- Department of Biological Science, The University of Tulsa, Tulsa, OK, United States
Aerotolerance in the microaerophilic species Campylobacter was previously reported and could increase bacterial survival and transmission in foods during stressful processing and storage conditions. In this study, 167 Campylobacter isolates (76 C. jejuni and 91 C. coli) were screened for aerotolerance; these strains were previously isolated from retail chicken meat, chicken livers, chicken gizzards, turkey, pork, and beef liver samples. Bacterial cultures were incubated aerobically in Mueller Hinton broth with agitation and viable cell counts were taken at 0, 6, 12, and 24 h. Approximately 47% of the screened Campylobacter isolates were aerotolerant (viable after a 12-h aerobic incubation period), whereas 24% were hyper-aerotolerant (viable after a 24-h aerobic incubation). A greater prevalence of aerotolerant strains (80%) was found among C. coli isolates as compared to C. jejuni isolates (6%). Differences in the oxidative stress response related genes were detected among C. jejuni and C. coli isolates when comparative genomics was used to analyze 17 Whole Genome Sequenced (WGS) strains from our laboratory. Genes encoding putative transcriptional regulator proteins and a catalase-like heme binding protein were found in C. coli genomes, but were absent in the genomes of C. jejuni. PCR screening showed the presence of a catalase-like protein gene in 75% (68/91) of C. coli strains, which was absent in all tested C. jejuni strains. While about 79% (30/38) of the hyper-aerotolerant C. coli strains harbored the catalase-like protein gene, the gene was also present in a number of the aerosensitive strains. The Catalase like protein gene was found to be expressed in both aerobic and microaerobic conditions with a 2-fold higher gene expression detected in aerobic conditions for an aerosensitive strain. However, the exact function of the gene remains unclear and awaits further investigation. In conclusion, aerotolerant Campylobacter strains (especially C. coli) are prevalent in various retail meats. Further studies are needed to investigate whether the genes encoding catalase-like heme binding protein and putative transcriptional regulators in C. coli strains are involved in stress response.
Introduction
Campylobacteriosis is a leading foodborne illness in developed countries, with symptoms including mild diarrhea and immunological disorders such as Guillian Barre syndrome (Dewey-Mattia et al., 2016). An increasing trend of Campylobacter infection has been reported in the USA from 2004 to 2012 at an annual rate of 11.4 cases per 100,000 individuals (Geissler et al., 2017). In 2014, 24 confirmed campylobacteriosis outbreaks with 324 confirmed illnesses were documented in the USA (Dewey-Mattia et al., 2016). C. jejuni accounts for more than 90% of clinical cases of campylobacteriosis, followed by C. coli with about 7% of clinical cases (Gillespie et al., 2002). Campylobacter is usually transmitted from poultry, but environmental sources also serve as transmission routes (Bronowski et al., 2014; Newell et al., 2017). Consumption of contaminated food products including retail meat, liver, dairy products, and water is commonly associated with clinical cases (Gillespie et al., 2002; Bronowski et al., 2014; Dewey-Mattia et al., 2016).
The prevalence of Campylobacter in retail meat and liver products has been reported (Noormohamed and Fakhr, 2012, 2013, 2014b; Huang et al., 2016). C. jejuni is predominant in retail meat products (mainly poultry products), whereas C. coli is prevalent in retail liver products and pork (Noormohamed and Fakhr, 2012, 2013, 2014b). C. coli strains from retail liver products were multidrug resistant and shared similar Sequence Type (ST) complexes with clinical isolates when subjected to Multilocus Sequence Typing (MLST) (Noormohamed and Fakhr, 2014a). The recent, increasing trend of antimicrobial resistance among Campylobacter strains indicates the potential threat of future outbreaks (Noormohamed and Fakhr, 2012, 2014b; Geissler et al., 2017).
Campylobacter is a microaerophilic, fastidious organism with an optimal growth temperature of approximately 42°C. Aerobic conditions, temperature variations, osmotic imbalances, and starvation are common stresses to Campylobacter during the processing and storage of retail meat and liver products (Bronowski et al., 2014; Bolton, 2015). The formation of viable but non-culturable (VBNC) state, biofilms, and aerotolerance are common strategies that enhance the viability of Campylobacter during stressful conditions (Bolton, 2015). Enhanced resistance to oxidative stress (Oh et al., 2015) and the production of oxidative stress response proteins (Rodrigues et al., 2016) are factors that likely increase the survival of Campylobacter exposed to aerobic conditions (Oh et al., 2015, 2017). A high incidence of aerotolerant C. jejuni from chicken was previously reported, with 35.7% of isolates identified as hyper-aerotolerant (HAT) (Oh et al., 2015). Furthermore, HAT strains had a higher prevalence of virulence genes than aerosensitive strains (Oh et al., 2017).
Most reports on the stress response of Campylobacter and gene expression analyses have been conducted with C. jejuni (Butcher et al., 2015; Handley et al., 2015). The availability of complete genome sequences for C. coli and C. jejuni from both retail meat and liver products (Marasini and Fakhr, 2016a,b,c, 2017a,b,c) has facilitated comparative genomic analyses. Furthermore, genomic differences in C. coli and C. jejuni strains (Fouts et al., 2005) might help to explain differences in aerotolerance (O'Kane and Connerton, 2017).
Previous reports from our laboratory showed high prevalence of C. coli and C. jejuni strains in retail liver products (Noormohamed and Fakhr, 2012, 2013, 2014b). Since the existence of aerotolerant strains would definitely enhance the survival of Campylobacter spp., we hypothesize that aerotolerant strains will be prevalent among those isolated from retail meats. The focus of the current study was to screen a large number of C. jejuni and C. coli strains from retail meat and liver products for aerotolerance. The presence of genes involved in the oxidative stress response were also explored among 17 C. coli and C. jejuni strains using sequence data previously generated in our laboratory.
Materials and Methods
Bacterial Strains and Growth Conditions
The C. jejuni (n = 76) and C. coli (n = 91) strains (Table S1) used in this study were previously isolated from retail chicken meat, chicken livers, chicken gizzards, turkey, pork, and beef livers (Noormohamed and Fakhr, 2012, 2013, 2014b). Campylobacter isolates were grown from stock cultures maintained at −70°C. Strains were inoculated to Mueller Hinton Agar (MHA) supplemented with 5% laked horse blood at 42°C for 48 h and incubated in microaerobic condition (6% O2, 13% CO2) in a water jacketed CO2 incubator (Thermo Scientific). Strains were transferred to fresh MHA with 5% laked horse blood and grown for 18 h prior to harvesting the cells for aerotolerance and hydrogen peroxide sensitivity assays.
Screening for Aerotolerant Campylobacter Strains
Aerotolerance was assayed as described previously (Oh et al., 2015) with slight modifications. Briefly, Campylobacter cells were harvested after an 18-h incubation and adjusted to OD600 = 0.5 in PBS (pH = 7.4). OD600 = 1 was used for 51 samples to ensure bacterial inoculum >107 CFU/ml). One ml of the Campylobacter cell suspension was then transferred to 9 ml of Mueller Hinton Broth (MHB) preincubated at 42°C in 50 ml Falcon tubes. Inoculated tubes with cracked open caps were incubated aerobically with agitation at 200 rpm in an incubator shaker with orbital diameter of 19 mm (New Brunswick I2400) at 42°C, and viable cell counts were obtained from 40 μl samples that were removed at 0, 6, 12, and 24 h. Aliquots (10 μl) from each dilution were spotted twice on MHA plates and incubated at 42°C for at least 48 h in microaerobic conditions (6% O2, 13% CO2). Each experiment was carried out in triplicate, and log CFU/ml values of viable cell counts were used for statistical analysis.
Comparative Genomic Analysis
Comparative genomic analyses among Campylobacter isolates sequenced in our laboratory (Table 1; Marasini and Fakhr, 2016a,b,c, 2017a,b,c) and reference genome sequences (GenBank) were performed with Rapid Annotation using Subsystem Technology (RAST) (Aziz et al., 2008) and NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Functional genomic comparisons were also performed for subsystems of the stress response and transcriptional regulators by RAST and BLAST. Multiple sequence alignment and phylogenetic analysis of nucleic acid sequences for catalase-like proteins from Campylobacter spp. from our study and GenBank (3/14/2017). (Table S2) were conducted using Geneious v. 11 (https://www.geneious.com).
