- 1Department of Microbiology and Cell Biology, Indian Institute of Science, Bengaluru, India
- 2Division of Biological Sciences, Indian Institute of Science, Bengaluru, India
- 3Undergraduate Studies, Indian Institute of Science, Bengaluru, India
- 4Centre for Biosystems Science and Engineering, Indian Institute of Science, Bengaluru, India
Enteropathogenic bacteria have been the cause of the majority of foodborne illnesses. Much of the research has been focused on elucidating the mechanisms by which these pathogens evade the host immune system. One of the ways in which they achieve the successful establishment of a niche in the gut microenvironment and survive is by a chain of elegantly regulated gene expression patterns. Studies have shown that this process is very elaborate and is also regulated by several factors. Pathogens like, enteropathogenic Escherichia coli (EPEC), Salmonella Typhimurium, Shigella flexneri, Yersinia sp. have been seen to employ various regulated gene expression strategies. These include toxin-antitoxin systems, quorum sensing systems, expression controlled by nucleoid-associated proteins (NAPs), several regulons and operons specific to these pathogens. In the following review, we have tried to discuss the common gene regulatory systems of enteropathogenic bacteria as well as pathogen-specific regulatory mechanisms.
Introduction
Bacteria with their supposedly primitive genomic architecture, have proven to be one of the smartest living things to have existed. Their success of survival depends on a chain of elegantly regulated gene expression patterns. This regulation becomes all the more important in the case of pathogens, which have to cleverly sabotage the host immune responses. For successful entry and survival in the host of an enteric pathogen, these bacteria have to cross the acidic pH of the stomach, find a way to cross the barrier of the existing intestinal microbiota, escape from the cationic anti-microbial peptides and immunoglobulins in the intestine. After breaching all of the above-mentioned barriers and entering a cell, survival becomes harder with the acidification of phagosomes, burst of reactive oxygen and reactive nitrogen species, iron deprivation by the cells, and autophagy. Enteropathogenic bacteria have been the cause of most of the foodborne illnesses. Diseases ranging from diarrhea to systemic fever like typhoid caused by the Salmonella Typhi to life-threatening sepsis are caused by these pathogens. In 2015, the World Health Organization (WHO) has reported an estimate of 582 million cases of 22 different foodborne enteric diseases and 351,000 associated deaths. Among these, 520,00 deaths were because of Salmonella Typhi and 370,00 deaths because of enteropathogenic Escherichia coli (EPEC). The disease burden is very high in Africa followed by southeastern Asia. Forty percent of the disease is reported in children under 5 years of age.
Pathogenesis of bacteria like EPEC and Salmonella Typhi have been fairly well characterized. However, with the emergence of multiple drug-resistant strains and the acquisition of many virulence genes by the bacteria with the horizontal gene transfer (HGT), control of disease progression has been difficult. Thorough understanding of the components dictating the life of these bacteria, inside and outside of the host is thus, vital. The following review discusses the gene regulation of some of the major enteric pathogens and some of the common themes of gene regulation in these bacteria.
Gene Regulatory Systems
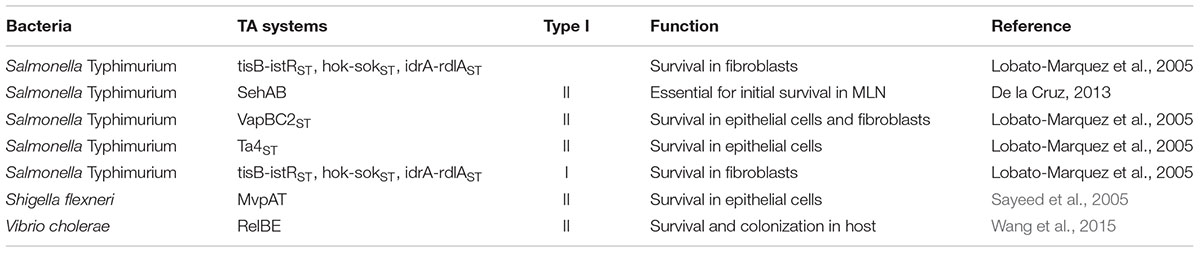
Regulation of Gene Expression by Toxin- Antitoxin (TA) Systems
The success of the pathogenic bacteria has been heavily reliant on the retention of virulence factors in mobile genetic elements (Cambray et al., 2010) like pathogenicity islands (PAI), virulence plasmids and conjugative integrons. The existence of such TA cascades require the continuous generation of the antitoxin molecule for their survival and thus help in the retention of the plasmid. There have been ever increasing roles of these TA modules emerging in bacterial pathogenesis.
Toxins are generally protein molecules and antitoxins are protein/RNA molecules that sequester the toxins. Induction of TA modules occur with the RNase or protease mediated antitoxin degradation of antitoxin molecule. This results in the release of toxin that kills the bacteria. TA modules have been classified into six types, depending on the mechanism by which the antitoxin inhibits the toxin activity (Page and Peti, 2016). Type I consists of the TA systems where the antitoxin is the antisense RNA that binds to the toxin mRNA and this heteroduplex formation inhibits translation of the toxin. Type II consists of the TA systems where antitoxin is also a protein molecule which forms a tight complex with the toxin, hiding its active site, thus resulting in inhibition. Type III consists of the TA systems where the antitoxin is a small RNA molecule that directly interacts with the toxin and inhibits activity. Type IV has TA systems in which the antitoxin protein molecule directly binds to the target of the toxin, thus inhibiting the toxin activity. TA systems in which the antitoxin molecule acts as a ribonuclease specific to the toxin mRNA constitute Type V and Type VI has the TA systems wherein the antitoxin brings about the proteolytic degradation of the toxin. Enteropathogenic bacteria have acquired many of these TA systems which reside in the virulence plasmids and horizontally acquired PAIs. Following are some of the TA systems existing in enteropathogenic bacteria which aid in their survival.
Vibrio cholerae have seven relBE Type II TA systems which are induced upon stress conditions experienced by the bacteria in the host. RelE is the toxin then cleaves mRNA leading to the inhibition of growth whose activity is inhibited in the presence of RelB antitoxin. Deletion of this TA system has been shown to hamper the ability of the pathogen to both survive and colonize in the host (Wang et al., 2015). Out of all the enteropathogenic bacteria, Salmonella species have the most number of well characterized TA systems. It possesses six putative Type I TA modules and seventeen Type II TA systems (Lobato-Marquez et al., 2015). One such Type II TA module present in one of the virulence plasmids, VapBC2ST helps the survival of Salmonella Typhimurium inside the host fibroblasts and epithelial cells (Lobato-Marquez et al., 2015). VapC being the toxin acts as an RNAse that cleaves the mRNA transcript independent of the ribosomes. Since the activation is cell type specific, activity of this particular TA system might be to dictate the outcome of the pathogenesis. This is highly homologous to mvpAT TA system located on the virulence plasmid of Shigella flexneri which is essential for the maintenance of the plasmid (Sayeed et al., 2005). Recent reports have also shown that positioning this TA system near to the 30 kb PAI leads to both global and local gene regulation. This also leads to the loss of expression of the Type III secretion system by the plasmid. Thus, altering the ability of the pathogen to invade (Pilla et al., 2017).
Another Type II TA system, SehAB is found to be important during the initial phases of infection of Salmonella. Especially in the survival of the pathogen in the mesenteric lymph nodes. This was demonstrated with the comparison of survival of a wild-type STM strain and a strain lacking SehAB. The mutant strains exhibited decreased virulence as compared to the wild-type when administered through the oral route whereas the virulence was not hindered when administered through intra-peritoneal route (De la Cruz et al., 2013). SehA, the toxin is the homolog of HigB, which is a ribosome-dependent endoribonuclease. In conditions that are known to increase the expression of genes involved in the intracellular proliferation, SehAB transcription is found to increase. Although SehAB transcription is found to be increased upon infection of the macrophages, it is not required for the replication. It is also not required for replication in bone marrow-derived macrophages or in epithelial cells like HeLa (Lobato-Marquez et al., 2015).
Toxin RelE was found to be important in the virulence as mutants that lacked RelE like toxin showed reduced virulence as compared to the wild-type. RelE toxin either directly or indirectly cleaves the mRNAs associated with the ribosomes. Toxins such as T4ST, T5ST, T2ST are found to be differentially expressed in fibroblasts as compared to the epithelial cells. Transcription of at least one of the three type I TA systems, namely, tisB-istRST, hok-sokST, idrA-rdlAST and two of the type II TAs, ta4ST and vapBC2ST, are found to be indispensable for the survival of STM inside fibroblasts (Lobato-Marquez et al., 2015). It is also observed that the type I and the type II TA systems differentially modulate survival in fibroblasts and epithelial cells. Thus, the existence of extrachromosomal genetic elements, harboring TA systems, play vital roles in the virulence of the enteropathogenic bacteria (Table 1).
Type IV TA system yeeUV in E. coli inhibits cell division. The antitoxin molecule YeeU helps in the polymerization of MerB and FtsZ by allowing them to bundle and thus protecting them from the action of the toxin YeeV (Masuda et al., 2012). The only reported Type V TA system is the ghoST system. GhoT is the toxin whose mRNA ids cleaved by the antitoxin GhoS. GhoT disrupts the membrane potential and thus, ATP synthesis (Wang et al., 2012). Interestingly, the ghoST TA system is recently found to be regulated by another TA system, mqsRA (Wang et al., 2013). MqsR toxin cleaves the ghoS transcript and thus, there is also cross regulation found among the existing TA systems.
The number of TA systems characterized to be exhibiting specific functions is miniscule when compared to the number of TA systems reported. Research devoted to characterization and a better understanding of these TA systems would open a wide arena for research wherein the TA systems can be employed to trick bacteria to negatively regulate its own virulence genes, as was done with the mvpAT of Shigella.
The induction of certain TA systems is also quorum dependent. One such chromosomal Type II TA system found in E. coli is mazEF which is regulated in a quorum dependent manner. The induction is brought about by the degradation of the antitoxin MazE by the Lon protease or other proteases depending on the stage of growth of the bacteria (Aizenman et al., 1996; Christensen et al., 2003). In the following section, we will see a more detailed quorum dependent gene regulatory systems.
Regulation of Gene Expression by Quorum Sensing
Quorum sensing (QS) represents the ability of bacteria to sense the population density in the surrounding environment. The way it does is through cell-to-cell signaling mechanism, by producing and/or responding to a chemical signal called autoinducer (AI). Gene expression is regulated depending on the surrounding concentration of AI. More than 45 years ago this phenomenon was discovered in the regulation of bioluminescence of Vibrio harveyi (Nealson et al., 1970) and later in Vibrio fischeri (Hastings and Nealson, 1977). One such diverse environment for bacterial crosstalk and signaling between beneficial microbiota, invading pathogen and the mammalian host is the large intestinal milieu. Inside intestinal milieu, beneficial microbiota helps the host in nutrient assimilation and immune competence (Hooper and Gordon, 2001). Pathogenic or opportunistic organisms can also occasionally establish their niche and thereby manifest a disease condition. QS is crucial because of the high density of microbes in the large intestine where interspecies, intraspecies and cross-kingdom coordination must be established. The enteric pathogens use mainly three types of QS systems: Lux R-I, LuxS/AI-2 and AI-3/epinephrine/norepinephrine. A thorough review of the kind of QS systems present and utilization of the same by pathogenic as well as commensal bacteria has been provided here. It will also shed some light on the kind of approach one can take to limit food borne pathogens disease manifestations.
Lux R-I
The regulation of bioluminescence in Vibrio sp. is through Lux R-I system. There are two regulatory proteins: Lux I, which is responsible for the production of N-acyl-homoserine-lactone (AHL) AI and Lux R, which get activated by AI thereby increasing transcription of the luciferase operon (Engebrecht et al., 1983; Engebrecht and Silverman, 1984). A similar regulation system has been discovered in gut-pathogen bacteria where homologs of LuxR-I operate and only difference lies in the downstream target gene regulation (de Kievit and Iglewski, 2000). Yersinia species also have LuxI and LuxR homologs including Yersinia enterocolitica which causes yersiniosis harbors only one pair of LuxRI (YenRI) in contrast to other species of Yersinia which harbor two pairs. yenI is involved in the synthesis of two short chains AHLs, along with three long-chain AHLs (Atkinson et al., 2006). One of the initial reports on Yersinia enterocolitica suggests that LuxR-I QS is involved in regulating the expression of the flagellar structural gene, fleB which in turn governs swimming motility (Atkinson et al., 2006). There are certain hypervirulent strain harboring a putative orphan luxR gene which is not linked to AHL synthase, ycoR. This QS system has been shown to regulate host cell attachment along with virulent plasmid pYve maintenance (Ng et al., 2018). Y. enterocolitica utilizes the type III secretion system (T3SS) to deliver multiple Yop effector proteins. It is regulated by Ca2+ ion concentration and temperature thereby maintain a tight control of phenotype (Cornelis et al., 1998; Cornelis, 2002; Trosky et al., 2008; Dewoody et al., 2013). This could probably improve the infectivity of the pathogen in terms of energy conservation and quick switch from not expressing virulent genes to expressing the same. Y. pseudotuberculosis and Y. pestis have convergently expressed two pairs of luxI/R orthologs. All the four major AHLs are produced by both the species.
On the other hand, E. coli, Salmonella and Shigella lack luxI gene, therefore, can’t synthesize AHLs. However, these organisms can respond to AHLs produced by other bacteria, with a receptor-like protein (SdiA). SdiA has an amino acid sequence homology with LuxR -type transcriptional factor (Yao et al., 2006). Yao et al. (2006) shows that the presence of several AHLs compounds is required for proper folding of SdiA. SdiA in E. coli K12 and EHEC was involved in cell division, increased quinolone resistance (Rahmati et al., 2002) and expression of virulence gene but only when SdiA is expressed in a high copy number plasmid. Chromosomally expressed SdiA regulate four genes involved in glutamate-dependent acid resistance and downregulate fliE in E. coli k12 and EHEC. Dyszel et al. (2010b) have thus concluded that gene regulation by SdiA in E. coli is only partially dependent on AHLs. The regulation through SdiA is only observed at 30°C also acid tolerance in E. coli is conferred by the same (Van Houdt et al., 2006). In E. coli, AHLs regulate adhesion to epithelial cells (Hep-2), biofilm formation (Lee et al., 2009) and acid tolerance (Van Houdt et al., 2006; Sharma and Bearson, 2013).
