
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 October 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02413
This article is part of the Research Topic New Knowledge of Food Microbiology in Asia View all 48 articles
 Junyan Liu1,2†
Junyan Liu1,2† Ling Yang3†
Ling Yang3† Yuchao Hou1
Yuchao Hou1 Thanapop Soteyome4
Thanapop Soteyome4 Bingbing Zeng5
Bingbing Zeng5 Jianyu Su1,6
Jianyu Su1,6 Lin Li1,6
Lin Li1,6 Bing Li1,6
Bing Li1,6 Dingqiang Chen7
Dingqiang Chen7 Yanyan Li8
Yanyan Li8 Aiwu Wu9*
Aiwu Wu9* Mark E. Shirtliff10
Mark E. Shirtliff10 Janette M. Harro10
Janette M. Harro10 Zhenbo Xu1,6,10,11*
Zhenbo Xu1,6,10,11* Brian M. Peters2
Brian M. Peters2Staphylococcus aureus is one of the representative foodborne pathogens which forms biofilm. Antibiotics are widely applied in livestock husbandry to maintain animal health and productivity, thus contribute to the dissemination of antimicrobial resistant livestock and human pathogens, and pose a significant public health threat. Effect of antibiotic pressure on S. aureus biofilm formation, as well as the mechanism, remains unclear. In this study, the regulatory mechanism of low concentration of ampicillin on S. aureus biofilm formation was elucidated. The viability and biomass of biofilm with and without 1/4 MIC ampicillin treatment for 8 h were determined by XTT and crystal violet straining assays, respectively. Transcriptomics analysis on ampicillin-induced and non-ampicillin-induced biofilms were performed by RNA-sequencing, differentially expressed genes identification and annotation, GO functional and KEGG pathway enrichment. The viability and biomass of ampicillin-induced biofilm showed dramatical increase compared to the non-ampicillin-induced biofilm. A total of 530 differentially expressed genes (DEGs) with 167 and 363 genes showing up- and down-regulation, respectively, were obtained. Upon GO functional enrichment, 183, 252, and 21 specific GO terms in biological process, molecular function and cellular component were identified, respectively. Eight KEGG pathways including “Microbial metabolism in diverse environments”, “S. aureus infection”, and “Monobactam biosynthesis” were significantly enriched. In addition, “beta-lactam resistance” pathway was also highly enriched. In ampicillin-induced biofilm, the significant up-regulation of genes encoding multidrug resistance efflux pump AbcA, penicillin binding proteins PBP1, PBP1a/2, and PBP3, and antimicrobial resistance proteins VraF, VraG, Dlt, and Aur indicated the positive response of S. aureus to ampicillin. The up-regulation of genes encoding surface proteins ClfB, IsdA, and SasG and genes (cap5B and cap5C) which promote the adhesion of S. aureus in ampicillin induced biofilm might explain the enhanced biofilm viability and biomass.
Animal protein demand for human consumption is rising worldwide. Daily animal protein intake in Asia increased from 7 grams/capita/day in 1960 to 25 grams/capita/day in 20131 (Guo et al., 2000). To meet the requirement, antibiotics are widely applied in livestock husbandry to maintain animal health and productivity, thus contribute to the dissemination of antimicrobial resistant livestock and human pathogens, and pose significant public health implications (Van Boeckel et al., 2015). The average annual consumption of antibiotics worldwide was estimated to be 172 mg/kg, 148 mg/kg, and 45 mg/kg for pigs, chicken, and cattle, respectively (Van Boeckel et al., 2015). In the United States, antimicrobial use in food animals is estimated to account for approximately 80% of the nation’s annual antimicrobial consumption (Food and Drug Administration, 2010). In 2010, China was the largest antimicrobial consumer for livestock and the consumption of antibiotics in China was estimated at 162,000 tons in 2013, with 52% applied in animals (Van Boeckel et al., 2015; Zhang et al., 2015). This widespread use of antibiotics in livestock contributes to the emergence of antimicrobial resistant pathogenic bacteria and lead to significant public health threats. Repeated exposure to low doses of antimicrobial agents builds optimum conditions for the emergence and dissemination of antimicrobial resistant bacteria in animals (You and Silbergeld, 2014; Lin et al., 2018). Animal origin antimicrobial resistant bacteria can be transmitted to agricultural workers by direct contact and to other humans through food products and environment (Price et al., 2005; Graham et al., 2009; Smith et al., 2013; Xu et al., 2017).
Staphylococcus aureus is one of the most common human and animal pathogens and one of the first strains characterized to be resistant to antimicrobials. Penicillin was first used in animal production in the late 1940s, and S. aureus with resistance to penicillin were observed in 1948 (Huttner et al., 2013). Nowadays, most of the S. aureus strains show resistance to penicillin by producing beta-lactams. In the past decade, S. aureus biofilms which are major cause for concern in multiple infections and are associated with chronic infections, have brought increased recognition (Conlon, 2014). Both in vitro and in vivo studies have showed biofilms pose challenges to medical and industrial due to their increased tolerance of antimicrobials and disinfectants (Bjarnsholt et al., 2013; Neopane et al., 2018). Biofilm development was classified into three stages: initial attachment, biofilm maturation, and dispersal and later proposed to include five stages: attachment, multiplication, exodus, maturation, and dispersal (O’Toole et al., 2000; Moormeier et al., 2014; Miao et al., 2017). Biofilm growth plays a major role during bacterial infection by the defense against several clearance mechanisms (Stewart and Costerton, 2001). Biofilm formation of S. aureus is also linked with irons, virulence factors, surface proteins and accessory gene regulator (agr) quorum-sensing system whose expression depends on environmental conditions (Cucarella et al., 2001; Yarwood et al., 2004; Poupel et al., 2018; Swarupa et al., 2018). In addition, biofilm cells display enhanced resistance to antibiotics (de la Fuente-Nunez et al., 2013; Lin et al., 2018). It has been reported that S. aureus isolated from hospitalized patients have high degree of biofilm-forming ability with high tendency to exhibit antimicrobial resistance, multidrug resistance and methicillin resistance (Neopane et al., 2018). Subinhibitory concentration of methicillin has been shown to lead to dramatic increase in biofilm formation of S. aureus which was dependent on autolysis activity linked to atl (Kaplan et al., 2012; Ranieri et al., 2018).
However, the effect of low-concentration of ampicillin pressure appeared in food products on S. aureus biofilm formation, as well as the mechanism, remains unclear. In this study, the regulatory mechanism of low concentration of ampicillin as a common antibiotic applied in livestock husbandry on S. aureus biofilm formation was elucidated by transcriptomics analysis.
Staphylococcus aureus strain FAHGMU10071 was isolated from a patient in the First Affiliated Hospital of Guangzhou Medical University in Guangzhou, China and was maintained as a glycerol stock stored at -80°C. A small amount of stock was spread onto Tryptone Soy agar (TSA) and incubated at 37°C for 24 h to obtain isolated colonies. A single colony was transferred to 2 mL of Tryptone Soy broth (TSB) and incubated at 37°C with shaking at 200 rpm overnight prior to further experiments. The minimal inhibitory concentration (MIC) was measured by broth microdilution method.
