
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 November 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02916
This article is part of the Research Topic The Role of Plant Hormones in Plant-Microbe Symbioses View all 15 articles
The ability of plant-associated microbes to produce gibberellin A (GA) phytohormones was first described for the fungal rice pathogen Gibberella fujikuroi in the 1930s. Recently the capacity to produce GAs was shown for several bacteria, including symbiotic alpha-proteobacteria (α-rhizobia) and gamma-proteobacteria phytopathogens. All necessary enzymes for GA production are encoded by a conserved operon, which appears to have undergone horizontal transfer between and within these two phylogenetic classes of bacteria. Here the operon was shown to be present and functional in a third class, the beta-proteobacteria, where it is found in several symbionts (β-rhizobia). Conservation of function was examined by biochemical characterization of the enzymes encoded by the operon from Paraburkholderia mimosarum LMG 23256T. Despite the in-frame gene fusion between the short-chain alcohol dehydrogenase/reductase and ferredoxin, the encoded enzymes exhibited the expected activity. Intriguingly, together these can only produce GA9, the immediate precursor to the bioactive GA4, as the cytochrome P450 (CYP115) that catalyzes the final hydroxylation reaction is missing, similar to most α-rhizobia. However, phylogenetic analysis indicates that the operon from β-rhizobia is more closely related to examples from gamma-proteobacteria, which almost invariably have CYP115 and, hence, can produce bioactive GA4. This indicates not only that β-rhizobia acquired the operon by horizontal gene transfer from gamma-proteobacteria, rather than α-rhizobia, but also that they independently lost CYP115 in parallel to the α-rhizobia, further hinting at the possibility of detrimental effects for the production of bioactive GA4 by these symbionts.
Gibberellin A (GA) was first discovered in the eponymous fungal rice pathogen Gibberella fujikuroi, which eventually enabled identification of these diterpenoids in plants where they serve as hormones regulating growth and development (Hedden and Sponsel, 2015). As suggested from their production by G. fujikuroi, these phytohormones also affect plant-microbe interactions (De Bruyne et al., 2014). However, in bacteria GA production was first reported from nitrogen-fixing rhizobial symbionts rather than plant pathogens (Bottini et al., 2004). More recently, bacterial GA biosynthesis has been elucidated (Morrone et al., 2009; Hershey et al., 2014; Lu et al., 2015; Tatsukami and Ueda, 2016; Nagel and Peters, 2017b; Nagel et al., 2017; Nett et al., 2017b), with all the necessary genes generally found in close association within a biosynthetic operon. This operon seems to only be found in plant-associated bacteria (Levy et al., 2017). However, beyond the symbiotic alpha-proteobacteria (α-rhizobia) where the operon was originally reported and characterized (Tully and Keister, 1993; Tully et al., 1998; Keister et al., 1999; Morrone et al., 2009; Hershey et al., 2014; Tatsukami and Ueda, 2016; Nett et al., 2017b), the operon also was found to be functionally present in a diverse group of plant pathogens from the gamma-proteobacteria class (Lu et al., 2015; Nagel and Peters, 2017b; Nagel et al., 2017). Indeed, there is greater phylogenetic diversity of the operon in these phytopathogens than rhizobia (Nagel and Peters, 2017b), suggesting that bacterial GA biosynthesis originally evolved in this distinct class of bacteria.
The GA biosynthetic operon typically contains genes encoding eight enzymes (Figure 1). First to be characterized were the pair of diterpene cyclases that produce ent-kaurene from the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGDP). This proceeds via sequential reactions, specifically through initial production of ent-copalyl diphosphate (ent-CDP) by a CDP synthase (CPS), followed by a subsequently acting ent-kaurene synthase (KS) (Morrone et al., 2009; Hershey et al., 2014; Lu et al., 2015). An isoprenyl diphosphate synthase (IDS) that produces GGDP from the common isoprenoid precursors isopentenyl diphosphate (IDP) and dimethylallyl diphosphate (DMADP) is also present (Hershey et al., 2014; Nagel and Peters, 2017b). In addition, there are typically at least three cytochromes P450 (CYPs), CYP112, CYP114 and CYP117, along with a ferredoxin (Fd) and short-chain alcohol dehydrogenase/reductase (SDR). The Fd is required for the ring-contraction reaction catalyzed by CYP114, which generates GA12-aldehyde that is further oxidized to GA12 by the SDR (Nagel and Peters, 2017b; Nagel et al., 2017; Nett et al., 2017b). Beyond these common/core genes, many copies of the operon contain an isopentenyl diphosphate isomerase (IDI) that interconverts IDP and DMADP (presumably to optimize production of GGDP), and/or an additional CYP, specifically CYP115 (Nagel and Peters, 2017b; Nagel et al., 2017; Nett et al., 2017a). However, while the core operon enables biosynthesis of GA9, this has not been shown to have classical hormonal activity, and CYP115 is required to catalyze a subsequent hydroxylation reaction to produce bioactive GA4 (Figure 1).
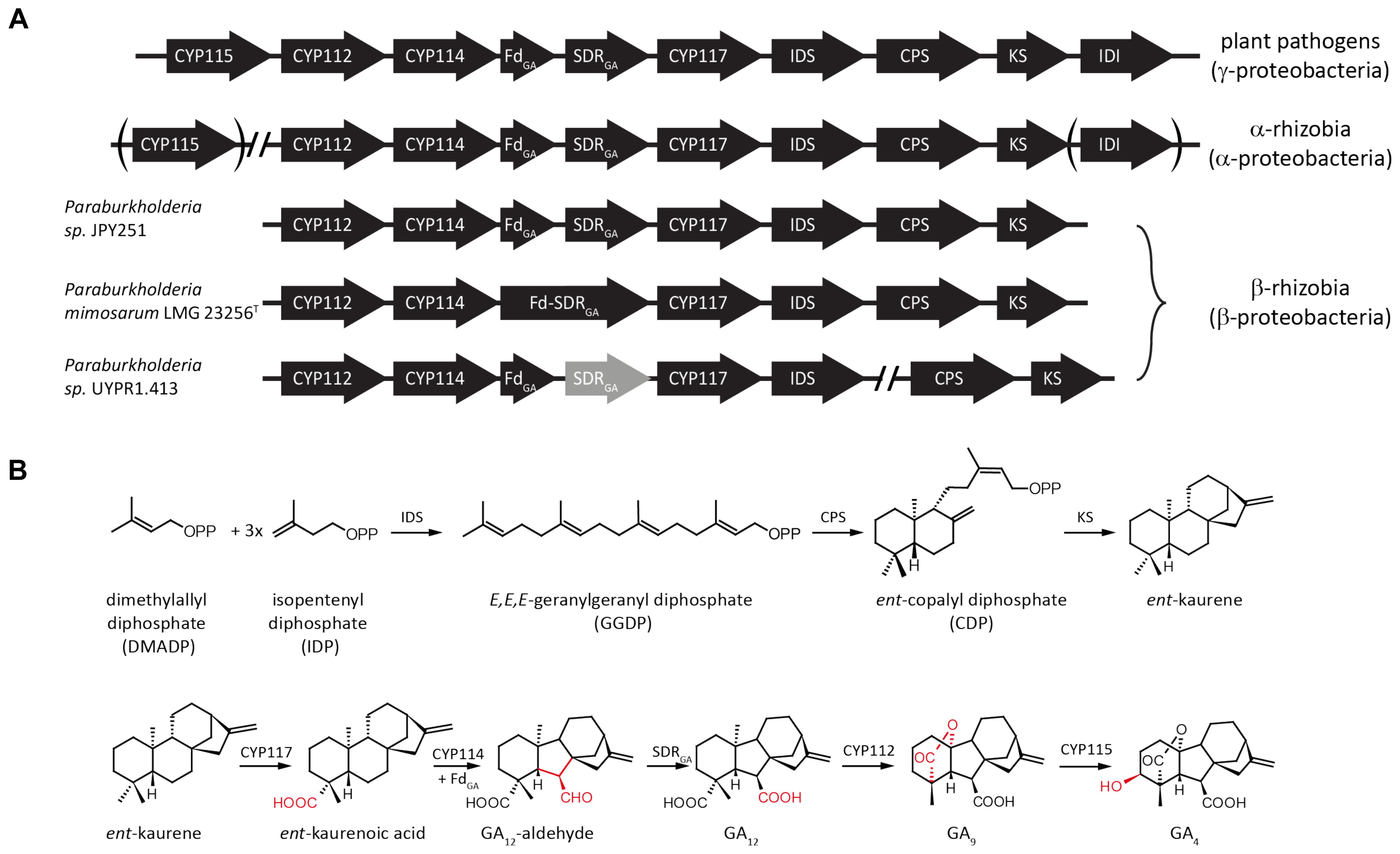
FIGURE 1. Schematic representation of bacterial gibberellin biosynthetic operon and encoded metabolic pathway. (A) Arrows indicate the direction of translation; abbreviations for the genes are CYP, cytochrome P450; Fd, ferredoxin; SDR, short-chain alcohol dehydrogenase/reductase; IDS, isoprenyl diphosphate synthase; CPS, copalyl diphosphate synthase; KS, ent-kaurene synthase; and IDI, isopentenyl diphosphate isomerase. (B) Reactions in the pathway catalyzed by the various enzymes encoded by individual genes from the operon.
