
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 01 November 2018
Sec. Microbiotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02625
This article is part of the Research Topic Microorganisms Degrading Organic Pollutants and Their Potential for the Bioremediation of Contaminated Environments View all 48 articles
Indole is long regarded as a typical N-heterocyclic aromatic pollutant in industrial and agricultural wastewater, and recently it has been identified as a versatile signaling molecule with wide environmental distributions. An exponentially growing number of researches have been reported on indole due to its significant roles in bacterial physiology, pathogenesis, animal behavior and human diseases. From the viewpoint of both environmental bioremediation and biological studies, the researches on metabolism and fates of indole are important to realize environmental treatment and illuminate its biological function. Indole can be produced from tryptophan by tryptophanase in many bacterial species. Meanwhile, various bacterial strains have obtained the ability to transform and degrade indole. The characteristics and pathways for indole degradation have been investigated for a century, and the functional genes for indole aerobic degradation have also been uncovered recently. Interestingly, many oxygenases have proven to be able to oxidize indole to indigo, and this historic and motivating case for biological applications has attracted intensive attention for decades. Herein, the bacteria, enzymes and pathways for indole production, biodegradation and biotransformation are systematically summarized, and the future researches on indole-microbe interactions are also prospected.
Indole, which was firstly isolated from indigo reduction process, is a typical nitrogen heterocyclic aromatic compound widespread in our daily products and natural environment. Indole ring is present in many alkaloids, phytohormones, plant flower oils, pigments and proteins (de Sá Alves et al., 2009; Arora et al., 2015). Since indole nucleus has a wide spectrum of biological activities, including anticancer, antiviral, antimicrobial, anti-inflammatory, antiHIV and antidiabetic activities, it is extensively used in the pharmaceutical industries (Gribble, 2010; Sharma et al., 2010). Plants and microorganisms can produce indole by tryptophanase or its analogs in the presence of indole-3-glycerol phosphate and tryptophan, thus indole is present in plants and bacteria-rich niches, such as soil, rhizosphere, sludge and intestinal tracts (Frey et al., 2000; Lee and Lee, 2010). In addition, relatively high concentration of indole has been found in coal tar and animal feces and sewage, constituting the major contributor to odor pollution (Yamamoto et al., 1991; Mackie et al., 1998).
Indole has long been regarded as a typical N-heterocyclic aromatic pollutant due to its toxicity and potential mutagenicity. Studies have shown that indole can cause animal hemolysi, hemoglobinuric nephrosis, temporary skin irritation, tumor formation and plant low pigmentation (Collin and Höke, 2000; Arora et al., 2015). For microorganisms, indole can induce bacterial membrane and oxidant toxicity, prevent cell division by modulating membrane potential, inhibit adenosine triphosphate production and protein folding, and cause reparable DNA damage (Osawa et al., 1983; Garbe et al., 2000; Chimerel et al., 2012; Kim J. et al., 2013). Therefore, the biodegradation and bioremediation of indole have been concerned over a century.
Recently, indole has been proven to be a versatile interspecies and interkingdom signaling molecule which plays important roles in prokaryotic and eukaryotic communities (Lee et al., 2015a). Indole can affect the spore formation, plasmid stability, cell division, antibiotic tolerance, biofilm formation and virulence of both indole-producing and non-indole-producing microorganisms (Lee and Lee, 2010). Indole can be produced by plants (e.g., maize) upon herbivore attack, which will act as the aerial priming agent to warn their non-attacked tissues and neighboring plants for the imminent attack (Erb et al., 2015). Indole can also control the behavior of insects. More importantly, increasing evidences indicate that indole is rich in human intestinal tracts and closely related with immune systems and several human diseases (Lee et al., 2015a). Overall, the function study of indole is still in initial stage and much is awaiting to be discovered and investigated.
Since indole is widely distributed in different environment matrices, it will encounter virous kinds of microorganisms. Many bacteria have acquired the ability to transform and degrade indole to improve their survival state. The researches on biotransformation and biodegradation of indole are of importance to realize environmental bioremediation and illuminate its biological functions. Herein, this review presents the history and recent advances regarding indole biotransformation and biodegradation, and the research trends in indole-microbe studies are also prospected.
Indole was firstly discovered in indigo reduction process by Baeyer in the year of 1866, which was then successfully synthesized by Baeyer (1866) and Fischer and Jourdan (1883). In 1897, it was found that bacterial strains, including Escherichia coli and Vibrio cholerae, could produce indole, and this feature has long been used for strain identification (Smith, 1897). The synthetic pathways for indole in bacteria are summarized in Figure 1. It is well known that biosynthesis of tryptophan is catalyzed by TrpABCDE encoded by the tryptophan operon. Tryptophan synthase is encoded by trpA and trpB, and catalyzes the hydrolysis of indole 3-glycerol phosphate to indole and the subsequent condensation of indole and L-serine to form L-tryptophan. The intermediate indole is transferred from the alpha-subunit to the beta-subunit and not released from the tryptophan synthase complex (Murdock et al., 1993). Another indole-related pathway is catalyzed by tryptophanase (TnaA), which can reversibly convert tryptophan to indole, pyruvate and ammonia (Watanabe and Snell, 1972). This is the sole indole-producing pathway in bacteria to date, and over 85 bacterial species (both Gram-negative and Gram-positive bacteria) can produce indole from tryptophan (Lee and Lee, 2010). The indole-producing bacteria occupy various environmental niches, suggesting indole can exist widespread such as in activated sludge, soil, plant rhizosphere, ocean, animals and livestock waste. It is noted that tryptophan can also be converted to indole-3-acetic acid and 3-methylindole (skatole), especially by gut bacteria under anoxic condition, whereas the mechanism and regulation of indole and skatole production remain unclear (Mackie et al., 1998; Deslandes et al., 2001).
Generally, the indole concentration can reach 0.5–0.6 mM when E. coli and V. cholerae are cultured in Luria-Bertani (LB) medium, and 0.25–1.1 mM in the mouse and human gut (Kobayashi et al., 2006; Mueller et al., 2009; Lee et al., 2015a). Several environmental and biological factors have been proven to affect the indole production level, of which the amount of exogenous tryptophan is the most important one. Li and Young (2013) found that the E. coli converted the tryptophan into an equal amount of indole up to 5 mM, and the synthesis process mainly depended on the tryptophan importer protein TnaB. However, further increase of tryptophan would not accumulate indole. The possible reason might be that excess indole inhibited TnaA enzyme activity and tryptophan transport process. High pH and temperature promoted indole production. Indole yield increased with pH value in the range of 4.0–9.0 since low pH inhibited TnaA expression, and TnaA was highly expressed at pH 9.0 (Han et al., 2011). Surprisingly, E. coli strain produced nearly 7 mM indole/cell OD at 50°C, 89-fold higher than that at 37°C, though it stopped growing at such high temperature. When E. coli strains exposed to antibiotics, such as ampicillin and kanamycin, the indole yield also significantly increased, which should be beneficial for the resistance and cell survival against antibiotics (Han et al., 2011).
Indole is also found in a variety of natural plant flower oils, such as jasmine and daffodils. It is reported that natural jasmine oils contain as high as 2.5% indole (Collin and Höke, 2000). Many plants, including cotton, corn, green beans, cucumbers and apple trees, can release volatile organic compounds to protect themselves from insect threats, and indole is one of the typical secondary metabolites (Erb et al., 2015). Three pathways relevant to indole production in plants have been reported as shown in Figure 2. The typical tryptophan biosynthetic route encoded by trp operon was also conserved in plants. Plant tryptophan synthase α (TSA) and tryptophan synthase β (TSB) showed high similarities with those in bacteria and catalyzed the conversion of indole-3-glycerol phosphate to tryptophan without indole release (Radwanski and Last, 1995). The enzymes encoded by gene cluster bx can catalyze the production of cyclic hydroxamic acid 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one (DIMBOA) from IGP in plants. Frey et al. (1997) demonstrated that the first enzyme BX1 in the cluster was a TSA homolog, and could directly catalyze the indole production from IGP independent of any TSB. Then another indole-producing enzyme, i.e., indole-3-glycerol phosphate lyase (IGL), was identified in maize (Frey et al., 2000). Sequence analysis indicated that both IGL and BX1 should be derived from TSA.
Anthropogenic activities significantly promote the distribution of indole in the environment. Indole is chemically synthesized in technical quantities as a feedstock or precursor in the production of tryptophan, plant growth regulators, dyes and pharmaceuticals (Barden, 2010). Indole has been found in coal tar over 100 years (Collin and Höke, 2000), and with the development of coal mining, petroleum processing and tobacco industries, plenty of indole-containing wastewater and smoke is produced and discharged into the environment. On the other hand, the increasing urbanization and intensification of animal production throughout the world have resulted in serious animal waste and odor nuisance. Indole and skatole produced by intestinal anaerobes from tryptophan have been implicated as a major factor in odor nuisance and livestock pollution, and they are released in the environment through gas transmission, livestock wastewater discharge and biosolids application to agricultural lands (Mackie et al., 1998; Kinney et al., 2006).