40 mL of overnight S. aureus culture was inoculated into 2 mL fresh TSB and incubated at 37°C with shaking at 200 rpm for 3 h to reach logarithmic growth phase. The logarithmic growth culture was then washed twice in sterile PBS and diluted to 107 cells/mL in TSB. 5 mL of the culture was added into a flat-bottomed 6-well plate (Trasadingen, Switzerland) and incubated at 37°C with gentle shaking (10–20 rpm) for 8 h to form biofilm. The biofilm viability and biomass were determined by XTT and Crystal Violet Staining assays, respectively (Christensen et al., 1985; Silva et al., 2008). The experiment was performed in triplicate and biofilm samples were collected at 8 h to be control groups (C1, C2, and C3) For ampicillin-induced groups (A1, A2, and A3), 1/4 MIC of ampicillin was added into the flat-bottomed 6-well plate at the meantime and biofilm samples were collected at 8 h.
Total RNA of S. aureus biofilm samples (A1, A2, A3, C1, C2, and C3) were extracted using the TRizol reagent (Sigma-Aldrich, United States) and Bacterial Total RNA Extraction kit (Sigma-Aldrich, United States) based on the manufacturer’s instruction (Liu et al., 2016a,b). The quality and quantity of the RNA samples were determined by Ultramicro spectrophotometer K5500 (Beijing Kaiao Technology Development Co., Ltd., China) and Agilent 2200 TapStation (Agilent Technologies, United States). To remove trace DNA and ribosomal RNAs, RNase-free DNase I (Ambion Inc., United States) and MICROBExpressTM kit (Ambion Inc., United States) were applied, respectively (Liu et al., 2016a). The mRNA was fragmented ultrasonically and was then reverse transcribed to cDNA (Liu et al., 2016a). RNA-seq libraries construction and RNA sequencing were performed in biological triplicate by RIBOBIO Biotechnology Company (Guangzhou, China) using the Illumina Paired End Sample Prep kit and the Illumina Hiseq 2500 platform, respectively (Liu et al., 2016a).
Raw reads were generated from image data and stored as FASTQ format. Raw data were filtered to remove adaptor contaminated and low quality sequences and obtain clean reads. Clean reads were quality examined by FastQC v.0.10.12 and aligned to reference genome of S. aureus NCTC 8325 (NCBI Reference Sequence: NC_007795) using TopHat (Trapnell et al., 2009). No more than 2 bases mismatch and read gap were allowed in the alignment. Gene coverage was calculated by the percentage of genes covered by reads and gene functional annotation was performed through ANNOVAR (Wang et al., 2010). Differentially expressed genes (DEGs) were identified using DEGseq panormalage (Anders, 2010; Anders and Huber, 2012) based on negative binomial distributions. The gene expression level was measured based on Reads Per Kilobase of transcript per Million mapped reads (RPKM) values and P values adjusted using the edgeR panormalage (Mortazavi et al., 2008; Robinson et al., 2010). Genes with an adjusted |log2(fold change)| > 1 and P-value < 0.05 were identified as DEGs. Kyoto encyclopedia of genes and genomes (KEGG) pathway and Gene Ontology (GO) annotation of DEGs were performed by kobas based on KEGG database (Kanehisa and Goto, 2000) and through GO database (Ashburner et al., 2000), respectively. The significantly enriched KEGG pathways and GO terms were identified by P-value < 0.05 in Fisher Exact Test and P-value < 0.01 in Hypergeometric Distribution, respectively, and adjusted by false discovery rates (FDR) (Storey, 2002; Rivals et al., 2007).
In order to validate the RNA-seq data, RT-PCR was performed to quantify the mRNA transcripts of 10 selected DEGs using the Light Cylcler 480 (Roche, Switzerland) according to the manufacturer’s instructions. Each RT-PCR reaction was performed in a final volume of 25 μL. The thermal cycling profile was as follows: 42°C for 60 min and 72°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 30 s, 72°C for 30 s, and a final extension of 68°C for 10 min. Negative control samples containing sterile water were also included. The cycle threshold values (CT) were determined and the relative fold differences were calculated by the 2-ΔΔCT method54 using 16S rRNA as the reference gene. Three independent experiments were run in triplicate.
Under the treatment of 1/4 MIC ampicillin for 8 h, the biofilm of S. aureus strain FAHGMU10071 showed dramatical increase in biomass and viability compared to the no treatment control (Figure 1; Xu et al., 2018). To gain further insight into the regulatory mechanism of low concentration antibiotic on S. aureus biofilm formation, RNA-seq analyses were performed on three biological replicates of S. aureus biofilm grown at 37°C for 8 h with (A1, A2, and A3) and without (C1, C2, and C3) 1/4 MIC of ampicillin. The cDNA libraries of the 6 S. aureus biofilm samples were constructed, sequenced and generated with a total of 29,684,588 to 40,756,932 reads which were mapped to the reference genome of S. aureus NCTC 8325 (NCBI Reference Sequence: NC_007795) (Supplementary Table S1). The overall gene expression levels of the three biological replicates of each group showed high similarity with each other, illustrating that the RNA-seq data were available for transcriptome analysis (NCBI SRA accession number: SRP154796). Gene expression levels determined by the average RPKM values demonstrated that 2840 total genes were expressed in ampicillin-induced and control groups.
Identified by adjusted |log2(fold change)| > 1 and P-value < 0.05 (Figure 2; Tan et al., 2015), 530 DEGs with 167 and 363 genes showing up- and down-regulation, respectively, were obtained (Supplementary Figure S1 and Supplementary Table S2). Considering the most differentially expressed genes, 6 of the significantly up-regulated genes showed |log2(fold change)| > 2 and 10 of the significantly down-regulated genes showed |log2(fold change)| > 4. The 6 up-regulated genes encode cation transporter E1-E2 family ATPase, argininosuccinate synthase, 6-phospho-beta-galactosidase, argininosuccinate lyase, PTS system lactose-specific transporter subunit IIBC, and riboflavin biosynthesis protein, respectively. The 10 down-regulated genes encode carbamate kinase, arginine/ornithine antiporter, 2-isopropylmalate synthase, ornithine carbamoyltransferase, phage head protein, 23S Ribosomal RNA, and 4 hypothetical proteins, respectively.
