- 1Institute of Infectious Disease, Beijing Ditan Hospital, Capital Medical University, Beijing, China
- 2Beijing Key Laboratory of Emerging Infectious Disease, Beijing, China
- 3Basic Research Laboratory, Frederick National Laboratory for Cancer Research, National Cancer Institute, Leidos Biomedical Research, Inc., Frederick, MD, United States
- 4Baruch S. Blumberg Institute, Hepatitis B Foundation, Doylestown, PA, United States
Interferon-induced transmembrane proteins (IFITMs) are a family of small proteins that localize in the plasma and endolysosomal membranes. IFITMs not only inhibit viral entry into host cells by interrupting the membrane fusion between viral envelope and cellular membranes, but also reduce the production of infectious virions or infectivity of progeny virions. Not surprisingly, some viruses can evade the restriction of IFITMs and even hijack the antiviral proteins to facilitate their infectious entry into host cells or promote the assembly of virions, presumably by modulating membrane fusion. Similar to many other host defense genes that evolve under the selective pressure of microorganism infection, IFITM genes evolved in an accelerated speed in vertebrates and many single-nucleotide polymorphisms (SNPs) have been identified in the human population, some of which have been associated with severity and prognosis of viral infection (e.g., influenza A virus). Here, we review the function and potential impact of genetic variation for IFITM restriction of viral infections. Continuing research efforts are required to decipher the molecular mechanism underlying the complicated interaction among IFITMs and viruses in an effort to determine their pathobiological roles in the context of viral infections in vivo.
Interferon-induced transmembrane proteins (IFITMs) are a family of small proteins that can be found in single cell organisms and are evolutionally conserved across vertebrates (Siegrist et al., 2011; Zhang et al., 2012). The human IFITM family comprises five members, including immune-related IFITM1, IFITM2, and IFITM3, as well as IFITM5 and IFITM10 with no known role in immunity. As key host defense genes, IFITMs evolved under the selective pressure of microorganism infection (Compton et al., 2016). IFITM proteins are involved in many aspects of virus–host interaction and play important roles in viral pathogenesis. Among the number of single-nucleotide polymorphisms (SNPs) in IFITM3 gene that have been identified in human populations, several are associated with disease severity and prognosis of influenza A virus (IAV) and other viral infections (Everitt et al., 2012; Zhang et al., 2015; Xu-Yang et al., 2016; Allen et al., 2017). Mechanistically, these SNPs either alter the expression of IFITM3 or result in expression of N-terminally truncated IFITM3 isoform, Δ21-IFITM3, with reduced antiviral activity against different viruses (Everitt et al., 2012; Allen et al., 2017). In this review, we will summarize the findings on human IFITM structural features related with antiviral activity, and impact of genetic variation on IFITM antiviral function in the control and pathogenesis of viral infections in humans.
IFITMs Restrict a Broad Spectrum of Viruses in Cultured Cells and In Vivo
To date, IFITMs have been shown to inhibit the infection of enveloped RNA viruses from 9 viral families (Perreira et al., 2013), non-enveloped RNA viruses, e.g., reovirus (Anafu et al., 2013) and foot-and-mouth disease virus (Xu et al., 2014), and several DNA viruses (Li et al., 2018). IFITMs efficiently inhibit a number of medically important human pathogenic viruses, including IAV (Brass et al., 2009; Bailey et al., 2012), dengue virus (DENV) (Brass et al., 2009; Jiang et al., 2010), West Nile virus (WNV) (Brass et al., 2009; Jiang et al., 2010), Zika Virus (ZIKV) (Savidis et al., 2016), Ebola virus (EBOV) (Huang et al., 2011; Wrensch et al., 2015), Marburg virus (MARV) (Huang et al., 2011), severe acute respiratory syndrome coronavirus (SARS-CoV) (Huang et al., 2011), Rift Valley fever virus (RVFV) (Mudhasani et al., 2013), Hantaan virus (HTNV) (Mudhasani et al., 2013; Xu-Yang et al., 2016), hepatitis C virus (HCV) (Wilkins et al., 2013), and human immunodeficiency virus (HIV) (Lu et al., 2011; Compton et al., 2014; Yu et al., 2015; Compton et al., 2016; Foster et al., 2016; Chesarino et al., 2017; Tartour et al., 2017; Wang et al., 2017). In vivo studies in IFITM3 knockout mice demonstrate the critical role of IFITM3 in restricting infection and reducing disease severity of infection by IAV (Bailey et al., 2012; Everitt et al., 2012), WNV (Gorman et al., 2016), Chikungunya virus and Venezuelan equine encephalitis virus (Poddar et al., 2016), and respiratory syncytial virus (Everitt et al., 2013). IFITM3 in mice not only protects lung epithelia cells from IAV infection, but it was also shown to restricts IAV infection of lung dendritic cells, which traffic to lymph nodes to prime CD8+ T cell anti-viral response (Infusini et al., 2015). Moreover, lung resident memory CD8+ T cells in mice were programmed to retain IFITM3 expression, facilitating their survival and protection from viral infection during subsequent exposures (Wakim et al., 2013).
IFITMs Mainly Restrict Virus Infection at Cell Entry
IFITM proteins localize at the plasma membrane as well as the membranes of endocytic vesicles and lysosomes (Bailey et al., 2014). IFITMs can also be incorporated into envelope membranes of many viruses (Yu et al., 2015; Tartour et al., 2017). Emerging evidence suggests that IFITM proteins on both viral and cellular membranes can restrict the infectious entry of diverse envelope viruses by inhibiting viral fusion at cell plasma or endolysosomal membranes, but IFITM does not impede endocytosis of virions into cells (Weidner et al., 2010; Li et al., 2013; Perreira et al., 2013; Compton et al., 2014; Desai et al., 2014). As extensively discussed in a previous review (Perreira et al., 2013), the potency of IFITM restriction of viral cell entry is generally correlated to the co-localization of IFITM proteins at the sites of viral fusion. For instance, IFITM1 more efficiently restricts the viruses that enter the cytoplasm via direct fusion with plasma membrane or via Rab-5 positive early endosomes, whereas IFITM3 more efficiently inhibits viruses that enter via Rab7-positive late endosomes or lysosomes. This rule is highlighted by the finding that mutation of IFITM3 endocytic signal results in its cell surface accumulation and gains a function to restrict the infection of human parainfluenza virus 3 (HPIV-3), which enters cells via direct fusion with the plasma membrane (Rabbani et al., 2016; Zhao et al., 2018). Another example is that the sensitivity of IAVs to IFITM3 appears to depend on the pH value at which the IAV hemagglutinin triggers membrane fusion and thus the endocytic compartments where the membrane fusion take place (Gerlach et al., 2017). However, exceptions of this rule do exist. For instance, it remains to know how Moloney leukemia virus (MLV) and Sendai virus that fuse at cell plasma membrane (Brass et al., 2009; Hach et al., 2013) as well as Lassa fever virus (LASV) and lymphocytic choriomeningitis virus (LCMV) that fuse at Rab7-positive late endosomes (Brass et al., 2009; Mudhasani et al., 2013) to escape IFITM1 and IFITM3 restriction, respectively.
The mechanism of IFITM inhibition of viral fusion and cell entry is not yet resolved. One study reported that IFITM inhibited Jaagsiekte sheep retrovirus envelope and IAV hemagglutinin fusion of viral envelope to cellular membranes prior to coalescence of lipid-bilayers, a process known as hemifusion (Li et al., 2013). However, another study using a direct virus-cell fusion assay in viable cells to investigate IAV entry found that overexpression of IFITM3 protein in late endosomes did not alter lipid mixing, but rather inhibited the release of viral contents into the cytoplasm, suggesting that IFITM3 inhibits the transition from hemifusion to full fusion of the respective lipid membranes (Desai et al., 2014). Others studies suggested that IFITM multimerization decreases membrane flexibility by altering membrane curvature, with the consequence of interruption of the virus-cell membrane fusion (John et al., 2013; Lin et al., 2013). Another group reported that IFITM expression increased cholesterol content in the endosome or lysosome via an interaction with vesicle-associated membrane protein-associated protein A (VAPA), which abrogated endolysosomal fusion with the viral envelop (Amini-Bavil-Olyaee et al., 2013). However, this later observation was not confirmed by other studies (Desai et al., 2014; Wrensch et al., 2014). In addition, IFITMs were reported to affect the trafficking of vacuolar ATPase (v-ATPase), implying that IFITMs may indirectly restrict viral entry by modulating endosomal acidity (Wee et al., 2012). It is also well documented that the antiviral potency of IFITM proteins varies among different cell types (Huang et al., 2011; Zhao et al., 2018), suggesting that IFITMs work together with other cellular proteins to modulate viral fusion. In support of this notion, zinc metallopeptidase STE24 (ZMPSTE24), a transmembrane metalloprotease localized in the inner nuclear membrane and cytoplasmic organelles, had been identified as a downstream partner of IFITM3 to restrict the entry of enveloped RNA and DNA viruses (Fu et al., 2017).
In addition to the protective function of IFITM proteins to reduce viral infection of host cells, IFITM proteins, particularly IFITM2 and 3, can lead to the production of virions that package IFITMs and display reduced entry into target cells (Compton et al., 2014; Tartour et al., 2014, 2017). One study found that IFITM proteins interact with HIV envelope protein in viral producer cells to disrupt envelope protein processing and virion incorporation, which impairs virion infectivity (Yu et al., 2015); however, another found that IFITM3 expression in producing cells did not affect the amount of envelope protein incorporation into progeny HIV-1 virions (Appourchaux et al., 2018). Both studies reported that the level of IFITM protein incorporation into progeny virions does not correlate with the extent of infectivity reduction.
