- 1Institute of Low Temperature Science, Hokkaido University, Sapporo, Japan
- 2Max Planck Institute for Terrestrial Microbiology, Marburg, Germany
- 3Research Faculty of Engineering, Hokkaido University, Sapporo, Japan
- 4Research Faculty of Agriculture, Hokkaido University, Sapporo, Japan
Even in the current era of metagenomics, the interpretation of nucleotide sequence data is primarily dependent on knowledge obtained from a limited number of microbes isolated in pure culture. Thus, it is of fundamental importance to expand the variety of strains available in pure culture, to make reliable connections between physiological characteristics and genomic information. In this study, two sulfur oxidizers that potentially represent two novel species were isolated and characterized. They were subjected to whole-genome sequencing together with 7 neutrophilic and chemolithoautotrophic sulfur-oxidizing bacteria. The genes for sulfur oxidation in the obtained genomes were identified and compared with those of isolated sulfur oxidizers in the classes Betaproteobacteria and Gammaproteobacteria. Although the combinations of these genes in the respective genomes are diverse, typical combinations corresponding to three types of core sulfur oxidation pathways were identified. Each pathway involves one of three specific sets of proteins, SoxCD, DsrABEFHCMKJOP, and HdrCBAHypHdrCB. All three core pathways contain the SoxXYZAB proteins, and a cytoplasmic sulfite oxidase encoded by soeABC is a conserved component in the core pathways lacking SoxCD. Phylogenetically close organisms share same core sulfur oxidation pathway, but a notable exception was observed in the family ‘Sulfuricellaceae’. In this family, some strains have either core pathway involving DsrABEFHCMKJOP or HdrCBAHypHdrCB, while others have both pathways. A proteomics analysis showed that proteins constituting the core pathways were produced at high levels. While hypothesized function of HdrCBAHypHdrCB is similar to that of Dsr system, both sets of proteins were detected with high relative abundances in the proteome of a strain possessing genes for these proteins. In addition to the genes for sulfur oxidation, those for arsenic metabolism were searched for in the sequenced genomes. As a result, two strains belonging to the families Thiobacillaceae and Sterolibacteriaceae were observed to harbor genes encoding ArxAB, a type of arsenite oxidase that has been identified in a limited number of bacteria. These findings were made with the newly obtained genomes, including those from 6 genera from which no genome sequence of an isolated organism was previously available. These genomes will serve as valuable references to interpret nucleotide sequences.
Introduction
Phylogenetically diverse bacteria have the capability of utilizing sulfur compounds as electron donors for respiration or phototrophic carbon fixation (Muyzer et al., 2013; Sorokin et al., 2013; Dahl, 2017). These sulfur-oxidizing bacteria have sulfur oxidation pathways consisting of various components (examples are shown in Figure 1), and distribution of the genes for sulfur oxidation has been investigated in diverse prokaryotic genomes (e.g., Meyer and Kuever, 2007; Meyer et al., 2007; Gregersen et al., 2011; Watanabe et al., 2014; Scott et al., 2018). These bacteria are also involved in the carbon and nitrogen cycles, playing crucial roles in natural environments (e.g., Mattes et al., 2013; Prokopenko et al., 2013; Herrmann et al., 2015; Dyksma et al., 2016; Lau et al., 2016). With their physiological functions, these bacteria have been intensively studied for applications such as water treatment, bioleaching and bioremediation (Pokorna and Zabranska, 2015; Lin et al., 2018). Recent advances in DNA sequencing technology have expanded our knowledge of uncultured microorganisms that are presumably oxidizing sulfur compounds (e.g., Kojima et al., 2015a; Mußmann et al., 2017; Tian et al., 2017; Hausmann et al., 2018). However, even in metagenomic studies, the interpretation of the resulting data is fundamentally dependent on knowledge obtained from studies on cultured organisms. In addition, pure culture-based experiments are also indispensable for verifying new concepts proposed by culture-independent studies. Because the majority of microorganisms remain unculturable, it is important to expand the variety of culturable isolates and to take full advantage of the available pure cultures.
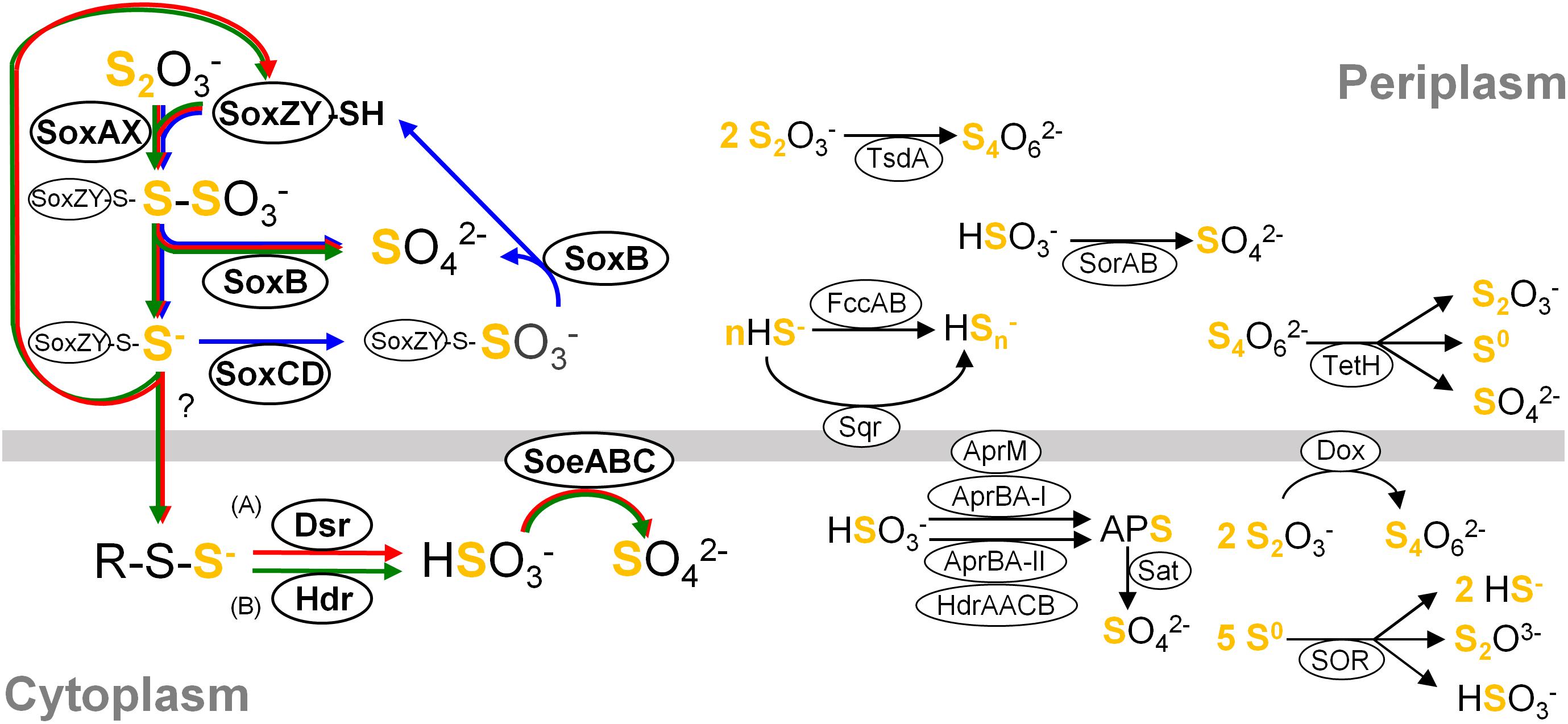
Figure 1. Simplified overview of major components constituting sulfur oxidation pathways in the classes Betaproteobacteria and Gammaproteobacteria. For clarity, reactions are not shown with exact stoichiometry. Pathways occurring in different organisms are summarized in this single picture, and a variety of pathways could consist of various combinations of these components connected by their reactants and products. Core pathways are highlighted with arrows in blue (cSox), red (Sox-Dsr-Soe) and green (Sox-Hdr-Soe), respectively. (A) The substrate for Dsr system is the persulfurated DsrC protein. (B) The substrate for HdrCBAHypHdrCB was postulated as persulfides (Koch and Dahl, 2018). Abbreviations are as follows: Sqr, sulfide:quinone oxidoreductase; Fcc, flavocytochrome c sulfide dehydrogenase; Sox, sulfur-oxidizing enzyme system; DoxDA, thiodulfate:quinone oxidoreductase; TetH, tetrathionate hydrolase; SOR, sulfur oxygenase reductase; Dsr, dissimilatory sulfite reductase; Apr, dissimilatory adenylylsulfate reductase; Hdr, heterodisulfide reductase; Sat, sulfate adenylyltransferase; Soe, sulfite-oxidizing enzyme; Sor, sulfur dehydrogenase. Details of these components are described in references cited in “Distribution of genes for sulfur oxidation” in main text.
