- 1Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
- 2Graduate School of Life and Environmental Sciences, University of Tsukuba, Ibaraki, Japan
- 3Research Center for Biology, Indonesia Institute of Sciences (LIPI), Cibinong, Indonesia
- 4Department of Biochemistry, School of Medicine, Keio University, Tokyo, Japan
- 5Clinical and Translational Research Center, School of Medicine, Keio University, Tokyo, Japan
Coenzyme A (CoA) is an essential cofactor for numerous cellular reactions in all living organisms. In the protozoan parasite Entamoeba histolytica, CoA is synthesized in a pathway consisting of four enzymes with dephospho-CoA kinase (DPCK) catalyzing the last step. However, the metabolic and physiological roles of E. histolytica DPCK remain elusive. In this study, we took biochemical, reverse genetic, and metabolomic approaches to elucidate role of DPCK in E. histolytica. The E. histolytica genome encodes two DPCK isotypes (EhDPCK1 and EhDPCK2). Epigenetic gene silencing of Ehdpck1 and Ehdpck2 caused significant reduction of DPCK activity, intracellular CoA concentrations, and also led to growth retardation in vitro, suggesting importance of DPCK for CoA synthesis and proliferation. Furthermore, metabolomic analysis showed that suppression of Ehdpck gene expression also caused decrease in the level of acetyl-CoA, and metabolites involved in amino acid, glycogen, hexosamine, nucleic acid metabolisms, chitin, and polyamine biosynthesis. The kinetic properties of E. histolytica and human DPCK showed remarkable differences, e.g., the Km values of E. histolytica and human DPCK were 58–114 and 5.2 μM toward dephospho-CoA and 15–20 and 192 μM for ATP, respectively. Phylogenetic analysis also supported the uniqueness of the amebic enzyme compared to the human counterpart. These biochemical, evolutionary features, and physiological importance of EhDPCKs indicate that EhDPCK represents the rational target for the development of anti-amebic agents.
Introduction
Coenzyme A (CoA) is an essential cofactor as an acyl group carrier and carbonyl activating group in metabolism that contributes to 9% out of 3,500 cellular activities.1 CoA is synthesized by five to six enzymatic reactions with pantothenic acid (vitamin B5), L-cysteine, and purine/pyrimidine nucleotides as substrates (Genschel et al., 1999). Pantothenate is either produced de novo, as in bacteria, archaea, mammals, and plants (Begley et al., 2001; Chakauya et al., 2008; Spry et al., 2008), or alternatively, scavenged by uptake (Saliba et al., 1998; Spry et al., 2008). It was shown that the first and last steps, catalyzed by pantothenate kinase (PanK) and dephospho-CoA kinase (DPCK, EC 2.7.1.24) are rate limiting and allosterically regulated (Leonardi et al., 2005; Hart et al., 2017). Both enzymes utilize ATP as a phosphate donor and are essential in many organisms (Hart et al., 2017).
Entamoeba histolytica is a parasitic protozoan, which causes amebiasis in humans. According to the World Health Organization, 50 million people, especially in the tropical countries, suffer from this infection, resulting in estimated 100,000 deaths annually (World Health Organization [WHO], 1997; Ximénez et al., 2009). No effective vaccine has yet been developed, and only metronidazole and its derivatives are the drugs of choice for treatment. It has been demonstrated that metronidazole targets pyruvate:ferredoxin oxidoreductase, which is involved in acetyl CoA production in central energy metabolism. However, their low efficacy against asymptomatic cyst carriers, cases of treatment failure (Hanna et al., 2000; Ali and Nozaki, 2007), and emergence of resistance (Orozco et al., 1985; Johnson, 1993; Samarawickrema et al., 1997) have been reported. Therefore, other rational targets need to be explored to develop new chemotherapeutics against amebiasis.
In E. histolytica, it has been previously shown that repression of panK gene causes defect in proliferation, and expression of dpck genes was transcriptionally upregulated by repression of panK gene expression (Nurkanto et al., 2018). This previous study reinforces the premise that the CoA biosynthetic pathway plays a pivotal role in trophozoite proliferation. While the first rate limiting enzyme was well characterized, the metabolic and physiological roles of the enzyme that catalyzes the last step of the pathway remain elusive. DPCK utilizes ATP to phosphorylate dephospho-CoA at the 3′-hydroxyl group of the ribose moiety to generate CoA. The E. histolytica genome apparently encodes two isotypes of DPCK (Nurkanto et al., 2018). In the present study, we conducted biochemical characterization of recombinant enzymes. We also undertook reverse genetic and metabolomic approaches to elucidate the physiological importance of EhDPCK in this parasitic protist.
Materials and Methods
Organisms, Cultivation, and Chemicals
Trophozoites of the E. histolytica clonal strain HM-1:IMSS cl 6 and G3 strains (Bracha et al., 2006) were maintained axenically in Diamond’s BI-S-33 medium at 35.5°C as described previously (Diamond et al., 1978). Trophozoites were continuously maintained in mid-log phase after inoculation of 1–30th to 1–12th of the total culture volume. Escherichia coli BL21 (DE3) strain was purchased from Invitrogen (Carlsbad, CA, United States). Lipofectamine and geneticin (G418) were also purchased from Invitrogen. Ni2+-NTA agarose was purchased from Novagen (Darmstadt, Germany). All other chemicals of analytical grade were purchased from Sigma-Aldrich (Tokyo, Japan) unless otherwise stated.
Phylogenetic Analyses of E. histolytica DPCK1 and DPCK2
By Basic Local Alignment Search Tool Protein (blastp) searches in public sequence databases, we collected 88 DPCK sequences including two E. histolytica sequences from non-redundant protein sequences (nr) database of National Center for Biotechnology Information (NCBI2) by using E. histolytica EhDPCK1 sequence (EHI_040840/XP_648971) as a query to obtain data from representative taxa (Supplementary Table S1). Hit sequences with E-value less than 1 × 10-10 were selected. The number of selected sequences in each blastp search for a representative taxon was determined based on the number of listed alignments. Sequences were aligned using Muscle program (Edgar, 2004) in SeaView package version 4.6.1 (Gouy et al., 2010). We selected 112 positions of unambiguously aligned by manual and used for phylogenetic analyses. The data matrices for phylogeny were subjected to IQTREE program (Nguyen et al., 2015) to select appropriate models for amino acid sequence evolution. LG + I + Ã4 and LG + Ã4 models were shown to be the best and the second-best models by using the information criterion. Maximum likelihood (ML) analysis implemented in the RAxML program version 7.2.6 (Stamatakis, 2006) was used to infer ML tree based on the LG + Ã4 model. In the bootstrap analysis, heuristic tree search was performed with a rapid bootstrap algorithm option (-f) for 100 bootstrap replicates. Bootstrap proportion (BP) values greater than 50 were indicated on the corresponding internal branches of the ML tree drawn by the use of FigTree program Version 1.4.2.3
Prediction of the Tertiary Structure of EhDPCK1 and EhDPCK2
The structure of EhDPCK1 and EhDPCK2 were predicted by homology modeling using Phyre 24 and Mus musculus COASY (PDB ID: 2F6R) as a template. Models were generated using one-to-one threading module of Phyre 2. All illustrations were prepared with EzMol version 1.21 (Reynolds et al., 2018).
Plasmid Construction for Recombinant EhDPCK1 and EhDPCK2
Plasmids to express recombinant EhDPCK1 and 2 proteins were constructed according to the procedures previously described (Sambrook and Russell, 2001). For PCR amplification of Ehdpck1 and 2 genes, total RNA was extracted from ∼106 trophozoites of Ehdpck1 and Ehdpck2 gene-silenced and control transformant strains using TRIzol (Ambion, Life Technologies) reagent as described in the previous study (Chomczynski and Mackey, 1995). DNase treatment was performed using DNase I (Invitrogen) to exclude genomic DNA. RNA quantity was determined by measuring the absorbance at 260 nm with NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). DNA fragments corresponding to the protein coding region of Ehdpck1 and Ehdpck2 gene were amplified from E. histolytica cDNA, using oligonucleotide primers: Ehdpck1 sense (5′-GCCGGGATCCATGAAAAAGATATTTGTTATTGGT-3′), antisense (5′-GCCGGTCGACTTAAAATTTATTTTCATTGAAGTCAA-3′) and Ehdpck2 sense (5′-GCCGGGATCCATGGTATTTGTTATTGGTATC-3′), antisense (5′-GCCGGTC GACTTAGTTTAAAGTAATTTTTAATTGTT-3′). Underlined letters indicate BamHI and SalI restriction sites. PCR was performed with primeSTAR HS DNA polymerase (Takara) and the following parameters: an initial incubation at 98°C for 30 s for denaturation; followed by the 30 cycles of denaturation at 98°C for 10 s; annealing at 55°C for 30 s; and elongation at 72°C for 1 min; and a final extension at 72°C for 7 min. The PCR fragments were digested with BamHI and SalI, purified with Wizard® SV gel and PCR clean-up system (Promega). The fragments were cloned into BamHI and SalI double digested pColdTM1 histidine-tag vector (Takara) to finally produce pColdTM1-Ehdpck1 and pColdTM1-Ehdpck2. The nucleotide sequences of the engineered plasmid were verified by sequencing.
Production and Purification of Recombinant EhDPCK1 and EhDPCK2
Plasmid pColdTM1-Ehdpck1 and pColdTM1-Ehdpck2 were introduced into E. coli BL21(DE3) cells by heat shock at 42°C for 1 min. E. coli BL21 (DE3) harboring pColdTM1-Ehdpck1 and pColdTM1-Ehdpck2 were grown at 37°C in 100 mL of Luria Bertani medium (Invitrogen) in the presence of 100 μg/mL ampicillin (Nacalai Tesque). The overnight culture was used to inoculate 500 mL of fresh medium, and the culture was further continued at 37°C with shaking at 180 rpm. When A600 reached 0.8, 0.5 mM isopropyl β-D-thio galactopyranoside (IPTG) was added, and cultivation was continued for another 24 h at 15°C. E. coli cells from the induced culture were harvested by centrifugation at 5,000 ×g for 20 min at 4°C. The cell pellet was washed with PBS, pH 7.4, re-suspended in 20 mL of the lysis buffer (50 mM Tris–HCl, pH 8.0, 300 mM NaCl, and 10 mM imidazole) containing 0.1% Triton X-100 (v/v), 100 μg/mL lysozyme, and 1 mM PMSF, and incubated at room temperature for 30 min, sonicated on ice and centrifuged at 25,000 ×g for 15 min at 4°C. The supernatant was mixed with 1.2 mL of 50% Ni2+-NTA His-bind slurry, incubated for 1 h at 4°C with mild shaking. The recombinant enzyme-bound resin in a column was washed three times with buffer A (50 mM Tris–HCl, pH 8.0, 300 mM NaCl, and 0.1% Triton X-100, v/v) containing 10–50 mM of imidazole. Bound protein was eluted with buffer A containing 100–300 mM imidazole. After the integrity and the purity of recombinant protein were confirmed with 12% SDS-PAGE analysis, followed by Coomassie Brilliant Blue (CBB) staining, the sample was dialyzed against a 300-fold volume of 50 mM Tris–HCl, 150 mM NaCl, pH 8.0 containing 10% glycerol (v/v) and the Complete Mini protease inhibitor cocktail (Roche, Mannheim, Germany) for 18 h at 4°C. The purified enzyme was stored at -80°C with 20% glycerol in small aliquots until use.
Enzymes Assay
EhDPCK activities of recombinant proteins and those in lysates were measured by a coupling assay using ADP HunterTM Plus Assay kit (DiscoverX, United States) according to the manufacturer’s instructions. Briefly, enzymatic assays were carried out using 50 ng of recombinant EhDPCK1 or EhDPCK2 and two kind of substrates (4–256 μM for dephospho-CoA, 5–100 μM ATP). All reactions were performed in assay buffer containing 15 mM Hepes, 20 mM NaCl, 1 mM EGTA, 0.02% Tween 20, 10 mM MgCl2, and 0.1% bovine gamma globulin in black microplate with 20 μL total volume. Plates were incubated for 60 min, added reagents A and B, and then re-incubated for 60 min in room temperature. After added with stop solution, The fluorescent intensity signal was measured using SpectraMax® Paradigm® (Molecular Devices, CA, United States) at excitation/emission wavelengths of 530/590 nm. The kinetic parameters were calculated using the non-linear regression function using the single saturating concentrations of substrates obtained from the GraphPad Prism software (GraphPad Software Inc., San Diego, CA, United States). The experiments were repeated three times in triplicate using protein preparations from three independent extractions, and kinetic values are presented as the means ± SE.
Production of Ehdpck1 and Ehdpck2 Gene-Silenced Strains
Small antisense RNA-mediated transcriptomic gene silencing (Bracha et al., 2006; Zhang et al., 2011) was used to repress gene expression of Ehdpck1 and Ehdpck2 genes with some modifications. Briefly, fragments corresponding to a 400 bp long of the open reading frame were amplified by PCR from cDNA using specific of oligonucleotides as follow: Ehdpck1 sense (5′-CAGAGGCCTATGAAAAAGATATTTGTTATTGGTAT-3′), antisense (5′-AATGAGCTCCAATGGCAATTTCAGGTGA-3′) and Ehdpck2 sense (5′-CAGAGGCCTATGGTATTTGTTATTGGTATCAC-3′), antisense (5′-AATGAGCTCTTCTAGTTTCCCAGATAACATTTA-3′). These oligonucleotides contained StuI and SacI restriction sites (shown in bold). PCR products were digested with StuI and SacI (New England BioLabs, MA, United States), and ligated into the StuI and SacI double digested psAP2-Gunma (Husain et al., 2011) to construct gene silencing plasmids. The trophozoites of G3 strain were transformed with the empty vector as a control and silencing plasmids by liposome-mediated transfection as previously described (Nozaki et al., 1999). Transformants were initially selected in the presence of 2 μg/mL geneticin with gradually increased up to 10 μg/mL.
Reverse Transcription PCR
Approximately 1 μg total RNA was used for cDNA synthesis using First-Stand cDNA Synthesis (Superscript® III, Invitrogen) with reverse transcriptase and oligo (dT) primers according to manufacturer’s instructions. The cDNA product was diluted 10-fold and PCR reactions were carried out in 50 μL, using primer pairs: Ehdpck1 sense (5′-ATGAAAAAGATATTTGTTATTGGT-3′), antisense (5′-TTAAAATTTATTTTCATTGAAGTCAA-3′) and Ehdpck2 sense (5′-ATGGTATTTGTTATTGGTATC-3′), antisense (5′-TTAGTTTAAAGTAATTTTTAATTGTT-3′). The PCR conditions were as follows: 98°C for 10 s, followed by 25 cycles of denaturation 98°C for 10 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. The PCR products obtained were resolved by agarose gel electrophoresis.