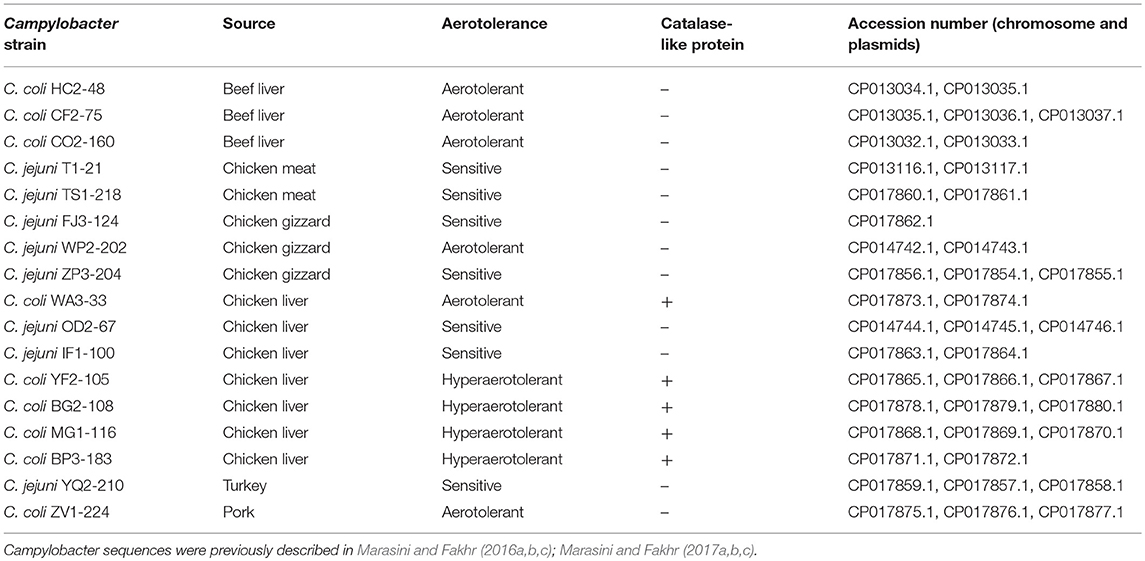
Table 1. Whole genome sequenced (WGS) Campylobacter strains (from our laboratory) used for comparative genomic analyses.
Screening for Genes Encoding Catalase-Like Proteins
Campylobacter strains from stock cultures (−70°C) were inoculated to MHA supplemented with 5% laked horse blood and incubated for 48 h in microaerobic conditions (6% O2, 13% CO2) in a water jacketed CO2 incubator (Thermo Scientific) at 42°C. Strains were screened for genes encoding catalase-like heme-binding proteins by PCR using the following primers: forward, 5′-TCAACTCAATGCGGATCCTAAA-3′ and reverse, 5′-AGCATAAGCCTCGTTTCTTACA-3′.
PCR reactions were conducted as described previously (Noormohamed and Fakhr, 2012). Briefly, DNA samples were prepared in single cell lysis buffer. PCR reactions contained 3 μl of each DNA sample, 12.5 μl GoTaq Green master Mix (Promega, Madison, WI, USA), 7.5 μl water, and 1 μl of each forward and reverse primer. PCR cycle conditions included 95°C for 3 min, and 30 cycles of the following: 95°C for 1 min, 50°C for 1 min, 72°C for 1 min, followed by a final extension for 10 min at 72°C and hold at 4°C. PCR products were subsequently analyzed using agarose gel electrophoresis.
Assay for Hydrogen Peroxide Sensitivity
Assay for hydrogen peroxide sensitivity was conducted as described previously with limited modifications (De Vries et al., 2017). Cell suspensions (OD600 = 0.15) were prepared in PBS (pH = 7.4) from 18 h cultures of 14 Campylobacter strains (nine C. coli and five C. jejuni isolates). Bacterial cell suspensions (4 ml) were mixed with 80 ml of MHA (~45°C), and 25 ml of each MHA-bacterial mixture was aliquoted into three, 90 mm petri dishes. Sterile filter paper discs (6 mm) containing 10 μl of H2O2 (0.05, 0.1, and 0.5 mM) were immediately placed onto the solid MHA-bacterial mixture, and inhibition zones were measured (n = 3) at 24 h.
Isolation of Total RNA
Three C. coli strains containing catalase like gene (P1-18, WA3-33, MG1-116) were cultured in MHA supplemented with 5% laked horse blood with antibiotics (cefoperazone 20 μg/ml, vancomycin 20 μg/ml, trimethoprim 20 μg/ml, and amphotericin B 10 μg/ml) for 48 h in microaerobic condition (6% O2, 13% CO2) in a water jacketed CO2 incubator (Thermo Scientific) at 42°C. After 48 h incubation, bacterial suspension was adjusted to OD600 = 0.5 in MH broth and diluted 1::10 in MH broth. Bacterial inoculum (80 μl) was added to 20 ml freshly prepared MH broth and incubated at 42°C for 16 h (log phase) in microaerobic condition. Then, bacterial culture was divided equally and incubated under two conditions (microaerobic and aerobic incubation) for 1 more hour. For aerobic incubation, the bacterial broth was incubated aerobically in 25 ml conical flask with agitation at 200 rpm in an incubator shaker with orbital diameter of 19 mm (New Brunswick I2400) at 42°C. After 1 h incubation, bacterial broths in both aerobic and microaerobic conditions were subjected to RNA isolation. Each experiment was conducted in triplicates.
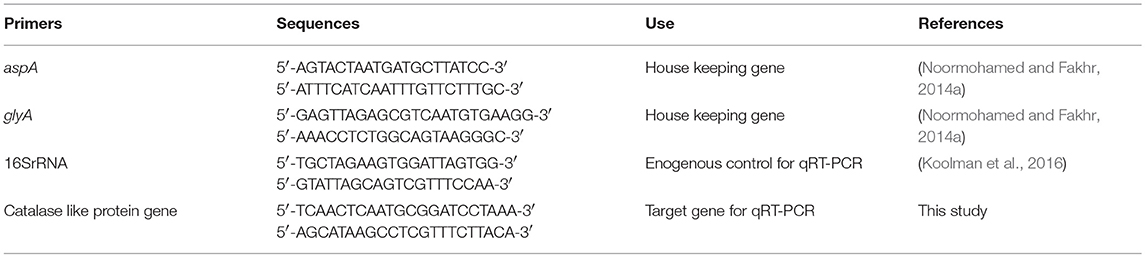
Multiple RNA samples were isolated for each strain and condition (microaerobic or aerobic) by using RNeasy Mini Kit (Qiagen) according to manufacturer's instruction. For total RNA isolation, 2 ml RNA protect bacteria reagent (Qiagen) was added to 1 ml of bacterial culture and vortexed for 5 s followed by incubation at room temperature for 5 min. Bacterial cell was harvested by centrifugation at 5,000 × g for 5 min at 4°C and cell pellet was resuspended in 700 μl of lysis buffer (RLT buffer, Qiagen) by vortexing for 10 s. Bacterial cell lysis was done by vortexing vigorously for 5 min in 2 ml safe lock tube containing ~30 mg acid washed glass beads (212–300 μm, Sigma, G1277). Lysate was centrifuged for 10 s at 13,800 × g (Heraeus Biofuge 13) and supernatant was transferred into a new tube. Equal volume of 70% ethanol was added to supernatant and mixed well by pipetting. Then, 700 μl of lysate was transferred to RNeasy spin column and centrifuged for 15 s at 13,800 × g. Flow through was discarded and remaining lysate solution was added and centrifuged in the same column. On-column DNA digestion with RNase Free DNase set (Qiagen) and RNA clean-up was done according to manufacturer's instructions. After washing and elution steps according to manufacturer's protocol, RNA samples were further subjected to DNA digestion in solution using RNase Free DNase set (Qiagen). DNA digestion process was carried out twice and cleaned up in new RNeasy spin columns. Quantity of total RNA was measured with NanoDrop™ 8,000 spectrophotometer. Absence of genomic DNA contamination in RNA samples was confirmed by PCR with primers of housekeeping genes glyA and aspA. All sequences of primers used in this study are listed in Table 2.
qRT-PCR
The qRT-PCR assay was carried out in 96 well plates (MicroAmp Fast 96 well reaction plate, Applied Biosystem) using QuantiTect SYBR Green RT-PCR kit (Qiagen) according to the manufacturer's instructions. One step qRT-PCR was run in StepOne Real-Time PCR system (Applied Biosystems). 16S rRNA gene was used as reference gene for relative quantification of expressed catalase like gene in different treatments (aerobic vs. microaerobic). 16S rRNA gene had been used as reference genes for various studies (Klančnik et al., 2006; Koolman et al., 2016). Primers for target and endogenous reference used in qRT-PCR are listed in Table 2. Standard curve for both catalase like gene and 16S rRNA was created to determine efficiency of qRT-PCR using 10−3 to 10−8 dilution series of amplified PCR products from cDNA templates. For each RNA sample, four replicates were included in this study. In 25 μl reaction mixture, 12.5 μl QuantiTect SYBR Green RT-PCR Master Mix, 1 μl each of forward primer and reverse primer, 0.25 μl of QuantiTect RT mix, 8.25 μl RNase free water, and 2 μl RNA sample were included. Negative control for each sample without RT mix was included. One step qRT-PCR conditions included 50°C for 30 min (reverse transcription step), 95°C for 15 min, 40 cycles of the following steps: 94°C for 15 s, 50°C for 30 s, 72°C for 30 s (data collection step), that was followed by melting curve analysis step with 0.03°C/s temperature rise up to 95°C. Amplification efficiency of target gene and endogenous control was determined to be in range of 1.944 to 2. So, calculation of fold change in transcription level for each strain in aerobic vs. microaerobic condition was done using the Pffafl method (Pfaffl, 2004) using mean CT values. Statistical analysis for relative comparison of transcription level was done using one-way ANOVA.