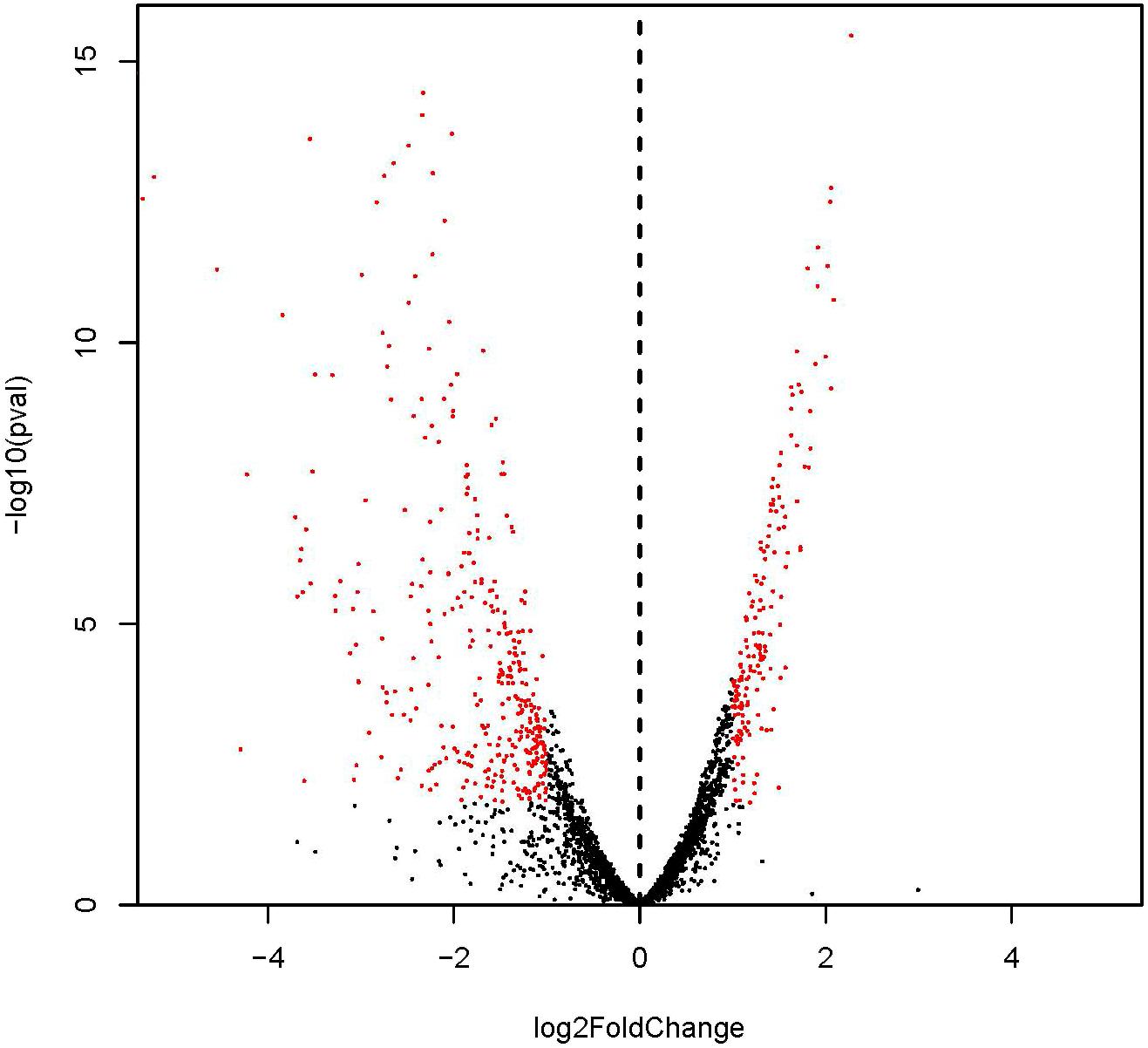
FIGURE 2. Differential expression level of ampicillin-induced (A1, A2, and A3) and control (C1, C2, and C3) groups identified by | log2(fold change)| > 1 and adjust P-value < 0.05.
Considering the S. aureus biofilm formation, DEGs in ampicillin-induced biofilm include some key genes encoding surface proteins, virulence factors, adhesins, proteases, and capsular polysaccharide proteins (Wang and Wood, 2011). Genes cap5B and cap5C mediating the capsular polysaccharide biosynthesis were significantly up-regulated. The S. sureus strain FAHGMU10071 might promote biofilm formation by the synthesis of capsular polysaccharides under low concentration of ampicillin. However, the expressions of genes encoding proteases ClpB and ClpC which are required for stress tolerance, intracellular replication and biofilm formation (Frees et al., 2004) and genes encoding adhesins SdrD and Fib significantly decreased. A newly reported virulence factor, spdC, which controls histidine kinase activity and the master regulator of cysteine metabolism, cymR, which plays a role in biofilm formation in S. aureus showed no significantly differential expression (Soutourina et al., 2009; Poupel et al., 2018).
To further understand the function of the DEGs underlying the effect of low concentration of ampicillin on S. aureus biofilm, GO enrichment analysis was performed with the 530 DEGs. Based on sequence homology, DEGs were assigned to one or more GO terms and categorized into 535, 536, and 277 secondary level GO terms in the three main categories (biological process, molecular function, and cellular component) of the GO function, respectively. Upon GO functional enrichment, 183, 252, and 21 specific GO terms in biological process, molecular function and cellular component were identified, respectively (Supplementary Table S3). Significantly enriched GO terms composed of the three specific categories were illustrated in Figure 3. Nine GO terms were enriched in the category of biological process, including “translation,” “regulation of transcription, DNA-templated,” “threonine biosynthetic process,” “cell division,” “carbohydrate transmembrane transport,” “carbohydrate metabolic process,” “phosphorelay signal transduction system,” “phosphoenolpyruvate-dependent sugar phosphotransferase system,” and “lysine biosynthetic process via diaminopimelate”. In the category of molecular function, “DNA binding,” “rRNA binding,” “structural constituent of ribosome,” “RNA binding,” “ATP binding,” “nickel cation binding,” and “metal ion binding” were significantly enriched. Only 2 GO terms “integral component of membrane” and “extracellular region” were enriched in the category of cellular component.
To identify the involved pathways, DEGs were mapped to the KEGG database, followed by KEGG pathway enrichment analysis (Supplementary Table S4). KEGG pathways “Glycine, serine and threonine metabolism,” “Phosphotransferase system (PTS),” “Arginine biosynthesis,” “Microbial metabolism in diverse environments,” “Staphylococcus aureus infection,” “Monobactam biosynthesis,” “Nitrogen metabolism,” and “Lysine biosynthesis” were significantly enriched (Figure 4).
Threonine which is an essential amino acid cannot be synthesized by animals, is derived from aspartate in bacteria and plants. In the “Glycine, serine and threonine metabolism” pathway (Figure 5), aspartate kinase (EC: 2.7.2.4), aspartate semialdehyde dehydrogenase (EC: 1.2.1.11), homoserine dehydrogenase (EC: 1.1.1.3), homoserine kinase (EC: 2.7.1.39), and threonine synthase (EC: 4.2.3.1) are functional during the threonine biosynthesis (aspartate - homoserine - threonine) (Rees and Hay, 1995; Gaupp et al., 2010). L-aspartate is phosphorylated by aspartate kinase to produce L-4-aspartylphosphate, which is reduced to L-aspartate-4-semialdehyde by aspartate semialdehyde dehydrogenase. The semialdehyde is further reduced by homoserine dehydrogenase to the non-protein amino acid homoserine. Homoserine kinase phosphorylates homoserine to produce O-phospho-L-homoserine, which is converted to threonine by threonine synthase (Rees and Hay, 1995). All the genes encoding the key functional enzymes in the threonine biosynthesis process showed significant down-regulation in the ampicillin-induced biofilm, indicating the attenuated activation of threonine biosynthesis pathway during the formation of S. aureus biofilm with antibiotic treatment. The functional enzymes including choline dehydrogenase (EC: 1.1.99.1) and betaine aldehyde dehydrogenase (EC: 1.2.1.8) during the betaine biosynthesis (choline - betaine) in the “Glycine, serine and threonine metabolism” pathway (Figure 5) were also significantly down-regulated in the ampicillin-induced biofilm.
