
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 February 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00092
Antimicrobial efflux is one of the important mechanisms causing multi-drug resistance (MDR) in bacteria. Chemosensitizers like 1-(1-naphthylmethyl)-piperazine (NMP) can inhibit an efflux pump and therefore can overcome MDR. However, secondary effects of NMP other than efflux pump inhibition are rarely investigated. Here, using phenotypic assays, phenotypic microarray and transcriptomic assays we show that NMP creates membrane destabilization in MDR Klebsiella pneumoniae MGH 78578 strain. The NMP mediated membrane destabilization activity was measured using β-lactamase activity, membrane potential alteration studies, and transmission electron microscopy assays. Results from both β-lactamase and membrane potential alteration studies shows that both outer and inner membranes are destabilized in NMP exposed K. pneumoniae MGH 78578 cells. Phenotypic Microarray and RNA-seq were further used to elucidate the metabolic and transcriptional signals underpinning membrane destabilization. Membrane destabilization happens as early as 15 min post-NMP treatment. Our RNA-seq data shows that many genes involved in envelope stress response were differentially regulated in the NMP treated cells. Up-regulation of genes encoding the envelope stress response and repair systems show the distortion in membrane homeostasis during survival in an environment containing sub-inhibitory concentration of NMP. In addition, the lsr operon encoding the production of autoinducer-2 responsible for biofilm production was down-regulated resulting in reduced biofilm formation in NMP treated cells, a phenotype confirmed by crystal violet-based assays. We postulate that the early membrane disruption leads to destabilization of inner membrane potential, impairing ATP production and consequently resulting in efflux pump inhibition.
Bacterial multi-drug resistance (MDR) is a serious threat to infectious disease therapy (Van Boeckel et al., 2015). Drug resistance is now recognized by the World Health Organization as one of the most significant challenges of the 21st century (World Health Organization, 2014). Currently, 25,000 people die each year in Europe alone due to multi-drug resistant (MDR) bacterial infections costing European Union € 1.5 billion annually (Davies et al., 2013). It is therefore imperative that new technologies be developed to mitigate drug resistance in bacteria.
The mechanisms used by bacteria to combat antimicrobial agents are (1) Reduced permeability of the antimicrobial compound; (2) Increased efflux of the molecule; (3) Gene mutation dependent modification of antimicrobial targets; and (4) Direct inactivation of drug molecules (Blair et al., 2015). The genes encoding proteins responsible for various antibiotic resistance mechanisms are frequently located on mobile genetic elements (MGE) such as plasmids, transposons, integrons and are disseminated via horizontal transfer (Carraro et al., 2014; Gillings, 2014).
Chromosomally encoded efflux pumps are membrane associated, ubiquitous in bacteria and act by extruding antimicrobial compounds from the bacteria – presenting a major challenge to antibiotic therapy (Anes et al., 2015; Blair et al., 2015). Antimicrobial concentration in the cytoplasm is thereby reduced leading to the development of resistance. Using an in vitro hollow-fiber infection model, it was shown that over-expression of efflux systems facilitates bacteria to initially adapt to antibiotic exposure, thereby acting as a primary mechanism of defense. This, in turn, provides time for the bacterium to evolve a mechanized framework of high-level resistance via target gene mutations (Singh et al., 2012). It is this initial tolerance in bacteria that precedes the fully expressed antimicrobial resistance phenotype as shown recently by in vitro evolution experiments (Levin-Reisman et al., 2017).
Five families of efflux pump transporters are recognized in bacteria, including those belonging to the RND (Resistance–Nodulation–Division) superfamily (Li et al., 2015). Structurally RND pumps are very complex with an internal membrane complex spanning inner membrane, periplasmic space, and outer membrane. This tripartite structure consists of an inner membrane efflux transporter with broad substrate specificity, outer membrane channel which extrudes the substrate across the outer membrane and the periplasmic adapter protein (Blair and Piddock, 2009; Nikaido and Pages, 2012). AcrAB-TolC is the most widely studied RND system in E. coli and it is often found overexpressed in MDR E. coli pathogens (Nikaido, 2011; Li et al., 2015). Resistance to various classes of antimicrobial agents have been associated with the overexpression of AcrB (Okusu et al., 1996; Li and Nikaido, 2009). The AcrAB-TolC pump has also been implicated in virulence and biofilm formation (Hirakata et al., 2009; Martinez et al., 2009; Baugh et al., 2014). RND pumps not only extrude antimicrobial compounds but also toxic cellular by-products (Okusu et al., 1996; Huguet et al., 2013; Ruiz and Levy, 2014) highlighting their important role in mediating bacterial stress responses and pathogenicity (Anes et al., 2015; Modarresi et al., 2015; Buckner et al., 2016).
The importance of efflux pumps in antimicrobial resistance has emphasized the utility of these pumps as primary targets for novel chemotherapeutic intervention strategies. Antimicrobial compounds when used in combination with efflux pump inhibitors (EPIs), or chemosensitizers, rendered drug resistant bacterial cells susceptible by facilitating a reduction in the minimum inhibitory concentration (MIC) of a given antimicrobial compound (Pages and Amaral, 2009; Song and Wu, 2016). In the past 20 years many chemosensitizers like PAβN, NMP, D13-9001, etc. have been developed (Li et al., 2015).
1-(1-Naphthylmethyl)-piperazine (NMP) is an arylpiperazine compound which is being investigated as a chemosensitizer. It was initially identified in a screen of N-heterocyclic organic compounds with activity against E. coli (Bohnert and Kern, 2005). Ethidium Bromide accumulation studies suggested that both AcrAB and AcrEF efflux pumps are inhibited by NMP in a dose dependent manner (Bohnert and Kern, 2005). NMP binds to the Phe residue rich lower pocket of AcrB binding site (Vargiu and Nikaido, 2012). Since this binding is weaker than normal AcrAB-TolC substrates, it was suggested by molecular docking experiments that NMP might act as a substrate of the AcrAB-TolC pump. Even though NMP acts as the substrate, during the process of pumping, NMP moves from the proximal to the distal pocket of AcrAB-TolC and instead of moving out they straddle the G-loop. This G-loop straddling interferes with normal substrate movement resulting in drug resistance reversal (Li et al., 2015).
1-(1-Naphthylmethyl)-piperazine was reported to reverse MDR in Acinetobacter baumannii (Pannek et al., 2006), Vibrio cholerae (Bina et al., 2009), Staphylococcus aureus (Li et al., 2011), E. coli (Kern et al., 2006), Citrobacter freundii, Enterobacter aerogenes, and Klebsiella pneumoniae (Schumacher et al., 2006). When used as an adjuvant, NMP reversed bacterial resistance against several compounds including dyes, phenicol-based drugs, fluoroquinolones, rifampicin, linezolid, tetracyclines, and others (Bohnert and Kern, 2005; Kern et al., 2006; Schumacher et al., 2006, 2007; Li et al., 2011; Marchetti et al., 2012). Furthermore, NMP was also reported to inhibit the secretion of two virulence factors, the cholera toxin and the toxin-co-regulated pilus in Vibrio cholerae (Bina et al., 2009). Molecular docking experiments reported that NMP binds to the distal pockets of the AcrB protein and competes with the antimicrobial agent for binding space within the efflux pump (Takatsuka et al., 2010; Vargiu and Nikaido, 2012; Schuster et al., 2014). Having greater affinity, NMP binds preferentially to the AcrB distal pocket than the antimicrobial agent (Poole and Lomovskaya, 2006) enabling the drug to concentrate in the bacterial cytoplasm and exert its bactericidal effect.
Phenotypically, efflux pump inhibition mediated by NMP is well-characterized (Takatsuka et al., 2010; Vargiu and Nikaido, 2012; Schuster et al., 2014). However, the secondary stress responses induced within bacteria following NMP exposure remain uncharacterized. Also, the cascade of phenotypic and genotypic events leading to efflux pump inhibition remains unknown. Taking cue from previous studies using PAβN (phenylalanine-arginine β naphthylamide) (Lamers et al., 2013; Misra et al., 2015) and PQQ4R -(4-(2-(piperazin-1-yl) ethoxy)-2-(4-propoxyphenyl) quinolone) (Machado et al., 2017) where efflux pump inhibition was preluded by widespread bacterial membrane destabilization, we hypothesized that the same could be true for NMP as well. To test this, we subjected NMP treated MDR K. pneumoniae MGH 78578 cells to a series of phenotypic assays testing bacterial membrane viability and found that NMP introduced widespread destabilization of the bacterial membrane. To further understand the genetic mechanisms underlying the membrane de-stabilization leading to efflux pump inhibition, we conducted RNA-seq on NMP treated bacterial cells to RNA-seq and transcriptional signals underlying the process were identified.
Multi-drug resistance K. pneumoniae MGH 78578 (ATCCTM700721) is a clinical sputum isolate originally isolated in 1994 and was purchased from the American Type Culture Collection. This strain was selected for this assay mainly because it is a multi-drug resistant type strain (Ogawa et al., 2005). Recently we elucidated the resistance profile of this organism and identified that strain is resistant to at least 15 antibiotics (Anes et al., 2017). Moreover, the efflux pumps present in this strain is well-characterized (Li et al., 2008; Ogawa et al., 2012). The main advantage was that the whole genome sequence of this is available in NCBI (Refence Sequence NC_009648.1) which made it easier for us to map our RNA-seq data. This bacterium was grown in Müeller-Hinton (MHB) broth and agar (MHA) (Sigma, Dublin, Ireland). The EPI compound 1-(1-naphthylmethyl)-piperazine (NMP) (Sigma) was prepared at defined concentrations according to the manufacturers’ instructions.
1-(1-Naphthylmethyl)-piperazine mediated destabilization of the cell wall was determined according to published protocols (Misra et al., 2015) with minor modifications. Briefly K. pneumoniae MGH 78578 were grown at 37°C in MHA to 108 bacterial cells/mL, washed twice, and resuspended in PBS. NMP (250 μg/mL) was added to washed bacterial cells resuspended in PBS containing nitrocefin (100 μg/mL). The assay was carried out in a 96-well plate using two technical replicates each with 100 μL. Absorbance was recorded at 492 nm every 5 min for a total of 100 min and results converted to nitrocefin hydrolysate produced (measured in μM). Polymyxin B (2 μg/mL) (corresponding to 2x minimum bactericidal concentration) was used as a positive control for membrane disruption. Hydrolysis rates were extracted from the exponential slope of the kinetic curves and reported as nitrocefin hydrolysate produced per minute. A standard curve was prepared from the complete hydrolysis of nitrocefin by resuspending 1 mL of an overnight culture of K. pneumoniae MGH 78578 at different concentrations of nitrocefin (3.125- to 100-μM) and incubated at 37°C for 20 min. This assay was performed in duplicate.
