
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 December 2018
Sec. Terrestrial Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.03122
This article is part of the Research Topic Landscape and Ecosystem Microbiology View all 10 articles
Nitrification driven by ammonia oxidizers is a key step of nitrogen removal in estuarine environments. Spatial distribution characteristics of ammonia-oxidizers have been well understood in mudflats, but less studied in the agricultural soils next to mudflats, which also play an important role in nitrogen cycling of the estuarine ecosystem. In the present research, we investigated ammonia oxidizers’ distributions along the Yangtze River estuary in Jiangsu Province, China, sampling soils right next to the estuary (mudflats) and the agricultural soils 100 m away. We determined the relationship between the abundance of amoA genes of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) and the potential nitrification rates of the mudflats and agricultural soils. We also identified the environmental variables that correlated with the composition of the ammonia oxidizers’ communities by 16S rRNA gene pyrosequencing. Results indicated that agricultural soils have significantly higher potential nitrification rates as well as the AOA abundance, and resulted in strong phylogenetic clustering only in AOA communities. The ammonia oxidizers’ community compositions differed dramatically among the mudflat and agricultural sites, and stochasticity played a dominant role. The AOA communities were dominated by the Group 1.1a cluster at the mudflat, whereas the 54D9 and 29i4 clusters were dominant in agriculture soils. The dominant AOB communities in the mudflat were closely related to the Nitrosospira lineage, whereas the agricultural soils were dominated by the Nitrosomonas lineage. Soil organic matter and salinity were correlated with the ammonia oxidizers’ community compositions.
Estuaries are mixing zones of coastal seawater and freshwater from continental runoff, acting as a conduit for the transport, transformation and production of organic carbon and nutrients. Nitrogen pollutants, emitted from both industrial and agricultural activities such as wastewater treatment plants, chemical and manufacturing industries, and livestock farming (Chung et al., 1996; Busca and Pistarino, 2003; Kim et al., 2007), are transferred from terrestrial ecosystems to aquatic ecosystems through mudflats by gravitational movement (Likens and Bormann, 1974). Coupled nitrification-denitrification can remove a substantial percentage (10∼80%) of anthropogenic nitrogen pollution in estuary ecosystems (Seitzinger, 2008), where nitrification converts ammonia to nitrite and nitrate, which are subsequently released to the atmosphere as N2 gas via denitrification or anammox. The removal of nitrogen pollutants in estuaries is a microbially mediated process. Because different nitrogen-cycling microbes own different activities, the niche separation of nitrogen-cycling microbes in estuaries are thus of great concern (Bernhard et al., 2007, 2010; Mosier and Francis, 2008; Moin et al., 2009; Jin et al., 2011; Damashek et al., 2015; Damashek and Francis, 2017; Zhang et al., 2018). In addition, increasing human disturbance in mudflats has a significant impact on changing soil properties, such as soil carbon and nitrogen contents (Wang et al., 2010), which are key environmental factors controlling the community composition of microorganisms involved the in nitrogen cycle. The ammonia oxidation process is the first and rate-limiting step of nitrification. It is necessary to assess the potential nitrification rates and niche differentiation of ammonia oxidizers in both multiflats and adjacent agricultural soils.
The study site of the current study was the Yangtze River, the third-largest river in the world, which has a high biogeochemical flux of nitrogen, discharging into the East China Sea with a nitrate load of 6.3 × 106 t/a. Intensive agricultural practice occurs in the upper estuary in Jiangsu Province, as shown in Figure 1. Ammonia oxidation is considered to be carried out in soils by ammonia oxidizing bacteria (AOB) (Kowalchuk and Stephen, 2001) and ammonia oxidizing archaea (AOA) (Leininger et al., 2006). Previous studies in open ocean environments indicated that AOA far outnumber AOB (Wuchter et al., 2006; Mincer et al., 2007; Beman et al., 2008), suggesting a high tolerance of AOA to salinity pressures (Santoro et al., 2008; Bernhard et al., 2010). However, Zhang et al. (2018) showed the equal numbers of AOA and AOB amoA genes at the soil/sediment interface, while AOB was more abundant than AOA in high salinity environments (Santoro et al., 2008). In addition, debatable results were also established in agricultural ecosystems. Many studies indicated the influence of ammonia concentration and fertilization on the niche separation of AOA and AOB, that AOB rather than AOA grew in soils treated with high levels of inorganic ammonium (Di et al., 2009, 2010; Jia and Conrad, 2009). In contrast, evidence from stable isotope probing (SIP) in paddy soils demonstrated that active AOA numerically outcompeted AOB (Wang et al., 2014). Thus, questions remain concerning their relative importance in mudflat and agricultural soils.
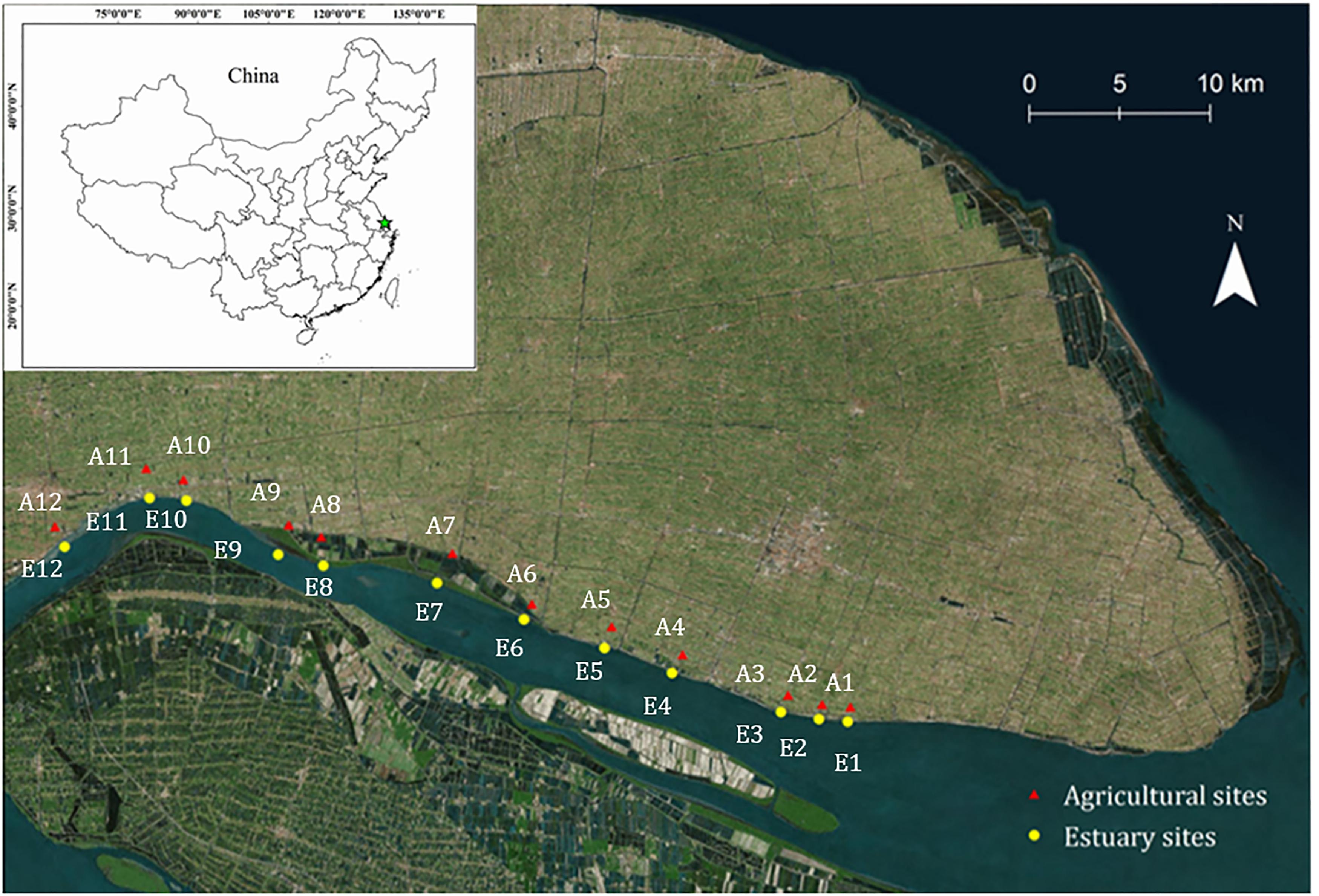
Figure 1. Map showing the location of sampling sites. Mudflat sites, E1–E12 (yellow circles); Agricultural sites, A1–A12 (red triangles).
The objectives of this study were to elucidate the differentiation of the potential nitrification rates and niche separation of community composition of ammonia oxidizers (AOA and AOB) between mudflat and adjacent agricultural soils and to investigate the potential links between the environmental variables and ammonia oxidizers’ community structure.
