FabB, which is a well studied 3-ketoacyl-ACP synthase for catalyzing the elongation reaction of fatty acid synthesis (Feng and Cronan, 2009), and additionally in FabA, which is hydroxyldecanoyl-ACP dehydratase/isomerase for the production of unsaturated fatty acids by many bacteria (Magnuson et al., 1993; Cronan and Rock, 1996). As demonstrated by Wang and Cronan (2004), FabF can functionally replace FabB, while FabZ adopts the function of FabA. It was also reported that expression of Lactococcus lactis FabF can functionally replace both FabB and FabF in E. coli (Morgan-Kiss and Cronan, 2008). Due to low sequence homologies, those enzymatic bi-functionalities are not predictable by genome analysis. Since both microorganisms grew to high cell densities in water kefir medium without any external fatty acids, those findings might also indicate the existence of other functional homologs for FabB and FabA in L. nagelii. Co-cultivation with S. cerevisiae does not alter the expression of any proteins involved in the fatty acid metabolism in both LAB (Supplementary Table S3). This indicates, that the beneficial effects of S. cerevisiae do not reside in a bilateral supply with unsaturated fatty acids. This situation resembles the one in L. hordei, which only lacks FabA but also should express functionally complementary alternatives (Xu et al., 2019a).
Moreover, there was a group of enzymes of L. nagelii, which showed decreased expression in response to the co-cultivation with S. cerevisiae, which are involved in the biosynthesis of riboflavin (as shown in Supplementary Figure S3). Riboflavin synthase (EC 2.5.1.9), 6,7-dimethyl-8-ribityllumazine synthase (EC 2.5.1.78), 5-amino-6-(5-phosphoribosylamine) uracil reductase (EC 1.1.1.193), GTP cyclohydrolase II (EC 3.5.4.25) and 3,4-dihydroxy-2-butanone 4-phosphate synthase (EC 4.1.99.12) were down-regulated in a range from -3.2 to -4.0 log2 fold. Those enzymes connect the purine metabolism and pentose phosphate pathway to synthesize riboflavin. Since generally yeast produce group B vitamins (Emery et al., 1946; Zeidler et al., 2002), and riboflavin production by some lactobacilli (such as L. plantarum and L. fermentum) was inducible (Burgess et al., 2006; Arena et al., 2014; Russo et al., 2014), this may be an evidence for the feeding of riboflavin from S. cerevisiae to L. nagelii, supporting its growth and leading to a stable water kefir consortium.
The label-free quantitative approach represents a powerful tool for the identification and quantification of proteins to study the bacteria–yeast interaction of microorganisms involved in food fermentation processes (Behr et al., 2007; Siragusa et al., 2014; Maeda et al., 2015). It may even be used to explore more complex combinations or the complete water kefir system. However, with several (closely related) lactobacilli/yeasts in the system the sorting of proteins to species along sequence homologies will probably be limited because of sequence similarities across species. So in turn one would probably not be able to see the specific L. nagelii/L. hordei responses to S. cerevisiae any more, which are markedly different. So the reduction of the system offers also some advantage for a deeper understanding.
The predicted functional genome and the differentially expressed proteins in the presence of S. cerevisiae TMW 3.221 depicted the adaption of L. nagelii TMW 1.1827 to the water kefir consortium and environment, although protein regulations were less distinct than in L. hordei TMW 1.1822 (Xu et al., 2019b). Both microorganisms are highly efficient in degrading sucrose by an extracellular glucansucrase and subsequent fructose uptake, which may then enter EMP, PKP or mannitol metabolism. As already described for L. hordei, also L. nagelii appears to favor PKP over EMP, indicating a metabolic switch induced by an altered redox potential in the presence of S. cerevisiae. While L. nagelii remained widely un-affected in its citrate metabolism, the yeast stimulated L. hordei to use citrate as additional carbon source and therefore, promoting its growth.
Both LAB profit from glutamine secreted by the yeast, whereas L. hordei also takes advantage of the provided glutamate. While L. hordei up-regulated all of its enzymes involved in the reduction of acid stress via ADI pathway, L. nagelii only altered the expression of OTC. It was obvious, that both microorganisms reduced external acid stress by switching from lactate and acetate production to butanediol formation in the case of L. hordei and ethanol production in the case of L. nagelii.
