
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 30 January 2019
Sec. Fungi and Their Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00099
This article is part of the Research TopicSensing and Signaling of the Environment by Fungi in Health and Disease View all 20 articles
Candida species, such as Candida albicans and Candida glabrata, cause infections at different host sites because they adapt their metabolism depending on the available nutrients. They are able to proliferate under both nutrient-rich and nutrient-poor conditions. This adaptation is what makes these fungi successful pathogens. For both species, sugars are very important nutrients and as the sugar level differs depending on the host niche, different sugar sensing systems must be present. Saccharomyces cerevisiae has been used as a model for the identification of these sugar sensing systems. One of the main carbon sources for yeast is glucose, for which three different pathways have been described. First, two transporter-like proteins, ScSnf3 and ScRgt2, sense glucose levels resulting in the induction of different hexose transporter genes. This situation is comparable in C. albicans and C. glabrata, where sensing of glucose by CaHgt4 and CgSnf3, respectively, also results in hexose transporter gene induction. The second glucose sensing mechanism in S. cerevisiae is via the G-protein coupled receptor ScGpr1, which causes the activation of the cAMP/PKA pathway, resulting in rapid adaptation to the presence of glucose. The main components of this glucose sensing system are also conserved in C. albicans and C. glabrata. However, it seems that the ligand(s) for CaGpr1 are not sugars but lactate and methionine. In C. glabrata, this pathway has not yet been investigated. Finally, the glucose repression pathway ensures repression of respiration and repression of the use of alternative carbon sources. This pathway is not well characterized in Candida species. It is important to note that, apart from glucose, other sugars and sugar-analogs, such as N-acetylglucosamine in the case of C. albicans, are also important carbon sources. In these fungal pathogens, sensing sugars is important for a number of virulence attributes, including adhesion, oxidative stress resistance, biofilm formation, morphogenesis, invasion, and antifungal drug tolerance. In this review, the sugar sensing and signaling mechanisms in these Candida species are compared to S. cerevisiae.
Over the last decades, the incidence of fungal infections has tremendously increased (Arendrup, 2013; Enoch et al., 2017). For every individual, there is a reasonable chance to suffer from a superficial fungal infection at least once during their lifetime. Due to organ transplants, catheter use and the redundant and prophylactic use of antibiotics, there is a higher number of at-risk people, which causes an increase in both superficial and systemic infection rate of these pathogenic fungi (Pfaller and Diekema, 2007; Arendrup, 2010; Yapar, 2014). From the pool of fungal infections, the most frequently isolated species are Candida sp. of which Candida albicans is the most prominent one (Guinea, 2014; Goemaere et al., 2018). Furthermore, the introduction of the antifungal fluconazole caused an increase in infections initiated by the inherently more fluconazole tolerant species, Candida glabrata (Pfaller et al., 2005; Diekema et al., 2012; Pfaller et al., 2014). C. glabrata is often found in the environment (Greppi et al., 2015; Morrison-Whittle et al., 2018), while C. albicans is only rarely found outside the mammalian host. Recently some studies showed its presence in chickens and turkeys and on old oaks (Bensasson et al., 2018; Liu et al., 2018; Sokół et al., 2018). The differences in lifestyle between C. albicans and C. glabrata are due to their different genomic evolution. C. albicans diverged from S. cerevisiae about 300 million years ago (Hedges et al., 2015). Therefore, these species are fairly different from each other with as most obvious example the different usage of the CUG codon (Santos and Tuite, 1995). It is estimated that about 100–150 million years ago, S. cerevisiae underwent a whole genome duplication (WGD) (Sugino and Innan, 2005). It is after this that C. glabrata has diverged from S. cerevisiae as this pathogen shows remnants of the WGD (Herrero, 2005). Therefore, C. glabrata and S. cerevisiae are phylogenetically much closer related to each other than to C. albicans. This difference in lifestyles also has an influence on their nutrient sensing mechanisms. C. glabrata is able to sense the broad range of nutrient availability which varies tremendously between host and environment. In the human host, the fungal cells proliferate under both nutrient-rich and nutrient-poor conditions; however, the range of nutrient concentrations is smaller (Figure 1). The adaptation to different environments is the key to successful survival of these pathogens (Brock, 2009; Fleck et al., 2011).
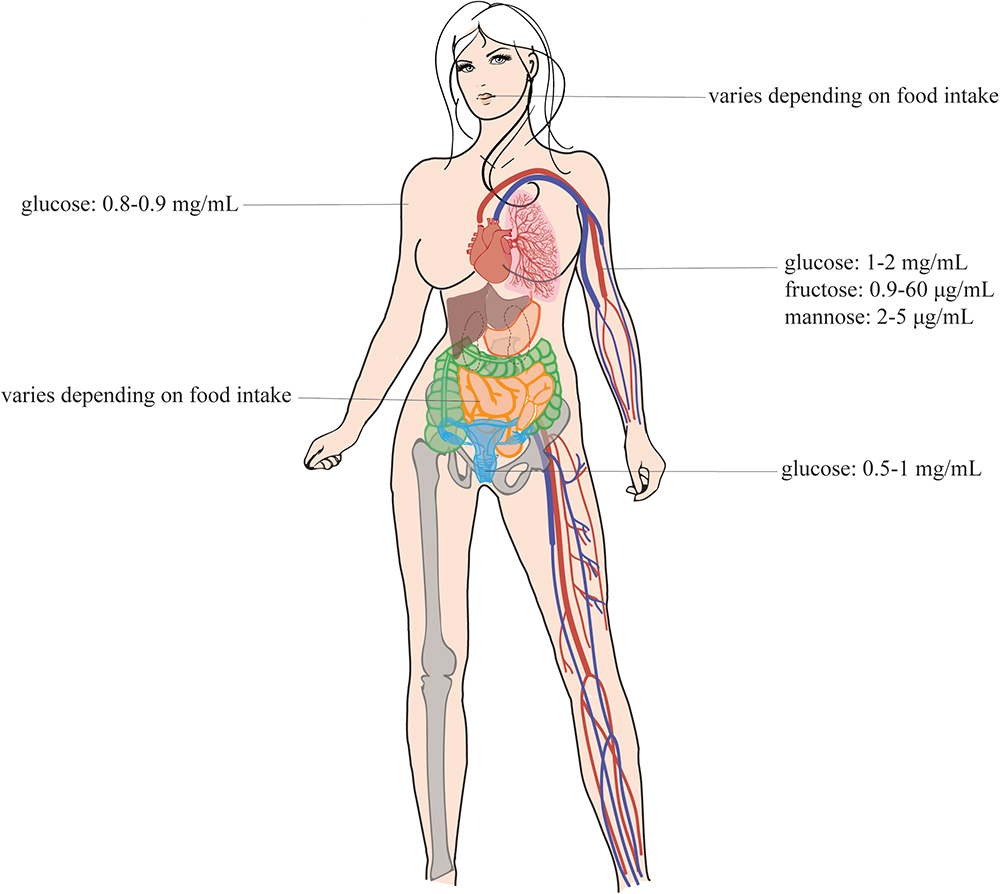
Figure 1. Sugar concentrations in the human body. This figure shows the sugar concentrations at the different infection sites of the Candida species. These pathogens are present at different sites of the human host, namely, in the mouth, in the gut, in the vagina, in the blood, and on the skin (Sone et al., 2003; Ehrstrom et al., 2006; Groenendaal et al., 2010; Laughlin, 2014). This figure is adapted from the review of Gulati and Nobile (2016). Permission was given to reuse this figure.
To elucidate the nutrient-induced signal transduction pathways in human fungal pathogens, the model yeast species S. cerevisiae has been used extensively. Three different glucose-sensing pathways have been described in S. cerevisiae. First, glucose binds to either the high affinity sensor ScSnf3 or the low affinity sensor ScRgt2, which results in the induction of hexose transporter (HXT) genes (Ozcan et al., 1996). Second, glucose also binds to the G-protein coupled receptor (GPCR) ScGpr1 resulting in the activation of its G-protein ScGpa2 and subsequent activation of adenylate cyclase (ScCyr1). This leads to an increase in cAMP levels and the activation of protein kinase A (PKA; Rolland et al., 2000; Thevelein et al., 2000). Full activation of this pathway also requires sugar phosphate-induced activation of ScCdc25 and ScRas (Toda et al., 1985; Rolland et al., 2000; Thevelein et al., 2000; Peeters et al., 2017). Once inside the cells, glucose causes repression of genes responsible for gluconeogenesis and respiration via the glucose repression pathway (Kayikci and Nielsen, 2015). This last one also has a negative effect on the expression of genes involved in the use of alternative carbon sources. For example, the GAL genes are repressed by the transcriptional inhibitor ScMig1, which is activated by high levels of glucose. This hinders the use of galactose as a carbon source when glucose is present in the medium. Candida species show many similarities with these pathways found in S. cerevisiae suggesting a same ancestor. However, also many differences in sensing and signaling have been described. This is to be expected, as Candida species have the ability to turn virulent whereas baker’s yeast generally does not. For example, glucose functions as a morphogen to influence yeast-to-hyphae transition in C. albicans (Maidan et al., 2005b). Additionally, glucose and other sugars have an influence on the ability of Candida cells to form a biofilm (Ng et al., 2016; Pemmaraju et al., 2016). This is a “city of microbes” where microorganisms live densely populated and surrounded by an excreted extracellular matrix as protection. Furthermore, sensing sugars is important for other virulence attributes, including adhesion, oxidative stress resistance, invasion, and antifungal drug tolerance (Vargas et al., 1993; Jin et al., 2004; Rodaki et al., 2009; Manns et al., 2014). In this review, we describe the different sugar sensing mechanisms in C. albicans and C. glabrata and compare them to the situation in S. cerevisiae. This is shown in Figure 2 for S. cerevisiae and in Figures 3, 4 for C. albicans and C. glabrata, respectively.
