
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 November 2018
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02704
This article is part of the Research Topic Carbapenem-Resistant Enterobacteriaceae in the Asia Pacific and Beyond View all 13 articles
The continuous emergence of carbapenem-resistant Escherichia coli (CRECO) presents a great challenge to public health. New Delhi metallo-lactamase (NDM) variants are widely disseminated in China, so the research on the prevalence and transmission of diverse blaNDM variants is urgently needed. In the present study, 54 CRECO isolates were collected from 1,185 Escherichia coli isolates in five hospitals in Northern Jiangsu Province, China from September 2015 to August 2016. Antimicrobial susceptibility tests, PCR detection of resistance determinants, multi-locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) were performed to characterize these strains. Plasmid conjugation experiments were carried out to determine the transferability of resistant genes from selected isolates. PCR-based replicon typing (PBRT), S1 nuclease-PFGE, and Southern blotting were conducted for plasmid profiling. Carbapenemase genes were detectable in all CRECO isolates, among which thirty-one CRECO isolates were found to carry blaNDM−5 (54.7%), while, blaNDM−1, blaNDM−7, blaNDM−4, blaNDM−9, and blaKPC−2 were identified in 14, five, two, one, and one isolates, respectively. MLST results revealed 15 different STs and four new STs were first reported to be linked with NDM-producing isolates. PFGE typing showed that no more than two isolates with the same ST appeared to the same band pattern except three ST410 isolates. Twenty-six selected NDM-producing isolates were successfully transferred to E. coli J53 by conjugation experiments. Notably, 50.0% (13/26) of blaNDM variants were found to be carried by ~55 kb IncX3 plasmid. Our study reported a high prevalence of blaNDM variants, especially blaNDM−5, in Northern Jiangsu province, China. Diverse blaNDM variants were mainly carried by ~55 kb IncX3 plasmids, suggesting that the fast evolution and high transferability of this kind of plasmid promote the high prevalence of blaNDM variants. Therefore, large-scale surveillance and effective infection control measures are also urgently needed to prevent diverse blaNDM variants from becoming epidemic in the future.
Carbapenem, a β-lactam that is highly potent against Gram-negative bacteria, has been recognized as a last resort for treating of infections caused by multidrug-resistant bacteria. However, the increasing number of carbapenem-resistant Enterobacteriaceae (CRE) is unexpected despite infection control efforts, and it poses a great challenge to clinic (Zilberberg and Shorr, 2013). Carbapenem resistance is predominantly attributed to the presence of carbapenemases, among which Class A (blaKPC), Class B (blaNDM, blaVIM, blaIMP), and Class D (blaOXA−48) types are most common for Enterobacteriaceae (Walsh, 2010; Albiger et al., 2015). An emerging carbapenemase, New Delhi metallo-lactamase (NDM), was first reported in a Swedish patient with a hospitalization history in India, it exhibited resistant to all β-lactams except for monobactams, and has great potential to cause global health crisis (Yong et al., 2009). Initially, blaNDM gene was endemic to India subcontinent and NDM-producing isolates tested worldwide have geographical links with these high prevalence areas. However, an increasing number of regions worldwide have reported that patients with blaNDM−positive isolates have never been abroad, indicating that blaNDM genes are also associated with some special clones (Leverstein-Van et al., 2010).
In China, since the first report of blaNDM gene in four carbapenem-resistant Acinetobacter baumannii isolates (Chen et al., 2011), increasing Enterobacteriaceae have been identified as carriers of the blaNDM gene. Escherichia coli, an important member of Enterobacteriaceae, are often spread globally through some epidemiological lineages. Although the prevalence of NDM-producing CRE strains is low, outbreaks caused by blaNDM-positive isolates have been identified in several regions of China, indicating high transferability of the blaNDM gene and the severity of infections caused by blaNDM−1-positive organisms (Wang et al., 2014; Jin et al., 2015; Yu et al., 2016). Furthermore, Kaase et al. (2011) first reported a novel blaNDM variant, blaNDM−2, which differs by one amino acid substitution (Pro28Ala) from blaNDM−1, and the subsequent discovery of other blaNDM variants highlights the rapid evolution of this multi-drug resistance gene. In 2012, the NDM enzyme reservoir, India, first reported diverse blaNDM variants among Enterobacteriaceae and blaNDM variants exhibited higher minimum inhibitory concentration (MIC) levels of carbapenem compared with blaNDM−1 (Rahman et al., 2014). Although the blaNDM gene is continuously recoverable in China, data on the prevalence and characteristics of blaNDM variants among Enterobacteriaceae are still needed for preventing its transmission. Notably, a study conducted by Hu et al. (2017) have discovered that various species of bacteria harbored several kinds of blaNDM variants in China, which were mainly carried by diverse plasmids with different sizes. In the present study, we reported a high prevalence of blaNDM variants among E. coli from five hospitals in Northern Jiangsu Province, China. Moreover, these diverse blaNDM variants were mainly located on the same plasmid.
From September 2015 to July 2016, five hospitals (two in Xuzhou, two in Suqian, and one in Lianyungang) in Northern Jiangsu Province of China collected 1,185 E. coli isolates to examine the prevalence and molecular epidemiology of carbapenem-resistance isolates. Initial species identification and antimicrobial susceptibility testing was performed by the Vitek 2 system (bioMe'rieux, France) and MALDI-TOF MS (Bruker Microflex LT, Bruker Daltonik GmbH, Bremen, Germany) according to the manufacturer's instructions.
Initial susceptibility testing was examined by Vitek 2 system. Further MICtesting was conducted by agar dilution method for cefoxitin, ceftriaxone, ceftazidime, cefepime, aztreonam, amikacin, ciprofloxacin, tigecycline, and piperacillin/tazobactam. The MICs of imipenem, meropenem, and ertapenem were determined by E-tests. For colistin, MIC values were tested by broth microdilution method. The agar dilution method and E-test were performed according to the standard Clinical and Laboratory Standards Institute guideline (M100-S26) (CLSI, 2017). The breakpoints of Food and Drug Administration (FDA) and European Committee on Antimicrobial Susceptibility Testing were used for tigecycline and polymyxin, respectively.
