- 1Institut des Géosciences de l’Environnement, Univ. Grenoble Alpes, CNRS, IRD, Grenoble INP, Grenoble, France
- 2Institut de Chimie de Clermont-Ferrand, UMR6096 CNRS–Université Clermont Auvergne-Sigma, Clermont-Ferrand, France
- 3CNRS UMR 5005, Environmental Microbial Genomics, Laboratoire Ampère, École Centrale de Lyon, Université de Lyon, Ecully, France
- 4Laboratory for Meteorological Physics (LaMP), Université Clermont Auvergne, Clermont-Ferrand, France
- 5Géosciences Environnement Toulouse, Centre National de la Recherche Scientifique, Institut de Recherche pour le Développement, Université de Toulouse, Toulouse, France
- 6Laboratory for Atmospheric Physics, Institute for Physics Research, Universidad Mayor de San Andrés, La Paz, Bolivia
- 7Department of Atmospheric and Oceanic Sciences, University of Maryland, College Park, MD, United States
- 8South African Weather Service, Stellenbosch, South Africa
- 9Key Laboratory of Tibetan Environment Changes and Land Surface Processes, Institute of Tibetan Plateau Research, Chinese Academy of Sciences, Beijing, China
The interplay between microbes and atmospheric physical and chemical conditions is an open field of research that can only be fully addressed using multidisciplinary approaches. The lack of coordinated efforts to gather data at representative temporal and spatial scales limits aerobiology to help understand large scale patterns of global microbial biodiversity and its causal relationships with the environmental context. This paper presents the sampling strategy and analytical protocols developed in order to integrate different fields of research such as microbiology, –omics biology, atmospheric chemistry, physics and meteorology to characterize atmospheric microbial life. These include control of chemical and microbial contaminations from sampling to analysis and identification of experimental procedures for characterizing airborne microbial biodiversity and its functioning from the atmospheric samples collected at remote sites from low cell density environments. We used high-volume sampling strategy to address both chemical and microbial composition of the atmosphere, because it can help overcome low aerosol and microbial cell concentrations. To account for contaminations, exposed and unexposed control filters were processed along with the samples. We present a method that allows for the extraction of chemical and biological data from the same quartz filters. We tested different sampling times, extraction kits and methods to optimize DNA yield from filters. Based on our results, we recommend supplementary sterilization steps to reduce filter contamination induced by handling and transport. These include manipulation under laminar flow hoods and UV sterilization. In terms of DNA extraction, we recommend a vortex step and a heating step to reduce binding to the quartz fibers of the filters. These steps have led to a 10-fold increase in DNA yield, allowing for downstream omics analysis of air samples. Based on our results, our method can be integrated into pre-existing long-term monitoring field protocols for the atmosphere both in terms of atmospheric chemistry and biology. We recommend using standardized air volumes and to develop standard operating protocols for field users to better control the operational quality.
Introduction
Biological particles are known to represent a significant fraction (∼20–70%) of the total number of aerosols > 0.2 μm, with large spatial and temporal variations (Matthias-Maser and Jaenicke, 1995; Graham et al., 2003; Jaenicke, 2005; Huffman et al., 2012). Among these, microorganisms are of particular interest in fields as diverse as epidemiology, including phytopathology (Morris et al., 2007), bioterrorism, forensic science and public health (Galán Soldevilla et al., 2007), and environmental sciences, like microbial ecology (Monteil et al., 2014; Mayol et al., 2017; Michaud et al., 2018), meteorology and climatology (Sesartic et al., 2012; Pouzet et al., 2017). More precisely concerning the latter, airborne microorganisms contribute to the pool of particles nucleating the condensation and crystallization of water and they are thus potentially involved in cloud formation and in the triggering of precipitation (Morris et al., 2014; Fröhlich-Nowoisky et al., 2016). Additionally, viable microbial cells act as chemical catalyzers interfering with atmospheric chemistry (e.g., Vaïtilingom et al., 2013). The constant flux of bacteria from the atmosphere to the Earth’s surface due to precipitation and dry deposition can also affect global biodiversity, but they are rarely taken into account when conducting ecological surveys (Hughes and Convey, 2010; Bar-On et al., 2018; Leyronas et al., 2018; Reche et al., 2018). As stressed by these studies attempting to decipher and understand the spread of microbes over the planet (e.g., Burrows et al., 2009a; Bowers et al., 2013; Morris and Sands, 2017; Šantl-Temkiv et al., 2018), concerted data are needed for documenting the abundance and distribution of airborne microorganisms, including at remote and altitudes sites.
Airborne bacteria are emitted by most Earth surfaces (plants, oceans, land, and urban areas) to the atmosphere via a variety of mechanical processes such as aeolian soil erosion, sea spray production, or mechanical disturbances including anthropogenic activities (e.g., Joung et al., 2017; Michaud et al., 2018). Due to their relatively small size (the median aerodynamic diameter of bacteria-containing particles is around 2–4 μm (Despres et al., 2012), these can then be transported upward by turbulent fluxes (Carotenuto et al., 2017) and carried by wind to long distances. As a consequence, bacteria are present in the air up to at least the lower stratosphere (Wainwright et al., 2006; DeLeon-Rodriguez et al., 2013; Smith et al., 2018). Given that the atmosphere is a large conveyor belt that moves air over thousands of kilometers, microorganisms are disseminated globally (Smith et al., 2012, 2013; Griffin et al., 2017). Airborne transport of microbes is therefore likely pervasive at the global scale, yet there have been only a limited number of studies that have looked at the spatial distribution of microbes across different geographical regions (e.g., Barberán et al., 2015; Griffin et al., 2017). One of the main difficulties is linked with the low microbial biomass associated with a high diversity existing in the atmosphere outdoor (∼102–105 cells/m3; e.g., Burrows et al., 2009b; Bowers et al., 2013; Amato et al., 2017), thus requiring reliable sampling procedures and controls. Furthermore, the site location and its environmental specificities have to be accounted for to some extent by considering chemical and meteorological variables (Deguillaume et al., 2014).
While these studies have led to novel findings regarding the link that may exist between airborne bacteria and their source and receptacle environments, the lack of uniform sampling and analysis methodology weaken the conclusions that can be drawn from independent studies. Here we aimed to provide sample collection and preparation methods intended to generate reproducible data, applicable to most sampling locations. This should allow the investigation of long-range transport, surface ecosystem interconnectivity and distribution of microorganisms in relation to meteorological and chemical contexts. Here, we present a method that allows simultaneous -sampling for chemical and microbiological characterization of aerosols, which can be deployed for long term monitoring at atmospheric observation sites throughout the planet. This has only been carried out previously in urban areas and the methods were not developed or optimized for non-urban environments (Bertolini et al., 2013) or for subsequent chemical analysis (Jiang et al., 2015; Luhung et al., 2015). The main objectives were to: (1) define appropriate sampling methods and duration (2) set up quality controls in order to improve the detection limit for various chemical species (3) improve DNA extraction methods from low biomass samples (4) ensure data intercomparability and (5) develop simplified experimental workflows that can easily be carried out by non-specialist onsite technical staff. These protocols were tested at 10 distinct sites covering various geographic regions of the globe. The need for a coordinated network for global monitoring of aerobiology has recently been identified in a number of recently published studies (Smith, 2013; Pearce et al., 2016; Cáliz et al., 2018), however, a standardized sampling method has yet to be proposed. Based on our results, our method can be integrated into pre-existing long-term monitoring field protocols for the atmosphere both in terms of atmospheric chemistry and biology, and could be included in future projects.
Methods
Experimental Strategy
A variety of methodologies for bioaerosol sampling, including passive sampling, filtration and impaction techniques exist (Haddrell and Thomas, 2017), however these have yet to be harmonized for concerted studies. In order to deal with the equipment available at most international monitoring stations, a sampling protocol that could be carried out by non-specialized personnel using on-site sampling equipment needed to be designed; these conditions constrained the choice of our bioaerosol sampling strategy toward high-volume samplers (high air flow-rate) on large diameter/size quartz fiber filters. Sampling time as well as DNA extraction protocols were improved for ensuring obtaining sufficient biological material for analyses from these types of filters, while maintaining the sampling time as short as possible for allowing detecting variations in connection with environmental variables. The sampling strategy and protocol development are outlined in the following section.
