- 1State Key Laboratory for Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 2Nanshan Center for Disease Control and Prevention, Shenzhen, China
- 3Shunyi District Center for Disease Control and Prevention, Beijing, China
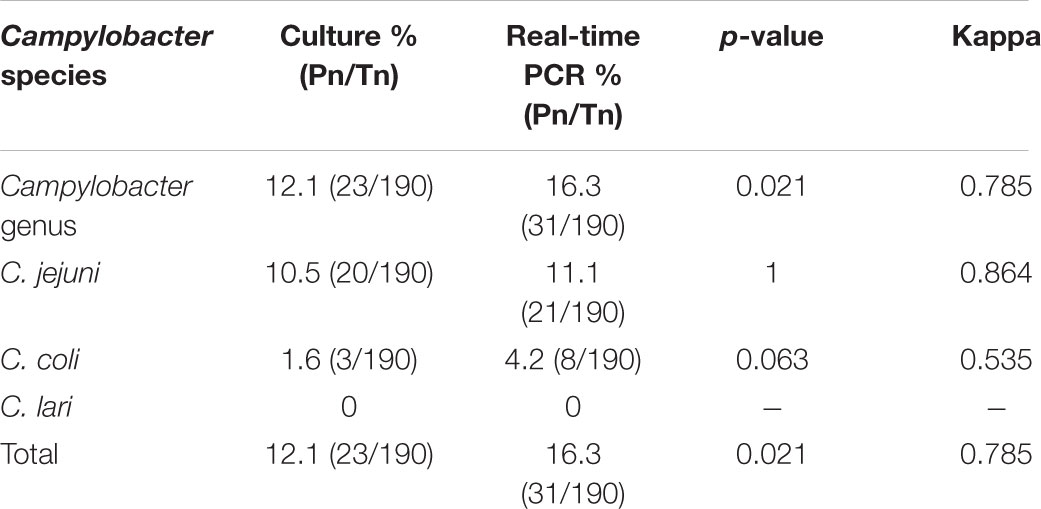
Campylobacter is one of the most common pathogens leading to the bacterial diarrheal illness. In order to set up one effective culture independent assay for the screen of the Campylobacter infection in the diarrheal patients, the quadruple real-time PCR method comparing to the culture based on the enriched filtration method which was recognized as the most effective isolation method was assessed for 190 stool samples from the diarrheal patients collected during the Foodborne Diseases Active Surveillance Network in Beijing. This multiple real-time PCR was designed to identify the Campylobacter genus, C. jejuni, C. coli, and C. lari simultaneously. With the enrichment culture method, 23 (12.1%, 23/190) Campylobacter isolates were obtained (20 C. jejuni and 3 C. coli), however, 31 samples (16.3%, 31/190) were detected positively with the real-time PCR (21 C. jejuni, 8 C. coli, and 2 Campylobacter genus only). With the comparison, the real-time-PCR method is more sensitive than the enrichment filtration method (16.3 vs. 12.1%, p = 0.021). Among the culture-positive samples, 95.7% (22/23) were detected positively by PCR which indicate the specificity of this method was higher. These two methods were consistent well (Kappa = 0.785, p < 0.05). Comparing to the culture methods, the result of the multiple real-time PCR method is sensitive, reliable and rapid. The present study indicated this multiple real-time PCR can be used both for the surveillance network and the preceding screen for bacteria isolation. This is first comparative study between the culture and multiple real-time PCR method for Campylobacter identification in stool specimens from the diarrheal patients.
Introduction
Foodborne Campylobacter is recognized as the leading causes of the bacterial diarrheal illness in both developing and developed countries (Coker et al., 2002; EFSA, 2015; Marder et al., 2017; Li et al., 2018; Natsos et al., 2018). One recent report showed that the isolation ratio of Campylobacter ranked the third among the major bacterial pathogen in the diarrheal cases in Beijing (Li et al., 2018).
Bacteria culture has been recognized as the “Gold standard” for the diagnosis of the Campylobacter infection (de Boer et al., 2015). However, the isolation result could be affected with many factors including the culture method and the medium. Culture-independent diagnostic tests (CIDTs) are increasingly used to diagnose Campylobacter infection in the Foodborne Diseases Active Surveillance Network (FoodNet) in United States and other countries (Bessède et al., 2011). The incidence of the culture confirmed and CIDT positive infections was highest for Campylobacter among bacterial and parasitic infections (confirmed = 11.79; CIDT positive–only = 17.43) per 100,000 persons in United States (Marder et al., 2017).