The effect(s) of NMP on membrane integrity were evaluated using the BacLight Bacterial Membrane Potential kit (Thermo Fisher, Dublin, Ireland) following manufacturer’s instructions. First an overnight culture of K. pneumoniae MGH78578 in MHB was sub-cultured into fresh 50 mL MHB and grown at 37°C until OD600nm 0.3 was reached. The culture was then diluted 10 times in PBS containing NMP at 125- and 250-μg/mL in the presence and absence of glucose (50 mM) at room temperature. Carbonyl cyanide m-chlorophenylhydrazone (CCCP) was also added to the cells diluted in PBS to a final concentration of 100- and 50-μM in the presence and absence of glucose to serve as a depolarized (positive) control. Following the addition of the testing compounds, 10 μL of 3 mM DiOC2(3) (3,30- diethyloxacarbocyanine iodide) was added to each sample and incubated at room temperature for 20 min. Samples were run in a BD Accuri C6 flow cytometer (BD, Oxford, England) using a 488 nm laser. Emission fluorescence was detected using the green fluorescence filter 533/30 and the red fluorescence filter 670LP.
Data obtained for all samples were analyzed by using the BD Accuri C6 software V. 1.0.264.21. Unstained control samples and NMP solutions were used to locate the bacterial population, and the compound signal in the forward scatter channel, respectively (Supplementary Figure S1). Bacterial cells devoid of compound and DiOC2(3) were used to select the cells in the forward and side scatter. Depolarized samples with CCCP were used to identify the bacterial cells with an altered membrane potential. These data were reanalyzed using the FCS Express 6 Plus beta research edition (De Novo software). All results were expressed as an average ratio of red fluorescence to green fluorescence of three biological replicates. One-way ANOVA was used to compare the results against the control sample, cells with DiOC2(3).
The effect of NMP on the metabolism of K. pneumoniae MGH 78578 was evaluated using the OmniLog (Biolog, Inc., Hayward, CA, United States) phenotypic microarray platform, testing 10 microplates named PM1 through PM10 except for PM5. All plates contained defined several carbon-, nitrogen-, sulfur- and phosphorous-substrates, ions, and osmolytes at different concentrations and pH (Bochner et al., 2001). K. pneumoniae MGH 78578 was grown at 37°C on MHA. Several colonies were picked with a sterile cotton swab and suspended in 15 mL physiological saline containing DMSO or NMP at 0.5% (v/v) and 250 μg/mL, respectively, until a cell density of 42% transmittance (T) was reached in a turbidimeter (Biolog, Inc., Hayward, CA, United States). Each 15 mL suspension was then added to 75 mL of physiological saline containing dye A, along with either DMSO or NMP at the concentrations mentioned previously. PM plates 1–2 were inoculated with the previous mixture. Plates PM 3–8 were inoculated with inoculating fluid-0 (IF-0) containing sodium pyruvate as carbon source. Finally plates PM 9–10 were inoculated with the inoculating fluid-10 (IF-10). One hundred microliters of each mixture were inoculated into each well of the microplates. All PM microplates were incubated at 37°C in an OmniLog reader and monitored for 72 h. Data was analyzed using DuctApe software v 0.17.4 (Galardini et al., 2014). Each isolate was analyzed in duplicate (Supplementary Table S1; WS 1).
The effects of NMP on the growth of K. pneumoniae MGH 78578 was carried out in MHB in 250 mL conical flasks with 200 rpm at 37°C. Bacterial cells were inoculated at OD 600 nm 0.005 and allowed to grow until OD 600 nm 0.6. Here NMP and DMSO were added at 250 μg/mL and 0.5% (v/v) respectively. Bacterial growth was enumerated followed by serial dilution and plating onto MHA. Samples were further taken for RNAseq and TEM imaging at time points 15 min, 3 and 21 h post-NMP exposure.
RNA was isolated from NMP treated K. pneumoniae MGH 78578 using RNAeasy extraction kit (Qiagen, Hilden, Germany) following manufacture’s instructions. Samples of the bacterial culture were taken at 15 min and 21 h post-NMP exposure and were then combined with two volumes of RNAprotect (Qiagen, Hilden, Germany). Bacterial cultures treated with DMSO for the same two-time points served as the negative control. The NMP/DMSO treatments and RNA isolations were carried out in biological replicates amounting to a total of eight libraries. The isolated RNA was then treated with Turbo DNase (Ambion, Foster City, CA, United States) to remove any contaminating DNA. DNAse treated RNA samples were subject to integrity testing and quantification using both the Bioanalyzer 2100 RNA 6000 nanochip (Agilent Technologies, Santa Clara, CA, United States) and the ND-1000 (Nanodrop Technologies, Wilmington, DE, United States), respectively. Purified RNA samples were then preserved at −80°C.
The library preparation and subsequent sequencing were conducted commercially at the Eurofins Genomics (Ebersberg, Germany). Ribo-Zero rRNA removal Kit for Bacteria (Illumina, San Diego, CA, United States) was used to carry out the depletion of ribosomal DNA according to the manufacturer’s instructions. Libraries were created using NEBNext® UltraTM Directional RNA Library Prep Kit (New England BioLabs, Frankfurt, Germany). Pooled libraries were loaded on the cBot (Illumina, San Diego, CA, United States) and cluster generation was performed according to manufacturer’s instructions. Single-end sequencing using 125 bp read length was performed on a HiSeq2500 machine (HiSeq Control Software 2.2.38) using HiSeq Flow Cell v4 and TruSeq SBS Kit v4 (Illumina). Raw sequencing read data was processed using RTA version 1.18.61 and CASAVA 1.8.4 to generate FASTQ-files. Genomic cDNA libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit (Illumina, San Diego, CA, United States) with Ribo-Zero to deplete ribosomal RNA (rRNA). An average of 1.48 Gb of raw sequence data was obtained per sample, in 125 bp single end reads.
Sequence reads were mapped against the K. pneumoniae MGH 78578 (NC_009648) reference genome using Segemehl (Hoffmann et al., 2009) with an accuracy set to 100%. To increase mapping quality, unmapped reads were sequentially truncated by one nucleotide beginning from the 3′-end of each read and then re-mapped. This process was iterated until a given read mapped to a single location on the bacterial chromosome, or a minimal read length of 20 nucleotides was reached. Only uniquely mapped reads were used for downstream computational analysis (Supplementary Table S1; WS 2).
Computational analysis of RNA-seq data was performed using R (version 3.2.41). To calculate the expression level of genes, the raw read counts were normalized using the VOOM function (Law et al., 2014) in the limma package (Ritchie et al., 2015). More specifically, counts were converted to log2 counts per mllion (log2 CPM), quantile normalized, and precision weighted using the VOOM function. A linear model was then fitted to each gene, empirical Bayes moderated t-statistics and its corresponding P-values were used to assess differences in expression (Smyth, 2004; Law et al., 2014). To account for multiple comparisons, Benjamini–Hochberg corrected P-values were computed. As reads for duplicated coding genes (paralogs) or duplicated small RNAs cannot be mapped unequivocally, these genes appear in the analysis as unmapped. The sequence reads can be visualized in Integrated Genome Browser (version 9.0.0) (Nicol et al., 2009). The read depth was adjusted in relation to the cDNA library with the lowest number of reads (Skinner et al., 2009).
The RNA-seq data generated from this study are deposited in NCBI-GEO and are available under the accession number GSE122651.
Quantitative Reverse Transcriptase PCR (qRT-PCR) experiments were performed to validate the RNA-seq data. RNA purified from the time points described previously was converted into cDNA using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher, Dublin, Ireland). Target genes were designed with IDT PrimeTime qPCR assays that included 6-FAM/ZEN/IBFQ double-quenched probes. Primers (Supplementary Table S1; WS 4) were synthesized by Integrated DNA Technologies (IDT, Leuven, Belgium). qRT-PCR was performed using the synthesized cDNA along with the PrimeTime qPCR assays using the PrimeTime Gene Expression Master Mix in an Eppendorf Mastercycler realplex ep gradient S (Eppendorf, Arlington, United Kingdom) according to the manufacturer’s instructions. Samples were run for 3 biological replicates each with three technical replicates. Data were analyzed using realplex software. The relative fold-increases in expression levels (ΔCt) were normalized based on the gene expression levels of the housekeeping gene rpoB. Comparative quantification was carried out using the ΔΔCt approach in the control (DMSO) versus the comparator (NMP) samples.
Klebsiella pneumoniae MGH 78578 was observed in the TEM in the presence of NMP (250 μg/mL) and DMSO 5% (v/v) following incubation in MHB at 37°C. NMP/DMSO treated bacterial cells were harvested at the corresponding time points described previously, centrifuged (at 13,000 rpm, 5 min, 4°C) and washed three times with cold PBS. Samples were fixed in PBS with 2.5% (v/v) glutaraldehyde and incubated overnight at 4°C. The fixed cells were treated with Sørensen’s phosphate buffer and 1% (w/v) osmium tetroxide (TAAB Laboratories Equipment, Aldermaston, United Kingdom) for 60 min at room temperature, followed by a washing step with Sørensen buffer. Cells were then centrifuged, and the pellets recovered were dehydrated in ethanol solutions of increasing concentrations and embedded in propylene oxide for 30 min. A mixture of propylene oxide – epoxy resin (Epon 812; Tousimis Research, Corp., Rockville, MD, United States) at a ratio of 1:1 was added to the pelleted cells and incubated at room temperature for 1 h. The epoxy resin was then embedded into the pellet, which was polymerized by incubation for 2 h at 37°C. After this step, cells were subjected to centrifugation and the pellet was re-suspended in epoxy resin which was polymerized by incubation at 60°C overnight. Ultrathin sections were collected on copper grids and stained with uranyl acetate and lead citrate. Observations were made using a FEI Tecnai transmission electron microscope (Tecnai, Co., Huston, TX, United States) operating at 120 kV. Electron microscope observations were documented through the acquisition of representative microphotographs for each time point.
The effect of NMP on the efficiency of K. pneumoniae MGH 78578 to develop a biofilm was tested by growing the bacterial cells in M9 minimal media (Neidhardt et al., 1974) with NMP. Briefly, an overnight culture of K. pneumoniae MGH 78578 was sub-cultured and grown until the culture reached an OD600 nm 0.3 in M9 minimal media (NH4Cl [1.9 mM], Na2HPO4 [42.3 mM], KH2PO4 [22 mM], NaCl [8.56 mM], MgSO4 [2 mM], CaCl2 [0.1 mM], and glucose [0.1% w/v]) in the presence and absence of NMP (250 μg/mL). Diluted cells were dispensed (as 200 μL volumes) across a 96-well microtiter plate and incubated statically for 24 h at 37°C.
Crystal violet (Sigma) was applied to stain the total biomass formed (Stepanović et al., 2007). Bacterial cells were washed with PBS and stained with crystal violet 0.4% (w/v) and incubated at room temperature for 15 min. After that period, cells were washed once more and fixed with 200 μL 33% [v/v] acetic acid. Absorbance was recorded in a MultiskanTM FC Microplate Photometer (Thermo Fisher Scientific, Dublin, Ireland) at 570 nm. This assay was repeated three times for each isolate and each assay was carried out over 10 technical replicates. Results were compared against the control well-containing fresh M9 minimal media alone. Biofilm data was processed individually for each condition and comparisons between conditions were performed using Prism 6 (GraphPad) using Student’s t-test analysis. P-values of < 0.05 were considered significant.