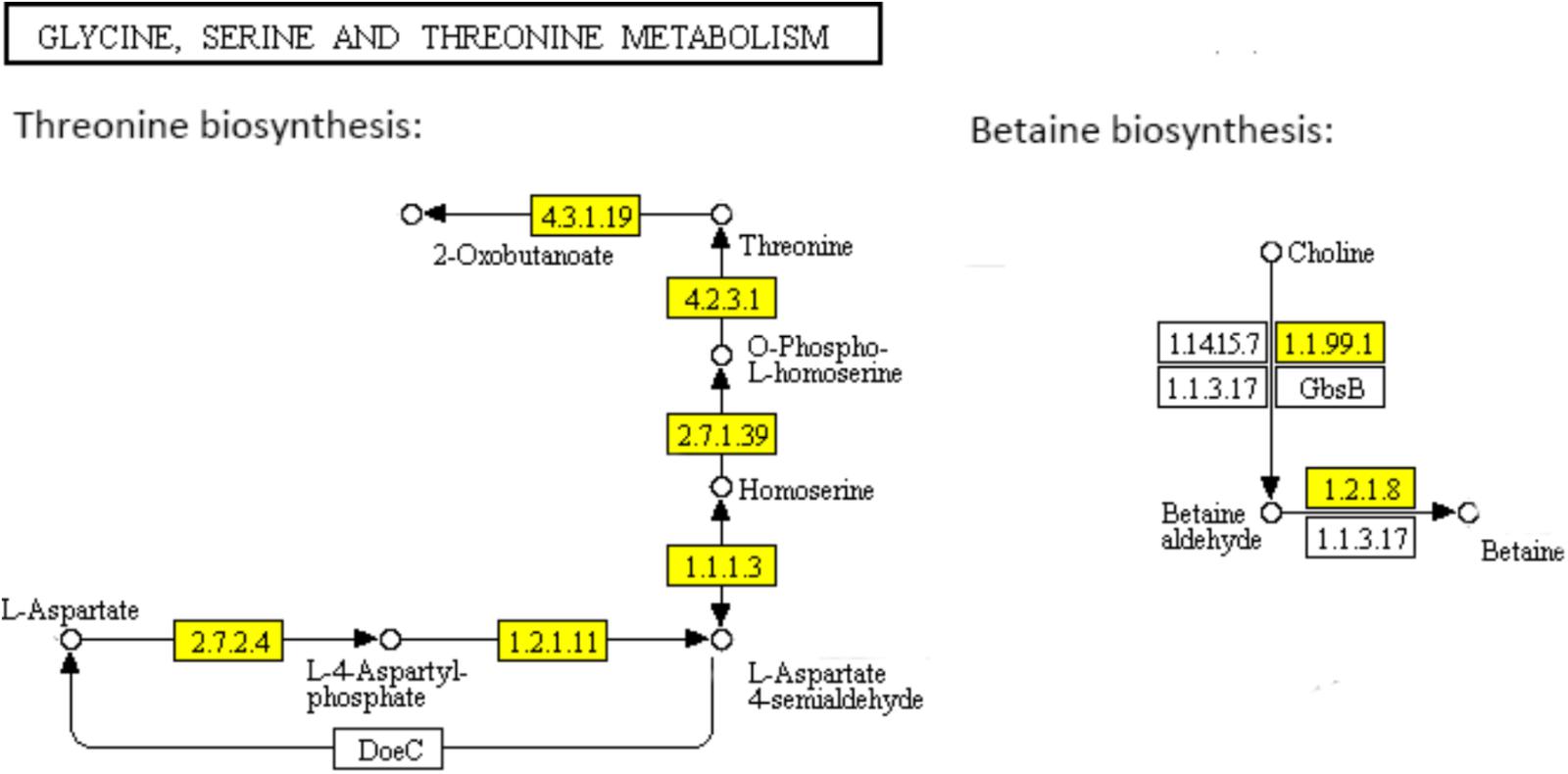
FIGURE 5. Significantly enriched KEGG pathway “Glycine, serine and threonine metabolism” (Yellow highlights represent down-regulation and green highlights represent up-regulation) (Kanehisa and Goto, 2000; Kanehisa et al., 2017).
The PTS has been characterized as a major bacterial mechanism for uptake of carbohydrates, especially hexoses, hexitols, and disaccharides, which are referred to PTS carbohydrate (Kotrba et al., 2001). The uptake of carbohydrates and their conversion are catalyzed into the respective phosphoesters by PTS during transport process (Tchieu et al., 2001). Four successive phosphoryl transfers appear in the PTS (Tchieu et al., 2001). Using phosphoenolpyruvate (PEP) as a substrate, initial autophosphorylation of enzyme I (EI) is followed by transfer of the phosphoryl group from EI to histidine phosphocarrier protein (HPr) (Tchieu et al., 2001). The self-phosphoryl transfer from HPr, after which the phosphoryl group is transferred to histidine or cysteine residues of EIIB, are catalyzed by enzyme II (EII) A (Tchieu et al., 2001). The specific saccharide is transported through the membrane-bound EIIC and is phosphorylated by the appropriate saccharide-specific EIIB (Tchieu et al., 2001). In the L-ascorbate family, the genes encoding EIIABC (UlaA, UlaB, and UlaC) which catalyzed, transported, and phosphorylated L-ascorbate to L-arsorbate 6-phosphate, were significantly down-regulated. It suggested the lower uptake of L-ascorbate in ampicillin-induced biofilm (Figure 6). The up-regulation of genes encoding EIIABC in the Lactose family (LacE and LacF) and Fructose family (MtlA, FruA and FruB) revealed the diminishing uptake of lactose and fructose in ampicillin-induced biofilm (Figure 6).
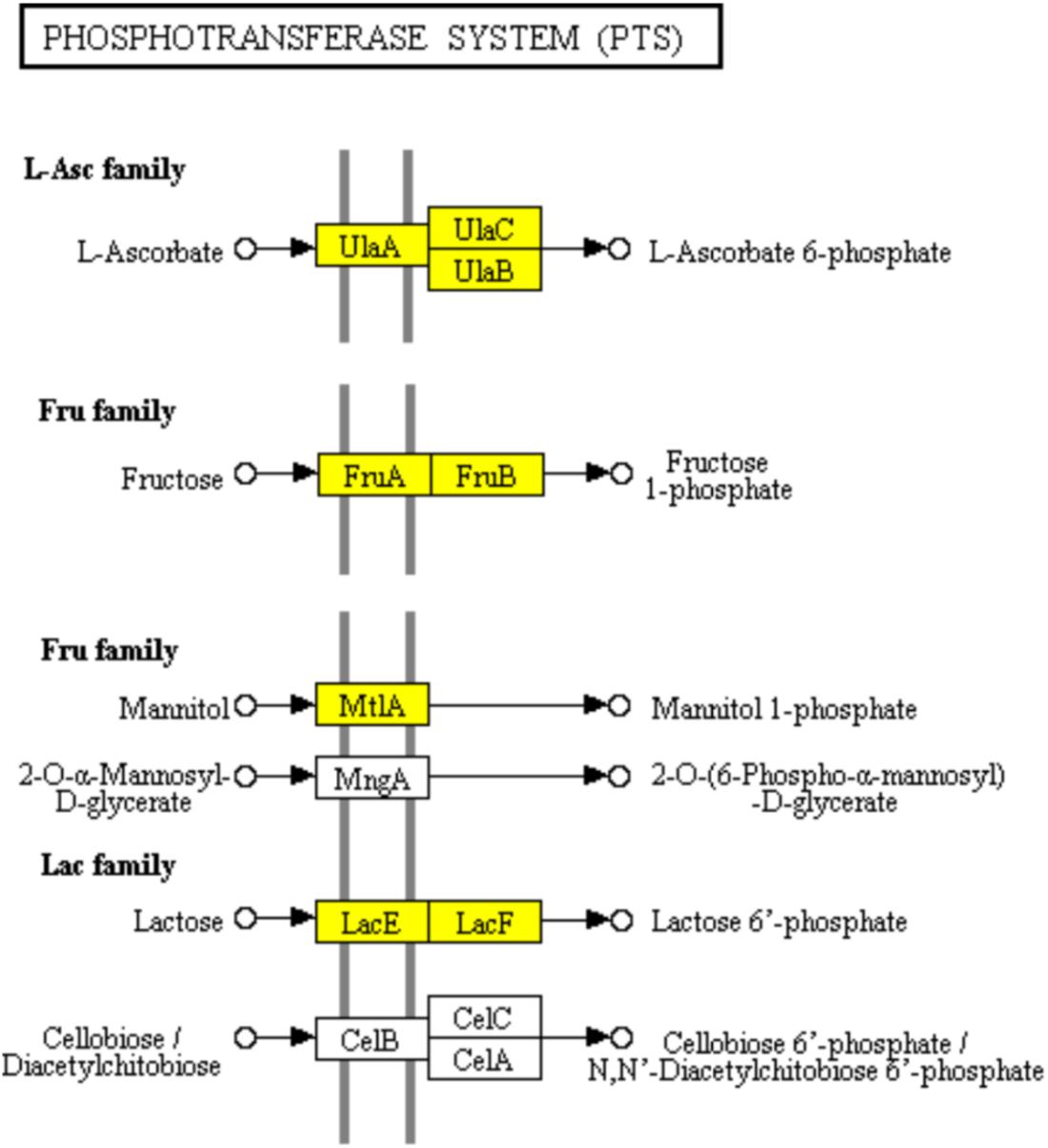
FIGURE 6. Significantly enriched KEGG pathway “Phosphotransferase system (PTS)” (Yellow highlights represent down-regulation and green highlights represent up-regulation) (Kanehisa and Goto, 2000; Kanehisa et al., 2017).