Filter Selection and Development of Sterilization Protocols
The analysis for chemical compounds and elements [e.g., elemental carbon (EC) and organic carbon (OC)] requires the combustion of a quartz fiber filter, which constrained the choice of sampling material. Depending on the sampler model, two filters sizes were used (5.9′′ round filter and 8′′ × 10′′ rectangular types) with filtration surface areas of 163 and 526 cm2, respectively. Several sterilization methods were tested to improve the biological quality of the filter without altering the detection limits of the chemical parameters. A standard method for atmospheric chemistry protocols is to dry heat the quartz filters at 500°C for at least a few hours (Jaffrezo et al., 2005). We tested additional heating and sterilization steps in laminar flow hoods or UV-exposure (10 min) on both filters and storage material. By adding these supplementary steps, we were able to significantly reduce the background OC concentrations of our blank filters as compared to the standard method (standard method OC concentration = 0.93 ± 0.35 μg/cm2 of filter, new method OC concentration = 0.55 ± 0.26 μg/cm2, p = 0.0009, Student T-test).
Based on our results, the following protocol was defined: filters were heated to 500°C for 8 h in order to remove traces of organic carbon including DNA. Filters were then handled within a laminar flow hood (UV sterilized, 254 nm, PSM – ESI FLUFRANCE BIOCYT 120, 10 min on each filter side) and individually stored in a folded aluminum foil and a thermally-sealed plastic (PE) bag or a zip-lock bag. All the material including foils, plastic bags, tweezers that would be in contact with the filters was UV-sterilized (2 J/cm2 for 2 min, 254 nm, CrossLinker, Bio-Link BLX). The filter holders were also UV-sterilized and stored individually in sterile bags. At the sampling sites, the field operators were trained and were given a detailed protocol (see Supplementary Information for detailed protocol) in order to replace and handle the filters properly at defined sampling times. After collection, filters were sealed in a folded sterile aluminum foil and plastic bags and stored at -20°C. At most sites, no microbiological safety cabinet was available; thus clean benches were made using pre-UV-sterilized plastic sheets in order to minimize contamination. All the sterile material was provided in sufficient quantity to the field operators. After sampling, filters (exposed and controls) were shipped to France for analysis from each sampling site at below zero temperature.
Optimization of DNA Extraction
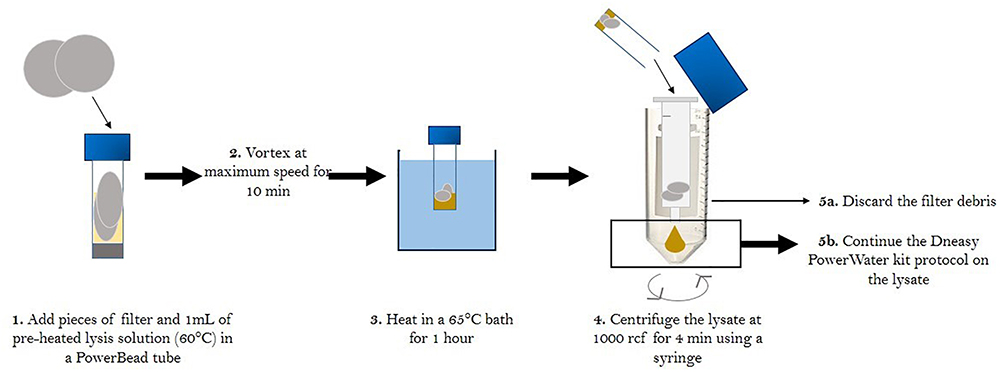
While quartz filters have been used for microbial studies in the air (Després et al., 2007; Smith et al., 2012; Jiang et al., 2015), limitations exist regarding the integrity of the samples collected [DNA degradation, cell mortality (Luhung et al., 2015)]. In addition, classic DNA extraction methods need to be improved for investigations in remote sites with low biomass. Since we were constrained by the choice of quartz filters for chemical analysis, we tested different extraction protocols in order to optimize DNA extraction yield from these filters. We tested different DNA extraction kits developed for environmental samples (e.g., DNeasy PowerWater, DNeasy PowerSoil and DNeasy Blood and Tissue kits from Qiagen). To do so, we set up a size selective high volume air sampling instrument (DIGITEL) equipped with a PM10 size-selective inlet in order to collect airborne particulate matter smaller than 10 μm (cut-off aerodynamic diameter) from the roof of the laboratory in Grenoble (France) to mimic field conditions. An atmospheric sample was collected for 24 h on a large filter. Nine sub-samples were collected from this filter and DNA was extracted according to the manufacturer’s instructions with the following modification: after the lysis step, the lysate was centrifuged in a syringe for 4 min at 1000 rcf to drain filter debris that tended to absorb the lysis solution. This additional centrifugation step increased lysate volume recovery by more than five-fold, potentially increasing DNA recovery. DNA concentrations were compared following quantification with a fluorometric method (Qubit dsDNA HS Assay Kit from Thermo Fisher Scientific, manufacturer’s instructions on 10 μL of sample) and 16S rRNA gene copy numbers were compared following quantification using qPCR. Briefly, the V3 region of the 16S rRNA gene was amplified using the SensiFast SYBR No-Rox kit (Bioline) and the following primers sequences: Eub 338f 5′-ACTCCTACGGGAGGCAGCAG-3′ as the forward primer and Eub 518r 5′-ATTACCGCGGCTGCTGG-3′ as the reverse primer (Fierer et al., 2005) on a Rotorgene 3000 machine (Qiagen). The reaction mixture of 20 μL contained 10 μL of SYBR master mix, 2 μL of DNA and RNAse-free water to complete the final 20 μL volume. The 2-step qPCR program consisted of an initial step at 95°C for 2 min for enzyme activation, then 35 cycles of 5 s at 95°C and 20 s at 60°C for hybridization and elongation, respectively. A final step was added to obtain a denaturation from 55 to 95°C with increments of 1°C. The amplicon length was around 200 bp. PCR products obtained from DNA from a pure culture of E. coli were cloned in a plasmid (pCRTM2.1-TOPO® vector, Invitrogen) and used as standard after quantification with the Broad-Range Qubit Fluorometric Quantification (Thermo Fisher Scientific).
The DNA concentrations measured were not significantly different among the three DNA extraction kits (Soil 0.008 ± 0.005 ng/μL, Water 0.010 ± 0.005 ng/μL, Tissue 0.012 ± 0.002 ng/μL, p > 0.22, One-way ANOVA, Tukey tests). However, the number of 16S rRNA gene copies per cubic meter of air was on average ten times higher with DNeasy PowerWater and PowerSoil kits than it was with DNeasy Blood & Tissue kit. Differences in extraction efficiency have been previously observed for several metagenomic and taxonomic studies and it is suggested that a variety of methods be tested to optimize results before studying new environments (Delmont et al., 2012; Zielińska et al., 2017). Based on these results and for practical reasons, we chose to use the DNeasy PowerWater kit. The DNeasy PowerBead tubes of the DNeasy PowerWater kit are 5 mL tubes (compared to the 2 mL PowerBead tubes of the DNeasy PowerSoil kit) allowing extraction of a larger filter surface.
The second series of extraction tests were carried out to determine whether the filter handling protocol had an effect on extraction efficiency. We tested four different treatments in triplicate: blank filters (processed and brought to the field but unexposed), filters that were passively exposed to the atmosphere for 5 min, filters that collected atmospheric samples during 24 h without heat treatment and filters that collected atmospheric samples during 24 h with heat treatment. DNA was extracted from filters using the protocol outlined above and quantified using Qubit. Based on the results of our test, heating the quartz filters at 500°C for 8 h before sampling has a significant impact on DNA extraction efficiency and reduced yield by up to 10-fold (24 h no heating 0.33 ± 0.16 ng/μL, n = 3, 24 h heating 0.038 ± 0.005 ng/μL, n = 3, p < 0.05, student t-test), but this is a critical step to ensure that trace carbon is removed from the filters prior to sampling. Therefore, we needed to further optimize our method to increase DNA extraction efficiency from heat sterilized quartz filters. Different options were tested, such as modifying the pH by addition of 1 M CaCO3 and shaking, as described in Bertolini et al. (2013), as well as adding a sonication step (Luhung et al., 2015), Neither of these methods increased yield significantly and we preferred to avoid adding a solution to our samples. Considering that high temperatures help desorbing DNA from the silica in the quartz filter (Vandeventer et al., 2013), we also tested the efficiency of a 1-h thermal treatment at 65°c during the lysis step (Jiang et al., 2015;Luhung et al., 2015) on DNA yield using eleven samples. We obtained four to five times higher DNA concentrations and 100–1000 times higher 16s rRNA gene copies per cubic meter of air using this optimized lysis technique. The final protocol is summarized in Figure 1 and was applied to all the filters collected during the sampling campaign.