Results
Prevalence of Aerotolerant C. coli Strains
Campylobacter spp. (76 C. jejuni and 91 C. coli strains; Table S1) were screened for aerotolerance; 46.7% (78/167) were aerotolerant (viable after a 12-h incubation in aerobic conditions), whereas 23.9% (40/167) were hyper-aerotolerant (viable after a 24-h incubation in aerobic conditions) (Figure 1). Among the 76 C. jejuni strains, 6.6% (5/76) were aerotolerant and two strains from chicken meat and chicken liver were hyper-aerotolerant. A greater incidence of aerotolerant strains (80.2%, 73/91) was observed for C. coli; 100% of isolates from chicken gizzards (3/3), turkey (2/2) and pork samples (2/2), and 85.9% (49/57) of chicken liver isolates were aerotolerant. Similarly, 41.7% of all C. coli isolates were hyper-aerotolerant, and 49.1% (28/57) of chicken liver isolates could survive up to 24 h of aerobic incubation (Table 3).
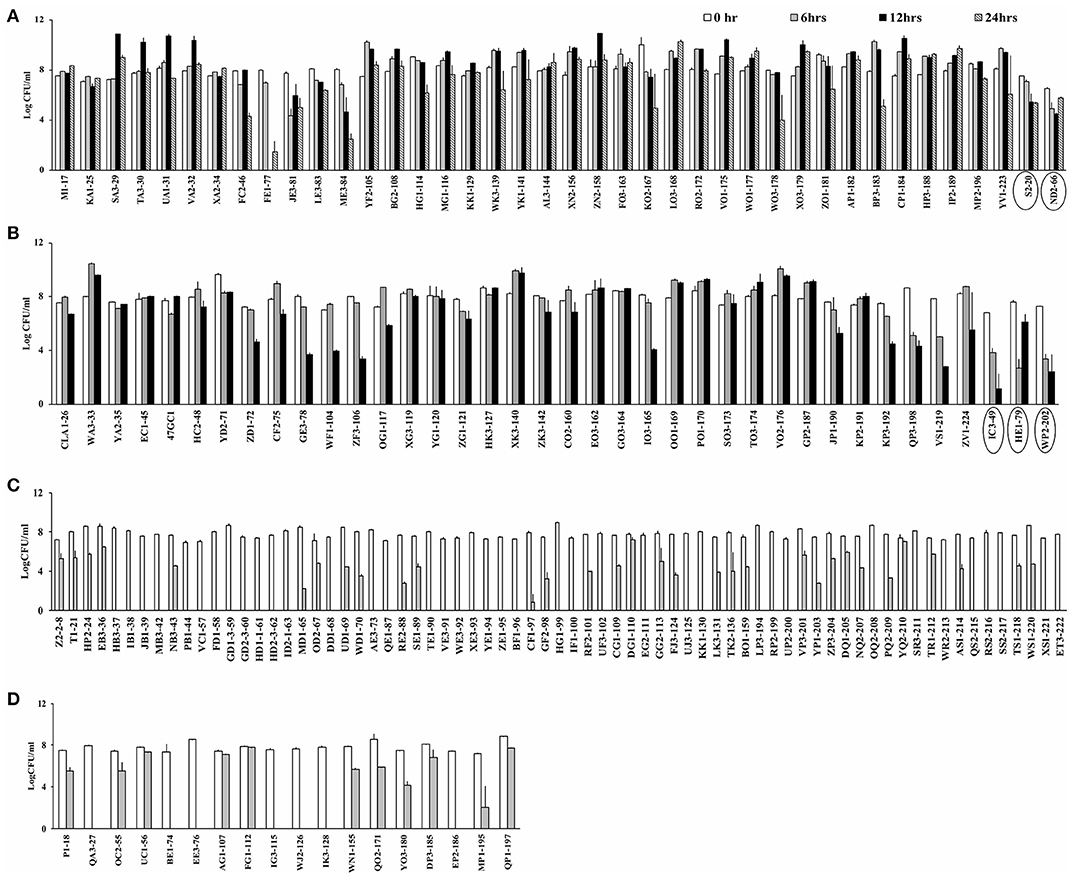
Figure 1. Screening for aerotolerant Campylobacter strains by incubating in MHB aerobically with agitation. (A) Hyper-aerotolerant C. coli and C. jejuni (circled) strains surviving a 24 h exposure to aerobic growth conditions. (B) Aerotolerant C. coli and C. jejuni (circled) strains surviving a 12 h exposure to aerobic conditions. Aerosensitive (C) C. jejuni and (D) C. coli strains not surviving the 12 h exposure to aerobic conditions. Error bars represent the mean standard error of triplicate experiments.
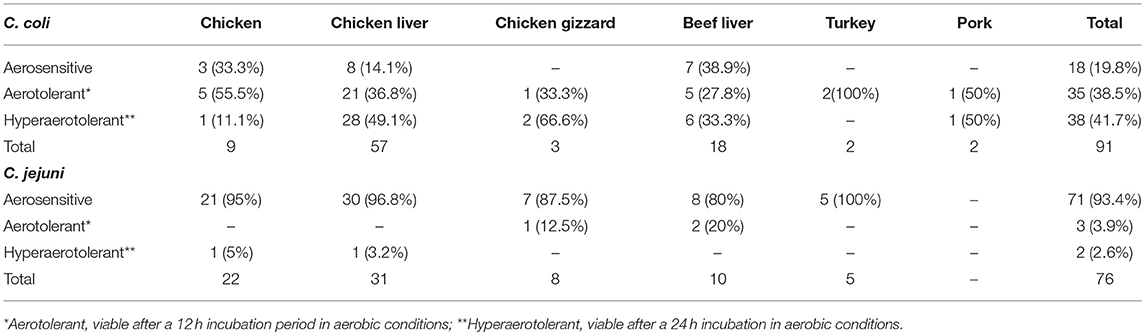
Table 3. Screening C. coli and C. jejuni strains from retail meat and liver sources for aerotolerance.