Research on indole degradation initiated from the 1920s and the reported indole-degrading strains are summarized in Table 1. Raistrick and Clark (1921) firstly proved that Bacillus pyocyaneus, B. fluorescens and B. prodigiosus could attack indole nucleus and release ammonia, and Supniewski (1923) further stated that B. pyocyaneus could degrade indole in a low rate. Gray (1928) obtained the Pseudomonas indoloxidans strain from soil which produced indigo in the process of indole decomposition, but it could not utilize indole as the sole carbon source. Detailed analysis of indole aerobic degradation was started by Sakamoto et al. (1953), who firstly proposed a reasonable indole degradation pathway as indole-indoxyl-isatin-N-formylanthranilate-anthranilate-salicylate-catechol by a Gram-negative bacterium. Fujioka and Wada (1968) then isolated a Gram-positive bacterium and obtained a dihydroxyindole oxygenase which directly catalyzed dihydroxyindole to anthranilic acid. Results also showed that the strain could not decompose N-formylanthranilate, isatin and 2-oxindole, indicating it might convert dihydroxyindole to anthranilate via a single enzyme step. However, the enzyme catalyzing the first step of indole oxidation was not obtained, which was associated with cellular debris after high-speed centrifugation. In Claus and Kutzner (1983), isolated an Alcaligenes spec. In3 and proposed the indole-indoxyl-isatin-anthranilate-gentisate pathway, while the gentisate formation process from anthranilate remained to be elucidated. In 1997, P. sp. ST-200 was reported to be able to assimilate high concentration of indole dissolved in organic solvent, and isatin and isatic acid were detected as the main intermediates (Doukyu and Aono, 1997). Strain ST-200 could not utilize isatic acid as the growth substrate, suggesting it assimilated indole in another degradation pathway. Another pseudomonad strain GS was isolated from mangrove sediment and produced two novel metabolites without further identification (Yin et al., 2005).
As the biological functions of indole have been extensively reported in the last decade, indole biotransformation studies have received more attention. Recently, Fukuoka et al. (2015) characterized the indole biotransformation pathway in Cupriavidus sp. KK10. It could degrade indole not only via N-heterocyclic ring cleavage pathway but also carbocyclic-aromatic ring cleavage pathway with initial attack at C4 and C5 positions. We have recently obtained two Cupriavidus strains named IDO and SHE from indole acclimated samples. They could utilize indole as sole carbon source and efficiently remove 100 mg/L indole within a day when cultured in simple mineral salt medium at 30°C and 150 r/min (Ma et al., 2015a; Qu et al., 2015). Intermediate analyses indicated that strain SHE produced isatin, isatic acid, and anthranilate in indole degradation process (Qu et al., 2017). As for Acinetobacter, Lin et al. (2015) demonstrated that strain A. baumannii ATCC19606 could detoxify indole by converting it to non-toxic indigo, and the functional genes responsible for indole oxidation were firstly identified and characterized. Subsequently, Sadauskas et al. (2017) reported that strain A. sp. O153 degraded indole via the indole-2,3-dihydrodiol, 3-hydroxyindolin-2-one, and anthranilate step through a similar iif gene cluster with that in strain ATCC19606. Yang et al. (2017) isolated an A. pittii L1 from coking wastewater and identified the 4-(3-hydroxy-1H-pyrrol-2-yl)-2-oxo-but-3-enoic acid and isatin as the intermediates, suggesting that strain L1 could attack both the carbocyclic and N-heterocyclic rings, similar with that in strain C. sp. KK10. In addition to Cupriavidus and Acinetobacter, other recently reported indole-degrading strains included Arthrobacter sp., Alcaligenes sp., Burkholderia unamae, P. aeruginosa, and Agrobacterium tumefaciens (Lee et al., 2009, 2015b; Kim D. et al., 2013; Arora et al., 2015). It is noted that some clinically important bacterial species (e.g., Providencia) can produce and degrade indole simultaneously, and the metabolic process and biological significance of this phenomenon need further investigation (Müller, 1986).
The indole degradation pathways are summarized in Figure 3, and it can be seen that isatin and anthranilate are the key intermediates in several studies (Qu et al., 2017). Isatic acid, the hydrolase product of isatin, was detected in P. sp. ST-200 and C. sp. SHE (Doukyu and Aono, 1997; Qu et al., 2015), further suggesting that isatin participated in indole degradation in these strains. However, studies in A. sp. O153 and a Gram-positive coccus suggested that anthranilate was directly formed from dihydroxyindole or 3-hydroxyindolin-2-one bypass isatin (E pathway in Figure 3; Fujioka and Wada, 1968; Sadauskas et al., 2017). Therefore, the roles of isatin in indole degradation should be further confirmed. Indole degradation via the carbocyclic ring cleavage pathway has been recently identified in C. sp. KK10 and A. pittii L1 (F pathway in Figure 3; Fukuoka et al., 2015; Yang et al., 2017), and the frequency of this pathway is worthy of further investigation. Moreover, the biotransformation of anthranilate, the key intermediate linking indole upstream and downstream metabolism, is diverse in different strains, and the understanding of anthranilate metabolic pathway is crucial to fully clarify the fate of indole in the microbial communities (Qu et al., 2017; Sadauskas et al., 2017).
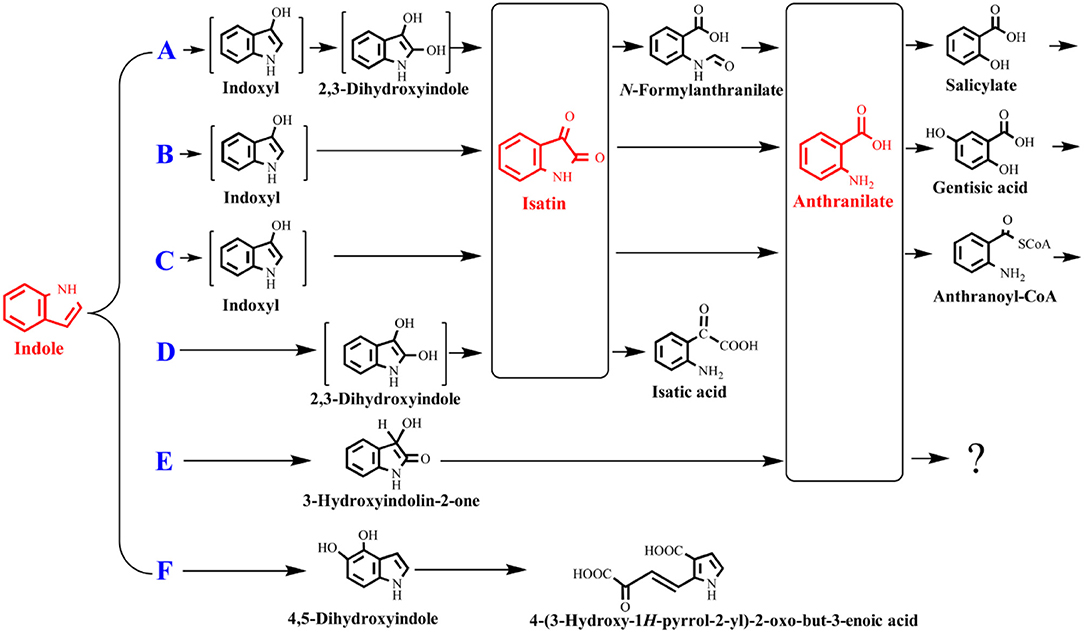
Figure 3. Summary of indole aerobic degradation pathways. A, a Gram-negative bacterium isolated from tap water (Sakamoto et al., 1953); B, Alcaligenes spec strain In 3 (Claus and Kutzner, 1983); C, C. sp. SHE (Qu et al., 2017); D, P. sp. ST-200 (Doukyu and Aono, 1997); E, a Gram-positive coccus (Fujioka and Wada, 1968) and A. sp. O153 (Sadauskas et al., 2017); F, C. sp. KK10 and A. pittii L1 (Fukuoka et al., 2015; Yang et al., 2017).
Identification of indole degradation related genes and gene clusters would undoubtedly enhance our understanding of its transformation process. Moreover, it can be inferred that indole oxygenase which utilized indole as the primary substrate may be advantageous over other aromatic oxygenases in indole biotransformation to form indigo. However, the functional genes responsible for indole degradation have remained a black box for a century. Many oxidoreductases, such as naphthalene dioxygenase, biphenyl dioxygenase, phenol hydroxylase and cytochrome P450 hydroxylase, could oxidize indole to indoxyl, which could be further dimerized into indigoids, while no evidences were shown that indole was the inherent substrate for these enzymes (Ensley et al., 1983; O'Connor et al., 1997; Kim et al., 2005). Fujioka and Wada (1968) have tried to purify the indole oxygenase, the key enzyme that catalyzed the first step reaction of indole. The target protein was not solubilized and the catalytic activity could not be determined since bacterial strain or cell debris oxidized indole to anthranilate directly. A dihydroxyindole oxygenase was unexpectedly obtained and it was proven to catalyze dihydroxyindole to anthranilate without formation of other intermediates, which should be the adjacent enzyme of indole oxygenase. Similarly, Kamath and Vaidyanathan (1990) tried to determine the indole oxygenase activity in Aspergillus niger under a series of conditions but ended in failure.
Very recently, Sadauskas et al. (2017) and Qu et al. (2017) simultaneously reported the genetic determinants for indole degradation in Acinetobacter and Cupriavidus, respectively (Figure 4). They both identified an iif (or ind) gene cluster, which was widespread in a range of bacteria, responsible for indole upstream metabolism to produce anthranilate. The oxygenase and flavin reductase units encoded by iifC and iifD, which were firstly identified and characterized by Lin et al. (2015), constituted the two-component indole oxygenase system responsible for indole oxidation to cis-2,3-dihydrodiol. The short-chain dehydrogenase IifB converted indole-2,3-dihydrodiol to 3-hydroxyindolin-2-one. And then a cofactor-independent oxygenase IifA was proven to produce anthranilate directly from 3-hydroxyindolin-2-one. The role of iifE in the cluster was ruled out due to its unchanged expression level and unclear function in indole transformation process.