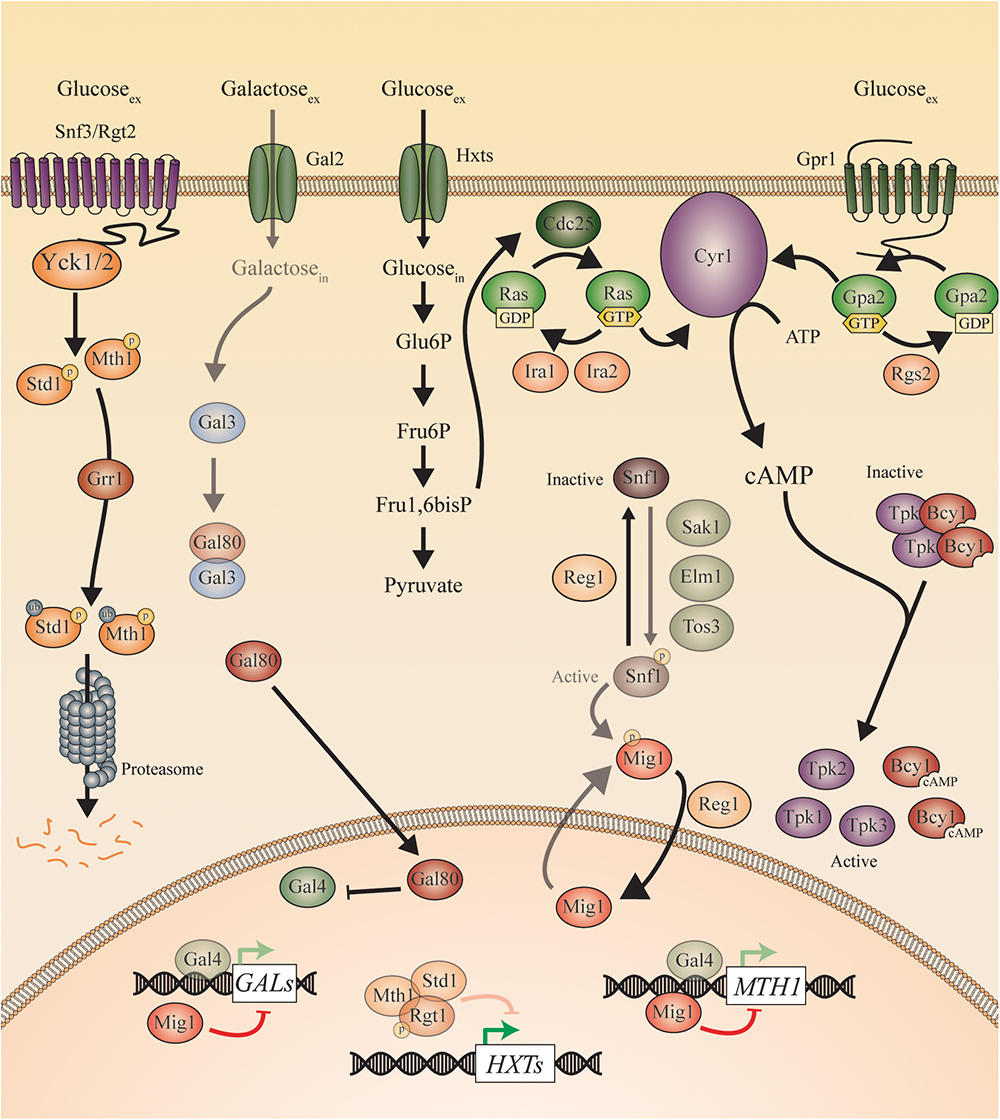
Figure 2. Sugar sensing and signaling in S. cerevisiae. The sugar-sensing network in S. cerevisiae. The faded arrows and proteins represent the situation when sugars are absent. All proteins are represented as circles. Activation is represented with green arrows, while red arrows indicate inhibition. When the ScSnf3/ScRgt2 receptors sense glucose, ScYck1 phosphorylates ScStd1 and ScMth1. These are subsequently ubiquitinated by ScGrr1 and broken down by the proteasome. In the absence of glucose, they function together with ScRgt1 to repress transcription of the ScHXT genes. The ScHxt transporters allow glucose to enter the cells after which this monosaccharide is converted to pyruvate during glycolysis. One of the intermediates of this pathway is fructose 1,6 bisphosphate which is found to activate ScCdc25. In this way, ScCdc25 allows the exchange of GDP to GTP on the ScRas proteins. ScRas is now activated and stimulates ScCyr1. Next to activation by ScRas, ScCyr1 needs the stimulus coming from ScGpr1 to be fully activated. ScGpr1 is activated by sensing extracellular glucose which allows the exchange of GDP to GTP on the ScGpa2 proteins. ScGpa2-GTP activates ScCyr1. ATP is converted into cAMP after complete activation of ScCyr1. cAMP binds to the regulatory subunits of PKA (Bcy1) thereby causing them to release the catalytic subunits (ScTpk1, 2, and 3) causing PKA activation. The absence of glucose triggers the phosphorylation of ScSnf1 by ScSak1, ScElm1, and ScTos3. The activated ScSnf1 protein phosphorylates ScMig1, preventing this protein to enter the nucleus. In the presence of glucose, both ScSnf1 and ScMig1 are dephosphorylated by ScReg1, allowing ScMig1 to enter the nucleus and function as a transcriptional repressor of the ScGAL genes and ScMTH1. When galactose is present (in the absence of glucose), it can enter the cells via the ScGal2 permease. Galactose can bind to ScGal3, which sequesters ScGal80 and prevents the latter to bind ScGal4. ScGal4 can thereby actively function in the nucleus as a transcription factor, inducing the expression of the ScGAL genes and of ScMTH1.
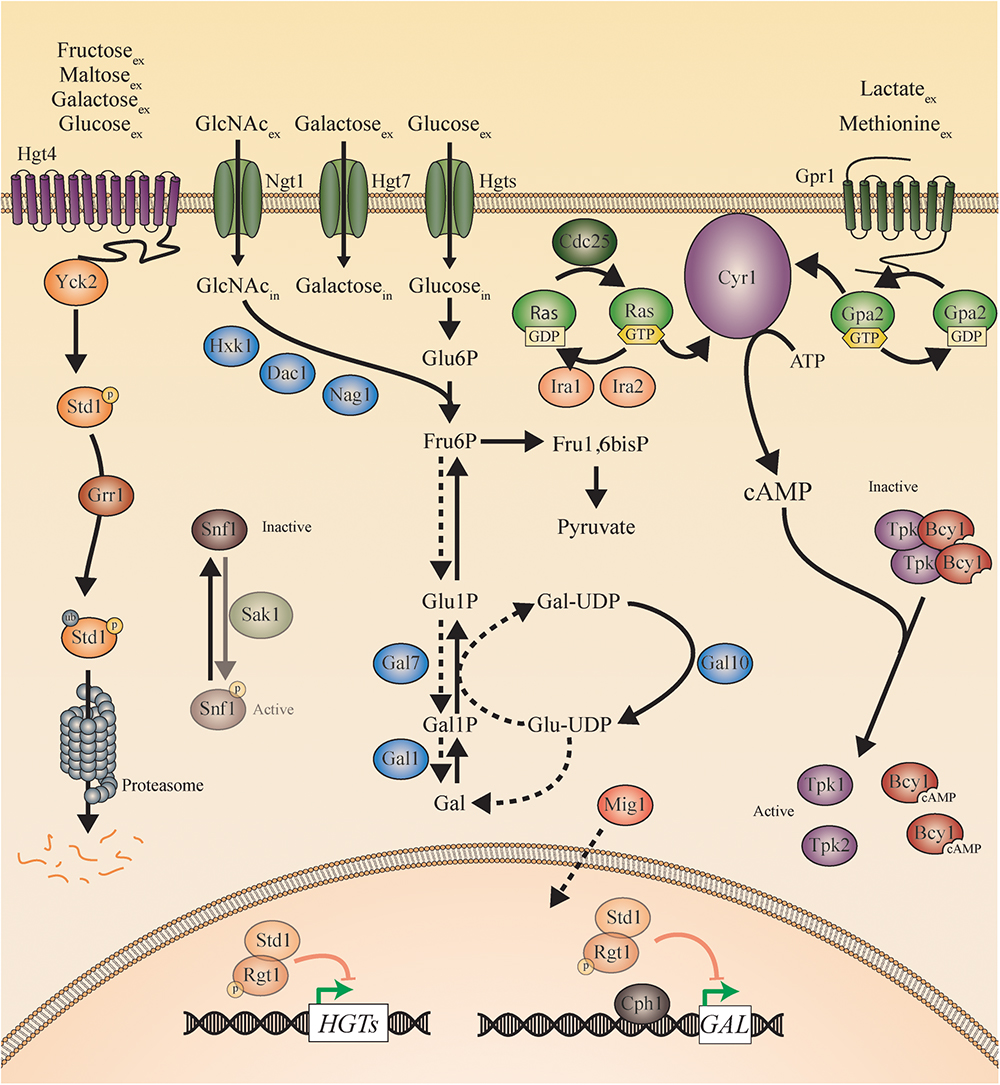
Figure 3. Sugar sensing and signaling in C. albicans. The sugar-sensing network in C. albicans. The faded arrows and proteins represent the situation when sugars are absent. All proteins are represented as circles. Activation is represented with green arrows, while red arrows indicate inhibition. The dashed lines represent the hypothesis. When the CaHgt4 receptor senses glucose, galactose, maltose, or fructose, CaYck2 phosphorylates CaStd1. This is subsequently ubiquitinated by CaGrr1 and broken down by the proteasome. In the absence of a carbon source, CaStd1 functions together with CaRgt1 to repress transcription of the CaHGT genes. The CaHgt transporters allow sugars to enter the cells. When glucose enters the cells, it is converted to pyruvate during glycolysis. In the presence of extracellular glucose, an active CaCdc25 allows the exchange of GDP to GTP on the CaRas proteins. CaRas is now activated and stimulates CaCyr1. Next to activation by CaRas, CaCyr1 needs the stimulus coming from CaGpr1 to be fully activated. CaGpr1 is activated by sensing extracellular molecules, presumably methionine or lactate, which allows the exchange of GDP to GTP on the CaGpa2 proteins. CaGpa2-GTP activates CaCyr1. ATP is converted into cAMP after complete activation of CaCyr1 by CaRas and by CaGpa2. cAMP binds to the regulatory subunits of PKA (CaBcy1) thereby causing them to release the catalytic subunits (CaTpk) causing PKA activation. When N-acetylglucosamine enters the cells via CaNgt1, it can be metabolized to fructose-6-phosphate by CaHxk1, CaDac1, and CaNag1. The hypothesis is that fructose-6-phosphate can be converted into galactose by the action of the CaGal enzymes. The galactose metabolism in C. albicans remains a mystery, but the expression of the CaGAL genes is found to be regulated by CaCph1 and CaRgt1. CaSnf1 can be phosphorylated by CaSak1 but no more in this pathway has been revealed.
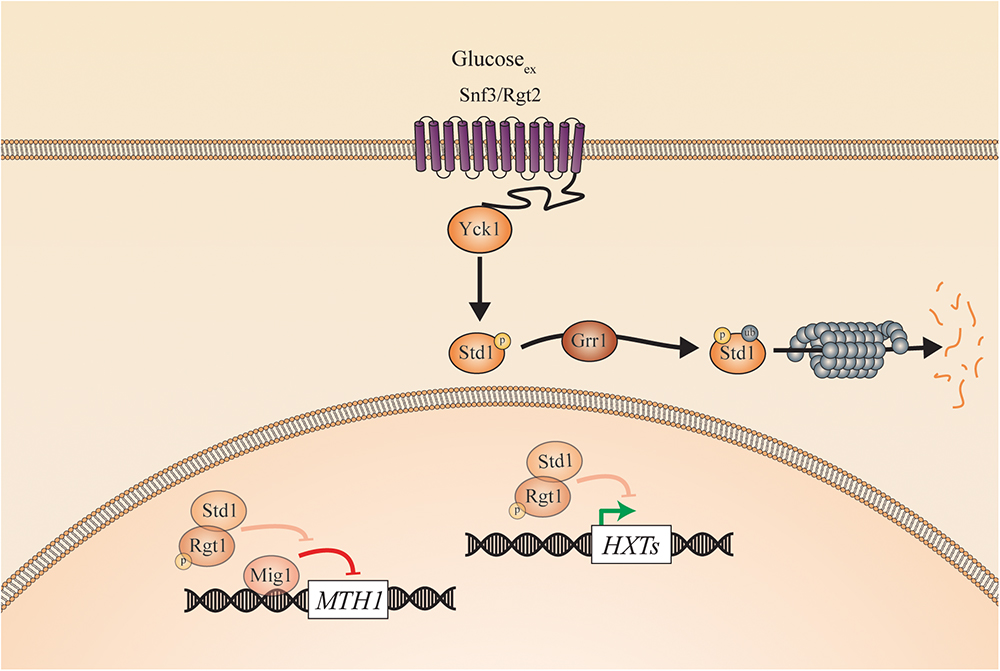
Figure 4. Sugar sensing and signaling in C. glabrata. The sugar-sensing network in C. glabrata. The faded arrows and proteins represent the situation when sugars are absent. All proteins are represented as circles. Activation is represented with green arrows, while red arrows indicate inhibition. When the CgSnf3/CgRgt2 receptors sense glucose CgYck1 phosphorylates CgStd1. This is subsequently ubiquitinated by CgGrr1 and broken down by the proteasome. In the absence of glucose, CgStd1 functions together with CgRgt1 to repress transcription of the CgHXT genes. The CgHxt transporters allow glucose to enter the cells.
As glucose is the most abundant hexose sugar on earth, it is the preferred carbon source for many species. Due to its central role in metabolism, glucose functions both as a ligand for receptors and as a substrate for transporters (Towle, 2005). As a result, glucose serves as starting material for the biosynthesis of other biomolecules as well as for energy production via fermentation or respiration (Verduyn, 1991; Rodrigues et al., 2006).
Saccharomyces cerevisiae encodes 17 hexose transporters which all have different affinity and kinetic properties in response to glucose (Ozcan and Johnston, 1995). Furthermore, they all consist out of 12 transmembrane domains (TMDs; Ozcan and Johnston, 1999). Two of these proteins, ScSnf3 and ScRgt2, are not able to transport glucose, but sense this sugar. Therefore, they have an additional long C-terminal extension compared to the sugar transporters (Ozcan and Johnston, 1999). ScSnf3 is the high-affinity sensor while ScRgt2 responds to higher levels of glucose (Ozcan et al., 1996; Boles, 2003). In the presence of glucose, the transcription factors, ScStd1 and ScMth1, are recruited to the cytoplasmic tail of ScSnf3 or ScRgt2 where they are phosphorylated by the yeast casein kinase ScYck1 (Schmidt et al., 1999; Lafuente et al., 2000). The phosphorylated ScMth1 and ScStd1 are subsequently ubiquitinated by ScGrr1 leading to degradation by the proteasome (Moriya and Johnston, 2004). This degradation prevents ScRgt1-ScMth1/ScStd1 complex formation and relieves the repression of the glucose transporter and metabolism genes. In the absence of glucose, ScMth1 and ScStd1 interact with ScRgt1 to repress genes involved in the glucose metabolism (e.g., HXTs) (see Figure 2). Interestingly, the Snf3/Rgt2 glucose sensors and the Rgt1-Mth1/Std1 transcriptional repression complex seem to be fungal specific (Todd and Andrianopoulos, 1997; Ozcan et al., 1998).
