- Laboratory of Microbiology, Parasitology and Hygiene, Department of Pharmaceutical Sciences, University of Antwerp, Wilrijk, Belgium
Streptococcus pneumoniae is the leading cause of bacterial pneumonia. Infection is linked to high morbidity and mortality rates and antibiotic resistance within this pathogen is on the rise. Therefore, there is a need for novel antimicrobial therapies. To lower the time and costs of the drug discovery process, alternative in vivo models should be considered. As such, Galleria mellonella larvae can be of great value. The larval immunity consisting of several types of haemocytes is remarkably similar to the human innate immune system. Furthermore, these larvae don’t require specific housing, are cheap and are easy to handle. In this study, the use of a G. mellonella infection model to study early pneumococcal infections and treatment is proposed. Firstly, the fitness of this model to study pneumococcal virulence factors is confirmed using streptococcal strains TIGR4, ATCC®49619, D39 and its capsule-deficient counterpart R6 at different inoculum sizes. The streptococcal polysaccharide capsule is considered the most important virulence factor without which streptococci are unable to sustain an in vivo infection. Kaplan–Meier survival curves showed indeed a higher larval survival after infection with streptococcal strain R6 compared to strain D39. Then, the infection was characterized by determining the number of haemocytes, production of oxygen free radicals and bacterial burden at several time points during the course of infection. Lastly, treatment of infected larvae with the standard antibiotics amoxicillin and moxifloxacin was evaluated. Treatment has proven to have a positive outcome on the course of infection, depending on the administered dosage. These data imply that G. mellonella larvae can be used to evaluate antimicrobial therapies against S. pneumoniae, apart from using the larval model to study streptococcal properties. The in-depth knowledge acquired regarding this model, makes it more suitable for use in future research.
Introduction
Streptococcus pneumoniae, also known as pneumococcus, is the most common cause of bacterial pneumonia, and can cause otitis media and meningitis in children, the elderly and immunocompromised patients. In a 2013 European study, an increase in the incidence of streptococcal bacteremia was reported. According to this study, S. pneumoniae was responsible for 11% of all bacteremia reports (de Kraker et al., 2013). In a 2018 European study, the incidence of pneumococcal community acquired pneumonia was estimated to be as high as 166 per 100,000 in Spain in elderly patients, with the incidence of invasive pneumococcal disease (IPD) to be 60 per 100,000. For all-cause pneumonia, the numbers rise to 7,000 per 100,000 in some countries (Torres et al., 2018). Overall, streptococcal infections are linked to high morbidity and mortality rates, even with the introduction of several vaccines. Antibiotic resistance highly depends on the antibiotic class and geographical region, reaching over 80% penicillin resistance in some parts of the African and Asian continents (Rennie, 2012).
While there is an urgent need for new antimicrobial therapies to combat streptococcal infections, this process is very time consuming and costly. One of the steps in this process is testing the potential novel compounds in in vivo models. Currently, rodents are the most commonly used animal models. However, these vertebrate models are often expensive, time consuming to maintain and ethically challenging (Graham and Prescott, 2015). Therefore, there is a need for new in vivo models for future research that can replace—or in any case significantly lower the amount of—vertebrates used in scientific research by bridging the gap between less complex in vitro models and vertebrate in vivo models. Over the past years, several alternative in vivo models have been proposed.
Caenorhabditis elegans is often used as a model due to its low cost, simple growth conditions and short generation time. Also, is has a completely sequenced genome and its immune system shows similarities with the human innate immune system (Schulenburg and Ewbank, 2007; López Hernández et al., 2015). However, this model poses limitations as well. The nematodes can only survive between 15 and 25°C, while human pathogens live at body temperatures. Also, its usefulness is limited by the lack of an adaptive immune system and of intracellular replication of pathogens (Squiban and Kurz, 2011). Adult zebrafish (Danio rerio) can also be used to study infectious diseases involving both the innate and adaptive immune system, while in embryos only the innate immunity is present. The embryos are transparent during the first days after fertilization, making it possible to follow an infection in real-time using fluorescent pathogens (Duggan and Mostowy, 2018). Also for this organism, the full genome is available (Chakraborty et al., 2009; Meijer and Spaink, 2011; López Hernández et al., 2015). Zebrafish can only survive at about 28°C, which limits their use for the study of human pathogens (Meeker and Trede, 2008).
Galleria mellonella larvae are larvae of the greater wax moth with a length of approx. 20 mm. These larvae can be obtained at commercial angler’s stores. Currently, there is only one company providing lab-grade larvae and genotyping has recently been carried out (Champion et al., 2016; Lange et al., 2018). Overall, G. mellonella larvae are cheap, do not require any specific caging and are easy to handle. Therefore, costs of this model are significantly lower compared to other ones (López Hernández et al., 2015; Tsai et al., 2016). Additionally, larvae can be maintained at 37°C, holding a big advantage over the C. elegans model when studying human pathogens (Loh et al., 2013; Champion et al., 2016). Wax moth larvae do not possess an adaptive immune system, but they do have an immune system analogous to the human innate immune system, making this model suitable to study early host-pathogen interactions and infections not involving adaptive immune responses. The haemolymph of these larvae consists of at least 8 types of haemocytes, of which plasmatocytes and granular cells possess macrophage-like functionalities, being involved in phagocytosis and encapsulation (Tsai et al., 2016). G. mellonella larvae have been used in previous research to study fungal and bacterial infections. It has been shown to be possible to study the virulence of different Candida albicans strains, and also to evaluate antifungal compounds and study biofilm formation in this model (Dunphy et al., 2003; Borghi et al., 2014; Borowiecki et al., 2018). For bacterial infections, it has been possible to infect larvae with Listeria monocytogenes and Legionella pneumophila. Also antimicrobial therapies against Pseudomonas aeruginosa have already been tested (Joyce and Gahan, 2010; Harding et al., 2013; Hill et al., 2014).
In this study, we propose the use of a G. mellonella infection model to study early pneumococcal infections and treatment. We determined the pathogenicity of several S. pneumoniae serotypes and thereby confirmed the suitability of this infection model (Evans and Rozen, 2012). Additionally, we further characterized the infection by determining the number of haemocytes, production of oxygen free radicals and bacterial burden in the haemolymph at several time points during the course of infection. Overall, our data demonstrate that G. mellonella larvae can be used to evaluate antimicrobial therapies for S. pneumoniae and give rise to more in-depth knowledge regarding this model, making it more suitable for use in future research.
Materials and Methods
G. mellonella Larvae and Bacterial Strains
G. mellonella last-instar larvae were purchased from a local vendor (Anaconda Reptiles, Kontich) and stored in wood chips at 15°C before use. Four hours before use, larvae were put at 4°C. Only healthy-looking larvae with no signs of melanization were used in the experiments. S. pneumoniae strains are listed in Table 1. Bacteria were cultured in brain-heart infusion (BHI) broth (LabM) or on 5% sheep blood agar plates (Tryptic Soy Agar from LabM, defibrinated sheep blood from Oxoid) at 37°C and 5% CO2. Agar plates or broth were supplemented with 1 μg/mL tetracycline (Sigma) when required. To obtain heat-killed streptococci, TIGR4 bacteria grown to mid-log phase were incubated for 30 min at 56°C. Killing was confirmed by plating out the suspension. To allow bacterial enumeration from G. mellonella haemolymph, streptococcal mutant TIGR4/10 (tetracycline resistant) was obtained through integration of plasmid pKB01_tagRFP into the streptococcal genome, as described previously. pKB01_tagRFP was a gift from Jan-Willem Veening (Addgene plasmid #69103) (Martin et al., 1995; Beilharz et al., 2015). Briefly, plasmid pKB01_tagRFP was purified using a NucleoSpin Plasmid EasyPure kit (Macherey-Nagel) and transformed into S. pneumoniae TIGR4 using competence-stimulating peptide 2 (CSP-2). Bacteria were grown in CAT-medium [1.0% casitone (BD Biosciences), 0.5% tryptone (Sigma-Aldrich), 0.1% yeast extract (Sigma-Aldrich), 0.5% NaCl (Merck), 0.2% glucose (Sigma-Aldrich), 16.7 M K2HPO4 (Roth)] until an OD600nm of 0.5 was reached. Bacteria were diluted 50 times in CTM-1 medium [CAT-medium supplemented with 1 mM CaCl2 (Merck), 0.2% bovine serum albumin (Sigma-Aldrich), pH 7.2] until OD600nm 0.15. Bacteria were then concentrated 10 times in CTM-1 medium and 15% glycerol was added. Bacteria were kept at –80°C until use. When needed, bacteria were thawed on ice and diluted 10 times in CTM-2 medium (CTM-1 medium, pH 7.9). 2% (v/v) 500 μg/mL CSP-2 stock was added to 200 μL aliquots and bacteria were incubated for 20 min at 37°C. Plasmid DNA was added and bacteria were incubated again for 20 min at 30°C to allow the uptake of the DNA. Bacteria were plated out on blood agar plates containing 1 μg/mL tetracycline and incubated overnight at 37°C and 5% CO2.