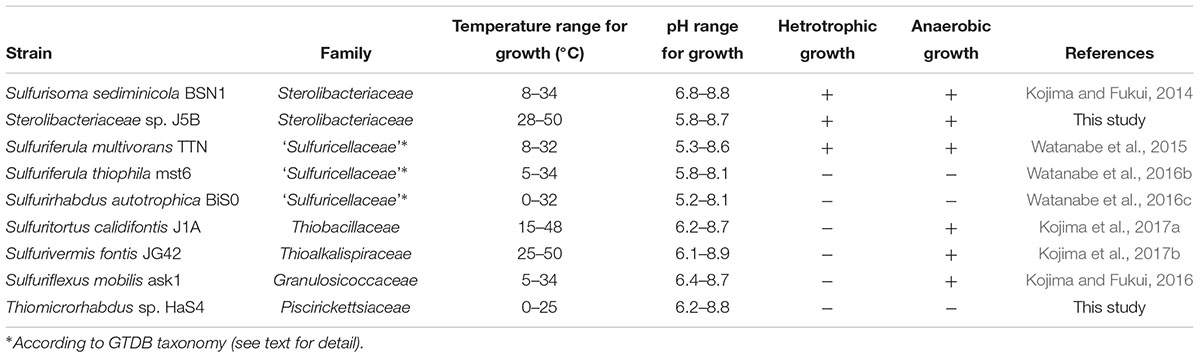
Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H are sulfur-oxidizing bacteria that were isolated from a same freshwater lake using the same defined medium (Kojima and Fukui, 2010, 2011). The isolation of these strains marked the beginning of a series of studies performed to obtain pure cultures of novel species of neutrophilic sulfur-oxidizing bacteria. In these studies, variations of the same medium were used with minor changes in composition to cultivate diverse sulfur oxidizers, primarily from freshwater environments. The use of this approach led to descriptions of 11 new species, which established 10 genera in the classes Betaproteobacteria and Gammaproteobacteria (Kojima and Fukui, 2010, 2011, 2014, 2016; Kojima et al., 2015b, 2016, 2017a,b; Watanabe et al., 2015, 2016b,c). The description of these genera resulted in some proposals for reclassification at higher taxonomic levels (Watanabe et al., 2014, 2015; Kojima et al., 2015b, 2017b). Among these sulfur oxidizers, complete genome sequences have been reported for the type strains of four species, Sulfuricella denitrificans, Sulfuritalea hydrogenivorans, Sulfurifustis variabilis, and Sulfuricaulis limicola (Watanabe et al., 2014; Umezawa et al., 2016).
In addition to the impact made on taxonomy, the isolation and characterization of these sulfur-oxidizing bacteria have contributed to a better understanding of the structure and function of microbial communities. Their gene sequences have served as reliable references to interpret nucleotide sequences retrieved from environments. As reviewed and demonstrated in previous studies, 16S rRNA gene sequences of the genera Sulfuricella and Sulfuritalea have been detected in various natural and engineered freshwater environments (Watanabe et al., 2012, 2014, 2016a,c, 2017). The relatives of Sulfuricella and Sulfuritalea have also been detected based on detection of sequences related to other genes of these genera (Watanabe et al., 2013; Kojima et al., 2014; Herrmann et al., 2015, 2017; Lau et al., 2016; Kumar et al., 2017, 2018; Feng et al., 2018; Luo et al., 2018). This is also the case for the other genome-sequenced species of Sulfurifustis and Sulfuricaulis (Watanabe et al., 2016a; Herrmann et al., 2017; Kumar et al., 2018; Włodarczyk et al., 2018). Furthermore, detailed inspections of the genomes of Sulfuricella and Sulfuritalea revealed the presence of characteristic genes for arsenic metabolism, arxAB and arrAB (Watanabe et al., 2014, 2017). These findings motivated studies regarding the previously unrecognized functions of these organisms. Consequently, arsenate respiration by Sulfuritalea hydrogenivorans sk43H was demonstrated (Watanabe et al., 2017), and arxA gene sequences closely related to that of Sulfuricella denitrificans skB26 were detected from freshwater environments (Ospino et al., 2018). Genome sequences also provide a basis for proteomic analyses. Based on the genome sequence of Sulfuricella denitrificans skB26, expression pattern of proteins involved in sulfur oxidation was investigated in this approach (Watanabe et al., 2012).
As shown by the previous studies mentioned above, the genome sequences of isolated and characterized sulfur oxidizers provide a great deal of valuable information for microbiology and related fields. In this study, the genomes of the remaining 7 species were sequenced along with those of two newly isolated sulfur oxidizers. These organisms represent 8 genera in the classes Betaproteobacteria and Gammaproteobacteria, including 6 genera whose genomes have not been available until now. The obtained sequences were used for comparative genomics with other sulfur oxidizers to obtain insights into the mechanisms of their sulfur metabolism. Furthermore, the expression of some notable genes was confirmed in proteomic experiments.
Materials and Methods
Isolation and Characterization of Novel Sulfur-Oxidizing Bacteria
Sulfur-oxidizing bacteria capable of chemolithoautotrophic growth were newly isolated and characterized. One of the novel strains, strain HaS4, was isolated from the water of Lake Harutori (Kubo et al., 2014). The other strain, strain J5B, was isolated from Jozankei hot spring via an enrichment culture established in a previous study (Kojima et al., 2017a). Prior to the detailed characterization of these microbes, the phylogenetic positions of the isolates were identified by sequencing their PCR-amplified 16S rRNA genes. The physiological characterizations were made in consideration of the features of their close relatives that were identified by the phylogenetic analysis. More detailed procedures for the isolation and characterization of these strains are described in the Supplementary Material.
Genome Sequencing
The newly isolated strains were subjected to whole-genome sequencing, along with other strains that have been isolated and maintained in our laboratory (Table 1). Genomic DNA from each strain was extracted with a Wizard Genomic DNA Purification Kit (Promega). For sequencing, different methods were used depending on the strains, as described below and summarized in Table 2.
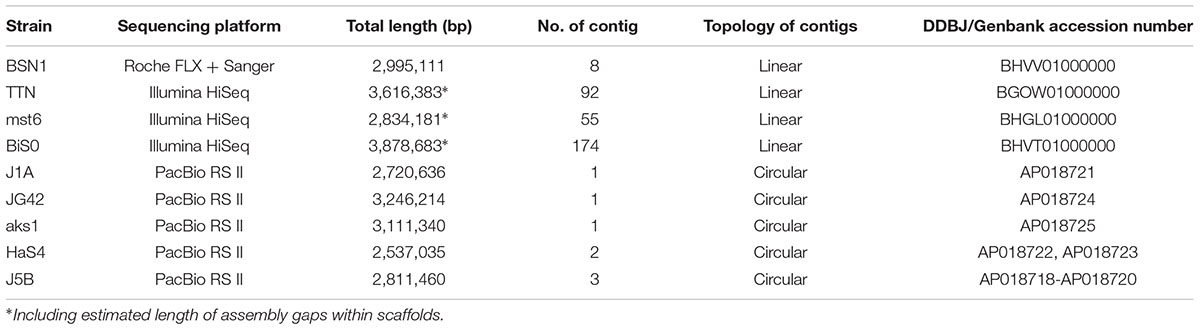
Table 2. General features of genome sequences obtained in this study (deposited under BioProject PRJDB7001).
The genome of Sulfurisoma sediminicola BSN1 was sequenced with the Genome Sequencer FLX System (Roche). The library preparation, FLX sequencing and subsequent PCR-based gap closing were performed as described previously (Watanabe et al., 2012). The genomes of three strains of the genera Sulfuriferula and Sulfurirhabdus were sequences using an Illumina HiSeq platform. These genera were formerly classified as members of the family ‘Sulfuricellaceae’ (Watanabe et al., 2014, 2015), which has subsequently been integrated with the family Gallionellaceae (Boden et al., 2017a). However, in this study, the term ‘Sulfuricellaceae’ is used to refer to the lineage consisting of these genera and Sulfuricella. The presence of the family ‘Sulfuricellaceae’ independent from Gallionellaceae is shown in the GTDB taxonomy1, which is based on extensive phylogenetic analysis using whole genomes (Parks et al., 2018). The genome sequences of Sulfuriferula and Sulfurirhabdus were obtained by paired-end sequencing, and the outputs of the Velvet assembler were directly used for further analyses. The genome sequences of the other strains were obtained using the PacBio RS II system with essentially the same methods as described previously (Umezawa et al., 2016). From the resulting linear contigs, circular contigs were manually constructed by connecting both ends of each linear contig based on duplicated sequences that appeared at the terminal regions.
Comparative Genome Analysis and the Identification of Genes for Sulfur Oxidation
For the comparative genome analysis, the genome sequences of sulfur oxidizers belonging to the classes Betaproteobacteria and Gammaproteobacteria were obtained from National Center for Biotechnology Information (as of June 2017). The organisms were selected for their ability for growth via the oxidation of inorganic sulfur compounds, as demonstrated by experiments with pure cultures. For the newly sequenced genomes, protein-coding sequences were identified using the RAST server (Aziz et al., 2008). In the newly and previously sequenced genomes, the genes encoding for proteins involved in sulfur oxidation (Sqr, FccAB, SoxXYZABCD, DoxDA, TsdA, TetH, Sor, DsrABEFHCMKJOP, AprBA, AprM, HdrAACB, Sat, HdrCBAHypHdrCB, SreABC, SoeABC, and SorAB) were identified based on sequence similarity, with the proteins listed in Supplementary Table S1 used as queries.
Proteomic Analysis
A proteomic analysis was performed for the strains Sulfuriferula thiophila mst6, Sulfurirhabdus autotrophica BiS0, and Sulfurifustis variabilis skN76. These strains were cultured in a bicarbonate-buffered medium that contained thiosulfate as the sole electron donor for chemolithoautotrophic growth. The growth of the strains was monitored by measuring the absorbance at 600 nm and by determining the concentrations of thiosulfate and sulfate. The cells were harvested by centrifugation and the proteins were extracted. The protein extracts were reduced with dithiothreitol, alkylated with iodoacetamide, and then digested with trypsin. The resulting peptide samples were analyzed by nanoscale liquid chromatography coupled to tandem mass spectrometry (nanoLC-MS/MS) using an Easy nLC1000 liquid chromatography system coupled to a Q-Exactive plus Orbitrap mass spectrometer (Thermo Fisher Scientific, IL, United States). The proteins were identified from the obtained mass spectra using Proteome Discoverer 2.0 (Thermo Fisher Scientific) and in-house databases constructed from the genomes of the respective strains. The abundance of proteins was estimated as the exponentially modified protein abundance index (emPAI) (Ishihama et al., 2005). For each strain, two sets of proteomic data were obtained from independent cultures, and the averaged emPAI values of the respective proteins were calculated from the two datasets. The detected proteins were sorted by the averaged emPAI value and were grouped into four categories based on the following ranking: within the top 2%, 2–10%, 10–30%, and below 30%. More detailed procedures are described in the Supplementary Material.