Moreover, recent studies have also shown that IFITMs can modulate viral infection and pathogenesis via mechanisms that are not directly related to the restriction of virus entry. For example, given the resistance of human papillomaviruses (HPV) to IFITMs, HPV infection of keratinocytes inhibits the expression of IFITM1 and RIPK3 to escape from IFN-γ and TNF-α-mediated antiproliferation and necroptosis, which is essential for establishing a persistent infection (Ma et al., 2016). In addition, it was demonstrated in IFITM3 knockout mice that IFITM3 limited murine CMV (MCMV) pathogenesis without directly preventing virus replication (Stacey et al., 2017). Instead, IFITM3 contributed to the antiviral cellular immunity by abrogating inflammatory cytokine-driven lymphopenia including apoptosis-independent NK cell death and T cells depletion (Stacey et al., 2017).
Viral Counteraction for Evading IFITMs
Not surprisingly, in the arms race between pathogen and host, many viruses have evolved strategies to evade the antiviral function of host restriction proteins. For instance, transmitted founder HIV-1 strains establishing de novo infection are generally capable of evading IFITM restriction (Foster et al., 2016; Wang et al., 2017). Mutations allow the virus to escape adaptive immune responses, and/or switches in the HIV-1 co-receptor tropism from CCR5 to CXCR4 increases viral sensitivity to inhibition by IFITM2 and IFITM3 in endosomal compartments (Foster et al., 2016). Moreover, IAV facilitates its infection by activating p53 or degrading eukaryotic translation initiation factor 4B (eIF4B) to inhibit the expression of IFITM proteins (Wang S. et al., 2014; Wang et al., 2018). In contrast, human coronavirus OC43 (HCoV-OC43) and human cytomegalovirus (HCMV) hijack IFITM3 to promote its infectious entry and progeny virions assembly, respectively (Zhao et al., 2014; Xie et al., 2015).
Structure and Polymorphism of IFITM Gene Locus
The human IFITM locus, approximately 18kb long, is located on chromosome 11 and comprises five genes: IFITM1, IFITM2, IFITM3, IFITM5, and IFITM10 (Figure 1). As an IFN-stimulated gene (ISG), IFITM1, IFITM2, IFITM3 genes each has an interferon stimulated response element (ISRE) in its promoter region besides the additional gamma-activated sequence (GAS) in the promoter region of IFITM1 gene (Siegrist et al., 2011). However, the protein expression of IFITM5 and IFITM10 are not induced by IFNs (Zhang et al., 2012). Though IFITM1-3 proteins are ubiquitously expressed in human tissues in the absence of IFN induction, they can be robustly up-regulated by all three types of IFNs (Zhao et al., 2014). The IFITM3 promoter has binding sites for dozens of transcription factors, including POLR2A, MYC, ELF1, PHF8, CHD1, TAF1, REST, SIN3AK20, SIN3A, IRF1, STAT1, TBP, STAT3, STAT2, ZBTB7A, and CTCF, some of which may affect IFITM3 expression (Allen et al., 2017).
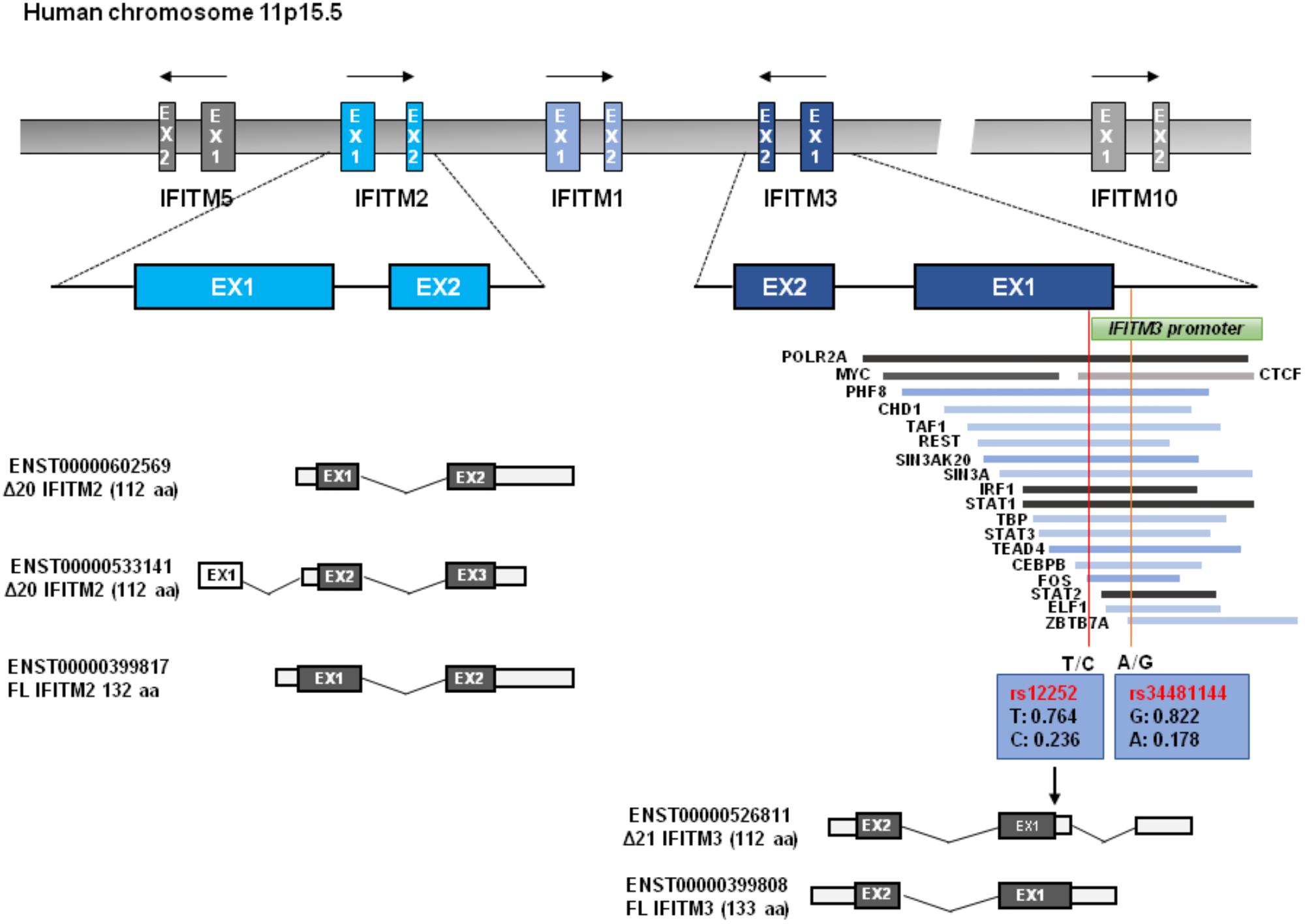
Figure 1. IFITM gene structure. Human IFITM genes located in chromosome 11p15.5 are indicated, and the exons of immunity-related IFITM genes (IFITM1, IFITM2, and IFITM3) are indicated in blue color. The putative transcripts of IFITM2 and IFITM3 are also shown below. The transcription factor binding regions derived from UCSC Genome Browser are shown, which is verified by Chip-seq from the 161 proteins tested through ENCODE; GRCh37/hg19. Two red vertical lines across transcription factor binding sites represent the Influenza associated SNPs of rs12252 and rs34481144 with allele frequencies, respectively. N-terminally truncated IFITM3 isoform (Δ21 IFITM3) that is presumably generated by rs12252 is indicated.
As illustrated in Figure 1, all IFITM genes contain two coding exons interspersed by one intron. Both IFITM2 and IFITM3 were predicated to encode a wild typed full-length form and a truncated isoform with 20/21 amino acid residues deletion from N-terminus. Most vertebrate animals have two or more IFITM genes. In many primate species, gene duplication, and divergence of IFITM3 has been identified (Zhang et al., 2012; Compton et al., 2016). IFITM locus in many modern primate species contains multiple copies of IFITM3-like genes due to gene duplication (Compton et al., 2016). For instance, Marmoset, Macaque, and Africa Green Monkey (AGM) each has five, six and eight copies of IFITM3 genes, respectively (Compton et al., 2016). Comparative genomics studies indicate that IFITM3 is the most ancient member in the IFITM family and IFITM2 emerges as a rather recent genetic event in human, chimpanzee, and gorilla (Compton et al., 2016). The multiplicity and diversity of IFITM2/3 indicates that there is a positive selection in IFITM evolution, which is consistent with their role in restricting pathogen invasion (Compton et al., 2016).
Structural Function Relationship of IFITM Proteins
IFITM proteins have several topologies that may affect their function. As shown in Figure 2A, although IFITM3 may adopt three different membrane topologies (Bailey et al., 2014), it exists predominantly as a type II transmembrane protein with the N-terminus in cytosol and the short C-terminus exposed to cellular exterior or in the lumen of endolysosome (Bailey et al., 2013). Although IFITM1 was also reported to predominately adopt a type II transmembrane topology, other membrane topologies may exist (Figure 2B; Li et al., 2015).