Even though glucose is of major importance for C. glabrata, the sugar receptor-repressor (SRR) pathway has not yet been investigated in detail in this species (Ng et al., 2016). Only 11 Hxts are identified in C. glabrata compared to 17 in S. cerevisiae and 20 in C. albicans. It is, however, possible that clinical C. glabrata isolates express more hexose transporters than the CBS138 laboratory strain because they have evolved within the human host. This hypothesis is based on recent findings regarding the emergence of antifungal resistance genes and the number of adhesins present in C. glabrata clinical isolates (Vale-Silva et al., 2017; Salazar et al., 2018). The glucose sensor CgSnf3 was first characterized by the group of Than as the most important and high affinity sensor of environmental glucose concentrations (Ng et al., 2015). CgSnf3 and CgRgt2 are 59 and 60% similar to their orthologs in S. cerevisiae, respectively. These putative glucose sensors also consist out of 12 TMDs and have a long cytoplasmic tail. In comparison to the situation in S. cerevisiae, CgSnf3 is important for growth on low physiological glucose levels (0.01–0.1%). The sensor is less important for growth on high glucose levels (1–2%) as it is probably not expressed under these conditions. Deletion of CgSNF3 results in downregulation of four out of nine hexose transporter genes (Ng et al., 2015). Furthermore, the disruption of CgSNF3 leads to the downregulation of CgYCK1, CgYCK2, and CgSTD1. This suggests that they work downstream of this high affinity glucose sensor. A Cgsnf3Δ mutant is also more susceptible to phagocytosis. This indicates that this receptor is important in host niches with low glucose levels, like inside macrophages (Ng et al., 2015). Whether this is due to the fact that some high affinity glucose transporters or other downstream genes are not expressed remains to be investigated. However, the Cgsnf3Δ mutant has no defect in the formation of biofilms. This suggests that other sensors must be necessary to sense the nutrient flow in this environment or that the glucose level is still sufficient (Ng et al., 2015). CgRGT2 is responsive to high surrounding glucose concentrations, indicating its low affinity, similar to ScRgt2 (see Figure 4). These findings suggest a conserved glucose signaling pathway between S. cerevisiae and C. glabrata. From the little knowledge there is, it is speculated that this pathway plays an important role in the virulence of this pathogenic fungus.
In C. albicans, the glucose transporter gene family was first studied by the group of Shen (Fan et al., 2002). The 12 TMDs of S. cerevisiae are conserved in C. albicans. Twenty high affinity glucose transporters are identified in the latter of which CaHGT9, CaHXT10, CaHGT12, and CaHGT17 are strongly expressed upon low glucose levels (0.2%) and repressed at 5% glucose. The other CaHGT genes are expressed in YPD medium (2% glucose). These transporter-like proteins are divided into two groups by performing a bootstrap neighbor-joining analysis: CaHXT (CaHGT6, CaHGT7, CaHGT8, and CaHGT11) and CaSNF3 (CaHGT4 and CaHGT12) (Fan et al., 2002). Even though it seems to be clear that CaHgt4 is a sugar sensor, similar to Snf3 in S. cerevisiae and C. glabrata, it is not yet clear whether CaHgt12 is a sensor and/or transporter (Fan et al., 2002; Brown et al., 2006). CaHgt4 has 56% identical and 73% similar residues compared to its orthologs ScSnf3/ScRgt2. Also, CaHgt12 shows high similarity to these latter ones (48% identical and 68% similar residues) but lacks the C-terminal extension which is a hallmark for the sensors (Helmerhorst et al., 2002). In order to define the protein responsible for sugar sensing in C. albicans, both CaHGT4 and CaHGT12 were disrupted. The Cahgt12Δ mutant shows no growth defect on sugar media containing glucose, fructose, or mannose. On the other hand, the Cahgt4Δ mutant shows a severe growth defect on fructose containing media and has problems growing on low concentrations of glucose and mannose (<0.2%) (Brown et al., 2006). The glucose concentrations which induce expression of CaHGT4 are comparable with glucose concentrations present in human serum. Furthermore, the Cahgt4Δ mutant is also affected in its growth on glucose containing media in the presence of Antimycin A, a compound that inhibits respiration (Helmerhorst et al., 2002). These data suggest that CaHgt4 is a protein essential for growth on low levels of fermentable carbon sources and is important during hypoxic conditions (Brown et al., 2006). Upon sensing glucose, CaHgt4 signals downstream toward the transcription of several CaHGTs. In the Cahgt4Δ mutant, CaHGT12, CaHGT7, and CaHXT10 are 10 times less expressed compared to their expression level in the CaHgt4 constitutively signaling strain (Brown et al., 2006). The promoters of these genes all contain six CaRgt1 binding sites causing constitutive repression when CaHGT4 is deleted. The Cargt1Δ mutant shows a fivefold increase in expression of these hexose transporter genes (Sexton et al., 2007). In the Cahgt4Δ Cargt1Δ double mutant, the expression of CaHGT12, CaHXT10 and CaHGT7 is restored. This shows, similar to the situation in S. cerevisiae, that CaRgt1 is operating downstream of CaHgt4, although it has only limited sequence similarity (Brown et al., 2006). Most of the similar residues are found in the zinc-cluster DNA binding domain in the N-terminus of the protein (Sexton et al., 2007). As low levels of glucose are known to induce morphogenesis, it is expected that disrupting CaHGT4 affects this virulence trait (Hudson et al., 2004). Indeed, the Cahgt4Δ mutant has a defect in filamentation, whereas the constitutively signaling form of CaHgt4 results in hyper filamentation (Brown et al., 2006). Furthermore, mice infected with the Cahgt4Δ mutant survive longer than mice infected with the wild type C. albicans strain. This shows the importance of this glucose sensor for virulence. As CaRgt1 signals downstream of CaHgt4, it is likely that repression of the hexose transporter genes by CaRgt1 also has an effect on filamentation (Maidan et al., 2005a). Indeed, the double Cahgt4Δ Cargt1Δ mutant shows hyper filamentation, similar to the phenotype observed in the strain containing the constitutively signaling form of Hgt4 (Brown et al., 2006; Sexton et al., 2007). Regarding the ScMth1 and ScStd1 proteins, only one ortholog is present in C. albicans: CaSTD1, which is regulated in a similar way as in S. cerevisiae (phosphorylation-dependent proteolytic degradation). This suggests that ScMTH1 occurred in S. cerevisiae by WGD after the split of the CUG clade and the Saccharomyces lineages (Brown et al., 2009).
Comparing the Cgsnf3Δ mutant with the C. albicans Cahgt4Δ mutant, the effect for growth on low glucose concentrations is different. The CaHgt4 sensor is important for fermentative growth while CgSnf3 is essential under both respiratory and fermentative conditions (Brown et al., 2006; Ng et al., 2015). Therefore, the group of Than suggests that this high affinity sensor is more essential in C. glabrata than in C. albicans. This can be due to the fact that C. glabrata is Crabtree positive and prefers fermentation over respiration and produces ethanol in the presence of oxygen (Van Urk et al., 1990). As fermentation is the less favorable way to provide the cells with energy (fermentation: 2 ATP/glucose; respiration: 38 ATP/glucose), it is obvious that C. glabrata needs two specific glucose sensors to increase the glucose uptake. Thereby, it is able to differentiate the expression of sugar transporters and fulfill its ATP demand (Ng et al., 2015). Also, the differences in lifestyles between C. glabrata and C. albicans are a possible explanation for their different glucose sensors. C. albicans may have more co-evolved with the human host and as no high levels of glucose are present inside the human body, the low-affinity glucose sensor seems unnecessary. C. glabrata is more similar to S. cerevisiae, as these species are phylogenetically more closely related and they experience high and low glucose levels in nature. However, since both C. glabrata and C. albicans occur as human fungal pathogens, it is expected that more similarities in their sugar sensing mechanisms will be found. To elucidate this, further research on the SRR pathway in C. glabrata is necessary.
A second mechanism by which S. cerevisiae is able to sense glucose and subsequently activate PKA is via the GPCR system, consisting of ScGpr1 and ScGpa2. The ScGpr1 receptor is able to sense both sucrose and glucose. Its affinity for glucose is very low, namely, a Ka of 75 mM (Rolland et al., 2000). This is related to the function of this GPCR in stimulating the switch between respirative/gluconeogenic growth and fermentative growth which only happens at glucose concentrations above 20 mM (Thevelein, 1991; Rolland et al., 2000). Other fermentable sugars like fructose, mannose, and galactose do not activate ScGpr1. However, mannose has an antagonistic effect on this sensor. Glucose, sucrose, and mannose all have very similar structures, so probably the position of the hydroxyl group on the second carbon atom is crucial for the activation of ScGpr1 (Lemaire et al., 2004). When ScGpr1 senses glucose or sucrose, it is able to activate ScGpa2. In its GTP bound state, ScGpa2 is able to activate ScCyr1, which is peripherally bound to the plasma membrane and converts ATP into cAMP (Uno et al., 1985; Nakafuku et al., 1988; Mitts et al., 1990; Kraakman et al., 1999). Increased cAMP levels subsequently activate PKA by binding to the regulatory subunit ScBcy1 releasing the catalytic subunits ScTpk1, 2, and 3. However, full activation of this pathway by glucose also requires glucose transport and its phosphorylation by one of the hexose kinases (Rolland et al., 2000; Thevelein et al., 2000). The molecular mechanism of this activation remained unclear until it was recently shown that the glycolytic intermediate, fructose-1,6-bisphosphate (FbP), is able to bind ScCdc25 thereby activating Ras (Peeters et al., 2017). The level of FbP is known to be controlled by multiple pathways indicating that this metabolite is important for the response of the cells to different levels of glucose (Hers, 1984). Activated GTP-bound ScRas binds to ScCyr1, leading to the activation of PKA (Toda et al., 1985; Jacquet, 1997). Both pathways upstream of ScCyr1 are required for full activation of PKA (Figure 2).
In the C. glabrata genome, orthologs of ScGpr1 and ScGpa2 are present, but so far, they are not characterized. Recently, a FRET system has been developed to measure PKA activity as well as cAMP levels in C. glabrata cells. Here, it is shown, that after addition of glucose to derepressed cells, both the intracellular cAMP level increases and PKA is activated. These findings suggest that, similar to the situation in S. cerevisiae, glucose stimulates the activation of PKA via cAMP (Demuyser et al., 2018).
The GPCR system of ScGpr1 and ScGpa2 is also present in C. albicans, where CaGpr1 has 43% sequence identity with its S. cerevisiae ortholog. Furthermore, the main structures like the seven transmembrane regions and the second half of the third cytosolic loop are conserved (Miwa et al., 2004). However, it seems that in this species the CaGpr1 receptor does not sense sugars as the receptor is dispensable for the glucose-induced increase in cAMP levels. CaGpr1 is clearly not required for the glucose-induced cAMP increase. This increase depends on the CaCdc25 and CaRas2 protein (Maidan et al., 2005a). However, the GPCR system seems to be upstream of the PKA pathway as addition of exogenous cAMP restores the hyphal defect of the Cagpr1Δ mutant on solid hyphae-inducing media. This morphogenesis defect is also restored by expression of a constitutively active CaGPA2 or by overexpression of CaTPK1, a catalytic subunit of PKA, indicating that CaGpr1 and CaGpa2 are upstream of the PKA pathway (Figure 3). This is further supported by the fact that the Cagpr1Δ mutant and the Cagpa2Δ mutant show the same morphogenesis phenotype as the Cagpr1Δ Cagpa2Δ double mutant (Maidan et al., 2005a). This raises the question about which ligand is able to activate CaGpr1. The lab of Van Dijck showed that methionine might be the ligand of CaGpr1 as this receptor is required for the methionine-induced and PKA mediated morphogenesis (Maidan et al., 2005a; Schrevens et al., 2018). Furthermore, it was shown that CaGpr1 internalizes in the presence of methionine, indicating ligand-induced internalization (Maidan et al., 2005a). Finally, physiological concentrations of methionine provide the strongest filamentation phenotype (Maidan et al., 2005b). However, more recently, it was shown that lactate may be the ligand of CaGpr1 and causes the internalization of this receptor (Ballou et al., 2016). More detailed receptor–ligand interactions need to be performed to elucidate the ligands of this receptor in C. albicans. To conclude, the exact mechanism by which glucose, methionine, and lactate integrate at the level of the Ras-cAMP/PKA pathway and how glycolysis may link with the cAMP/PKA pathway in regulating virulence remains to be investigated. Nevertheless, it was previously shown that glycolysis is upregulated during infections and that it plays an important role during pathogenicity of the fungus (Barelle et al., 2006). Since PKA controls different virulence factors, it is possible that the effect of glycolysis on the virulence factors is signaled via the cAMP/PKA pathway.
