
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 February 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00276
 Christian Schweizer1,2†
Christian Schweizer1,2† Peter Bischoff3*†
Peter Bischoff3*† Jennifer Bender4†
Jennifer Bender4† Axel Kola3
Axel Kola3 Petra Gastmeier3
Petra Gastmeier3 Manfred Hummel1
Manfred Hummel1 Frank-Rainer Klefisch1
Frank-Rainer Klefisch1 Felix Schoenrath5,6
Felix Schoenrath5,6 Andre Frühauf4
Andre Frühauf4 Yvonne Pfeifer4
Yvonne Pfeifer4Carbapenem-resistant Enterobacteriaceae (CRE) cause health care-associated infections worldwide, and they are of severe concern due to limited treatment options. We report an outbreak of KPC-2-producing CRE that was caused by horizontal transmission of a promiscuous plasmid across different genera of bacteria and hospitals in Germany. Eleven isolates (8 Citrobacter freundii, 2 Klebsiella oxytoca, and 1 Escherichia coli) were obtained from seven critically ill patients during the six months of the outbreak in 2016. One patient developed a CRE infection while the other six patients were CRE-colonized. Three patients died in the course of the hospital stay. Six of the seven patients carried the same C. freundii clone; one K. oxytoca clone was found in two patients, and one patient carried E. coli and C. freundii. Molecular analysis confirmed the presence of a conjugative, blaKPC-2-carrying 70 kb-IncN plasmid in C. freundii and E. coli and an 80 kb-IncN plasmid in K. oxytoca. All transconjugants harbored either the 70 or 80 kb plasmid with blaKPC-2, embedded within transposon variant Tn4401g. Whole genome sequencing and downstream bioinformatics analyses of all plasmid sequences showed an almost perfect match when compared to a blaKPC-2-carrying plasmid of a large outbreak in another German hospital two years earlier. Differences in plasmid sizes and open reading frames point to the presence of inserted mobile genetic elements. There are few outbreak reports worldwide on the transmission of blaKPC-2-carrying plasmids across different bacterial genera. Our data suggest a regional and supraregional spread of blaKPC-2-carrying IncN-plasmids harboring the Tn4401g isoform in Germany.
“Klebsiella pneumoniae carbapenemase” (KPC) was first described in carbapenem-resistant K. pneumoniae isolates from the United States in the 1990s. According to the NCBI database more than 30 different KPC variants are known today but KPC-2 and KPC-3 are the most prevalent enzymes worldwide (Tzouvelekis et al., 2012; Mathers et al., 2015). Multilocus sequence typing (MLST) analyses have confirmed the worldwide emergence of the successful clonal group CG258 of K. pneumoniae, which includes different sequence types (STs), e.g., ST258, ST512, ST11, ST340, and ST437. KPC-2 or KPC-3-producing K. pneumoniae-ST258 are spread worldwide, and over the last 10 years the number of infections has increased dramatically in some countries, for instance the United States, Israel, Greece, and Italy (Mathers et al., 2015; Wyres and Holt, 2016). The blaKPC gene is embedded in transposon Tn4401, which is located on plasmids of different sizes and replicon type, or is integrated into the bacterial chromosome (Mathers et al., 2017). As a result of its plasmid location, blaKPC can be transferred to species other than K. pneumoniae, e.g., Enterobacter spp., Citrobacter spp., or E. coli; interspecies transmission during outbreaks has been reported (Mathers et al., 2011, 2017; Tzouvelekis et al., 2012).
Plasmids of the incompatibility group N (IncN) are prevalent in Enterobacteriaceae and are known to carry various resistance determinants including extended-spectrum β-lactamases (ESBLs) and carbapenemases like KPC. The 119 IncN plasmids currently deposited in the plasmid MLST database1 represent the high plasticity of this plasmid type. Of special interest are IncN plasmids that harbor blaKPC-2 and the β-lactamase gene blaTEM-1 in the truncated transposon isoform Tn4401g. These plasmids appear to be very “promiscuous” and have been described in different Enterobacteriaceae species in Israel and Germany (Chmelnitsky et al., 2014; Yao et al., 2014). A reported outbreak with KPC-2-producing Enterobacteriaceae (including Citrobacter spp., Klebsiella oxytoca, Enterobacter spp., E. coli) in Germany in 2014 left 132 patients rectally colonized but without clinical symptoms. The kitchen in the hospital was identified as the probable source of the outbreak after detection of KPC-2 producers (C. freundii and K. oxytoca) in different foods (Robert Koch-Institut, 2014). The present study analyzed KPC-2-producing isolates of different species from patients in two other German hospitals for the presence of plasmids as vehicles for transmission of carbapenem resistance.