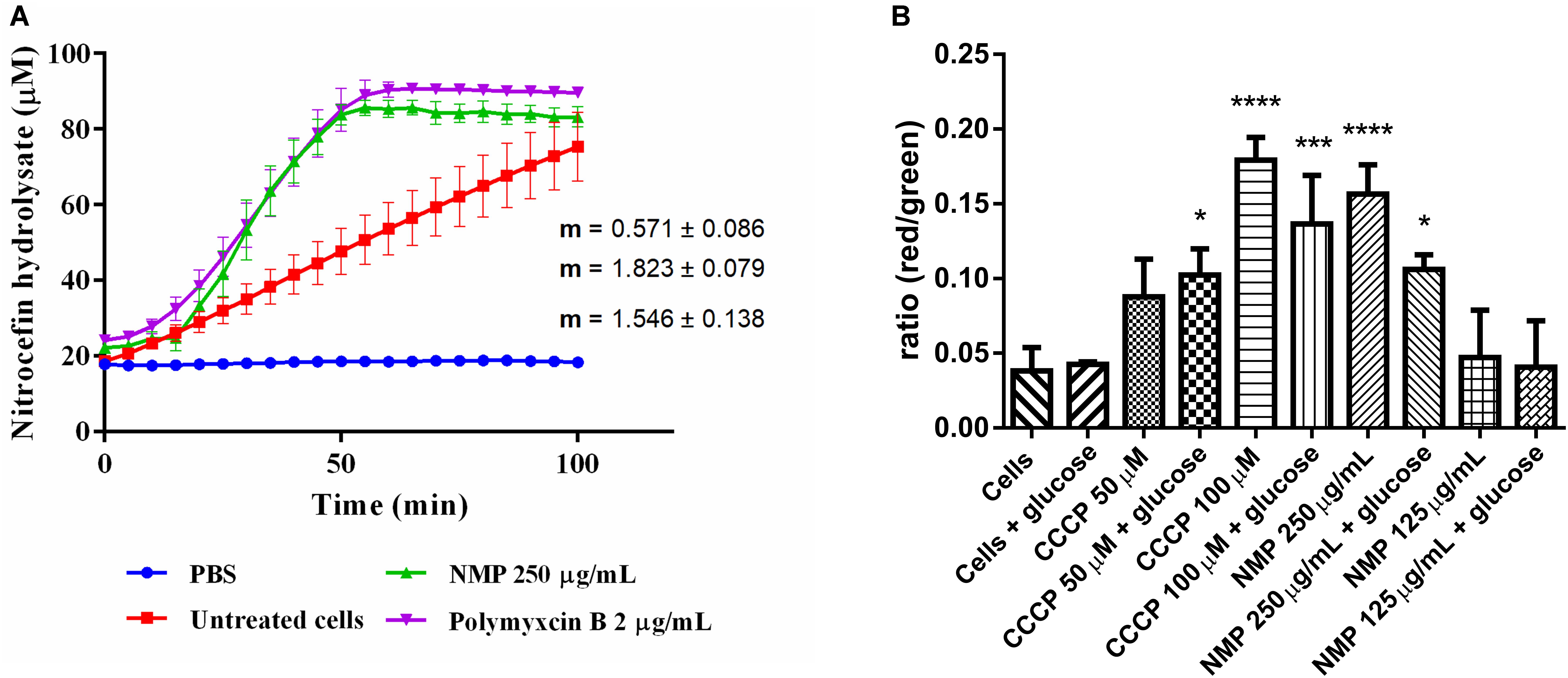
Figure 1. (A) Measurements of β-lactamase enzymatic activity in Klebsiella pneumoniae MGH 78578, in the presence of 250 μg/mL NMP. Activity was measured over 100 min at 37°C. Slope values of the activity curves represent the rate of nitrocefin hydrolysis/min. Polymyxin B was used as an internal control for cell wall disruption at a concentration of 2 μg/mL. Background absorbance values for nitrocefin, nitrocefin with NMP and nitrocefin with polymyxin B were subtracted from the corresponding samples. Slopes and standard deviations were calculated from three individual experiments with two technical replicates each. (B) Effect of NMP on the membrane potential of K. pneumoniae MGH 78578. Bacterial membrane potential was measured by flow cytometry using the Baclight kit (Invitrogen). Bacterial cells were diluted in PBS (control) and in PBS containing defined concentrations of the proton-motive force inhibitor CCCP and NMP in presence and absence of glucose. DiOC2(3) was added to all samples and fluorescence emission data collected in the green and red channels. Variations in the membrane potential cause a change in fluorescence of DiOC2(3) from green to red. Results were compared to the control sample [Cells + DiOC2(3)] by one-way ANOVA statistical test – ∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Since chemosensitizers like PQQ4R and PAβN were observed to have a membrane destabilization effect in several bacteria (Lamers et al., 2013; Misra et al., 2015; Machado et al., 2017), our first hypothesis was that NMP might have similar effects on K. pneumoniae MGH78578. Previously we had determined the MIC of NMP on multi-drug resistant K. pneumoniae MGH78578 as 500 μg/mL (Anes et al., 2018). Here, we subjected K. pneumoniae MGH78578 to a β-lactamase assay in the presence of 250 μg/mL (0.5x MIC), defined as a sub-inhibitory concentration of NMP. β-Lactamase is a periplasmic enzyme, its extracellular release correlates with disruption of the outer membrane (OM) which can be assayed by the hydrolysis of the chromogenic substrate nitrocefin (O’Callaghan et al., 1972). Polymyxin B, an antibiotic, known to disrupt the bacterial membrane (Storm et al., 1977), was used as the positive control and untreated K. pneumoniae MGH78578 cells acted as the negative control. NMP treated cells were determined to have a β-lactamase activity of 1.8 μM hydrolyzed nitrocefin/min, similar to that of polymyxin B (1.5 μM/min) (Figure 1A). This assay showed that the OM of K. pneumoniae MGH78578 was disrupted following treatment with sub-inhibitory concentrations of NMP.
Any agent that damages the cell membrane should alter membrane permeability (MP). Therefore, to further confirm the NMP-mediated disruption of the bacterial cell membrane we assayed the fluctuations in the MP during NMP treatment. Here, we used the Baclight kit (Invitrogen) to assay the MP of NMP treated K. pneumoniae MGH 78578 (Spindler et al., 2011). This kit measures the accumulation of the fluorescent dye DiO2C(3) (3,30- diethyloxacarbocyanine iodide) within a cell with intact cell membrane measured as a shift from green to red fluorescence. The DiO2C(3) accumulation within a cell with depolarized membrane is indicated by a shift in fluorescence from red to green (Spindler et al., 2011). The uncoupling agent CCCP which is known to depolarize the bacterial membrane was used as the positive control at concentrations of 50 and 100 μM.
To standardize the protocol, bacterial autofluorescence was first checked by gating unstained bacteria, followed by the gating of the bacterial population using side scatter versus forward scatter (Supplementary Figure S1). Next NMP was gated to ensure that the background signal noise from the compound did not influence the emission readings being taken. Subsequently DiOC2(3)-, NMP-treated and untreated K. pneumoniae MGH78578 were assayed for their membrane potential using the 488 nm blue light laser (Texas Red filter – LP 670 nm and Green filter – BP 530/30 nm). Treatment with NMP reduced the membrane potential to levels similar to CCCP treated cells and significantly different from untreated cells (Figure 1B). Since membrane potential is associated with inner membrane, the DiO2C(3) accumulation assay indicates that inner membrane is destabilized. However, the effect of NMP on membrane potential was concentration dependent. While at 250 μg/mL (0.5 MIC) there was a significant reduction in bacterial membrane potential, at 125 μg/mL (0.25 MIC) the reduction was negligible. Stabilization of membrane potential was noted when NMP treated cells were grown in the presence of glucose confirming an earlier observation that glucose restores membrane potential by activating ATP dependent mechanisms (Paixao et al., 2009). Results from β-lactamase and DiO2C(3) accumulation assays indicate that both outer and inner membranes of K. pneumoniae MGH 78578 cells are destabilized during NMP exposure.
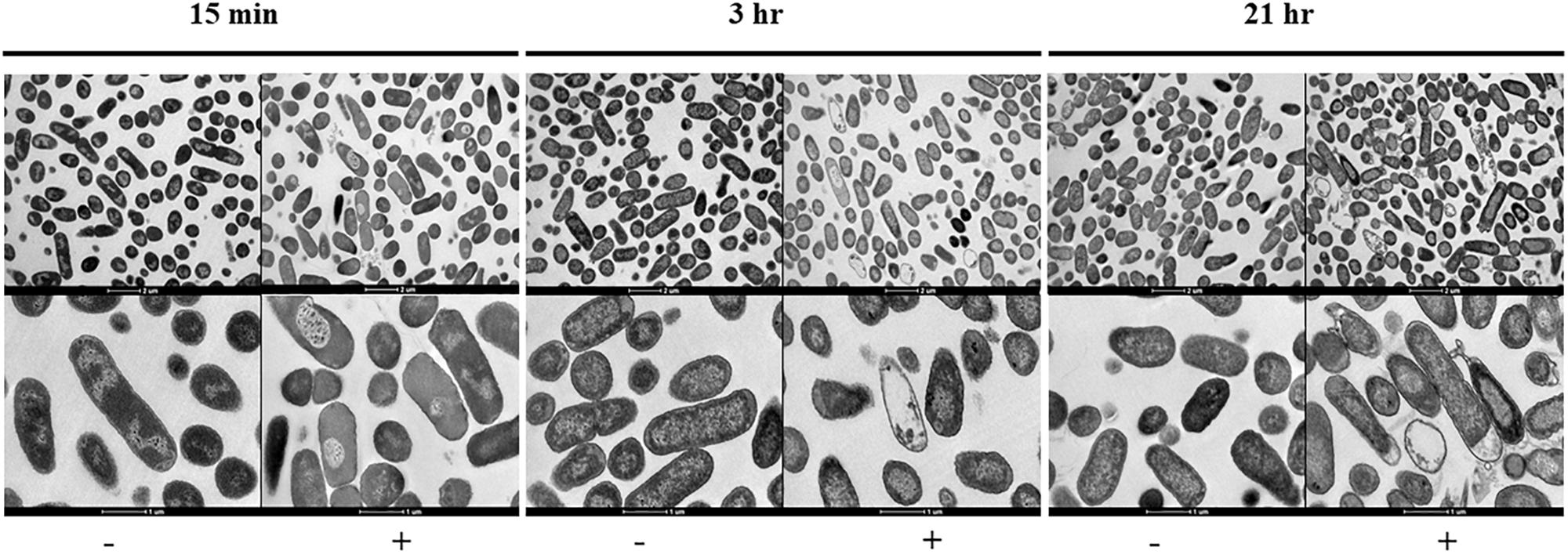
Figure 2. Transmission electron micrographs sowing the morphological changes observed on NMP treated K. pneumoniae MGH 78578. Images were collected after 15 min, 3- and 21-h of exposure to NMP. The images in the top panel were magnified 6,000X, bottom panel images shown were magnified 16,500X. The sign ‘–’ denotes DMSO and ‘+’ denotes DMSO plus NMP.
