
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
GENERAL COMMENTARY article
Front. Microbiol., 01 February 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00059
This article is a commentary on:
Study the Features of 57 Confirmed CRISPR Loci in 38 Strains of Staphylococcus aureus
A Commentary on
Study the Features of 57 Confirmed CRISPR Loci in 38 Strains of Staphylococcus aureus
by Zhao, X., Yu, Z., and Xu, Z. (2018). Front. Microbiol. 9:1591. doi: 10.3389/fmicb.2018.01591
As mentioned in the article by Xihong Zhao, Zhixue Yu and Zhenbo Xu, bioinformatics analysis provides data support for bacterial typing, traceability analysis, and exploration of clustered regularly interspaced short palindromic repeats (CRISPR) (Zhao et al., 2018). However, there are relatively few studies on the CRISPR of Staphylococcus aureus. For example, Yang et al. (2015) analyzed the features of 45 identified CRISPR loci in 32 S. aureus. Zhang and Ye (2017) studied the CRISPR like elements in S. aureus. These articles have contributed greatly to our understanding of the distribution, architecture, function, and evolution of S. aureus CRISPR. At present, the research about the characteristics of CRISPR has made some headway. For instance, based on the specificity of virulence genes and CRISPR, Liu et al. (2011) have developed a sequence typing scheme (designated CRISPR–MVLST) to subtype Salmonella enterica isolated from different sources, Shariat et al. (2013) combined the method with multiple site sequence typing to distinguish the outbreak isolates during pathogen outbreaks. Furthermore, with the development of CRISPR-mediated genome editing and engineering, these CRISPR-based tools will have major implications for both basic and applied research (Galizi and Jaramillo, 2018).
In Zhao et al. paper, they described data sources as “the different S. aureus strain genomes were searched by the National Center for Biotechnology Information (NCBI) nucleotide database (http://www.ncbi.nlm.nih.gov/) with default parameters; then S. aureus CRISPR loci were searched by the CRISPR Finder server (E-value ≤ 0.001) (http://crispr.i2bc.paris-saclay.fr/Server/) (Last updated on May 9, 2017).” They found 22 strains of S. aureus contained one CRISPR locus, 14 strains of S. aureus contained 2 CRISPR loci, and the other 2 strains contained 3 and 4 CRISPR loci. We downloaded the genomes of 38 strains from NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/) according to the GenBank ID they provided and then searched the CRISPR loci by CRISPR Finder. However, we found different results from their article description. As shown in Figure 1, only 5 strains were detected carrying the confirmed CRISPR locus, three of which contained one CRISPR locus, and the other 2 strains contained 2 and 3 CRISPR loci. Especially, the last strain, MSHR1132, is not a strain of S. aureus, but Staphylococcus argenteus. S. argenteus was given status as a separate species distinct from S. aureus in 2015 (Tong et al., 2015). According to the confirmed CRISPR locus, Cas proteins were searched by CRISPRCas Finder. We found all 4 strains of S. aureus contained Cas3 and only S. aureus 08BA02176 contained diverse Cas proteins that belongs to subtype III-A.
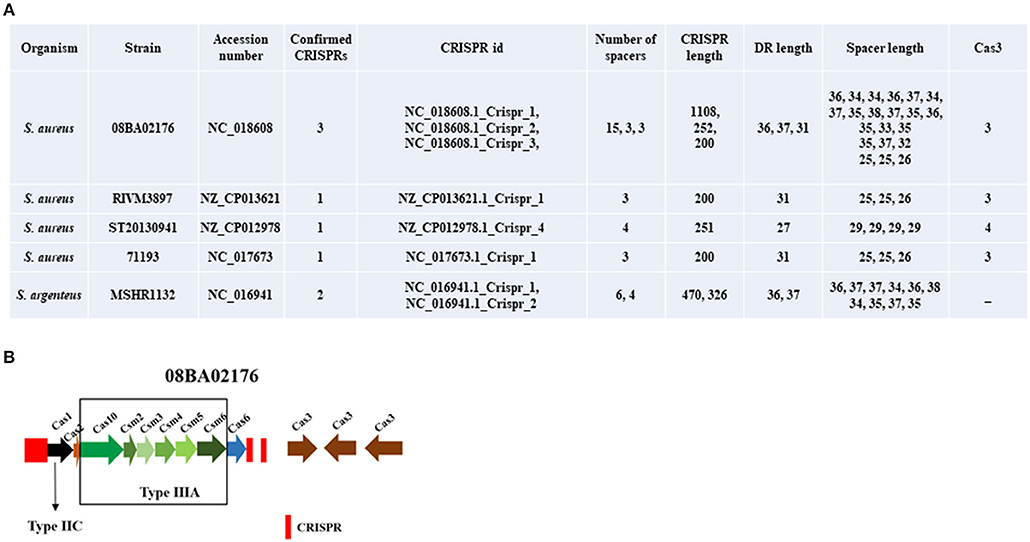
Figure 1. The features of confirmed CRISPR loci. (A) The statistical table of 8 CRISPR loci of strains. (B) The distribution of CRISPR and cas genes in S. aureus 08BA02176.