Optimization of Sampling Time for DNA Analysis
Once the filter treatment and DNA extraction protocols were validated, we carried out different tests to optimize sample collection duration. We used the same sampling set-up on the roof of the laboratory in Grenoble to collect atmosphere samples. Several different sampling times were considered: 5 min, 1, 24, and 72 h. DNA was extracted from filters using the protocol outlined above and quantified using Qubit. Filters analyzed in triplicate at 5 min and 1 h where found to be below the detection limit of the apparatus (0.01 ng/μL). Filters collected at 24 h showed a significantly lower yield than those collected at 72 h (24 h = 0.02 ± 0.01 ng/μL, 72 h = 0.08 ± 0.03 ng/μL, n = 3, p < 0.05, Student t-test), which suggests that sampling duration impacts DNA yield. Based on the results of this test and considering the remoteness of the sites, we decided to sample continuously for 7 days.
Protocol Testing and Deployment
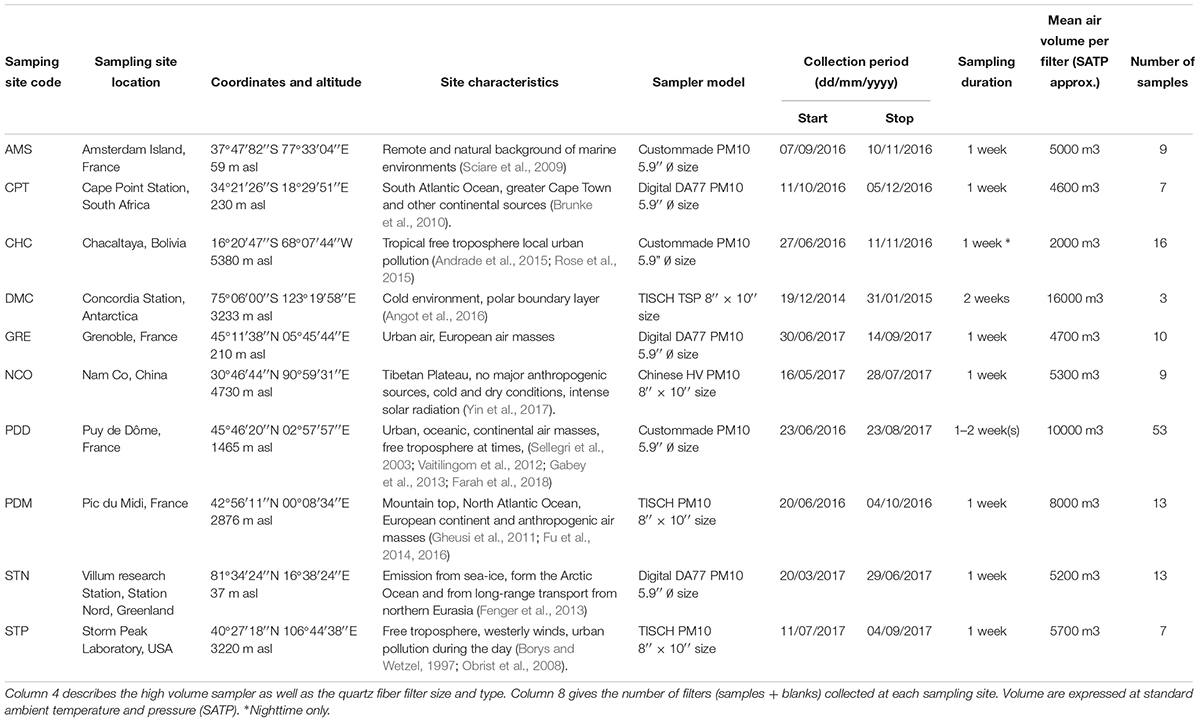
Ten sites were chosen based on latitudinal positions, known chemical characteristics, historic atmospheric data and logistical support. The characteristics for each site are described in Table 1 and the geographic distribution can be seen in Figure 2. Sites included Arctic and Antarctic stations as well as mid-latitude stations. In order to access information on long range transport of aerosols, dusts and airborne microorganisms, three sites that are frequently in the free troposphere were selected. Temporal variability is an important but poorly understood factor of microbial community diversity (e.g., Bertolini et al., 2013), so we completed the dataset with continuous sampling for more than one entire year at a 1-week resolution at a single reference site: puy de Dôme, France. To avoid snap-shot sampling, each site was sampled for a minimum of 2 months, so manned research sites were selected. At all sampling sites, meteorological parameters, such as wind speed and direction, rainfall, temperature, humidity, air pressure, and solar radiation were systematically recorded. Depending on the site, continuous measurements providing additional information for data interpretation were also collected. These included aerosol properties (size, concentration) and gas concentrations (ambient O3, nitrogen species, CO, CO2, CH4, H2O, O2, and volatile organic compounds (VOCs). Finally, total gaseous mercury (TGM) concentration was also continuously monitored by our collaborators at Cape Point Station, Nam Co, Amsterdam Island and Villum Research Station.
Several brands of size selective high volume air sampling instruments (TISCH, DIGITEL, home-made) were used in the present study. All these samplers are based on the same physical principle. Air was drawn into the sampler and through a large quartz fiber filter by means of a powerful pump, so that particulate material impacted the filter surface. All the samplers, but one (at Concordia station in Antarctica -DMC), were equipped with a PM10 size-selective inlet in order to collect particulate matter smaller than 10 μm (cut-off aerodynamic diameter). The use of the PM10 inlet was an important aspect in order to guarantee that both chemical and microbial data could be compared among the sites and to prevent rain and hydrometeors from reaching the filter and modifying its porosity and air flow rate properties. At DMC, total suspended particles were collected (median aerodynamic diameter of 20 μm approx). Sampling air flow rates were between 30 and 70 m3/h (±2%) and collected volumes ranged from 2000 to 10000 m3 over a 1-week period (except at DMC, where the samples were collected over a 2-week period). Flow regulation of the pump was controlled with a flow-meter that was regularly checked and calibrated. The total volume of air was subsequently corrected to standard ambient temperature and pressure (SATP, 298K, 101.325 kPa) to standardize air collection at all the sites (Table 1). For some mountain sites (PDM, CHC, STP), we generally sampled at nights in order to limit the sampling of the planetary boundary layer air.
Quality Control
In addition to the 140 samples, we also collected 38 blank filters named “transportation blanks” (TB, 18 filters) and “field blanks” (FB, 20 filters) in order to monitor and check the quality of the sampling protocol (see Figure 3). The transportation blanks were filters shipped back and forth to the sampling sites but without any manipulation. The field blanks were exposed for 24–72 h without switching on the high-volume sampler and then processed and stored similarly as the actual samples. The field blanks had significantly higher OC concentrations as compared to the transportation blanks, but these were in the same range as the values obtained for blank filters using the standard sterilization technique (without subsequent sterilization steps).
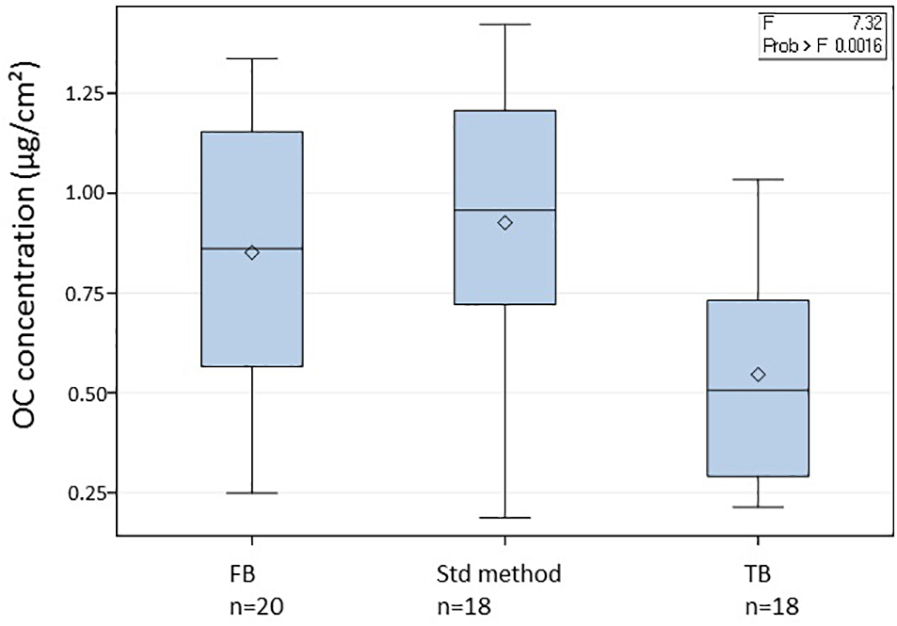
Figure 3. Organic concentrations on different types of filters: field blanks (FB), transport blanks (TB) and filters treated using the standard method (Std method). Significance was tested using One-way ANOVA and Tukey tests.