Oxidative Stress Subsystem and Transcriptional Regulators
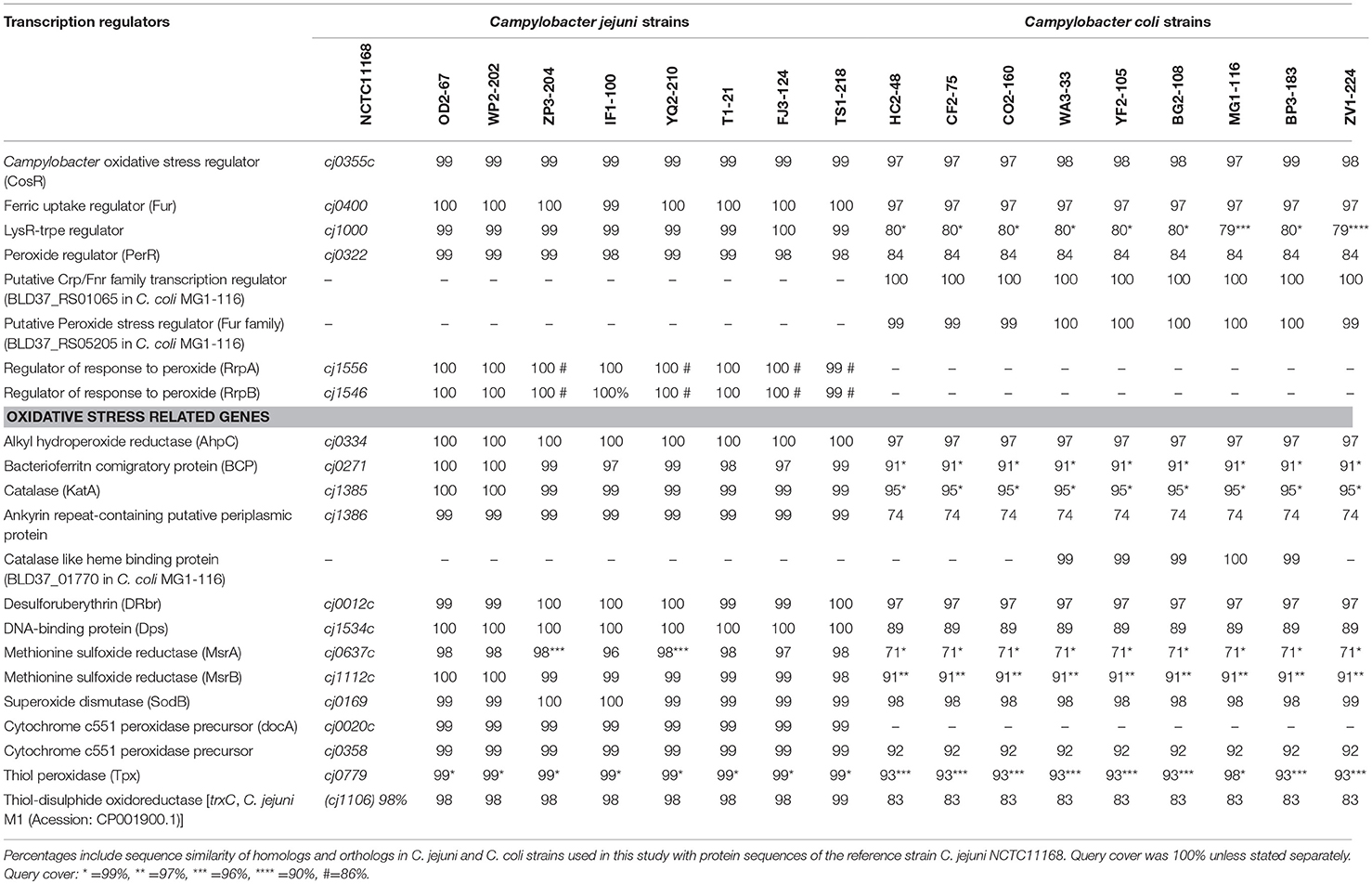
A total of 17 WGS Campylobacter strains (8 C. jejuni, 9 C. coli strains) from our laboratory were used for comparative genomic analysis. Among WGS strains, only one C. jejuni WP2-202 but all WGS C. coli strains showed less sensitivity to aerobic conditions (Table 1). C. coli strains YF2-105, BG2-108, MG1-116, and BP3-183 from chicken liver were hyper-aerotolerant. Functional subsystem comparison of WGS Campylobacter strains (Table 1) by RAST and BLAST revealed relatively few genomic differences between C. jejuni and C. coli strains with respect to genes involved in oxidative stress (Table 4). Homologs for oxidative stress-related genes and transcriptional regulators were present in all sequenced C. jejuni and C. coli strains (Table 4). Transcriptional regulators for responsiveness to peroxide, e.g., RrpA (encoded by cj1546) and RrpB (cj1556), were previously shown to function in the aerobic stress response (Gundogdu et al., 2015); however, the sequenced C. coli strains in our study lacked homologs or orthologs for RrpA and RrpB (Table 4). Genes encoding the cytochrome c551 peroxidase precursor (cj0020c and cj0358) play a role in Campylobacter colonization (Bingham-Ramos and Hendrixson, 2008), and homologs for both genes were present in all WGS C. jejuni strains. Interestingly, C. coli strains lacked a homologs or orthologous sequence for cj0020c, which is involved in colonization (Bingham-Ramos and Hendrixson, 2008).
A putative transcriptional regulator sequence in the Crp/Fnr family (BLD37_RS01065, Table 5) was present in C. coli MG1-116, but was not identified in the WGS C. jejuni strains or the reference strain, C. jejuni NCTC11168. However, several C. jejuni strains associated with Guillian Barre syndrome (GenBank accession numbers: CP012689.1, CP012689.1, CP002029.1) and C. jejuni strains in the ST 677 clonal complex isolated from human feces and blood (Bioproject PRJNA268846) harbor orthologous sequences (~58% sequence similarity) of putative CRP/Fnr family transcriptional regulators. Similarly, another sequence for a putative peroxide stress regulator/ferric uptake regulation protein (Fur family) [(BLD37_RS05205) in C. coli MG1-116] (Table 5) was identified in all C. coli strains, but was absent in C. jejuni strains.
Catalase-Like Heme Binding Protein
Comparative genomic analysis revealed that a gene encoding a catalase-like heme binding protein (BLD37_RS01770 in C. coli MG1-116, Table 5) was present in aerotolerant C. coli strains from chicken liver (e.g., strains WA3-33, YF2-105, BG2-108, MG1-116, and BP3-183; Table 4). However, WGS C. coli strains from other sources and all C. jejuni strains lacked the catalase-like gene sequence. We subsequently screened all 167 Campylobacter strains for the gene encoding the catalase-like heme binding protein by PCR analyses. PCR revealed an 844-bp product encoding the catalase-like protein in 74.7% (68/91) of all C. coli strains; this product was absent in 76 C. jejuni strains (Table 6, Figure 2). Most C. coli strains from poultry contained the gene encoding catalase-like heme-binding protein; these included strains from chicken meat (88.9%), chicken liver (94.7%), chicken gizzard (100%), and turkey, 100%. The incidence of the PCR product encoding the catalase-like gene was low to negligible in C. coli strains from beef liver (5.6%) and pork (0%) (Table 6).
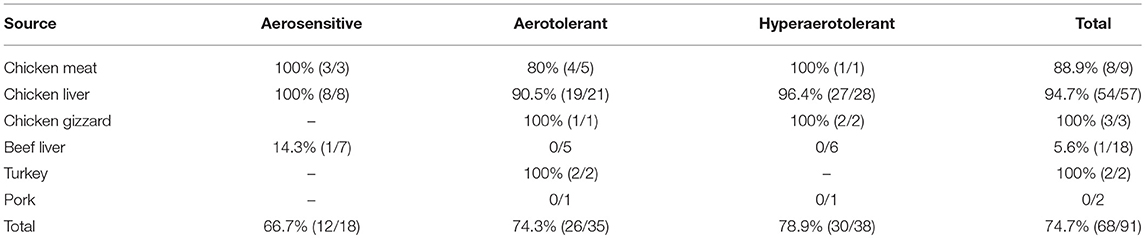
Table 6. C. coli strains testing positive in PCR analysis for the gene encoding the catalase-like heme binding protein.
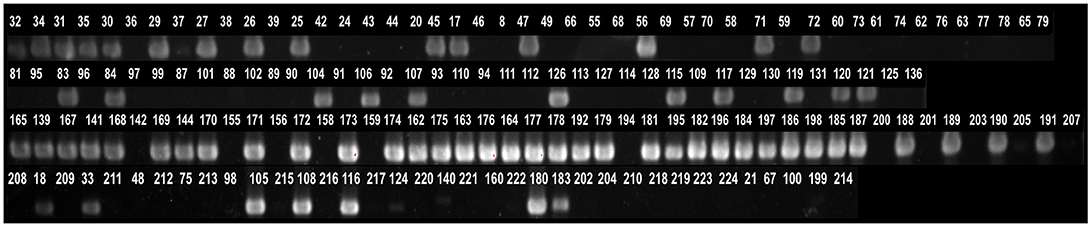
Figure 2. Detection of genes encoding a catalase-like heme binding protein by PCR. A 844-bp PCR product representing the gene was detected in many Campylobacter strains. The numbers above lanes represent the Campylobacter strain number (Table S1).
Genes encoding catalase-like heme-binding proteins were previously reported in urease-positive C. lari (Nakajima et al., 2016) and C. jejuni CFSAN032806 from chicken breast (GenBank accession no. CP023543.1). The genomic arrangement of the region containing the catalase-like gene in C. coli MG1-116 was compared with C. jejuni CFSAN032806 and C. lari UPTC CF 89-12 (Figure 3). The arsenate resistance operon was upstream of the catalase-like protein in C. coli MG1116 and divergently transcribed (Figure 3A, genes e-i). In C. lari UPTC CF 89-12, the catalase-like gene mapped adjacent to mreB and mreC, which are involved in determining morphological shape of the bacterial cell (Figure 3C, genes 8–9) (Nakajima et al., 2016). The gene encoding the catalase-like protein in C. jejuni CFSAN032806 is flanked by genes similar to those in C. coli, but lacks the genes encoding Acr3 and arsenate reductase (Figures 3A,B). Nucleotide similarity was highest between strains of the same Campylobacter species, and differences were greater between species (Table 7). Phylogenetic analyses indicated that the genes encoding the catalase-like proteins were more similar between C. coli and C. jejuni, which grouped in a different clade than C. lari and C. volucris (Figure 4).
