
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 February 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00283
 Chengsi Li1†
Chengsi Li1† Haiyan Zeng1†
Haiyan Zeng1† Jumei Zhang1†
Jumei Zhang1† Wenjing He1
Wenjing He1 Na Ling1
Na Ling1 Moutong Chen1
Moutong Chen1 Shi Wu1
Shi Wu1 Tao Lei1
Tao Lei1 Haoming Wu1
Haoming Wu1 Yingwang Ye1
Yingwang Ye1 Yu Ding2
Yu Ding2 Juan Wang3
Juan Wang3 Xianhu Wei1
Xianhu Wei1 Youxiong Zhang1
Youxiong Zhang1 Qingping Wu1*
Qingping Wu1*Cronobacter spp. are foodborne pathogens that can infect and cause life-threatening diseases in all age groups, particularly in infants and immunocompromised elderly. This study aimed to investigate the prevalence, antibiotic susceptibility, and molecular characteristics of Cronobacter spp. isolates in edible mushrooms collected from 44 cities in China. In total, 668 edible mushrooms were collected from traditional retail markets and supermarkets and were analyzed by quantitative methods, PCR-based serotyping, multilocus sequence typing (MLST), and antibiotic susceptibility testing. Among the 668 samples tested, 89 (13.32%) were positive for Cronobacter spp., and the contamination levels exceeded the 110 most probable number (MPN)/g in 13.48% (12/89) of the samples. Flammulina velutipes samples had the highest contamination rate of 17.54% (37/211), whereas Hypsizygus marmoreus samples had the lowest contamination rate of 3.28% (2/61). Ten serotypes were identified among 115 isolates, of which the C. sakazakii serogroup O1 (n = 32) was the primary serotype. MLST indicated that there was quite high genetic diversity in Cronobacter spp. and 72 sequence types were identified, 17 of which were new. Notably, C. sakazakii ST148 (n = 10) was the most prevalent, followed by C. malonaticus ST7 (n = 5). Antibiotic susceptibility testing revealed that the majority of Cronobacter spp. strains were susceptible to the 16 antibiotics tested. However, a portion of isolates exhibited relatively high resistance to cephalothin, with resistance and intermediate rates of 93.91 and 6.09%, respectively. One isolate (cro300A) was multidrug-resistant, with resistance to five antibiotics. Overall, this large-scale study revealed the relatively high prevalence and high genetic diversity of Cronobacter spp. on edible mushrooms in China, indicating a potential public health concern. To our knowledge, this is the first large-scale and systematic study on the prevalence of Cronobacter spp. on edible mushrooms in China, and the findings can provide valuable information that can guide the establishment of effective measures for the control and precaution of Cronobacter spp on edible mushrooms during production processes.
Cronobacter spp. is a gram-negative, motile, rod-shaped, non-sporulating pathogenic bacterium belonging to the family Enterobacteriaceae (Barron and Forsythe, 2007). Currently, the genus comprises seven species: C. sakazakii, C. malonaticus, C. turicensis, C. universalis, C. muytjensii, C. dublinensis, and C. condimenti (Iversen and Forsythe, 2007; Joseph and Forsythe, 2012; Forsythe et al., 2014). Of these, C. sakazakii, C. malonaticus, and C. turicensis have been proven relevant to human infections (Liu et al., 2013; Forsythe et al., 2014), and outbreaks have been reported in some countries (Caubilla-Barron et al., 2007; Patrick et al., 2014). Cronobacter infections occur in all age groups, although with a greater incidence in the very young and elderly, who are typically more immunocompromised (Forsythe S., 2018). Clinical symptoms of infection in infants include meningitis, bacteraemia, and necrotizing enterocolitis, with mortality rates of 40–80% (Friedemann, 2009). Infections in the adults show a wide range of symptoms: conjunctivitis, biliary sepsis, urosepsis, wound infections, appendicitis, and pneumonia (Lai, 2001; Patrick et al., 2014). Cronobacter spp. has been isolated not only from clinical samples, but also from various foods, including cereals, meat, herbs, spices, salads, fruits, and vegetables, as well as their as well as derivative food products (Alsonosi et al., 2015; Ueda, 2017). Cronobacter spp. infecting infants are generally believed to be derived from infant milk formulas (van Acker et al., 2001), although other food sources, such as breast milk, have been suspected in some cases (Stoll et al., 2004). The source of infections in adults is still unidentified (Joseph and Forsythe, 2012). Some research reports indicated that the principal sources of this organism are associated with soil, water, and vegetables (Ueda, 2017; Zeng et al., 2018b). However, a clear understanding about the epidemiology and reservoirs of Cronobacter spp. is still lacking (Holý and Forsythe, 2014). An in-depth understanding of the genetic diversity of Cronobacter spp. can effectively promote reliable source tracking of contaminated foods and enhance the resolution of surveillance.