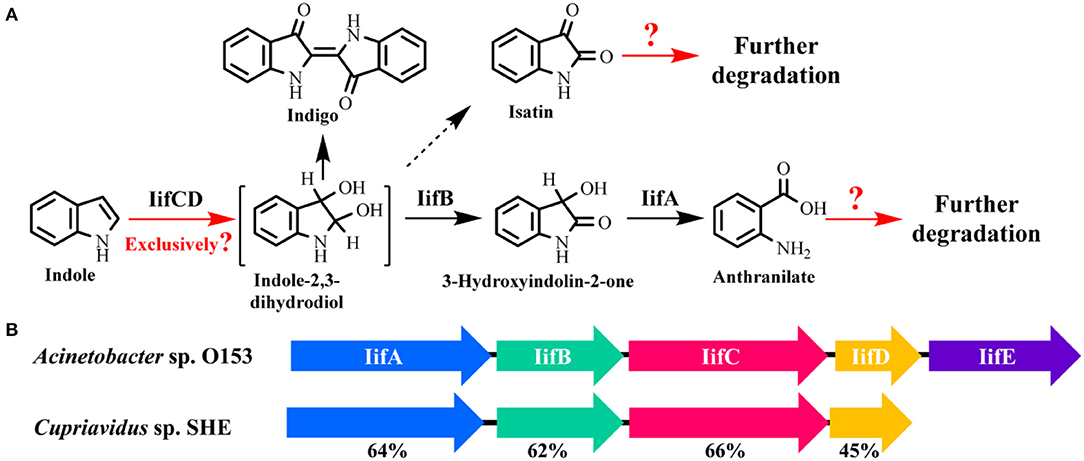
Figure 4. Summary of indole aerobic degradation functional enzymes. (A) overall indole degradation pathway and corresponding enzymes; (B) iif gene cluster organization in strain A. sp. O153 and C. sp. SHE.
Several key questions still remain in this research. (i) Is iif the exclusive gene cluster for indole degradation? Answer should be no. One typical example is that the type strain P. aeruginosa PAO1 can degrade indole effectively, while genomic analysis indicates that this strain lacks iif related genes (Lee et al., 2009). In the iif-dependent degradation bacteria, more than one copy of iif gene cluster exists in some strains, such as C. sp. IDO (Ma et al., 2015a), thus the role of these clusters should be resolved. (ii) Is isatin involved in indole degradation? It was shown that there was no gene in the iif cluster associated with isatin production or transformation, thus Sadauskas et al. (2017) suggested that isatin should be a dead-end product or even an experimental artifact. Nevertheless, isatic acid was detected in some studies, indicating that the isatin was formed and further metabolized possibly by an isatin hydrolase. Further BLAST analysis indicates that the isatin hydrolase gene is spread in numerous strains, including indole-degrading genera Cupriavidus, Burkholderia and Azoarcus, and functional identification of this gene is worth investigation (Bjerregaard-Andersen et al., 2014). (iii) What is the functional gene cluster for anthranilate metabolism in indole degradation process? Anthranilate can be further degraded by four pathways, i.e., catechol pathway catalyzed by anthranilate-1,2-dioxygenase, gentisate pathway catalyzed by an unreported enzyme, 3-hydroxyanthranilate pathway catalyzed by anthranilate hydroxylase, and 2-aminobenzoyl-CoA pathway catalyzed by 2-aminobenzoayl-CoA ligase and 2-aminobenzoyl-CoA monooxygenase/reductase (Langkau et al., 1995; Arora, 2015). Sadauskas et al. (2017) and Lee et al. (2009) speculated that anthranilate might be degraded by the anthranilate dioxygenase by genomic and transcriptomic analyses. However, we proved that a 2-aminobenzoayl-CoA gene cluster was vital for anthranilate degradation by proteomic and genetic verification (Qu et al., 2017). Moreover, gentisate was detected as the intermediate in indole degradation (Claus and Kutzner, 1983), and the functional gene for gentisate pathway has not yet been revealed to date. (iv) How is the iif gene cluster regulated? Previous study indicated that an IifR protein was required for indole oxygenase expression in strain A. baumannii ATCC19606 (Lin et al., 2015). However, IifR was constitutively expressed irrespective of indole concentrations and the regulation mechanism was not further clarified.
Apart from pure culture studies, indole degradation by aerobic microbial communities was rarely investigated. In this respect, we have systematically explored microbial community composition and structure using flask and sequencing batch reactor systems by Illumina high throughput sequencing technique (Ma et al., 2015b; Zhang et al., 2015). Azorcus and Thauera were the dominant genera in the microbial community with indole and glucose as the carbon source. When indole was utilized as sole carbon and/or nitrogen source, Alcaligenes, Burkholderia, and Comamonas were the main compositions. Several strains from these genera have been reported previously, which should be valuable microbial resources for indole remediation. However, it was noted that indole degradation intermediates could be the carbon sources for many strains, thus the functional populations toward indole should be further affirmed. Besides, the functional genes in the microbial community for indole transformation have not been concerned.
Indole anaerobic degradation studies were concentrated on microbial communities and the initial studies could date back to 1980s. Balba and Evans (1980) investigated the methanogenic fermentation of tryptophan and indole was detected as the key intermediate, which was further converted to anthranilate, salicylate and benzoate. Wang et al. (1984) showed that indole could be degraded under methanogenic conditions and mineralized to methane and carbon dioxide. However, these results were inconclusive due to the sorption phenomena. Berry et al. (1987) confirmed the mineralization of indole to methane with the inoculum of sewage sludge, and oxindole was carefully identified as the degradation intermediate. Further experiment indicated that with the decrease of temperature and concentration of sewage sludge inoculum, oxindole could reach higher concentration and persisted longer under methanogenic conditions (Madsen et al., 1988). Complete mineralization of indole has also been observed under sulfate-reducing conditions, which shared similar pathway with that under methanogenic conditions (Shanker and Bollag, 1990). Gu and Berry (1991) has shown that the substituted groups at indole would affect its degradation rates and pathway. Specially, a methyl group at C1 (1-methylindole) or C2 (2-methylindole) inhibited its hydroxylation and degradation, and 3-methylindole could only be hydroxylated in the 2-position resulting in 3-methyloxindole formation without further mineralization under methanogenic conditions. Indole degradation under denitrifying conditions was carefully conducted by Madsen et al. (1988) with nitrate-reducing sewage sludge. Oxindole appeared only in trace amounts without accumulation possibly due to the exceeded oxindole elimination rate over production rate. High-performance liquid chromatography, thin-layer chromatography, and mass spectrometry analyses identified four key intermediates, i.e., oxindole, isatin, dioxindole and anthranilate, in indole mineralization process in the nitrate-reducing sewage (Figure 5; Madsen and Bollag, 1989).
Pathways for indole aerobic and anaerobic degradation share certain similarities. Hydroxylation of indole is believed to be the essential step in its metabolism. Isatin, dihydroxyindole and anthranilate are the common intermediates detected under both conditions. The obvious difference is that indole is often aerobically oxidized at C3 or C2/C3 position as the first step, which, however, is generally hydroxylated at C2 position leading to 2-oxindole production under anaerobic condition. Under methanogenic and sulfate-reducing conditions, 2-oxindole can be accumulated and easily detected as main intermediate, while it is effectively converted under nitrifying conditions, which may be the reason for the high indole degradation efficiency (Madsen et al., 1988; Shanker and Bollag, 1990). All studies have indicated that benzene ring of indole remained intact and hydroxylation only occurred at pyrrole ring. It can be speculated that functional enzyme for indole anaerobic degradation is distinct from the aerobic ones. The source of oxygen used in the enzyme-mediated oxidation should be from water rather than molecular oxygen, and anaerobic enzyme regioselectively attacked C2 rather than C3 position.
Indole could be degraded under various anaerobic conditions, while the functional microbes have been sporadically referred. Hong et al. (2010) firstly explored the microbial communities using the 16S rRNA clone library method. It was shown that Alicycliphilus, Alcaligenes, and Thauera were dominant genera in the denitrifying bioreactor. The major genera were similar with those of aerobic degradation bacterial community, suggesting they were the facultative and functionally important bacteria in indole degradation process. Under sulfate-reducing condition, bacterial community was significantly distinct, and Clostridia and Actinobacteria were the major classes, which should be resulted from the different electron acceptors of these two systems. Recently, indole degradation in microbial fuel cell (MFC) was achieved with electricity generation (Luo et al., 2010). Up to 88% indole (250 mg/L) could be removed within 3 h, whose rate was much superior to microbial reactions. MFC system provided another avenue for indole-containing wastewater treatment, while the microbial community on the anodes, possibly Desulfovibrio related genera, required further investigation.
Desulfobacterum indolicum In04 was the first and only isolated bacterium which could degrade indole under strictly anaerobic conditions up to date (Bak and Widdel, 1986). It could completely convert indole to CO2 using sulfate as the electron acceptor. The indole degradation intermediates by strain In04 were oxindole, isatin and anthranilate, same with those detected in community systems. Three P. putida strains were also isolated from activated sludges that could use indole as sole carbon source under anoxic condition (Li and Young, 2013). Metabolite analyses verified that indole was degraded via the traditional anaerobic pathway, i.e., oxindole, isatin and anthracitic acid. Similar with Pseudomonas, Thauera was also reported to be able to degrade indole under both aerobic and anaerobic conditions (Mao et al., 2010). This genus was detected as the main population in the denitrifying bioreactor and acerbic degradation community, further highlighting its versatile indole degradation capacity.