We found that the confirmed CRISPR loci searched by CRISPR Finder according to genomes from NCBI are different from the information in CRISPR database, although the CRISPR Finder and the CRISPR database are connected to each other. So, we should be cautious of the data sources when we use online database for analysis.
In summary, bioinformatics of CRISPR study would provide new ideas on the phylogenetic distribution and potential role of CRISPR-Cas systems in shaping the S. aureus accessory genome and antibiotic resistance elements.
TM and HY designed the study. TM and JL analyzed data and wrote the paper. GD offered some good advice. All authors read and approved the final manuscript.
This study received financial support from the Henan Province University Science and Technology Innovation Talent Projects (17HASTIT045).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Galizi, R., and Jaramillo, A. (2018). Engineering CRISPR guide RNA riboswitches for in vivo applications Roberto Galizi1 and Alfonso Jaramillo2,3,4. Curr. Opin. Biotechnol. 55, 103–113. doi: 10.1016/j.copbio.2018.08.007
Liu, F., Kariyawasam, S., Jayarao, B. M., Barrangou, R., Gerner-Smidt, P., Ribot, E. M., et al. (2011). Subtyping Salmonella enterica Serovar Enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs). Appl. Environ. Microb. 77, 4520–4526. doi: 10.1128/AEM.00468-11
Shariat, N., Kirchner, M. K., Sandt, C. H., Trees, E., Barrangou, R., and Dudley, E. G. (2013). Subtyping of Salmonella enterica Serovar newport outbreak isolates by CRISPR-MVLST and determination of the relationship between CRISPR-MVLST and PFGE results. J. Clin. Microbiol. 51, 2328–2336. doi: 10.1128/JCM.00608-13
Tong, S. Y. C., Schaumburg, F., Ellington, M. J., Corander, J., Pichon, B., Leendertz, F., et al. (2015). Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int. J. Syst. Evol. Micr. 65, 15–22. doi: 10.1099/ijs.0.062752-0
Yang, S., Liu, J., Shao, F., Wang, P., Duan, G., and Yang, H. (2015). Analysis of the features of 45 identified CRISPR loci in 32 Staphylococcus aureus. Biochem. Bioph. Res. Co. 464, 894–900. doi: 10.1016/j.bbrc.2015.07.062
Zhang, Q., and Ye, Y. (2017). Not all predicted CRISPR–Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinformatics 18:4. doi: 10.1186/s12859-017-1512-4
Keywords: CRISPR, Staphylococcus aureus, Staphylococcus argenteus, bioinformatics, data sources
Citation: Mao T, Long J, Duan G and Yang H (2019) Commentary: Study the Features of 57 Confirmed CRISPR Loci in 38 Strains of Staphylococcus aureus. Front. Microbiol. 10:59. doi: 10.3389/fmicb.2019.00059
Received: 23 October 2018; Accepted: 14 January 2019;
Published: 01 February 2019.
Edited by:
Teresa Zotta, Italian National Research Council (CNR), ItalyReviewed by:
Mario Andrea Marchisio, Tianjin University, ChinaCopyright © 2019 Mao, Long, Duan and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyan Yang, eWh5QHp6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.