Results and Discussion
Characteristics of Newly Isolated Strains
Two novel sulfur oxidizers, designated as strains HaS4 and J5B, were obtained from lake water and a hot spring microbial mat, respectively. The 16S rRNA gene sequence analysis revealed that strain HaS4 belongs to the genus Thiomicrorhabdus in the class Gammaproteobacteria, but it is distinct from the existing species in this genus, with a sequence similarity lower than 96% (Figure 2). All known Thiomicrorhabdus species are obligately chemolithoautotrophic and oxidize inorganic sulfur compounds (Boden et al., 2017b,c). It was also revealed that strain J5B belongs to the family Sterolibacteriaceae in the class Betaproteobacteria (Figure 2) and potentially represents a novel genus, because none of previously described genera can accommodate this strain (Supplementary Figure S1). The family Sterolibacteriaceae was defined on the basis of 16S rRNA gene sequences and currently consists of 6 monospecific genera (Boden et al., 2017a). Among these genera, Sulfuritalea and Sulfurisoma have been described as facultatively autotrophic sulfur oxidizers which utilize some organic acids.
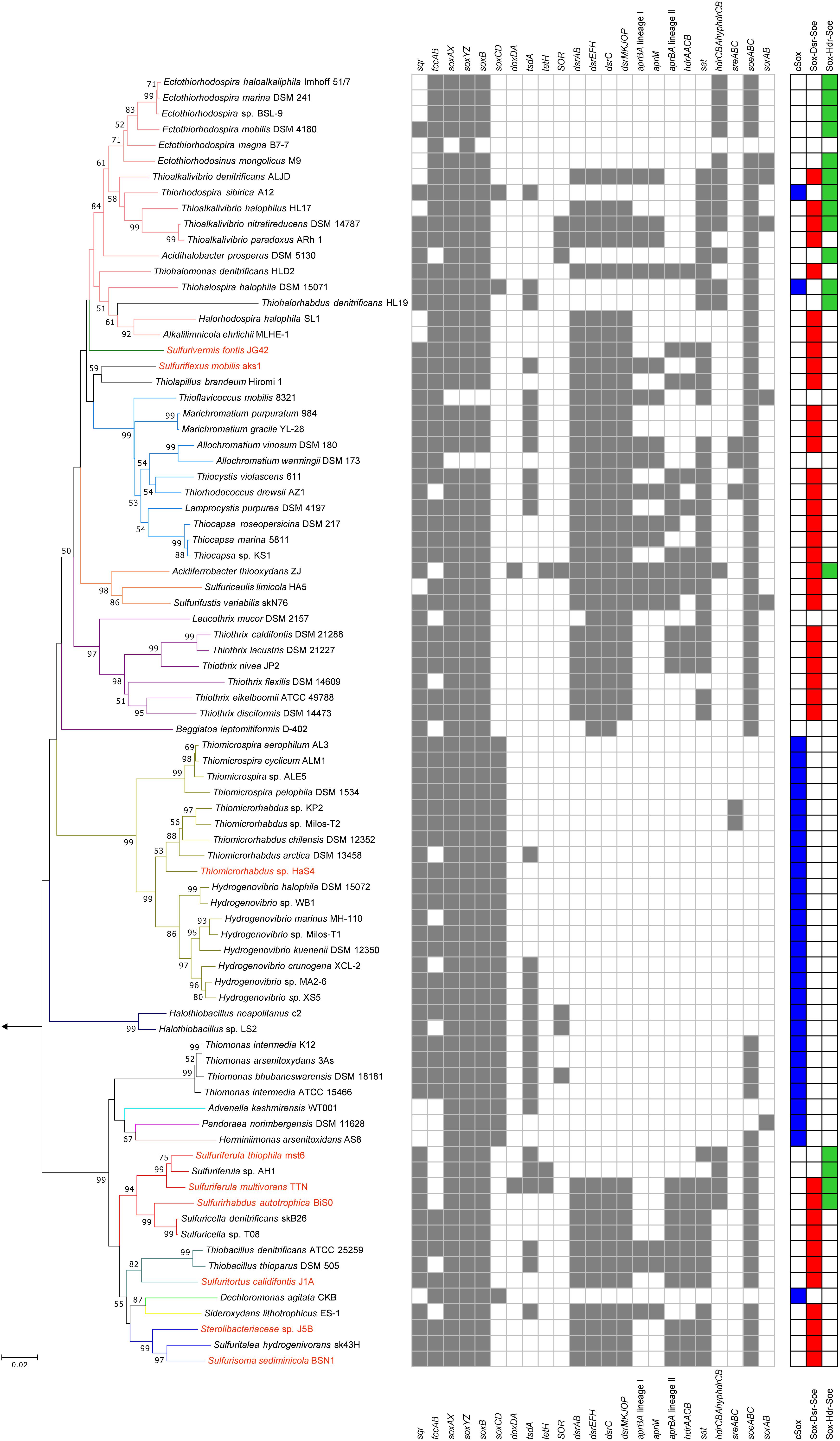
Figure 2. Phylogeny of sulfur-oxidizing bacteria and distribution of genes for sulfur oxidation. Phylogenetic tree was constructed by neighbor-joining method with 16S rRNA gene sequences aligned by ClustalW. The number of final comparable positions was 1224. Bootstrap values (50% >) from 1,000 replicates are shown next to branches. Branches are colorized on the basis of the family-level taxonomy. Strains shown in red are organisms whose genomes were sequenced in this study. The tree was rooted with the 16S rRNA sequence of Sulfurimonas autotrophica OK10. Three right-most columns represent distribution of the core sulfur oxidation pathways.
Strain HaS4 grew at a temperature range of 0–25°C, with optimum growth observed at 22°C. The growth of this bacterium was observed at a pH range of 6.2–8.8, with an optimum range of 6.6–7.4. Strain HaS4 grew chemolithoautotrophically on thiosulfate, tetrathionate, elemental sulfur and sulfide, but not on hydrogen gas. The following organic substrates did not support growth of the strain: lactate, acetate, formate, fumarate, glucose, maltose, fructose, N-acetyl-D-glucosamine, sucrose, and cellobiose.
Strain J5B grew at temperature range of 28–55°C, with optimum growth observed at 45–50°C. The growth of this strain was observed at a pH range of 5.8–8.7, with an optimum range of 6.7–7.4. Under nitrate-reducing conditions, strain J5B grew chemolithoautotrophically on thiosulfate and elemental sulfur, but not on sulfide, tetrathionate, or hydrogen gas. Strain J5B grew anaerobically on the following organic substrates in the presence of nitrate: pyruvate, lactate, acetate, propionate, succinate, fumarate, malate, and butyrate. The following substrates could not support anaerobic growth of strain J5B: benzoate, isobutyrate, methanol, ethanol, formate, citrate, glucose, xylose, phenol, o-cresol, and m-cresol.
Sequencing and Assembly of Genomes
The basic characteristics of the newly sequenced genomes are summarized in Table 2. The draft genome sequence of Sulfurisoma sediminicola BSN1 was obtained with a combination of GS FLX and Sanger sequencing for gap closing. By closing 46 gaps within the scaffolds, 8 contigs were finally obtained. For the Sulfuriferula species strains, draft genome sequences were obtained using a HiSeq platform. By assembling paired-end reads, 87 and 48 scaffolds were constructed for the strains TTN and mst6, respectively. Some of these scaffolds are segmented into two or more contigs separated by unclosed gaps, the lengths of which were estimated. Similarly, 178 contigs in 83 scaffolds were assembled for Sulfurirhabdus autotrophica BiS0. For the other 5 strains analyzed with PacBio, single circular chromosomes were successfully constructed. Additionally, small circular contigs were obtained for strains J5B and HaS4, suggesting the presence of plasmids in these strains. These results indicated the advantage of using the long-reads produced by PacBio sequencing to obtain complete genome sequences.
Distribution of Genes for Sulfur Oxidation
The genes for sulfur oxidation identified in the newly sequenced genomes were compared with those of other sulfur oxidizers, and summarized in Figure 2. For the comparative analysis, the genomes of 73 strains isolated in pure culture were obtained as verifiable references, and genes encoding the following proteins were identified in the genomes: Sqr (Griesbeck et al., 2002; Marcia et al., 2009), FccAB (Chen et al., 1994), SoxXZYABCD (Friedrich et al., 2000, 2001), DsrAB (Pott and Dahl, 1998), DsrEFH (Stockdreher et al., 2012), DsrC (Stockdreher et al., 2012), DsrMKJOP (Grein et al., 2010), HdrCBAHypHdrCB (Cao et al., 2018; Koch and Dahl, 2018), SoeABC (Dahl et al., 2013), SorAB (Kappler et al., 2000; Kappler and Bailey, 2005), AprBA (Lyric and Suzuki, 1970; Fritz et al., 2002), AprM (Pires et al., 2003; Meyer and Kuever, 2007; Parey et al., 2013), HdrAACB/QmoABHdrCB (Meyer and Kuever, 2007; Ramos et al., 2012; Watanabe et al., 2014), TsdA (Denkmann et al., 2012; Brito et al., 2015), SOR (Kletzin, 1989, 1992), TetH (De Jong et al., 1997), DoxDA (Müller et al., 2004), and SreABC (Laska et al., 2003). Except for the SreABC, functions of these proteins in sulfur oxidation have been examined in the previous studies listed above. The involvement of SreABC in the reverse reaction of persulfide reductase was suggested in a green sulfur bacterium but has not yet been examined (Eddie and Hanson, 2013).
Core Sulfur Oxidation Pathways
Although combinations of sulfur oxidation genes are highly diverse among the genomes, typical combinations were identified as a genetic basis for three types of core sulfur oxidation pathways that consisted of the following different sets of enzymes: (1) the “cSox” pathway, with SoxXYZABCD; (2) the “Sox-Dsr-Soe” pathway, with SoxXYZAB, DsrABEFHCMKJOP, and SoeABC; and (3) the “Sox-Hdr-Soe” pathway, with SoxXYZAB, HdrCBAHypHdrCB, and SoeABC (Figures 1, 2). All these core pathways contain SoxXYZAB, and their structural genes were the most commonly observed in the sulfur-oxidizing bacteria analyzed. Wide distribution of soxXYZAB has also been shown in a previous study (Meyer et al., 2007). SoxAX catalyzes oxidative conjugation of thiosulfate to cysteine residue on SoxY of the SoxYZ complex, and the sulfonate group is removed by hydrolysis via SoxB with the generation of sulfate and SoxYZ with the sulfane sulfur (Friedrich et al., 2000, 2001; Bamford et al., 2002; Sauvé et al., 2007, 2009). It was also reported that a mixture of the purified SoxAX, SoxYZ, and SoxB enzymes catalyzes sulfite oxidation (Friedrich et al., 2000). Such functional versatility of Sox complex may be related to its wide distribution.