Between February 2016 and December 2017, a total of 13 carbapenem-resistant isolates (8 C. freundii, 2 K. oxytoca, and 3 E. coli) were obtained from seven patients hospitalized in two different hospitals in Berlin, Germany. Species identification and determination of antibiotic susceptibilities was performed using an automated system (VITEK 2, AST-N248, bioMérieux, Nürthingen, Germany). The presence of carbapenemase genes (blaNDM-like, blaOXA-48-like, blaV IM-like, blaIMP-like, and blaKPC-like) and further β-lactamase genes (blaTEM-like, blaSHV -like, blaCTX-M-like, blaOXA-1-like, blaOXA-2-like, blaOXA-9-like, blaOXA-10-like, blaCMY, and blaDHA) was confirmed by PCR as previously described (Gröbner et al., 2009; Pfeifer et al., 2011). Screening for plasmid-mediated quinolone resistance (PMQR) genes [qnrA/B/S; aac(6’)Ib-cr] was additionally performed using primers published in other studies (Park et al., 2006; Pfeifer et al., 2009). The genetic environment of blaKPC was analyzed using previously described primers AM07/AM08 (Mathers et al., 2011) and primers TEMg 5′-AGGCAACTATGGATGAACGA-3′ and KPCg 5′-AGAGACAAGACAGCAGAACTAGAC-3′, designed by the National Reference Laboratory for multidrug-resistant Gram-negative bacteria in Bochum, Germany, for rapid identification of the Tn4401g isoform (Chmelnitsky et al., 2014).
In addition, the genetic relationship of the isolates was analyzed by pulsed-field gel electrophoresis (PFGE) using XbaI-restricted whole genomic DNA. Macrorestriction patterns were interpreted based on the criteria of Tenover et al. (1995). E. coli isolates were assigned to the four main phylogenetic groups (A, B1, B2, and D) and clonal lineage O25b:H4-ST131 using PCR-based assays (Clermont et al., 2000; Blanco et al., 2009).
Transfer of β-lactam resistance was tested for all isolates in broth mating assays using a sodium azide-resistant E. coli J53 Azir recipient (Clowes and Rowley, 1954). Transconjugants were selected on Luria-Bertani agar plates with 200 mg/L sodium azide and 50 mg/L ampicillin. Plasmid content and the plasmid size of clinical isolates and transconjugants were determined by means of S1-nuclease restriction and PFGE as described by Barton et al. (1995). PCR-based replicon typing (“PBRT KIT,” DIATHEVA, Fano, Italy) was performed using plasmid DNA of transconjugants (Carattoli et al., 2005). For comparison of plasmid restriction patterns, EcoRI restriction was performed after plasmid DNA isolation from transconjugants using the Qiagen Plasmid Mini Kit.
After performance and evaluation of PFGE-based bacterial strain typing and conjugation assays four isolates (2 C. freundii, 1 E. coli, and 1 K. oxytoca) and their respective KPC-2-positive transconjugants were selected for WGS analysis (Table 1). These eight isolates were cultivated in Brain Heart Infusion (BHI) broth. DNA was extracted from overnight cultures using the MagAttract Kit (Qiagen) and the DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s instructions. A Qubit dsDNA HS Assay Kit (Invitrogen) was used for DNA quantification. Sequencing libraries were prepared by applying the Nextera XT DNA Library Prep Kit (Illumina). They were sequenced on an Illumina Miseq using a v3 chemistry (2 × 300 bp) as described in the manufacturer’s protocol. The quality of fastq data obtained was assessed and de novo assembly was performed by the in-house developed pipelines QCumber 2.0.02 and batch Assembly 2.1.2. The latter implements the assembler a5-miseq or SPAdes, respectively (Coil et al., 2015)3.
Contigs, obtained after de novo assembly, were submitted to the CGE Finder Series (Centre for Genomic Epidemiology, Technical University of Denmark (DTU)4, pubmlst.org (University of Oxford, Great Britain) and to the SeqSphere+ software suite (Ridom GmbH, Muenster, Germany). Different analysis tools (MLST and pMLST from pubmlst.org, cgMLST, or ResFinder 3.0) were applied to extract the multilocus ST as described by (Wirth et al., 2006) the plasmid multilocus sequence type (pST), and information on genes that mediate resistance to β-lactams, fluoroquinolones and other antibiotics (Larsen et al., 2012; Zankari et al., 2012; Carattoli et al., 2014; Joensen et al., 2014, 2015).
The BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011) was used to align all assembled reads of the strains sequenced to the blaKPC-2-carrying C. freundii plasmid pCF8698 [accession no. LN610760, 54,036 bp (Yao et al., 2014)]. In addition, mapping of selected transconjugant reads to C. freundii pCF8698 or to the transposon Tn4401g carrying blaKPC-2 (Chmelnitsky et al., 2014) was performed using Geneious 10.0.5.
Raw read data was assigned SRA Accession No. SRP153806.
No informed consent or ethical approval was required since all isolates were generated and analyzed as part of microbiological diagnostics (therapeutic purposes) and/or infection prevention and control requirements and measures.