Quantitative Real-Time (qRT) PCR
Relative levels of mRNA of the Ehpank (EHI_183060), Ehdpck1 and Ehdpck2 (EHI_040840 and EHI_155780, respectively) were measured using qRT-PCR. RNA polymerase II gene (EHI_056690) was used as a reference. Each 20 μL reaction mixture contained 10 μL of 2× Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA, United States), 0.6 μL each of 10 μM sense and antisense primers, 5 μL 10× diluted cDNA, and nuclease-free water. PCR was performed using StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) with the cycling conditions: enzyme activation at 95°C for 20 s, followed by 40 cycles of denaturation at 95°C for 3 s, and annealing extension at 60°C for 30 s. All reactions were carried out in triplicate, including cDNA-minus controls. The amount of the steady state mRNA of each target gene was determined by the ΔCt method with RNA polymerase II as a reference gene (Livak and Schmittgen, 2001). The mRNA expression level of each gene in the transformants was expressed relative to that in the control transfected with psAP2.
Growth Kinetics
Trophozoite cultures were continuously maintained in mid-log phase as described above. Glass tubes (6-mL, Pyrex) containing approximately 5 × 105 cells were placed on ice for 5 min to detach cells from the glass surface. Centrifugation at 500 × g for 5 min at room temperature was conducted to collect cells. After discarding the spent medium, the pellet was re-suspended in 1 mL of BI-S-33 medium. Cell densities were estimated on a hemocytometer. Approximately 10,000 trophozoites were inoculated in 6 mL fresh BI-S-33 medium. Cell density of the cultures were estimated on a hemocytometer every 24 h for 5 days. Cell viability was also measured using WST-1 (Roche, Mannheim, Germany). Two hundred microliter aliquots of cultures were collected at every time point of incubation and transferred to the 96-well plate. The plate was incubated under anaerobic conditions at 35.5°C for 1 h. After the medium was removed, 100 μL of pre-warmed Opti-MEM I (Life Technologies, Grand Islands, NY, United States) containing 10% volume of WST-1 was added to each well, followed by further incubation at 35.5°C for 30 min. The viability of trophozoites was estimated by measuring absorbance at 450 nm by SpectraMax® Paradigm® (Molecular Devices, CA, United States).
Cell Fractionation and Immunoblot Analysis to Detect the Localization of EhDPCK
Trophozoites of the amoeba transformant expressing EhDPCK1 or 2 fused to human influenza virus hemagglutinin (HA) and the mock transformant, transfected with pEhEx-HA, were washed three times with PBS containing 2% glucose. Cells were homogenized and disrupted in homogenization buffer (50 mM Tris–HCl, pH 7.5, 250 mM sucrose, 50 mM NaCl, and 0.5 mg/mL E-64 protease inhibitor) followed by centrifugation at 500 ×g for 5 min, and the supernatant was collected to remove unbroken cells in the pellet. The supernatant fraction was centrifuged at 5,000 ×g for 10 min to isolate pellet and supernatant fractions. The 5,000 ×g supernatant fraction was further centrifuged at 100,000 ×g for 60 min to produce a 100,000 ×g supernatant and pellet fractions. The pellets at each step were further washed twice with homogenization buffer and re-centrifuged at 100,000 ×g for 10 min. Immunoblot analysis was performed using the fractions and anti-HA mouse monoclonal IgG. Anti-CPBF1 (cysteine protease-binding family protein 1) and anti-CS1 (cysteine synthase 1) rabbit antisera were used as organelle membrane and cytosolic markers, respectively.
Enzyme Activity and CoA Determination From Cell Lysates
Approximately 106 cells cultivated in 25 cm2 flasks (Iwaki, Fukushima, Japan) for 48 h were harvested, and the cell number was estimated as described above. The cell pellet was resuspended and homogenized in homogenization buffer (50 mM Tris–HCl, pH 7.5, 250 mM sucrose, and 50 mM NaCl) supplemented with 1 mM PMSF and 0.5 mg/mL E-64 (Peptide Institute, Osaka, Japan) by mechanical homogenization with a glass homogenizer. After kept on ice for 30 min, the suspension was centrifuged at 500 ×g for 30 min at 4°C to remove unbroken cells. Enzyme activity in the supernatant was used for enzymatic assay.
Concentrations of CoA in cell lysates were measured using the CoA assay kit (BioVision, CA, United States) according to the manufacturer’s instructions. CoA at 0.05–1 nmol was used to produce a standard curve to determine the CoA concentrations in lysates. Experiments were conducted in triplicate and repeated three times on three different days.
Intracellular Metabolite Extraction
Intracellular metabolites from 1 × 107 Ehdpck1 and Ehdpck2 gene silencing cells were extracted according to the protocol previously described (Jeelani et al., 2012). Internal standards, 200 μM of 2-(N-morpholino)-ethanesulfonic acid and methionine sulfone, were added to every sample to ensure that experimental artifacts such as ion suppression did not lead to misinterpretation of metabolite levels.
Instrumentation of Capillary Electrophoresis–Mass Spectrometry (CE-MS)
Capillary electrophoresis–mass spectrometry was performed as previously described (Kinoshita et al., 2007; Yamamoto et al., 2014). The instrument used was an Agilent CE Capillary Electrophoresis System equipped with an air pressure pump, an Agilent 1100 series isocratic high-performance liquid chromatography pump and an Agilent 1100 series MSD mass spectrometer. We also used a G1603A Agilent CE-MS adapter kit, and a G1607A Agilent CE-MS sprayer kit (Agilent Technologies). G2201AA Agilent ChemStation software for CE-MSD was used for system control and data acquisition.
CE-MS Conditions for Cationic, Anionic Compounds, and Nucleotides
Cationic nucleotides compounds were carried out on a gas chromatograph capillary, polydimethylsiloxane (DB-1) (50-μm inner diameter × 100-cm total length) (Agilent Technologies). CE separation was using 50 mM ammonium acetate, pH 7.5 as the electrolyte. Samples solution (∼3 nL) was injected at 50 mbar for 30 s, and a positive voltage of 30 kV was applied. All other conditions were the same as in the anionic metabolite analysis. Anions were coated in SMILE (+) capillary 50-μm inner diameter × 100-cm total length obtained from Nacalai Tesque (Kyoto, Japan). The electrolyte was 50 mM ammonium acetate solution (pH 8.5). Sample solution (∼30 nL) was injected at 50 mbar for 30 s and a negative voltage of 30 kV. The capillary voltage was set at 3.5 kV for ESI-MS in the negative ion mode. Deprotonated [M-H] ions were monitored for anionic metabolites of interest.
Statistical Analysis of Metabolomic Data and Pathway Analysis
For each experimental condition, three independent biological replicates were made and for each biological replicate, two technical replicates were made. All data are shown as means ± SE or the indicated number of experiments. Statistical comparisons were made by Student’s t-test. Pathway analysis was conducted using Kyoto Encyclopedia of Genes and Genomes database (KEGG5).
Results
Identification and Features of Two EhDPCK Isotypes
Two genes encoding EhDPCK were identified from the genome database of E. histolytica HM-1:IMSS (HM-1) reference strain6: EHI_040840 and EHI_155780, herein designated as Ehdpck1 and Ehdpck2 genes, respectively. The open reading frames of the two genes are 621 and 615 bp in length and encode 206- and 204-amino acid long proteins with the calculated molecular mass of 23.9 and 23.1 kDa, respectively. The two proteins exhibit 38% mutual similarity. EhDPCK1 shows 31, 29, and 26% percentage similarity to orthologs from human, Arabidopsis thaliana, and E. coli, whereas EhDPCK2 shows 34, 29, and 28% identity to the orthologs from human, A. thaliana, and E. coli, respectively. Human, A. thaliana, and E. coli only have one functional copy of DPCK and seem to be slightly closer to EhDPCK2 than to EhDPCK1.
DPCKs are a member of the family of P-loop kinases with an overall topology similar to that of nucleotide monophosphate-binding kinases (Vonrhein et al., 1995; Leipe et al., 2003). DPCKs comprise three domains: the nucleotide-binding domain in parallel β-sheet, the substrate-binding domain in α-helices, and the LID domain. All of these DPCK features are present in EhDPCK (Figures 1A,B). DPCKs generally contain five parallel β-strands flanked by α-helices with ATP-binding site and CoA-binding domain (Obmolova et al., 2001; O’Toole et al., 2003). Based on secondary structure prediction, each monomer of EhDPCKs consist of five to six parallel β-strands flanked by 11 α-helices, which, the domain configuration, differs from 10 α-helices in both E. coli and H. influenza counterparts. EhDPCK1 and 2 contain the highly conserved P-loop or Walker A sequence motif (GXXXXGKT/S, where X is any residue), which is reported to be involved in the nucleotide binding (Obmolova et al., 2001; Supplementary Figure S1). EhDPCKs P-loop motifs are located in a region between β1 and α1. The amino acid residues involved in the CoA binding (T10, D35, W90, A117, L118, and Q163), and in the ATP binding (G11, I13, G16, K17, R144, and T179) are conserved in EhDPCKs.
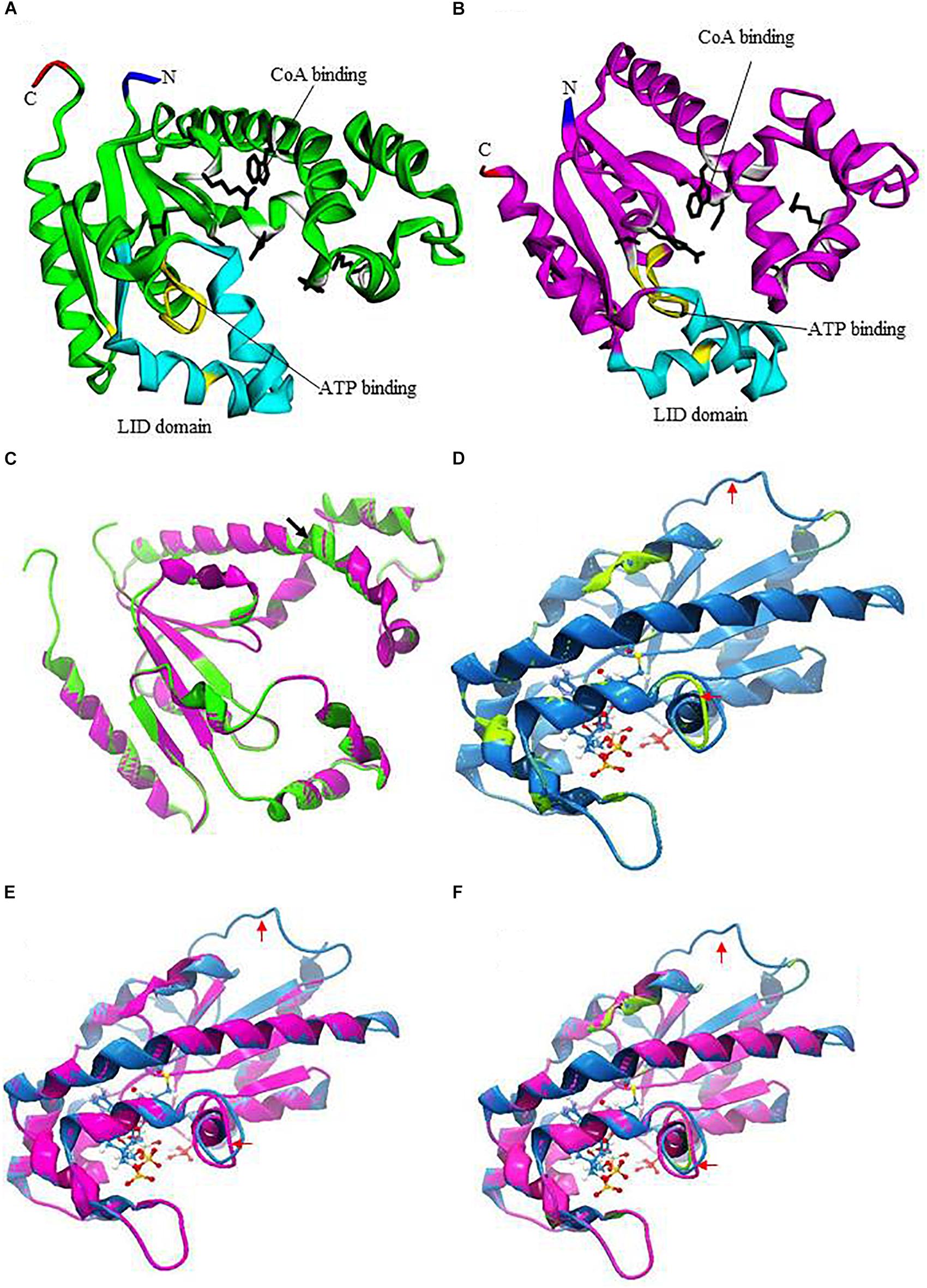
FIGURE 1. Prediction of three-dimensional structures of EhDPCKs using Mus musculus COASY as the Pyre 2 homology model. (A) A predicted structure of EhDPCK1 (accession number: XP_648971). The region implicated in the ATP binding is marked in yellow; the side chains of the residues involved in the CoA binding are marked with white ribbons with black sticks; and LID domain is depicted with cyan ribbons. The amino and carboxyl termini are marked in blue and red, respectively. The remaining regions are shown in green. (B) A predicted structure of EhDPCK2 (accession number: XP_655761). The region implicated in the ATP binding is marked in yellow; side chains of the residues involved in the CoA binding are marked with white ribbons with black sticks; and LID domain is depicted with cyan ribbons. The amino and carboxyl termini are marked in green and red, respectively. The remaining regions are shown in purple. (C) Superimposition of EhDPCK1 (green) and EhDPCK2 (pink). Structures shown in (C–F) are rotated 180° around the axis shown in (A) of the structure shown in (A,B). Black arrowhead indicated an additional β-sheet in EhDPCK1, but the 3-residue beta sheet is flanked by two alpha helices and thus it is not obvious. (D) Superimposition of the EhDPCK1 (green) on M. musculus COASY (PDB ID: 2F6R) (blue). ATP is shown with sticks and balls. (E) Superimposition of the EhDPCK2 (pink) on M. musculus COASY (blue). (F) Superimposition of the EhDPCK1 (green) and EhDPCK2 (pink) on M. musculus COASY (blue). Red arrows indicate the different area between EhDPCKs and M. musculus COASY.
