
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 January 2019
Sec. Evolutionary and Genomic Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.03205
 Fabio Faria da Mota1,2
Fabio Faria da Mota1,2 Daniele Pereira Castro2,3
Daniele Pereira Castro2,3 Cecilia Stahl Vieira3
Cecilia Stahl Vieira3 Marcia Gumiel3
Marcia Gumiel3 Julia Peixoto de Albuquerque4
Julia Peixoto de Albuquerque4 Nicolas Carels5*
Nicolas Carels5* Patricia Azambuja2,3
Patricia Azambuja2,3Serratia marcescens is a bacterium with the ability to colonize several niches, including some eukaryotic hosts. S. marcescens have been recently found in the gut of hematophagous insects that act as parasite vectors, such as Anopheles, Rhodnius, and Triatoma. While some S. marcescens strains have been reported as symbiotic or pathogenic to other insects, the role of S. marcescens populations from the gut microbiota of Rhodnius prolixus, a vector of Chagas’ disease, remains unknown. Bacterial colonies from R. prolixus gut were isolated on BHI agar. After BOX-PCR fingerprinting, the genomic sequences of two isolates RPA1 and RPH1 were compared to others S. marcescens from the NCBI database in other to estimate their evolutionary divergence. The in vitro trypanolytic activity of these two bacterial isolates against Trypanosoma cruzi (DM28c clone and Y strain) was assessed by microscopy. In addition, the gene expression of type VI secretion system (T6SS) was detected in vivo by RT-PCR. Comparative genomics of RPA1 and RPH1 revealed, besides plasmid presence and genomic islands, genes related to motility, attachment, and quorum sensing in both genomes while genes for urea hydrolysis and type II secretion system (T2SS) were found only in the RPA1 genome. The in vitro trypanolytic activity of both S. marcescens strains was stronger in their stationary phases of growth than in their exponential ones, with 65–70 and 85–90% of epimastigotes (Dm28c clone and Y strain, respectively) being lysed after incubation with RPA1 or RPH1 in stationary phase. Although T6SS transcripts were detected in guts up to 40 days after feeding (DAF), R. prolixus morbidity or mortality did not appear to be affected. In this report, we made available two trypanolytic S. marcescens strains from R. prolixus gut to the scientific community together with their genomic sequences. Here, we describe their genomic features with the purpose of bringing new insights into the S. marcescens adaptations for colonization of the specific niche of triatomine guts. This study provides the basis for a better understanding of the role of S. marcescens in the microbiota of R. prolixus gut as a potential antagonist of T. cruzi in this complex system.
Trypanosoma cruzi is a protozoan parasite that causes Chagas disease (also known as American Trypanosomiasis), a neglected disease that affects six to seven million people worldwide and is transmitted by triatomines, which are insect vectors from the family of Reduviidae (Coura, 2015; World Health Organization [WHO], 2018). T. cruzi, after been ingested by a triatomine vector, multiplies as epimastigotes within its midgut and finally in the rectum where it differentiates into infective metacyclic trypomastigotes, which can be eliminated through feces during the triatomine blood meal on the vertebrate host (Cortez et al., 2012; Ferreira et al., 2016). Although T. cruzi transmission through triatomine vectors is considered the principal infection mechanism, the oral route transmission through food contaminated by infected triatomines is increasing, especially in the Amazon region, including Brazil, Colombia, Venezuela, French Guyana, and Bolivia (Noya and González, 2015).
Chagas disease presents an acute phase with mild or no symptoms and a chronic phase during which parasites can be hidden mainly in the heart and digestive muscles (Coura and Viñas, 2010). As the infection proceeds, it leads to sudden heart failure due to the progressive destruction of nervous connections. Chagas disease has been reported mainly in Latin America where it is endemic in 21 countries, but over the past decades, it has also been detected in the United States, Canada, Europe, and some Western Pacific countries (Coura and Viñas, 2010; World Health Organization [WHO], 2018) due to infected people emigration and blood transfusion (Hernández-Romano et al., 2015). Because of the lack of a vaccine, vector control plays a key role in the prevention of Chagas disease, which is performed through the use of insecticides. However, insecticide resistance has been extensively detected over the last 15 years (Vassena et al., 2000) and deserves attention because of its increasing impacts on costs of vector control (Germano et al., 2012) and of its interference with the dynamic of triatomine population in situ. As a result, the re-infestation by triatomines of different species, such as Triatoma brasiliensis and Triatoma pseudomaculata, infected with T. cruzi, has been notified in endemic areas to replace Triatoma infestans, the original triatomine vector (Coura and Viñas, 2010; Pessoa et al., 2015; Barbosa-Silva et al., 2016).
A sustainable alternative strategy to insecticide is biological control. Fungi (Vázquez-Martínez et al., 2014; Forlani et al., 2015; Garcia et al., 2016), and toxin-producing bacteria such as Bacillus thuringiensis are already largely used as commercial biopesticides. The biological control by symbiotic bacteria, such as Wolbachia sp., has also been demonstrated to be effective in the control of Aedes aegypti (McMeniman et al., 2009; Bian et al., 2010; Hancock et al., 2016).
Since 2002, Kondo et al. (2002) demonstrated that the biological control of host’s reproduction with Wolbachia is a successful alternative strategy to transgenic approaches. In mosquitos, Wolbachia can be transferred between insect species and rapidly spread through natural populations of A. aegypti (McMeniman et al., 2009). However, exclusion of Wolbachia from the reproductive organs of mosquito vectors by other bacteria such as those from the genus Asaia may prevent its control effectiveness (Rossi et al., 2015). In addition, antibiotics produced by the native microbiota of triatomine digestive tract (TDT) also could disturb Wolbachia colonization in new vector species (Rossi et al., 2015). Consequently, it is important to know the members of TDT microbiota and their respective functionalities in that environment.
Rhodococcus rhodnii, a bacterial symbiont of Rhodnius prolixus reported in some specimens bred in insectaries, was suggested to provide the vector with vitamins (Pachebat et al., 2013). Paratransgenic approaches proposed that R. rhodnii could be genetically modified and reintroduced in the TDT microbiota to control Chagas disease vectors (Hurwitz et al., 2011) through (i) systemic RNAi (Taracena et al., 2015; Whitten et al., 2016), (ii) the expression of antimicrobial molecules, such as cecropin A (Durvasula et al., 1997; Hurwitz et al., 2012), or (iii) functional antibody fragment (Durvasula et al., 1999).
Gumiel et al. (2015) observed that the TDT microbiota might differ between triatomines from insectary and from the wild. Although actinobacteria such as R. rhodnii were reported in some traditional Rhodnius insectary colonies (Eichler and Schaub, 2002; Dias et al., 2015), these have been only sporadically observed in specimens from field capture. On the other hand, Serratia species has been predominantly diagnosed in the TDT microbiota of wild Triatoma specimens, by high-throughput sequencing of ribosomal gene (Gumiel et al., 2015), and in Rhodnius from other insectaries by Sanger sequencing (da Mota et al., 2012) as well as by 454 pyrosequencing of 16S rRNA (Vieira et al., 2015). Globally, it seems that bacteria with GC-rich genomes are the dominant microbial components in the ecological niche of TDT (Carels et al., 2017).
Serratia sp. is ubiquitous and found widespread around the globe. Members of Serratia genus exist in a wide range of habitats, including water, soil, and the digestive tracts of various animals, showing extremely diverse ecological adaptations (Petersen and Tisa, 2013). Serratia includes insect symbionts (Manzano-Marín et al., 2016), insect pathogens (Grimont et al., 1979), nematode pathogens (Schulenburg and Ewbank, 2004), plant pathogens (Zhang et al., 2003), plant growth-promoting rhizobacterium (Berg, 2000), coral pathogens (Patterson et al., 2002), nosocomial human pathogens (Mahlen, 2011), and bacterial antagonists (Thomson et al., 2000; Masschelein et al., 2013).
While R. rhodnii has been cited as a symbiotic member of TDT, the role of Serratia in the TDT environment and in vector homeostasis is not clear. However, there are some pieces of evidence that S. marcencens interferes with the success rate of digestive tract colonization by T. cruzi in R. prolixus (Azambuja et al., 2004; Castro et al., 2012; Vieira et al., 2016). In addition, an entomopathogenic S. liquefaciens Strain FK01 (Taira et al., 2014), which was originally isolated from the ant lion, was shown to be highly virulent to the American cockroach and the silkworm (Egami et al., 2009). Ultrastructures showing the lysis of the human parasite Leishmania (Leishmania) chagasi by S. marcescens were also reported by electron microscopy (Moraes et al., 2008).
Different secretions systems were reported in S. marcescens isolated from other environments (Alcoforado-Diniz and Coulthurst, 2015). These secretions systems could export toxins or other molecules with high antagonistic potential against other bacterial members of TDT microbiota community or against eukaryotic cells, such as fungi, protozoa, or epithelial cells from the vector itself (Li et al., 2015). Among these bacterial secretion systems, the type VI secretion system (T6SS) fires toxic proteins into target cells like a nanomachine. The target structure can vary depending on the different subassemblies of the T6SS machinery (Gerc et al., 2015). The T6SS represents a means by which bacteria interact with their host or attack competitors organisms by contact with an attacking neighbor cell. Azambuja et al. (2004) suggested that prodigiosin could be an important factor contributing to the trypanolytic action of Serratia. Castro et al. (2007a) also suggested that bacterial fimbriae could participate to the lysis of protozoan parasites induced by S. marcescens, once the trypanolytic effect of S. marcescens in vitro was dependent on D-mannose and distinct from the hemolytic activity. However, the genomic diversity of Serratia populations in TDT, as well as the ability of S. marcescens strains to express their antagonistic mechanisms in vivo, remain unknown. Therefore, the aim of this study was (i) to obtain S. marcescens isolates from R. prolixus TDT microbiota, (ii) to evaluate their in vitro trypanolytic activity, (iii) to acquire their draft genomic sequences, (iv) to suggest genetic determinants in their genomes with antagonist potential against T. cruzi, and (v) to detect T6SS transcripts in R. prolixus TDT.
Specimens of R. prolixus were maintained, under controlled temperature and humidity, in a colony at Laboratório de Bioquímica e Fisiologia de Insetos, Instituto Oswaldo Cruz. The triatomines were fed through an artificial apparatus with defibrinated rabbit blood provided by the Instituto de Ciência e Tecnologia em Biomodelos (ICTB) (Azambuja and Garcia, 1997). The rabbit blood was obtained according to the ethical principles in animal experimentation approved by the Comissão de Ética no Uso de Animais from Oswaldo Cruz Foundation (CEUA/FIOCRUZ) under the protocol number LW019/17 following the recommendations of Ministério da Ciência, Tecnologia e Inovação/Conselho Nacional de Controle de Experimenyação Animal (MCTI/CONCEA) available at http://pages.cnpem.br/ceua/wp-content/uploads/sites/56/2015/06/DBCA.pdf, which is approved by the Federation of European Laboratory Animal Science Associations (FELASA), the American Association for Animal Science (AAAS), the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC), and the International Council for Animal Science (ICLAS).
The T. cruzi Dm28c clone (Contreras et al., 1988) and T. cruzi Y strain (Silva and Nussenzweig, 1953), previously classified as TcI and TcII, respectively (Zingales et al., 2009), were supplied by Dr. Otacílio Moreira (Laboratório de Biologia Molecular de Doenças Endêmicas, Fiocruz, Brazil). Epimastigotes of T. cruzi were grown in vitro in brain heart infusion (BHI) media (Sigma-Aldrich) containing folic acid (30 mg/L), hemin (25 mg/L), and supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 28°C (Azambuja and Garcia, 1997). The epimastigotes parasites in the exponential growth phase were used for further in vivo and in vitro assays. The number of parasites was quantified using an optical microscope in a Neubauer chamber and diluted to concentrations of 2.5 × 106 epimastigotes/mL for in vitro assay and 1 × 107 epimastigotes/mL for triatomine infection (in vivo assay). The Mann–Whitney statistical test was used to verify whether there were any statistically significant differences in the parasite number from TDTs.
Fifth instar nymphs were randomly picked up and fed with defibrinated rabbit blood containing T. cruzi epimastigotes of the Y strain or Dm28c clone. The blood complement system was previously heat-inactivated by centrifugation at 1890 × g for 15 min at 4°C and plasma (supernatant) incubated for 30 min at 55°C. Subsequently, erythrocytes previously washed with phosphate buffered saline (PBS; 0.15 NaCl in 0.01 M sodium phosphate buffer, pH 7.2) were mixed with plasma treated as described above and T. cruzi were added at a final concentration of 1 × 107 epimastigotes/mL to the reconstituted blood. Uninfected individuals (control) were fed only on inactivated blood without parasites. Only fully engorged fifth instar R. prolixus nymphs were used for the experiments.
At 1, 2, 7, 15, 30, and 40 days after feeding (DAF), fifth instar nymphs of R. prolixus were washed to eliminate surface bacteria with 70% ethanol and several times with sterile distilled water. All dissections were performed under aseptic conditions. For parasite quantification in insect digestive tract, the abdomen cuticle was removed and the contents of the whole digestive tract were collected in sterile Eppendorf tubes, containing PBS at a proportion of five stomachs or intestines/500 μL PBS. To perform RNA extraction, three pools containing each one five (stomach) or posterior (intestine) midgut samples were collected and stored in sterile 1.5 mL microtubes at -70°C (Vieira et al., 2016).
In order to obtain single colonies, the content of stomach lumens was serially diluted, spread onto the surface of BHI agar, and incubated for 48 h at 28°C. Thirty colonies were randomly picked up and subcultured at 28°C for 48 h on BHI agar.
Genomic DNAs were isolated using a Wizard genomic DNA purification kit, according to the manufacturer’s instructions. For each bacterial isolate, the cells from 10 mL cultures grown in BHI medium at 28°C for 24 h were centrifuged at 10,000 × g for 10 min.
Amplification reactions with BOXA1R primer were performed as described by da Mota et al. (2002) in a mix containing 600 μM of each dNTP, 3.75 mM MgCl2, 50 mM KCl, 20 mM Tris–HCl (pH 8.4), 1 μM of the primer BOXA1R (5′-CTACGGCAAGGCGACGCTGACG-3′), about 50 ng of genomic DNA and 1.25 U of Taq DNA polymerase. The PCR conditions were comprised of an initial denaturation step for 7 min at 95°C followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 53°C for 1 min, and extension at 65°C for 8 min, with a final elongation step of 16 min at 65°C. The PCR products were analyzed by electrophoresis in 1.4% agarose gel in Tris–borate–EDTA buffer for 3 h 30 min, at 90 V and room temperature.