In the urea cycle of “Arginine biosynthesis” pathway, genes encoding major enzymes including ornithine carbamoyltransferase (EC: 2.1.3.3), argininosuccinate synthase (EC: 6.3.4.5), argininosuccinate lyase (EC: 4.3.2.1), arginase (EC: 3.5.3.1), and arginine deiminase (EC: 3.5.3.6) showed significant down-regulation (Figure 7). However, the genes encoding bifunctional ornithine acetyltransferase/N-acetylglutamate synthase argJ (EC: 2.3.1.1) acetylglutamate kinase (EC: 1.2.1.38), and N-acetyl-gamma-glutamyl-phosphate reductase argC (EC: 2.7.2.8) which catalyze glutamate to N-acetylglutamate semialdehyde were significantly up-regulated (Figure 7A). The diaminopimelic acid pathway (DAP) in “Lysine biosynthesis” pathway has been reported to be related to arginine metabolism (Velasco et al., 2002). Similar to the genes involved in “Arginine biosynthesis” pathway, the genes encoding enzymes [homoserine dehydrogenase (EC: 1.1.1.3), aspartate kinase (EC: 2.7.2.4, EC: 2.7.2.4), aspartate semialdehyde dehydrogenase (EC: 1.2.1.11), 4-hydroxy-tetrahydrodipicolinate synthase (EC: 4.3.3.7), and 4-hydroxy-tetrahydrodipicolinate reductase (EC: 1.17.1.8)] involved in DAP were significantly down-regulated (Figure 7B).
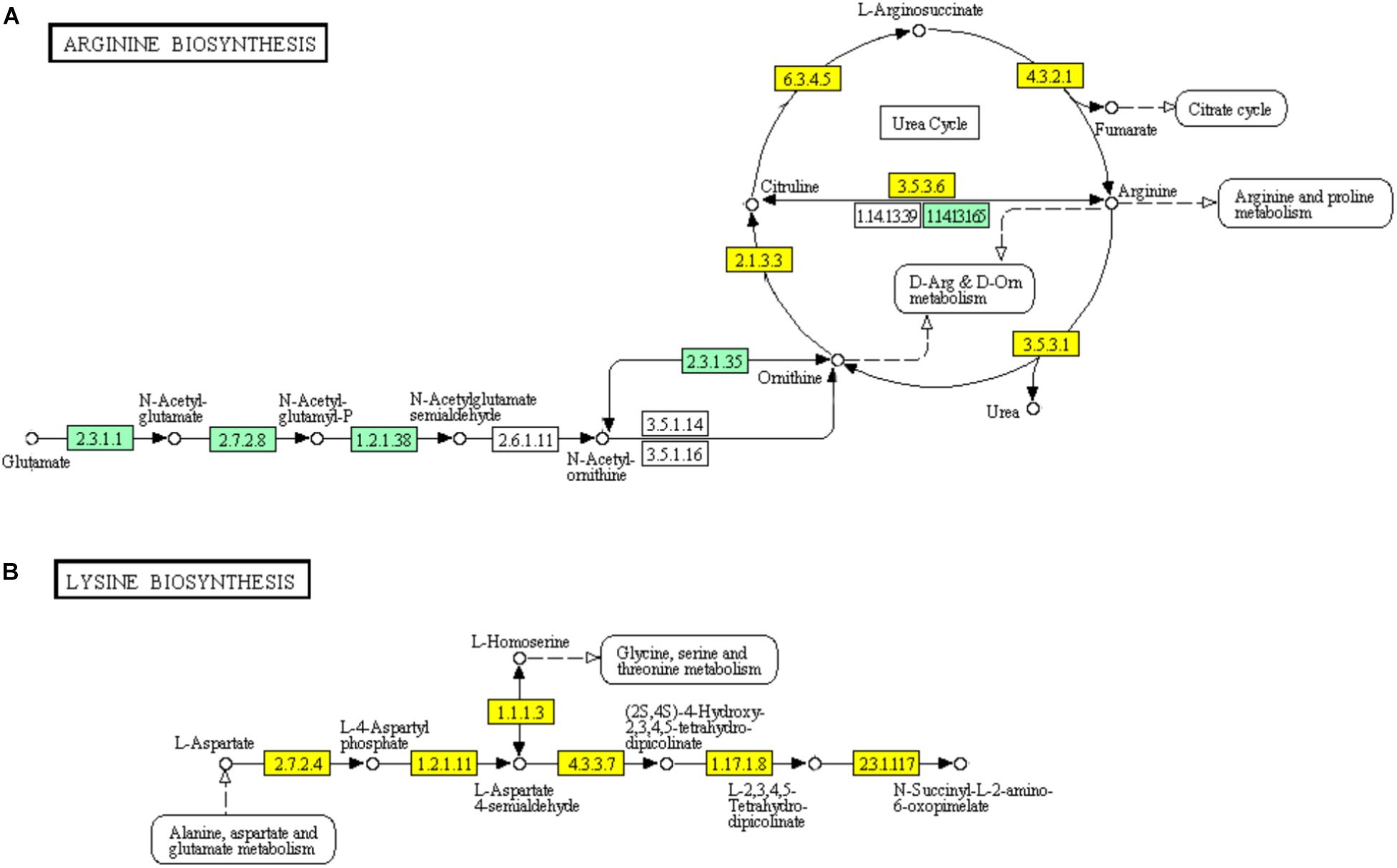
FIGURE 7. Significantly enriched KEGG pathways “Arginine biosynthesis” (A) and “Lysine biosynthesis” (B) (Yellow highlights represent down-regulation and green highlights represent up-regulation) (Kanehisa and Goto, 2000; Kanehisa et al., 2017).