PCR-based O-antigen serotyping methods for identifying isolates in epidemiological studies of bacteria have been successfully developed (Jones et al., 2012). To date, 24 serogroups of Cronobacter spp. have been identified (BlaŽková et al., 2015; Forsythe S. J., 2018). Furthermore, multilocus sequence typing (MLST) based on seven housekeeping genes (atpD, fusA, glnS, gltB, gyrB, infB, and ppsA) has been established in the genus Cronobacter. The Cronobacter PubMLST database (http://pubmlst.org/Cronobacter/) is an open access, sequence-based repository in which 633 sequence types (STs) are stored. MLST has been proven a powerful tool to effectively identify and discriminate different Cronobacter species, and its application has accelerated our understanding of the STs and studies on outbreaks of these bacteria (Joseph et al., 2012b; Xu et al., 2015). C. sakazakii ST4 is the predominant ST in the clinical setting (Joseph and Forsythe, 2011; Hariri et al., 2013). C. malonaticus ST7 is significantly associated with adult infections, although the source has not yet been determined (Joseph and Forsythe, 2012). Till now, some novel STs were also reported to cause infant meningitis (Cui et al., 2017; Chaves et al., 2018; Zeng et al., 2018a).
Currently, antimicrobial resistance, particularly multidrug resistance, poses a public health concern because certain pathogenic bacteria are not disrupted by conventional treatment, which results in prolonged illness and a greater risk of death. Most Cronobacter spp. isolates are susceptible to commonly used antibiotic agents; however, prolonged and extensive antibiotic use has led to the emergence of single- and multidrug resistant strains. Of late, some Cronobacter spp. isolates from food have been reported to be resistant to cephalothin, cefotaxime, streptomycin, ampicillin, penicillin G, and amoxicillin-clavulanate (Chon et al., 2012; Pan et al., 2014; Fei et al., 2017a). Therefore, it is necessary analyze the potential association between antibiotic resistance and the source of Cronobacter spp. isolates, especially the clinical setting and various foods.
In China, edible mushrooms are popular foods that are frequently used as raw materials in other foods owing to their high nutritional value, delicacy, and good chewiness (Chang and Buswell, 1996; Bao et al., 2013; Ye et al., 2014). Although infants are not fed with mushroom, the elderly (or adults) will eat it, despite the potential risk (Chon et al., 2012; Aksu et al., 2016; Chitrakar et al., 2018). A recent study reported an acute gastroenteritis outbreak by the infection of C. sakazakii which occurred in a local senior high school of China, although without any specification about the food (Yong et al., 2018). In addition, China's share in global mushroom production increased from 5.7% in 1978 to 80% in 2011 (Zhang et al., 2014). However, our previous study showed that edible mushrooms are often contaminated with foodborne pathogens, such as Listeria monocytogenes, Staphylococcus aureus, and Cronobacter (Chen et al., 2014; Ye et al., 2014; Wu et al., 2015; Huang et al., 2018). These findings highlight the importance of evaluating the microbiological profile of edible mushrooms for food safety. Therefore, this study aimed to investigate the contamination levels of edible mushrooms in China with Cronobacter spp., to identify molecular features of the isolates by O-antigen serotyping and MLST analysis, and to evaluate antibiotic resistance patterns.
In total, 668 edible mushroom samples, including Flammulina velutipes (n = 211), Lentinus edodes (n = 114), Pleurotus ostreatus (n = 104), Pleurotus eryngii (n = 98); Hypsizygus marmoreus (n = 80), and other species (n = 61) were collected from traditional retail markets and supermarkets during July 2011 and June 2016 (Table 1). The samples were obtained from 44 cities geographically spread over China (Figure 1). The samples were placed in sterile sealed plastic bags, transferred under cold conditions (below 4°C) to the laboratory, and analyzed immediately.
Figure 1. The sampling locations of the edible mushrooms for this study in China, including 44 cities, covering most cities of China.
Quantitative detection was conducted by an isolation and enrichment method according to the National Food Safety Standard of China Food microbiological examination: Enterobacter sakazakii (GB 4789.40-2010; National Standard of the People's Republic of China, 2010) for powdered infant formula (PIF), as described previously, with some modifications (Xu et al., 2015; Ling et al., 2018). We modified the method based on the analysis of PIF to mushroom for Cronobacter. The nine-tube method was applied to calculate the most probable number (MPN). The MPN was determined on the basis of the number of positive tube(s) in each of the three sets and the MPN table (GB 4789.7-2013, National Standard of the People's Republic of China, 2013). Green or blue-green colonies in chromogenic Enterbacter sakazakii Agar Plate were considered as presumptive Cronobacter spp. and were selected for analysis using API 20E diagnostic strips (BioMérieux, Marcy-l'Étoile, France). Species identification of these isolates was conducted by fusA sequencing (Joseph and Forsythe, 2012).
Genomic DNA was extracted with the HiPure Bacterial DNA Kit (Magen Technologies, Guangzhou, China). The serotypes of Cronobacter spp. isolates were identified according to previously reported Cronobacter molecular serotyping schemes, including C. sakazakii O1 to O4, O6, and O7 (Jarvis et al., 2011; Sun et al., 2011, 2012), C. malonaticus O1 to O4 (BlaŽková et al., 2015), and C. dublinensis O1 to O4 (Jarvis et al., 2011).