Inceptive studies on Salmonella have shown that SdiA was involved in regulation of exclusively one gene rck which functions to impart resistance against human complement. The SdiA in Salmonella is induced at low pH (pH 4) during aerobic conditions. Although SdiA mutant, can survive in Luria-Bertani medium minimum for a week at pH 4 (Rychlik and Barrow, 2005). The SdiA of Salmonella Typhimurium was activated during transit through the gastrointestinal tract of turtle colonized with Aeromonas hydrophila, capable of AHL synthesis (Smith et al., 2008). Also, report from Dyszel et al. (2010a)shows that Salmonella Typhimurium SdiA was activated during the transit through mice gut colonized by Yersinia enterocolitica, which is also capable of synthesis of AHLs. In the same study, they have shown that cloning luxI of Yersinia into Salmonella gave a benefit of fitness to the pathogen in mice model (Dyszel et al., 2010a), these studies suggest that even though Salmonella lacks AHL producing capabilities it can very well respond to it via orphan receptors such as SdiA. This will not only help Salmonella to crosstalk but could be one of the mechanisms to establish co-infection.
In Salmonella, the AHLs also regulate survival in rabbit and guinea pig serum apart from anchoring and invasion in HeLa and HEp-2 cells respectively (Nesse et al., 2011; Liu et al., 2014; Campos-Galvao et al., 2016). During micro-aerobiosis and anaerobiosis conditions the gene expression through LuxS and SdiA is also reduced (Lamas et al., 2016). Therefore, it is evident that entero-pathogens regulate their virulence factors at transcriptional level itself via LuxRI systems. These virulence factors are majorly associated with establishment of infection. This often serves them with an upper hand in a competitive environment such as intestinal milieu.
LuxS/AI-2
Most of the sequenced bacterial genomes have luxS QS system (Waters and Bassler, 2005) and hence it is a universal signaling molecule which serves the purpose of interspecies communication. Subsequently, it was found that the enzyme luxS which is responsible for AI-2 synthesis is extremely conserved and globally expressed among a variety of bacterial species (Han and Lu, 2009; Pereira et al., 2013; Even-Tov et al., 2016) Primarily it was identified in V. harveyi for regulation of bioluminescence (Surette et al., 1999). The key enzyme in this sensing is LuxS which has a role in S-adenosyl methionine metabolism. LuxS catalyzes the reaction of S-ribosyl-homocysteine into homocysteine and highly unstable 4,5- Dihydroxy 2,3 pentanedione, which cyclizes to form furanones (Schauder et al., 2001; Winzer et al., 2002a; Sperandio et al., 2003), serving as a precursor of AI-2 compound(Schauder et al., 2001). AI-2 is imported inside the cell by specific receptor transport system present on bacterial membrane. Several previous works have identified various AI-2 receptors in diverse bacterial species. LuxP was discovered in V. harveyi, LsrB in Samonella Typhimurium, and RbsB in Haemophilus influenzae (Armbruster et al., 2011). The LuxS/AI-2 type of QS system has been found to be involved in the regulation of bacterial bioluminescence, competence, biofilm formation, antibiotic resistance (Xue et al., 2013), sporulation, and virulence factor secretion (Novotny et al., 2015), type three secretion system (Han et al., 2013, 2015a,b). In Salmonella, it was initially reported that AI-2 only regulated gene expression of ABC transporter named Lsr (LuxS regulated). AI-2 is a transcriptional activator of lsr operon which consists of seven genes (lsrACDBFGE) (Taga et al., 2001). The Lsr transporter was also reported in E. coli and in both bacteria have homology with sugar transporter. Once AI-2 is inside the cell it is further phosphorylated thereby interacts with LsrR which is a SorC-like transcription factor that represses lsr operon (Taga et al., 2001; Taga et al., 2003). The homology between lsr transporter and sugar importer lead to suggesting that the function of AI-2 is basically metabolic (Winzer et al., 2002b, 2003; Vendeville et al., 2005) The AI-2 import is tightly regulated and remains shut off in presence of glucose because of repression mediated by cAMP-catabolite activator protein (CAP) (Wang et al., 2005; Xavier and Bassler, 2005). Microarray based study has shown that luxS mutation leads to multiple metabolic changes, especially in processes involving nitrogen and carbon metabolism (Walters et al., 2006). AI-2 mediated signaling has also been shown to modulate the gut microbiota to facilitate colonization of a Firmicutes while hindering the Bacteroidetes during antibiotic mediated dysbiosis (Thompson et al., 2015). It is also reported for identification of AI-2 receptor in recombinant E. coli background. AI-2 interacts with LsrB in a temperature dependent manner (Zhang et al., 2017). Recent work on S. Dublin also shows that AI-2 is involved in biofilm formation and antibiotic resistance (Ju et al., 2018). Even though the genetic regulation via AI-2 system majorly involves metabolic control and interspecies crosstalk, there is some host protective role of commensals via AI-2 signaling to maintain the quorum of enteric microbiota. Therefore, AI-2 system can be said as a two-way sword which helps the host and can be also utilized by pathogens to suppress the inhibition by commensals to establish successful infection in host.
AI-3/Cross-Kingdom Quorum Sensing
The third and most diverse type QS involves with AI-3 compound which facilitates cross-kingdom signaling between interspecies and inter-kingdom (host–microbes). Mammalian host produces a variety of hormones to maintain it’s signaling these include peptide and steroidal hormones. These hormones are then sensed by microflora residing in our system and serves as a cue for modulating their gene expression. The AI-3 compound is different from AI-2 and the former is capable of binding C-18 column and elution is possible only with methanol (Sperandio et al., 2003). The AI-3 is involved in quorum-sensing mechanisms to regulate the genes associated with EHEC LEE, flagellar and motility (Sperandio et al., 1999, 2002). AI-3 is produced by commensal organisms of gut microflora or by the host (epinephrine/norepinephrine). Mass spectroscopy analysis has shown that there are structural similarities between AI-3 and host epinephrine/norepinephrine, therefore, it is hypothesized as an antagonist of adrenergic receptors would also block QS through AI-3(Sperandio et al., 2003). During EHEC infection, the host adrenaline/ noradrenaline (NA) can’t cross the cellular membrane, thus it employs a two-component systems (TCSs) QseBC and QseEF to carry the signal inside bacteria. The two-component system has a membrane-bound histidine kinase which senses the signal and phosphorylates downstream cytoplasmic response regulator (RR) that results in the regulation of target genes (Jung et al., 2012). In the case of EHEC, genes regulated by QseC via adrenaline and NA are most often associated with flagella (Clarke and Sperandio, 2005b), motility (Reading et al., 2009), stress responses, ion uptake, osmolarity regulation, attaching and effacing lesions formation on enterocytes (Nakashima et al., 1992; Sperandio et al., 2002; Clarke and Sperandio, 2005a; Reading et al., 2007; Hughes et al., 2009; Njoroge et al., 2012). QseC is indispensable for EHEC virulence, and deletion leads to attenuation in rabbit and bovine models. QseEF regulates the gene which is involved in Shiga toxins synthesis, SOS responses, and other TCSs, which includes RcsBC, PhoPQ (Reading et al., 2009, 2010; Njoroge and Sperandio, 2012). Similarly, other hosts signaling molecules are also sensed by gut microflora which includes peptide hormones such as gastrin (Chowers et al., 1999), EGF, natriuretic peptides (Veron et al., 2007; Blier et al., 2011) and opioid hormones (Beury-Cirou et al., 2013).
Apart from these host-microbe interaction bacteria have also ability to sense host active defense/immune components such as cytokines (Porat et al., 1991; Luo et al., 1993; Zav’yalov et al., 1995; Wu et al., 2005), antimicrobial peptides (Miller et al., 1989; Bader et al., 2005) along with various host membrane component such as phosphatidylethanolamine (Bertin et al., 2011; Thiennimitr et al., 2011; Kendall et al., 2012; Luzader et al., 2013; Gonyar and Kendall, 2014), sugars (Pacheco et al., 2012) and host antioxidants, glutathione (Reniere et al., 2015; Kendall and Sperandio, 2016). Sensing the host intestinal environment and counter responding to the same is one of the preliminary strategies to evade host immune system and establishing the infection. AI-3 QS being the major crosstalk between host–pathogen rather helps the pathogen to comprehend the host anti-pathogenic retaliations.
There are several other enteric bacterial genera such as Shigella, Salmonella, Klebsiella, Enterobacter, and Citrobacter which produces the AI-3 compound and are used for inter-species communication (Walters et al., 2006). In enteric bacteria genera Shigella, Salmonella, and Yersinia it has been reported to have amino acid sequence conservation for QseC sensors (Clarke et al., 2006). Combining all the type of QS system we have tried to summarize in schematic (Figure 1). Targeting these molecules could be one of the smart ways for future therapeutics for food and water-borne diseases.
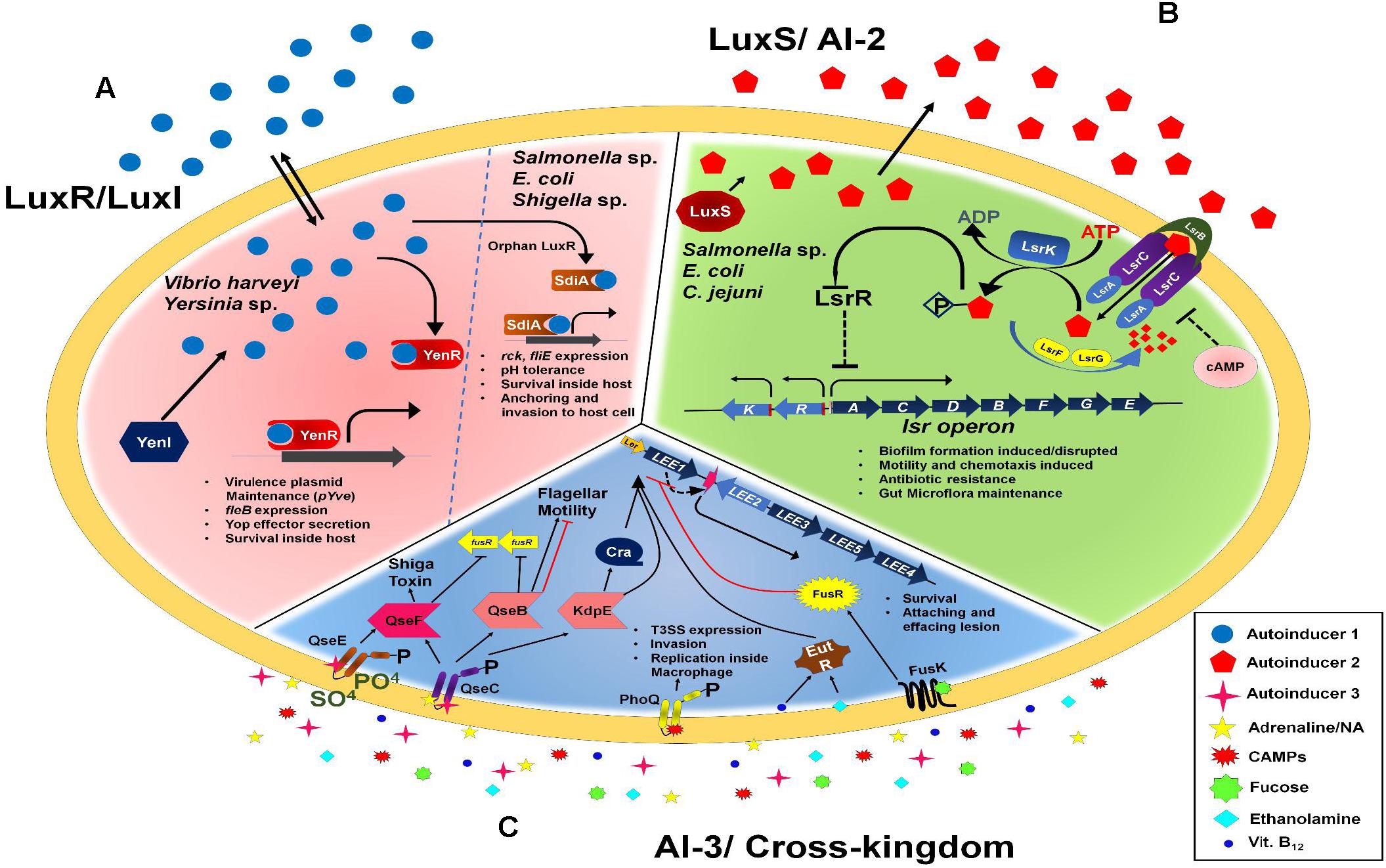
Figure 1. Quorum sensing in an overview, (A) describes the AI-1 mediated Quorum sensing and its downstream target gene regulation, (B) describes the AI-2 mediated gene regulation through lsr operon, (C) Cross-kingdom communication to facilitate the bacteria to sense the cues from the host and thereby establish successful infection.
Regulation of Gene Expression by Nucleoid-Associated Proteins (NAPs)
Nucleoid-associated proteins are proteins in non-eukaryotes involved in the compact storage of genetic material. They are responsible for altering the direction of DNA in the nucleoid, i.e., the configuration of the DNA in three-dimensional space by bending, wrapping and bridging it. They are known to regulate gene expression at the transcriptional-level (Dillon and Dorman, 2010).
Actively growing bacteria are probably shown more stable looped chromosomal domains in line with higher transcriptional activity such as synthesis of rRNA and other structures involved in translation (Postow et al., 2004; Deng et al., 2005; Stein et al., 2005). NAPs such as H-NS and factor of inversion (Fis), are found to bind to genomes (Grainger et al., 2006; Lucchini et al., 2006; Navarre et al., 2006; Noom et al., 2007). The binding locations of H-NS are believed to function as chromosomal domain loop boundaries (Noom et al., 2007). Proteins that bridge DNA are conjectured to be the best candidates for the formation of domain loop boundaries (Dillon and Dorman, 2010). Interestingly, nucleoid structure is found to be maintained even in the absence of Fis and H-NS among other NAPs (Zimmerman, 2006).
DNA-protein-DNA bridges are believed to be capable of either enabling or disabling the RNA polymerase to bind to DNA (Dame et al., 2002; Grainger et al., 2006). StpA and Sfh are H-NS paralogs and can form heteromers with H-NS (Dorman, 2004). The change in the numbers of the multimeric NAPs is shown to change gene expression patterns (Muller et al., 2006). Haemolysin expression modulating (Hha)/YdgT protein’s interaction with H-NS influences gene regulation activity of H-NS (Madrid et al., 2007). H-NS also plays a key role in the regulation of certain virulence genes in response to temperature (Rowe et al., 2000).
HU is another NAP. It has two sub-units, Huα and Huβ. In E. coli HU exists as either heterodimer or homodimer according to the stage of growth (Claret and Rouviere-Yaniv, 1997). HU influences the expression of a broad spectrum of genes in E. coli which includes genes involved in central metabolism and respiration (Oberto et al., 2009). It interacts with topoisomerase I and induces changes in the super-helicity of the DNA, thereby influencing gene expression (Broyles and Pettijohn, 1986). In several instances, the activity of promoters can be increased or decreased in response to super-helical variation (Dorman, 2006). HU also induces DNA bending which enables the formation of DNA loops which influences gene regulation and at the same time H-NS has the opposite effect (Becker et al., 2007).
Unlike HU, IHF binds to a well-conserved nucleotide sequence eventually causing the DNA to take a U-turn centered at the binding site of the protein to the DNA (Swinger and Rice, 2004; Dillon and Dorman, 2010). In E. coli and its relatives, IHF consists of an alpha subunit and a beta subunit. The alpha-beta heterodimer is the predominant active form, but it has been shown that the alpha–alpha and beta–beta homodimers are also biologically active in Salmonella enterica subspecies enterica serovar Typhimurium (Mangan et al., 2006). IHF has an influence on the global transcription in E. coli (Arfin et al., 2000) and Salmonella Typhimurium (Mangan et al., 2006). IHF recruits σ54-containing RNA polymerase to promoters (Macchi et al., 2003). The ilvPG promoter of the ilvGMEDA operon of E. coli is activated by IHF through the release of free energy that is normally stored in the IHF-binding site, making this energy available to assist with the formation of the open transcription complex at the promoter (Sheridan et al., 1998).
Besides the mentioned NAPs there are several more with functions involved in the structuring of the DNA, gene expression and DNA replication. Being directly associated with the DNA give these set of proteins a direct advantage and a major chance of being involved in the regulation of gene expression.
Now that we have summarized some of the broad gene regulatory mechanisms that the enteropathogenic bacteria employ, we will discuss some of the pathogen specific mechanisms.
Enteric Pathogens
Gene Regulation in Escherichia coli
Escherichia coli is the predominant commensal found to be residing in the mucous layer of the mammalian colon. The bacteria and the host, by large, co-exist with mutual benefits. The pathology associated with these bacteria is the result of the immunocompromised state of the host where conditions favor the establishment of the disease. Among the pathotypes reported, the following have been well characterized, Enteropathogenic E. coli (EPEC), Enterohemorrhagic E. coli (EHEC), Enteroaggregative E. coli (EAEC), Enteroinvasive E. coli (EIEC), and Diffusely adherent E. coli (DAEC). Also, urinary tract infections are the most common extra-intestinal E. coli infection caused by the uropathopathogenic E. coli (UPEC).
The genes responsible for the attaching and effacing (A/E) phenotype which is the primary histopathological characteristic of EPEC infections reside in a locus called the locus of enterocyte effacement (LEE) located on a 35 kb PAI (McDaniel et al., 1995). These PAIs are typically associated with tRNA genes and possess a different GC content as compared to the genomic DNA of the bacteria. Integration of these happens at selC tRNA genes which have proven to be the “hot-spots for mobile gene insertions. They are also adjacent to other virulence genes, most of which code for the structural and effector proteins of the type three secretion system (T3SS) (Tauschek et al., 2002). There are five major polycistronic regions designated as LEE 1-5 comprising of 41 open reading frames (ORFs) (Kenny et al., 1997) LEE1, LEE2, LEE3 encode for T3SS apparatus that span the inner and outer membranes (Yerushalmi et al., 2014), they also code for the outer membrane porin, EscC and an ATPase called EscN. LEE4 encodes a necessary protein EspA (Knutton et al., 1998) which forms the filament structure over the secretion needle upon polymerization that is necessary for injection of effectors into the host cell, EspB and EspD which aid in pore formation in the host cell membrane (Ide et al., 2001) and EspF which is injected into the host cell that targets the mitochondria triggering the activation of cell death pathways (McNamara et al., 2001). EspF also causes redistribution of an important tight junction protein called the occludin, which results in the loss of trans-epithelial membrane resistance. LEE5 codes for intimin and Tir proteins and also CesT which is the Tir chaperone (Elliott et al., 1999).
The first open reading frame, LEE1 encodes for the LEE-encoded regulator (Ler) which counteracts the inhibitory effect of H-NS which is a DNA binding protein that inhibits transcription of LEE ORFs (Winardhi et al., 2014). At a temperature of 27°C, LEE1 is repressed whereas it gets activated at 37°C (Rosenshine et al., 1996). Ler protein shares amino acid sequence similarity with the H-NS (majorly the C terminal domain which binds to the DNA). Ler activates the transcription of LEE2-5 (Sperandio et al., 2000), map, the gene product is known to target mitochondria of the host cell and espC (Stein et al., 1996). EspC is encoded by a second PAI of EPEC which is translocated through pinocytosis and causes disruption of host cell cytoskeleton. Since Ler regulates the transcription of espC that does not reside in the LEE locus, it is termed as the global regulator of virulence gene expression in EPEC. In EHEC, the stcE gene on the plasmid pO157 is also regulated by Ler (Grys et al., 2005). Although it is shown that ler mutants are severely attenuated in their ability to form A/E lesions (Zhu et al., 2006), it is observed that ler is necessary only in the initial stages of infection and later represses the transcription of LEE1 (Zhu et al., 2006). Other generic E. coli proteins, such as, Fis, integration host factor (IHC) and BipA positively regulate LEE1 expression whereas Hha, which is a regulator of α-hemolysin directly binds to LEE1 region and negatively regulates ler expression (Hong et al., 2010).
Other regulatory systems like RcsC-RcsD-RcsB phosphorelay system and EHEC specific GrvA protein is known to control the expression of ler (Tobe et al., 2005). Transcription of the ler encoding LEE1 operon is found to be enhanced with the global response to starvation, entry into the stationary growth phase and is included in any form of stringent response requiring relA and spoT (Nakanishi et al., 2006).
Transcription of LEE operons, however, is not under the sole control of Ler. In EHEC, two novel regulators, YhiF and YhiE are known to regulate the secretion of EspB, EspD and Tir proteins (Tatsuno et al., 2003) which shows they regulate the transcription of LEE2 and LEE4 operons. These proteins share sequence similarity with the luxR family of proteins (Tatsuno et al., 2003). Also, in EHEC, two genes in the cryptic T3SS of the Sakai 813 strain, namely, etrA or eivF led to the increased secretion of EspA, EspB, Tir and also pO157 encoded StcE and EspP (Zhang et al., 2004). Also, mitomycin C treatment is also known to induce expression of LEE2 and LEE3 in recA and lexA dependent manner (Mellies et al., 2007). LexA was found to bind to the SOS box located in between the overlapping promoter regions of LEE2 and LEE3.
The Ler transcription itself is regulated by the action of proteins encoded by the plasmid-encoded regulator (Per) locus found on a plasmid called the EPEC adherence factor (EAF) plasmid which is 70–100 kb in size and is a characteristic of typical EPEC strains (Bieber et al., 1998). The Per locus on the EAF plasmid has three regions, namely PerA, PerB and PerC (Porter et al., 2004). PerA controls the bundle Forming Pilus (BFP) expression that also helps in adhesion. PerA has amino acid sequence similarity to AraC family of proteins, which is also closely related to VirF protein of Shigella (Dorman and Porter, 1998). PerA also exhibits autoregulation (Ibarra et al., 2003). BFP expression is also regulated by the Cpx two-component system (TCS). This TCS comprising of the sensor kinase CpxA and response regulator CpxR is responsive to lethal damage to the cells. BFP is not assembled until this TCS is activated (Nevesinjac and Raivio, 2005). BFP also exhibits maximum expression at 37°C (Puente et al., 1996). bfpA gene is also under NH4+ ion regulation. Increased NH4+ is found to repress the expression of this gene (Puente et al., 1996). PerC regulates Ler expression by directly binding to the region upstream to the LEE1 leading to transcriptional repression (Kimmitt et al., 2000). In acidic conditions, the Per genes are repressed by the action of GadX which also activates GadAB genes that bring about resistance to acidic environment (Tramonti et al., 2002). This acts as a key step in regulating the virulence gene expression as the expression can only be activated once the bacteria pass through the acidic environment of the stomach and reach the alkaline environment of the small intestine.
Another protein that acts as an adhesive is a large 385 kDa protein called lymphostatin (LifA) that is encoded by the region adjacent to PAI (outside of LEE). This protein inhibits lymphocyte activation. Apart from these, the porcine A/E associated gene (paa) and Long Polar Fimbriae (LPF) also aid in adhesion. Once the expression of these virulence genes has been activated, the release of effector proteins by the T3SS into the host cells leads to the disease manifestation.
Till date, many pathotypes of E. coli have been identified and the list keeps growing. Additionally, with the abuse of antibiotics in the recent past, the emergence of drug-resistant bacteria is also on the rise. Thus, there is a need to better characterize the virulence gene expressions and their triggers before the number of pathotypes become overwhelming.
Gene Regulation in Salmonella
Salmonella is a Gram-negative rod-shaped pathogen, bearing peritrichous motility and has been known to cause a pathogenicity in a wide array of hosts spanning from cold-blooded animals to humans. In humans, it causes various diseases ranging from occasionally fatal systemic fever to self-limiting diarrhea. Regardless of current medical advancement, Salmonella remains a major cause of morbidity and mortality in developing countries. Salmonella Typhimurium has a plethora of genes that are strictly regulated and thereby facilitates adaptation in various environments. There are at least 23 Salmonella pathogenicity islands (SPI, which are the molecular toolbox for Salmonella pathogenicity) in SPI pan-genome (Fookes et al., 2011; Hayward et al., 2013). In response to environmental signals such as temperature, redox state, pH, osmolarity, antimicrobial peptides and nutrient deprivation Salmonella achieves adaptation via well-coordinated gene regulation. The pathogenicity is caused by two steps encoded by two different T3SS, SPI-1 and SPI-2. The SPI-1 encoded T3SS acts during initial contact with the host cells, it then injects bacterial effector protein into the host cell and mediates uptake in non-phagocytic cells. To survive inside the phagosome/vacuole it then expresses SPI-2 T3SS and its effectors are then injected into the host cell. Major regulation of the gene in the bacterial domain is via transcription regulation. One prominent example is five alternative sigma factors designated σE (σ24), σF (σ28), σH (σ32), σS (σ38), and σN (σ54) which are activated differentially under stress or changes in growth conditions (Gruber and Gross, 2003). Upon general stress such as starvation and environmental stress response, σS upregulates approx. 500 genes (Hengge, 2009). The SPIs are AT-rich regions of the genome encoding genes required to induced phagocytosis in non-phagocytic cells (Lostroh and Lee, 2001; Patel and Galan, 2005; Fink and Cookson, 2007), negative regulator of SPI-1 is hilE, which has three promoters, controlled by Mlc repressor (response to glucose) (Lim et al., 2007), FimZ activator also reported to regulate Salmonella attachment and adherence (Baxter and Jones, 2005). Also, a key regulatory factor for SPI-1 regulation is HilD whose expression is in control of std fimbrial operon gene via a DNA adenine methylation pathway (Lopez-Garrido and Casadesus, 2012). In a recent report, it has been shown that in Salmonella the Hfq dependent sRNA Spot 42 is transcriptionally regulated by the CRP-cAMP regulatory circuit. In the same study, it was shown the upon relieving repression of Spot 42 is up-regulated hilD by binding to 3′UTR unstructured regions of hilD transcripts (El Mouali et al., 2018). Two-component systems also regulate Salmonella response to external environmental stimuli. One of the major two components in Salmonella is PhoP/Q system which functions in acid tolerance response. Classical response regulators can bind the RNA polymerase directly. Mechanism of transcriptional regulation also involves repressor DNA binding protein which remains bound to DNA and suppresses transcription unless an inducer molecule bind.
Two-component systems which is involved in combating oxidative and nitrosative stress is SoxRS, the SoxR has one (2Fe–2S) for sensing superoxide and nitric oxide radical, upon sensing it gets activated and further upregulated the expression of SoxS, which then further induces genes responsible for oxidative stress (Nunoshiba et al., 1993; Ding and Demple, 2000). The regulator of cellular NO response is NsrR, which is again an iron-sulfur cluster containing NO stress response regulator. NsrR is a transcriptional repressor and its regulon’s most conserved genes hmp encodes for a flavohaemoprotein which can detoxify NO in both anaerobic and aerobic conditions. It has been also reported that hmp expression can, in turn, exacerbate the oxidative stress by promoting Fenton Chemistry (Bang et al., 2006). The transcriptional regulator OxyR belonging to LysR family is a non-metal containing proteins reported to activate transcription via a redox and nitrosative stress sensitive Cys199 of the genes involved in oxidative stress (Kim et al., 2002; Paget and Buttner, 2003). Apart from this one more fascinating example is Salmonella is mgtA which is responsive to low magnesium. Also, a proline-rich region in the open reading frame also located within the leader mRNA of mgtA, named as mgtL. It has been demonstrated to confer responsiveness against proline and osmolarity (Park et al., 2010).
In Salmonella, the role of H-NS, NAP in global silencing of genes that are acquired via HGT (Lucchini et al., 2006; Navarre et al., 2006). Usually, the gene acquired has lower GC content than the resident gene, based on this difference the H-NS specifically binds to the high AT-rich region and thereby silence the acquired gene in this process (Navarre et al., 2006; Pacheco et al., 2012). Following this, the subsequent role is played by an evolutionary modification that further incorporates acquired genes into regulatory networking. To express silenced genes Salmonella incorporated mechanism of counter silencing such as: the antagonist binding to H-NS subsequently disrupting the multiunit complex of H-NS, competing with H-NS to bind DNA, alternative sigma factor assigned which demonstrates higher affinity for A-T rich sequence, and last is changing geometry of the promoter such that H-NS can no longer bind (Navarre et al., 2006; Stoebel et al., 2008). This type of counter silencing regulation is mostly seen in genes associated with virulence of the pathogen. One example of this kind of regulation is biofilm formation in Salmonella, biofilm formation helps to colonize the host and protects cells outside of the host too. In absence of H-NS the csgBAC genes can be even transcribed by σ70 but in presence of H-NS, it requires an alternative sigma factor σS (Olsen et al., 1993). On the other hand, the master regulator csgD is also in turn regulated by OmpR, IHF, H-NS and MlrA by bind to upstream of the csgD promoter (Gerstel et al., 2003).
The SPIs are also silenced by H-NS until it is sensed by external stimuli from the host and counter silenced by a transcriptional factor. There are various transcription factors such as HilE, HilA (Lostroh and Lee, 2001). Under low osmolarity condition hilA is silenced until H-NS mediated the repression is relieved by two other factor HilC and HilD encoded by SPI-1 (Schechter et al., 2003).
Apart from these regulations, there is other gene regulation which is imparted by proteolytic signaling cascade and small non-coding RNA which are typically involved in the immediate gene expression/repression. One excellent example is the upregulation of σ S via σ E (Bang et al., 2005; McCullen et al., 2010) rpoH and hfq. Hfq promotes translation of σS by binding to small RNAs DsrA and RprA thereby stabilizing their interaction with rpoS mRNA (Sledjeski et al., 1996; McCullen et al., 2010; Soper et al., 2010). The outer membrane proteins (OMPs) also participate in sensing and gene regulation. Misfolded OMPs can activate σE (Alba and Gross, 2004; Ades, 2008), under non-stressed conditions the σE is bound to RseA an anti-sigma factor and thus remains sequestered at the inner membrane. But it has also been shown in Salmonella that activation of σE can be independent of misfolded OMPs. Inside acidified phagosomes of host macrophages, the low pH triggers the DegS independent activation of σE, which is essential for Salmonella survival (Muller et al., 2009). As a result, it was concluded that different environmental stimuli can activate common stress response regulator σE. Salmonella can also sense and utilize intestinal metabolites such as ethanolamine. It is sensed by EutR receptor and upregulate genes which facilitate utilization of ethanolamine during gut-inflammation scenario thereby providing it with an upper hand of survival to the commensal microbiota (Thiennimitr et al., 2011). Apart from in general gene regulation Salmonella maintains transcriptional regulation directly at the RNA polymerase machinery level. It has assigned different sigma factor for different environmental cues to regulate the subsequent downstream genetic locus.
Gene Regulation in Yersinia
Three species of Yersinia are pathogenic to humans among them Y. enterocolitica and Y. pseudotuberculosis, represents gastrointestinal pathogens and it adopts the fecal-oral route of transmission (Rich et al., 1990). Y. pestis is a causative organism of bubonic plague. To establish infection in mammalian host successfully, it must adapt and combat multiple numbers of environmental stress from the host. Therefore, Yersinia regulates their gene expression in a tight fashion and only express the gene required for survival at any moment. Bacteria, in general, regulate gene expression in the level of transcription, for example, sigma factor, promoter sequence, and transcriptional activator and repressor all respond and act to environmental cues and bacterial need (Cornelis et al., 1989; Iriarte et al., 1995a,b; Trulzsch et al., 2001; Brzostek et al., 2007; Gao et al., 2008; Raczkowska et al., 2011). It is well studied that there is an intricate layer of regulation for the expression of several survival and virulence gene. One instance is the expression of Yop-Ysc type III secretion system and its effector proteins are required for virulence in mammalian system at the same time it is essential to shut off this machinery to escape host immune responses (Woestyn et al., 1994; Cornelis and Wolf-Watz, 1997; Cornelis et al., 1998; Shao, 2008). One of the excellent reviews that give enormous knowledge on transcriptional regulation in Yersinia is by Marceau (2005). While transcriptional control is a broader form of regulation, post-transcriptional/translational gene regulation research is on higher interest recently. The post-transcriptional research provides fine tuning of gene regulations and facilitates more rapid adaptation ability in the fleeting environment. Yersinia possesses various RNA binding regulatory protein which either stabilizes or degrades target mRNA thereby regulating them. yopD is one such effector protein a part of the T3SS translocon, in addition, it binds to 5′ untranslated region of yopQ (effector protein gene) mRNA along with its chaperone lcrH and mediates decay of the transcript (Anderson et al., 2002). The regulation of yopQ is also via class II regulators (yopD, lcrH, and Yop secretion factors yscM1/yscM2) which completes with the ribosome 30S and there downregulates yopQ translation (Kopaskie et al., 2013).
Another prominent regulator of gene expression in Yersinia is carbon storage regulator A (CsrA), there are various well-established mechanistic models which suggests that depending upon the binding site on the target gene, translation is either activated or repressed. CsrA binds to 5′-UTR of target mRNA and either alter translation, mRNA turnover or transcription elongation (Vakulskas et al., 2015). The function of CsrA is in turn regulated by two non-coding RNA molecules termed as CsrB and CsrC which sequester CsrA thereby regulating the amount of free CsrA present to function (Liu et al., 1997; Romeo, 1998) (Weilbacher et al., 2003; Vakulskas et al., 2015). CsrA also regulates the global transcriptional regulator RovA in Y. pseudotuberculosis thus controlling the expression of genes involved in virulence indirectly (Cathelyn et al., 2006) (Heroven et al., 2008). CsrA stabilizes the master regulator of flagellum biosynthesis and chemotaxis, flhDC by preventing cleavage of the same by cellular RNase E (Heroven et al., 2008). Recent reports suggest that CsrA also involved in fine-tuning of expression of two T3SS genetic regulatory cascades Ysa, Ysc, flagellar T3SS and biofilm formation in response to distinct environmental cues (Willias et al., 2015; Ozturk et al., 2017). The small protein SmpB functioning along with small RNA SsrA to rescue stalled ribosomes on incomplete mRNA transcripts (Komine et al., 1994; Keiler et al., 1996; Karzai et al., 2000) are also involved in regulating transcription of the master regulator gene of virulence lcrF/virF (Okan et al., 2006). Apart from this SsrA is important for Y. pestis virulence via intranasal and intravenous routes and ΔssrA strains were also used as candidate vaccine in the same study (Okan et al., 2010). Another protein which plays a central part in the regulation of a large number of genes as much as 243 genes in Yersinia (Geng et al., 2009) is a pleiotropic small RNA chaperone protein Hfq. The Hfq is 11 KDa in Yersinia, forms homo-hexameric ring complex that allows it to bind more than one RNA molecule at any given point (Kajitani and Ishihama, 1991; Vytvytska et al., 1998; Sauter et al., 2003). The site of binding of Hfq to small RNA and target mRNA determines whether the transcript will be stabilized or degraded (Vytvytska et al., 1998; Masse et al., 2003b; Meibom et al., 2009). The Hfq regulates genes involved in virulence, metabolism (carbohydrates, nitrogen, iron, fatty acids, and ATP), siderophore (Kakoschke et al., 2014) and surface pathogenicity factor, and stress response (Kakoschke et al., 2016). Some of the genes regulate are plasminogen activator protease gene pla, the F1 antigen gene cafI, the diguanylate cyclase gene hmsT and 50% gene of T3SS. Hfq also participates in the repression of non-flagellar dependent swarming motility (Geng et al., 2009) and biofilm formation in vitro (Kakoschke et al., 2014). On the other hand, it is also required for the formation of biofilm inside proventriculus of flea thereby maintaining the life cycle in flea vector (Rempe et al., 2012). Hfq deficient strains showed growth impairment, higher sensitive to environmental stresses and antibiotics, along with disability in QS, and host invasion. One of the recent reports suggests that various Hfq-dependent phenotypic alteration in Y. enterocolitica is mediated by depression of the transcriptional regulator RovM (master regulator of motility, chemotaxis and flagella synthesis) (Leskinen et al., 2017).
A different set of elegant regulation is mediated by small RNAs in Yersinia, majorly regulating genes which are required to express/repress immediately upon a change in environment. In response to temperature shift from 26 to 37°C (host temperature), there is differential sRNA expression in Y. pestis which enhances host infection (Beauregard et al., 2013; Schiano et al., 2014), it was also reported in infected mice’s lungs (Yan et al., 2013). One of the candidates sRNAs, ysr35, deletion resulted in attenuation of Y. pestis in mice models (Koo et al., 2011). There are small RNAs such as ysr141 expressed on pCD1 plasmid that encodes T3SS effectors injected into the host system to escape host immune system and establish infection successfully (Schiano et al., 2014). Ysr141 targets 5′ UTR of yopJ (T3SS effector) and activates its synthesis. Also, in Y. pestis sRNA HmsB has been shown to upregulate biofilm formation (Fang et al., 2014). One report suggests that upon knocking-down of ysr170 there was a significant decrease in infection efficiency of pathogenic Yersinia resulting in higher production of the proinflammatory cytokines TNF-alpha, IL-8 and EGR1. In the same study, ysr170 was proposed to the global regulator of virulence and metabolism in Yersinia (Li et al., 2016). Another mode of regulation in Yersinia involves RNase E mediated degradation along with Hfq of cellular mRNA of T3SS effector YopE (Yang et al., 2008) in post-translational step, similar regulations have also been reported for PNPase (Rosenzweig et al., 2005; Rosenzweig and Schesser, 2007). There is moreover a tight regulation maintained during the change in surrounding temperature. A report which states that over 400 genes are differentially regulated at two different temperature 26 and 37°C. Out of which 61% of them are downregulated, whereas 39% were upregulated (Han et al., 2004). Many of these genes are associated with T3SS which gets activated exclusively inside mammalian host at 37°C by de-repressing the transcription of lcrF, encoding for AraC like transcription factor. Therefore, it has been proposed that lcrF is thermo-sensor and mutation of the same, critically hampers the virulence of the pathogen (Hoe and Goguen, 1993). The secondary structure of lcrF also forms motif homologous to heat shock protein in eubacteria (Kortmann and Narberhaus, 2012; Schumann, 2012). Regarding riboswitches the mechanism underlying is yet to be discovered, prediction of one of the bioinformatics studies show there are a few switches like FMN, cobalamin, yybP-ykoY element, Moco which are yet to be characterized. One of the well-characterized riboswitches is mgtA, which is a Mg2+ responsive (Korth and Sigel, 2012). Among all the enteropathogens, most diverse set of gene regulation is shown by Yersinia. There is a growing need for more research in this field as this would lead us to some very important clues to how these deadly pathogens regulate their genetic locus to effectively establish the infection with high rates of mortality and morbidity. Genetic regulations in Yersinia also gives us insights that not only it invade the host system but there are specific set of regulations performed which mask them from host immune system and thereby escaping the same.
Gene Regulation in Shigella
Shigella causes bacterial dysentery in humans. The disease progresses in the lower part of the digestive tract. The bacteria invade colonic epithelial cells, reproduces within them and proceeds to other cells. The genes necessary for Shigella virulence are present on a 230-kbp plasmid and these are clustered in a 31 kbp portion (Dorman and Porter, 1998). S. flexneri virulence genes belong to the large family of H-NS repressed genes (Dorman and Porter, 1998). In addition to H-NS and StpA, Shigella flexneri 2a encodes Sfh, a third member of H-NS like family (Gore and Payne, 2010).
CsrA and Cra have antagonist effects on glycolysis and gluconeogenesis and it affects epithelial cell invasion in early stages (Gore and Payne, 2010). pfkA is a gene required for glycolysis. CsrA positively regulates pfkA expression (Sabnis et al., 1995) while Cra negatively regulates the same (Chin et al., 1989). Gore and Payne (2010) showed that expression of virulence gene regulators virF and virB were reduced in pfkA and csrA mutants (Gore and Payne, 2010).
The VirF-VirB modulatory mechanism controls the expression of ipa genes. This regulation of the expression of ipa (invasion plasmid antigens) genes is subject to environmental factors like osmolarity, temperature and pH (Bernardini et al., 1990; Porter and Dorman, 1997a; Jennison and Verma, 2004). Transcription of the virulence genes on the plasmid has shown to require virF and virB genes (Adler et al., 1989).
VirF is a member of AraC family of transcription regulators that exerts influence on gene expression usually in response to temperature (Gallegos et al., 1997). IHF is necessary for the complete activation of virF and virB (Porter and Dorman, 1997a) while H-NS represses expression of virB (Tobe et al., 1993). Transcription of the virF gene is under pH control. But thermal regulation can supersede pH control at genes downstream in the regulatory cascade (Porter and Dorman, 1997a,b). In S. sonnei, and almost certainly also in S. flexneri, this pH control depends on the cpxRA gene located on the chromosome. cpxR, the gene encoding the response regulator, binds at virF and regulates it in response to pH (Nakayama and Watanabe, 1995).
The Hfq protein is an RNA binding protein. It performs several physiological roles in bacteria (Hayashi-Nishino et al., 2012; Kakoschke et al., 2014; Vogt and Raivio, 2014). Hfq accommodates for environmental stress and affects virulence in S. flexneri (Yang et al., 2015). Hfq augments acid resistance in S. flexneri by regulating acid resistance genes-gadB, gadA, hdeB, hdeA, and hdeD. Hfq on top plays a significant role in the virulence of Shigella by positively regulating T3SS gene expression (Yang et al., 2015).
In Shigella, when the iron is replete, the transcription factor Fur binds to iron and Fe-Fur restricts the expression of iron transport genes (De Lorenzo et al., 1988). ryhB encodes a small RNA that promotes degradation of transcripts for iron storage, oxidative metabolism and stress proteins and Fe-Fur represses expression of rhyB too (Masse et al., 2003a, 2005) Iron availability modulates both transcription of fur and the activity of the Fur protein (De Lorenzo et al., 1988; Delany et al., 2002; Sala et al., 2003; Hernandez et al., 2006). ArcA represses expression of fur and it is equally involved in the regulation of feo and siderophore biosynthesis and transport genes (Boulette and Payne, 2007). ArcA (aerobic respiration control A) is a global transcription factor and feo is an iron transport system encoded by S. flexneri which transports ferrous ion. Oxygen availability also plays an important role as a general signal for regulation of iron acquisition (Boulette and Payne, 2007).
SlyA in S. flexneri is involved in the regulation of virulence-associated genes and in acid resistance. In S. flexneri, the slyA promoter activity is most during the stationary phase (Weatherspoon-Griffin and Wing, 2016). slyA promoter is negatively autoregulated and positively regulated by PhoP (Norte et al., 2003; Santiago et al., 2016).
The Shigella virulence gene regulon is an example of a complex gene control system. However, a detailed characterization of the gene regulatory mechanisms needs to be discovered.
Conclusion and Future Direction
In the review, we have tried to summarize some of the common as well as pathogen specific gene regulatory mechanisms employed by enteropathogenic bacteria. Since the amount of literature available with respect to gene regulation in these bacteria is huge, we have tried to include some of the major and elegantly regulated mechanisms. For example, in QS, AI 1, AI 2, AI 3 mediated genetic control, different types of TA systems induced gene regulation and other direct transcription regulators like NAPs. Emerging literature on NAPs are suggesting that there might be a very thin line between the conventional transcriptional regulators and NAPs. We have also summarized the pathogen specific genetic regulation in E. coli, Salmonella sp., Yersinia sp., Shigella sp. which majorly comprise the class of entero-pathogenic bacteria.
Recently, with the advent of bioinformatics, studying gene regulation has become far simpler. Prediction, detection and designation of functions of virulence-associated genes (like TA systems) have become a matter of few hours. Since the rate of failure of antibiotics is increasing with the emergence of multiple drug resistance strains, we are in dire need of novel strategies to target these pathogens. Studies on NAPs and other novel proteins that directly regulate the survival of bacteria are on the rise, which might prove to be successful in being targeted for therapeutics. Also, the design of small molecule inhibitors of PAMPs, nanoparticle-based drug delivery systems has been proven to be successful. Although these pathogens have been associated with causing foodborne illness since decades, our failure in being able to control them is the proof that there is a gap to be filled in the knowledge of our understanding their survival strategies. It should also be considered that the need of therapeutics is majorly in economically backward countries in Africa and south-eastern Asia and thus the novel strategies to control disease should be feasible for use in these countries for bringing down the numbers in the statistics of morbidity and mortality associated with such illnesses.
Author Contributions
RC, MS, RS, and DC designed and prepared the manuscript. DC supervised the work and all the authors were involved in the final editing of the manuscript.
Funding
DAE Outstanding research fellow (DAE-SRC), DBT-IISc partnership program, and Infosys Foundation grant for advanced research in biological sciences to DC is highly acknowledged. Infrastructure support from ICMR (Center for Advanced Study in Molecular Medicine), DST (FIST), and UGC-CAS (special assistance) is acknowledged. DC received DAE SRC outstanding Investigator award and funds. RC duly acknowledge CSIR-JRF for funding.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ades, S. E. (2008). Regulation by destruction: design of the sigmaE envelope stress response. Curr. Opin. Microbiol. 11, 535–540. doi: 10.1016/j.mib.2008.10.004
Adler, B., Sasakawa, C., Tobe, T., Makino, S., Komatsu, K., and Yoshikawa, M. (1989). A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3, 627–635. doi: 10.1111/j.1365-2958.1989.tb00210.x
Aizenman, E., Engelberg-Kulka, H., and Glaser, G. (1996). An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U.S.A. 93, 6059–6063. doi: 10.1073/pnas.93.12.6059
Alba, B. M., and Gross, C. A. (2004). Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52, 613–619. doi: 10.1111/j.1365-2958.2003.03982.x
Anderson, D. M., Ramamurthi, K. S., Tam, C., and Schneewind, O. (2002). YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J. Bacteriol. 184, 1287–1295. doi: 10.1128/JB.184.5.1287-1295.2002
Arfin, S. M., Long, A. D., Ito, E. T., Tolleri, L., Riehle, M. M., Paegle, E. S., et al. (2000). Global gene expression profiling in Escherichia coli K12, the effects of integration host factor. J. Biol. Chem. 275, 29672–29684. doi: 10.1074/jbc.M002247200
Armbruster, C. E., Pang, B., Murrah, K., Juneau, R. A., Perez, A. C., Weimer, K. E., et al. (2011). RbsB (NTHI_0632) mediates quorum signal uptake in nontypeable Haemophilus influenzae strain 86-028NP. Mol. Microbiol. 82, 836–850. doi: 10.1111/j.1365-2958.2011.07831.x
Atkinson, S., Chang, C. Y., Sockett, R. E., Camara, M., and Williams, P. (2006). Quorum sensing in Yersinia enterocolitica controls swimming and swarming motility. J. Bacteriol. 188, 1451–1461. doi: 10.1128/JB.188.4.1451-1461.2006
Bader, M. W., Sanowar, S., Daley, M. E., Schneider, A. R., Cho, U., Xu, W., et al. (2005). Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122, 461–472. doi: 10.1016/j.cell.2005.05.030
Bang, I. S., Frye, J. G., McClelland, M., Velayudhan, J., and Fang, F. C. (2005). Alternative sigma factor interactions in Salmonella: sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol. Microbiol. 56, 811–823. doi: 10.1111/j.1365-2958.2005.04580.x
Bang, I. S., Liu, L., Vazquez-Torres, A., Crouch, M. L., Stamler, J. S., and Fang, F. C. (2006). Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J. Biol. Chem. 281, 28039–28047. doi: 10.1074/jbc.M605174200
Baxter, M. A., and Jones, B. D. (2005). The fimYZ genes regulate Salmonella enterica Serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect. Immun. 73, 1377–1385. doi: 10.1128/IAI.73.3.1377-1385.2005
Beauregard, A., Smith, E. A., Petrone, B. L., Singh, N., Karch, C., McDonough, K. A., et al. (2013). Identification and characterization of small RNAs in Yersinia pestis. RNA Biol. 10, 397–405. doi: 10.4161/rna.23590
Becker, N. A., Kahn, J. D., and Maher, L. J. III. (2007). Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 35, 3988–4000. doi: 10.1093/nar/gkm419
Bernardini, M. L., Fontaine, A., and Sansonetti, P. J. (1990). The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J. Bacteriol. 172, 6274–6281. doi: 10.1128/jb.172.11.6274-6281.1990
Bertin, Y., Girardeau, J. P., Chaucheyras-Durand, F., Lyan, B., Pujos-Guillot, E., Harel, J., et al. (2011). Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13, 365–377. doi: 10.1111/j.1462-2920.2010.02334.x
Beury-Cirou, A., Tannieres, M., Minard, C., Soulere, L., Rasamiravaka, T., Dodd, R. H., et al. (2013). At a supra-physiological concentration, human sexual hormones act as quorum-sensing inhibitors. PLoS One 8:e83564. doi: 10.1371/journal.pone.0083564
Bieber, D., Ramer, S. W., Wu, C. Y., Murray, W. J., Tobe, T., Fernandez, R., et al. (1998). Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280, 2114–2118. doi: 10.1126/science.280.5372.2114
Blier, A. S., Veron, W., Bazire, A., Gerault, E., Taupin, L., Vieillard, J., et al. (2011). C-type natriuretic peptide modulates quorum sensing molecule and toxin production in Pseudomonas aeruginosa. Microbiology 157(Pt 7), 1929–1944. doi: 10.1099/mic.0.046755-0
Boulette, M. L., and Payne, S. M. (2007). Anaerobic regulation of Shigella flexneri virulence: ArcA regulates Fur and iron acquisition genes. J. Bacteriol. 189, 6957–6967. doi: 10.1128/JB.00621-07
Broyles, S. S., and Pettijohn, D. E. (1986). Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol. 187, 47–60. doi: 10.1016/0022-2836(86)90405-5
Brzostek, K., Brzostkowska, M., Bukowska, I., Karwicka, E., and Raczkowska, A. (2007). OmpR negatively regulates expression of invasin in Yersinia enterocolitica. Microbiology 153(Pt 8), 2416–2425. doi: 10.1099/mic.0.2006/003202-0
Cambray, G., Guerout, A. M., and Mazel, D. (2010). Integrons. Annu. Rev. Genet. 44, 141–166. doi: 10.1146/annurev-genet-102209-163504
Campos-Galvao, M. E., Ribon, A. O., Araujo, E. F., and Vanetti, M. C. (2016). Changes in the Salmonella enterica enteritidis phenotypes in presence of acyl homoserine lactone quorum sensing signals. J. Basic Microbiol. 56, 493–501. doi: 10.1002/jobm.201500471
Cathelyn, J. S., Crosby, S. D., Lathem, W. W., Goldman, W. E., and Miller, V. L. (2006). RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. U.S.A. 103, 13514–13519. doi: 10.1073/pnas.0603456103
Chin, A. M., Feldheim, D. A., Saier, M. H. Jr. (1989). Altered transcriptional patterns affecting several metabolic pathways in strains of Salmonella typhimurium which overexpress the fructose regulon. J. Bacteriol. 171, 2424–2434. doi: 10.1128/jb.171.5.2424-2434.1989
Chowers, M. Y., Keller, N., Tal, R., Barshack, I., Lang, R., Bar-Meir, S., et al. (1999). Human gastrin: a Helicobacter pylori–specific growth factor. Gastroenterology 117, 1113–1118. doi: 10.1016/S0016-5085(99)70396-3
Christensen, S. K., Pedersen, K., Hansen, F. G., and Gerdes, K. (2003). Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332, 809–819. doi: 10.1016/S0022-2836(03)00922-7
Claret, L., and Rouviere-Yaniv, J. (1997). Variation in HU composition during growth of Escherichia coli: the heterodimer is required for long term survival. J. Mol. Biol. 273, 93–104. doi: 10.1006/jmbi.1997.1310
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10420–10425. doi: 10.1073/pnas.0604343103
Clarke, M. B., and Sperandio, V. (2005a). Transcriptional autoregulation by quorum sensing Escherichia coli regulators B and C (QseBC) in enterohaemorrhagic E. coli (EHEC). Mol. Microbiol. 58, 441–455. doi: 10.1111/j.1365-2958.2005.04819.x
Clarke, M. B., and Sperandio, V. (2005b). Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 57, 1734–1749. doi: 10.1111/j.1365-2958.2005.04792.x
Cornelis, G., Sluiters, C., de Rouvroit, C. L., and Michiels, T. (1989). Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171, 254–262. doi: 10.1128/jb.171.1.254-262.1989
Cornelis, G. R. (2002). The Yersinia Ysc-Yop virulence apparatus. Int. J. Med. Microbiol. 291, 455–462. doi: 10.1078/1438-4221-00153
Cornelis, G. R., Boland, A., Boyd, A. P., Geuijen, C., Iriarte, M., Neyt, C., et al. (1998). The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62, 1315–1352.
Cornelis, G. R., and Wolf-Watz, H. (1997). The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23, 861–867. doi: 10.1046/j.1365-2958.1997.2731623.x
Dame, R. T., Wyman, C., Wurm, R., Wagner, R., and Goosen, N. (2002). Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277, 2146–2150. doi: 10.1074/jbc.C100603200
de Kievit, T. R., and Iglewski, B. H. (2000). Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68, 4839–4849. doi: 10.1128/IAI.68.9.4839-4849.2000
De la Cruz, M. A., Zhao, W., Farenc, C., Gimenez, G., Raoult, D., Cambillau, C., et al. (2013). A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 9:e1003827. doi: 10.1371/journal.ppat.1003827
De Lorenzo, V., Herrero, M., Giovannini, F., and Neilands, J. B. (1988). Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. Eur. J. Biochem. 173, 537–546. doi: 10.1111/j.1432-1033.1988.tb14032.x
Delany, I., Spohn, G., Pacheco, A. B., Ieva, R., Alaimo, C., Rappuoli, R., et al. (2002). Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol. Microbiol. 46, 1107–1122. doi: 10.1046/j.1365-2958.2002.03227.x
Deng, S., Stein, R. A., and Higgins, N. P. (2005). Organization of supercoil domains and their reorganization by transcription. Mol. Microbiol. 57, 1511–1521. doi: 10.1111/j.1365-2958.2005.04796.x
Dewoody, R. S., Merritt, P. M., and Marketon, M. M. (2013). Regulation of the Yersinia type III secretion system: traffic control. Front. Cell Infect. Microbiol. 3:4. doi: 10.3389/fcimb.2013.00004
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Ding, H., and Demple, B. (2000). Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. U.S.A. 97, 5146–5150. doi: 10.1073/pnas.97.10.5146
Dorman, C. J. (2004). H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2, 391–400. doi: 10.1038/nrmicro883
Dorman, C. J. (2006). DNA supercoiling and bacterial gene expression. Sci. Prog. 89(Pt 3–4), 151–166. doi: 10.3184/003685006783238317
Dorman, C. J., and Porter, M. E. (1998). The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29, 677–684. doi: 10.1046/j.1365-2958.1998.00902.x
Dyszel, J. L., Smith, J. N., Lucas, D. E., Soares, J. A., Swearingen, M. C., Vross, M. A., et al. (2010a). Salmonella enterica serovar Typhimurium can detect acyl homoserine lactone production by Yersinia enterocolitica in mice. J. Bacteriol. 192, 29–37. doi: 10.1128/JB.01139-09
Dyszel, J. L., Soares, J. A., Swearingen, M. C., Lindsay, A., Smith, J. N., and Ahmer, B. M. (2010b). E. coli K-12 and EHEC genes regulated by SdiA. PLoS One 5:e8946. doi: 10.1371/journal.pone.0008946
El Mouali, Y., Gaviria-Cantin, T., Sanchez-Romero, M. A., Gibert, M., Westermann, A. J., Vogel, J., et al. (2018). CRP-cAMP mediates silencing of Salmonella virulence at the post-transcriptional level. PLoS Genet. 14:e1007401. doi: 10.1371/journal.pgen.1007401
Elliott, S. J., Hutcheson, S. W., Dubois, M. S., Mellies, J. L., Wainwright, L. A., Batchelor, M., et al. (1999). Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33, 1176–1189. doi: 10.1046/j.1365-2958.1999.01559.x
Engebrecht, J., Nealson, K., and Silverman, M. (1983). Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32, 773–781. doi: 10.1016/0092-8674(83)90063-6
Engebrecht, J., and Silverman, M. (1984). Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U.S.A. 81, 4154–4158. doi: 10.1073/pnas.81.13.4154
Even-Tov, E., Bendori, S. O., Valastyan, J., Ke, X., Pollak, S., Bareia, T., et al. (2016). Social evolution selects for redundancy in bacterial quorum sensing. PLoS Biol. 14:e1002386. doi: 10.1371/journal.pbio.1002386
Fang, N., Qu, S., Yang, H., Fang, H., Liu, L., Zhang, Y., et al. (2014). HmsB enhances biofilm formation in Yersinia pestis. Front. Microbiol. 5:685. doi: 10.3389/fmicb.2014.00685
Fink, S. L., and Cookson, B. T. (2007). Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol. 9, 2562–2570. doi: 10.1111/j.1462-5822.2007.01036.x
Fookes, M., Schroeder, G. N., Langridge, G. C., Blondel, C. J., Mammina, C., Connor, T. R., et al. (2011). Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog. 7:e1002191. doi: 10.1371/journal.ppat.1002191
Gallegos, M. T., Schleif, R., Bairoch, A., Hofmann, K., and Ramos, J. L. (1997). Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410.
Gao, H., Zhou, D., Li, Y., Guo, Z., Han, Y., Song, Y., et al. (2008). The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 190, 3063–3075. doi: 10.1128/JB.01910-07
Geng, J., Song, Y., Yang, L., Feng, Y., Qiu, Y., Li, G., et al. (2009). Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS One 4:e6213. doi: 10.1371/journal.pone.0006213
Gerstel, U., Park, C., and Romling, U. (2003). Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49, 639–654. doi: 10.1046/j.1365-2958.2003.03594.x
Gonyar, L. A., and Kendall, M. M. (2014). Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 82, 193–201. doi: 10.1128/IAI.00980-13
Gore, A. L., and Payne, S. M. (2010). CsrA and Cra influence Shigella flexneri pathogenesis. Infect. Immun. 78, 4674–4682. doi: 10.1128/IAI.00589-10
Grainger, D. C., Hurd, D., Goldberg, M. D., and Busby, S. J. (2006). Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 34, 4642–4652. doi: 10.1093/nar/gkl542
Gruber, T. M., and Gross, C. A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466. doi: 10.1146/annurev.micro.57.030502.090913
Grys, T. E., Siegel, M. B., Lathem, W. W., and Welch, R. A. (2005). The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect. Immun. 73, 1295–1303. doi: 10.1128/IAI.73.3.1295-1303.2005
Han, X., Bai, H., Liu, L., Dong, H., Liu, R., Song, J., et al. (2013). The luxS gene functions in the pathogenesis of avian pathogenic Escherichia coli. Microb Pathog. 55, 21–27. doi: 10.1016/j.micpath.2012.09.008
Han, X., Bai, H., Tu, J., Yang, L., Xu, D., Wang, S., et al. (2015a). Deletion of luxS further attenuates the virulence of the avian pathogenic Escherichia coli aroA mutant. Microb. Pathog. 88, 39–47. doi: 10.1016/j.micpath.2015.08.003
Han, X., Liu, L., Fan, G., Zhang, Y., Xu, D., Zuo, J., et al. (2015b). Riemerella anatipestifer lacks luxS, but can uptake exogenous autoinducer-2 to regulate biofilm formation. Res. Microbiol. 166, 486–493. doi: 10.1016/j.resmic.2015.06.004
Han, X., and Lu, C. (2009). Biological activity and identification of a peptide inhibitor of LuxS from Streptococcus suis serotype 2. FEMS Microbiol. Lett. 294, 16–23. doi: 10.1111/j.1574-6968.2009.01534.x
Han, Y., Zhou, D., Pang, X., Song, Y., Zhang, L., Bao, J., et al. (2004). Microarray analysis of temperature-induced transcriptome of Yersinia pestis. Microbiol. Immunol. 48, 791–805. doi: 10.1111/j.1348-0421.2004.tb03605.x
Hastings, J. W., and Nealson, K. H. (1977). Bacterial bioluminescence. Annu. Rev. Microbiol. 31, 549–595. doi: 10.1146/annurev.mi.31.100177.003001
Hayashi-Nishino, M., Fukushima, A., and Nishino, K. (2012). Impact of hfq on the intrinsic drug resistance of Salmonella enterica serovar typhimurium. Front. Microbiol. 3:205. doi: 10.3389/fmicb.2012.00205
Hayward, M. R., Jansen, V., and Woodward, M. J. (2013). Comparative genomics of Salmonella enterica serovars Derby and Mbandaka, two prevalent serovars associated with different livestock species in the UK. BMC Genomics 14:365. doi: 10.1186/1471-2164-14-365
Hengge, R. (2009). Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res. Microbiol. 160, 667–676. doi: 10.1016/j.resmic.2009.08.014
Hernandez, J. A., Muro-Pastor, A. M., Flores, E., Bes, M. T., Peleato, M. L., and Fillat, M. F. (2006). Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J. Mol. Biol. 355, 325–334. doi: 10.1016/j.jmb.2005.10.079
Heroven, A. K., Bohme, K., Rohde, M., and Dersch, P. (2008). A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol. 68, 1179–1195. doi: 10.1111/j.1365-2958.2008.06218.x
Hoe, N. P., and Goguen, J. D. (1993). Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175, 7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993
Hong, S. H., Lee, J., and Wood, T. K. (2010). Engineering global regulator Hha of Escherichia coli to control biofilm dispersal. Microb. Biotechnol. 3, 717–728. doi: 10.1111/j.1751-7915.2010.00220.x
Hooper, L. V., and Gordon, J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115–1118. doi: 10.1126/science.1058709
Hughes, D. T., Clarke, M. B., Yamamoto, K., Rasko, D. A., and Sperandio, V. (2009). The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog. 5:e1000553. doi: 10.1371/journal.ppat.1000553
Ibarra, J. A., Villalba, M. I., and Puente, J. L. (2003). Identification of the DNA binding sites of PerA, the transcriptional activator of the bfp and per operons in enteropathogenic Escherichia coli. J. Bacteriol. 185, 2835–2847. doi: 10.1128/JB.185.9.2835-2847.2003
Ide, T., Laarmann, S., Greune, L., Schillers, H., Oberleithner, H., and Schmidt, M. A. (2001). Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell Microbiol. 3, 669–679. doi: 10.1046/j.1462-5822.2001.00146.x
Iriarte, M., Stainier, I., and Cornelis, G. R. (1995a). The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect. Immun. 63, 1840–1847.
Iriarte, M., Stainier, I., Mikulskis, A. V., and Cornelis, G. R. (1995b). The fliA gene encoding sigma 28 in Yersinia enterocolitica. J. Bacteriol. 177, 2299–2304. doi: 10.1128/jb.177.9.2299-2304.1995
Jennison, A. V., and Verma, N. K. (2004). Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol. Rev. 28, 43–58. doi: 10.1016/j.femsre.2003.07.002
Ju, X., Li, J., Zhu, M., Lu, Z., Lv, F., Zhu, X., et al. (2018). Effect of the luxS gene on biofilm formation and antibiotic resistance by Salmonella serovar Dublin. Food Res. Int. 107, 385–393. doi: 10.1016/j.foodres.2018.02.039
Jung, K., Fried, L., Behr, S., and Heermann, R. (2012). Histidine kinases and response regulators in networks. Curr. Opin. Microbiol. 15, 118–124. doi: 10.1016/j.mib.2011.11.009
Kajitani, M., and Ishihama, A. (1991). Identification and sequence determination of the host factor gene for bacteriophage Q beta. Nucleic Acids Res. 19, 1063–1066. doi: 10.1093/nar/19.5.1063
Kakoschke, T., Kakoschke, S., Magistro, G., Schubert, S., Borath, M., Heesemann, J., et al. (2014). The RNA chaperone Hfq impacts growth, metabolism and production of virulence factors in Yersinia enterocolitica. PLoS One 9:e86113. doi: 10.1371/journal.pone.0086113
Kakoschke, T. K., Kakoschke, S. C., Zeuzem, C., Bouabe, H., Adler, K., Heesemann, J., et al. (2016). The RNA chaperone Hfq is essential for virulence and modulates the expression of four adhesins in Yersinia enterocolitica. Sci. Rep. 6:29275. doi: 10.1038/srep29275
Karzai, A. W., Roche, E. D., and Sauer, R. T. (2000). The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7, 449–455. doi: 10.1038/75843
Keiler, K. C., Waller, P. R., and Sauer, R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993. doi: 10.1126/science.271.5251.990
Kendall, M. M., Gruber, C. C., Parker, C. T., and Sperandio, V. (2012). Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. MBio 3:e00050-12. doi: 10.1128/mBio.00050-12
Kendall, M. M., and Sperandio, V. (2016). What a dinner party! Mechanisms and functions of interkingdom signaling in host-pathogen associations. MBio 7:e01748. doi: 10.1128/mBio.01748-15
Kenny, B., DeVinney, R., Stein, M., Reinscheid, D. J., Frey, E. A., and Finlay, B. B. (1997). Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520. doi: 10.1016/S0092-8674(00)80437-7
Kim, S. O., Merchant, K., Nudelman, R., Beyer, W. F. Jr., Keng, T., et al. (2002). OxyR: a molecular code for redox-related signaling. Cell 109, 383–396. doi: 10.1016/S0092-8674(02)00723-7
Kimmitt, P. T., Harwood, C. R., and Barer, M. R. (2000). Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 6, 458–465. doi: 10.3201/eid0605.000503
Knutton, S., Rosenshine, I., Pallen, M. J., Nisan, I., Neves, B. C., Bain, C., et al. (1998). A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17, 2166–2176. doi: 10.1093/emboj/17.8.2166
Komine, Y., Kitabatake, M., Yokogawa, T., Nishikawa, K., and Inokuchi, H. (1994). A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 91, 9223–9227. doi: 10.1073/pnas.91.20.9223
Koo, J. T., Alleyne, T. M., Schiano, C. A., Jafari, N., and Lathem, W. W. (2011). Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia -specific small, noncoding RNAs required for virulence. Proc. Natl. Acad. Sci. U.S.A.108, E709–E717. doi: 10.1073/pnas.1101655108
Kopaskie, K. S., Ligtenberg, K. G., and Schneewind, O. (2013). Translational regulation of Yersinia enterocolitica mRNA encoding a type III secretion substrate. J. Biol. Chem. 288, 35478–35488. doi: 10.1074/jbc.M113.504811
Korth, M. M., and Sigel, R. K. (2012). Unusually high-affinity Mg(2+) binding at the AU-rich sequence within the antiterminator hairpin of a Mg(2+) riboswitch. Chem. Biodivers. 9, 2035–2049. doi: 10.1002/cbdv.201200031
Kortmann, J., and Narberhaus, F. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265. doi: 10.1038/nrmicro2730
Lamas, A., Miranda, J. M., Vazquez, B., Cepeda, A., and Franco, C. M. (2016). Biofilm formation, phenotypic production of cellulose and gene expression in Salmonella enterica decrease under anaerobic conditions. Int. J. Food Microbiol. 238, 63–67. doi: 10.1016/j.ijfoodmicro.2016.08.043
Lee, J., Maeda, T., Hong, S. H., and Wood, T. K. (2009). Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl. Environ. Microbiol. 75, 1703–1716. doi: 10.1128/AEM.02081-08
Leskinen, K., Pajunen, M. I., Varjosalo, M., Fernandez-Carrasco, H., Bengoechea, J. A., and Skurnik, M. (2017). Several Hfq-dependent alterations in physiology of Yersinia enterocolitica O:3 are mediated by derepression of the transcriptional regulator RovM. Mol. Microbiol. 103, 1065–1091. doi: 10.1111/mmi.13610
Li, N., Hennelly, S. P., Stubben, C. J., Micheva-Viteva, S., Hu, B., Shou, Y., et al. (2016). Functional and structural analysis of a highly-expressed Yersinia pestis small RNA following infection of cultured macrophages. PLoS One 11:e0168915. doi: 10.1371/journal.pone.0168915
Lim, S., Yun, J., Yoon, H., Park, C., Kim, B., Jeon, B., et al. (2007). Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res. 35, 1822–1832. doi: 10.1093/nar/gkm060
Liu, M. Y., Gui, G., Wei, B., Preston, J. F. III, Oakford, L., Yuksel, U., et al. (1997). The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272, 17502–17510. doi: 10.1074/jbc.272.28.17502
Liu, Z., Que, F., Liao, L., Zhou, M., You, L., Zhao, Q., et al. (2014). Study on the promotion of bacterial biofilm formation by a Salmonella conjugative plasmid and the underlying mechanism. PLoS One 9:e109808. doi: 10.1371/journal.pone.0109808
Lobato-Marquez, D., Moreno-Cordoba, I., Figueroa, V., Diaz-Orejas, R., and Garcia-del Portillo, F. (2015). Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep. 5:9374. doi: 10.1038/srep09374
Lopez-Garrido, J., and Casadesus, J. (2012). Crosstalk between virulence loci: regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by products of the std fimbrial operon. PLoS One 7:e30499. doi: 10.1371/journal.pone.0030499
Lostroh, C. P., and Lee, C. A. (2001). The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3, 1281–1291. doi: 10.1016/S1286-4579(01)01488-5
Lucchini, S., Rowley, G., Goldberg, M. D., Hurd, D., Harrison, M., and Hinton, J. C. (2006). H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. doi: 10.1371/journal.ppat.0020081
Luo, G., Niesel, D. W., Shaban, R. A., Grimm, E. A., and Klimpel, G. R. (1993). Tumor necrosis factor alpha binding to bacteria: evidence for a high-affinity receptor and alteration of bacterial virulence properties. Infect. Immun. 61, 830–835.
Luzader, D. H., Clark, D. E., Gonyar, L. A., and Kendall, M. M. (2013). EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 195, 4947–4953. doi: 10.1128/JB.00937-13
Macchi, R., Montesissa, L., Murakami, K., Ishihama, A., De Lorenzo, V., and Bertoni, G. (2003). Recruitment of sigma54-RNA polymerase to the Pu promoter of Pseudomonas putida through integration host factor-mediated positioning switch of alpha subunit carboxyl-terminal domain on an UP-like element. J. Biol. Chem. 278, 27695–27702. doi: 10.1074/jbc.M303031200
Madrid, C., Balsalobre, C., Garcia, J., and Juarez, A. (2007). The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol. Microbiol. 63, 7–14. doi: 10.1111/j.1365-2958.2006.05497.x
Mangan, M. W., Lucchini, S., Danino, V., Croinin, T. O., Hinton, J. C., and Dorman, C. J. (2006). The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 1831–1847. doi: 10.1111/j.1365-2958.2006.05062.x
Marceau, M. (2005). Transcriptional regulation in Yersinia: an update. Curr. Issues Mol. Biol. 7, 151–177.
Masse, E., Escorcia, F. E., and Gottesman, S. (2003a). Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374–2383. doi: 10.1101/gad.1127103
Masse, E., Majdalani, N., and Gottesman, S. (2003b). Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6, 120–124. doi: 10.1016/S1369-5274(03)00027-4
Masse, E., Vanderpool, C. K., and Gottesman, S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005
Masuda, H., Tan, Q., Awano, N., Yamaguchi, Y., and Inouye, M. (2012). A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiol. Lett. 328, 174–181. doi: 10.1111/j.1574-6968.2012.02496.x
McCullen, C. A., Benhammou, J. N., Majdalani, N., and Gottesman, S. (2010). Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J. Bacteriol. 192, 5559–5571. doi: 10.1128/JB.00464-10
McDaniel, T. K., Jarvis, K. G., Donnenberg, M. S., and Kaper, J. B. (1995). A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 92, 1664–1668. doi: 10.1073/pnas.92.5.1664
McNamara, B. P., Koutsouris, A., O’Connell, C. B., Nougayrede, J. P., Donnenberg, M. S., and Hecht, G. (2001). Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Invest. 107, 621–629. doi: 10.1172/JCI11138
Meibom, K. L., Forslund, A. L., Kuoppa, K., Alkhuder, K., Dubail, I., Dupuis, M., et al. (2009). Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect. Immun. 77, 1866–1880. doi: 10.1128/IAI.01496-08
Mellies, J. L., Haack, K. R., and Galligan, D. C. (2007). SOS regulation of the type III secretion system of enteropathogenic Escherichia coli. J. Bacteriol. 189, 2863–2872. doi: 10.1128/JB.01859-06
Miller, S. I., Kukral, A. M., and Mekalanos, J. J. (1989). A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. U.S.A. 86, 5054–5058. doi: 10.1073/pnas.86.13.5054
Muller, C., Bang, I. S., Velayudhan, J., Karlinsey, J., Papenfort, K., Vogel, J., et al. (2009). Acid stress activation of the sigma(E) stress response in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 71, 1228–1238. doi: 10.1111/j.1365-2958.2009.06597.x
Muller, C. M., Dobrindt, U., Nagy, G., Emody, L., Uhlin, B. E., and Hacker, J. (2006). Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 188, 5428–5438. doi: 10.1128/JB.01956-05
Nakanishi, N., Abe, H., Ogura, Y., Hayashi, T., Tashiro, K., Kuhara, S., et al. (2006). ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61, 194–205. doi: 10.1111/j.1365-2958.2006.05217.x
Nakashima, K., Sugiura, A., Momoi, H., and Mizuno, T. (1992). Phosphotransfer signal transduction between two regulatory factors involved in the osmoregulated kdp operon in Escherichia coli. Mol. Microbiol. 6, 1777–1784. doi: 10.1111/j.1365-2958.1992.tb01350.x
Nakayama, S., and Watanabe, H. (1995). Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei vir F gene. J. Bacteriol. 177, 5062–5069. doi: 10.1128/jb.177.17.5062-5069.1995
Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238. doi: 10.1126/science.1128794
Nealson, K. H., Platt, T., and Hastings, J. W. (1970). Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322.
Nesse, L. L., Berg, K., Vestby, L. K., Olsaker, I., and Djonne, B. (2011). Salmonella Typhimurium invasion of HEp-2 epithelial cells in vitro is increased by N-acylhomoserine lactone quorum sensing signals. Acta Vet. Scand. 53:44. doi: 10.1186/1751-0147-53-44
Nevesinjac, A. Z., and Raivio, T. L. (2005). The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J. Bacteriol. 187, 672–686. doi: 10.1128/JB.187.2.672-686.2005
Ng, Y. K., Grasso, M., Wright, V., Garcia, V., Williams, P., and Atkinson, S. (2018). The quorum sensing system of Yersinia enterocolitica 8081 regulates swimming motility, host cell attachment, and virulence plasmid maintenance. Genes (Basel) 9:E307. doi: 10.3390/genes9060307
Njoroge, J., and Sperandio, V. (2012). Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect. Immun. 80, 688–703. doi: 10.1128/IAI.05921-11
Njoroge, J. W., Nguyen, Y., Curtis, M. M., Moreira, C. G., and Sperandio, V. (2012). Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. MBio 3:e280-12. doi: 10.1128/mBio.00280-12
Noom, M. C., Navarre, W. W., Oshima, T., Wuite, G. J., and Dame, R. T. (2007). H-NS promotes looped domain formation in the bacterial chromosome. Curr. Biol. 17, R913–R914. doi: 10.1016/j.cub.2007.09.005
Norte, V. A., Stapleton, M. R., and Green, J. (2003). PhoP-responsive expression of the Salmonella enterica serovar Typhimurium slyA gene. J. Bacteriol. 185, 3508–3514. doi: 10.1128/JB.185.12.3508-3514.2003
Novotny, L. A., Jurcisek, J. A., Ward, M. O. Jr., Jordan, Z. B., Goodman, S. D., et al. (2015). Antibodies against the majority subunit of type IV Pili disperse nontypeable Haemophilus influenzae biofilms in a LuxS-dependent manner and confer therapeutic resolution of experimental otitis media. Mol. Microbiol. 96, 276–292. doi: 10.1111/mmi.12934
Nunoshiba, T., deRojas-Walker, T., Wishnok, J. S., Tannenbaum, S. R., and Demple, B. (1993). Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc. Natl. Acad. Sci. U.S.A. 90, 9993–9997. doi: 10.1073/pnas.90.21.9993
Oberto, J., Nabti, S., Jooste, V., Mignot, H., and Rouviere-Yaniv, J. (2009). The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One 4:e4367. doi: 10.1371/journal.pone.0004367
Okan, N. A., Bliska, J. B., and Karzai, A. W. (2006). A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2:e6. doi: 10.1371/journal.ppat.0020006
Okan, N. A., Mena, P., Benach, J. L., Bliska, J. B., and Karzai, A. W. (2010). The smpB-ssrA mutant of Yersinia pestis functions as a live attenuated vaccine to protect mice against pulmonary plague infection. Infect. Immun. 78, 1284–1293. doi: 10.1128/IAI.00976-09
Olsen, A., Arnqvist, A., Hammar, M., Sukupolvi, S., and Normark, S. (1993). The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol. Microbiol. 7, 523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x
Ozturk, G., LeGrand, K., Zheng, Y., and Young, G. M. (2017). Yersinia enterocolitica CsrA expression of the Ysa and Ysc type 3 secretion system in unique ways. FEMS Microbiol. Lett. 364:fnx204. doi: 10.1093/femsle/fnx204
Pacheco, A. R., Curtis, M. M., Ritchie, J. M., Munera, D., Waldor, M. K., Moreira, C. G., et al. (2012). Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117. doi: 10.1038/nature11623
Page, R., and Peti, W. (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214. doi: 10.1038/nchembio.2044
Paget, M. S., and Buttner, M. J. (2003). Thiol-based regulatory switches. Annu. Rev. Genet. 37, 91–121. doi: 10.1146/annurev.genet.37.110801.142538
Park, S. Y., Cromie, M. J., Lee, E. J., and Groisman, E. A. (2010). A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell 142, 737–748. doi: 10.1016/j.cell.2010.07.046
Patel, J. C., and Galan, J. E. (2005). Manipulation of the host actin cytoskeleton by Salmonella–all in the name of entry. Curr. Opin. Microbiol. 8, 10–15. doi: 10.1016/j.mib.2004.09.001
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Pilla, G., McVicker, G., and Tang, C. M. (2017). Genetic plasticity of the Shigella virulence plasmid is mediated by intra- and inter-molecular events between insertion sequences. PLoS Genet. 13:e1007014. doi: 10.1371/journal.pgen.1007014
Porat, R., Clark, B. D., Wolff, S. M., and Dinarello, C. A. (1991). Enhancement of growth of virulent strains of Escherichia coli by interleukin-1. Science 254, 430–432. doi: 10.1126/science.1833820
Porter, M. E., and Dorman, C. J. (1997a). Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol. Gen. Genet. 256, 93–103. doi: 10.1007/s004380050550
Porter, M. E., and Dorman, C. J. (1997b). Positive regulation of Shigella flexneri virulence genes by integration host factor. J. Bacteriol. 179, 6537–6550. doi: 10.1128/jb.179.21.6537-6550.1997
Porter, M. E., Mitchell, P., Roe, A. J., Free, A., Smith, D. G., and Gally, D. L. (2004). Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 54, 1117–1133. doi: 10.1111/j.1365-2958.2004.04333.x
Postow, L., Hardy, C. D., Arsuaga, J., and Cozzarelli, N. R. (2004). Topological domain structure of the Escherichia coli chromosome. Genes Dev. 18, 1766–1779. doi: 10.1101/gad.1207504
Puente, J. L., Bieber, D., Ramer, S. W., Murray, W., and Schoolnik, G. K. (1996). The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20, 87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x
Raczkowska, A., Brzostkowska, M., Kwiatek, A., Bielecki, J., and Brzostek, K. (2011). Modulation of inv gene expression by the OmpR two-component response regulator protein of Yersinia enterocolitica. Folia Microbiol. (Praha) 56, 313–319. doi: 10.1007/s12223-011-0054-9
Rahmati, S., Yang, S., Davidson, A. L., and Zechiedrich, E. L. (2002). Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol. Microbiol. 43, 677–685. doi: 10.1046/j.1365-2958.2002.02773.x
Reading, N. C., Rasko, D., Torres, A. G., and Sperandio, V. (2010). A transcriptome study of the QseEF two-component system and the QseG membrane protein in enterohaemorrhagic Escherichia coli O157: H7. Microbiology 156(Pt 4), 1167–1175. doi: 10.1099/mic.0.033027-0
Reading, N. C., Rasko, D. A., Torres, A. G., and Sperandio, V. (2009). The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 5889–5894. doi: 10.1073/pnas.0811409106
Reading, N. C., Torres, A. G., Kendall, M. M., Hughes, D. T., Yamamoto, K., and Sperandio, V. (2007). A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J. Bacteriol. 189, 2468–2476. doi: 10.1128/JB.01848-06
Rempe, K. A., Hinz, A. K., and Vadyvaloo, V. (2012). Hfq regulates biofilm gut blockage that facilitates flea-borne transmission of Yersinia pestis. J. Bacteriol. 194, 2036–2040. doi: 10.1128/JB.06568-11
Reniere, M. L., Whiteley, A. T., Hamilton, K. L., John, S. M., Lauer, P., Brennan, R. G., et al. (2015). Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517, 170–173. doi: 10.1038/nature14029
Rich, E. J., McDonald, R. A., and Christie, D. L. (1990). Yersinia pseudotuberculosis: report of a case with endoscopic findings. J. Pediatr. Gastroenterol. Nutr. 10, 413–415. doi: 10.1097/00005176-199004000-00025
Romeo, T. (1998). Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29, 1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x
Rosenshine, I., Ruschkowski, S., and Finlay, B. B. (1996). Expression of attaching/effacing activity by enteropathogenic Escherichia coli depends on growth phase, temperature, and protein synthesis upon contact with epithelial cells. Infect. Immun. 64, 966–973.
Rosenzweig, J. A., and Schesser, K. (2007). Polynucleotide phosphorylase and the T3SS. Adv. Exp. Med. Biol. 603, 217–224. doi: 10.1007/978-0-387-72124-8_19
Rosenzweig, J. A., Weltman, G., Plano, G. V., and Schesser, K. (2005). Modulation of Yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 280, 156–163. doi: 10.1074/jbc.M405662200
Rowe, S., Hodson, N., Griffiths, G., and Roberts, I. S. (2000). Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J. Bacteriol. 182, 2741–2745. doi: 10.1128/JB.182.10.2741-2745.2000
Rychlik, I., and Barrow, P. A. (2005). Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 29, 1021–1040. doi: 10.1016/j.femsre.2005.03.005
Sabnis, N. A., Yang, H., and Romeo, T. (1995). Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 270, 29096–29104. doi: 10.1074/jbc.270.49.29096
Sala, C., Forti, F., Di Florio, E., Canneva, F., Milano, A., Riccardi, G., et al. (2003). Mycobacterium tuberculosis FurA autoregulates its own expression. J. Bacteriol. 185, 5357–5362. doi: 10.1128/JB.185.18.5357-5362.2003
Santiago, A. E., Yan, M. B., Tran, M., Wright, N., Luzader, D. H., Kendall, M. M., et al. (2016). A large family of anti-activators accompanying XylS/AraC family regulatory proteins. Mol. Microbiol. 101, 314–332. doi: 10.1111/mmi.13392
Sauter, C., Basquin, J., and Suck, D. (2003). Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 31, 4091–4098. doi: 10.1093/nar/gkg480
Sayeed, S., Brendler, T., Davis, M., Reaves, L., and Austin, S. (2005). Surprising dependence on postsegregational killing of host cells for maintenance of the large virulence plasmid of Shigella flexneri. J. Bacteriol. 187, 2768–2773. doi: 10.1128/JB.187.8.2768-2773.2005
Schauder, S., Shokat, K., Surette, M. G., and Bassler, B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41, 463–476. doi: 10.1046/j.1365-2958.2001.02532.x
Schechter, L. M., Jain, S., Akbar, S., and Lee, C. A. (2003). The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 71, 5432–5435. doi: 10.1128/IAI.71.9.5432-5435.2003
Schiano, C. A., Koo, J. T., Schipma, M. J., Caulfield, A. J., Jafari, N., and Lathem, W. W. (2014). Genome-wide analysis of small RNAs expressed by Yersinia pestis identifies a regulator of the Yop-Ysc type III secretion system. J. Bacteriol. 196, 1659–1670. doi: 10.1128/JB.01456-13
Schumann, W. (2012). Thermosensor systems in eubacteria. Adv. Exp. Med. Biol. 739, 1–16. doi: 10.1007/978-1-4614-1704-0_1
Shao, F. (2008). Biochemical functions of Yersinia type III effectors. Curr. Opin. Microbiol. 11, 21–29. doi: 10.1016/j.mib.2008.01.005
Sharma, V. K., and Bearson, S. M. (2013). Evaluation of the impact of quorum sensing transcriptional regulator SdiA on long-term persistence and fecal shedding of Escherichia coli O157:H7 in weaned calves. Microb. Pathog. 57, 21–26. doi: 10.1016/j.micpath.2013.02.002
Sheridan, S. D., Benham, C. J., and Hatfield, G. W. (1998). Activation of gene expression by a novel DNA structural transmission mechanism that requires supercoiling-induced DNA duplex destabilization in an upstream activating sequence. J. Biol. Chem. 273, 21298–21308. doi: 10.1074/jbc.273.33.21298
Sledjeski, D. D., Gupta, A., and Gottesman, S. (1996). The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15, 3993–4000. doi: 10.1002/j.1460-2075.1996.tb00773.x
Smith, J. N., Dyszel, J. L., Soares, J. A., Ellermeier, C. D., Altier, C., Lawhon, S. D., et al. (2008). SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS One 3:e2826. doi: 10.1371/journal.pone.0002826
Soper, T., Mandin, P., Majdalani, N., Gottesman, S., and Woodson, S. A. (2010). Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. U.S.A. 107, 9602–9607. doi: 10.1073/pnas.1004435107
Sperandio, V., Mellies, J. L., Delahay, R. M., Frankel, G., Crawford, J. A., Nguyen, W., et al. (2000). Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38, 781–793. doi: 10.1046/j.1365-2958.2000.02168.x
Sperandio, V., Mellies, J. L., Nguyen, W., Shin, S., and Kaper, J. B. (1999). Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 15196–15201. doi: 10.1073/pnas.96.26.15196
Sperandio, V., Torres, A. G., Jarvis, B., Nataro, J. P., and Kaper, J. B. (2003). Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U.S.A. 100, 8951–8956. doi: 10.1073/pnas.1537100100
Sperandio, V., Torres, A. G., and Kaper, J. B. (2002). Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43, 809–821. doi: 10.1046/j.1365-2958.2002.02803.x
Stein, M., Kenny, B., Stein, M. A., and Finlay, B. B. (1996). Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J. Bacteriol. 178, 6546–6554. doi: 10.1128/jb.178.22.6546-6554.1996
Stein, R. A., Deng, S., and Higgins, N. P. (2005). Measuring chromosome dynamics on different time scales using resolvases with varying half-lives. Mol. Microbiol. 56, 1049–1061. doi: 10.1111/j.1365-2958.2005.04588.x
Stoebel, D. M., Free, A., and Dorman, C. J. (2008). Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154(Pt 9), 2533–2545. doi: 10.1099/mic.0.2008/020693-0
Surette, M. G., Miller, M. B., and Bassler, B. L. (1999). Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. U.S.A. 96, 1639–1644. doi: 10.1073/pnas.96.4.1639
Swinger, K. K., and Rice, P. A. (2004). IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 14, 28–35. doi: 10.1016/j.sbi.2003.12.003
Taga, M. E., Miller, S. T., and Bassler, B. L. (2003). Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50, 1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x
Taga, M. E., Semmelhack, J. L., and Bassler, B. L. (2001). The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42, 777–793. doi: 10.1046/j.1365-2958.2001.02669.x
Tatsuno, I., Nagano, K., Taguchi, K., Rong, L., Mori, H., and Sasakawa, C. (2003). Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71, 2598–2606. doi: 10.1128/IAI.71.5.2598-2606.2003
Tauschek, M., Strugnell, R. A., and Robins-Browne, R. M. (2002). Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44, 1533–1550. doi: 10.1046/j.1365-2958.2002.02968.x
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108, 17480–17485. doi: 10.1073/pnas.1107857108
Thompson, J. A., Oliveira, R. A., Djukovic, A., Ubeda, C., and Xavier, K. B. (2015). Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10, 1861–1871. doi: 10.1016/j.celrep.2015.02.049
Tobe, T., Ando, H., Ishikawa, H., Abe, H., Tashiro, K., Hayashi, T., et al. (2005). Dual regulatory pathways integrating the RcsC-RcsD-RcsB signalling system control enterohaemorrhagic Escherichia coli pathogenicity. Mol. Microbiol. 58, 320–333. doi: 10.1111/j.1365-2958.2005.04828.x
Tobe, T., Yoshikawa, M., Mizuno, T., and Sasakawa, C. (1993). Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J. Bacteriol. 175, 6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993
Tramonti, A., Visca, P., De Canio, M., Falconi, M., and De Biase, D. (2002). Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184, 2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002
Trosky, J. E., Liverman, A. D., and Orth, K. (2008). Yersinia outer proteins: Yops. Cell Microbiol. 10, 557–565. doi: 10.1111/j.1462-5822.2007.01109.x
Trulzsch, K., Roggenkamp, A., Pelludat, C., Rakin, A., Jacobi, C., and Heesemann, J. (2001). Cloning and characterization of the gene encoding periplasmic 2′,3′-cyclic phosphodiesterase of Yersinia enterocolitica O:8. Microbiology 147(Pt 1), 203–213. doi: 10.1099/00221287-147-1-203
Vakulskas, C. A., Potts, A. H., Babitzke, P., Ahmer, B. M., and Romeo, T. (2015). Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 79, 193–224. doi: 10.1128/MMBR.00052-14
Van Houdt, R., Aertsen, A., Moons, P., Vanoirbeek, K., and Michiels, C. W. (2006). N-acyl-L-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol. Lett. 256, 83–89. doi: 10.1111/j.1574-6968.2006.00103.x
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M., and Hardie, K. R. (2005). Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3, 383–396. doi: 10.1038/nrmicro1146
Veron, W., Lesouhaitier, O., Pennanec, X., Rehel, K., Leroux, P., Orange, N., et al. (2007). Natriuretic peptides affect Pseudomonas aeruginosa and specifically modify lipopolysaccharide biosynthesis. FEBS J. 274, 5852–5864. doi: 10.1111/j.1742-4658.2007.06109.x
Vogt, S. L., and Raivio, T. L. (2014). Hfq reduces envelope stress by controlling expression of envelope-localized proteins and protein complexes in enteropathogenic Escherichia coli. Mol. Microbiol. 92, 681–697. doi: 10.1111/mmi.12581
Vytvytska, O., Jakobsen, J. S., Balcunaite, G., Andersen, J. S., Baccarini, M., and von Gabain, A. (1998). Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc. Natl. Acad. Sci. U.S.A. 95, 14118–14123. doi: 10.1073/pnas.95.24.14118
Walters, M., Sircili, M. P., and Sperandio, V. (2006). AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188, 5668–5681. doi: 10.1128/JB.00648-06
Wang, L., Hashimoto, Y., Tsao, C. Y., Valdes, J. J., and Bentley, W. E. (2005). Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187, 2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005
Wang, X., Lord, D. M., Cheng, H. Y., Osbourne, D. O., Hong, S. H., Sanchez-Torres, V., et al. (2012). A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 8, 855–861. doi: 10.1038/nchembio.1062
Wang, X., Lord, D. M., Hong, S. H., Peti, W., Benedik, M. J., Page, R., et al. (2013). Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 15, 1734–1744. doi: 10.1111/1462-2920.12063
Wang, Y., Wang, H., Hay, A. J., Zhong, Z., Zhu, J., and Kan, B. (2015). Functional RelBE-family toxin-antitoxin pairs affect biofilm maturation and intestine colonization in Vibrio cholerae. PLoS One 10:e0135696. doi: 10.1371/journal.pone.0135696
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell. Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Weatherspoon-Griffin, N., and Wing, H. J. (2016). Characterization of SlyA in Shigella flexneri identifies a novel role in virulence. Infect. Immun. 84, 1073–1082. doi: 10.1128/IAI.00806-15
Weilbacher, T., Suzuki, K., Dubey, A. K., Wang, X., Gudapaty, S., Morozov, I., et al. (2003). A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48, 657–670. doi: 10.1046/j.1365-2958.2003.03459.x
Willias, S. P., Chauhan, S., Lo, C. C., Chain, P. S., and Motin, V. L. (2015). CRP-Mediated carbon catabolite regulation of Yersinia pestis biofilm formation is enhanced by the carbon storage regulator protein, CsrA. PLoS One 10:e0135481. doi: 10.1371/journal.pone.0135481
Winardhi, R. S., Gulvady, R., Mellies, J. L., and Yan, J. (2014). Locus of enterocyte effacement-encoded regulator (Ler) of pathogenic Escherichia coli competes off histone-like nucleoid-structuring protein (H-NS) through noncooperative DNA binding. J. Biol. Chem. 289, 13739–13750. doi: 10.1074/jbc.M113.545954
Winzer, K., Hardie, K. R., Burgess, N., Doherty, N., Kirke, D., Holden, M. T., et al. (2002a). LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148(Pt 4), 909–922. doi: 10.1099/00221287-148-4-909
Winzer, K., Hardie, K. R., and Williams, P. (2002b). Bacterial cell-to-cell communication: sorry, can’t talk now – Gone to lunch! Curr. Opin. Microbiol. 5, 216–222. doi: 10.1016/S1369-5274(02)00304-1
Winzer, K., Hardie, K. R., and Williams, P. (2003). LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv. Appl. Microbiol. 53, 291–396. doi: 10.1016/S0065-2164(03)53009-X
Woestyn, S., Allaoui, A., Wattiau, P., and Cornelis, G. R. (1994). YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol. 176, 1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994
Wu, L., Estrada, O., Zaborina, O., Bains, M., Shen, L., Kohler, J. E., et al. (2005). Recognition of host immune activation by Pseudomonas aeruginosa. Science 309, 774–777. doi: 10.1126/science.1112422
Xavier, K. B., and Bassler, B. L. (2005). Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187, 238–248. doi: 10.1128/JB.187.1.238-248.2005
Xue, T., Zhao, L., and Sun, B. (2013). LuxS/AI-2 system is involved in antibiotic susceptibility and autolysis in Staphylococcus aureus NCTC 8325. Int. J. Antimicrob Agents 41, 85–89. doi: 10.1016/j.ijantimicag.2012.08.016
Yan, Y., Su, S., Meng, X., Ji, X., Qu, Y., Liu, Z., et al. (2013). Determination of sRNA expressions by RNA-seq in Yersinia pestis grown in vitro and during infection. PLoS One 8:e74495. doi: 10.1371/journal.pone.0074495
Yang, G., Wang, L., Wang, Y., Li, P., Zhu, J., Qiu, S., et al. (2015). hfq regulates acid tolerance and virulence by responding to acid stress in Shigella flexneri. Res. Microbiol. 166, 476–485. doi: 10.1016/j.resmic.2015.06.007
Yang, J., Jain, C., and Schesser, K. (2008). RNase E regulates the Yersinia type 3 secretion system. J. Bacteriol. 190, 3774–3778. doi: 10.1128/JB.00147-08
Yao, Y., Martinez-Yamout, M. A., Dickerson, T. J., Brogan, A. P., Wright, P. E., and Dyson, H. J. (2006). Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J. Mol. Biol. 355, 262–273. doi: 10.1016/j.jmb.2005.10.041
Yerushalmi, G., Litvak, Y., Gur-Arie, L., and Rosenshine, I. (2014). Dynamics of expression and maturation of the type III secretion system of enteropathogenic Escherichia coli. J. Bacteriol. 196, 2798–2806. doi: 10.1128/JB.00069-14
Zav’yalov, V. P., Chernovskaya, T. V., Navolotskaya, E. V., Karlyshev, A. V., MacIntyre, S., Vasiliev, A. M., et al. (1995). Specific high affinity binding of human interleukin 1 beta by Caf1A usher protein of Yersinia pestis. FEBS Lett. 371, 65–68. doi: 10.1016/0014-5793(95)00878-D
Zhang, L., Chaudhuri, R. R., Constantinidou, C., Hobman, J. L., Patel, M. D., Jones, A. C., et al. (2004). Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72, 7282–7293. doi: 10.1128/IAI.72.12.7282-7293.2004.
Zhang, Y., Qi, K., Jing, Y., Zuo, J., Hu, J., Lv, X., et al. (2017). LsrB-based and temperature-dependent identification of bacterial AI-2 receptor. AMB Express 7:188. doi: 10.1186/s13568-017-0486-y.
Zhu, C., Feng, S., Thate, T. E., Kaper, J. B., and Boedeker, E. C. (2006). Towards a vaccine for attaching/effacing Escherichia coli: a LEE encoded regulator (ler) mutant of rabbit enteropathogenic Escherichia coli is attenuated, immunogenic, and protects rabbits from lethal challenge with the wild-type virulent strain. Vaccine 24, 3845–3855. doi: 10.1016/j.vaccine.2005.07.019.
Keywords: enteropathogen, quorum sensing, E. coli, Shigella, Salmonella
Citation: Chatterjee R, Shreenivas MM, Sunil R and Chakravortty D (2019) Enteropathogens: Tuning Their Gene Expression for Hassle-Free Survival. Front. Microbiol. 9:3303. doi: 10.3389/fmicb.2018.03303
Received: 26 September 2018; Accepted: 19 December 2018;
Published: 09 January 2019.
Edited by:
Xihui Shen, Northwest A&F University, ChinaReviewed by:
Bei Han, Xi’an Jiaotong University, ChinaWeipeng Zhang, Guangzhou HKUST Fok Ying Tung Research Institute, China
Yunlong Li, Wadsworth Center, United States
Copyright © 2019 Chatterjee, Shreenivas, Sunil and Chakravortty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dipshikha Chakravortty, ZGlwYUBpaXNjLmFjLmlu
 Ritika Chatterjee
Ritika Chatterjee Meghanashree M. Shreenivas1,2,3
Meghanashree M. Shreenivas1,2,3 Rohith Sunil
Rohith Sunil Dipshikha Chakravortty
Dipshikha Chakravortty