Indigo is one of the oldest pigments used in dying, food, pharmaceutical and semiconductor industries, and it is known for the application of producing the popular blue denim. Indigo is traditionally extracted from plants, while the indigo market has been soon taken up by chemical production after Baeyer synthesized the coloring matter in the lab (Huxtable, 2001). However, chemical synthesis will unavoidably use toxic compounds (e.g., aniline) as the substrates and produce industrial wastewater, causing environmental and public health concerns. Microbial production of indigo has been attracting a huge attention in the last few decades due to its environment-benign and mild condition requirements. The formation of indigo was firstly reported in the study of indole degradation process by P. indoloxidans (Gray, 1928; Oshima et al., 1965). Two molecules of indole were oxidized to one molecule of indigo and indoxyl was detected as the intermediate in indigo formation reaction. Up to date, there have been amounts of bacterial strains reported with the ability of indigo formation from indole as shown in Table 2.
Pseudomonas, a typical type of strain for organic compounds biodegradation, was the most reported indigo-producing genus. O'Connor and Hartmans (1998) investigated five Pseudomonas strains and found that styrene-induced P. putida S12 and CA-3 produced indigo at high rates of 5–10 mg/L in 30 min, while toluene- and naphthalene-induced P. putida F1 and PpG7 produced 1.3–1.6 mg/L indigo in 30 min. Afterwards, Bhushan et al. (2000) found that naphthalene-induced P. putida PKJ1 could also produce indigo at a relative low rate around ~2.0 mg/L in 30 min. Pathak and Madamwar (2010) isolated an efficient naphthalene degrading strain P. sp. HOB1, and it could produce as high as 246 mg/L indigo. We have previously reported that phenol-induced P. sp. QM and PI1 could also produce indigo, and other red and purple indigoids were also generated (Ma et al., 2012; Wang et al., 2015).
Acinetobacter was another famous indigo-producing genus. Doukyu et al. (2002) obtained A. sp. ST-550 from soil sample and it could produce 292 mg/L indigo in the diphenylmethane-water two phase system. Ishikawa et al. (2014) introduced the bacterionanofiber gene ataA into strain ST-550 to improve its adhesiveness, and the recombinant strain could be easily immobilized on a polyurethane support. The immobilized strain could tolerant higher concentration of indole and produce more indigo, and this strategy provided a smart way for efficient indigo production and recovery. A. sp. strain PP-2 was previously obtained in our lab and could produce indigo induced by different aromatics, such as phenol, naphthalene and xylene (Qu et al., 2010). The indigo formation conditions were optimized and the yield could reach the highest of 202 mg/L when induced by phenol.
Indoxyl is the precursor of indigo, thus indole oxidation to indoxyl is the key step for indigo production. Many oxygenases have been reported to be able to oxidize indole, and these enzymes could be mainly divided into dioxygenase and monooxygenase. The indigo formation enzymes have been summarized in Table 3, and typical oxygenases include naphthalene dioxygenase (NDO), multicomponent phenol hydroxylase (mPH), cytochrome P450 monooxygenase and flavin monooxygenase (FMO). Since indigo pigment produced by E. coli strains is visible and can be easily screened from clone libraries, the indigo-screening method has been widely used for isolation of oxygenases both from genome and metagenome libraries (Guan et al., 2007; Van Hellemond et al., 2007; Singh et al., 2010). Although high and pure indigo could be produced by microbial process, the eco-efficiency analysis indicated that bio-indigo production is less efficient than chemical processes (Philp et al., 2013). Nevertheless, this novel and green biological process has attracted numerous attention, and related studies continue today.
The first as well as the most reported oxygenase in indigo production field is NDO. Ensley et al. (1983) cloned an NDO gene from P. putida PpG7 and expressed it in E. coli. The recombinant strain was able to produce indigo, an unnatural end product of E. coli, in nutrient medium. Analysis indicated that E. coli could convert tryptophan to indole by tryptophanase, which was further oxidized to indoxyl by NDO, leading to the production of indigo (Figure 6). This study initiated the indigo bioproduction history. Ten years later, Amgen industry successfully constructed the first metabolic pathway catalyzing indigo biosynthesis from glucose in E. coli (Murdock et al., 1993; Bialy, 1997). The activity and half-life time of NDO was significantly improved by promoter and ferredoxin gene component modification, and then high level of indole was produced from glucose in E. coli by inactivation of the β subunit of tryptophan synthetase. However, the indigo yield, 135 mg/L in lab experiment, was not satisfactory for commercial application. Almost another 10 years later, scientists from Genencor International reconstructed the fermentation process based on metabolic engineering to improve the production of bio-indigo (Berry et al., 2002). A high-level indigo producing strain was developed designated as FM5/pBMW, pTacFd911. The production of indigo was further enhanced by improving 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) formation via several strategies, reaching 23 g/L in 42 h. Indirubin was a common byproduct usually co-produced in indigo production process. It was proposed that indoxyl and isatin/2-hydroxyindole were the substrate for indirubin formation. The serious and common by-product problem was resolved by incorporation of an isatin hydrolase, which could open the pyrrole ring of isatin and block indirubin production. In addition to indole, it has been proved that NDO could convert various indole derivatives, including methylindoles, methoxyindoles, nitroindoles, chloroindioles, and bromoindoles, to different indigoid pigments (Kim et al., 2003; Zhang et al., 2013).
Cytochrome P450 enzymes have been found in all kingdoms of life, including bacteria, fungi, plants, animals and human beings. They are among the most versatile proteins with the capability of catalyzing C-hydroxylation, heteroatom oxygenation, heteroatom release, and epoxide formation, and have been widely applied in chiral compounds production, drug development and biodegradation fields (Urlacher and Girhard, 2012). Guengerich group firstly reported and undertook massive work in the indigo formation by cytochrome P450 enzymes (Gillam et al., 1999). P450 2A6 was active in indole biotransformation and produced indigo, indirubin and a new product 6H-oxazolo[3,2-a:4,5-b′]diindole (Figure 7) (Gillam et al., 2000). Random mutagenesis of P450 2A6 resulted in some mutants with higher indigo production capacities, and the mutants could convert substituted indoles to different indigoids. The 5-methylindole transformation product, i.e., 5-methylindirubin, 5′-methylindirubin and 5,5′-dimethoxyindirubin, showed highest inhibition activity toward human cyclin-dependent kinases (CDK)-1 and - 5 and glycogen synthase kinase-3 (GSK3), facilitating the research in drug development (Guengerich et al., 2004). Lu and Mei (2007) co-expressed the P450 BM3 and glucose dehydrogenase and improved the indigo yield. Random and saturation mutagenesis produced several mutants with higher indole oxidation performance, and the mutant D168W could produce indirubin as the main product (90%) rather than indigo by the wild P450 BM3 enzyme, providing novel information for indirubin bio-production (Hu et al., 2010).
The functional enzyme mPH was firstly identified in P. sp. CF600 in phenol and (di)methylphenol degradation process, which was encoded by six genes dmpKLMNOP (Nordlund et al., 1990). Doukyu et al. (2003) has cloned the mPH gene from A. sp. ST-550 and co-expressed with NAD-dependent formate dehydrogenase gene, and the host cell could produce 52 mg/L indigo. Kim et al. (2005) expressed two different mPHs from P. sp. KL33 and P. sp. KL28. The mPHKL28 mainly produced indigoid pigments from indole and could catalyze the formation of various dyestuffs from a wide range of indoles. However, the mPHKL33 converted indole to 7-hydroindole. Our previous study indicated that the mPH from Arthrobacter sp. W1 could transform indole to three pigments, i.e., indigo (blue), indirubin (red) and 2-(7-oxo-1H-indol-6(7H)-ylidene) indolin-3-one (purple) (Qu et al., 2012a). It was reported that many other phenol-degrading bacteria, including P. sp. CF600, P. sp. KL33, and P. sp. QM, converted indole to deep purple or red-purple pigments (Kim et al., 2005; Ma et al., 2012). 7-Hydroindole was identified as the intermediate in these strains, suggesting the purple products might contained 2-(7-oxo-1H-indol-6(7H)-ylidene) indolin-3-one, which was derived from indoxyl and 7-hydroindole. The proposed indole transformation pathway by mPH should cover three pathways, i.e., C2, C3, and C7, distinct from other oxygenases (Figure 8) (Qu et al., 2012a).
The newly reported indigo-producing enzyme FMO was identified from strain Methylophaga aminisulfidivorans MPT (Choi et al., 2003). The recombinant E. coli strain expressing fmo gene could originally produce 160 mg/L indigo in tryptophan medium. Subsequently, the upstream sequence of fmo and the culture medium composition were optimized by surface methodology method. The maximum yield of bio-indigo reached as high as 920 mg/L in the 5-L and 3000-L fermentation system under optimal conditions, the highest yield in batch fermentation system up to date (Han et al., 2008). In addition, Han et al. (2013) found that addition of cysteine could lead to the 2-hydroxyindole formation from indole, which significantly enhanced indirubin production (Figure 9). This finding provided a new and simple approach for the production of valuable indirubin.
Microbial production of indole has solved the problem of hazardous and toxic reagents used in indigo chemical synthesis, while the indigo dying process is still environmentally unfriendly. Indigo is insoluble in water and excess reducing agent, such as sodium dithionite, must be added to covert indigo to leuco form before dying. Hsu et al. (2018) has developed a two-step sustainable biochemical route to solve the indigo reducing problem, as shown in Figure 10. They firstly obtained a glucosyltransferase from Japanese indigo plant Polygonum tinctorium. Expression of the glucosyltransferase gene and fmo gene in E. coli could successfully lead to indicant production rather than the unstable indoxyl. The glucosyl group in indicant could be enzymatically hydrolyzed by a β-glucosidase to produce indoxyl and leucoindigo whenever needed, thus the dying process could avoid the reducing step. Compared with indoxyl, indican is stable, water soluble, easily concentrated and suitable for long storage. Although some challenges still exist for practical dying process, this indoxyl glucosylation method is historic and remarkably promotes bio-indigo application.