The outbreak investigation was conducted in accordance with article 25, section 1 of the German Infection Protection Act of 2001.
Between February and July 2016, cases, all cardiosurgical patients with major comorbidities, were identified in two separate intensive care units (ICUs) and one peripheral ward all spread across two different hospitals in the same city. Unveiling this epidemiological link was possible due to close cooperation of the local infection prevention and control teams. A carbapenem-resistant C. freundii was isolated from both urine and tracheobronchial secretion in the index patient (#1) in hospital 2 (Figure 1). Both contact patients (#2 and #3) in the 3-bed room were found to be positive for carbapenem-resistant C. freundii by rectal screening over the course of two weeks. Prior to the initial pathogen detection, the index patient had been hospitalized for about six weeks, four of those in hospital 1. Notably, none of the previous screenings (on average two per patient) had yielded a carbapenem-resistant strain in any of these three patients. About four weeks after the contact patients tested positive, a carbapenem-resistant E. coli, along with C. freundii, was found by rectal screening of patient #3.
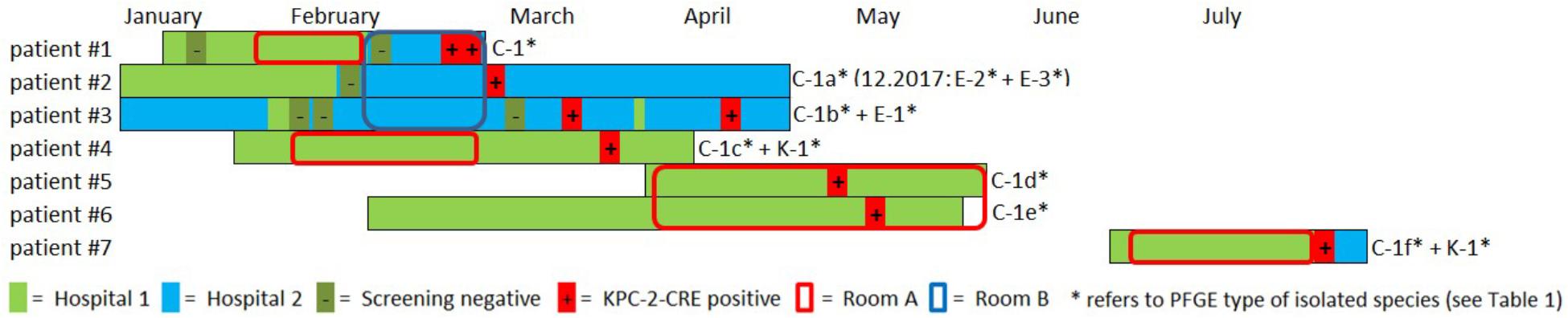
Figure 1. Overview and description of the outbreak in 2016. Colored bars represent the patient’s stay in hospital 1 (light green) and hospital 2 (light blue). Circles indicate the stay in a certain ICU room (red = room A, blue = room B). Negative and positive microbiological results (screening or clinical specimen) for each patient is indicated with (–) and (+), respectively. A carbapenem-resistant C. freundii (PFGE type C1) was isolated from both urine and tracheobronchial secretion in the index patient (#1) in hospital 2. Both contact patients (#2 and #3) in the 3-bed-room (room B) were positive for carbapenem-resistant C. freundii over the course of two weeks (rectal screening). Both a KPC-2-producing C. freundii (rectal) and a KPC-2-producing K. oxytoca (urine) were isolated from patient #4 on a peripheral ward in hospital 1. Patient #1 and patient #4 had shared a 2-bed-room in the ICU of hospital 1 five weeks earlier. Nine weeks after patients #1 and #4 had left, KPC-2-producing C. freundii were detected in other patients in this room. For patient #5, a positive result was obtained from tracheobronchial secretion and the contact patient #6 had a rectal and pharyngeal colonization. Patient #7 was admitted to the same ICU 2-bed-room in hospital 1 about one month later and occupied the room for about five weeks. Only after discharge and during admission screening in hospital 2 both a KPC-2-producing C. freundii and K. oxytoca (both rectal) were isolated. In patient #2 two KPC-2 producing E. coli morphotypes were isolated upon re-admission 20 months later.
The National Reference Laboratory for multidrug-resistant Gram-negative bacteria in Bochum, Germany, identified carbapenemase gene blaKPC-2 in all the isolates as the cause of resistance. Seemingly independent of this situation, both a KPC-2-producing C. freundii (rectal swab) and a KPC-2-producing K. oxytoca (urine) were isolated from another patient (#4) on a peripheral ward in hospital 1. However, epidemiological investigation revealed that patient #1 and patient #4 had shared a 2-bed room in the ICU of hospital 1 five weeks earlier. Nine weeks after patients #1 and #4 had left, KPC-2-producing C. freundii were detected in other patients in this room. In patient #5, a positive result was obtained from a tracheobronchial secretion and the contact patient (#6) exhibited rectal and pharyngeal colonization. The room was thoroughly disinfected and all materials there were discarded, as far as possible.