DNA templates were prepared by alkaline lysis method using the kit (MoBio, USA). Carbapenemase genes (blaKPC blaNDM, blaSME, blaGES,blaVIM, blaIMP, and blaOXA−48) (Senda et al., 1996; Queenan et al., 2000; Poirel et al., 2004; Endimiani et al., 2008; Yang et al., 2012; Pereira et al., 2015; Al-Agamy et al., 2017), extended spectrumβ-lactamase genes (blaSHV, blaTEM, blaCTX−M−1group, blaCTX−M−2group, blaCTX−M−8group, and blaCTX−M−9group) (Schmitt et al., 2007; Yu et al., 2007), and plasmid-mediated AmpC genes(blaACC, blaFOX, blaMOX, blaDHA, blaCIT/SPM, and blaEBC) (Pérez-Pérez and Hanson, 2002) were examined by PCR. E. coli ATCC25922 was used for the quality control. Positive amplifications were subject to Sanger sequencing (GENEWIZ Company, Suzhou, China).
Molecular typing of 54 NDM-producing E. coli isolates was performed by pulsed-field gel electrophoresis (PFGE). The plugs containing genomic DNA were prepared according to the procedure described by Pereira et al. (2015). The DNA fragments digested with restriction endonuclease XbaI (TaKaRa Biotechnology, Dalian, China) were separated by PFGE on 1% SeaKem Gold agarose (Lonza, Rockland, ME, USA) using the CHEF Mapper XA PFGE system (Bio-Rad, USA) for 18 h at 14°C. The electrophoretic switch times were 6.8–35.4 s. Salmonella H9812 was used as reference marker. Dice coefficients was used to calculate the similarity of PFGE patterns. Dendrograms were constructed by the unweighted pair group method with arithmetic averages (UPGMA) using BioNumerics software version 5.10. Isolates were categorized to be of the same cluster when their dice similarity index was ≥85%. Multi-locus sequence typing of E. coli was conducted by PCR as previously described (Wirth et al., 2006). The allelic profiles and sequence types were identified by amplifying and sequencing the seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, recA) according to the reference website (https://enterobase.warwick.ac.uk/species/index/ecoli). A minimum spanning tree of 54 blaNDM-positive isolates was also constructed by BioNumerics software version 5.10.
The conjugation experiment was implemented by mix broth mating among 26 selected isolates. The donor (clinical strains harboring the blaNDM gene) and recipient (sodium azide-resistant E. coli J53) were mixed and cultured in broth at 37°C overnight. The transconjugants were selected on MH agar with sodium-azide (180 μg/mL) and imipenem (1 μg/mL). Initial species identification was conducted by Vitek MS system. Transformants were regarded as transconjugants when it exhibited resistance to carbapenem and harbored the blaNDM gene.
Twenty-six selected isolates, including blaNDM-5 and blaNDM-1 gene with different STs from the aforementioned five different hospitals and all blaNDM-4 (one isolates was lost in transit), blaNDM-7, and blaNDM-9-positive isolates, were subjected to further plasmid analysis. Incompatibility groups of plasmids extracted from transconjugants were determined by PCR-based replicon typing as described previously (Carattoli et al., 2005; Johnson et al., 2012). S1-PFGE and Southern blotting were conducted to isolate and locate resistance plasmids. Briefly, the gel plugs embedded with blaNDM-positive isolates were digested with S1 nuclease (TaKaRa Biotechnology, Dalian, China) and linear plasmids were separated by CHEF-Mapper XA PFGE system (Bio-Rad) as described above. The universal primers (F: GAAGCTGAGCACCGCATTAG; R: GGGCCGTATGAGTGATTGC) were used for probe synthesis. The plasmid DNA were transferred to positive-charged nylon membranes (Millipore, USA), and DIG-labeled blaNDM−specific probe served to hybridize plasmids according to the instructions of the DIG High Prime DNA Labeling and Detection Starter Kit (Roche, USA).
A total of 54 (4.56%, 54/1185) non-duplicate E. coli isolates that exhibited resistance to imipenem or meropenem were obtained from five hospitals in Northern Jiangsu Province, China. The Affiliated Hospital of Xuzhou Medical University (Hosptial A, n = 18), the Children's Hospital of Xuzhou (Hosptial B, n = 13), the People's Hospital of Suqian (Hosptial C, n = 11), the First People's Hospital of Suqian (Hosptial D, n = 6), and the Second People's Hospital of Lianyungang (Hosptial E, n = 6) were included. Among 54 CRECO isolates, 53 (98.1%, 53/54) were found to be blaNDM-positive and 1 was blaKPC−2-positive. Interestingly, five different blaNDM variants were identified in this collection (Figure 1). Among them, the blaNDM−5 was the prevailing variant, accounting for 58.5% (31/53) of blaNDM-positive isolates, followed by blaNDM−1 (26.4%, 14/53). Moreover, blaNDM−7, blaNDM−4, and blaNDM−9 genes were also identified in 5, 2, and 1 isolates, respectively. NDM variations in amino acid substitutions at various positions are shown in Table 1. The distribution of the blaNDM variants in hospitals is depicted in Figure 1.