The function study of indole as an interspecies and interkingdom signaling molecule has been a hot topic, especially its impact on bacterial antibiotic resistance, virulence, and persister formation (Lee and Lee, 2010; Lee et al., 2015a). Bacteria, plants and anthropic activities can produce considerable amounts of indole and release it into environment. Bacteria will inevitably encounter indole, while the interactions between indole and its surrounding environments largely remain uncovered. The following aspects on indole-microbe interactions beyond signaling research are worthy of further investigation (Figure 11).
(1) Industrial application of indigoids bio-production. More efficient enzymes and pathways for indole biotransformation are to be constructed, the downstream procession of bio-indigo requires attention, and the challenge of indirubin biosynthesis needs in-depth investigation.
(2) Molecular mechanisms for indole degradation. The enzymes besides indole oxygenase for indole oxidation in some bacteria remain unknow. Formation and transformation of isatin and anthranilate in indole degradation may be strain-dependent and remain controversial, and the regulation of indole metabolic genes are rarely referred. Furthermore, the indole degradation mechanism under anaerobic conditions has still been a black box.
(3) Impacts of indole on human health. Indole, derived from tryptophan in food via microbial transformation, is rich in animal and human gut (Lee et al., 2015a). However, the substrate tryptophan can be also converted to indole-3-acetic acid and skatole (Deslandes et al., 2001). The functional strains and regulation of the transformation process in intestinal tracts are still unknown. Indole may impact several diseases such as inflammatory bowel diseases and diabetes (Bansal et al., 2010; Chimerel et al., 2014). However, solid evidences of indole roles should be further provided and the functional mechanism urgently needs to be solved. Indole has been proven to be able to extend healthspan (remain healthy and free of age-related infirmities) in animals by interacting with the aryl hydrocarbon receptor (Sonowal et al., 2017), highlighting the potential of indoles as new therapeutics to improve life quality and reduce frailty.
(4) Functions of indole on marine organisms. Marine bacteria can produce indole, while the researches of indole production by marine bacteria are numbered (Takao et al., 1994; Lee and Lee, 2010). Practically in marine aquaculture industries, tryptophan is abundant in aquaculture feed, and the indole-producing Vibrio strains are the most common pathogenic marine bacteria. Thus, it can be inferred that indole is spread in aquaculture systems. Indole has been found to control the biofilm and virulence of Vibrio strains, and it may also regulate the antibiotic resistance and virulence of other indole-producing and non-indole-producing marine strains (Mueller et al., 2009). Therefore, indole may contribute to, negatively or positively, the aquaculture diseases, antibiotic resistance gene (ARG) expression and propagation.
(5) Impacts of indole on the waste treatment process. Treatment of indole-containing wastewater is always accompanied by indigoids formation, thus isolation of non-indigo production indole-degrading bacteria or genetically modification of bacterial metabolic pathways are feasible solutions (Lee et al., 2015a). Indole as a signaling molecule can coordinate cell-cell communication in microbial communities. Quorum sensing (QS), the most well studied cell-cell communication mechanism, has been found to play crucial roles in wastewater treatment process via control of microbial adhesion, colonization, and biofilm formation, while bacterial QS of indole in these systems has yet been concerned (Yeon et al., 2008; Feng et al., 2013). Indole and skatole are the major contributor to boar taint and the malodor emission from livestock waste and human feces (Mackie et al., 1998; Hales et al., 2012), and development of microbial agents and enzymes will have a broad application prospect in livestock industry. More importantly, livestock wastes are the major sources of ARG which may pose potential worldwide human health risks (Zhu et al., 2013). Indoles in livestock wastes can affect the antibiotic resistance and virulence of several pathogens, influencing the ARG richness and antibiotic usage. Indoles-containing wastewater and manure will enter river, soil and farmland, further interacting with ARG, and the impacts deserve to be investigated.
QM wrote the paper, and YQ and XZ revised the paper. All authors approved it for publication.
The study was supported by the National Natural Science Foundation of China (No. 31800091, No. 51508068), and the Fundamental Research Funds for the Central Universities (No. 3132018173, No. 3132016330).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alemayehu, D., Gordon, L. M., O'Mahony, M. M., O'Leary, N. D., and Dobson, A. D. (2004). Cloning and functional analysis by gene disruption of a novel gene involved in indigo production and fluoranthene metabolism in Pseudomonas alcaligenes PA-10. FEMS Microbiol. Lett. 239, 285–293. doi: 10.1016/j.femsle.2004.08.046
America, S. P. L., Jung, H. S., Kim, H. S., Han, S. S., Kim, H. S., and Lee, J. H. (2015). Characterization of a flavin-containing monooxygenase from Corynebacterium glutamicum and its application to production of indigo and indirubin. Biotechnol. Lett. 37, 1637–1644. doi: 10.1007/s10529-015-1824-2
Arora, P. K. (2015). Bacterial degradation of monocyclic aromatic amines. Front. Microbiol. 6:820. doi: 10.3389/fmicb.2015.00820
Arora, P. K., and Bae, H. (2014). Identification of new metabolites of bacterial transformation of indole by gas chromatography-mass spectrometry and high performance liquid chromatography. Int. J. Anal. Chem. 2014:239641. doi: 10.1155/2014/239641
Arora, P. K., Sharma, A., and Bae, H. (2015). Microbial degradation of indole and its derivatives. J. Chem. 2015:129159. doi: 10.1155/2015/129159
Baeyer, A. (1866). Ueber die reduction aromatischer verbindungen mittelst zinkstaub. Eur. J. Inorg. Chem. 140, 295–296. doi: 10.1002/jlac.18661400306
Bak, F., and Widdel, F. (1986). Anaerobic degradation of indolic compounds by sulfate-reducing enrichment cultures, and description of Desulfobacterium indolicum gen. nov., sp. nov. Arch. Microbiol. 146, 170–176. doi: 10.1007/BF00402346
Balba, M. T., and Evans, W. C. (1980). Methanogenic fermentation of the naturally occurring aromatic amino acids by a microbial consortium. Biochem. Soc. Trans. 8, 625–627. doi: 10.1042/bst0080625
Bansal, T., Alaniz, R. C., Wood, T. K., and Jayaraman, A. (2010). The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 228–233. doi: 10.1073/pnas.0906112107
Barden, T. C. (2010). “Indoles: industrial, agricultural and over-the-counter uses,” in Heterocyclic Scaffolds II:. Topics in Heterocyclic Chemistry, Vol. 26, ed G. Gribble (Berlin; Heidelberg: Springer), 31–46.
Berry, A., Dodge, T. C., Pepsin, M., and Weyler, W. (2002). Application of metabolic engineering to improve both the production and use of biotech indigo. J. Ind. Microbiol. Biotechnol. 28, 127–133. doi: 10.1038/sj/jim/7000228
Berry, D. F., Madsen, E. L., and Bollag, J. M. (1987). Conversion of indole to oxindole under methanogenic conditions. Appl. Environ. Microbiol. 53, 180–182.
Bhushan, B., Samanta, S. K., and Jain, R. K. (2000). Indigo production by naphthalene-degrading bacteria. Lett. Appl. Microbiol. 31, 5–9. doi: 10.1046/j.1472-765x.2000.00754.x
Bialy, H. (1997). Biotechnology, bioremediation, and blue genes. Nat. Biotechnol. 15:110. doi: 10.1038/nbt0297-110
Bjerregaard-Andersen, K., Sommer, T., Jensen, J. K., Jochimsen, B., Etzerodt, M., and Morth, J. P. (2014). A proton wire and water channel revealed in the crystal structure of isatin hydrolase. J. Biol. Chem. 289, 21351–21359. doi: 10.1074/jbc.M114.568824
Boyd, C., Larkin, M. J., Reid, K. A., Sharma, N. D., and Wilson, K. (1997). Metabolism of naphthalene, 1-naphthol, indene, and indole by Rhodococcus sp. strain NCIMB 12038. Appl. Environ. Microbiol. 63, 151–155.
Cheng, L., Yin, S., Chen, M., Sun, B., Hao, S., and Wang, C. (2016). Enhancing indigo production by over-expression of the styrene monooxygenase in Pseudomonas putida. Curr. Microbiol. 73, 248–254. doi: 10.1007/s00284-016-1055-3
Chimerel, C., Emery, E., Summers, D. K., Keyser, U., Gribble, F. M., and Reimann, F. (2014). Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 9, 1202–1208. doi: 10.1016/j.celrep.2014.10.032
Chimerel, C., Field, C. M., Piñero-Fernandez, S., Keyser, U. F., and Summers, D. K. (2012). Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim. Biophys. Acta 1818, 1590–1594. doi: 10.1016/j.bbamem.2012.02.022
Choi, H. S., Kim, J. K., Cho, E. H., Kim, Y. C., Kim, J. I., and Kim, S. W. (2003). A novel flavin-containing monooxygenase from Methylophaga sp. strain SK1 and its indigo synthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 306, 930–936. doi: 10.1016/S0006-291X(03)01087-8
Claus, G., and Kutzner, H. J. (1983). Degradation of indole by Alcaligenes spec. Syst. Appl. Microbial. 4, 169–180. doi: 10.1016/S0723-2020(83)80046-0
Collin, G., and Höke, H. (2000). Ullmann's Encyclopedia of Industrial Chemistry. John Wiley & Sons, 19, 75–77. doi: 10.1002/14356007.a14_167
de Sá Alves, F. R., Barreiro, E. J., and Fraga, C. A. (2009). From nature to drug discovery: the indole scaffold as a ‘privileged structure’. Mini. Rev. Med. Chem. 9, 782–793. doi: 10.2174/138955709788452649
Deslandes, B., Gariépy, C., and Houde, A. (2001). Review of microbiological and biochemical effects of skatole on animal production. Livest. Prod. Sci. 71, 193–200. doi: 10.1016/S0301-6226(01)00189-0
Doukyu, N., and Aono, R. (1997). Biodegradation of indole at high concentration by persolvent fermentation with Pseudomonas sp. ST-200. Extremophiles 1, 100–105. doi: 10.1007/s007920050021
Doukyu, N., Arai, T., and Aono, R. (1998). Effects of organic solvents on indigo formation by Pseudomonas sp. strain ST-200 grown with high levels of indole. Biosci. Biotechnol. Biochem. 62, 1075–1080.