A third important pathway in glucose sensing is the glucose repression pathway. This pathway has a negative effect on the use of alternative carbon sources and respiration (Gancedo, 1998; Carlson, 1999; Hedbacker and Carlson, 2008). S. cerevisiae has a very efficient glucose repression system. When glucose is present, the utilization of alternative carbon sources is completely blocked (Gancedo, 1992). For example, the gene expression of enzymes working in the gluconeogenic pathway is repressed (Mercado et al., 1991; Mercado and Gancedo, 1992). In addition, also the degradation of mRNAs of those genes accelerates (Mercado et al., 1994; Yin et al., 1996). In this way, the only used carbon source is glucose. An important player in this pathway is ScSnf1. This is a serine threonine protein kinase and the yeast ortholog of the mammalian AMPK protein. Upon glucose depletion, this protein is activated due to phosphorylation by three upstream kinases (ScSak1, ScTos3, and ScElm1) (Ludin et al., 1998). Phosphorylated ScSnf1 has an indirect effect on the use of alternative carbon sources, by phosphorylating ScMig1, whereby the latter can no longer translocate to the nucleus. This causes the derepression of genes involved in the utilization of other sugars. In the presence of glucose, the dephosphorylation and subsequent inactivation of ScSnf1 is stimulated by ScReg1 (Ludin et al., 1998). At the same time, ScMig1 can be dephosphorylated by ScReg1 and migrates from the cytoplasm to the nucleus causing repression of certain genes (Treitel and Carlson, 1995; Hardie et al., 1998; Cottier et al., 2017; Shashkova et al., 2017; Figure 2). ScMig1 has, for example, an inhibitory effect on ScSUC2, ScGAL1, ScGAL4 and on the gluconeogenic genes (Nehlin and Ronne, 1990; Griggs and Johnston, 1991; DeVit and Johnston, 1999). ScSnf1 and ScMig1 also have an effect on the expression of the high affinity glucose sensor ScSnf3 and the high affinity transporters ScHxt2 and ScHxt4 (Ozcan and Johnston, 1999; Kaniak et al., 2004). There is no expression of these two transporters in a Scsnf1Δ mutant due to the constant repression by ScMig1 (Ozcan and Johnston, 1995). This indicates that ScSnf1 is essential for the derepression of high affinity transport (Bisson, 1988).
Also C. glabrata has a ScSnf1 ortholog with high similarity to its orthologs in S. cerevisiae and C. albicans (Celenza and Carlson, 1986; Petter et al., 1997). The deletion of CgSNF1 causes the inability to use trehalose as a carbon source. A typical character trait for C. glabrata is that it solely uses glucose and trehalose as fermentable carbon sources. This finding indicates that also here, compared to the situation in S. cerevisiae, the use of alternative sugars is repressed in a CgSnf1-dependent manner (Petter and Kwon-Chung, 1996).
In contrast to S. cerevisiae, C. albicans is able to use alternative carbon sources in the presence of glucose. This is due to posttranscriptional rewiring during the evolution of this pathogen (Sandai et al., 2012). Glucose still causes the degradation of certain gene transcripts involved in the metabolism of these other carbon sources, but the protein levels stay at a constant level. For example, isocitrate lyase (Icl1), a protein working in the glyoxylate cycle, is not ubiquitinated in the presence of glucose. On the other hand, when ScICL1 is expressed in C. albicans, ScIcl1 is degraded upon glucose addition. This indicates that the ubiquitin-dependent catabolite inactivation machinery is still present in C. albicans. However, enzymes working in the gluconeogenic and glyoxylate pathway of this pathogen are missing this ubiquitination site present in their orthologs in S. cerevisiae. Therefore, they are not degraded when glucose is present (Sandai et al., 2012). From further research, it became clear that the absence of this ubiquitination site is important for the virulence of C. albicans. When such a site is added to CaIcl1, it is degraded in the presence of glucose and the cells have a reduced ability to colonize the gastrointestinal (GI) tract and to cause a systemic infection in mice. This indicates that the colonization and infection of the human host are enhanced by flexible carbon utilization (Childers et al., 2016). An important player in this glucose repression pathway is CaSnf1. This protein is essential for viability of this pathogen, which is not the case for baker’s yeast and C. glabrata (Petter and Kwon-Chung, 1996; Petter et al., 1997). CaSnf1 is highly upregulated during stress conditions and plays a role in drug resistance, indicating its possible importance for virulence (Orlova et al., 2008). CaSnf1 is activated via phosphorylation by CaSak1, an enzyme essential for metabolic adaptation and in vivo fitness. This indicates that the other two upstream kinases may not be present or active in C. albicans. Indeed, there is no ortholog of ScTos3 and an inactivating mutation of CaELM1 does not reduce CaSnf1 phosphorylation. This indicates that CaSak1 is most likely the only dedicated CaSnf1 activating kinase (Ramirez-Zavala et al., 2017; Figure 3). In contrast to the situation in S. cerevisiae, CaMig1 has no consensus sequence for phosphorylation by CaSnf1. This suggests that the activity of CaMig1 may be differently controlled than ScMig1 or that it has a different role in C. albicans. This is strengthened by the fact that CaMig1 is not differentially expressed on different carbon sources (Zaragoza et al., 2000). However, CaMig1 does have a differential effect on the expression of other genes in the presence of different carbon sources. This is shown by disruption of CaMig1, leading to a higher sensitivity to weak organic acids when grown on YPD compared to maltose containing media. This is probably due to the lack of repression of several transporter genes (Cottier et al., 2017). Nevertheless, CaMIG1 disruption is found to have no effect on the expression of the GAL regulon, which is later refuted by the group of Pavelka, who shows an increased CaGAL1 expression in the Camig1Δ mutant (Zaragoza et al., 2000; Cottier et al., 2017). In S. cerevisiae, ScMig1 recruits the ScTup1-ScCyc8 complex to repress glucose-sensitive promoters (Tzamarias and Struhl, 1995). Furthermore, ScTup1 also functions independently from ScMig1 (Tzamarias and Struhl, 1995). This seems to be similar in C. albicans. Here, CaMig1 functions both dependently and independently from CaTup1 (Murad et al., 2001). This is strengthened by the fact that the Camig1Δ mutant shows a different phenotype than the Catup1Δ mutant. A disruption in CaTUP1 causes constitutive filamentation whereas the Camig1Δ mutant shows normal morphogenesis (Tzamarias and Struhl, 1995; Zaragoza et al., 2000). CaMig1 probably also translocates to the nucleus as it has a KMPPK sequence, which is also present in the N-terminal region of ScHxk2. This region is involved in catabolite repression and targeting of the ScHxk2 protein to the nucleus (Herrero et al., 1998; Zaragoza et al., 2000).
As mentioned above, there are similarities as well as differences in the glucose sensing and signaling pathways between S. cerevisiae and Candida species. The SRR pathway seems to be very similar, while the glucose repression pathway seems to work differently. Also, for the activation of the PKA pathway, there are both similarities and differences observed. In C. albicans, it seems that other nutrients are also able to activate PKA using the same sensor. This is probably due to the fact that the Candida species are pathogenic fungi and need to use all the available carbon sources to become a successful pathogen. It is not a surprise to observe these differences between signal transduction pathways in S. cerevisiae and C. albicans as similar rewiring has been described for a number of pathways including for example the galactose metabolism circuitry and the regulation of stress response (Martchenko et al., 2007a; Brown et al., 2014).
Galactose metabolism is strongly regulated by the type of carbon source present in the environment. Most of the genes from this pathway are clustered in the ScGAL regulon, which is a genetic switch for the utilization of galactose in the absence of glucose (Lohr et al., 1995; Bhat and Murthy, 2001; Malakar and Venkatesh, 2014). S. cerevisiae encodes three regulatory ScGAL genes (ScGAL4, ScGAL80, ScGAL3) and five structural ScGAL genes (ScGAL1, ScGAL2, ScGAL7, ScGAL10, ScMEL1), all belonging to the Leloir pathway (Leloir, 1951; Holden et al., 2003). In the absence of galactose, the transcriptional activation domain of ScGal4 is bound by ScGal80, thereby preventing the induction of downstream genes. When galactose enters the cells via the ScGal2 permease, it results in the binding of ScGal3 with ScGal80 and thereby relieving the inhibitory activity of ScGal80 on ScGal4. This results in the induction of the galactose metabolism genes leading to proper galactose utilization (Laughon and Gesteland, 1982; West et al., 1984; Johnston, 1987; Lohr et al., 1995; Platt and Reece, 1998; Peng and Hopper, 2002). ScGal4 also induces expression of ScMTH1, encoding a co-repressor necessary for ScRgt1 function. In this way, galactose inhibits glucose assimilation by repressing the ScHXT genes (Ren et al., 2000). On the other hand, the ScGAL genes are also responsive to glucose repression via the ScMig1 repressor. However, as stated above, when glucose levels are low, ScMig1 is phosphorylated by ScSnf1 and leaves the nucleus to be broken down by the proteasome, relieving its repression of the ScGAL genes (Nehlin et al., 1991; Flick and Johnston, 1992; Figure 2).
The regulation of galactose metabolism in C. albicans seems to be rewired compared to the one seen in S. cerevisiae. Whereas most of the galactose structural genes (CaGAL1, CaGAL10, CaGAL7, CaGAL2) are conserved between C. albicans and S. cerevisiae, this is not the case for the galactose regulatory genes. No ScGAL3 or ScGAL80 orthologs seem to be present in the C. albicans genome. A strong conservation is observed for CaGal4, which shares 86% sequence identity with ScGal4. However, the activation domain and the negatively charged region, necessary for interaction with Gal80, is missing in C. albicans (Martchenko et al., 2007a,b). In contrast to the situation in S. cerevisiae, the disruption of CaGAL4 does not change the expression of the CaGAL genes. Surprisingly, the CaHgt4 sensor is required as a low affinity galactose sensor that begins to respond to galactose at 0.6% (Brown et al., 2006, 2009). Since both glucose and galactose are sensed by the same sensor, intracellular signaling must be strictly regulated. However, it is found for wild type C. albicans cells that 94% of the genes induced in response to galactose are also induced in response to low or high levels of glucose (Brown et al., 2009). This result may be explained by the fact that heterologous expression of CaHgt4 in S. cerevisiae results in galactose-induced expression of ScHXT genes. This indicates that CaHgt4 is able to couple to the downstream signal transduction pathway as the endogenous ScSnf3/ScRgt2 receptors are unable to sense galactose (Brown et al., 2009). The addition of galactose to C. albicans wild type cells induces expression of several genes listed in Supplementary Table S1. Addition of 2% galactose to glycerol grown C. albicans cells results in expression of the Leloir enzyme encoding genes (CaGAL1, CaGAL7, CaGAL10), genes involved in glucose metabolism (CaHGT7, CaHXK2, CaMTH1), and many others (CaQDR1, CaAOX2, CaCMK1) (Martchenko et al., 2007a; Brown et al., 2009). Most of the galactose-induced genes contain a CaRgt1-consensus site (5′-CGGANNA-3′) and show lower expression in a Cacph1Δ mutant, suggesting a coordinated regulation by CaRgt1 and CaCph1 (ortholog of Ste12 in S. cerevisiae) (Martchenko et al., 2007a). The expression of CaSTD1 is unaffected by galactose, but is by glucose. This implies that galactose sensing is different between C. albicans and S. cerevisiae while glucose sensing is similar in both species (CaHgt4-CaStd1-CaRgt1 or ScSnf3-ScStd1/ScMth1-ScRgt1). Galactose sensing in C. albicans is possibly due to a broad unspecific hexose sensor (CaHgt4) and an absent Gal4 pathway (Brown et al., 2009).