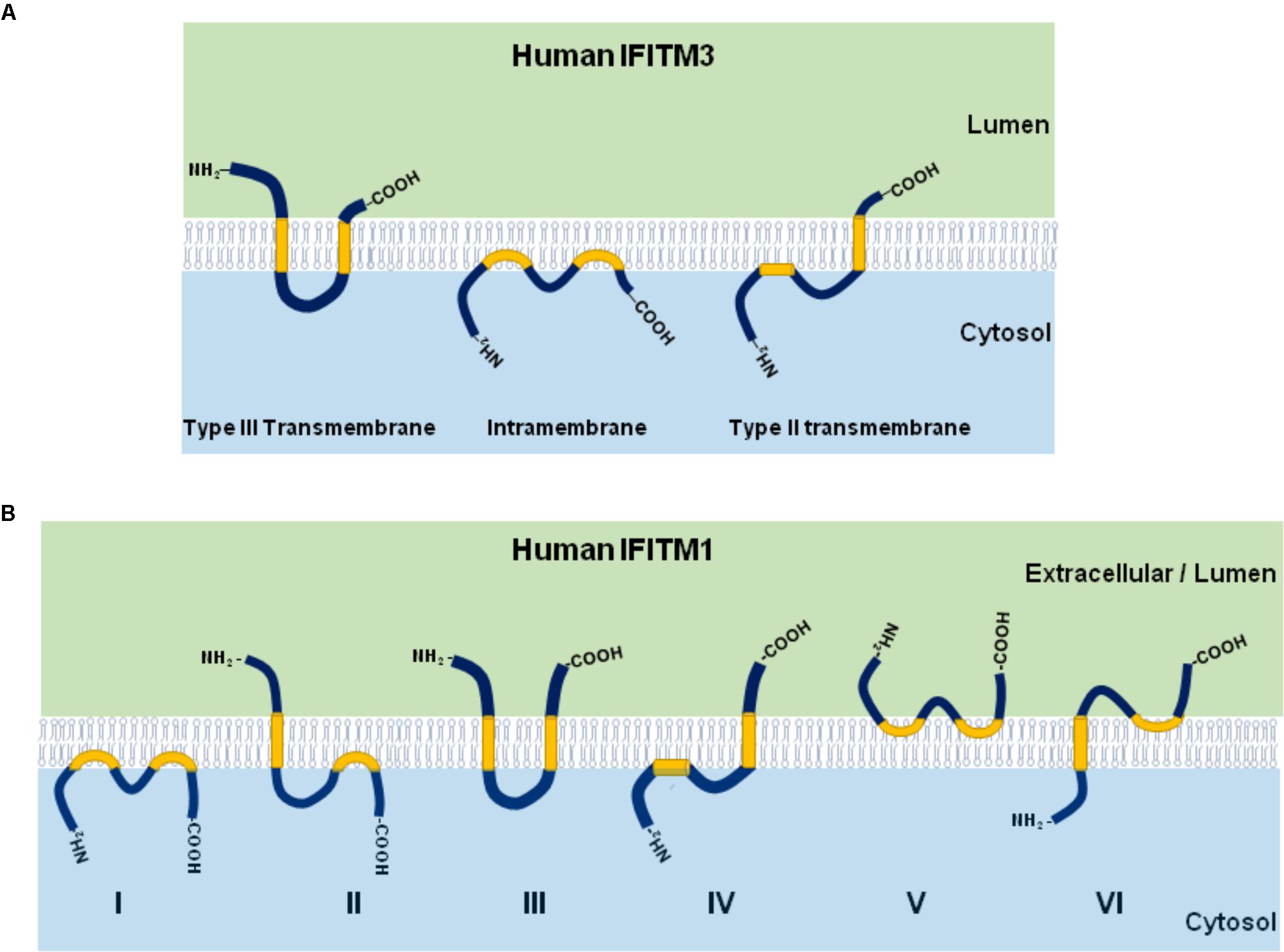
Figure 2. Illustration of IFITM protein membrane topology. The three possible membrane topologies of IFITM3 (A) and six possible membrane topological forms of IFITM1 (B) are presented. The two transmembrane (or intramembrane) regions were highlighted in yellow.
As depicted in Figure 3, IFITMs consist of intramembrane (IMD) and transmembrane (TMD) domains separated by an intracellular loop (CIL) and variable N and C terminal domains (NTD and CTD), respectively (Bailey et al., 2013, 2014; Chesarino et al., 2017). While the NTD and CTD are highly variable in length and sequence among IFITM orthologs and paralogs, the canonical CD225 domain spanning IMD to CIL domains is evolutionally more conserved (Bailey et al., 2014). Critical structure motifs and amino acid residues undergoing post-translational modifications required for IFITM oligomerization as well as their biological and antiviral functions are discussed below.
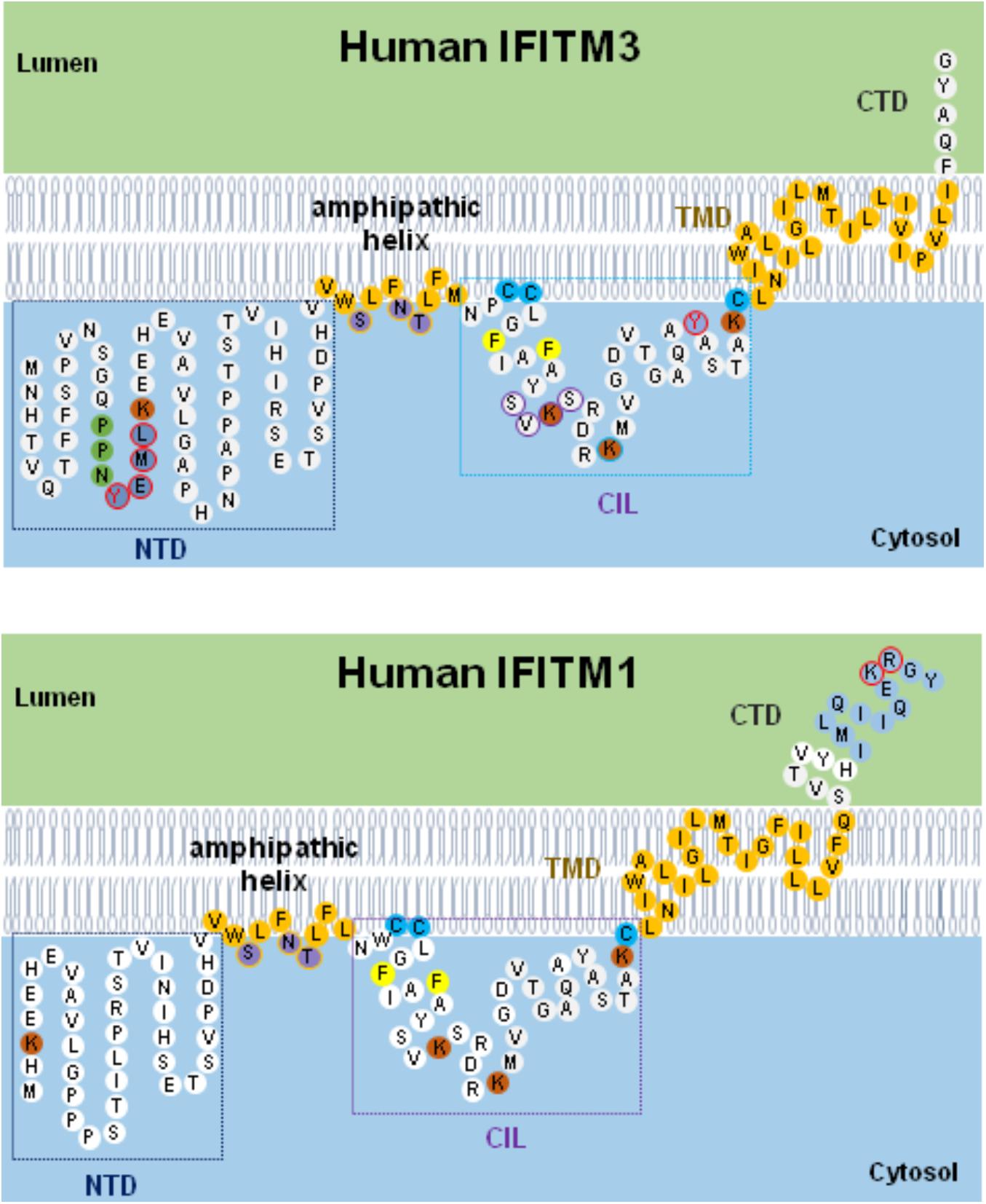
Figure 3. Schematic diagram of IFITM1/3 topology and structural determinants essential for their modulation of viral entry. Five structural domains, including the N-terminal domain (NTD), amphipathic helix domain (AHD), intracellular loop (CIL), transmembrane domain (TMD), and C-terminal domain (CTD), are illustrated. Two membrane-associated domains (AHD and TMD) are embodied in light orange. In IFITM3, three hydrophilic residues (S61, N64, and T65) in amphipathic helix are labeled in violet. In NTD domain, 20YEML23 motif required for IFITM3 endocytosis are depicted in blue with red circle and 17PPNY20 motif recruiting NEDD4 E3 ligase are shown in green. In CIL domain, 81SVKS84 motif required for IFITM3 to inhibit IAV, DENV, and LLOV infection is indicated with purple circle. The residues with post-translational modification, including phosphorylation (Y20 and Y99 labeled with red circle), palmitoylation (C71, C72, and C105 marked with light blue), and ubiquitination (highlighted in orange), are indicated. In addition, two residues (F75 and F78) essential for oligomerization are marked with yellow. In IFITM1, C-terminal 12-aa residues critical for modulating human coronaviruses entry is indicated in blue, and 122KR123 dibasic motif constituted as IFITM1 sorting signal is depicted with red circle.