Bacterial isolates with different fingerprinting profiles were previously cultured in liquid BHI medium for 18 h at 30°C. Then, 10 microliters of each culture were aseptically sub cultured into 5 mL of sterile BHI and incubated at 30°C for 2 h (exponential phase) or 20 h (stationary phase) and 90 rpm. Since the antagonistic effect of S. marcescens over T. cruzi occurs through trypanolysis, aliquots collected from these bacterial cultures were used in a trypanolytic assay in vitro with T. cruzi Dm28c and Y epimastigotes, as previously described by Azambuja et al. (2004) and Castro et al. (2007b). Bacterial suspensions in the concentration of 1 × 108 CFU/mL were added to 80 μL of T. cruzi Dm28c clone and Y strains epimastigote suspensions at the final concentration of 2.5 × 106 parasites/mL in Eppendorf tubes, and incubated at 30°C for 120 min. The whole procedure was carried out under aseptic conditions and incubated for 2 h at 30°C. In each assay, the living parasites, predictable by their apparently un-affected morphology and at least some slight flagellar movements, were counted using a light microscope and a Neubauer hemocytometer chamber. The control of each assay was obtained by adding 10 μL of PBS in the medium instead of a bacterial suspension. The experiment was repeated twice with around ten incubations for each group. The trypanolytic activity was expressed as the ratio (%) of lysed parasites to the number of parasites in the control after 2 h of incubation. The one-way ANOVA statistical test was used to determine whether there were any statistically significant differences between the samples incubated with bacteria or without bacteria (control). Moreover, we tested the statistical significance of differences between averages through t test.
The trypanolytic strains, RPA1 and RPH1 had two different BOX fingerprinting profiles and were deposited in the Coleção de Enterobacterias – CENT at the Fundação Oswaldo Cruz (Fiocruz), Brazil. Whole-genome sequencing (WGS) of these strains were performed using an Illumina HiSeq 2500 sequencer from the high-throughput sequencing platform of Fiocruz. Reads were de novo assembled using the SPADES (version 3.1.1) assembler. Gene prediction and annotation were performed with the Prokaryotic Genome Automatic Annotation Pipeline (version 1.11). The Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank under the accession NCQJ00000000 and NCQI00000000. The versions described in this paper are versions NCQJ01000000 and NCQI01000000, respectively, for RPA1 and RPH1 strains.
The average nucleotide identity (ANIm) was calculated using the NUCleotide MUMmer software (version 3.1) for RPA1, RPH1, and others 377 genomic sequences of Serratia marcescens available at RefSeq NCBI https://www.ncbi.nlm.nih.gov/genome/genomes/1112, accessed on 05/29/2018.
The BLAST Ring Image Generator (BRIG) software was used to generate genomic maps that show multiple prokaryote genome comparisons among the RPA1, RPH1, and their closest genomes based on their ANIm. The closest genomic sequences for the RPA1 strain were sicaria-Ss1, ADJS-2D_White, SOLR4, and RSC-14, while for the RPH1 strain they were 19F, S2I7, WW4, and EGD-HP20, as shown in the spreadsheet called “ANIm Serratia strains” from the Supplementary Material.
Antimicrobial resistance genes were searched in draft genomic sequences of S. marcescens RPA1 and RPH1 using KmerResistance (Clausen et al., 2016) while the putative genomic islands related to antibiotic resistance, metabolism, pathogenicity, or symbiotic lifestyle were found using GIPSy (Soares et al., 2016). Genomic islands and operons were visualized on genome sequences with BRIG and ARTEMIS, respectively.
All protein sequences predicted from coding sequences (CDS) from draft genomes of S. marcescens RPA1 and RPH1 strains were compared to nr database (non-redundant protein NCBI database, release 72) and analyzed by MEGAN (version 5.11.3) in order to identify the proteins related to bacterial secretion systems.
The sequences of ClpV (a gene involved in T6SS recycling, Douzi et al., 2016), Hcp1 (a key hexameric protein of the tubular T6SS secretion system, Brunet et al., 2014), and DotU1 (encoding homologs of T4SS stabilizing proteins, Bröms et al., 2012) that encode key proteins of T6SS were used to design primers, as shown in the spreadsheet called “SecretionSystems” from the Supplementary Material.
Triatomines at 1, 7, 15, and 40 DAF (T. cruzi infected and non-infected) were dissected to prepare pools of five stomachs and intestines, as previously described (Vieira et al., 2016). Total RNA was obtained using the NucleoSpin®RNA II Kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions and quantified using a NanoDrop 2000 Spectrophotometer (Thermo Scientific, Waltham, MA, United States). Synthesis of cDNA was performed with a First-Strand cDNA Synthesis Kit (GE Healthcare, Buckinghamshire, United Kingdom) following the manufacturer’s protocol using 2.5 μg of total RNA and the pd(N)6 primer. cDNA was quantified by fluorescence using a Qubit Fluorimeter (Life Technologies) with the ssDNA assay kit. Here, we analyzed the gene expression of three genes for T6SS of S. marcescens (ClpV, Hcp, and DotU) and the constitutive rpoB gene, which encodes the β subunit of bacterial RNA polymerase. The following primers were designed based on the sequences of RPA1 and RPH1 genomes for the amplification of the gene fragments: ClpV_F1692-1718(5′-GGT GkT GkC sGA yTG GAC CGG CAT CCC-3′) and ClpV_R2158-2183(5′-CCG TCy TCC ATC mmG CCC TTG TCG AA-3′); DotU_F264-290(5′-CGG GCT GGT GAC CTT CCA TAA CGA-3′) and DotU_R792-815(5′-CAC GCT TTG CTG CGC TTC GTC GCG-3′); Hcp_F26-51(5′-TGT TCC TGA AAG TTG AAG GTG CCA GC-3′); Hcp_R489-515(5′-CGG TTT TCC TTG ATG TTC CAA CCT GCG-3′). Previously designed primers by Dahllöf et al. (2000) were used for rpoB constitutive gene, rpoB_1698f (5′-AAC ATC GGT TTG ATC AAC-3′) and rpoB_2041r (5′-CGT TGC ATG TTG GTA CCC AT-3′). Each singleplex reaction was run for each pool of triatomines (n = 3) and contained 10 ng cDNA, primer pairs (0.2 μM), MgCl2 (2.5 mM), dNTPs (200 μM each), buffer and GoTaq®DNA polymerase (0.6 U) from Promega at a final volume of 25 μl. The cDNA was amplified at 95°C for 5 min followed by 38 cycles of 95°C for 40 s, 52°C for 1 min and 72°C for 40 s, with a final extension for 7 min at 72°C. Negative controls of PCR reactions were carried out without cDNA template in order to assess dimer formation between primers or reaction contaminations by foreign DNA. The agarose gel electrophoresis of PCR products was performed with 1.4% agarose in Tris–borate–EDTA buffer at 100 V for 2 h at room temperature.
We observed distinct types of morphologies as well as colors (white and red) among the colonies isolated from the R. prolixus gut microbiota and sub cultured on BHI agar plate (data not shown). Molecular fingerprinting of their genomic DNA revealed that these isolates belong to at least two distinct genomic profiles (Figure 1). BOXA1R profiles similar to those of RPA1 and RPH1, corresponded to about 73 and 26% of isolates, respectively. RPA1 and RPH1 isolates also showed differences in the morphologies of their bacterial colonies on BHI agar. While RPA1 presented white colonies, RPH1 showed strong red colonies, suggesting the synthesis of prodigiosin by this strain.
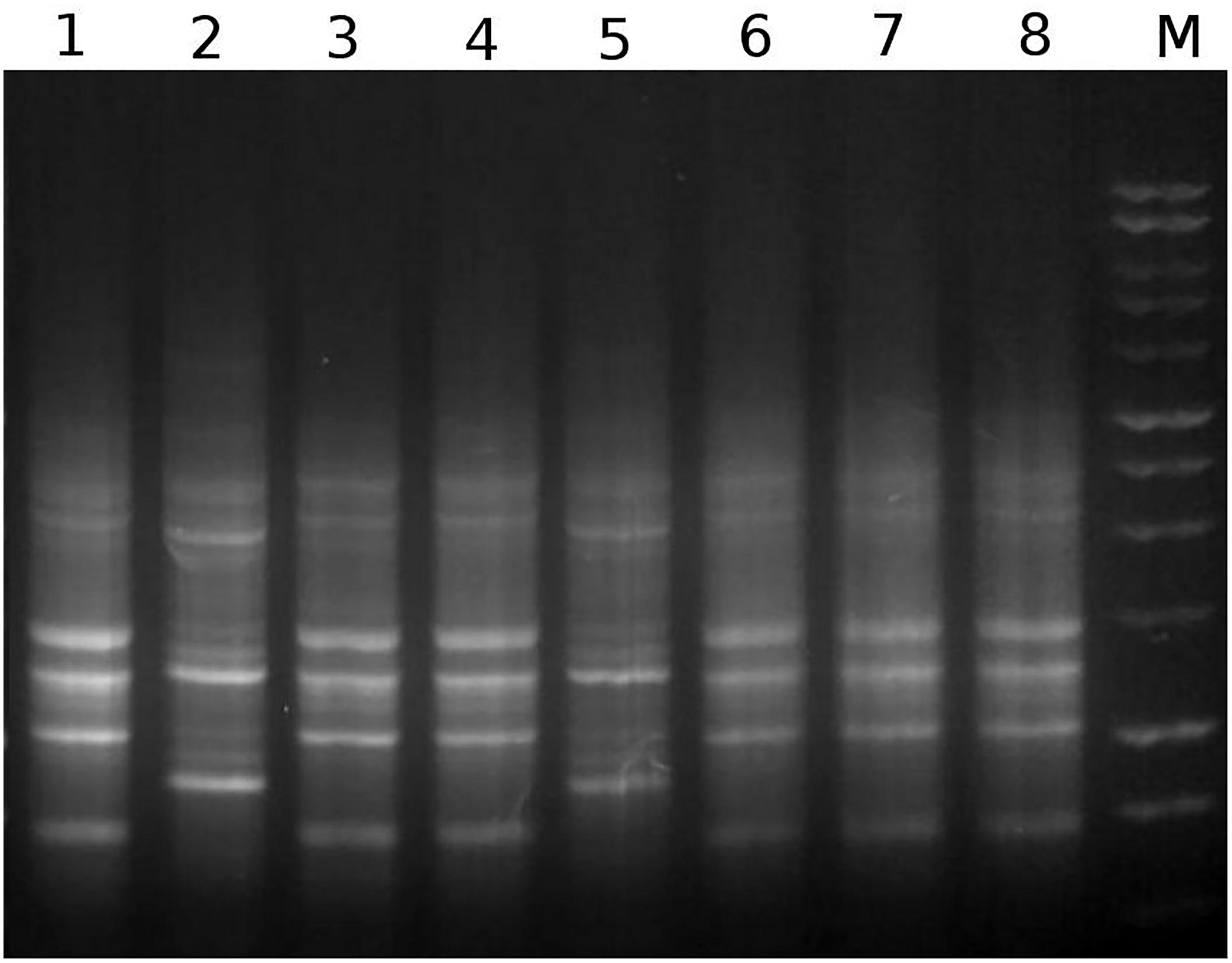
Figure 1. A representative example of genomic fingerprints generated by separation of BOXA1R amplicons through agarose gel electrophoresis. The numbers 1–8 represent BOXA1R profiles obtained with the isolates RPA1, RPH1, RPD1, RPE1, RPG11, RPC11, RPD4, and RPB1, respectively. M represents the 1 kb DNA ladder from Promega.
The trypanolytic activity of both RPA1 and RPH1 S. marcescens strains was stronger in their stationary phases of growth than in their exponential ones. In exponential phase (Figure 2A, blue), the percentage of T. cruzi Dm28c lysis was significantly higher under incubation with RPA1 (50%) than with RPH1 (7%), on average (Figure 2A; p = 0.0003; F = 1.358). In their exponential phase, RPA1 and RPH1 isolates showed similar lysis percentages for T. cruzi Y, which was about 30–40% (Figure 2A, green). In stationary phase, the percentage of epimastigotes of T. cruzi Y that were lysed by both RPH1 and RPA1 was significantly higher with an average of 84 and 88%, respectively (Figure 2B, green). The percentage of epimastigotes of T. cruzi Dm28c that were lysed was also significantly higher, on average, when incubated with RPH1 (67%) and RPA1 (73%) in stationary phase (Figure 2B, blue). T. cruzi Y was significantly more sensitive to RPH1 in the stationary phase than T. cruzi Dm28c (Figure 2B; p = 0.0035; F = 2.377).
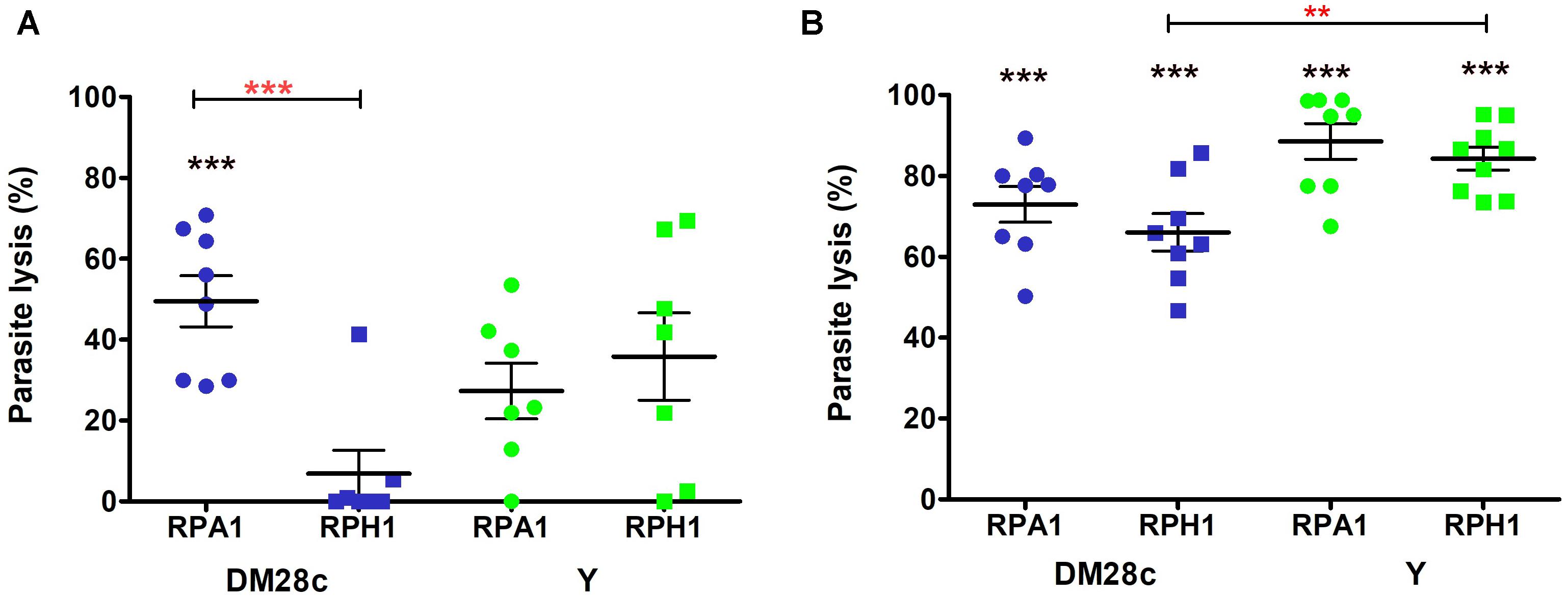
Figure 2. Trypanolytic activity during the exponential (A) and stationary (B) phases of S. marcescens cultures (RPA1 and RPH1 are represented by circles and squares, respectively) with respect to epimastigotes of T. cruzi Dm28c (blue) and Y strains (green) after incubation for 2 h at 30°C. The vertical axis represents the percentage of lysed parasites/mL in comparison to the controls without the bacteria (0%). The red asterisks above the bars indicate statistical significances at p = 0.0035 (∗∗) and F = 2.377 or p = 0.0003 (∗∗∗) and F = 1.358 differences between averages using t test. The black asterisks (∗∗∗) indicate statistical significances at p < 0.001 for average differences between the control without bacteria and the group in one-way ANOVA (see details in Supplementary Material).