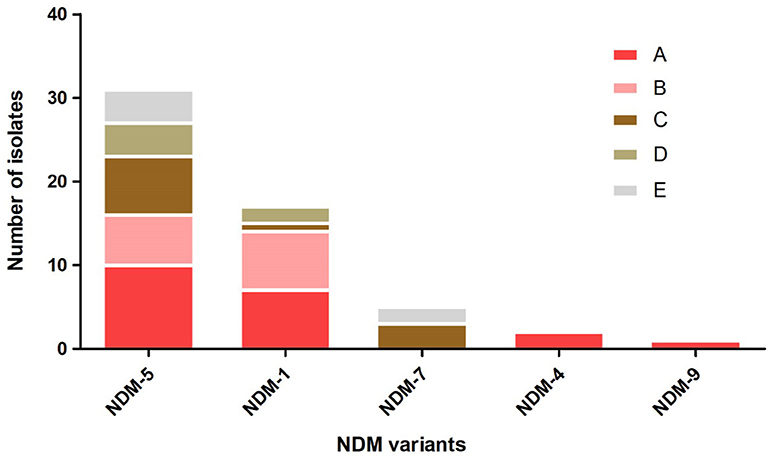
Figure 1. The distribution of blaNDM variants among different hospitals. Hospital A, the Affiliated Hospital of Xuzhou Medical University; Hospital B, the Children's Hospital of Xuzhou; Hospital C, the People's Hospital of Suqian; Hospital D, the First People's Hospital of Suqian; Hospital E, the Second People's Hospital of Lianyungang.
Fifty-three NDM-producing CRECO isolates were resistant to all cephalosporins (cefoxitin, ceftriaxone, ceftazidime, and cefepime) and enzyme inhibitors (piperacillin/tazobactam) but remained susceptible to colistin and tigecycline. The resistance rates to aztreonam, amikacin, and ciprofloxacin were 84.9, 22.6, and 92.4%, respectively. As shown in Table 2, among 31 blaNDM−5-positive isolates, 93.5% were resistant to ciprofloxacin, 74.2% to aztreonam, and 32.2% to amikacin. Resistance to aztreonam and ciprofloxacin were 85.7 and 92.9% among 14 blaNDM−1-producing isolates and were susceptible to amikacin. As for blaNDM−7-positive isolates, susceptibility was only found for amikacin. The blaNDM−4 and blaNDM−9 isolates were resistant to all antibiotics tested in this study except for colistin and tigecycline. One blaKPC−2-positive isolate was resistant to amikacin, colistin, and tigecycline. Molecular features revealed that most CRECO isolates carried the ESBLs gene, AmpC gene, or both. Overall, blaCTX−M, blaSHV, and blaTEM were identified in 42, 11, and 32 isolates, respectively. blaCTX−M-15 was the most common ESBLs gene in our study, accounting for 26.4% followed by blaCTX−M-55 (n = 9), blaCTX−M-65 (n = 8), blaCTX−M-14 (n = 8). blaCTX−M-64 (n = 1), blaCTX−M-90 (n = 1), and blaCTX−M-123 (n = 1). All blaTEM positive isolates were identified as blaTEM−1. Moreover, 6 blaCMY−2, 5 blaCMY−42, and 1 blaDHA−1 were also identified.
A total of 15 STs were identified in 54 CRECO isolates (Figure 2). Among 53 blaNDM-producing isolates, ST167 was the most prevalent, accounting for 35.8% (19/53), followed by ST410 (16.9%, 9/53), ST617 (13.2%, 7/53), ST405 (5.6%, 3/53), ST155 (3.8%, 2/53), ST156 (3.8%, 2/53), ST361 (3.8%, 2/53), and ST2659 (3.8%, 2/53). ST90, ST224, ST46, ST648, ST2376, and ST2083 were identified in one isolate. ST167 and ST617 are different by one allele and both correspond to clonal complex CC10. Moreover, ST410 and ST90 belong to clonal complex CC23. One KPC-2-producing isolate belong to ST131. PFGE typing revealed that no more than two isolates with the same ST type appeared to the same band pattern except for three ST410 isolates from hospital B (Figure 3). Surprisingly, the two isolates (E11 and E28) from hospital A and B shared the same patterns, indicating the occurrence of cross-transmission between hospitals.
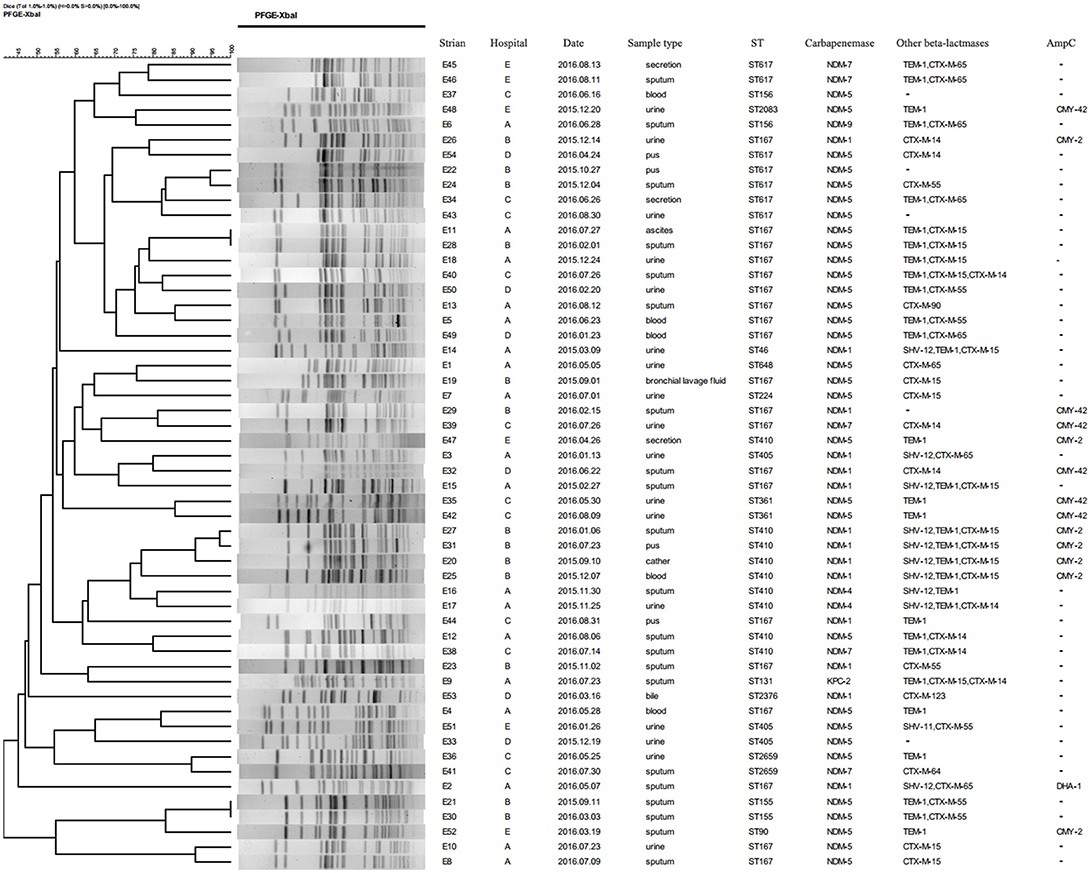
Figure 3. Dendrogram of PFGE profiles of 54 CRECO isolates. The UPGMA algorithm was used to construct dendrogram based on the dice similarity coefficient. Isolates were categorized to be of the same cluster when their dice similarity index was ≥ 85%.