The galactose-sensing signal transduction pathway in C. albicans was one of the first pathways where a clear rewiring was observed at the transcriptional level when compared to the situation in S. cerevisiae. The difference in regulation of the Leloir genes in both fungi may be due to the fact that galactose plays an important role in certain virulence processes of C. albicans, namely, adhesion and biofilm formation, which are less important in S. cerevisiae (Jin et al., 2004; Martchenko et al., 2007a). In contrast to baker’s yeast, C. albicans uses glucose and galactose at the same time, which may be of importance when colonizing the GI tract of the human host in early stages of life (Cox, 1986; Sabina and Brown, 2009). At this time, glucose and galactose are present in high amounts due to milk intake (Grenov et al., 2016). C. glabrata is unable to metabolize galactose and does not encode any gene of the GAL regulon (Hittinger et al., 2004).
The amino-sugar N-acetylglucosamine (GlcNAc), which is present in the human host in mucosal layers, is used as a carbon and nitrogen source by C. albicans. GlcNAc is an important signaling molecule and is also the monosaccharide of chitin. Therefore, GlcNAc forms part of the fungal cell wall. GlcNAc is transported into the cells by the CaNgt1 transporter and is metabolized by CaNag1, CaHxk1, and CaDac1 into fructose-6-phosphate, which is further metabolized to pyruvate (Simonetti et al., 1974; Cardinali et al., 1997; Kumar et al., 2000; Leberer et al., 2001; Yamada-Okabe et al., 2001; Alvarez and Konopka, 2007). It is intriguing that next to galactose, GlcNAc also induces the expression of CaGAL1, CaGAL7, and CaGAL10 (Kamthan et al., 2013). A disruption of CaGAL10 shows a severely reduced expression of CaGAL1 and CaGAL7 in the presence of GlcNAc. This finding suggests an important role for CaGAL10 in the GlcNAc mediated upregulation of the Leloir pathway genes. The induction of these genes peaks 90 min after the addition of GlcNAc whereas this expression maximum is already visualized 15 min after galactose addition (Kamthan et al., 2013). These data suggest that GlcNAc has an indirect effect on the induction of the CaGAL genes. Interestingly, intracellular galactose levels increase in cells growing in GlcNAc containing media which implies that GlcNAc induces the synthesis of the internal galactose, a trait already reported in Escherichia coli and Kluyveromyces lactis (Wu and Kalckar, 1966; Cardinali et al., 1997). This may then also explain the delayed and possibly indirect induction of the CaGAL genes. Even though CaCph1 is responsible for galactose induced expression, the transcriptional effect of GlcNAc remains unchanged in a Cacph1Δ mutant (Sexton et al., 2007; Kamthan et al., 2013; Figure 3).
Apart from the genes described above, many other genes are induced in the presence of GlcNAc (Supplementary Table S1) (Gunasekera et al., 2010). Several of them are involved in virulence (Shepherd et al., 1980; Singh et al., 2001; Huang et al., 2010; Noble et al., 2010). For example, GlcNAc seems to activate the cAMP/PKA pathway as this pathway is required for the GlcNAc-induced morphogenesis. C. albicans grows on GlcNAc as a sole carbon source which needs to be internalized to cause filamentation (Singh and Datta, 1979; Kumar et al., 2000; Alvarez and Konopka, 2007). On the other hand, the catabolism of GlcNAc is not required to induce morphogenesis, as the Cahxk1Δ Canag1Δ Cadag1Δ triple mutant is efficiently stimulated by GlcNAc to form hyphae (Naseem et al., 2011). Interestingly, GlcNAc induces a phenomenon called sugar-induced cell death (SICD; Du et al., 2015). This phenomenon was first identified in S. cerevisiae where the induction of glucose in water-cultured cells “tricks” the cells to enter an active cell cycle and leave the protective stationary phase. However, since there is a lack of other nutrients, they rapidly lose viability and undergo programmed cell death (apoptosis) (Granot and Snyder, 1991). C. albicans also undergoes apoptosis when it encounters different environmental stresses. The fact that GlcNAc can also induce SICD shows that this amino-sugar regulates multiple cellular programs in a coordinated manner and thereby maximizing the efficiency of nutrient use (Du et al., 2015). S. cerevisiae strains do not grow on GlcNAc as sole carbon source, but introducing the C. albicans GlcNAc catabolic genes allows baker’s yeast to use GlcNAc as a sole carbon source. This strain is useful in the research toward a renewable way of biofuel production (Wendland et al., 2009). This GlcNAc metabolism is not present in C. glabrata.
Together with glucose, mannose and fructose belong to the top three of most preferred sugars for yeast species. In S. cerevisiae, fructose and mannose have similar sensing and transport mechanisms as glucose via the SRR pathway. Both sugars are sensed by ScSnf3/ScRgt2 and the extracellular fructose/mannose level has an effect on the expression of the hexose transporter genes (Dietvorst et al., 2010). All hexose transporter genes which are induced by the ScSnf3/ScRgt2 signaling pathway are able to transport fructose and mannose (Boles and Hollenberg, 1997). Seven transporters (ScHxt1–ScHxt7) are essential for growth on fructose/mannose at any concentration tested (Reifenberger et al., 1995; Liang and Gaber, 1996; Reifenberger et al., 1997).
This broad substrate sensing is conserved for the C. albicans CaHgt4 sensor. Apart from glucose, it also senses fructose and mannose as a Cahgt4Δ mutant is strongly impaired for growth on either sugar, with a more severe effect on fructose. The sensing of fructose and mannose induces the expression of seven of the 20 hexose transporters genes under which, CaHGT12 and CaHGT7 (Brown et al., 2006; Luo et al., 2007; Brown et al., 2009). This last one is activated by low levels (0.04%) of fructose and mannose (Brown et al., 2009). It suggests that these transporters are more critical for fructose transport in comparison to their relevance for glucose and mannose transport (Brown et al., 2006). As a final remark, C. glabrata is unable to use fructose or mannose as a carbon source.
Taken together, both similarities and differences are found in the sugar sensing and signaling mechanisms of S. cerevisiae and the Candida species. The most important observation is that C. albicans is able to use multiple carbon sources at the same time, while glucose represses the use of all other carbon sources in S. cerevisiae (Gancedo, 1992; Sandai et al., 2012). Probably, this is due to the metabolic flexibility of this pathogen, which is necessary to colonize the human body. The response of the cells to the nutrients present in the microenvironment is essential to become a successful pathogen (Rodaki et al., 2009).
Glucose has an effect on different virulence traits of both C. albicans and C. glabrata. One of the most important virulence factors of these pathogens is their ability to form biofilms. In this heterogenic community, the cells show a differentially active metabolism. Those which are in contact with glucose have an upregulation of glycolysis while others have an upregulated gluconeogenesis (Rodaki et al., 2009). During biofilm maturation, C. albicans forms hyphae, which are important for structural support, adherence to other cells and fulfillment of their nutrient needs (Chandra et al., 2001; Xu et al., 2014). Upon high levels of glucose, morphogenesis is repressed whereas hyphae formation is strongly induced from sugar levels lower than 0.25% (Maidan et al., 2005b). This indicates that sugar levels in biofilms are presumably below 0.25%. Depending on the environmental conditions, Candida cells disperse from the biofilm. During rich sugar conditions, there is more dispersion observed than under low sugar availability (Uppuluri et al., 2010). This suggests that biofilms are a survival strategy during poor nutrient conditions. Especially the carbon source present in the surroundings has a major effect on this process as the presence of glucose induces more dispersion than the presence of galactose (Uppuluri et al., 2010).
Second, another important virulence factor of Candida species is their high stress tolerance. Increasing glucose concentrations (0.01–1%) lead to upregulation of stress resistance genes in C. albicans, causing an increased resistance to azole antifungal drugs and osmotic stress (Rodaki et al., 2009; Ene et al., 2012). This is remarkable as S. cerevisiae downregulates its stress response genes in the presence of increasing concentrations of glucose (Rodaki et al., 2009). More comparable to the situation in S. cerevisiae, C. glabrata has a lower resistance against antifungal drugs upon increasing glucose levels. After treatment with amphotericin B, there was 50% less survival of the C. glabrata cells in the presence of 1% glucose then when less glucose (0.2%) was present (Ng et al., 2016). This indicates that this pathogen is more resistant due to a slower growth rate in the presence of low glucose levels (Ng et al., 2016). Additionally, C. albicans and C. glabrata are less susceptible for oxidative stress in the presence of increasing concentrations of glucose. C. glabrata is even more resistant than C. albicans as it tolerates 0.05 M of hydrogen peroxide, while this concentration damages the latter in the presence of 1% glucose (Rodaki et al., 2009; Ng et al., 2016). The mechanisms which link glucose to oxidative and osmotic stress resistance may be of importance during the infection process of C. albicans and C. glabrata. When entering the bloodstream, the cells are suddenly exposed to glucose, possibly leading to a fast induction of oxidative and osmotic stress resistance genes. This may be important for survival as, in the bloodstream, Candida cells are attacked by neutrophils, which try to kill off the pathogens by oxidative burst and low nutrient availability (Rodaki et al., 2009).
The glucose sensing pathways of C. albicans and C. glabrata described above are of importance for systemic infections. Many involved mutants have been tested in systemic mice models showing various results. It is known that the Cahgt4Δ mutant is not able to form hyphae. Consequently, these cells have a delayed virulence in mice. Where wild type cells can kill 90% of the mice after 5 days, the Cahgt4Δ mutants were only able to do this after 14 days (Brown et al., 2006). This indicates that the lack of this sugar sensor is not sufficient to render the cells completely avirulent. The Cahgt4Δ Cargt1Δ mutant shows a hyper-filamentous phenotype (Brown et al., 2006). This block in morphogenesis indicates that the double mutant will be impaired in virulence during a systemic infection. This has not yet been tested. Different components of the sugar signaling pathway toward PKA have been tested for their effect during systemic infections. Both CaGpr1 and CaGpa2 do not have any effect on the host survival. No difference in the mice survival rate was detected when mice were infected with the Cagpa2Δ mutant and the wild type. Mice infected with the Cagpr1Δ mutant showed a slight increase in survival of 20% compared to the wild type (Miwa et al., 2004; Maidan et al., 2005a). Moreover, it seems that CaCyr1 is very important during systemic infections since all mice infected with the Cacyr1Δ mutant could survive (Rocha et al., 2001). Finally, until now no components of the glucose repression pathway were tested for an effect on systemic infections.
From the findings presented above, it is clear that blood sugar levels have an effect on virulence by Candida albicans cells (Lorenz et al., 2004; Rodaki et al., 2009). Furthermore, C. glabrata cells isolated from the blood have a higher tendency toward biofilm formation than other cells (Ng et al., 2016). Also sugar related diseases have an effect on the virulence of these pathogens. For example, patients with diabetes have a higher chance of getting a systemic Candida infection compared to non-diabetic people (Rodaki et al., 2009). Furthermore, also a higher amount of oral Candida infections occur in people suffering from diabetes mellitus compared to healthy individuals (Zomorodian et al., 2016). This indicates that colonization and invasion by Candida species is enhanced via dietary glucose (Vargas et al., 1993). Constituently, we can conclude that the different sugar sensing and signaling pathways described above are important factors for both virulence and pathogenesis of C. albicans and C. glabrata.
As Candida species are an increasing problem, it is important to investigate their main virulence factors in order to find proper treatments. We focused on C. albicans and C. glabrata, because they are the two most isolated Candida species. However, in the last 10 years, there has been a big increase in other Candida species like Candida auris and Candida krusei (Jeffery-Smith et al., 2018). The problem with these species is that they show high antifungal resistance toward the current antifungal drugs which leads to major concerns. Further, C. glabrata is found to be inherently more resistant to azole drugs (Whaley and Rogers, 2016). Finally, more and more C. albicans clinical isolates show antifungal drug resistance (Whaley et al., 2016). This indicates the need for new antifungal drug targets and corresponding drugs. In this review, we show that sugar sensing and signaling are important for Candida species to turn virulent. Therefore, better understanding of the sugar sensing and metabolism pathways offer valuable antifungal drug targets. As stated, many of the players in these pathways are conserved between S. cerevisiae, C. albicans, and C. glabrata. Targeting these players result in a broad-spectrum antifungal drug against fungi. This kind of drug is desired by clinics and pharmaceutical companies, as nowadays one of the biggest issues with fungal infections is the diagnosis of the species (Perfect, 2017). For example, targeting fungal Gpr1 proteins will not lead to a fungistatic or fungicidal drug. However, this protein is important for morphogenesis and blocking this protein prevents the cells to turn virulent. This may be a better strategy for commensal fungi which are also present in a healthy human body, as it does not block normal cell growth (Brown et al., 2018). Other proteins may be less desired as many of them are highly conserved in human cells and thereby the corresponding drugs may have toxic effects.