G. mellonella Killing Assay
A sterile 20 μL Hamilton syringe was used to inject 10 μL aliquots of bacterial suspensions in PBS into the hindmost left proleg of G. mellonella. Larvae were infected with 104, 105, or 106 CFU/larvae, with a minimum of 15 larvae per group. The control group was injected with 10 μL phosphate buffered saline (PBS) (Life Technologies). Following the injections, larvae were incubated at 37°C in the dark for several days to allow the progression of the pneumococcal infection. Every 24 h, larvae were scored as dead or alive. Larvae were determined dead when no signs of movement could be observed in response to external stimuli and when the larvae showed dark pigmentation due to melanization. For each strain, 4 independent repeats were carried out.
G. mellonella Bacterial Burden
To determine the bacterial burden, larvae were infected with streptococcal strain TIGR4/10, with a minimum of 15 larvae per time point. At fixed time points after infection (1, 24, 48, and 72 h p.i.), alive larvae were bled into ice-cold microcentrifuge tubes and haemolymph of 5 larvae was pooled together and further processed as one sample. Bacterial burden was determined using the viable plate count method with blood agar plates containing 1 μg/mL tetracycline to inhibit growth of native larval flora and allow streptococcal selection. For each time point, 5 independent repeats were carried out, with preferably two pooled samples per repeat.
G. mellonella Treatment Assay
Larvae infected with streptococcal strain TIGR4/10, D39, or ATCC 49619 at a concentration of 106 CFU/larvae were treated with 10 μL amoxicillin (Sigma-Aldrich), moxifloxacin (Fluka) or tetracycline (Sigma-Alrich) 1 h p.i. at different concentrations (0.1, 1, or 10 μg/mL for amoxicillin and tetracycline and 1, 10, or 100 μg/mL for moxifloxacin) through injection into the hindmost right proleg. After treatment, larvae were incubated and scored as dead or alive as described previously. For each strain, at least 3 independent repeats were carried out with a minimum of 10 larvae per group. To determine the bacterial load in the haemolymph after treatment, bacterial burden was determined as described above. For each time point, 3 independent repeats were carried out with two samples per repeat.
Preparation and Counting of Larval Haemocytes
At fixed time points p.i. (1, 24, 48, and 72 h p.i.), alive larvae infected with several streptococcal strains were disinfected using 70% ethanol after which they were bled into ice-cold microcentrifuge tubes containing 50 μL of insect physiological solution (IPS) [150 mM NaCl, 5 mM KCl (Sigma-Aldrich), 10 mM tris HCl (Sigma-Aldrich), pH 6.9] containing 30 mM EDTA (Sigma-Aldrich) and 30 mM sodium citrate (VWR) as anticoagulants. Haemolymph of 5 larvae was pooled together into 1 tube and further processed as one sample. Cells were diluted ten times in PBS and counted using KOVA®Glasstic®Slide 10 counting chambers (Thermo Fisher Scientific). For each time point, 4 independent repeats were carried out, with preferably two pooled samples per repeat.
Detection of Oxygen Free Radicals
Electron paramagnetic resonance spectroscopy (EPR) was used to measure the formation of oxygen free radicals. At different time points p.i. (1, 24, 48, and 72 h p.i.), alive larvae were bled out into ice-cold microcentrifuge tubes. Haemolymph of 5 larvae was pooled together and further processed as one sample. The haemolymph was diluted 100 × in 1 mL Krebs-Hepes Buffer (KHB, pH 7.4) before adding 1-hydroxy-3-methoxycarbonyl-2,2,5,5-tetramethylpyrrolidine (CM-H) as a spin probe. After incubation for 90 min at 37°C, 50 μL of the suspension was sampled into a capillary tube for measurement. All EPR measurements were performed on a Magnettech MiniScope MS 200 spectrometer as follows: frequency, 9.4 GHz; power: 5 dBm (3.16 mW); modulation frequency, 100 kHz; modulation amplitude, 0.1 mT; sweep time, 30 s; time constant, 0.1; sweep width, 10 mT; and number of scans, 1. For the measurements, the analyzed samples (50 μl) were contained in glass capillaries (Hirschmann). Data were analyzed using the SpecView software. Peak height was measured and reported in arbitrary units (a.u.). For each time point, at least 4 independent repeats were carried out, with preferably two pooled samples per repeat.
Statistical Analyses
Data were analyzed for statistical significance using GraphPad Prism Version 7. Continuous variables were compared by two-way Anova, unpaired t-test or survival analysis. Statistical significance was defined as p < 0.05. Statistical significance between two groups is visualized as asterisks in figures (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001).
Results
Survival of Larvae Was Dependent on MOI, Serotype, and Presence of Virulence Factors
The outcome of infection in the G. mellonella model was – as it is in vertebrate models – dependent on both MOI (TIGR4 p < 0.0001; R6 p = 0.0044; D39 p < 0.0001; ATCC 49619 p < 0.0001; Survival analysis, Log-rank Mantel-Cox test) and streptococcal strain (Figure 1). For all strains, administering 104 CFU/larvae hardly led to any larval death, while the effect of administering 106 CFU/larvae was variable. For strains D39 and TIGR4 – which differ in serotype – there was no difference in virulence (p = 0.8961; Survival analysis, Log-rank Mantel-Cox test). It should be noted that, while there was no difference in larval survival, the visual pathology of the larvae infected with strain D39 seemed worse than that of strain TIGR4. The melanization process in larvae infected with strain D39 occurred both faster and more intense and their mobility steadily decreased. For larvae infected with strain TIGR4, a more diverse result was observed. Despite the similarity in survival curves, strain D39 was considered more virulent compared to strain TIGR4. Polysaccharide capsule deficient strain R6, which originates from strain D39, showed a clearly attenuated virulence compared to strain D39 (p = 0.0008; Survival analysis, Log-rank Mantel-Cox test). The polysaccharide capsule is known as one of the most important virulence factors, which is in accordance with the loss of virulence in this model when the capsule is not present (Kadioglu et al., 2008; Preston and Dockrell, 2008). While strains TIGR4 and D39 showed no difference in survival, there was a significant difference between those strains and strain ATCC 49619, which proved to be the most virulent (D39 and ATCC 49619: p = 0.0006; TIGR4 and ATCC 49619: p < 0.0001; Survival analysis, Log-rank Mantel-Cox test).
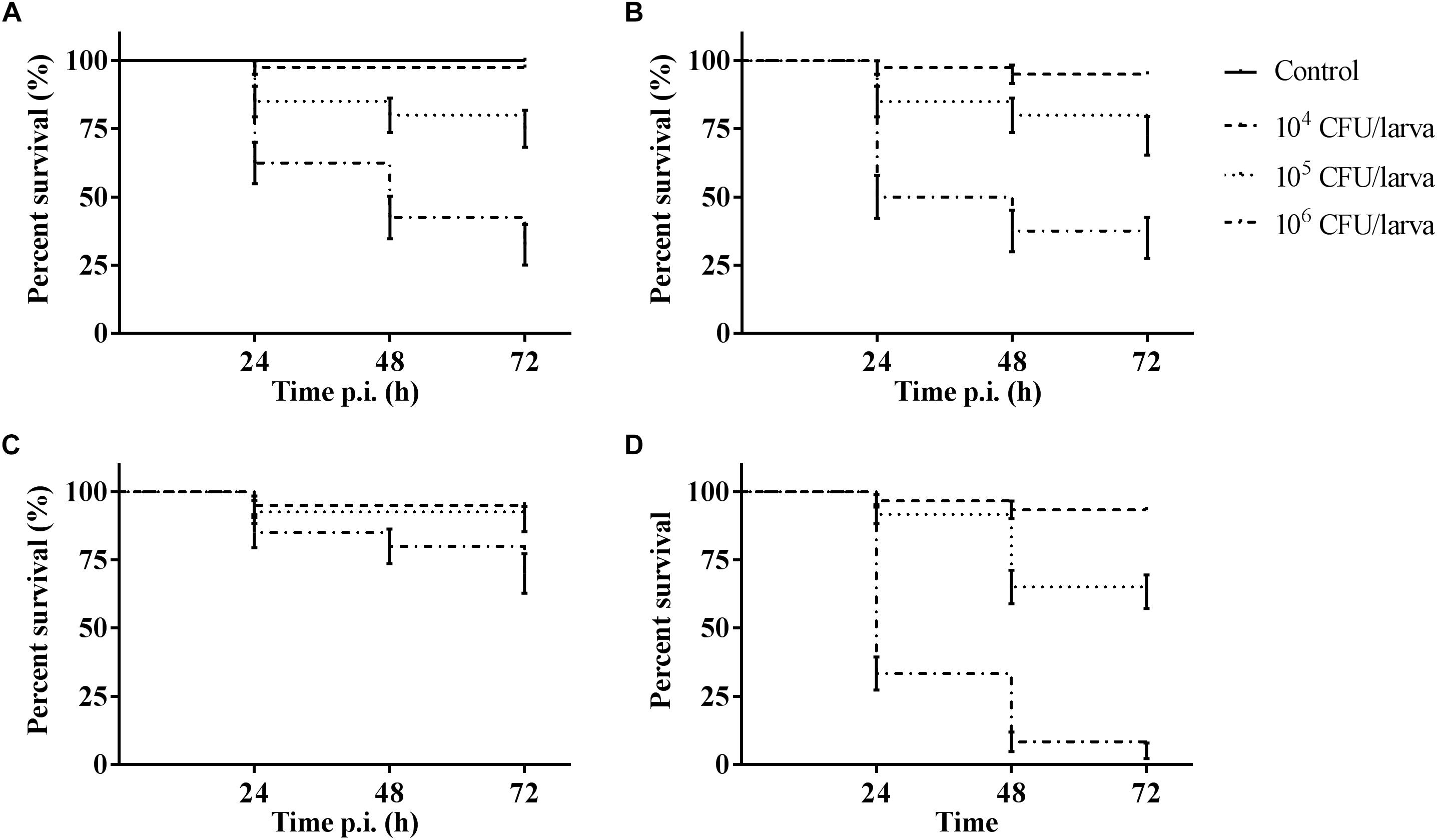
Figure 1. Kaplan–Meier survival curves of G. mellonella larvae after infection with S. pneumoniae at different inoculum sizes (104 CFU/larva, 105 CFU/larva and 106 CFU/larva). (A) Infection with strain TIGR4; (B) Infection with strain D39; (C) Infection with strain R6; (D) Infection with strain ATCC 49619. Control group is injected with PBS. Error bars represent SE (standard error) (n = 15 × 4).