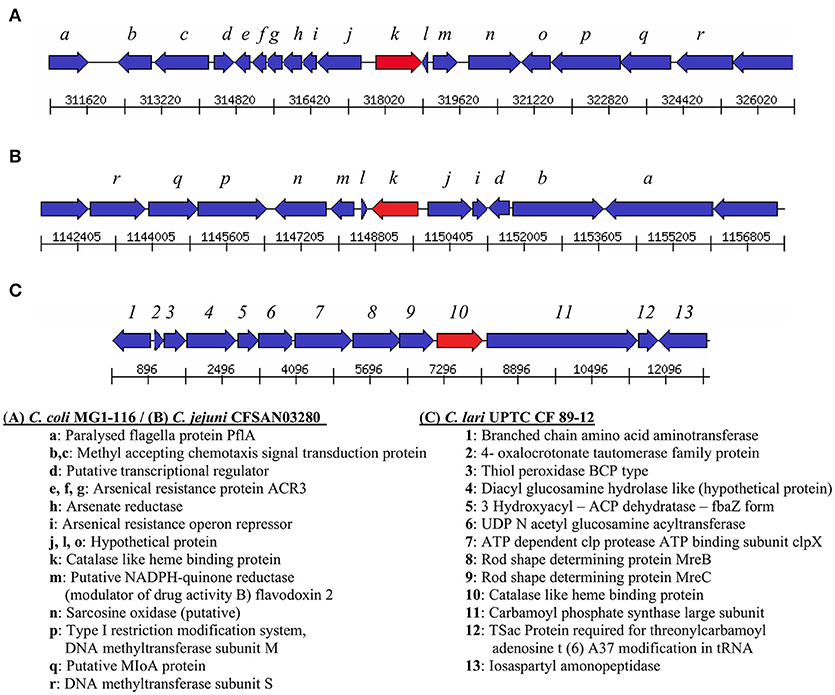
Figure 3. Arrangement of genes flanking the catalase-like heme binding protein (indicated by the red arrow) in (A) C. coli MG1-116 (B) C. jejuni CFSAN032806, and (C) C. lari UPTC CF 89-12.
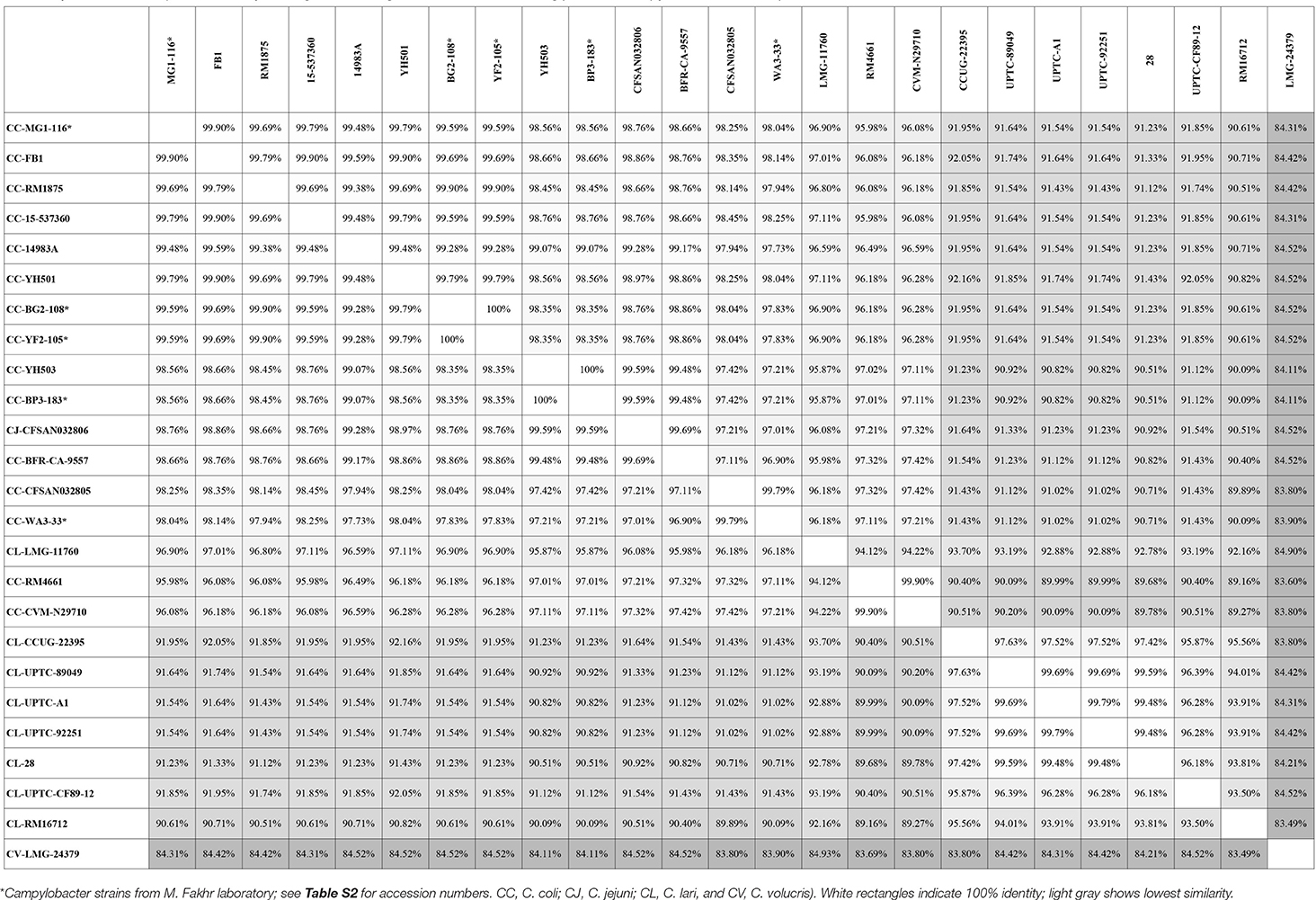
Table 7. Nucleic acid sequence similarity of the gene encoding catalase-like heme binding protein in Campylobacter strains deposited in GenBank.
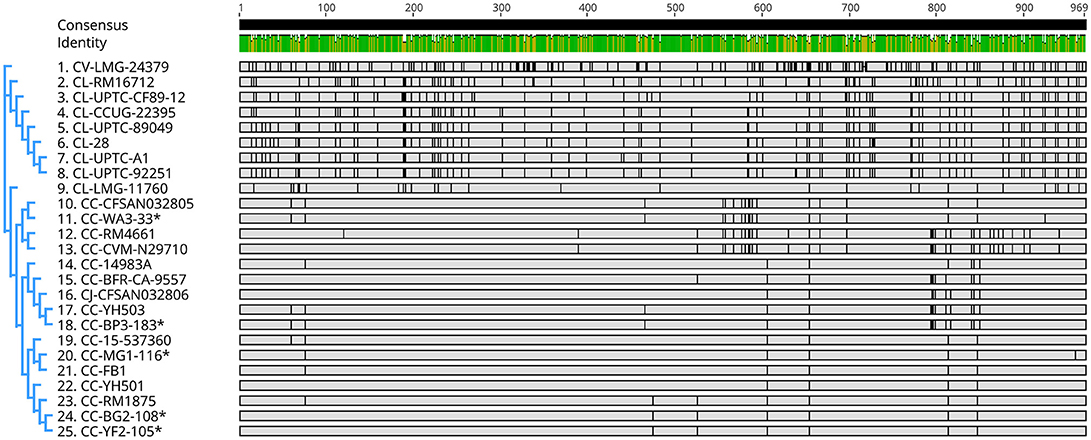
Figure 4. Nucleotide sequence similarity and phylogenetic analysis of genes encoding catalase-like heme binding proteins in Campylobacter (Table S2). Graphics were constructed using the neighbor-joining method (Tamura-Nei model) with Geneious V. 11. C. coli strains from the author's laboratory are marked with asterisks. Cc, C. coli; Cj, C. jejuni; Cl, C. lari; and Cv, C. volucris.