Patient #7 was admitted to the same ICU 2-bed-room in hospital 1 about one month later and occupied the room for about five weeks. Only after discharge and upon admission screening (rectal swabs) in hospital 2, both KPC-2-producing C. freundii and K. oxytoca were isolated. Three of the patients (#1, #5, and #6) died as a result of their underlying diseases. One patient developed a CRE infection (#1), while the other six patients remained colonized during their stay and at discharge. Information on the previous travel history of the patients was not available. All carbapenem-resistant isolates identified were sent to the Robert Koch Institute (RKI) for further analysis.
Outbreak investigation identified stay in the same ICU room as the most obvious epidemiological link (room A+B; Figure 1). Moreover, one particular ICU room was found to be the common source for CRE acquisition for five patients who had all had stays in that room at separate times over the course of a six months period (patients #1,#4,#5,#6,#7, room A; Figure 1). Environmental investigation for CRE was not carried out during the patient stays. After the discharge of the last patient, the ICU ward in hospital 1 was completely renovated and no new cases have occurred ever since. After 20 months, one of the CRE carrying patients (patient #2) was admitted to the hospital again when screening revealed colonization with two carbapenem-resistant E. coli morphotypes.
Between February and July 2016, 11 carbapenem-resistant isolates (8 C. freundii, 2 K. oxytoca, and 1 E. coli) were obtained from seven patients in two different hospitals. The isolates were resistant to imipenem, meropenem, piperacillin/tazobactam, cefotaxime, ceftazidime, aztreonam, ciprofloxacin, moxifloxacin, gentamicin, tobramycin, and trimethoprim/sulfamethoxazole. Susceptibility to amikacin, colistin, tigecyline, and fosfomycin was confirmed in all isolates.
XbaI-macrorestriction patterns of C. freundii isolated from six patients in the period February–May 2016 varied in 1–4 bands, which confirmed the presence of the same clone (PFGE types C1a-e, Table 1). Two C. freundii isolates of patient #7 isolated at the end of July 2016 showed a difference in 6–7 bands in their macrorestriction pattern compared to the previous isolates. According to the criteria of Tenover et al. (1995) and Mathers et al. (2011), this difference is to be interpreted as showing a possible relation or no relation of these isolates. Because of the epidemiological link of the patients and WGS-based assignment of both isolates to ST98, we decide to assign both isolates to the same clone as all the other C. freundii isolates. Furthermore, PFGE analysis confirmed the presence of one K. oxytoca clone in two patients, and three E. coli clones were identified (patients #2 and #3; Supplementary Figure S1). Finally, utilizing WGS and the programs MLST and cgMLST of pubmlst.org and SeqSphere+, respectively, both C. freundii isolates were typed ST98, the K. oxytoca clone was typed ST29 and the E. coli clone as a member of ST369 and complex type CT789 (Table 1).
PCR and sequencing confirmed the presence of the carbapenemase gene blaKPC-2 in all 13 isolates (Table 1). This gene showed a unique genetic environment, the Tn4401g transposon variant with blaTEM-1 next to blaKPC-2, as described previously in Enterobacteriaceae isolates from Israel and in outbreak isolates from Germany in 2014 (Chmelnitsky et al., 2014; Robert Koch-Institut, 2014; Yao et al., 2014). Broth mating successfully transferred the blaKPC-2-carrying plasmids of approx. 70–80 kb and IncN group. All transconjugants harbored only one plasmid which were either 70 kb (E. coli and C. freundii donor isolates) or 80 kb (K. oxytoca donor) plasmids (Table 1 and Supplementary Figure S2). All transconjugants were positive for β-lactamase genes blaKPC-2, blaOXA-1, and blaTEM-1, and PMQR gene aac(6’)Ib-cr. PMQR gene qnrB2 was additionally present in transconjugants and their respective clinical isolates (C. freundii from three patients and the two K. oxytoca isolates (Table 1).
Simple EcoR1-restriction of the 70 and 80 kb plasmids revealed similar restriction patterns (Supplementary Figure S3). Use of WGS and the tool pMLST, identified the IncN plasmids of all transconjugants as pST15. The pMLST database5 contains only three entries of IncN-pST15 plasmids, identified in E. coli and K. pneumoniae in Israel in 2005–2006 (Goren et al., 2010; Leavitt et al., 2010). A BRIG analysis of all plasmid sequences showed an almost perfect match with the blaKPC-2-carrying 54 kb-IncN plasmid pCF8698_KPC2 (accession no. LN610760). This plasmid was isolated from C. freundii from a KPC-mediated outbreak in a German hospital in 2014 (Yao et al., 2014). Minor deviations in coverage and gaps from the sequence of pCF8698_KPC2 were noted in a region comprising five hypothetical proteins (Figure 2). In accordance with differences in plasmid sizes in pCF8698 and the ones investigated here, it was presumed that insertion of mobile genetic elements (MGEs) or deletion of certain plasmid fragments had occurred. Therefore, paired-end reads were aligned to the reference plasmid pCF8698_KPC2, followed by a MAUVE alignment of contigs of interest. MGEs possibly inserted in the region of the five hypothetical proteins could not be determined. However, we detected a 777 bp insertion in gene secA in all isolates of the present outbreak that encodes an IS1 family transposase (BLASTn results, data not shown).