The core nitrogen cycle in “Nitrogen metabolism” pathway involves four reduction pathways and two oxidation pathways. In ampicillin induced S. aureus biofilm, 6 genes encoding nitrate reductase NarGHIJ, NirBD, and NxrAB involved in the dissimilatory nitrate reduction, denitrification, and nitrification were significantly down-regulated (Figure 8). It has been reported that nitrogen metabolism is involved in antibiotic resistance (Borriello et al., 2004; Yan et al., 2013). Thus, the significant enrichment of “Nitrogen metabolism” pathway in ampiciliin induced biofilm group indicated the response of S. aureus biofilm to ampicillin. Besides, a total of 44 genes involved in “Microbial metabolism in diverse environments” were significantly up- or down-regulated, demonstrating the response of S. aureus biofilm to ampicillin.
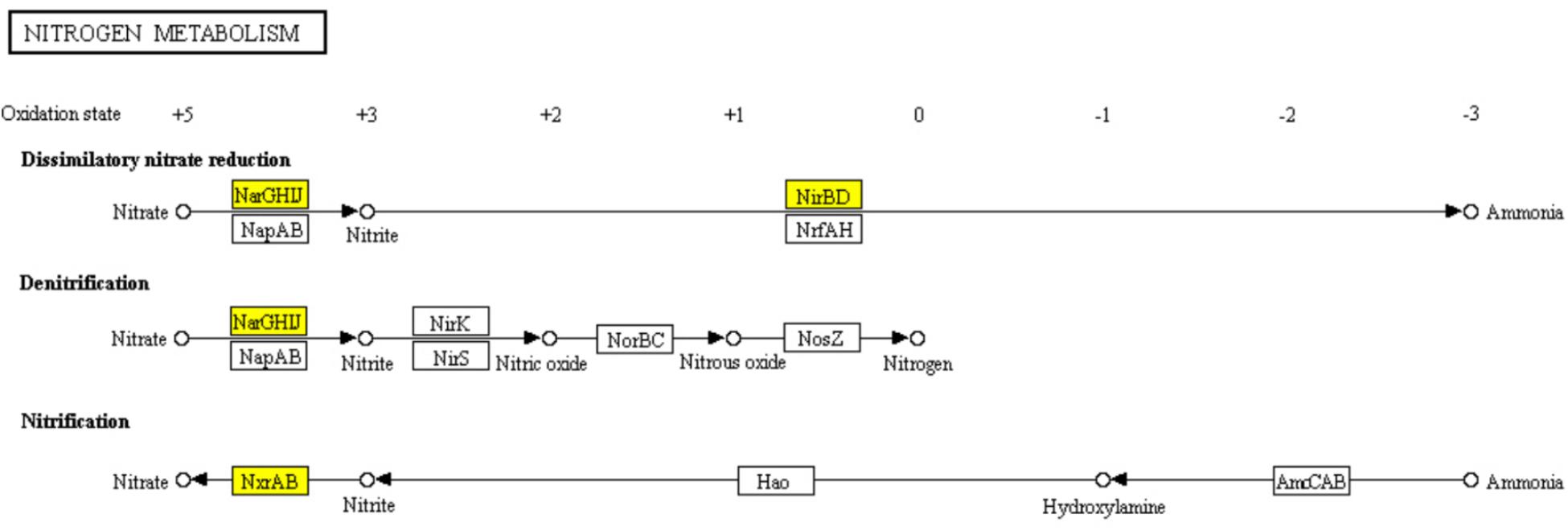
FIGURE 8. Significantly enriched KEGG pathway “Nitrogen metabolism” (Yellow highlights represent down-regulation and green highlights represent up-regulation) (Kanehisa and Goto, 2000; Kanehisa et al., 2017).
Infections including superficial skin infections, food poisoning infections, and life-threatening infections can be caused by S. aureus, which diverts the effectiveness of the immune system in several ways: modulating the cationic antimicrobial peptides sensitivity by increasing the cytoplasmic membrane positive net charge, superantigens expression which prevent common immune response development or cause emetic response when ingested, and secreting immune modulating proteins which inhibit complement activation and neutrophil chemotaxis or lysis (Foster, 2009; Kanehisa et al., 2017). The ability of S. aureus to colonize the host and to form biofilm is likely to be partially determined by its adhesion ability (Foulston et al., 2014; Scherr et al., 2014). Surface proteins including clumping factor B (ClfB), iron-regulated surface determinant (IsdA), SdrC, SdrD, and S. aureus surface protein (SasG) have been reported to be able to promote adhesion in vitro (Foster, 2009). SasG-expressing strains of S. aureus were reported to form biofilm independently of the polysaccharide intercellular adhesin (PIA) (Corrigan et al., 2007). SasG is able to mask S. aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) binding to their ligands and to improve biofilm formation (Corrigan et al., 2007). The up-regulation of genes encoding surface proteins ClfB, IsdA, and SasG which promote the adhesion of S. aureus in ampicillin induced biofilm explained the enhanced biofilm viability and biomass (Figure 9). Clumping factor A (ClfA) which is the major fibrinogen-binding protein on the surface of cells from the stationary phase of growth, has been reported to act as a protective antigen and a virulence determinant (Josefsson et al., 2001). The up-regulation of clfA gene implied the protection of S. aureus encountering antibiotic stress by ClfA (Figure 9). Obviously, the significantly up-regulated genes encoding VraF, VraG, Dlt, Aur proteins sharing antimicrobial resistance activities indicated the positively response of S. aureus to ampicillin (Figure 9; Foster, 2009).