All of plasmids haboring blaNDM gene from 26 selected CRECO isolates were successfully transferred to E. coli J53, and transconjugants exhibited resistance to carbapenem, cephalosporins and enzyme inhibitors (Table 3). As shown in Figure 4, S1-PFGE and Southern blotting revealed that all blaNDM−5 genes were located on the same size (~55 kb) plasmids, which was associated with IncX3 (n = 5), IncFI (n = 3), IncFII (n = 1), and untypeable replicon (n = 3). The blaNDM−1 genes were carried by 55~210 kb plasmids, including IncX3 (n = 5), IncFI (n = 1), and IncFII (n = 1) replicon types. Among five blaNDM−7 positive isolates, four harbored ~55 kb IncX3 plasmids with DIG-labeled blaNDM−7, and the remaining one was carried by ~110 kb IncFI plasmid. The blaNDM−4 and blaNDM−9 genes were located on ~55 kb IncX3 and ~110 kb IncI1 plasmids, respectively. Surprisingly, E5 isolates harbored three blaNDM−5−positive plasmids of ~55, ~105, and ~320 kb in size, suggesting high insertion efficiency of the blaNDM gene. Further plasmid sequencing from E5 isolates revealed that the respective GenBank accession numbers for ~55 and ~105 kb plasmids were NC_022740.1 and AP018144.1, however, the nucleotide sequences of ~320 kb plasmid could not be completely obtained.
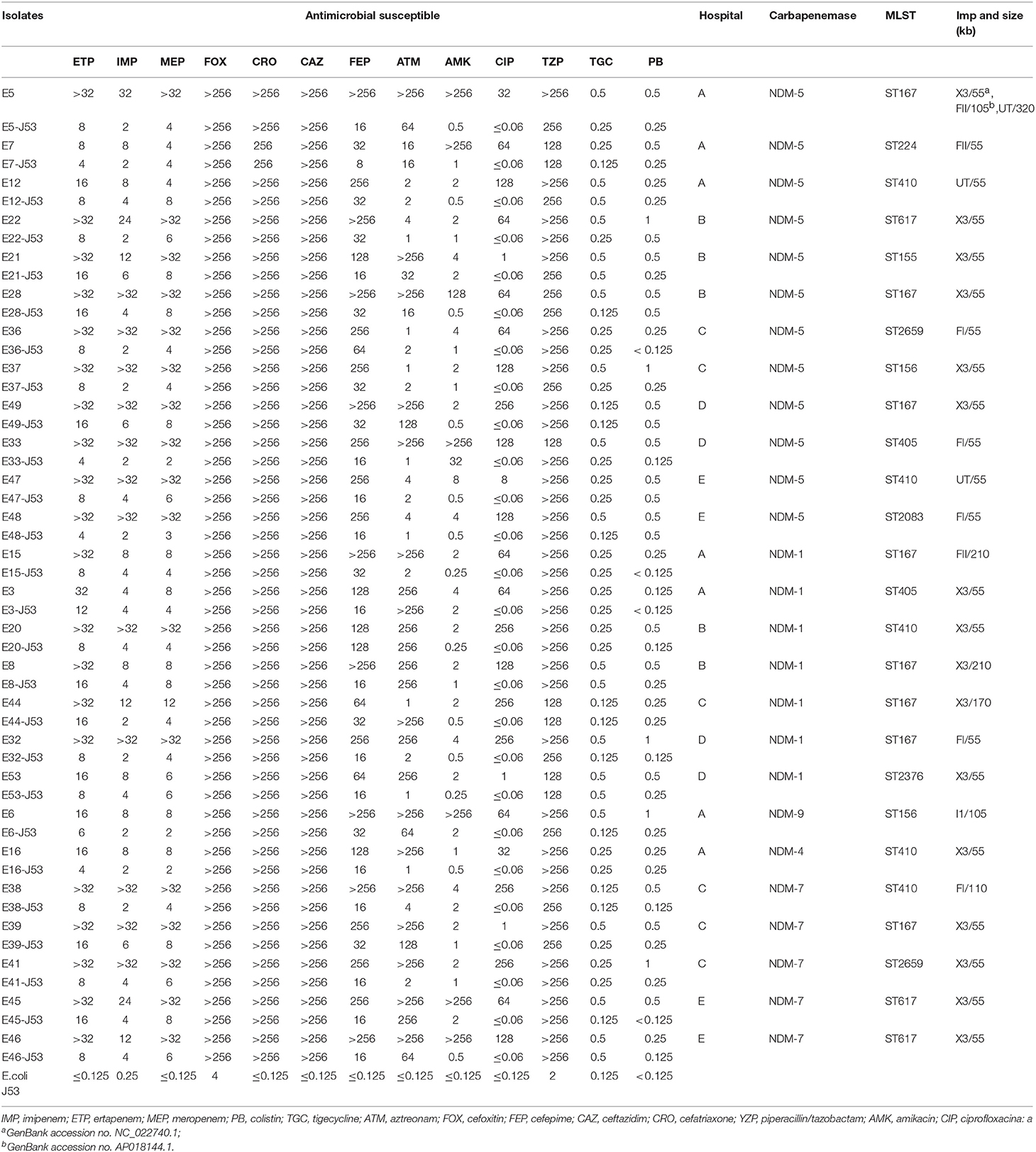
Table 3. Antimicrobial susceptible patterns and characteristics of 26 selected NDM-producing Escherichia coli (μg/mL).