To visually interpret the effect of NMP on K. pneumoniae MGH 78578, we carried out transmission electron microscopy (TEM) on NMP and DMSO-only treated cells. K. pneumoniae MGH 78578 was exposed to 250 μg/mL NMP at three different time points – 15 min, 3- and 21-h. TEM images confirmed that for NMP treated K. pneumoniae MGH 78578, these cells underwent discernible morphological changes in their bacterial membrane (Figure 2). Cytoplasmic condensation and cell content leakage were noted following exposure to NMP as early as 15 min. Prolonged exposure exacerbated the above effects with more evident loss of cell content and coagulation of cytoplasmic material observed post 21 h NMP treatment. All these observations confirmed that NMP specifically affected cell membrane integrity.
Biochemical composition of the bacterial membrane lipids is dependent on cell growth conditions and subsequent metabolic variations (Sohlenkamp and Geiger, 2016). To understand the metabolic variations in NMP treated K. pneumoniae MGH 78578 compared to untreated cells, we assessed the ability of NMP treated cells to metabolize several substrates using a phenotypic microarray (Biolog). This assay measures the respiration of bacterial cells, by detecting colorimetric changes in the presence of a tetrazolium dye. Since DMSO is the solvent used for NMP, DMSO treated cells were used as control to elicit any possible metabolic responses.
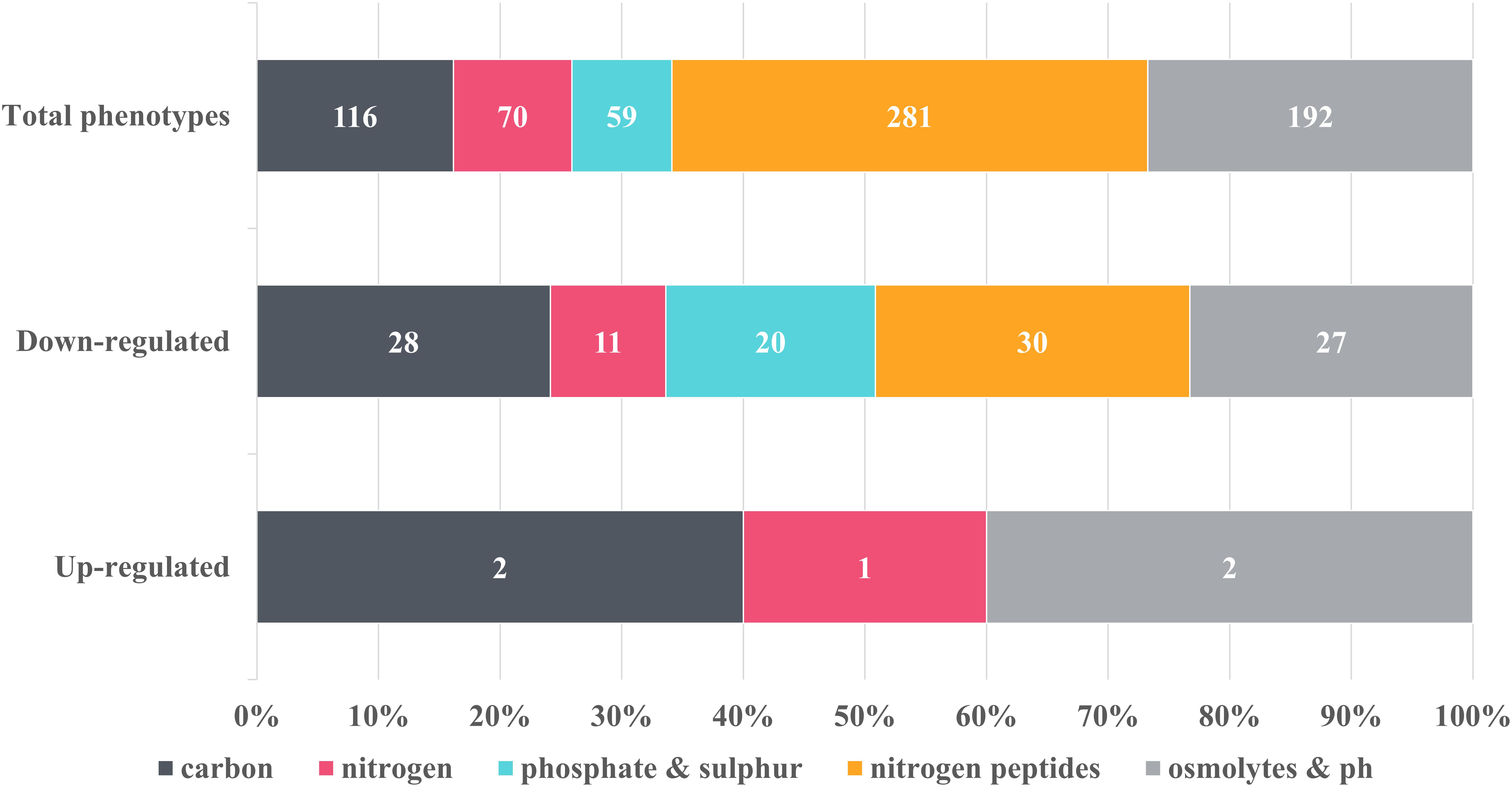
Figure 3. Overview of the metabolic changes detected in NMP-treated K. pneumoniae MGH 78578 as observed in the phenotypic microarray platform. Phenotypes were grouped into classes and the number of representatives contained within each class was indicated on the graph. Control phenotypes were assessed in the presence of DMSO, the solvent used to solubilize NMP. Raw data was analyzed using DuctApe software (v 0.17.4) and significant changes were considered for Δ activities lower and higher than –2 and 2 respectively. Five metabolic pathways were considered to be up-regulated in the presence of 250 μg/mL NMP with 116 being down-regulated.
Of the total 597 substrates measured, NMP treated K. pneumoniae MGH 78578 showed decreased respiration on 116 substrates and increased respiration on five substrates (Figure 3). Increased respiration was noted on substrates like glycerol and D-tagatose, suggesting that NMP stressed cells improved glycerol uptake similar to Saccharomyces cerevisae during hyperosmotic stress (Petelenz-Kurdziel et al., 2013). NMP treated K. pneumoniae MGH 78578 also exhibited an increase in respiration rate in sodium nitrate. Since sodium nitrate is widely used as an electron acceptor during anaerobic respiration (Stewart and Cali, 1990), increased respiration rate suggests that NMP treated cells can endure anaerobic or microaerophilic conditions better than DMSO treated control cells. Of the 116 substrates on which NMP treated K. pneumoniae MGH 78578 exhibited a decreased respiration rate, the most significant was alkaline pH. The piperazine ring of NMP has a pKa of 9.73. So, at alkaline pH the ring will be protonated on the nitrogen positions (Wishart et al., 2018) making the compound highly toxic to the bacterial cells thereby reducing the respiration rate. Overall these data showed that K. pneumoniae MGH 78578 could adapt to metabolic pathways that contributed to the bacterial recovery during NMP stress.
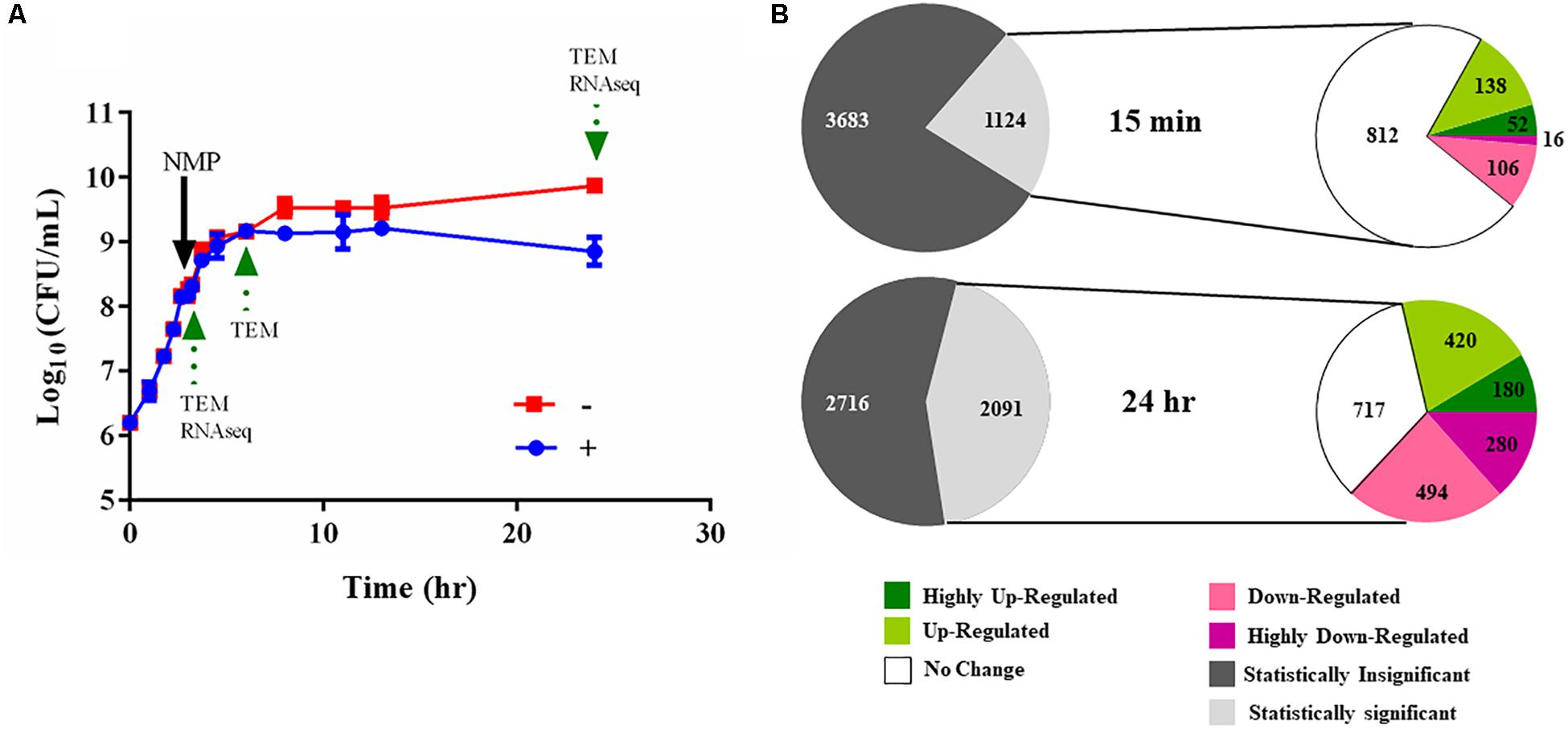
Figure 4. Experimental design of the RNA-seq experiments. (A) Typical bacterial growth curve measured at 37°C in MH broth in the presence of the 250 μg/mL NMP and the control solvent DMSO. NMP was added when OD600 nm of the culture reached mid-log phase (0.6 approximately after 3 h of growth). The sign ‘–’ denotes DMSO and ‘+’ denotes DMSO plus NMP. Samples were taken for transmission electron microscopy (TEM) and for RNA purification as indicated by green arrows at time points 15 min, 3 and 21 h following exposure. (B) Overview of the number of the differentially regulated genes identified using RNA-seq. The gray and white colored pie chart [upper] depicts the number of genes for which statistically significant data (Voom, P < 0.05) was obtained from the two time points. The second pie chart [lower] depicts the number of up- and down-regulated genes at the respective time point. The color of each pie represents the relative gene expression based on the color scale given below.