Chemical Analyses
Elemental carbon (EC), organic carbon (OC), sugar anhydrides and alcohols, major soluble anions and cations as described by Waked et al. (2014) as well as total mercury were systematically analyzed in all the quartz fiber filters (including blank filters). EC and OC were analyzed from a 1.5 cm2 punch sample using a thermo optical transmission method on a Sunset Lab analyzer (Birch and Cary, 1996). Punches of 38 mm-diameter from each samples were extracted using ultrapure water under mechanical agitation for a period of 30 min. The extracts were then filtered with 0.22 μm Nucleopore filters before injection in the instruments (Piot et al., 2012). The extracts were used for quantification of sugar anhydrides and alcohols (levoglucosan, mannosan, galactosan, inositol, glycerol, erythriol, xylitol, arabitol, sorbitol, mannitol, trehalose, rhamnose, glucose, fructose, and sucrose) by HPLC-PAD using a set of Methrom columns (MetroSep A Supp 15 and Metrosep Carb1) in a Thermo ScientificTM DionexTM ICS-5000+ Capillary HPICTM system. Soluble anions (MSA, SO42-, NO3-, Cl-, Ox) and cations (Na+, NH4+, K+, Mg2+, Ca2+) were analyzed by ion chromatography (IC, Dionex ICS3000) on the same extracts. AS/AG 11HC and CS/CG 12A columns were used for anions and cations analyses, respectively. Finally, twenty-five low molecular weight organic acids (C3-C9) were analyzed from the same extracts by LC-MS (DX500 – LCQ Fleet with an inverse phase C18 column). On some of the filters, more than twenty-five organic components were detected (glycolic acid, glyoxylic acid, tartaric acid, malic acid, lactic acid, malonic acid, succinic acid, hydroxybutyric acid, methylmalonic acid, fumaric acid, ketobutyric acid, maleic acid, glutaric acid, oxoheptanedioic acid, citraconic acid, methlysuccinic acid, methylglutaric acid, adipic acid, pimelic acid, phtalic acid, pinic acid, isophtalic acid, suberic acid, benzoic acid, azelaic acid, and sebacic acid). Total mercury measurements from filter samples were performed with a DMA-80 (Milestone) analytical system based on the principles of sample thermal decomposition, mercury amalgamation and atomic absorption detection. Additionally, filters collected at AMS, CPT, PDD, and PDM sampling sites were analyzed by LC-MS technique. Except for total mercury measurements (which were performed at GET, Toulouse), all analyses were performed at the AirOSol chemical analytical platform facility at IGE, Grenoble, France.
Results
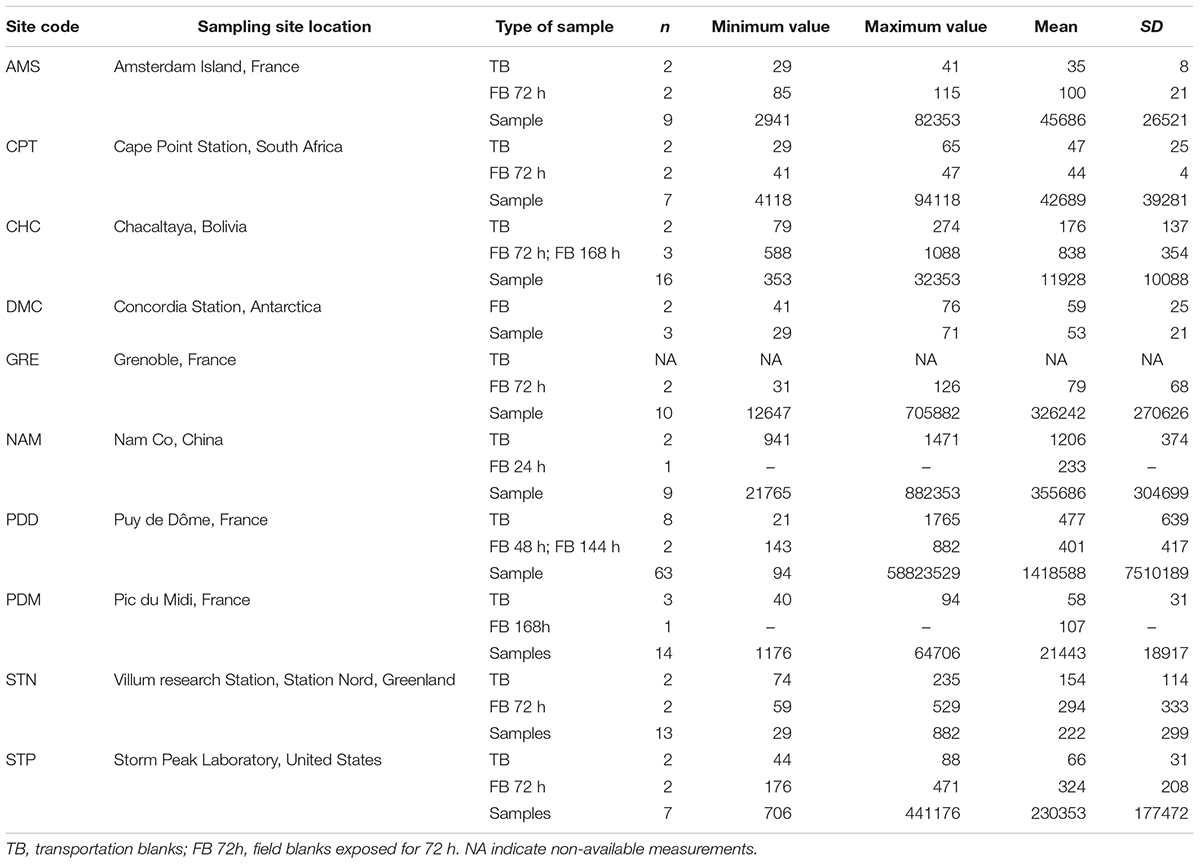
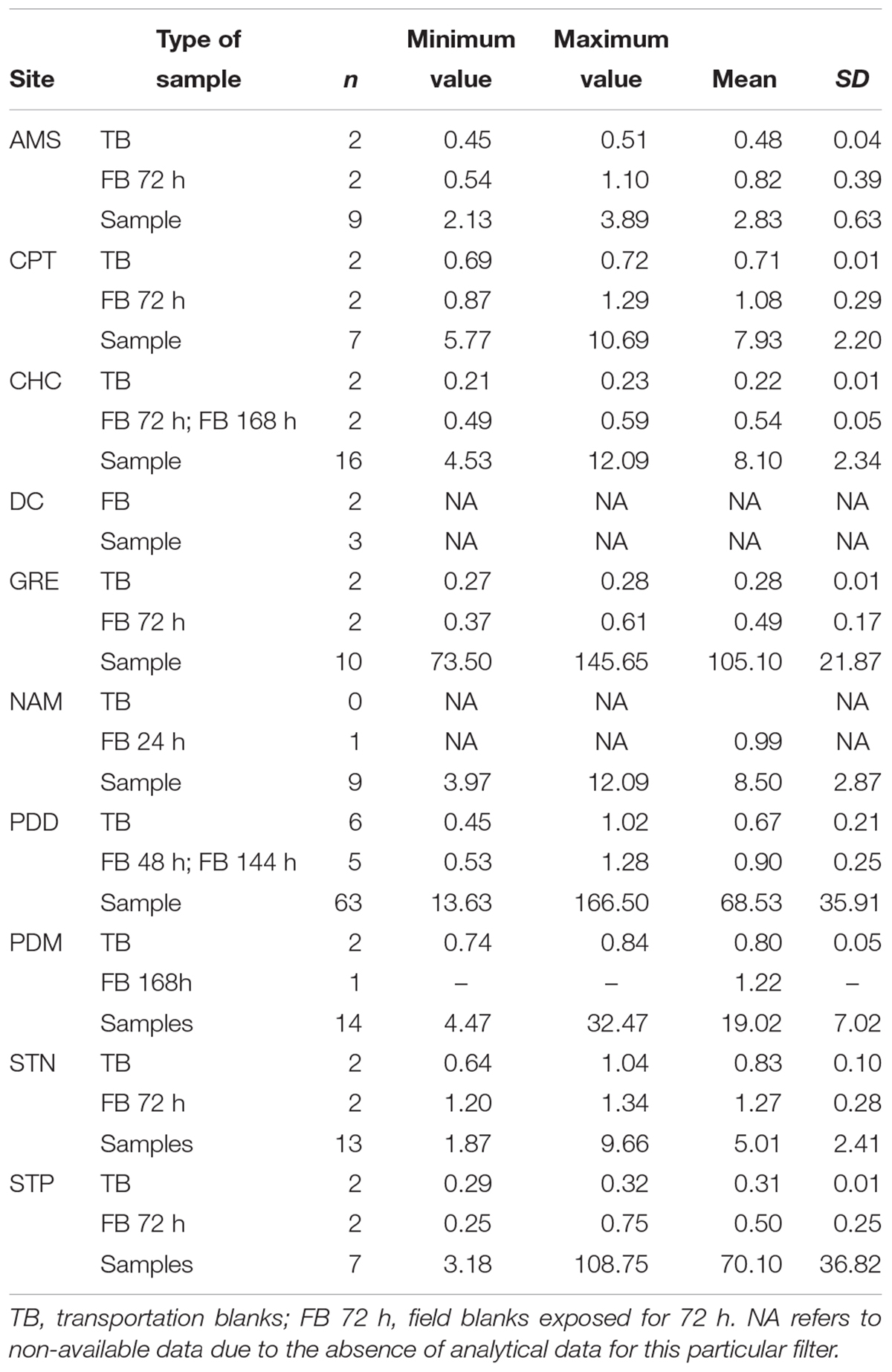
For most of the sites, the TB were in the range of 101–102 16S rRNA copies per mm2 (Table 2). A few high outliers remained and they could be attributed to the cleaning and packing procedures, and to the DNA extraction (including possible cross contamination during the subsampling phase). FB consisting of filters exposed to the atmosphere for up to 1 week but with no air forced to pass through, had, as expected, 16S rRNA gene concentrations one-to five-fold higher than the TB. These combined the passive contribution of the atmospheric environment and the DNA contamination occurring during the different phases of filter handling in the field. Except for the polar sites and CHC, the concentration of 16S rRNA gene copies in blank samples were < 0.3% that in the corresponding atmospheric samples. The blanks at CHC were up to 7% of the average number of copies in the atmospheric samples, due to the low concentrations of DNA sampled from air at this high altitude site. At both polar sites (DMC and Villum) the 16S rRNA gene concentrations were similar to controls, indicating very low biomass.