In order to explore the effective methods used for Campylobacter surveillance in China, in this study, we compared the quadruple real-time PCR method which could simultaneously identify the Campylobacter genus and three major foodborne Campylobacter spp., C. jejuni, C. coli, and C. lari to the culturing method from the stool specimens collected from the diarrheal patients in the Foodborne Diseases Active Surveillance Network (FoodNet) in Shunyi District, Beijing.
Materials and Methods
Sample Collection and Patients
A total of 190 stool samples from diarrhea patients were collected in Shunyi and Konggang Hospitals in Shunyi district, Beijing from July 2017 to February 2018. 190 specimens were collected from 190 cases. Among the 190 cases, 121 cases were male and 69 cases were female. The age of the patients ranged from 15 to 85 years old. The clinical character of the patients were summarized in Table 1. Five mg fresh stool samples were collected from each patient and transported in Cary-Blair medium at 4°C to the laboratory within 24 h.
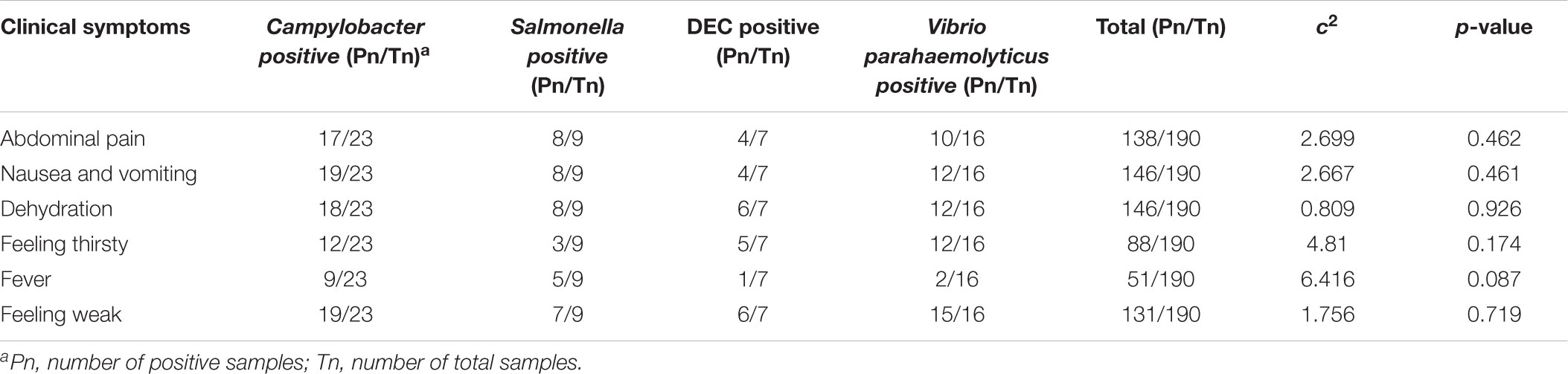
TABLE 1. Clinical characteristics of the entire tested cases and the characteristics of the culture-positive cases of each pathogen.
Bacteria Isolation and Identification
Campylobacter isolation was performed using the Campylobacter isolation kit (ZC-CAMPY-002, Qingdao Sinova Biotechnology Co., Ltd., Qingdao, China) according to the manufacturer’s instructions (Li et al., 2018) and the suspected isolates were picked on the Karmali agar in a microaerophilic atmosphere (5% O2,10% CO2, and 85% N2) at 42°C for 48 h. The species of Campylobacter were identified by multiple PCR (Wang et al., 2002). Meanwhile, the isolation for other four bacterial pathogens were also performed such as Salmonella, Shigella, Diarrheagenic E. coli [DEC, including Escherichia coli (EPEC), enterotoxigenic E. coli (ETEC), enterohemorrhagic E. coli (EHEC), enteroinvasive E. coli (EIEC), and enteroaggregative E. coli (EAEC)] and V. parahaemolyticus by traditional culture methods as described previously (Li et al., 2018).