MLST was used for molecular typing of the Cronobacter isolates, as previously reported (Joseph et al., 2012a). The designation of new alleles and STs was verified by Stephen J. Forsythe (https://pubmlst.org/cronobacter/), the MLST database curator. Neighbor-joining trees were constructed based on the seven MLST loci (concatenated length of 3,036 bp) of the Cronobacter spp. using MEGA (version 5.05) as previous study (Ling et al., 2018).
The susceptibility profiles of Cronobacter spp. isolates were determined by antimicrobial dilution and disk susceptibility testing using Mueller-Hinton agar (Huankai, Guangzhou, China), following the protocols of the Clinical and Laboratory Standards Institute (Jorgensen, 2015). Sixteen antimicrobials (AMs) (Oxoid, Hampshire, United Kingdom) recommended for Enterobacteriaceae were tested, as previously reported (Ling et al., 2018).
As shown in Table 1, the 668 samples were isolated from six mushroom species, and 89 (13.32%) samples tested positive for Cronobacter spp. The prevalence rate of Cronobacter spp. varied among the different mushroom species: 17.54% (37/211) were obtained from F. velutipes, 5.26% (6/114) from L. edodes, 14.42% (15/104) from P. ostreatus, 10.20% (10/98) from P. eryngii, 5.00% (4/80) from H. marmoreus, and 27.87% (17/61) from other species. Based on the MPN analysis, the contamination level of Cronobacter spp. was <10 MPN/g in 77.53% (69/89) of the samples, ranged between 10 and 110 MPN/g in eight samples, and exceeded 110 MPN/g in 12 samples, five of which were F. velutipes samples. Moreover, the positive samples, including H. marmoreus (55.46 MPN/g), V. volvacea (50.29 MPN/g), and F. velutipes (21.43 MPN/g) showed a higher mean contamination level than other species of mushroom. The mean contamination level of L. edodes, P. ostreatus, and P. eryngii was lower than 2 MPN/g. Isolates from the same sample belonging to the same ST and serotype were considered clonal. Thus, the 115 Cronobacter isolates from the 89 positive samples were identified as three species (Table 2). The majority (60.87%, 70/115) of the isolates were identified as C. sakazakii, followed by C. malonaticus (29.57%, 34/115), and C. dublinensis (9.57%, 11/115).
O-antigen serotyping according to the size of the target gene was employed to evaluate the distribution of O-antigen serotypes among the 115 Cronobacter spp. isolates from edible mushrooms. As shown in Table 2, all C. sakazakii serotypes were detected among the 70 isolates. Among them, O1 was the dominant serotype (32 isolates), followed by serotype O2 (20 isolates). In addition, 34 C. malonaticus isolates were classified into serotypes O1 (10 isolates), O2 (15 isolates), and O3 (9 isolates). Nine C. dublinensis serotype O1 and 2 serotype O2 isolates were identified.
A phylogenetic tree was constructed by the neighbor-joining method using the concatenated sequences of seven housekeeping genes (3,036 bp) for the 115 Cronobacter isolates. As shown in Figure 2, the tree showed a clear relatedness between the 115 Cronobacter spp. isolates, which were grouped into 72 STs, 17 of which were novel, and divided into three clusters representing the three species The new nucleotide sequences have been deposited in the PubMLST database under IDs 2457 to 2474 (http://pubmlst.org/Cronobacter/). Fifty-two of the 72 STs were unique to only one isolate, and the remaining 20 STs covered two to 10 isolates each. There were 44 STs including seven new STs in C. sakazakii isolates. ST148 was the dominant ST (n = 10), followed by ST4 (n = 4), and ST40 (n = 4). Eighteen STs were included in C. malonaticus isolates and ST7 (n = 5) was the dominant ST, followed by ST60 (n = 4). In C. dublinensis isolates, the newly identified ST658 (n = 2) was the dominant ST, the other isolates had unique STs. Furthermore, ST1 was found only in F. velutipes and P. eryngii; ST4 was found in F. velutipes, P. ostreatus, and H. marmoreus; ST7 was found in F. velutipes, P. eryngii, and V. volvacea; ST8 was found in F. velutipes and V. volvacea; ST60 was found in F. velutipes, P. ostreatus, and Agaricus bisporus, and ST64 was found in F. velutipes and P. ostreatus. Several pathogenic STs were found in F. velutipes.