Organic carbon results (Table 3) also confirmed that the sampling protocols were adequately designed with a mean OC value of 0.55 ± 0.26 μg/cm2 for all the transportation blanks, and of 0.85 ± 0.32 μg/cm2 for the field blanks. In the case of the PDD samples, TB or FB represented less than 1.5% of the OC content in a sample. At remote sites with a very low OC concentration such as at AMS, the FB fraction reached up to 30% of the value in samples, but was thus still clearly distinct. Based on our experience in atmospheric chemistry field programs (Sprovieri et al., 2016; Daellenbach et al., 2017; Pandolfi et al., 2018), these are low concentrations for blank series. As shown in Figure 3, our protocols significantly lower the transportation blank regarding OC. Inevitably, the handling of the filter clearly induces some contamination. This contamination can be significantly reduced when using a laminar flow hood, as in Grenoble (field blank of 0.49 μg/cm2) and PDD.
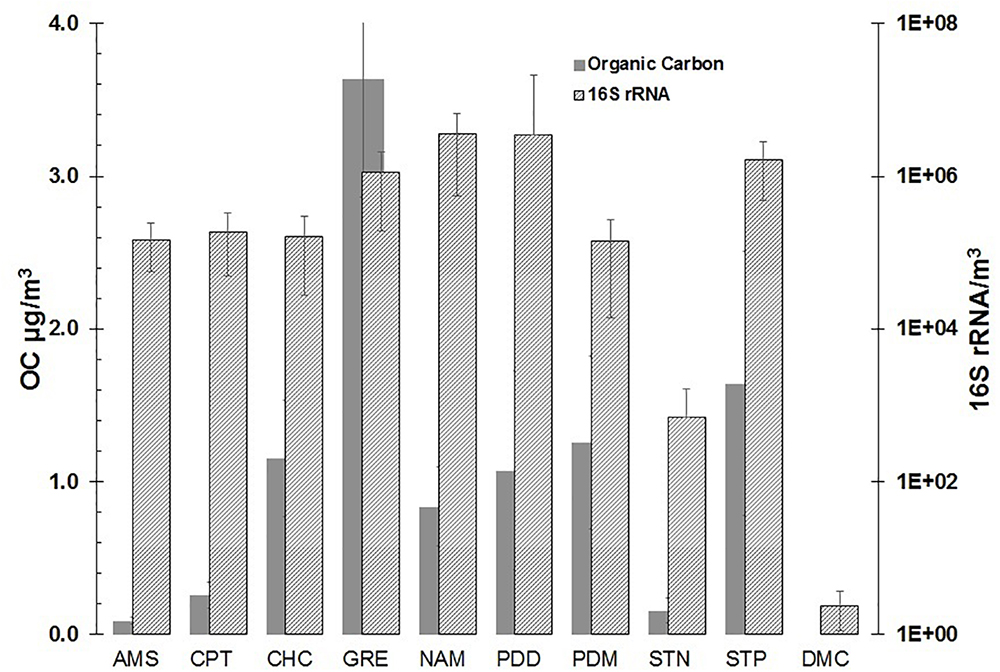
Figure 4 reports the range of OC and 16S rRNA gene concentrations measured at the different sampling sites. As expected, the urban site (Grenoble) had the highest OC values (3.63 ± 0.78 μg/m3) and 16S rRNA gene concentrations, of around 106 copies/m3 of air (1.1 ± 0.9 × 106). Comparable gene copies concentrations were observed at Storm Peak (1.6 ± 1.1) 106 copies/m3, Nam Co (3.6 ± 3.1) 106 copies/m3, but lower OC values comparatively to the Grenoble site.
This indicates that DNA material has, at least in part, sources distinct from OC. Puy de Dome station, where 53 samples were collected over 1 year, showed a range of gene copies from 104 to 108 copies/m3 ((3.4 ± 18) 106 copies/m3 in average (±standard deviation) illustrating the great temporal variability of airborne biological material in the air at a single site. This is to be related with the wide diversity of air masses and meteorological conditions that can occur at a given site over a year (see for example Deguillaume et al., 2014). More remote sites had all 16S gene concentrations one order of magnitude lower than at PDD, such as Chacaltaya (1.6 ± 1.3) 105 copies/m3 (night samples only), Cape Point (1.9 ± 1.4) 105 copies/m3, Pic du Midi (1.4 ± 1.3) 105 copies/m3 and Amsterdam Island (1.5 ± 1.0) 105 copies/m3. In turn, the OC concentrations were clearly distinct between the sites, with Amsterdam Island showing the lowest OC levels of this study (0.09 ± 0.02) μg/m3. The arctic site had a very low 16S rRNA gene concentration of 102/m3 for corresponding average OC concentrations of 1.6 ± 0.9 μg/m3, while the Antarctic site on the plateau showed gene copies similar to the blank.
Conclusion
We developed suitable easy-to-use and standardized protocols that generated consistent microbial and chemical datasets from the same samples at concentrations high enough for confident analysis. The validity of these protocols was then tested by deploying them on 10 sites over the globe and collect samples to explore atmospheric bacteria, fungi, or viruses over large spatial scales. For large scale studies coupling chemical measurements to biological measurements, we were able to demonstrate that quartz filters can be used, but that the extraction protocols must be optimized to maximize DNA yield. In addition to using the protocol outlined here, we also recommend the following:
• carefully prepare the filter and all the material using a combination of heating (500°C) and UV treatment (254 nm).
• provide detailed protocols intended to limit contamination (SOP, see SI for an example) to field users.
• include enough control samples (including transportation and field work blanks) to monitor the quality of the sampling procedure.