One of the strains isolated in this study, strain HaS4, possesses the cSox pathway consisting of SoxXYZABCD (Figure 2). This pathway is completely conserved in sulfur oxidizers of the family Piscirickettsiaceae. Genomic features of these sulfur oxidizers were closely inspected in a recent study, which revealed prevalence of the SoxCD among them (Scott et al., 2018). It also pointed out that genes encoding SoeABC are absent from their genomes. The comparative analysis in the present study revealed that sulfur oxidizers with the cSox pathway generally lack SoeABC (Figure 2), one of the most common sulfite-oxidizing enzyme complexes in the analyzed strains. The SoxCD complex oxidizes the sulfane sulfur of SoxZY-Cys-S- derived from the SoxXYZAB reaction using water molecules to yield the sulfonate group as SoxZY-Cys-SO3- (Quentmeier et al., 2000; Zander et al., 2010), which is further hydrolyzed to free sulfate ion by SoxB. In a revised model of Sox reaction recently proposed, sulfur carrier for the SoxCD reaction is SoxZY-Cys-S(n)-S- rather than SoxZY-Cys-S- (Grabarczyk and Berks, 2017). The cytoplasmic sulfite oxidase SoeABC may not be important for organisms with SoxCD because SoxXYZABCD completely oxidizes thiosulfate to sulfate in the periplasm (cSox means “complete Sox system”). Interestingly, it turned out that Thiorhodospira sibirica A12 and Thiohalospira halophila DSM 15071 have soeABC in addition to soxCD (Figure 2). These sulfur oxidizers also possess the hdrCBAhyphdrCB gene cluster, which is always accompanied by soeABC, as discussed below.
The Sox-Dsr-Soe pathway is the most common core sulfur oxidation pathway in the newly sequenced genomes from this study (Figure 2). In this pathway, sulfane sulfur derived from thiosulfate via SoxXYZAB is transported to the cytoplasm presumably in the form of persulfides, as suggested by a phototrophic sulfur oxidizer (Frigaard and Dahl, 2008). Persulfide sulfur is then transferred to DsrC via Rhd, TusA, and DsrEFH (Cort et al., 2008; Stockdreher et al., 2012, 2014). The persulfurated form of DsrC is considered to be the substrate for DsrAB, as shown by the crystal structure of the DsrABC complex from Desulfovibrio vulgaris (Oliveira et al., 2008). The siroheme-containing cytoplasmic enzyme DsrAB is involved in the reverse reaction of reduction of sulfite to sulfide as demonstrated in sulfate-reducing prokaryotes (Schedel et al., 1979; Pott and Dahl, 1998); generates sulfite and disulfide bond in DsrC, which is reduced to free thiols by the DsrMKJOP transmembrane complex for restart of the sulfur relay system (Dahl et al., 2005; Pires et al., 2006; Grein et al., 2010). The resulting sulfite is oxidized to sulfate by the cytoplasmic enzyme complex SoeABC (Dahl et al., 2013). In this study, it was found that the genes encoding the SoeABC complex are completely conserved in the dsr-positive sulfur oxidizers (Figure 2). The reactions catalyzed by Dsr and SoeABC both occur in the cytoplasm (Figure 1), and this colocalization may be one of the reasons for the coexistence of the genes encoding Dsr and SoeABC in the genomes. In theory, sulfite generated by DsrAB can be oxidized by Sox proteins (Friedrich et al., 2000), but transport of the sulfite across the cytoplasmic membrane would be required in this case since the Sox reaction occurs in the periplasm (Figure 1).
The other core sulfur oxidation pathway is Sox-Hdr-Soe. This pathway essentially consists of the proteins of the Sox-Dsr-Soe pathway, but Dsr proteins are replaced with Hdr proteins that are encoded in the gene cluster hdrCBAhyphdrCB (Figure 1). Recently, the role of these Hdr proteins in sulfur oxidation was genetically investigated with Hyphomicrobium denitrificans, a bacterium which degrades dimethylsulfide (DMS) (Koch and Dahl, 2018). The mutagenesis of hdr genes resulted in loss of ability to metabolize DMS and a lower rate of the sulfate formation from thiosulfate than wild type under chemoorganoheterotrophic growth conditions. Although the reaction catalyzed by HdrCBAHypHdrCB has not yet been revealed, involvement of these proteins in the sulfite generation from persulfides was proposed based on sequence similarities of hdr genes with archaeal counterparts along with mutagenesis experiments. In thiosulfate metabolism, HdrCBAHypHdrCB might functionally substitute for Dsr system as shown in Figure 1. As is the case with the dsr genes, all the analyzed genomes with hdrCBAhyphdrCB also harbor soeABC genes.
The majority of the strains analyzed have one of the three core pathways for sulfur oxidation, although some exceptions were noted, as described below. Among the 82 strains included in the comparative analysis (shown in Figure 2), three strains lack some or all of the genes encoding SoxXYZAB, the common component of the three core pathways. The strains Ectothiorhodospira magna B7-7, Thioflavicoccus mobilis 8321, and Allochromatium warmingii DSM173 are sulfur oxidizers that utilize sulfur compounds other than thiosulfate. Leucothrix mucor DSM2157 and Beggiatoa leptomitiformis D-402 harbor the genes encoding SoxXYZAB and SoeABC but lack those encoding conserved proteins in the core pathways. In contrast to these 5 strains lacking components for the core pathways, some other strains were observed to have full gene sets for two core pathways. It has been noted that the dsr and hdrCBAhyphdrCB genes are almost exclusive to each other, but a small number of organisms have both genes in their genomes (Koch and Dahl, 2018). This notable overlap was observed in the genomes of Sulfurirhabdus autotrophica BiS0 and Sulfuriferula multivorans TTN, which were sequenced in this study. In addition, a recent study revealed that three Acidiferrobacter strains have dsr and hdrCBAhyphdrCB along with soeABC (Issotta et al., 2018). These findings indicate that these strains have two core pathways, Sox-Dsr-Soe and Sox-Hdr-Soe. As mentioned above, Thiorhodospira sibirica A12 and Thiohalospira halophila DSM 15071 harbor both soxCD and hdrCBAhyphdrCB, representing organisms with cSox and Sox-Hdr-Soe pathways. In this study, no organism possessing both the cSox and Sox-Dsr-Soe pathways was identified. The soxCD and dsr genes are well known to be mutually exclusive (Meyer and Kuever, 2007; Frigaard and Dahl, 2008; Gregersen et al., 2011; Lenk et al., 2012).
Variations in Sulfur Oxidation Pathways Within the Family ‘Sulfuricellaceae’
The patterns for the occurrence of the core sulfur oxidation pathways were generally consistent with the 16S rRNA gene-based phylogeny (Figure 2). In other words, phylogenetically close organisms share a similar genetic basis for sulfur oxidation. However, there were some notable deviations from this tendency. In particular, considerable variations were observed among the genomes of ‘Sulfuricellaceae’ strains. In this family, 6 genomes of isolated strains were available for the analysis, including three genomes obtained in this study. Sulfuriferula thiophila mst6 and Sulfuriferula sp. AH1 lack the dsr operon, which is present in all the other members of the family. In contrast, two Sulfuricella strains lack the hdrCBAhyphdrCB cluster, which all the other strains possess. Consequently, 6 ‘Sulfuricellaceae’ strains were classified into three types, two strains with the Sox-Dsr-Soe pathway, two strains with the Sox-Hdr-Soe pathway, and two strains with both pathways. A close inspection of the genomes of these bacteria revealed that the hdrCBAhyphdrCB cluster is located between genomic regions that are conserved among these strains, and these conserved regions are directly adjacent to each other in the genomes of Sulfuricella strains lacking this gene cluster (Figure 3). This observation may indicate that a common ancestor of ‘Sulfuricellaceae’ had hdrCBAhyphdrCB and that Sulfuricella strains lost this cluster during evolution. The coexistence of the hdrCBAhyphdrCB and dsr genes in Sulfuriferula multivorans TTN and Sulfurirhabdus autotrophica BiS0 may represent a transition state, and one of them may be selected from the genomes in the future.