EhDPCK1 and 2 show striking similarity based on three-dimensional structure predicted by homology modeling using Phyre 2 (also see below, Figure 1C). EhDPCK1 differs from EhDPCK2 by an additional β-sheet from residue Ile88 and Trp90, indicated with a black arrowhead (Figure 1C). When compared to E. coli DPCK, almost all key residues implicated for CoA binding are conserved in both EhDPCK1 and EhDPCK2 (O’Toole et al., 2003). The only exceptions are the substitution of Val473 in human ortholog with Leu118 and Leu117 in EhDPCK1 and EhDPCK2, respectively. Gly56 and Gly55 in EhDPCK1 and EhDPCK2, respectively, located between helices α3 and α4 are predicted to be important to maintain structural integrity. Briefly, based on our in silico analysis, E. histolytica has two genes encoding EhDPCK with relatively low similarity of amino acids identity but share highly conserved amino acids in term of substrates binding.
Phylogenetic Analysis of EhDPCK
Eighty-eight DPCK sequences from broad taxonomic groups were obtained, aligned, and analyzed (Supplementary Table S1 and Supplementary Figure S1). All major groups of Eukaryota possess putative homologs of DPCK. There are only few other eukaryotes that possess more than one copy of DPCK homologs (e.g., trypanosomes). However, we did not perform further searches for duplicated homologs in other eukaryotic species. While most of the sequences were well aligned throughout the proteins, three eukaryotic homologs (Homo sapiens, M. musculus, and D. melanogaster) possess an approximately 350 amino acids long N-terminal extension, which corresponds to phosphopantetheine adenylyltransferase domain of the phosphopantetheine adenylyltransferase/DPCK bifunctional enzyme. As shown in an alignment of the selected DPCK sequences from representative taxa (Supplementary Figure S1), the amino acid residues involved in the CoA binding and in the ATP binding are well conserved.
Phylogenetic analyses of 88 sequences on the basis of 112 aligned positions inferred an optimal maximum likelihood (ML) tree shown in Supplementary Figure S2. As a whole, DPCK sequences are highly divergent suggested by long branches. Consequently, no resolution has been inferred for deep branching patterns across all parts of the tree. Neither was inferred the origin of the Entamoeba homologs. However, in the genus Entamoeba, all DPCK sequences including two isotypes for each species (E. histolytica, E. nuttalli, E. dispar, and Entamoeba invadens) are monophyletic, although the bootstrap support value was not high (66%). Furthermore, monophyly of copy 1 or copy 2 isotype among the four Entamoeba species was supported with 87 and 88% BP values, respectively, suggesting that DPCK gene duplication likely occurred in the common ancestor of four Entamoeba species, followed by speciation of the four species.
Structural Comparison of EhDPCK and Its Mammalian Counterpart
We utilized M. musculus COASY as a representative mammalian DPCK (the Protein Data Bank ID: 2F6R), as a crystal structure of human DPCK was not available. Human and M. musculus COASY are closely related with 87% identity. Molecular modeling using Phyre 2 (see text footnote 4) indicates overall similarities between EhDPCK1 and 2 and also between amebic DPCKs and mammalian counterpart despite low amino acid identity (<24%) (Figures 1D–F). Several areas are remarkably different in structures such as the loop region indicated by red arrows, some residues in CoA binding, and LID domain, suggesting the potential for designing of specific inhibitors against amebic DPCKs.
Biochemical Characterization of Recombinant EhDPCK1 and EhDPCK2
EhDPCK1 and EhDPCK2 recombinant enzymes were successfully produced using E. coli expression system and purified for enzymological characterization. As demonstrated by SDS-PAGE analysis under reducing conditions, followed by CBB staining and immunoblot assay, both enzymes were detected in both the soluble and insoluble fractions. Purified proteins were obtained with a relatively low yield: typically around 0.3 mg of protein, purified from a 500 mL E. coli culture (Supplementary Table S2. The purity of recombinant EhDPCK1 and 2 was estimated to be 90∼95% by densitometric scanning of the CBB stained gels after SDS-PAGE (Supplementary Figure S3). Immunoblots also indicate only negligible truncated forms of both EhDPCK1 and 2 were present in the final preparations. The apparent molecular mass of the purified proteins was consistent with the molecular mass of 23.9 and 23.1 kDa for native EhDPCK1 and EhDPCK2, respectively, plus 2.6 kDa corresponding to the histidine tag.
The specific activity of the purified enzymes was estimated as 2.13 ± 0.15 and 2.54 ± 0.26 μmole/min/mg (mean ± SE of triplicates) for EhDPCK1 and EhDPCK2, respectively. EhDPCK1 and EhDPCK2 showed a broad pH optimum between 7 and 9 with the maximum activity at 8 (Supplementary Figure S4). No difference was observed in the pH dependency between EhDPCK1 and EhDPCK2. Kinetic constants for EhDPCK1 and EhDPCK2 were determined by measuring the initial rates obtained with different concentrations of ATP and dephospho-CoA (Table 1). Both ATP and dephospho-CoA exhibited hyperbolic saturation kinetics when assayed over the range of 4–128 μM dephospho-CoA in the presence of 100 μM ATP and in the range of 1–100 μM ATP substrate in the presence of 128 μM dephospho-CoA (data not shown). EhDPCK1 showed the Km values of 114 ± 19 and 19.6 ± 1.2 μM for dephospho-CoA and ATP, respectively. EhDPCK2 showed lower Km values with 57.9 ± 6.1 and 15.0 ± 2.4 μM for dephospho-CoA and ATP, respectively. Neither EhDCPK1 nor 2 utilized pantothenate as a substrate when the assay was conducted with 100 μM of pantothenate and 80 μM of ATP (<0.02 μmole/min/mg; data not show in tables).
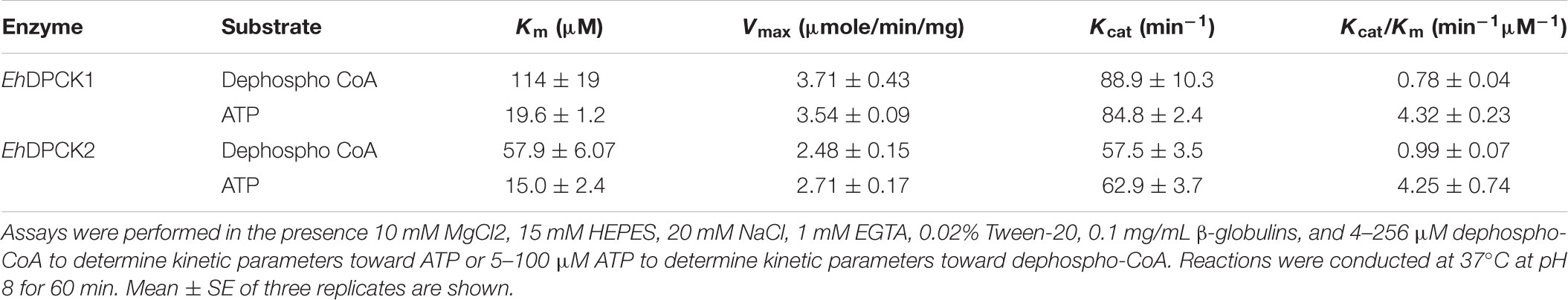
TABLE 1. Kinetic parameters of E. histolytica dephospho CoA kinase 1 (EhDPCK1) and dephospho CoA kinase 2 (EhDPCK2).
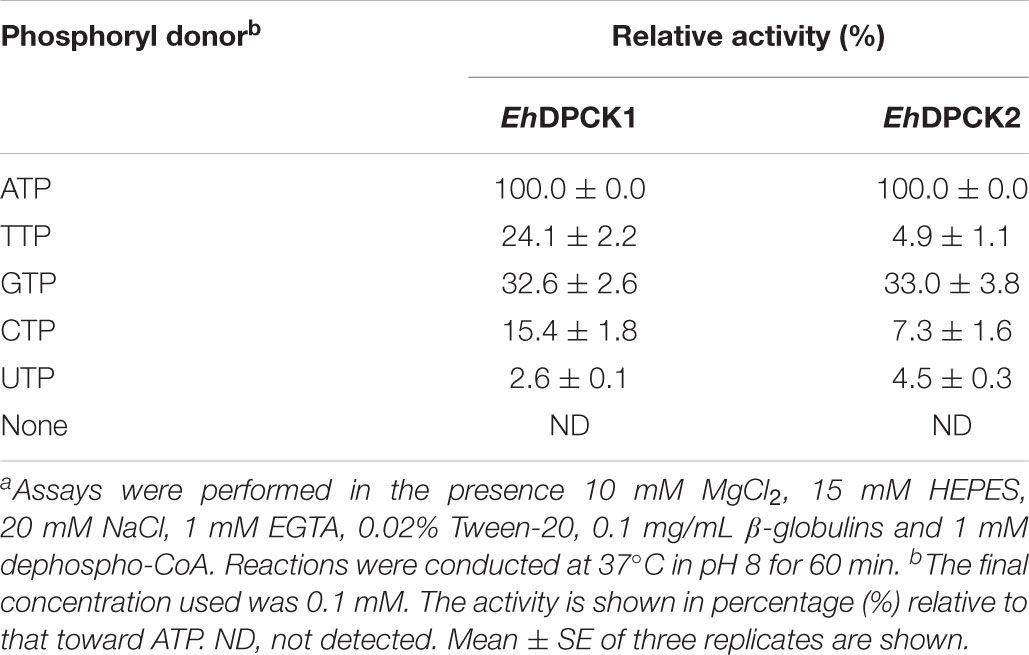
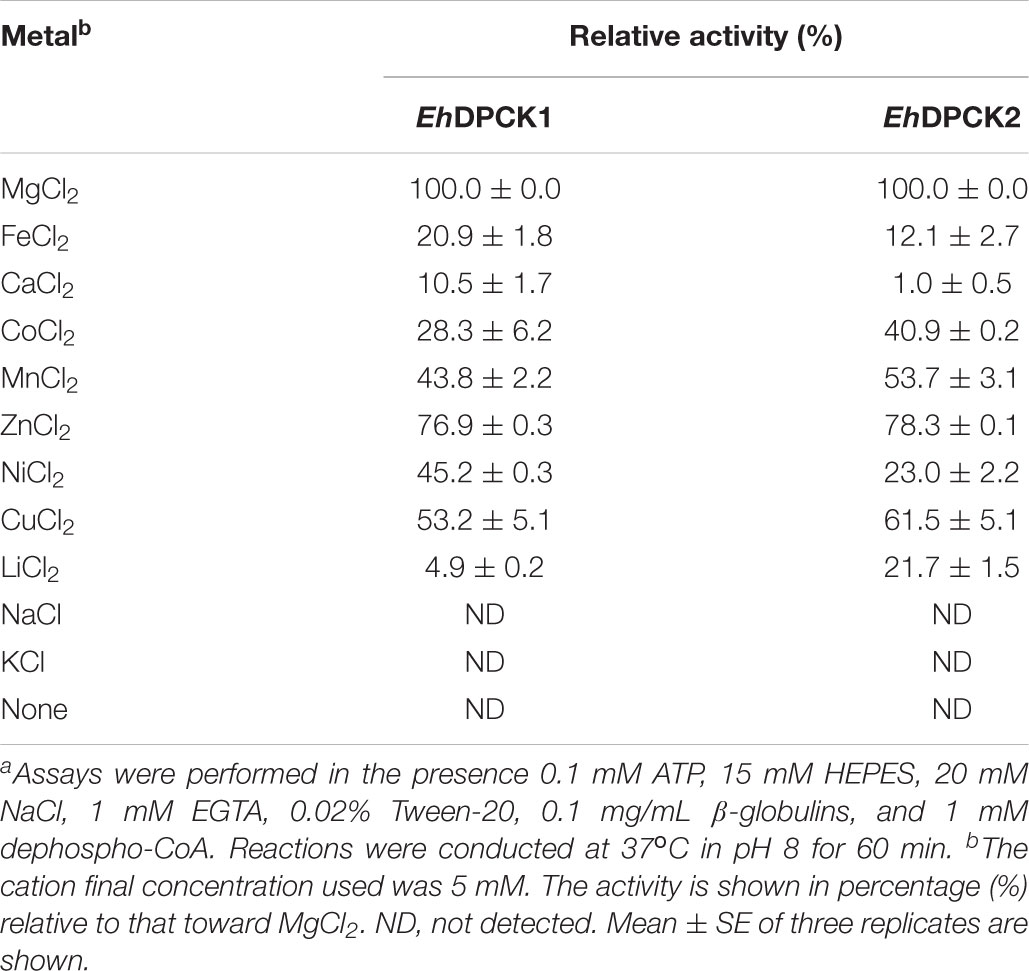
Recombinant EhDPCK1 and EhDPCK2 could use various nucleoside triphosphates, such as ATP, CTP, GTP, and UTP, as phosphoryl donors (Table 2). GTP could partially replace ATP for EhDPCK1 and EhDPCK2 (33% of the activity relative to that with ATP). This is a unique feature of EhDPCKs as in general only a few kinases exhibit dual usage of ATP or GTP (Niefind et al., 1999; Becher et al., 2013). EhDPCK1 and EhDPCK2 showed noticeably difference in nucleotide triphosphate preference toward TTP (24.1 and 4.9%, respectively). The effects of metal ions were examined by assaying the activity after the addition of metal salts to the standard reaction mixture (Table 3). The data also suggest that both EhDPCK1 and EhDPCK2 activities are Mg2+-dependent. Furthermore, some cations (Zn2+ and Cu2+) could replace Mg2+ with relative activity higher than 50%. No activity was detected when Na+ or K+ was used instead of Mg2+. Both EhDPCK1 and EhDPCK2 showed similar profiles of inhibition by CoA, acetyl-CoA, and malonyl-CoA at relatively high concentrations (Supplementary Figure S5). Taken together from enzymological characterization including kinetic parameters, phosphoryl donor specificities, metal dependence, and inhibitors, EhDPCK1 and 2 showed are both active and show noticeable biochemical differences.
Cellular Localization of EhDPCK
Cellular fractionation of the transformant cells expressing HA-tagged EhDPCK2, followed by immunoblot analysis with anti-HA antibody was performed in order to determine localization of EhDPCK2. EhDPCK2 was detected in a wide range of fractions, namely 5,000 ×g supernatant and pellet and 100,000 ×g supernatant and pellet fractions with the highest amount being detected in the 100,000 ×g supernatant fraction, suggesting that EhDPCK2 is located in both cytosol and organelles (or associated with the membrane) (Supplementary Figure S6C). Despite repeated trials, EhDPCK1 with epitope-tags (i.e., HA, Myc, and Flag) were unable to be expressed.
