- 1College of Veterinary Medicine, Mississippi State University, Starkville, MS, United States
- 2Bioinformatics and Systems Biology, Justus-Liebig-University Giessen, Giessen, Germany
Virulent Aeromonas hydrophila causes severe motile Aeromonas septicemia in warmwater fishes. In recent years, channel catfish farming in the U.S.A. and carp farming in China have been affected by virulent A. hydrophila, and genome comparisons revealed that these virulent A. hydrophila strains belong to the same clonal group. Bacterial secretion systems are often important virulence factors; in the current study, we investigated whether secretion systems contribute to the virulent phenotype of these strains. Thus, we conducted comparative secretion system analysis using 55 A. hydrophila genomes, including virulent A. hydrophila strains from U.S.A. and China. Interestingly, tight adherence (TaD) system is consistently encoded in all the vAh strains. The majority of U.S.A. isolates do not possess a complete type VI secretion system, but three core elements [tssD (hcp), tssH, and tssI (vgrG)] are encoded. On the other hand, Chinese isolates have a complete type VI secretion system operon. None of the virulent A. hydrophila isolates have a type III secretion system. Deletion of two genes encoding type VI secretion system proteins (hcp1 and vgrG1) from virulent A. hydrophila isolate ML09-119 reduced virulence 2.24-fold in catfish fingerlings compared to the parent strain ML09-119. By determining the distribution of genes encoding secretion systems in A. hydrophila strains, our study clarifies which systems may contribute to core A. hydrophila functions and which may contribute to more specialized adaptations such as virulence. Our study also clarifies the role of type VI secretion system in A. hydrophila virulence.
Introduction
Aeromonas hydrophila is common in freshwater environments and causes disease in fish, reptiles, amphibians, and humans (Janda and Abbott, 2010; Tomás, 2012). The U.S.A. and China aquaculture industries have had significant losses due to A. hydrophila disease (Nielsen et al., 2001). In the southeastern U.S.A., severe A. hydrophila outbreaks began impacting the catfish aquaculture industry in 2009 and are caused by a clonal group of strains named virulent A. hydrophila (vAh) (Hemstreet, 2010; Hossain et al., 2014).
Comparative genomics methods have helped identify taxonomically mislabeled A. hydrophila genomes in Genbank (Beaz-Hidalgo et al., 2015). The same methods also revealed that the U.S.A. vAh clonal lineage is similar to a clonal lineage of A. hydrophila that is responsible for significant economic losses in the Chinese aquaculture industry (Griffin et al., 2013; Hossain et al., 2013; Zhang et al., 2014; Pang et al., 2015). Both clonal groups are categorized as sequence type ST251 (Rasmussen-Ivey et al., 2016). It has been theorized that the U.S.A. vAh originated from transport of carrier fish from Asia (Hossain et al., 2014).
Comparative genomics has revealed that the vAh clonal group has unique characteristics. Some of the unique biochemical pathways include sialic acid biosynthesis, myo-inositol utilization, and L-fucose metabolism. They also have unique O-antigen biosynthesis and characteristic mobilome elements and secretion systems (Hossain et al., 2013; Pang et al., 2015). Intriguingly, Asian vAh isolates encode all the core components of type VI secretion system (T6SS), whereas most of the U.S.A. vAh isolates carry remnants of the T6SS (Rasmussen et al., 2016).
Several virulence mechanisms of A. hydrophila including secretion systems, motility, toxins, tissue-destructive enzymes, iron acquisition, and S-layer have been studied (Tomás, 2012). Secretion systems are used by bacteria to interact with the environment, including host adaptation and competing against other bacteria (Cianfanelli et al., 2016). A thorough investigation of secretion systems distribution in A. hydrophila, including vAh, other fish disease strains, and environmental strains, has not been conducted. Hence, in this study, we analyzed 55 A. hydrophila genomes from distinct geographical origins and hosts. We also evaluated type 4 pili (T4P), tight adherence systems (Tad), and flagella components due to their sequence similarity to secretion systems. Potential host-pathogen interactions of the identified secretion system proteins were evaluated.
In the current study, we conducted comparative genomics of secretion systems encoded by A. hydrophila. Our analysis showed that all the evaluated A. hydrophila genomes encode the whole operon or remnants of T6SS. To clarify the function of T6SS genes in vAh, we mutated two T6SS genes in strain ML09-119, and we determined the virulence of mutant strains in catfish fingerlings. Overall, the comparative genomics and mutational analyses reported here clarify the distribution of various secretion systems in A. hydrophila and provide functional information on the role of T6SS components in vAh.
Materials and Methods
Genome Sequences and Annotation
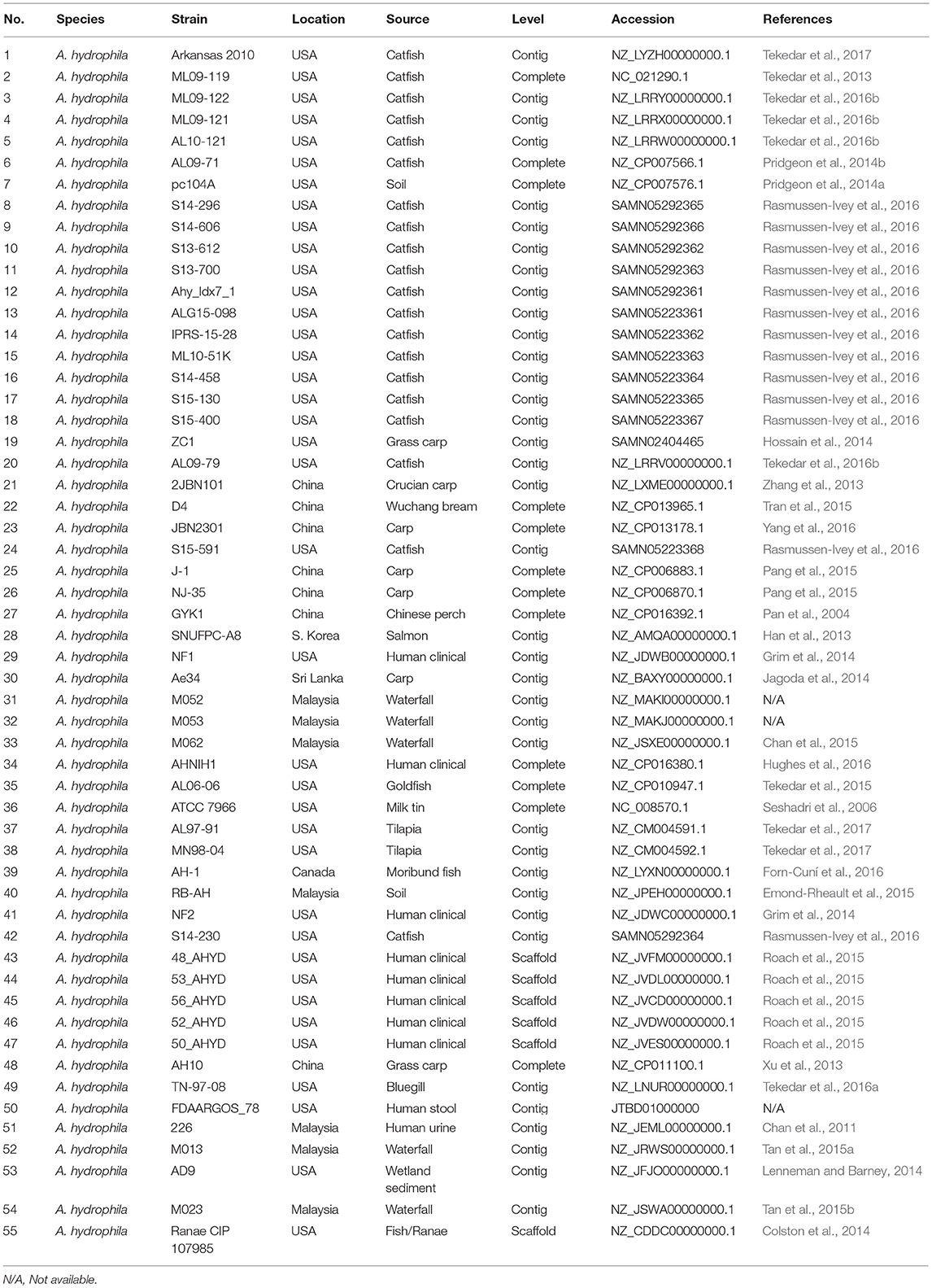
The genome sequences (including complete sequences, draft assemblies, and raw reads) of 55 A. hydrophila strains were downloaded from the National Center for Biotechnology Information (Table 1). Raw data were assembled using CLC workbench 6.5.1 after trimming sequence reads, followed by error correction and contig creation. All unannotated genomes were annotated by RAST (Brettin et al., 2015). All selected genomes had at least 95% average nucleotide identity (ANI).