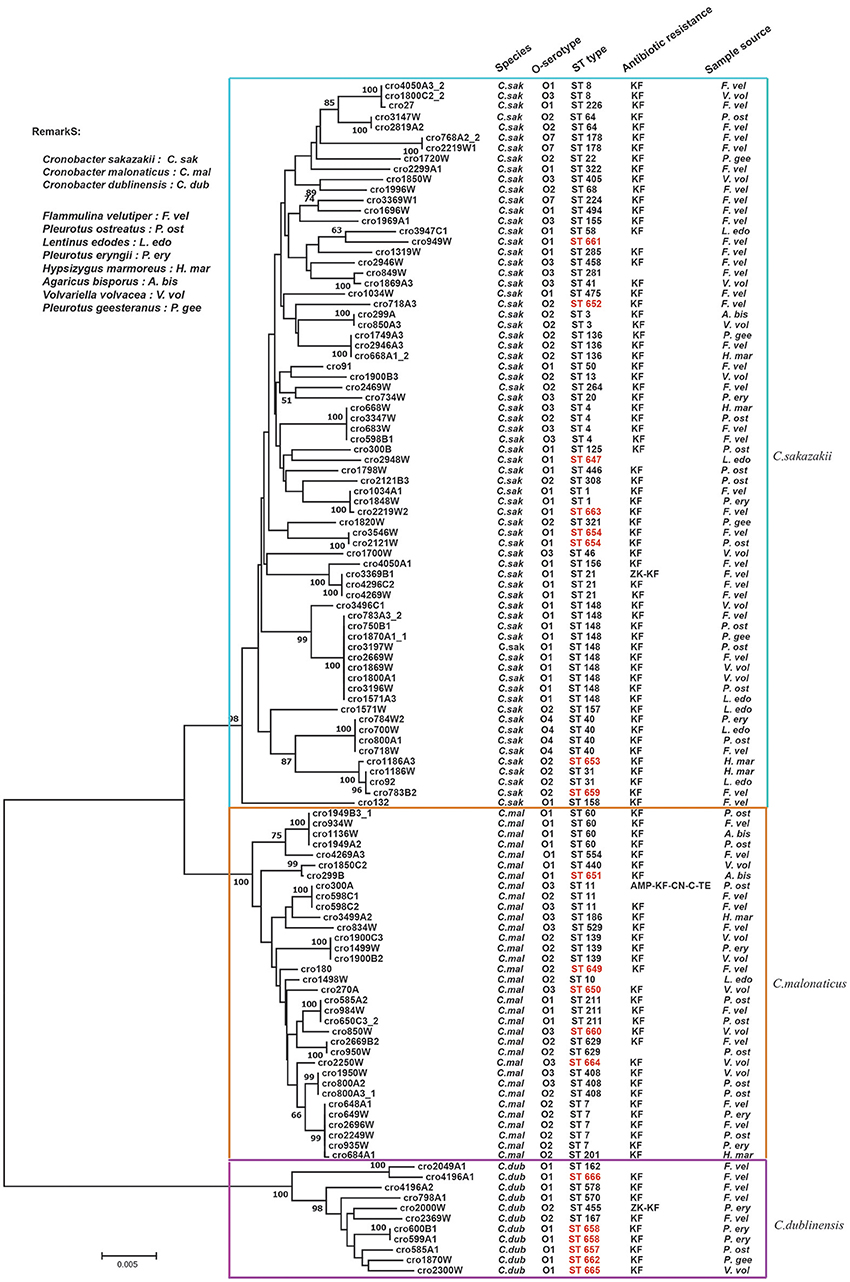
Figure 2. Neighbor-joining phylogenetic tree based on the seven MLST loci (3,036 base pair concatenated length) of Cronobacter spp. isolates. This tree was generated using MEGA (version 5.05) with 1,000 bootstrap replicates.
The 115 Cronobacter spp. isolates were subjected to 16 antimicrobial susceptibility tests (Table 3). All isolates were susceptible to TOB, SAM, FEP, CRO, CIP, IPM, SXT, ATM, and AMC, and there was no significant difference among C. sakazakii, C. malonaticus, and C. dublinensis isolates. The majority of isolates were susceptible to AMP, KZ, CN, AK, C, and TE, with sensitivity rates of 99.13, 72.17, 99.13, 97.39, 99.13, and 99.13%, respectively. The tests revealed that 93.91 and 6.09% of isolates exhibited resistance and intermediate to KF, respectively. One C. malonaticus isolate (cro300A) was detected as multidrug-resistant strains with resistance to five AMs (AMP-KF-CN-C-TE).
Cronobacter spp. are well known to be opportunistic foodborne pathogens that can cause infections and life-threatening diseases in all age groups, especially in infants and the elderly, who are immunocompromised. In this long-term and large-scale study, 668 edible mushroom samples were analyzed, and the overall prevalence of Cronobacter spp. was determined to be 13.32% (89/668). Based on comparison with other large-scale and systematic investigations in China, the prevalence of Cronobacter spp. on edible mushrooms is lower than that in ready-to-eat foods (18.6%, 52/280) (Xu et al., 2015) and raw vegetables (30.27%, 122/403) (Ling et al., 2018). The prevalence of Cronobacter spp. in F. velutipes was very high 17.54% (37/211). In our previous study, we frequently detected Listeria monocytogenes on edible mushrooms, particularly in F. velutipes (Chen et al., 2014), for which the contamination rate was 55.6% (Wu et al., 2015). Therefore, it is necessary to pay close attention to this correlation between Cronobacter spp. and F. velutipes. The contamination rate in the other mushroom species was not analyzed because of the limited number of samples for each species, but it is worth noting the high contamination rate of V. volvacea (11/17) (detail not shown). Usually, edible mushrooms require relatively high temperature (28–35°C) and high humidity to grow; conditions that permit bacterial growth (Bao et al., 2013). V. volvacea were largely cultivated in soil, whereas the other mushroom species were mainly grown in plastic bags and thus had no direct contact with the soil (Chang, 1978; Zhang et al., 2014). Some studies have shown that the soil maybe a principal source of Cronobacter spp. (Ueda, 2017). Moreover, the base growth material for V. volvacea was generally not sterilized. Furthermore, V. volvacea usually was stored in bulk at ambient temperature, which promotes the growth of pathogens (Bao et al., 2013). These might be reasons for the high contamination rate of Cronobacter spp. in V. volvacea. Future studies should include a larger sample size of V. volvacea and establish a continuous surveillance of Cronobacter spp. for reliable determination of the contamination status. Even low levels of Cronobacter spp. in powdered infant formula are considered to be a risk factor, which suggests that our result also indicate a potential risk for the immunocompromised. Although edible mushrooms are not ready-to-eat foods, they can potentially cause cross contamination of the environment and other foods (Kilonzo-Nthenge et al., 2012). To date, no standard permissible limit of Cronobacter spp. on edible mushrooms has been established in China. The establishment of a definite microbiological standard to ensure the quality of edible mushrooms products is essential for food safety in China.