At first glance, the fatty acid metabolism of both microorganisms appears to be impaired by the lack of one or more genes coding for key fatty acid biosynthesis enzymes. As it was already reported for other bacteria (Wang and Cronan, 2004), it is likely, that the functional role of those enzymes may be undertaken by other enzymes of the fatty acid biosynthesis gene cluster. This would explain, why both, L. hordei and L. nagelii, grew to high cell densities while facing an environment insufficient in unsaturated fatty acids. While S. cerevisiae TMW 3.221 modulated the protein expression of L. hordei TMW 1.1822 mainly in its carbohydrate metabolism, L. nagelii TMW 1.1827 seems to profit from secreted riboflavin. With respect to the establishment of a consortium maintaining physical proximity of lactobacilli and yeasts L. hordei appears to have a more prominent role as compared to L. nagelii as a result of its unique dextran causing yeast aggregation and proteins involved in adhesion functions.
The datasets generated for this study can be found in GenBank, CP018180–CP018183, and the ProteomeXchange via the PRIDE partner repository with the dataset identifier PXD012513.
DX conducted the wet lab experiments and performed the primary data analysis. CL helped with proteomic data analyses and deposition. JlB conducted detailed analysis and metabolic predictions. JgB supervised data analyses. DX and JlB wrote the first draft of manuscript. JgB and RV established general layout of experimental approach, supervised DX and JlB and did final discussions and shaping of the manuscript.
This study was supported by the China Scholarship Council in grant no. 201306820010, and the German Ministry of Economics and Technology (via AiF) and the WiFö (Wissenschaftsförderung der Deutschen Brauwirtschaft e.V., Berlin) in project AiF 19180 N.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00325/full#supplementary-material
FIGURE S1 | Genomic atlas of L. nagelii TMW 1.1827. Forward CDS (red), reverse CDS (blue), pseudogenes on both strands (black), tRNA and rRNA (dark green), % GC plot (yellow, high GC spike and green, low GC spike), GC skew [(G - C)/(G + C)] (gray).
FIGURE S2 | Overview of enzymatic activity of L. nagelii TMW 1.1827 in the complete metabolic and other pathways in presence of S. cerevisiae TMW 3.221: the nodes colored in bold red represent up-regulated, in bold blue represent down-regulated enzymes or proteins according to proteomic data, while nodes colored in thin red represent all the rest enzymes or proteins according to genomic annotation data presented in iPath 3.0.
FIGURE S3 | Overview of riboflavin metabolism of L. nagelii TMW 1.1827 generated in KEGG mapper. The EC numbers colored in red show up-regulated enzymes of L. nagelii in the presence of S. cerevisiae.
TABLE S1 | Annotated key features of the L. nagelii TMW 1.1827 genome.
TABLE S2 | List of annotated enzymes by NCBI and RAST involved in carbohydrate metabolism (glycolysis, pentose phosphate pathway, pyruvate metabolism, and TCA cycle).
TABLE S3 | List of enzymes involved in fatty acid biosynthesis.
Alikhan, N. F., Petty, N. K., Zakour, N. L. B., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Arena, M. P., Russo, P., Capozzi, V., López, P., Fiocco, D., and Spano G. (2014). Probiotic abilities of riboflavin-overproducing Lactobacillus strains: a novel promising application of probiotics. Appl. Microbiol. Biotechnol. 98, 7569–7581. doi: 10.1007/s00253-014-5837-x
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Behr, J., Geissler, A. J., Schmid, J., Zehe, A., and Vogel, R. F. (2016). The identification of novel diagnostic marker genes for the detection of beer spoiling Pediococcus damnosus strains using the BlAst diagnostic gene findEr. PLoS One 11:e0152747. doi: 10.1371/journal.pone.0152747
Behr, J., Israel, L., Gänzle, M. G., and Vogel, R. F. (2007). Proteomic approach for characterization of hop-inducible proteins in Lactobacillus brevis. Appl. Environ. Microbiol. 73, 3300–3306. doi: 10.1128/AEM.00124-127
Bernard, S. M., and Habash, D. Z. (2009). The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182, 608–620. doi: 10.1111/j.1469-8137.2009.02823.x
Burgess, C. M., Smid, E. J., Rutten, G., and van Sinderen, D. (2006). A general method for selection of riboflavin-overproducing food grade micro-organisms. Microb. Cell Fact. 5:24. doi: 10.1186/1475-2859-5-24
Carver, T., Thomson, N., Bleasby, A., Berriman, M., and Parkhill, J. (2008). DNAplotter: circular and linear interactive genome visualization. Bioinformatics 25, 119–120. doi: 10.1093/bioinformatics/btn578
Cox, J., Neuhauser, N., Michalski, A., Scheltema, R. A., Olsen, J. V., and Mann, M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. doi: 10.1021/pr101065j
Cronan, J. E., and Rock, C. O. (1996). Escherichia coli and Salmonella typhimurium. Cell. Mol. Biol. 1, 612–636.