In the future, these pathways should be unraveled in different pathogenic fungi and similarities should be marked. Similar players in different fungal species which are not conserved in humans are the desired drug targets nowadays. Furthermore, connections between the different pathways should be elucidated to better understand sugar signaling in pathogenic fungi and to find appropriate drug targets.
All authors contributed to the writing of the manuscript.
SW was supported by a Strategic Basic (SB) Research grant from the Fund for Scientific Research-Flanders (FWO; Grant No. 1S91118N). This work was further supported by a grant from the KU Leuven Research Fund (C14/17/063).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a past co-authorship with one of the authors PV.
We would like to acknowledge Nico Vangoethem and Frederik Van Leemputte for their help with the figures. We also thank Griet Van Zeebroeck to proofread this review.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00099/full#supplementary-material
Alvarez, F. J., and Konopka, J. B. (2007). Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol. Biol. Cell 18, 965–975. doi: 10.1091/mbc.e06-10-0931
Arendrup, M. C. (2010). Epidemiology of invasive candidiasis. Curr. Opin. Crit. Care 16, 445–452. doi: 10.1097/MCC.0b013e32833e84d2
Arendrup, M. C. (2013). Candida and candidaemia. Susceptibility and epidemiology. Dan. Med. J. 60:B4698.
Ballou, E. R., Avelar, G. M., Childers, D. S., Mackie, J., Bain, J. M., Wagener, J., et al. (2016). Lactate signalling regulates fungal beta-glucan masking and immune evasion. Nat. Microbiol. 2:16238. doi: 10.1038/nmicrobiol.2016.238
Barelle, C. J., Priest, C. L., Maccallum, D. M., Gow, N. A., Odds, F. C., and Brown, A. J. (2006). Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 8, 961–971. doi: 10.1111/j.1462-5822.2005.00676.x
Bensasson, D., Dicks, J., Ludwig, J. M., Bond, C. J., Elliston, A., Roberts, I. N., et al. (2018). Diverse lineages of Candida albicans live on old oaks. Genetics doi: 10.1534/genetics.118.301482 [Epub ahead of print].
Bhat, P. J., and Murthy, T. V. (2001). Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: mechanism of galactose-mediated signal transduction. Mol. Microbiol. 40, 1059–1066. doi: 10.1046/j.1365-2958.2001.02421.x
Bisson, L. F. (1988). High-affinity glucose transport in Saccharomyces cerevisiae is under general glucose repression control. J. Bacteriol. 170, 4838–4845. doi: 10.1128/jb.170.10.4838-4845.1988
Boles, E. (2003). “Yeast as a model system for studying glucose transport,” in Transmembrane Transporters, ed. M. W. Quick (Hoboken, NJ: Wiley).
Boles, E., and Hollenberg, C. P. (1997). The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev. 21, 85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x
Brock, M. (2009). Fungal metabolism in host niches. Curr. Opin. Microbiol. 12, 371–376. doi: 10.1016/j.mib.2009.05.004
Brown, A. J., Budge, S., Kaloriti, D., Tillmann, A., Jacobsen, M. D., Yin, Z., et al. (2014). Stress adaptation in a pathogenic fungus. J. Exp. Biol. 217(Pt 1), 144–155. doi: 10.1242/jeb.088930
Brown, N. A., Schrevens, S., Van Dijck, P., and Goldman, G. H. (2018). Fungal G-protein-coupled receptors: mediators of pathogenesis and targets for disease control. Nat. Microbiol. 3, 402–414. doi: 10.1038/s41564-018-0127-5
Brown, V., Sabina, J., and Johnston, M. (2009). Specialized sugar sensing in diverse fungi. Curr. Biol. 19, 436–441. doi: 10.1016/j.cub.2009.01.056
Brown, V., Sexton, J. A., and Johnston, M. (2006). A glucose sensor in Candida albicans. Eukaryot. Cell 5, 1726–1737. doi: 10.1128/EC.00186-06
Cardinali, G., Vollenbroich, V., Jeon, M. S., de Graaf, A. A., and Hollenberg, C. P. (1997). Constitutive expression in gal7 mutants of Kluyveromyces lactis is due to internal production of galactose as an inducer of the Gal/Lac regulon. Mol. Cell Biol. 17, 1722–1730. doi: 10.1128/MCB.17.3.1722
Carlson, M. (1999). Glucose repression in yeast. Curr. Opin. Microbiol. 2, 202–207. doi: 10.1016/S1369-5274(99)80035-6
Celenza, J. L., and Carlson, M. (1986). A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233, 1175–1180. doi: 10.1126/science.3526554
Chandra, J., Kuhn, D. M., Mukherjee, P. K., Hoyer, L. L., McCormick, T., and Ghannoum, M. A. (2001). Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183, 5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001
Childers, D. S., Raziunaite, I., Mol Avelar, G., Mackie, J., Budge, S., Stead, D., et al. (2016). The rewiring of ubiquitination targets in a pathogenic yeast promotes metabolic flexibility, host colonization and virulence. PLoS Pathog. 12:e1005566. doi: 10.1371/journal.ppat.1005566
Cottier, F., Tan, A. S. M., Yurieva, M., Liao, W., Lum, J., Poidinger, M., et al. (2017). The transcriptional response of Candida albicans to weak organic acids, carbon source, and MIG1 inactivation unveils a role for HGT16 in mediating the fungistatic effect of acetic acid. G3 7, 3597–3604. doi: 10.1534/g3.117.300238
Cox, F. (1986). Candida albicans adherence in newborn infants. J. Med. Vet. Mycol. 24, 121–125. doi: 10.1080/02681218680000181
Demuyser, L., Van Genechten, W., Mizuno, H., Colombo, S., and Van Dijck, P. (2018). Introducing fluorescence resonance energy transfer-based biosensors for the analysis of cAMP-PKA signalling in the fungal pathogen Candida glabrata. Cell Microbiol. 20:e12863. doi: 10.1111/cmi.12863
DeVit, M. J., and Johnston, M. (1999). The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9, 1231–1241. doi: 10.1016/S0960-9822(99)80503-X
Diekema, D., Arbefeville, S., Boyken, L., Kroeger, J., and Pfaller, M. (2012). The changing epidemiology of healthcare-associated candidemia over three decades. Diagn. Microbiol. Infect. Dis. 73, 45–48. doi: 10.1016/j.diagmicrobio.2012.02.001
Dietvorst, J., Karhumaa, K., Kielland-Brandt, M. C., and Brandt, A. (2010). Amino acid residues involved in ligand preference of the Snf3 transporter-like sensor in Saccharomyces cerevisiae. Yeast 27, 131–138. doi: 10.1002/yea.1737
Du, H., Guan, G., Li, X., Gulati, M., Tao, L., Cao, C., et al. (2015). N-acetylglucosamine-induced cell death in Candida albicans and its implications for adaptive mechanisms of nutrient sensing in yeasts. mBio 6:e01376-15. doi: 10.1128/mBio.01376-15
Ehrstrom, S., Yu, A., and Rylander, E. (2006). Glucose in vaginal secretions before and after oral glucose tolerance testing in women with and without recurrent vulvovaginal candidiasis. Obstet. Gynecol. 108, 1432–1437. doi: 10.1097/01.AOG.0000246800.38892.fc
Ene, I. V., Adya, A. K., Wehmeier, S., Brand, A. C., MacCallum, D. M., Gow, N. A., et al. (2012). Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol. 14, 1319–1335. doi: 10.1111/j.1462-5822.2012.01813.x
Enoch, D. A., Yang, H., Aliyu, S. H., and Micallef, C. (2017). The changing epidemiology of invasive fungal infections. Methods Mol. Biol. 1508, 17–65. doi: 10.1007/978-1-4939-6515-1_2
Fan, J., Chaturvedi, V., and Shen, S. H. (2002). Identification and phylogenetic analysis of a glucose transporter gene family from the human pathogenic yeast Candida albicans. J. Mol. Evol. 55, 336–346. doi: 10.1007/s00239-002-2330-4
Fleck, C. B., Schöbel, F., and Brock, M. (2011). Nutrient acquisition by pathogenic fungi: nutrient availability, pathway regulation, and differences in substrate utilization. Int. J. Med. Microbiol. 301, 400–407. doi: 10.1016/j.ijmm.2011.04.007
Flick, J. S., and Johnston, M. (1992). Analysis of URSG-mediated glucose repression of the GAL1 promoter of Saccharomyces cerevisiae. Genetics 130, 295–304.
Gancedo, J. M. (1992). Carbon catabolite repression in yeast. Eur. J. Biochem. 206, 297–313. doi: 10.1111/j.1432-1033.1992.tb16928.x
Goemaere, B., Becker, P., Van Wijngaerden, E., Maertens, J., Spriet, I., Hendrickx, M., et al. (2018). Increasing candidaemia incidence from 2004 to 2015 with a shift in epidemiology in patients preexposed to antifungals. Mycoses 61, 127–133. doi: 10.1111/myc.12714
Granot, D., and Snyder, M. (1991). Glucose induces cAMP-independent growth-related changes in stationary-phase cells of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 88, 5724–5728. doi: 10.1073/pnas.88.13.5724
Grenov, B., Briend, A., Sangild, P. T., Thymann, T., Rytter, M. H., Hother, A. L., et al. (2016). Undernourished children and milk lactose. Food Nutr. Bull. 37, 85–99. doi: 10.1177/0379572116629024
Greppi, A. Ł. K., Costantini, A., Rantsiou, K., Hounhouigan, D. J., Arneborg, N., Cocolin, L., et al. (2015). Phytase-producing capacity of yeasts isolated from traditional African fermented food products and PHYPk gene expression of Pichia kudriavzevii strains. Int. J. Food Microbiol. 205, 81–89. doi: 10.1016/j.ijfoodmicro.2015.04.011
Griggs, D. W., and Johnston, M. (1991). Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc. Natl. Acad. Sci. U.S.A. 88, 8597–8601. doi: 10.1073/pnas.88.19.8597
Groenendaal, W., von Basum, G., Schmidt, K. A., Hilbers, P. A., and van Riel, N. A. (2010). Quantifying the composition of human skin for glucose sensor development. J. Diabetes Sci. Technol. 4, 1032–1040. doi: 10.1177/193229681000400502
Guinea, J. (2014). Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 20(Suppl. 6), 5–10. doi: 10.1111/1469-0691.12539
Gulati, M., and Nobile, C. J. (2016). Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 18, 310–321. doi: 10.1016/j.micinf.2016.01.002
Gunasekera, A., Alvarez, F. J., Douglas, L. M., Wang, H. X., Rosebrock, A. P., and Konopka, J. B. (2010). Identification of GIG1, a GlcNAc-induced gene in Candida albicans needed for normal sensitivity to the chitin synthase inhibitor nikkomycin Z. Eukaryot. Cell 9, 1476–1483. doi: 10.1128/ec.00178-10
Hardie, D. G., Carling, D., and Carlson, M. (1998). The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67, 821–855. doi: 10.1146/annurev.biochem.67.1.821
Hedbacker, K., and Carlson, M. (2008). SNF1/AMPK pathways in yeast. Front. Biosci. 13, 2408–2420. doi: 10.2741/2854
Hedges, S. B., Marin, J., Suleski, M., Paymer, M., and Kumar, S. (2015). Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845. doi: 10.1093/molbev/msv037
Helmerhorst, E. J., Murphy, M. P., Troxler, R. F., and Oppenheim, F. G. (2002). Characterization of the mitochondrial respiratory pathways in Candida albicans. Biochim. Biophys. Acta 1556, 73–80. doi: 10.1016/S0005-2728(02)00308-0
Herrero, E. (2005). Evolutionary relationships between Saccharomyces cerevisiae and other fungal species as determined from genome comparisons. Rev. Iberoam. Micol. 22, 217–222. doi: 10.1016/S1130-1406(05)70046-2
Herrero, P., Martinez-Campa, C., and Moreno, F. (1998). The hexokinase 2 protein participates in regulatory DNA-protein complexes necessary for glucose repression of the SUC2 gene in Saccharomyces cerevisiae. FEBS Lett. 434, 71–76. doi: 10.1016/S0014-5793(98)00872-2
Hers, H. G. (1984). The discovery and the biological role of fructose 2,6-bisphosphate. Biochem. Soc. Trans. 12, 729–735. doi: 10.1042/bst0120729
Hittinger, C. T., Rokas, A., and Carroll, S. B. (2004). Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc. Natl. Acad. Sci. U.S.A. 101, 14144–14149. doi: 10.1073/pnas.0404319101
Holden, H. M., Rayment, I., and Thoden, J. B. (2003). Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 278, 43885–43888. doi: 10.1074/jbc.R300025200
Huang, G., Yi, S., Sahni, N., Daniels, K. J., Srikantha, T., and Soll, D. R. (2010). N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6:e1000806. doi: 10.1371/journal.ppat.1000806
Hudson, D. A., Sciascia, Q. L., Sanders, R. J., Norris, G. E., Edwards, P. J., Sullivan, P. A., et al. (2004). Identification of the dialysable serum inducer of germ-tube formation in Candida albicans. Microbiology 150(Pt 9), 3041–3049. doi: 10.1099/mic.0.27121-0
Jacquet, M. (1997). [Ras proteins in Saccharomyces cerevisiae, their partners and their activation]. C. R. Seances Soc. Biol. Fil. 191, 221–235.