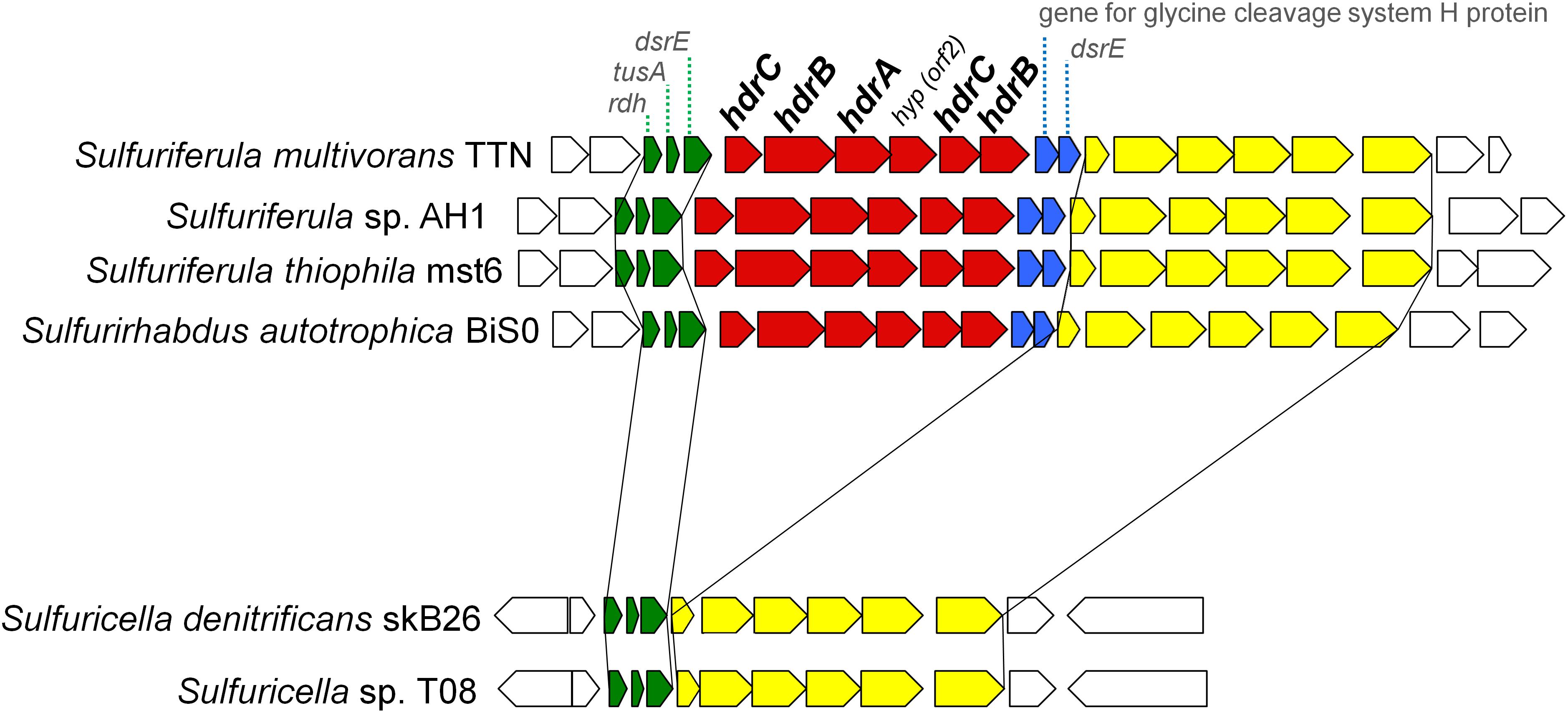
Figure 3. Gene arrangement around hdrCBAhyphdrCB gene cluster in the genomes of ‘Sulfuricellaceae’. Genes of hdrCBAhyphdrCB cluster are shown in red. Genes shown in blue are conserved along with the hdr genes in the ‘Sulfuricellaceae’. Genes shown in green and yellow are conserved in the all ‘Sulfuricellaceae’ genomes in the same order and direction.
Another example of differences observed among organisms in this family is that Sulfuriferula multivorans TTN has doxDA gene encoding a membrane-bound thiosulfate:quinone oxidoreductase, which couples oxidation of thiosulfate to reduction of quinone (Müller et al., 2004). DoxDA is one of the most minor proteins involved in sulfur oxidation in the reference strains (Figure 2). The gene encoding DoxDA was only identified in the acidophilic gammaproteobacteria Acidihalobacter ferrooxidans and Acidiferrobacter thiooxidans (Valdés et al., 2008; Quatrini et al., 2009; Issotta et al., 2018).
Expression of Proteins Involved in Sulfur Oxidation
The expression of the proteins required for sulfur oxidation was investigated for the whole proteomes of three sulfur oxidizers with different sets of the core pathways, including Sulfuriferula thiophila mst6 with Sox-Hdr-Soe, Sulfurifustis variabilis skN76 with Sox-Dsr-Soe, and Sulfurirhabdus autotrophica BiS0 with both. In the proteomes of the strains grown on thiosulfate, proteins corresponding 41–48% of the total protein-coding genes predicted in the corresponding genomes were detected. In all the strains, the summed abundance of the top 10% most abundant proteins represented more than 90% of the total protein abundance (Supplementary Figure S2). As shown in Figure 4, almost all the proteins involved in sulfur oxidation were detected in the proteomes. Majority of the Sox, Dsr, and Hdr proteins were among the top 10% most abundant proteins. In addition to these major components of the core pathways, some additional proteins were also detected among the most abundant proteins in strain skN76.
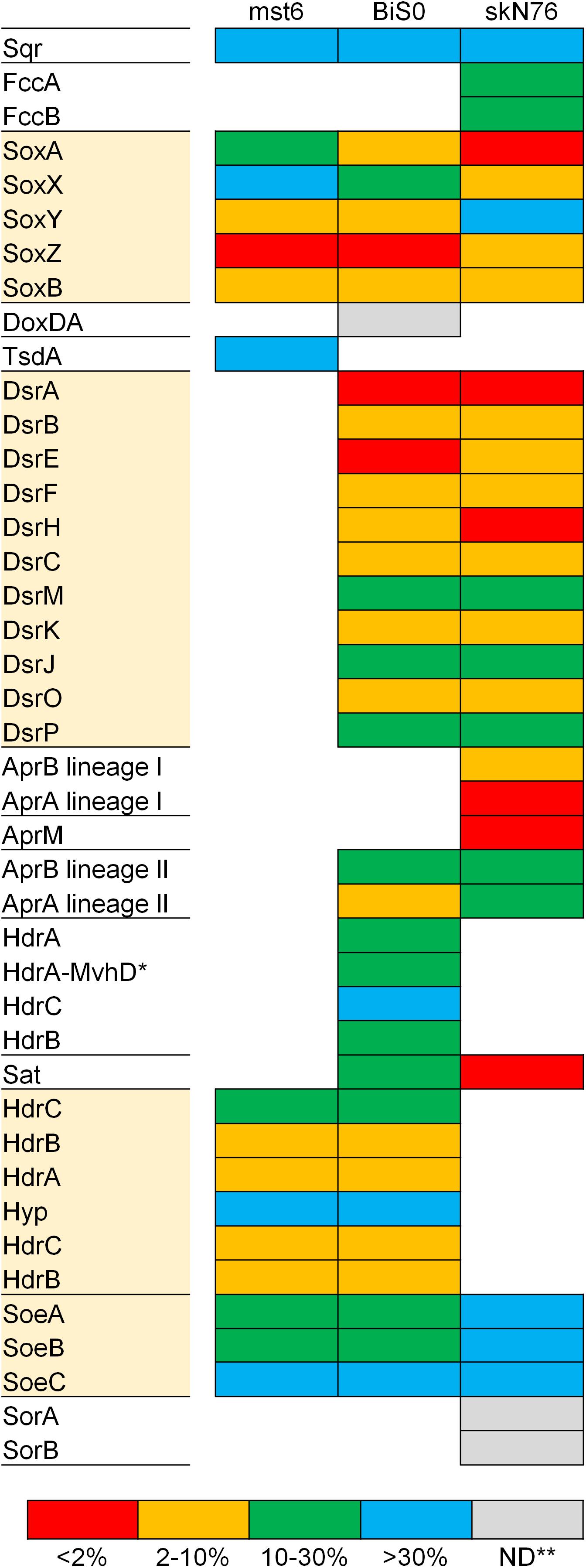
Figure 4. Expression levels of proteins for sulfur oxidation estimated with proteomic analysis of Sulfuriferula thiophila mst6, Sulfurirhabdus autotrophica BiS0 and Sulfurifustis variabilis skN76 grown on thiosulfate. The detected proteins were grouped into four categories based on ranking of abundance (see text for detail). Proteins highlighted with shaded boxes are components of the core sulfur oxidation pathway. ∗HdrA-MvhD, HdrA fused with MvhD; ∗∗ND, not detected.
Compared to the Sox and Dsr proteins, evidence for the essential contribution of the Hdr proteins encoded by hdrCBAhyphdrCB in sulfur oxidation had been limited. The involvement of this gene cluster in sulfur oxidation was previously investigated by transcriptomic approaches using Acidithiobacillus strains (Quatrini et al., 2009; Chen et al., 2012). In the transcriptomes of these strains fed elemental sulfur or tetrathionate, the transcripts from this gene cluster were detected except for that of orf2 (referred to as hyp in this study) located between hdrCBA and hdrCB. A recent study proposed the involvement of HdrCBAHypHdrCB in generation of sulfate from thiosulfate via the sulfite formation from persulfides (Koch and Dahl, 2018). In the present study, the products of this gene cluster were detected in Sulfuriferula thiophila mst6 and Sulfurirhabdus autotrophica BiS0 grown on thiosulfate. These results support the involvement of these proteins in thiosulfate-dependent chemolithoautotrophic growth.
Sulfurirhabdus autotrophica BiS0 has both the Sox-Dsr-Soe and Sox-Hdr-Soe pathways. A previous study of Hyphomicrobium suggested that HdrCBAHypHdrCB catalyzes the oxidation of protein-bound persulfide sulfur to generate sulfite and that this reaction can functionally substitute for that catalyzed by the Dsr proteins (Koch and Dahl, 2018). In the Sulfurirhabdus autotrophica BiS0 proteome, the products of both the dsr and hdrCBAhyphdrCB operons were detected at a relatively high abundance (Figure 4). These results suggest that strain BiS0 uses both systems to generate sulfite. The biological importance of the apparent simultaneous expression of these proteins is currently unclear. This subject may be an important key to understanding the evolution of sulfur-oxidizing systems, because many sulfur oxidizers have two or more genes performing the same physiological function. Such functional overlap was prominently observed in the genome and proteome of Sulfurifustis variabilis skN76. This strain has two copies each of dsrAB, aprBA and fccAB genes (Umezawa et al., 2016). In its proteome, encoded products of these genes were all detected (Figure 4).
Genes for Arsenite Oxidation and Respiratory Arsenate Reduction
Some prokaryotes, including sulfur oxidizers, are known to have genes encoding either of the two types of arsenite oxidases, Aio or Arx (van Lis et al., 2013). The arxAB genes, which encode catalytic units of the Arx, have been identified in a limited number of bacteria that primarily belong to the class Gammaproteobacteria (Zargar et al., 2012; Ospino et al., 2018). In this study, the genomes of Sulfuritortus calidifontis J1A and strain J5B were observed to harbor the arxAB genes. Among sulfur oxidizers, the arxAB genes have been only identified in members of the family Ectothiorhodospiraceae isolated from salty alkaline environments. As an exception, the genes were identified in a plasmid from Sulfuricella denitrificans skB26, which is a neutrophilic betaproteobacterium belonging to the family ‘Sulfuricellaceae’ (Watanabe et al., 2014). The strains J1A and J5B are neutrophilic betaproteobacteria isolated from a same microbial mat, but they belong to the families Thiobacillaceae and Sterolibacteriaceae, respectively. In the present study, the genomes of four other strains belonging to the families ‘Sulfuricellaceae’ and Sterolibacteriaceae were also sequenced (Table 1) but the arxAB genes were identified in none of them. In contrast to strain skB26, the arxAB genes of strains J1A and J5B are located in their chromosomes. Interestingly, strain J1A also has the aioAB genes encoding catalytic units of Aio in its genome. To the best of our knowledge, this is the first report describing the coexistence of aioAB and arxAB genes in single bacterial strain isolated in pure culture. It will be subject of future work to identify roles of these two oxidases in arsenic metabolism of the strain J1A.