Phenotypes Caused by Gene Silencing of Ehdpck1 and Ehdpck2
To better understand the role of EhDPCK in E. histolytica, we created a transformant strain where dpck gene expression was selectively repressed by small antisense RNA-mediated transcriptional gene silencing. A 400-bp long region of Ehdpck1 and Ehdpck2 genes that corresponds to the amino-terminal portion of the protein was used to design gene silencing constructs. Despite low identity at nucleotide and amino acid levels between the two isotypes, gene silencing of Ehdpck1 gene also affected Ehdpck2 gene and vice versa (Figures 2A,B, 3A,B). Gene silencing of expression Ehdpck1 gene caused 77 ± 1.2 and 46 ± 18.9% reduction of Ehdpck1 and Ehdpck2 transcripts, respectively, while gene silencing of Ehdpck2 gene caused 41 ± 14.8 and 88 ± 2.9% reduction of Ehdpck1 and Ehdpck2 transcripts, respectively. There was no change in the transcript level of irrelevant genes such as NAD(H) kinase (EHI_151920), mitosomal membrane protein-Tom40 (EHI_104420), and cysteine protease binding protein 2 (EHI_087660) (Supplementary Figure S7), suggesting the specificity of Ehdpcks silencing. Despite only 45% amino acid identity at the amino-terminal portion of EhDPCK1 and 2, the first 400 bp of Ehdpck1 and 2 genes show 53% identity at the nucleotide level. Moreover, most of the conserved domains including the ATP-binding domain is located within the first 100 amino acids in both EhDPCK1 and 2. Therefore, this high similarity probably caused cross-repression of gene expression by antisense small RNA-mediated transcriptional silencing in our experiment. Interestingly, gene silencing of either Ehdpck1 or Ehdpck2 gene caused upregulation of EhPanK transcript (Figures 2B, 3B), which may indicate compensation for Ehdpck gene silencing.
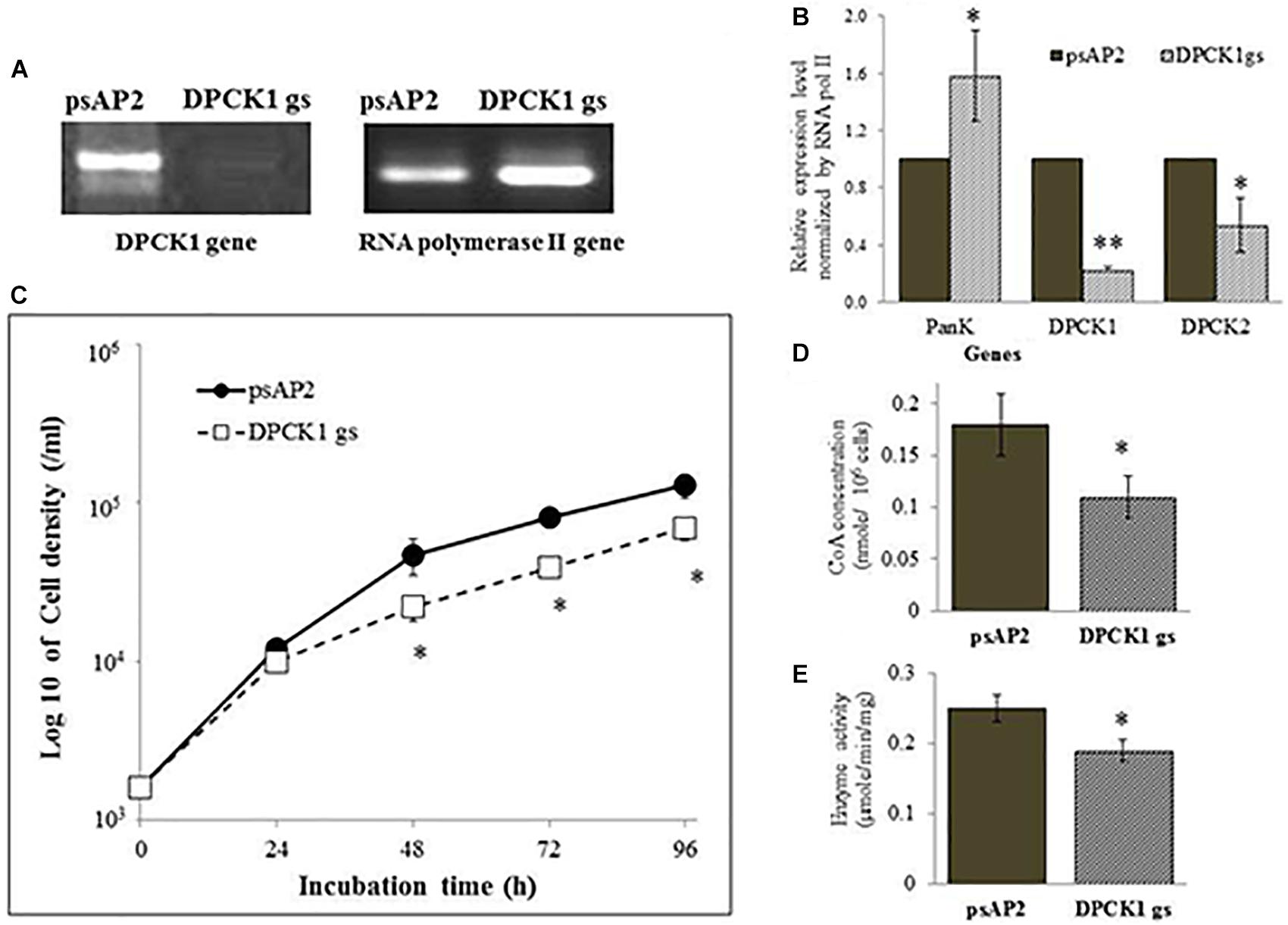
FIGURE 2. The dpck gene expression, cell growth, CoA concentration, and DPCK activity in Ehdpck1 gene-silenced (gs) strain. (A) RT-PCR of Ehdpck1 and RNA polymerase II genes from cDNA of E. histolytica transformants. “psAP2” indicates G3 strain transfected with mock psAP-2-Gunma vector, and “DPCK1 gs” indicates GS strain transfected with Ehdpck1 gene silencing plasmid. (B) Relative levels of gene transcripts encoding EhPanK, EhDPCK1, and EhDPCK2 involved in CoA biosynthesis, estimated by qRT-PCR analysis in psAP2 and Ehdpck1 gs transformants. qRT-PCR data were normalized against RNA polymerase II, and are shown in percentage relative to the transcript level of each gene in psAP2 control. (C) Growth kinetics of E. histolytica transformants in BI-S-33 medium. (D) CoA concentrations in cell lysates. (E) DPCK activity in cell lysates of psAP2 and Ehdpck1 gs transformants. Data shown in mean ± SE. Statistical comparison is made by Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
Ehdpck1 and Ehdpck2 gene-silenced (gs) strains showed significant growth defect in normal growth medium (Figures 2C, 3C and Supplementary Figures S6A,B, by cell count and WST-1 growth assay, respectively). The level of growth inhibition by Ehdpck2 gene silencing was more severe compared to that by Ehdpck1 gene silencing; the percentage growth at 96 h of Ehdpck1 gs and Ehdpck2 gs trophozoites was 53.8 ± 9.2% and 23.1 ± 6.2% (mean ± SE of triplicates) of the control transformant, respectively. Cells lysates from Ehdpck1 gs strain contained 38.9 ± 7.1% lower level of CoA (Figure 2D) and approximately 24.0 ± 2.0% less EhDPCK activity (Figure 2E). The effects of Ehdpck2 gene silencing on the reduction of DPCK activity and CoA concentrations in cell lysates were more severe than those by Ehdpck1 gene silencing: 61.1 ± 7.1% decrease in the CoA concentration (Figure 3D) and 44.0 ± 6.2% decrease in EhDPCK2 activity (Figure 3E) by Ehdpck2 gene silencing. Altogether, these results suggest that both EhDPCK isotypes play important roles in cell growth and the role played by EhDPCK2 is more important than that of EhDPCK1. In our study, the repression level of both Ehdpck1 and Ehdpck2 gene transcripts by gene silencing was not complete; residual expression was also observed in the silenced strains. This likely contributes the poor but viable cultures of these gene-silenced strains. The fact that we also failed to create Ehdpcks gene-silenced lines in three additional attempts is also consistent with the presumed essentiality of these genes.
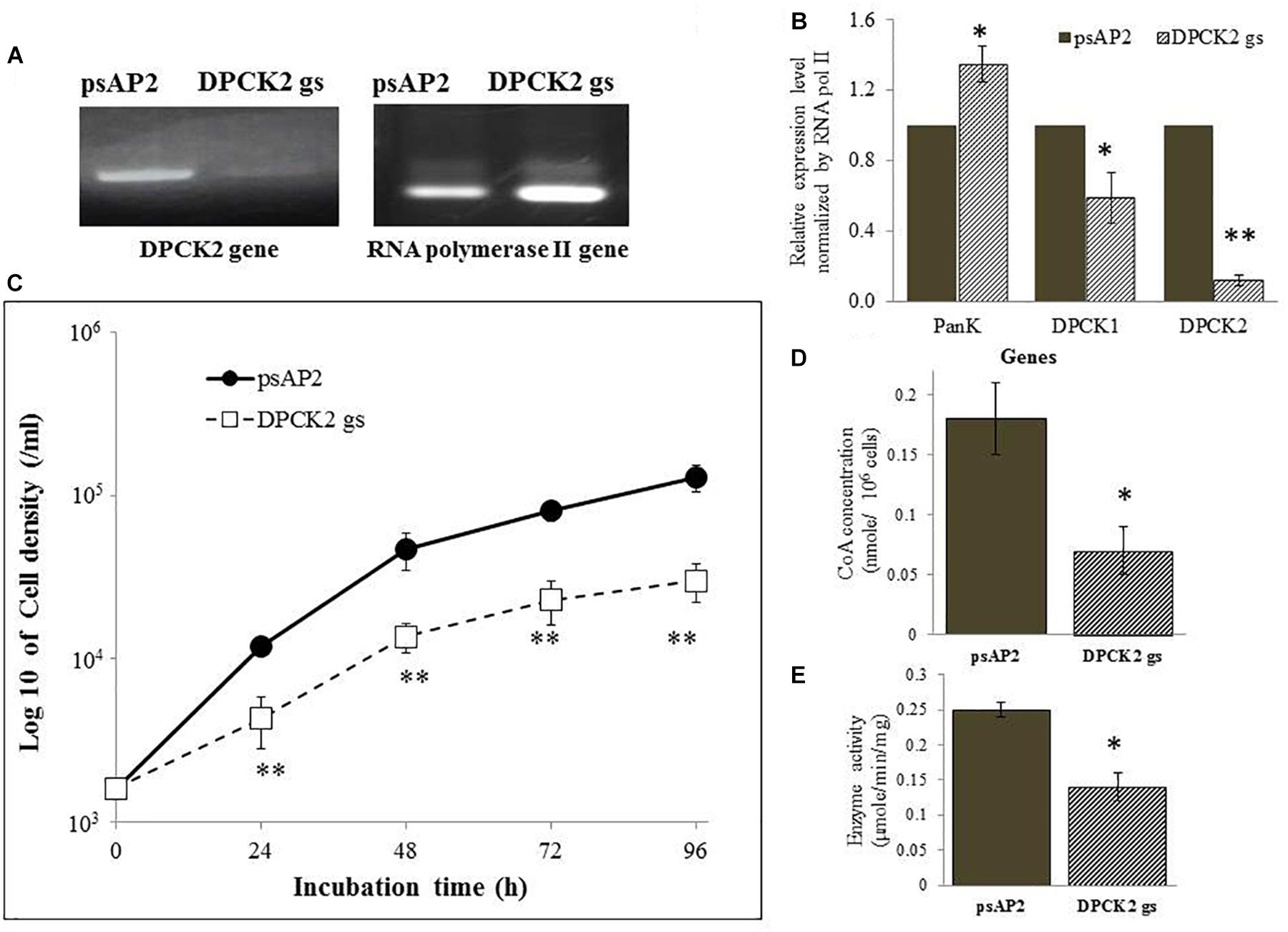
FIGURE 3. DPCK gene expression, cell growth, CoA concentration, and DPCK activity in Ehdpck2 gene-silenced strain. (A) RT-PCR of Ehdpck2 and RNA polymerase II genes from cDNA of E. histolytica transformants. (B) Relative levels of gene transcripts encoding EhPanK, EhDPCK1, and EhDPCK2, estimated by qRT-PCR analysis using RNA from psAP2 and Ehdpck2 gs transformants. qRT-PCR data were normalized against RNA polymerase II, and are shown in percentage relative to the transcript level of each gene in psAP2 control. (C) Growth kinetic of E. histolytica transformants in BI-S-33 medium. (D) CoA concentrations in cell lysates. (E) DPCK activity in cell lysates of psAP2 and Ehdpck2 gs transformants. Labels and abbreviations are as in Figure 1.
Metabolomic Analyses of Ehdpck1 and Ehdpck2 Gene-Silenced Strains
We performed metabolomics analyses of the Ehdpck1 and Ehdpck2 gs strains. The capillary electrophoresis–mass spectrometry (CE-MS)-based quantitation systems were used both in cation and anion modes to identify >110 metabolites which include precursors, intermediates, and end products of central carbon metabolism, biosynthetic and catabolic intermediates of amino acids, sugars, nucleic acids, and lipids. All data were presented as normalized by cell number (per 1 × 106 cells), as commonly accepted and used in many studies. The levels of some metabolites were changed in both directions (i.e., increase and decrease) in response to Ehdpck1 and Ehdpck2 gene silencing (Supplementary Table S3). However, all of the statistically significant changes in the concentrations of metabolites in both Ehdpck1 and Ehdpck2 gene silencing were decrease, not increase, relative to the metabolite levels in the control strain.
Besides decrease in the CoA concentration mentioned above, Ehdpck1 gene silencing caused decrease only in citrate, ornithine, some nucleic acids, and S-adenosyl-L-methionine (Figure 4). On the other hand, Ehdpck2 gene silencing caused decrease in a wider range of metabolites than Ehdpck1, such as pantothenate, acetyl-CoA, glutamate, ornithine, putrescine, spermidine, methionine, and S-adenosyl-L-methionine, which were mapped to metabolites involved in ornithine and polyamine biosynthesis in KEGG maps (Figure 5). Ehdpck2 gene silencing also caused decrease in some (five of nine metabolites measured) metabolites involved in glycogen/glucuronate and chitin metabolism, such as UDP-glucose, glutamate, and N-acetylglucosamine 6-phosphate (Figure 6). Interestingly, some (six of 14 metabolites measured) of the intermediate metabolites in purine metabolism were also decreased by Ehdpck2 gene silencing (Figure 7): in particular adenine, hypoxanthine, xanthine, GDP, GTP, and ATP. Metabolite changes caused by Ehdpck1 gene silencing in ornithine and polyamine biosynthesis, glycogen/glucuronate, chitin, and purine metabolisms were presented in Supplementary Figures S8–S10. Taken together, our metabolomics data showed that Ehdpck2 gene silencing more severe affected overall metabolisms than that of Ehdpck1.