In this study, 10 Cronobacter spp. serogroups were identified in the 115 isolates. C. sakazakii serotype O1 (32/70 isolates) was the most prevalent, followed by C. sakazakii serotype O2 (20/70 isolates). This finding was in agreement with our previous survey in raw vegetables (Ling et al., 2018), but in disagreement with previous studies, in which C. sakazakii serotype O2 was the most dominant serotype in ready-to-eat foods in China (Xu et al., 2015) and powdered infant formula (PIF) (Fei et al., 2017b). It is worth noting that both serotypes O1 and O2 have been associated with clinical sources (Scharinger et al., 2017).
Approximately 30% of isolates in the Cronobacter PubMLST database are from China (Forsythe et al., 2014); the identification of 17 new STs in this study indicates that further studies on genetic characteristic of Cronobacter in China are necessary, especially those from infections with unknown sources. This would help developing control measures for this pathogen and future epidemiological studies. Notably, C. sakazakii ST148 was the most dominant ST. Although ST148 have not reported to associated with human infection, one C. sakazakii ST148 strain was isolated from blood of a 64-year-old in Denmark in 2009 according to PubMLST database (http://pubmlst.org/Cronobacter/). Some pathogenic STs (e.g., ST1, ST4, ST7, ST8, ST60, and ST64) were also identified in this study.
Antibiotic susceptibility tests revealed that the isolates from edible mushrooms were susceptible to most antibiotics. However, a portion of isolates exhibited high or intermediate resistance to cephalothin, cefazolin, cephalothin, and amikacin, which was basically consistent with previous reports (Lai, 2001; Molloy et al., 2009; Chon et al., 2012). It has been reported that some isolates that exhibit intermediate resistance can become resistant under certain circumstances (Ruiz-Bolivar et al., 2011). β-Lactamases and extended spectrum-β-lactamases (ESBL)-related genes have already been reported in Cronobacter spp. (Girlich et al., 2001; Caubilla-Barron et al., 2007; Müller et al., 2014). ESBL confer resistance to penicillins, third-generation cephalosporins, and monobactams (Vasconcellos et al., 2018). Most clinical Cronobacter spp. isolates are multidrug-resistant (Cui et al., 2017; Shi et al., 2017; Zeng et al., 2018a), whereas isolates from foods generally have low drug resistance. It is still unclear whether multi-drug resistance is acquired by Cronobacter before or after infections in the adaptation process. Therefore, a comprehensive study of the molecular mechanisms of antibiotic resistance and correlation analysis of isolates from various environments, foods, and clinical settings should be explored. Clearly, over- and misuse of antimicrobial agents should be avoided, and the transmission and transfer of resistance genes should be closely evaluated.
In this study, it is necessary to pay more attention to the correlation between Cronobacter spp. and F. velutipes or V. volvacea with the high contamination rates of Cronobacter spp. Detection of pathogenic Cronobacter STs strongly implicates that this could be a potential risk to humans. Although edible mushrooms are not ready-to-eat foods, they can potentially cause cross contamination of the environment and other foods during transport, sales, and food preparation processes (Kilonzo-Nthenge et al., 2012). The correlation analysis among foodborne bacteria isolates from environments, foods, and clinics need to be established for identifying the route of transmission and surveillance of multi-drug resistance of Cronobacter spp.
CL, HZ, and JZ contributed to this work equally. QW and HZ conceived and designed the experiments. CL, WH, NL, MC, SW, HW, YY, XW, and YZ performed the experiments. CL, HZ, JZ, YD, and JW analyzed the data. HZ and WH drafted the manuscript. QW supervised the project. All authors read and approved the final manuscript.