Sulfuritalea hydrogenivorans sk43H is the first betaproteobacterium for which the ability for arsenate respiration was demonstrated, and it has arrAB genes encoding respiratory arsenate reductase (Watanabe et al., 2017). In this study, the genomes of its relatives within the same family were sequenced. Although Sulfurisoma sediminicola BSN1 and strain J5B have physiological traits that are similar to those of strain sk43H (e.g., facultatively anaerobic, facultatively autotrophic, neutrophilic), the arrAB genes were not observed in their genomes. These observations of the arx and arr indicate that the presence of these genes in genomes cannot be predicted by phylogenetic proximity.
Conclusion
In this study, the genomes of 9 sulfur-oxidizing bacteria were sequenced. These sulfur oxidizers belong to 8 genera, including 6 for which no genome sequence of a cultured organism was available. In the comparative genome analysis, typical suites of genes were identified for core sulfur oxidation pathways. The results of the analysis suggested the crucial importance of the cytoplasmic sulfite oxidase encoded by soeABC in the sulfur oxidizers without soxCD. In addition, large variations in the sulfur oxidation pathways were observed among members of the family ‘Sulfuricellaceae’. Furthermore, the arx genes were discovered in the families Thiobacillaceae and Sterolibacteriaceae. These findings provide some insights into the mechanism and evolution of sulfur metabolism and expand knowledge of arsenite oxidases. The primary significance of this study may lie in providing the genome sequences of 9 sulfur oxidizers, which are certainly linked to the identity and physiology of the respective organisms. These genomes will serve as valuable references for various kinds of sequence-based analyses including amplicon sequencing of various genes, metagenomics, metatranscriptomics and metaproteomics.
Author Contributions
TW, HK, and MF designed the study. HK and YK isolated and characterized strains. TW and HK performed experiments for genome sequencing. TW, KU, CH, and TT carried out proteomic experiments. TW conducted data analyses. TW and HK wrote the manuscript.
Funding
This study was supported by JSPS KAKENHI Grant No. 15K07209. TW was supported by the Alexander von Humboldt Foundation and by Japan Society for the Promotion of Science.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Arisa Shinohara and Riho Tokizawa for their technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00316/full#supplementary-material
Footnotes
References
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bamford, V. A., Bruno, S., Rasmussen, T., Appia-Ayme, C., Cheesman, M. R., Berks, B. C., et al. (2002). Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J. 21, 5599–5610. doi: 10.1093/emboj/cdf566
Boden, R., Hutt, L. P., and Rae, A. W. (2017a). Reclassification of Thiobacillus aquaesulis (Wood & Kelly, 1995) as Annwoodia aquaesulis gen. nov., comb. nov., transfer of Thiobacillus (Beijerinck, 1904) from the Hydrogenophilales to the Nitrosomonadales, proposal of Hydrogenophilalia class. nov. within the ‘Proteobacteria’, and four new families within the orders Nitrosomonadales and Rhodocyclales. Int. J. Syst. Evol. Microbiol. 67, 1191–1205. doi: 10.1099/ijsem.0.001927
Boden, R., Scott, K. M., Rae, A. W., and Hutt, L. P. (2017b). Reclassification of Thiomicrospira hydrogeniphila (Watsuji et al. 2016) to Thiomicrorhabdus hydrogenophila comb. nov., with emended description of Thiomicrorhabdus (Boden, 2017). Int. J. Syst. Evol. Microbiol. 67, 4205–4209. doi: 10.1099/ijsem.0.002279
Boden, R., Scott, K. M., Williams, J., Russel, S., Antonen, K., Rae, A. W., et al. (2017c). An evaluation of Thiomicrospira, Hydrogenovibrio and Thioalkalimicrobium: reclassification of four species of Thiomicrospira to each Thiomicrorhabdus gen. nov. and Hydrogenovibrio, and reclassification of all four species of Thioalkalimicrobium to Thiomicrospira. Int. J. Syst. Evol. Microbiol. 67, 1140–1151. doi: 10.1099/ijsem.0.001855
Brito, J. A., Denkmann, K., Pereira, I. A., Archer, M., and Dahl, C. (2015). Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: structural and functional insights into thiosulfate oxidation. J. Biol. Chem. 290, 9222–9238. doi: 10.1074/jbc.M114.623397
Cao, X., Koch, T., Steffens, L., Finkensieper, J., Zigann, R., Cronan, J. E., et al. (2018). Lipoate-binding proteins and specific lipoate-protein ligases in microbial sulfur oxidation reveal an atpyical role for an old cofactor. Elife 7:e37439. doi: 10.7554/eLife.37439
Chen, L., Ren, Y., Lin, J., Liu, X., Pang, X., and Lin, J. (2012). Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS One 7:e39470. doi: 10.1371/journal.pone.0039470
Chen, Z. W., Koh, M., Van Driessche, G., Van Beeumen, J. J., Bartsch, R. G., Meyer, T. E., et al. (1994). The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science 266, 430–432. doi: 10.1126/science.7939681
Cort, J. R., Selan, U., Schulte, A., Grimm, F., Kennedy, M. A., and Dahl, C. (2008). Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation. J. Mol. Biol. 382, 692–707. doi: 10.1016/j.jmb.2008.07.022
Dahl, C. (2017). “Sulfur metabolism in phototrophic bacteria,” in Modern Topics in the Phototrophic Prokaryotes, ed. P. Hallenbeck (Cham: Springer), 27–66. doi: 10.1007/978-3-319-51365-2_2
Dahl, C., Engels, S., Pott-Sperling, A. S., Schulte, A., Sander, J., Lubbe, Y., et al. (2005). Novel genes of the dsr gene cluster and evidence for close interaction of dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 187, 1392–1404. doi: 10.1128/JB.187.4.1392-1404.2005
Dahl, C., Franz, B., Hensen, D., Kesselheim, A., and Zigann, R. (2013). Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159, 2626–2638. doi: 10.1099/mic.0.071019-0
De Jong, G. A., Hazeu, W., Bos, P., and Kuenen, J. G. (1997). Isolation of the tetrathionate hydrolase from Thiobacillus acidophilus. Eur. J. Biochem. 243, 678–683. doi: 10.1111/j.1432-1033.1997.00678.x
Denkmann, K., Grein, F., Zigann, R., Siemen, A., Bergmann, J., van Helmont, S., et al. (2012). Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ. Microbiol. 14, 2673–2688. doi: 10.1111/j.1462-2920.2012.02820.x
Dyksma, S., Bischof, K., Fuchs, B. M., Hoffmann, K., Meier, D., Meyerdierks, A., et al. (2016). Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 10, 1939–1953. doi: 10.1038/ISMEJ.2015.257
Eddie, B. J., and Hanson, T. E. (2013). Chlorobaculum tepidum TLS displays a complex transcriptional response to sulfide addition. J. Bacteriol. 195, 399–408. doi: 10.1128/JB.01342-12
Feng, G., Xie, T., Wang, X., Bai, J., Tang, L., Zhao, H., et al. (2018). Metagenomic analysis of microbial community and function involved in cd-contaminated soil. BMC Microbiol. 18:11. doi: 10.1186/s12866-018-1152-5
Friedrich, C. G., Quentmeier, A., Bardischewsky, F., Rother, D., Kraft, R., Kostka, S., et al. (2000). Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 182, 4677–4687. doi: 10.1128/JB.182.17.4677-4687.2000
Friedrich, C. G., Rother, D., Bardischewsky, F., Quentmeier, A., and Fischer, J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67, 2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001
Frigaard, N.-U., and Dahl, C. (2008). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200. doi: 10.1016/S0065-2911(08)00002-7
Fritz, G., Roth, A., Schiffer, A., Buchert, T., Bourenkov, G., Bartunik, H. D., et al. (2002). Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 99, 1836–1841. doi: 10.1073/pnas.042664399
Grabarczyk, D. B., and Berks, B. C. (2017). Intermediates in the Sox sulfur oxidation pathway are bound to a sulfane conjugate of the carrier protein SoxYZ. PLoS One 12:e0173395. doi: 10.1371/journal.pone.0173395
Gregersen, L. H., Bryant, D. A., and Frigaard, N. U. (2011). Mechanisms and evolution of oxidative sulphur metabolism in green sulphur bacteria. Front. Microbiol. 2:116. doi: 10.3389/fmicb.2011.00116
Grein, F., Pereira, I. A. C., and Dahl, C. (2010). Biochemical characterization of individual components of the Allochromatium vinosum DsrMKJOP transmembrane complex aids understanding of complex function in vivo. J. Bacteriol. 24, 6369–6377. doi: 10.1128/JB.00849-10
Griesbeck, C., Schütz, M., Schödl, T., Bathe, S., Nausch, L., Mederer, N., et al. (2002). Mechanism of sulfide-quinone reductase investigated using site-directed mutagenesis and sulfur analysis. Biochemistry 41, 11552–11565. doi: 10.1021/bi026032b
Hausmann, B., Pelikan, C., Herbold, C. W., Köstlbacher, S., Albertsen, M., Eichorst, S. A., et al. (2018). Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J. 12, 1729–1742. doi: 10.1038/s41396-018-0077-1