Phylogenetic Tree Creation
A phylogenetic tree was built from the complete core genomes of 55 A. hydrophila strains, which included 115,335 coding sequences (2,097/genome) with 101,851,090 amino acid residues (1,851,838/genome). The gene sets of the core genome were aligned one by one using MUSCLE (Edgar, 2004), and alignments were concatenated. This alignment was used to compute a Kimura distance matrix, which was used as input for the Neighbor-Joining algorithm as implemented in PHYLP (Felsenstein, 1989). The resulting tree was verified by bootstrapping with 250 iterations.
ANI and AAI Calculation
Average nucleotide identity and average amino acid identity (AAI) values (Konstantinidis and Tiedje, 2005a,b; Konstantinidis et al., 2006) were calculated using EDGAR (Konstantinidis and Tiedje, 2005b). Briefly, the average amino acid identities were based on all protein sequences encoded by genes in the core genome (2,097 per genome). Percent identity values were extracted from BLASTP (Altschul et al., 1990) results that are stored in the EDGAR database, summed up, and averaged for every pair of genomes. ANI using BLAST (ANIb) was based on BLASTN results as described (Goris et al., 2007) using the same cutoffs as JSpeciesWS (Richter and Rossello-Mora, 2009).
Identification of Secretion Systems
MacSyFinder was used with default features to identify secretion systems from the A. hydrophila genomes. The “unordered” type of dataset option was chosen because the majority of the evaluated genomes were draft genomes. The topology of the replicon was linear/circular, maximal E-value was 1.0, maximal independent E-value was 0.001, and minimal profile coverage was 0.5. Both mandatory genes and accessory genes were identified (Abby et al., 2014, 2016).
Bacterial Strains and Plasmids
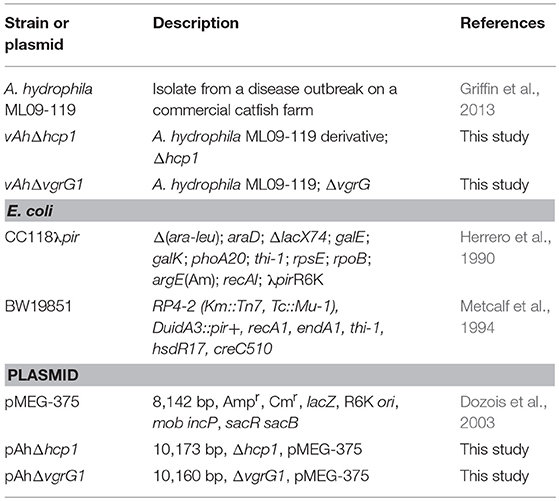
Bacterial strains and plasmids used in this study are listed in Table 2. Aeromonas hydrophila strain ML09-119 represents the vAh clonal group impacting U.S.A. channel catfish aquaculture. The strain was grown on brain heart infusion (BHI) agar or broth (Difco, Sparks, MD, USA) and incubated at 37°C. Escherichia coli strain CC118 λpir was used for cloning, and strain BW19851 was used for transferring suicide plasmid pMEG-375 into A. hydrophila by conjugation. Escherichia coli strains were cultured in Luria–Bertani (LB) agar and broth (Difco) and incubated at 37°C. The following antibiotics and reagents (Sigma-Aldrich, Saint Louis, MN, USA) were used when needed: ampicillin (100 μg/ml), chloramphenicol (10–25 μg/ml), colistin (12.5 μg/ml), sucrose (5%), and mannitol (0.35%).
In-frame Deletion of A. hydrophila Genes
Two chromosomal in-frame deletion mutants of type six secretion system (T6SS) effector genes hcp1 (AHML_05970) and vgrG1 (AHML_05975) were constructed by allelic exchange and homologous recombination using suicide plasmid pMEG-375 containing the counter-selectable marker sacB (Dozois et al., 2003). Recombinant DNA and mutant construction procedures were completed as described previously (Abdelhamed et al., 2013). Briefly, four primers (A, B, C, and D) were designed for each gene using Primer3 (Untergasser et al., 2012) (Table 3). Compatible restriction enzyme sites were embedded in A and D primers (bold line in primers A and D) for cloning, and the reverse complement of primer B was added to the 5′ end of primer C (underlined letters in primers C) to allow fusion of PCR fragments by overlap extension PCR (Horton et al., 1989). The upstream (fragment AB) and downstream (fragment CD) of each gene was amplified using two sets of primers. PCR fragments AB and CD were annealed at the overlapping regions and were amplified as a single fragment using primers A and D. The fusion products were purified, digested, ligated into digested pMEG-375, electroporated into E. coli CC118λpir, and spread on LB agar plus ampicillin.
The resulting plasmids were purified from E. coli CC118λpir and transferred into A. hydrophila ML09-119 by conjugation using E. coli BW19851. Transconjugants were selected on plates containing chloramphenicol and colistin; chloramphenicol was used to select the integration of pMEG-375 in A. hydrophila chromosome while colistin was used as counter-selection against E. coli. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA. After sucrose treatment, transconjugants that were colistin resistant and chloramphenicol sensitive were selected, and the deletion was confirmed by colony PCR using A and D primers. Mutant validation was done by sequencing of AD fragments amplified from chloramphenicol sensitive mutants using hcp1Seq and vgrG1Seq primers (Table 2). The A. hydrophila mutants were designated vAhΔhcp1 and vAhΔvgrG1.
Virulence of vAh Mutants in Catfish Fingerlings
All fish experiments were conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee at Mississippi State University. Virulence of vAhΔhcp1 and vAhΔvgrG1 was compared to A. hydrophila wild-type (WT) strain ML09-119 by immersion route of exposure as described (Abdelhamed et al., 2016). Briefly, 120 6-month-old specific-pathogen-free (SPF) channel catfish fingerlings (18.10 ± 0.56 cm, 50.90± 3.76 g) were stocked into twelve 40-liter flow-through tanks (10 fish/tank) and acclimated for a week. Tanks were assigned randomly to four treatment groups: vAhΔhcp1, vAhΔvgrG1, vAh WT, and BHI (sham). Each group included three replicate tanks. Water temperature was maintained at 32°C (±2) throughout the experiments. Fish were fed twice a day with a commercial catfish feed. On the challenge day, the water levels in each tank were decreased to 10 L, and 100 mL of overnight culture was added directly to each tank (1.02 × 1010 CFU/mL water). Negative control tanks were exposed to 100 mL of sterile BHI broth. During immersion, water was well aerated. After 6 h, water flow was restored, and fish were maintained as usual. Fish mortalities were recorded daily for a total of 21 days, and percent mortality was calculated for each group. Protection against vAh WT challenge was determined in fingerlings that survived infection by the vAhΔhcp1 and vAhΔvgrG1 mutants. Briefly, at 21 days post-infection, catfish fingerlings were re-challenged by vAh WT by immersion (2.21 × 1010 CFU/ml water), and mortalities were recorded daily for 14 days. At the end of the experiments, mean percent survival was calculated for each treatment.
Statistical Analysis
Mean percent mortality data were arcsine transformed, and analysis of variance (ANOVA) was applied using PROC GLM in SAS for Windows v9.4 (SAS Institute, Inc., Cary, NC) to assess significance. An alpha level of 0.05 was used in all analyses.
Host-Pathogen Interaction Network
Protein-protein interactions between A. hydrophila secretion system proteins and catfish proteins (accession: PRJNA281269) were determined using the Host-Pathogen Interaction Database (Ammari et al., 2016). For pathogen sequences, default upload options were: database search: bacterial pathogens, matrix: Blosum62, E-value: 0.00001, pathogen percent identity: 30, and query coverage filter: 50%. For host sequences, selected animal protein options were: for the database search matrix: Blosum62, E-value cutoff: 0.00001, percent identity and query coverage filter: 70% (Ammari et al., 2016).
Results
Genome Features
The 55 genome sequences included in the current study are A. hydrophila isolates from different geographical locations and hosts (Table 1). Of these, our group sequenced vAh strains ML09-119, ML09-121, ML09-122, AL09-79, AL10-121, and Arkansas 2010. We also sequenced A. hydrophila strains AL06-06, AL97-91, MN98-04, and TN97-08. Additionally, we assembled and annotated 12 draft vAh strain genome sequences released in 2016 (strains Ahy_Idx71, ALG15-098, IPRS15-28, ML10-51K, S13-612, S13-700, S14-296, S14-458, S14-606, S15-130, S15-400, and S15-591) and one non-vAh strain genome (S14-230) for inclusion in our analysis. Genome size of the 55 strains ranged from ~4.67 to 5.28 Mb, and G+C ratio of the genomes ranged from 60.47 to 61.60.
Average Nucleotide Identities (ANI) and Phylogenetic Tree Creation
A phylogenetic tree based on the complete core genome of 55 A. hydrophila strains shows the 27 vAh strains forming a highly conserved branch separated clearly from the other strains. The separation of the vAh cluster from the rest of the tree showed 100% branch conservation. These findings were confirmed by ANI as well as Average AAI (Supplementary File 1). ANI and AAI values within the cluster of 27 strains were above 99.88% (ANI) and 99.89% (AAI), respectively (Figure 1).
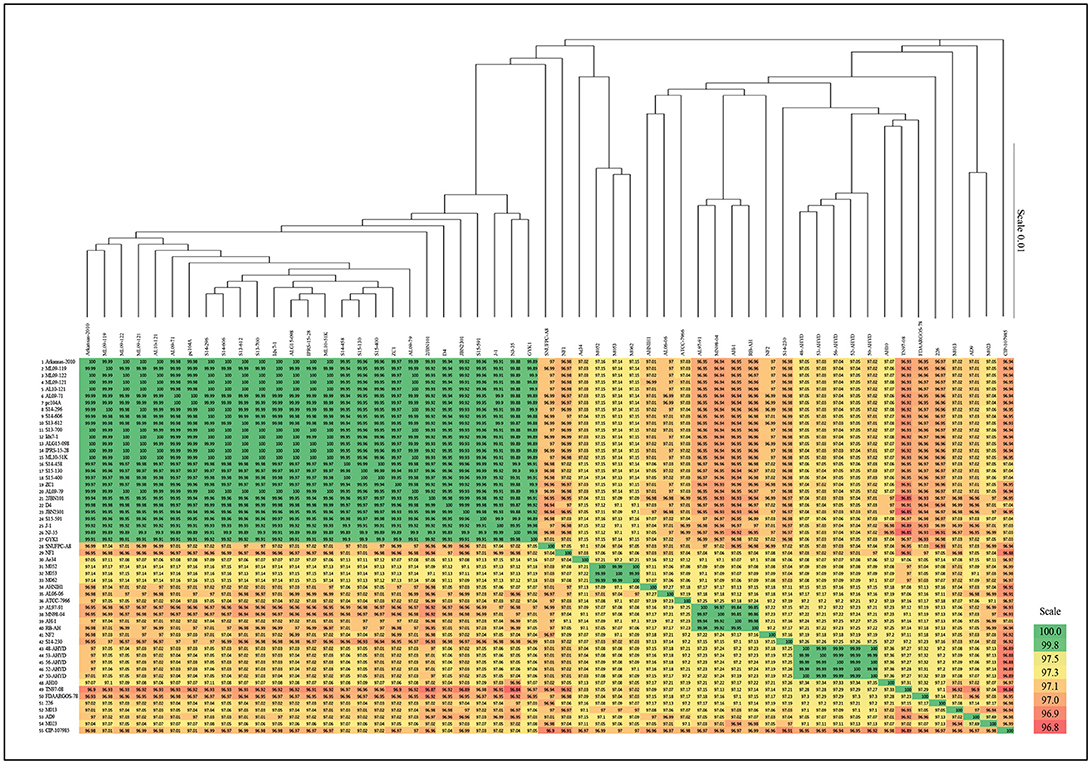
Figure 1. Average nucleotide identities (ANI) of A. hydrophila genomes and phylogenetic tree based on core genome. Note that branch lengths of the phylogenetic tree were reduced to fit the image.
Secretion Systems in Aeromonas hydrophila Genomes
In our in silico secretion systems analysis, we identified that most of the U.S.A. and Chinese vAh isolates tend to encode more T1SS core components, for instance ATP-binding cassette (abc) and mfp genes, compared to environmental isolates. Additionally, the genome of human strain FDAARGOS_78 encodes more abc and outer membrane factor (omf) genes than the other 54 genomes (Figure 2).
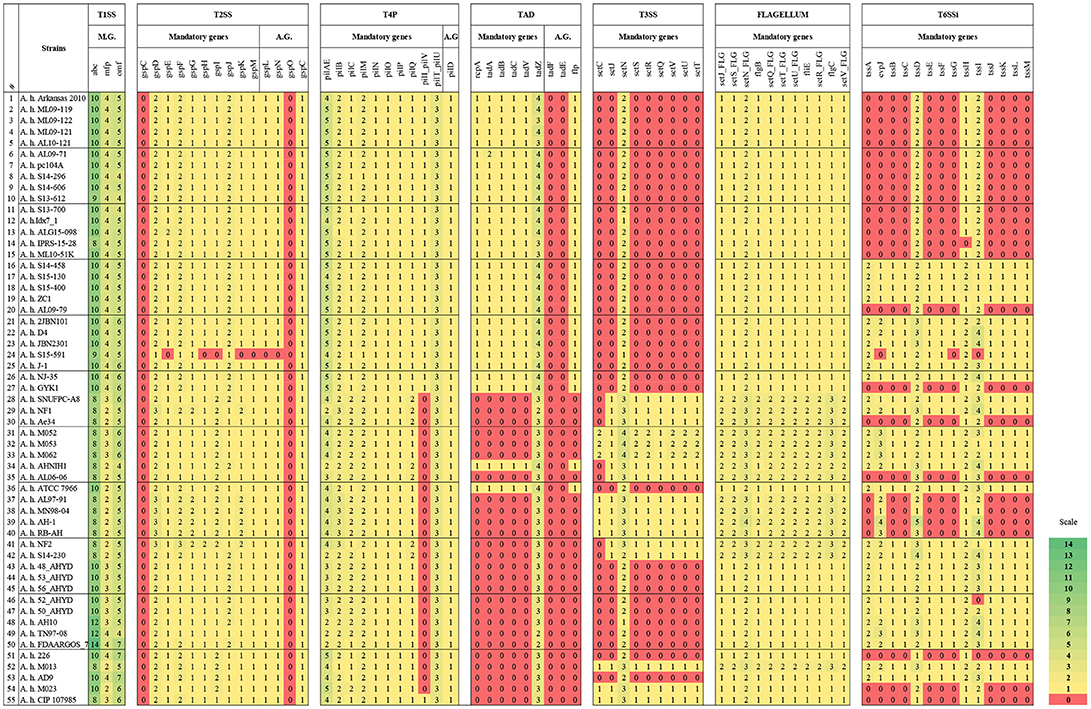
Figure 2. Core and accessory proteins of secretion systems, T4P, Tad, and flagella encoded in A. hydrophila genomes. Numbers and color represent the number of copies of each listed gene. The strains are listed in the same order as Table 1 and Figure 1 (the first 27 strains are vAh strains). tssD is also known as hcp and tssI is also known as vgrG. A.G. indicates Accessory genes.
All of the evaluated A. hydrophila genomes encode a T2SS system except one: strain S15-591. However, this strain does not have a completed genome sequence; it is a draft assembly with a large number of contigs. Therefore, it is possible that the genes may not have been detected due to the large number of gaps in the genome (Figure 2).
All the vAh strains in the current study encode mandatory and accessory genes of type 4 pilus (T4P). By contrast, non-vAh strains from different origins and locations lacked the pilQ gene. One non-vAh strain (Ranae CIP 107985, which was isolated from a frog) encodes all the T4P elements (Figure 2).
Intriguingly, only one gene (tadZ) from the Tad system is present in all the evaluated A. hydrophila genomes. On the other hand, all the vAh strains as well as two non-vAh strains (ATCC 7966 and AHNIH1) encode this system (Figure 2).
The majority of vAh strains from the U.S.A. (except strain S14-230) and Chinese isolates do not carry all of the mandatory T3SS genes in their genomes (Figure 2). The mandatory T3SS gene cluster is composed of sctU, sctJ, sctN, sctS, sctR, sctQ, sctV, sctU, and sctT. Interestingly, only the sctN gene is present in all of the evaluated A. hydrophila genomes. Only two of the eight human isolates encode T3SS except for the sctC gene. A. hydrophila ATCC 7966 does not encode T3SS, but it has two copies of the sctN gene. By contrast, most of the environmental isolates carry T3SS. Interestingly, A. hydrophila strains that encode more T1SS components tend to have fewer or no genes encoding T3SS (Figure 2).
Some T3SS genes are similar to flagella genes. Therefore, we used MacSyfinder to discriminate between T3SS and flagella genes. Of the 55 A. hydrophila genomes we evaluated, all carry the mandatory flagella genes (Figure 2).
All of the evaluated A. hydrophila genomes encode either the entire operon or remnants of the T6SSi. Most of the U.S.A. vAh isolates have only three T6SSi genes: hcp1 tssH, and vgrG. By contrast, almost all the China isolates encode the entire T6SSi. The exception was strain GYK1 from China, which has the same three T6SSi genes as the U.S.A. vAh isolates. Additionally, fish isolate Ae34 from Sri Lanka, four non-vAh isolates from the U.S.A. (AL06-06, MN98-04, AL97-91, and Ranae CIP 107985), and one fish isolate (AH-1) from Canada do not encode the entire T6SSi elements but have the same three genes as the U.S.A. vAh strains. Three of the Malaysian isolates (M023, RB-AH, and 226) encode the same three elements from T6SSi, whereas four Malaysian isolates (M013, M052, M053, and M054) encode the entire T6SSi.
Only one gene encoding a T9SS-like protein (sprA) was identified in the A. hydrophila genomes. This gene is encoded by all the evaluated A. hydrophila genomes.
Construction and Virulence of Mutant Strains
In the present study, we successfully introduced in-frame deletions in two genes encoding T6SS effectors: hcp1 and vgrG1 (Table 4). The Δhcp1 mutation has a deletion of 537 bp out of 564 bp (95.21%), and ΔvgrG1 has a deletion of 2,739 bp out of 2,781 bp (98.49%).

Table 4. The sizes of upstream (USF), downstream (DSF), and in-frame fused fragments (FF), deleted region (DR), and undeleted region (UD) by base pair (bp).
Results of the immersion challenge in catfish fingerlings indicated that the mortality rate was significantly lower (p < 0.05) in vAhΔhcp1 and vAhΔvgrG1 compared with parent vAh strain ML09-119 (33.33 and 33.33% mortality vs. 60% mortality) (Figure 3A). Fingerlings surviving infection with vAhΔhcp1 and vAhΔvgrG1 had 91.67 and 100.00% percent survival, respectively, compared to 60.00% survival in the sham-infected control group (Figure 3B). In both experimental infections, all mortalities occurred within 72 h post-infection.
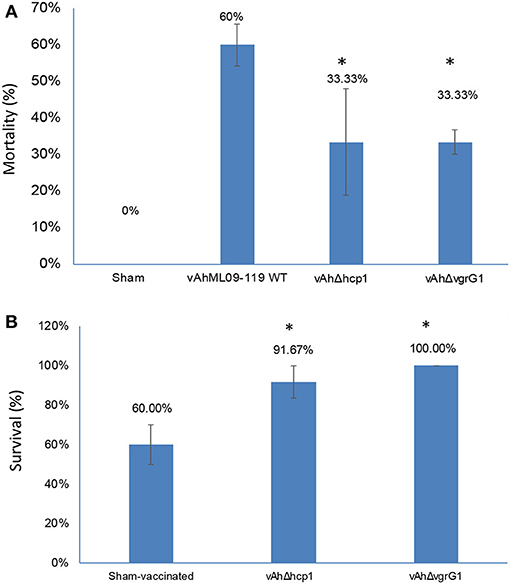
Figure 3. Virulence of vAh T6SS mutants in channel catfish fingerlings. (A) Percent mortalities in catfish fingerlings experimentally infected with vAh T6SS mutants and vAh wild type (WT) strain ML09-119. (B) Percent survival in catfish fingerlings surviving infection with T6SS mutants and re-challenged with vAh WT at 21 d post-infection. Data are the mean ± SE of three replicate tanks. Significant differences between challenged and non-vaccinated treatments are indicated with asterisks (p < 0.05).
Host-Pathogen Interaction
Using HPIDB, we predicted the interaction of identified A. hydophila secretion system components with host channel catfish (Ictalurus punctatus) proteins. We identified 333 catfish proteins that potentially interact with 30 different components of the A. hydrophila secretion systems (Supplementary File 2).
Discussion
In this study, our goal was to compare the distribution of secretion systems in A. hydrophila genomes using comparative genomics. We found that some of the secretion systems commonly involved in pathogenesis of Gram-negative bacterial infections are not consistently present in the U.S.A. vAh isolates. However, there are three secretion systems (T1SS, T2SS, and T4P) present in all A. hydrophila strains we analyzed, and one system (Tad) that is present almost specifically in vAh strains. We determined that genes hcp1 (tssD) and vgrG1 (tssI) contribute to vAh virulence in catfish despite the absence of a complete T6SS.
Phylogenetic ANI analysis based on the complete core genome of the 55 strains in our study confirmed their classification as A. hydrophila, and it showed that the 27 vAh strains formed a highly conserved branch that is clearly separated from the other A. hydrophila strains. Also, ANI analysis showed that the U.S.A. vAh isolates and Chinese epidemic isolates were derived from the same monophyletic clade.
Aeromonas hydrophila secretes a wide range of extracellular enzymes and toxins. Type I secretion systems are capable of secreting exotoxins and enzymes by a one-step process from cytoplasm to outer membrane. T1SS consists of three main components: ATP-binding cassette (ABC) transporters, membrane fusion protein (MFP), and outer membrane factor (OMF) (Green and Mecsas, 2016). All of the evaluated A. hydrophila genomes carry core components of the T1SS. However, most of the vAh isolates and some of the other isolates encode additional copies of some genes encoding core components of the T1SS. A domain search analysis (data not shown) for all the evaluated 55 genomes revealed that the vast majority of the vAh isolates encode RTX toxins, which are cytotoxins that potentially cause host cell rounding and apoptotic death. In Vibrio, RTX toxin is secreted by T1SS (Boardman and Satchell, 2004). Presence of T1SS increases virulence of Vibrio cholerae (Dolores et al., 2015) and Serratia marcescens (Létoffé et al., 1996).
Not surprisingly, all the evaluated A. hydrophila genomes possess a T2SS. This system is capable of secreting enzymes such as proteases, phosphatases, and lipases (Korotkov et al., 2012; Green and Mecsas, 2016); in A. hydrophila, it is also well known for exporting cytotoxic enterotoxin (Act), which has hemolytic and cytotoxic activities (Chopra et al., 2000; Galindo et al., 2004; Korotkov et al., 2012). T2SS is a large, trans-envelope apparatus encoded by a set of 12-16 core genes. It is located in the outer membrane, and it transports folded proteins from periplasm into the extracellular environment. T2SS differs from T1SS, which releases proteins to the outer medium, and T3SS, T4SS, and T6SS, which are contact-dependent (Hayes et al., 2010). T2SS secretes specific toxins, effectors, and large proteins that could not be secreted to the host or competitor bacteria otherwise (Rondelet and Condemine, 2013; Rosenzweig and Chopra, 2013). T2SS has sequence similarity with the type 4 pilus (T4P) system, which is responsible for motility, signaling, and adhesion (Nivaskumar and Francetic, 2014). T4P has not been studied extensively in A. hydrophila. T4P and T2SS show a high degree of similarity in their components, and one of the genes encoding a T2SS component, gspO, is located in the T4P-encoding locus (Nivaskumar and Francetic, 2014). Our secretion system analysis assigned the A. hydrophila gspO gene as pilD, which is one of the accessory genes of T4P. A. hydrophila gspC gene is listed as a missing mandatory gene in Figure 2, but it is present as an accessory gene (Figure 2). In E. coli, T2SS and T4P are important for persistent infection (Kulkarni et al., 2009).
The tight adherence (Tad) system contributes to biofilm formation, colonization, and virulence of several pathogens (Tomich et al., 2007). The Tad system is similar to T2SS systems (Peabody et al., 2003; Tomich et al., 2007). Intriguingly, our results showed that a complete Tad secretion system is available in vAh strains, whereas the majority of the other evaluated A. hydrophila genomes do not encode a Tad system except for two strains, one of which is human clinical isolate (strain AHNIH1), and the other is from milk (strain ATCC 7966). Interestingly, only one gene (tadZ) from this system is available in all the evaluated A. hydrophila genomes. TadZ is encoded by one of the mandatory genes of the Tad system and plays a major role in mediating polar localization of the Tad secretion system (Perez-Cheeks et al., 2012).