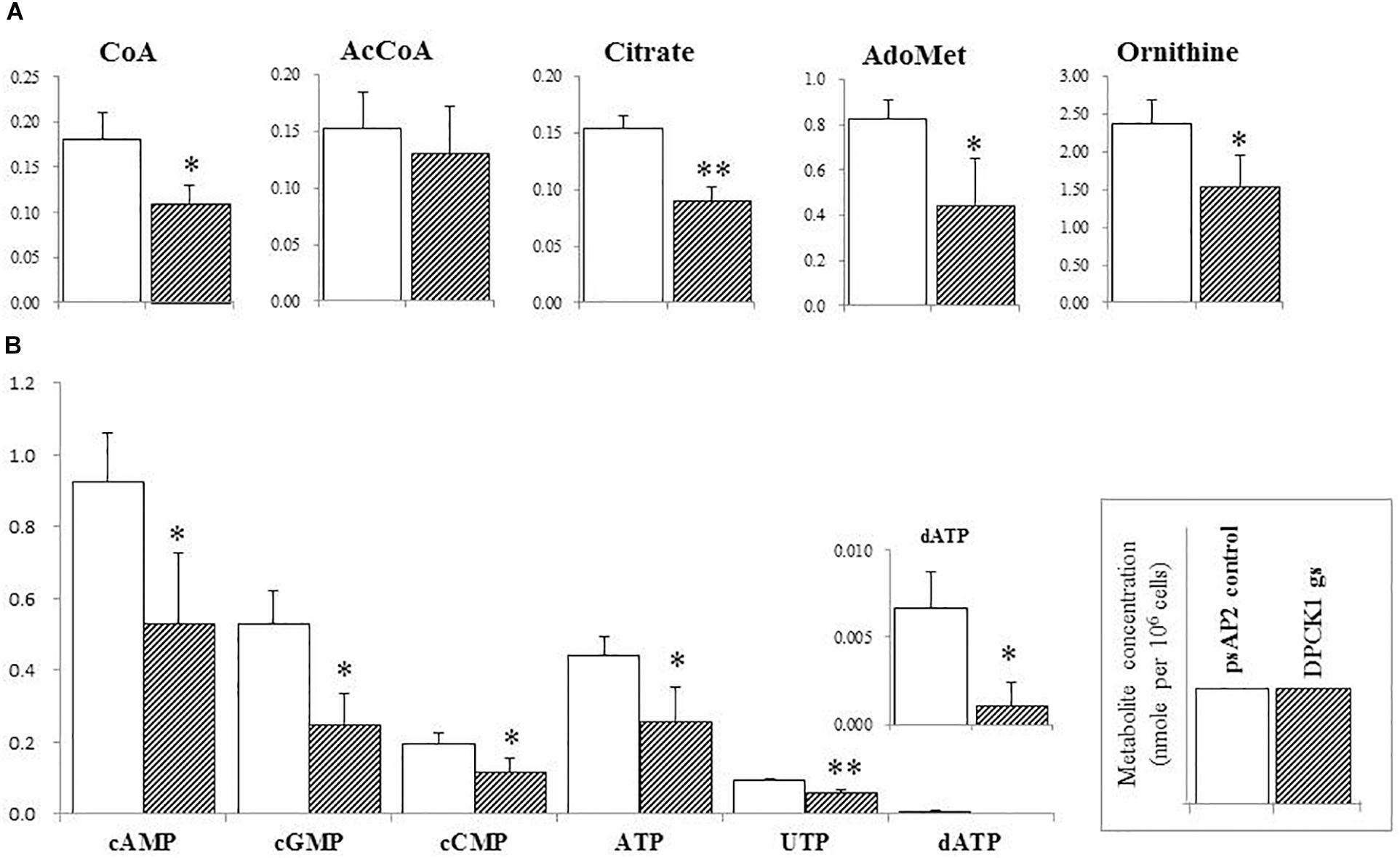
FIGURE 4. The levels of representative intermediary metabolites affected by Ehdpck1 gene silencing. (A) Note decrease in the concentrations of coenzyme A (CoA), acetyl-CoA (AcCoA), citrate, ornithine, and S-adenosyl-L-methionine (AdoMet). (B) Depletion of nucleotides. Data are shown in mean ± SE. Statistical comparison is made by Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
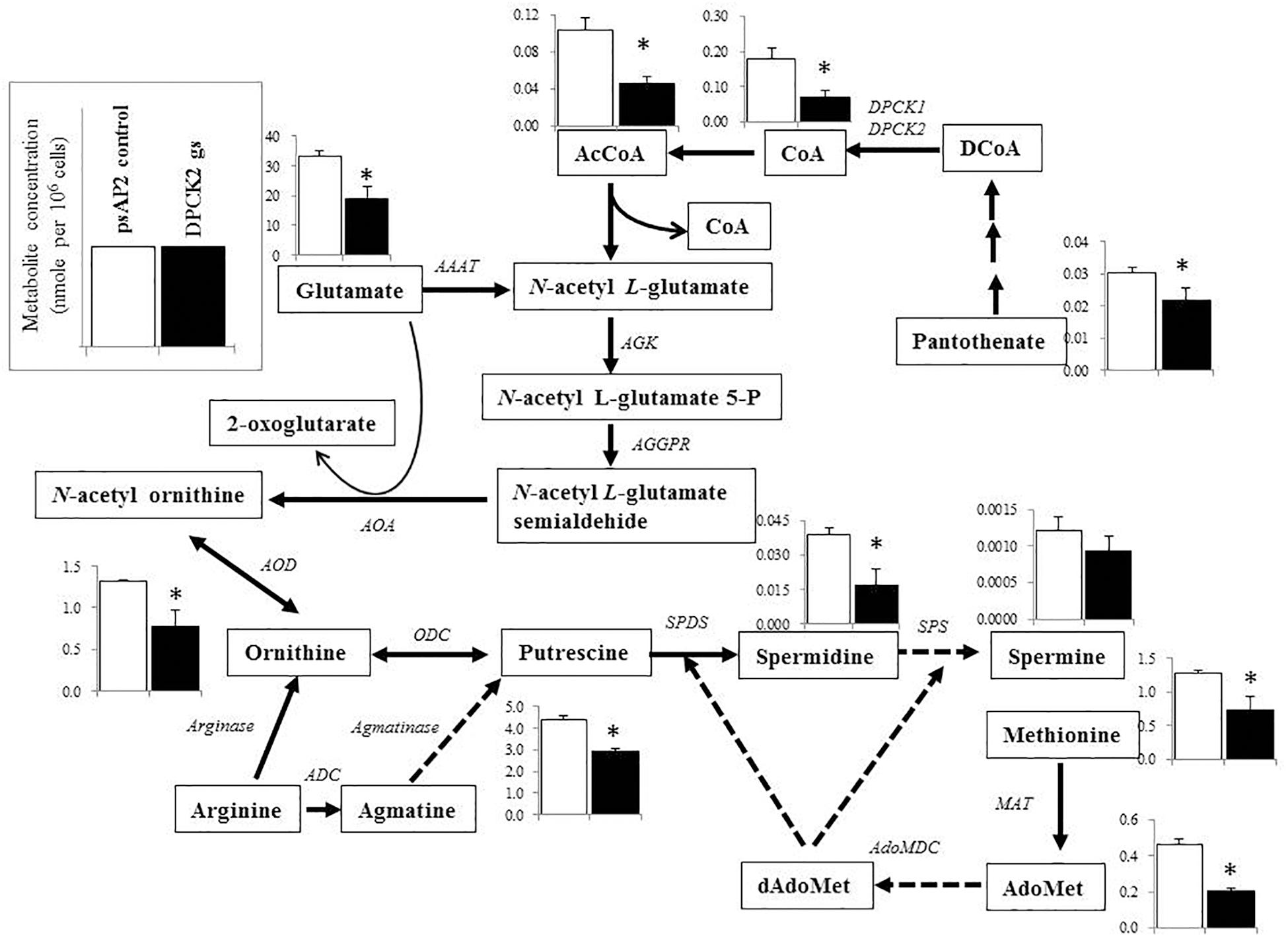
FIGURE 5. Metabolic profiles of amino acids, polyamines, and associated metabolites affected by Ehdpck2 gene silencing. Broken lines indicate genes whose encoding enzymes known to catalyze the steps are likely absent in the genome. Abbreviations: ND, not detected; CS, citrate synthase; AAAT, amino-acid N-acetyltransferase; AGK, acetylglutamate kinase; AGGPR, N-acetyl-L-glutamyl-phosphate reductase; AOA, acetylornithine aminotransferase; AOD, acetylornithine deacetylase; ODC, ornithine decarboxylase; ADC, arginine decarboxylase; MAT, methionine adenosyltransferase, AdoMet, S-adenosyl-L-methionine; dAdoMet, S-adenosylmethioninamine; MAO, monoamine oxidase; AdoMDC, S-adenosylmethionine decarboxylase; SPDS, spermidine synthase; SPS, spermine synthase. Short and successive arrows indicated multiple steps in the corresponding pathway. Data are shown in mean ± SE. Statistical comparison is made by Student’s t-test (∗P < 0.05).
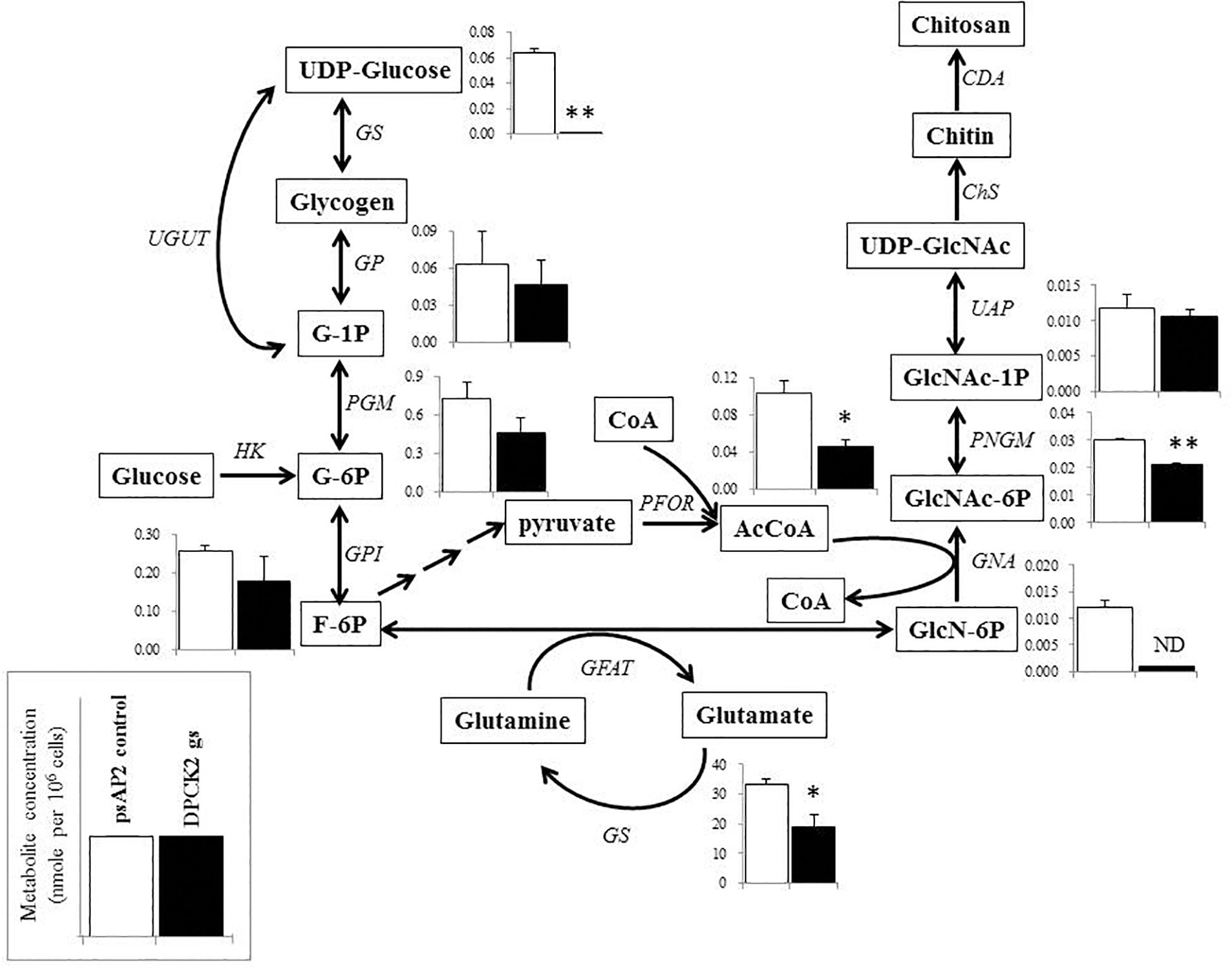
FIGURE 6. Metabolic profiles of hexose phosphates, glycogen metabolism, chitin biosynthesis, and associated metabolism affected by Ehdpck2 gene silencing. Abbreviations: G-1P, glucose 1-phosphate; G-6P, glucose 6-phosphate; F-6P, fructose 6-phosphate; GlcN-6P, glucosamine 6-phosphate; GlcNAc-6P, N-acetylglucosamine 6-phosphate; GlcNAc-1P, N-acetyl glucosamine 1-phosphate; UDP-GlcNAc, UDP-N-acetylglucosamine; GS, glycogen synthase; GP, glycogen phosphorylase; UGUT, UTP-glucose-1-phosphate uridyltransferase; PGM, phosphoglucomutase; GPI, glucose-6-phosphate isomerase; HK, hexokinase; GFAT, glutamine:fructose-6-phosphate aminotransferase; GS, glutamine synthetase; PFOR, pyruvate ferredoxin oxidoreductase; GNA, glucosamine-phosphate N-acetyltransferase; PNGM, phosphoacetylglucosamine mutase; UAP, UDP-N-acetylgalactosamine diphosphorylase; ChS, chitin synthase; CDA, chitin deacetylase; ND, not detected. Short and successive arrows indicated multiple steps in the corresponding pathway. Data are shown in mean ± SE. Statistical comparison is made by Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
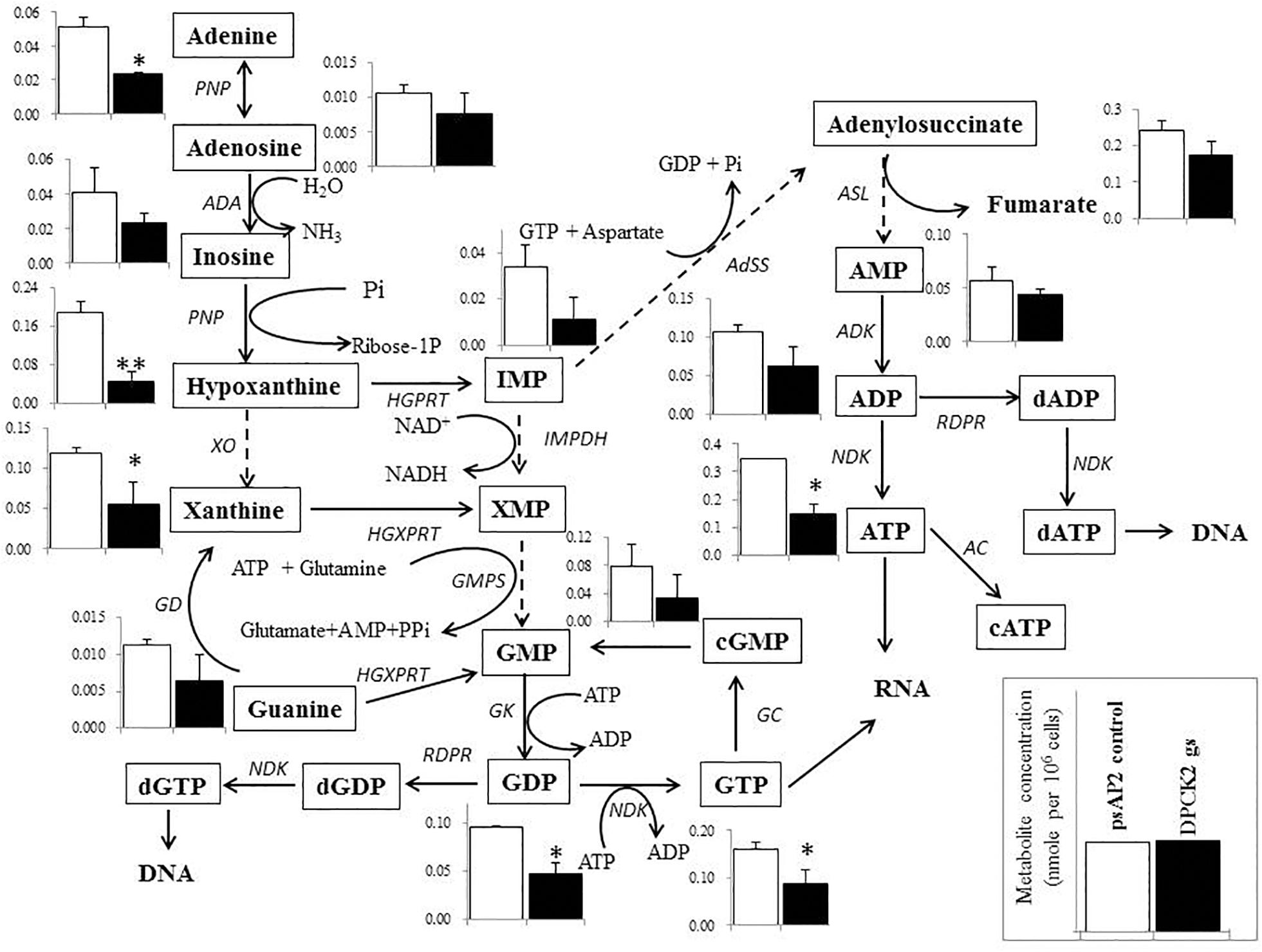
FIGURE 7. Metabolic profiles of purine metabolism affected by Ehdpck2 gene silencing. Broken lines indicate genes whose encoding enzymes known to catalyze the steps are likely absent in the genome. Abbreviations: PNP, purine nucleoside phosphorylase; ADA, adenosine deaminase; XO, xanthine oxidase; HGPRT, hypoxanthine-guanine phosphoribosyltransferase; HGXPRT, hypoxanthine-guanine-xanthine phosphoribosyltransferase; IMPDH, inosine 5’-monophosphate dEhydrogenase; GMPS, guanosine monophosphate synthetase; GK, guanylate kinase; NDK, nucleoside diphosphate kinase; RDPR, ribonucleoside diphosphate reductase; GC, guanylate cyclase; AdSS, adenylosuccinate synthetase; ASL, adenylosuccinate lyase; ADK, adenylate kinase; AC, adenylate cyclase. Data are shown in mean ± SE. Statistical comparison is made by Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
Discussion
Suggested Specific Roles of EhDPCK Isotypes
Our previous study on E. histolytica PanK, the enzyme that catalyzes the first step of CoA biosynthesis, clearly demonstrated that this enzyme was important for growth of E. histolytica trophozoites. In this study, we have shown that both of two EhDPCKs, EhDPCK1 and EhDPCK2, play an indispensable, likely non-overlapping role in CoA biosynthesis. A line of evidence supports that both enzymes are involved in CoA biosynthesis in E. histolytica. First, E. coli-produced recombinant enzymes show comparable specific activity and kinetic parameters. Second, Ehdpck1 and Ehdpck2 gene silencing caused decrease in EhDPCK activity and intracellular CoA concentrations, as well as decrease in cell growth. Most eukaryotic organisms possess a single copy of dpck gene with some exceptions including Trypanosoma (TriTrypDB7) and Entamoeba (this study). In a rodent Plasmodium species, which has only one copy of DPCK, it has recently been reported that DPCK is essential and indispensable for development of blood stage parasites (Hart et al., 2017). Human infecting Plasmodium species (P. falciparum and P. vivax) also have a single copy of dpck gene.
In this study, we have shown that Entamoeba, where two copies of dpck are present, gene silencing of Ehdpck2 produced more severe effects on both the metabolite profile and growth, compared to that of Ehdpck1. Our previous microarray analysis of E. histolytica trophozoites showed that the Ehdpck2 transcript level is higher than that of EhDPCK1, approximately fivefold in HM-1:IMSS cl6 strain (Penuliar et al., 2012, 2015) and 11-fold in G3 strain (Furukawa et al., 2012, 2013; Nakada-Tsukui et al., 2012). Furthermore, based on our previous transcriptomic analysis of encystation (differentiation of the trophozoite into the cyst) of E. invadens (De Cádiz et al., 2013), which is the amebic encystation model (Donaldson et al., 1975; Kojimoto et al., 2001; Stanley, 2003; Chia et al., 2009), we found that both of the E. invadens genes encoding DPCK1 and DPCK2 were expressed during encystation (Supplementary Figure S11). E. invadens dpck2 gene was expressed at comparable levels throughout encystation. In contrast, E. invadens dpck1 transcript was dramatically increased (∼30-fold) during encystation. These data suggest that these isoenzymes play specific roles during proliferation and stage conversion. dpck1 is likely involved in encystation in E. invadens. However, gene silencing of Ehdpck1 and Ehdpck2 caused comparable, though slightly more pronounced in Ehdpck2 gs strain, growth and metabolic defects. Our results indicate that EhDPCK2 plays a primary role in the proliferative trophozoite stage and EhDPCK1 may have a specific role during developmental stage transition, which needs to be examined in the future.
Uniqueness of EhDPCK