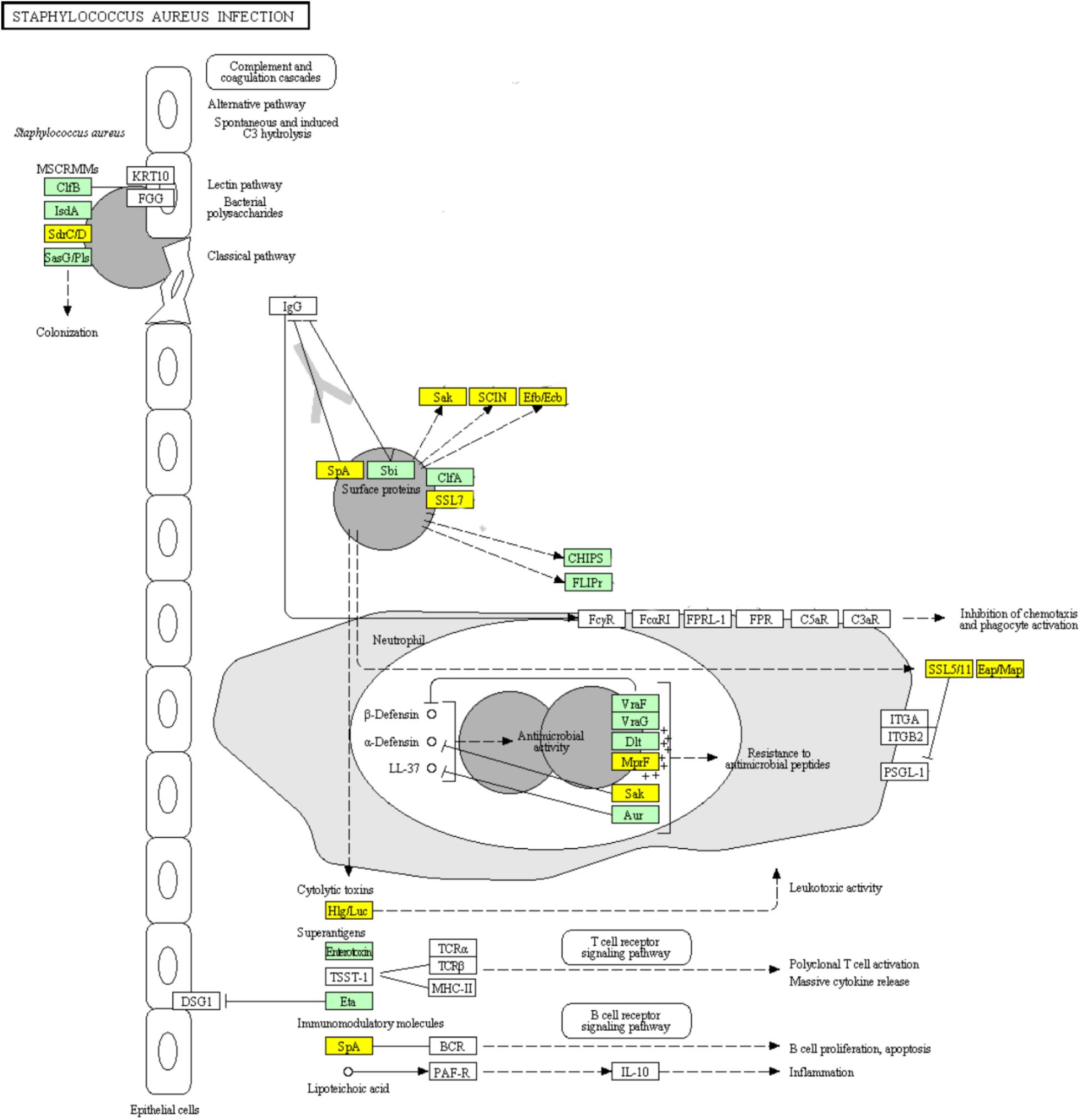
FIGURE 9. Significantly enriched KEGG pathway “Staphylococcus aureus infection” (Yellow highlights represent down-regulation and green highlights represent up-regulation) (Kanehisa and Goto, 2000; Kanehisa et al., 2017).
Concerning the toxins involved in this pathway, genes encoding cytolytic toxins Hlg/Luc and immunomodulatory module Spa showed significant down-regulation, while genes encoding superantigens enterotoxin and exfoliative toxin Eta were significantly up-regulated during ampicillin-induced biofilm formation (Figure 9). The genes encoding superantigenic toxic shock syndrome toxin-1 (TSST-1) did not show significantly differential expression.
Monobactams are beta-lactam antibiotics containing a monocyclic beta-lactam nucleus, that is different from penicillin and cephalosporin core structures with another fused ring (Hamed et al., 2013). The genes encoding aspartate kinase (EC: 2.7.2.4), aspartate semialdehyde dehydrogenase (EC: 1.2.1.11), 4-hydroxy-tetrahydrodipicolinate synthase (EC: 4.3.3.7), and 4-hydroxy-tetrahydrodipicolinate reductase (EC:1.17.1.8) which involved in part of the “Monobactam biosynthesis” pathway showed significant down-regulation (Figure 10A).
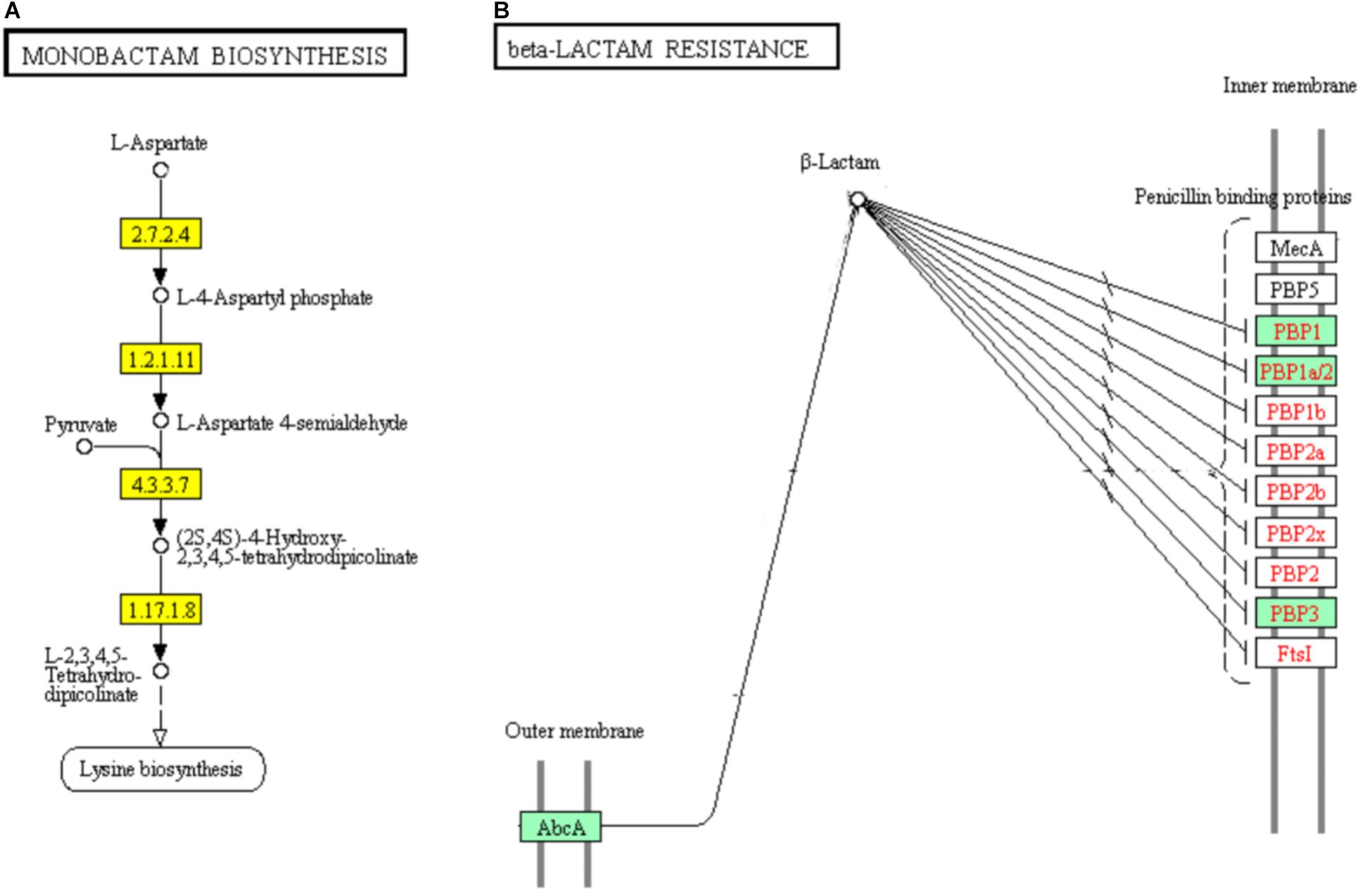
FIGURE 10. Significantly enriched KEGG pathways “Monobactam biosynthesis” (A) and “beta-lactam resistance” (B) (Yellow highlights represent down-regulation and green highlights represent up-regulation) (Kanehisa and Goto, 2000; Kanehisa et al., 2017).