NTD of IFITM2 and IFITM3
Compared to IFITM1, IFITM2 and IFITM3 have N-terminal 20-aa or 21-aa extension which was previously regarded to be absent in rs12252-C encoding IFITM3 isoform. Although deletion of this N-terminal 21-aa region significantly impaired its ability to inhibit the infection by IAV, VSV, and DENV, the deletion apparently did not affect its ability to enhance HCoV-OC43pp infection (Weidner et al., 2010; John et al., 2013). Interestingly, Δ21-IFITM3 enhanced inhibition of HIV-1 fusion (Compton et al., 2016). Moreover, Δ20-IFITM2, a N-terminal 20-aa truncated isoform of IFITM2, which is derived from an alternatively initiated RNA transcript (Figure 1), demonstrated a more potent suppression on HIV-1 infection than that by full-length wild-type IFITM2 (Wu et al., 2017). In fact, a recent study argues that the Δ20-IFITM2, rather than the full-length IFITM2 and IFITM3, is the effective restriction factor of HIV-1 with CXCR4-tropism (Wu et al., 2017).
The 20YXXΦ23 motif of IFITM3 is an endocytic signal essential for endocytosis and localization of IFITM3 to endocytic vesicles and lysosomes (Jia et al., 2014). Artificial mutations of 20YEML23 motif (Y20D, Y20A, or L23A) result in an accumulation of IFITM3 in the plasma membrane and reduced inhibition of viruses that enter the cytoplasm at endocytic vesicles/lysosomes, such as IAV, human coronaviruses NL63 and -229E (Chesarino et al., 2014a; Jia et al., 2014; Williams et al., 2014; Zhao et al., 2018), but enhanced inhibition of viruses that directly enter cells at the plasma membranes, such as HPIV-3 and HIV-1 (Jia et al., 2012; Compton et al., 2016; Rabbani et al., 2016). Intriguingly, replacement of IFITM3 tyrosine (Y) 20 with either alanine (A) or aspartic acid (D) to mimic unphosphorylated or phosphorylated IFITM3 converted the antiviral protein to enhance the entry of SARS-CoV and MERS-CoV (Zhao et al., 2018). These studies suggest that the NTD, particularly 20YXXΦ23 motif, plays important roles in IFITM2/3 subcellular localization and antiviral activity. IFITM3 are phosphorylated by the protein-tyrosine kinase Fyn on tyrosine 20 (Y20), which results in its plasma membrane accumulation and decreased antiviral activity against influenza viruses (Jia et al., 2012, 2014; Chesarino et al., 2014a). This result is consistent with the mutagenesis studies of 20YXXΦ23 motif, where Y20 is part of an endocytosis signal that can be blocked by phosphorylation (Chesarino et al., 2014a). Additionally, phosphorylation of IFITM3 by Fyn and mutagenesis of Y20 also lead to decreased IFITM3 ubiquitination, suggesting modification of Y20 as an important mechanism to control IFITM3 trafficking and degradation (Chesarino et al., 2014a).
CTD of IFITM1
Compared to IFITM2 and IFITM3, human IFITM1 has a relatively long C-terminal region of 18 amino acid residues. The 122KRXX125 motif serves as a sorting signal for IFITM1. Substitution of two basic residues 122KR123 with alanine (KR/AA) in IFITM1 reduced its distribution in LAMP1-positive lysosomes but enriched its localization in CD63-positive multivesicular bodies (Li et al., 2015). The KR/AA mutant IFITM1 executed increased activity to inhibit the infection by Jaagsiekte sheep retrovirus (JSRV) and 10A1 amphotropic murine leukemia virus (MLV) (Li et al., 2015). Interestingly, although SARS-CoV and HCoV-NL63 share the ACE2 receptor for infection of host cells, deletion of the C-terminal 3, 6, or 9 amino acids did not apparently affect the activity of IFITM1 to inhibit SARS-CoV entry but enhanced the activity of IFITM1 to inhibit the entry of HCoV-NL63 (Zhao et al., 2018). However, further deletion of the C-terminal 12, 15, or 18 amino acids significantly attenuated or even abolished the ability of IFITM1 to inhibit the entry of SARS-CoV, but did not apparently affect the entry of NL63 (Zhao et al., 2018). More strikingly, deletion of C-terminal 12-aa or 18-aa converted IFITM1 to a potent enhancer of MERS-CoV and HCoV-OC43 entry (Zhao et al., 2014, 2018). A recent mutagenesis study indicated that amino acid residues 127–132 in the CTD domain of IFITM3 differentially modulates its activities in target cell protection and negative imprinting of progeny HIV-1 infectivity (Appourchaux et al., 2018). These studies clearly indicate that the CTD of IFITMs contains multiple structure motifs that regulate the subcellular localization and antiviral functions.
CD225 Domain
The canonical CD225 domain comprises IMD to CIL domains and is evolutionally conserved (Bailey et al., 2014). IMD domain (residues 58–80) is a hydrophobic region and possesses an amphipathic alpha helix spanning residues 59–68 (Chesarino et al., 2017). It was demonstrated recently that either deletion of this alpha helices or mutations altering amphipathicity largely impaired or even abolished IFITM antiviral activity, suggesting a critical role in IFITM antiviral function, presumably through affecting membrane physical properties (Chesarino et al., 2017). In addition, the 81SVKS84 motif residing in CIL domain is essential for IFITM3 to inhibit IAV and DENV infection, but replacement of this motif with four residues of alanine promoted cellular entry driven by Lloviu virus (LLOV) glycoprotein (Wrensch et al., 2015).
Palmitoylation of IFITM
IFITM proteins can be palmytoylated at three cystines in CD225 domain by multiple zinc finger DHHC domain-containing palmitoyltransferases (ZDHHCs) (McMichael et al., 2017). While Cys105 localizes close to the amphipathic alpha helix in the IMD domain, Cys71 and Cys72 are adjacent to the TMD domain. Mutagenesis studies indicated that substitution of those three cysteine residues with alanine alters the IFITM3 distribution from punctate clusters in the cellular membrane to a more diffused pattern and the resulting palmitoylation-deficient mutant showed impaired or even abolished antiviral activity (Yount et al., 2010). Interestingly, when other lipid modification sites, such as myristoylation and prenylation, were introduced in IFITM NTD or CTD domains, the antiviral function of palmitoylation-deficient IFITM3 mutant could be restored. These findings imply that anchoring IFITM protein to membranes, but not structure alteration by S-palmitoylation, is important for IFITM restriction of virus entry (Yount et al., 2012; Chesarino et al., 2014b).
Ubiquitination of IFITM
Human IFITM proteins possess four conserved lysine residues (Figure 2) that can be ubiquitinated by E3 ubiquitin ligase such as NEDD4 (Yount et al., 2012; Chesarino et al., 2015). Previous studies demonstrated that each lysine residue can be modified with mono- and poly-ubiquitination through Lys-48 and Lys-63 linkages (Yount et al., 2012). By replacing all four residues of lysine with alanine, the mutant IFITM3 demonstrated endolysosomal distribution and execute hyperactivity to restrict IAV infection than wild type (Yount et al., 2012). However, our studies demonstrated that ubi-deficient IFITM3 lost antiviral activity against all the viruses tested, including IAV (Zhao et al., 2014, 2018). The discrepancy of those studies is not clear. Importantly, ubiquitination and endocytosis of IFITM3 is required for mTOR inhibitor-induced degradation of IFITM3 (Shi et al., 2018). In addition, IFITM3 K88 can be monomethylated by lysine methyltransferase SET7 and demethylated by histone demethylase LSD1 (Shan et al., 2013, 2017). While vesicular stomatitis virus (VSV) and IAV infection increased IFITM3-K88me1 levels by promoting the interaction between IFITM3 and SET7 and disassociation from LSD1 to attenuate IFITM3 antiviral activity, IFN-α reduced IFITM3-K88me1 levels and increased its antiviral activity (Shan et al., 2013, 2017).
Oligomerization of IFITMs
IFITM proteins function as homo- or hetero-oligomers. Two phenylalanine residues (F75 and F78) are essential for IFITM homo- and hetero-oligomerization (John et al., 2013). IFITM3 bearing F75A and F78A mutations (IFITM3/2FA) showed reduced ability to inhibit the entry of HCoV-NL63, but lost ability to inhibit or enhance the entry of all other tested viruses (Zhao et al., 2014, 2018).