The WGS reads were de novo assembled using the SPADES (version 3.1.1) assembler, which yielded a dataset composed of 60 contigs, totaling ∼5.3 Mb, with an average G+C content of 59.32% for strain RPA1 and 35 contigs, totaling ∼5.1 Mb, with an average G+C content of 59.63% for strain RPH1.
In order to globally evaluate the genomic status of RPA1 and RPH1 genomes according to all genome sequences of S. marcescens available in the NCBI repository, we calculated the ANIm for 379 genomes (Supplementary Material, ANI Serratia strains). The ANIm analysis indicated up to six clades of S. marcescens (red squares, ANIm ≥95%) along the heatmap diagonal (Figure 3). The RPA1 and RPH1 strains (isolated from R. prolixus gut) are found in distinct red clades, suggesting that both are S. marcescens, but from distinct evolutionary origins and refuting the clonal hypothesis for these strains. The RPA1 genome was closer to sicaria-Ss1 (Apis mellifera), ADJS-2D_White (Scapteriscus borellii), SOLR4 (Solanacea), and RSC-14 (Solanum nigrum), while RPH1 strain was closer to 19F (Atelopus zeteki), S2I7 (oil contaminated soil), WW4 (paper machine), and EGD-HP20 (wastwater), according to their ANIm percentage identity. Two strains (ano1 and ano2) previously isolated from the gut lumen of adult female Anopheles stephensi mosquitoes were found in another distinct clades as shown by arrows in Figure 3.
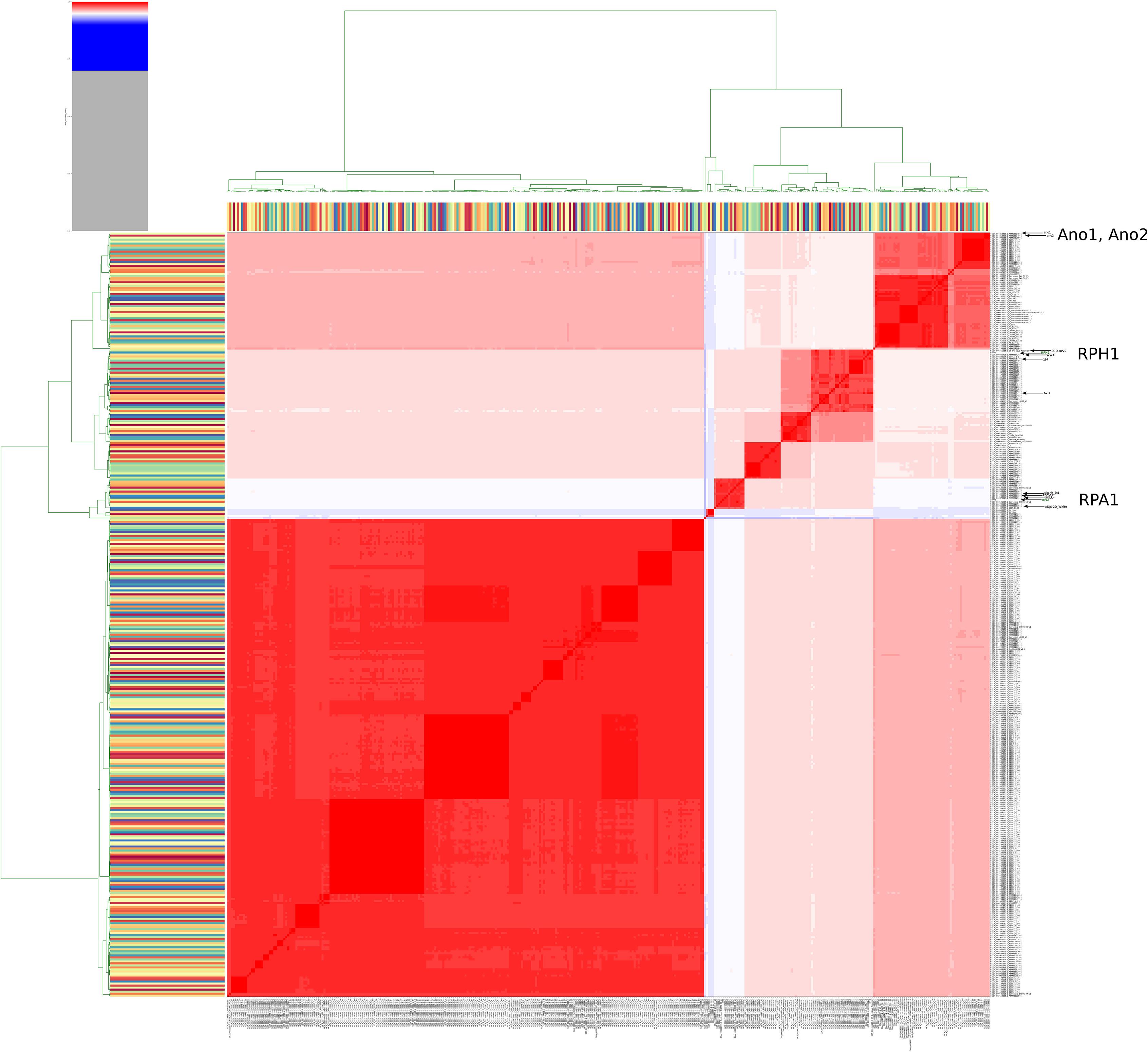
Figure 3. Heatmap of ANIm percentage identity for 379 genomes of S. marcescens obtained from RefSeq. White and red cells in the heatmap corresponded to ANIm sequence identity higher than 95% and therefore to the same species (Richter and Rosselló-Móra, 2009), while blue cells correspond to ANIm lower than 95%. The hierarchical clustering follows a two dimension dendrograms organized according to simple linkage of ANIm percentage identities. The analysis indicates up to six strains clades along the heatmap diagonal. The RPA1 and RPH1 strains (isolated from R. prolixus gut) are found in different clades, suggesting distinct evolutionary origins and refuting the hypothesis of clonality for these strains. The arrows show the strains that are the closest to RPA1 (sicaria-Ss1, ADJS-2D_White, SOLR4, and RSC-14), RPH1 (19F, S2I7, WW4, and EGD-HP20), and ano1 according to ANIm percentage identity. Ano1 and Ano2 were previously isolated from the gut lumen of adult females of A. stephensi mosquitoes (see details and high resolution image in Supplementary Material).
Genomic maps of RPA1 and RPH1 strains showed that although they have a high identity with other RefSeq strains, each strain has unique gene islands (Figure 4). Genomic mapping also revealed that strains associated to plants and animals were closer to RPA1 than those associated to free living ones. Some gene islands shared by strains RPA1 and RPH1, suggests that these strains with different evolutionary origins could have recently acquired these gene islands, possibly after niche adaptation. RPA1 has a plasmid highly similar to that of RPH1 (indicated by the brown arrows of Figure 4), which has genes also found in animal-associated strains (ADJS-2D_White, sicaria-Ss1, 19F, ano1, and ano2) but not in plant-associated strains (SOLR4 and RSC-14).
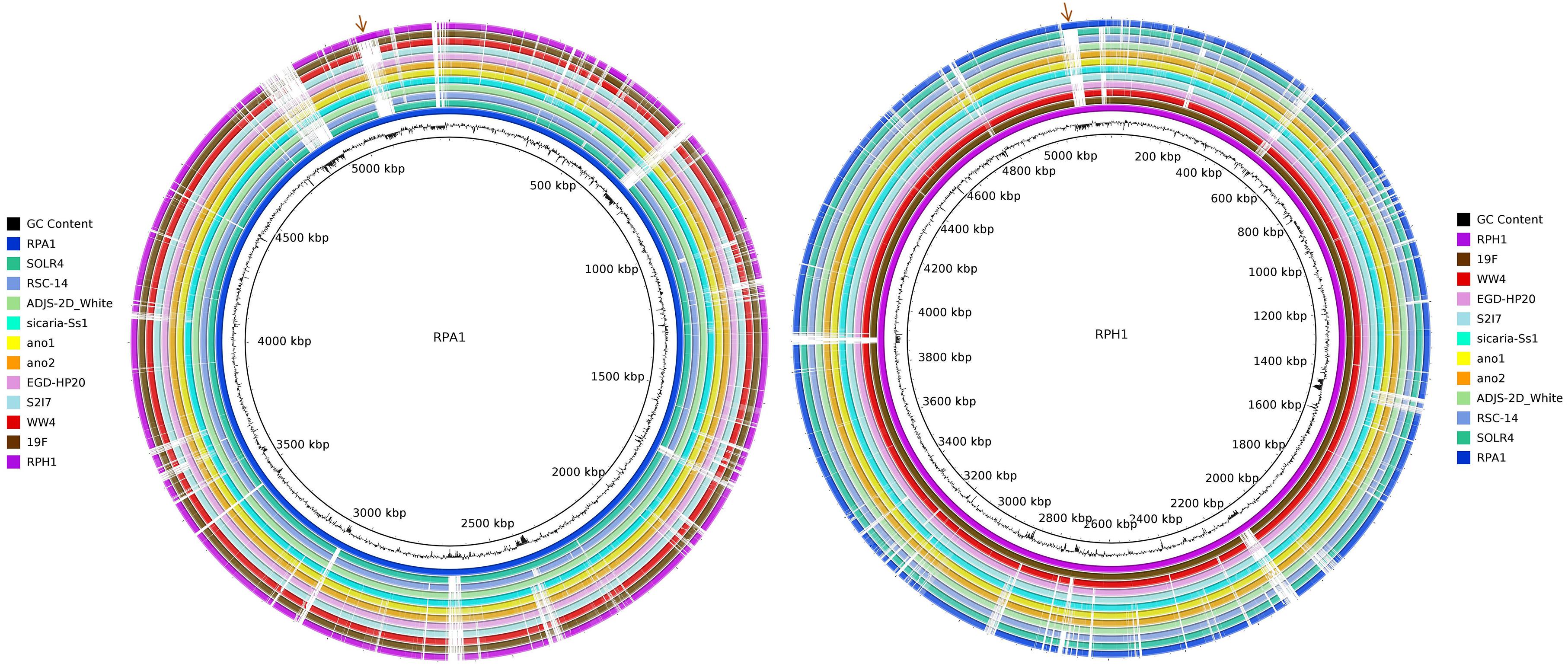
Figure 4. Circular representation of two S. marcescens genomes. The CG content bias was plotted as the second internal circle for RPA1 and RPH1 genomes. The brown arrows represent a plasmid shared by RPA1 and RPH1 that includes genes also found in other animal-associated strains. The stronger colors represent the higher blastn identity for the draft genome sequences of RPA1 and RPH1 strains with their closest complete genomes in RefSeq-NCBI based on ANIm, i.e., sicaria-Ss1 (A. mellifera), ADJS-2D_White (S. borellii), SOLR4 (Solanacea), RSC-14 (Solanum nigrum) for RPA1, 19F (skin of A. zeteki), S2I7 (soil), WW4 (paper mill process waters), and EGD-HP20 (wastwater) for RPH1, as well as the ano1 and ano2 strains previously isolated from the gut lumen of adult female of A. stephensi mosquitoes.
The identification of CDSs revealed the presence of at least 4,969 putative CDSs, regions of DNA that can potentially be translated into proteins in RPA1 (locus tag prefix B7L32_) while 4,816 putative CDSs were found in RPH1 genome (locus tag prefix B7L62_). Plasmidial CDSs were found in both genomes, RPA1 (B7L32_24230 to B7L32_24595, B7L32_25505 to B7L32_25515 and B7L32_25585) and RPH1 (B7L62_23970 to B7L62_24190, B7L62_24560 to B7L62_24710 and B7L62_24795). Plasmid stability is probably maintained due to some RelE toxin genes found in the chromosome of each strain (B7L32_21785, B7L32_16920, and B7L32_11945 for RPA1; B7L62_19950, B7L62_13775, and B7L62_06530 for RPH1), while the gene for RelB antitoxin (B7L32_24540 and B7L62_24620) was found only in their plasmids. In addition, a copy of RelE toxin gene (B7L32_25460) was also found in the RPA1 plasmid. Moreover, genes for other complete system toxin–antitoxin (PemK toxins: B7L32_24575 and B7L62_24585 as well as their PemL antitoxins: B7L32_24580 and B7L62_24580) were observed in their plasmids. Among plasmidial CDSs, we also found conjugation genes, including tra genes.
Most of GIs found in the RPA1 and RPH1 genomes corresponded to regions with a GC bias (Figure 5), suggesting that they could have been acquired on a recent genomic timescale. These GIs included genes related to biologic functions such as antibiotic resistance, metabolism, pathogenicity, and symbiosis.
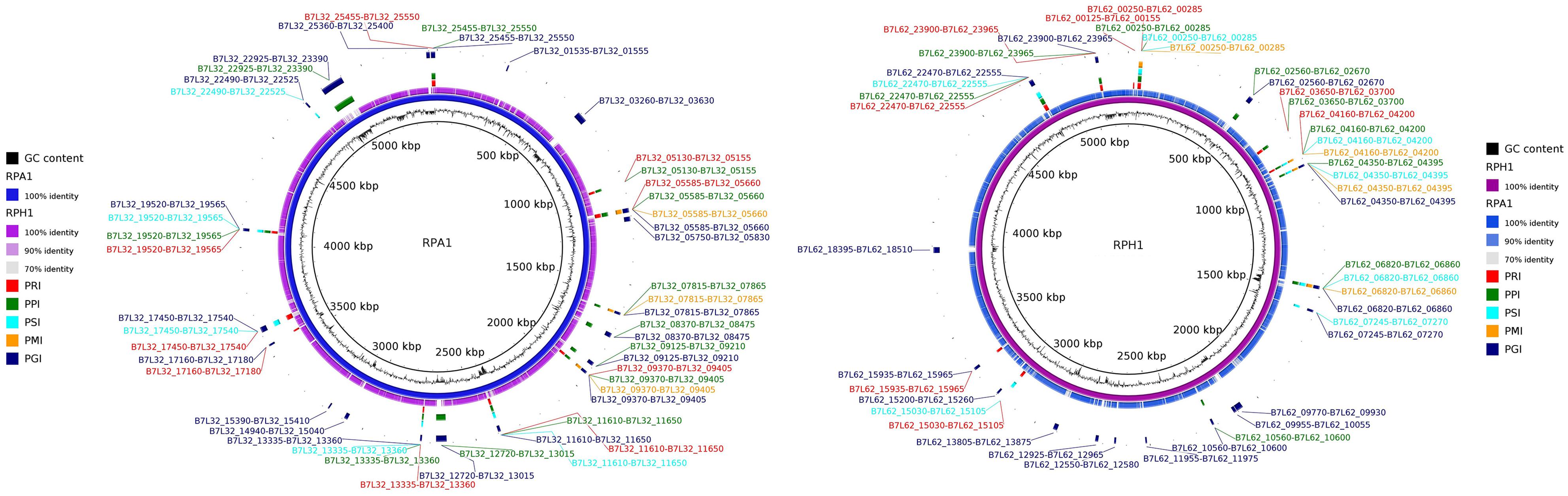
Figure 5. Genomic Islands on RPA1 and RPH1 S. marcescens draft genomes and their corresponding genes. The putative phenotypes were predicted by GIPSy software, [Putative Resistance Island (PRI, red), Putative Pathogenic Islands (PPI, green), Putative Symbiotic Islands (PSI, aqua), Putative Metabolic Island (PMI, orange), Putative Genomic Island (PGI, navy)]. The CG content bias was plotted in internal circle of each genome (black).