DNA Extraction and the Quadruple Real-Time PCR Analysis
The DNA of the stool samples were extracted using the QIAamp DNA Stool Mini Kit (Qiagen) according to the manufacturer’s protocol. All of the DNA samples were triplicate and stored at -80°C until use. The quadruple real-time PCR commercial kit (Qingdao Sinova Biotechnology Co., Ltd., Qingdao, China) was used for detecting Campylobacter genus, C. jejuni, C. coli, and C. lari. As the description of manufacturer’s instructions, the primers and fluorophore-linked probes for genus Campylobacter were targeted on the 16s rRNA gene, which can detect the major foodborne Campylobacter spp. including C. jejuni, C. coli, C. lari, C. upsaliensis, and C. hyointestinalis. Moreover, the target genes of C. jejuni, C. coli, and C. lari are gyrA, ceuE, and glyA, respectively. We used a real-time PCR commercial kit in this study so the exact primer and probe sequences are not available. The fluorescence labeled for the probes of the Campylobacter genus, C. jejuni, C. coli, and C. lari were FAM, HEX, Cy5, and Quasar 705 with the quencher dye BHQ1, BHQ1, BHQ3, and BHQ3, respectively.
The quadruple real-time PCR was carried out on a CFX96 (Bio-Rad, United States) thermal cycler, with the TransStartTM Probe real-time PCR SuperMix (TransGen Biotech, China). For this real-time PCR, a 20 μl reaction was performed contained 3 μl of template DNA, 10 μl TransStartTM Probe real-time PCR SuperMix, 0.3 μmol l-1 of each amplification primer and 0.15 μmol l-1 of each probe. The thermal cycle protocol consisted of initial denaturation at 94°C for 30 s, followed by 40 cycles of 94°C for 15 s and 60°C for 40 s. The result data of real-time PCR was analyzed with CFX manager software (version 2.0). Quantitative results of real-time PCR were based on log10 value transformed and threshold cycle (Ct). All the samples with Ct < 30 were considered positive. The Ct > 35 was considered as negative and 30 < Ct < 35 was defined as suspected with three times repeat.
The three species of Campylobacter strains stored in our laboratory were used as the positive reference strains. Several kinds of other genus bacteria stored in our laboratory was used as negative controls such as Helicobacter pylori, E. coli, Streptococcus pneumoniae, etc.
Statistical Analysis
Significance of differences (p < 0.05) between culture and the real-time PCR was analyzed using chi-squared test and correlations between culture and detection was tested using Kappa test. The statistical analysis was performed using SPSS (version 22.0).
Ethics Statement
There was no tissue or blood samples were involved in this study. All the bacteria were isolated from stool specimens from the patients. The purpose of the study was informed to the participants and written consent was given to all of them. This project was approved by the ethics committee of the China CDC and the academic committee in the National Institute for Communicable Disease Control and Prevention. All relevant documents were recorded in China CDC (ICDC-2014012).
Results
Bacterial Isolation
Among the 190 specimens, 54 samples were positive for the tested five major enteric pathogens and two of them were mixed infection. In one sample, C. jejuni and ETEC were isolated and in another one, C. coli and Shigella were isolated. Campylobacter (12.1%, 23/190) was the most common pathogen. 20 samples (10.5%, 20/190) were positive for C. jejuni and 3 (1.5%, 3/190) samples were positive for C. coli. The isolation of other pathogens was listed in Table 2. Nine samples were Salmonella positive, one Shigella isolate was obtained from 1 sample, 16 samples were Vibrio parahaemolyticus positive and 7 samples were DEC positive, respectively. The clinical symptoms of Campylobacter infection patients are not statistically different (see Table 1, p > 0.05) with other pathogens infection.
Real-Time PCR Results and Comparison With Bacterial Culture
The entire 190 samples were performed by real-time PCR to detect the genus and identify the species of Campylobacter simultaneously. The results of the real-time PCR are shown in Table 3. Twenty-one C. jejuni (11%, 21/190) and eight C. coli (4.2%, 8/190) were detected in all samples. Among the 21 C. jejuni PCR positive samples, 18 (86%, 18/21) of them were culture positive but only 3 of the 8 C. coli PCR positive samples were culture positive (37.5%, 3/8). Only one culture-positive sample was not detected by this PCR method. There were two samples had positively result only on the genus Campylobacter but negative for the tested three species, which indicated the possibility of the infection with C. upsaliensis and C. hyointestinalis and need further confirmation.