Many Gram-negative pathogens use type III secretion systems, which delivers effector proteins directly into host cells. Many components of this system are homologous to flagellum proteins. T3SS is an important contributor to pathogenesis of some A. hydrophila strains (Vilches et al., 2004; Yu et al., 2004); however, our comparative genomics analysis showed that 27 vAh strains lack genes encoding T3SS (except for the sctN gene, which encodes a highly conserved ATPase that contributes to energy metabolism and provides recognition capability for T3SS effectors and other virulence factors) (Zarivach et al., 2007). Most of the non-vAh isolates in our study encode T3SS, but the majority of these are environmental isolates from outside the U.S.A. On the other hand, U.S.A. environmental isolates (soil and wetland sediment) do not encode a T3SS, and they also lack Tad systems. Our results showing absence of T3SS in vAh strains are consistent with a previous smaller-scale comparative genomics study (Pang et al., 2015). Therefore, presence of genes encoding a T3SS may not a good indicator of virulence potential for A. hydrophila strains in fish. Similarly, the majority of human clinical isolates (seven out of ten) do not encode T3SS.
Interestingly, there is an inverse relationship between presence of a Tad system and a T3SS in many of the A. hydrophila genomes we analyzed. The Tad system is encoded in the vAh isolates, but they do not encode T3SS. On the other hand, almost all of the non-vAh A. hydrophila strains do not encode a Tad system, but many of these genomes encode T3SS (Figure 2). Ten strains have neither Tad nor T3SS systems, and only one strain (human isolate AHNIH1) encodes both systems. Therefore, the three secretion systems consistently encoded in vAh strains are T2SS, T4P, and Tad.
Flagella are important in motility and often in attachment to the host. They are linked with biofilm formation, which contributes to persistent infection (Tomás, 2012). In eels, an A. hydrophila polar flagellum mutant had decreased survival and adherence to eel macrophages (Qin et al., 2014). Because flagella proteins are similar to T3SS proteins (Nguyen et al., 2000; Gophna et al., 2003), we included them in our comparative genomics analysis. All the evaluated A. hydrophila genomes encode mandatory flagella genes. In some bacteria, T3SS components play a role in flagellar rotation (Diepold and Armitage, 2015), but in A. hydrophila, there is only one T3SS gene (sctN) shared by all the evaluated A. hydrophila genomes. In Sodalis glossinidius, SctN mediated entry into tsetse fly cells (Dale et al., 2001). An A. hydrophila master regulator of T3SS (ExsA) negatively affects the lateral flagella (Zhao and Shaw, 2016), so it is possible that T3SS and flagella proteins interact in A. hydrophila strains encoding both systems.
T6SS is widely distributed in Gram-negative bacteria, and it contributes to bacterial fitness in specific niches (Cianfanelli et al., 2016). In particular, it delivers secreted proteins into competitor bacteria or host cells (Zoued et al., 2014). T6SS is categorized into three phylogenetic subtypes (T6SSi, T6SSii, T6SSiii) (Russell et al., 2014). All of the A. hydrophila genomes we evaluated encode the entire T6SSi operon or remnants of the T6SSi. Some of the U.S.A. vAh strains have only three genes (hcp, tssH, and vgrG) of T6SS, while other vAh strains from U.S.A. and China encode all the core genes of T6SS. We extended our research to understand the role of these remnants in the pathogenicity of A. hydrophila.
Strain ML09-119 encodes two hemolysin co-regulated proteins (Hcp) (AHML_05970 and AHML_10025) and two valine-glycine repeat G (VgrG) proteins (AHML_05975 and AHML_10030). The hcp genes are located adjacent to the vgrG genes in strain ML09-119; the hcp1 gene is adjacent to vgrG1 gene, and hcp2 gene is adjacent to the vgrG2 gene. Multiple copies of hcp and vgrG genes are commonly seen in several bacterial species that possess a T6SS, including V. cholerae, Pseudomonas aeruginosa, A. hydrophila SSU, and A. hydrophila ATCC 7966T (Mougous et al., 2006; Podladchikova et al., 2011; Sha et al., 2013).
Hcp and VgrG are effector proteins of T6SS (Cascales, 2008). However, structural analysis of Hcp and VgrG from P. aeruginosa and V. cholerae showed that these proteins independently formed a transportation channel between the inner and outer membranes through which other effector molecules can be transported to the host cell (Leiman et al., 2009; Pell et al., 2009). Thus, Hcp and VgrG could also be part of the secretion apparatus. Hcp and VgrG contribute to pathogenesis of several Gram-negative species, including E. coli (Dudley et al., 2006), P. aeruginosa (Hood et al., 2010), Edwardsiella tarda (Rao et al., 2004), and Aeromonas (Sha et al., 2013). In V. cholera, an hcp1/hcp2 mutant is avirulent, whereas individual hcp1 or hcp2 mutants retain virulence. Therefore, at least one Hcp protein is required and sufficient for virulence (Pukatzki et al., 2006).
Secretion systems and effector proteins of A. hydrophila strain SSU have been studied extensively. It has been proposed that strain SSU be reclassified as Aeromonas dhakensis (Beaz-Hidalgo et al., 2015), but being a closely related species to A. hydrophila, SSU provides valuable comparative information. Strain SSU encodes a full T6SS, and its components are capable of translocating effector protein Hcp into eukaryotic cells (Suarez et al., 2008). Hcp modulates the activation of macrophages during infection in a mouse model (Suarez et al., 2010b). Effector protein VgrG is responsible for inducing host cell toxicity by ADP ribosylation of actin (Suarez et al., 2010a). In an intraperitoneal murine model of infection, all Hcp and VgrG paralogues were required for optimal A. hydrophila SSU virulence and dissemination to mouse peripheral organs (Sha et al., 2013).
vAh strain NJ-35 also encodes a functional T6SS that is located on a genomic island (Pang et al., 2015). This strain encodes three Hcp proteins. Hcp1 is responsible for T6SS assembly and inhibiting bacterial competition, Hcp2 negatively impacts biofilm formation and bacterial adhesion, and Hcp3 positively contributes to bacterial adhesion and biofilm formation (Wang et al., 2018). In NJ-35, all three genes contribute significantly to virulence, but a hcp2 mutant had greater attenuation than hcp1 and hcp3 mutants (7-fold increase in LD50 for hcp2 compared to 2-fold increase in LD50 for hcp1 and hcp3).
In our study, deletion of the hcp1 and vgrG1 genes in vAh strain ML09-119 affected virulence significantly (Figure 3A). This finding is consistent with those reported for A. hydrophila strain SSU and vAh strain NJ-35, but both of these strains encode a functional T6SS, while strain ML09-119 does not. So what is the role of Hcp and VrgG in vAh strains that do not encode a functional T6SS? Our virulence data substantiates they could have similar roles in pathogenesis as Hcp and VgrG proteins in strains SSU and NJ-35. However, there is another intriguing possibility. It has been hypothesized that putative effector islands could be translocated by Hcp and VgrG (De Maayer et al., 2011), and it is worth noting that T6SS is encoded on a genomic island in vAh strain NJ-35. Therefore, it is possible that Hcp and VgrG mobilize effector islands in A. hydrophila and are responsible for the genomic variation in T6SS encoded in the species.
With the goal of developing an effective vaccine to protect catfish from MAS caused by vAh, we determined the level of protection provided by the Δhcp1 and ΔvgrG1 mutants. Both mutants provided significant protection. The Δhcp1 and ΔvgrG1 mutants are not safe enough for use as vaccines, but our results validate our approach of using comparative genomics to identify candidate virulence genes. Our results also indicate that deletion of virulence genes is a valid approach for live attenuated vaccine development against vAh.
T9SS is typically only found in some species in the Bacteroidetes phylum, so it is not surprising that only one gene (sprA) encoding a protein similar to T9SS is present in all the evaluated A. hydrophila genomes. T9SS functions as a secretion system but also enables gliding motility (McBride and Zhu, 2013; Sato et al., 2013; McBride and Nakane, 2015). In Flavobacterium johnsoniae, SprA is responsible (along with SprE and SprT) for secretion of SprB (Shrivastava et al., 2013).
Due to their role in secreting proteins involved in pathogenesis of multiple bacterial species, it is not surprising that 30 of the A. hydrophila secretion system proteins have predicted interactions with channel catfish proteins. We chose channel catfish as the host species for this analysis because of its importance as an aquaculture species in the U.S.A. and due to the impacts and known virulence of vAh strains on this species. These results confirm the multiple interactions between A. hydrophila secretion systems and channel catfish, adding additional evidence to their potential roles in A. hydrophila virulence.
In summary, our analysis indicates that vAh strains do not encode two of the contact-dependent secretion systems commonly involved in virulence of many Gram-negative pathogens, T3SS and T6SS. In fact, the T3SS is missing in all vAh strains and many other A. hydrophila strains. This suggests that vAh utilizes other systems to secrete effectors, toxins, and large secreted proteins. T1SS, T2SS, and T4P systems are encoded in all the A. hydrophila strains we sequenced, and these systems likely secrete several virulence-related proteins. Interestingly, the Tad system is present in all the vAh strains we sequenced, but it is only present in two of the non-vAh strains we analyzed. It is possible that the Tad system is one of the vAh-specific adaptations that make this clade of A. hydrophila more virulent.
Although only some A. hydrophila have a complete T6SS, all of the strains in our analysis encode three T6SS proteins. We determined that two of these genes, Δhcp1 and ΔvgrG1, contribute significantly to channel catfish virulence. Further investigation of the role of these T6SS genes in A. hydrophila is warranted, including the effects of deleting all the hcp and vrgG alleles on A. hydrophila virulence.
Author Contributions
HT, HA, AK, and ML designed and conceived the analysis and experiments. HT, HA, JB, and SK performed experiments and analyzed the data. HT, HA, AK, and ML wrote the manuscript. All authors read and approved the final manuscript.
Funding
This project was supported by Agriculture and Food Research Initiative competitive grant no. 2013-67015-21313 from the USDA National Institute of Food and Agriculture.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Laboratory Animal Resources and Care at the College of Veterinary Medicine for providing the SPF channel catfish.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03216/full#supplementary-material
Supplementary File 1. AAI distances.
Supplementary File 2. Host-Pathogen interactions.
References
Abby, S. S., Cury, J., Guglielmini, J., Neron, B., Touchon, M., and Rocha, E. P. (2016). Identification of protein secretion systems in bacterial genomes. Sci. Rep. 6:23080. doi: 10.1038/srep23080
Abby, S. S., Neron, B., Menager, H., Touchon, M., and Rocha, E. P. (2014). MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS ONE 9:e110726. doi: 10.1371/journal.pone.0110726
Abdelhamed, H., Lu, J., Shaheen, A., Abbass, A., Lawrence, M. L., and Karsi, A. (2013). Construction and evaluation of an Edwardsiella ictaluri fhuC mutant. Vet. Microbiol. 162, 858–865. doi: 10.1016/j.vetmic.2012.11.006
Abdelhamed, H., Nho, S. W., Turaga, G., Banes, M. M., Karsi, A., and Lawrence, M. L. (2016). Protective efficacy of four recombinant fimbrial proteins of virulent Aeromonas hydrophila strain ML09-119 in channel catfish. Vet. Microbiol. 197, 8–14. doi: 10.1016/j.vetmic.2016.10.026
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ammari, M. G., Gresham, C. R., McCarthy, F. M., and Nanduri, B. (2016). HPIDB 2.0: a curated database for host-pathogen interactions. Database 2016:baw103. doi: 10.1093/database/baw103
Beaz-Hidalgo, R., Hossain, M. J., Liles, M. R., and Figueras, M. J. (2015). Strategies to avoid wrongly labelled genomes using as example the detected wrong taxonomic affiliation for aeromonas genomes in the GenBank database. PLoS ONE 10:e0115813. doi: 10.1371/journal.pone.0115813
Boardman, B. K., and Satchell, K. J. (2004). Vibrio cholerae strains with mutations in an atypical type I secretion system accumulate RTX toxin intracellularly. J. Bacteriol. 186, 8137–8143. doi: 10.1128/JB.186.23.8137-8143.2004
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Cascales, E. (2008). The type VI secretion toolkit. EMBO Rep. 9, 735–741. doi: 10.1038/embor.2008.131
Chan, K. G., Puthucheary, S. D., Chan, X. Y., Yin, W. F., Wong, C. S., Too, W. S., et al. (2011). Quorum sensing in Aeromonas species isolated from patients in Malaysia. Curr. Microbiol. 62, 167–172. doi: 10.1007/s00284-010-9689-z
Chan, K. G., Tan, W. S., Chang, C. Y., Yin, W. F., and Mumahad Yunos, N. Y. (2015). Genome sequence analysis reveals evidence of quorum-sensing genes present in Aeromonas hydrophila strain M062, isolated from freshwater. Genome Announc. 3:e00100–15. doi: 10.1128/genomeA.00100-15
Chopra, A. K., Xu, X., Ribardo, D., Gonzalez, M., Kuhl, K., Peterson, J. W., et al. (2000). The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect. Immun. 68, 2808–2818. doi: 10.1128/IAI.68.5.2808-2818.2000
Cianfanelli, F. R., Monlezun, L., and Coulthurst, S. J. (2016). Aim, Load, Fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62. doi: 10.1016/j.tim.2015.10.005
Colston, S. M., Fullmer, M. S., Beka, L., Lamy, B., Gogarten, J. P., and Graf, J. (2014). Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. MBio 5:e02136. doi: 10.1128/mBio.02136-14
Dale, C., Young, S. A., Haydon, D. T., and Welburn, S. C. (2001). The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. U.S.A. 98, 1883–1888. doi: 10.1073/pnas.98.4.1883
De Maayer, P., Venter, S. N., Kamber, T., Duffy, B., Coutinho, T. A., and Smits, T. H. (2011). Comparative genomics of the type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics 12:576. doi: 10.1186/1471-2164-12-576
Diepold, A., and Armitage, J. P. (2015). Type III secretion systems: the bacterial flagellum and the injectisome. Philos. Trans. R. Soc. Lond B Biol. Sci. 370:1679. doi: 10.1098/rstb.2015.0020
Dolores, J. S., Agarwal, S., Egerer, M., and Satchell, K. J. (2015). Vibrio cholerae MARTX toxin heterologous translocation of beta-lactamase and roles of individual effector domains on cytoskeleton dynamics. Mol. Microbiol. 95, 590–604. doi: 10.1111/mmi.12879
Dozois, C. M., Daigle, F., and Curtiss, R. III. (2003). Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. U.S.A. 100, 247–252. doi: 10.1073/pnas.232686799
Dudley, E. G., Thomson, N. R., Parkhill, J., Morin, N. P., and Nataro, J. P. (2006). Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 61, 1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Emond-Rheault, J. G., Vincent, A. T., Trudel, M. V., Brochu, F., Boyle, B., Tanaka, K. H., et al. (2015). Variants of a genomic island in Aeromonas salmonicida subsp. salmonicida link isolates with their geographical origins. Vet. Microbiol. 175, 68–76. doi: 10.1016/j.vetmic.2014.11.014
Forn-Cuní, G., Tomas, J. M., and Merino, S. (2016). Whole-genome sequence of Aeromonas hydrophila strain AH-1 (serotype O11). Genome Announc. 4:e00920–16. doi: 10.1128/genomeA.00920-16
Galindo, C. L., Fadl, A. A., Sha, J., Gutierrez, C. Jr., Popov, V. L., Boldogh, I., et al. (2004). Aeromonas hydrophila cytotoxic enterotoxin activates mitogen-activated protein kinases and induces apoptosis in murine macrophages and human intestinal epithelial cells. J. Biol. Chem. 279, 37597–37612. doi: 10.1074/jbc.M404641200
Gophna, U., Ron, E. Z., and Graur, D. (2003). Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312, 151–163. doi: 10.1016/S0378-1119(03)00612-7
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57(Pt 1), 81–91. doi: 10.1099/ijs.0.64483-0
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems: an overview. Microbiol. Spectr. 4, 215–239. doi: 10.1128/microbiolspec.VMBF-0012-2015
Griffin, M. J., Goodwin, A. E., Merry, G. E., Liles, M. R., Williams, M. A., Ware, C., et al. (2013). Rapid quantitative detection of Aeromonas hydrophila strains associated with disease outbreaks in catfish aquaculture. J. Vet. Diagn. Invest. 25, 473–481. doi: 10.1177/1040638713494210
Grim, C. J., Kozlova, E. V., Ponnusamy, D., Fitts, E. C., Sha, J., Kirtley, M. L., et al. (2014). Functional genomic characterization of virulence factors from necrotizing fasciitis-causing strains of Aeromonas hydrophila. Appl. Environ. Microbiol. 80, 4162–4183. doi: 10.1128/AEM.00486-14
Han, J. E., Kim, J. H., Choresca, C., Shin, S. P., Jun, J. W., and Park, S. C. (2013). Draft genome sequence of a clinical isolate, Aeromonas hydrophila SNUFPC-A8, from a moribund cherry salmon (Oncorhynchus masou masou). Genome Announc. 1:e00133–12. doi: 10.1128/genomeA.00133-12
Hayes, C. S., Aoki, S. K., and Low, D. A. (2010). Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 44, 71–90. doi: 10.1146/annurev.genet.42.110807.091449
Hemstreet, W. B. (2010). An update on Aeromonas hydrophila from a fish health specialist for summer 2010. Catfish J. 24:4.