Two geographic sampling regions were selected in our study: the mudflat sites (sites E1–E12) and agricultural sites (sites A1–A12) (Figure 1). The main crop is rapeseed, which is the world’s second most important oilseed crop. Soil samples were collected in April 2017 to a depth of 0–5 cm. At each sampling location, 3 replicates of soil samples were collected. Each replicate was arranged into two randomized blocks and then mixed. All the sampled soils were sieved (<2 mm) to remove aboveground plant materials, roots, and stones. The fresh soil samples were stored in -20°C for the analysis of nitrification potential and ammonia oxidizers’ community, while dried and ground samples were used for the analysis of soil chemical properties.
Soil properties were determined as previously described (Zhou et al., 2015). Soil pH and salinity were determined in soil samples of a 1:2.5 (weight: volume) ratio with distilled water using pH and conductivity probes (Jiang et al., 2007). Soil organic matter was determined by the Walkley–Black acid digestion method, which uses chromic acid to measure the oxidizable organic carbon in the soil (Walkley and Black, 1934). NH4+-N and NO3--N were extracted with 2M KCl at a soil/solution ratio of 1:5 by shaking at 200 rpm for 60 min and determined by a Continuous Flow Analyzer (San++System, Skalar, Holland). Soil chemistry measures normalized to dry or wet soil weight.
Potential nitrification rates were measured in triplicates by the shaken-slurry method as described previously (Yao et al., 2011). Ten grams of soil from each sample were preincubated at room temperature for 7 days and then mixed with 100 ml of 1.5 mM ammonium sulfate. After incubation for 0, 3, and 7 days, 10-ml slurry samples were centrifuged and the supernatant was filtered through a 0.45-μm-pore-size membrane. NO3--N content in the supernatant was immediately analyzed and, as described above for KCl-extracted NO3--N. NO3-N, concentration increased linearly and nitrification potential was calculated from the rate of increase in NO3-–N concentration over time in the slurry using linear regression.
The net nitrification rates (n) were calculated by the formula presented by Persson and Wirén (1995) as follows:
where (NO3--N)/t2 and (NO3--N)/t1 are the concentrations of NO3--N in the soil at time t2 and time t1, respectively, and t is the number of days between t2 and t1.
DNA was extracted from 0.5 g of each homogenized sample using the FastDNA spin for Soil Kit (Qbiogene, Inc., Irvine, CA, United States). The methods followed the manufacturer’s protocol. The quality and abundance of the DNA extractions were measured by gel electrophoresis (0.8% agarose) and NanoDrop spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, United States), and stored at -80°C.
Functional marker genes encoding amoA of AOA and AOB were quantified by real-time PCR (q-PCR) using the primer pairs of Arch-amoAF/Arch-amoAR (Francis et al., 2005) and amoA-1F/amoA-2R (Rotthauwe et al., 1997), respectively (Supplementary Table S2). Real-time quantitative PCR was carried out on a CFX96 Optical Real-Time Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, United States). Quantitative PCR reactions were performed in 20-μL reaction mixtures containing 10 μL 2x SYBR Premix Ex Taq (Takara Biotech, Dalian, China), 200 nM of each primer, and 20 ng DNA template. The thermal program for the real-time PCR assay is shown in Supplementary Table S2. Blanks were run with water, instead of soil DNA extract, as the template to serve as the negative control. To ensure that there was no significant effect of PCR inhibition on the q-PCR data, a dilution series of templates, encompassing the working concentration, from each site were assayed using the archaeal amoA qPCR assay. A linear correlation was established between the dilution and copy number (R > 0.99), indicating no significant effect of PCR inhibitors on gene quantification. Standards for qPCR were generated using a serial dilution of known copies of PCR fragments of the respective functional gene generated using M13 PCR from clones. Standard curves, spanning 107-101, were constructed by a dilution series of plasmids harboring the amoA gene. The plasmid was extracted by the Plasmid Purification Kit (Takara) and the concentration was measured by a NanoDrop ND-1000 UV-Vis Spectrophotometer and used for the calculation of standard copy numbers. The amplification efficiency ranged from 98 to 110% with R2 values of approximately 0.99 in each reaction. Special amplification of amoA genes was confirmed by agarose gel electrophoresis and melting curve analysis.
Universal primer pair Tag-515F and 907R (Turner et al., 1999) were used to amplify soil bacterial 16S rRNA gene fragments for the MiSeq Illumina Sequencing Platform. PCR was carried out in 50-μl reaction mixtures containing each deoxynucleoside triphosphate at a concentration of 1.25 mM, 1 μl of forward and reverse primers (20 mM), 2U of Taq DNA polymerase (TaKaRa, Japan), and 50 ng of DNA. The following cycling parameters were used: 5 min of incubation at 94°C, followed by 32 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s, and a 10 min extension at 72°C (Supplementary Table S2). Triplicate reaction mixtures per sample were pooled together and purified using an agarose gel DNA purification kit (TaKaRa), and quantified using NanoDrop ND-1000 (Thermo Scientific, United States). The bar-coded PCR products were pooled in equimolar amounts (10 pg for each sample) before sequencing as described in Xiang et al. (2016). Sequences were merged by FLASH (Mago and Salzberg, 2011) and then processed using sequence data through the program AmpliconNoise, version 1.24, for the detection and correction of probable errors (Quince et al., 2011), obtainable from Quantitative Insights Into Microbial Ecology (QIIME1) (Caporaso et al., 2010). Poor-quality sequences (below an average quality score of 25) and short sequences (<200 bp) were removed (Supplementary Table S3). Sequences of each replicate were merged, and then clustered into Operational Taxonomic Units (OTUs) using a 97% identity threshold (default QIIME settings) by UCLUST (Edgar, 2010) and all singleton OTUs were deleted. The ammonia oxidizers’ sequences were screened at the phylum level for Thaumarchaea and at the genus levels for both AOB (Nitrosospira and Nitrosomonas), and the representative sequences were extracted for phylogenetic analysis (Supplementary Figures S3, S4).
Phylogenetic analysis of the Thaumarchaea 16S rRNA gene sequenced in this study was performed using the Molecular Evolutionary Genetics Analysis (MEGA 4.0) software package (Kumar et al., 2004). The basic tree of sequences from known AOA and AOB cultures was constructed through a neighbor-joining algorithm. The tree topology was checked using the neighbor-joining algorithm and the minimum evolution method. The nucleotide sequences of the representative OTUs were deposited in the NCBI (National Center for Biotechnology Information) under the accession numbers MH115263 to MH115276 (AOA) and MH115277 to MH115284 (AOB) of the 16S rRNA genes, respectively. The pyrosequencing reads of the total 16S rRNA genes were deposited in the NCBI under accession number SRP136463.
Sequences with 97% similarity were clustered into operational taxonomic units (OTUs) before matching the resulting consensus sequence of each OTU to known sequences in the SILVA database. Each OTU was then used as a proxy for representing a microbial species present in a sample, where the relative abundance of an OTU was directly proportional to the abundance of the microbial taxa that this OTU describes.
Non-metric multidimensional scaling analyses (NMDS) were performed with Bray-Curtis dissimilarity for the microbial community data. Canonical Correspondence Analysis (CCA) and BIO-ENV were used to identify the relationship between the ammonia oxidizers’ community structure and environmental parameters. The NMDS, CCA and BIO-ENV analysis were conducted using the “vegan” R package2.
In order to evaluate the phylogenetic community composition within each sample, the net relatedness index (NRI) was calculated by the mean phylogenetic distance (MPDs) among all co-occurring individuals, indicating the ‘basal’ dispersion of the lineages within the community. The nearest taxon index (NTI) measured the mean nearest taxon distance (MNTD) among individuals, and estimated the ‘terminal’ phylogenetic dispersion of the community (Webb, 2000). For a single community, NRI or NTI greater than +2 or less than -2 indicated that coexisting taxa were more or less related than expected by chance (phylogenetic clustering), respectively. A positive mean NRI and NTI of all communities suggested that communities were more affected by phylogenetically clustering than dispersal limitation, and a negative mean implied that communities were phylogenetically over-dispersed. NRI and NTI were calculated in the R “picante” package (Purcell et al., 2007). Beta nearest taxon index (betaNTI) was the number of standard deviations of the observed betaMNTD from the mean of null distribution (Stegen et al., 2012). If betaNTI values were below -2 or above +2, deterministic processes were dominant in shaping the community composition across all sites, and if betaNTI values were between -2 and 2, stochastic processes played an important role. The betaNTI was calculated in phylocom4.2 (Hardy, 2008). The potential relationship between amoA gene abundance and 16S rRNA relative abundance of AOA and AOB and environmental characteristics was analyzed by SPSS version 10.0 (IBM Co., Armonk, NY, United States), and multicollinearity was ruled out. Differences at P < 0.05 were considered statistically significant.
The basic physio-chemical parameters of the sites are given in Figure 2 and Supplementary Table S1. The sites were defined as mudflat (prefix E) or agriculture (prefix A); the agricultural sites are about 100 meters away from the mudflat. A salinity gradient from 243 to 3000 μS/cm was observed from the upper estuary (E12) to the lower estuary (E1). The salinity ranged from 46.2 to 226 μS cm-1 in the agriculture sites and no spatial heterogeneity was found. A similar distribution pattern was observed for the concentration of NH4+, which increased from the upper estuary (4.2 mg kg-1) to the lower estuary (11.4 mg kg-1), with no significant difference between the agriculture sites (P > 0.05). The salinity was positively correlated with the concentration of NH4+ (P < 0.05). The organic matter content ranged from 8.08 to 16.8% along the estuarine sites (mean 11.5%), whereas the mean organic matter content for the agricultural sites was 21.0%. There was no significant difference in soil pH between the mudflat (pH 8.3–9.3) and agricultural (pH 7.9–9.4) sites.
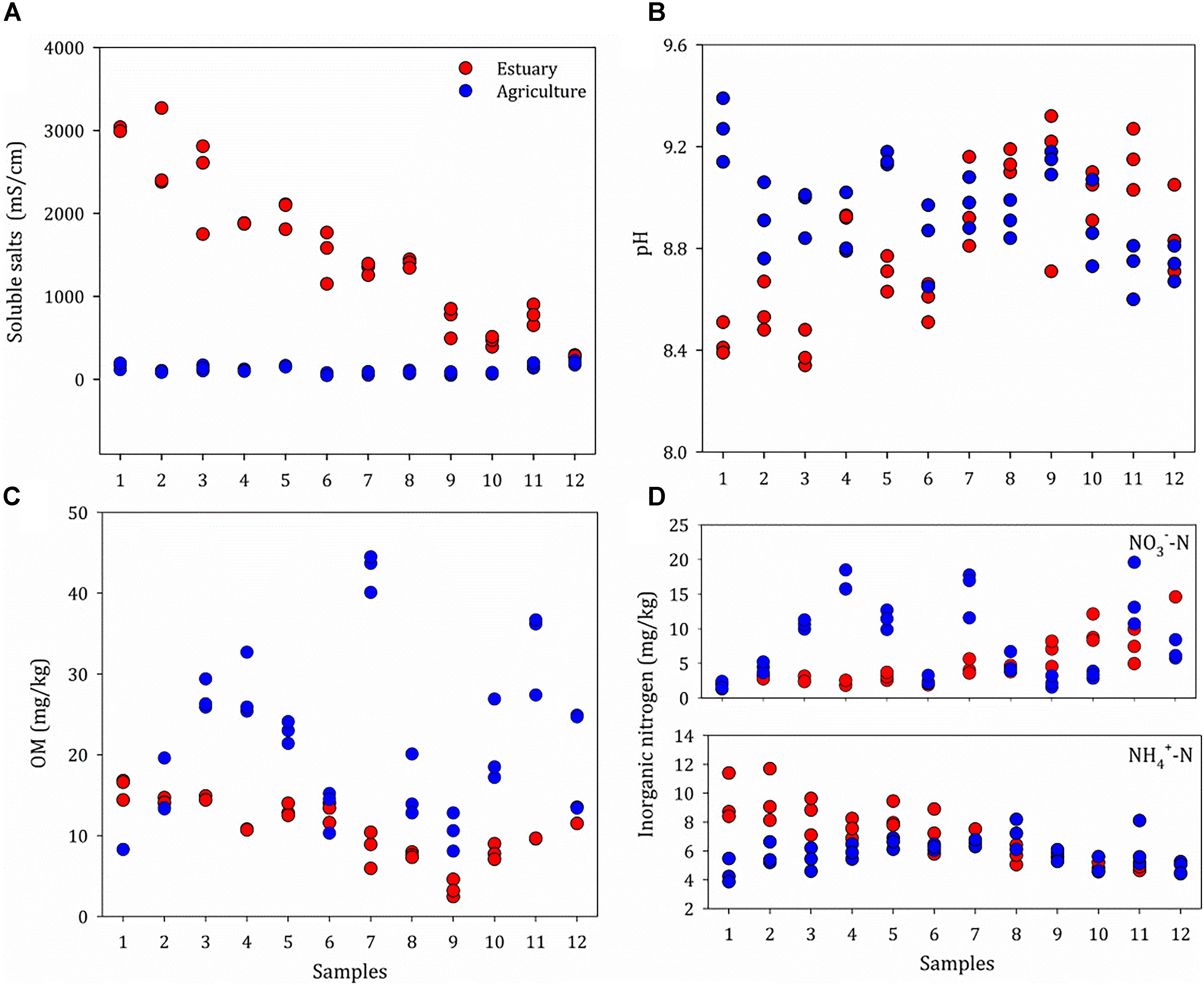
Figure 2. Properties of soils at the mudflat (Estuary) and agricultural (Agriculture) sites, including Soluble salts (A), pH (B), OM (C) and Inorganic nitrogen (D). Original values are given in Supplementary Table S1. OM, organic matter.
The rate of nitrification at the agricultural sites ranged from 2.0 to 65.1 μg g-1 day-1 and was generally higher than the rates at the mudflat sites (0–10.3 μg g-1 day-1) (Figure 3A). The soil nitrification rates at the upper estuary sites (E7–E12) were significantly higher than those in the lower estuary sites (E1–E6). Strong positive correlations between the amount of soil organic matter and the potential nitrification rates were observed among the agricultural soils (P < 0.05), indicating that organic matter was the key factor influencing the potential nitrification rate in agricultural sites. Contrarily, no correlation was observed between environmental factors and the rates of nitrification at the mudflat sites.
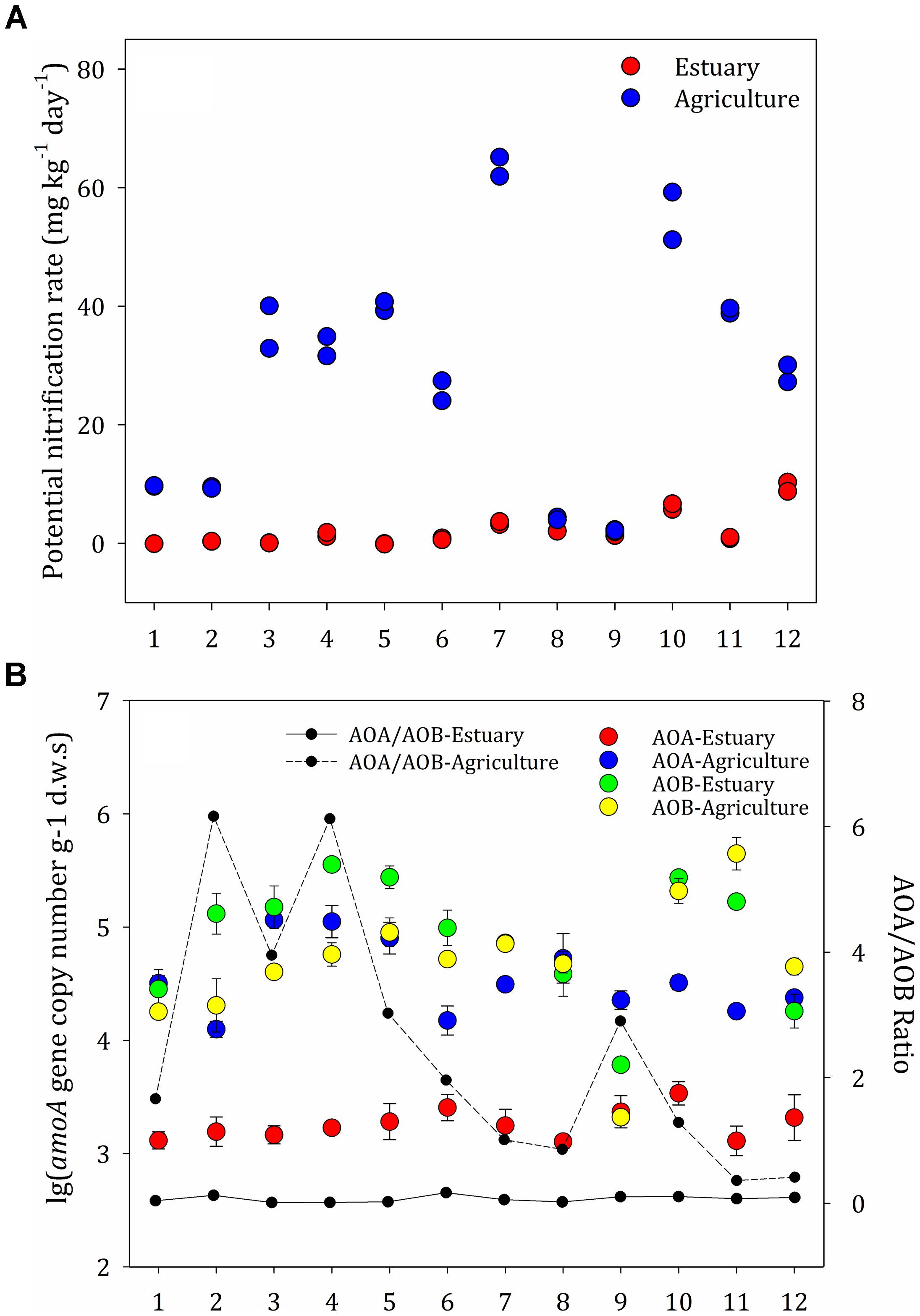
Figure 3. (A) Potential nitrification rates at the mudflat and agricultural sites. (B) Abundance of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) along the mudflat and at the agricultural sites based on the amoA copy numbers per gram dry weight of soil. The ratios of AOA to AOB amoA gene copies are shown as black (mudflat samples) and dotted (agricultural samples) lines. The error bars indicate the standard deviation.
The distribution pattern for the ammonia oxidizers was significantly different between the mudflat and agricultural soils (Figure 3B). Across all mudflat sites, the ammonia oxidizers’ communities were dominated by AOB (inferred from the amoA genes from AOB, ranging from 1.3 × 104 to 1.2 × 105 copies g-1 d.w.s), which were significantly more abundant than AOA (1.3 × 103–3.4 × 103 copies g-1 d.w.s). At the majority of the agricultural sites (A1, A2, A3, A4, A5, A6, A7, A9, and A10), the abundance of archaeal amoA genes (6.1 × 103–3.6 × 105 copies g-1 d.w.s) was significantly higher than the abundance of bacterial amoA genes (2.1 × 103–2.1 × 105 copies g-1 d.w.s) (P < 0.05), whereas the abundance of bacterial amoA genes was higher than the abundance of archaeal amoA genes at the other agricultural sites. The AOA:AOB ratios were between 0.01 and 0.17 at the mudflat sites and between 0.36 and 6.17 at the agricultural sites. There was no correlation between the abundance of archaea organisms and the potential nitrification rates.
The composition of the phylotypes of the ammonia oxidizers was determined by pyrosequencing samples taken from each of the sites. The 16S rRNA sequences of the ammonia oxidizers showed the presence of AOA (14 OTUs) (Figure 4) and AOB (8 OTUs) (Figure 5). The AOA OTUs could be classified into six clusters: four (OTUs 1, 2, 3, and 4) were related to the Group 1.1a cluster; one (OTU 5) was related to the Group 1.1a-associated cluster; and nine OTUs were related to the Group 1.1b cluster, including two (OTUs 6 and 7) related to the 54D9 cluster, four (OTUs 7, 8, 9, 10, and 11) related to the 29i4 cluster, one (OTU 12) falling within the 29i4-related cluster and two (OTUs 13 and 14) that most likely matched the Nitrososphaera viennensis cluster. The majority of AOA communities in the mudflat sites were in Group 1.1a (including OTUs 1 and 2), a cluster well-known in marine environments, with relative abundances ranging from 67.7 to 98.4%. In contrast, the dominant AOA phylotype in agricultural soils fell within clusters 54D9 (OTU 6) and 29i4 (OTU 8), with abundances ranging from 42.8 to 68.4% and 10.5 to 39.6%, respectively.
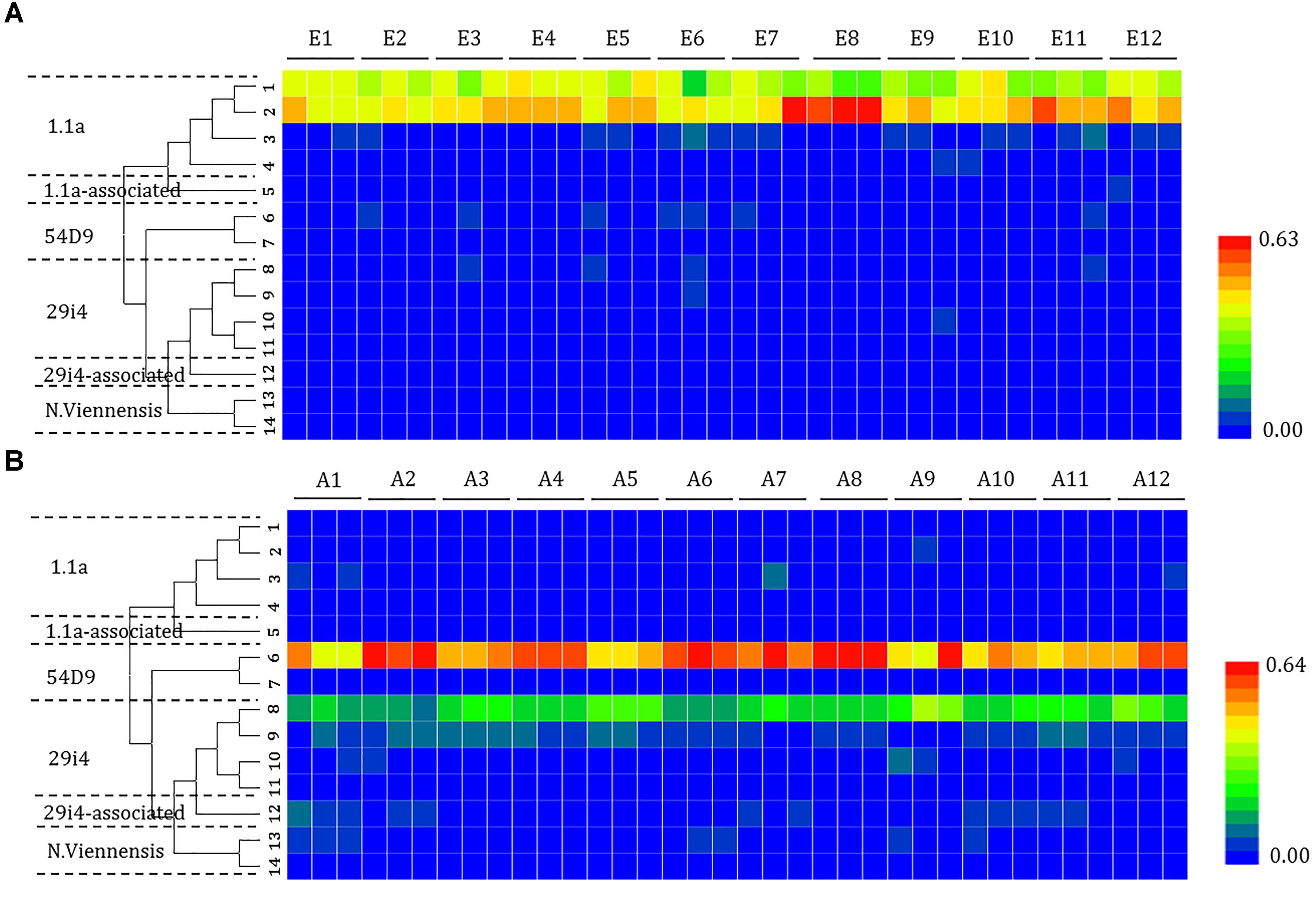
Figure 4. Heat map showing the relatedness of the ammonia-oxidizing archaea (AOA) community structures in the analysis of the (A) mudflat and (B) agricultural sites based on the relative abundance of distinct lineages of ammonia-oxidizing archaea 16S rRNA genes analyzed using pyrosequencing. The relative abundance of each lineage in each soil is displayed as a heat map representation. The phylogenetic position of each representative operational taxonomic unit is shown in the phylogenetic tree on the left-hand side.
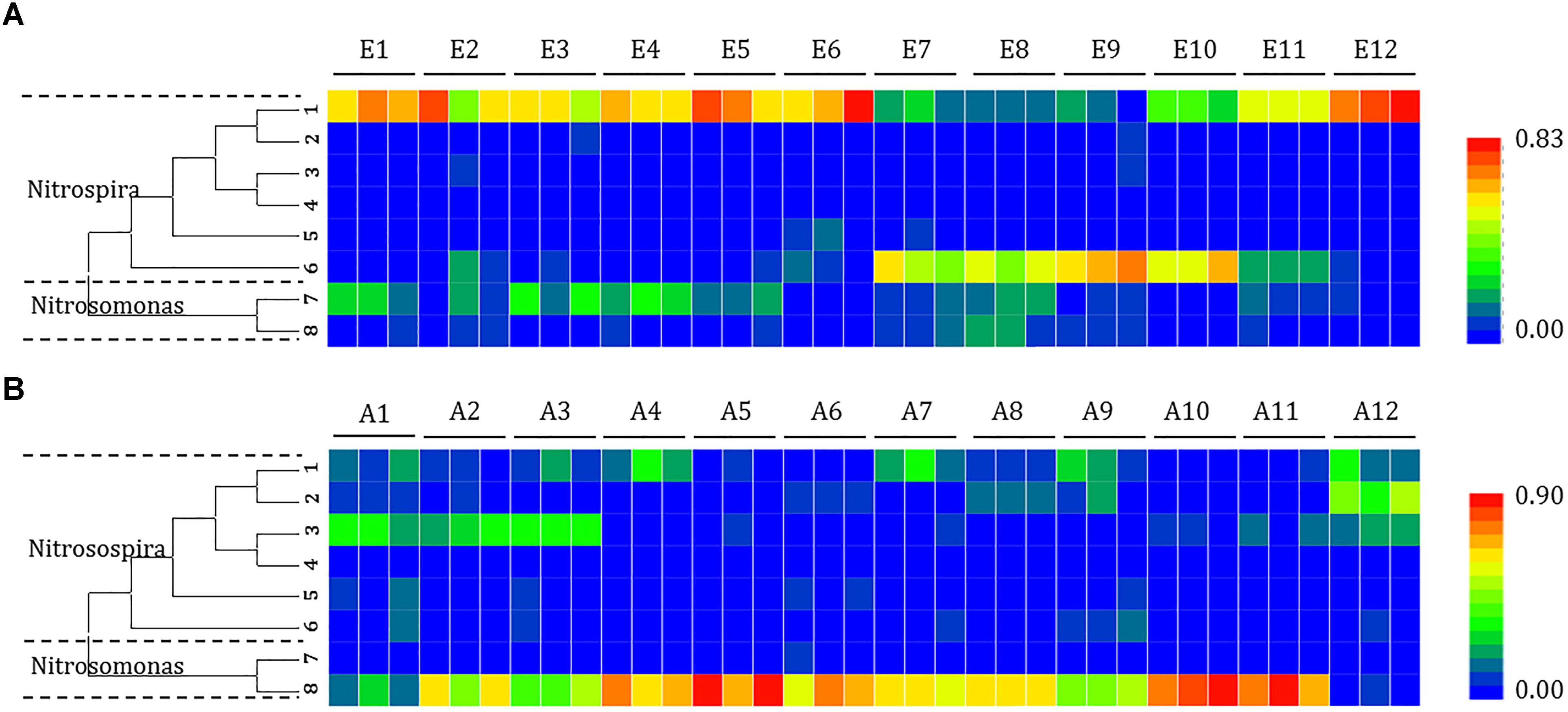
Figure 5. Heat map showing the relatedness of ammonia-oxidizing bacteria community structures in the analysis of the (A) mudflat and (B) agricultural sites based on the relative abundance of distinct lineages of ammonia-oxidizing bacteria 16S rRNA genes analyzed using pyrosequencing. The relative abundance of each lineage in each soil is displayed as a heat map representation. The phylogenetic position of each representative operational taxonomic unit is shown in the phylogenetic tree on the left-hand side.
Changes in the distribution of phylotypes were also detected in the AOB communities. All the sequences of the AOB matched those of the well-described AOB in the Nitrosospira (OTUs 1, 2, 3, 4, 5, and 6) and Nitrosomonas (OTUs 7 and 8) lineages. A significant portion of the OTUs (OTU 1 at sites E1, E2, E3, E4, E5, E6, E11, and E12; OTU 6 at sites E7, E8, E9, and E10) detected at the mudflat sites belonged to the AOB Nitrosospira (ranging from 41.9 to 88.4% and 42.7 to 72.9%, respectively), whereas the AOB Nitrosomonas OTU 8 was dominant at the agricultural sites (ranging from 42.9 to 96.7%), except at site A1 (OTU 3) and A12 (OTU 2), where Nitrosospira was the major lineage.
Non-metric multidimensional scaling (NMDS) analysis was used to demonstrate the structural divergence of the community composition and to show the community differentiation of AOA and AOB between the mudflat sites and the agricultural sites (Figure 6).
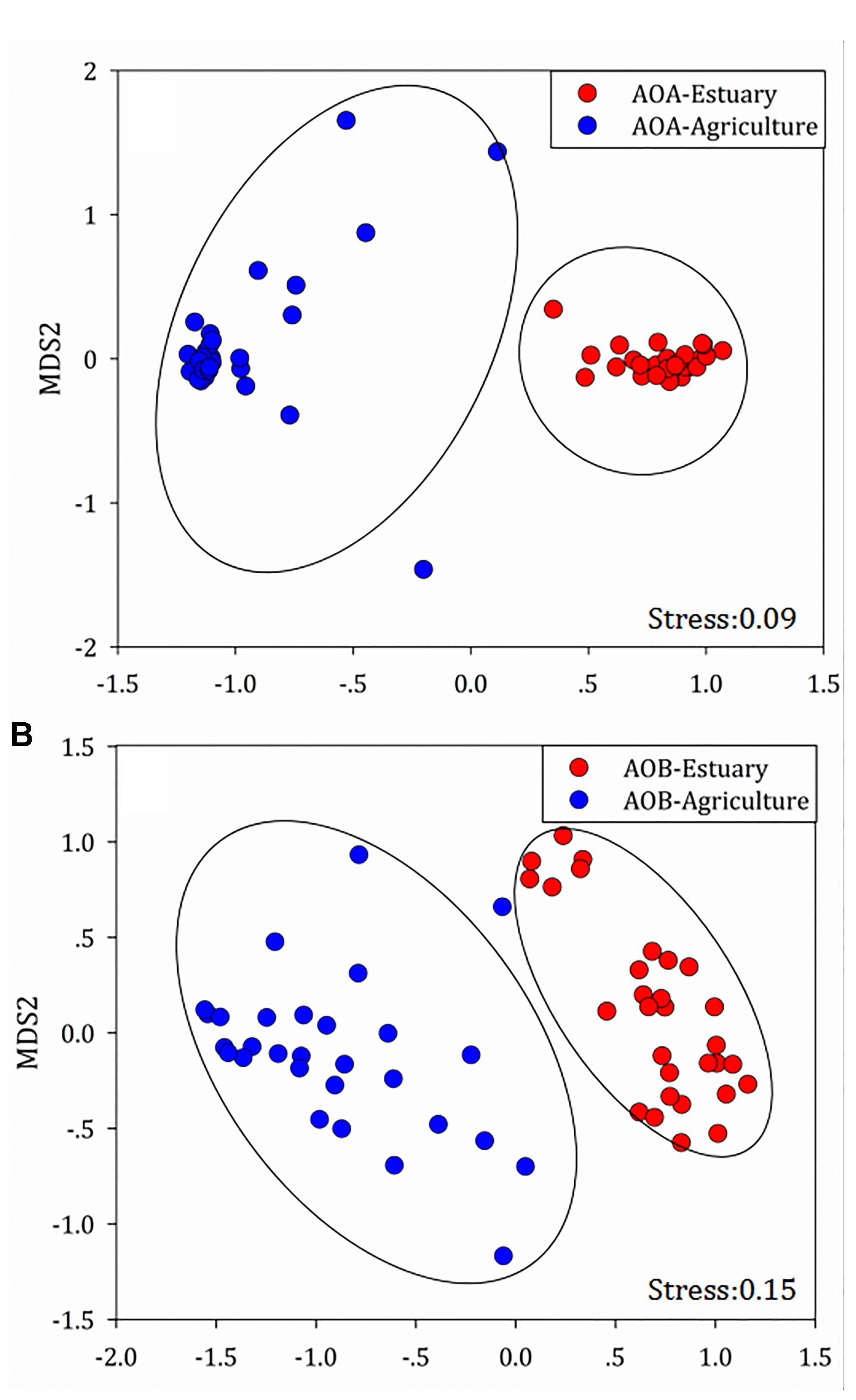
Figure 6. Compositional structure of (A) ammonia-oxidizing archaea (AOA) and (B) ammonia-oxidizing bacteria (AOB) 16S rRNA genes among the mudflat and agricultural sites as described by non-metric multidimensional scaling plots.
To test whether dispersal limited or niche-based mechanisms better explained the assembly of ammonia oxidizers’ community composition in mudflat and agricultural sites, we calculated the NRI (Figure 7) and NTI (Supplementary Figure S1) for each single community.
For the AOA and AOB, we found that the mean NRI and mean NTI were positive (Figure 7 and Supplementary Figure S1, P < 0.05), suggesting AOA and AOB communities in both mudflat and agricultural soils were more influenced by niche processes than dispersal limitation. For the AOA, the mean value of NRI in agricultural soils was higher than that in mudflat sites, whereas no significant difference of the mean NTI value was observed between mudflat and agricultural soils. In addition, the mean NRI value of AOA was above 2 in agricultural soils. For AOB, no differentiation of either the mean NRI or NTI value was observed between mudflat and agricultural soils, and the NRI and NTI scores in all samples were between -2 and 2.
In order to establish which process controlled community dynamics at different spatial scales, βNTI values were determined. The results showed that a stochastic process was dominant at all the sites in our study (Supplementary Figure S2; value > -2 and <2).
There was no correlation between the abundance of amoA genes and environmental parameters at any of the mudflat or agricultural soils. Potential relationships between the composition of the ammonia oxidizers’ community and the soil physico-chemical properties were inferred through unconstrained (BIO-ENV) (Clarke and Ainsworth, 1993) and constrained (CCA) multivariate methods based on the relative abundance of the OTUs of the ammonia oxidizers (Figure 8). The BIO-ENV analysis showed that soil salinity had the best individual associations with the composition of AOA, whereas soil organic matter explained the composition of AOB (Supplementary Tables S4, S5). Two axes of the CCA explained 38.6% of the total variance in the AOA 16S rRNA genotype composition and 73.9% of the cumulative variance of the genotype–environment relationship. Only soil salinity and organic matter significantly influenced the distribution of the AOA 16S rRNA genotypes (r2 = 0.52, P < 0.001; r2 = 0.23, P < 0.001, respectively, 999 permutations). For the AOB, two axes of the CCA explained 34.7% of the total variance in the AOB genotype composition and 64.8% of the cumulative variance of the genotype–environment relationship. The salinity (r2 = 0.55, P < 0.001, 999 permutations) and amount of organic matter (r2 = 0.24, P < 0.001, 999 permutations) significantly affected the 16S rRNA gene composition of AOB.
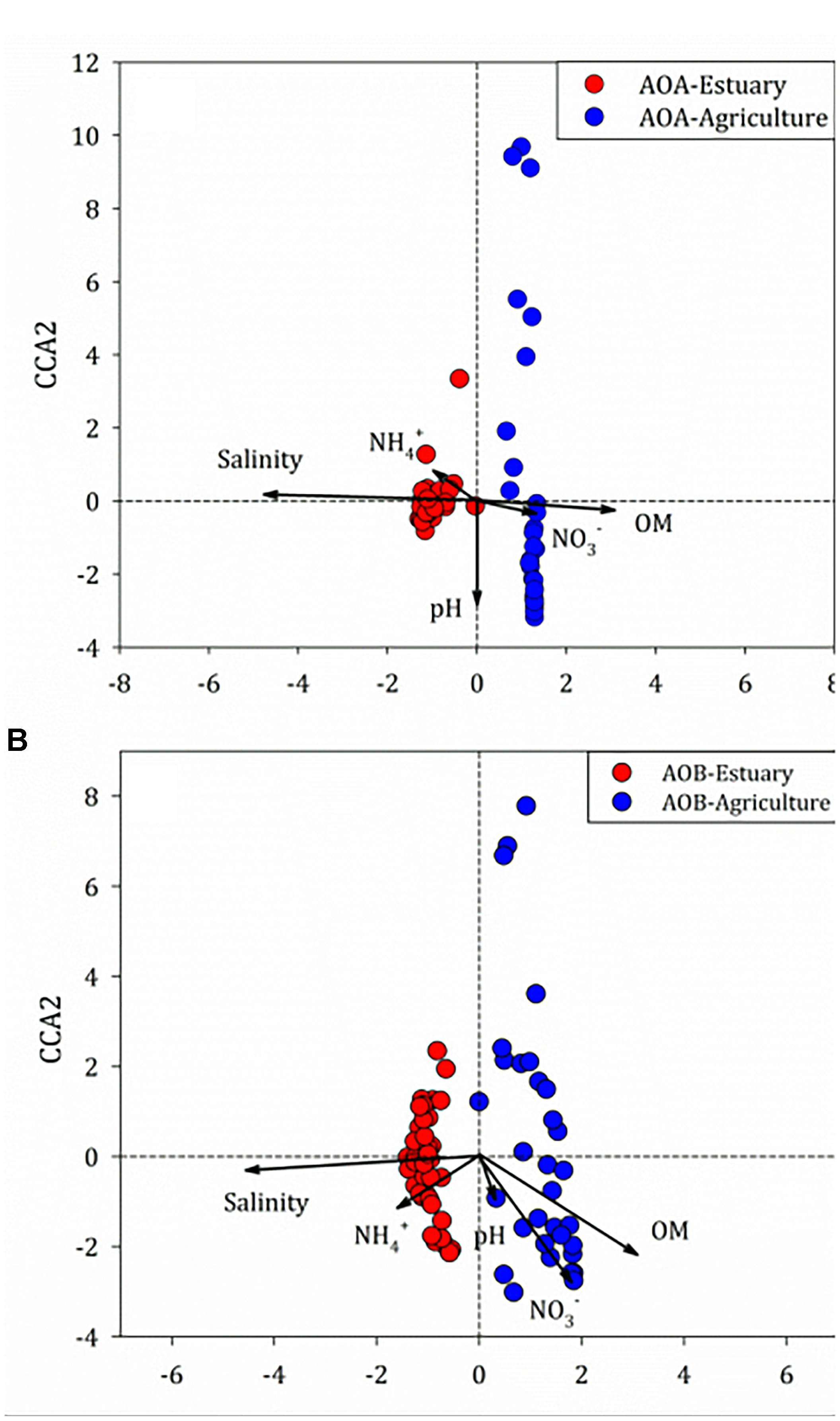
Figure 8. Canonical correspondence analysis of environmental variables and (A) ammonia-oxidizing archaea (AOA) and (B) ammonia-oxidizing bacteria (AOB) 16S rRNA genes among the mudflat and agricultural sites.
Agricultural practice increased the soil potential nitrification rates. The nitrification potential rates were considerably higher in the agricultural soils than those at the mudflat sites, except for sites E8 and E9, suggesting the higher organic matter concentration and lower salinity might both contribute to the increase of potential nitrification rates in agricultural soils. This is in agreement with publications reported previously, in which salinity and organic matter were important in controlling the rates of nitrification (Jiang et al., 2007; Zhou et al., 2015) as the nitrification rates generally decreased with the increasing salinity and external nitrogen addition stimulated nitrification (Offre et al., 2009; Gubry-Rangin et al., 2010; Zhang et al., 2010; He et al., 2012) by regulating the nitrifying community (Bernhard and Bollmann, 2010). Spatial differences in the nitrification potential rates were also observed in mudflat sites. The nitrification rates at the low-salinity sites (E7–E12) were significantly higher than those at high-salinity sites (E1–E6), further confirming the negative effects of salinity on nitrification rates.
It is important to note that the nitrification potential rates were determined with samples that were frozen at -20°C after thawing. Freezing/thawing will change the community structure of active microbes and activity potentials due to the disruption of cells. Soil freezing has been suggested to destroy microbes physically, releasing nutrients from the destroyed cells (Schimel and Clein, 1996; Sharma et al., 2006). The thaw-related flush of nutrients increases in activity and biomass of those microbes that survive such soil freeze (Skogland et al., 1988). However, the response pattern of AOA and AOB to thaw still evaded understanding. The amoA gene abundance significantly increased from -4°C to 5°C (Sophie et al., 2012), while quantities of amoA gene expression did not significantly change over the spring thaw (Németh et al., 2014). Thus, whether freezing/thawing soil accurately reflects soil nitrification potential rates still requires further research.
We observed stronger phylogenetic clustering in the AOA community of agricultural sites compared with the AOA community in mudflat sites (Figure 7). Although MNTD analysis also demonstrated the phylogenetic clustering effects in AOB communities of all the sites, no difference of the effects was observed between mudflat and agricultural sites, suggesting agricultural practice had stronger effects on the AOA community structure than the AOB.
We also found that the ammonia oxidizers’ community of both agricultural and mudflat sites were more strongly influenced by stochastic factors such as ecological drift, mutation, and random growth and deaths (Kembel, 2009; Fine and Kembel, 2011; Stegen et al., 2012) rather than by deterministic processes. These findings are consistent with publications reported previously, in which deterministic influences were more likely to dominate at large scales, and stochasticity trended to dominate at local scale (Shi et al., 2018).
Much of what we know about marine AOA has come from studies in open ocean environments, where AOA far outnumber AOB (Wuchter et al., 2006; Mincer et al., 2007; Beman et al., 2008), suggesting the high tolerance of AOA to salinity pressures (Santoro et al., 2008; Bernhard et al., 2010). In the mudflat, the dominant cluster was Group 1.1a and was highly related to Nitrosopumilus maritimus AOA, which has been isolated from marine environments (Konneke et al., 2005) and has a much higher tolerance of high salinity than other AOA. Based on the abundance of archaeal and bacterial amoA genes and AOA/AOB ratios, our study showed that the population size of AOA remained stable along the changing salinity gradient of mudflat sites. On the contrary, the abundance of AOB amoA gene were much higher in the high-salinity mudflat sites than those in low-salinity mudflat sites, which is consistent with previous results showing that AOB amoA gene abundances are greater and increase with salinity (Mosier and Francis, 2008; Santoro et al., 2008; Zhang et al., 2018), suggesting the potentially dominant contribution of AOB in mudflats. A recent study also established that the AOB community was first regulated by salinity (Zhang et al., 2018). In addition, the large shifts and contribution in the composition of the 16S rRNA and amoA clone libraries of AOB communities have been reported for the Schelde estuary in the Netherlands (De Bie et al., 2001; Bollmann and Laanbroek, 2002) and Waquoit Bay in the United States (Bernhard et al., 2005), indicating that salinity may affect the niche separation of AOB. However, no significantly distinct community structure of AOB was observed along the mudflat in this study. Sequences relating to Nitrosospira spp. were dominant at the mudflat sites ranging from low salinity to high salinity. Publications previously reported a selection for Nitrosospira spp. with increasing salinity in estuarine systems (Bernhard et al., 2007; Sahan and Muyzer, 2008). However, no statistical correlation was observed between nitrification rates and AOB population size, which might be attributed to the uncoupling between specific activity and the abundance of ammonia oxidizers.
Although agricultural practice in neutral soils has been widely reported with AOB activity, as AOB rather than AOA dominate oxidation of added ammonium (Xia et al., 2011; Prosser and Nicol, 2012), AOA outnumbered AOB amoA genes in the agricultural soils of this study, which might be attributed to the accumulation of organic matter rather than ammonia content. In fact, the significant growth of thaumarchaeota was also observed by root exudation (Chen et al., 2008) and mature slurry (Zhou et al., 2015), but less affected by the addition of inorganic fertilizers (Lehtovirta-Morley et al., 2011), indicating the importance of organic matter in the AOA growth. In contrast, AOB cannot grow on organic compounds (Krümmel and Harms, 1982). The dominant OTUs in the AOA guild for the agricultural sites were related to the 54D9 and 29i4 clusters, which have been observed to be the predominant AOA community in agricultural systems with high organic content (Bernhard and Bollmann, 2010; Zhou et al., 2015). The proposed mechanisms include that (1) mineralized ammonia was the major substrate source for thaumarchaeal nitrification processes (Stopnišek et al., 2010), (2) thaumarchaea had a mixotrophic lifestyle for assimilation of organic matters (Levičnik-Höfferle et al., 2011). The potentially hetetrophic pathway of AOA was also consistent with the positive correlation between OM and nitrification rates in agricultural sites.
In summary, both potential nitrification rates and AOA abundance were higher in the agricultural soils than those in mudflat, indicating the importance of 29i4 and 54D9 AOA in agricultural soils, while AOB was dominant in mudflats. Stronger phylogenetic clustering was only observed in the AOA community under agricultural practices, suggesting the importance of AOA in agricultural soils. Our study also revealed that stochastic processes dominated in AOA and AOB communities in both mudflat and agricultural sites, suggesting stochastic processes played an important role at the small scale (22 km) of our study. Both organic matter and salinity contributed to the distinct community compositions of the AOA and AOB. These results expand our understanding of ammonia oxidizers’ biogeographic distribution in both mudflat and agricultural soils of estuarine ecosystems. To date, recent discovery of complete ammonia oxidizer (comammox) organisms, i.e., bacteria that completely oxidize ammonia to nitrate (Daims et al., 2016), within the genus Nitrospira has led to a dramatic shift in the current model of nitrification. The niche differentiation of comammox in the mudflat is worth being explored further. We should also notice that environmental parameters not measured in the study, such as oxygen concentration and metal ion concentration, are also likely to influence the community structure.
Our study assessed the niche differentiation of ammonia oxidizers and potential nitrification rates in estuarine ecosystems in both mudflat and adjacent agricultural soils.
XZ and CL designed the experiments. XZ, BL, and ZG carried out the experiments and performed the analyses. XZ, CL, ZW, and JL substantially contributed to the interpretation of the results and writing of the paper.
CL acknowledges financial support from the National Natural Science Foundation of China (51679067) and the 111 project (B17015). ZW acknowledges financial support from the National Key Research Project (2017YFC0404501). XZ acknowledges financial support of the China Postdoctoral Science Foundation (2017M611683) and National Natural Science Foundation of China (Grant No. 41807029).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03122/full#supplementary-material
Beman, J. M., Popp, B. N., and Francis, C. A. (2008). Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2, 429–441. doi: 10.1038/ismej.2007.118
Bernhard, A. E., and Bollmann, A. (2010). Estuarine nitrifiers: new palyers, patterns and processes. Estuarine Coastal Shelf Sci. 88, 1–11. doi: 10.1016/j.ecss.2010.01.023
Bernhard, A. E., Donn, T., Giblin, A. E., and Stahl, D. A. (2005). Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7, 1289–1297. doi: 10.1111/j.1462-2920.2005.00808.x
Bernhard, A. E., Landry, Z. C., Blevins, A., de la Torre, J. R., Giblin, A. E., and Stahl, D. A. (2010). Abundance of ammonia-oxidizing archaea and bacteria along an estuarine salinity gradient in relation to potential nitrification rates. Appl. Environ. Microbiol. 76:1285. doi: 10.1128/AEM.02018-09
Bernhard, A. E., Tucker, J., Giblin, A. E., and Stahl, D. A. (2007). Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ. Microbiol. 9, 1439–1447. doi: 10.1111/j.1462-2920.2007.01260.x
Bollmann, A., and Laanbroek, H. J. (2002). Influence of oxygen partial pressure and salinity on the community composition of ammonia-oxidizing bacteria in the Schelde estuary. Aqu. Microb. Ecol. 28, 239–247. doi: 10.3354/ame028239
Busca, G., and Pistarino, C. (2003). Abatement of ammonia and amines from waste gases: a summary. J. Loss Prev. Proc. Ind. 16, 157–163. doi: 10.1016/S0950-4230(02)00093-1
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, X.-P., Zhu, Y.-G., Xia, Y., Shen, J.-P., and He, J.-Z. (2008). Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10, 1978–1987. doi: 10.1111/j.1462-2920.2008.01613.x
Chung, Y. C., Huang, C., and Tseng, C. (1996). Reduction of H2S/NH3 production from pig feces by controlling environmental conditions. Environ. Lett. 31, 139–155.
Clarke, K., and Ainsworth, M. (1993). A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. 92, 205–219. doi: 10.1016/j.biortech.2014.12.081
Daims, H., Lücker, S., and Wagner, M. (2016). A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712. doi: 10.1016/j.tim.2016.05.004
Damashek, J., and Francis, C. A. (2017). Microbial nitrogen cycling in estuaries: from genes to ecosystem processes. Estuaries Coasts 41, 1–35.
Damashek, J., Smith, J. M., Mosier, A. C., and Francis, C. A. (2015). Benthic ammonia oxidizers differ in community structure and biogeochemical potential across a riverine delta. Front. Microbiol. 5:743. doi: 10.3389/fmicb.2014.00743
De Bie, M. J. M., Speksnijder, A. G. C. L., Kowalchuk, G. A., Schuurman, T., Zwart, G., Stephen, J. R., et al. (2001). Shifts in the dominant populations of ammonia-oxidizing Ã y-subclass proteobacteria along the eutrophic Schelde estuary. Aquatic Microb. Ecol. 23, 225–236. doi: 10.3354/ame023225
y-subclass proteobacteria along the eutrophic Schelde estuary. Aquatic Microb. Ecol. 23, 225–236. doi: 10.3354/ame023225
Di, H., Cameron, K., Shen, J. P., Winefield, C., O’Callaghan, M., Bowatte, S., et al. (2009). Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat. Geosci. 2, 621–624. doi: 10.1038/ngeo613
Di, H. J., Cameron, K. C., Shen, J.-P., Winefield, C. S., O’Callaghan, M., Bowatte, S., et al. (2010). Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol. Ecol. 72, 386–394. doi: 10.1111/j.1574-6941.2010.00861.x
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Fine, P. V. A., and Kembel, S. W. (2011). Phylogenetic community structure and phylogenetic turnover across space and edaphic gradients in western Amazonian tree communities. Ecography 34, 552–565. doi: 10.1111/j.1600-0587.2010.06548.x
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102, 14683–14688. doi: 10.1073/pnas.0506625102
Gubry-Rangin, C., Nicol, G. W., and Prosser, J. I. (2010). Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol. Ecol. 74, 566–574. doi: 10.1111/j.1574-6941.2010.00971.x
Hardy, O. J. (2008). Testing the spatial phylogenetic structure of local communities: statistical performances of different null models and test statistics on a locally neutral community. J. Ecol. 96, 914–926. doi: 10.1111/j.1365-2745.2008.01421.x
He, J.-Z., Hu, H.-W., and Zhang, L.-M. (2012). Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol. Biochem. 55, 146–154. doi: 10.1016/j.soilbio.2012.06.006
Jia, Z., and Conrad, R. (2009). Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11, 1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x
Jiang, H., Dong, H., Zhang, G., Yu, B., Chapman, L. R., and Fields, M. W. (2007). Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl. Environ. Microbiol. 72, 3832–3845. doi: 10.1128/AEM.02869-05
Jin, T., Zhang, T., Ye, L., Lee, O. O., Yue, H. W., and Qian, P. Y. (2011). Diversity and quantity of ammonia-oxidizing archaea and bacteria in sediment of the pearl river estuary. China. Appl. Microbiol. Biotechnol. 90, 1137–1145. doi: 10.1007/s00253-011-3107-8
Kembel, S. W. (2009). Disentangling niche and neutral influences on community assembly: assessing the performance of community phylogenetic structure tests. Ecol. Lett. 12, 949–960. doi: 10.1111/j.1461-0248.2009.01354.x
Kim, J. H., Rene, E. R., and Park, H. S. (2007). Performance of an immobilized cell biofilter for ammonia removal from contaminated air stream. Chemosphere 2007, 274–280. doi: 10.1016/j.chemosphere.2006.12.100
Konneke, M., Bernhard, A., de la Torre, J., Walker, C., Waterbury, J., and Stahl, D. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Kowalchuk, G. A., and Stephen, J. R. (2001). Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55, 485–529. doi: 10.1146/annurev.micro.55.1.485
Krümmel, A., and Harms, H. (1982). Effect of organic matter on growth and cell yield of ammonia-oxidizing bacteria. Arch. Microbiol. 133, 50–54. doi: 10.1007/BF00943769
Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004, 150–163. doi: 10.1093/bib/5.2.150
Lehtovirta-Morley, L. E., Stoecker, K., Vilcinskas, A., Prosser, J. I., and Nicol, G. W. (2011). Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. 108, 15892–15897. doi: 10.1073/pnas.1107196108
Leininger, S., Urich, T., Schloter, M., Schwark, L., Qi, J., Nicol, G., et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809. doi: 10.1038/nature04983
Levičnik-Höfferle,Š, Nicol, G. W., Ausec, L., Mandić-Mulec, I., and Prosser, J. I. (2011). Stimulation of thaumarchaeal ammonia oxidation by ammonia derived from organic nitrogen but not added inorganic nitrogen. FEMS Microbiol. Ecol. 80, 114–123. doi: 10.1111/j.1574-6941.2011.01275.x
Likens, G. E., and Bormann, F. H. (1974). Linkages between terrestrial and aquatic ecosystems. Bioscience 24, 447–456. doi: 10.2307/1296852
Mago, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mincer, T. J., Church, M. J., Taylor, L. T., Preston, C., Karl, D. M., and Delong, E. F. (2007). Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9, 1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x
Moin, N. S., Nelson, K. A., Bush, A., and Bernhard, A. E. (2009). Distribution and diversity of archaeal and bacterial ammonia oxidizers in salt marsh sediments. Appl. Environ. Microbiol. 75, 7461–7468. doi: 10.1128/AEM.01001-09
Mosier, A. C., and Francis, C. A. (2008). Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002. doi: 10.1111/j.1462-2920.2008.01764.x
Németh, D. D., Wagner-Riddle, C., and Dunfield, K. E. (2014). Abundance and gene expression in nitrifier and denitrifier communities associated with a field scale spring thaw N 2 O flux event. Soil Biol. Biochem. 73, 1–9. doi: 10.1016/j.soilbio.2014.02.007
Offre, P., Prosser, J. I., and Nicol, G. W. (2009). Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol. Ecol. 70, 99–108. doi: 10.1111/j.1574-6941.2009.00725.x
Persson, T., and Wirén, A. (1995). Nitrogen mineralization and potential nitrification at different depths in acid forest soils. Plant Soil 16, 55–65. doi: 10.1007/BF00029313
Prosser, J. I., and Nicol, G. W. (2012). Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 20, 523–531. doi: 10.1016/j.tim.2012.08.001
Purcell, D., Sompong, U., Yim, L. C., Barraclough, T. G., Peerapornpisal, Y., and Pointing, S. B. (2007). The effects of temperature, pH and sulphide on the community structure of hyperthermophilic streamers in hot springs of northern Thailand. FEMS Microbiol. Ecol. 60, 456–466. doi: 10.1111/j.1574-6941.2007.00302.x
Quince, C., Lanzen, A., Davenport, R. J., and Turnbaugh, P. J. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. doi: 10.1186/1471-2105-12-38
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712.
Sahan, E., and Muyzer, G. (2008). Diversity and spatio-temporal distribution of ammonia-oxidizing Archaea and Bacteria in sediments of the Westerschelde estuary. FEMS Microbiol. Ecol. 64, 175. doi: 10.1111/j.1574-6941.2008.00462.x
Santoro, A. E., Francis, C. A., De Sieyes, N. R., and Boehm, A. B. (2008). Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 10, 1068–1079. doi: 10.1111/j.1462-2920.2007.01547.x
Schimel, J. P., and Clein, J. S. (1996). Microbial response to freeze-thaw cycles in tundra and taiga soils. Soil Biol. Biochem. 28, 1061–1066. doi: 10.1016/0038-0717(96)00083-1
Sharma, S., Szele, Z., Schilling, R., Munch, J. C., and Schloter, M. (2006). Influence of freeze-thaw stress on the structure and function of microbial communities and denitrifying populations in soil. Appl. Environ. Microbiol. 72, 2148–2154. doi: 10.1128/AEM.72.3.2148-2154.2006
Shi, Y., Li, Y., Xiang, X., Sun, R., Yang, T., He, D., et al. (2018). Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome 6:27. doi: 10.1186/s40168-018-0409-4
Skogland, T., Lomeland, S., and Goksøyr, J. (1988). Respiratory burst after freezing and thawing of soil: experiments with soil bacteria. Soil Biol. Biochem. 20, 851–856. doi: 10.1016/0038-0717(88)90092-2
Sophie, W., Claudia, G., Zebarth, B. J., Burton, D. L., Enrico, T., Chantigny, M. H., et al. (2012). Effects of temperatures near the freezing point on N2O emissions, denitrification and on the abundance and structure of nitrifying and denitrifying soil communities. FEMS Microbiol. Ecol. 83, 242–254. doi: 10.1111/j.1574-6941.2012.01468.x
Stegen, J. C., Lin, X., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Stopnišek, N., Gubry-Rangin, C., Höfferle,Š., Nicol, G. W., Mandič-Mulec, I., and Prosser, J. I. (2010). Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl. Environ. Microbiol. 76, 7626–7634. doi: 10.1128/AEM.00595-10
Turner, S., Pryer, K. M., Miao, V. P., and Palmer, J. D. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 46, 327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x
Walkley, A. J., and Black, I. A. (1934). An examination of the degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 37, 29–38. doi: 10.1097/00010694-193401000-00003
Wang, B., Zhao, J., Guo, Z., Ma, J., Xu, H., and Jia, Z. (2014). Differential contributions of ammonia oxidizers and nitrite oxidizers to nitrification in four paddy soils. ISME J. 9, 1062–1075. doi: 10.1038/ismej.2014.194
Wang, W. Q., Zeng, C. S., Zhong, C. Q., and Tong, C. (2010). Effect of human disturbance on ecological stoichiometry characteristics of soil carbon, nitrogen and phosphorus in Minjiang River estuarine wetland. Huan Jing Ke Xue 31, 2411–2416.
Webb, C. O. (2000). Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 156, 145–155. doi: 10.1086/303378
Wuchter, C., Abbas, B., Coolen, M. J. L., Herfort, L., Bleijswijk, J. V., Timmers, P., et al. (2006). Archaeal nitrification in the ocean. Proc. Nat. Acad. Sci. U.S.A. 103, 12317–12322. doi: 10.1073/pnas.0600756103
Xia, W., Zhang, C., Zeng, X., Feng, Y., Weng, J., Lin, X., et al. (2011). Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 5, 1226–1236. doi: 10.1038/ismej.2011.5
Xiang, X., Gibbons, S. M., He, J.-S., Wang, C., He, D., Li, Q., et al. (2016). Rapid response of arbuscular mycorrhizal fungal communities to short-term fertilization in an alpine grassland on the Qinghai-Tibet Plateau. PeerJ 4:e2226. doi: 10.7717/peerj.2226
Yao, Y. G., Graeme, W., Nicol, Colin, D., Campbell, C. D., Prosser, J. I., et al. (2011). Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl. Environ. Microbiol. 77, 4618–4625. doi: 10.1128/AEM.00136-11
Zhang, L. M., Duff, A. M., and Smith, C. J. (2018). Community and functional shifts in ammonia oxidizers across terrestrial and marine (soil/sediment) boundaries in two coastal Bay ecosystems. Environ. Microbiol. 20, 2834–2853. doi: 10.1111/1462-2920.14238
Zhang, L.-M., Offre, P. R., He, J.-Z., Verhamme, D. T., Nicol, G. W., and Prosser, J. I. (2010). Autotrophic ammonia oxidation by soil thaumarchaea. Proc. Natl. Acad. Sci. 107, 17240–17245. doi: 10.1073/pnas.1004947107
Keywords: agricultural soil, mudflat, ammonia-oxidizing archaea, ammonia-oxidizing bacteria, agricultural practice, MNTD
Citation: Zhou X, Li B, Guo Z, Wang Z, Luo J and Lu C (2018) Niche Separation of Ammonia Oxidizers in Mudflat and Agricultural Soils Along the Yangtze River, China. Front. Microbiol. 9:3122. doi: 10.3389/fmicb.2018.03122
Received: 13 August 2018; Accepted: 03 December 2018;
Published: 18 December 2018.
Edited by:
Andreas Ulrich, Leibniz-Zentrum für Agrarlandschaftsforschung (ZALF), GermanyReviewed by:
Marcus A. Horn, Leibniz University Hannover, GermanyCopyright © 2018 Zhou, Li, Guo, Wang, Luo and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue Zhou, eHVlemhvdXNub3d5QGhvdG1haWwuY29t Chunhui Lu, Y2x1QGhodS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.