Bacterial Burden Decreased Over Time
One hour post-infection, larval burden of S. pneumoniae strain TIGR4 decreased with 1 log compared to the initial inoculum of (1.37 ± 0.64) ∗ 108 CFU/mL to (1.04 ± 0.07) ∗ 107 CFU/mL (Figure 2). After 24 h, the burden increased to (1.26 ± 0.2) ∗ 108 CFU/mL 24 h p.i. Larvae that outlived the infection over 24 h, seemed to start clearing the streptococci [(9.38 ± 0.93) ∗ 106 CFU/mL 48 h p.i.; (1.08 ± 0.33) ∗ 106 CFU/mL 72 h p.i.]. This is probably due to the efficient phagocytosis mechanism of larval haemocytes (Kavanagh and Reeves, 2004; Wojda, 2017). It should be noted that 72 h post-infection, the presence of streptococci in haemolymph could no longer be determined for 60% of all samples. Of note, there was no difference in survival between infection with strain TIGR4 and tetracycline-resistant strain TIGR4/10 (Supplementary Figure 1).
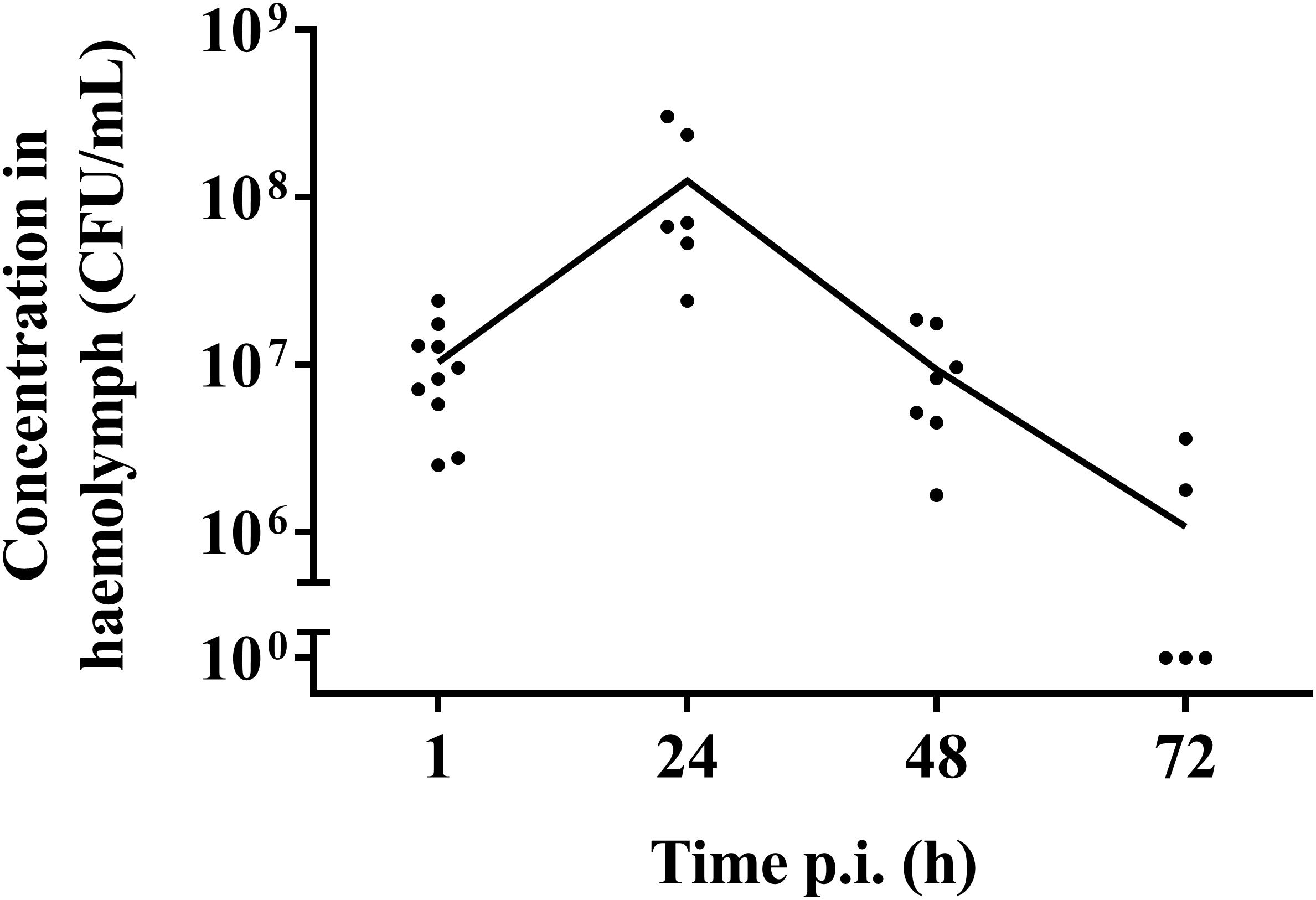
Figure 2. Streptococcal burden in a G. mellonella infection model using streptococcal strain TIGR4/10. After a rapid bacterial multiplication (1–24 h), larvae that survived showed a decrease in bacterial burden over the course of the infection (48–72 h). The mean is shown (n = 2 × 5).
Number of Haemocytes Did Not Differ Over Time
There was no difference in the number of cells between the control group and infected larvae (p = 0.3940; two-way Anova) (Figure 3). This suggests that there was no influx of haemocytes that can be accredited to the bacterial infection, nor do haemocytes of infected larvae go into apoptosis or necrosis more quickly. Overall, an average of (3.72 ± 0.68) ∗ 105 cells/larvae was counted.