Antiviral Activity of Human IFITM Variants
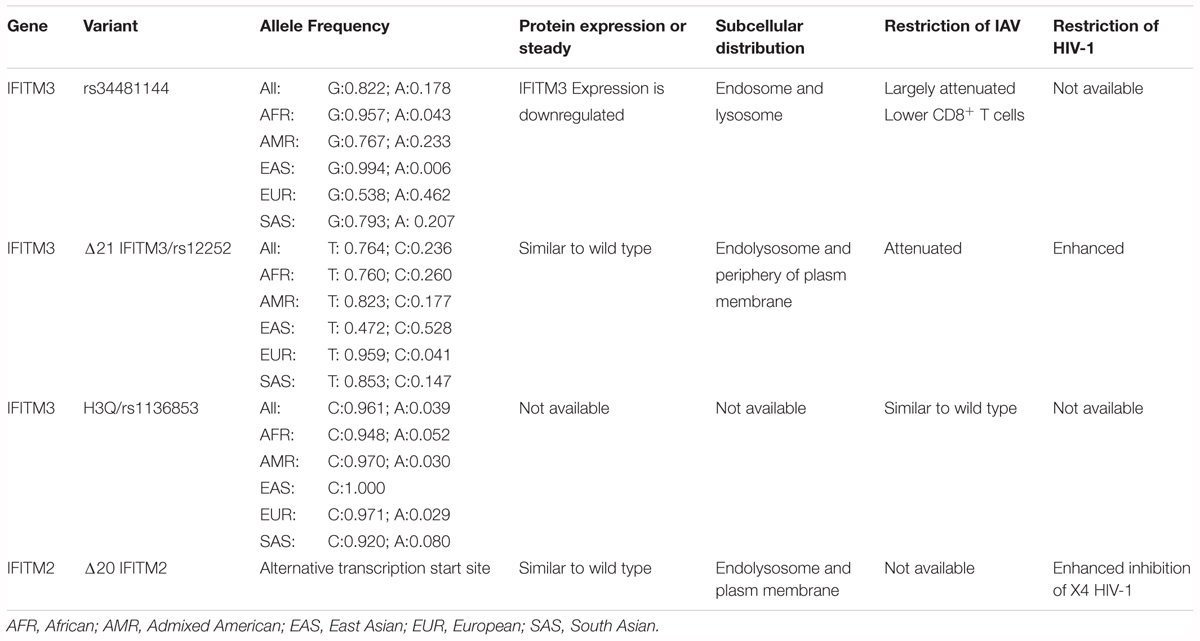
The antiviral activity of many naturally existing human IFITM variants has been tested in cell cultures. As shown in Table 1 and mentioned in previous sections, although SNP rs12252 is predicted to encode a splice variant specifying a N-terminally truncated isoform IFITM3 (Everitt et al., 2012), the putative Δ21-IFITM3 protein has not been detected in the tissues or cells of affected subjects (Wu et al., 2017; Makvandi-Nejad et al., 2018). However, subcellular localization and antiviral activity of genetically engineered Δ21-IFITM3 have been extensively investigated in cultured cells (John et al., 2013; Compton et al., 2016). Interestingly, IFITM2 was reported recently to have a similar N-terminally truncated isoform (Δ20-IFITM2) expressed in human innate immune cells and CD4+ T cells (Wu et al., 2017). Compared with the full-length wild-type IFITM2, Δ20-IFITM2 demonstrated an increased plasma membrane accumulation and enhanced antiviral activity against HIV-1, particularly, HIV-1 with CXCR4 tropism (Compton et al., 2016; Wu et al., 2017). In addition, expression and antiviral activity of some human nonsynonymous IFITM3 variants have also been investigated in cell cultures (John et al., 2013). While many of the SNPs results in undetectable levels of IFITM3 in the transfected cells, presumably due to the reduced stability of mutant proteins, and thus loss of antiviral activity against IAV, a few SNPs did not apparently alter the amount of IFITM proteins but impaired the antiviral activity against IAV. It will be interesting to test all the IFITM variants against a group of viruses and identify the SNPs that potentially impact the control and pathogenesis of specific viral infections in humans.
IFITM Variants and Diseases
IFITM Variants in Human Populations
As a potent viral restriction factor, IFITMs play a pivotal role in limiting the infection by multiple viruses and any genetic variation affecting the IFITM expression or function might contribute to viral pathogenesis. Thus, natural variation in IFITM genes and its association with illness severity has been extensively investigated. Currently, a dozen of SNP in IFITM3 have been reported, some of which may modulate IFITM expression, affect RNA splicing, or result in nonsynonymous or synonymous variants (Table 1). Several SNPs affecting function of IFITMs have been investigated for their association with morbidity, severity and prognosis of microorganism infection (Everitt et al., 2012; Shen et al., 2013; Zhang et al., 2013, 2015; Wang Z. et al., 2014; Allen et al., 2017).
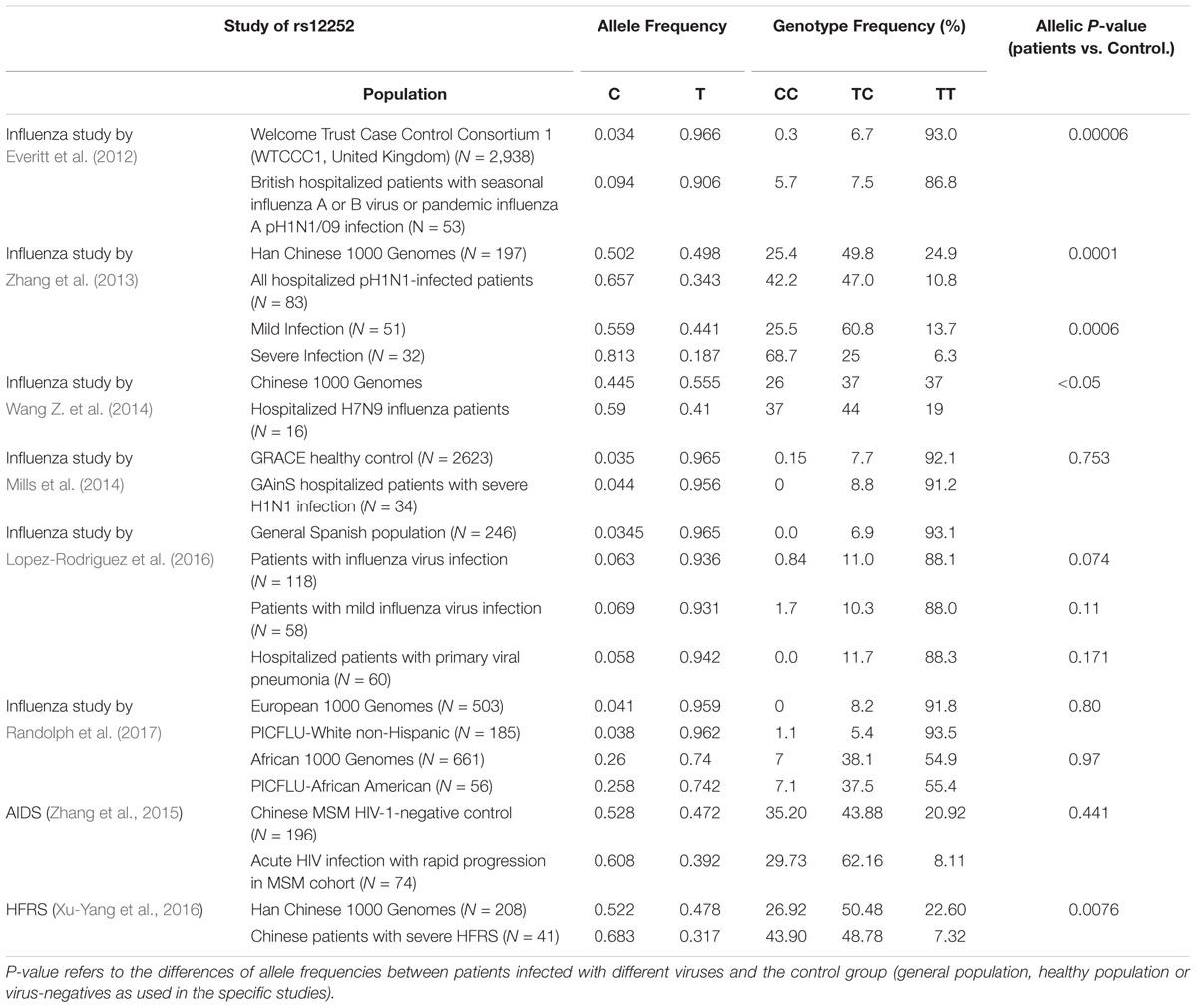
The most studied SNP associated with severe outcomes of IAV infection is SNP rs12252, which is a nonsynonymous variation in the first exon of IFITM3 (Everitt et al., 2012). The substitution of the major allele of T with alternative allele of C was predicted to alter IFITM3 mRNA splicing and generate a N-terminally truncated variant of IFITM3 with 21 amino acid residues deletion (Δ21-IFITM3) (Everitt et al., 2012). The SNP rs12252 has a higher prevalence in the population of East Asia than Europe (0.528 vs. 0.041) (Everitt et al., 2012). Moreover, the homozygous CC genotype and heterozygous TC genotype have significantly higher frequency in East Asia than that in Europe (0.300 vs. 0.000 and 0.456 vs. 0.082, respectively) (Everitt et al., 2012).
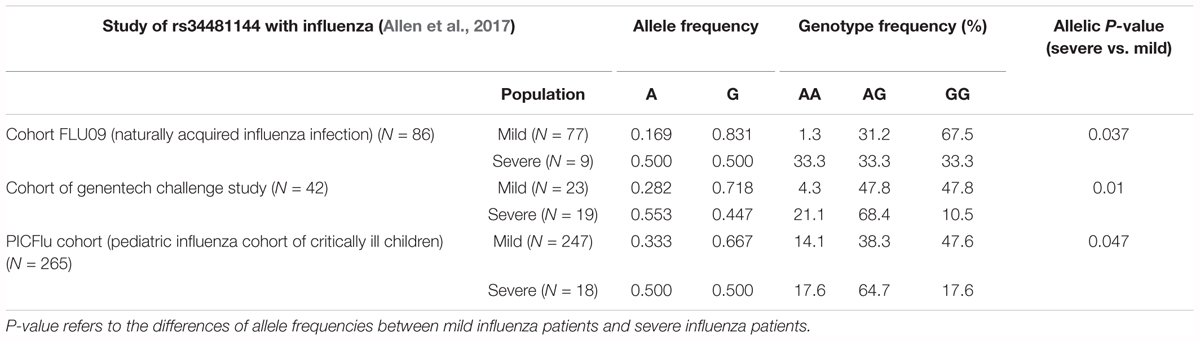
rs34481144 located at the IFITM3 promoter is associated with severe influenza in three human cohorts (Allen et al., 2017). SNP rs34481144, wherein the majority G allele is replaced with a minor A allele, controls IFITM3 promoter activity and determines the expression level. Compared to the G allele, A allele has activities of decreased IRF3 binding and increased CTCF binding, thus resulting in lower IFITM3 expression. SNP rs34481144 has diverse allele and genotypic frequencies in different human populations (Table 1). The allele frequency of rs34481144-G is higher in the population of East Asia and Africa than that in Europe (0.957 and 0.994 vs. 0.538, respectively) (Allen et al., 2017). Also, the homozygous GG genotype has significantly higher frequency in East Asia than Europe (0.988 vs. 0.294) (Allen et al., 2017).