Intriguingly, while all phytopathogens contain CYP115 in their operons, almost all α-rhizobia only have a non-functional fragment remaining, and of the less than 20% with the GA operon that also contain a full-length CYP115 gene, in every case but one it is not located in the operon (Nett et al., 2017a). Thus, most α-rhizobia can only produce the penultimate intermediate GA9, as has been demonstrated for Bradyrhizobium japonicum, although given that the operon is only expressed in nodules and the inability to otherwise distinguish between plant and bacteria produced GAs, this required isolation of bacteroids from nodules and radio-isotope feeding studies (Mendez et al., 2014). By contrast, phytopathogens can produce the bioactive GA4. This difference in final GA product presumably reflects their distinct relationships with plant hosts. While it has been noted that a symbiotic beta-proteobacteria (β-rhizobia) also contains the operon (Nagel and Peters, 2017b), this has not otherwise been investigated. The genome of Paraburkholderia mimosarum LMG 23256T contains the core operon necessary for GA biosynthesis. This β-rhizobium was isolated from Mimosa pigra and shown to form indeterminate nodules with its host (Chen et al., 2005, 2006). Here the enzymes encoded by the operon from P. mimosarum LMG 23256T were characterized and shown to be functionally conserved, with further phylogenetic analysis indicating not only independent acquisition of the operon from gamma-proteobacteria, but also loss of CYP115, which supports the hypothesis that direct production of bioactive GA4 may have a deleterious effect in the symbiotic relationship between rhizobia and their leguminous host plants.
To find copies of the operon from newly sequenced bacterial genomes, BLAST searches were carried out using individual genes from the most divergent copy of the operon, namely that previously characterized from Erwinia tracheiphila (Nagel and Peters, 2017b). The results for examples found outside of the α-rhizobia are summarized in Supplementary Table S2 (along with that for the α-rhizobia used in the phylogenetic analyses reported here). Note that the bacterial genus Burkholderia was recently reclassified as Paraburkholderia (Sawana et al., 2014; Dobritsa and Samadpour, 2016).
P. mimosarum LMG 23256T (Willems et al., 2014) was obtained from the German Collection of Microorganisms and Cell Cultures (LMG 23256T = DSM21841T). Genes from the P. mimosarum operon were cloned from genomic DNA with Q5 Hot Start High-Fidelity DNA polymerase (New England Biolabs) according to the product manual with 5 μl of the high-GC-content enhancer and gene specific primers (Supplementary Table S1). The IDS, KS, CPS, and CYP112 genes were cloned into pET100/D-TOPO (Invitrogen). CYP117 was cloned into pET101/D-TOPO (Invitrogen) including a stop codon to omit the C-terminal His-Tag of the vector. CYP114 was cloned in tandem with either the Fd-SDR fusion or only the Fd into pET100. For the later construct the amino acid sequence after position 86 was changed from ET to that of the Fd from Paraburkholderia sp. JPY251 with the sequence ADDEAT followed by a stop codon. The CPS also was obtained as a synthetic codon optimized gene (Invitrogen) and similarly cloned into pET100. Recombinant expression, protein purification and in vitro enzyme assays or recombinant feeding studies, as well as organic extraction and GC-MS analysis were performed as previously described (Nagel and Peters, 2017b). Briefly, all proteins were expressed in E. coli BL21(Star). Cells expressing PmIDS were centrifuged to collect cells, which were homogenized in MOPSO buffer pH 7.2 with 10% glycerol and 10 mM MgCl2. The cell lysate was incubated with 1 ml Ni-NTA Agarose (Qiagen) and PmIDS was eluted with imidazole and 20 μg protein was used in 0.5 ml in vitro assays with 50 mM IDP and DMADP each (Sigma-Aldrich). Assays were dephosphorylated using alkaline phosphatase and extracted 3 times with equal volumes of pentane. PmCPS and PmKS were expressed in a 50 ml metabolic engineering setting – i.e., with an IDS from Abies grandis that produces GGDP and either AtKS or AtCPS, respectively, just as previously described for analysis of other bacterial CPSs and KSs (Morrone et al., 2009; Hershey et al., 2014; Lu et al., 2015) – for 3 days at 18°C together with pIRS, to increase flux to isoprenoids (Morrone et al., 2010). Note that neither CPS or KS alone in this setting produce ent-kaurene. PmCYP117, PmCYP114 + Fd, PmCYP114 + Fd-SDR and PmCYP112 were expressed alone and their respective substrates were added to the 25 ml culture and the culture was incubated at 18 °C for 3 days under shaking at 180 rpm. Cultures were extracted with 3 times with equal volumes of hexanes or ethyl acetate in case of PmCYP114 + Fd, PmCYP114 + Fd-SDR and PmCYP112. Extracts were partially purified using silica gel columns developed with hexanes and eluted with increasing concentrations of ethyl acetate. Products of PmCYP117, PmCYP114 + Fd, PmCYP114 + Fd-SDR and PmCYP112 were methylated with diazomethane and quantified by GC-MS using a Varian GC-MS with a HP5-MS column (Agilent). The injector temperature was set at 250 °C with a helium column flow of 1.2 ml/min and 1 μl injections in split-less mode. The initial temperature of the GC oven was 50°C which was held for 3 min and increased by either 15°C/min for products of PmIDS, PmCPS and PmKS or by 10°C/min for all other enzymes, until a temperature of 300 °C was reached, which was held for 3 min.