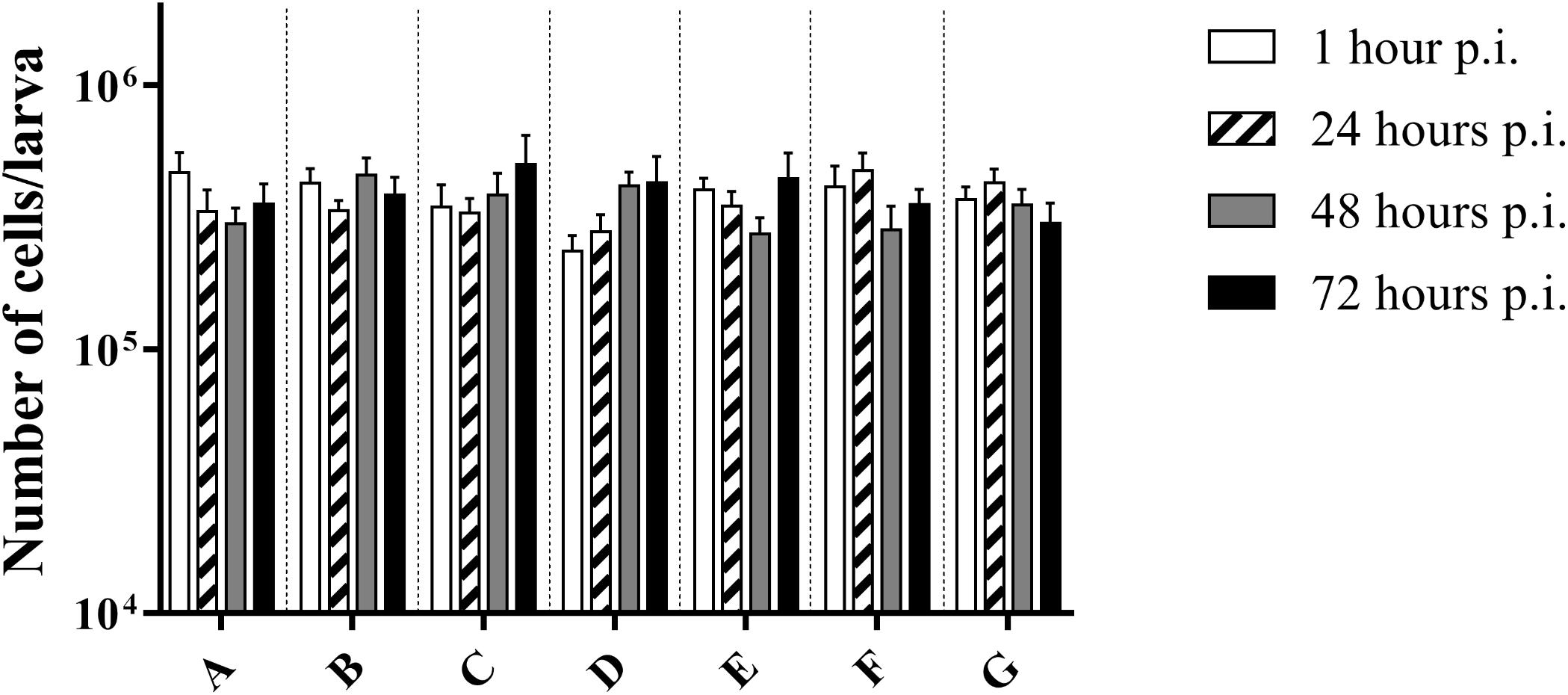
Figure 3. Number of cells in haemolymph of G. mellonella at different time points post-infection. (A) Non-injected control group; (B) PBS-injected control group; (C) Infection with heat-killed bacteria; (D) Infection strain TIGR4; (E) Infection with strain ATCC 49619; (F) Infection with strain D39; (G) Infection with strain R6. Error bars represent SE (standard error). There are no statistical differences between infected and control groups (Two-way Anova, p < 0.05) (n = 2 × 4).
Production of Oxygen Free Radicals Differed During the Course of Infection
Haemocytes are known to produce oxygen free radicals as a way of battling infections. This can be monitored using the spin probe CM-H (Slepneva et al., 1999). Directly after infection, an increase in production of oxygen free radicals could be observed between the control group and infected larvae for strains TIGR4 (p = 0.0100, Unpaired t-test) (Figure 4A). Afterward, there was no detectable difference between this strain and the control group (24 h p.i. p = 0.1731; 48 h p.i. p = 01314; 72 h p.i. p = 0.1517; Unpaired t-test). This concurs with previous results (Figure 2), where also the streptococcal burden was lowered after 24 h, presuming the infection was being cleared.
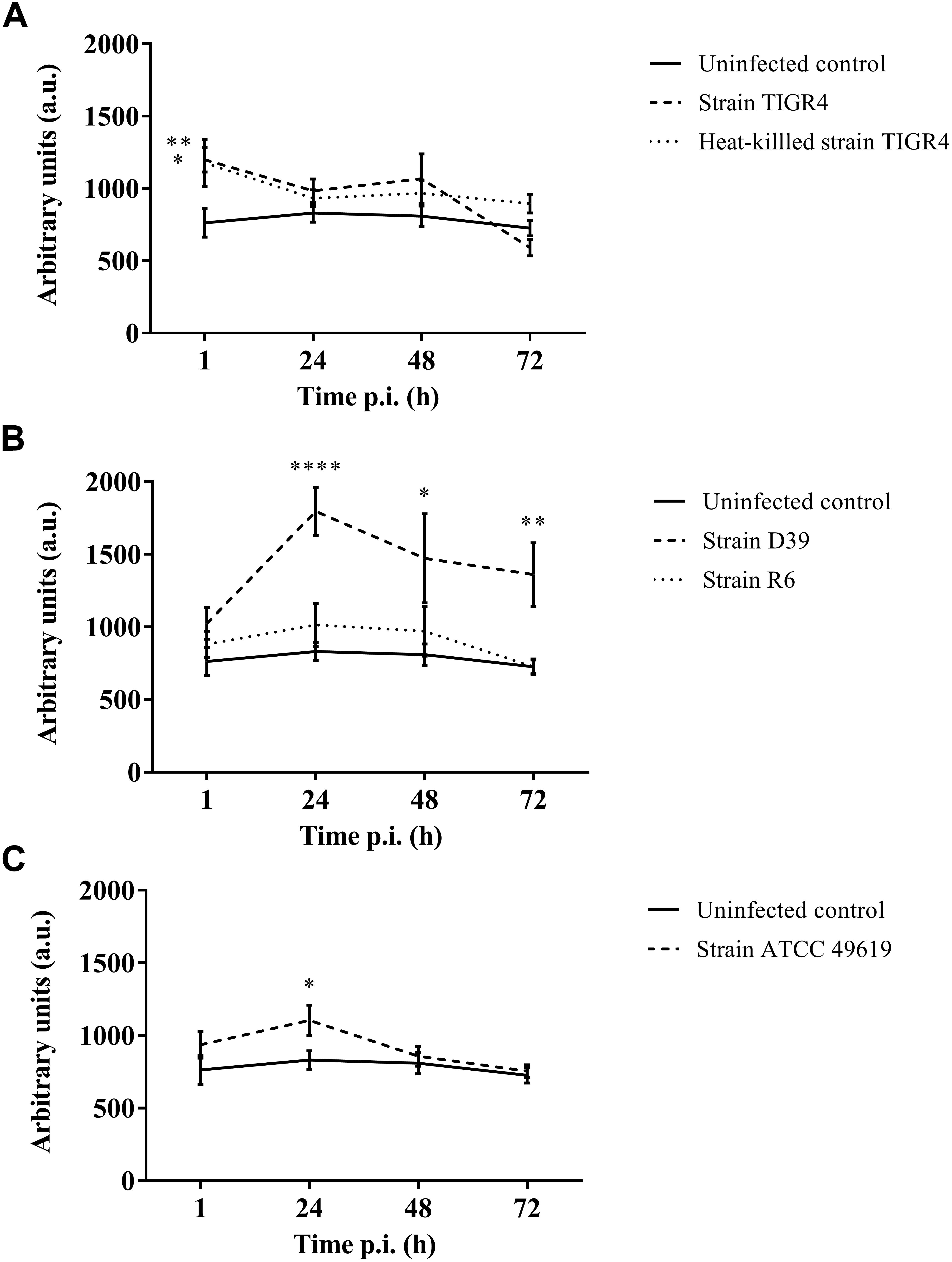
Figure 4. Production of oxygen free radicals of haemocytes of G. mellonella at different time points post-infection. Amount of oxygen radicals captured by CM-H is calculated by measuring the peak height after 90 min and expressed in arbitrary units. (A) Infection with Strain TIGR4 and heat-killed bacteria; (B) Infection with strain D39 and R6; (C) Infection with strain ATCC 49619. Error bars represent SE (standard error). Asterisks represent statistical differences between infected groups and the uninfected control group (Unpaired t-test, p ≤ 0.05, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001) (n = 2 × 4).
Furthermore, for strain D39 (Figure 4B), a clear increase in oxygen free radicals could be observed during the course of infection compared to the control group (1 h p.i. p = 0.1067; 24 h p.i. p < 0.0001; 48 h p.i. p = 0.0242; 72 h p.i. p = 0.0045, Unpaired t-test). The increase in oxygen free radical is also significantly higher for infection with D39 as it is for infection with all other strains (TIGR4 p = 0.0394; R6 p = 0.0155; ATCC 49619 p = 0.0195, One-way Anova). It should be noted that—as stated earlier—the visual pathology of larvae infected with strain D39 was also worse than that of larvae infected with strains TIGR4 or ATCC 49619. Infection with strain R6 showed no increase in production of oxygen free radicals compared to the control group (1 h p.i. p = 0.3951; 24 h pi. p = 0.1427; 48 h p.i. p = 0.4017; 72 h p.i. p = 0.9986, Unpaired t-test). This complies with earlier results, as strain R6 was the least virulent strain (Figure 1C). For infection with strain ATCC 49619 (Figure 4C) no increase in oxygen free radicals was observed 1 h p.i. (p = 0.2291, Unpaired t-test), but there was an increase 24 h p.i. (p = 0.0380, Unpaired t-test). However, this increase was not observed 48 or 72 h p.i. (48 h p.i. p = 0.3521; 72 h p.i. p = 0.7146, Unpaired t-test). At 24 h p.i. survival of larvae is approx. 33%. This number further lowers to approx. 8% at 48 h p.i. (Figure 1D). This implies most larvae are dying at 24 h p.i., which could explain the temporary increase in oxygen free radicals.
Lastly, in case of infection with heat-killed streptococci (Figure 4A), only directly after infection a difference between infected larvae and control larvae could be observed (1 h p.i. p = 0.0494; 24 h p.i. p = 0.2665; 48 h p.i. p = 0.1941; 72 h p.i. p = 0.0679, Unpaired t-test).
Treatment With Amoxicillin or Moxifloxacin Enhanced the Survival of Larvae
It was possible to treat infected larvae with amoxicillin. For strain TIGR4/10 (Figure 5A), there was no difference between untreated controls and treatment with 0.1 μg/mL amoxicillin (p = 0.9725, Survival analysis, Log-rank Mantel-Cox test), but there was a significant difference between untreated controls and treatment with 1 or 10 μg/mL amoxicillin (p = 0.0022 and p < 0.0001 respectively, Survival analysis, Log-rank Mantel-Cox test). In addition, there was no difference between treatment with 1 or 10 μg/mL amoxicillin (p = 0.1179, Survival analysis, Log-rank Mantel-Cox test). The bacterial burden in the haemolymph of larvae infected with strain TIGR4/10 decreased over time for all amoxicillin concentrations used (Figure 6). In concordance with the larval survival results, bacterial burden remained highest in groups treated with the lowest antibiotic concentration.
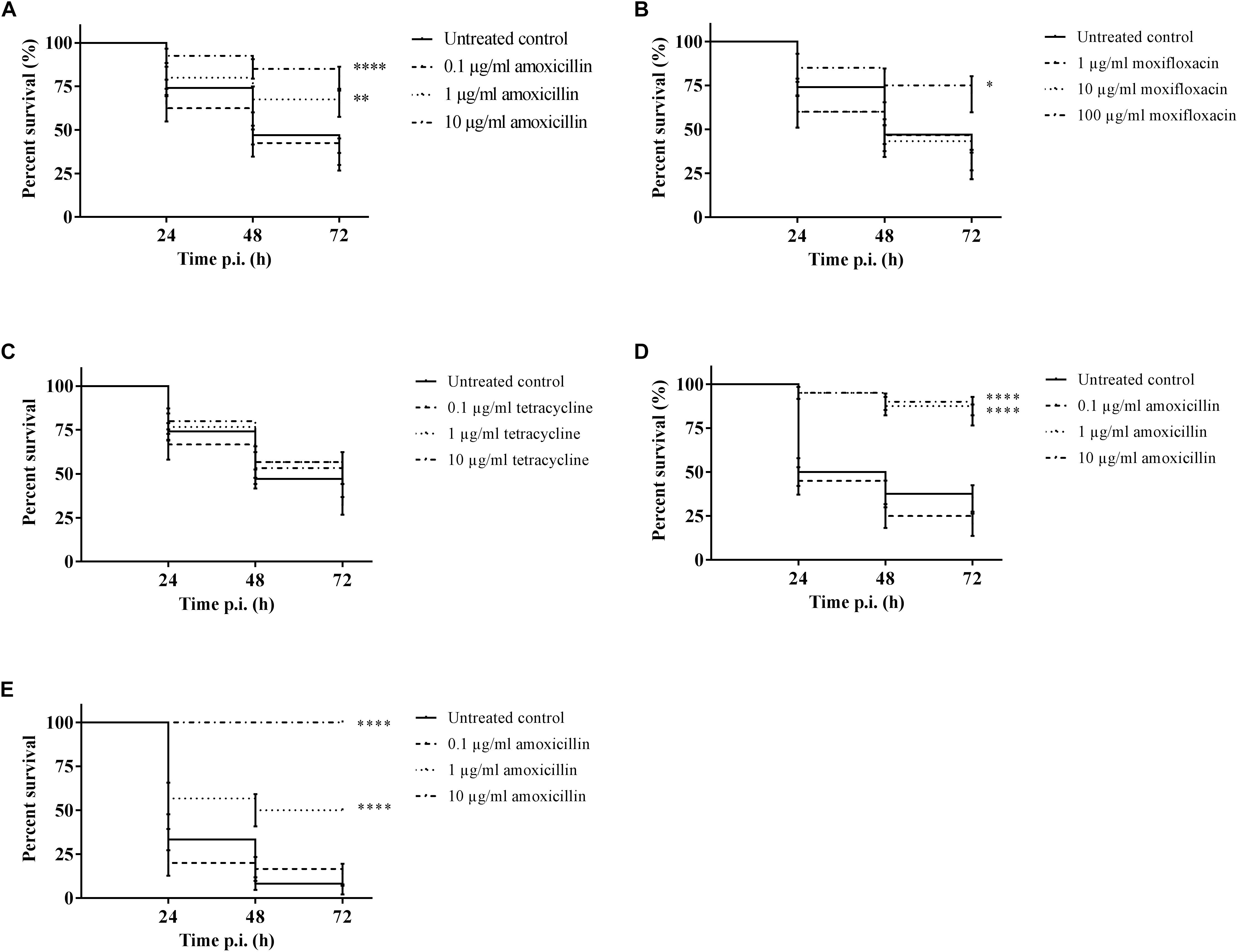
Figure 5. Survival of infected larvae after treatment with different concentrations of amoxicillin, moxifloxacin and tetracycline. (A) Survival of larvae treated with amoxicillin after TIGR4/10 infection; (B) Survival of larvae treated with moxifloxacin after TIGR4/10 infection; (C) Survival of larvae treated with tetracycline after TIGR4/10 infection; (D) Survival of larvae treated with amoxicillin after D39 infection; (E) Survival of larvae treated with amoxicillin after ATCC 49619 infection. Asterisks represent statistical differences between untreated control groups and treated groups (Survival analysis, p ≤ 0.05, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001) (n = 10 × 3).
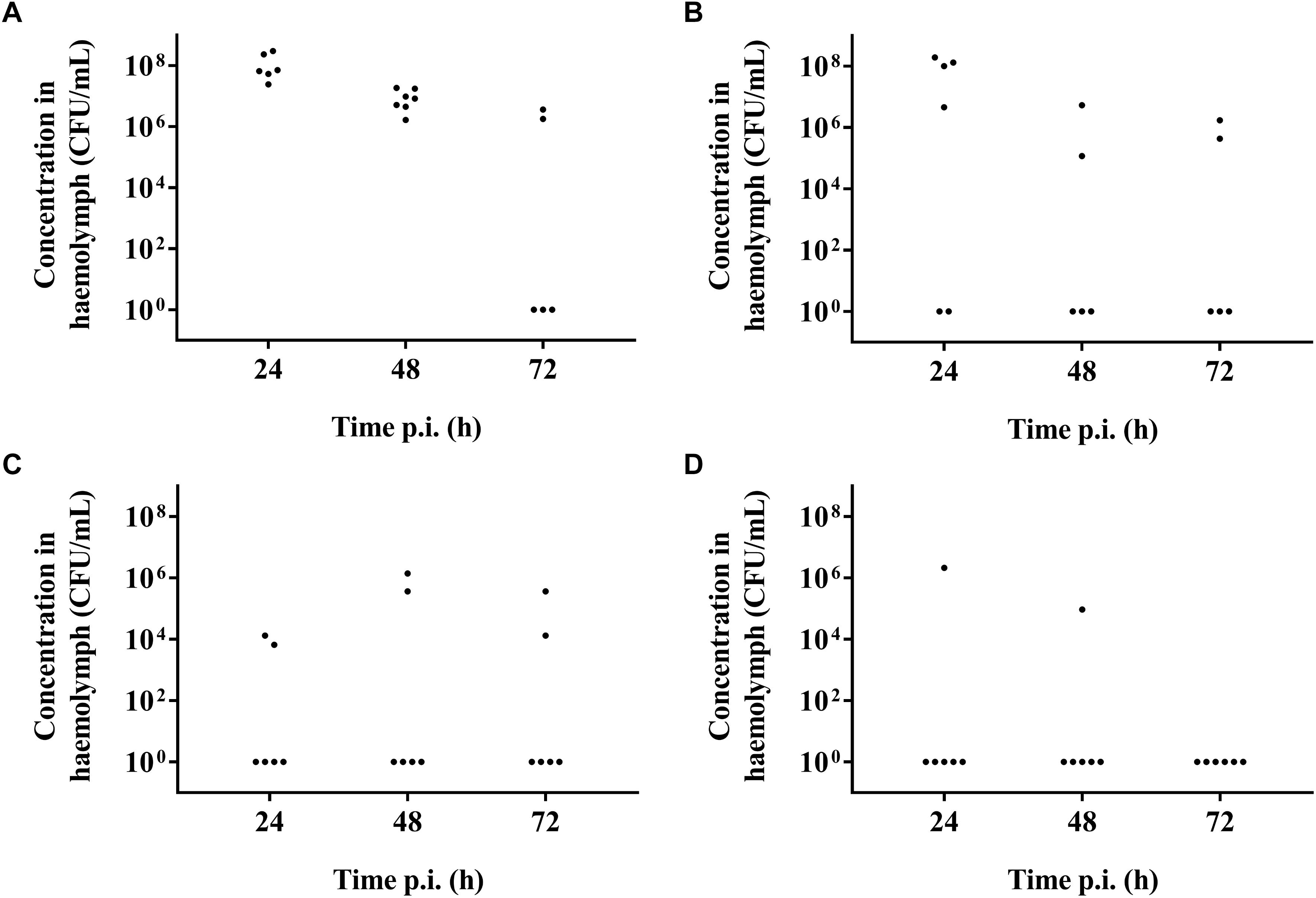
Figure 6. Bacterial burden of larval haemolymph 24, 48, and 72 h post-infection and post-treatment. (A) Burden in untreated control larvae; (B) Burden after treatment with 0.1 μg/mL amoxicillin; (C) Burden after treatment with 1 μg/mL amoxicillin; (D) Burden after treatment with 10 μg/mL amoxicillin. Each burden was determined on a pool of 3–5 living larvae (n = 2 × 3).