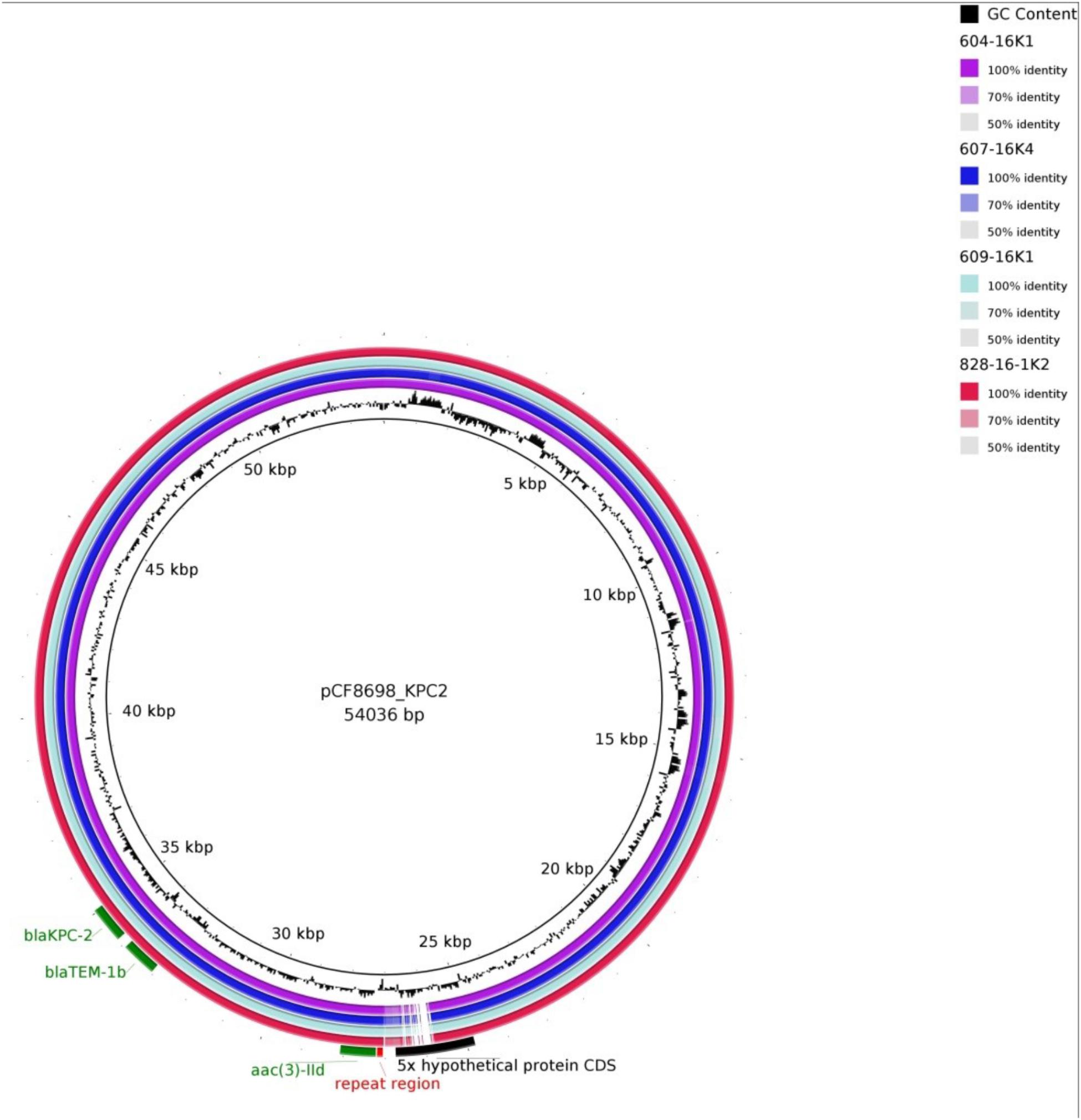
Figure 2. BLAST Ring Image Generator analysis of plasmid sequences. Plasmid sequences of four transconjugants were compared with plasmid pCF8698_KPC2 (Yao et al., 2014). violett, 604-16K1 E. coli K12J53 blaKPC-2, blaTEM-1, blaOXA-1, aac(6’)Ib-cr (patient #1); dark blue, 607-16K4 E. coli K12J53 blaKPC-2, blaTEM-1, blaOXA-1, aac(6’)Ib-cr (patient #3); light blue, 609-16K1 E. coli K12J53 blaKPC-2, blaTEM-1, blaOXA-1, aac(6’)Ib-cr, qnrB2 (patient #4); red, 828-16-1K2 E. coli K12J53 blaKPC-2, blaTEM-1, blaOXA-1, aac(6’)Ib-cr, qnrB2 (patient #7); important resistance genes are labeled in green.