Phylogenetic analyses focused on a representative set of species and utilized nucleotide sequences spanning the core operon (i.e., from CYP112 to the KS, with CYP115 and IDI not included due to their absence in most rhizobia), including the intergenic regions. This was carried out with the nucleotide sequence of the operon, instead of the previously used concatenated protein sequences of the same region (Nagel and Peters, 2017b), as this allowed inclusion of operons with inactivating mutations (e.g., premature stop codons or frame shift mutations) as well as intergenic regions. Sequences were aligned using the Muscle algorithm in MEGA 7 (Kumar et al., 2016), and phylogenetic trees were then constructed and tested with the Maximum Likelihood Neighbor Joining and Minimum Evolution algorithms. The Tamura 3-parameter model was used with inclusion of a gamma distribution. For the Maximum Likelihood algorithm all sites were used, while for the Neighbor Joining and Minimum Evolution algorithms positions with less than 50% coverage were eliminated. The accuracy of the tree was tested via bootstrap testing with 1000 replicates each.
Beyond the core operon previously reported from Paraburkholderia sp. JPY251 (Nagel and Peters, 2017b), BLAST searches found two other copies in β-rhizobia. Another complete copy of the core operon was found in P. mimosarum LMG 23256T, while an incomplete copy is present in Paraburkholderia sp. UYPR1.413, where the operon is split between two contigs, with a transposable element found adjacent to the 3’ end of the IDS, and the SDR contains inactivating frame-shift mutations (see Figure 1A). As previously reported for Paraburkholderia sp. JPY251 (Nagel and Peters, 2017b), CYP115 is not found in the operon, nor elsewhere in the genome, in P. mimosarum or Paraburkholderia sp. UYPR1.413.
To determine if the operon found in the beta-proteobacteria is fully functional – i.e., enables GA production – that from P. mimosarum was biochemically characterized by recombinant expression in Escherichia coli and examination of enzymatic activity. PmIDS was assessed via in vitro assays, which readily demonstrated the expected production of GGDP from IDP and DMADP (Figure 2 and Supplementary Figure S1). PmCPS was not well-expressed in E. coli and no activity was observed with the purified protein in vitro. Activity was then assessed via a metabolic engineering approach, involving co-expression with AtKS, the ent-CDP specific KS from Arabidopsis thaliana, along with a GGDP producing IDS from Abies grandis (AgGGPS), as previously described (Morrone et al., 2009; Hershey et al., 2014; Lu et al., 2015). While the native gene for PmCPS further did not exhibit activity in this setting either, use of a synthetic gene codon-optimized for expression in E. coli enabled the expected production of ent-kaurene (Figure 2 and Supplementary Figure S1). PmKS also was assessed via a metabolic engineering approach, via co-expression with AgGGPS and AtCPS, the ent-CDP producing CPS from A. thaliana, again as previously described (Morrone et al., 2009; Hershey et al., 2014; Lu et al., 2015). These cultures similarly produced the expected ent-kaurene (Figure 2 and Supplementary Figure S1).
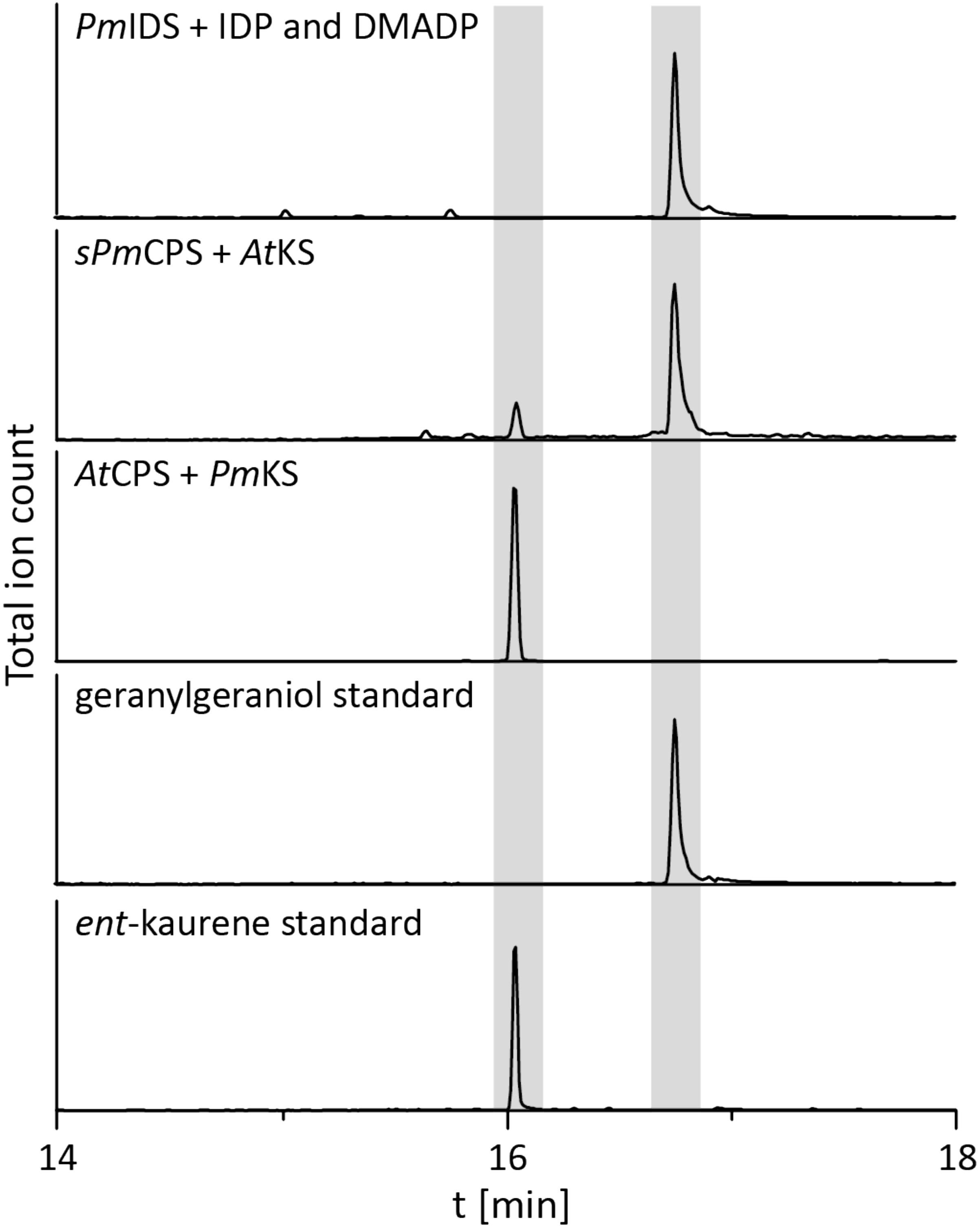
FIGURE 2. IDS, CPS, and KS enzyme activity. GC-MS chromatogram from an in vitro enzyme assay of PmIDS with dimethyl allyl diphosphate (DMADP) and isopentenyl diphosphate (IDP) as substrates, with subsequent dephosphorylation with alkaline calf intestine phosphatase. GC-MS chromatograms of organic extracts from E. coli cultures expressing PmKS or synthetic sPmCPS along with either AtCPS or AtKS, with additional engineering for GGDP production. GC-MS chromatograms of geranylgeraniol (dephosphorylated GGDP) and ent-kaurene standards are included for reference.
The remaining oxidative enzymes were investigated by whole-cell feeding studies. Accordingly, ent-kaurene was added to E. coli expressing PmCYP117, leading to the expected conversion to ent-kaurenoic acid, with the intermediates ent-kaurenol and ent-kaurenal not observed (Figure 3 and Supplementary Figure S1), consistent with the previously investigated mechanism for this reaction (Nagel and Peters, 2017a). Notably, in P. mimosarum the genes for the Fd and SDR are fused (in-frame). Given the previously reported requirement for the Fd to enable full activity with CYP114 (Nagel and Peters, 2017b; Nagel et al., 2017; Nett et al., 2017b), two constructs were generated to evaluate the implications of this fusion for activity. First, PmCYP114 was cloned into pET100 together with the fused PmFd-SDR, including the native intergenic region between CYP114 and the Fd-SDR fusion. The second construct consisted of PmCYP114 and only PmFd, with introduction of a stop codon based on the sequence of the Fd in Paraburkholderia sp. JPY251 where the Fd and SDR are not fused. Both constructs were expressed in E. coli and ent-kaurenoic acid was added to the resulting cultures. Cultures expressing PmCYP114 + PmFd (i.e., without the SDR) produced a mixture of GA12-aldehyde and GA12, while the PmCYP114 + PmFd-SDR construct exclusively produced GA12, with the putative intermediate ent-7α-hydroxykaurenoic acid not observed in either case (Figure 4 and Supplementary Figure S1), consistent with previous mechanistic investigation of this reaction (Nett et al., 2016). This demonstrated the ability of the Fd to enable full CYP114 activity as a fusion protein with the SDR, as well as the expected more efficient oxidation of GA12-aldehyde to GA12 by the SDR. Finally, E. coli expressing PmCYP112 and fed GA12 efficiently converted this to GA9, with only small amounts of the intermediate GA15 observed and none of the intermediate GA24 or the side product GA25 (Figure 5 and Supplementary Figure S1), again consistent with the previously investigated mechanism for this reaction (Nagel and Peters, 2018a,b).
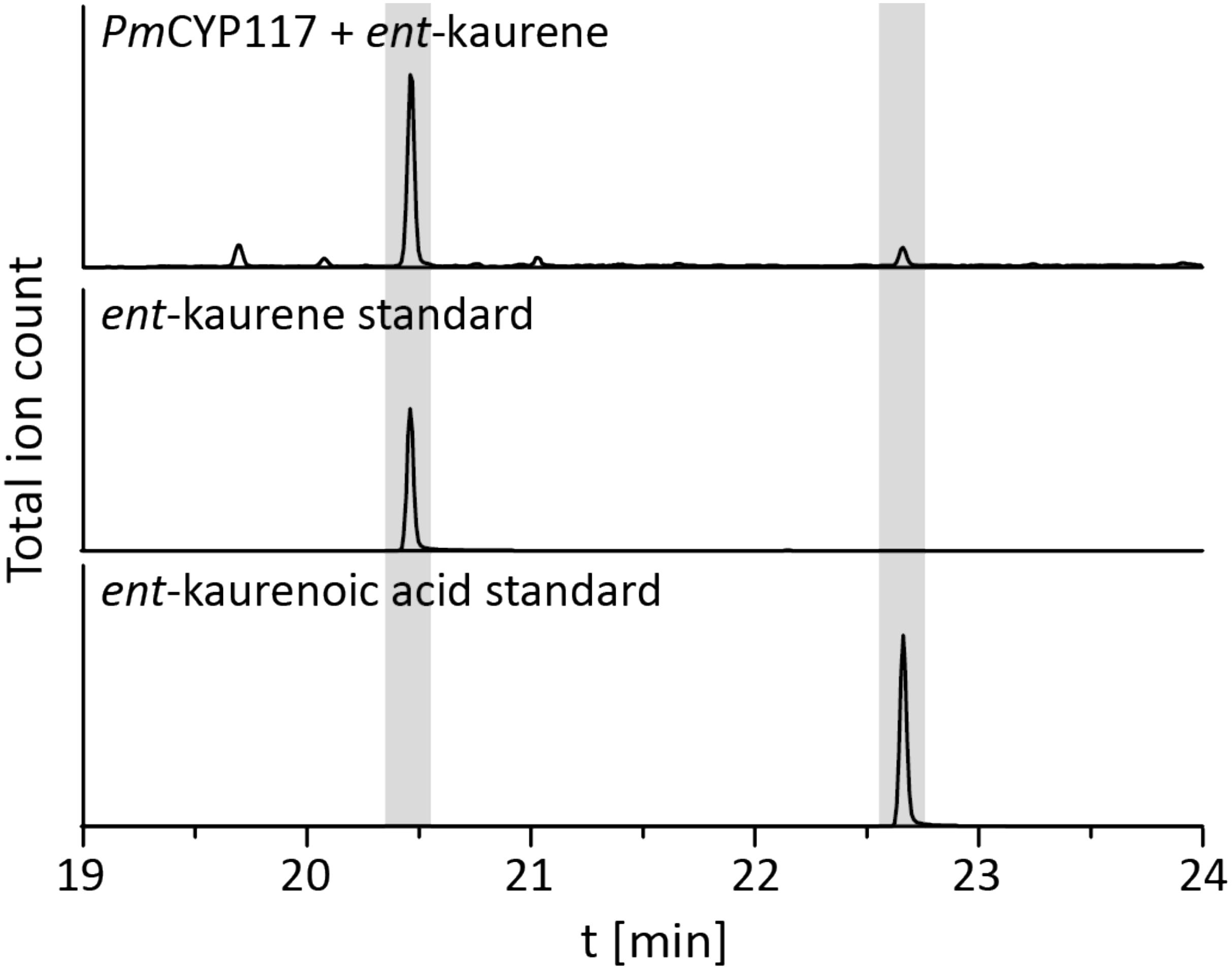
FIGURE 3. CYP117 enzyme activity. GC-MS chromatogram of the organic extract of an E. coli culture expressing PmCYP117 and fed ent-kaurene. GC-MS chromatograms of ent-kaurene and ent-kaurenoic acid standards are included for reference.
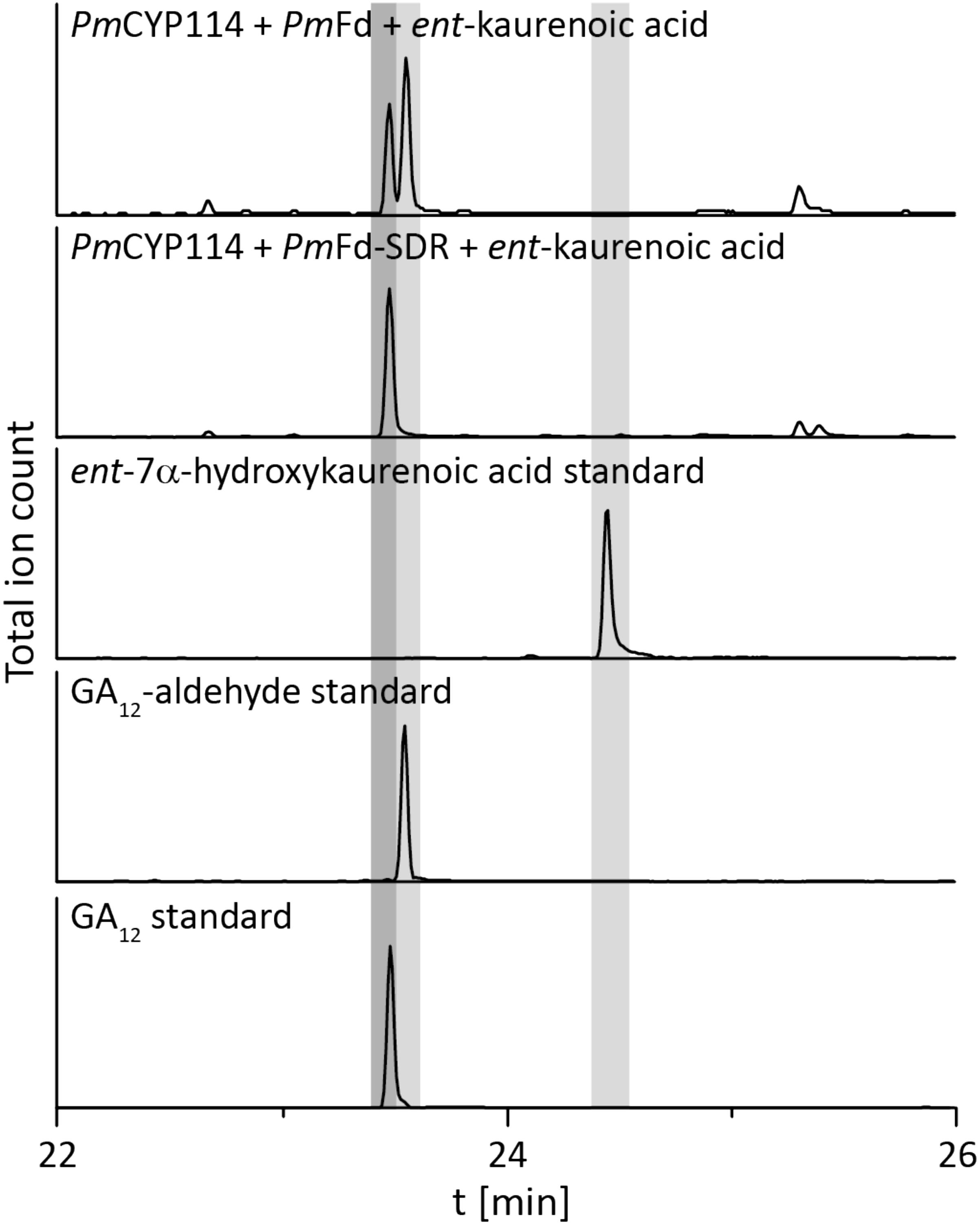
FIGURE 4. CYP114+Fd and CYP114 + Fd-SDR enzyme activity. GC-MS chromatograms from organic extracts of E. coli cultures expressing PmCYP114 either with the artificial PmFd or the PmFd-SDR fusion protein found in P. mimosarum and fed ent-kaurenoic acid. GC-MS chromatograms of ent-7a-hydroxykaurenoic acid, GA12-aldehyde and GA12 standards are included for reference.
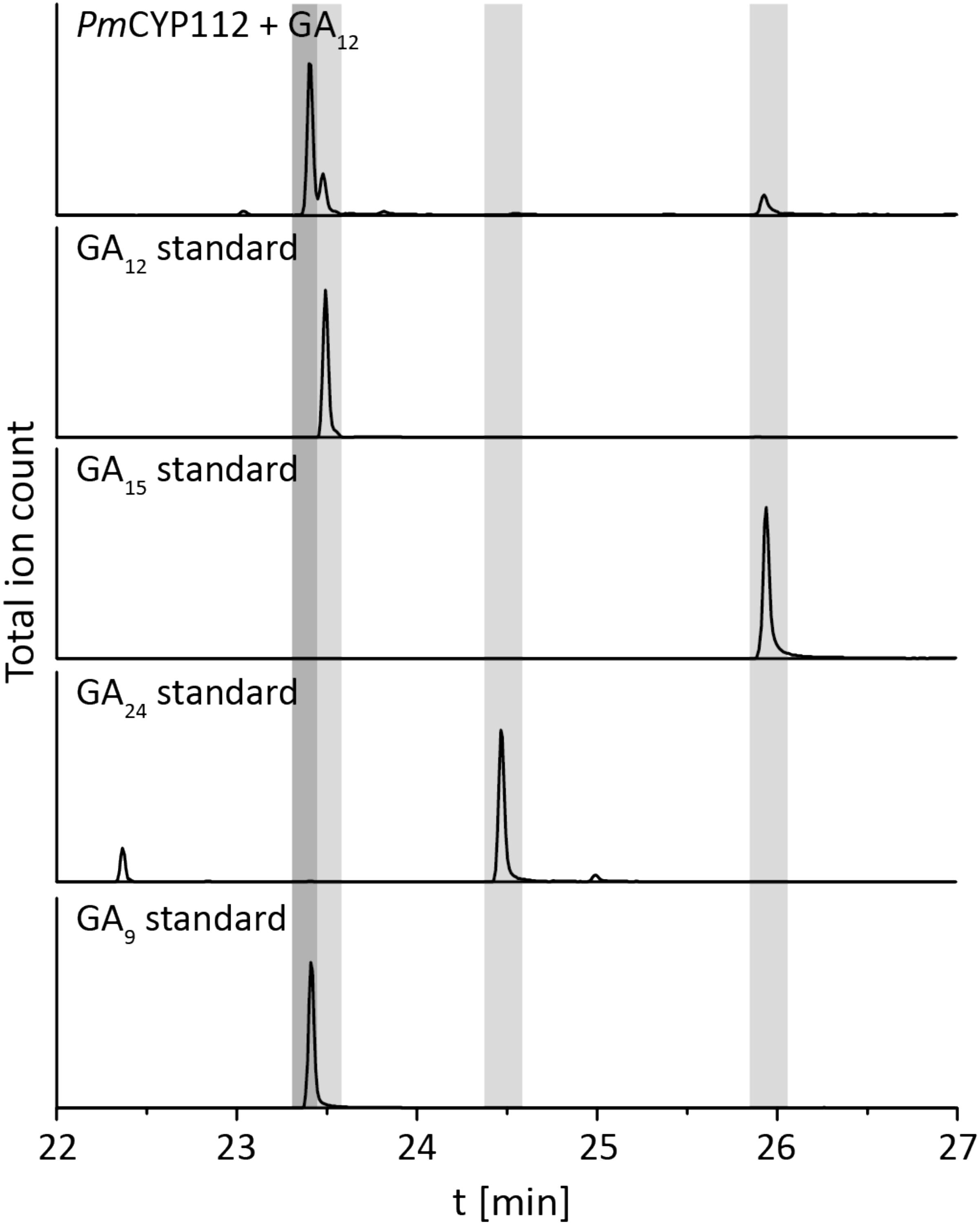
FIGURE 5. CYP112 enzyme activity. GC-MS chromatograms from organic extract of an E. coli culture expressing PmCYP112 and fed GA12. Chromatograms of GA12, GA15, GA24, and GA9 standards are included for reference.