We used RNA-seq to elucidate the transcriptional landscape of NMP stressed K. pneumoniae MGH 78578. Here, total RNA was isolated from bacterial cells grown in the presence of NMP (250 μg/mL) or DMSO (5%) for 15 min and 21 h (Figure 4A). The 15-min time point was used to simulate an NMP shock condition to identify early transcriptional signals following NMP exposure while the 21-h assay was selected to identify the adaptive responses to prolonged exposure – NMP adaptive response. About 62 million reads mapped uniquely across eight libraries making an average of 7.75 million reads/library, sufficient for robust transcriptomic analysis (Haas et al., 2012). The expression levels and differential regulation of the 4,809 K. pneumoniae MGH 78578 genes during NMP stress were calculated using the Voom approach (limma package). To confirm the reproducibility of the RNA-seq data, the correlation coefficients (Spearman) were calculated for normalized read counts and between two biological replicates, correlation co-efficient was ∼0.99 (Supplementary Figure S2), indicating significant statistical correlation between replicates. Statistically significant gene expression data was obtained for 1,124 genes at 15 min post-NMP exposure (NMP shock) and 2,091 genes at 21 h post-exposure (NMP adaptive response) (Figure 4B). Of those genes detected, during the NMP shock, 190 genes were up-regulated and 122 were down-regulated (>2 fold). During the NMP adaptive response, 600 genes were up-regulated while 774 genes were down-regulated.
AcrAB-TolC is an important RND efflux system in all Gram-negative bacteria (Li et al., 2015). Genes acrAB and tolC were up-regulated (threefold) (Supplementary Table S1; WS 3) in NMP stressed K. pneumoniae MGH78578 as observed earlier in Salmonella Typhimurium (Lawler et al., 2013). Similarly, the genes marAB (AraC family transcription regulator operon) were up-regulated 3.5 fold and ramA (a transcription regulator protein) had a 9.95 fold up-regulation in NMP exposed K. pneumoniae. This NMP mediated up-regulation of genes agreed with the expression patterns previously reported in the literature confirming the robustness of our analysis. qRT-PCR was conducted on representative differentially regulated genes to validate our RNA-seq based transcriptomic data. For all tested genes, the qRT-PCR data reflected the observations from our RNA-seq data further supporting the robustness of our RNA-seq based transcriptomic analysis (Figure 5).
Important examples of up-regulated genes identified during both NMP shock and NMP adaptive responses were nhaA, degP, ramA, spy, pspABCD, pmrHFIK, mgtE, etc. – showing bacterial response to maintain homeostasis during NMP stress. For example, NhaA is an integral membrane protein which is crucial for the maintenance of cytosolic pH and Na+ concentrations in S. Typhimurium (Lentes et al., 2014). The up-regulation of mgtE is to maintain Mg2+ homeostasis as reported earlier (Hattori et al., 2009). Lipid A modification plays a key role in the resistance to factors that compromise bacterial cell membrane (Moreira et al., 2013). Many lipid A modification genes were up-regulated in NMP stressed K. pneumoniae MGH 78578. The up-regulation of eptB [Kdo(2)-lipid A phosphoethanolamine 7-transferase which adds phosphatidylethanolamine to the outer Kdo residue of Lipid A], lpxO (α-keto glutarate dependent dioxygenase enzyme that hydroxylates the secondary acyl chain of Kdo2-lipid A in the presence of O2), pmrHFIK (biosynthesis of L-Ara4N of Lipid A) (Raetz et al., 2007) etc., are responsible for an overall reduction in the negative charge on lipid A. Exogenic stress factors like heat shock, envelope stress, etc. generally result in the accumulation of toxic misfolded proteins in the bacterial periplasm, as observed in S. Typhimurium when exposed to ethanol and polymyxin B – induced the expression of the spy gene encoded chaperonic spheroplast protein y (Spy) (Jeong et al., 2017). In NMP stressed K. pneumoniae MGH 78578, continuous up-regulation of spy was noted in both the NMP shock and NMP adaptive response datasets. A similar up-regulation was noted for degP, chaperon degrading heat stress associated misfolded periplasmic proteins during heat stress, as reported previously in E. coli (Kim and Sauer, 2014). NMP exposed K. pneumoniae MGH 78578 also up-regulated periplasmic peptidylprolyl cis–trans isomerases which catalyzes the isomerization of peptidyl-prolyl bonds, a rate limiting step during protein synthesis (Obi et al., 2011). The up-regulation of cpxP shows that the protein represses CpxA inactivating the conjugative pilus system as seen in envelope stressed E. coli (Weatherspoon-Griffin et al., 2014). The phage shock protein operon (pspABCD and pspG) encoding responsive switch (Flores-Kim and Darwin, 2016) during membrane stress was also up-regulated during NMP exposure. All these transcriptomic responses show continuous efforts by the bacterium to maintain membrane homeostasis during NMP stress.
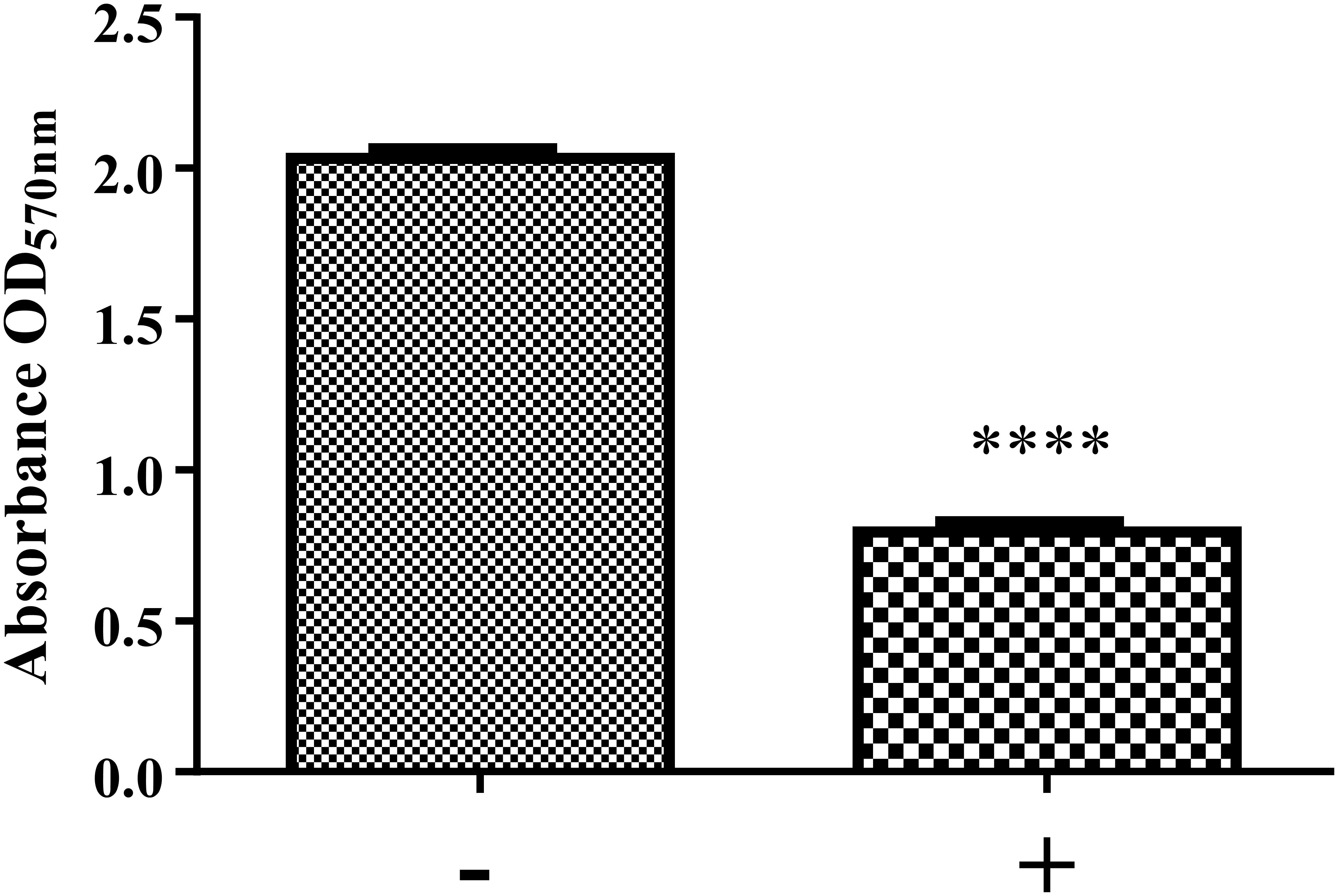
Figure 6. Biofilm formation of K. pneumoniae MGH 78578 in the presence and absence of NMP. Bacterial cells were grown statically in M9 minimal media in the presence of NMP at 250 μg/mL for 24 h at 37°C. The sign ‘–’ denotes DMSO and ‘+’ denotes DMSO plus NMP. Biofilm formation was measured by crystal violet method and compared between NMP treated and untreated cultures (∗∗∗∗P < 0.05).