The comparison of the PCR and the culturing results is shown in Figure 1. The coherence analysis of these two methods was Kappa = 0.785 (p < 0.05), which means the two methods are consistent well. According to the result of chi-squared test, the two methods in detecting Campylobacter are statistically different (p = 0.021) and the positive ratio of real-time PCR is higher than the culture (16.3 to 12.1%). For C. jejuni and C. coli, the difference of these two methods were not statistically significant (p = 1 and p = 0.063, respectively).

FIGURE 1. Venn-diagram summarizing the results of the culturing and the multiple real-time PCR detection method. The numbers in blue modules represent the number of culturing positive samples. The numbers in light yellow modules represent the number of culturing negative samples. The numbers in pink modules represent the number of real-time PCR negative samples. The numbers in green modules represent the number of real-time PCR positive samples.
Discussion
Bacterial diarrhea is a serious public health problem worldwide, especially Campylobacter spp. considered as a leading cause (Lynch et al., 2011; Wei et al., 2014; Nohra et al., 2016; Pavlova et al., 2016). In this study, the isolation of Campylobacter is also being the most common pathogen among the tested five enteric bacterial pathogens with the ratio of 12.1% (23/190). The Campylobacter isolation from the diarrheal patients using selective medium is variable. The considerable difference of the infection ratio in these reports was observed (Zhu et al., 2016; Chang et al., 2017; Zhang et al., 2017). Picking out the suspected Campylobacter colonies on the selective medium was laboratory experience depending and become more and more difficult with the increased drug-resistant microorganisms in the samples which reduced the selection capacity of the selective medium in these days in China. The enrichment with filtration method was recognized as the most effective isolation method for Campylobacter from the stool samples from the diarrheal patients (Li et al., 2018).
The use of CIDTs is increasing in the surveillance network in the developed counties (Cronquist et al., 2012; Babady et al., 2018). Stool antigen tests for Campylobacter from the stool samples was rapid but still with the problems of specificity and positive predictive value (Giltner et al., 2013). The method of multiple real-time PCR is widely used in detecting and identifying Campylobacter (Rudi et al., 2004; Ivanova et al., 2014). In this study, we used quadruple real-time PCR method which can detect the major foodborne Campylobacter spp. in one tube simultaneously. Comparing to the filtration method (culture with enrichment), this quadruple real-time PCR is more sensitive and reliable (16.3 to 12.1%, p = 0.021) and the whole PCR process was only take about 40 min to get the result. In this study, the Campylobacter was detected as the most common pathogen among the 190 diarrheal patients, which was consisting with the result of the FoodNet in United States (Marder et al., 2017).
Among the 31 PCR positive samples (16.3%, 31/190), the isolation rate is 71.0% (22/31). In addition to the sensitivity problem of the method (de Boer et al., 2015), there were other reasons may cause the result of 9 samples uniquely had PCR positive: the quality of the samples or the proceed of the sample transport caused the Campylobacter in a non-cultivable state. Certainly, the false positive results of the PCR might happened. PCR detection has the capacity to detect the DNA of dead bacteria or non-cultivable bacteria which could also indicated the recent infection. Only one sample was culture positive with C. jejuni but the CT value of the genus was more than 35 with three times repeats. The reason might be the amount of the pathogen is not enough or there was lost during the DNA extraction.
There are numbers of previous study indicated that the sensitivity of the real-time PCR was less than the culture method (Lund et al., 2004; Hong et al., 2007; Mily et al., 2011). Our study indicated the two methods for each of C. coli and C. jejuni is agreed with almost perfect agreement (Kappa = 0.864) and moderate agreement (Kappa = 0.535), respectively, and the difference was not statistically significant. This is first comparative study between the culture and multiple real-time PCR method for Campylobacter identification in stool specimens from the diarrheal patients.
To date, the surveillance for Campylobacter infection in the diarrheal patients were only carried out in parts of China. According to some previous publication (Zhu et al., 2016; Chang et al., 2017; Zhang et al., 2017), the campylobacteriosis in China was low and it might because of the detection method was inefficient. From this study, the results using the multiple- real time PCR indicated the Campylobacter infection accounts for a high proportion in diarrheal patients in Beijing. With the effective screening method, we could get the reliable data more quickly and it will specifically benefit the food safety assessment plan in China.