Detmers, F. J., Kunji, E. R., Lanfermeijer, F. C., Poolman, B., and Konings, W. N. (1998). Kinetics and specificity of peptide uptake by the oligopeptide transport system of Lactococcus lactis. Biochemistry 37, 16671–16679. doi: 10.1021/bi981712t
Dincturk, H. B., Cunin, R., and Akce, H. (2011). Expression and functional analysis of glutamate synthase small subunit-like proteins from archaeon Pyrococcus horikoshii. Microbiol. Res. 166, 294–303. doi: 10.1016/j.micres.2010.03.006
Emery, W. B., McLeod, N., and Robinson, F. A. (1946). Comparative microbiological assays of members of the vitamin B complex in yeast and liver extracts. Biochem. J. 40, 426–432. doi: 10.1042/bj0400426
Fels, L., Jakob, F., Vogel, R. F., and Wefers, D. (2018). Structural characterization of the exopolysaccharides from water kefir. Carbohydr. Polym. 189, 296–303. doi: 10.1016/j.carbpol.2018.02.037
Feng, Y. J., and Cronan, J. E. (2009). Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J. Biol. Chem. 284, 29526–29535. doi: 10.1074/jbc.M109.023440
Fernández, M., and Zúñiga, M. (2006). Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 32, 155–183. doi: 10.1080/10408410600880643
Gardy, J. L., Laird, M. R., Chen, F., Rey, S., Walsh, C. J., Ester, M., et al. (2004). PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21, 617–623. doi: 10.1093/bioinformatics/bti057
Gulitz, A., Stadie, J., Wenning, M., Ehrmann, M. A., and Vogel, R. F. (2011). The microbial diversity of water kefir. Int. J. Food Microbiol. 151, 284–288. doi: 10.1016/j.ijfoodmicro.2011.09.016
Horisberger, M. (1969). Structure of the dextran of the tibi grain. Carbohydr. Res. 10, 379–385. doi: 10.1016/S0008-6215(00)80897-6
Kainulainen, V., Loimaranta, V., Pekkala, A., Edelman, S., Antikainen, J., Kylväjä, R., et al. (2012). Glutamine synthetase and glucose-6-phosphate isomerase are adhesive moonlighting proteins of Lactobacillus crispatus released by cathelicidin LL-37. J. Bacteriol. 194, 2509–2519. doi: 10.1128/JB.06704-11
Kleerebezem, M., Boekhorst, J., Van Kranenburg, R., Molenaar, D., Kuipers, O. P., Leer, R., et al. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. PNAS 100, 1990–1995. doi: 10.1073/pnas.0337704100
Kleerebezem, M., and Hugenholtz, J. (2003). Metabolic pathway engineering in lactic acid bacteria. Curr. Opin. Biotechnol. 14, 232–237. doi: 10.1016/S0958-1669(03)00033-8
Laureys, D., Aerts, M., Vandamme, P., and De Vuyst, L. (2018). Oxygen and diverse nutrients influence the water kefir fermentation process. Food Microbiol. 73, 351–361. doi: 10.1016/j.fm.2018.02.007
Laureys, D., and De Vuyst, L. (2014). Microbial species diversity, community dynamics, and metabolite kinetics of water kefir fermentation. Appl. Environ. Microbiol. 80, 2564–2572. doi: 10.1128/AEM.03978-13
Letort, C., Nardi, M., Garault, P., Monnet, V., and Juillard, V. (2002). Casein utilization by Streptococcus thermophilus results in a diauxic growth in milk. Appl. Environ. Microbiol. 68, 3162–3165. doi: 10.1128/AEM.68.6.3162-3165.2002
Maeda, K., Nagata, H., Ojima, M., and Amano, A. (2015). Proteomic and transcriptional analysis of interaction between oral microbiota Porphyromonas gingivalis and Streptococcus oralis. J. Proteome Res. 14, 82–94. doi: 10.1021/pr500848e
Magnuson, K., Jackowski, S., and Rock, C. O. (1993). Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57, 522–542.