IFITM3 Variants and Influenza A Virus Infection
Several groups previously reported that IFITM3 SNP of rs12252 is associated with the susceptibility and severity of patients with seasonal influenza and pandemic H1N1 and H7N9 IAV infection (Everitt et al., 2012; Zhang et al., 2013; Wang Z. et al., 2014; Pan et al., 2017). As summarized in Table 2, Everitt et al. (2012) first discovered that a higher allelic frequency of rs12252-C exists in Caucasian hospitalized influenza patients than healthy control. Moreover, they observed a remarkably higher frequency of the homologous CC genotype in hospitalized influenza patients compared to the normal European population (5.7% vs. 0.3%) (Everitt et al., 2012). Importantly, another group reported that the minor C allele of rs12252 in the Caucasian population is much more prevalent in Han Chinese and Japanese and is associated with disease severity in patients with pH1N1/09 IAV infection (Zhang et al., 2013). In line with this observation, results obtained from study of H7N IAV infection revealed that H7N9 infected patients with rs12252-CC genotype exhibited accelerated disease progression and increased mortality rate than patients with the TC and TT genotype (Wang Z. et al., 2014). These studies collectively suggest that rs12252-C is a risk allele associated with severe IAV infection. However, several recent studies from European or Africa-American ethnic groups did not support the association of rs12252 with severe influenza infection (Mills et al., 2014; Lopez-Rodriguez et al., 2016; Randolph et al., 2017). The rare frequency of rs12252-C in European population may account for this discrepancy. Through meta-analysis of 11 studies, rs12252 T > C was associated with risk to severe influenza infection with odds ratio of 1.69 (95% CI 1.23, 2.33) in both European and East Asian populations, but for the mild infection, the results remained uncertain (Prabhu et al., 2018).
Although the association of rs12252 with the susceptibility to influenza infection and disease severity was observed by many studies, the mechanism for this association is largely unknown. As mentioned above, although rs12252-C is speculated to encode a N-terminal truncated IFITM3 with attenuated antiviral activity to impede IAV entry, the truncated variant has not been detected so far (Everitt et al., 2012; Makvandi-Nejad et al., 2018). Moreover, the homozygous CC genotype was demonstrated to only express full-length IFITM3 at a similar level to TT genotype (Wu et al., 2017). Thus rs12252-C genetic variant does not appear to affect the biochemical nature and expression of IFITM3. Obviously, further investigation to determine whether rs12252-C influences IFITM3 gene splicing or protein levels in a cell type-specific manner and whether this SNP co-segregates with a different causative allele is warranted.
A recent study found that another IFITM3 SNP rs34481144 has a strong association with disease severity in three influenza cohorts (Allen et al., 2017). rs34481144 is located in the promoter region of IFITM3 (Figure 1). The substitution of the majority allele of G with minority allele of C reduces IFITM3 expression level and consequently results in the reduced number of antiviral CD8+ T cells in lung tissue upon IAV infection (Allen et al., 2017). In cohort FLU09, a cohort of naturally acquired influenza infection, a higher frequency of homozygosity of risk A allele was observed in patients with severe illness than the mild cases (33.3% vs. 1.3%) (Table 3; Allen et al., 2017). Also, an increased frequency of the A allele was found in patients who suffered with severe influenza in other two cohorts (Table 3; Allen et al., 2017). Thus, rs34481144-A is a risk allele associated with severe IAV infection and its association with other viral infection disease deserves to be investigated in future.
IFITM3 Variants and Other Viral Infections
In addition to influenza, SNP of rs12252 was also observed to have association with development of AIDS and Hantaan virus associated hemorrhagic fever with renal syndrome (HFRS) (Zhang et al., 2015; Xu-Yang et al., 2016). Zhang et al. (2015) reported that rs12252 is associated with rapid progression of AIDS, but not the susceptibility to HIV infection. Patients of AIDS rapid progressors had a higher frequency of rs12252 CC and CT genotype than non-progressors (Zhang et al., 2015). Compared with TT genotype, more patients with CC/CT genotypes were characterized with higher viremia level and more significantly reduced CD4 T+ cells (Zhang et al., 2015). In addition, the association of rs12252-C and homozygous CC genotype with disease severity in HFRS patients infected by Hantaan virus was reported recently (Xu-Yang et al., 2016). A higher allelic frequency of rs12252-C was observed in severe HFRS patients hospitalized than healthy Han Chinese controls (68.29% vs. 52.16%). Also, severe HFRS patients had a higher frequency of rs12252 CC than patients with mild HFRS and healthy Han Chinese. Along these lines, this group also reported that the viral titer detected in the plasma of HFRS patients with rs12252-CC genotype was more significantly alleviated than those with the TC and TT genotype (Xu-Yang et al., 2016). Thus, rs12252 C is a risk allele associated with severity of AIDS and HFRS, indicating that SNP rs12252 may have a profound impact on the pathogenesis of multiple viral diseases.
Future Directions
IFITMs are a group of small fusogenic proteins that restrict the infection of broad spectrum of viruses by inhibiting the fusion of viral and cellular membranes. However, whether IFITMs inhibit hemifusion, the process whereby the outer, but not the inner, leaflet of the viral and cellular membranes merge, or the transition from hemifusion to pore formation remains to be rigorously determined. It is also important to note that IFITMs do not always inhibit virus entry, but may promote membrane fusion under selected conditions, such as in the case of HCoV-OC43 infection (Zhao et al., 2014). Moreover, our recent studies indicated that specific mutations can flip the biological activity of IFITMs, from inhibiting to promoting the infection of selected human coronaviruses (Zhao et al., 2018). Those later findings strongly suggest that the fusogenic activity of IFITMs might be bidirectional and could be regulated by viral and host factors. The elucidation of molecular mechanisms underlying IFITM-mediated innate immunity via modulation of viral entry into host cells as well as the negative imprinting of progeny virions (virions incorporating IFITMs display decreased infectivity) will open new avenues for future research. We have only just begun to appreciate the role of IFITM proteins in innate antiviral defenses. Genetic association studies of rare and common variants may explain population-specific and individual variance in susceptibility to common viral pathogens and identify specific IFITM – virus interactions. Expansion of genetic studies incorporating gene knock-down or knock-off screens, single cell transcriptomics, epigenetic modification, and targeted sequencing for different viral infections will provide needed insights into cellular mechanisms and pathways involved in IFITM-mediated host response to viral exposure and infection.
Author Contributions
All the authors participated in the draft and revision of this review article.
Funding
The project was supported by grants from the U.S. National Institutes of Health (AI113267), the National Natural Science Foundation of China (81571976 and 81772173), the Commonwealth of Pennsylvania through the Hepatitis B Foundation as well as federal funds from the National Cancer Institute, National Institutes of health, under contract HHSN26120080001E. This project was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor do mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We want to thank the critical comments and suggestions by Dr. Jinhong Chang and Julia Ma on this manuscript.
References
Allen, E. K., Randolph, A. G., Bhangale, T., Dogra, P., Ohlson, M., Oshansky, C. M., et al. (2017). SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat. Med. 23, 975–983. doi: 10.1038/nm.4370
Amini-Bavil-Olyaee, S., Choi, Y. J., Lee, J. H., Shi, M., Huang, I. C., Farzan, M., et al. (2013). The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13, 452–464. doi: 10.1016/j.chom.2013.03.006
Anafu, A. A., Bowen, C. H., Chin, C. R., Brass, A. L., and Holm, G. H. (2013). Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 288, 17261–17271. doi: 10.1074/jbc.M112.438515
Appourchaux, R., Delpeuch, M., Zhong, L., Burlaud-Gaillard, J., Tartour, K., Savidis, G., et al. (2018). Functional mapping of regions involved in the negative imprinting of virion particles infectivity and in target cell protection by the Interferon-Induced Transmembrane Protein 3 (IFITM3) against HIV-1. J. Virol. doi: 10.1128/JVI.01716-18 [Epub ahead of print].