This work was supported by grants from the National Key R&D Program of China (2017YFC1601200), the National Natural Science Foundation of China (31601571), Local Innovative and Research Teams Project of Guangdong PEARL River Talents Program (2017BT01S174), Pearl River S&T Nova Program of Guangzhou (201806010062), and GDAS' Special Project of Science and Technology Development (2017GDASCX-0201).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aksu, F., Sandikçi Altunatmaz, S., Issa, G., Özmen Togay, S., and Aksu, H. (2016). Prevalence and identification by multiplex polymerase chain reaction patterns of Cronobacter spp. isolated from plant-based foods. Food Sci. Technol. 36, 730–736. doi: 10.1590/1678-457x.16916
Alsonosi, A., Hariri, S., Kajsík, M., Oriešková, M., Hanulík, V., Röderová, M., et al. (2015). The speciation and genotyping of Cronobacter isolates from hospitalised patients. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1979–1988. doi: 10.1007/s10096-015-2440-8
Bao, D., Gong, M., Zheng, H., Chen, M., Zhang, L., Wang, H., et al. (2013). Sequencing and comparative analysis of the straw mushroom (Volvariella volvacea) genome. PLoS ONE 8:e58294. doi: 10.1371/journal.pone.0058294
Barron, J. C. and Forsythe, S. J. (2007). Dry stress and survival time of Enterobacter sakazakii and other Enterobacteriaceae in dehydrated powdered infant formula. J. Food Prot. 70, 2111–2117. doi: 10.1111/j.1550-7408.2007.00286.x.
BlaŽková, M., Javurková, B., Vlach, J., Göselová, S., Karamonová, L., Ogrodzki, P., et al. (2015). Diversity of O Antigens within the Genus Cronobacter: from disorder to order. Appl. Environ. Microbiol. 81, 5574–5582. doi: 10.1128/AEM.00277-15
Caubilla-Barron, J., Hurrell, E., Townsend, S., Cheetham, P., Loc-Carrillo, C., Fayet, O., et al. (2007). Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J. Clin. Microbiol. 45, 3979–3985. doi: 10.1128/JCM.01075-07
Chang, S. T. (1978). Volvariella volvacea - The Biology and Cultivation of Edible Mushrooms - 27. Biology & Cultivation of Edible Mushrooms, 573–603. doi: 10.1016/B978-0-12-168050-3.50033-5
Chang, S. T., and Buswell, J. A. (1996). Mushroom nutriceuticals. World J. Microbiol. Biotechnol. 12, 473–476. doi: 10.1007/BF00419460
Chaves, C. E. V., Brandão, M. L. L., Lacerda, M., Rocha, C., Leone de Oliveira, S., Parpinelli, T. C., et al. (2018). Fatal Cronobacter sakazakii sequence type 494 meningitis in a Newborn, Brazil. Emerg. Infect. Dis. 24, 1948–1950. doi: 10.3201/eid2410.180373
Chen, M., Wu, Q., Zhang, J., Guo, W., Wu, S., and Yang, X. (2014). Prevalence and contamination patterns of Listeria monocytogenes in Flammulina velutipes plants. Foodborne Pathog. Dis. 11, 620–627. doi: 10.1089/fpd.2013.1727
Chitrakar, B., Zhang, M., and Adhikari, B. (2018). Dehydrated foods: are they microbiologically safe? Crit. Rev. Food Sci. Nutr. 19, 1–12. doi: 10.1080/10408398.2018.1466265
Chon, J. W., Song, K. Y., Kim, S. Y., Hyeon, J. Y., and Seo, K. H. (2012). Isolation and characterization of Cronobacter from desiccated foods in Korea. J. Food Sci. 77, M354–358. doi: 10.1111/j.1750-3841.2012.02750.x
Cui, J. H., Yu, B., Xiang, Y., Zhang, Z., Zhang, T., Zeng, Y. C., et al. (2017). Two cases of multi-antibiotic resistant Cronobacter spp. Infections of infants in China. Biomed. Environ. Sci. 30, 601–605. doi: 10.3967/bes2017.079
Fei, P., Jiang, Y., Feng, J., Forsythe, S. J., Li, R., Zhou, Y., et al. (2017a). Antibiotic and desiccation resistance of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and processing environments. Front. Microbiol. 8:316. doi: 10.3389/fmicb.2017.00316
Fei, P., Jiang, Y., Jiang, Y., Yuan, X., Yang, T., Chen, J., et al. (2017b). Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter sakazakii isolates from powdered infant formula collected from chinese retail markets. Front. Microbiol. 8:2026. doi: 10.3389/fmicb.2017.02026
Forsythe, S. (2018). Microbial source tracking of Cronobacter spp. Adv. Appl. Microbiol. 103, 49–101. doi: 10.1016/bs.aambs.2018.01.004
Forsythe, S. J. (2018). Updates on the Cronobacter Genus. Annu. Rev. Food Sci. Technol. 9, 23–44. doi: 10.1146/annurev-food-030117-012246
Forsythe, S. J., Dickins, B., and Jolley, K. A. (2014). Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics 15:1121. doi: 10.1186/1471-2164-15-1121
Friedemann, M. (2009). Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. Eur. J. Clin. Microbiol. Infect. Dis. 28, 1297–1304. doi: 10.1007/s10096-009-0779-4
Girlich, D., Poirel, L., Leelaporn, A., Karim, A., Tribuddharat, C., Fennewald, M., et al. (2001). Molecular epidemiology of the integron-located VEB-1 extended-spectrum beta-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39, 175–182. doi: 10.1128/JCM.39.1.175-182.2001
Hariri, S., Joseph, S., and Forsythe, S. J. (2013). Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerg. Infect. Dis. 19, 175–177. doi: 10.3201/eid1901.120649
Holý, O., and Forsythe, S. (2014). Cronobacter spp. as emerging causes of healthcare-associated infection. J. Hosp. Infect. 86, 169–177. doi: 10.1016/j.jhin.2013.09.011
Huang, J., Shi, W. U., Zhang, F., Zhang, J., and Qingping, W. U. (2018). Prevalence, antibiotic resistance,and enterotoxin gene detection of Staphylococcus aureus isolated from retail edible mushrooms in China. J. Food Sci. Technol. 36, 25–32. doi: 10.3969/j.issn.2095-6002.2018.03.004.