Two amebic DPCK isotypes, EhDPCK1 and EhDPCK2, showed comparable specific activity and substrate affinity despite only 38% mutual identity. When the Km values of amebic DPCKs were compared with the human counterpart, phosphopantetheine adenylyltransferase and DPCK bifunctional enzyme (Aghajanian and Worrall, 2002) from human has an approximately 10-fold higher Km value toward dephospho-CoA and a 10-fold lower Km value toward ATP (5.2 and 192 μM, respectively). These data indicate significant biochemical and structural differences exist between amebic and human DPCK and also suggest a possibility of discovering or designing parasite-specific DPCK inhibitors. ATP production in E. histolytica relies solely on substrate-level phosphorylation during glycolysis because this parasite lacks the enzymes and machineries involved in the Krebs cycle and oxidative phosphorylation and thus is unable to produce ATP as efficiently as aerobic organisms (McLaughlin and Aley, 1985; Reeves, 1985). It is thus plausible that DPCK from E. histolytica is able to have higher affinity to ATP than human counterpart. Moreover, our results suggest that both EhDPCK1 and EhDPCK2 activity was regulated biochemically not only by the end products of this pathway, CoA, but also its thioesters, acetyl CoA and malonyl-CoA through feedback inhibition.
Domain Structures
Dephospho-CoA kinase has three domains: the nucleotide binding, the dephospho-CoA binding, and the LID domains (Obmolova et al., 2001). In E. coli, two highly conserved prolines (Pro90 and Pro134) are predicted to be essential for the lid flexibility. These residues are located between the nucleotide binding and dephospho-CoA binding domains. However, these residues are only partially conserved in EhDPCK1 (Pro138) and EhDPCK2 (Pro90), indicating that the lid flexibility of DPCK is different between E. histolytica and its bacterial ortholog. A lysine residue in the GXXXGKS motif is also one of the conserved motifs of DPCKs among organisms, and known to be essential for the binding of ATP. Thr10 for EhDPCK1 and Thr8 for EhDPCK2 are probably involved in dephospho-CoA binding, whereas Asp35 in EhDPCK1 and Asp33 for EhDPCK2 are involved in activation of the 3′-OH group of the ribose for the attack on the β-phosphate of ATP, as previously reported in bacteria (Obmolova et al., 2001; O’Toole et al., 2003). Leu85 in EhDPCK1 and Ile84 in EhDPCK2, both of which are also predicted to be localized to the CoA-binding site, are also conserved. Asn91 in EhDPCK1 is replaced with Pro90 in EhDPCK2; the corresponding residue Pro90 in E. coli is located at a hinge and implicated for the movement of the CoA-binding domain during catalysis, suggesting a potential difference in a catalytic process between EhDPCK1 and 2.
Metabolic Disturbance Caused by Ehdpck Gene Silencing
Metabolomics analysis has been widely used to complement genomics, transcriptomics, and proteomics analyses (Dumas, 2012; Castro-Santos et al., 2015), and to provide a comprehensive view on the responses to the environmental changes (Fuhrer and Zamboni, 2015), a metabolic-responsive profile (Holmes et al., 2008; Husain et al., 2011), and also to validate target for drug development (Krug and Müller, 2014; Mastrangelo et al., 2014). As expected, dpck gene silencing (particularly gene silencing targeting Ehdpck2) caused decrease in the level of CoA and acetyl-CoA levels. Acetyl-CoA occupies a crucial position in metabolisms for all living organisms, and serves as an essential metabolic intermediate, a precursor of many anabolic reactions, also a donor of protein acetylation and allosteric enzymatic regulator (Choudhary et al., 2014). Our metabolome data have also shown the disturbance, mostly decrease, in the broad range of related metabolites, in particular those involved in ornithine and polyamine biosynthesis. Ornithine is an important precursor of polyamines and essential for proliferation (Bacchi and Yarlett, 1993; Müller et al., 2001; Heby et al., 2003; Wallace et al., 2003). Polyamine metabolism was targeted to develop drugs against Trypanosoma (Begolo et al., 2014) and Leishmania (Das et al., 2016). In P. falciparum, it has been known that many cellular processes, such as DNA replication, transcription and translation are polyamine dependent and the cellular levels of polyamines reach their peak during the intra-erythrocytic development (Assaraf et al., 1987). Our metabolomics analysis has shown that Ehdpck2 gene silencing significantly reduced the levels of many intermediates in the polyamine pathway, while Ehdpck1 gene silencing showed decrease to a lesser extent (not statistically significant). Citrate was significantly decreased by Ehdpck1 gene silencing, which was not found in Ehdpck2 gs strain. Citrate is an important intermediate metabolite derived from the condensation of acetyl CoA and oxaloacetate in the Krebs cycle. However, since Krebs cycle is absent in E. histolytica, the role of citrate remains elusive and needs future investigation.
The levels of metabolites in hexose metabolism were also decreased by Ehdpck2 gene silencing (Figure 6). In particular, metabolites involved in the hexosamine-amino acid amide transfer and chitin biosynthesis, such as glucosamine 6-phosphate and N-acetyl-D-glucosamine 6-phosphate, significantly decreased compared to control (P = 0.008). However, it is not well understood how glucosamine 6-phosphate was completely deprived by Ehdpck2 gene silencing. The decrease in glucosamine 6-phosphates was apparently associated with the decreased level of glutamate in Ehdpck2 gs strain, via the reaction converting D-fructose-6-phosphate/L-glutamate and D-glucosamine-6-phosphate and L-glutamine. On the other hand, the decrease in N-acetyl-D-glucosamine-6-phosphate was likely caused by the decrease in acetyl-CoA as N-acetyl-D-glucosamine-6-phosphate is generated from a combination of D-glucosamine-6-phosophate and acetyl-CoA. Glucosamine 6-phosphate and N-acetyl-D-glucosamine 6-phosphate are also important intermediates for chitin biosynthesis. Chitin is important and present as the major component of cell wall in cyst and oocyst stages of many protozoan parasites (Chávez-Munguía et al., 2007) including E. histolytica (Arroyo-Begovich and Carabez-Trejo, 1982). Chitin biosynthesis was also proposed as a parasite-specific drug target as it is absent in humans (Spindler et al., 1990). Interestingly, reduction in the intermediates in the chitin biosynthetic pathway was observed only in Ehdpck2 gs strain, but not in Ehdpck1 gs strain. Although statistically insignificant, several metabolites in the pathway were increased in Ehdpck1 gs strain: glucose-1-phosphate, glucose-6-phosphate, N-acetyl-D-glucosamine-1-phosphate, and N-acetyl-D-glucosamine-6-phosphate, suggesting that the roles of EhDPCK1 and EhDPCK2 in chitin metabolism are different.