Bailey, C. C., Huang, I. C., Kam, C., and Farzan, M. (2012). Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 8:e1002909. doi: 10.1371/journal.ppat.1002909
Bailey, C. C., Kondur, H. R., Huang, I. C., and Farzan, M. (2013). Interferon-induced transmembrane protein 3 is a type II transmembrane protein. J. Biol. Chem. 288, 32184–32193. doi: 10.1074/jbc.M113.514356
Bailey, C. C., Zhong, G., Huang, I. C., and Farzan, M. (2014). IFITM-family proteins: the cell’s first line of antiviral defense. Ann. Rev. Virol. 1, 261–283. doi: 10.1146/annurev-virology-031413-085537
Brass, A. L., Huang, I. C., Benita, Y., John, S. P., Krishnan, M. N., Feeley, E. M., et al. (2009). The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254. doi: 10.1016/j.cell.2009.12.017
Chesarino, N. M., Compton, A. A., McMichael, T. M., Kenney, A. D., Zhang, L., Soewarna, V., et al. (2017). IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep. 18, 1740–1751. doi: 10.15252/embr.201744100
Chesarino, N. M., McMichael, T. M., Hach, J. C., and Yount, J. S. (2014a). Phosphorylation of the antiviral protein interferon-inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J. Biol. Chem. 289, 11986–11992. doi: 10.1074/jbc.M114.557694
Chesarino, N. M., McMichael, T. M., and Yount, J. S. (2014b). Regulation of the trafficking and antiviral activity of IFITM3 by post-translational modifications. Future Microbiol. 9, 1151–1163. doi: 10.2217/fmb.14.65
Chesarino, N. M., McMichael, T. M., and Yount, J. S. (2015). E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 11:e1005095. doi: 10.1371/journal.ppat.1005095
Compton, A. A., Bruel, T., Porrot, F., Mallet, A., Sachse, M., Euvrard, M., et al. (2014). IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe 16, 736–747. doi: 10.1016/j.chom.2014.11.001
Compton, A. A., Roy, N., Porrot, F., Billet, A., Casartelli, N., Yount, J. S., et al. (2016). Natural mutations in IFITM3 modulate post-translational regulation and toggle antiviral specificity. EMBO Rep. 17, 1657–1671. doi: 10.15252/embr.201642771
Desai, T. M., Marin, M., Chin, C. R., Savidis, G., Brass, A. L., and Melikyan, G. B. (2014). IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 10:e1004048. doi: 10.1371/journal.ppat.1004048
Everitt, A. R., Clare, S., McDonald, J. U., Kane, L., Harcourt, K., Ahras, M., et al. (2013). Defining the range of pathogens susceptible to Ifitm3 restriction using a knockout mouse model. PLoS One 8:e80723. doi: 10.1371/journal.pone.0080723
Everitt, A. R., Clare, S., Pertel, T., John, S. P., Wash, R. S., Smith, S. E., et al. (2012). IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523. doi: 10.1038/nature10921
Foster, T. L., Wilson, H., Iyer, S. S., Coss, K., Doores, K., Smith, S., et al. (2016). Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe 20, 429–442. doi: 10.1016/j.chom.2016.08.006
Fu, B., Wang, L., Li, S., and Dorf, M. E. (2017). ZMPSTE24 defends against influenza and other pathogenic viruses. J. Exp. Med. 214, 919–929. doi: 10.1084/jem.20161270
Gerlach, T., Hensen, L., Matrosovich, T., Bergmann, J., Winkler, M., Peteranderl, C., et al. (2017). pH optimum of hemagglutinin-mediated membrane fusion determines sensitivity of influenza a viruses to the interferon-induced antiviral state and IFITMs. J. Virol. 91:e00246-17. doi: 10.1128/JVI.00246-17
Gorman, M. J., Poddar, S., Farzan, M., and Diamond, M. S. (2016). The interferon-stimulated gene ifitm3 restricts west nile virus infection and pathogenesis. J. Virol. 90, 8212–8225. doi: 10.1128/JVI.00581-16
Hach, J. C., McMichael, T., Chesarino, N. M., and Yount, J. S. (2013). Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J. Virol. 87, 9923–9927. doi: 10.1128/JVI.00621-13
Huang, I. C., Bailey, C. C., Weyer, J. L., Radoshitzky, S. R., Becker, M. M., Chiang, J. J., et al. (2011). Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7:e1001258. doi: 10.1371/journal.ppat.1001258
Infusini, G., Smith, J. M., Yuan, H., Pizzolla, A., Ng, W. C., Londrigan, S. L., et al. (2015). Respiratory DC Use IFITM3 to avoid direct viral infection and safeguard virus-specific CD8+ T cell priming. PLoS One 10:e0143539. doi: 10.1371/journal.pone.0143539
Jia, R., Pan, Q., Ding, S., Rong, L., Liu, S. L., Geng, Y., et al. (2012). The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 86, 13697–13707. doi: 10.1128/JVI.01828-12
Jia, R., Xu, F., Qian, J., Yao, Y., Miao, C., Zheng, Y. M., et al. (2014). Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell. Microbiol. 16, 1080–1093. doi: 10.1111/cmi.12262
Jiang, D., Weidner, J. M., Qing, M., Pan, X. B., Guo, H., Xu, C., et al. (2010). Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J. Virol. 84, 8332–8341. doi: 10.1128/JVI.02199-09
John, S. P., Chin, C. R., Perreira, J. M., Feeley, E. M., Aker, A. M., Savidis, G., et al. (2013). The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J. Virol. 87, 7837–7852. doi: 10.1128/JVI.00481-13
Li, C., Du, S., Tian, M., Wang, Y., Bai, J., Tan, P., et al. (2018). The host restriction factor interferon-inducible transmembrane protein 3 inhibits Vaccinia virus infection. Front. Immunol. 9:228. doi: 10.3389/fimmu.2018.00228
Li, K., Jia, R., Li, M., Zheng, Y. M., Miao, C., Yao, Y., et al. (2015). A sorting signal suppresses IFITM1 restriction of viral entry. J. Biol. Chem. 290, 4248–4259. doi: 10.1074/jbc.M114.630780
Li, K., Markosyan, R. M., Zheng, Y. M., Golfetto, O., Bungart, B., Li, M., et al. (2013). IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9:e1003124. doi: 10.1371/journal.ppat.1003124
Lin, T. Y., Chin, C. R., Everitt, A. R., Clare, S., Perreira, J. M., Savidis, G., et al. (2013). Amphotericin B increases influenza A virus infection by preventing IFITM3-mediated restriction. Cell Rep. 5, 895–908. doi: 10.1016/j.celrep.2013.10.033
Lopez-Rodriguez, M., Herrera-Ramos, E., Sole-Violan, J., Ruiz-Hernandez, J. J., Borderias, L., Horcajada, J. P., et al. (2016). IFITM3 and severe influenza virus infection. No evidence of genetic association. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1811–1817. doi: 10.1007/s10096-016-2732-7
Lu, J., Pan, Q., Rong, L., He, W., Liu, S. L., and Liang, C. (2011). The IFITM proteins inhibit HIV-1 infection. J. Virol. 85, 2126–2137. doi: 10.1128/JVI.01531-10
Ma, W., Tummers, B., van Esch, E. M., Goedemans, R., Melief, C. J., Meyers, C., et al. (2016). Human papillomavirus downregulates the expression of IFITM1 and RIPK3 to escape from IFNgamma- and TNFalpha-mediated antiproliferative effects and necroptosis. Front. Immunol. 7:496. doi: 10.3389/fimmu.2016.00496
Makvandi-Nejad, S., Laurenson-Schafer, H., Wang, L., Wellington, D., Zhao, Y., Jin, B., et al. (2018). Lack of truncated IFITM3 transcripts in cells homozygous for the rs12252-C variant that is associated with severe influenza infection. J. Infect. Dis. 217, 257–262. doi: 10.1093/infdis/jix512
McMichael, T. M., Zhang, L., Chemudupati, M., Hach, J. C., Kenney, A. D., Hang, H. C., et al. (2017). The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity. J. Biol. Chem. 292, 21517–21526. doi: 10.1074/jbc.M117.800482
Mills, T. C., Rautanen, A., Elliott, K. S., Parks, T., Naranbhai, V., Ieven, M. M., et al. (2014). IFITM3 and susceptibility to respiratory viral infections in the community. J. Infect. Dis. 209, 1028–1031. doi: 10.1093/infdis/jit468
Mudhasani, R., Tran, J. P., Retterer, C., Radoshitzky, S. R., Kota, K. P., Altamura, L. A., et al. (2013). IFITM-2 and IFITM-3 but not IFITM-1 restrict Rift Valley fever virus. J. Virol. 87, 8451–8464. doi: 10.1128/JVI.03382-12
Pan, Y., Yang, P., Dong, T., Zhang, Y., Shi, W., Peng, X., et al. (2017). IFITM3 Rs12252-C variant increases potential risk for severe influenza virus infection in chinese population. Front. Cell Infect. Microbiol. 7:294. doi: 10.3389/fcimb.2017.00294
Perreira, J. M., Chin, C. R., Feeley, E. M., and Brass, A. L. (2013). IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 425, 4937–4955. doi: 10.1016/j.jmb.2013.09.024
Poddar, S., Hyde, J. L., Gorman, M. J., Farzan, M., and Diamond, M. S. (2016). The interferon-stimulated gene IFITM3 restricts infection and pathogenesis of arthritogenic and encephalitic alphaviruses. J. Virol. 90, 8780–8794. doi: 10.1128/JVI.00655-16