Iversen, C., and Forsythe, S. J. (2007). Comparison of media for the isolation of Enterobacter sakazakii. Appl. Environ. Microbiol. 73, 48–52. doi: 10.1128/AEM.01562-06
Jarvis, K. G., Grim, C. J., Franco, A. A., Gopinath, G., Sathyamoorthy, V., Hu, L., et al. (2011). Molecular characterization of Cronobacter lipopolysaccharide O-antigen gene clusters and development of serotype-specific PCR assays. Appl. Environ. Microbiol. 77, 4017–4026. doi: 10.1128/AEM.00162-11
Jones, J. L., Lüdeke, C. H., Bowers, J. C., Garrett, N., Fischer, M., Parsons, M. B., et al. (2012). Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 50, 2343–2352. doi: 10.1128/JCM.00196-12
Jorgensen, J. H. (2015). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. Approved Guideline. Wayne, PA: Clinical and Laboratory Standards Institute.
Joseph, S., Desai, P., Ji, Y., Cummings, C. A., Shih, R., Degoricija, L., et al. (2012a). Comparative analysis of genome sequences covering the seven cronobacter species. PLoS ONE 7:e49455. doi: 10.1371/journal.pone.0049455
Joseph, S., and Forsythe, S. J. (2011). Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg. Infect. Dis. 17, 1713–1715. doi: 10.3201/eid1709.110260
Joseph, S., and Forsythe, S. J. (2012). Insights into the emergent bacterial pathogen Cronobacter spp., generated by multilocus sequence typing and analysis. Front. Microbiol. 3:397. doi: 10.3389/fmicb.2012.00397
Joseph, S., Sonbol, H., Hariri, S., Desai, P., McClelland, M., and Forsythe, S. J. (2012b). Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J. Clin. Microbiol. 50, 3031–3039. doi: 10.1128/JCM.00905-12
Kilonzo-Nthenge, A., Rotich, E., Godwin, S., Nahashon, S., and Chen, F. (2012). Prevalence and antimicrobial resistance of Cronobacter sakazakii isolated from domestic kitchens in middle Tennessee, United States. J. Food Prot. 75, 1512–1517. doi: 10.4315/0362-028X.JFP-11-442
Lai, K. K. (2001). Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine 80, 113–122. doi: 10.1097/00005792-200103000-00004
Ling, N., Li, C., Zhang, J., Wu, Q., Zeng, H., He, W., et al. (2018). Prevalence and molecular and antimicrobial characteristics of Cronobacter spp. Isolated from raw vegetables in China. Front. Microbiol. 9:1149. doi: 10.3389/fmicb.2018.01149
Liu, H., Cui, J. H., Cui, Z. G., Hu, G. C., Yang, Y. L., Li, J., et al. (2013). Cronobacter carriage in neonate and adult intestinal tracts. Biomed. Environ. Sci. 26, 861–864. doi: 10.3967/bes2013.011
Molloy, C., Cagney, C., O'Brien, S., Iversen, C., Fanning, S., and Duffy, G. (2009). Surveillance and characterisation by pulsed-field gel electrophoresis of Cronobacter spp. in farming and domestic environments, food production animals and retail foods. Int. J. Food Microbiol. 136, 198–203. doi: 10.1016/j.ijfoodmicro.2009.07.007
Müller, A., Hächler, H., Stephan, R., and Lehner, A. (2014). Presence of AmpC Beta-Lactamases, CSA-1, CSA-2, CMA-1, and CMA-2 conferring an unusual resistance phenotype in Cronobacter sakazakii and Cronobacter malonaticus. Microb. Drug Resist. 20, 275–80. doi: 10.1089/mdr.2013.0188
Pan, Z., Cui, J., Lyu, G., Du, X., Qin, L., Guo, Y., et al. (2014). Isolation and molecular typing of Cronobacter spp. in commercial powdered infant formula and follow-up formula. Foodborne Pathog. Dis. 11, 456–461. doi: 10.1089/fpd.2013.1691
Patrick, M. E., Mahon, B. E., Greene, S. A., Rounds, J., Cronquist, A., Wymore, K., et al. (2014). Incidence of Cronobacter spp. infections, United States, 2003-2009. Emerg. Infect. Dis. 20, 1520–1523. doi: 10.3201/eid2009.140545
Ruiz-Bolivar, Z., Neuque-Rico, M. C., Poutou-Piñales, R. A., Carrascal-Camacho, A. K., and Mattar, S. (2011). Antimicrobial susceptibility of Listeria monocytogenes food isolates from different cities in Colombia. Foodborne Pathog. Dis. 8, 913–919. doi: 10.1089/fpd.2010.0813
Scharinger, E. J., Dietrich, R., Wittwer, T., Märtlbauer, E., and Schauer, K. (2017). Multiplexed lateral flow test for detection and differentiation of Cronobacter sakazakii serotypes O1 and O2. Front. Microbiol. 8:1826. doi: 10.3389/fmicb.2017.01826