In conclusion, with the comparison of the real-time PCR method to the bacteria culture with enrichment in the Campylobacter detection from the stool sample, this quadruple real-time PCR method is a reliable and sensitive. This method is suitable to be used as a rapid screening method. Besides, the result can be used as guide for the pathogen culture which can get the isolates in the surveillance work in the future.
Author Contributions
HL, YD, ZW, and YG performed the molecular test and data analysis. YL was involved in the collection of samples and DNA extraction. HL and MZ drafted and revised this manuscript.
Funding
This work was supported by the Sanming Project of Medicine in Shenzhen (SZSM201803081) and the Major State Basic Research Development Program (2013CB127204).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the colleagues form Shunyi CDC, Shunyi Hospital, and Konggang Hospital for collecting the samples.
References
Babady, N. E., England, M. R., Jurcic Smith, K. L., He, T., Wijetunge, D. S., Tang, Y. W., et al. (2018). Multicenter evaluation of the ePlex respiratory pathogen panel for the detection of viral and bacterial respiratory tract pathogens in nasopharyngeal swabs. J. Clin. Microbiol. 56:e01658-17. doi: 10.1128/JCM.01658-17
Bessède, E., Delcamp, A., Sifré, E., Buissonnière, A., and Mégraud, F. (2011). New methods for detection of campylobacters in stool samples in comparison to culture. J. Clin. Microbiol. 49, 941–944. doi: 10.1128/JCM.01489-10
Chang, H., Zhang, L., Ge, Y., Cai, J., Wang, X., Huang, Z., et al. (2017). A hospital-based case-control study of diarrhea in children in Shanghai. Pediatr. Infect. Dis. J. 36, 1057–1063. doi: 10.1097/INF.0000000000001562
Coker, A. O., Isokpehi, R. D., Thomas, B. N., Amisu, K. O., and Obi, C. L. (2002). Human campylobacteriosis in developing countries1. Emerg. Infect. Dis. 8, 237–244. doi: 10.3201/eid0803.010233
Cronquist, A. B., Mody, R. K., Atkinson, R., Besser, J., D’Angelo, M. T., Hurd, S., et al. (2012). Impacts of culture-independent diagnostic practices on public health surveillance for bacterial enteric pathogens. Clin. Infect. Dis. 54(Suppl._5), S432–S439. doi: 10.1093/cid/cis267
de Boer, P., Rahaoui, H., Leer, R. J., Montijn, R. C., and van der Vossen, J. M. (2015). Real-time PCR detection of Campylobacter spp.: a comparison toclassic culturing and enrichment. Food Microbiol. 51, 96–100. doi: 10.1016/j.fm.2015.05.006
EFSA (2015). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 13:4329.
Giltner, C. L., Saeki, S., Bobenchik, A. M., and Humphries, R. M. (2013). Rapid detection of Campylobacter antigen by enzyme immunoassay leads to increased positivity rates. J. Clin. Microbiol. 51, 618–620. doi: 10.1128/JCM.02565-12
Hong, J., Jung, W. K., Kim, J. M., Kim, S. H., Koo, H. C., Ser, J., et al. (2007). Quantification and differentiation of Campylobacter jejuni and Campylobacter coli in raw chicken meats using a real-time PCR method. J. Food Prot. 70, 2015–2022. doi: 10.4315/0362-028X-70.9.2015
Ivanova, M., Singh, R., Dharmasena, M., Gong, C., Krastanov, A., and Jiang, X. (2014). Rapid identification of Campylobacter jejuni from poultry carcasses and slaughtering environment samples by real-time PCR. Poult. Sci. 93:1587. doi: 10.3382/ps.2013-03736
Li, Y., Zhang, S., He, M., Zhang, Y., Fu, Y., Liang, H., et al. (2018). Prevalence and Molecular Characterization of Campylobacter spp. Isolated from patients with diarrhea in Shunyi, Beijing. Front. Microbiol. 9:52. doi: 10.3389/fmicb.2018.00052