Prabhu, S. S., Chakraborty, T. T., Kumar, N., and Banerjee, I. (2018). Association between IFITM3 rs12252 polymorphism and influenza susceptibility and severity: a meta-analysis. Gene 674, 70–79. doi: 10.1016/j.gene.2018.06.070
Rabbani, M. A., Ribaudo, M., Guo, J. T., and Barik, S. (2016). Identification of interferon-stimulated gene proteins that inhibit human parainfluenza virus type 3. J. Virol. 90, 11145–11156. doi: 10.1128/JVI.01551-16
Randolph, A. G., Yip, W. K., Allen, E. K., Rosenberger, C. M., Agan, A. A., Ash, S. A., et al. (2017). Evaluation of IFITM3 rs12252 association with severe pediatric influenza infection. J. Infect. Dis. 216, 14–21. doi: 10.1093/infdis/jix242
Savidis, G., Perreira, J. M., Portmann, J. M., Meraner, P., Guo, Z., Green, S., et al. (2016). The IFITMs Inhibit Zika Virus Replication. Cell Rep. 15, 2323–2330. doi: 10.1016/j.celrep.2016.05.074
Shan, J., Zhao, B., Shan, Z., Nie, J., Deng, R., Xiong, R., et al. (2017). Histone demethylase LSD1 restricts influenza A virus infection by erasing IFITM3-K88 monomethylation. PLoS Pathog. 13:e1006773. doi: 10.1371/journal.ppat.1006773
Shan, Z., Han, Q., Nie, J., Cao, X., Chen, Z., Yin, S., et al. (2013). Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation. J. Biol. Chem. 288, 35093–35103. doi: 10.1074/jbc.M113.511949
Shen, C., Wu, X. R., Jiao, W. W., Sun, L., Feng, W. X., Xiao, J., et al. (2013). A functional promoter polymorphism of IFITM3 is associated with susceptibility to pediatric tuberculosis in Han Chinese population. PLoS One 8:e67816. doi: 10.1371/journal.pone.0067816
Shi, G., Ozog, S., Torbett, B. E., and Compton, A. A. (2018). mTOR inhibitors lower an intrinsic barrier to virus infection mediated by IFITM3. Proc. Natl. Acad. Sci. U.S.A. 115, E10069–E10078. doi: 10.1073/pnas.1811892115
Siegrist, F., Ebeling, M., and Certa, U. (2011). The small interferon-induced transmembrane genes and proteins. J. Interferon Cytokine Res. 31, 183–197. doi: 10.1089/jir.2010.0112
Stacey, M. A., Clare, S., Clement, M., Marsden, M., Abdul-Karim, J., Kane, L., et al. (2017). The antiviral restriction factor IFN-induced transmembrane protein 3 prevents cytokine-driven CMV pathogenesis. J. Clin. Invest. 127, 1463–1474. doi: 10.1172/JCI84889
Tartour, K., Appourchaux, R., Gaillard, J., Nguyen, X. N., Durand, S., Turpin, J., et al. (2014). IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology 11:103. doi: 10.1186/s12977-014-0103-y
Tartour, K., Nguyen, X. N., Appourchaux, R., Assil, S., Barateau, V., Bloyet, L. M., et al. (2017). Interference with the production of infectious viral particles and bimodal inhibition of replication are broadly conserved antiviral properties of IFITMs. PLoS Pathog. 13:e1006610. doi: 10.1371/journal.ppat.1006610
Wakim, L. M., Gupta, N., Mintern, J. D., and Villadangos, J. A. (2013). Enhanced survival of lung tissue-resident memory CD8(+) T cells during infection with influenza virus due to selective expression of IFITM3. Nat. Immunol. 14, 238–245. doi: 10.1038/ni.2525
Wang, B., Lam, T. H., Soh, M. K., Ye, Z., Chen, J., and Ren, E. C. (2018). Influenza A virus facilitates its infectivity by activating p53 to inhibit the expression of interferon-induced transmembrane proteins. Front. Immunol. 9:1193. doi: 10.3389/fimmu.2018.01193
Wang, S., Chi, X., Wei, H., Chen, Y., Chen, Z., Huang, S., et al. (2014). Influenza A virus-induced degradation of eukaryotic translation initiation factor 4B contributes to viral replication by suppressing IFITM3 protein expression. J. Virol. 88, 8375–8385. doi: 10.1128/JVI.00126-14
Wang, Y., Pan, Q., Ding, S., Wang, Z., Yu, J., Finzi, A., et al. (2017). The V3 Loop of HIV-1 Env determines viral susceptibility to ifitm3 impairment of viral infectivity. J. Virol. 91:e02441-16. doi: 10.1128/JVI.02441-16
Wang, Z., Zhang, A., Wan, Y., Liu, X., Qiu, C., Xi, X., et al. (2014). Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc. Natl. Acad. Sci. U.S.A. 111, 769–774. doi: 10.1073/pnas.1321748111
Wee, Y. S., Roundy, K. M., Weis, J. J., and Weis, J. H. (2012). Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate Immun. 18, 834–845. doi: 10.1177/1753425912443392
Weidner, J. M., Jiang, D., Pan, X. B., Chang, J., Block, T. M., and Guo, J. T. (2010). Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84, 12646–12657. doi: 10.1128/JVI.01328-10
Wilkins, C., Woodward, J., Lau, D. T., Barnes, A., Joyce, M., McFarlane, N., et al. (2013). IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology 57, 461–469. doi: 10.1002/hep.26066
Williams, D. E., Wu, W. L., Grotefend, C. R., Radic, V., Chung, C., Chung, Y. H., et al. (2014). IFITM3 polymorphism rs12252-C restricts influenza A viruses. PLoS One 9:e110096. doi: 10.1371/journal.pone.0110096
Wrensch, F., Karsten, C. B., Gnirss, K., Hoffmann, M., Lu, K., Takada, A., et al. (2015). Interferon-induced transmembrane protein-mediated inhibition of host cell entry of ebolaviruses. J. Infect. Dis. 212(Suppl. 2), S210–S218. doi: 10.1093/infdis/jiv255
Wrensch, F., Winkler, M., and Pohlmann, S. (2014). IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses 6, 3683–3698. doi: 10.3390/v6093683
Wu, W. L., Grotefend, C. R., Tsai, M. T., Wang, Y. L., Radic, V., Eoh, H., et al. (2017). Delta20 IFITM2 differentially restricts X4 and R5 HIV-1. Proc. Natl. Acad. Sci. U.S.A. 114, 7112–7117. doi: 10.1073/pnas.1619640114
Xie, M., Xuan, B., Shan, J., Pan, D., Sun, Y., Shan, Z., et al. (2015). Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment. J. Virol. 89, 3049–3061. doi: 10.1128/JVI.03416-14
Xu, J., Qian, P., Wu, Q., Liu, S., Fan, W., Zhang, K., et al. (2014). Swine interferon-induced transmembrane protein, sIFITM3, inhibits foot-and-mouth disease virus infection in vitro and in vivo. Antiviral Res. 109, 22–29. doi: 10.1016/j.antiviral.2014.06.008
Xu-Yang, Z., Pei-Yu, B., Chuan-Tao, Y., Wei, Y., Hong-Wei, M., Kang, T., et al. (2016). Interferon-induced transmembrane protein 3 inhibits hantaan virus infection, and its single nucleotide polymorphism rs12252 influences the severity of hemorrhagic fever with renal syndrome. Front. Immunol. 7:535. doi: 10.3389/fimmu.2016.00535
Yount, J. S., Karssemeijer, R. A., and Hang, H. C. (2012). S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 287, 19631–19641. doi: 10.1074/jbc.M112.362095
Yount, J. S., Moltedo, B., Yang, Y. Y., Charron, G., Moran, T. M., López, C. B., et al. (2010). Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6, 610–614. doi: 10.1038/nchembio.405
Yu, J., Li, M., Wilkins, J., Ding, S., Swartz, T. H., Esposito, A. M., et al. (2015). IFITM proteins restrict HIV-1 infection by antagonizing the envelope glycoprotein. Cell Rep. 13, 145–156. doi: 10.1016/j.celrep.2015.08.055
Zhang, Y., Makvandi-Nejad, S., Qin, L., Zhao, Y., Zhang, T., Wang, L., et al. (2015). Interferon-induced transmembrane protein-3 rs12252-C is associated with rapid progression of acute HIV-1 infection in Chinese MSM cohort. AIDS 29, 889–894. doi: 10.1097/QAD.0000000000000632
Zhang, Y. H., Zhao, Y., Li, N., Peng, Y. C., Giannoulatou, E., Jin, R. H., et al. (2013). Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 4:1418. doi: 10.1038/ncomms2433
Zhang, Z., Liu, J., Li, M., Yang, H., and Zhang, C. (2012). Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One 7:e49265. doi: 10.1371/journal.pone.0049265
Zhao, X., Guo, F., Liu, F., Cuconati, A., Chang, J., Block, T. M., et al. (2014). Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. U.S.A. 111, 6756–6761. doi: 10.1073/pnas.1320856111
Keywords: host susceptibility, interferon-induced transmembrane proteins, IFITM, single nucleotide polymorphisms, viral infection
Citation: Zhao X, Li J, Winkler CA, An P and Guo J-T (2019) IFITM Genes, Variants, and Their Roles in the Control and Pathogenesis of Viral Infections. Front. Microbiol. 9:3228. doi: 10.3389/fmicb.2018.03228
Received: 08 October 2018; Accepted: 12 December 2018;
Published: 08 January 2019.
Edited by:
Jianrong Li, The Ohio State University, United StatesReviewed by:
Arnaud Moris, Center for the National Scientific Research (CNRS), FranceShitao Li, Oklahoma State University, United States
Copyright © 2019 Zhao, Li, Winkler, An and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ju-Tao Guo, anUtdGFvLmd1b0BiYmx1bWJlcmcub3Jn
 Xuesen Zhao
Xuesen Zhao Jiarui Li1,2
Jiarui Li1,2 Ping An
Ping An Ju-Tao Guo
Ju-Tao Guo