Jeffery-Smith, A., Taori, S. K., Schelenz, S., Jeffery, K., Johnson, E. M., Borman, A., et al. (2018). Candida auris: a review of the literature. Clin. Microbiol. Rev. 31:e00029-17. doi: 10.1128/CMR.00029-17
Jin, Y., Samaranayake, L. P., Samaranayake, Y., and Yip, H. K. (2004). Biofilm formation of Candida albicans is variably affected by saliva and dietary sugars. Arch. Oral Biol. 49, 789–798. doi: 10.1016/j.archoralbio.2004.04.011
Johnston, M. (1987). A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 51, 458–476.
Kamthan, M., Kamthan, A., Ruhela, D., Maiti, P., Bhavesh, N. S., and Datta, A. (2013). Upregulation of galactose metabolic pathway by N-acetylglucosamine induced endogenous synthesis of galactose in Candida albicans. Fungal Genet. Biol. 54, 15–24. doi: 10.1016/j.fgb.2013.02.006
Kaniak, A., Xue, Z., Macool, D., Kim, J. H., and Johnston, M. (2004). Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 3, 221–231. doi: 10.1128/EC.3.1.221-231.2004
Kayikci,Ö., and Nielsen, J. (2015). Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 15:fov068. doi: 10.1093/femsyr/fov068
Kraakman, L., Lemaire, K., Ma, P., Teunissen, A. W., Donaton, M. C., Van Dijck, P., et al. (1999). A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32, 1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x
Kumar, M. J., Jamaluddin, M. S., Natarajan, K., Kaur, D., and Datta, A. (2000). The inducible N-acetylglucosamine catabolic pathway gene cluster in Candida albicans: discrete N-acetylglucosamine-inducible factors interact at the promoter of NAG1. Proc. Natl. Acad. Sci. U.S.A. 97, 14218–14223. doi: 10.1073/pnas.250452997
Lafuente, M. J., Gancedo, C., Jauniaux, J. C., and Gancedo, J. M. (2000). Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol. 35, 161–172. doi: 10.1046/j.1365-2958.2000.01688.x
Laughlin, M. R. (2014). Normal roles for dietary fructose in carbohydrate metabolism. Nutrients 6, 3117–3129. doi: 10.3390/nu6083117
Laughon, A., and Gesteland, R. F. (1982). Isolation and preliminary characterization of the GAL4 gene, a positive regulator of transcription in yeast. Proc. Natl. Acad. Sci. U.S.A. 79, 6827–6831. doi: 10.1073/pnas.79.22.6827
Leberer, E., Harcus, D., Dignard, D., Johnson, L., Ushinsky, S., Thomas, D. Y., et al. (2001). Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42, 673–687. doi: 10.1046/j.1365-2958.2001.02672.x
Leloir, L. F. (1951). The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch. Biochem. Biophys. 33, 186–190. doi: 10.1016/0003-9861(51)90096-3
Lemaire, K., Van de Velde, S., Van Dijck, P., and Thevelein, J. M. (2004). Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16, 293–299. doi: 10.1016/j.molcel.2004.10.004
Liang, H., and Gaber, R. F. (1996). A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol. Biol. Cell 7, 1953–1966. doi: 10.1091/mbc.7.12.1953
Liu, J., Liu, H., Yan, J., Liu, N., Zhang, H., Zhao, C., et al. (2018). Molecular typing and genetic relatedness of 72 clinical Candida albicans isolates from poultry. Vet. Microbiol. 214, 36–43. doi: 10.1016/j.vetmic.2017.11.030
Lohr, D., Venkov, P., and Zlatanova, J. (1995). Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 9, 777–787. doi: 10.1096/fasebj.9.9.7601342
Lorenz, M. C., Bender, J. A., and Fink, G. R. (2004). Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3, 1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004
Ludin, K., Jiang, R., and Carlson, M. (1998). Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 95, 6245–6250. doi: 10.1073/pnas.95.11.6245
Luo, L., Tong, X., and Farley, P. C. (2007). The Candida albicans gene HGT12 (orf19.7094) encodes a hexose transporter. FEMS Immunol. Med. Microbiol. 51, 14–17. doi: 10.1111/j.1574-695X.2007.00274.x
Maidan, M. M., De Rop, L., Serneels, J., Exler, S., Rupp, S., Tournu, H., et al. (2005a). The G protein-coupled receptor Gpr1 and the Galpha protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16, 1971–1986. doi: 10.1091/mbc.e04-09-0780
Maidan, M. M., Thevelein, J. M., and Van Dijck, P. (2005b). Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem. Soc. Trans. 33(Pt 1), 291–293. doi: 10.1042/bst0330291
Malakar, P., and Venkatesh, K. V. (2014). GAL regulon of Saccharomyces cerevisiae performs optimally to maximize growth on galactose. FEMS Yeast Res. 14, 346–356. doi: 10.1111/1567-1364.12109
Manns, D. C., Churey, J. J., and Worobo, R. W. (2014). Nutrient-dependent efficacy of the antifungal protein YvgO correlates to cellular proliferation rate in Candida albicans 3153A and Byssochlamys fulva H25. Probiotics Antimicrob. Proteins 6, 198–207. doi: 10.1007/s12602-014-9167-1
Martchenko, M., Levitin, A., Hogues, H., Nantel, A., and Whiteway, M. (2007a). Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr. Biol. 17, 1007–1013. doi: 10.1016/j.cub.2007.05.017
Martchenko, M., Levitin, A., and Whiteway, M. (2007b). Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryot. Cell 6, 291–301. doi: 10.1128/EC.00183-06
Mercado, J. J., and Gancedo, J. M. (1992). Regulatory regions in the yeast FBP1 and PCK1 genes. FEBS Lett. 311, 110–114. doi: 10.1016/0014-5793(92)81379-Z
Mercado, J. J., Smith, R., Sagliocco, F. A., Brown, A. J., and Gancedo, J. M. (1994). The levels of yeast gluconeogenic mRNAs respond to environmental factors. Eur. J. Biochem. 224, 473–481. doi: 10.1111/j.1432-1033.1994.00473.x
Mercado, J. J., Vincent, O., and Gancedo, J. M. (1991). Regions in the promoter of the yeast FBP1 gene implicated in catabolite repression may bind the product of the regulatory gene MIG1. FEBS Lett. 291, 97–100. doi: 10.1016/0014-5793(91)81112-L
Mitts, M. R., Grant, D. B., and Heideman, W. (1990). Adenylate cyclase in Saccharomyces cerevisiae is a peripheral membrane protein. Mol. Cell Biol. 10, 3873–3883. doi: 10.1128/MCB.10.8.3873
Miwa, T., Takagi, Y., Shinozaki, M., Yun, C. W., Schell, W. A., Perfect, J. R., et al. (2004). Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3, 919–931. doi: 10.1128/ec.3.4.919-931.2004
Moriya, H., and Johnston, M. (2004). Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. U.S.A. 101, 1572–1577. doi: 10.1073/pnas.0305901101
Morrison-Whittle, P., Lee, S. A., Fedrizzi, B., and Goddard, M. R. (2018). Co-evolution as tool for diversifying flavor and aroma profiles of wines. Front. Microbiol. 9:910. doi: 10.3389/fmicb.2018.00910
Murad, A. M., d’Enfert, C., Gaillardin, C., Tournu, H., Tekaia, F., Talibi, D., et al. (2001). Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42, 981–993. doi: 10.1046/j.1365-2958.2001.02713.x
Nakafuku, M., Obara, T., Kaibuchi, K., Miyajima, I., Miyajima, A., Itoh, H., et al. (1988). Isolation of a second yeast Saccharomyces cerevisiae gene (GPA2) coding for guanine nucleotide-binding regulatory protein: studies on its structure and possible functions. Proc. Natl. Acad. Sci. U.S.A. 85, 1374–1378. doi: 10.1073/pnas.85.5.1374
Naseem, S., Gunasekera, A., Araya, E., and Konopka, J. B. (2011). N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. J. Biol. Chem. 286, 28671–28680. doi: 10.1074/jbc.M111.249854
Nehlin, J. O., Carlberg, M., and Ronne, H. (1991). Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 10, 3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x
Nehlin, J. O., and Ronne, H. (1990). Yeast MIG1 repressor is related to the mammalian early growth response and Wilms’ tumour finger proteins. EMBO J. 9, 2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x
Ng, T. S., Chew, S. Y., Rangasamy, P., Mohd Desa, M. N., Sandai, D., Chong, P. P., et al. (2015). SNF3 as high affinity glucose sensor and its function in supporting the viability of Candida glabrata under glucose-limited environment. Front. Microbiol. 6:1334. doi: 10.3389/fmicb.2015.01334
Ng, T. S., Desa, M. N. M., Sandai, D., Chong, P. P., and Than, L. T. L. (2016). Growth, biofilm formation, antifungal susceptibility and oxidative stress resistance of Candida glabrata are affected by different glucose concentrations. Infect. Genet. Evol. 40, 331–338. doi: 10.1016/j.meegid.2015.09.004
Noble, S. M., French, S., Kohn, L. A., Chen, V., and Johnson, A. D. (2010). Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42, 590–598. doi: 10.1038/ng.605
Orlova, M., Barrett, L., and Kuchin, S. (2008). Detection of endogenous Snf1 and its activation state: application to Saccharomyces and Candida species. Yeast 25, 745–754. doi: 10.1002/yea.1628
Ozcan, S., Dover, J., and Johnston, M. (1998). Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17, 2566–2573. doi: 10.1093/emboj/17.9.2566
Ozcan, S., Dover, J., Rosenwald, A. G., Wölfl, S., and Johnston, M. (1996). Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. U.S.A. 93, 12428–12432. doi: 10.1073/pnas.93.22.12428
Ozcan, S., and Johnston, M. (1995). Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell Biol. 15, 1564–1572. doi: 10.1128/MCB.15.3.1564
Ozcan, S., and Johnston, M. (1999). Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63, 554–569.
Peeters, K., Van Leemputte, F., Fischer, B., Bonini, B. M., Quezada, H., Tsytlonok, M., et al. (2017). Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras. Nat. Commun. 8:922. doi: 10.1038/s41467-017-01019-z
Pemmaraju, S. C., Pruthi, P. A., Prasad, R., and Pruthi, V. (2016). Modulation of Candida albicans biofilm by different carbon sources. Mycopathologia 181, 341–352. doi: 10.1007/s11046-016-9992-8
Peng, G., and Hopper, J. E. (2002). Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc. Natl. Acad. Sci. U.S.A. 99, 8548–8553. doi: 10.1073/pnas.142100099
Perfect, J. R. (2017). The antifungal pipeline: a reality check. Nat. Rev. Drug Discov. 16, 603–616. doi: 10.1038/nrd.2017.46
Petter, R., Chang, Y. C., and Kwon-Chung, K. J. (1997). A gene homologous to Saccharomyces cerevisiae SNF1 appears to be essential for the viability of Candida albicans. Infect. Immun. 65, 4909–4917.
Petter, R., and Kwon-Chung, K. J. (1996). Disruption of the SNF1 gene abolishes trehalose utilization in the pathogenic yeast Candida glabrata. Infect. Immun. 64, 5269–5273.