Among the gene islands of the RPA1 genome were included genes encoding for (i) filamentous hemagglutinin, (ii) hemolysin transporter protein (ShlB) precursor, (iii) phospholipase D, (iv) teichoic acids translocation permease export system (TagGH), (v) teichuronic acid biosynthesis glycosyltransferase (TuaH), (vi) modular non-ribosomal peptide synthase related to linear gramicidin synthase, (vii) chitinases, (viii) pullulanase secretion protein (PulS), (ix) type 4 prepilin-like proteins leader, (x) type II secretion system MLKJIHGFED, (xi) invasion protein regulator, (xii) firmbria A, (xiii) pilus, (xiv) exotoxin, (xv) endolysin, and (xvi) non-ribosomal peptide synthase, among others (see RPA1genesInGeneIslands and RPH1genesInGeneIslands spreadsheets of the Supplementary Material).
In silico KmerResistance and Resfinder methods for resistance genes prediction revealed the presence of different antibiotic classes: aminoglycosides [aadA1, aac(6′)-Ib, and aac(6′)-Ic], tetracyclines [tet(41)], and beta-lactams (blaOXA-9, blaTEM-1A, and blaSST-1) in the RPA1 genome sequence. On the other hand, blaOXA-9 and aadA1 genes were not found in the RPH1 genome (Supplementary Material). However, the genes aadA1, aac(6′)-Ib, blaTEM-1A, and blaOXA-9 were found in plasmids (see the ResistanceGenes spreadsheet of the Supplementary Material).
Ninety genes from the RPA1 genome were absent in RPH1, when annotated by reference to KEGG. We found that they included genes (eight) belonging to urease operon (Figure 6) and fimbriae (see the SharedOrExclusiveKOrthologous spreadsheet of the Supplementary Material). These recent gene acquisitions should be further studied once these genes could bring advantages to Serratia considering the specific features of the adaptation niche of hematophagous insects such as R. prolixus. On the other hand, RPH1 exhibited 104 genes, including some belonging to multidrug efflux systems, absent in RPA1.
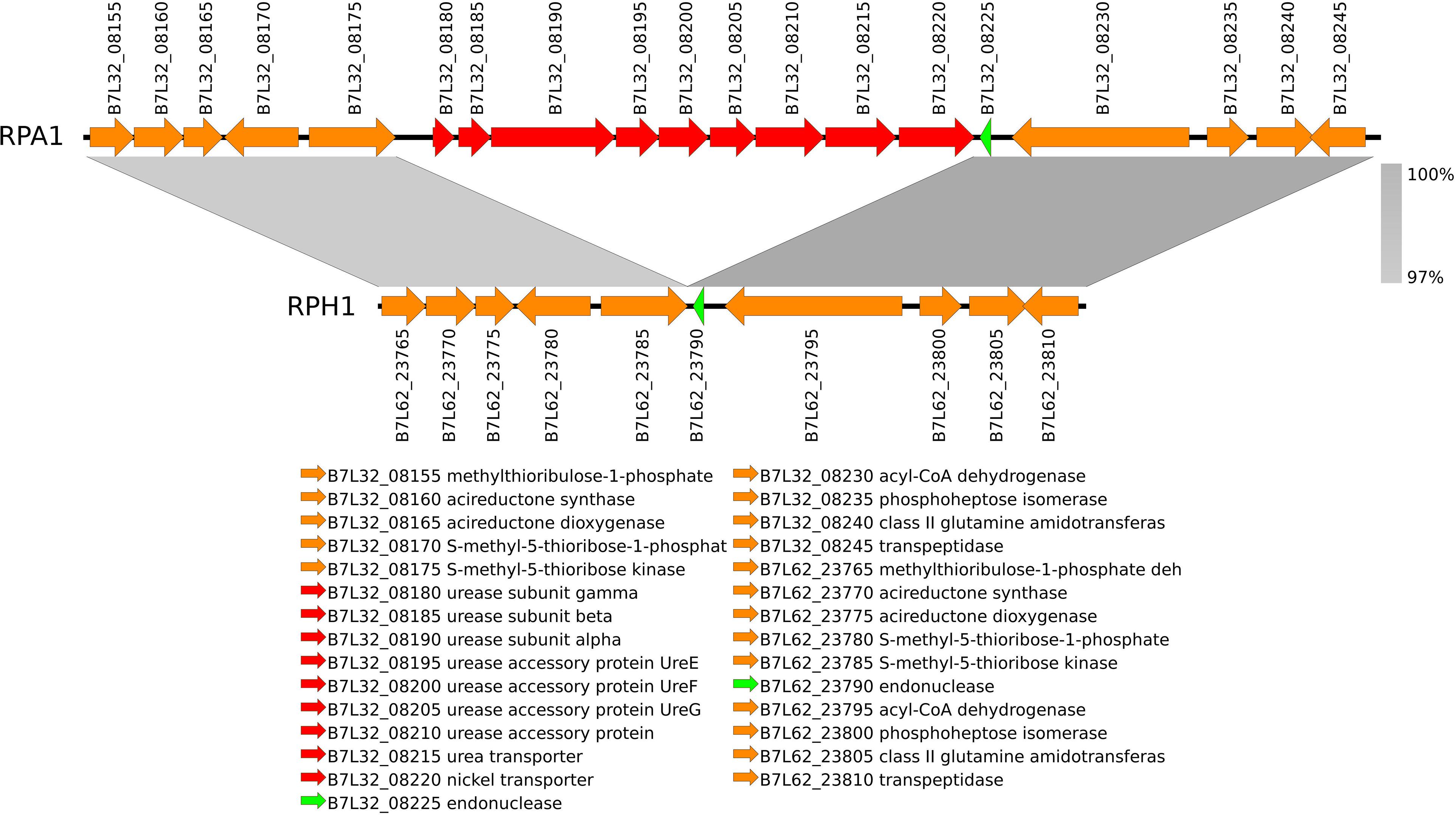
Figure 6. The nickel-dependent urease (EC 3.5.1.5) and accessory genes (red) are included in a gene island found in RPA1 genome, but not in RPH1 strain. This gene island is located between two conserved syntenic flanking regions (gray) characterized by nucleotide similarities higher than 97%.
Another difference between RPA1 and RPH1 is the colicin V/pyocin bacteriocin operon, which is present only in RPH1. In RPH1, the colicin/pyocin operon includes cda1, which is a gene encoding colicin-D absent from the WW4 strain (red arrow in Figure 7). Colicin V (ColV) is an antibiotic peptide secreted by some members of the Enterobacteriaceae that enable them to kill closely related bacteria, thereby reducing competition for essential nutrients.
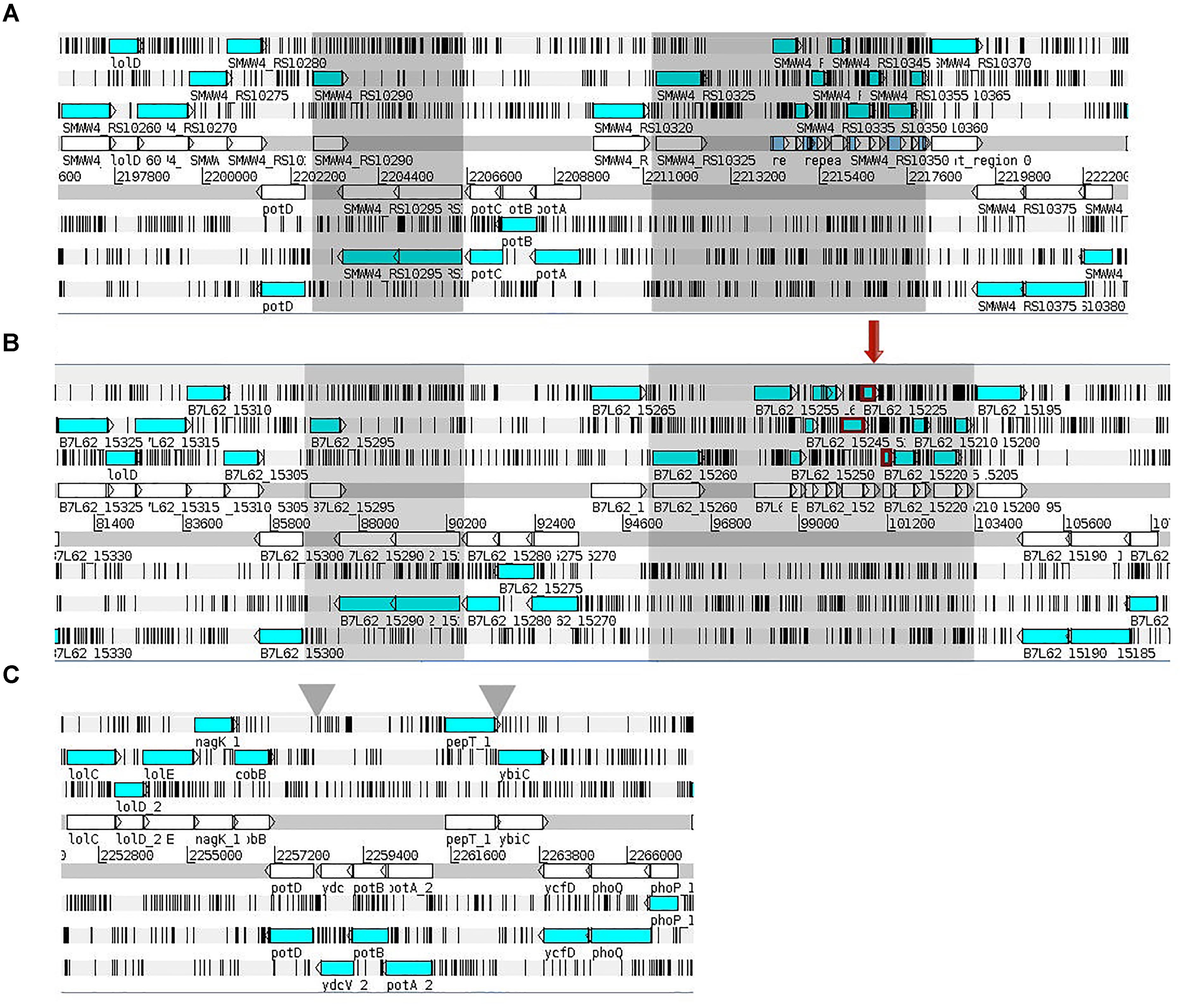
Figure 7. The two genomic regions of S. marcescens highlighted by gray rectangles in the WW4 (A) and RPH1 (B) strains are absent in the RPA1 strain (C). They are localized inside the pot operon (spermidine/putrescine transport system) and between the pepT and ybiC genes. These regions include genes coding for the PTS system maltose-specific EIICB component and the colicin/pyocin bacteriocin operon. The colicin/pyocin bacteriocin operon in RPH1 (B) includes a whole pyocin (B7L62_15230), which is partial in WW4; the colicin immunity protein (B7L62_15225) present in RPH1 is absent in WW4 (red arrow). The genes of the two-component regulatory system known as PhoP/PhoQ were present in all strains (A–C).
Among the genes shared by RPA1 and RPH1 are those of chitinases, esterases, phospholipid/cholesterol/gamma-HCH transport system, exonucleases, heme acquisition and exporter proteins, iron uptake, transport and chelatases, bicyclomycin/chloramphenicol resistance, microcin C resistance, catalase, peroxidase, chloroperoxidase, nitronate monoxygenase, superoxide dismutases, enterobactin synthetase, pyochelin synthetase, filamentous hemagglutinin, flagellar, secretion system proteins and serralysin, bacillolysin, aquaporin Z, hemolysin, hemolysin III, and virulence factors (see the SharedOrExclusiveKOrthologous spreadsheet of the Supplementary Material).
Some bacterial secretion systems were observed in both RPA1 and RPH1 genomes, including Sec-SRP, twin-arginine targeting (Tat), T6SS, and some other genes, such as TolC (an outer membrane protein required for several efflux systems), ShlA (a hemolysin precursor), and ShlB (a hemolysin transporter protein) (Figure 8), while type II system (T2SS) was observed only in RPA1. T6SS is widely used throughout Gram-negative bacteria to inject effector proteins and some toxins in a one-step mechanism directly into the cytosol of the eukaryotic cells being targeted since Hcp proteins can cross the plasmatic membrane of eukaryotic cells (Figure 8) and possibly that of T. cruzi.
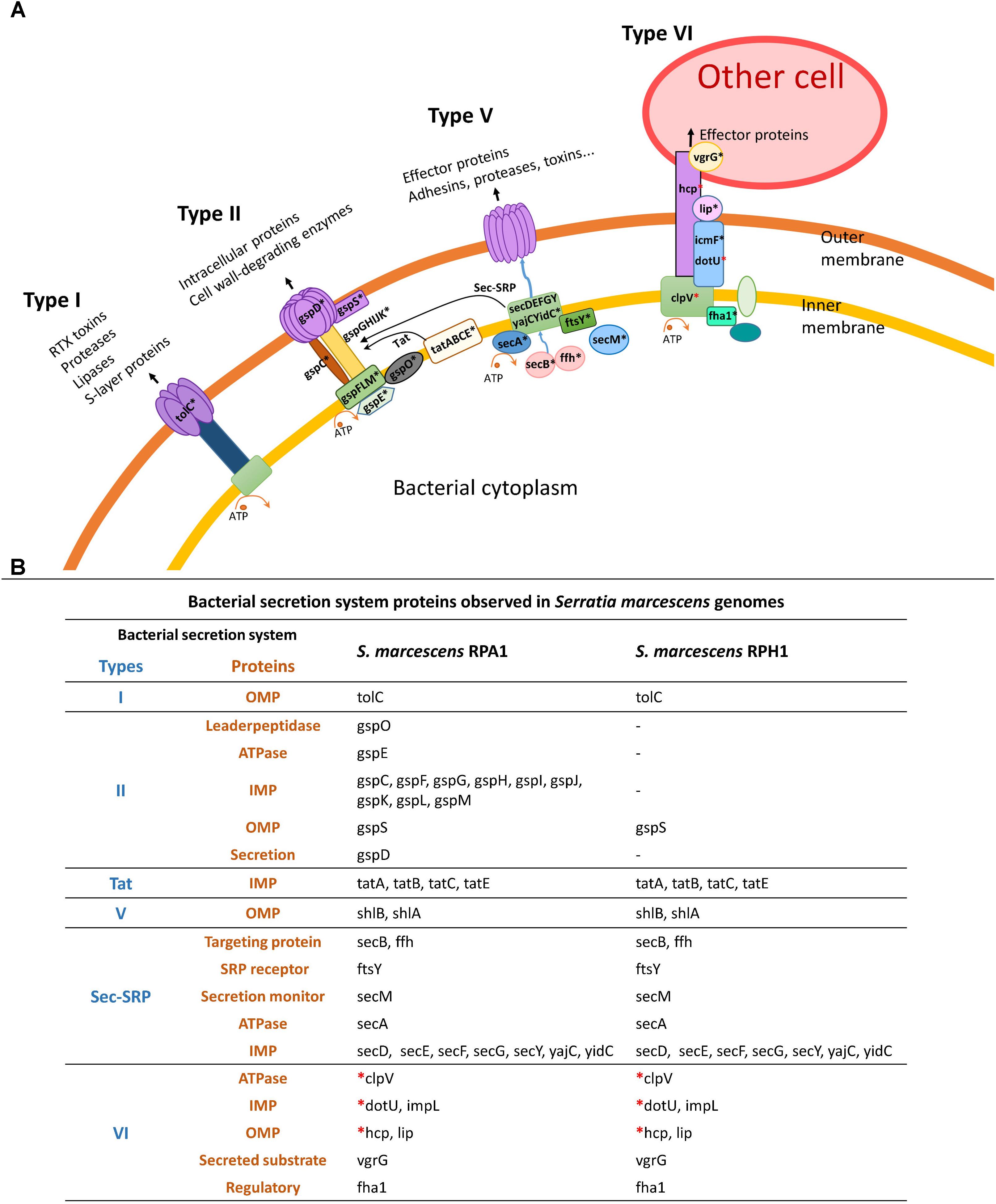
Figure 8. Bacterial secretion systems and their proteins observed in S. marcescens RPA1 and RPH genomes isolated from R. prolixus gut. (A) Schematic illustration of bacterial secretion systems type I, type II, type Vb, and type VI. (B) Proteins found in the bacterial secretion systems of S. marcescens RPA1 and RPH1 genomes. The red asterisks indicate the genes used for primer design for gene expression analysis in vivo. OMP, outer membrane protein; IMP, inner membrane protein; Tat, twin-arginine targeting.
In other to confirm if T6SS genes of S. marcescens could be transcripted in vivo in the digestive tract of R. prolixus infected with T. cruzi, the tissues were analyzed by RT-PCR from 1 to 40 DAF. Transcripts encoding for T6SSs proteins were detected in triatomines infected or not with T. cruzi DM28c clone or Y strains. The in vivo gene expression of T6SS was observed by RT-PCR in R. prolixus gut microbiota of uninfected stomachs Figures 9A–C, intestines Figure 9D, as well as in stomachs infected with T. cruzi DM28c clone Figures 9E–G and Y strain Figures 9H,I. Apparently, samples from triatomines infected with T. cruzi showed an earlier and stronger transcription of T6SSs when compared to those from the uninfected group, especially in the case of DM28c clone (7 and 15 DAF, Figures 9F,G, respectively). On the other hand, mRNA for the T6SSs proteins was still detected in stomachs at 15 DAF and intestines at 40 DAF of uninfected triatomines, as showed in Figures 9C,D, respectively.
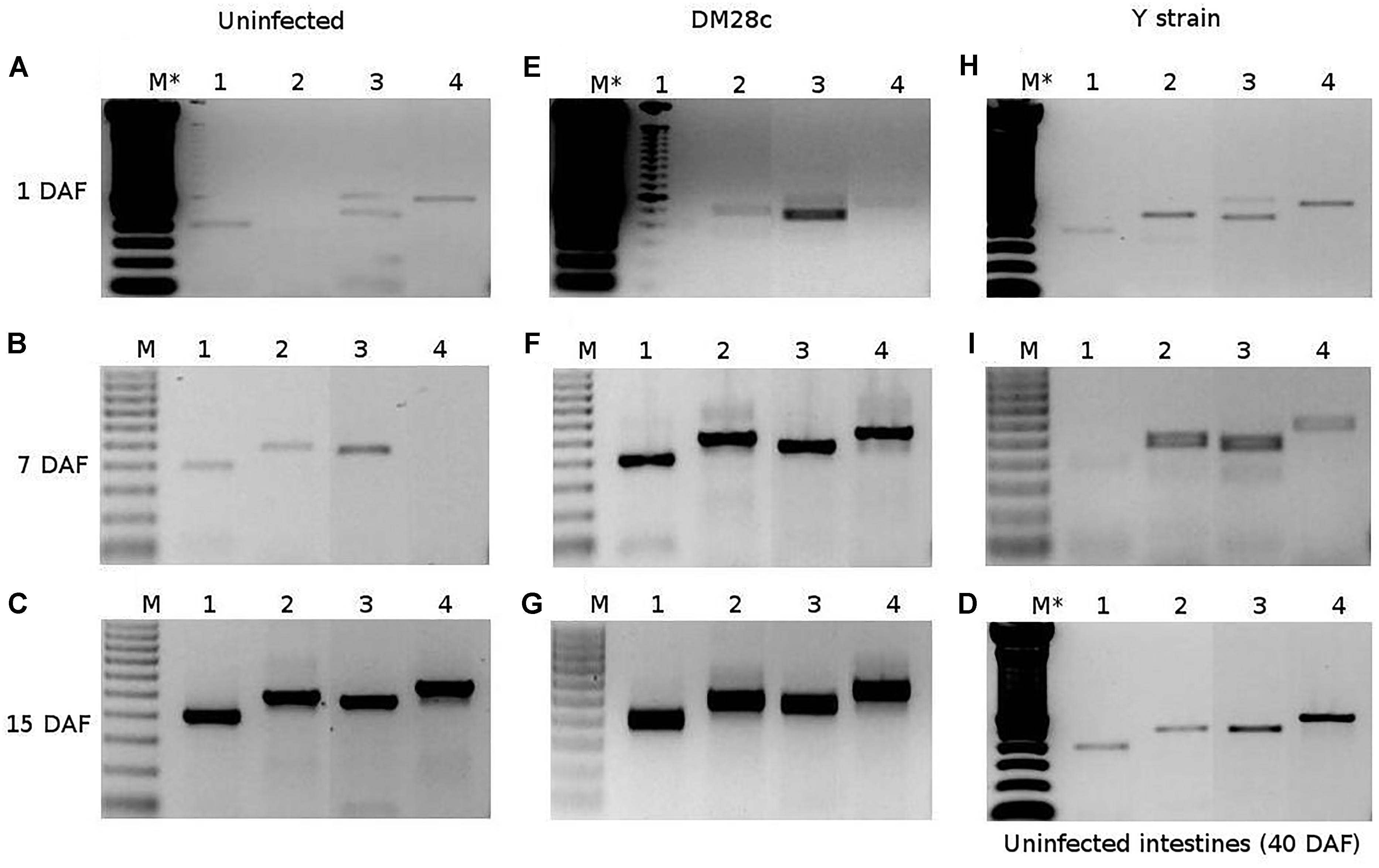
Figure 9. RT-PCR assessment of in vivo gene expression of R. prolixus gut microbiota in uninfected stomachs (A–C), intestines (D), as well as in stomachs infected with T. cruzi DM28c clone (E–G) or Y strain (H,I). The samples were collected at 1 (A,E,H), 7 (B,F,I), 15 (C,G), and 40 (D) days after feeding. The numbers represent amplicons for a constitutive gene rpoB (1), and genes for T6SSs proteins (ClpV, Hcp1, and DotU1) of S. marcescens (2–4, respectively). The letter M represents the 0.5 μg of Thermo Scientific GeneRuler 100 bp DNA ladder, and the symbol (∗) indicates the double DNA ladder amount.
When counting the number of T. cruzi DM28c parasites in the R. prolixus TDT, we found it significantly higher compared to that of the Y strain, especially at 7 and 30 DAF (Figure 10, p = 0.0079; p = 0.0019, respectively).
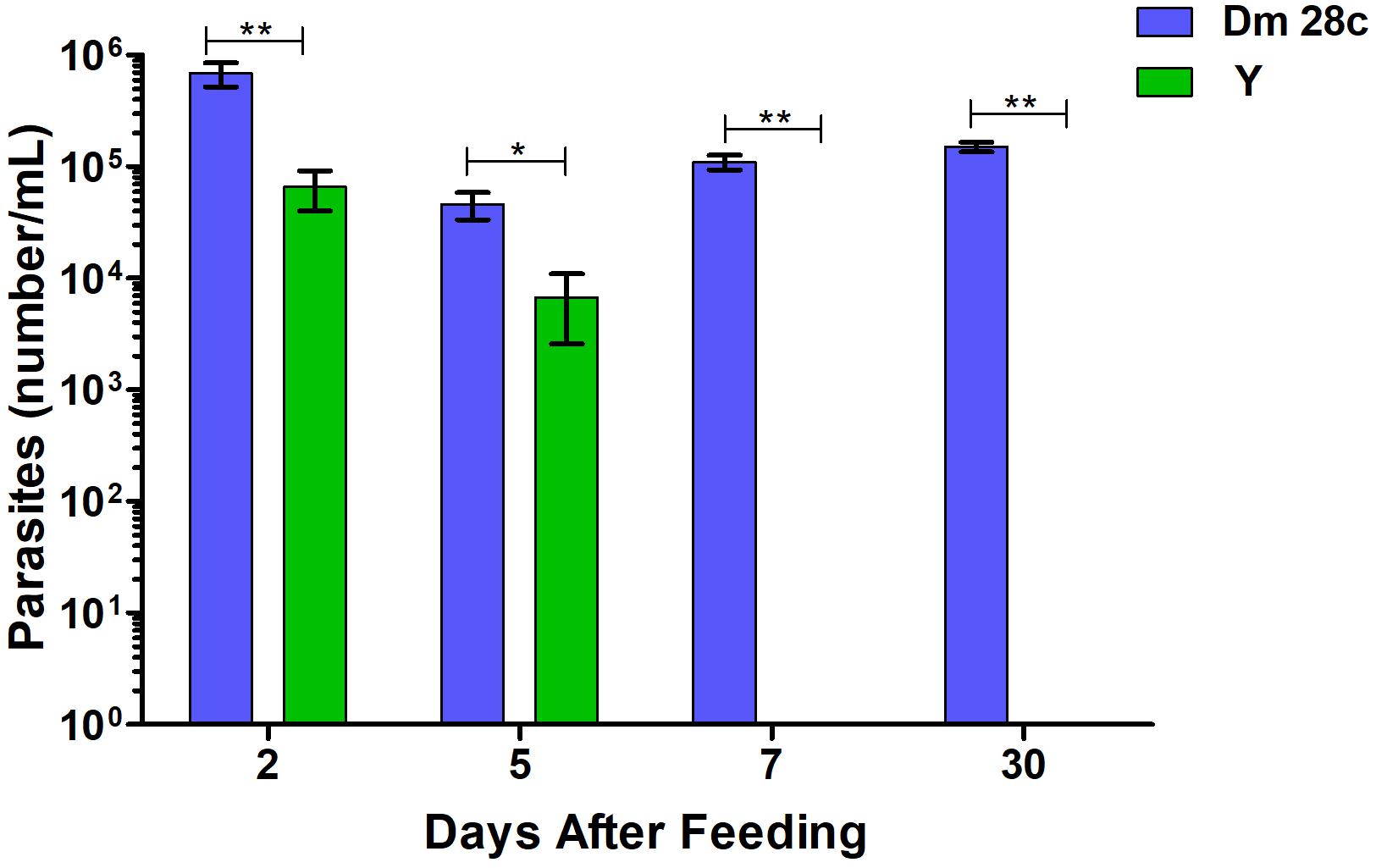
Figure 10. The number of parasites per mL quantified by optical microscope in R. prolixus gut in 2, 5, 7, and 30 days after feeding with blood infected with Trypanosoma cruzi Dm28c clone (blue) and Y strain (green). Parasites could not be observed in the gut 7 or 30 days after feeding with Y strain. Bars represent the average of parasites in insect digestive tract. Averages were compared using Mann–Whitney test (∗p < 0.05, ∗∗p < 0.01).
In this study, we reported on the genomes of two S. marcescens strains isolated from the gut microbiota of R. prolixus and made available publicly through DDBJ/ENA/GenBank. We analyzed the genomic features of these strains and demonstrated theirs in vitro trypanolytic activity against T. cruzi as well as the expression of the T6SS in vivo.
The study of the intestinal microbiota of insect vectors has focused on the influence of some resident bacteria to interrupt the parasite development within the invertebrate host gut (Azambuja et al., 2004; Cirimotich et al., 2011; Weiss and Aksoy, 2011; Geiger et al., 2013). In Diptera, the bacteria S. marcescens isolated from the digestive tract of different species has been proposed as a strategy to prevent Plasmodium and T. brucei transmission to vertebrates (Rahul et al., 2015; Wang et al., 2017). S. marcescens isolated from R. prolixus gut possess an in vitro hemolytic activity against T. cruzi (Azambuja et al., 2004). Here, we found that both S. marcescens strains (RPA1 and RPH1) possess genes related to the bacterial hemolytic activity, which could benefit the blood digestion by triatomines. Actually, the trypanolytic activity of S. marcescens with regard to T. cruzi was previously demonstrated in vitro (Azambuja et al., 2004; Castro et al., 2007a,b). Moreover, feeding experiments in which R. prolixus was infected with blood containing antibiotics resulted in the reduction of the bacterial population together with the increase of parasites number in the midgut (Castro et al., 2012; Vieira et al., 2016), which indicated the antagonistic effect of S. marcescens on T. cruzi in vivo (Vieira et al., 2016).
The growth dynamics of bacterial microbiota from R. prolixus in liquid medium reaches its maximum around the eighth DAF (Castro et al., 2012). Later, it was shown that the bacterial proliferation activates a basal immune response in R. prolixus midgut corresponding to the increase of genes expression of antimicrobial peptides (Vieira et al., 2014), which control the over-proliferation of microorganisms. The trypanolytic activity of S. marcescens was previously reported (Azambuja et al., 2004; Genes et al., 2011). Moreover, it was demonstrated that the S. marcescens trypanolytic activity is dependent on its adhesion to T. cruzi through mannose sensitive fimbriae (Castro et al., 2007a,b; Moraes et al., 2009). Here, we characterized the trypanolytic activity of S. marcescens in vitro for the two strains RPA1 and RPH1 with regard to the Dm 28c and Y strains of T. cruzi. The prodigiosin (red pigment) production in vitro does not seems to be a key determinant of S. marcescens antagonism against T. cruzi since the lysis induced by RPA1 (non-pigmented strain) was larger than the one induced by RPH1 (pigmented strain).