Secondly, larvae were infected with streptococcal strain D39. While there was no difference between untreated controls and treatment with 0.1 μg/mL amoxicillin for strain D39 (Figure 5D) (p = 0.1951, Survival analysis, Log-rank Mantel-Cox test), there was a difference between controls and treatment with 1 or 10 μg/mL amoxicillin (p < 0.0001 in both cases, Survival analysis, Log-rank Mantel-Cox test). Also for this strain, there was no difference between treatment with 1 or 10 μg/mL amoxicillin (p = 0.5469, Survival analysis, Log-rank Mantel-Cox test). Lastly, larvae were infected with streptococcal strain ATCC 49619 before amoxicillin treatment. Again, there was no difference between untreated controls and treatment with 0.1 μg/mL amoxicillin (Figure 5E) (p = 0.8283, Survival analysis, Log-rank Mantel-Cox test), but there was a difference between controls and treatment with 1 or 10 μg/mL amoxicillin (p < 0.0001 in both cases, Survival analysis, Log-rank Mantel-Cox test). However, there was also a difference between treatment with 1 or 10 μg/mL amoxicillin (p < 0.0001, Survival analysis, Log-rank Mantel-Cox test). This indicates that the dosage highly affected the outcome of infection.
For moxifloxacin (Figure 5B), only the highest concentration of moxifloxacin had an effect on the outcome of infection (p = 0.0117, Survival analysis, Log-rank Mantel-Cox test). For 1 and 10 μg/mL no differences in survival compared to the untreated controls could be observed (p = 0.6460 and p = 0.5833 respectively, Survival analysis, Log-rank Mantel-Cox test). Since TIGR4/10 harbors a tetracycline resistance gene, tetracycline was not expected to influence the course of infection (Figure 5C). Indeed, there was no difference between untreated controls and treatment with 0.1, 1, or 10 μg/mL tetracycline (p = 0.1309, p = 0.0864, and p = 0.0815 respectively, Survival analysis, Log-rank Mantel-Cox test).
Discussion
In accordance with the “Three R’s” principle (Replacement, Reduction, Refinement), alternative in vivo models to vertebrate models have been proposed to lower the costs of drug development experiments and gain data more rapidly (Graham and Prescott, 2015; López Hernández et al., 2015). While these models are often quite different from humans, they do provide valuable information regarding host-microbe interactions and are a useful in-between step in drug development (Ermolaeva and Schumacher, 2014; Trofimov et al., 2017). To study lung infections different animal models without a pulmonary system, such as C. elegans, zebrafish, or G. mellonella, are currently used (López Hernández et al., 2015). These models are used to investigate interactions with immune cells, and the model of choice is often based on the specific research need of immune cells. For all alternative models, the benefits of low costs, easy handling, lower ethical barriers and fast results often outweigh the differences in immunity between human hosts and these models (López Hernández et al., 2015).
In this study, more in-depth knowledge of a streptococcal infection was acquired. In a previous study using G. mellonella larvae, and to the best of our knowledge the only study using streptococci, the effect of a deletion in the polysaccharide capsule of streptococci was determined. The sole outcome parameter of these experiments was the survival of the larvae (Evans and Rozen, 2012). In this study, we consolidated the use of this model to study streptococcal virulence factors by using different streptococcal strains. While there was a difference in survival between serotypes, there was also a difference in survival within the same serotype with or without the polysaccharide capsule. These findings concur with data from other pathogens, such as Vibrio paraphaemolyticus, Streptococcus pyogenes, and the fungus C. albicans, where G. mellonella larvae were used to assess the effects of various virulence factors (Tsai et al., 2016; Pérez-Reytor and García, 2018; Rossoni et al., 2018). In our study, streptococcal serotype 19F was the most virulent one, as opposed to most mice models where serotype 19 is commonly less virulent compared to serotypes 2 or 4 (Briles et al., 1992; Chiavolini et al., 2008). However, serotype 19F has been reported previously to be more likely to cause invasive disease in small children compared to in older individuals (Kalin, 1998). This variation could be due to the immaturity of the adaptive immune system of small children. Since adaptive immunity is absent in G. mellonella larvae, this could explain the differences in serotype virulence compared to mice. In addition, it is worth noting virulence in mice doesn’t necessarily concur to virulence in humans. Previous research showed clinical isolates of serotype 19F were avirulent in mice, despite having their origin as human disease-causing pathogens (Briles et al., 1992). Furthermore, analogous to vertebrate models and other G. mellonella infection models, the inoculum size determines the outcome of infection (MacGowan et al., 2001; Tsai et al., 2016). An inoculum size of 106 CFU/mL was best to observe differences in virulence or to evaluate treatment efficiency. In a previous study, production of oxygen free radicals was investigated and was found to be increased in haemolymph of larvae stimulated with dihydroxyphenylalanine (DOPA) (Slepneva et al., 1999). However, these findings have never been confirmed using bacterial agents. The working mechanism of plasmatocytes and granulocytes is not fully understood, but it is known to involve a burst in oxidative metabolism (Kavanagh and Reeves, 2004). Here, we demonstrated an increase in oxygen free radicals directly after infection with strain TIGR4, yet we did not observe this effect after 24 h of infection or beyond. These data complied with the bacterial burden data, where the burden clearly decreased during the course of infection. This suggests that the haemocytes were partially capable of clearing the infection through the production of oxygen free radicals in a macrophage-like manner rather than oxygen free radicals being produced by the streptococci as a way of battling the host immune system (Slepneva et al., 1999; Bergin et al., 2005). This is confirmed by infecting the larvae with heat-killed streptococci, which are unable to produce hydrogen peroxide. Contrarily, the amount of oxygen free radicals produced during infection with strain ATCC 49619 only increases 24 h p.i., at the time point where most larvae are severely ill or moribund. Hydrogen peroxide is known to be produced by streptococci to combat the immune system and sustain an infection (Preston and Dockrell, 2008; Yesilkaya et al., 2013). For both ATCC 49619 and D39, the increase of oxygen free radicals could be appointed to this. However, if oxygen free radicals were produced by streptococci, haemocytes would start to go into apoptosis and necrosis and numbers would lower, which was not observed (Rai et al., 2015). Nevertheless, numbers might stabilize due to an equal influx of cells to combat the infection and streptococcal killing of cells.
After thoroughly assessing the model, we determined the possibility of antimicrobial treatment. In previous research studies, several therapies against Gram-negative agents, such as Pseudomonas aeruginosa, were evaluated using G. mellonella larvae. For example, it was recently shown that a cocktail of four novel bacteriophages can cure infections in both mice and wax moth larvae (Forti et al., 2018). For an in-depth overview of evaluation of antibacterial agents using the G. mellonella model and a list of pathogens which have already been studied, see the review of Tsai et al. (2016). Treatment of fungal infections has also been proven possible. Recently, the use of probiotic Lactobacillus spp. for C. albicans infections has been studied and proven effective (Santos et al., 2018). Furthermore, in the first 6 months of 2018 alone, 5 novel research studies were published evaluating both prophylactic and therapeutic treatment options for C. albicans in G. mellonella—showing a clear interest and application for this model (Liu et al., 2017; Borowiecki et al., 2018; Chupáčová et al., 2018; Gu et al., 2018; Staniszewska et al., 2018). Here, we assessed the functionality of 2 standard antibiotics, amoxicillin and moxifloxacin. Amoxicillin is a bactericidal beta-lactam antibiotic commonly used in the first-line treatment of lower pulmonary infections in both children and adults. Moxifloxacin is a fluoroquinolone used when amoxicillin is contra-indicated due to a penicillin allergy in adults (Belgische Commissie voor de Coördinatie van het Antibioticabeleid, 2012). We observed a clear dose-response effect in our in vivo model for both amoxicillin and moxifloxacin and observed a decrease in bacterial burden after amoxicillin usage compared to a non-treated group.
There are several limitations to the wax moth model. Firstly, in contrast to mammalian models, G. mellonella larvae do not possess an adaptive immune system (Wojda, 2017). However, the innate immune system is the first-line defense in human lungs through the presence of alveolar macrophages (Marriott and Dockrell, 2007; Camberlein et al., 2015). Therefore, the larval model – with its phagocytic haemocytes – can be used as a substitution for more complex vertebrate models. Larval haemocytes possess a similar mechanism for phagocytosis compared to human macrophages, also using reactive oxygen species through activation of the NADPH oxidase complex. In this larval complex, immunologically related proteins to human p47phox and p67phox have been observed (Bergin et al., 2005). While the differences in immunity between G. mellonella larvae and vertebrates limit the possibility of replacing vertebrate in vivo models, this model can be used as an in-between step between in vitro and complex in vivo models (Tsai et al., 2016).