Marsh, A. J., O’sullivan, O., Hill, C., Ross, R. P., and Cotter, P. D. (2013). Sequence-based analysis of the microbial composition of water kefir from multiple sources. FEMS Microbiol. Lett. 348, 79–85. doi: 10.1111/1574-6968.12248
Martínez-Torres, A., Gutiérrez-Ambrocio, S., Heredia-Del-Orbe, P., Villa-Tanaca, L., and Hernández-Rodríguez, C. (2017). Inferring the role of microorganisms in water kefir fermentations. Int. J. Food Sci. Technol. 52, 559–571. doi: 10.1111/ijfs.13312
Morgan-Kiss, R. M., and Cronan, J. E. (2008). The Lactococcus lactis FabF fatty acid synthetic enzyme can functionally replace both the FabB and FabF proteins of Escherichia coli and the FabH protein of Lactococcus lactis. Arch. Microbiol. 190, 427–437. doi: 10.1007/s00203-008-0390-6
Nepomuceno, R., Tavares, M., Lemos, J., Griswold, A., Ribeiro, J., Balan, A., et al. (2007). The oligopeptide (opp) gene cluster of Streptococcus mutans: identification, prevalence, and characterization. Oral Microbiol. Immunol. 22, 277–284. doi: 10.1111/j.1399-302X.2007.00368.x
Neve, H., and Heller, K. J. (2002). The microflora of water kefir: a glance by scanning electron microscopy. Kiel Milchwirtsch Forschungsber 54, 337–349.
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2013). The seed and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pidoux, M. (1989). The microbial flora of sugary kefir grain (the gingerbeer plant): biosynthesis of the grain from Lactobacillus hilgardii producing a polysaccharide gel. MIRCEN J. Appl. Microbiol. Biotechnol. 5, 223–238. doi: 10.1007/BF01741847
Pidoux, M., Brillouet, J. M., and Quemener, B. (1988). Characterization of the polysaccharides from a Lactobacillus brevis and from sugary kefir grains. Biotechnol. Lett. 10, 415–420. doi: 10.1007/BF01087442
Pidoux, M., De Ruiter, G. A., Brooker, B. E., Colquhoun, I. J., and Morris, V. J. (1990). Microscopic and chemical studies of a gelling polysaccharide from Lactobacillus hilgardii. Carbohydr. Polym. 13, 351–362. doi: 10.1016/0144-8617(90)90035-Q
Rimaux, T., Vrancken, G., Pothakos, V., Maes, D., De Vuyst, L., and Leroy, F. (2011). The kinetics of the arginine deiminase pathway in the meat starter culture Lactobacillus sakei CTC 494 are pH-dependent. Food Microbiol. 28, 597–604. doi: 10.1016/j.fm.2010.11.016
Russo, P., Capozzi, V., Arena, M. P., Spadaccino, G., Dueñas, M. T., López, P., et al. (2014). Riboflavin-overproducing strains of Lactobacillus fermentum for riboflavin-enriched bread. Appl. Microbiol. Biotechnol. 98, 3691–3700. doi: 10.1007/s00253-013-5484-7
Siragusa, S., De Angelis, M., Calasso, M., Campanella, D., Minervini, F., Di Cagno, R., et al. (2014). Fermentation and proteome profiles of Lactobacillus plantarum strains during growth under food-like conditions. J. Proteomics 96, 366–380. doi: 10.1016/j.jprot.2013.11.003
Smeianov, V. V., Wechter, P., Broadbent, J. R., Hughes, J. E., Rodríguez, B. T., Christensen, T. K., et al. (2007). Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 73, 2661–2672. doi: 10.1128/AEM.00005-07
Stadie, J., Gulitz, A., Ehrmann, M. A., and Vogel, R. F. (2013). Metabolic activity and symbiotic interactions of lactic acid bacteria and yeasts isolated from water kefir. Food Microbiol. 35, 92–98. doi: 10.1016/j.fm.2013.03.009
Stolz, P., Vogel, R. F., and Hammes, W. P. (1995). Utilization of electron acceptors by lactobacilli isolated from sourdough. Z. Lebensm. Unters. Forsch. 201, 402–410. doi: 10.1007/BF01193208