Many nucleic acids were also decreased in both Ehdpck1 and Ehdpck2 gs strains (Figures 4, 7 and Supplementary Table S3). This may be explained at least in part by the decrease in polyamine synthesis (Figure 5). Polyamines can interact with macromolecules such as nucleic acids and are required during nucleic acids packaging (Ruan et al., 1994). Also, the interaction between polyamines and nucleic acids can affect structure and stability of DNA (Gosule and Schellman, 1976; Raspaud et al., 1999; van Dam et al., 2002). Moreover, most of the intermediary metabolites in purine metabolism in Ehdpck2 gs strain were also reduced (Figure 7). Importantly, these changes were specific to Ehdpck2 gene silencing. In Ehdpck1 gs strain, several purines and intermediates including adenine, guanine, IMP, and ADP, were increased compared to control. Purine metabolism is very important for DNA and RNA synthesis, nucleotide-dependent enzyme reactions, as chemical energy sources (e.g., ATP, GTP), intracellular second messengers (e.g., cAMP), and switch signals (e.g., GDP, GTP) for metabolic and gene regulation (Hyde, 2007). However, a majority of parasitic protists including E. histolytica (Lo and Wang, 1985) are incapable of de novo synthesis of purines and rely on salvage from the host. Hypoxanthine, guanine, xanthine, adenine, and adenosine must be imported from the hosts or media via nucleoside transporters described in P. falciparum (Carter et al., 2000), Toxoplasma gondii (De Koning et al., 2003), and Trypanosoma brucei (Sanchez et al., 2002). Interestingly, while most of the above-mentioned purines were decreased in Ehdpck2 gs strain, hypoxanthine was most dramatically decreased (Figure 7). Hypothanthine is regarded as the key precursor of the other purines. For instance, degradation of hypoxanthine by added xanthine oxidase in the culture medium strongly inhibits in vitro growth of P. falciparum (Berman et al., 1991). Purine metabolic pathways were proposed as promising targets for novel drug development against infections caused by P. falciparum (Cassera et al., 2011), T. gondii (De Koning et al., 2003), and Mycobacterium tuberculosis (Parker and Long, 2007).
Regulation of CoA Biosynthesis
The Ehdpck2 gene silencing caused reduction of CoA and also led to decrease in pantothenate, which can be explained by either high consumption of pantothenate or decrease in pantothenate uptake. Most eukaryotes including E. histolytica are unable to synthesize pantothenate and rely on uptake of exogenous pantothenate. Our qRT-PCR data indicate that the decreased level of CoA by Ehdpck1 or Ehdpck2 gene silencing led to the increase in steady-state mRNA level of Ehpank gene encoding the protein that catalyzes conversion of pantothenate to 4′-phosphopantothenate. This was also confirmed by the observation that PanK activity increased in Ehdpck1 and Ehdpck2 gs strains. Although none of the intermediates, 4′-phosphopantothenate, 4′-phosphopantothenoyl-L-cysteine, pantetheine 4-phosphate, and dephospho-CoA was measured in our metabolomics analysis, it is conceivable that these metabolites accumulated intracellularly as a consequence of lack of EhDPCK and upregulation of EhPanK. These data also reinforce allosteric feedback of EhPanK by the CoA regulatory network of CoA biosynthesis, which was demonstrated in our previous study (Nurkanto et al., 2018).
Finally, together with our previous report on EhPanK, which catalyzes the first step of CoA biosynthesis (Nurkanto et al., 2018), the two key enzymes of CoA biosynthesis, EhPanK and EhDPCKs have been proven to be attractive and rational targets for the development of anti-amebic agents. However, EhPanK and EhDPCKs need to be further validated for suitability as drug target in the future by screening chemical and microbial libraries to discover specific inhibitors.
Conclusion
We have demonstrated that EhDPCK is the important enzyme involved in CoA biosynthesis and its repression causes significant disturbance in the level of some intermediates in polyamine, hexose, and nucleic acid metabolisms in the proliferative trophozoite stage of E. histolytica. EhDPCK2 plays a predominant role compared to EhDPCK1 in trophozoites stage. Notable differences in kinetic parameters of DPCK between Entamoeba and its host suggest that amebic DPCK is the rational target for the development of anti-amebic agents.
Author Contributions
AN, GJ, YN, MS, TH, and TN designed the experiments and analyzed the data. AN, GJ, TY, YN, and THI performed the experiments. AN, TH, and TN wrote the manuscript. TN conceived the project and supervised the study. All authors reviewed the results and approved the final version of the manuscript.
Funding
This work was supported in part by a grant for Science and Technology Research Partnership for Sustainable Development (SATREPS) from Japan Agency for Medical Research and Development (AMED) and Japan International Cooperation Agency (JICA), a grant for Research on Emerging and Re-emerging Infectious Diseases from AMED, and Grants-in-Aid for Challenging Research (Exploratory) (17K19416) for Scientific Research (15H04406) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. AN was supported by a fellowship from Riset-Pro, The Ministry of Research and Higher Education (Kemenristek-DIKTI), Republic of Indonesia.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of Nozaki Lab, particularly, Kumiko Nakada-Tsukui, Yumiko Saito-Nakano, Takeshi Annoura, Herbert J. Santos, Yuki Hanadate, and Ratna Wahyuni.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02902/full#supplementary-material
FIGURE S1 | Alignment of DPCK protein sequences from E. histolytica and other organisms. The sequences were aligned using Muscle program (Edgar, 2004) in SeaView package version 4.6.1 (Gouy et al., 2010). Identical residues in these conserved domains are denoted by asterisks (∗). The GXXXXGKS/T p-loop consensus sequence, which is an ATP/GTP-binding motif, is shown in gray background. The lysine residues are highlighted with white letters on black background. Human DPCK is located at the carboxyl terminus of a fused bifunctional enzyme of phosphopantetheine adenylyltransferase and dephospho-CoA kinase (COASY), which catalyzes the last two steps of CoA biosynthesis.
FIGURE S2 | Phylogenetic tree of dephospho-CoA Kinase (DPCK) proteins from E. histolytica and other organisms. Optimal ML tree inferred by RAxML program with LG + Ã4 model was shown from 112 DPCK positions were used in this analysis. The numbers at the nodes represent the bootstrap values as a percentage of 100 replicates. The scale bar indicates 0.3 substitutions at each amino acid position.
FIGURE S3 | Expression and purification of recombinant EhDPCK1 (A,B) and EhDPCK2 (C,D) | (A,C) Protein samples of recombinant EhDPCK1 (A) and EhDPCK2 (C) at each step of purification were subjected to 15% SDS-PAGE under reducing conditions and then stained with Coomassie Brilliant Blue R250. (B,D) Immunoblot analysis of purified recombinant EhDPCK1 (B) and EhDPCK2 (D) using anti-His antibody.
FIGURE S4 | Optimum pH of the recombinant EhDPCK1 (A) and EhDPCK2 (B) | Enzymes activities were measured at 37°C at the various pHs indicated in the figure. Assays were performed as described in Section “Experimental procedures,” with 50 ng of EhDPCK1 or EhDPCK2 recombinant enzyme, 15 mM HEPES, 20 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 0.02% Tween-20, 0.1 mg/mL β-globulins, 100 μM ATP, and 200 μM dephospho-CoA at 37°C. Data are shown in mean ± SE of three replicates.
FIGURE S5 | Inhibition activities of the EhDPCK1 (A) and EhDPCK2 (B) by CoA, acetyl-CoA, and malonyl-CoA. The assays were carried out three times independently. Data are shown in mean ± SE.
FIGURE S6 | Growth kinetic of E. histolytica transformants during 96 h incubation in BI-S-33 medium observed using WST1 (A,B) and the cellular fractionation and immunoblot analysis of EhDPCK2 (C) | Data are shown in mean of absorbance 450 nm ± SE of three replicates (A,B) as described in Section “Materials and Methods.” Figure (C) is immunoblot analysis using anti-HA monoclonal antibody, anti-CPBF1, and anti-CS1 polyclonal antisera, where CPBF1 and CS1 serve as a control for organelle and cytosolic proteins, respectively. Whole lysate of control strain was also subjected to analysis.
FIGURE S7 | Relative levels of gene transcripts encoding Ehdpck1 (A) and Ehdpck2(B) and its related and unrelated genes in CoA biosynthesis pathway. Data were normalized against RNA polymerase II, and are shown in percentage relative to the transcript level of each gene in psAP2 control. NADHK, Tom-40, and CPBF2 are corresponding to NAD(H) kinase (EHI_151920), mitosomal membrane protein-Tom40 (EHI_104420), and cysteine protease binding protein 2 (EHI_087660), respectively. Data are shown in mean ± SE. Statistical comparison is made by Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
FIGURE S8 | Metabolic profiles of amino acids (ornithine, glutamate, and methionine), polyamines, and associated metabolites affected by Ehdpck1 gene silencing. Broken lines indicate genes and their encoded enzymes are likely absent in the genome. Labels and abbreviations are as in Figure 4. Data are shown in mean ± SE. Statistical comparison is made by Student’s t-test (∗P < 0.05).
FIGURE S9 | Metabolic profiles of hexose phosphates, glycogen metabolism, chitin biosynthesis, and associated metabolism affected by Ehdpck1 gene silencing. Labels and abbreviations are as in Figure 5. Data are shown in mean ± SE.
FIGURE S10 | Metabolic profiles of purine metabolism affected by Ehdpck2 gene silencing. Broken lines indicate genes whose encoding enzymes known to catalyze the steps are likely absent in the genome. Labels and abbreviations are as in Figure 6. Data are shown in mean ± SE. Statistical comparison is made by Student’s t-test (∗P < 0.05).
FIGURE S11 | Relative levels of dpck1 and 2 gene transcripts in E. invadens encystation stage. Data was generated from our previous publication (De Cádiz et al., 2013).
TABLE S1 | Sequence data of dephospho-CoA kinase (DPCK) used in phylogenetic analysis.
TABLE S2 | Protein concentrations, activities, specific activities, percentage yield, and fold purification of recombinant EhDPCK.
TABLE S3 | Metabolite profiles of Ehdpck1 and Ehdpc2 gene silencing strain obtained from CE-MS analysis. All data are shown as means ± SE from three independent biological replicates, and for each biological replicate, two technical replicates were made.
Footnotes
- ^http://www.brenda-enzymes.info/
- ^http://www.ncbi.nlm.nih.gov/
- ^http://tree.bio.ed.ac.uk/software/figtree/
- ^http://www.sbg.bio.ic.ac.uk/phyre2
- ^https://www.genome.jp/kegg
- ^http://amoebadb.org/amoeba/
- ^http://tritrypdb.org/tritrypdb/
References
Aghajanian, S., and Worrall, D. M. (2002). Identification and characterization of the gene encoding the human phosphopantetheine adenylyltransferase and dephospho-CoA kinase bifunctional enzyme (CoA synthase). Biochem. J. 365, 13–18. doi: 10.1042/BJ20020569
Ali, V., and Nozaki, T. (2007). Current therapeutics, their problems, and sulfur-containing-amino-acid metabolism as a novel target against infections by “amitochondriate” protozoan parasites. Clin. Microbiol. Rev. 20, 164–187. doi: 10.1128/CMR.00019-06
Arroyo-Begovich, A., and Carabez-Trejo, A. (1982). Location on chitin in the cyst wall of Entamoeba invadens with colloidal gold tracers. J. Parasitol. 68, 253–258. doi: 10.2307/3281183
Assaraf, Y. G., Abu-Elheiga, L., Spira, D. T., Desser, H., and Bachrach, U. (1987). Effect of polyamine depletion on macromolecular synthesis of the malarial parasite, Plasmodium falciparum, cultured in human erythrocytes. Biochem. J. 242, 221–226. doi: 10.1042/bj2420221
Bacchi, C. J., and Yarlett, N. (1993). Effects of antagonists of polyamine metabolism on African trypanosomes. Acta Trop. 54, 225–236. doi: 10.1016/0001-706X(93)90095-S
Becher, I., Savitski, M. M., Savitski, M. F., Hopf, C., Bantscheff, M., and Drewes, G. (2013). Affinity profiling of the cellular kinome for the nucleotide cofactors ATP, ADP, and GTP. ACS Chem. Biol. 8, 599–607. doi: 10.1021/cb3005879
Begley, T. P., Kinsland, C., and Strauss, E. (2001). The biosynthesis of coenzyme A in bacteria. Vitam. Horm. 61, 157–171. doi: 10.1016/s0083-6729(01)61005-7
Begolo, D., Erben, E., and Clayton, C. (2014). Drug target identification using a trypanosome overexpression library. Antimicrob. Agents Chemother. 58, 6260–6264. doi: 10.1128/AAC.03338-14
Berman, P. A., Human, L., and Freese, J. A. (1991). Xanthine oxidase inhibits growth of Plasmodium falciparum in human erythrocytes in vitro. J. Clin. Invest. 88, 1848–1855. doi: 10.1172/JCI115506
Bracha, R., Nuchamowitz, Y., Anbar, M., and Mirelman, D. (2006). Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2:e48. doi: 10.1371/journal.ppat.0020048
Carter, N. S., Ben Mamoun, C., Liu, W., Silva, E. O., Landfear, S. M., Goldberg, D. E., et al. (2000). Isolation and functional characterization of the PfNT1 nucleoside transporter gene from Plasmodium falciparum. J. Biol. Chem. 275, 10683–10691. doi: 10.1074/jbc.275.14.10683
Cassera, M. B., Zhang, Y., Hazleton, K. Z., and Schramm, V. L. (2011). Purine and pyrimidine pathways as targets in Plasmodium falciparum. Curr. Top. Med. Chem. 11, 2103–2115. doi: 10.2174/156802611796575948
Castro-Santos, P., Laborde, C. M., and Díaz-Peña, R. (2015). Genomics, proteomics and metabolomics: their emerging roles in the discovery and validation of rheumatoid arthritis biomarkers. Clin. Exp. Rheumatol. 33, 279–286.
Chakauya, E., Coxon, K. M., Wei, M., MacDonald, M. V., Barsby, T., Abell, C., et al. (2008). Towards engineering increased pantothenate (vitamin B5) levels in plants. Plant Mol. Biol. 68, 493–503. doi: 10.1007/s11103-008-9386-5
Chávez-Munguía, B., Omaña-Molina, M., González-Lázaro, M., González-Robles, A., Cedillo-Rivera, R., Bonilla, P., et al. (2007). Ultrastructure of cyst differentiation in parasitic protozoa. Parasitol. Res. 100, 1169–1175. doi: 10.1007/s00436-006-0447-x
Chia, M.-Y., Jeng, C.-R., Hsiao, S.-H., Lee, A.-H., Chen, C.-Y., and Pang, V. F. (2009). Entamoeba invadens myositis in a common water monitor lizard (Varanus salvator). Vet. Pathol. 46, 673–676. doi: 10.1354/vp.08-VP-0224-P-CR
Chomczynski, P., and Mackey, K. (1995). Modification of the TRI Reagent(TM) procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques 19, 942–945. doi: 10.2144/000113156
Choudhary, C., Weinert, B. T., Nishida, Y., Verdin, E., and Mann, M. (2014). The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550. doi: 10.1038/nrm3841
Das, M., Singh, S., and Dubey, V. K. (2016). Novel Inhibitors of Ornithine Decarboxylase of Leishmania Parasite (LdODC): the parasite resists ldodc inhibition by overexpression of spermidine synthase. Chem. Biol. Drug Des. 87, 352–360. doi: 10.1111/cbdd.12665
De Cádiz, A. E., Jeelani, G., Nakada-Tsukui, K., Caler, E., and Nozaki, T. (2013). Transcriptome analysis of encystation in Entamoeba invadens. PLoS One 8:e74840. doi: 10.1371/journal.pone.0074840
De Koning, H. P., Al-Salabi, M. I., Cohen, A. M., Coombs, G. H., and Wastling, J. M. (2003). Identification and characterisation of high affinity nucleoside and nucleobase transporters in Toxoplasma gondii. Int. J. Parasitol. 33, 821–831. doi: 10.1016/S0020-7519(03)00091-2
Diamond, L. S., Harlow, D. R., and Cunnick, C. C. (1978). A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72, 431–432. doi: 10.1016/0035-9203(78)90144-X
Donaldson, M., Heyneman, D., Dempster, R., and Garcia, L. (1975). Epizootic of fatal amebiasis among exhibited snakes: epidemiologic, pathologic, and chemotherapeutic considerations. Am. J. Vet. Res. 36, 807–817.