Concerning adhesive molecules, we found several genes associated to fimbriae, pili, and hemagglutinins in the genomes of both S. marcescens strains, which are supposed to be related to their trypanolytic activity. However, only S. marcescens RPA1 possessed fimbriae type I (fimB) and type IV (fimA), which are known to be associated to adherence, invasion, and virulence (Kennan et al., 2001; Pusz et al., 2014). The presence of these genes in RPA1 strain could be related to its stronger trypanolytic activity when compared to RPH1 strain. Moreover, it was found that both S. marcescens strains have a PTS-mannose specific related to the uptake of this carbohydrate from the extracellular environment (Postma et al., 1993; Uchiyama et al., 2003). As described in previous publications, the trypanosomatids possess several mannose residues on their membrane surfaces (Bonay et al., 2001; Barboza et al., 2005; Moraes et al., 2008, 2009). In addition, the reported mannose sensitive fimbriae of S. marcescens mediate adhesion to eukaryotic cells (Reid and Sobel, 1987). Having taken these pieces of evidence together, we propose that the S. marcescens fimbriae play a role in the protozoan recognition by S. marcescens, which is supposed to activate a mechanism of incompatibility through the induction of T6SS or other secretion systems. However, further studies are necessary in order to evaluate this cause–effect association.
Type VI secretion system represents an efficient means by which bacteria interact with host organisms or attack competitors. The T6SS of S. marcescens Db10, a white entomopathogenic strain, which if directly introduced into the body cavity of Drosophila is insensitive to the host’s systemic immune response and kills flies in a day, is active and expressed constitutively under normal growth conditions (Nehme et al., 2007). The T6SS of Db10 strain can assemble itself and be fired without needing to be triggered by cell contact, which broadens its range of cell targets and suggests that the activation of the T6SS is tailored to survival in specific niches (Gerc et al., 2015). Moreover, a multi-stage and dynamic assembly process was observed in Db10 with different subassemblies occurring simultaneously (Gerc et al., 2015). Although some T6SS transcripts have been detected in vivo, neither morbidity nor mortality of R. prolixus was observed in this study (data not shown). Further studies are, therefore, necessary to clarify if correct assemblies or activations are occurring in R. prolixus gut and evaluate whether T6SS is, indeed, involved in the trypanolytic activity observed in vivo.
In addition, there have been several recent reports stating that some T6SSs can target bacteria rather than eukaryotic cells (Murdoch et al., 2011). The T6SS has been shown to be involved in the bacteria–bacteria competition of S. marcescens with other opportunistic bacterial pathogens, but the role of T6SS of S. marcescens in the R. prolixus gut is still not completely clear. Although T6SS has been implicated in the virulence of some mammalian pathogens, it is not required for the virulence of S. marcescens Db10 strain in three non-mammalian virulence models (Murdoch et al., 2011). It does, however, exhibit dramatic killing activity against several other bacterial species and is required for S. marcescens to persist in mixed culture with the opportunist pathogen Enterobacter cloacae, which is occasionally found in triatomine gut (Murdoch et al., 2011; Gumiel et al., 2015).
Although the trypanolytic activity of S. marcescens was reported in in vitro conditions, it was found in this study and others (Azambuja et al., 2004; Genes et al., 2011) that some T. cruzi strains successfully colonized triatomine gut even in the presence of S. marcescens. Observation from the wild showed that Serratia, Dietzia, Gordonia, Mycobacterium, Corynebacterium, and Rhodococcus do not ensure protection to prevent triatomines from infection by T. cruzi alone or in combination (Gumiel et al., 2015). In fact, bacterial cytotoxic gene expression could vary under distinct physiological conditions, such as digestive tract compartment colonization.
Differences between the two populations of S. marcescens isolated from the digestive tract of Rhodnius spp. were previously observed using the analysis of 16S rDNA (da Mota et al., 2012) and confirmed here with the genome sequences of two bacterial strains, RPA1 and RPH1. The RPA1 genome shows a high level of similarity with SOLR4 and RSC-14 (two strains associated to plants) as well as ADJS-2D_White (a strain isolated from a phytophagous insect), whereas RPH1 has identity with the aquatic strain 19F (isolated from the skin of A. zeteki), WW4 (paper mills use water in their manufacturing process), and EGD-HP20 (from wastewater). The evolutionary divergences between RPA1 and RPH1 isolates were highlighted by comparative genomics and trypanolytic activity as well.
Serratia marcescens SOLR4 and RSC-14 are bacteria associated with plants; the first one isolated from an unclassified Solanaceae from Brazil (SAMN08287009) and the second from Solanum nigrum (Khan et al., 2015). Besides the hematophagic and coprophagic habits of triatomines (Wigglesworth, 1974; Sandoval et al., 2000), Díaz-Albiter et al. (2016) have also identified the alternative phytophagic behavior of R. prolixus, which may feed on Solanum lycopersicum fruit and sucrose solution in case of starvation. This finding could explain the close phylogenetic relationship between the strains RPA1 and plant-associated S. marcescens strains. Phytophagy by triatomines might enable them to avoid death from starvation in dry environments but might also be the source of endophytic bacteria in their guts.
Similarly, to the digestive enzymes of hematophagous insect, plants include the production of reactive oxygen species (ROS) and reactive nitrogen species, such as nitric oxide (NO) in their defense arsenal (Nascimento et al., 2018). Thus, to be able to colonize internal plant tissues, bacterial endophytes need to be able to cope with these stressful conditions (Nascimento et al., 2018). S. marcescens genomes from R. prolixus gut encode for various enzymes related to ROS detoxification, including superoxide dismutases, catalases, alkyl peroxidase (only RPA1), peroxidases, peroxiredoxins, organic hydroperoxide, peroxide stress protein YaaA, resistance gene ohrB, and biofilm peroxide resistance protein (BsmA). In addition, many glutathione S-transferase genes, the glutathione ABC transporters, glutathione peroxidases, glutathione synthetases, glutathione reductases, and glutaredoxin genes are found in the RPA1 and RPH1 genomes. A NO dioxygenase gene is present only in RPA1 genome and may account for the strain ability to cope with nitrosative stress. On the other hand, alkylhydroperoxidase and lipid hydroperoxide peroxidase were found only in RPH1.
Gut colonization by symbiotic bacteria requires multiple genomic adaptations since uncontrolled growth could lead to tissue damage and host death. Although the function of pRPA1 and pRPH1 plasmids remains unknown, a previous study showed that the S. marcescens colonies maintaining plasmids grow at a slower rate than the plasmid-free once (Platt and Chesham, 1990). Both S. marcescens pRPA1 and pRPH1 plasmids share many genes with a high level of identity, including some genes also found in other S. marcescens strains isolated from animals (mainly insects), but not observed in plant-associated strains (SOLR4 and RSC-14), suggesting a recent host adaptation niche, despite the evolutionary divergence between RPA1 and RPH1 chromosomes.
The investigation of Serratia genomics performed here revealed its adaptation to the life in community with multi-species interactions. The genes involved in motility, chemotaxis, and attachment that have been suggested to play an important role in endophytic bacterial colonization could act in a similar way in the settlement of Serratia in triatomine guts. Flagella, fimbria, LPS, and quorum-sensing genes are also abundant in Serratia genomes, which is consistent with its ability to sense and colonize several niches, including plants, insects, and nematode (Nascimento et al., 2018).
As previously reported, secretion systems and toxins (Sarris et al., 2014) were found to be encoded by GIs in RPA1 and RPH1 strains. The distinct composition in secretion systems between S. marcescens strains may explain their differences in trypanolytic activities. T2SS was observed only in RPA1, which could explain its higher trypanolytic activity compared to that of RPH1. This secretion system was also detected in S. marcescens strains Ano1 and Ano2, from A. stephensi (Chen et al., 2017), although the mechanism that could interfere with the Plasmodium development in the mosquito midgut has not been described yet.
The expression of the T6SS transcripts, found in both RPA1 and RPH1 genome, was also detected in the R. prolixus digestive tract. However, the T6SS transcription appeared earlier and stronger in R. prolixus infected with T. cruzi Dm 28c. The expression of the T6SS was also reported in the S. marcescens strain WW4 (Chen et al., 2017).
The role played by chitinases in the metabolism of S. marcescens is still unclear, but they were detected in RPA1 as well as in RPH1. Partially purified chitinases of S. marcescens PRNK-1, an isolated from cockroach, strongly inhibited the fungal growth of Rhizoctonia solani and Fusarium oxysporum in vitro (Moon et al., 2017). Chitinases secreted in the TDT could be especially important to protect its microbiota from infection by entomopathogenic fungi, i.e., Beauveria bassiana, Gliocladium virens, Metarhizium anisopliae, and Isaria fumosorosea (Vázquez-Martínez et al., 2014; Forlani et al., 2015; Huang et al., 2016). Although S. marcescens possess chitinolytic activity over the fungal wall, it is unknown if it affects the perimicrovillar membrane of TDT which also contains chitin (Alvarenga et al., 2016). The FS14 strain significantly suppressed the growth of phytopathogenic fungi through non-contact inhibition, which might be attributed to the extracellular secretion of chitinases.
Colicins and pyocins are bacteriocins with antibiotic activity secreted by some members of the Enterobacteriaceae that enable them to kill closely related bacterial cells, thereby reducing competition for essential nutrients. Although the antagonistic potential against bacteria has not been evaluated in vitro in this study, a whole colicin/pyocin bacteriocin operon including a colicin immunity protein was found in RPH1 (but not in RPA1) next to a PhoP/PhoQ system genes present in all strains (A, B, and C). PhoP/PhoQ is a two-component regulatory system that has been reported to be critical in the virulence control of S. marcescens (Barchiesi et al., 2012).
Microbial urease presents the enzymatic property of yielding ammonia, but also carbamate, which is very toxic to the host cells. Urease has been considered as a virulent factor not only for host cells but also for fungi and yeast (Giannouli et al., 2014). In presence of the S. marcescens urease, the urea from ingested mammalian blood might generate ammonia, which might contribute to nitrogen bioavailability and increase pH in midguts of hematophagous insects. This mechanism of pH control was proposed in A. stephensi (Chen et al., 2017) and might be expanded to other insect vectors such as R. prolixus. In the present study, a complete cassette encoding genes for urease was detected in S. marcescens RPA1 and RPH1 genomes.
The acquisition of plasmids is a major factor in the ability of bacteria to exploit new environments and hosts, including human and Hemipteran insects (Ricci and Hernandez, 2000; Douidah et al., 2014; Perilla-Henao and Casteel, 2016). Some Serratia strains are considered opportunist pathogens for human and some virulence genes are frequently described in their genomes, mainly in plasmids (Hurst et al., 2000; Iguchi et al., 2014; Gruber et al., 2015). pKPC-56ce, a plasmid similar to those of pRPA1 and pRPH1, was found in the SSNIH1 strain of Serratia sp. isolated in 2015 from a hospital tubing in the United States (Weingarten et al., 2018). Fortunately, pRPA1 and pRPH1 plasmids do not include the integron found in pKPC-56ce and its resistance genes for quinolones (QnrB19, C3F38_25970), betalactams extended-spectrum (SHV-12, C3F38_25945), and carbapenems (KPC-2, C3F38_25920). On the other hand, pRPA1 plasmid encodes the aadA1 gene (B7L32_25510) for aminoglycoside resistance also found in pKPC-56ce, but not in pRPH1.
The coprophagic behavior of triatomines might explain the variability of S. marcescens populations in the digestive tract of specimens from the wild. Gumiel et al. (2015), analyzing the microbiota diversity in the digestive tract of T. brasiliensis and T. pseudomaculata, also observed the presence of S. marcescens in all specimens captured in the endemic northeastern region of Brazil. In addition, Gumiel et al. (2015) also reported the sporadic presence of Rhodococcus, which belong to the Corynebacterineae suborder as well as other bacterium genera. Moreover, Carels et al. (2017) noted that all these bacterial species share one common feature: bacteria with GC-rich genomes overcome those with GC-poor genomes. These authors attributed the comparative success of GC-rich bacteria in the TDT niche to their increased ability to process complex substrates.
In this study, Actinobacteria such as the symbiont R. rhodnii were not detected, probably due to the short time of incubation on BHI agar. In fact, the TDT microbiota diversity is probably underrepresented in this study due to limitations of in vitro culture and the fastest growth of S. marcescens. Despite its apparent predominance, the limitations on molecular fingerprint methods could sub represent the genetic diversity of Serratia populations, since only one genetic marker (one fingerprint primer) was applied to classify the isolates (da Mota et al., 2004, 2012). Interestingly, the general distribution of S. marcescens in triatomines (da Mota et al., 2012; Gumiel et al., 2015) and mosquitoes (Wang et al., 2017) seems to be a favorable factor to propose it as a suitable competitor species in these systems. However, S. marcescens does not seems to be antagonistic of Wolbachia sp., since this bacterium is intracellular and has already been observed in co-colonization with Serratia sp. in a sylvatic adult specimen of Rhodnius sp. from Amazon, Brazil (da Mota et al., 2012). At least two types of Wolbachia sp. were also detected in specimens of Rhodnius pallescens raised in insectarium and captured in the field in Republic of Panama (Espino et al., 2009).
Although this study presents the first pieces of evidence of an antagonistic potential of S. marcescens with regard to T. cruzi in R. prolixus based on the genomic characterization of RPA1 and RPH1 strains, further investigations are needed to improve our understanding of this system in triatomines.
FM, DC, CV, MG, and PA conceived and designed the experiments. FM, DC, CV, and MG performed the experiments. FM, DC, CV, JA, NC, and PA analyzed the data. FM, DC, CV, and PA contributed reagents, materials, and analysis tools. FM, DC, CV, JA, NC, and PA wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Deyse Vallim and Rodrigo Lisboa for their assistance in the bacteria isolation and the Instituto Oswaldo Cruz and Fundação Oswaldo Cruz for funding as well as for allowing the use of its facilities of high-throughput sequencing and bioinformatics. We also thank the fundings provided by PAPES and PROEP programs, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular (INCT-EM).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03205/full#supplementary-material
TABLE S1 | Average nucleotide identity between Serratia genomes, bacterial secretion systems, KEGG and Ano1 orthologous, gene islands, antibiotic resistance genes and one way ANOVA of trypanolytic activity.