• carefully design atmospheric sampling at peak stations in order to take into account vertical turbulent mixing, and night-time hours (in general) should be preferred to avoid the influence of local sources of aerosols.
• Correct sampling times for the remoteness of the sites and for the measurements to be carried out. For example, a 1 week sampling with a volume of around 5000 (normalized) m3 is sufficient for amplicon and metagenomic sequencing for most remote sites, except Antarctica, where the biomass is too low for DNA investigations even from total filtered volumes of 16000 m3.
• Collected volumes should be normalized using STP or SATP standards.
One of the main disadvantages of a weekly sampling is the loss of information regarding rapid atmospheric chemistry processes and rapid changes in terms of aerosol sources. In turn, it has the advantage to smooth the data and avoid the stochastic-like behavior of biological content in the air often observed (Bowers et al., 2009, etc.). With the development of better extraction protocols and more sensitive sequencing techniques, this limitation could be overcome, allowing for daily sampling in the future.
Author Contributions
AD, CathL, TV, and PA conceived the study. OM implemented the technical phase and protocols together with AT and RT-P. RT-P performed the biomolecular work. J-LJ supervised the analytical work. MJ, LB, KS, JS, MA, IM, CasL, LM, and QZ helped and participated to the implementation of the field experiments.
Funding
This program is funded by ANR-15-CE01-0002 – INHALE. RT-P’s doctoral grant is funded by Région Auvergne – Rhöne Alpes. Part of the analytical equipment was supported at IGE by the AirOSol platform within Labex OSUG@2020 (ANR10 LABX56). AD and QZ were also supported by the program CAMPUS France program XU GUANGQI (38718ZC INHALE_NAMCO). Financial support for AMS, DMC and Villum field campaigns was provided by the French Polar Institute IPEV (programs 1028 and 399).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank IPEV GMOStral 1028 and ALCHIMI 399 program. We are very grateful to Gannet Hallar and Ian McCubbin at Storm Peak, Joel Savarino, Nicolas Caillon, Roxanne Jacob for DMC samples, Benny and Jesper for the sampling at Villum Research Station, and to Bjarne Jensen and Henrik Skov for their support, Isabelle Jouvie at Amsterdam Island. We deeply thank Ross Edwards at Curtin University and James Schauer and Christopher Worley at University of Wisconsin–Madison for lending us a high-volume sampler. We would like to thank Alessia Nicosia, Mickael Ribeiro (OPGC/LaMP), Ludovic Besaury (FRE), and the UMS 831 Pic du Midi Observatory team for help with sample collection. We thank Manuel Rocca and Fernando Velarde for help at CHC. Alexandre Poulain is deeply acknowledged for his ideas and thoughts on the experimental design and strategy. AD would also like to thank the FFCR and the University of Ottawa (Department of Biology) for hosting him. We also acknowledge Vincent Lucaire, Laure Jullien, and Antony Vella for the lab work in the AirOsol platform.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00243/full#supplementary-material
References
Amato, P., Joly, M., Besaury, L., Oudart, A., Taib, N., Moné, A. I., et al. (2017). Active microorganisms thrive among extremely diverse communities in cloud water. PLoS One 12:e0182869. doi: 10.1371/journal.pone.0182869
Andrade, M., Zaratti, F., Forno, R., Gutiérrez, R., Moreno, I., Velarde, F., et al. (2015). Puesta en marcha de una nueva estación de monitoreo climático en los andes centrales de Bolivia: la estación Gaw/Chacaltaya Bolivia: the Gaw/Chacaltaya station set to work of a new climate monitoring station in the central andes of Bolivia: the Gaw/Chacal. Revista Boliviana De Física 26, 6–15.
Angot, H., Magand, O., Helmig, D., Ricaud, P., Quennehen, B., Gallée, H., et al. (2016). New insights into the atmospheric mercury cycling in central Antarctica and implications on a continental scale. Atmos. Chem. Phys. 16, 8249–8264. doi: 10.5194/acp-16-8249-2016
Barberán, A., Ladau, J., Leff, J. W., Pollard, K. S., Menninger, H. L., Dunn, R. R., et al. (2015). Continental-scale distributions of dust-associated bacteria and fungi. Proc. Natl. Acad. Sci.U.S.A. 112, 5756–5761. doi: 10.1073/pnas.1420815112
Bar-On, Y. M., Phillips, R., and Milo, R. (2018). The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511. doi: 10.1073/pnas.1711842115
Bertolini, V., Gandolfi, I., Ambrosini, R., Bestetti, G., Innocente, E., Rampazzo, G., et al. (2013). Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Appl. Microbiol. Biotechnol. 97, 6561–6570. doi: 10.1007/s00253-012-4450-0
Birch, M. E., and Cary, R. A. (1996). Elemental carbon-based method for monitoring occupational exposures to particulate diesel exhaust. Aerosol Sci. Technol. 25, 221–241. doi: 10.1080/02786829608965393
Borys, R. D., and Wetzel, M. A. (1997). Storm peak laboratory: a research, teaching, and service facility for the atmospheric sciences. Bull. Am. Meteorol. Soc. 78, 2115–2124. doi: 10.1175/1520-0477(1997)078<2115:SPLART>2.0.CO;2
Bowers, R. M., Clements, N., Emerson, J. B., Wiedinmyer, C., Hannigan, M. P., and Fierer, N. (2013). Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environ. Sci. Technol. 47, 12097–12106. doi: 10.1021/es402970s
Bowers, R. M., Lauber, C. L., Wiedinmyer, C., Hamady, M., Hallar, A. G., Fall, R., et al. (2009). Characterization of airborne microbial communities at a high-elevation site and their potential to act as atmospheric ice nuclei. Appl. Environ. Microbiol. 75, 5121–5130. doi: 10.1128/AEM.00447-09
Brunke, E. G., Labuschagne, C., Ebinghaus, R., Kock, H. H., and Slemr, F. (2010). Gaseous elemental mercury depletion events observed at cape point during 2007-2008. Atmos. Chem. Phys. 10, 1121–1131. doi: 10.5194/acp-10-1121-2010
Burrows, S. M., Butler, T., Jöckel, P., Tost, H., Kerkweg, A., Pöschl, U., et al. (2009a). Bacteria in the global atmosphere – part 2: modeling of emissions and transport between different ecosystems. Atmos. Chem. Phys. 9, 9281–9297. doi: 10.5194/acp-9-9281-2009
Burrows, S. M., Elbert, W., Lawrence, M. G., and Pöschl, U. (2009b). Bacteria in the global atmosphere – part 1: review and synthesis of literature data for different ecosystems. Atmos. Chem. Phys. 9, 9263–9280. doi: 10.5194/acp-9-9263-2009
Cáliz, J., Triadó-Margarit, X., Camarero, L., and Casamayor, E. O. (2018). A long-term survey unveils strong seasonal patterns in the airborne microbiome coupled to general and regional atmospheric circulations. Proc. Natl. Acad. Sci. U.S.A. 115, 12229-12234. doi: 10.1073/pnas.1812826115
Carotenuto, F., Georgiadis, T., Gioli, B., Leyronas, C., Morris, C. E., Nardino, M., et al. (2017). Measurements and modeling of surface–atmosphere exchange of microorganisms in mediterranean grassland. Atmos. Chem. Phys. 17, 14919–14936. doi: 10.5194/acp-17-14919-2017
Daellenbach, K. R., Stefenelli, G., Bozzetti, C., Vlachou, A., Fermo, P., Gonzalez, R., et al. (2017). Long-term chemical analysis and organic aerosol source apportionment at nine sites in central europe: source identification and uncertainty assessment. Atmos. Chem. Phys. 17, 13265–13282. doi: 10.5194/acp-17-13265-2017