Sun, Z., Harris, H. M., Mccann, A., Guo, C., Argimón, S., Zhang, W., et al. (2015). Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 6:8322. doi: 10.1038/ncomms9322
Tonon, T., and Lonvaud-Funel, A. (2002). Arginine metabolism by wine Lactobacilli isolated from wine. Food Microbiol. 19, 451–461. doi: 10.1006/fmic.2002.0502
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., et al. (2016). The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 13, 731–740. doi: 10.1038/NMETH.3901
Tynkkynen, S., Buist, G., Kunji, E., Kok, J., Poolman, B., Venema, G., et al. (1993). Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J. Bacteriol. 175, 7523–7532. doi: 10.1128/jb.175.23.7523-7532.1993
Waldherr, F. W., Doll, V. M., Meißner, D., and Vogel, R. F. (2010). Identification and characterization of a glucan-producing enzyme from Lactobacillus hilgardii TMW 1.828 involved in granule formation of water kefir. Food Microbiol. 27, 672–678. doi: 10.1016/j.fm.2010.03.013
Wang, H., and Cronan, J. E. (2004). Functional replacement of the FabA and FabB proteins of Escherichia coli fatty acid synthesis by Enterococcus faecalis FabZ and FabF homologues. J. Biol. Chem. 279, 34489–34495. doi: 10.1074/jbc
Ward, H. M. (1891). The’ginger-beer plant,’and the organisms composing it: a contribution to the study of fermentation-yeasts and bacteria. Proc. R. Soc. Lond. 50, 261–265.
Xu, D., Bechtner, J., Behr, J., Eisenbach, L., Geißler, A. J., and Vogel, R. F. (2019a). Lifestyle of Lactobacillus hordei isolated from water kefir based on genomic, proteomic and physiological characterization. Int. J. Food. Microbiol. 290, 141–149. doi: 10.1016/j.ijfoodmicro.2018.10.004
Xu, D., Behr, J., Geißler, A. J., Bechtner, J., Ludwig, C., and Vogel, R. F. (2019b). Label-free quantitative proteomic analysis reveals the lifestyle of Lactobacillus hordei in the presence of Sacchromyces cerevisiae. Int. J. Food Microbiol. 294, 18–26. doi: 10.1016/j.ijfoodmicro.2019.01.010
Xu, D., Fels, L., Wefers, D., Behr, J., Jakob, F., and Vogel, R. F. (2018). Lactobacillus hordei dextrans induce Saccharomyces cerevisiae aggregation and network formation on hydrophilic surfaces. Int. J. Biol. Macromol. 115, 236–242. doi: 10.1016/j.ijbiomac.2018.04.068
Yamada, T., Letunic, I., Okuda, S., Kanehisa, M., and Bork, P. (2011). iPath2. 0: interactive pathway explorer. Nucleic Acids Res. 39, W412–W415. doi: 10.1093/nar/gkr313
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Keywords: Lactobacillus nagelii, Lactobacillus hordei, functional genome prediction, proteomic analysis, metabolism
Citation: Bechtner J, Xu D, Behr J, Ludwig C and Vogel RF (2019) Proteomic Analysis of Lactobacillus nagelii in the Presence of Saccharomyces cerevisiae Isolated From Water Kefir and Comparison With Lactobacillus hordei. Front. Microbiol. 10:325. doi: 10.3389/fmicb.2019.00325
Received: 19 December 2018; Accepted: 07 February 2019;
Published: 28 February 2019.
Edited by:
Giuseppe Spano, University of Foggia, ItalyReviewed by:
Analia Graciela Abraham, National University of La Plata, ArgentinaCopyright © 2019 Bechtner, Xu, Behr, Ludwig and Vogel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rudi F. Vogel, cnVkaS52b2dlbEB0dW0uZGU=; cnVkaS52b2dlbEB3encudHVtLmRl
†These authors have contributed equally to this work as joint first authors
‡Present address: Jürgen Behr, Leibniz-Institut für Lebensmittel-Systembiologie an der Technischen Universität München, Freising, Germany
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.