Alcoforado-Diniz, J., and Coulthurst, S. J. (2015). Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J. Bacteriol. 197, 2350–2360. doi: 10.1128/JB.00199-15
Alvarenga, E. S., Mansur, J. F., Justi, S. A., Figueira-Mansur, J., Dos Santos, V. M., Lopez, S. G., et al. (2016). Chitin is a component of the Rhodnius prolixus midgut. Insect Biochem. Mol. Biol. 69, 61–70. doi: 10.1016/j.ibmb.2015.04.003
Azambuja, P., Feder, D., and Garcia, E. S. (2004). Isolation of Serratia marcescens in the midgut of Rhodnius prolixus: impact on the establishment of the parasite Trypanosoma cruzi in the vector. Exp. Parasitol. 107, 89–96. doi: 10.1016/j.exppara.2004.04.007
Azambuja, P., and Garcia, E. S. (1997). “Care and maintenance of triatomine colonies,” in Molecular Biology of Insect Disease Vectors: a Methods Manual, eds J. M. Crampton, C. B. Beard, and C. Louis (London: Chapman and Hall), 56–64. doi: 10.1007/978-94-009-1535-0_6
Barbosa-Silva, A. N., Câmara, A. C., Martins, K., Nunes, D. F., Oliveira, P. I., Azevedo, P. R., et al. (2016). Characteristics of Triatomine infestation and natural Trypanosoma cruzi infection in the State of Rio Grande do Norte, Brazil. Rev. Soc. Bras. Med. Trop. 49, 57–67. doi: 10.1590/0037-8682-0300-2015
Barboza, M., Duschak, V. G., Fukuyama, Y., Nonami, H., Erra-Balsells, R., Cazzulo, J. J., et al. (2005). Structural analysis of the N-glycans of the major cysteine proteinase of Trypanosoma cruzi. Identification of sulfated high-mannose type oligosaccharides. FEBS J. 272, 3803–3815. doi: 10.1111/j.1742-4658.2005.04787.x
Barchiesi, J., Castelli, M. E., Di Venanzio, G., Colombo, M. I., and García Véscovi, E. (2012). The PhoP/PhoQ system and its role in Serratia marcescens pathogenesis. J. Bacteriol. 194, 2949–2961. doi: 10.1128/JB.06820-11
Berg, G. (2000). Diversity of antifungal and plant-associated Serratia plymuthica strains. J. Appl. Microbiol. 88, 952–960. doi: 10.1046/j.1365-2672.2000.01064.x
Bian, G., Xu, Y., Lu, P., Xie, Y., and Xi, Z. (2010). The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 6:e1000833. doi: 10.1371/journal.ppat.1000833
Bonay, P., Molina, R., and Fresno, M. (2001). Binding specificity of mannose-specific carbohydrate-binding protein from the cell surface of Trypanosoma cruzi. Glycobiology 11, 719–729. doi: 10.1093/glycob/11.9.719
Bröms, J. E., Meyer, L., Lavander, M., Larsson, P., and Sjöstedt, A. (2012). DotU and VgrG, core components of type VI secretion systems, are essential for Francisella LVS pathogenicity. PLoS ONE 7:e34639. doi: 10.1371/journal.pone.0034639
Brunet, Y. R., Hénin, J., Celia, H., and Cascales, E. (2014). Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 15, 315–321. doi: 10.1002/embr.201337936
Carels, N., Gumiel, M., da Mota, F. F., de Carvalho Moreira, C. J., and Azambuja, P. (2017). A metagenomic analysis of bacterial microbiota in the digestive tract of triatomines. Bioinform. Biol. Insights 11:1177932217733422. doi: 10.1177/1177932217733422
Castro, D., Moraes, C., Gonzalez, M., Ratcliffe, N., Azambuja, P., and Garcia, E. (2012). Trypanosoma cruzi immune response modulation decreases microbiota in Rhodnius prolixus gut and is crucial for parasite survival and development. PLoS One 7:e36591. doi: 10.1371/journal.pone.0036591
Castro, D. P., Moraes, C. S., Garcia, E. S., and Azambuja, P. (2007a). Inhibitory effects of d-mannose on trypanosomatid lysis induced by Serratia marcescens. Exp. Parasitol. 115, 200–204. doi: 10.1016/j.exppara.2006.08.001
Castro, D. P., Seabra, S. H., Garcia, E. S., de Souza, W., and Azambuja, P. (2007b). Trypanosoma cruzi: ultrastructural studies of adhesion, lysis and biofilm formation by Serratia marcescens. Exp. Parasitol. 117, 201–207. doi: 10.1016/j.exppara.2007.04.014
Chen, S., Blom, J., and Walker, E. D. (2017). Genomic, physiologic, and symbiotic characterization of Serratia marcescens strains isolated from the mosquito Anopheles stephensi. Front. Microbiol. 8:1483. doi: 10.3389/fmicb.2017.01483
Cirimotich, C. M., Dong, Y., Clayton, A. M., Sandiford, S. L., Souza-Neto, J. A., Mulenga, M., et al. (2011). Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332, 855–858. doi: 10.1126/science.1201618
Clausen, P. T., Zankari, E., Aarestrup, F. M., and Lund, O. (2016). Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J. Antimicrob. Chemother. 71, 2484–2488. doi: 10.1093/jac/dkw184
Contreras, V. T., Araújo-Jorge, T. C., Bonaldo, M. C., Thomaz, N., Barbosa, H. S., Meirelles, M., et al. (1988). Biological aspects of the Dm 28c clone of a Trypanosoma cruzi after metacyclogenesis in chemically defined media. Mem. Inst. Oswaldo Cruz 83, 123–133. doi: 10.1590/S0074-02761988000100016
Cortez, M. R., Provencano, A., Silva, C. E., Mello, C. B., Zimmermann, L. T., Schaub, G. A., et al. (2012). Trypanosoma cruzi: effects of azadirachtin and ecdysone on the dynamic development in Rhodnius prolixus larvae. Exp. Parasitol. 131, 363–371. doi: 10.1016/j.exppara.2012.05.005
Coura, J. R. (2015). The main sceneries of Chagas disease transmission. The vectors, blood and oral transmissions–a comprehensive review. Mem. Inst. Oswaldo Cruz 110, 277–282. doi: 10.1590/0074-0276140362
Coura, J. R., and Viñas, P. A. (2010). Chagas disease: a new worldwide challenge. Nature 465, S6–S7. doi: 10.1038/nature09221
da Mota, F. F., Gomes, E. A., Paiva, E., Rosado, A. S., and Seldin, L. (2004). Use of rpoB gene analysis for identification of nitrogen-fixing Paenibacillus species as an alternative to the 16S rRNA gene. Lett. Appl. Microbiol. 39, 34–40. doi: 10.1111/j.1472-765X.2004.01536.x
da Mota, F. F., Marinho, L. P., Moreira, C. J., Lima, M. M., Mello, C. B., Garcia, E. S., et al. (2012). Cultivation-independent methods reveal differences among bacterial gut microbiota in triatomine vectors of Chagas disease. PLoS Negl. Trop. Dis. 6:e1631. doi: 10.1371/journal.pntd.0001631
da Mota, F. F., Nóbrega, A., Marriel, I. E., Paiva, E., and Seldin, L. (2002). Genetic diversity of Paenibacillus polymyxa populations isolated from the rhizosphere of four cultivars of maize (Zea Mays) planted in Cerrado Soil. Appl. Soil Ecol. 20, 119–132. doi: 10.1016/s0929-1393(02)00016-1
Dahllöf, I., Baillie, H., and Kjelleberg, S. (2000). rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66, 3376–3380. doi: 10.1128/AEM.66.8.3376-3380.2000
Dias, F. A., Guerra, B., Vieira, L. R., Perdomo, H. D., Gandara, A. C., Amaral, R. J., et al. (2015). Monitoring of the parasite load in the digestive tract of Rhodnius prolixus by combined qPCR analysis and imaging techniques provides new insights into the Trypanosome life cycle. PLoS Negl. Trop. Dis. 9:e0004186. doi: 10.1371/journal.pntd.0004186
Díaz-Albiter, H. M., Ferreira, T. N., Costa, S. G., Rivas, G. B., Gumiel, M., Cavalcante, D. R., et al. (2016). Everybody loves sugar: first report of plant feeding in triatomines. Parasit. Vectors 9:114. doi: 10.1186/s13071-016-1401-0
Douidah, L., De Zutter, L., Van Nieuwerburgh, F., Deforce, D., Ingmer, H., Vandenberg, O., et al. (2014). Presence and analysis of plasmids in human and animal associated Arcobacter species. PLoS One 9:e85487. doi: 10.1371/journal.pone.0085487
Douzi, B., Brunet, Y. R., Spinelli, S., Lensi, V., Legrand, P., Blangy, S., et al. (2016). Structure and specificity of the Type VI secretion system ClpV-TssC interaction in enteroaggregative Escherichia coli. Sci. Rep. 6:34405. doi: 10.1038/srep34405
Durvasula, R. V., Gumbs, A., Panackal, A., Kruglov, O., Aksoy, S., Merrifield, R. B., et al. (1997). Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proc. Natl. Acad. Sci. U.S.A. 94, 3274–3278. doi: 10.1073/pnas.94.7.3274
Durvasula, R. V., Gumbs, A., Panackal, A., Kruglov, O., Taneja, J., Kang, A. S., et al. (1999). Expression of a functional antibody fragment in the gut of Rhodnius prolixus via transgenic bacterial symbiont Rhodococcus rhodnii. Med. Vet. Entomol. 13, 115–119. doi: 10.1046/j.1365-2915.1999.00175.x
Egami, I., Iiyama, K., Zhang, P., Chieda, Y., Ino, N., Hasegawa, K., et al. (2009). Insecticidal bacterium isolated from an ant lion larva from Munakata, Japan. J. Appl. Entomol. 133, 117–124. doi: 10.1111/j.1439-0418.2008.01329.x
Eichler, S., and Schaub, G. A. (2002). Development of symbionts in triatomine bugs and the effects of infections with trypanosomatids. Exp. Parasitol. 100, 17–27. doi: 10.1006/expr.2001.4653
Espino, C. I., Gómez, T., González, G., do Santos, M. F., Solano, J., Sousa, O., et al. (2009). Detection of Wolbachia bacteria in multiple organs and feces of the triatomine insect Rhodnius pallescens (Hemiptera, Reduviidae). Appl. Environ. Microbiol. 75, 547–550. doi: 10.1128/AEM.01665-08
Ferreira, R. C., Kessler, R. L., Lorenzo, M. G., Paim, R. M., Ferreira, L., De, L., et al. (2016). Colonization of Rhodnius prolixus gut by Trypanosoma cruzi involves an extensive parasite killing. Parasitology 14, 434–443. doi: 10.1017/S0031182015001857
Forlani, L., Pedrini, N., Girotti, J. R., Mijailovsky, S. J., Cardozo, R. M., Gentile, A. G., et al. (2015). Vector Triatoma infestans with the Entomopathogenic Fungus Beauveria bassiana combined with an aggregation cue: field, laboratory and mathematical modeling assessment. PLoS Negl. Trop. Dis. 9:e0003778. doi: 10.1371/journal.pntd.0003778
Garcia, A. R. M., Rocha, A. P., Moreira, C. C., Rocha, S. L., Guarneri, A. A., and Elliot, S. L. (2016). Screening of fungi for biological control of a triatomine vector of chagas disease: temperature and Trypanosome infection as factors. PLoS Negl. Trop. Dis. 10:e0005128. doi: 10.1371/journal.pntd.0005128
Geiger, A., Fardeau, M. L., Njiokou, F., and Ollivier, B. (2013). Glossina spp. Gut bacterial flora and their putative role in fly-hosted trypanosome development. Front. Cell. Infect. Microbiol. 3:34. doi: 10.3389/fcimb.2013.00034
Genes, C., Baquero, E., Echeverri, F., Maya, J. D., and Triana, O. (2011). Mitochondrial dysfunction in Trypanosoma cruzi: the role of Serratia marcescens prodigiosin in the alternative treatment of Chagas disease. Parasit. Vectors 4:66. doi: 10.1186/1756-3305-4-66
Gerc, A. J., Diepold, A., Trunk, K., Porter, M., Rickman, C., Armitage, J. P., et al. (2015). Visualization of the serratia type VI secretion system reveals unprovoked attacks and dynamic assembly. Cell Rep. 12, 2131–2142. doi: 10.1016/j.celrep.2015.08.053
Germano, M. D., Santo-Orihuela, P., Roca-Acevedo, G., Toloza, A. C., Vassena, C., Picollo, M. I., et al. (2012). Scientific evidence of three different insecticide-resistant profiles in Triatoma infestans (Hemiptera: Reduviidae) populations from Argentina and Bolivia. J. Med. Entomol. 49, 1355–1360. doi: 10.1603/ME12070
Giannouli, M., Palatucci, A. T., Rubino, V., Ruggiero, G., Romano, M., Triassi, M., et al. (2014). Use of larvae of the wax moth Galleria mellonella as an in vivo model to study the virulence of Helicobacter pylori. BMC Microbiol. 14:228. doi: 10.1186/s12866-014-0228-0
Grimont, P. A., Grimont, F., Le Minor, S., Davis, B., and Pigache, F. (1979). Compatible results obtained from biotyping and serotyping in Serratia marcescens. J. Clin. Microbiol. 10, 425–432.
Gruber, T. M., Göttig, S., Mark, L., Christ, S., Kempf, V. A., Wichelhaus, T. A., et al. (2015). Pathogenicity of pan-drug-resistant Serratia marcescens harbouring blaNDM-1. J. Antimicrob. Chemother. 70, 1026–1030. doi: 10.1093/jac/dku482
Gumiel, M., da Mota, F. F., Rizzo, V. S., Sarquis, O., Castro, D. P., Lima, M. M., et al. (2015). Characterization of the microbiota in the guts of Triatoma brasiliensis and Triatoma pseudomaculata infected by Trypanosoma cruzi in natural conditions using culture independent methods. Parasit. Vectors 8:245. doi: 10.1186/s13071-015-0836-z
Hancock, P. A., White, V. L., Ritchie, S. A., Hoffmann, A. A., and Godfray, H. C. (2016). Predicting Wolbachia invasion dynamics in Aedes aegypti populations using models of density-dependent demographic traits. BMC Biol. 14:96. doi: 10.1186/s12915-016-0319-5
Hernández-Romano, P., Cámara-Contreras, M., Bravo-Sarmiento, E., and López-Balderas, N. (2015). Prevalence of Trypanosoma cruzi antibodies in blood donors from Veracruz State, Mexico. Transfusion 55, 647–656. doi: 10.1111/trf.12860
Huang, Z., Hao, Y., Gao, T., Huang, Y., Ren, S., and Keyhani, N. O. (2016). The Ifchit1 chitinase gene acts as a critical virulence factor in the insect pathogenic fungus Isaria fumosorosea. Appl. Microbiol. Biotechnol. 100, 5491–5503. doi: 10.1007/s00253-016-7308-z
Hurst, M. R., Glare, T. R., Jackson, T. A., and Ronson, C. W. (2000). Plasmid-located pathogenicity determination of Serratia entomophila, the causal agent of amber diseases of grass grub, showing similarity to the insecticidal toxins of Photorhabdus luminescens. J. Bacteriol. 182, 5127–5138. doi: 10.1128/JB.182.18.5127-5138.2000
Hurwitz, I., Fieck, A., and Durvasula, R. (2012). Antimicrobial peptide delivery strategies: use of recombinant antimicrobial peptides in paratransgenic control systems. Curr. Drug Targets 13, 1173–1180. doi: 10.2174/138945012802002366
Hurwitz, I., Fieck, A., Read, A., Hillesland, H., Klein, N., Kang, A., et al. (2011). Paratransgenic control of vector borne diseases. Int. J. Biol. Sci. 7, 1334–1344. doi: 10.7150/ijbs.7.1334
Iguchi, A., Nagaya, Y., Pradel, E., Ooka, T., Ogura, Y., Katsura, K., et al. (2014). Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol. Evol. 6, 2096–2110. doi: 10.1093/gbe/evu160
Kennan, R. M., Dhungyel, O. P., Whittington, R. J., Egerton, J. R., and Rood, J. I. (2001). The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 183, 4451–4458. doi: 10.1128/JB.183.15.4451-4458.2001
Khan, A. R., Ullah, I., Khan, A. L., Park, G. S., Waqas, M., Hong, S. J., et al. (2015). Improvement in phytoremediation potential of Solanum nigrum under cadmium contamination through endophytic-assisted Serratia sp. RSC-14 inoculation. Environ. Sci. Pollut. Res. Int. 22, 14032–14042. doi: 10.1007/s11356-015-4647-8
Kondo, N., Nikoh, N., Ijichi, N., Shimada, M., and Fukatsu, T. (2002). Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc. Natl. Acad. Sci. U.S.A. 99, 14280–14285. doi: 10.1073/pnas.222228199
Li, P., Kwok, A. H., Jiang, J., Ran, T., Xu, D., Wang, W., et al. (2015). Comparative genome analyses of Serratia marcescens FS14 reveals its high antagonistic potential. PLoS One 10:e0123061. doi: 10.1371/journal.pone.0123061
Mahlen, S. D. (2011). Serratia infections: from military experiments to current practice. Clin. Microbiol. Rev. 24, 755–791. doi: 10.1128/CMR.00017-11
Manzano-Marín, A., Simon, J. C., and Latorre, A. (2016). Reinventing the wheel and making it round again: evolutionary convergence in Buchnera-Serratia symbiotic consortia between the distantly related lachninae aphids Tuberolachnus salignus and Cinara cedri. Genome Biol. Evol. 8, 1440–1458. doi: 10.1093/gbe/evw085
Masschelein, J., Mattheus, W., Gao, L. J., Moons, P., Van Houdt, R., Uytterhoeven, B., et al. (2013). A PKS/NRPS/FAS hybrid gene cluster from Serratia plymuthica RVH1 encoding the biosynthesis of three broad spectrum, zeamine-related antibiotics. PLoS One 8:e54143. doi: 10.1371/journal.pone.0054143
McMeniman, C. J., Lane, R. V., Cass, B. N., Fong, A. W., Sidhu, M., Wang, Y. F., et al. (2009). Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 323, 141–144. doi: 10.1126/science.1165326
Moon, C., Seo, D. J., Song, Y. S., Hong, S. H., Choi, S. H., and Jung, W. J. (2017). Antifungal activity and patterns of N-acetyl-chitooligosaccharide degradation via chitinase produced from Serratia marcescens PRNK-1. Microb. Pathog. 113, 218–224. doi: 10.1016/j.micpath.2017.10.039
Moraes, C. S., Seabra, S. H., Albuquerque-Cunha, J. M., Castro, D. P., Genta, F. A., de Souza, W., et al. (2009). Prodigiosin is not a determinant factor in lysis of Leishmania (Viannia) braziliensis after interaction with Serratia marcescens D-mannose sensitive fimbriae. Exp. Parasitol. 122, 84–90. doi: 10.1016/j.exppara.2009.03.004
Moraes, C. S., Seabra, S. H., Castro, D. P., Brazil, R. P., de Souza, W., Garcia, E. S., et al. (2008). Leishmania (Leishmania) chagasi interactions with Serratia marcescens: ultrastructural studies, lysis and carbohydrate effects. Exp. Parasitol. 118, 561–568. doi: 10.1016/j.exppara.2007.11.015
Murdoch, S., Trunk, K., English, G., Fritsch, M., Pourkarimi, E., and Coulthurst, S. (2011). The opportunistic pathogen Serratia marcescens utilizes Type VI secretion to target bacterial competitors. J. Bacteriol. 193, 6057–6069. doi: 10.1128/JB.05671-11
Nascimento, F., Vicente, C., Cock, P., Tavares, M., Rossi, M., Hasegawa, K., et al. (2018). From plants to nematodes: Serratia grimesii BXF1 genome reveals an adaptation to the modulation of multi-species interactions. Microb. Genom 4:e000178. doi: 10.1099/mgen.0.000178
Nehme, N. T., Liégeois, S., Kele, B., Giammarinaro, P., Pradel, E., Hoffmann, J. A., et al. (2007). A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 3:e173. doi: 10.1371/journal.ppat.0030173
Noya, B. A., and González, O. N. (2015). An ecological overview on the factors that drives to Trypanosoma cruzi oral transmission. Acta Trop. 151, 94–102. doi: 10.1016/j.actatropica.2015.06.004
Pachebat, J. A., van Keulen, G., Whitten, M. M., Girdwood, S., Del Sol, R., Dyson, P. J., et al. (2013). Draft genome sequence of Rhodococcus rhodnii strain LMG5362, a symbiont of Rhodnius prolixus (Hemiptera, Reduviidae, Triatominae), the Principle Vector of Trypanosoma cruzi. Genome Announc. 20:e00329-13. doi: 10.1128/genomeA.00329-13
Patterson, K. L., Porter, J. W., Ritchie, K. B., Polson, S. W., Mueller, E., Peters, E. C., et al. (2002). The etiology of white pox, a lethal disease of the Caribbean elkhorn coral, Acropora palmata. Proc. Natl. Acad. Sci. U.S.A. 99, 8725–8730. doi: 10.1073/pnas.092260099
Perilla-Henao, L. M., and Casteel, C. L. (2016). Vector-borne bacterial plant pathogens: interactions with hemipteran insects and plants. Front. Plant Sci. 7:1163. doi: 10.3389/fpls.2016.01163
Pessoa, G. C., Trevizani, N. A., Dias, L. S., Bezerra, C. M., Melo, B. V., and Diotaiut, L. (2015). Toxicological profile of deltamethrin in Triatoma brasiliensis (Hemiptera: Reduviidae) in State of Ceará, Northeastern Brazil. Ver. Soc. Bras. Med. Trop. 48, 39–43. doi: 10.1590/0037-8682-0010-2015
Petersen, L. M., and Tisa, L. S. (2013). Friend or foe? A review of the mechanisms that drive Serratia towards diverse lifestyles. Can. J. Microbiol. 59, 627–640. doi: 10.1139/cjm-2013-0343
Platt, D. J., and Chesham, J. S. (1990). Growth depression of Serratia marcescens by plasmids that belong to incompatibility group P. Microbios 63, 151–157.
Postma, P. W., Lengeler, J. W., and Jacobson, G. R. (1993). Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594.
Pusz, P., Bok, E., Mazurek, J., Stosik, M., and Baldy-Chudzik, K. (2014). Type 1 fimbriae in commensal Escherichia coli derived from healthy humans. Acta Biochim. Pol. 61, 389–392.
Rahul, S., Chandrashekhar, P., Hemant, B., Bipinchandra, S., Mouray, E., Grellier, P., et al. (2015). In vitro antiparasitic activity of microbial pigments and their combination with phytosynthesized metal nanoparticles. Parasitol. Int. 64, 353–356. doi: 10.1016/j.parint.2015.05.004
Reid, G., and Sobel, J. D. (1987). Bacterial adherence in the pathogenesis of urinary tract infection: a review. Rev. Infect. Dis. 9, 470–487. doi: 10.1093/clinids/9.3.470
Ricci, J. C. D., and Hernandez, M. E. (2000). Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 20, 79–108. doi: 10.1080/07388550008984167
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rossi, P., Ricci, I., Cappelli, A., Damiani, C., Ulissi, U., Mancini, M. V., et al. (2015). Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasit. Vectors 8:278. doi: 10.1186/s13071-015-0888-0
Sandoval, C. M., Joya, M. I., Gutierez, R., and Ângulo, V. M. (2000). Cleptohaematophagy of the triatomine bug Belminus herreri. Med. Vet. Entomol. 14, 100–101. doi: 10.1046/j.1365-2915.2000.00210.x
Sarris, P. F., Ladoukakis, E. D., Panopoulos, N. J., and Scoulica, E. V. (2014). A phage tail-derived element with wide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol. Evol. 6, 1739–1747. doi: 10.1093/gbe/evu136
Schulenburg, H., and Ewbank, J. J. (2004). Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol. Biol. 4:49. doi: 10.1186/1471-2148-4-49
Silva, L. H. P., and Nussenzweig, V. (1953). Sôbre uma cepa de Trypanosoma cruzi altamente virulenta para o camundongo branco. Folia Clin. Biol. 20, 191–208.
Soares, S. C., Geyik, H., Ramos, R. T., de Sá, P. H., Barbosa, E. G., Baumbach, J., et al. (2016). GIPSy: genomic island prediction software. J. Biotechnol. 232, 2–11. doi: 10.1016/j.jbiotec.2015.09.008
Taira, E., Iiyama, K., Mon, H., Mori, K., Akasaka, T., Tashiro, K., et al. (2014). Draft genome sequence of entomopathogenic Serratia liquefaciens strain FK01. Genome Announc. 2:e00609-14. doi: 10.1128/genomeA.00609-14
Taracena, M. L., Oliveira, P. L., Almendares, O., Umaña, C., Lowenberger, C., Dotson, E. M., et al. (2015). Genetically modifying the insect gut microbiota to control Chagas disease vectors through systemic RNAi. PLoS Negl. Trop. Dis. 9:e0003358. doi: 10.1371/journal.pntd.0003358
Thomson, N. R., Crow, M. A., McGowan, S. J., Cox, A., and Salmond, G. P. (2000). Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 36, 539–556. doi: 10.1046/j.1365-2958.2000.01872.x
Uchiyama, T., Kaneko, R., Yamaguchi, J., Inoue, A., Yanagida, T., Nikaidou, N., et al. (2003). Uptake of N, N’-diacetylchitobiose [(GlcNAc)2] via the phosphotransferase system is essential for chitinase production by Serratia marcescens 2170. J. Bacteriol. 185, 1776–1782. doi: 10.1128/JB.185.6.1776-1782.2003
Vassena, C. V., Picollo, M. I., and Zerba, E. N. (2000). Insecticide resistance in Brazilian Triatoma infestans and Venezuelan Rhodnius prolixus. Med. Vet. Entomol. 14, 51–55. doi: 10.1046/j.1365-2915.2000.00203.x
Vázquez-Martínez, M. G., Cirerol-Cruz, B. E., Torres-Estrada, J. L., and López, M. H. (2014). Potential for entomopathogenic fungi to control Triatoma dimidiata (Hemiptera: Reduviidae), a vector of Chagas disease in Mexico. Rev. Soc. Bras. Med. Trop. 47, 716–722. doi: 10.1590/0037-8682-0193-2014
Vieira, C. S., Mattos, D. P., Waniek, P. J., Santangelo, J. M., Figueiredo, M. B., Gumiel, M., et al. (2015). Rhodnius prolixus interaction with Trypanosoma rangeli: modulation of the immune system and microbiota population. Parasit. Vectors 8:135. doi: 10.1186/s13071-015-0736-2
Vieira, C. S., Waniek, P. J., Castro, D. P., Mattos, D. P., Moreira, O. C., and Azambuja, P. (2016). Impact of Trypanosoma cruzi on antimicrobial peptide gene expression and activity in the fat body and midgut of Rhodnius prolixus. Parasit. Vectors 9:119. doi: 10.1186/s13071-016-1398-4
Vieira, C. S., Waniek, P. J., Mattos, D. P., Castro, D. P., Mello, C. B., Ratcliffe, N. A., et al. (2014). Humoral responses in Rhodnius prolixus: bacterial feeding induces differential patterns of antibacterial activity and enhances mRNA levels of antimicrobial peptides in the midgut. Parasit. Vectors 7:232. doi: 10.1186/1756-3305-7-232
Wang, S., Dos-Santos, A. L. A., Huang, W., Liu, K. C., Oshaghi, M. A., Wei, G., et al. (2017). Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 357, 1399–1402. doi: 10.1126/science.aan5478
Weingarten, R. A., Johnson, R. C., Conlan, S., Ramsburg, A. M., Dekker, J. P., Lau, A. F., et al. (2018). Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. doi: 10.1128/mBio.02011-17
Weiss, B., and Aksoy, S. (2011). Microbiome influences on insect host vector competence. Trends Parasitol. 27, 514–522. doi: 10.1016/j.pt.2011.05.001
Whitten, M. M., Facey, P. D., Del Sol, R., Fernández-Martínez, L. T., Evans, M. C., Mitchell, J. J., et al. (2016). Symbiont-mediated RNA interference in insects. Proc. Biol. Sci. 283:20160042. doi: 10.1098/rspb.2016.0042
Wigglesworth, V. B. (1974). The Principles of Insect Physiology, 7th Edn. London: Chapman and Hall. doi: 10.1007/978-1-4899-3202-0
World Health Organization [WHO] (2018). Chagas Disease (American Trypanosomiasis). Fact Sheet N° 340. Geneva: World Health Organization.
Zhang, Q., Weyant, R., Steigerwalt, A. G., White, L. A., Melcher, U., Bruton, B. D., et al. (2003). Genotyping of Serratia marcescens strains associated with cucurbit yellow vine disease by repetitive elements-based polymerase chain reaction and DNA-DNA hybridization. Phytopathology 93, 1240–1246. doi: 10.1094/PHYTO.2003.93.10.1240
Keywords: Serratia marcescens, Rhodnius prolixus, Trypanosoma cruzi, trypanocidal activity, antagonistic genes
Citation: da Mota FF, Castro DP, Vieira CS, Gumiel M, de Albuquerque JP, Carels N and Azambuja P (2019) In vitro Trypanocidal Activity, Genomic Analysis of Isolates, and in vivo Transcription of Type VI Secretion System of Serratia marcescens Belonging to the Microbiota of Rhodnius prolixus Digestive Tract. Front. Microbiol. 9:3205. doi: 10.3389/fmicb.2018.03205
Received: 28 August 2018; Accepted: 11 December 2018;
Published: 24 January 2019.
Edited by:
Frank T. Robb, University of Maryland, Baltimore, United StatesReviewed by:
Darlene Miller, University of Miami, United StatesCopyright © 2019 da Mota, Castro, Vieira, Gumiel, de Albuquerque, Carels and Azambuja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicolas Carels, bmljb2xhcy5jYXJlbHNAY2R0cy5maW9jcnV6LmJy; bmljb2xhcy5jYXJlbHNAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.