Despite the lack of CYP115 in the GA biosynthetic operon from β-rhizobia, which nominally resembles the operons found in α-rhizobia, it has been suggested that β-rhizobia independently obtained the operon via horizontal gene transfer from gamma-proteobacteria (Nagel and Peters, 2017b). However, this hypothesis was proposed by phylogenetic analyses limited by the single examples then available – i.e., that from Paraburkholderia JPY251 and the most closely related copy from a gamma-proteobacteria, Pseudomonas psychrotolerans (Ps. psychrotolerans) NS274, which includes CYP115. Given that increased numbers of species with the operon are now available, including the multiple copies noted above for Paraburkholderia and additional strains of Ps. psychrotolerans, each of which contain CYP115 (Supplementary Table S2), this phylogenetic analysis was repeated here. The results are consistent with the hypothesis that the operon in β-rhizobia is most closely related to that from Ps. psychrotolerans (Figure 6 and Supplementary Figure S2).
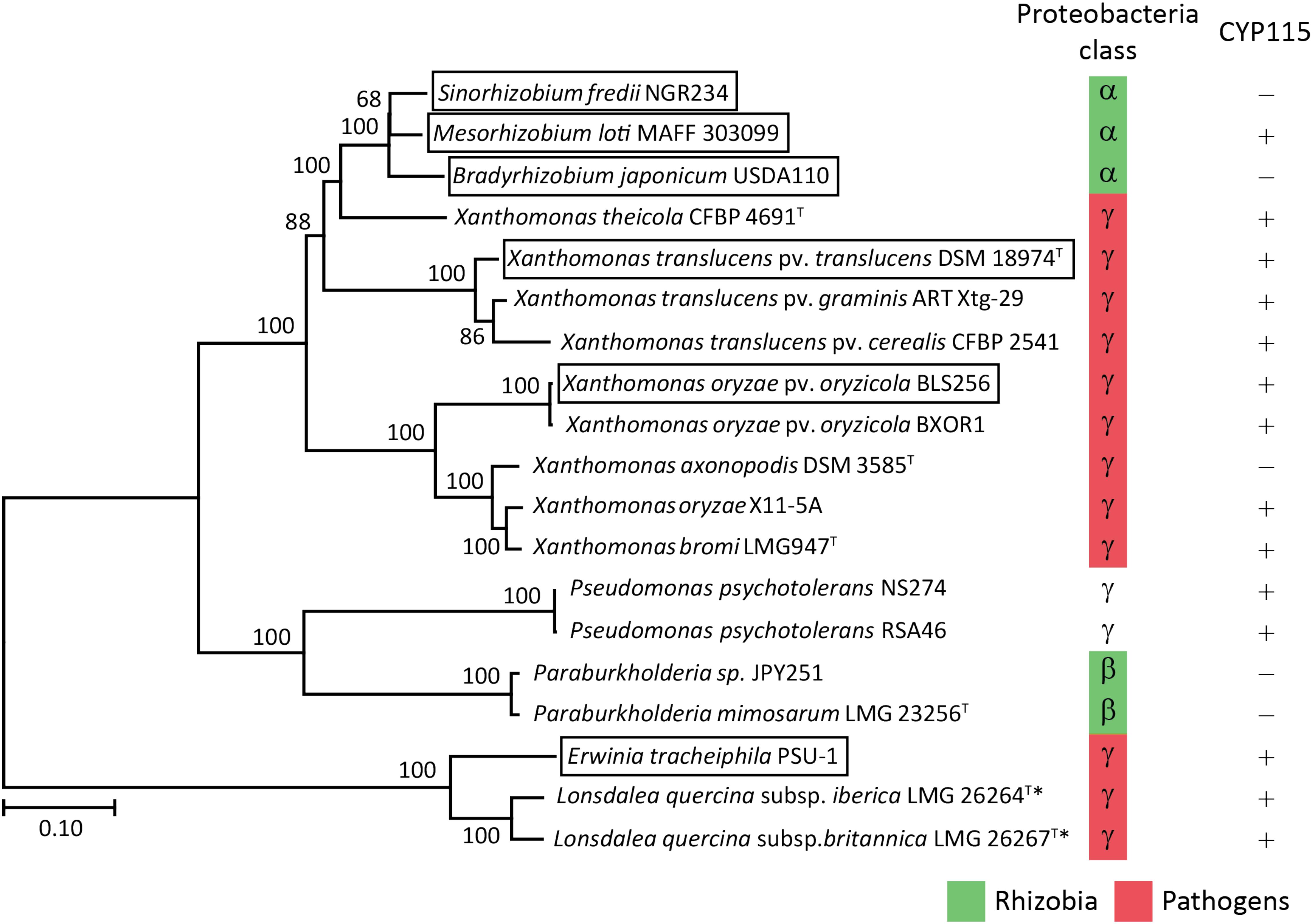
FIGURE 6. Phylogeny of the GA biosynthetic operon. Unrooted Maximum Likelihood phylogenetic tree of the operon nucleotide sequence spanning CYP112 to KS including intergenic regions. Bootstrap values were determined with 1000 replicates using MEGA 7. The scale bar represents substitutions per site and the asterisk (∗) indicates operons with inactivating mutations in at least one gene. The GA operon genes found within boxed species have been functionally characterized. The GeneBank accession numbers of the contig containing the operon for each of the presented bacteria can be found in Supplementary Table S2.
The biochemical results reported here demonstrate that the GA biosynthetic operon is functionally present in β-rhizobia, representing the third class of proteobacteria in which this operon can be found. However, the absence of CYP115 limits the β-rhizobia to production of the penultimate intermediate GA9 rather than bioactive GA4. While this nominally resembles previous findings in α-rhizobia, phylogenetic analysis strongly implies that the β-rhizobia independently obtained their copy of the operon from gamma-proteobacteria, as these are most closely related to the operon found in Ps. psychrotolerans, where CYP115 is invariably present. This further indicates that β-rhizobia also independently lost CYP115. In α-rhizobia with a full assembled genome the GA operon is invariably located within the symbiotic island or plasmid that also contains the necessary genes for nitrogen fixation (Perret et al., 1999; Gottfert et al., 2001; Sullivan et al., 2002; Gonzalez et al., 2003; Uchiumi et al., 2004; Nett et al., 2017a). However, it is unknown if the operon in beta-proteobacteria also is associated with the symbiotic island as the genome of all three species are not completely assembled and the contigs with the operon have few adjoining genes.