Herrero, M., de Lorenzo, V., and Timmis, K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172, 6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990
Hood, R. D., Singh, P., Hsu, F., Guvener, T., Carl, M. A., Trinidad, R. R., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K., and Pease, L. R. (1989). Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68. doi: 10.1016/0378-1119(89)90359-4
Hossain, M. J., Sun, D., McGarey, D. J., Wrenn, S., Alexander, L. M., Martino, M. E., et al. (2014). An Asian origin of virulent Aeromonas hydrophila responsible for disease epidemics in United States-farmed catfish. MBio 5:e00848–14. doi: 10.1128/mBio.00848-14
Hossain, M. J., Waldbieser, G. C., Sun, D., Capps, N. K., Hemstreet, W. B., Carlisle, K., et al. (2013). Implication of lateral genetic transfer in the emergence of Aeromonas hydrophila isolates of epidemic outbreaks in channel catfish. PLoS ONE 8:e80943. doi: 10.1371/journal.pone.0080943
Hughes, H. Y., Conlan, S. P., Lau, A. F., Dekker, J. P., Michelin, A. V., Youn, J. H., et al. (2016). Detection and whole-genome sequencing of carbapenemase-producing Aeromonas hydrophila isolates from routine perirectal surveillance culture. J. Clin. Microbiol. 54, 1167–1170. doi: 10.1128/JCM.03229-15
Jagoda, S. S., Tan, E., Arulkanthan, A., Kinoshita, S., Watabe, S., and Asakawa, S. (2014). Draft Genome sequence of Aeromonas hydrophila strain Ae34, isolated from a septicemic and moribund koi carp (Cyprinus carpio koi), a freshwater aquarium fish. Genome Announc. 2:e00572–14. doi: 10.1128/genomeA.00572-14
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Konstantinidis, K. T., Ramette, A., and Tiedje, J. M. (2006). The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1929–1940. doi: 10.1098/rstb.2006.1920
Konstantinidis, K. T., and Tiedje, J. M. (2005a). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Konstantinidis, K. T., and Tiedje, J. M. (2005b). Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005
Korotkov, K. V., Sandkvist, M., and Hol, W. G. (2012). The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10, 336–351. doi: 10.1038/nrmicro2762
Kulkarni, R., Dhakal, B. K., Slechta, E. S., Kurtz, Z., Mulvey, M. A., and Thanassi, D. G. (2009). Roles of putative type II secretion and type IV pilus systems in the virulence of uropathogenic Escherichia coli. PLoS ONE 4:e4752. doi: 10.1371/journal.pone.0004752
Leiman, P. G., Basler, M., Ramagopal, U. A., Bonanno, J. B., Sauder, J. M., Pukatzki, S., et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159. doi: 10.1073/pnas.0813360106
Lenneman, E. M., and Barney, B. M. (2014). Draft genome sequences of the alga-degrading bacteria Aeromonas hydrophila strain AD9 and Pseudomonas pseudoalcaligenes strain AD6. Genome Announc. 2:e00709–14. doi: 10.1128/genomeA.00709-14
Létoffé, S., Delepelaire, P., and Wandersman, C. (1996). Protein secretion in gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 15, 5804–5811. doi: 10.1002/j.1460-2075.1996.tb00967.x
McBride, M. J., and Nakane, D. (2015). Flavobacterium gliding motility and the type IX secretion system. Curr. Opin. Microbiol. 28, 72–77. doi: 10.1016/j.mib.2015.07.016
McBride, M. J., and Zhu, Y. (2013). Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J. Bacteriol. 195, 270–278. doi: 10.1128/JB.01962-12
Metcalf, W. W., Jiang, W., and Wanner, B. L. (1994). Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138, 1–7. doi: 10.1016/0378-1119(94)90776-5
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Nguyen, L., Paulsen, I. T., Tchieu, J., Hueck, C. J., and Saier, M. H. Jr. (2000). Phylogenetic analyses of the constituents of type III protein secretion systems. J. Mol. Microbiol. Biotechnol. 2, 125–144.
Nielsen, M. E., Hoi, L., Schmidt, A. S., Qian, D., Shimada, T., Shen, J. Y., et al. (2001). Is Aeromonas hydrophila the dominant motile Aeromonas species that causes disease outbreaks in aquaculture production in the Zhejiang Province of China? Dis. Aquat. Org. 46, 23–29. doi: 10.3354/dao046023
Nivaskumar, M., and Francetic, O. (2014). Type II secretion system: a magic beanstalk or a protein escalator. Biochim. Biophys. Acta 1843, 1568–1577. doi: 10.1016/j.bbamcr.2013.12.020
Pan, H. J., Wu, S. Q., Dong, C. F., Shi, C. B., Ye, M. X., Lin, T. L., et al. (2004). Identification, virulence, hemolytic activity of GYK1, a strain of pathogenic Aeromonas hydrophila isolated from mandarinfish. J. Shanghai Fish. Univ. 13, 23–29.
Pang, M., Jiang, J., Xie, X., Wu, Y., Dong, Y., Kwok, A. H., et al. (2015). Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics. Sci. Rep. 5:9833. doi: 10.1038/srep09833
Peabody, C. R., Chung, Y. J., Yen, M. R., Vidal-Ingigliardi, D., Pugsley, A. P., and Saier, M. H. Jr. (2003). Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149(Pt 11), 3051–3072. doi: 10.1099/mic.0.26364-0
Pell, L. G., Kanelis, V., Donaldson, L. W., Howell, P. L., and Davidson, A. R. (2009). The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165. doi: 10.1073/pnas.0900044106
Perez-Cheeks, B. A., Planet, P. J., Sarkar, I. N., Clock, S. A., Xu, Q., and Figurski, D. H. (2012). The product of tadZ, a new member of the parA/minD superfamily, localizes to a pole in Aggregatibacter actinomycetemcomitans. Mol. Microbiol. 83, 694–711. doi: 10.1111/j.1365-2958.2011.07955.x
Podladchikova, O., Antonenka, U., Heesemann, J., and Rakin, A. (2011). Yersinia pestis autoagglutination factor is a component of the type six secretion system. Int. J. Med. Microbiol. 301, 562–569. doi: 10.1016/j.ijmm.2011.03.004
Pridgeon, J. W., Zhang, D., and Zhang, L. (2014a). Complete genome sequence of a moderately virulent Aeromonas hydrophila strain, pc104A, isolated from soil of a catfish pond in west Alabama. Genome Announc. 2:e00554–14. doi: 10.1128/genomeA.00554-14
Pridgeon, J. W., Zhang, D., and Zhang, L. (2014b). Complete genome sequence of the highly virulent Aeromonas hydrophila AL09-71 isolated from diseased channel catfish in west Alabama. Genome Announc. 2:e00450–14. doi: 10.1128/genomeA.00450-14
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Qin, Y., Lin, G., Chen, W., Huang, B., Huang, W., and Yan, Q. (2014). Flagellar motility contributes to the invasion and survival of Aeromonas hydrophila in Anguilla japonica macrophages. Fish Shellfish Immunol. 39, 273–279. doi: 10.1016/j.fsi.2014.05.016
Rao, P. S., Yamada, Y., Tan, Y. P., and Leung, K. Y. (2004). Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53, 573–586. doi: 10.1111/j.1365-2958.2004.04123.x
Rasmussen, M., Olsen, M. S., Sunde, L., Pedersen, L., and Petersen, O. B. (2016). Positive predictive value and completeness of prenatally assigned International Classification of Disease-10 kidney anomaly diagnoses in the Danish National Patient Registry. Clin. Epidemiol. 8, 9–14. doi: 10.2147/CLEP.S94394
Rasmussen-Ivey, C. R., Hossain, M. J., Odom, S. E., Terhune, J. S., Hemstreet, W. G., Shoemaker, C. A., et al. (2016). Classification of a hypervirulent Aeromonas hydrophila pathotype responsible for epidemic outbreaks in warm-water fishes. Front. Microbiol. 7:1615. doi: 10.3389/fmicb.2016.01615
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Roach, D. J., Burton, J. N., Lee, C., Stackhouse, B., Butler-Wu, S. M., Cookson, B. T., et al. (2015). A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet. 11:e1005413. doi: 10.1371/journal.pgen.1005413