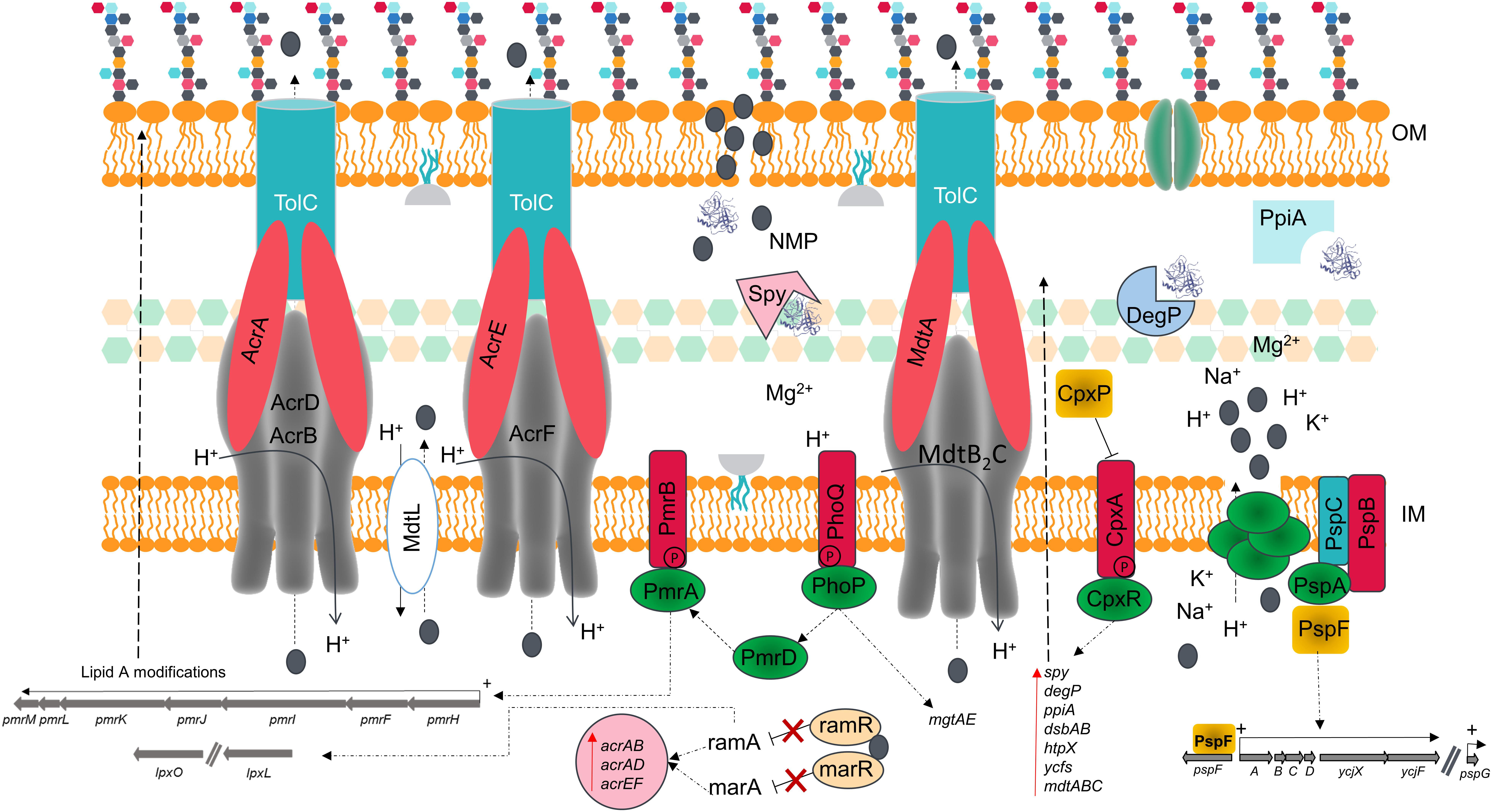
Figure 7. Schematic model summarizing the effects of NMP of the membrane of K. pneumoniae MGH 78578. The chemosensitizer NMP (indicated by the gray colored circle) crosses the cell membrane by disrupting the integrity of the lipid bilayer [both outer membrane (OM) and inner membrane (IM)]. This disruption causes the leakage of essential ions and cell content components leading to several structural and metabolic changes in a manner reminiscent of the action of similar cationic antimicrobial peptides.
Some genes were differentially regulated only during adaptive response. Important examples include rpoH (encodes σ32), phoPQ (two- component regulatory system) and pmrD. The up-regulation of rpoH had been noted in membrane stress associated K. pneumoniae (Ramos et al., 2016) and temperature stressed E. coli (Nonaka et al., 2006). Similarly, phoPQ was reported to be up-regulated when RNA-seq was used to study the transcriptional response of K. pneumoniae during exposure to colistin (Wright et al., 2015). A PhoP dependent gene pmrD [promoting lipid A modifications increasing bacterial resistance to cationic antimicrobial peptides (Rubin et al., 2015)] was also up-regulated 4.5 fold during adaptive NMP response. Genes encoding chaperone DnaK and co-chaperone DnaJ are also highly up-regulated (7 to 8 fold) during adaptive response to NMP. DnaK is known to directly interact with lipids and forms dimers with the membrane bilayer (Lopez et al., 2016). Moreover, when transcriptional response of a double deletion of dnaKJ was studied on a microarray, the expression of many inner membrane proteins were down-regulated (Fan et al., 2016). Overall many genes involved in the maintenance of the normal cell membrane were up-regulated during NMP exposure, a feature which shows that NMP mediated cell membrane destabilization causes the expected bacterial response, to maintain membrane homeostasis.
1-(1-Naphthylmethyl)-piperazine is well-known as an EPI (Kern et al., 2006; Schumacher et al., 2006; Schuster et al., 2014). However, NMP treated K. pneumoniae MGH 78578 significantly up-regulated the expression of two major AraC-transcriptional global regulator genes, ramA and marA. NMP-mediated overexpression of ramA was reported previously in S. Typhimurium using promoter-gfp fusion experiments (Lawler et al., 2013). Molecular dynamics simulation experiments show that NMP functions as a substrate for the AcrAB-TolC pump and during pumping NMP straddles the G-loop, effectively blocking the efflux pump (Li et al., 2015). This leads to increased accumulation of the antibiotic in the bacterial cytoplasm reversing the resistance in multi-drug resistant bacteria (Vargiu and Nikaido, 2012). Global regulators like RamR can bind to some substrates of the AcrAB-TolC pump (Yamasaki et al., 2013). We postulate that NMP could bind to RamR, reducing its DNA binding efficiency subsequently increasing the expression of ramA. However, our postulation that NMP could bind RamR is hypothetical and will need further experiments for confirmation.
Biofilm associated cells exhibit significantly higher tolerance/resistance to antibiotics (Hoiby et al., 2010). Increased expression of efflux pumps is one of the mechanisms that confer increased antibiotic resistance to biofilm associated cells (Gilbert et al., 2002). Therefore inactivation of efflux pumps is seen as a strategy to eradicate biofilm formation in bacteria (Baugh et al., 2014). Gene deletion based inactivation of efflux pumps showed inhibition of biofilm formation in S. Typhimurium (Baugh et al., 2014). Addition of EPIs also reduced biofilm formation but the rate of reduction varied with the type of EPI used. While Thoridazine and PAβN did significantly reduce the biofilm formation in Staphylococcus aureus and Pseudomonas putida, application of NMP did not produce significant reduction in biofilm formation in both these pathogens (Kvist et al., 2008). Interestingly, application of PAβN reduced the MBEC (minimum biofilm eradication concentration) of Ceftazidime and Doxycycline against Burkholderia pseudomallei biofilms (Sirijant et al., 2016), indicating compromised biofilm formation capability in PAβN treated bacterial cells.
The preparation and maintenance of biofilms is mainly coordinated by quorum sensing (QS), where signaling molecules like autoinducers (AIs) coordinate various bacterial behaviors in relation to bacterial cell density. Our RNA-seq data showed that the lsr operon encoding autoinducer-2 (AI-2) (Vendeville et al., 2005; Brito et al., 2013) was down-regulated during exposure to NMP (Supplementary Table S1; WS 3). This led us to hypothesize that the down-regulation of the lsr operon might reduce the production of AI-2, leading to a consequent reduction in the biofilm formation ability of NMP treated K. pneumoniae MGH78578. Our crystal violet staining analysis showed that treatment with NMP reduced the ability to form biofilms by 60% compared to untreated cells (Figure 6). When PQQ4R was used as the EPI, membrane destabilization along with impairment in the production of intracellular ATP was recorded, ultimately leading to the inhibition of efflux pump (Machado et al., 2017). In this case too membrane destabilization coupled with efflux pump inhibition could result in the inhibition of biofilm formation in NMP treated K. pneumoniae MGH78578.
The alarming increase in antimicrobial resistance has driven the need for alternative approaches to control the spread of MDR bacteria. The use of chemosensitizers like NMP may contribute to the development of a new antimicrobial therapy, by allowing the use of older antimicrobial drugs in combination with NMP-like drugs, thereby increasing their efficacy. Even though efflux pump inhibition is the main theme of NMP action, we propose that membrane destabilization is an additional effect of NMP on bacterial cells. We confirm this using β-lactamase, membrane potential fluctuation and TEM assays in K. pneumoniae MGH78578. This effect is observable as early as 15 min post-NMP treatment. Our investigations using phenotypic microarray and RNA-seq revealed that cells actively synthesize proteins required to repair and maintain membrane homeostasis. This disruption of the bacterial membrane could lead to the loss of intracellular content changing the proton gradient, confirmed by the changes in the membrane potential seen in the experiments and by microscopy. acrB is found up-regulated in the presence of NMP and NMP is found to bind to the distal pocket of AcrB (Schuster et al., 2014; Vargiu et al., 2014). The up-regulation of other efflux systems such as AcrD, AcrF, and MdtB2C could be an alternative to the blocked AcrB efflux pump. The loss of membrane homeostasis caused the activation of PhoPQ which in turn activates PmrD, a regulator of the PmrAB two-component system (Cheng et al., 2010; Rubin et al., 2015). Activation of the PmrAB system induces the up-regulation of several LPS-based modification genes via the operon pmrMLKIFH (Farizano et al., 2012). Induction of RamA can in turn induce different LPS modification enzymes (Schneiders et al., 2003). These modifications are expected to set the bacterial membrane net charge toward a predominantly positive charge and prevent further interaction of NMP with the cell wall (Figure 7). This paper therefore provides unique insights in to the effects of NMP on a bacterial cell.
JA, SF, and SS designed the experiments. JA performed the experiments. JA and SS analyzed the data. DMM performed the qRT-PCR. SKS performed the RNA-seq analysis. JA, SF, and SS wrote and revised the manuscript into which DMM, SKS commented.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZS declared a shared affiliation, with no collaboration, with one of the authors, SKS, to the handling Editor at the time of the review.
We kindly acknowledge Enterprise Ireland funded Sequencing Alliance for Food Environment (SAFE) project for funding both JA and SS. JA would also like to acknowledge the financial support through the research grant 11/F/051 provided by the Department of Agriculture, Food and the Marine (DAFM), Ireland.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00092/full#supplementary-material
FIGURE S1 | Sequential gating strategy applied to characterize K. pneumoniae MGH 78578 membrane potential shifts in the presence of the chemosensitizer NMP. The bacterial population was first identified with an unstained sample to check for autofluorescence using 633 nm laser with the filter 675/25 nm; gated samples (singlets 1) were next analyzed based on the forward scatter and gated as singlets 2. Finally, bacterial population was gated using the side scatter versus forward scatter. In order to ensure that the signal from the compounds did not affect the gating, a sample of NMP was used to exclude the signal in the Scatter gate. A stained sample with DiOC2(3) was used to assess the bacterial membrane potential using the 488 nm blue light laser using the Texas red filter (LP 670 nm) and the green filter (BP 530/30 nm). The ionophore CCCP and the chemosensitizer NMP caused a disruption/blockage on the membrane potential of K. pneumoniae resulting in a shift in the fluorescence.