Deguillaume, L., Charbouillot, T., Joly, M., Vaïtilingom, M., Parazols, M., Marinoni, A., et al. (2014). Classification of clouds sampled at the puy de Dôme (France) based on 10 yr of monitoring of their physicochemical properties. Atmos. Chem. Phys. 14, 1485–1506. doi: 10.5194/acp-14-1485-2014
DeLeon-Rodriguez, N., Lathem, T. L., Rodriguez-R, L. M., Barazesh, J. M., Anderson, B. E., Beyersdorf, A. J., et al. (2013). Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc. Natl. Acad. Sci. U.S.A. 110, 2575–2580. doi: 10.1073/pnas.1212089110
Delmont, T. O., Prestat, E., Keegan, K. P., Faubladier, M., Robe, P., Clark, I. M., et al. (2012). Structure, fluctuation and magnitude of a natural grassland soil metagenome. Isme J. 6, 1677–1687. doi: 10.1038/ismej.2011.197
Despres, V. R., Huffman, J. A., Burrows, S. M., Hoose, C., Safatov, A. S., Buryak, G., et al. (2012). Primary biological aerosol particles in the atmosphere: a review. Tellus B Chem. Phys. Meteorol. 64, doi: 10.3402/tellusb.v64i0.15598
Després, V. R., Nowoisky, J. F., Klose, M., Conrad, R., Andreae, M. O., and Pöschl, U. (2007). Characterization of primary biogenic aerosol particles in urban, rural, and high-alpine air by DNA sequence and restriction fragment analysis of ribosomal RNA genes. Biogeosciences 4, 1127–1141. doi: 10.5194/bg-4-1127-2007
Farah, A., Freney, E., Chauvigné, A., Baray, J.-L., Rose, C., Picard, D., et al. (2018). Seasonal variation of aerosol size distribution data at the Puy de Dôme station with emphasis on the boundary layer/free troposphere segregation. Atmosphere 9:244. doi: 10.3390/atmos9070244
Fenger, M., Sørensen, L. L., Kristensen, K., Jensen, B., Nguyen, Q. T., Nøjgaard, J. K., et al. (2013). Sources of anions in aerosols in northeast Greenland during late winter. Atmos. Chem. Phys. 13, 1569–1578. doi: 10.5194/acp-13-1569-2013
Fierer, N., Jackson, J. A., Vilgalys, R., and Jackson, R. B. (2005). Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71, 4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005
Fröhlich-Nowoisky, J., Kampf, C. J., Weber, B., Huffman, J. A., Pöhlker, C., Andreae, M. O., et al. (2016). Bioaerosols in the earth system: climate, health, and ecosystem interactions. Atmos. Res. 182, 346–376. doi: 10.1016/j.atmosres.2016.07.018
Fu, X., Heimbürger, L.-E., and Sonke, J. E. (2014). Collection of atmospheric gaseous mercury for stable isotope analysis using iodine- and chlorine-impregnated activated carbon traps. J. Anal. Atom. Spectrom. 29, 841–841. doi: 10.1039/c3ja50356a
Fu, X., Marusczak, N., Wang, X., Gheusi, F., and Sonke, J. E. (2016). Isotopic composition of gaseous elemental mercury in the free troposphere of the pic du midi observatory, france. Environ. Sci. Technol. 50, 5641–5650. doi: 10.1021/acs.est.6b00033
Gabey, A. M., Vaitilingom, M., Freney, E., Boulon, J., Sellegri, K., Gallagher, M. W., et al. (2013). Observations of fluorescent and biological aerosol at a high-altitude site in central France. Atmos. Chem. Phys. 13, 7415–7428. doi: 10.5194/acp-13-7415-2013
Galán Soldevilla, C., Cariñanos González, P., Alcázar Teno, P., and Domínguez Vilches, E. (2007). Spanish Aerobiology Network (REA): Management and Quality Manual. Cordoba: Servicio de Publicaciones.
Gheusi, F., Ravetta, F., Delbarre, H., Tsamalis, C., Chevalier-Rosso, A., Leroy, C., et al. (2011). Pic 2005, a field campaign to investigate low-tropospheric ozone variability in the Pyrenees. Atmos. Res. 101, 640–665. doi: 10.1016/j.atmosres.2011.04.014
Graham, B., Guyon, P., Maenhaut, W., Taylor, P. E., Ebert, M., Matthias-Maser, S., et al. (2003). Composition and diurnal variability of the natural Amazonian aerosol. J. Geophys. Res. 108, 4765–4765. doi: 10.1029/2003JD004049
Griffin, D., Gonzalez-Martin, C., Hoose, C., and Smith, D. (2017). “Global-Scale Atmospheric Dispersion of Microorganisms,” in Microbiology of Aerosols, eds A. M. Delort and P. Amato (Hoboken, NJ: John Wiley & Sons).
Haddrell, A. E., and Thomas, R. J. (2017). Aerobiology: experimental considerations, observations, and future tools. Appl. Environ. Microbiol. 83:e809–17. doi: 10.1128/AEM.00809-17
Huffman, J. A., Sinha, B., Garland, R. M., Snee-Pollmann, A., Gunthe, S. S., Artaxo, P., et al. (2012). Size distributions and temporal variations of biological aerosol particles in the amazon rainforest characterized by microscopy and real-time UV-APS fluorescence techniques during AMAZE-08. Atmos. Chem. Phys. 12, 11997–12019. doi: 10.5194/acp-12-11997-2012
Hughes, K. A., and Convey, P. (2010). The protection of Antarctic terrestrial ecosystems from inter- and intra-continental transfer of non-indigenous species by human activities: a review of current systems and practices. Glob. Environ. Change 20, 96–112. doi: 10.1016/j.gloenvcha.2009.09.005
Jaenicke, R. (2005). Abundance of cellular material and proteins in the atmosphere. Science 308, 73–73. doi: 10.1126/science.1106335
Jaffrezo, J. L., Aymoz, G., and Cozic, J. (2005). Size distribution of EC and OC in the aerosol of alpine valleys during summer and winter. Atmos. Chem. Phys. 5, 2915–2925. doi: 10.5194/acp-5-2915-2005
Jiang, W., Liang, P., Wang, B., Fang, J., Lang, J., Tian, G., et al. (2015). Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat. Protoc. 10, 768–779. doi: 10.1038/nprot.2015.046
Joung, Y. S., Ge, Z., and Buie, C. R. (2017). Bioaerosol generation by raindrops on soil. Nat. Commun. 8:14668. doi: 10.1038/ncomms14668
Leyronas, C., Morris, C. E., Choufany, M., and Soubeyrand, S. (2018). Assessing the aerial interconnectivity of distant reservoirs of sclerotinia sclerotiorum. Front. Microbiol. 9:2257. doi: 10.3389/fmicb.2018.02257
Luhung, I., Wu, Y., Ng, C. K., Miller, D., Cao, B., and Chang, V. W.-C. (2015). Protocol improvements for low concentration DNA-based bioaerosol sampling and analysis. PLoS One 10:e0141158. doi: 10.1371/journal.pone.0141158
Matthias-Maser, S., and Jaenicke, R. (1995). The size distribution of primary biological aerosol particles with radii 0.2 μm in an urban/rural influenced region. Atmos. Res. 39, 279–286. doi: 10.1016/0169-8095(95)00017-8
Mayol, E., Arrieta, J. M., Jiménez, M. A., Martínez-Asensio, A., Garcias-Bonet, N., Dachs, J., et al. (2017). Long-range transport of airborne microbes over the global tropical and subtropical ocean. Nat. Commun. 8:201. doi: 10.1038/s41467-017-00110-9
Michaud, J. M., Thompson, L. R., Kaul, D., Espinoza, J. L., Richter, R. A., Xu, Z. Z., et al. (2018). Taxon-specific aerosolization of bacteria and viruses in an experimental ocean-atmosphere mesocosm. Nat. Commun. 9:2017. doi: 10.1038/s41467-018-04409-z
Monteil, C. L., Bardin, M., and Morris, C. E. (2014). Features of air masses associated with the deposition of Pseudomonas syringae and Botrytis cinerea by rain and snowfall. Isme J. 8:2290. doi: 10.1038/ismej.2014.55
Morris, C. E., Conen, F., Huffman, J. A., Phillips, V., Poschl, U., and Sands, D. C. (2014). Bioprecipitation: a feedback cycle linking earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere. Glob. Change Biol. 20, 341–351. doi: 10.1111/gcb.12447
Morris, C. E., Kinkel, L. L., Xiao, K., Prior, P., and Sands, D. C. (2007). Surprising niche for the plant pathogen Pseudomonas syringae. Infect. Genet. Evol. 7, 84–92. doi: 10.1016/j.meegid.2006.05.002
Morris, C. E., and Sands, D. C. (2017). “Impacts of Microbial Aerosols on Natural and Agro-ecosystems: Immigration, Invasions, and their Consequences,” in Microbiology of Aerosols, eds A. M. Delort and P. Amato (Hoboken, NJ: John Wiley & Sons).