In addition, 5 of the 9 DEGs were involved in “beta-lactam resistance” pathway and showed significantly up-regulated in ampicillin-induced group compared to control group (Figure 10B). The up-regulation of “beta-lactam resistance” related genes indicated the strain was response to beta-lactam (ampicillin) stress. The beta-lactam antibiotics expand their effect in bacterial cell walls by interfering with the structural crosslinking of peptidoglycans. Bacterial beta-lactam antibiotic resistance can be acquired by strategies including the production of inactivated beta-lactamases, changing the beta-lactam targets for penicillin-binding proteins (PBPs), reducing the transport of beta-lactams into the periplasmic space through porin changes, and exclusion of beta-lactams via efflux pumps (Zapun et al., 2008; Kanehisa et al., 2017). In ampicillin-induced biofilm, genes encoding multidrug resistance efflux pump AbcA and penicillin binding proteins PBP1, PBP1a/2, and PBP3 were significantly up-regulated. The low concentration ampicillin resistance mechanism of S. aureus strain FAHGMU10071 might be the activation of multidrug resistance efflux pump AbcA and penicillin binding proteins PBP1, PBP2, and PBP3. PBPs which are enzymes involved in the last stages of peptidoglycan biosynthesis, have been intense studied especially considering the role of some important pathogens including S. sureus in the beta-lactams resistance. Four native PBPs (PBP1 to PBP4) and an methicillin-resistant S. aureus (MRSA) specific PBP2a have been identified in S. aureus. PBPs 1, 2 and 3 of S. aureus are essential for viability and are the targets of beta-lactam antibiotics (Waxman and Strominger, 1983). PBP1 of S. aureus has also been reported to be essential for growth and cell division (Pereira et al., 2007).
Ten genes with different expression profiles were selected to validate the results of RNA-seq. The 10 genes were selected based on three criteria: (i) Gene function. The 10 genes were DEGs from different significantly enriched GO terms, COG categories and KEGG pathways. (ii) Expression level. Among the 10 genes, 5 were up-regulated and the others down-regulated. (iii) Gene position in the genome. The selected 10 genes were located at different positions in the genome. The mRNA levels of the 10 selected genes determined by qRT-PCR were consistent with those from the RNA-Seq analysis.
In this study, the regulatory mechanism of low concentration of ampicillin on S. aureus biofilm formation was elucidated. The viability and biomass of ampicillin-induced biofilm showed dramatical increase compared to the non-ampicillin-induced biofilm. A total of 530 DEGs with 167 and 363 genes showing up- and down-regulation, respectively, were obtained by RNA-sequencing. Upon GO functional enrichment, 183, 252, and 21 specific GO terms in biological process, molecular function and cellular component were identified, respectively. Eight KEGG pathways including “Microbial metabolism in diverse environments,” “Staphylococcus aureus infection,” and “Monobactam biosynthesis” were significantly enriched. In addition, “beta-lactam resistance” pathway was also highly enriched. In ampicillin-induced biofilm, the significant up-regulation of genes encoding multidrug resistance efflux pump AbcA, penicillin binding proteins PBP1, PBP1a/2, and PBP3, and antimicrobial resistance proteins VraF, VraG, Dlt, and Aur indicated the positively response of S. aureus to ampicillin. The up-regulation of genes encoding surface proteins ClfB, IsdA, and SasG and genes (cap5B and cap5C) which promote the adhesion of S. aureus in ampicillin induced biofilm might explain the enhanced biofilm viability and biomass.
ZX and AW conceived of the study, participated in its design and coordination. YH, LY, and DC carried out the strains collection and samples preparation. LL and BL conducted the RNA extraction, library construction and RNA-seq. JL and BP performed the DEGs identification and annotation. JS, TS, JH, and MS carried out the bioinformatics analyses. JL, BZ, and YL performed the pathway analyses. All authors reviewed and approved the final manuscript.
This work was supported by the National Key Research and Development Program of China (2016YFD04012021), the Scientific and Technological Program of Guangzhou City (201510010241), Guangdong Special Support Program (2016TQ03N682), Pearl River S&T Nova Program of Guangzhou (201710010061), Science and Technology Planning Project of Guangdong Province (2017A050501007), National Outstanding Doctoral Dissertation Funding (201459), the Fundamental Research Funds for the Central Universities (2017ZD092), the 111 Project (B17018), and the Natural Science Foundation of Guangdong Province (Nos. 2018A030310170 and 2018A030313279).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02413/full#supplementary-material
FIGURE S1 | The expression level of differentially expressed genes.
TABLE S1 | Overview of the RNA-seq statistics.
TABLE S2 | Differentially expressed genes of ampicillin-induced (A1, A2, and A3) and control (C1, C2, and C3) groups.
TABLE S3 | GO enrichment analysis (Biological Process) of differentially expressed genes in ampicillin-induced (A1, A2, and A3) group compared to control (C1, C2, and C3).
TABLE S4 | KEGG pathway enrichment analysis of differentially expressed genes in ampicillin-induced (A1, A2, and A3) group compared to control (C1, C2, and C3) group.
Anders, S. (2010). Analysing RNA-seq data with the DESeq package. Mol. Biol. 2010, 1–17. doi: 10.7717/peerj.576
Anders, S., and Huber, W. (2012). Differential Expression of RNA-Seq Data at the Gene Level-the DESeq Package. Heidelberg: EMBL.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bjarnsholt, T., Alhede, M., Alhede, M., Eickhardt-Sørensen, S. R., Moser, C., Kühl, M., et al. (2013). The in vivo biofilm. Trends Microbiol. 21, 466–474. doi: 10.1016/j.tim.2013.06.002
Borriello, G., Werner, E., Roe, F., Kim, A. M., Ehrlich, G. D., and Stewart, P. S. (2004). Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 48, 2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004
Christensen, G. D., Simpson, W. A., Younger, J. J., Baddour, L. M., Barrett, F. F., Melton, D. M., et al. (1985). Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22, 996–1006.
Conlon, B. P. (2014). Staphylococcus aureus chronic and relapsing infections: evidence of a role for persister cells: an investigation of persister cells, their formation and their role in S. aureus disease. Bioessays 36, 991–996. doi: 10.1002/bies.201400080
Corrigan, R. M., Rigby, D., Handley, P., and Foster, T. J. (2007). The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153, 2435–2446. doi: 10.1099/mic.0.2007/006676-0
Cucarella, C., Solano, C., Valle, J., Amorena, B., Lasa, I., and Penadés, J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001
de la Fuente-Nunez, C., Reffuveille, F., Fernandez, L., and Hancock, R. E. (2013). Bacterial biofilm development as amulticellular adaptation: antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 16, 580–589. doi: 10.1016/j.mib.2013.06.013
Food and Drug Administration (2010)). CVM Updates - CVM Reports on Antimicrobials Sold or Distributed for Food-Producing Animals. Available at: www.fda.gov/AnimalVeterinary/NewsEvents/CVMUpdates/ucm236143.htm
Foster, T. J. (2009). Colonization and infection of the human host by staphylococci: adhesion, survival and immune evasion. Vet. