
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 12 December 2018
Sec. Infectious Agents and Disease
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.03101
This article is part of the Research Topic Novel Antichlamydial Treatment Strategies and Resistance Mechanisms View all 15 articles
The Chlamydiaceae comprise a group of highly adapted bacterial pathogens sharing a unique intracellular lifestyle. Three Chlamydia species are pathogenic to humans: Chlamydia trachomatis, Chlamydia pneumoniae, and Chlamydia psittaci. C. trachomatis is the leading bacterial cause of sexually-transmitted infections and infectious blindness worldwide. Chlamydia pneumoniae is a major cause of community-acquired atypical pneumonia. C. psittaci primarily affects psittacine birds and can be transmitted to humans causing psittacosis, a potentially fatal form of pneumonia. As opposed to other bacterial pathogens, the spread of clinically relevant antimicrobial resistance genes does not seem to be a major problem for the treatment of Chlamydia infections. However, when exposed to stressing conditions, like those arising from exposure to antimicrobial stimuli, these bacteria undergo a temporary interruption in their replication cycle and enter a viable but non-cultivable state known as persistence. When the stressing conditions are removed, Chlamydia resumes replication and generation of infectious particles. This review gives an overview of the different survival strategies used by Chlamydia to evade the deleterious effects of penicillin and IFNγ, with a focus on the different models used to study Chlamydia persistence, their contribution to elucidating the molecular basis of this complex phenomenon and their potential implications for studies in animal models of infection.
The Chlamydiaceae are a family of Gram-negative obligate intracellular bacteria comprising 11 species pathogenic for a variety of animals (Elwell et al., 2016), 3 of which are human pathogens: Chlamydia trachomatis, Chlamydia pneumoniae, and C. psitacci. The main impact on human health is caused by C. trachomatis, which leads to a variety of oculo-genital and perinatal infections. Based on the antigenic properties of the major outer membrane protein MOMP, C. trachomatis strains can be classified into different serovars (Stephens et al., 1987; Baehr et al., 1988; Gomes et al., 2007). Serovars A-C are the etiologic agents of trachoma, the leading cause of infectious blindness worldwide (Stocks et al., 2014). With an estimate of 131 million new cases per year, C. trachomatis serovars D-K are the main bacterial cause of sexually-transmitted infections (STI) globally (Newman et al., 2015). Notably, up to 70–90% of these infections may be asymptomatic and long-lasting (Stamm, 1999; Gonzales et al., 2004), leading to serious complications such as pelvic inflammatory disease, ectopic pregnancy, and infertility in women (Lan et al., 1995; Westrom, 1995; Haggerty et al., 2010). C. trachomatis serovars D-K also cause inclusion conjunctivitis in adults, and perinatal infections such us ophthalmia neonatorum and chlamydial pneumonia in infants (Rönnerstam et al., 1985; Schachter et al., 1986; Stenberg and Mardh, 1991; Darville, 2005; Hammerschlag, 2011). The more invasive serovars L1-L3 are the cause of a less frequent form of STI called lymphogranuloma venereum, a systemic illness characterized by inguinal lymphadenopathy and/or severe proctitis/proctocolitis (Herring and Richens, 2006; White, 2009). C. pneumoniae is a widespread pathogen of the human respiratory tract, causing bronchitis, community-acquired atypical pneumonia and asthma exacerbation (Grayston et al., 1993; Asner et al., 2014). C. psitacci is the etiologic cause of psittacosis, a zoonotic disease that can be transmitted to humans upon close contact with a variety of birds, most frequently Psittacidae (cockatoos, parrots, parakeets and lories) or Columbiformes (pigeons) and recently, exposure to equine placental material has also been found as a risk factor for transmission (Polkinghorne and Greub, 2017). Clinical symptoms of psittacosis include high fever, chills, headache, myalgia, non-productive coughing, respiratory distress and may be fatal if untreated (Beeckman and Vanrompay, 2009).
All members of the Chlamydiaceae are highly successful intracellular parasites that undergo a propagation cycle involving developmental forms with clearly different morphological and functional properties. The elementary bodies (EBs) are very small (ca. 0.2 μm in diameter), infectious, environmentally resistant and non-replicative. The reticulate bodies (RBs), on the other hand, are larger (ca. 0.8 μm in diameter), non-infectious, labile in the extracellular environment and actively replicative. EBs are pre-loaded with virulence factors including type III secretion effectors involved in internalization into the host cell, while RBs are enriched in proteins involved in nutrient uptake, ATP generation and translation (Saka et al., 2011). Upon attachment to epithelial cells, EBs are internalized and confined into a vacuole, termed an inclusion. At early times post-infection, EBs differentiate into RBs, which start replicating as the inclusion gradually expands. During this process, Chlamydia manipulates host cell pathways to acquire essential nutrients and avoid interaction with degradative organelles like lysosomes. At mid-cycle, some RBs begin to differentiate back into EBs asynchronously. At late stages post-infection the inclusion contains mostly EBs, which are finally released to the extracellular environment either by host cell lysis or extrusion of inclusions. In the extracellular environment, EBs can infect neighboring cells and continue propagating (Saka and Valdivia, 2010; Elwell et al., 2016). If during replication Chlamydia encounter non bacteriocidal stress conditions, like those arising from exposure to certain antibiotics, cytokines or nutrient deprivation, the bacteria respond by markedly reducing RBs division and production of infectious particles, being able to rapidly resume to normal replication once the stressing conditions are removed (Figure 1A). This response is referred to as chlamydial persistence or chlamydial stress response, as it allows these bacteria to survive for long periods of time in cell culture in presence of unfavorable growth conditions (Schoborg, 2011; Byron, 2012; Elwell et al., 2016).
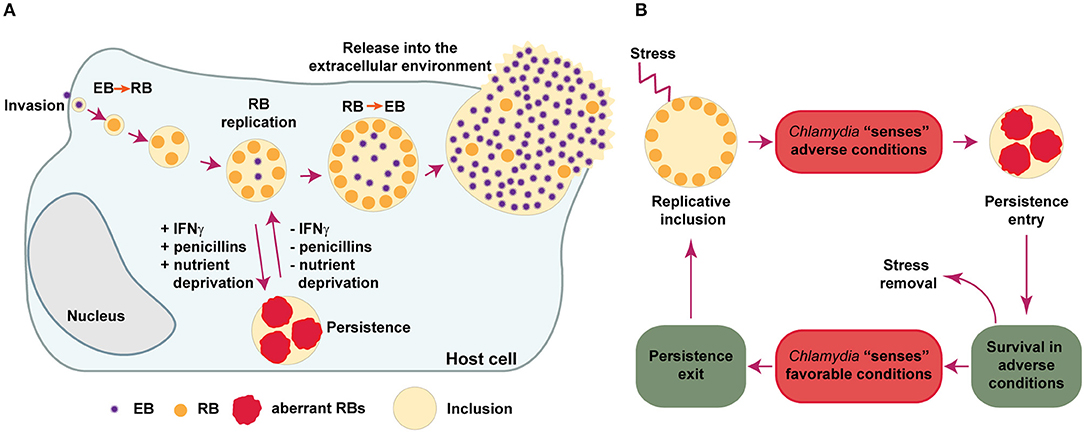
Figure 1. Chlamydia developmental cycle. (A) Chlamydia are obligate intracellular bacteria that undergo multiple developmental forms with distinct morphological and functional properties. The infectious elementary bodies (EB) are internalized in the host cells and are confined to a membrane-bound vacuole, termed an “inclusion.” Soon after invasion, EBs differentiate into replicative but non-infectious reticulate bodies (RBs), which actively divide. Around mid-cycle, RBs begin to asynchronously differentiate back into EBs and are finally released (by cell lysis or extrusion of intact inclusions) into the extracellular environment, where they can infect neighboring cells. If during replication Chlamydia is exposed to stressing conditions, like those caused by gamma-interferon (IFNγ), penicillins or deprivation of essential nutrients, the bacteria enter into a long-lasting, viable but non-cultivable state known as “Chlamydia persistence,” which is typically associated to the presence of enlarged, aberrant RBs. When conditions are again favorable, the persistence state is reversed and normal replication ensues. (B) This scheme summarizes different events that Chlamydia may use in order to successfully evade antimicrobial effects triggered during infection in-vitro. First, Chlamydia “senses” different types of stresses and then respond by entering into a temporary, reversible interruption in the replication cycle (termed “Chlamydia persistence” or “Chlamydia stress response”). In this state, Chlamydia adapts to adverse conditions by prioritizing cell functions required for long-lasting survival. When the bacteria sense that the stressing condition has ceased, they exit from persistence and resume normal replication and generation of infectious progeny. The Chlamydia factors and the molecular mechanisms required for the successful execution of each one of these steps remain poorly elucidated.
As opposed to other bacterial pathogens, for which acquisition of resistance genes by lateral gene transfer confer resistance to antimicrobial drugs and represent a major concern for public health (Chatterjee et al., 2018), Chlamydia have so far remained susceptible to all anti-chlamydial antibiotics. An exception to this widespread susceptibility is the well-characterized tetracycline resistance island in the pig pathogen Chlamydia suis, which so far has not spread to Chlamydia species affecting humans (Seth-Smith et al., 2017). In fact, there are no reports of naturally-acquired, stable antibiotic resistance in Chlamydia strains recovered from human samples (Sandoz and Rockey, 2010). Interestingly, despite wide availability of effective drugs and apparent lack of antibiotic resistance mechanisms, Chlamydia continue to be widespread pathogens, notorious for their ability to cause long-lasting, persistent infections (Byron, 2012). Perhaps as a result of their long evolutionary history as obligate intracellular parasites, Chlamydia seem to have evolved unique mechanisms to resist antimicrobial effects triggered by the host innate and adaptive immune responses. In this review, we will address the current knowledge about the strategies used by these microorganisms to resist antimicrobial stimuli triggered by penicillins and IFNγ in cell culture or in-vivo models of infection, with a focus in Chlamydia persistence.
The view that Chlamydia only alternates between EB and RB developmental forms is an oversimplified view of its life cycle. Already in 1950, Weiss discovered that Chlamydia muridarum and C. felis (then known as the murine and feline pneumonitis viruses, respectively), displayed an enlarged, abnormal morphology upon exposure to the antibiotic penicillin (Weiss, 1950). Similar observations were reported for C. trachomatis LGV and C. psittaci (by then named lymphogranuloma and meningopneumonitis viruses, respectively) (Hurst et al., 1953; Tamura and Manire, 1968; Matsumoto and Manire, 1970). A seminal report from Galasso and Manire (1961), showed that penicillin was able to reduce the generation of C. psittaci infectious progeny in HeLa cells to values as low as 0.1% of the original titer. Surprisingly, these investigators found that normal replication rates recovered quickly upon penicillin removal, even after 3 and a half months of continuous presence of the antibiotic in cell culture. Morphological analysis by transmission electron microscopy, demonstrated that in the presence of penicillin, C. psittaci inclusions contained greatly enlarged RBs, while normal morphology was recovered upon removal of the antibiotic (Matsumoto and Manire, 1970). We now know that those early observations are in line with the presence of enlarged, aberrant RBs (aRBs), typically found when Chlamydia undergo nutrient deprivation, exposure to certain antibiotics, INFγ or other compounds/conditions that impose a stress to these microorganisms (Byron, 2012). The aRBs observed in-vitro, may be considered the morphological manifestation of Chlamydia entry into a persistent state, characterized by lack of cultivability while conditions are unfavorable, and followed by a quick return to normal replication upon removal of the stressor. In this context, the persistent state may function as a resilience pathway triggered in Chlamydia to deal with stressing conditions elicited by antimicrobial stimuli. Thus, it is reasonable to imagine that in order to execute such response, these bacteria should follow a sequence of events involving: (i) sensing of unfavorable conditions, (ii) entry into a persistent state, (iii) survival in adverse conditions, (iv) sensing of favorable conditions, (iv) exit from persistence, and (v) resume to normal production of infectious progeny (Figure 1B). The mechanistic and molecular basis underlying each one of the mentioned events, remain poorly understood. However, we know that under the persistent state, Chlamydia slows down DNA replication and continues to transcribe genes, but stops dividing, becoming viable but non-cultivable (Ouellette et al., 2006; Muramatsu et al., 2016). This is frequently accompanied by the presence of enlarged aRB forms, which retain their ability to transition into infectious EBs once conditions are again favorable. It is important to mention, however, that a functional approach to the definition of Chlamydia persistence as a reversible interruption of the productive cycle (Byron, 2012), is probably more appropriate than a morphological definition, since aRBs are not always observed (Schoborg, 2011).
A variety of stimuli induce Chlamydia persistence including several antibiotics, gamma-interferon (IFNγ), deprivation of essential nutrients (i.e., iron, amino acids, glucose), heat-shock, components found on cigarette smoke, exposure to adenosine, infection with chlamydiaphage, co-infection with Herpes Simplex Virus or Porcine Epidemic Diarrhea Virus (Beatty et al., 1994b; Hsia et al., 2000; Wiedeman et al., 2005; Huston et al., 2008; Pettengill et al., 2009; Vanover et al., 2010; Schoborg, 2011; Prusty et al., 2012; Schoborg and Borel, 2014). The diversity of stimuli that can elicit persistence, suggests that Chlamydia does not respond equally to all of them. This notion is supported by several studies showing that the transcriptional responses triggered are significantly different according to the persistence model used (Mathews et al., 2001; Belland et al., 2003; Gérard et al., 2004; Goellner et al., 2006; Ouellette et al., 2006; Mäurer et al., 2007; Brinkworth et al., 2018). Among the different models of Chlamydia persistence, we will focus on the two that have been most thoroughly studied: the beta-lactam antibiotic penicillin and IFNγ, a cytokine that is critical for innate and adaptive immunity.
As a member of the beta-lactams family of antibiotics, penicillins exert their antimicrobial activity by blocking the cross-linking of the peptidoglycan, a key step in cell wall synthesis in bacteria. The general mechanism of action for beta-lactam antibiotics is the inactivation of penicillin binding proteins (PBPs) by targeting their transpeptidase domain. PBPs catalyze the last step in peptidoglycan synthesis, which is the cross-linking of pentapeptide sidechains by a transpeptidation reaction. Due to the structural similarity of beta-lactams with the terminal D-Ala-D-Ala of the pentapeptide precursor, which is the natural substrate of PBPs, these antibiotics form a stable acyl-enzyme covalent bond with PBPs abolishing their peptidoglycan cross-linking activity (Sauvage et al., 2008). As these cross-links are pivotal for conferring strength and rigidity to both, Gram-positive and Gram-negative bacterial cell walls, beta-lactams usually lead to bacteria lysis due to high internal osmotic pressures.
Exposure to penicillin in cell culture provided the first evidence for Chlamydia persistence and the first in-vitro models to study this phenomenon (reviewed in Beatty et al., 1994b). Under these conditions, EB to RB transition is not prevented but once in the RB stage, cell division is blocked, inclusions are small and populated by only a few enlarged RBs (aRBs) and there is a halt in RB to EB differentiation until the antibiotic is removed (Galasso and Manire, 1961; Matsumoto and Manire, 1970; Skilton et al., 2009). Another feature of penicillin-induced persistence is that the inhibition of cell division is accompanied by accumulation of 16s rRNA and only a partial reduction in replication of genomic or plasmidic DNA (Lambden et al., 2006; Ouellette et al., 2006; Skilton et al., 2009; Kintner et al., 2014). Remarkably, a detailed video-microscopy analysis of C. trachomatis development during penicillin-induced persistence showed that in the recovery phase, even though aRBs remain observable inside the inclusion, normal RBs are generated from them in an asynchronous manner through a budding-resembling process (Skilton et al., 2009).
A relatively recent study carried out a careful analysis of the effect caused by beta-lactams on C. trachomatis serovar E (Kintner et al., 2014). In this study, HeLa cells monolayers were infected with C. trachomatis exposed to a variety of beta-lactam antibiotics at physiologically relevant concentrations, including 6 different penicillins (penicillin G, penicillin V, amoxicillin, ampicillin, carbenicillin, and piperacillin). Addition of any of the penicillins tested at the time of infection, did not prevent inclusion formation even at concentrations 100 times higher than the physiological serum concentration. However, inclusions were small and contained aRBs. Also, Kintner et al. demonstrated that if penicillins are added at 24 h post-infection, when inclusions are already established, the formation of aRBs is induced and production of infectious progeny is reduced by 95%. Noticeably, beta-lactams not belonging to the penicillin group, like the monobactam aztreonam and the cephalosporins ceftriaxone or cefotaxime, did not seem to affect inclusion size, morphology or infectious progeny generation, indicating that not all beta-lactams trigger the same response (Kintner et al., 2014). The relevance of these observations is that they could mimic clinically important scenarios that occur when patients asymptomatically infected with Chlamydia, are prescribed with beta-lactams due to other concomitant infections.
Chlamydia encode peptidoglycan-synthesizing high molecular weight PBPs (PBP-2 and PBP-3) and produce peptidoglycan, indicative that they possess the natural target of beta-lactam antibiotics (Barbour et al., 1982; Ouellette et al., 2012; Jacquier et al., 2014; Packiam et al., 2015). Thus, inhibition of peptidoglycan production, leading to a disruption in cell wall synthesis, may be a stressing condition that triggers Chlamydia persistence in presence of these antibiotics.
The molecular basis of Chlamydia response to stress caused by exposure to penicillins is still poorly characterized (Figure 2A). According to a transcriptional analysis carried out by Dr. Byrne's group in C. pneumoniae during penicillin-induced persistence, the late genes omcB and hctB, involved in RB to EB differentiation, are not induced even at 48 h post-infection, strongly suggesting that in this condition the expression profile resembles that of the RB stage (Ouellette et al., 2006). Another work revealed that late gene transcription is also downregulated during penicillin-induced persistence in C. psittaci, together with upregulation of the stress response genes grpE and groES (Goellner et al., 2006). Additionally, the stress response protease/chaperone HtrA and its gene, which is conserved across Chlamydia species, have been reported to be increased during penicillin-induced persistence (Huston et al., 2008; Di Pietro et al., 2012). In line with this, treatment with the HtrA inhibitor JO146 severely impairs production of infectious progeny during recovery from penicillin-induced persistence in C. trachomatis, suggesting that this protease plays a role in the reversion process (Ong et al., 2013).
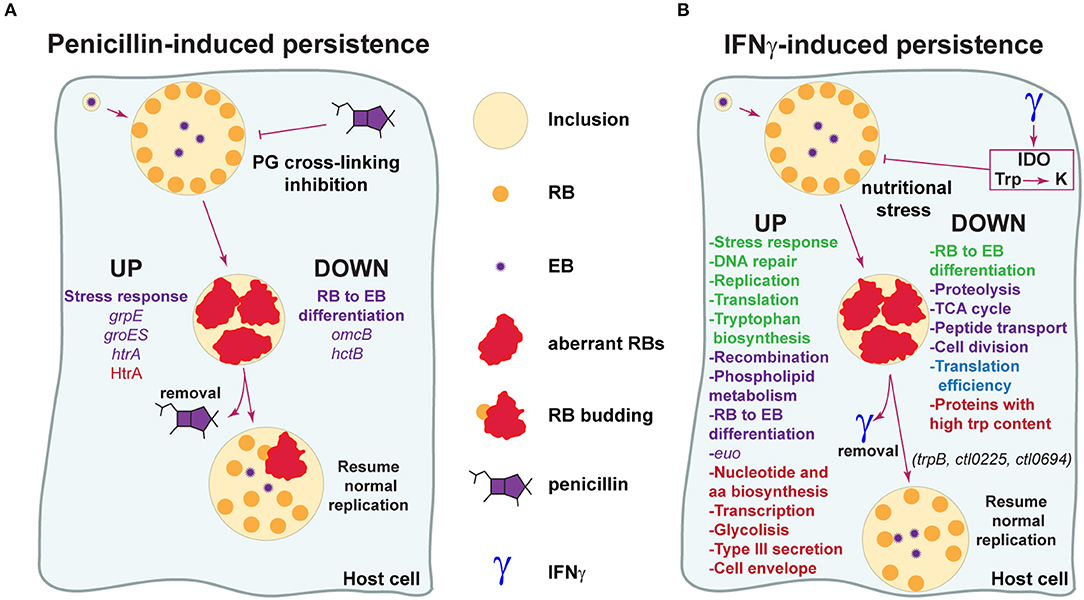
Figure 2. Model for the molecular basis of Chlamydia evasion of antimicrobial activity elicited by penicillins and IFNγ. (A) Penicillins inhibit crosslinking of the peptidoglycan (PG), leading to disruption of cell wall synthesis. Chlamydia respond to this stress by downregulating genes required for RB to EB transition (omcB, hctB), halting cell division and adopting an aberrant, enlarged morphology (aberrant RB) (Goellner et al., 2006; Ouellette et al., 2006). Genes associated to stress responses are upregulated (grpE, groES, htrA) (Goellner et al., 2006; Di Pietro et al., 2012). During this stage, the bacteria is able to “persist” for long periods of time in a viable but non-cultivable state and infectious progeny generation is abrogated. When penicillin is removed, Chlamydia exit persistence and resume generation of normal RBs, which may originate from aRBs through a budding-like process. The protease/chaperone protein HtrA may participate in reversion from penicillin-induced persistence (Huston et al., 2008; Ong et al., 2013). Reported processes/genes/proteins up- and down-regulated during penicillin-induced persistence are indicated. Data based on transcriptional or protein studies are highlighted in purple and red, respectively. (B) IFNγ is a cytokine with key roles in host defense against pathogens, including Chlamydia. IFNγ activates indoleamine-2,3-dioxygenase (IDO), causing the degradation of tryptophan (Trp) into kynurenine (K) and triggering nutritional stress in Chlamydia. In response to this stress, Chlamydia enters into a persistent state analogous to what is observed with penicillin. In this state, Chlamydia alters gene transcription and expression in order to modify key biological processes and warrant survival. Reported processes/genes/proteins up- and down-regulated during IFNγ-induced persistence are indicated. Data based on transcriptional studies (Byrne et al., 2001; Belland et al., 2003; Goellner et al., 2006; Ouellette et al., 2006) is indicated in purple. Data based on protein studies (Molestina et al., 2002; Mukhopadhyay et al., 2006; Ouellette et al., 2006; Østergaard et al., 2016) is shown in red. Data supported by both transcriptional and protein studies is indicated in green. Data based on global activity of the translation pathway is shown in blue. Note that RB to EB differentiation transcripts have been found increased and decreased in different studies, as discussed in the manuscript. trpB (tryptophan synthase), ctl0225 (a predicted small neutral amino acid transporter) and ctl0694 (a hypothetical oxidoreductase) have been found to play a role in reversion from IFNγ-induced persistence in C. trachomatis (Muramatsu et al., 2016).
A relevant question is: can observations emerged from in-vitro models of penicillin-induced persistence be extrapolated to events occurring in infected humans? Dr. Schoborg's group reported the first animal model to study Chlamydia persistence (Phillips Campbell et al., 2012). By infecting the female genital tract of amoxicillin-treated BALB/c mice with C. muridarum, these investigators made several significant findings: (i) C. muridarum enters a viable non-infectious state in the genital tract; (ii) accumulation of chlamydial pre-16s rRNA is not affected; (iii) aRBs are observed in the infected tissue and (iv) shedding of infectious particles peaks upon termination of amoxicillin treatment. These findings indicate that Chlamydia persistence, as defined in cell culture, can occur in-vivo upon amoxicillin treatment. Another relevant observation is that amoxicillin-induced persistence in mice infected intravaginally with C. muridarum, results in increased failure of subsequent treatment with the first choice anti-chlamydial antibiotic azithromycin (Phillips-Campbell et al., 2014). These observations are consistent with previous results obtained in cell culture infection models (Wyrick and Knight, 2004). Then, it seems a reasonable clinical consideration that penicillin-induced Chlamydia persistence may confer increased resistance to other antibiotics, including first choice anti-Chlamydia drugs.
IFNγ is a pleiotropic cytokine secreted primarily by T-lymphocytes and NK cells, with key roles in regulation of immune responses and in host defense against pathogens, including Chlamydia (McClarty et al., 2007; Billiau and Matthys, 2009). Initially identified as an antiviral factor (Wheelock, 1965), IFNγ exerts antimicrobial effects on Chlamydia (reviewed in Beatty et al., 1994b). Byrne et al. (1986) found that IFNγ restricted C. psittaci growth in human uroepithelial T24 cells and that this effect was due to increased catabolism of the essential amino acid tryptophan within the host cell. They proposed that depletion of intracellular pools of tryptophan was achieved by IFNγ-mediated increased activity of indoleamine-2,3-dioxygenase (IDO), a catabolic enzyme that degrades tryptophan into N-formylkynurenine and kynurenine. Indeed, increased IDO activity and tryptophan depletion are responsible for IFNγ-induced growth inhibition of C. trachomatis serovar B in human conjunctival epithelial cells (Rapoza et al., 1991). Soon after, a seminal paper from Beatty and collaborators discovered that in presence of IFNγ, C. trachomatis leads to a persistent infection “characterized by the development of noninfectious atypical chlamydial forms, from which infectious progeny could be recovered only when IFNγ was removed from the culture system” (Beatty et al., 1993). A follow-up study confirmed that IDO-mediated deprivation of tryptophan was responsible for IFNγ induction of a persistent C. trachomatis form in cell culture (Beatty et al., 1994a). Similar observations have been made in C. pneumoniae (Pantoja et al., 2001). All these findings are consistent with the idea that Chlamydia undergo persistence to evade the antimicrobial effects caused by IFNγ-mediated starvation of an amino acid for which these bacteria are auxotrophs. Independently of tryptophan starvation, antimicrobial effects triggered by IFNγ during Chlamydia infection include GTPases of the Immunity Related GTPases (IRG) and Guanylate Binding Proteins (GBP), which can restrict the replication of C. trachomatis in mouse and human cells, respectively (Bernstein-Hanley et al., 2006; Tietzel et al., 2009).
It is of great interest to understand the molecular basis of Chlamydia persistence in response to IFNγ (Figure 2B). Belland et al. cataloged C. trachomatis serovar D transcripts that were up- and down-regulated in an IFNγ-induced persistence and reactivation model of infection in HeLa cells (Belland et al., 2003). These investigators showed that in presence of IFNγ, Chlamydia genes involved in tryptophan biosynthesis, DNA repair and recombination, phospholipid metabolism, stress response, and protein translation were up-regulated. On the other hand, genes related to proteolysis, peptide transport, the TCA cycle, cell division and RB to EB differentiation were down-regulated. In addition, the early gene euo, which has been proposed as a repressor of late genes transcription (Rosario and Tan, 2012), was found highly upregulated during IFNγ-induced persistence both in C. trachomatis and C. pneumoniae (Belland et al., 2003; Ouellette et al., 2006). Notably, the transcriptome of C. trachomatis during IFNγ-induced persistence is in agreement with well-established properties attributed to aRBs: a halt in cell division and infectious progeny generation. Similarly, work from Byrne et al. revealed that genes involved in chromosome replication, repair and recombination are transcribed during IFNγ-induced persistence in C. pneumoniae-infected HEp2 cells, while genes related to cytokinesis are downregulated (Byrne et al., 2001). One transcriptional study reported that genes involved in RB to EB differentiation were upregulated in C. pneumoniae in presence of IFNγ (Ouellette et al., 2006). Even though this is not in full agreement with previous reports (Belland et al., 2003; Goellner et al., 2006), the apparent inconsistencies may be due to different normalization methods (i.e., 16s rRNA vs. gyrA vs. DNA content) used for transcription data analysis (Schoborg, 2011).
Because transcriptional data not always correlate with actual protein expression profiles, proteomic studies can be very useful tools to investigate the molecular basis of Chlamydia persistence. As shown in Figure 2B, early proteomic analysis carried out in C. pneumoniae during IFNγ-induced persistence, found increased levels of proteins involved in stress response, nucleotide and amino acid biosynthesis, DNA replication, transcription, translation, glycolysis, type III secretion, and cell envelope (Molestina et al., 2002; Mukhopadhyay et al., 2006). More, a recent paper used a label-free proteomics approach and carried out the first comprehensive comparison of C. trachomatis serovar D EBs, RBs and IFNγ-induced aRBs (Østergaard et al., 2016). Remarkably, this study uncovered that the proteome of aRBs was very similar to that of the RBs, except that they expressed very high levels of tryptophan synthase (subunits TrpA and TrpB), which can transform indol into tryptophan. This finding is in agreement with previously mentioned transcriptomic data (Belland et al., 2003). Interestingly, it has been proposed that tryptophan synthase, not functional in ocular serovars, could provide a salvage pathway against tryptophan depletion for C. trachomatis genital serovars, given their co-existence with indol-producing microbiota in the female genital tract (Caldwell et al., 2003; Aiyar et al., 2014). This is in line with evidence showing that TrpB from C. trachomatis genital serovars, is capable of utilizing indole for the biosynthesis of tryptophan (Fehlner-Gardiner et al., 2002). Indeed, a trpB null mutant of C. trachomatis serovar D can be fully recovered from IFNγ-induced persistence upon addition of tryptophan but not indole (Kari et al., 2011). Another notable finding of the mentioned proteomic study, was that aRBs expressed overall lower levels of proteins with high tryptophan content, reflecting at a proteome level, the struggle to resist tryptophan restriction imposed by IFNγ (Østergaard et al., 2016). This is in agreement with the observation that the Chlamydiaceae have undergone evolutionary selection for proteins with lower-than-average tryptophan content (Down-Trp selection), which may operate during the persistent state triggered by tryptophan starvation (Bonner et al., 2014). Ouellette and coworkers found that during IFNγ-induced tryptophan restriction, C. pneumoniae accumulates Trp-codon rich transcripts, presumably as a result of ribosome stalling on Trp-codons (Ouellette et al., 2016, 2018). This could partially explain a previous report showing that in response to tryptophan restriction, C. pneumoniae undergoes increased transcription but inefficient translation, pointing out to an apparent uncoupling of these two pathways (Ouellette et al., 2006).
Because Chlamydia have been historically refractory to genetic manipulation, the role of individual genes involved in persistence is ill-defined. Only recently, tools have become available for Chlamydia genetic analysis, though they are still limited compared to other bacteria (reviewed in Bastidas and Valdivia, 2016). To date, there is only one report of a systematic approach to elucidate the genetic basis of Chlamydia persistence (Muramatsu et al., 2016). Muramatsu et al. used a collection of ~2,000 GFP-labeled, chemically-mutagenized C. trachomatis LGV-L2 strains, to screen for mutants exhibiting a reduced ability to reactivate from IFNγ-induced stress. The screening consisted on infecting in parallel HeLa cells pre-treated or not for 24 h with 10 ng/mL, with each individual mutant. The untreated condition was fixed at 24 h post-infection, while de IFNγ-treated were instead washed off the cytokine and left to recover for additional 24 h in culture media supplemented with indole before fixation. By comparing the ratio of the number of inclusions formed in untreated/recovery for each mutant, 6 mutants sensitive to IFNγ-induced persistence were identified and for 3 of them the causative mutation was elucidated. These were missense mutations in TrpB (subunit of tryptophan synthase), CTL0225 (a predicted small neutral amino acid transporter), and CTL0694 (a hypothetical oxidoreductase). Interestingly, all of these mutants were able to enter into a persistent state in presence of IFNγ and failed to reactivate upon addition of tryptophan or indole, yet displayed different sensitivities to IFNγ (Muramatsu et al., 2016), highlighting the relative impact of the mutations on protein function and the potential involvement of different Chlamydia genes in persistence.
A general principle for the biomedical relevance of cell culture experiments, is that conditions should resemble the in-vivo situation as much as possible. In this context, a study investigated the influence of oxygen levels on IFNγ-induced persistence (Roth et al., 2010). Roth and collaborators reasoned that in the female genital tract, C. trachomatis must survive under hypoxia (oxygen concentrations ≤5%), while most of the in-vitro experiments are carried out in normoxia. According to this study, switching the oxygen levels from normoxia to hypoxia in human fallopian tube cells in-vitro and ex-vivo, correlated with a reduction in the anti-chlamydial activity of IFNγ against genital but not ocular serovars of C. trachomatis and with decreased levels of IDO under hypoxia (Roth et al., 2010). Another study from the same group later evaluated the activity of first-line anti-chlamydial antibiotics against C. trachomatis L2 in HeLa cells during IFNγ-induced persistence in hypoxia vs. normoxia (Shima et al., 2013), and observed that the efficacy of subinhibitory concentrations of azithromycin against C. trachomatis was moderately reduced under hypoxia. Considering that the cellular concentrations of oxygen were not measured and thus distinctions between anoxic and hypoxic states within the cells cannot be made, the physiological relevance of these observations is not clear. However, these studies highlight that experimental conditions used to investigate Chlamydia persistence in-vitro should be carefully considered, as they may have profound effects on the results, their interpretation and extrapolation for the in-vivo situation.
During infection of their hosts Chlamydia encounters stress imposed by immune responses or eventually by administration of antimicrobial agents. In light of the experimental findings discussed above, it is reasonable to speculate that Chlamydia can enter a persistent state in-vivo. However, the evidence is so far indirect. Evidence that persistence may occur in-vivo, comes from direct observation of aRBs in infected tissues. For instance, by means of immunogold electron microscopy, aRBs have been observed for C. pneumoniae in atherosclerotic tissue from human patients (Borel et al., 2008). Similarly, aRBs were detected for C. suis by immunohistochemistry and immunogold electron microscopy in the gut of naturally infected pigs (Pospischil et al., 2009). Also, C. muridarum aRBs were present in endocervical cells using a female genital tract model of infection in mice (Rank et al., 2011; Phillips Campbell et al., 2012). Moreover, Dr. Quayle group identified the presence of aRBs in endocervical cells obtained from a woman with C. trachomatis cervicitis (Lewis et al., 2014). While the observation of aRBs in infected tissues does not necessarily indicate a persistent infection, the widely demonstrated ability of Chlamydia to cause persistent, asymptomatic, chronic infections and apparent reactivations, is strongly suggestive. A clinical observation that further support the case for Chlamydia persistence in-vivo is post-gonococcal urethritis. A proportion of patients treated with penicillin for gonorrhoeae in the 60's and the 70's developed post-gonococcal urethritis caused by C. trachomatis (Richmond et al., 1972). It has been reported that C. trachomatis is recovered in 11–50% of patients with gonococcal urethritis, and 75–100% of these patients may develop post-gonococcal urethritis after treatment with an antibiotic ineffective for Chlamydia, like the beta-lactams penicillin or cephalosporins (Augenbraun and McCormack, 2015). While beta-lactams may succeed in clearance of Neisseria gonorrhoeae, they may only induce a persistent state in Chlamydia. Therefore, post-gonococcal urethritis could be conceived as the clinical manifestation of Chlamydia reactivation after completion of the beta-lactam regime in gonorrhoeae patients co-infected with C. trachomatis. Another indirect evidence of reactivation is the repeated observation of apparently healed immigrants from endemic areas undergoing active trachoma decades after, in absence of an identifiable re-exposure (Thygeson, 1963). Similarly, based on mice models of infection, C. pneumoniae can be reactivated in the lungs weeks after the initial inoculation upon treatment with cortisone, but not in immunocompetent animals, strongly suggesting that anti-Chlamydia immune responses trigger these bacteria to enter into a persistent state in the infected tissues (Malinverni et al., 1995; Laitinen et al., 1996). Also, the detection of Chlamydia antigens, DNA or RNA in clinical specimens in the absence of cultivability, is consistent with a persistent state occurring in-vivo (reviewed in Hogan et al., 2004). For instance, C. trachomatis antigens or DNA were identified in biopsy samples from culture-negative women with post-infectious tubal infertility, even after antibiotic treatment (Patton et al., 1994). In addition, C. trachomatis DNA and RNA have been detected weeks after cultures were already negative in a trachoma model of infection based on cynomolgus monkeys (Holland et al., 1992). However, it should be noted that detection of relatively stable molecules such as DNA or protein antigens does not necessarily indicate the presence of viable organisms and thus is not conclusive evidence of a persistent phenotype in-vivo. Importantly, a recent whole-genome sequence analysis from clinical samples/isolates presented compelling evidence that the same strain of C. trachomatis can persist for 3–5 years in the genital tract of women, regardless of regular antibiotic treatment and with none to few genomic mutations accumulated (Suchland et al., 2017). This indicates that Chlamydia's strategy to persist in the host cell is not mutational, but may instead rely on the orchestration of a specific response oriented to avoid pathogen elimination.
If persistence occurs in-vivo, a likely scenario is that a fraction of Chlamydia bacteria enters into a persistent state in the host in response to unfavorable growth conditions, like those arisen from antibiotic treatment or from IFNγ produced by immune cells in response to infection. Chlamydia persistence may then contribute to the establishment of chronic and minimally symptomatic infections that are considered critical for long term tissue damage and pathogenesis of chlamydial diseases.
The ability of Chlamydia to enter a persistent state in response to antimicrobials in cell culture has been studied for decades. This persistent state invariably includes a reversible interruption in the productive cycle, but the precise mechanism required to achieve this vary with the different inducers. Experimental models of Chlamydia persistence in cell culture are helpful only if they can be extrapolated to in-vivo infections. To date the in-vivo occurrence of a persistent state(s) in Chlamydia resembling that observed in cell culture is consistent with morphological and clinical observations. For activation of a persistence program in Chlamydia, the pathogen should undergo a series of events including: sensing the stress condition, activation of gene expression programs that enable entry into persistence, staying persistent, and ultimately sensing favorable conditions that enable exit from persistence to resume normal replication and transition into infectious EBs. Many critical questions regarding each of these steps remain. How is damage sensed? How is persistence regulated? How does Chlamydia enter into and exit from persistence? What are the genetic basis for persistence in the context of different stimuli and interactions with the host? What chlamydial genes are required for persistence and how do they participate? Can we intervene in order to prevent Chlamydia entry or exit from persistence? What are the consequences of such interventions for Chlamydia pathogenesis in-vivo? Emerging tools for genetic manipulations in Chlamydia have started a new and exciting time for Chlamydia research, opening the possibility to address those and other relevant questions about Chlamydia persistence.
HS, RV, and MP wrote the paper. HS and RV contributed with funding support.
MP is a CONICET doctoral fellow. HS is an investigator of CONICET and a Professor at Universidad Nacional de Cordoba. RV is a Professor at Duke University School of Medicine and his work is supported by grants from the National Institute of Allergy and Infectious Diseases (NIAID). Work in the Saka lab is supported by the grants Mario Hirsch from Bunge & Born Foundation, PICT2014-2238-Prestamo BID from the Ministry of Science and Technology of Argentina, SECyT-UNC Tipo A 2016 from Universidad Nacional de Córdoba and GRFT 2017 from the Ministry of Science and Technology of Cordoba.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aiyar, A., Quayle, A. J., Buckner, L. R., Sherchand, S. P., Chang, T. L., Zea, A. H., et al. (2014). Influence of the tryptophan-indole-IFNγ axis on human genital Chlamydia trachomatis infection: role of vaginal co-infections. Front. Cell. Infect. Microbiol. 4:72. doi: 10.3389/fcimb.2014.00072
Asner, S. A., Jaton, K., Kyprianidou, S., Nowak, A. M., and Greub, G. (2014). Chlamydia pneumoniae: possible association with asthma in children. Clin. Infect. Dis. 58, 1198–1199. doi: 10.1093/cid/ciu034
Augenbraun, M. H., and McCormack, W. M. (2015). “Urethritis,” in Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases 8th edn, eds J. E. Bennett, R. Dolin, and M. J. Blaser (Philadelphia, PA: Elsevier Saunders), 1349–1357.
Baehr, W., Zhang, Y. X., Joseph, T., Su, H., Nano, F. E., Everett, K. D., et al. (1988). Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. U.S.A. 85, 4000–4004. doi: 10.1073/pnas.85.11.4000
Barbour, A. G., Amano, K., Hackstadt, T., Perry, L., and Caldwell, H. D. (1982). Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J. Bacteriol. 151, 420–428.
Bastidas, R. J., and Valdivia, R. H. (2016). Emancipating Chlamydia: advances in the genetic manipulation of a recalcitrant intracellular pathogen. Microbiol. Mol. Biol. Rev. 80, 411–427. doi: 10.1128/MMBR.00071-15
Beatty, W. L., Belanger, T. A., Desai, A. A., Morrison, R. P., and Byrne, G. I. (1994a). Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 62, 3705–3711.
Beatty, W. L., Byrne, G. I., and Morrison, R. P. (1993). Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 90, 3998–4002. doi: 10.1073/pnas.90.9.3998
Beatty, W. L., Morrison, R. P., and Byrne, G. I. (1994b). Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58, 686–699.
Beeckman, D. S., and Vanrompay, D. C. (2009). Zoonotic Chlamydophila psittaci infections from a clinical perspective. Clin. Microbiol. Infect. 15, 11–17. doi: 10.1111/j.1469-0691.2008.02669.x
Belland, R. J., Nelson, D. E., Virok, D., Crane, D. D., Hogan, D., Sturdevant, D., et al. (2003). Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. U.S.A. 100, 15971–15976. doi: 10.1073/pnas.2535394100
Bernstein-Hanley, I., Coers, J., Balsara, Z. R., Taylor, G. A., Starnbach, M. N., and Dietrich, W. F. (2006). The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proc. Natl. Acad. Sci. U.S.A. 103, 14092–14097. doi: 10.1073/pnas.0603338103
Billiau, A., and Matthys, P. (2009). Interferon-gamma: a historical perspective. Cytokine Growth Factor Rev. 20, 97–113. doi: 10.1016/j.cytogfr.2009.02.004
Bonner, C. A., Byrne, G. I., and Jensen, R. A. (2014). Chlamydia exploit the mammalian tryptophan-depletion defense strategy as a counter-defensive cue to trigger a survival state of persistence. Front. Cell. Infect. Microbiol. 4:17. doi: 10.3389/fcimb.2014.00017
Borel, N., Summersgill, J. T., Mukhopadhyay, S., Miller, R. D., Ramirez, J. A., and Pospischil, A. (2008). Evidence for persistent Chlamydia pneumoniae infection of human coronary atheromas. Atherosclerosis 199, 154–161. doi: 10.1016/j.atherosclerosis.2007.09.026
Brinkworth, A. J., Wildung, M. R., and Carabeo, R. A. (2018). Genomewide transcriptional responses of iron-starved Chlamydia trachomatis reveal prioritization of metabolic precursor synthesis over protein translation. mSystems 3:e00184–17. doi: 10.1128/mSystems.00184-17
Byrne, G. I., Lehmann, L. K., and Landry, G. J. (1986). Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect. Immun. 53, 347–351.
Byrne, G. I., Ouellette, S. P., Wang, Z., Rao, J. P., Lu, L., Beatty, W. L., et al. (2001). Chlamydia pneumoniae expresses genes required for DNA replication but not cytokinesis during persistent infection of HEp-2 cells. Infect. Immun. 69, 5423–5429. doi: 10.1128/IAI.69.9.5423-5429.2001
Byron, E. B. (2012). “Chlamydia infection and epidemiology,” in Intracellular Pathogens I: Chlamydiales, eds M. Tan and P. Bavoil (Washington, DC: ASM Press), 1–26.
Caldwell, H. D., Wood, H., Crane, D., Bailey, R., Jones, R. B., Mabey, D., et al. (2003). Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 111, 1757–1769. doi: 10.1172/JCI17993
Chatterjee, A., Modarai, M., Naylor, N. R., Boyd, S. E., Atun, R., Barlow, J., et al. (2018). Quantifying drivers of antibiotic resistance in humans: a systematic review. Lancet Infect. Dis. 18, e368–e378. doi: 10.1016/S1473-3099(18)30296-2
Darville, T. (2005). Chlamydia trachomatis infections in neonates and young children. Semin. Pediatr. Infect. Dis. 16, 235–244. doi: 10.1053/j.spid.2005.06.004
Di Pietro, M., Tramonti, A., De Santis, F., De Biase, D., Schiavoni, G., Filardo, S., et al. (2012). Analysis of gene expression in penicillin G induced persistence of Chlamydia pneumoniae. J. Biol. Regul. Homeost. Agents 26, 277–284.
Elwell, C., Mirrashidi, K., and Engel, J. (2016). Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14, 385–400. doi: 10.1038/nrmicro.2016.30
Fehlner-Gardiner, C., Roshick, C., Carlson, J. H., Hughes, S., Belland, R. J., Caldwell, H. D., et al. (2002). Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277, 26893–26903. doi: 10.1074/jbc.M203937200
Galasso, G. J., and Manire, G. P. (1961). Effect of antiserum and antibiotics on persistent infection of HeLa cells with meningopneumonitis virus. J. Immunol. 86, 382–385.
Gérard, H. C., Whittum-Hudson, J. A., Schumacher, H. R., and Hudson, A. P. (2004). Differential expression of three Chlamydia trachomatis hsp60-encoding genes in active vs. persistent infections. Microb. Pathog. 36, 35–39. doi: 10.1016/j.micpath.2003.08.005
Goellner, S., Schubert, E., Liebler-Tenorio, E., Hotzel, H., Saluz, H. P., and Sachse, K. (2006). Transcriptional response patterns of Chlamydophila psittaci in different in vitro models of persistent infection. Infect. Immun. 74, 4801–4808. doi: 10.1128/IAI.01487-05
Gomes, J. P., Bruno, W. J., Nunes, A., Santos, N., Florindo, C., Borrego, M. J., et al. (2007). Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 17, 50–60. doi: 10.1101/gr.5674706
Gonzales, G. F., Muñoz, G., Sánchez, R., Henkel, R., Gallegos-Avila, G., Diaz-Gutierrez, O., et al. (2004). Update on the impact of Chlamydia trachomatis infection on male fertility. Andrologia 36, 1–23. doi: 10.1046/j.0303-4569.2003.00594.x
Grayston, J. T., Aldous, M. B., Easton, A., Wang, S. P., Kuo, C. C., Campbell, L. A., et al. (1993). Evidence that Chlamydia pneumoniae causes pneumonia and bronchitis. J. Infect. Dis. 168, 1231–1235. doi: 10.1093/infdis/168.5.1231
Haggerty, C. L., Gottlieb, S. L., Taylor, B. D., Low, N., Xu, F., and Ness, R. B. (2010). Risk of sequelae after Chlamydia trachomatis genital infection in women. J. Infect. Dis. 201 (Suppl. 2), S134–S155. doi: 10.1086/652395
Hammerschlag, M. R. (2011). Chlamydial and gonococcal infections in infants and children. Clin. Infect. Dis. 53 (Suppl. 3), S99–S102. doi: 10.1093/cid/cir699
Herring, A., and Richens, J. (2006). Lymphogranuloma venereum. Sex. Transm. Infect. 82 (Suppl. 4), iv23–iv25. doi: 10.1136/sti.2006.023143
Hogan, R. J., Mathews, S. A., Mukhopadhyay, S., Summersgill, J. T., and Timms, P. (2004). Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 72, 1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004
Holland, S. M., Hudson, A. P., Bobo, L., Whittum-Hudson, J. A., Viscidi, R. P., Quinn, T. C., et al. (1992). Demonstration of chlamydial RNA and DNA during a culture-negative state. Infect. Immun. 60, 2040–2047.
Hsia, R., Ohayon, H., Gounon, P., Dautry-Varsat, A., and Bavoil, P. M. (2000). Phage infection of the obligate intracellular bacterium, Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect. 2, 761–772. doi: 10.1016/S1286-4579(00)90356-3
Hurst, E. W., Landquist, J. K., Melvin, P., Peters, J. M., Senior, N., Silk, J. A., et al. (1953). The therapy of experimental psittacosis and lymphogranuloma venereum (inguinale) II. The activity of quinoxaline-1:4-dioxide and substituted and related compounds, with a note on the morphological changes induced in lymphogranuloma virus by these compounds and by antibiotics. Br. J. Pharmacol. Chemother. 8, 297–305. doi: 10.1111/j.1476-5381.1953.tb00798.x
Huston, W. M., Theodoropoulos, C., Mathews, S. A., and Timms, P. (2008). Chlamydia trachomatis responds to heat shock, penicillin induced persistence, and IFN-gamma persistence by altering levels of the extracytoplasmic stress response protease HtrA. BMC Microbiol. 8:190. doi: 10.1186/1471-2180-8-190
Jacquier, N., Frandi, A., Pillonel, T., Viollier, P. H., and Greub, G. (2014). Cell wall precursors are required to organize the chlamydial division septum. Nat. Commun. 5:3578. doi: 10.1038/ncomms4578
Kari, L., Goheen, M. M., Randall, L. B., Taylor, L. D., Carlson, J. H., Whitmire, W. M., et al. (2011). Generation of targeted Chlamydia trachomatis null mutants. Proc. Natl. Acad. Sci. U.S.A. 108, 7189–7193. doi: 10.1073/pnas.1102229108
Kintner, J., Lajoie, D., Hall, J., Whittimore, J., and Schoborg, R. V. (2014). Commonly prescribed β-lactam antibiotics induce C. trachomatis persistence/stress in culture at physiologically relevant concentrations. Front. Cell. Infect. Microbiol. 4:44. doi: 10.3389/fcimb.2014.00044
Laitinen, K., Laurila, A. L., Leinonen, M., and Saikku, P. (1996). Reactivation of Chlamydia pneumoniae infection in mice by cortisone treatment. Infect. Immun. 64, 1488–1490.
Lambden, P. R., Pickett, M. A., and Clarke, I. N. (2006). The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology 152(Pt 9), 2573–2578. doi: 10.1099/mic.0.29032-0
Lan, J., van den Brule, A. J., Hemrika, D. J., Risse, E. K., Walboomers, J. M., Schipper, M. E., et al. (1995). Chlamydia trachomatis and ectopic pregnancy: retrospective analysis of salpingectomy specimens, endometrial biopsies, and cervical smears. J. Clin. Pathol. 48, 815–819. doi: 10.1136/jcp.48.9.815
Lewis, M. E., Belland, R. J., AbdelRahman, Y. M., Beatty, W. L., Aiyar, A. A., Zea, A. H., et al. (2014). Morphologic and molecular evaluation of Chlamydia trachomatis growth in human endocervix reveals distinct growth patterns. Front. Cell. Infect. Microbiol. 4:71. doi: 10.3389/fcimb.2014.00071
Malinverni, R., Kuo, C. C., Campbell, L. A., and Grayston, J. T. (1995). Reactivation of Chlamydia pneumoniae lung infection in mice by cortisone. J. Infect. Dis. 172, 593–594. doi: 10.1093/infdis/172.2.593
Mathews, S., George, C., Flegg, C., Stenzel, D., and Timms, P. (2001). Differential expression of ompA, ompB, pyk, nlpD and Cpn0585 genes between normal and interferon-gamma treated cultures of Chlamydia pneumoniae. Microb. Pathog. 30, 337–345. doi: 10.1006/mpat.2000.0435
Matsumoto, A., and Manire, G. P. (1970). Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J. Bacteriol. 101, 278–285.
Mäurer, A. P., Mehlitz, A., Mollenkopf, H. J., and Meyer, T. F. (2007). Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 3:e83. doi: 10.1371/journal.ppat.0030083
McClarty, G., Caldwell, H. D., and Nelson, D. E. (2007). Chlamydial interferon gamma immune evasion influences infection tropism. Curr. Opin. Microbiol. 10, 47–51. doi: 10.1016/j.mib.2006.12.003
Molestina, R. E., Klein, J. B., Miller, R. D., Pierce, W. H., Ramirez, J. A., and Summersgill, J. T. (2002). Proteomic analysis of differentially expressed Chlamydia pneumoniae genes during persistent infection of HEp-2 cells. Infect. Immun. 70, 2976–2981. doi: 10.1128/IAI.70.6.2976-2981.2002
Mukhopadhyay, S., Miller, R. D., Sullivan, E. D., Theodoropoulos, C., Mathews, S. A., Timms, P., et al. (2006). Protein expression profiles of Chlamydia pneumoniae in models of persistence versus those of heat shock stress response. Infect. Immun. 74, 3853–3863. doi: 10.1128/IAI.02104-05
Muramatsu, M. K., Brothwell, J. A., Stein, B. D., Putman, T. E., Rockey, D. D., and Nelson, D. E. (2016). Beyond tryptophan synthase: identification of genes that contribute to Chlamydia trachomatis survival during Gamma Interferon-induced persistence and reactivation. Infect. Immun. 84, 2791–2801. doi: 10.1128/IAI.00356-16
Newman, L., Rowley, J., Vander Hoorn, S., Wijesooriya, N. S., Unemo, M., Low, N., et al. (2015). Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS ONE 10:e0143304. doi: 10.1371/journal.pone.0143304
Ong, V. A., Marsh, J. W., Lawrence, A., Allan, J. A., Timms, P., and Huston, W. M. (2013). The protease inhibitor JO146 demonstrates a critical role for CtHtrA for Chlamydia trachomatis reversion from penicillin persistence. Front. Cell. Infect. Microbiol. 3:100. doi: 10.3389/fcimb.2013.00100
Østergaard, O., Follmann, F., Olsen, A. W., Heegaard, N. H., Andersen, P., and Rosenkrands, I. (2016). Quantitative protein profiling of Chlamydia trachomatis growth forms reveals defense strategies against tryptophan starvation. Mol. Cell. Proteomics 15, 3540–3550. doi: 10.1074/mcp.M116.061986
Ouellette, S. P., Hatch, T. P., AbdelRahman, Y. M., Rose, L. A., Belland, R. J., and Byrne, G. I. (2006). Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNgamma-mediated host cell tryptophan starvation. Mol. Microbiol. 62, 1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x
Ouellette, S. P., Karimova, G., Subtil, A., and Ladant, D. (2012). Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol. Microbiol. 85, 164–178. doi: 10.1111/j.1365-2958.2012.08100.x
Ouellette, S. P., Messerli, P. R., Wood, N. A., and Hajovsky, H. (2018). Characterization of Chlamydial rho and the role of rho-mediated transcriptional polarity during interferon gamma-mediated tryptophan limitation. Infect. Immun. 86:e00240–18. doi: 10.1128/IAI.00240-18
Ouellette, S. P., Rueden, K. J., and Rucks, E. A. (2016). Tryptophan codon-dependent transcription in Chlamydia pneumoniae during gamma interferon-mediated tryptophan limitation. Infect. Immun. 84, 2703–2713. doi: 10.1128/IAI.00377-16
Packiam, M., Weinrick, B., Jacobs, W.R. Jr, and Maurelli, A. T. (2015). Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly”. Proc. Natl. Acad. Sci. U.S.A. 112, 11660–11665. doi: 10.1073/pnas.1514026112
Pantoja, L. G., Miller, R. D., Ramirez, J. A., Molestina, R. E., and Summersgill, J. T. (2001). Characterization of Chlamydia pneumoniae persistence in HEp-2 cells treated with gamma interferon. Infect. Immun. 69, 7927–7932. doi: 10.1128/IAI.69.12.7927-7932.2001
Patton, D. L., Askienazy-Elbhar, M., Henry-Suchet, J., Campbell, L. A., Cappuccio, A., Tannous, W., et al. (1994). Detection of Chlamydia trachomatis in fallopian tube tissue in women with postinfectious tubal infertility. Am. J. Obstet. Gynecol. 171, 95–101. doi: 10.1016/S0002-9378(94)70084-2
Pettengill, M. A., Lam, V. W., and Ojcius, D. M. (2009). The danger signal adenosine induces persistence of chlamydial infection through stimulation of A2b receptors. PLoS ONE 4:e8299. doi: 10.1371/journal.pone.0008299
Phillips Campbell, R., Kintner, J., Whittimore, J., and Schoborg, R. V. (2012). Chlamydia muridarum enters a viable but non-infectious state in amoxicillin-treated BALB/c mice. Microbes Infect. 14, 1177–1185. doi: 10.1016/j.micinf.2012.07.017
Phillips-Campbell, R., Kintner, J., and Schoborg, R. V. (2014). Induction of the Chlamydia muridarum stress/persistence response increases azithromycin treatment failure in a murine model of infection. Antimicrob. Agents Chemother. 58, 1782–1784. doi: 10.1128/AAC.02097-13
Polkinghorne, A., and Greub, G. (2017). A new equine and zoonotic threat emerges from an old avian pathogen, Chlamydia psittaci. Clin. Microbiol. Infect. 23, 693–694. doi: 10.1016/j.cmi.2017.05.025
Pospischil, A., Borel, N., Chowdhury, E. H., and Guscetti, F. (2009). Aberrant chlamydial developmental forms in the gastrointestinal tract of pigs spontaneously and experimentally infected with Chlamydia suis. Vet. Microbiol. 135, 147–156. doi: 10.1016/j.vetmic.2008.09.035
Prusty, B. K., Bohme, L., Bergmann, B., Siegl, C., Krause, E., Mehlitz, A., et al. (2012). Imbalanced oxidative stress causes chlamydial persistence during non-productive human herpes virus co-infection. PLoS ONE 7:e47427. doi: 10.1371/journal.pone.0047427
Rank, R. G., Whittimore, J., Bowlin, A. K., and Wyrick, P. B. (2011). In vivo ultrastructural analysis of the intimate relationship between polymorphonuclear leukocytes and the chlamydial developmental cycle. Infect. Immun. 79, 3291–3301. doi: 10.1128/IAI.00200-11
Rapoza, P. A., Tahija, S. G., Carlin, J. P., Miller, S. L., Padilla, M. L., and Byrne, G. I. (1991). Effect of interferon on a primary conjunctival epithelial cell model of trachoma. Invest. Ophthalmol. Vis. Sci. 32, 2919–2923.
Richmond, S. J., Hilton, A. L., and Clarke, S. K. (1972). Chlamydial infection. Role of Chlamydia subgroup A in non-gonococcal and post-gonococcal urethritis. Br. J. Vener. Dis. 48, 437–444. doi: 10.1136/sti.48.6.437
Rönnerstam, R., Persson, K., Hansson, H., and Renmarker, K. (1985). Prevalence of chlamydial eye infection in patients attending an eye clinic, a VD clinic, and in healthy persons. Br. J. Ophthalmol. 69, 385–388. doi: 10.1136/bjo.69.5.385
Rosario, C. J., and Tan, M. (2012). The early gene product EUO is a transcriptional repressor that selectively regulates promoters of Chlamydia late genes. Mol. Microbiol. 84, 1097–1107. doi: 10.1111/j.1365-2958.2012.08077.x
Roth, A., Konig, P., van Zandbergen, G., Klinger, M., Hellwig-Burgel, T., Daubener, W., et al. (2010). Hypoxia abrogates antichlamydial properties of IFN-gamma in human fallopian tube cells in vitro and ex vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 19502–19507. doi: 10.1073/pnas.1008178107
Saka, H. A., Thompson, J. W., Chen, Y. S., Kumar, Y., Dubois, L. G., Moseley, M. A., et al. (2011). Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol. Microbiol. 82, 1185–1203. doi: 10.1111/j.1365-2958.2011.07877.x
Saka, H. A., and Valdivia, R. H. (2010). Acquisition of nutrients by Chlamydiae: unique challenges of living in an intracellular compartment. Curr. Opin. Microbiol. 13, 4–10. doi: 10.1016/j.mib.2009.11.002
Sandoz, K. M., and Rockey, D. D. (2010). Antibiotic resistance in Chlamydiae. Future Microbiol. 5, 1427–1442. doi: 10.2217/fmb.10.96
Sauvage, E., Kerff, F., Terrak, M., Ayala, J. A., and Charlier, P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258. doi: 10.1111/j.1574-6976.2008.00105.x
Schachter, J., Grossman, M., Sweet, R. L., Holt, J., Jordan, C., and Bishop, E. (1986). Prospective study of perinatal transmission of Chlamydia trachomatis. JAMA 255, 3374–3377. doi: 10.1001/jama.1986.03370240044034
Schoborg, R. V. (2011). Chlamydia persistence – a tool to dissect chlamydia–host interactions. Microbes Infect. 13, 649–662. doi: 10.1016/j.micinf.2011.03.004
Schoborg, R. V., and Borel, N. (2014). Porcine epidemic diarrhea virus (PEDV) co-infection induced chlamydial persistence/stress does not require viral replication. Front. Cell. Infect. Microbiol. 4:20. doi: 10.3389/fcimb.2014.00020
Seth-Smith, H. M., Wanninger, S., Bachmann, N., Marti, H., Qi, W., Donati, M., et al. (2017). The Chlamydia suis genome exhibits high levels of diversity, plasticity, and mobile antibiotic resistance: comparative genomics of a recent livestock cohort shows influence of treatment regimes. Genome Biol. Evol. 9, 750–760. doi: 10.1093/gbe/evx043
Shima, K., Klinger, M., Solbach, W., and Rupp, J. (2013). Activities of first-choice antimicrobials against gamma interferon-treated Chlamydia trachomatis differ in hypoxia. Antimicrob. Agents Chemother. 57, 2828–2830. doi: 10.1128/AAC.02211-12
Skilton, R. J., Cutcliffen, L. T., Barlow, D., Wang, Y., Salim, O., Lambden, P. R., et al. (2009). Penicillin induced persistence in Chlamydia trachomatis: high quality time lapse video analysis of the developmental cycle. PLoS ONE 4:e7723. doi: 10.1371/journal.pone.0007723
Stamm, W.E. (1999). Chlamydia trachomatis infections: progress and problems. J. Infect. Dis. 179 (Suppl. 2), S380–S383. doi: 10.1086/513844
Stenberg, K., and Mardh, P. A. (1991). Genital infection with Chlamydia trachomatis in patients with chlamydial conjunctivitis: unexplained results. Sex. Transm. Dis. 18, 1–4. doi: 10.1097/00007435-199101000-00001
Stephens, R. S., Sanchez-Pescador, R., Wagar, E. A., Inouye, C., and Urdea, M. S. (1987). Diversity of Chlamydia trachomatis major outer membrane protein genes. J. Bacteriol. 169, 3879–3885. doi: 10.1128/jb.169.9.3879-3885.1987
Stocks, M. E., Ogden, S., Haddad, D., Addiss, D. G., McGuire, C., and Freeman, M. C. (2014). Effect of water, sanitation, and hygiene on the prevention of trachoma: a systematic review and meta-analysis. PLoS Med. 11:e1001605. doi: 10.1371/journal.pmed.1001605
Suchland, R. J., Dimond, Z. E., Putman, T. E., and Rockey, D. D. (2017). Demonstration of persistent infections and genome stability by whole-genome sequencing of repeat-positive, same-serovar Chlamydia trachomatis collected from the female genital tract. J. Infect. Dis. 215, 1657–1665. doi: 10.1093/infdis/jix155
Tamura, A., and Manire, G. P. (1968). Effect of penicillin on the multiplication of meningopneumonitis organisms (Chlamydia psittaci). J. Bacteriol. 96, 875–880.
Thygeson, P. (1963). Epidemiologic observations on Trachoma in the United States. Invest. Ophthalmol. 2, 482–489.
Tietzel, I., El-Haibi, C., and Carabeo, R. A. (2009). Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS ONE 4:e6499. doi: 10.1371/journal.pone.0006499
Vanover, J., Kintner, J., Whittimore, J., and Schoborg, R. V. (2010). Interaction of herpes simplex virus type 2 (HSV-2) glycoprotein D with the host cell surface is sufficient to induce Chlamydia trachomatis persistence. Microbiology 156(Pt 5), 1294–1302. doi: 10.1099/mic.0.036566-0
Weiss, E. (1950). The effect of antibiotics on agents of the psittacosis-lymphogranuloma group: I. the effect of penicillin. J. Infect. Dis. 87, 249–263. doi: 10.1093/infdis/87.3.249
Wheelock, E. F. (1965). Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin. Science 149, 310–311. doi: 10.1126/science.149.3681.310
White, J. A. (2009). Manifestations and management of lymphogranuloma venereum. Curr. Opin. Infect. Dis. 22, 57–66. doi: 10.1097/QCO.0b013e328320a8ae
Wiedeman, J. A., Kaul, R., Heuer, L. S., Thao, N. N., Pinkerton, K. E., and Wenman, W. M. (2005). Tobacco smoke induces a persistent, but recoverable state in Chlamydia pneumoniae infection of human endothelial cells. Microb. Pathog. 39, 197–204. doi: 10.1016/j.micpath.2005.09.001
Keywords: Chlamydia persistence, penicillin-induced persistence, gamma interferon-induced persistence, Chlamydia evasion of antimicrobial stimuli, aberrant reticulate bodies, in-vivo implications of Chlamydia persistence, Chlamydia persistence inducers
Citation: Panzetta ME, Valdivia RH and Saka HA (2018) Chlamydia Persistence: A Survival Strategy to Evade Antimicrobial Effects in-vitro and in-vivo. Front. Microbiol. 9:3101. doi: 10.3389/fmicb.2018.03101
Received: 26 September 2018; Accepted: 30 November 2018;
Published: 12 December 2018.
Edited by:
Robert V. Schoborg, East Tennessee State University, United StatesReviewed by:
Jennifer H. Shaw, Oklahoma State University, United StatesCopyright © 2018 Panzetta, Valdivia and Saka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hector Alex Saka, YWxleC5zYWthQHVuYy5lZHUuYXI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.