Approximately 78.9% (30/38) of the hyper-aerotolerant, 74.3% (26/35) aerotolerant, and 66.7% (12/18) of aerosensitive C. coli strains contain the gene encoding the catalase-like heme binding protein (Table 6). Among 14 strains tested for hydrogen peroxide sensitivity, seven of C. coli strains harbored catalase like gene. Similarly, six of strains (including both C. jejuni and C. coli) were hyper-aerotolerant and four of them were aerotolerant. However, significant differences in H2O2 sensitivity among tested strains were neither correlated with aerotolerancy nor with the presence of the catalase-like gene (Figure 5). All three C. coli strains (P1-18, WA3-33, and MG1-116) used for gene expression study showed transcription of catalase like gene in both aerobic and microaerobic conditions. Hence, relative quantification of catalase like gene transcript level in aerobic condition vs. microaerobic condition was carried out for the three tested C. coli strains using 16S rRNA gene as endogenous control. No significant difference of transcript level was seen in aerobic vs. microaerobic condition for hyperaerotolerant C. coli strain MG1-116 and aerotolerant strain WA3-33 [Fold change: MG1-116 = 0.6973 (p > 0.05), WA3-33 = 0.696 (p > 0.05)] (Figure 6). Interestingly, the aerosensitive strain C. coli P1-18 had 2.0526-fold (p < 0.001) higher transcript level in aerobic condition when compared to microaerobic condition (Figure 6). However, in microaerobic condition, no significant difference of transcript level for the catalase like gene was seen between the hyperaerotolerant C. coli strain MG1-116, the aerotolerant strain WA3-33, and the aerosensitive strain P1-18 when normalized against the transcript level of the aerosensitive strain. In other words, the catalase like gene transcript level was relatively similar among the three strains when tested in microaerobic condition.
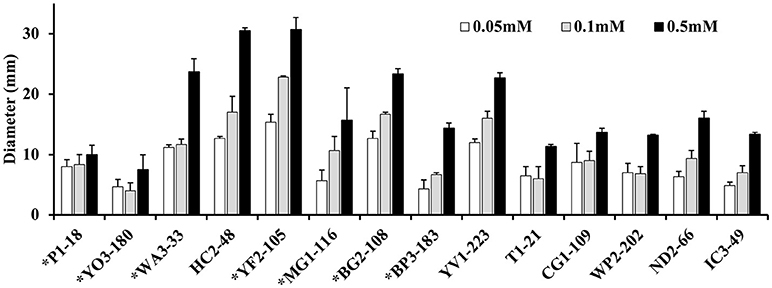
Figure 5. Assay for hydrogen peroxide sensitivity in C. coli strains P1-18 (CC18), YO3-180, WA3-33, YF2-105, MG1-116, BG2-108, BP3-183, HC2-48, and YV1-223 and C. jejuni strains T1-21, CG1-109, WP2-202, ND2-66, and IC3-49. Bacterial cells were exposed to different concentrations of H2O2 (0.05, 0.1, and 0.5 mM) and the diameter of diffusion zones was measured. C. coli strains with the gene encoding the catalase-like heme binding protein are indicated with asterisks. Vertical bars represent the standard error for the mean values from triplicate experiments.
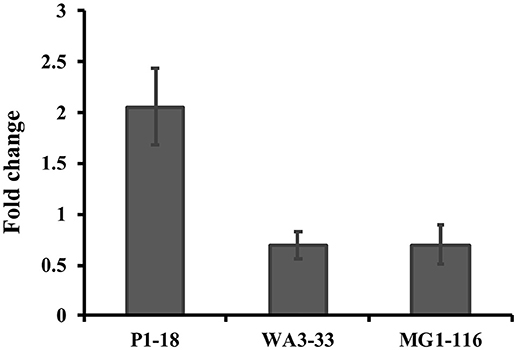
Figure 6. Relative quantification of catalase like protein gene expression in C. coli strains in aerobic condition compared to microaerobic condition. Hyperaerotolerant C. coli strain MG1-116, aerotolerant WA3-33, and aerosensitive P1-18 were used in this study.
Discussion
Campylobacter is an important foodborne pathogen that is transmitted from contaminated food products and water (Newell et al., 2017). Despite being microaerophilic and fastidious organism, many previous reports have reported the isolation of aerotolerant Campylobacter strains that could survive and grow in aerobic conditions (Rodrigues et al., 2015; Oh et al., 2017; O'Kane and Connerton, 2017). In our study, 46.7% (78/167) of tested Campylobacter strains were aerotolerant. Aerotolerant Campylobacter strains had shown prolonged survival during oxidative stress conditions (Oh et al., 2015, 2017; O'Kane and Connerton, 2017), increased biofilm formation (Bronnec et al., 2016b) and a high incidence of virulence genes (Oh et al., 2017) which help to survive in harsh environmental conditions. Even at low-temperature storage conditions, aerotolerant strains survive better than aerosensitive strains in different gaseous atmospheres (Oh et al., 2017). Hence, the high prevalence of Campylobacter reported in retail meat and liver products (Noormohamed and Fakhr, 2012, 2013, 2014b) seems to be associated with higher prevalence of aerotolerant Campylobacter strains found in this study. Our previous study reported the higher prevalence of Campylobacter in retail meat and liver products, where more than 75% of beef and chicken liver were contaminated with C. coli (Noormohamed and Fakhr, 2012, 2013). In the current study, 80.2% of the C. coli isolates from retail meat and liver products were aerotolerant; however, only 6.6% of C. jejuni were aerotolerant. The increased incidence of aerotolerant C. coli might be a contributing factor to the prevalence of C. coli in retail meat and liver products. Aerotolerant C. jejuni were not common in our study; however, Oh et al. found that 71% of C. jejuni strains from retail chicken were aerotolerant and 37% were hyper-aerotolerant (Oh et al., 2015). Although C. coli is the causal agent in only 7% of human clinical cases of campylobacteriosis (Gillespie et al., 2002), aerotolerant C. coli strains with a ST complex similar to clinical isolates (Noormohamed and Fakhr, 2014a) might result in additional clinical cases.
Various genes in Campylobacter function in the oxidative stress response (Flint et al., 2016), and most studies relevant to the oxidative stress response in Campylobacter have been conducted with C. jejuni (Butcher et al., 2015; Handley et al., 2015; Rodrigues et al., 2016). The oxidative stress response is modulated by many transcriptional regulators (PerR, Fur, RrpA, RrpB, CosR, CsrA) (Fields and Thompson, 2008; Hwang et al., 2011; Gundogdu et al., 2015; Handley et al., 2015; Flint et al., 2016), and some of these also function in iron transport (van Vliet et al., 2002; Holmes et al., 2005). Genes previously identified in the regulation of the oxidative stress response in C. jejuni (RrpA and RrpB) (Gundogdu et al., 2015, 2016) and colonization (cj0020c) (Bingham-Ramos and Hendrixson, 2008) were absent in the C. coli strains in our study. Mutagenesis study for MarR like transcriptional regulators RrpA and RrpB genes in C. jejuni strains have shown their role in oxidative and aerobic stress response. Mutants of these genes showed reduced survival under aerobic stress (Gundogdu et al., 2015, 2016). However, the absence of RrpA and RrpB seems to be distinctive genomic characteristics among C. coli strains (Gundogdu et al., 2016; O'Kane and Connerton, 2017). Comparative analysis of >4,000 Campylobacter genome sequences had shown the lack of RrpA and RrpB in C. coli strains (Gundogdu et al., 2016). A link between presence of transcriptional regulator like RrpB in C. jejuni strains with adaptation and survival capability of these strains in variable environmental conditions has been proposed (Gundogdu et al., 2016). Thus, it is likely that unique genomic traits and various putative transcription regulators found in C. coli strains might play a role in differential adaptation to oxidative stress. Presence of putative Crp/Fnr family transcriptional regulators in C. coli and some clinical C. jejuni strains might also indicate a possible association with virulence. The functional analyses of the transcriptional regulators identified in C. coli strains and their role in oxidative stress is underway in our laboratory.
In our study, comparative genomic analyses and PCR screening indicated that most C. coli isolates contained the gene encoding the catalase-like heme binding protein. The fact that none of the C. jejuni strains screened in our study possessed the gene is not surprising since out of all sequenced C. jejuni genomes in Genbank, only one C. jejuni CFSAN032806 genome harbors the catalase-like gene. Interestingly, C. jejuni CFSAN032806 and most of the C. coli isolates with the catalase-like protein gene originated from chicken or turkey, which might explain the greater prevalence of the catalase like protein gene in poultry isolates in the current study. A previously published report has shown presence of catalase-like gene in urease-positive C. lari strains (Nakajima et al., 2016). Sequence data from GenBank showed presence of catalase like gene in various species of Campylobacter. The high degree of sequence similarity in the catalase-like genes suggests the recent dissemination of this gene between strains; however, comparison of similar genes in C. coli, C. jejuni, and C. lari revealed inter-species variation in this region.