This model has previously been evaluated for its use in toxicity studies and antimicrobial susceptibility testing using standard antibiotics and several Gram-positive and Gram-negative strains. A correlation between toxicity in wax moth larvae and rodents could be observed. Furthermore, the therapeutic doses for human use of the tested antibiotics could be translated to the doses in larvae (Ignasiak and Maxwell, 2017). Also, the model is currently being used for the evaluation of potential novel antimicrobial strategies against pathogens such as C. albicans, Burkholderia cenocepacia, Porphyromonas gingivalis, and Klebsiella pneumoniae (dos Santos et al., 2017; Van den Driessche et al., 2017; Nath et al., 2018; Staniszewska et al., 2018). The implementation of this model leads to a lower number of compounds going into further in vivo trials, thereby reducing the number of vertebrates (Ignasiak and Maxwell, 2017). While the determination of streptococcal bacterial burden currently requires larval death, bioluminescence could be a valuable addition to the model. Use of bioluminescence for in vivo imaging in G. mellonella has been previously assessed using lux tagged promotors of virulence factors in L. monocytogenes infections (Joyce and Gahan, 2010). In fungal infections, bioluminescence has been used to study C. albicans burden both ex vivo and in vivo (Delarze et al., 2015). More recently, bioluminescence has been applied to study in vivo infections with Mycobacterium abscessus (Meir et al., 2018).
Conclusion
In conclusion, our results strengthen the idea of implementing the G. mellonella model in the drug discovery process on a standardized basis. It is possible to evaluate host parameters, such as haemocyte count and production of oxygen free radicals, and to monitor bacterial burden during the course of infection. This model could provide valuable information regarding potential novel antimicrobial compounds, without the use of expensive and ethically challenging vertebrate models. While this model cannot replace the vertebrate models currently used, it can be a valuable addition in the transition from in vitro to in vivo data.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
FC, BV, and PC conceived the presented idea. FC carried out the experiments. ET, JA, BV, DC, PD, and PC contributed to interpretation of results. FC wrote the manuscript with support of ET, JA, BV, DB, LM, and GC. PD and PC helped supervise the project. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was partially supported by the European Training Network (MSCA-ITN-ETN #722467, PRINT-AID project).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00281/full#supplementary-material
References
Beilharz, K., van Raaphorst, R., Kjos, M., and Veening, J.-W. (2015). Red fluorescent proteins for gene expression and protein localization studies in Streptococcus pneumoniae and efficient transformation with DNA assembled via the gibson assembly method. Appl. Environ. Microbiol. 81, 7244–7252. doi: 10.1128/AEM.02033-15
Belgische Commissie voor de Cordinatie van het Antibioticabeleid (2012). Belgische gids voor anti-infectieuze behandeling in de ambulante praktijk. Available at: http://www.bcfi.be/legacy_assets/antibioticagids-nl.pdf [Accessed June 20, 2018].
Bergin, D., Reeves, E. P., Renwick, J., Wientjes, F. B., and Kavanagh, K. (2005). Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect. Immun. 73, 4161–4170. doi: 10.1128/IAI.73.7.4161-4170.2005
Borghi, E., Romagnoli, S., Fuchs, B. B., Cirasola, D., Perdoni, F., Tosi, D., et al. (2014). Correlation between Candida albicans biofilm formation and invasion of the invertebrate host Galleria mellonella. Future Microbiol. 9, 163–173. doi: 10.2217/fmb.13.159
Borowiecki, P., Wińska, P., Bretner, M., Gizińska, M., Koronkiewicz, M., and Staniszewska, M. (2018). Synthesis of novel proxyphylline derivatives with dual anti-Candida albicans and anticancer activity. Eur. J. Med. Chem. 150, 307–333. doi: 10.1016/j.ejmech.2018.02.077
Briles, D. E., Crain, M. J., Gray, B. M., Forman, C., and Yother, J. (1992). Strong association between capsular type and virulence for mice among human isolates of Streptococcus pneumoniae. Infect. Immun. 60, 111–116.
Camberlein, E., Cohen, J. M., José, R., Hyams, C. J., Callard, R., Chimalapati, S., et al. (2015). Importance of bacterial replication and alveolar macrophage-independent clearance mechanisms during early lung infection with Streptococcus pneumoniae. Infect. Immun. 83, 1181–1189. doi: 10.1128/IAI.02788-14
Chakraborty, C., Hsu, C. H., Wen, Z. H., Lin, C. S., and Agoramoorthy, G. (2009). Zebrafish: a complete animal model for in vivo drug discovery and development. Curr. Drug Metab. 10, 116–124. doi: 10.2174/138920009787522197
Champion, O. L., Wagley, S., and Titball, R. W. (2016). Galleria mellonella as a model host for microbiological and toxin research. Virulence 7, 840–845. doi: 10.1080/21505594.2016.1203486
Chiavolini, D., Pozzi, G., and Ricci, S. (2008). Animal models of Streptococcus pneumoniae disease. Clin. Microbiol. Rev. 21, 666–685. doi: 10.1128/cmr.00012-08
Chupáčová, J., Borghi, E., Morace, G., Los, A., and Bujdáková, H. (2018). Anti-biofilm activity of antibody directed against surface antigen complement receptor 3-related protein—comparison of Candida albicans and Candida dubliniensis. Pathog. Dis. 76:ftx127. doi: 10.1093/femspd/ftx127
de Kraker, M. E. A., Jarlier, V., Monen, J. C. M., Heuer, O. E., van de Sande, N., and Grundmann, H. (2013). The changing epidemiology of bacteraemias in Europe: trends from the european antimicrobial resistance surveillance system. Clin. Microbiol. Infect. 19, 860–868. doi: 10.1111/1469-0691.12028
Delarze, E., Ischer, F., Sanglard, D., and Coste, A. T. (2015). Adaptation of a Gaussia princeps luciferase reporter system in Candida albicans for in vivo detection in the Galleria mellonella infection model. Virulence 6, 684–693. doi: 10.1080/21505594.2015.1081330
dos Santos, J. D., de Alvarenga, J. A., Rossoni, R. D., García, M. T., Moraes, R. M., Anbinder, A. L., et al. (2017). Immunomodulatory effect of photodynamic therapy in Galleria mellonella infected with Porphyromonas gingivalis. Microb. Pathog. 110, 507–511. doi: 10.1016/j.micpath.2017.07.045
Duggan, G. M., and Mostowy, S. (2018). Use of zebrafish to study Shigella infection. Dis. Model. Mech. 11, dmm032151. doi: 10.1242/dmm.032151
Dunphy, G. B., Oberholzer, U., Whiteway, M., Zakarian, R. J., and Boomer, I. (2003). Virulence of Candida albicans mutants toward larval Galleria mellonella (insecta, lepidoptera, galleridae). Can. J. Microbiol. 49, 514–524. doi: 10.1139/w03-064
Ermolaeva, M. A., and Schumacher, B. (2014). Insights from the worm: the C. elegans model for innate immunity. Semin. Immunol. 26, 303–309. doi: 10.1016/j.smim.2014.04.005
Evans, B. A., and Rozen, D. E. (2012). A Streptococcus pneumoniae infection model in larvae of the wax moth Galleria mellonella. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2653–2660. doi: 10.1007/s10096-012-1609-7
Forti, F., Roach, D. R., Cafora, M., Pasini, M. E., Horner, D. S., Fiscarelli, E. V., et al. (2018). Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 62, e2573-17. doi: 10.1128/AAC.02573-17
Graham, M. L., and Prescott, M. J. (2015). The multifactorial role of the 3Rs in shifting the harm-benefit analysis in animal models of disease. Eur. J. Pharmacol. 759, 19–29. doi: 10.1016/j.ejphar.2015.03.040
Gu, W., Yu, Q., Yu, C., and Sun, S. (2018). In vivo activity of fluconazole/tetracycline combinations in Galleria mellonella with resistant Candida albicans infection. J. Glob. Antimicrob. Resist. 13, 74–80. doi: 10.1016/j.jgar.2017.11.011
Harding, C. R., Schroeder, G. N., Collins, J. W., and Frankel, G. (2013). Use of Galleria mellonella as a model organism to study Legionella pneumophila infection. J. Vis. Exp. 81:e50964. doi: 10.3791/50964