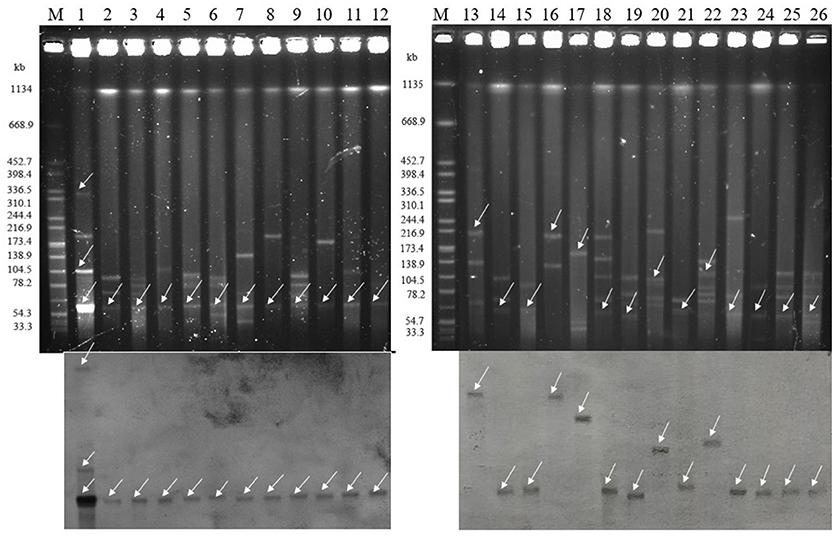
Figure 4. Isolation and determination of plasmids harboring the blaNDM gene in 26 clinical and transconjugants (Top: S1-nuclease PFGE patterns; Bottom: Southern blotting with a blaNDM specific probe). The arrows represent thelocation of linear plasmid DNA hybridized with the blaNDM specific probe. M: Braenderup H9812 Marker; lane 1: E5; lane 2: E7; lane 3: E12; lane4: E22; lane 5: E21; lane 6: E28; lane 7: E36; lane 8: E37; lane 9: E49; lane 10: E33; lane 11: E47; lane 12: E48; lane 13: E15; lane 14: E3; lane 15: E20; lane 16: E8; lane 17: E44; lane18: E32; lane 19: E53; lane 20: E6; lane 21: E16; lane 22: E38; lane 23: E39; lane 24: E41; lane 25: E45; lane 26: E46.
Metallo-lactamase, NDM, is an emerging carbapenem-resistant β-lactamase that is of major public concern due to its high medical and economic burden (Otter et al., 2017), especially for developing countries such as India, Pakistan, and the Balkan countries. As the most populous country in the world, there are major difficulties in preventing the dissemination of multidrug resistant genes in China. Therefore, comprehensive, extensive studies on diverse blaNDM variant-positive E. coli are needed to provide clear information to optimize antibiotic policy in endemic areas.
Generally, the prevalence of the blaNDM gene has continuously increased worldwide. As of now, the NDM enzyme has been identified in almost all of the world, including many countries in Asia, Africa, Europe, the Americas, and Australia (Berrazeg et al., 2014). A study from India also analyzed the occurrence of the blaNDM gene among carbapenem resistant isolates, and it accounted for 45.4% of them (Rahman et al., 2018). Recently, a survey from the French National Reference Center revealed that among 140 carbapenem-resistant isolates 21% were NDM producer (Gauthier et al., 2018). In 2017, a nationwide study of clinical CRE strains in China demonstrated that 49% were NDM producer among carbapenem-resistant E. coli (Zhang et al., 2017). Furthermore, a multicenter study of the China CRE network revealed that among 39 carbapenem-resistant E. coli isolates, 74.4% were NDM producer, suggesting that there is a serious challenge in combating infections caused by this “superbug” in China (Zhang et al., 2018). In the present study, we identified 53 blaNDM-carrying isolates among 54 CRECO, which is much higher than in any other region of China (Wang S. et al., 2016; Hu et al., 2017; Liang et al., 2017). To the best of our knowledge, this is also the first report on the blaNDM gene in Northern Jiangsu Province. Moreover, the emergence of such a high prevalence of blaNDM variants indicates that the blaNDM gene is increasing in this area.
Since its first identification in 2009, the blaNDM gene has evolved at a fast pace during the past 10 years. Twenty-one blaNDM variants have been identified in different countries, all of which are archived at http://www.lahey.org/Studies/other.asp. Khan et al. (2017) reported that the Asian continent, especially China and India, was a reservoir of NDM producers, in which about a 58.2% abundance of the blaNDM−1 variants was found. Among these blaNDM variants, the blaNDM−1 gene has been reported as the most prevalent type worldwide (Nordmann and Poirel, 2014). In our study, five blaNDM variants were identified as responsible for MBL production, with the blaNDM−5 gene being the most prevalent. Compared with NDM-1, NDM-5 producers exhibited higher hydrolytic activity and toxicity toward carbapenem and cephalosporin (Mei et al., 2017). The blaNDM−5 gene is an emerging blaNDM variants and has tended to surpass blaNDM−1 recently, and it differs from the NDM-1 enzyme at two amino acid substitutions, exhibited increased carbapenem resistance (Zhang et al., 2016). The blaNDM−4 and blaNDM−7 genes have been discovered among E. coli in China with relatively low prevalence, involving in 4 and 6 patients, respectively. Moreover, clinical blaNDM−9-positive E. coli has only been found in Taiwan (Lai et al., 2017), with this being the first report on the mainland China. Moreover, not merely NDM-5, amino acid substitutions in NDM-4 (M154L), NDM-7 (D130A), and NDM-9 (E152A) could also result in high levels of carbapenem resistance (Düzgün, 2018). Stewart et al. (2017) reported that the substitution in M154L, which is found in most blaNDM variants, could enhance resistance to ampicillin at low zinc(II) concentrations relevant to infection sites.
blaNDM variant-positive isolates exhibited multi-drug resistance. Variable resistances to aztreonam, amikacin, and ciprofloxacin were identified in NDM-1 and NDM-5-producing E. coli. The NDM-7, NDM-4, and NDM-9-producing isolates exhibited pan-resistant phenotypes, showing resistance to almost all commonly used clinical antibiotics except for colistin and tigecycline. In view of this, the expert recommended polymyxins was regarded as the last resort for NDM-producing isolates (Yamamoto and Pop-Vicas, 2014).