Although, a previous report (Oh et al., 2015) showed enhanced resistance toward hydrogen peroxide in aerotolerant strains, significant differences among hydrogen peroxide sensitivity could not be correlated among tested Campylobacter strains with their aerotolerancy in this study. Likewise, no significant difference in hydrogen peroxide sensitivity was observed between strains containing the catalase-like gene, and the gene was prevalent in both aerotolerant and aerosensitive strains. Nakajima et al. had also reported variable levels of catalase activity in C. lari strains that were independent of the presence of this gene (Nakajima et al., 2016). In a previous study, an aerotolerant C. jejuni strain showed higher transcript level for genes related to oxidative stress response in aerobic condition when compared to microaerobic condition (Rodrigues et al., 2016). A higher catalase equivalent activity in microaerobic condition was also reported in aerotolerant C. jejuni strains when compared to aerosensitive ones (Rodrigues et al., 2016). This was not the case in our study since the catalase like gene transcript level was relatively similar among the three hyperaerotolerant, aerotolerant, and the aerosensitive strains when tested in microaerobic condition. However, in the current study, higher expression of catalase like gene in aerobic condition was seen in aerosensitive C. coli strain P1-18 than aerotolerant strains (MG1-116 and WA3-33). Thus, it remains plausible that this gene may not be directly involved in conferring aerotolerancy but might help aerosensitive strains to slightly cope with oxidative stress. The absence of this gene in C. jejuni despite the presence of aerotolernat C. jejuni strains indicates that another mechanism might be involved in conferring aerotolerancy in C. jejuni and C. coli strains. The exact function of the gene encoding catalase-like heme binding protein remains unclear and awaits further investigation using mutagenesis and complementation.
Some prominent genomic differences were observed among the WGS strains, and these might contribute to discrepancies in aerotolerance as well as survival. A Type VI secretion system (T6SS) in Campylobacter had an enhanced hemolytic effect on blood cells and functioned in virulence (Lertpiriyapong et al., 2012; Bleumink-Pluym et al., 2013; Marasini, 2016). WGS strains C. coli ZV1-224 and C. jejuni (OD2-67, IF1-100, WP2-202, ZP3-204, YQ2-210, and TS1-218) harbor sequences for a T6SS (Marasini and Fakhr, 2016b, 2017a,b,c). A T6SS was also present in the previously reported aerotolerant C. jejuni Bf strain (Bronnec et al., 2016a,b) and C. coli OR12 strain (O'Kane and Connerton, 2017). In this study, C. coli ZV1-224 and C. jejuni WP2-202 were aerotolerant, but all other Campylobacter strains with putative T6SSs were aerosensitive. Hence, it is unlikely that the T6SS is a contributing factor for enhanced aerotolerance, a conclusion supported by a recent study (O'Kane and Connerton, 2017). The presence of a functional Entner Doudoroff (ED) pathway could enhance survival and biofilm formation in Campylobacter (Vegge et al., 2016). Thus, the ED pathway encoded by the C. coli ZV1-224 genome could contribute to enhanced aerotolerance, but further validation is needed.
O'Kane and Connerton recently demonstrated that relatively few genomic differences and mutations can create aerotolerance in a wild-type aerosensitive Campylobacter strain (O'Kane and Connerton, 2017). Hence, genomic differences between Campylobacter spp. (Fouts et al., 2005) might play a role in the differential aerotolerance. It is also possible that transcriptional and translational modifications might be sufficient to facilitate aerotolerance without significant genetic differences in genomic structure (Bronnec et al., 2016a,b).
In conclusion, aerotolerant C. coli strains are highly prevalent in retail meat and liver products. Aerotolerant C. coli strains with antimicrobial resistance and ST complexes similar to clinical strains pose a risk towards emerging clinical cases. Some genes encoding transcriptional regulators and a catalase-like protein are present in C. coli strains which are missing in C. jejuni strains. Although the catalase like gene is being transcribed in C. coli strains, its exact function in stress response or virulence is still not explored. Mutagenesis studies are currently underway in our laboratory to investigate the potential role of this gene in C. coli.
Author Contributions
AK and MF research design and manuscript preparation. AK, DM, CO, and KM experimental procedures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge financial support from the Research Office of The University of Tulsa (Tulsa, OK, USA) for granting CO a student research grant. The authors would also like to thank the Beta Beta Beta National Biological Honor Society for granting KM a student research grant.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02951/full#supplementary-material
References
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75. doi: 10.1186/1471-2164-9-75
Bingham-Ramos, L. K., and Hendrixson, D. R. (2008). Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect. Immun. 76, 1105–1114. doi: 10.1128/IAI.01430-07
Bleumink-Pluym, N. M., van Alphen, L. B., Bouwman, L. I., Wösten, M. M., and van Putten, J. P. (2013). Identification of a functional type VI secretion system in Campylobacter jejuni conferring capsule polysaccharide sensitive cytotoxicity. PLoS Pathog. 9:e1003393. doi: 10.1371/journal.ppat.1003393
Bolton, D. J. (2015). Campylobacter virulence and survival factors. Food Microbiol. 48, 99–108. doi: 10.1016/j.fm.2014.11.017
Bronnec, V., Haddad, N., Cruveiller, S., Hernould, M., Tresse, O., and Zagorec, M. (2016a). Draft genome sequence of Campylobacter jejuni Bf, an a typical strain able to grow under aerobiosis. Genome Announc. 4, e00120–e00116. doi: 10.1128/genomeA.00120-16
Bronnec, V., Turonová, H., Bouju, A., Cruveiller, S., Rodrigues, R., Demnerova, K., et al. (2016b). Adhesion, biofilm formation, and genomic features of Campylobacter jejuni Bf, an atypical strain able to grow under aerobic conditions. Front. Microbiol. 7:1002. doi: 10.3389/fmicb.2016.01002
Bronowski, C., James, C. E., and Winstanley, C. (2014). Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol. Lett. 356, 8–19. doi: 10.1111/1574-6968.12488
Butcher, J., Handley, R. A., van Vliet, A. H., and Stintzi, A. (2015). Refined analysis of the Campylobacter jejuni iron-dependent/independent Fur- and PerR-transcriptomes. BMC Genomics 16:498. doi: 10.1186/s12864-015-1661-7
De Vries, S. P. W., Gupta, S., Baig, A., Wright, E., Wedley, A., Jensen, A. N., et al. (2017). Genome-wide fitness analyses of the foodborne pathogen Campylobacter jejuni in in vitro and in vivo models. Sci. Rep. 7, 1–17. doi: 10.1038/s41598-017-01133-4
Dewey-Mattia, D., Manikonda, K., and Vieira, A. (2016). Surveillance for Foodborne Disease Outbreaks United States, 2014: Annual Report (Atlanta: US Department of Health and Human Services, CDC), 1–24.
Fields, J. A., and Thompson, S. A. (2008). Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 190, 3411–3416. doi: 10.1128/JB.01928-07
Flint, A., Stintzi, A., and Saraiva, L. M. (2016). Oxidative and nitrosative stress defences of Helicobacter and Campylobacter species that counteract mammalian immunity. FEMS Microbiol. Rev. 40, 938–960. doi: 10.1093/femsre/fuw025
Fouts, D. E., Mongodin, E. F., Mandrell, R. E., Miller, W. G., Rasko, D. A., Ravel, J., et al. (2005). Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:15. doi: 10.1371/journal.pbio.0030015
Geissler, A. L., Bustos Carrillo, F., Swanson, K., Patrick, M. E., Fullerton, K. E., Bennett, C., et al. (2017). Increasing Campylobacter infections, outbreaks, and antimicrobial resistance in the United States, 2004-2012. Clin. Infect. Dis. 65, 1624–1631. doi: 10.1093/cid/cix624
Gillespie, I. A., O'Brien, S. J., Frost, J. A., Adak, G. K., Horby, P., Swan, A. V., et al. (2002). A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8, 937–942. doi: 10.3201/eid0809.010187
Gundogdu, O., da Silva, D. T., Mohammad, B., Elmi, A., Mills, D. C., Wren, B. W., et al. (2015). The Campylobacter jejuni MarR-like transcriptional regulators RrpA and RrpB both influence bacterial responses to oxidative and aerobic stresses. Front. Microbiol. 6:724. doi: 10.3389/fmicb.2015.00724
Gundogdu, O., da Silva, D. T., Mohammad, B., Elmi, A., Wren, B. W., van Vliet, A. H., et al. (2016). The Campylobacter jejuni oxidative stress regulator RrpB is associated with a genomic hypervariable region and altered oxidative stress resistance. Front. Microbiol. 7:2117. doi: 10.3389/fmicb.2016.02117
Handley, R. A., Mulholland, F., Reuter, M., Ramachandran, V. K., Musk, H., Clissold, L., et al. (2015). PerR controls oxidative stress defence and aerotolerance but not motility-associated phenotypes of Campylobacter jejuni. Microbiology 161, 1524–1536. doi: 10.1099/mic.0.000109
Holmes, K., Mulholland, F., Pearson, B. M., Pin, C., McNicholl-Kennedy, J., Ketley, J. M., et al. (2005). Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151, 243–257. doi: 10.1099/mic.0.27412-0
Huang, J., Zong, Q., Zhao, F., Zhu, J., and Jiao, X.-an. (2016). Quantitative surveys of Salmonella and Campylobacter on retail raw chicken in Yangzhou, China. Food Control 59, 68–73. doi: 10.1016/j.foodcont.2015.05.009
Hwang, S., Kim, M., Ryu, S., and Jeon, B. (2011). Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PLoS ONE 6:22300. doi: 10.1371/journal.pone.0022300
Klancnik, A., Botteldoorn, N., Herman, L., and Mozina, S. S. (2006). Survival and stress induced expression of groEL and rpoD of Campylobacter jejuni from different growth phases. Int. J. Food Microbiol. 112, 200–207. doi: 10.1016/j.ijfoodmicro.2006.03.015
Koolman, L., Whyte, P., Burgess, C., and Bolton, D. (2016). Virulence gene expression, adhesion and invasion of Campylobacter jejuni exposed to oxidative stress (H2O2). Int. J. Food Microbiol. 220, 33–38. doi: 10.1016/j.ijfoodmicro.2016.01.002
Lertpiriyapong, K., Gamazon, E. R., Feng, Y., Park, D. S., Pang, J., Botka, G., et al. (2012). Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS ONE 7:42842. doi: 10.1371/journal.pone.0042842
Marasini, D. (2016). Molecular Characterization of Megaplasmids in Campylobacter jejuni and Campylobacter coli Isolated from Retail Meats. Available online at: http://search.proquest.com/openview/fa0fa3228bd0e7c21703fd18fba37f7b/1?pq-origsite=gscholar&cbl=18750&diss=y (Accessed January 23, 2018).