Obrist, D., Hallar, A. G., Mccubbin, I., Stephens, B. B., and Rahn, T. (2008). Atmospheric mercury concentrations at storm peak laboratory in the rocky mountains: evidence for long-range transport from asia, boundary layer contributions, and plant mercury uptake. Atmos. Environ. 42, 7579–7589. doi: 10.1016/j.atmosenv.2008.06.051
Pandolfi, M., Alados-Arboledas, L., Alastuey, A., Andrade, M., Angelov, C., Artiñano, B., et al. (2018). A european aerosol phenomenology – 6: scattering properties of atmospheric aerosol particles from 28 ACTRIS sites. Atmos. Chem. Phys. 18, 7877–7911. doi: 10.5194/acp-18-7877-2018
Pearce, D. A., Alekhina, I. A., Terauds, A., Wilmotte, A., Quesada, A., Edwards, A., et al. (2016). Aerobiology over antarctica - a new initiative for atmospheric ecology. Front. Microbiol. 7:16. doi: 10.3389/fmicb.2016.00016
Piot, C., Jaffrezo, J. L., Cozic, J., Pissot, N., El Haddad, I., Marchand, N., et al. (2012). Quantification of levoglucosan and its isomers by high performance liquid chromatography - electrospray ionization tandem mass spectrometry and its applications to atmospheric and soil samples. Atmos. Measur. Tech. 5, 141–148. doi: 10.5194/amt-5-141-2012
Pouzet, G., Peghaire, E., Aguès, M., Baray, J.-L., Conen, F., and Amato, P. (2017). Atmospheric processing and variability of biological ice nucleating particles in precipitation at opme, France. Atmosphere 8:229. doi: 10.3390/atmos8110229
Reche, I., D’orta, G., Mladenov, N., Winget, D. M., and Suttle, C. A. (2018). Deposition rates of viruses and bacteria above the atmospheric boundary layer. ISME J. 12, 1154–1162. doi: 10.1038/s41396-017-0042-4
Rose, C., Sellegri, K., Velarde, F., Moreno, I., Ramonet, M., Weinhold, K., et al. (2015). Frequent nucleation events at the high altitude station of Chacaltaya (5240 m a.s.l.), Bolivia. Atmos. Environ. 102, 18–29. doi: 10.1016/j.atmosenv.2014.11.015
Šantl-Temkiv, T., Gosewinkel, U., Starnawski, P., Lever, M., and Finster, K. (2018). Aeolian dispersal of bacteria in southwest greenland: their sources, abundance, diversity and physiological states. FEMS Microbiol. Ecol. 94:fiy031. doi: 10.1093/femsec/fiy031
Sciare, J., Favez, O., Sarda-Estève, R., Oikonomou, K., Cachier, H., and Kazan, V. (2009). Long-term observations of carbonaceous aerosols in the austral ocean atmosphere: evidence of a biogenic marine organic source. J. Geophys. Res. 114, D15302–D15302. doi: 10.1029/2009JD011998
Sellegri, K., Laj, P., Marinoni, A., Dupuy, R., Legrand, M., and Preunkert, S. (2003). Contribution of gaseous and particulate species to droplet solute composition at the Puy de Dôme, France. Atmos. Chem. Phys. 3, 1509–1522. doi: 10.5194/acp-3-1509-2003
Sesartic, A., Lohmann, U., and Storelvmo, T. (2012). Bacteria in the ECHAM5-HAM global climate model. Atmos. Chem. Phys. 12, 8645–8661. doi: 10.5194/acp-12-8645-2012
Smith, D. J. (2013). Aeroplankton and the need for a global monitoring network. Bioscience 63, 515–516. doi: 10.1525/bio.2013.63.7.3
Smith, D. J., Jaffe, D. A., Birmele, M. N., Griffin, D. W., Schuerger, A. C., Hee, J., et al. (2012). Free tropospheric transport of microorganisms from asia to north america. Microb. Ecol. 64, 973–985. doi: 10.1007/s00248-012-0088-9
Smith, D. J., Ravichandar, J. D., Jain, S., Griffin, D. W., Yu, H., Tan, Q., et al. (2018). Airborne Bacteria in Earth’s lower stratosphere resemble taxa detected in the troposphere: results from a new NASA Aircraft Bioaerosol Collector (ABC). Front. Microbiol. 9:1752. doi: 10.3389/fmicb.2018.01752
Smith, D. J., Timonen, H. J., Jaffe, D. A., Griffin, D. W., Birmele, M. N., Perry, K. D., et al. (2013). Intercontinental dispersal of bacteria and archaea by transpacific winds. Appl. Environ. Microbiol. 79, 1134–1139. doi: 10.1128/AEM.03029-12
Sprovieri, F., Pirrone, N., Bencardino, M., D’amore, F., Carbone, F., Cinnirella, S., et al. (2016). Atmospheric mercury concentrations observed at ground-based monitoring sites globally distributed in the framework of the GMOS network. Atmos. Chem. Phys. 16, 11915–11935. doi: 10.5194/acp-16-11915-2016
Vaitilingom, M., Attard, E., Gaiani, N., Sancelme, M., Deguillaume, L., Flossmann, A. I., et al. (2012). Long-term features of cloud microbiology at the puy de Dome (France). Atmos. Environ. 56, 88–100. doi: 10.1016/j.atmosenv.2012.03.072
Vaïtilingom, M., Deguillaume, L., Vinatier, V., Sancelme, M., Amato, P., Chaumerliac, N., et al. (2013). Potential impact of microbial activity on the oxidant capacity and organic carbon budget in clouds. Proc. Natl. Acad. Sci.U.S.A. 110, 559–564. doi: 10.1073/pnas.1205743110
Vandeventer, P. E., Mejia, J., Nadim, A., Johal, M. S., and Niemz, A. (2013). DNA adsorption to and elution from silica surfaces: influence of amino acid buffers. J. Phys. Chem. 117, 10742–10749. doi: 10.1021/jp405753m
Wainwright, M., Wickramasinghe, N. C., Narlikar, J. V., and Rajaratnam, P. (2006). Microorganisms cultured from stratospheric air samples obtained at 41 km. FEMS Microbiol. Lett. 218, 161–165. doi: 10.1111/j.1574-6968.2003.tb11513.x
Waked, A., Favez, O., Alleman, L. Y., Piot, C., Petit, J. E., Delaunay, T., et al. (2014). Source apportionment of PM10 in a north-western Europe regional urban background site (Lens, France) using positive matrix factorization and including primary biogenic emissions. Atmos. Chem. Phys. 14, 3325–3346. doi: 10.5194/acp-14-3325-2014
Yin, X., Kang, S., De Foy, B., Cong, Z., Luo, J., Zhang, L., et al. (2017). Surface ozone at Nam Co in the inland Tibetan Plateau: variation, synthesis comparison and regional representativeness. Atmos. Chem. Phys. 17, 11293–11311. doi: 10.5194/acp-17-11293-2017
Keywords: atmosphere, microorganisms biodiversity, aerobiology, biogeography, protocols, methods, aerosols
Citation: Dommergue A, Amato P, Tignat-Perrier R, Magand O, Thollot A, Joly M, Bouvier L, Sellegri K, Vogel T, Sonke JE, Jaffrezo J-L, Andrade M, Moreno I, Labuschagne C, Martin L, Zhang Q and Larose C (2019) Methods to Investigate the Global Atmospheric Microbiome. Front. Microbiol. 10:243. doi: 10.3389/fmicb.2019.00243
Received: 12 July 2018; Accepted: 29 January 2019;
Published: 21 February 2019.
Edited by:
Thulani Peter Makhalanyane, University of Pretoria, South AfricaReviewed by:
David Anthony Pearce, Northumbria University, United KingdomDavid G. Schmale, Virginia Tech, United States
Copyright © 2019 Dommergue, Amato, Tignat-Perrier, Magand, Thollot, Joly, Bouvier, Sellegri, Vogel, Sonke, Jaffrezo, Andrade, Moreno, Labuschagne, Martin, Zhang and Larose. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catherine Larose, Q2F0aGVyaW5lLkxhcm9zZUBlYy1seW9uLmZy
 Aurelien Dommergue
Aurelien Dommergue Pierre Amato
Pierre Amato Romie Tignat-Perrier1,3
Romie Tignat-Perrier1,3 Olivier Magand
Olivier Magand Muriel Joly
Muriel Joly Timothy Vogel
Timothy Vogel Jeroen E. Sonke
Jeroen E. Sonke Isabel Moreno
Isabel Moreno Casper Labuschagne
Casper Labuschagne Lynwill Martin
Lynwill Martin Catherine Larose
Catherine Larose