ST167 was confirmed as significant carrier in the present study, and is reported to be associated with the production of the NDM enzyme, especially NDM-5 (Chen et al., 2016). Notably, four blaNDM-positive ST617 strains were identified in the world, but we identified 6 isolates in the present study. Moreover, a novel new variant of blaNDM, blaNDM−21, belonged to ST617, which should deserve more attention (Liu et al., 2018). Furthermore, four new STs ST361, ST46, ST2376, and ST2083 were firstly reported to be linked with NDM-producing isolates.
The high efficiency of transfer renders the NDM producer ubiquitous throughout the world. Torres-González et al. (2015) reported an outbreak caused by blaNDM−1-carrying plasmid, which is easily transferred between E. coli and Klebsiella pneumoniae. Similarly, in the present study, the plasmids of 26 selected CRECO isolates were also tested for transfer. All selected blaNDM−5 genes were identified to be located on 55 kb plasmid, among which IncX3 was the most common replicon type. Notably, blaNDM−5 gene was frequently reported to be carried by the IncX3 plasmid of 55 kb in size, which was widely reported in China (Yang et al., 2014), Inidia (Krishnaraju et al., 2015), Australia (Wailan et al., 2015), and Damark (Hammerum et al., 2015). Moreover, Li et al. (2018) reported that IncX3 type plasmids play an important role in the transmission of the blaNDM−5 gene in Enterobacteriaceae and this kind of plasmid occurred in different species. Further illustrated the challenge of preventing the dissemination of the blaNDM−5 gene. The widespread blaNDM−1-carrying plasmid has been found to be associated with multiple replicon types, including IncX3, IncF, and IncA/C etc. In agreement with previous studies (Göttig et al., 2013; Wang L. H. et al., 2016), four blaNDM−7-positive plasmids were identified as ~55 kb IncX3 type, while the blaNDM−7 gene was also detected in the IncFI plasmid, which have been identified in India (Rahman et al., 2014). As for blaNDM−9, to the best of our knowledge, this is the first report on ~105 kb IncI1 type plasmid harboring blaNDM−9 gene. Overall, most plasmids harboring blaNDM variants were identified as 55 kb IncX3 types, hinting that amino acid mutations might occur in the process of plasmid transfer, resulting in the emergence of blaNDM variants. The high prevalence of the blaNDM genes due in part to plasmid transfer, meanwhile, the fast evolution of this multidrug resistance gene also favors the persistence of such bacteria harboring it.
In summary, the present study reported high prevalence of blaNDM variants, especially blaNDM−5, among carbapenem-resistant E. coli in Northern Jiangsu Province. The presence of five different variants further increases the threat to public health because of the limited treatment options. Notably, diverse blaNDM variants were mainly located on ~55 kb IncX3 plasmids, indicating that the fast evolution and high transferability of this kind of plasmid has led to the high prevalence of blaNDM variants. Timely detection of NDM enzyme and antimicrobial susceptibility testing are necessary so that infections caused by NDM producers receive appropriate and effective therapy. Similarly, large-scale surveillance and effective infection control measures are also urgently needed to prevent diverse blaNDM variants from becoming epidemics in the future.
The laboratory measurements were performed by RB, ZK, and HQ. BG and PM participated in experimental design and manuscript revision. Data analysis were implemented by RB, ZK, HQ, FJ, and HK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported by the National Natural Science Foundation of China (81471994), Jiangsu Provincial Natural Science Foundation (BK20151154), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six Talent Peaks Project of Jiangsu Province (WSN-135), Advanced Health Talent of Six-One Project of Jiangsu Province (LGY2016042), and Jiangsu Provincial Commission of Health and Family Planning Research Project (H201631).
Al-Agamy, M. H., Jeannot, K., El-Mahdy, T. S., Shibl, A. M., Kattan, W., Plésiat, P., et al. (2017). First detection of GES-5 carbapenemase-producing Acinetobacter baumannii isolate. Microb. Drug Resist. 23, 556–562. doi: 10.1089/mdr.2016.0152
Albiger, B., Glasner, C., Struelens, M. J., Grundmann, H., and Monnet, D. L. (2015). Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro. Surveill. 20:30062. doi: 10.2807/1560-7917.ES.2015.20.45.30062
Berrazeg, M., Diene, S., Medjahed, L., Parola, P., Drissi, M., Raoult, D., et al. (2014). New Delhi Metallo-beta-lactamase around the world: an eReview using Google Maps. Euro. Surveill. 19:20809.