Beyond such implications for the origin of the operon in β-rhizobia, a number of other observations were derived from the bioinformatic analyses carried out here. For example, copies were found in Lonsdalea quercina subsp. britanica and Lonsdalea quercina subsp. iberica, which are close relatives of Erwinia tracheiphila, and are pathogens of oak trees (Brady et al., 2012). The Lonsdalea operons are also most closely related to that from E. tracheiphila (Figure 6 and Supplementary Figure S2), which has already been shown to be fully functional (Nagel and Peters, 2017b). However, the operons from Lonsdalea all are disrupted by an inactivating frame-shift mutation in CYP112, with several exhibiting additional inactivating mutations in other genes (Supplementary Table S2), suggesting that these are no longer functional, presumably reflecting a loss of selective pressure for GA production in these phytopathogens.
In the Xanthomonas genus, consistent with a previous report (Nagel et al., 2017), the operon is selectively present in certain pathovars of X. oryzae, namely Xanthomonas oryzae pv. oryzicola (all 21 sequenced genomes), but not in the other major pathovar Xanthomonas oryzae pv. oryzae, where it is not found in any of the 380 genomes currently available. By contrast, the operon is widespread in X. translucens, where it is present in all 48 sequenced genomes, which cover a range of pathovars, although there appears to be a premature stop codon in CYP115 in 3 of the 5 sequenced strains from Xanthomonas translucens pv. poae (Supplementary Table S2). These form a distinct cluster within the relevant clade from X. translucens (Langlois et al., 2017), perhaps indicating some loss of selective pressure for production of bioactive GA4 relative to the immediate precursor GA9 in this group of non-cereal phytopathogens. Regardless, it seems clear that X. translucens acquired the operon early, with the selective advantage provided by the production of GA4 leading to its widespread retention (i.e., vertical descent).
In addition, the operon was found in two new Xanthomonas species, X. axonopodis and X. theicola, although the operon in X. axonopodis appears to be missing CYP115 (Supplementary Table S2). While the division between the operons in X. translucens and X. theicola relative to X. oryzae, X. bromi and X. axonopodis reflects their broader phylogenic relationship (Merda et al., 2017), the numerous other species within the Xanthomonas genus that completely lack the operon indicate that these individually acquired the operon – i.e., via horizontal gene transfer. Consistent with this hypothesis, X. translucens and X. theicola represent a quite divergent group even within the broader Xanthomondaceae family (Naushad et al., 2015). Moreover, although X. oryzae, X. bromi and X. axonopodis all come from the same clade within this genus they do not otherwise group together (Merda et al., 2017), and the selective presence of the operon in X. oryzae pv. oryzicola versus X. oryzae pv. oryzae, as well as in Xanthomonas axonopodis DSM 3585 versus other strains of X. axonopodis (several of which have genome sequences available), suggests that even within this clade the operon also may be acquired by horizontal gene transfer rather than vertical descent.
Interestingly, the operon from X. theicola is more closely related to those from α-rhizobia than even those from X. translucens (Figure 6 and Supplementary Figure S2). Accordingly, while the absence of CYP115 in X. axonopodis nominally resembles the operon structure in α-rhizobia, this appears to represent an independent gene loss event. In particular, the presence of CYP115 in the much more closely related X. theicola operon supports the hypothesis that α-rhizobia originally acquired a full operon with subsequent early loss of CYP115, which contrasts with the complete gene loss observed in β-rhizobia, in that a non-functional fragment remains in almost all α-rhizobia. This further implies that direct production of bioactive GA4 is generally selected against in the symbiotic relationship between these rhizobia and their leguminous hosts. However, at least one α-rhizobia GA biosynthetic operon retains CYP115 (i.e., in the same position as found in gamma-proteobacteria copies), and this appears to have undergone independent gene transfer into a subset of other α-rhizobia (Nett et al., 2017a), indicating that such direct hormone biosynthesis does provide a selective advantage for at least the α-rhizobia under certain circumstances.
In conclusion, the results reported here extend our understanding of the phylogenetic range for functional acquisition of the GA biosynthetic operon beyond those previously characterized from alpha- and gamma- into beta- proteobacteria as well. Moreover, the phylogenetic analysis not only supports the hypothesis that the operon arose in the gamma-proteobacteria, but also the previously advanced hypothesis that this was acquired by independent horizontal gene transfer by both α- and β-rhizobia (Nagel and Peters, 2017b), as well as suggesting more specific origins. Strikingly, this further supports independent loss of CYP115 in both classes of rhizobia, implying that direct production of bioactive GA4 relative to the immediate precursor GA9 generally (although not universally) has deleterious effects in such symbiotic relationships. While this has been suggested to stem from suppression of the host plant defense response against microbial pathogens (Nett et al., 2017a), the actual selective pressure against retention of CYP115 remains unclear, representing an avenue for future investigation.
RN and RP designed the research and wrote the manuscript. RSN, JB, and MS-D revised the manuscript. RN, JB, MS-D, and RSN performed the research and analyzed the data.
This work was supported by grants from the NIH (GM109773) and NSF (CHE-1609917) to RP, along with a postdoctoral fellowship to RN from the Deutsche Forschungsgemeinschaft (DFG) NA 1261/1-2.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02916/full#supplementary-material
Bottini, R., Cassan, F., and Piccoli, P. (2004). Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 65, 497–503. doi: 10.1007/s00253-004-1696-1
Brady, C. L., Cleenwerck, I., Denman, S., Venter, S. N., Rodríguez-Palenzuela, P., Coutinho, T. A., et al. (2012). Proposal to reclassify Brenneria quercina (Hildebrand and Schroth 1967) Hauben 1999 into a new genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., descriptions of Lonsdalea quercina subsp. quercina comb. nov., Lonsdalea quercina subsp. iberica subsp. nov. and Lonsdalea quercina subsp. britannica subsp. nov., emendation of the description of the genus Brenneria, reclassification of Dickeya dieffenbachiae as Dickeya dadantii subsp. dieffenbachiae comb. nov., and emendation of the description of Dickeya dadantii. Int. J. Syst. Evol. Microbiol. 62, 1592–1602. doi: 10.1099/ijs.0.035055-0
Chen, W. M., James, E. K., Chou, J. H., Sheu, S. Y., Yang, S. Z., and Sprent, J. I. (2005). Beta-rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol. 168, 661–675. doi: 10.1111/j.1469-8137.2005.01533.x