Doukyu, N., Nakano, T., Okuyama, Y., and Aono, R. (2002). Isolation of an Acinetobacter sp. ST-550 which produces a high level of indigo in a water-organic solvent two-phase system containing high levels of indole. Appl. Microbiol. Biotechnol. 58, 543–546. doi: 10.1007/s00253-001-0919-y
Doukyu, N., Toyoda, K., and Aono, R. (2003). Indigo production by Escherichia coli carrying the phenol hydroxylase gene from Acinetobacter sp. strain ST-550 in a water-organic solvent two-phase system. Appl. Microbiol. Biotechnol. 60, 720–725. doi: 10.1007/s00253-002-1187-1
Drewlo, S., Brämer, C. O., Madkour, M., Mayer, F., and Steinbüchel, A. (2001). Cloning and expression of a Ralstonia eutropha HF39 gene mediating indigo formation in Escherichia coli. Appl. Environ. Microbiol. 67, 1964–1969. doi: 10.1128/AEM.67.4.1964-1969.2001
Ensley, B. D., Ratzkin, B. J., Osslund, T. D., Simon, M. J., Wackett, L. P., and Gibson, D. T. (1983). Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science 222, 167–169. doi: 10.1126/science.6353574
Erb, M., Veyrat, N., Robert, C. A., Xu, H., Frey, M., Ton, J., et al. (2015). Indole is an essential herbivoreinduced volatile priming signal in maize. Nat. Commun. 6:6273. doi: 10.1038/ncomms7273
Feng, L., Wu, Z., and Yu, X. (2013). Quorum sensing in water and wastewater treatment biofilms. J. Environ. Biol. 34, 437–444. Available online at: http://www.jeb.co.in/journal_issues/201304_apr13_supp/paper_20.pdf
Fischer, E., and Jourdan, F. (1883). Ueber die hydrazine der brenztraubensäure. Eur. J. Inorg. Chem. 16, 2241–2245. doi: 10.1002/cber.188301602141
Frey, M., Chomet, P., Glawischnig, E., Stettner, C., Grün, S., Winklmair, A., et al. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277, 696–699. doi: 10.1126/science.277.5326.696
Frey, M., Stettner, C., Pare, P. W., Schmelz, E. A., Tumlinson, J. H., and Gierl, A. (2000). An herbivore elicitor activates the gene for indole emission in maize. Proc. Natl. Acad. Sci. U. S. A. 97, 14801–14806. doi: 10.1073/pnas.260499897
Fujioka, M., and Wada, H. (1968). The bacterial oxidation of indole. Biochim. Biophys. Acta 158, 70–78. doi: 10.1016/0304-4165(68)90073-1
Fukuoka, K., Tanaka, K., Ozeki, Y., and Kanaly, R. A. (2015). Biotransformation of indole by Cupriavidus sp. strain KK10 proceeds through N-heterocyclic-and carbocyclic-aromatic ring cleavage and production of indigoids. Int. Biodeter. Biodegr. 97, 13–24. doi: 10.1016/j.ibiod.2014.11.007
Garbe, T. R., Kobayashi, M., and Yukawa, H. (2000). Indole-inducible proteins in bacteria suggest membrane and oxidant toxicity. Arch. Microbiol. 173, 78–82. doi: 10.1007/s002030050012
Gillam, E. M., Aguinaldo, A. M., Notley, L. M., Kim, D., Mundkowski, R. G., Volkov, A. A., et al. (1999). Formation of indigo by recombinant mammalian cytochrome P450. Biochem. Biophys. Res. Commun. 265, 469–472.
Gillam, E. M., Notley, L. M., Cai, H., DeVoss, J. J., and Guengerich, F. P. (2000). Oxidation of indole by cytochrome P450 enzymes. Biochemistry 39, 13817–13824. doi: 10.1021/bi001229u
Gray, P. H. H. (1928). The formation of indigotin from indol by soil bacteria. Proc. R. Soc. Lond. B 102, 263–280. doi: 10.1098/rspb.1928.0003
Gribble, G. W. (2010). Heterocyclic Scaffolds II: Reactions and Applications of Indoles. Heidelberg: Springer.
Gu, J. D., and Berry, D. F. (1991). Degradation of substituted indoles by an indole-degrading methanogenic consortium. Appl. Environ. Microbiol. 57, 2622–2627.
Guan, C., Ju, J., Borlee, B. R., Williamson, L. L., Shen, B., Raffa, K. F., et al. (2007). Signal mimics derived from a metagenomic analysis of the gypsy moth gut microbiota. Appl. Environ. Microbiol. 73, 3669–3676. doi: 10.1128/AEM.02617-06
Guengerich, F. P., Sorrells, J. L., Schmitt, S., Krauser, J. A., Aryal, P., and Meijer, L. (2004). Generation of new protein kinase inhibitors utilizing cytochrome P450 mutant enzymes for indigoid synthesis. J. Med. Chem. 47, 3236–3241. doi: 10.1021/jm030561b
Hales, K. E., Parker, D. B., and Cole, N. A. (2012). Potential odorous volatile organic compound emissions from feces and urine from cattle fed corn-based diets with wet distillers grains and solubles. Atmos. Environ. 60, 292–297. doi: 10.1016/j.atmosenv.2012.06.080
Han, G. H., Gim, G. H., Kim, W., Seo, S. I., and Kim, S. W. (2013). Enhanced indirubin production in recombinant Escherichia coli harboring a flavin-containing monooxygenase gene by cysteine supplementation. J. Biotechnol. 164, 179–187. doi: 10.1016/j.jbiotec.2012.08.015
Han, G. H., Shin, H. J., and Kim, S. W. (2008). Optimization of bio-indigo production by recombinant E. coli harboring fmo gene. Enzyme Microb. Technol. 42, 617–623. doi: 10.1016/j.enzmictec.2008.02.004
Han, T. H., Lee, J. H., Cho, M. H., Wood, T. K., and Lee, J. (2011). Environmental factors affecting indole production in Escherichia coli. Res. Microbiol. 162, 108–116. doi: 10.1016/j.resmic.2010.11.005
Hong, X., Zhang, X., Liu, B., Mao, Y., Liu, Y., and Zhao, L. (2010). Structural differentiation of bacterial communities in indole-degrading bioreactors under denitrifying and sulfate-reducing conditions. Res. Microbiol. 161, 687–693. doi: 10.1016/j.resmic.2010.06.010
Hsu, T. M., Welner, D. H., Russ, Z. N., Cervantes, B., Prathuri, R. L., Adams, P. D., et al. (2018). Employing a biochemical protecting group for a sustainable indigo dyeing strategy. Nat. Chem. Biol. 14, 256–261. doi: 10.1038/nchembio.2552
Hu, S., Huang, J., Mei, L., Yu, Q., Yao, S., and Jin, Z. (2010). Altering the regioselectivity of cytochrome P450 BM-3 by saturation mutagenesis for the biosynthesis of indirubin. J. Mol. Catal. B Enzym. 67, 29–35. doi: 10.1016/j.molcatb.2010.07.001
Huxtable, R. J. (2001). The mutability of blue. Mol. Interv. 1, 141–144. Available online at: http://triggered.stanford.clockss.org/ServeContent?url=http%3A%2F%2Fmolinterv.aspetjournals.org%2Fcontent%2F1%2F3%2F141.full
Ishikawa, M., Shigemori, K., and Hori, K. (2014). Application of the adhesive bacterionanofiber AtaA to a novel microbial immobilization method for the production of indigo as a model chemical. Biotechnol. Bioeng. 111, 16–24. doi: 10.1002/bit.25012
Kamath, A. V., and Vaidyanathan, C. S. (1990). “New pathway for the biodegradation of indole,” in Aspergillus niger. Appl. Environ. Microbiol. 56, 275–280.