FIGURE S2 | Confirmation of the reproducibility of the RNA-seq data calculated as Spearman correlation coefficients from the normalized read counts obtained from two biological replicates.
TABLE S1; WS 1 | Metabolic analysis – phenotypic Microarray analysis results on the NMP-mediated variations in K. pneumoniae metabolism.
TABLE S1; WS 2 | RNA-seq metadata – details of the reads mapping against the genome of K. pneumoniae MGH 78578.
TABLE S1; WS 3 | NMP RNA-seq – NMP based gene expression profiling of K. pneumoniae MGH 78578 during exposure to NMP.
TABLE S1; WS 4 | qRT-PCR primers – sequences of qRT-PCR primers used in this study.
Anes, J., Hurley, D., Martins, M., and Fanning, S. (2017). Exploring the genome and phenotype of multi-drug resistant Klebsiella pneumoniae of clinical origin. Front. Microbiol. 8:1913. doi: 10.3389/fmicb.2017.01913
Anes, J., Martins, M., and Fanning, S. (2018). Reversing antimicrobial resistance in multidrug-resistant Klebsiella pneumoniae of clinical origin using 1-(1-Naphthylmethyl)-Piperazine. Microb. Drug Resist. doi: 10.1089/mdr.2017.0386 [Epub ahead of print].
Anes, J., Mccusker, M. P., Fanning, S., and Martins, M. (2015). The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 6:587. doi: 10.3389/fmicb.2015.00587
Baugh, S., Phillips, C. R., Ekanayaka, A. S., Piddock, L. J., and Webber, M. A. (2014). Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J. Antimicrob. Chemother. 69, 673–681. doi: 10.1093/jac/dkt420
Bina, X. R., Philippart, J. A., and Bina, J. E. (2009). Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J. Antimicrob. Chemother. 63, 103–108. doi: 10.1093/jac/dkn466
Blair, J. M., and Piddock, L. J. (2009). Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin. Microbiol. 12, 512–519. doi: 10.1016/j.mib.2009.07.003
Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Bochner, B. R., Gadzinski, P., and Panomitros, E. (2001). Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11, 1246–1255. doi: 10.1101/gr.186501
Bohnert, J. A., and Kern, W. V. (2005). Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 49, 849–852. doi: 10.1128/AAC.49.2.849-852.2005
Brito, P. H., Rocha, E. P., Xavier, K. B., and Gordo, I. (2013). Natural genome diversity of AI-2 quorum sensing in Escherichia coli: conserved signal production but labile signal reception. Genome Biol. Evol. 5, 16–30. doi: 10.1093/gbe/evs122
Buckner, M. M., Blair, J. M., La Ragione, R. M., Newcombe, J., Dwyer, D. J., Ivens, A., et al. (2016). Beyond antimicrobial resistance: evidence for a distinct role of the acrd efflux pump in Salmonella biology. mBio 7:e1916-16. doi: 10.1128/mBio.01916-16
Carraro, N., Matteau, D., Luo, P., Rodrigue, S., and Burrus, V. (2014). The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet 10:e1004714. doi: 10.1371/journal.pgen.1004714
Cheng, H. Y., Chen, Y. F., and Peng, H. L. (2010). Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J. Biomed. Sci. 17:60. doi: 10.1186/1423-0127-17-60
Davies, S. C., Fowler, T., Watson, J., Livermore, D. M., and Walker, D. (2013). Annual report of the chief medical officer: infection and the rise of antimicrobial resistance. Lancet 381, 1606–1609. doi: 10.1016/S0140-6736(13)60604-2
Fan, D., Liu, C., Liu, L., Zhu, L., Peng, F., and Zhou, Q. (2016). Large-scale gene expression profiling reveals physiological response to deletion of chaperone dnaKJ in Escherichia coli. Microbiol. Res. 186–187, 27–36. doi: 10.1016/j.micres.2016.03.001
Farizano, J. V., Pescaretti Mde, L., Lopez, F. E., Hsu, F. F., and Delgado, M. A. (2012). The PmrAB system-inducing conditions control both lipid a remodeling and o-antigen length distribution, influencing the Salmonella Typhimurium-host interactions. J. Biol. Chem. 287, 38778–38789. doi: 10.1074/jbc.M112.397414
Flores-Kim, J., and Darwin, A. J. (2016). The phage shock protein response. Annu. Rev. Microbiol. 70, 83–101. doi: 10.1146/annurev-micro-102215-095359
Galardini, M., Mengoni, A., Biondi, E. G., Semeraro, R., Florio, A., Bazzicalupo, M., et al. (2014). DuctApe: a suite for the analysis and correlation of genomic and OmniLog phenotype microarray data. Genomics 103, 1–10. doi: 10.1016/j.ygeno.2013.11.005
Gilbert, P., Allison, D. G., and Mcbain, A. J. (2002). Biofilms in vitro and in vivo: do singular mechanisms imply cross-resistance? J. Appl. Microbiol. 92, 98S–110S. doi: 10.1046/j.1365-2672.92.5s1.5.x
Gillings, M. R. (2014). Integrons: past, present, and future. Microbiol. Mol. Biol. Rev. 78, 257–277. doi: 10.1128/MMBR.00056-13
Haas, B. J., Chin, M., Nusbaum, C., Birren, B. W., and Livny, J. (2012). How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics 13:734. doi: 10.1186/1471-2164-13-734
Hattori, M., Iwase, N., Furuya, N., Tanaka, Y., Tsukazaki, T., Ishitani, R., et al. (2009). Mg(2+)-dependent gating of bacterial MgtE channel underlies Mg(2+) homeostasis. EMBO J. 28, 3602–3612. doi: 10.1038/emboj.2009.288
Hirakata, Y., Kondo, A., Hoshino, K., Yano, H., Arai, K., Hirotani, A., et al. (2009). Efflux pump inhibitors reduce the invasiveness of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 34, 343–346. doi: 10.1016/j.ijantimicag.2009.06.007
Hoffmann, S., Otto, C., Kurtz, S., Sharma, C. M., Khaitovich, P., Vogel, J., et al. (2009). Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 5:e1000502. doi: 10.1371/journal.pcbi.1000502
Hoiby, N., Bjarnsholt, T., Givskov, M., Molin, S., and Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Huguet, A., Pensec, J., and Soumet, C. (2013). Resistance in Escherichia coli: variable contribution of efflux pumps with respect to different fluoroquinolones. J. Appl. Microbiol. 114, 1294–1299. doi: 10.1111/jam.12156
Jeong, S. M., Lee, H. J., Park, Y. M., Kim, J. S., Lee, S. D., and Bang, I. S. (2017). Inducible spy transcription acts as a sensor for envelope stress of Salmonella typhimurium. Korean J. Food Sci. Anim. Resour. 37, 134–138. doi: 10.5851/kosfa.2017.37.1.134
Kern, W. V., Steinke, P., Schumacher, A., Schuster, S., Von Baum, H., and Bohnert, J. A. (2006). Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J. Antimicrob. Chemother. 57, 339–343. doi: 10.1093/jac/dki445
Kim, S., and Sauer, R. T. (2014). Distinct regulatory mechanisms balance DegP proteolysis to maintain cellular fitness during heat stress. Genes Dev. 28, 902–911. doi: 10.1101/gad.238394.114
Kvist, M., Hancock, V., and Klemm, P. (2008). Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 74, 7376–7382. doi: 10.1128/AEM.01310-08
Lamers, R. P., Cavallari, J. F., and Burrows, L. L. (2013). The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAbetaN) permeabilizes the outer membrane of gram-negative bacteria. PLoS One 8:e60666. doi: 10.1371/journal.pone.0060666
Law, C. W., Chen, Y., Shi, W., and Smyth, G. K. (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29. doi: 10.1186/gb-2014-15-2-r29
Lawler, A. J., Ricci, V., Busby, S. J., and Piddock, L. J. (2013). Genetic inactivation of acrAB or inhibition of efflux induces expression of ramA. J. Antimicrob. Chemother. 68, 1551–1557. doi: 10.1093/jac/dkt069
Lentes, C. J., Mir, S. H., Boehm, M., Ganea, C., Fendler, K., and Hunte, C. (2014). Molecular characterization of the Na+/H+-antiporter NhaA from Salmonella Typhimurium. PLoS One 9:e101575. doi: 10.1371/journal.pone.0101575
Levin-Reisman, I., Ronin, I., Gefen, O., Braniss, I., Shoresh, N., and Balaban, N. Q. (2017). Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830. doi: 10.1126/science.aaj2191
Li, D. W., Onishi, M., Kishino, T., Matsuo, T., Ogawa, W., Kuroda, T., et al. (2008). Properties and expression of a multidrug efflux pump AcrAB-KocC from Klebsiella pneumoniae. Biol. Pharm. Bull. 31, 577–582. doi: 10.1248/bpb.31.577
Li, L., Li, Z., Guo, N., Jin, J., Du, R., Liang, J., et al. (2011). Synergistic activity of 1-(1-naphthylmethyl)-piperazine with ciprofloxacin against clinically resistant Staphylococcus aureus, as determined by different methods. Lett. Appl. Microbiol. 52, 372–378. doi: 10.1111/j.1472-765X.2011.03010.x
Li, X. Z., and Nikaido, H. (2009). Efflux-mediated drug resistance in bacteria: an update. Drugs 69, 1555–1623. doi: 10.2165/11317030-000000000-00000
Li, X. Z., Plesiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Lopez, V., Cauvi, D. M., Arispe, N., and De Maio, A. (2016). Bacterial Hsp70 (DnaK) and mammalian Hsp70 interact differently with lipid membranes. Cell Stress Chaperones 21, 609–616. doi: 10.1007/s12192-016-0685-5
Machado, D., Fernandes, L., Costa, S. S., Cannalire, R., Manfroni, G., Tabarrini, O., et al. (2017). Mode of action of the 2-phenylquinoline efflux inhibitor PQQ4R against Escherichia coli. PeerJ 5:e3168. doi: 10.7717/peerj.3168
Marchetti, M. L., Errecalde, J., and Mestorino, N. (2012). Effect of 1-(1-naphthylmethyl)-piperazine on antimicrobial agent susceptibility in multidrug-resistant isogenic and veterinary Escherichia coli field strains. J. Med. Microbiol. 61, 786–792. doi: 10.1099/jmm.0.040204-0
Martinez, J. L., Sanchez, M. B., Martinez-Solano, L., Hernandez, A., Garmendia, L., Fajardo, A., et al. (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33, 430–449. doi: 10.1111/j.1574-6976.2008.00157.x
Misra, R., Morrison, K. D., Cho, H. J., and Khuu, T. (2015). Importance of real-time assays to distinguish multidrug efflux pump-inhibiting and outer membrane-destabilizing activities in Escherichia coli. J. Bacteriol. 197, 2479–2488. doi: 10.1128/JB.02456-14
Modarresi, F., Azizi, O., Shakibaie, M. R., Motamedifar, M., Valibeigi, B., and Mansouri, S. (2015). Effect of iron on expression of efflux pump (adeABC) and quorum sensing (luxI, luxR) genes in clinical isolates of Acinetobacter baumannii. APMIS 123, 959–968. doi: 10.1111/apm.12455
Moreira, C. G., Herrera, C. M., Needham, B. D., Parker, C. T., Libby, S. J., Fang, F. C., et al. (2013). Virulence and stress-related periplasmic protein (VisP) in bacterial/host associations. Proc. Natl. Acad. Sci. U.S.A. 110, 1470–1475. doi: 10.1073/pnas.1215416110
Neidhardt, F. C., Bloch, P. L., and Smith, D. F. (1974). Culture medium for enterobacteria. J. Bacteriol. 119, 736–747.