Rondelet, A., and Condemine, G. (2013). Type II secretion: the substrates that won't go away. Res. Microbiol. 164, 556–561. doi: 10.1016/j.resmic.2013.03.005
Rosenzweig, J. A., and Chopra, A. K. (2013). Modulation of host immune defenses by Aeromonas and Yersinia species: convergence on toxins secreted by various secretion systems. Front. Cell. Infect. Microbiol. 3:70. doi: 10.3389/fcimb.2013.00070
Russell, A. B., Wexler, A. G., Harding, B. N., Whitney, J. C., Bohn, A. J., Goo, Y. A., et al. (2014). A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236. doi: 10.1016/j.chom.2014.07.007
Sato, K., Yukitake, H., Narita, Y., Shoji, M., Naito, M., and Nakayama, K. (2013). Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol. Lett. 338, 68–76. doi: 10.1111/1574-6968.12028
Seshadri, R., Joseph, S. W., Chopra, A. K., Sha, J., Shaw, J., Graf, J., et al. (2006). Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188, 8272–8282. doi: 10.1128/JB.00621-06
Sha, J., Rosenzweig, J. A., Kozlova, E. V., Wang, S., Erova, T. E., Kirtley, M. L., et al. (2013). Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiology 159(Pt 6), 1120–1135. doi: 10.1099/mic.0.063495-0
Shrivastava, A., Johnston, J. J., van Baaren, J. M., and McBride, M. J. (2013). Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J. Bacteriol. 195, 3201–3212. doi: 10.1128/JB.00333-13
Suarez, G., Sierra, J. C., Erova, T. E., Sha, J., Horneman, A. J., and Chopra, A. K. (2010a). A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168. doi: 10.1128/JB.01260-09
Suarez, G., Sierra, J. C., Kirtley, M. L., and Chopra, A. K. (2010b). Role of Hcp, a type 6 secretion system effector, of Aeromonas hydrophila in modulating activation of host immune cells. Microbiology 156(Pt 12), 3678–3688. doi: 10.1099/mic.0.041277-0
Suarez, G., Sierra, J. C., Sha, J., Wang, S., Erova, T. E., Fadl, A. A., et al. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44, 344–361. doi: 10.1016/j.micpath.2007.10.005
Tan, W. S., Yin, W. F., and Chan, K. G. (2015a). Insights into the quorum-sensing activity in Aeromonas hydrophila strain M013 as revealed by Whole-Genome sequencing. Genome Announc. 3:e01372–14. doi: 10.1128/genomeA.01372-14
Tan, W. S., Yin, W. F., Chang, C. Y., and Chan, K. G. (2015b). Whole-genome sequencing analysis of quorum-sensing Aeromonas hydrophila strain M023 from freshwater. Genome Announc. 3:e01548–14. doi: 10.1128/genomeA.01548-14
Tekedar, H. C., Karsi, A., Akgul, A., Kalindamar, S., Waldbieser, G. C., Sonstegard, T., et al. (2015). Complete genome sequence of fish pathogen Aeromonas hydrophila AL06-06. Genome Announc. 3:e00368–15. doi: 10.1128/genomeA.00368-15
Tekedar, H. C., Kumru, S., Kalindamar, S., Karsi, A., Waldbieser, G. C., Sonstegard, T., et al. (2017). Draft genome sequences of three Aeromonas hydrophila isolates from catfish and tilapia. Genome Announc. 5:e01509–16. doi: 10.1128/genomeA.01509-16
Tekedar, H. C., Kumru, S., Karsi, A., Waldbieser, G. C., Sonstegard, T., Schroeder, S. G., et al. (2016a). Draft genome sequence of Aeromonas hydrophila TN97-08. Genome Announc. 4:e00436–16. doi: 10.1128/genomeA.00436-16
Tekedar, H. C., Kumru, S., Karsi, A., Waldbieser, G. C., Sonstegard, T., Schroeder, S. G., et al. (2016b). Draft genome sequences of four virulent Aeromonas hydrophila strains from catfish aquaculture. Genome Announc. 4:e00860–16. doi: 10.1128/genomeA.00860-16
Tekedar, H. C., Waldbieser, G. C., Karsi, A., Liles, M. R., Griffin, M. J., Vamenta, S., et al. (2013). Complete genome sequence of a channel catfish epidemic isolate, Aeromonas hydrophila strain ML09-119. Genome Announc. 1:e00755–13. doi: 10.1128/genomeA.00755-13
Tomás, J. M. (2012). The main Aeromonas pathogenic factors. ISRN Microbiol. 2012:256261. doi: 10.5402/2012/256261
Tomich, M., Planet, P. J., and Figurski, D. H. (2007). The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5, 363–375. doi: 10.1038/nrmicro1636
Tran, N. T., Gao, Z. X., Zhao, H. H., Yi, S. K., Chen, B. X., Zhao, Y. H., et al. (2015). Transcriptome analysis and microsatellite discovery in the blunt snout bream (Megalobrama amblycephala) after challenge with Aeromonas hydrophila. Fish Shellfish Immunol. 45, 72–82. doi: 10.1016/j.fsi.2015.01.034
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Vilches, S., Urgell, C., Merino, S., Chacon, M. R., Soler, L., Castro-Escarpulli, G., et al. (2004). Complete type III secretion system of a mesophilic Aeromonas hydrophila strain. Appl. Environ. Microbiol. 70, 6914–6919. doi: 10.1128/AEM.70.11.6914-6919.2004
Wang, N., Liu, J., Pang, M., Wu, Y., Awan, F., Liles, M. R., et al. (2018). Diverse roles of Hcp family proteins in the environmental fitness and pathogenicity of Aeromonas hydrophila Chinese epidemic strain NJ-35. Appl. Microbiol. Biotechnol. 102:7083–7095. doi: 10.1007/s00253-018-9116-0
Xu, L., Wang, H., Yang, X., and Lu, L. (2013). Integrated pharmacokinetics/pharmacodynamics parameters-based dosing guidelines of enrofloxacin in grass carp Ctenopharyngodon idella to minimize selection of drug resistance. BMC Vet. Res. 9:126. doi: 10.1186/1746-6148-9-126
Yang, W., Li, N., Li, M., Zhang, D., and An, G. (2016). Complete genome sequence of fish pathogen Aeromonas hydrophila JBN2301. Genome Announc. 4:e01615–15. doi: 10.1128/genomeA.01615-15
Yu, H. B., Rao, P. S., Lee, H. C., Vilches, S., Merino, S., Tomas, J. M., et al. (2004). A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 72, 1248–1256. doi: 10.1128/IAI.72.3.1248-1256.2004
Zarivach, R., Vuckovic, M., Deng, W., Finlay, B. B., and Strynadka, N. C. (2007). Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 14, 131–137. doi: 10.1038/nsmb1196
Zhang, X., Yang, W., Wu, H., Gong, X., and Li, A. (2014). Multilocus sequence typing revealed a clonal lineage of Aeromonas hydrophila caused motile Aeromonas septicemia outbreaks in pond-cultured cyprinid fish in an epidemic area in central China. Aquaculture 432, 1–6. doi: 10.1016/j.aquaculture.2014.04.017
Zhang, X. J., Yang, W. M., Li, T. T., and Li, A. H. (2013). Research on the genetic diversity and virulence characteristics of Aeromonas hydrophila isolated from fishponds with disease outbreaks in Hubei province. Acta Hydrobiol. 3, 458–466.
Zhao, Y. H., and Shaw, J. G. (2016). Cross-talk between the Aeromonas hydrophila type III secretion system and lateral flagella system. Front. Microbiol. 7:1434. doi: 10.3389/fmicb.2016.01434
Keywords: Aeromonas hydrophila, comparative genomics, secretion systems, T6SS, Hcp, VgrG
Citation: Tekedar HC, Abdelhamed H, Kumru S, Blom J, Karsi A and Lawrence ML (2018) Comparative Genomics of Aeromonas hydrophila Secretion Systems and Mutational Analysis of hcp1 and vgrG1 Genes From T6SS. Front. Microbiol. 9:3216. doi: 10.3389/fmicb.2018.03216
Received: 30 May 2018; Accepted: 11 December 2018;
Published: 09 January 2019.
Edited by:
Martin Stephen Llewellyn, University of Glasgow, United KingdomReviewed by:
Ana Cristina Esteves, University of Aveiro, PortugalHetron Mweemba Munang'andu, Norwegian University of Life Sciences, Norway
Copyright © 2019 Tekedar, Abdelhamed, Kumru, Blom, Karsi and Lawrence. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark L. Lawrence, bGF3cmVuY2VAY3ZtLm1zc3RhdGUuZWR1
†These authors have contributed equally to this work
 Hasan C. Tekedar
Hasan C. Tekedar Hossam Abdelhamed
Hossam Abdelhamed Salih Kumru
Salih Kumru Jochen Blom
Jochen Blom Attila Karsi
Attila Karsi Mark L. Lawrence
Mark L. Lawrence