Marasini, D., and Fakhr, M. K. (2016a). Complete genome sequences of Campylobacter jejuni strains OD267 and WP2202 isolated from retail chicken livers and gizzards reveal the presence of novel 116-kilobase and 119-kilobase megaplasmids with type VI secretion systems. Genome Announc. 4:e01060–16. doi: 10.1128/genomeA.01060-16
Marasini, D., and Fakhr, M. K. (2016b). Complete genome sequences of the plasmid-bearing campylobacter coli strains HC2-48, CF2-75, and CO2-160 isolated from retail beef liver. Genome Announc. 4:01004–16. doi: 10.1128/genomeA.01004-16
Marasini, D., and Fakhr, M. K. (2016c). Whole-genome sequencing of a Campylobacter jejuni strain isolated from retail chicken meat reveals the presence of a megaplasmid with mu-like prophage and multidrug resistance genes. Genome Announc. 4:00460–16. doi: 10.1128/genomeA.00460-16
Marasini, D., and Fakhr, M. K. (2017a). Complete genome sequences of Campylobacter jejuni strains isolated from retail chicken and chicken gizzards. Genome Announc. 5, e01351–17. doi: 10.1128/genomeA.01351-17
Marasini, D., and Fakhr, M. K. (2017b). Complete genome sequences of plasmid-bearing Campylobacter coli and Campylobacter jejuni strains isolated from retail chicken liver. Genome Announc. 5:e01350–17. doi: 10.1128/genomeA.01350-17
Marasini, D., and Fakhr, M. K. (2017c). Complete genome sequences of plasmid-bearing multidrug-resistant Campylobacter jejuni and Campylobacter coli strains with type VI secretion systems, isolated from retail turkey and pork. Genome Announc. 5:e01360–17. doi: 10.1128/genomeA.01360-17
Nakajima, T., Kuribayashi, T., Moore, J. E., Millar, B. C., Yamamoto, S., and Matsuda, M. (2016). Molecular identification and characterisation of catalase and catalase-like protein genes in urease-positive thermophilic Campylobacter (UPTC). Br. J. Biomed. Sci. 73, 56–66. doi: 10.1080/09674845.2016.1156867
Newell, D. G., Mughini-Gras, L., Kalupahana, R. S., and Wagenaar, J. A. (2017). “Campylobacter epidemiology-sources and routes of transmission for human infection,” in Campylobacter: Features, Detection, and Prevention of Foodborne Disease, ed G. Klein (Cambridge, MA: Academic Press), 85–110. doi: 10.1016/B978-0-12-803623-5.00005-8
Noormohamed, A., and Fakhr, M. K. (2012). Incidence and antimicrobial resistance profiling of Campylobacter in retail chicken livers and gizzards. Foodborne Pathog. Dis. 9, 617–624. doi: 10.1089/fpd.2011.1074
Noormohamed, A., and Fakhr, M. K. (2013). A higher prevalence rate of Campylobacter in retail beef livers compared to other beef and pork meat cuts. Int. J. Environ. Res. Public Health 10, 2058–2068. doi: 10.3390/ijerph10052058
Noormohamed, A., and Fakhr, M. (2014a). Molecular typing of Campylobacter jejuni and Campylobacter coli isolated from various retail meats by MLST and PFGE. Foods 3, 82–93. doi: 10.3390/foods3010082
Noormohamed, A., and Fakhr, M. K. (2014b). Prevalence and antimicrobial susceptibility of Campylobacter spp. in Oklahoma conventional and organic retail poultry. Open Microbiol. J. 8, 130–137. doi: 10.2174/1874285801408010130
Oh, E., McMullen, L., and Jeon, B. (2015). High prevalence of hyper-aerotolerant Campylobacter jejuni in retail poultry with potential implication in human infection. Front. Microbiol. 6:1263. doi: 10.3389/fmicb.2015.01263
Oh, E., McMullen, L. M., Chui, L., and Jeon, B. (2017). Differential survival of hyper-aerotolerant Campylobacter jejuni under different gas conditions. Front. Microbiol. 8:954. doi: 10.3389/fmicb.2017.00954
O'Kane, P. M., and Connerton, I. F. (2017). Characterisation of aerotolerant forms of a robust chicken colonizing Campylobacter coli. Front. Microbiol. 8:513. doi: 10.3389/fmicb.2017.00513
Pfaffl, M. (2004). “Quantification strategies in real-time PCR,” in A-Z Quant. PCR, ed S. A. Bustin (La Jolla, CA: International University Line), 87–112.
Rodrigues, R. C., Haddad, N., Chevret, D., Cappelier, J. M., and Tresse, O. (2016). Comparison of proteomics profiles of Campylobacter jejuni strain Bf under microaerobic and aerobic conditions. Front. Microbiol. 7:1596. doi: 10.3389/fmicb.2016.01596
Rodrigues, R. C., Pocheron, A. L., Hernould, M., Haddad, N., Tresse, O., and Cappelier, J. M. (2015). Description of Campylobacter jejuni Bf, an atypical aero-tolerant strain. Gut Pathog. 7:30. doi: 10.1186/s13099-015-0077-x
van Vliet, A. H., Ketley, J. M., Park, S. F., and Penn, C. W. (2002). The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26, 173–186. doi: 10.1016/S0168-6445(02)00095-5
Vegge, C. S., Jansen van Rensburg, M. J., Rasmussen, J. J., Maiden, M. C., Johnsen, L. G., Danielsen, M., et al. (2016). Glucose metabolism via the entner-doudoroff pathway in Campylobacter: a rare trait that enhances survival and promotes biofilm formation in some isolates. Front. Microbiol. 7:1877. doi: 10.3389/fmicb.2016.01877
Keywords: aerotolerance, hyper-aerotolerant, Campylobacter, retail liver, retail meat, oxidative stress, transcriptional regulators, catalase
Citation: Karki AB, Marasini D, Oakey CK, Mar K and Fakhr MK (2018) Campylobacter coli From Retail Liver and Meat Products Is More Aerotolerant Than Campylobacter jejuni. Front. Microbiol. 9:2951. doi: 10.3389/fmicb.2018.02951
Received: 24 February 2018; Accepted: 16 November 2018;
Published: 12 December 2018.
Edited by:
Giovanna Suzzi, Università degli Studi di Teramo, ItalyReviewed by:
Odile Tresse, INRA Centre Angers-Nantes Pays de la Loire, FranceBeatrix Stessl, Veterinärmedizinische Universität Wien, Austria
Copyright © 2018 Karki, Marasini, Oakey, Mar and Fakhr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamed K. Fakhr, bW9oYW1lZC1mYWtockB1dHVsc2EuZWR1
 Anand B. Karki
Anand B. Karki Daya Marasini
Daya Marasini Clark K. Oakey
Clark K. Oakey Mohamed K. Fakhr
Mohamed K. Fakhr