Shi, L., Liang, Q., Zhan, Z., Feng, J., Zhao, Y., Chen, Y., et al. (2017). Co-occurrence of 3 different resistance plasmids in a multi-drug resistant Cronobacter sakazakii isolate causing neonatal infections. Virulence 9, 110–120. doi: 10.1080/21505594.2017.1356537
Stoll, B. J., Hansen, N., Fanaroff, A. A., and Lemons, J. A. (2004). Enterobacter sakazakii is a rare cause of neonatal septicemia or meningitis in VLBW infants. J. Pediatr. 144, 821–823. doi: 10.1016/j.jpeds.2004.02.045
Sun, Y., Wang, M., Liu, H., Wang, J., He, X., Zeng, J., et al. (2011). Development of an O-antigen serotyping scheme for Cronobacter sakazakii. Appl. Environ. Microbiol. 77, 2209–2214. doi: 10.1128/AEM.02229-10
Sun, Y., Wang, M., Wang, Q., Cao, B., He, X., Li, K., et al. (2012). Genetic analysis of the Cronobacter sakazakii O4 to O7 O-antigen gene clusters and development of a PCR assay for identification of all C. sakazakii O serotypes. Appl. Environ. Microbiol. 78, 3966–3974. doi: 10.1128/AEM.07825-11
Ueda, S. (2017). Occurrence of Cronobacter spp. in dried foods, fresh vegetables and soil. Biocontrol. Sci. 22, 55–59. doi: 10.4265/bio.22.55
van Acker, J., de Smet, F., Muyldermans, G., Bougatef, A., Naessens, A., and Lauwers, S. (2001). Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39, 293–297. doi: 10.1128/JCM.39.1.293-297.2001
Vasconcellos, L., Carvalho, C. T., Tavares, R. O., de Mello Medeiros, V., de Oliveira Rosas, C., Silva, J. N., et al. (2018). Isolation, molecular and phenotypic characterization of Cronobacter spp. in ready-to-eat salads and foods from Japanese cuisine commercialized in Brazil. Food Res. Int. 107, 353–359. doi: 10.1016/j.foodres.2018.02.048
Wu, S., Wu, Q., Zhang, J., Chen, M., Yan, Z. A., and Hu, H. (2015). Listeria monocytogenes prevalence and characteristics in retail raw foods in China. PLoS ONE 10:e0136682. doi: 10.1371/journal.pone.0136682
Xu, X., Li, C., Wu, Q., Zhang, J., Huang, J., and Yang, G. (2015). Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter spp. in Chinese ready-to-eat foods. Int. J. Food Microbiol. 204, 17–23. doi: 10.1016/j.ijfoodmicro.2015.03.003Ye
Ye, Y., Li, H., Wu, Q., Chen, M., Lu, Y., and Yan, C. (2014). Isolation and phenotypic characterization of Cronobacter from dried edible macrofungi samples. J. Food Sci. 79, M1382–1386. doi: 10.1111/1750-3841.12513
Yong, W., Guo, B., Shi, X., Cheng, T., Chen, M., Jiang, X., et al. (2018). An investigation of an acute gastroenteritis outbreak: Cronobacter sakazakii, a potential cause of food-borne illness. Front. Microbiol. 9:2549. doi: 10.3389/fmicb.2018.02549
Zeng, H., Lei, T., He, W., Zhang, J., Liang, B., Li, C., et al. (2018a). Novel multidrug-resistant Cronobacter sakazakii causing meningitis in neonate, China, 2015. Emerg. Infect. Dis. 24, 2121–2124. doi: 10.3201/eid2411.180718
Zeng, H., Zhang, J., Wu, Q., He, W., Wu, H., Ye, Y., et al. (2018b). Reconstituting the history of cronobacter evolution driven by differentiated CRISPR activity. Appl. Environ. Microbiol. 84, e00267–e00218. doi: 10.1128/AEM.00267-18
Keywords: Cronobacter, prevalence, edible mushrooms, multilocus sequence typing, O-antigen serotyping, antibiotic susceptibility test
Citation: Li C, Zeng H, Zhang J, He W, Ling N, Chen M, Wu S, Lei T, Wu H, Ye Y, Ding Y, Wang J, Wei X, Zhang Y and Wu Q (2019) Prevalence, Antibiotic Susceptibility, and Molecular Characterization of Cronobacter spp. Isolated From Edible Mushrooms in China. Front. Microbiol. 10:283. doi: 10.3389/fmicb.2019.00283
Received: 30 October 2018; Accepted: 04 February 2019;
Published: 26 February 2019.
Edited by:
Julio Parra-Flores, University of the Bío-Bío, ChileReviewed by:
Juan Aguirre, Universidad de Chile, ChileCopyright © 2019 Li, Zeng, Zhang, He, Ling, Chen, Wu, Lei, Wu, Ye, Ding, Wang, Wei, Zhang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, d3VxcDIwM0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.