Nicol, J. W., Helt, G. A., Blanchard, S. G. Jr., Raja, A., and Loraine, A. E. (2009). The integrated genome browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730–2731. doi: 10.1093/bioinformatics/btp472
Nikaido, H. (2011). Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 77, 1–60.
Nikaido, H., and Pages, J. M. (2012). Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 36, 340–363. doi: 10.1111/j.1574-6976.2011.00290.x
Nonaka, G., Blankschien, M., Herman, C., Gross, C. A., and Rhodius, V. A. (2006). Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20, 1776–1789. doi: 10.1101/gad.1428206
Obi, I. R., Nordfelth, R., and Francis, M. S. (2011). Varying dependency of periplasmic peptidylprolyl cis-trans isomerases in promoting Yersinia pseudotuberculosis stress tolerance and pathogenicity. Biochem. J. 439, 321–332. doi: 10.1042/BJ20110767
O’Callaghan, C. H., Morris, A., Kirby, S. M., and Shingler, A. H. (1972). Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1, 283–288. doi: 10.1128/AAC.1.4.283
Ogawa, W., Li, D. W., Yu, P., Begum, A., Mizushima, T., Kuroda, T., et al. (2005). Multidrug resistance in Klebsiella pneumoniae MGH78578 and cloning of genes responsible for the resistance. Biol. Pharm. Bull. 28, 1505–1508. doi: 10.1248/bpb.28.1505
Ogawa, W., Onishi, M., Ni, R., Tsuchiya, T., and Kuroda, T. (2012). Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene 498, 177–182. doi: 10.1016/j.gene.2012.02.008
Okusu, H., Ma, D., and Nikaido, H. (1996). AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178, 306–308. doi: 10.1128/jb.178.1.306-308.1996
Pages, J. M., and Amaral, L. (2009). Mechanisms of drug efflux and strategies to combat them: challenging the efflux pump of Gram-negative bacteria. Biochim. Biophys. Acta 1794, 826–833. doi: 10.1016/j.bbapap.2008.12.011
Paixao, L., Rodrigues, L., Couto, I., Martins, M., Fernandes, P., De Carvalho, C. C., et al. (2009). Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 3:18. doi: 10.1186/1754-1611-3-18
Pannek, S., Higgins, P. G., Steinke, P., Jonas, D., Akova, M., Bohnert, J. A., et al. (2006). Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide. J. Antimicrob. Chemother. 57, 970–974. doi: 10.1093/jac/dkl081
Petelenz-Kurdziel, E., Kuehn, C., Nordlander, B., Klein, D., Hong, K. K., Jacobson, T., et al. (2013). Quantitative analysis of glycerol accumulation, glycolysis and growth under hyper osmotic stress. PLoS Comput. Biol. 9:e1003084. doi: 10.1371/journal.pcbi.1003084
Poole, K., and Lomovskaya, O. (2006). Can efflux inhibitors really counter resistance? Drug Discov. Today Ther. Strateg. 3, 145–152. doi: 10.1016/j.ddstr.2006.05.005
Raetz, C. R., Reynolds, C. M., Trent, M. S., and Bishop, R. E. (2007). Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329. doi: 10.1146/annurev.biochem.76.010307.145803
Ramos, P. I., Custodio, M. G., Quispe Saji, G. D., Cardoso, T., Da Silva, G. L., Braun, G., et al. (2016). The polymyxin B-induced transcriptomic response of a clinical, multidrug-resistant Klebsiella pneumoniae involves multiple regulatory elements and intracellular targets. BMC Genomics 17:737. doi: 10.1186/s12864-016-3070-y
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Rubin, E. J., Herrera, C. M., Crofts, A. A., and Trent, M. S. (2015). PmrD is required for modifications to Escherichia coli endotoxin that promote antimicrobial resistance. Antimicrob. Agents Chemother. 59, 2051–2061. doi: 10.1128/AAC.05052-14
Ruiz, C., and Levy, S. B. (2014). Regulation of acrAB expression by cellular metabolites in Escherichia coli. J. Antimicrob. Chemother. 69, 390–399. doi: 10.1093/jac/dkt352
Schneiders, T., Amyes, S. G., and Levy, S. B. (2003). Role of AcrR and ramA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob. Agents Chemother. 47, 2831–2837. doi: 10.1128/AAC.47.9.2831-2837.2003
Schumacher, A., Steinke, P., Bohnert, J. A., Akova, M., Jonas, D., and Kern, W. V. (2006). Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J. Antimicrob. Chemother. 57, 344–348. doi: 10.1093/jac/dki446
Schumacher, A., Trittler, R., Bohnert, J. A., Kummerer, K., Pages, J. M., and Kern, W. V. (2007). Intracellular accumulation of linezolid in Escherichia coli, Citrobacter freundii and Enterobacter aerogenes: role of enhanced efflux pump activity and inactivation. J. Antimicrob. Chemother. 59, 1261–1264. doi: 10.1093/jac/dkl380
Schuster, S., Kohler, S., Buck, A., Dambacher, C., Konig, A., Bohnert, J. A., et al. (2014). Random mutagenesis of the multidrug transporter AcrB from Escherichia coli for identification of putative target residues of efflux pump inhibitors. Antimicrob. Agents Chemother. 58, 6870–6878. doi: 10.1128/AAC.03775-14
Singh, R., Swick, M. C., Ledesma, K. R., Yang, Z., Hu, M., Zechiedrich, L., et al. (2012). Temporal interplay between efflux pumps and target mutations in development of antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 56, 1680–1685. doi: 10.1128/AAC.05693-11
Sirijant, N., Sermswan, R. W., and Wongratanacheewin, S. (2016). Burkholderia pseudomallei resistance to antibiotics in biofilm-induced conditions is related to efflux pumps. J. Med. Microbiol. 65, 1296–1306. doi: 10.1099/jmm.0.000358
Skinner, M. E., Uzilov, A. V., Stein, L. D., Mungall, C. J., and Holmes, I. H. (2009). JBrowse: a next-generation genome browser. Genome Res. 19, 1630–1638. doi: 10.1101/gr.094607.109
Smyth, G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:3. doi: 10.2202/1544-6115.1027
Sohlenkamp, C., and Geiger, O. (2016). Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159. doi: 10.1093/femsre/fuv008
Song, L., and Wu, X. (2016). Development of efflux pump inhibitors in antituberculosis therapy. Int. J. Antimicrob. Agents 47, 421–429. doi: 10.1016/j.ijantimicag.2016.04.007
Spindler, E. C., Hale, J. D., Giddings, T. H. Jr., Hancock, R. E., and Gill, R. T. (2011). Deciphering the mode of action of the synthetic antimicrobial peptide Bac8c. Antimicrob. Agents Chemother. 55, 1706–1716. doi: 10.1128/AAC.01053-10
Stepanović, S., Vuković, D., Hola, V., Di Bonaventura, G., Djukić, S., Ćirković, I., et al. (2007). Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115, 891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x
Stewart, V., and Cali, B. M. (1990). Genetic evidence that NarL function is not required for nitrate regulation of nitrate assimilation in Klebsiella pneumoniae M5al. J. Bacteriol. 172, 4482–4488. doi: 10.1128/jb.172.8.4482-4488.1990
Storm, D. R., Rosenthal, K. S., and Swanson, P. E. (1977). Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46, 723–763. doi: 10.1146/annurev.bi.46.070177.003451
Takatsuka, Y., Chen, C., and Nikaido, H. (2010). Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 6559–6565. doi: 10.1073/pnas.1001460107
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U.S.A. 112, 5649–5654. doi: 10.1073/pnas.1503141112
Vargiu, A. V., and Nikaido, H. (2012). Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 109, 20637–20642. doi: 10.1073/pnas.1218348109
Vargiu, A. V., Ruggerone, P., Opperman, T. J., Nguyen, S. T., and Nikaido, H. (2014). Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob. Agents Chemother. 58, 6224–6234. doi: 10.1128/AAC.03283-14
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M., and Hardie, K. R. (2005). Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3, 383–396. doi: 10.1038/nrmicro1146
Weatherspoon-Griffin, N., Yang, D., Kong, W., Hua, Z., and Shi, Y. (2014). The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J. Biol. Chem. 289, 32571–32582. doi: 10.1074/jbc.M114.565762
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082. doi: 10.1093/nar/gkx1037
World Health Organization (2014). Antimicrobial Resistance Global Report on surveillance. Geneva: World Health Organization.
Wright, M. S., Suzuki, Y., Jones, M. B., Marshall, S. H., Rudin, S. D., Van Duin, D., et al. (2015). Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. doi: 10.1128/AAC.04037-14
Keywords: Klebsiella pneumoniae, chemosensitizers, 1-(1-naphthylmethyl)-piperazine, membrane destabilization, NMP, secondary effects
Citation: Anes J, Sivasankaran SK, Muthappa DM, Fanning S and Srikumar S (2019) Exposure to Sub-inhibitory Concentrations of the Chemosensitizer 1-(1-Naphthylmethyl)-Piperazine Creates Membrane Destabilization in Multi-Drug Resistant Klebsiella pneumoniae. Front. Microbiol. 10:92. doi: 10.3389/fmicb.2019.00092
Received: 27 September 2018; Accepted: 16 January 2019;
Published: 13 February 2019.
Edited by:
Yuji Morita, Meiji Pharmaceutical University, JapanReviewed by:
Zhangqi Shen, China Agricultural University, ChinaCopyright © 2019 Anes, Sivasankaran, Muthappa, Fanning and Srikumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Séamus Fanning, c2Zhbm5pbmdAdWNkLmll Shabarinath Srikumar, c3Jpa3VtYXIuc2hhYmFyaW5hdGhAdWNkLmll
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.