Herrmann, M., Opitz, S., Harzer, R., Totsche, K., and Küsel, K. (2017). Attached and suspended denitrifier communities in pristine limestone aquifers harbor high fractions of potential autotrophs oxidizing reduced iron and sulfur compounds. Microb. Ecol. 74, 264–277. doi: 10.1007/s00248-017-0950-x
Herrmann, M., Rusznyak, A., Akob, D. M., Schulze, I., Opitz, S., Totsche, K. U., et al. (2015). Large fractions of CO2-fixing microorganisms in pristine limestone aquifers appear to be involved in the oxidation of reduced sulfur and nitrogen compounds. Appl. Environ. Microbiol. 81, 2384–2394. doi: 10.1128/AEM.03269-14
Ishihama, Y., Oda, Y., Tabata, T., Sato, T., Nagasu, T., Rappsilber, J., et al. (2005). Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 4, 1265–1272. doi: 10.1074/mcp.M500061-MCP
Issotta, F., Moya-Beltrán, A., Mena, C., Covarrubias, P. C., Thyssen, C., Bellenberg, S., et al. (2018). Insights into the biology of acidophilic members of the Acidiferrobacteraceae family derived from comparative genomic analyses. Res. Microbiol. 169, 608–617. doi: 10.1016/j.resmic.2018.08.001
Kappler, U., and Bailey, S. (2005). Molecular basis of intramolecular electron transfer in sulfite-oxidizing enzymes is revealed by high resolution structure of a heterodimeric complex of the catalytic molybdopterin subunit and a c-type cytochrome subunit. J. Biol. Chem. 280, 24999–25007. doi: 10.1074/jbc.M503237200
Kappler, U., Bennett, B., Rethmeier, J., Schwarz, G., Deutzmann, R., McEwan, A. G., et al. (2000). Sulfite:cytochrome c oxidoreductase from Thiobacillus novellus – purification, characterization, and molecular biology of a heterodimeric member of the sulfite oxidase family. J. Biol. Chem. 275, 13202–13212. doi: 10.1074/jbc.275.18.13202
Kletzin, A. (1989). Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. J. Bacteriol. 171, 1638–1643. doi: 10.1128/jb.171.3.1638-1643.1989
Kletzin, A. (1992). Molecular characterization of the sor gene, which encodes the sulfur oxygenase/reductase of the thermoacidophilic Archaeum Desulfurolobus ambivalens. J. Bacteriol. 174, 5854–5859. doi: 10.1128/jb.174.18.5854-5859.1992
Koch, T., and Dahl, C. (2018). A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J. 12, 2479–2491. doi: 10.1038/s41396-018-0209-7
Kojima, H., and Fukui, M. (2010). Sulfuricella denitrificans gen. nov., sp. nov., a sulfur-oxidizing autotroph isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 60, 2862–2866. doi: 10.1099/ijs.0.016980-0
Kojima, H., and Fukui, M. (2011). Sulfuritalea hydrogenivorans gen. nov., sp. nov., a facultative autotroph isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 61, 1651–1655. doi: 10.1099/ijs.0.024968-0
Kojima, H., and Fukui, M. (2014). Sulfurisoma sediminicola gen. nov., sp. nov., a facultative autotroph isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 64, 1587–1592. doi: 10.1099/ijs.0.057281-0
Kojima, H., and Fukui, M. (2016). Sulfuriflexus mobilis gen. nov., sp. nov., a sulfur-oxidizing bacterium isolated from a brackish lake sediment. Int. J. Syst. Evol. Microbiol. 66, 3515–3518. doi: 10.1099/ijsem.0.001227
Kojima, H., Ogura, Y., Yamamoto, N., Togashi, T., Mori, H., Watanabe, T., et al. (2015a). Ecophysiology of Thioploca ingrica as revealed by the complete genome sequence supplemented with proteomic evidence. ISME J. 9, 1166–1176. doi: 10.1038/ismej.2014.209
Kojima, H., Shinohara, A., and Fukui, M. (2015b). Sulfurifustis variabilis gen. nov., sp. nov., a sulfur oxidizer isolated from a lake, and proposal of Acidiferrobacteraceae fam. nov. and Acidiferrobacterales ord. nov. Int. J. Syst. Evol. Microbiol. 65, 3709–3713. doi: 10.1099/ijsem.0.000479
Kojima, H., Watanabe, M., and Fukui, M. (2017a). Sulfuritortus calidifontis gen. nov., sp. nov., a sulfur oxidizer isolated from a hot spring microbial mat. Int. J. Syst. Evol. Microbiol. 67, 1355–1358. doi: 10.1099/ijsem.0.001813
Kojima, H., Watanabe, M., and Fukui, M. (2017b). Sulfurivermis fontis gen. nov., sp. nov., a novel sulfur-oxidizing autotroph, and proposal of Thioprofundaceae fam. nov. Int. J. Syst. Evol. Microbiol. 67, 3458–3461. doi: 10.1099/ijsem.0.002137
Kojima, H., Watanabe, T., and Fukui, M. (2016). Sulfuricaulis limicola gen. nov., sp. nov., a sulfur oxidizer isolated from a lake. Int. J. Syst. Evol. Microbiol. 66, 266–270. doi: 10.1099/ijsem.0.000709
Kojima, H., Watanabe, T., Iwata, T., and Fukui, M. (2014). Identification of major planktonic sulfur oxidizers in stratified freshwater lake. PLoS One 9:e93877. doi: 10.1371/journal.pone.0093877
Kubo, K., Kojima, H., and Fukui, M. (2014). Vertical distribution of major sulfate-reducing bacteria in a shallow eutrophic meromictic lake. Syst. Appl. Microbiol. 37, 510–519. doi: 10.1016/j.syapm.2014.05.008
Kumar, S., Herrmann, M., Blohm, A., Hilke, I., Frosch, T., Trumbore, S. E., et al. (2018). Thiosulfate-and hydrogen-driven autotrophic denitrification by a microbial consortium enriched from groundwater of an oligotrophic limestone aquifer. FEMS Microbiol. Ecol. 94:fiy141. doi: 10.1093/femsec/fiy141
Kumar, S., Herrmann, M., Thamdrup, B., Schwab, V. F., Geesink, P., Trumbore, S. E., et al. (2017). Nitrogen loss from pristine carbonate-rock aquifers of the Hainich Critical Zone Exploratory (Germany) is primarily driven by chemolithoautotrophic anammox processes. Front. Microbiol. 8:1951. doi: 10.3389/fmicb.2017.01951
Laska, S., Lottspeich, F., and Kletzin, A. (2003). Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology 149, 2357–2371. doi: 10.1099/mic.0.26455-0
Lau, M. C. Y., Kieft, T. L., Kuloyo, O., Linage-Alvarez, B., van Heerden, E., Lindsay, M. R., et al. (2016). An oligotrophic deep-subsurface community dependent on syntrophy is dominated by sulfur-driven autotrophic denitrifiers. Proc. Natl. Acad. Sci. U.S.A. 113, E7927–E7936. doi: 10.1073/pnas.1612244113
Lenk, S., Moraru, C., Hahnke, S., Arnds, J., Richter, M., Kube, M., et al. (2012). Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J. 6, 2178–2187. doi: 10.1038/ismej.2012.66
Lin, S., Mackey, H. R., Hao, T., Guo, G., van Loosdrecht, M. C., and Chen, G. (2018). Biological sulfur oxidation in wastewater treatment: a review of emerging opportunities. Water Res. 143, 339–415. doi: 10.1016/j.watres.2018.06.051
Luo, J., Tan, X., Liu, K., and Lin, W. (2018). Survey of sulfur-oxidizing bacterial community in the Pearl River water using soxB, sqr, and dsrA as molecular biomarkers. 3 Biotech 8:73. doi: 10.1007/s13205-017-1077-y
Lyric, R. M., and Suzuki, I. (1970). Enzymes involved in the metabolism of thiosulfate by Thiobacillus thioparus. II. Properties of adenosine-5’-phosphosulfate reductase. Can. J. Biochem. 48, 344–354. doi: 10.1139/o70-057
Marcia, M., Ermler, U., Peng, G., and Michel, H. (2009). The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc. Natl. Acad. Sci. U.S.A. 106, 9625–9630. doi: 10.1073/pnas.0904165106
Mattes, T. E., Nunn, B. L., Marshall, K. T., Proskurowski, G., Kelley, D. S., Kawka, O. E., et al. (2013). Sulfur oxidizers dominate carbon fixation at a biogeochemical hot spot in the dark ocean. ISME J. 7, 2349–2360. doi: 10.1038/ismej.2013.113
Meyer, B., Imhoff, J. F., and Kuever, J. (2007). Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria – evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 9, 2957–2977. doi: 10.1111/j.1462-2920.2007.01407.x
Meyer, B., and Kuever, J. (2007). Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5’-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 153, 3478–3498. doi: 10.1099/mic.0.2007/008250-0
Müller, F. H., Bandeiras, T. M., Urich, T., Teixeira, M., Gomes, C. M., and Kletzin, A. (2004). Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53, 1147–1160. doi: 10.1111/j.1365-2958.2004.04193.x
Muyzer, G., Kuenen, J. G., and Robertson, L. A. (2013). “Colorless sulfur bacteria,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 555–588. doi: 10.1007/978-3-642-30141-4_78
Mußmann, M., Pjevac, P., Krüger, K., and Dyksma, S. (2017). Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J. 11, 1276–1281. doi: 10.1038/ismej.2016.185
Oliveira, T. F., Vonrhein, C., Matias, P. M., Venceslau, S. S., Pereira, I. A. C., and Archer, M. (2008). Purification, crystallization and preliminary crystallographic analysis of a dissimilatory DsrAB sulfite reductase in complex with DsrC. J. Struct. Biol. 164, 236–239. doi: 10.1016/j.jsb.2008.07.007
Ospino, M. C., Kojima, H., Watanabe, T., Iwata, T., and Fukui, M. (2018). Diversity of anaerobic arsenite-oxidizing bacteria in low-salt environments analyzed with a newly developed PCR-based method. Limnology 19, 177–183. doi: 10.1007/s10201-018-0539-1