Another 13.202 bp insertion in the gene umuC was present only in the 80 kb blaKPC-2-carrying plasmid in K. oxytoca. BLASTn analysis of the inserted sequence revealed regions homologous to plasmid pKPN-edaa (accession no. NZ_CP026398.1) from K. pneumoniae isolated in an ICU piping system of an United States hospital in 2017 (Weingarten et al., 2018). The sequence encodes hypothetical proteins, a membrane anchor protein, a Tat signal sequence, an MFS transporter, and the nickel/cobalt efflux protein RcnA. The size of the inserted sequence corresponds consistently to the size difference of the K. oxytoca plasmid demonstrated by S1-nuclease PFGE (Supplementary Figure S2).
It must be noted that transconjugants TC 604-16K1 of C. freundii and TC 607-16K4 of E. coli demonstrate a 3rd insertion site in a hypothetical protein located around nucleotide position 29.460. The fragment revealed homologies to regions from plasmids of various gram-negative pathogens and comprises a DDE transposase and genes belonging to arsenical efflux (data not shown).
The transconjugants harboured blaKPC, further β-lactamase and PMQR genes and in addition a number of other antibiotic resistance genes. Using ResFinder with a threshold of 90% identity and a minimum length of 60%, acquired resistance genes that confer resistance to rifampicin, phenicols, aminoglycosides, trimethoprim, sulfonamide, and macrolides were extracted, e.g., catB3, arr-3, strA, dfrA18, sul1, and mph(A) (Supplementary Table S1).
In Germany, the first outbreak with KPC-2-producing K. pneumoniae was detected in 2007–2008 followed by another large outbreak in 2010/13 (Wendt et al., 2010; Lübbert et al., 2014). In both outbreaks admission of patients previously hospitalized in Greece were identified as possible index patients. Transmission of blaKPC-2 to other species, e.g., E. coli, was only detected in a single case in the outbreak 2007–2008. Subsequently performed analyses in the RKI revealed another genetic environment of this gene (istB gene next to blaKPC-2, Mathers et al., 2011) which is in contrast to the isolates of the present study. This MGE was integrated into a conjugative 100 kb plasmid. Unlike the worldwide spread of KPC-producing K. pneumoniae clones, e.g., ST258, there are several outbreak reports on the transmission of promiscuous blaKPC-2-carrying plasmids across different bacterial species and genera (Mathers et al., 2011; Chmelnitsky et al., 2014; Robert Koch-Institut, 2014; Huang et al., 2018; Kukla et al., 2018; Li et al., 2018).
The location of blaKPC-2 on conjugative IncN plasmids in different bacterial species from patients and environmental samples has been described worldwide, and blaKPC-2 was embedded primarily in the Tn4401b isoform (Pereira et al., 2015; Eilertson et al., 2017; Nascimento et al., 2017; Weingarten et al., 2018). The transfer of these different plasmids between adapted species (e.g., Klebsiella spp., Citrobacter spp., and Enterobacter spp.) in biofilms of the aquatic environment in hospitals (e.g., sinks, pipes) suggests that these are an important reservoir for resistance genes (Weingarten et al., 2018). In the present study blaKPC-2 was embedded in the Tn4401g isoform in an IncN-pST15 plasmid that had been described only in Enterobacteriaceae isolates from Israel and Germany (Chmelnitsky et al., 2014; Yao et al., 2014). Consistent with the isolates of the outbreak in Hesse, Germany, in 2014, the isolates in present outbreak were primarily of species C. freundii and K. oxytoca. Furthermore, patients with co-colonization indicate possible events of plasmid transfer to commensal E. coli in the gut or in the hospital environment. According to the MLST database, the E. coli-ST369 identified in this study seems to be widespread. ST369 was identified previously in poultry, food and some clinical samples in Germany, the Netherlands, the United States and Japan6 (Ozawa et al., 2010).
WGS analyses showed a high degree of similarity of the blaKPC-2-carrying IncN-plasmids of the 2016 outbreak in Berlin to these of the 2014 outbreak in Hesse, Germany. Nevertheless, the sizes of the blaKPC-2-carrying plasmids varied, which is caused primarily by insertions of various MGEs. Because short read sequencing is limited with respect to sequence reconstruction, we were not able to identify all inserted MGEs or to complete the entire plasmid sequences. This is a limitation of the present study.