Kim, D., Rahman, A., Sitepu, I. R., and Hashidoko, Y. (2013). Accelerated degradation of exogenous indole by Burkholderia unamae strain CK43B exposed to pyrogallol-type polyphenols. Biosci. Biotechnol. Biochem. 77, 1722–1727. doi: 10.1271/bbb.130282
Kim, J., Hong, H., Heo, A., and Park, W. (2013). Indole toxicity involves the inhibition of adenosine triphosphate production and protein folding in Pseudomonas putida. FEMS Microbiol. Lett. 343, 89–99. doi: 10.1111/1574-6968.12135
Kim, J. Y., Kim, J. K., Lee, S. O., Kim, C. K., and Lee, K. (2005). Multicomponent phenol hydroxylase-catalysed formation of hydroxyindoles and dyestuffs from indole and its derivatives. Lett. Appl. Microbiol. 41, 163–168. doi: 10.1111/j.1472-765X.2005.01734.x
Kim, J. Y., Lee, K., Kim, Y., Kim, C. K., and Lee, K. (2003). Production of dyestuffs from indole derivatives by naphthalene dioxygenase and toluene dioxygenase. Lett. Appl. Microbiol. 36, 343–348. doi: 10.1046/j.1472-765X.2003.01279.x
Kim, M., Lee, J. H., Kim, E., Choi, H., Kim, Y., and Lee, J. (2016). Isolation of indole utilizing bacteria Arthrobacter sp. and Alcaligenes sp. from livestock waste. Indian J. Microbiol. 56, 158–166. doi: 10.1007/s12088-016-0570-z
Kinney, C. A., Furlong, E. T., Zaugg, S. D., Burkhard, M. R., Werner, S. L., Cahill, J. D., et al. (2006). Survey of organic wastewater contaminants in biosolids destined for land application. Environ. Sci. Technol. 40, 7207–7215. doi: 10.1021/es0603406
Kobayashi, A., Hirakawa, H., Hirata, T., Nishino, K., and Yamaguchi, A. (2006). Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J. Bacteriol. 188, 5693–5703. doi: 10.1128/JB.00217-06
Kwon, N. R., Chae, J. C., Choi, K. Y., Yoo, M., Zylstra, G. J., Kim, Y. M., et al. (2008). Identification of functionally important amino acids in a novel indigo-producing oxygenase from Rhodococcus sp. strain T104. Appl. Microbiol. Biotechnol. 79, 417–422. doi: 10.1007/s00253-008-1445-y
Langkau, B., Vock, P., Massey, V., Fuchs, G., and Ghisla, S. (1995). 2-Aminobenzoyl-CoA monooxygenase/reductase: evidence for two distinct loci catalyzing substrate monooxygenation and hydrogenation. Eur. J. Biochem. 230, 676–685. doi: 10.1111/j.1432-1033.1995.tb20609.x
Lee, J., Attila, C., Cirillo, S. L., Cirillo, J. D., and Wood, T. K. (2009). Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2, 75–90. doi: 10.1111/j.1751-7915.2008.00061.x
Lee, J. H., Kim, Y. G., Baek, K. H., Cho, M. H., and Lee, J. (2015a). The multifaceted roles of the interspecies signaling molecule indole in Agrobacterium tumefaciens. Environ. Microbiol. 17, 1234–1244. doi: 10.1111/1462-2920.12560
Lee, J. H., and Lee, J. (2010). Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 34, 426–444. doi: 10.1111/j.1574-6976.2009.00204.x
Lee, J. H., Wood, T. K., and Lee, J. (2015b). Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 23, 707–718. doi: 10.1016/j.tim.2015.08.001
Li, G., and Young, K. D. (2013). Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 159, 402–410. doi: 10.1099/mic.0.064139-0
Li, P., Tong, L., Liu, K., Wang, Y., and Wang, Y. (2009). Indole degrading of ammonia oxidizing bacteria isolated from swine wastewater treatment system. Water Sci. Technol. 59, 2405–2410. doi: 10.2166/wst.2009.312
Lim, H. K., Chung, E. J., Kim, J. C., Choi, G. J., Jang, K. S., Chung, Y. R., et al. (2005). Characterization of a forest soil metagenome clone that confers indirubin and indigo production on Escherichia coli. Appl. Environ. Microbiol. 71, 7768–7777. doi: 10.1128/AEM.71.12.7768-7777.2005
Lin, G. H., Chen, H. P., Huang, J. H., Liu, T. T., Lin, T. K., Wang, S. J., et al. (2012). Identification and characterization of an indigo-producing oxygenase involved in indole 3-acetic acid utilization by Acinetobacter baumannii. Antonie Van Leeuwenhoek 101, 881–890. doi: 10.1007/s10482-012-9704-4
Lin, G. H., Chen, H. P., and Shu, H. Y. (2015). Detoxification of indole by an indole-induced flavoprotein oxygenase from Acinetobacter baumannii. PLoS ONE 10:e0138798. doi: 10.1371/journal.pone.0138798
Lu, Y., and Mei, L. (2007). Co-expression of P450 BM3 and glucose dehydrogenase by recombinant Escherichia coli and its application in an NADPH-dependent indigo production system. J. Ind. Microbiol. Biotechnol. 34, 247–253. doi: 10.1007/s10295-006-0193-1
Luo, Y., Zhang, R., Liu, G., Li, J., Li, M., and Zhang, C. (2010). Electricity generation from indole and microbial community analysis in the microbial fuel cell. J. Hazard. Mater. 176, 759–764. doi: 10.1016/j.jhazmat.2009.11.100
Ma, Q., Qu, Y., Tang, H., Yu, H., Ma, F., Shi, S., et al. (2012). Genome sequence of a novel indigo-producing strain, Pseudomonas monteilii QM. J. Bacteriol. 194, 4459–4460. doi: 10.1128/JB.00867-12
Ma, Q., Qu, Y., Zhang, X., Liu, Z., Li, H., and Zhang, Z. (2015b). Systematic investigation and microbial community profile of indole degradation processes in two aerobic activated sludge systems. Sci. Rep. 5:17674. doi: 10.1038/srep17674
Ma, Q., Qu, Y., Zhang, Z., Li, P., and Tang, H. (2015a). Genome sequence of an efficient indole-degrading bacterium, Cupriavidus sp. strain IDO, with potential polyhydroxyalkanoate production applications. Genome. Announc. 3, e00102–e00115. doi: 10.1128/genomeA.00102-15
Mackie, R. I., Stroot, P. G., and Varel, V. H. (1998). Biochemical identification and biological origin of key odor components in livestock waste. J. Anim. Sci. 76, 1331–1342.
Madsen, E. L., and Bollag, J. M. (1989). Pathway of indole metabolism by a denitrifying microbial community. Arch. Microbiol. 151, 71–76.
Madsen, E. L., Francis, A. J., and Bollag, J. M. (1988). Environmental factors affecting indole metabolism under anaerobic conditions. Appl. Environ. Microbiol. 54, 74–78.
Mao, Y., Zhang, X., Xia, X., Zhong, H., and Zhao, L. (2010). Versatile aromatic compound-degrading capacity and microdiversity of Thauera strains isolated from a coking wastewater treatment bioreactor. J. Ind. Microbiol. Biotechnol. 37, 927–934. doi: 10.1007/s10295-010-0740-7
McClay, K., Boss, C., Keresztes, I., and Steffan, R. J. (2005). Mutations of toluene-4-monooxygenase that alter regiospecificity of indole oxidation and lead to production of novel indigoid pigments. Appl. Environ. Microbiol. 71, 5476–5483. doi: 10.1128/AEM.71.9.5476-5483.2005
Mercadal, J. P., Isaac, P., Siñeriz, F., and Ferrero, M. A. (2010). Indigo production by Pseudomonas sp. J26, a marine naphthalene-degrading strain. J. Basic. Microbiol. 50, 290–293. doi: 10.1002/jobm.200900276
Mermod, N., Harayama, S., and Timmis, K. N. (1986). New route to bacterial production of indigo. Nat. Biotechnol. 4, 321–324. doi: 10.1038/nbt0486-321
Meyer, A., Würsten, M., Schmid, A., Kohler, H. P. E., and Witholt, B. (2002). Hydroxylation of indole by laboratory-evolved 2-hydroxybiphenyl 3-monooxygenase. J. Biol. Chem. 277, 34161–34167. doi: 10.1074/jbc.M205621200
Moreno-Ruiz, E., Hernáez, M. J., Martínez-Pérez, O., and Santero, E. (2003). Identification and functional characterization of Sphingomonas macrogolitabida strain TFA genes involved in the first two steps of the tetralin catabolic pathway. J. Bacteriol. 185, 2026–2030. doi: 10.1128/JB.185.6.2026-2030.2003
Mueller, R. S., Beyhan, S., Saini, S. G., Saini, S. G., Yildiz, F. H., and Bartlett, D. H. (2009). Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 191, 3504–3516. doi: 10.1128/JB.01240-08
Müller, H. E. (1986). Production and degradation of indole by gram-negative bacteria. Zbl. Bakt. Hyg. A 261, 1–11. doi: 10.1016/S0176-6724(86)80058-X
Murdock, D., Ensley, B. D., Serdar, C., and Thalen, M. (1993). Construction of metabolic operons catalyzing the de novo biosynthesis of indigo in Escherichia coli. Nat. Biotechnol. 11, 381–386. doi: 10.1038/nbt0393-381
Nakamura, K., Martin, M. V., and Guengerich, F. P. (2001). Random mutagenesis of human cytochrome P450 2A6 and screening with indole oxidation products. Arch. Biochem. Biophys. 395, 25–31. doi: 10.1006/abbi.2001.2569
Nordlund, I., Powlowski, J., and Shingler, V. (1990). Complete nucleotide sequence and polypeptide analysis of multi component phenol hydroxylase from Pseudomonas sp. strain CF600. J. Bacteriol. 172, 6826–6833.
O'Connor, K. E., Dobson, A. D., and Hartmans, S. (1997). Indigo formation by microorganisms expressing styrene monooxygenase activity. Appl. Environ. Microbiol. 63, 4287–4291.
O'Connor, K. E., and Hartmans, S. (1998). Indigo formation by aromatic hydrocarbon-degrading bacteria. Biotechnol. Lett. 20, 219–223.
Osawa, T., Namiki, M., Suzuki, K., and Mitsuoka, T. (1983). Mutagen formation by intestinal bacteria. Mutat. Res. 122, 299–304. doi: 10.1016/0165-7992(83)90010-6
Oshima, T., Kawai, S., and Egami, F. (1965). Oxidation of indole to indigotin by Pseudomonas indoloxidans. J. Biochem. 58, 259–263.