Hill, L., Veli, N., and Coote, P. J. (2014). Evaluation of Galleria mellonella larvae for measuring the efficacy and pharmacokinetics of antibiotic therapies against Pseudomonas aeruginosa infection. Int. J. Antimicrob. Agents 43, 254–261. doi: 10.1016/j.ijantimicag.2013.11.001
Ignasiak, K., and Maxwell, A. (2017). Galleria mellonella (greater wax moth) larvae as a model for antibiotic susceptibility testing and acute toxicity trials. BMC Res. Notes 10:428. doi: 10.1186/s13104-017-2757-8
Joyce, S. A., and Gahan, C. G. M. (2010). Molecular pathogenesis of Listeria monocytogenes in the alternative model host Galleria mellonella. Microbiology 156, 3456–3468. doi: 10.1099/mic.0.040782-0
Kadioglu, A., Weiser, J. N., Paton, J. C., and Andrew, P. W. (2008). The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6, 288–301. doi: 10.1038/nrmicro1871
Kalin, M. (1998). Pneumococcal serotypes and their clinical relevance. Thorax 53, 159–162. doi: 10.1136/THX.53.3.159
Kavanagh, K., and Reeves, E. P. (2004). Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28, 101–112. doi: 10.1016/j.femsre.2003.09.002
Lange, A., Beier, S., Huson, D. H., Parusel, R., Iglauer, F., and Frick, J.-S. (2018). Genome sequence of galleria mellonella (greater wax moth). Genome Announc. 6, e1220-17. doi: 10.1128/genomeA.01220-17
Liu, X., Li, T., Wang, D., Yang, Y., Sun, W., Liu, J., et al. (2017). Synergistic antifungal effect of fluconazole combined with licofelone against resistant Candida albicans. Front. Microbiol. 8:2101. doi: 10.3389/fmicb.2017.02101
Loh, J. M., Adenwalla, N., Wiles, S., and Proft, T. (2013). Galleria mellonella larvae as an infection model for group a streptococcus. Virulence 4, 419–428. doi: 10.4161/viru.24930
López Hernández, Y., Yero, D., Pinos-Rodríguez, J. M., and Gibert, I. (2015). Animals devoid of pulmonary system as infection models in the study of lung bacterial pathogens. Front. Microbiol. 6:38. doi: 10.3389/fmicb.2015.00038
MacGowan, A., Rogers, C., and Bowker, K. (2001). In vitro models, in vivo models, and pharmacokinetics: what can we learn from in vitro models? Clin. Infect. Dis. 33, S214–S220. doi: 10.1086/321850
Marriott, H. M., and Dockrell, D. H. (2007). The role of the macrophage in lung disease by bacteria. Exp. Lung Res. 33, 493–505. doi: 10.1080/01902140701756562
Martin, B., García, P., Castanié, M. P., and Claverys, J. P. (1995). The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol. Microbiol. 15, 367–379. doi: 10.1111/j.1365-2958.1995.tb02250.x
Meeker, N. D., and Trede, N. S. (2008). Immunology and zebrafish: spawning new models of human disease. Dev. Comp. Immunol. 32, 745–757. doi: 10.1016/J.DCI.2007.11.011
Meijer, A. H., and Spaink, H. P. (2011). Host-pathogen interactions made transparent with the zebrafish model. Curr. Drug Targets 12, 1000–1017. doi: 10.2174/138945011795677809
Meir, M., Grosfeld, T., and Barkan, D. (2018). Establishment and validation of galleria mellonella as a novel model organism to study Mycobacterium abscessus infection, pathogenesis, and treatment. Antimicrob. Agents Chemother. 62, e2539-17. doi: 10.1128/AAC.02539-17
Nath, S., Moussavi, F., Abraham, D., Landman, D., and Quale, J. (2018). In vitro and in vivo activity of single and dual antimicrobial agents against KPC-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 431–436. doi: 10.1093/jac/dkx419
Pérez-Reytor, D., and García, K. (2018). Galleria mellonella: a model of infection to discern novel mechanisms of pathogenesis of non-toxigenic Vibrio parahaemolyticus strains. Virulence 9, 22–24. doi: 10.1080/21505594.2017.1388487
Preston, J. A., and Dockrell, D. H. (2008). Virulence factors in pneumococcal respiratory pathogenesis. Future Microbiol. 3, 205–221. doi: 10.2217/17460913.3.2.205
Rai, P., Parrish, M., Tay, I. J. J., Li, N., Ackerman, S., He, F., et al. (2015). Streptococcus pneumoniae secretes hydrogen peroxide leading to DNA damage and apoptosis in lung cells. Proc. Natl. Acad. Sci. U.S.A. 112, E3421–E3430. doi: 10.1073/pnas.1424144112
Rennie, R. P. (2012). Current and future challenges in the development of antimicrobial agents. Handb. Exp. Pharmacol. 211, 45–65. doi: 10.1007/978-3-642-28951-4_4
Rossoni, R. D., dos Santos Velloso, M., Figueiredo, L. M. A., Martins, C. P., Jorge, A. O. C., and Junqueira, J. C. (2018). Clinical strains of Lactobacillus reduce the filamentation of Candida albicans and protect Galleria mellonella against experimental candidiasis. Folia Microbiol. 63, 307–314. doi: 10.1007/s12223-017-0569-9
Santos, R. B., Scorzoni, L., Namba, A. M., Rossoni, R. D., Jorge, A. O. C., and Junqueira, J. C. (2018). Lactobacillus species increase the survival of Galleria mellonella infected with Candida albicans and non–albicans Candida clinical isolates. Med. Mycol. doi: 10.1093/mmy/myy032 [Epub ahead of print].
Schulenburg, H., and Ewbank, J. J. (2007). The genetics of pathogen avoidance in Caenorhabditis elegans. Mol. Microbiol. 66, 563–570. doi: 10.1111/j.1365-2958.2007.05946.x
Slepneva, I. A., Glupov, V. V., Sergeeva, S. V., and Khramtsov, V. V. (1999). EPR detection of reactive oxygen species in hemolymph of Galleria mellonella and Dendrolimus superans sibiricus (Lepidoptera) larvae. Biochem. Biophys. Res. Commun. 264, 212–215. doi: 10.1006/bbrc.1999.1504
Squiban, B., and Kurz, C. L. (2011). C. elegans: an all in one model for antimicrobial drug discovery. Curr. Drug Targets 12, 967–977. doi: 10.2174/138945011795677854
Staniszewska, M., Gizińska, M., Mikulak, E., Adamus, K., Koronkiewicz, M., and Łukowska-Chojnacka, E. (2018). New 1,5 and 2,5-disubstituted tetrazoles-dependent activity towards surface barrier of Candida albicans. Eur. J. Med. Chem. 145, 124–139. doi: 10.1016/j.ejmech.2017.11.089
Torres, A., Cillóniz, C., Blasi, F., Chalmers, J. D., Gaillat, J., Dartois, N., et al. (2018). Burden of pneumococcal community-acquired pneumonia in adults across Europe: a literature review. Respir. Med. 137, 6–13. doi: 10.1016/J.RMED.2018.02.007
Trofimov, V., Costa-Gouveia, J., Hoffmann, E., and Brodin, P. (2017). Host–pathogen systems for early drug discovery against tuberculosis. Curr. Opin. Microbiol. 39, 143–151. doi: 10.1016/j.mib.2017.11.017
Tsai, C. J., Loh, J. M., and Proft, T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7, 214–229. doi: 10.1080/21505594.2015.1135289
Van den Driessche, F., Vanhoutte, B., Brackman, G., Crabbé, A., Rigole, P., Vercruysse, J., et al. (2017). Evaluation of combination therapy for Burkholderia cenocepacia lung infection in different in vitro and in vivo models. PLoS One 12:e0172723. doi: 10.1371/journal.pone.0172723
Wojda, I. (2017). Immunity of the greater wax moth Galleria mellonella. Insect Sci. 24, 342–357. doi: 10.1111/1744-7917.12325
Keywords: Streptococcus pneumoniae, Galleria mellonella, infection model, virulence, antibiotics, wax moth larvae
Citation: Cools F, Torfs E, Aizawa J, Vanhoutte B, Maes L, Caljon G, Delputte P, Cappoen D and Cos P (2019) Optimization and Characterization of a Galleria mellonella Larval Infection Model for Virulence Studies and the Evaluation of Therapeutics Against Streptococcus pneumoniae. Front. Microbiol. 10:311. doi: 10.3389/fmicb.2019.00311
Received: 16 November 2018; Accepted: 05 February 2019;
Published: 21 February 2019.
Edited by:
Grégory Resch, Université de Lausanne, SwitzerlandReviewed by:
Sabine Stegemann-Koniszewski, Universitätsklinikum Magdeburg, GermanyLance Edward Keller, University of Groningen, Netherlands
Copyright © 2019 Cools, Torfs, Aizawa, Vanhoutte, Maes, Caljon, Delputte, Cappoen and Cos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul Cos, cGF1bC5jb3NAdWFudHdlcnBlbi5iZQ==
†These authors have contributed equally to this work
 Freya Cools
Freya Cools Eveline Torfs
Eveline Torfs Juliana Aizawa
Juliana Aizawa Bieke Vanhoutte
Bieke Vanhoutte Louis Maes
Louis Maes Guy Caljon
Guy Caljon Peter Delputte
Peter Delputte Davie Cappoen
Davie Cappoen Paul Cos
Paul Cos