In summary, we have confirmed another multispecies-outbreak caused by a blaKPC-2-carrying Tn4401g transposon on IncN plasmids that occurred in two German hospitals two years after a large outbreak in a hospital located at a distance of 500 km. In the present outbreak, the hospital environment seemed to have played a particular role as a source of KPC-producing C. freundii. Over a course of six months five patients with consecutive, separate stays in the same hospital room acquired the same CRE clones. Although the surfaces and objects in the room had been thoroughly disinfected multiple times, the unknown source was only eliminated when a planned renovation of the ward was carried out. Transfer of colonized patients and contamination of the aquatic environment with KPC-producing bacteria might have facilitated the regional and supraregional spread of blaKPC-2-carrying plasmids.
CS and PB carried out the epidemiological investigations. AK, PG, MH, F-RK, and FS supervised and supported this work. JB, AF, and YP planned and carried out the experiments. CS, PB, JB, and YP drafted the manuscript and designed the figures. All authors contributed to the interpretation of the results, provided critical feedback and helped to finalize the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Sibylle Müller-Bertling, Kirstin Ganske, Christine Günther and Roland Pfüller for excellent technical assistance and Gerald Brennan for editing and proofreading the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00276/full#supplementary-material
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Blanco, M., Alonso, M. P., Nicolas-Chanoine, M. H., Dahbi, G., Mora, A., Blanco, J. E., et al. (2009). Molecular epidemiology of Escherichia coli producing extended-spectrum {beta}- lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 63, 1135–1141. doi: 10.1093/jac/dkp122
Carattoli, A., Bertini, A., Villa, L., Falbob, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Meth. 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chmelnitsky, I., Shklyar, M., Leavitt, A., Sadovsky, E., Navon-Venezia, S., Ben Dalak, M., et al. (2014). Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn. Microbiol. Infect. Dis. 79, 255–260. doi: 10.1016/j.diagmicrobio.2014.03.008
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
Clowes, R. C., and Rowley, D. (1954). Some observations on linkage effects in genetic recombination in Escherichia coli K-12. J. Gen. Microbiol. 11, 250–260. doi: 10.1099/00221287-11-2-250
Coil, D., Jospin, G., and Darling, A. E. (2015). A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31, 587–589. doi: 10.1093/bioinformatics/btu661
Eilertson, B., Chen, L., Chavda, K. D., and Kreiswirth, B. N. (2017). Genomic characterization of two KPC-producing klebsiella isolates collected in 1997 in New York City. Antimicrob Agents Chemother. 61:e2458-16. doi: 10.1128/AAC.02458-16
Goren, M. G., Navon-Venezia, S., Chmelnitsky, I., and Carmeli, Y. (2010). Carbapenem-resistant KPC-2-producing Escherichia coli in a tel aviv medical center, 2005 to 2008. Antimicrob Agents Chemother. 54, 2687–2691. doi: 10.1128/AAC.01359-09
Gröbner, S., Linke, D., Schutz, W., Fladerer, C., Madlung, J., Autenrieth, I. B., et al. (2009). Emergence of carbapenem-non-susceptible extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tubingen, Germany. J. Med. Microbiol. 58, 912–922. doi: 10.1099/jmm.0.005850-0
Huang, J., Ding, H., Shi, Y., Zhao, Y., Hu, X., Ren, J., et al. (2018). Further spread of a blaKPC-harboring untypeable plasmid in Enterobacteriaceae in China. Front. Microbiol. 9:1938. doi: 10.3389/fmicb.2018.01938
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Joensen, K. G., Tetzschner, A. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Kukla, R., Chudejova, K., Papagiannitsis, C. C., Medvecky, M., Habalova, K., Hobzova, L., et al. (2018). Characterization of KPC-encoding plasmids from Enterobacteriaceae isolated in a Czech Hospital. Antimicrob Agents Chemother. 62:e2152-17. doi: 10.1128/AAC.02152-1
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Leavitt, A., Carmeli, Y., Chmelnitsky, I., Goren, M. G., Ofek, I., and Navon-Venezia, S. (2010). Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob Agents Chemother. 54, 3002–3006. doi: 10.1128/AAC.01818-09
Li, B., Feng, J., Zhan, Z., Yin, Z., Jiang, Q., Wei, P., et al. (2018). Dissemination of KPC-2-encoding IncX6 plasmids among multiple Enterobacteriaceae species in a single Chinese hospital. Front. Microbiol. 9:478. doi: 10.3389/fmicb.2018.00478