Pathak, H., and Madamwar, D. (2010). Biosynthesis of indigo dye by newly isolated naphthalene-degrading strain Pseudomonas sp. HOB1 and its application in dyeing cotton fabric. Appl. Biochem. Biotechnol. 160, 1616–1626. doi: 10.1007/s12010-009-8638-4
Philp, J. C., Ritchie, R. J., and Allan, J. E. (2013). Biobased chemicals, the convergence of green chemistry with industrial biotechnology. Trends Biotechnol. 31, 219–222. doi: 10.1016/j.tibtech.2012.12.007
Qu, Y., Ma, Q., Liu, Z., Wang, W., Tang, H., Zhou, J., et al. (2017). Unveiling the biotransformation mechanism of indole in a Cupriavidus sp. strain. Mol. Microbiol. 106, 905–918. doi: 10.1111/mmi.13852
Qu, Y., Pi, W., Ma, F., Zhou, J., and Zhang, X. (2010). Influence and optimization of growth substrates on indigo formation by a novel isolate Acinetobacter sp. PP-2. Bioresour. Technol. 101, 4527–4532. doi: 10.1016/j.biortech.2010.01.033
Qu, Y., Shen, E., Ma, Q., Zhang, Z., Liu, Z., Shen, W., et al. (2015). Biodegradation of indole by a newly isolated Cupriavidus sp. SHE. J. Environ. Sci. 34, 126–132. doi: 10.1016/j.jes.2015.01.023
Qu, Y., Shi, S., Zhou, H., Ma, Q., Li, X., Zhang, X., et al. (2012a). Characterization of a novel phenol hydroxylase in indoles biotranformation from a strain Arthrobacter sp. W1. PLoS ONE 7:e44313. doi: 10.1371/journal.pone.0044313
Qu, Y., Xu, B., Zhang, X., Ma, Q., Zhou, H., Kong, C., et al. (2013). Biotransformation of indole by whole cells of recombinant biphenyl dioxygenase and biphenyl-2, 3-dihydrodiol-2, 3-dehydrogenase. Biochem. Eng. J. 72, 54–60. doi: 10.1016/j.bej.2012.12.021
Qu, Y., Zhang, X., Ma, Q., Ma, F., Zhang, Q., and Li, X. (2012b). Indigo biosynthesis by Comamonas sp. MQ. Biotechnol. Lett. 34, 353–357. doi: 10.1007/s10529-011-0778-2
Radwanski, E. R., and Last, R. L. (1995). Tryptophan biosynthesis and metabolism, biochemical and molecular genetics. Plant Cell 7, 921–934. doi: 10.1105/tpc.7.7.921
Raistrick, H., and Clark, A. B. (1921). Studies on the cycloclastic power of bacteria, Part II. A quantitative study of the aerobic decomposition of tryptophan and tyrosine by bacteria. Biochem. J. 15, 76–82.
Regar, R. K., Gaur, V. K., Mishra, G., Jadhao, S., Kamthan, M., and Manickam, N. (2016). Draft genome sequence of Alcaligenes faecalis strain IITR89, an indole-oxidizing bacterium. Genome. Announc. 4, e00067–e00016. doi: 10.1128/genomeA.00067-16
Rui, L., Reardon, K. F., and Wood, T. K. (2005). Protein engineering of toluene ortho-monooxygenase of Burkholderia cepacia G4 for regiospecific hydroxylation of indole to form various indigoid compounds. Appl. Microbiol Biotechnol. 66, 422–429. doi: 10.1007/s00253-004-1698-z
Sadauskas, M., Vaitekunas, J., Gasparaviciute, R., and Meškys, R. (2017). Indole biodegradation in Acinetobacter sp. strain O153, genetic and biochemical characterization. Appl. Environ. Microbiol. 83, e01453–e01417. doi: 10.1128/AEM.01453-17
Sakamoto, Y., Uchida, M., and Ichihara, K. (1953). The bacterial decomposition of indole. I. Studies on its metabolic pathway by successive adaptation. Med. J. Osaka Univ. 3, 477–486.
Shanker, R., and Bollag, J. M. (1990). Transformation of indole by methanogenic and sulfate-reducing microorganisms isolated from digested sludge. Microb. Ecol. 20, 171–183. doi: 10.1007/BF02543875
Sharma, V., Kumar, P., and Pathak, D. (2010). Biological importance of the indole nucleus in recent years: a comprehensive review. J. Heterocyclic Chem. 47, 491–502. doi: 10.1002/jhet.349
Singh, A., Singh Chauhan, N., Thulasiram, H. V., Taneja, V., and Sharma, R. (2010). Identification of two flavin monooxygenases from an effluent treatment plant sludge metagenomic library. Bioresour. Technol. 101, 8481–8484. doi: 10.1016/j.biortech.2010.06.025
Smith, T. (1897). A modification of the method for determining the production of indol by bacteria. J. Exp. Med. 2, 543–547.
Sonowal, R., Swimm, A., Sahoo, A., Luo, L., Matsunaga, Y., Wu, Z., et al. (2017). Indoles from commensal bacteria extend healthspan. Proc. Natl. Acad. Sci. U.S.A. 114, E7506–E7515. doi: 10.1073/pnas.1706464114
Supniewski, J. (1923). Transformation of carbon compounds by Bacillus pyocyaneus. Compt. Rend. Soc. Biol. 89, 1377–1379.
Takao, T., Kitatani, F., Watanab, N., Yagi, A., and Sakata, K. (1994). A simple screening method for antioxidants and isolation of several antioxidants produced by marine bacteria from fish and shellfish. Biosci. Biotechnol. Biochem. 5, 1780–1783. doi: 10.1271/bbb.58.1780
Urlacher, V. B., and Girhard, M. (2012). Cytochrome P450 monooxygenases, an update on perspectives for synthetic application. Trends Biotechnol. 30, 26–36. doi: 10.1016/j.tibtech.2011.06.012
Van Hellemond, E. W., Janssen, D. B., and Fraaije, M. W. (2007). Discovery of a novel styrene monooxygenase originating from the metagenome. Appl. Environ. Microbiol. 73, 5832–5839. doi: 10.1128/AEM.02708-06
Wang, J., Zhang, X., Fan, J., Zhang, Z., Ma, Q., and Peng, X. (2015). Indigoids biosynthesis from indole by two phenol-degrading strains, Pseudomonas sp. PI1 and Acinetobacter sp. PI2. Appl. Biochem. Biotechnol. 176, 1263–1276. doi: 10.1007/s12010-015-1644-9
Wang, Y. T., Suidan, M. T., and Pfeffer, J. T. (1984). Anaerobic biodegradation of indole to methane. Appl. Environ. Microbiol. 48, 1058–1060.
Watanabe, T., and Snell, E. E. (1972). Reversibility of the tryptophanase reaction, synthesis of tryptophan from indole, pyruvate, and ammonia. Proc. Natl. Acad. Sci. U.S.A. 69, 1086–1090. doi: 10.1073/pnas.69.5.1086
Yamamoto, Y., Sato, Y., Ebina, T., Yokoyama, C., Takahasi, S., Mito, Y., et al. (1991). Separation of high purity indole from coal tar by high pressure crystallization. Fuel 70, 565–566. doi: 10.1016/0016-2361(91)90039-D
Yang, Z., Zhou, J., Xu, Y., Zhang, Y., Luo, H., Chang, K., et al. (2017). Analysis of the metabolites of indole degraded by an isolated Acinetobacter pittii L1. Biomed. Res. Int. 2017:2564363. doi: 10.1155/2017/2564363
Yeon, K. M., Cheong, W. S., Oh, H. S., Lee, W. N., Hwang, B. K., Lee, C. H., et al. (2008). Quorum sensing, a new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ. Sci. Technol. 43, 380–385. doi: 10.1021/es8019275
Yin, B., Gu, J. D., and Wan, N. (2005). Degradation of indole by enrichment culture and Pseudomonas aeruginosa Gs isolated from mangrove sediment. Int. Biodeterior. Biodegrad. 56, 243–248. doi: 10.1016/j.ibiod.2005.10.001
Zhang, X., Qu, Y., Ma, Q., Zhang, Z., Li, D., Wang, J., et al. (2015). Illumina MiSeq sequencing reveals diverse microbial communities of activated sludge systems stimulated by different aromatics for indigo biosynthesis from indole. PLoS ONE 10:e0125732. doi: 10.1371/journal.pone.0125732
Zhang, X., Qu, Y., Ma, Q., Zhou, H., Li, X., Kong, C., et al. (2013). Cloning and expression of naphthalene dioxygenase genes from Comamonas sp. MQ for indigoids production. Process Biochem. 48, 581–587. doi: 10.1016/j.procbio.2013.02.008
Keywords: indole, signaling molecule, biodegradation, biotransformation, indigo, functional study
Citation: Ma Q, Zhang X and Qu Y (2018) Biodegradation and Biotransformation of Indole: Advances and Perspectives. Front. Microbiol. 9:2625. doi: 10.3389/fmicb.2018.02625
Received: 28 July 2018; Accepted: 15 October 2018;
Published: 01 November 2018.
Edited by:
Shaohua Chen, South China Agricultural University, ChinaReviewed by:
Willem J. H. Van Berkel, Wageningen University & Research, NetherlandsCopyright © 2018 Ma, Zhang and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiao Ma, eGlhb21hMDU1NkBkbG11LmVkdS5jbg==
Yuanyuan Qu, cXl5QGRsdXQuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.