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Chen, D., Gong, L., Walsh, T. R., Lan, R., Wang, T., Zhang, J., et al. (2016). Infection by and dissemination of NDM-5-producing Escherichia coli in China. J. Antimicrob. Chemother. 71, 563–565. doi: 10.1093/jac/dkv352
Chen, Y., Zhou, Z., Jiang, Y., and Yu, Y. (2011). Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66, 1255–1259. doi: 10.1093/jac/dkr082
Clinical and Laboratory Standards Institute [CLSI]. (2017). Performance Standards for antimicrobial Susceptibility Testing, 27th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
Düzgün, A. Ö. (2018). Effect of amino acid substitution in New Delhi metallo-β-lactamase on carbapenem susceptibility. Acta Microbiol. Immunol. Hung. 13, 1–9. doi: 10.1556/030.65.2018.022
Endimiani, A., Carias, L. L., Hujer, A. M., Bethel, C. R., Hujer, K. M., Perez, F., et al. (2008). Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae isolates possessing blaKPC in the United States. Antimicrob. Agents Chemother. 52, 2680–2682. doi: 10.1128/AAC.00158-08
Gauthier, L., Dortet, L., Cotellon, G., Creton, E., Cuzon, G., Ponties, V., et al. (2018). Diversity of Carbapenemase-producing Escherichia coli isolates in France in 2012-2013. Antimicrob Agents Chemother. 62:e00266–18. doi: 10.1128/AAC.00266-18
Göttig, S., Hamprecht, A. G., Christ, S., Kempf, V. A., and Wichelhaus, T. A. (2013). Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-β-lactamase with increased carbapenemase activity. J. Antimicrob. Chemother. 68, 1737–1740. doi: 10.1093/jac/dkt088
Hammerum, A. M., Hansen, F., Olesen, B., Struve, C., Holzknecht, B. J., Andersen, P. S., et al. (2015). Investigation of a possible outbreak of NDM-5-producing ST16 Klebsiella pneumoniae among patients in Denmark with no history of recent travel using whole-genome sequencing. J. Glob. Antimicrob. Resist. 3, 219–221. doi: 10.1016/j.jgar.2015.05.003
Hu, X., Xu, X., Wang, X., Xue, W., Zhou, H., Zhang, L., et al. (2017). Diversity of New Delhi metallo-beta-lactamase-producing bacteria in China. Int. J. Infect. Dis. 55, 92–95. doi: 10.1016/j.ijid.2017.01.011
Jin, Y., Shao, C., Li, J., Fan, H., Bai, Y., and Wang, Y. (2015). Outbreak of multidrug resistant NDM-1-producing Klebsiella pneumoniae from a neonatal unit in Shandong Province, China. PLoS ONE 10:e0119571. doi: 10.1371/journal.pone.0119571
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Kaase, M., Nordmann, P., Wichelhaus, T. A., Gatermann, S. G., Bonnin, R. A., and Poirel, L. (2011). NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66, 1260–1262. doi: 10.1093/jac/dkr135
Khan, A. U., Maryam, L., and Zarrilli, R. (2017). Structure, genetics and worldwide spread of New Delhi Metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 17:101. doi: 10.1186/s12866-017-1012-8
Krishnaraju, M., Kamatchi, C., Jha, A. K., Devasena, N., Vennila, R., Sumathi, G., et al. (2015). Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J. Med. Microbiol. 33, 30–38. doi: 10.4103/0255-0857.148373
Lai, C. C., Chuang, Y. C., Chen, C. C., and Tang, H. J. (2017). Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int. J. Antimicrob. Agents 49, 517–518. doi: 10.1016/j.ijantimicag.2017.02.001
Leverstein-Van, H. M. A., Stuart, J. C., Voets, G. M., Versteeg, D., Tersmette, T., and Fluit, A. C. (2010). Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect. Dis. 10, 830–831. doi: 10.1016/S1473-3099(10)70277-2
Li, X., Fu, Y., Shen, M., Huang, D., Du, X., Hu, Q., et al. (2018). Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control 7:59. doi: 10.1186/s13756-018-0349-6
Liang, W. J., Liu, H. Y., Duan, G. C., Zhao, Y. X., Chen, S. Y., Yang, H. Y., et al. (2017). Emergence and mechanism of carbapenem-resistant Escherichia coli in Henan, China, 2014. J. Infect. Public Health 11, 347–351. doi: 10.1016/j.jiph.2017.09.020
Liu, L., Feng, Y., McNally, A., and Zong, Z. (2018). blaNDM-21, a new variant of blaNDM in an Escherichia coli clinical isolate carrying blaCTX-M-55 and rmtB. J. Antimicrob. Chemother. 73, 2336–2339. doi: 10.1093/jac/dky226
Mei, Y. F., Liu, P. P., Wan, L. G., Liu, Y., Wang, L. H., Wei, D. D., et al. (2017). Virulence and genomic feature of a virulent Klebsiella pneumoniae sequence type 14 strain of serotype K2 harboring blaNDM-5 in China. Front. Microbiol. 8:335. doi: 10.3389/fmicb.2017.00335
Nordmann, P., and Poirel, L. (2014). The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 20, 821–830. doi: 10.1111/1469-0691.12719
Otter, J. A., Burgess, P., Davies, F., Mookerjee, S., Singleton, J., Gilchrist, M., et al. (2017). Counting the cost of an outbreak of carbapenemase-producing Enterobacteriaceae: an economic evaluation from a hospital perspective. Clin. Microbiol. Infect. 23, 188–196. doi: 10.1016/j.cmi.2016.10.005
Pereira, P. S., Borghi, M., de Araújo, C. F., Aires, C. A., Oliveira, J. C., Asensi, M. D., et al. (2015). Clonal Dissemination of OXA-370-producing Klebsiella pneumoniae in Rio de Janeiro, Brazil. Antimicrob. Agents Chemother. 59, 4453–4456. doi: 10.1128/AAC.04243-14
Pérez-Pérez, F. J., and Hanson, N. D. (2002). Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40, 2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002
Poirel, L., Héritier, C., Tolün, V., and Nordmann, P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 15–22. doi: 10.1128/AAC.48.1.15-22.2004
Queenan, A. M., Torres-Viera, C., Gold, H. S., Carmeli, Y., Eliopoulos, G. M., Moellering, R. C., et al. (2000). SME-type carbapenem-hydrolyzing class A beta-lactamases from geographically diverse Serratia marcescens strains. Antimicrob. Agents Chemother. 44, 3035–3039. doi: 10.1128/AAC.44.11.3035-3039.2000
Rahman, M., Mukhopadhyay, C., Rai, R. P., Singh, S., Gupta, S., Singh, A., et al. (2018). Novel variant NDM-11 and other NDM-1 variants in multidrug-resistant Escherichia coli from South India. J. Glob. Antimicrob. Resist. 14, 154–157. doi: 10.1016/j.jgar.2018.04.001
Rahman, M., Shukla, S. K., Prasad, K. N., Ovejero, C. M., Pati, B. K., Tripathi, A., et al. (2014). Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int. J. Antimicrob. Agents 44, 30–37. doi: 10.1016/j.ijantimicag.2014.03.003
Schmitt, J., Jacobs, E., and Schmidt, H. (2007). Molecular characterization of extended-spectrum beta-lactamases in Enterobacteriaceae from patients of two hospitals in Saxony, Germany. J. Med. Microbiol. 56(Pt 2), 241–249. doi: 10.1099/jmm.0.46670-0
Senda, K., Arakawa, Y., Ichiyama, S., Nakashima, K., Ito, H., Ohsuka, S., et al. (1996). PCR detection of metallo-beta-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum beta-lactams. J. Clin. Microbiol. 34, 2909–2913.
Stewart, A. C., Bethel, C. R., VanPelt, J., Bergstrom, A., Cheng, Z., Miller, C. G., et al. (2017). Clinical variants of New Delhi metallo-β-lactamase are evolving to overcome zinc scarcity. ACS Infect. Dis. 3, 927–940. doi: 10.1021/acsinfecdis.7b00128
Torres-González, P., Bobadilla-Del Valle, M., Tovar-Calderón, E., Leal-Vega, F., Hernández-Cruz, A., Martínez-Gamboa, A., et al. (2015). Outbreak caused by Enterobacteriaceae harboring NDM-1 metallo-β-lactamase carried in an IncFII plasmid in a tertiary care hospital in Mexico City. Antimicrob. Agents Chemother. 59, 7080–7083. doi: 10.1128/AAC.00055-15.
Wailan, A. M., Paterson, D. L., Caffery, M., Sowden, D., and Sidjabat, H. E. (2015). Draft genome sequence of NDM-5-producing Escherichia coli sequence Type 648 and genetic context of blaNDM-5 in Australia. Genome. Announc. 3:e00194–15. doi: 10.1128/genomeA.00194-15
Walsh, T. R. (2010). Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36(Suppl. 3), S8–S14. doi: 10.1016/S0924-8579(10)70004-2
Wang, L. H., Liu, P. P., Wei, D. D., Liu, Y., Wan, L. G., Xiang, T. X., et al. (2016). Clinical isolates of uropathogenic Escherichia coli ST131 producing NDM-7 metallo-β-lactamase in China. Int. J. Antimicrob. Agents 48, 41–45. doi: 10.1016/j.ijantimicag.2016.03.009
Wang, S., Zhao, S. Y., Xiao, S. Z., Gu, F. F., Liu, Q. Z., Tang, J., et al. (2016). Antimicrobial resistance and molecular epidemiology of Escherichia coli causing bloodstream infections in three hospitals in Shanghai, China. PLoS ONE 11:e0147740. doi: 10.1371/journal.pone.0147740
Wang, X., Xu, X., Li, Z., Chen, H., Wang, Q., Yang, P., et al. (2014). An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb. Drug Resist. 20, 144–149. doi: 10.1089/mdr.2013.0100
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Yamamoto, M., and Pop-Vicas, A. E. (2014). Treatment for infections with carbapenem-resistant Enterobacteriaceae: what options do we still have? Crit. Care 18:229. doi: 10.1186/cc13949
Yang, J., Chen, Y., Jia, X., Luo, Y., Song, Q., Zhao, W., et al. (2012). Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin. Microbiol. Infect. 18, E506–E513. doi: 10.1111/1469-0691.12035
Yang, P., Xie, Y., Feng, P., and Zong, Z. (2014). blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob. Agents Chemother. 58, 7548–7552. doi: 10.1128/AAC.03911-14
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Yu, J., Tan, K., Rong, Z., Wang, Y., Chen, Z., Zhu, X., et al. (2016). Nosocomial outbreak of KPC-2- and NDM-1-producing Klebsiella pneumoniae in a neonatal ward: a retrospective study. BMC Infect. Dis. 16:563. doi: 10.1186/s12879-016-1870-y
Yu, Y., Ji, S., Chen, Y., Zhou, W., Wei, Z., Li, L., et al. (2007). Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J. Infect. 54, 53–57. doi: 10.1016/j.jinf.2006.01.014
Zhang, F., Xie, L., Wang, X., Han, L., Guo, X., Ni, Y., et al. (2016). Further spread of blaNDM-5 in Enterobacteriaceae via IncX3 plasmids in Shanghai, China. Front. Microbiol. 7:424. doi: 10.3389/fmicb.2016.00424
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017). Nationwide surveillance of clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Zhang, Y., Wang, Q., Yin, Y., Chen, H., Jin, L., Gu, B., et al. (2018). Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE network. Antimicrob. Agents Chemother. 62. doi: 10.1128/AAC.01882-17
Keywords: carbapenem, Escherichia coli, blaNDM variants, diversity, plasmid
Citation: Bi R, Kong Z, Qian H, Jiang F, Kang H, Gu B and Ma P (2018) High Prevalence of blaNDM Variants Among Carbapenem-Resistant Escherichia coli in Northern Jiangsu Province, China. Front. Microbiol. 9:2704. doi: 10.3389/fmicb.2018.02704
Received: 26 June 2018; Accepted: 23 October 2018;
Published: 13 November 2018.
Edited by:
Liang Chen, Public Health Research Institute (PHRI), United StatesReviewed by:
Jian-Hua Liu, South China Agricultural University, ChinaCopyright © 2018 Bi, Kong, Qian, Jiang, Kang, Gu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Gu, Z2IyMDAzMTEyOUAxNjMuY29t
Ping Ma, NjcyNDQzMTkzQHFxLmNvbQ==
†Present Address: Ruru Bi, Department of Laboratory Medicine, Suzhou Science and Technology Town Hospital, Suzhou, China
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.