Dumas, M.-E. (2012). Metabolome 2.0: quantitative genetics and network biology of metabolic phenotypes. Mol. Biosyst. 8, 2494–2502. doi: 10.1039/c2mb25167a
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fuhrer, T., and Zamboni, N. (2015). High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 31, 73–78. doi: 10.1016/j.copbio.2014.08.006
Furukawa, A., Nakada-Tsukui, K., and Nozaki, T. (2012). Novel transmembrane receptor involved in phagosome transport of lysozymes and β-hexosaminidase in the enteric protozoan Entamoeba histolytica. PLoS Pathog. 8:e1002539. doi: 10.1371/journal.ppat.1002539
Furukawa, A., Nakada-Tsukui, K., and Nozaki, T. (2013). Cysteine protease-binding protein family 6 mediates the trafficking of amylases to phagosomes in the enteric protozoan Entamoeba histolytica. Infect. Immun. 81, 1820–1829. doi: 10.1128/IAI.00915-12
Genschel, U., Powell, C., Abell, C., and Smith, A. (1999). The final step of pantothenate biosynthesis in higher plants: cloning and characterization of pantothenate synthetase from Lotus japonicus and Oryza sativum (rice). Biochem. J. 678, 669–678. doi: 10.1042/0264-6021:3410669
Gosule, L. C., and Schellman, J. A. (1976). Compact form of DNA induced by spermidine. Nature 259, 333–335. doi: 10.1038/259333a0
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Hanna, R. M., Dahniya, M. H., Badr, S. S., and El-Betagy, A. (2000). Percutaneous catheter drainage in drug-resistant amoebic liver abscess. Trop. Med. Int. Heal. 5, 578–581. doi: 10.1046/j.1365-3156.2000.00586.x
Hart, R. J., Abraham, A., and Aly, A. S. I. (2017). Genetic characterization of coenzyme a biosynthesis reveals essential distinctive functions during malaria parasite development in blood and mosquito. Front. Cell. Infect. Microbiol. 7:260. doi: 10.3389/Fcimb.2017.00260
Heby, O., Roberts, S. C., and Ullman, B. (2003). Polyamine biosynthetic enzymes as drug targets in parasitic protozoa. Biochem. Soc. Trans. 31, 415–419. doi: 10.1042/BST0310415
Holmes, E., Wilson, I. D., and Nicholson, J. K. (2008). Metabolic phenotyping in health and disease. Cell 134, 714–717. doi: 10.1016/j.cell.2008.08.026
Husain, A., Jeelani, G., Sato, D., and Nozaki, T. (2011). Global analysis of gene expression in response to L-Cysteine deprivation in the anaerobic protozoan parasite Entamoeba histolytica. BMC Genomics 12:275. doi: 10.1186/1471-2164-12-275
Hyde, J. E. (2007). Targeting purine and pyrimidine metabolism in human apicomplexan parasites. Curr. Drug Targets 8, 31–47. doi: 10.2174/138945007779315524
Jeelani, G., Sato, D., Husain, A., Escueta-de Cadiz, A., Sugimoto, M., Soga, T., et al. (2012). Metabolic profiling of the protozoan parasite Entamoeba invadens revealed activation of unpredicted pathway during encystation. PLoS One 7:e37740. doi: 10.1371/journal.pone.0037740
Johnson, P. J. (1993). Metronidazole and drug resistance. Parasitol. Today 9, 183–186. doi: 10.1016/0169-4758(93)90143-4
Kinoshita, A., Tsukada, K., Soga, T., Hishiki, T., Ueno, Y., Nakayama, Y., et al. (2007). Roles of hemoglobin allostery in hypoxia-induced metabolic alterations in erythrocytes: Simulation and its verification by metabolome analysis. J. Biol. Chem. 282, 10731–10741. doi: 10.1074/jbc.M610717200
Kojimoto, A., Uchida, K., Horii, Y., Okumura, S., Yamaguch, R., and Tateyama, S. (2001). Amebiasis in four ball pythons, Python reginus. J. Vet. Med. Sci. 63, 1365–1368. doi: 10.1292/jvms.63.1365
Krug, D., and Müller, R. (2014). Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat. Prod. Rep. 31, 768–783. doi: 10.1039/c3np70127a
Leipe, D. D., Koonin, E. V., and Aravind, L. (2003). Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 333, 781–815. doi: 10.1016/j.jmb.2003.08.040
Leonardi, R., Zhang, Y. M., Rock, C. O., and Jackowski, S. (2005). Coenzyme a: back in action. Prog. Lipid Res. 44, 125–153. doi: 10.1016/j.plipres.2005.04.001
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real- time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lo, H. S., and Wang, C. C. (1985). Purine salvage in Entamoeba histolytica. J. Parasitol. 71, 662–669. doi: 10.2307/3281440
Mastrangelo, A., Armitage, E. G., García, A., and Barbas, C. (2014). Metabolomics as a tool for drug discovery and personalised medicine. A review. Curr. Top. Med. Chem. 14, 2627–2636. doi: 10.2174/1568026614666141215124956
McLaughlin, J., and Aley, S. (1985). The biochemistry and functional morphology of the Entamoeba. J. Eukaryot. Microbiol. 32, 221–240. doi: 10.1111/j.1550-7408.1985.tb03043.x
Müller, S., Coombs, G. H., and Walter, R. D. (2001). Targeting polyamines of parasitic protozoa in chemotherapy. Trends Parasitol. 17, 242–249. doi: 10.1016/S1471-4922(01)01908-0
Nakada-Tsukui, K., Tsuboi, K., Furukawa, A., Yamada, Y., and Nozaki, T. (2012). A novel class of cysteine protease receptors that mediate lysosomal transport. Cell. Microbiol. 14, 1299–1317. doi: 10.1111/j.1462-5822.2012.01800.x
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Niefind, K., Putter, M., Guerra, B., Issinger, O. G., and Schomburg, D. (1999). GTP plus water mimic ATP in the active site of protein kinase CK2. Nat. Struct. Biol. 6, 1100–1103. doi: 10.1038/70033
Nozaki, T., Asai, T., Sanchez, L. B., Kobayashi, S., Nakazawa, M., and Takeuchi, T. (1999). Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar. J. Biol. Chem. 274, 32445–32452. doi: 10.1074/jbc.274.45.32445
Nurkanto, A., Jeelani, G., Yamamoto, T., Naito, Y., Hishiki, T., Mori, M., et al. (2018). Characterization and validation of Entamoeba histolytica pantothenate kinase as a novel anti-amebic drug target. Int. J. Parasitol. Drugs Drug Resist. 8, 125–136. doi: 10.1016/j.ijpddr.2018.02.004
Obmolova, G., Teplyakov, A., Bonander, N., Eisenstein, E., Howard, A. J., and Gilliland, G. L. (2001). Crystal structure of dephospho-coenzyme A kinase from Haemophilus influenzae. J. Struct. Biol. 136, 119–125. doi: 10.1006/jsbi.2001.4428
Orozco, E., de la Cruz Hernández, F., and Rodríguez, M. A. (1985). Isolation and characterization of Entamoeba histolytica mutants resistant to emetine. Mol. Biochem. Parasitol. 15, 49–59. doi: 10.1016/0166-6851(85)90028-3
O’Toole, N., Barbosa, J. A., Li, Y., Hung, L.-W., Matte, A., and Cygler, M. (2003). Crystal structure of a trimeric form of dephosphocoenzyme A kinase from Escherichia coli. Protein Sci. 12, 327–336. doi: 10.1110/ps.0227803
Parker, W. B., and Long, M. C. (2007). Purine metabolism in Mycobacterium tuberculosis as a target for drug development. Curr. Pharm. Des. 13, 599–608. doi: 10.2174/138161207780162863
Penuliar, G. M., Furukawa, A., Nakada-Tsukui, K., Husain, A., Sato, D., and Nozaki, T. (2012). Transcriptional and functional analysis of trifluoromethionine resistance in Entamoeba histolytica. J. Antimicrob. Chemother. 67, 375–386. doi: 10.1093/jac/dkr484
Penuliar, G. M., Nakada-Tsukui, K., and Nozaki, T. (2015). Phenotypic and transcriptional profiling in Entamoeba histolytica reveal costs to fitness and adaptive responses associated with metronidazole resistance. Front. Microbiol. 6:354. doi: 10.3389/fmicb.2015.00354
Raspaud, E., Chaperon, I., Leforestier, A., and Livolant, F. (1999). Spermine-induced aggregation of DNA, nucleosome, and chromatin. Biophys. J. 77, 1547–1555. doi: 10.1016/S0006-3495(99)77002-5
Reeves, R. E. (1985). Metabolism of Entamoeba histolytica Schaudinn, 1903. Adv. Parasitol. 23, 105–142. doi: 10.1016/S0065-308X(08)60286-9
Reynolds, C. R., Islam, S. A., and Sternberg, M. J. E. (2018). EzMol: a web server wizard for the rapid visualisation and image production of protein and nucleic acid structures. J. Mol. Biol. 430, 2244–2248. doi: 10.1016/j.jmb.2018.01.013
Ruan, H., Hill, J. R., Fatemie-Nainie, S., and Morris, D. R. (1994). Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA: influence of the structure of the 5’ transcript leader on regulation by the upstream open reading frame. J. Biol. Chem. 269, 17905–17910.
Saliba, K. J., Horner, H. A., and Kirk, K. (1998). Transport and metabolism of the essential vitamin pantothenic acid in human erythrocytes infected with the malaria parasite Plasmodium falciparum. J. Biol. Chem. 273, 10190–10195. doi: 10.1074/jbc.273.17.10190
Samarawickrema, N. A., Brown, D. M., Upcroft, J. A., Thammapalerd, N., and Upcroft, P. (1997). Involvement of superoxide dismutase and pyruvate:ferredoxin oxidoreductase in mechanisms of metronidazole resistance in Entamoeba histolytica. J. Antimicrob. Chemother. 40, 833–840. doi: 10.1093/jac/40.6.833
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor. LaboratoryPress.
Sanchez, M., Tryon, R., Green, J., Boor, I., and Landfear, S. (2002). Six related nucleoside/nucleobase transporters from Trypanosoma brucei exhibit distinct biochemical functions. J. Biol. Chem. 277, 21499–21504. doi: 10.1074/jbc.M202319200
Spindler, K. D., Spindler-Barth, M., and Londershausen, M. (1990). Chitin metabolism: a target for drugs against parasites. Parasitol. Res. 76, 283–288. doi: 10.1007/BF00928180
Spry, C., Kirk, K., and Saliba, K. J. (2008). Coenzyme A biosynthesis: an antimicrobial drug target. FEMS Microbiol. Rev. 32, 56–106. doi: 10.1111/j.1574-6976.2007.00093.x
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
van Dam, L., Korolev, N., and Nordenskiöld, L. (2002). Polyamine-nucleic acid interactions and the effects on structure in oriented DNA fibers. Nucleic Acids Res. 30, 419–428. doi: 10.1093/nar/30.2.419
Vonrhein, C., Schlauderer, G. J., and Schulz, G. E. (1995). Movie of the structural changes during a catalytic cycle of nucleoside monophosphate kinases. Structure 3, 483–490. doi: 10.1016/S0969-2126(01)00181-2
Wallace, H. M., Fraser, A. V., and Hughes, A. (2003). A perspective of polyamine metabolism. Biochem. J. 376, 1–14. doi: 10.1042/BJ20031327
Ximénez, C., Morán, P., Rojas, L., Valadez, A., and Gómez, A. (2009). Reassessment of the epidemiology of amebiasis: state of the art. Infect. Genet. Evol. 9, 1023–1032. doi: 10.1016/j.meegid.2009.06.008
Yamamoto, T., Takano, N., Ishiwata, K., Ohmura, M., Nagahata, Y., Matsuura, T., et al. (2014). Reduced methylation of PFKFB3 in cancer cells shunts glucose towards the pentose phosphate pathway. Nat. Commun. 5:3480. doi: 10.1038/ncomms4480
Keywords: Entamoeba histolytica, coenzyme A, gene silencing, metabolome, drug development
Citation: Nurkanto A, Jeelani G, Yamamoto T, Hishiki T, Naito Y, Suematsu M, Hashimoto T and Nozaki T (2018) Biochemical, Metabolomic, and Genetic Analyses of Dephospho Coenzyme A Kinase Involved in Coenzyme A Biosynthesis in the Human Enteric Parasite Entamoeba histolytica. Front. Microbiol. 9:2902. doi: 10.3389/fmicb.2018.02902
Received: 03 August 2018; Accepted: 13 November 2018;
Published: 30 November 2018.
Edited by:
Guan Zhu, Texas A&M University, United StatesReviewed by:
Abigail Betanzos, Centro de Investigación y de Estudios Avanzados (CINVESTAV), MexicoSergio Adrian Guerrero, Universidad Nacional del Litoral, Argentina
Xianyong Liu, China Agricultural University, China
Copyright © 2018 Nurkanto, Jeelani, Yamamoto, Hishiki, Naito, Suematsu, Hashimoto and Nozaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoyoshi Nozak, bm96YWtpQG0udS10b2t5by5hYy5qcA==
 Arif Nurkanto
Arif Nurkanto Ghulam Jeelani
Ghulam Jeelani Takehiro Yamamoto
Takehiro Yamamoto Takako Hishiki4,5
Takako Hishiki4,5 Makoto Suematsu
Makoto Suematsu Tetsuo Hashimoto
Tetsuo Hashimoto Tomoyoshi Nozaki
Tomoyoshi Nozaki