Lübbert, C., Lippmann, N., Busch, T., Kaisers, U. X., Ducomble, T., Eckmanns, T., et al. (2014). Long-term carriage of Klebsiella pneumoniae carbapenemase-2-producing K. pneumoniae after a large single-center outbreak in Germany. Am. J. Infect. Control. 42, 376–380. doi: 10.1016/j.ajic.2013.12.001
Mathers, A. J., Cox, H. L., Kitchel, B., Bonatti, H., Brassinga, A. K., Carroll, J., et al. (2011). Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2:e204-11. doi: 10.1128/mBio.00204-11
Mathers, A. J., Peirano, G., and Pitout, J. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
Mathers, A. J., Stoesser, N., Chai, W., Carroll, J., Barry, K., Cherunvanky, A., et al. (2017). Chromosomal Integration of the Klebsiella pneumoniae carbapenemase gene, of blaKPC, in Klebsiella species is elusive but not rare. Antimicrob Agents Chemother. 61:e1823-16. doi: 10.1128/AAC.01823-16
Nascimento, T., Cantamessa, R., Melo, L., Fernandes, M. R., Fraga, E., Dropa, M., et al. (2017). International high-risk clones of Klebsiella pneumoniae KPC-2/CC258 and Escherichia coli CTX-M-15/CC10 in urban lake waters. Sci. Total Environ. 598, 910–915. doi: 10.1016/j.scitotenv.2017.03.207
Ozawa, M., Baba, K., and Asai, T. (2010). Molecular typing of avian pathogenic Escherichia coli O78 strains in Japan by using multilocus sequence typing and pulsed-field gel electrophoresis. J. Vet. Med. Sci. 72, 1517–1520. doi: 10.1292/jvms.10-0174
Park, C. H., Robicsek, A., Jacoby, G. A., Sahm, D., and Hooper, D. C. (2006). Prevalence in the United States of aac(6’)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50, 3953–3955. doi: 10.1128/AAC.00915-06
Pereira, P. S., Borghi, M., Albano, R. M., Lopes, J. C., Silveira, M. C., Marques, E. A., et al. (2015). Coproduction of NDM-1 and KPC-2 in Enterobacter hormaechei from Brazil. Microb. Drug Resist. 21, 234–236. doi: 10.1089/mdr.2014.0171
Pfeifer, Y., Matten, J., and Rabsch, W. (2009). Salmonella enterica serovar Typhi with CTX-M β-lactamase, Germany. Emerg. Infect. Dis. 15, 1533–1535. doi: 10.3201/eid1509.090567
Pfeifer, Y., Wilharm, G., Zander, E., Wichelhaus, T. A., Gottig, S., Hunfeld, K. P., et al. (2011). Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66, 1998–2001. doi: 10.1093/jac/dkr256
Robert Koch-Institut (2014). Available at: https://www.rki.de/DE/Content/Infekt/EpidBull/Archiv/2014/Ausgaben/47_14.pdf?__blob=publicationFile
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Weingarten, R. A., Johnson, R. C., Conlan, S., Ramsburg, A. M., Dekker, J. P., Lau, A. F., et al. (2018). NISC comparative sequencing program, Henderson DK, Palmore TN, Segre JA, Frank KM. genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e2011-17. doi: 10.1128/mBio.02011-17
Wendt, C., Schütt, S., Dalpke, A. H., Konrad, M., Mieth, M., Trierweiler-Hauke, B., et al. (2010). First outbreak of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae in Germany. Eur. J. Clin. Microbiol. Infect. Dis. 29, 563–570. doi: 10.1007/s10096-010-0896-0
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Wyres, K. L., and Holt, K. E. (2016). Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 24, 944–956. doi: 10.1016/j.tim.2016.09.007
Yao, Y., Imirzalioglu, C., Hain, T., Kaase, M., Gatermann, S., Exner, M., et al. (2014). Complete nucleotide sequence of a Citrobacter freundii plasmid carrying KPC-2 in a unique genetic environment. Genome Announc. 2:e1157-14. doi: 10.1128/genomeA.01157-14
Keywords: multidrug-resistance, Citrobacter freundii ST98, Klebsiella oxytoca ST29, Escherichia coli ST369, IncN, WGS
Citation: Schweizer C, Bischoff P, Bender J, Kola A, Gastmeier P, Hummel M, Klefisch F-R, Schoenrath F, Frühauf A and Pfeifer Y (2019) Plasmid-Mediated Transmission of KPC-2 Carbapenemase in Enterobacteriaceae in Critically Ill Patients. Front. Microbiol. 10:276. doi: 10.3389/fmicb.2019.00276
Received: 13 August 2018; Accepted: 01 February 2019;
Published: 19 February 2019.
Edited by:
Laurent Poirel, Université de Fribourg, SwitzerlandReviewed by:
Costas C. Papagiannitsis, University of Thessaly, GreeceCopyright © 2019 Schweizer, Bischoff, Bender, Kola, Gastmeier, Hummel, Klefisch, Schoenrath, Frühauf and Pfeifer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Bischoff, cGV0ZXIuYmlzY2hvZmZAY2hhcml0ZS5kZQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.