Lund, M., Nordentoft, S., Pedersen, K., and Madsen, M. (2004). Detection of Campylobacter spp. in Chicken fecal samples by real-time PCR. J. Clin. Microbiol. 42, 5125–5132. doi: 10.1128/JCM.42.11.5125-5132.2004
Lynch, O. A., Cagney, C., Mcdowell, D. A., and Duffy, G. (2011). Occurrence of fastidious Campylobacter spp. in fresh meat and poultry using an adapted cultural protocol. Int. J. Food Microbiol. 150, 171–177. doi: 10.1016/j.ijfoodmicro.2011.07.037
Marder, E. P., Cieslak, P. R., Cronquist, A. B., Dunn, J., Lathrop, S., Rabatskyehr, T., et al. (2017). Incidence and trends of infections with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance – foodborne diseases active surveillance network, 10 U.S. Sites, 2013-2016. MMWR 66:397. doi: 10.15585/mmwr.mm6615a1
Mily, L. M., François, B., Henri, S., Martine, D., and Catherine, B. (2011). Rapid identification and quantification of Campylobacter coli and Campylobacter jejuni by real-time PCR in pure cultures and in complex samples. BMC Microbiol. 11:113. doi: 10.1186/1471-2180-11-113
Natsos, G., Koutoulis, K. C., Sossidou, E., Chemaly, M., and Mouttotou, N. K. (2018). Campylobacter spp. infection in humans and poultry. J. Hellenic Vet. Med. Soc. 67, 65–82. doi: 10.12681/jhvms.15624
Nohra, A., Grinberg, A., Midwinter, A. C., Marshall, J. C., Collinsemerson, J. M., and French, N. P. (2016). Molecular epidemiology of Campylobacter coli strains isolated from different sources in new zealand between 2005 and 2014. Appl. Environ. Microbiol. 82, 4363–4370. doi: 10.1128/AEM.00934-16
Pavlova, M. R., Dobreva, E. G., Ivanova, K. I., Asseva, G. D., Ivanov, I. N., Petrov, P. K., et al. (2016). Multiplex PCR assay for Identifi cation and differentiation of Campylobacter jejuni and Campylobacter coli isolates. Folia Med. 58, 95–100. doi: 10.1515/folmed-2016-0016
Rudi, K., Høidal, H. K., Katla, T., Johansen, B. K., Nordal, J., and Jakobsen, K. S. (2004). Direct Real-Time PCR Quantification of Campylobacter jejuni in chicken fecal and cecal samples by integrated cell concentration and DNA purification. Appl. Environ. Microbiol. 70, 790–797. doi: 10.1128/AEM.70.2.790-797.2004
Wang, G., Clark, C. G., Taylor, T. M., Pucknell, C., Barton, C., Price, L., et al. (2002). Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 40, 4744–4747. doi: 10.1128/JCM.40.12.4744-4747.2002
Wei, B., Cha, S. Y., Kang, M., Roh, J. H., Seo, H. S., Yoon, R. H., et al. (2014). Antimicrobial susceptibility profiles and molecular typing of Campylobacter jejuni and Campylobacter coli isolates from ducks in South Korea. Appl. Environ. Microbiol. 80, 7604–7610. doi: 10.1128/AEM.02469-14
Zhang, Z., Lai, S., Yu, J., Geng, Q., Yang, W., Chen, Y., et al. (2017). Etiology of acute diarrhea in the elderly in China: a six-year observational study. PLoS One 12:e0173881. doi: 10.1371/journal.pone.0173881
Keywords: Campylobacter, culture, quadruple real-time PCR, diarrheal patients, comparison
Citation: Liang H, Wen Z, Li Y, Duan Y, Gu Y and Zhang M (2018) Comparison of the Filtration Culture and Multiple Real-Time PCR Examination for Campylobacter spp. From Stool Specimens in Diarrheal Patients. Front. Microbiol. 9:2995. doi: 10.3389/fmicb.2018.02995
Received: 14 August 2018; Accepted: 20 November 2018;
Published: 05 December 2018.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Zixin Peng, China National Center for Food Safety Risk Assessment, ChinaHongxia Jiang, South China Agricultural University, China
Copyright © 2018 Liang, Wen, Li, Duan, Gu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maojun Zhang, emhhbmdtYW9qdW5AaWNkYy5jbg==
 Hao Liang
Hao Liang Ziyu Wen2
Ziyu Wen2 Ying Li
Ying Li Yongxiang Duan
Yongxiang Duan Maojun Zhang
Maojun Zhang