Pfaller, M. A., Andes, D. R., Diekema, D. J., Horn, D. L., Reboli, A. C., Rotstein, C., et al. (2014). Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004-2008. PLoS One 9:e101510. doi: 10.1371/journal.pone.0101510
Pfaller, M. A., and Diekema, D. J. (2007). Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163. doi: 10.1128/CMR.00029-06
Pfaller, M. A., Diekema, D. J., Rinaldi, M. G., Barnes, R., Hu, B., Veselov, A. V., et al. (2005). Results from the ARTEMIS DISK global antifungal surveillance study: a 6.5-year analysis of susceptibilities of Candida and other yeast species to fluconazole and voriconazole by standardized disk diffusion testing. J. Clin. Microbiol. 43, 5848–5859. doi: 10.1128/JCM.43.12.5848-5859.2005
Platt, A., and Reece, R. J. (1998). The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 17, 4086–4091. doi: 10.1093/emboj/17.14.4086
Ramirez-Zavala, B., Mottola, A., Haubenreisser, J., Schneider, S., Allert, S., Brunke, S., et al. (2017). The Snf1-activating kinase Sak1 is a key regulator of metabolic adaptation and in vivo fitness of Candida albicans. Mol. Microbiol. 104, 989–1007. doi: 10.1111/mmi.13674
Reifenberger, E., Boles, E., and Ciriacy, M. (1997). Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur. J. Biochem. 245, 324–333. doi: 10.1111/j.1432-1033.1997.00324.x
Reifenberger, E., Freidel, K., and Ciriacy, M. (1995). Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol. Microbiol. 16, 157–167. doi: 10.1111/j.1365-2958.1995.tb02400.x
Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., et al. (2000). Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309. doi: 10.1126/science.290.5500.2306
Rocha, C. R., Schroppel, K., Harcus, D., Marcil, A., Dignard, D., Taylor, B. N., et al. (2001). Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12, 3631–3643. doi: 10.1091/mbc.12.11.3631
Rodaki, A., Bohovych, I. M., Enjalbert, B., Young, T., Odds, F. C., Gow, N. A., et al. (2009). Glucose promotes stress resistance in the fungal pathogen Candida albicans. Mol. Biol. Cell 20, 4845–4855. doi: 10.1091/mbc.E09-01-0002
Rodrigues, F., Ludovico, P., and Leão, C. (2006). Sugar Metabolism in Yeasts: an Overview of Aerobic and Anaerobic Glucose Catabolism. Berlin: Springer.
Rolland, F., De Winde, J. H., Lemaire, K., Boles, E., Thevelein, J. M., and Winderickx, J. (2000). Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38, 348–358. doi: 10.1046/j.1365-2958.2000.02125.x
Sabina, J., and Brown, V. (2009). Glucose sensing network in Candida albicans: a sweet spot for fungal morphogenesis. Eukaryot. Cell 8, 1314–1320. doi: 10.1128/EC.00138-09
Salazar, S. B., Wang, C., Münsterkötter, M., Okamoto, M., Takahashi-Nakaguchi, A., Chibana, H., et al. (2018). Comparative genomic and transcriptomic analyses unveil novel features of azole resistance and adaptation to the human host in Candida glabrata. FEMS Yeast Res. 18:fox079. doi: 10.1093/femsyr/fox079
Sandai, D., Yin, Z., Selway, L., Stead, D., Walker, J., Leach, M. D., et al. (2012). The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. mBio 3:e02489-14. doi: 10.1128/mBio.02489-14
Santos, M. A., and Tuite, M. F. (1995). The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 23, 1481–1486. doi: 10.1093/nar/23.9.1481
Schmidt, M. C., McCartney, R. R., Zhang, X., Tillman, T. S., Solimeo, H., Wölfl, S., et al. (1999). Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell Biol. 19, 4561–4571. doi: 10.1128/MCB.19.7.4561
Schrevens, S., Van Zeebroeck, G., Riedelberger, M., Tournu, H., Kuchler, K., and Van Dijck, P. (2018). Methionine is required for cAMP-PKA-mediated morphogenesis and virulence of Candida albicans. Mol. Microbiol. 108, 258–275. doi: 10.1111/mmi.13933
Sexton, J. A., Brown, V., and Johnston, M. (2007). Regulation of sugar transport and metabolism by the Candida albicans Rgt1 transcriptional repressor. Yeast 24, 847–860. doi: 10.1002/yea.1514
Shashkova, S., Wollman, A. J. M., Leake, M. C., and Hohmann, S. (2017). The yeast Mig1 transcriptional repressor is dephosphorylated by glucose-dependent and-independent mechanisms. FEMS Microbiol. Lett. 364:fnx133. doi: 10.1093/femsle/fnx133
Shepherd, M. G., Yin, C. Y., Ram, S. P., and Sullivan, P. A. (1980). Germ tube induction in Candida albicans. Can. J. Microbiol. 26, 21–26. doi: 10.1139/m80-004
Simonetti, N., Strippoli, V., and Cassone, A. (1974). Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature 250, 344–346. doi: 10.1038/250344a0
Singh, B., and Datta, A. (1979). Regulation of N-acetylglucosamine uptake in yeast. Biochim. Biophys. Acta 557, 248–258. doi: 10.1016/0005-2736(79)90107-X
Singh, P., Ghosh, S., and Datta, A. (2001). Attenuation of virulence and changes in morphology in Candida albicans by disruption of the N-acetylglucosamine catabolic pathway. Infect. Immun. 69, 7898–7903. doi: 10.1128/IAI.69.12.7898-7903.2001
Sokół, I., Gawel, A., and Bobrek, K. (2018). The prevalence of yeast and characteristics of the isolates from the digestive tract of clinically healthy turkeys. Avian Dis. 62, 286–290. doi: 10.1637/11780-121117-Reg.1
Sone, H., Shimano, H., Ebinuma, H., Takahashi, A., Yano, Y., Iida, K. T., et al. (2003). Physiological changes in circulating mannose levels in normal, glucose-intolerant, and diabetic subjects. Metabolism 52, 1019–1027. doi: 10.1016/S0026-0495(03)00153-7
Sugino, R. P., and Innan, H. (2005). Estimating the time to the whole-genome duplication and the duration of concerted evolution via gene conversion in yeast. Genetics 171, 63–69. doi: 10.1534/genetics.105.043869
Thevelein, J. M. (1991). Fermentable sugars and intracellular acidification as specific activators of the RAS-adenylate cyclase signalling pathway in yeast: the relationship to nutrient-induced cell cycle control. Mol. Microbiol. 5, 1301–1307. doi: 10.1111/j.1365-2958.1991.tb00776.x
Thevelein, J. M., Cauwenberg, L., Colombo, S., De Winde, J. H., Donation, M., Dumortier, F., et al. (2000). Nutrient-induced signal transduction through the protein kinase A pathway and its role in the control of metabolism, stress resistance, and growth in yeast. Enzyme Microb. Technol. 26, 819–825. doi: 10.1016/S0141-0229(00)00177-0
Toda, T., Uno, I., Ishikawa, T., Powers, S., Kataoka, T., Broek, D., et al. (1985). In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40, 27–36. doi: 10.1016/0092-8674(85)90305-8
Todd, R. B., and Andrianopoulos, A. (1997). Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21, 388–405. doi: 10.1006/fgbi.1997.0993
Towle, H. C. (2005). Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol. Metab. 16, 489–494. doi: 10.1016/j.tem.2005.10.003
Treitel, M. A., and Carlson, M. (1995). Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. U.S.A. 92, 3132–3136. doi: 10.1073/pnas.92.8.3132
Tzamarias, D., and Struhl, K. (1995). Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 9, 821–831. doi: 10.1101/gad.9.7.821
Uno, I., Mitsuzawa, H., Matsumoto, K., Tanaka, K., Oshima, T., and Ishikawa, T. (1985). Reconstitution of the GTP-dependent adenylate cyclase from products of the yeast CYR1 and RAS2 genes in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 82, 7855–7859. doi: 10.1073/pnas.82.23.7855
Uppuluri, P., Chaturvedi, A. K., Srinivasan, A., Banerjee, M., Ramasubramaniam, A. K., Kohler, J. R., et al. (2010). Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6:e1000828. doi: 10.1371/journal.ppat.1000828
Vale-Silva, L., Beaudoing, E., Tran, V. D. T., and Sanglard, D. (2017). Comparative genomics of two sequential Candida glabrata clinical isolates. G3 7, 2413–2426. doi: 10.1534/g3.117.042887
Van Urk, H., Voll, W. S., Scheffers, W. A., and Van Dijken, J. P. (1990). Transient-state analysis of metabolic fluxes in crabtree-positive and crabtree-negative yeasts. Appl. Environ. Microbiol. 56, 281–287.
Vargas, S. L., Patrick, C. C., Ayers, G. D., and Hughes, W. T. (1993). Modulating effect of dietary carbohydrate supplementation on Candida albicans colonization and invasion in a neutropenic mouse model. Infect. Immun. 61, 619–626.
Verduyn, C. (1991). Physiology of yeasts in relation to biomass yields. Antonie Van Leeuwenhoek 60, 325–353. doi: 10.1007/BF00430373
Wendland, J., Schaub, Y., and Walther, A. (2009). N-acetylglucosamine utilization by Saccharomyces cerevisiae based on expression of Candida albicans NAG genes. Appl. Environ. Microbiol. 75, 5840–5845. doi: 10.1128/AEM.00053-09
West, R. W. Jr., Yocum, R. R., and Ptashne, M. (1984). Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol. Cell Biol. 4, 2467–2478. doi: 10.1128/MCB.4.11.2467
Whaley, S. G., Berkow, E. L., Rybak, J. M., Nishimoto, A. T., Barker, K. S., and Rogers, P. D. (2016). Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front. Microbiol. 7:2173. doi: 10.3389/fmicb.2016.02173
Whaley, S. G., and Rogers, P. D. (2016). Azole resistance in Candida glabrata. Curr. Infect. Dis. Rep. 18:41. doi: 10.1007/s11908-016-0554-5
Wu, H. C., and Kalckar, H. M. (1966). Endogenous induction of the galactose operon in Escherichia coli K12. Proc. Natl. Acad. Sci. U.S.A. 55, 622–629. doi: 10.1073/pnas.55.3.622
Xu, H., Jenkinson, H. F., and Dongari-Bagtzoglou, A. (2014). Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol. Oral Microbiol. 29, 99–116. doi: 10.1111/omi.12049
Yamada-Okabe, T., Sakamori, Y., Mio, T., and Yamada-Okabe, H. (2001). Identification and characterization of the genes for N-acetylglucosamine kinase and N-acetylglucosamine-phosphate deacetylase in the pathogenic fungus Candida albicans. Eur. J. Biochem. 268, 2498–2505. doi: 10.1046/j.1432-1327.2001.02135.x
Yapar, N. (2014). Epidemiology and risk factors for invasive candidiasis. Ther. Clin. Risk Manag. 10, 95–105. doi: 10.2147/TCRM.S40160
Yin, Z., Smith, R. J., and Brown, A. J. (1996). Multiple signalling pathways trigger the exquisite sensitivity of yeast gluconeogenic mRNAs to glucose. Mol. Microbiol. 20, 751–764. doi: 10.1111/j.1365-2958.1996.tb02514.x
Zaragoza, O., Rodriguez, C., and Gancedo, C. (2000). Isolation of the MIG1 gene from Candida albicans and effects of its disruption on catabolite repression. J. Bacteriol. 182, 320–326. doi: 10.1128/JB.182.2.320-326.2000
Keywords: Candida albicans, Candida glabrata, Saccharomyces cerevisiae, sugar sensing, sugar transport, Snf3/Rgt2-Hgt4, Gpr1/Gpa2, Snf1/Mig1
Citation: Van Ende M, Wijnants S and Van Dijck P (2019) Sugar Sensing and Signaling in Candida albicans and Candida glabrata. Front. Microbiol. 10:99. doi: 10.3389/fmicb.2019.00099
Received: 30 November 2018; Accepted: 16 January 2019;
Published: 30 January 2019.
Edited by:
Neil Andrew Brown, University of Bath, United KingdomReviewed by:
Rebecca Anne Hall, University of Birmingham, United KingdomCopyright © 2019 Van Ende, Wijnants and Van Dijck. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick Van Dijck, cGF0cmljay52YW5kaWpja0BrdWxldXZlbi52aWIuYmU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.