Chen, W. M., James, E. K., Coenye, T., Chou, J. H., Barrios, E., De Faria, S. M., et al. (2006). Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 56, 1847–1851. doi: 10.1099/ijs.0.64325-0
De Bruyne, L., Hofte, M., and De Vleesschauwer, D. (2014). Connecting growth and defense: the emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol. Plant 7, 943–959. doi: 10.1093/mp/ssu050
Dobritsa, A. P., and Samadpour, M. (2016). Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 66, 2836–2846. doi: 10.1099/ijsem.0.001065
Gonzalez, V., Bustos, P., Ramirez-Romero, M. A., Medrano-Soto, A., Salgado, H., Hernandez-Gonzalez, I., et al. (2003). The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 4:R36. doi: 10.1186/gb-2003-4-6-r36
Gottfert, M., Rothlisberger, S., Kundig, C., Beck, C., Marty, R., and Hennecke, H. (2001). Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183, 1405–1412. doi: 10.1128/JB.183.4.1405-1412.2001
Hedden, P., and Sponsel, V. (2015). A century of gibberellin research. J. Plant Growth Regul. 34, 740–760. doi: 10.1007/s00344-015-9546-1
Hershey, D. M., Lu, X., Zi, J., and Peters, R. J. (2014). Functional conservation of the capacity for ent-kaurene biosynthesis and an associated operon in certain rhizobia. J. Bact. 196, 100–106. doi: 10.1128/JB.01031-13
Keister, D. L., Tully, R. E., and Van Berkum, P. (1999). A cytochrome P450 gene cluster in the Rhizobiaceae. J. Gen. Appl. Microbiol. 45, 301–303. doi: 10.2323/jgam.45.301
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Langlois, P. A., Snelling, J., Hamilton, J. P., Bragard, C., Koebnik, R., Verdier, V., et al. (2017). Characterization of the Xanthomonas translucens complex using draft genomes, comparative genomics, phylogenetic analysis, and diagnostic lamp assays. Phytopathology 107, 519–527. doi: 10.1094/PHYTO-08-16-0286-R
Levy, A., Salas Gonzalez, I., Mittelviefhaus, M., Clingenpeel, S., Herrera Paredes, S., Miao, J., et al. (2017). Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. doi: 10.1038/s41588-017-0012-9
Lu, X., Hershey, D. M., Wang, L., Bogdanove, A. J., and Peters, R. J. (2015). An ent-kaurene derived diterpenoid virulence factor from Xanthomonas oryzae pv. oryzicola. New Phytol.406, 295–302. doi: 10.1111/nph.13187
Mendez, C., Baginsky, C., Hedden, P., Gong, F., Caru, M., and Rojas, M. C. (2014). Gibberellin oxidase activities in Bradyrhizobium japonicum bacteroids. Phytochemistry 98, 101–109. doi: 10.1016/j.phytochem.2013.11.013
Merda, D., Briand, M., Bosis, E., Rousseau, C., Portier, P., Barret, M., et al. (2017). Ancestral acquisitions, gene flow and multiple evolutionary trajectories of the type three secretion system and effectors in Xanthomonas plant pathogens. Mol. Ecol. 26, 5939–5952. doi: 10.1111/mec.14343
Morrone, D., Chambers, J., Lowry, L., Kim, G., Anterola, A., Bender, K., et al. (2009). Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 583, 475–480. doi: 10.1016/j.febslet.2008.12.052
Morrone, D., Lowry, L., Determan, M. K., Hershey, D. M., Xu, M., and Peters, R. J. (2010). Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol. 85, 1893–1906. doi: 10.1007/s00253-009-2219-x
Nagel, R., and Peters, R. J. (2017a). 18O2 labeling experiments illuminate the oxidation of ent-kaurene in bacterial gibberellin biosynthesis. Org. Biomol. Chem. 15, 7566–7571. doi: 10.1039/c7ob01819c
Nagel, R., and Peters, R. J. (2017b). Investigating the phylogenetic range of Gibberellin biosynthesis in bacteria. Mol. Plant Microbe Interact. 30, 343–349. doi: 10.1094/MPMI-01-17-0001-R
Nagel, R., and Peters, R. J. (2018a). Diverging mechanisms: cytochrome-P450-catalyzed demethylation and gamma-lactone formation in bacterial gibberellin biosynthesis. Angew. Chem. Int. Ed. Engl. 57, 6082–6085. doi: 10.1002/anie.201713403
Nagel, R., and Peters, R. J. (2018b). Probing the specificity of CYP112 in bacterial gibberellin biosynthesis. Biochem. J. 475, 2167–2177. doi: 10.1042/BCJ20180317
Nagel, R., Turrini, P. C. G., Nett, R. S., Leach, J. E., Verdier, V., Van Sluys, M. A., et al. (2017). An operon for production of bioactive gibberellin A4 phytohormone with wide distribution in the bacterial rice leaf streak pathogen Xanthomonas oryzae pv. oryzicola. New Phytol. 214, 1260–1266. doi: 10.1111/nph.14441
Naushad, S., Adeolu, M., Wong, S., Sohail, M., Schellhorn, H. E., and Gupta, R. S. (2015). A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales, the family Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Antonie Van Leeuwenhoek 107, 467–485. doi: 10.1007/s10482-014-0344-8
Nett, R. S., Contreras, T., and Peters, R. J. (2017a). Characterization of CYP115, a gibberellin 3-oxidase, demonstrates that certain rhizobia produce bioactive gibberellin A4. ACS Chem. Biol. 12, 912–917. doi: 10.1021/acschembio.6b01038
Nett, R. S., Montanares, M., Marcassa, A., Lu, X., Nagel, R., Charles, T. C., et al. (2017b). Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat. Chem. Biol. 13, 69–74. doi: 10.1038/nchembio.2232
Nett, R. S., Dickschat, J. S., and Peters, R. J. (2016). Labeling studies clarify the committed step in bacterial Gibberellin biosynthesis. Org. Lett. 18, 5974–5977. doi: 10.1021/acs.orglett.6b02569
Perret, X., Freiberg, C., Rosenthal, A., Broughton, W. J., and Fellay, R. (1999). High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 32, 415–425. doi: 10.1046/j.1365-2958.1999.01361.x
Sawana, A., Adeolu, M., and Gupta, R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:429. doi: 10.3389/fgene.2014.00429
Sullivan, J. T., Trzebiatowski, J. R., Cruickshank, R. W., Gouzy, J., Brown, S. D., Elliot, R. M., et al. (2002). Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184, 3086–3095. doi: 10.1128/JB.184.11.3086-3095.2002
Tatsukami, Y., and Ueda, M. (2016). Rhizobial gibberellin negatively regulates host nodule number. Sci. Rep. 6:27998. doi: 10.1038/srep27998
Tully, R. E., and Keister, D. L. (1993). Cloning and mutagenesis of a Cytochrome P-450 Locus from Bradyrhizobium japonicum that is expressed anaerobically and symbiotically. Appl. Environ. Microbiol. 59, 4136–4142.
Tully, R. E., Van Berkum, P., Lovins, K. W., and Keister, D. L. (1998). Identifcation and sequencing of a cytochrome P450 gene cluster from Bradyrhizobium japonicum. Biochim. Biophys. Acta 1398, 243–255. doi: 10.1016/S0167-4781(98)00069-4
Uchiumi, T., Ohwada, T., Itakura, M., Mitsui, H., Nukui, N., Dawadi, P., et al. (2004). Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J. Bacteriol. 186, 2439–2448. doi: 10.1128/JB.186.8.2439-2448.2004
Keywords: symbiosis, gibberellin, rhizobia, legume (nodules), evolution
Citation: Nagel R, Bieber JE, Schmidt-Dannert MG, Nett RS and Peters RJ (2018) A Third Class: Functional Gibberellin Biosynthetic Operon in Beta-Proteobacteria. Front. Microbiol. 9:2916. doi: 10.3389/fmicb.2018.02916
Received: 14 September 2018; Accepted: 13 November 2018;
Published: 27 November 2018.
Edited by:
Eloise Foo, University of Tasmania, AustraliaReviewed by:
Maria Jose Soto, Spanish National Research Council (CSIC), SpainCopyright © 2018 Nagel, Bieber, Schmidt-Dannert, Nett and Peters. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reuben J. Peters, cmpwZXRlcnNAaWFzdGF0ZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.