Parey, K., Demmer, U., Warkentin, E., Wynen, A., Ermler, U., and Dahl, C. (2013). Structural, biochemical and genetic characterization of dissimilatory ATP sulfurylase from Allochromatium vinosum. PLoS One 8:e74707. doi: 10.1371/journal.pone.0074707
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Pires, R. H., Lourenço, A. I. C., Morais, F., Teixeira, M., Xavier, A. V., Saraiva, L. M., et al. (2003). A novel membrane-bound respiratory complex from Desulfovibrio desulfuricans ATCC 27774. Biochim. Biophys. Acta 1605, 67–82. doi: 10.1016/S0005-2728(03)00065-3
Pires, R. H., Venceslau, S. S., Morais, F., Teixeira, M., Xavier, A. V., and Pereira, I. A. C. (2006). Characterization of the Desulfovibrio desulfuricans ATCC 27774 DsrMKJOP complex – A membrane-bound redox complex involved in the sulfate respiratory pathway. Biochemistry 4, 249–262. doi: 10.1021/bi0515265
Pokorna, D., and Zabranska, J. (2015). Sulfur-oxidizing bacteria in environmental technology. Biotech. Adv. 33, 1246–1259. doi: 10.1016/j.biotechadv.2015.02.007
Pott, A. S., and Dahl, C. (1998). Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144, 1881–1894. doi: 10.1099/00221287-144-7-1881
Prokopenko, M. G., Hirst, M. B., De Brabandere, L., Lawrence, D. J. P., Berelson, W. M., Granger, J., et al. (2013). Nitrogen losses in anoxic marine sediments driven by Thioploca-anammox bacterial consortia. Nature 500, 194–198. doi: 10.1038/nature12365
Quatrini, R., Appia-Ayme, C., Denis, Y., Jedlicki, E., Holmes, D., and Bonnefoy, V. (2009). Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. doi: 10.1186/1471-2164-10-394
Quentmeier, A., Kraft, R., Kostka, S., Klockenkämper, R., and Friedrich, C. G. (2000). Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Arch. Microbiol. 173, 117–125. doi: 10.1007/s002039900118
Ramos, A. R., Keller, K. L., Wall, J. D., and Pereira, I. A. C. (2012). The membrane QmoABC complex interacts directly with the dissimilatory adenosine 5’-phosphosulfate reductase in sulfate reducing bacteria. Front. Microbiol. 3:137. doi: 10.3389/fmicb.2012.00137
Sauvé, V., Bruno, S., Berks, B. C., and Hemmings, A. M. (2007). The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J. Biol. Chem. 282, 23194–23204. doi: 10.1074/jbc.M701602200
Sauvé, V., Roversi, P., Leath, K. J., Garman, E. F., Antrobus, R., Lea, S. M., et al. (2009). Mechanism for the hydrolysis of a sulfur-sulfur bond based on the crystal structure of the thiosulfohydrolase SoxB. J. Biolog. Chem. 284, 21707–21718. doi: 10.1074/jbc.M109.002709
Schedel, M., Vanselow, M., and Trüper, H. G. (1979). Siroheme sulfite reductase isolated from Chromatium vinosum. Purification and investigation of some of its molecular and catalytic properties. Arch. Microbiol. 121, 29–36. doi: 10.1007/BF00409202
Scott, K. M., Williams, J., Porter, C. M., Russel, S., Harmer, T. L., Paul, J. H., et al. (2018). Genomes of ubiquitous marine and hypersaline Hydrogenovibrio, Thiomicrorhabdus and Thiomicrospira spp. encode a diversity of mechanisms to sustain chemolithoautotrophy in heterogeneous environments. Environ. Microbiol. 20, 2686–2708. doi: 10.1111/1462-2920.14090
Sorokin, D. Y., Banciu, H., Robertson, L. A., Kuenen, J. G., Muntyan, M. S., and Muyzer, G. (2013). “Halophilic and haloalkaliphilic sulfur-oxidizing bacteria,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 529–554. doi: 10.1007/978-3-642-30141-4_77
Stockdreher, Y., Sturm, M., Josten, M., Sahl, H.-G., Dobler, N., Zigann, R., et al. (2014). New proteins involved in sulfur trafficking in the cytoplasm of Allochromatium vinosum. J. Biol. Chem. 289, 12390–12403. doi: 10.1074/jbc.M113.536425
Stockdreher, Y., Venceslau, S. S., Josten, M., Sahl, H. G., Pereira, I. A., and Dahl, C. (2012). Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum: evidence for sulfur transfer from DsrEFH to DsrC. PLoS One 7:e40785. doi: 10.1371/journal.pone.0040785
Tian, R. M., Zhang, W., Cai, L., Wong, Y. H., Ding, W., and Qian, P. Y. (2017). Genome reduction and mricrobe-host interactions drive adaptation of a sulfur-oxidizing bacterium associated with a cold seep sponge. mSystems 2:e184-16. doi: 10.1128/mSystems.00184-16
Umezawa, K., Watanabe, T., Miura, A., Kojima, H., and Fukui, M. (2016). The complete genome sequences of sulfur-oxidizing Gammaproteobacteria Sulfurifustis variabilis skN76T and Sulfuricaulis limicola HA5T. Stand. Genomic. Sci. 11:71. doi: 10.1186/s40793-016-0196-0
Valdés, J., Pedroso, I., Quatrini, R., Dodson, R. J., Tettelin, H., and Blake, R. (2008). Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597. doi: 10.1186/1471-2164-9-597
van Lis, R., Nitschke, W., Duval, S., and Schoepp-Cothenet, B. (2013). Arsenics as bioenergetic substrates. Biochim. Biophys. Acta 1827, 176–188. doi: 10.1016/j.bbabio.2012.08.007
Watanabe, T., Kojima, H., and Fukui, M. (2012). Draft genome sequence of a psychrotolerant sulfur-oxidizing bacterium, Sulfuricella denitrificans skB26, and proteomic insights into cold adaptation. Appl. Environ. Microbiol. 78, 6545–6549. doi: 10.1128/AEM.01349-12
Watanabe, T., Kojima, H., and Fukui, M. (2014). Complete genomes of freshwater sulfur oxidizers Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H: genetic insights into the sulfur oxidation pathway of betaproteobacteria. Syst. Appl. Microbiol. 37, 387–395. doi: 10.1016/j.syapm.2014.05.010
Watanabe, T., Kojima, H., and Fukui, M. (2015). Sulfuriferula multivorans gen. nov., sp. nov., isolated from a freshwater lake, reclassification of ‘Thiobacillus plumbophilus’ as Sulfuriferula plumbophilus sp. nov., and description of Sulfuricellaceae fam. nov. and Sulfuricellales ord. nov. Int. J. Syst. Evol. Microbiol. 65, 1504–1508. doi: 10.1099/ijs.0.000129
Watanabe, T., Kojima, H., and Fukui, M. (2016a). Identity of major sulfur-cycle prokaryotes in freshwater lake ecosystems revealed by a comprehensive phylogenetic study of the dissimilatory adenylylsulfate reductase. Sci. Rep. 6:36262. doi: 10.1038/srep36262
Watanabe, T., Kojima, H., and Fukui, M. (2016b). Sulfuriferula thiophila sp. nov., a chemolithoautotrophic sulfur-oxidizing bacterium, and correction of the name Sulfuriferula plumbophilus Watanabe, Kojima and Fukui 2015 to Sulfuriferula plumbiphila corrig. Int. J. Syst. Evol. Microbiol. 66, 2041–2045. doi: 10.1099/ijsem.0.000988
Watanabe, T., Kojima, H., Shinohara, A., and Fukui, M. (2016c). Sulfurirhabdus autotrophica gen. nov., sp. nov., isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 66, 113–117. doi: 10.1099/ijsem.0.000679
Watanabe, T., Kojima, H., Takano, Y., and Fukui, M. (2013). Diversity of sulfur-cycle prokaryotes in freshwater lake sediments investigated using aprA as the functional marker gene. Syst. Appl. Microbiol. 36, 436–443. doi: 10.1016/j.syapm.2013.04.009
Watanabe, T., Miura, A., Iwata, T., Kojima, H., and Fukui, M. (2017). Dominance of Sulfuritalea species in nitrate-depleted water of a stratified freshwater lake and arsenate respiration ability within the genus. Environ. Microbiol. Rep. 9, 522–527. doi: 10.1111/1758-2229.12557
Włodarczyk, A., Lirski, M., Fogtman, A., Koblowska, M., Bidziński, G., and Matlakowska, R. (2018). The oxidative metabolism of fossil hydrocarbons and sulfide minerals by the lithobiontic microbial community inhabiting deep subterrestrial Kupferschiefer black shale. Front. Microbiol. 9:972. doi: 10.3389/fmicb.2018.00972
Zander, U., Faust, A., Klink, B. U., de Sanctis, D., Panjikar, S., Quentmeier, A., et al. (2010). Structural basis for the oxidation of protein-bound sulfur by the sulfur cycle molybdohemo-enzyme sulfane dehydrogenase SoxCD. J. Biol. Chem. 286, 8349–8360. doi: 10.1074/jbc.M110.193631
Keywords: sulfur-oxidizing bacteria, ‘Sulfuricellaceae’, Thiobacillaceae, Sterolibacteriaceae, comparative genomics
Citation: Watanabe T, Kojima H, Umezawa K, Hori C, Takasuka TE, Kato Y and Fukui M (2019) Genomes of Neutrophilic Sulfur-Oxidizing Chemolithoautotrophs Representing 9 Proteobacterial Species From 8 Genera. Front. Microbiol. 10:316. doi: 10.3389/fmicb.2019.00316
Received: 26 October 2018; Accepted: 06 February 2019;
Published: 25 February 2019.
Edited by:
Monika Bright, University of Vienna, AustriaReviewed by:
Kathleen Scott, University of South Florida, United StatesStefan M. Sievert, Woods Hole Oceanographic Institution, United States
Copyright © 2019 Watanabe, Kojima, Umezawa, Hori, Takasuka, Kato and Fukui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hisaya Kojima, a29qaW1haEBwb3AubG93dGVtLmhva3VkYWkuYWMuanA=
 Tomohiro Watanabe
Tomohiro Watanabe Hisaya Kojima
Hisaya Kojima Kazuhiro Umezawa
Kazuhiro Umezawa Chiaki Hori
Chiaki Hori Taichi E. Takasuka
Taichi E. Takasuka Yukako Kato
Yukako Kato Manabu Fukui
Manabu Fukui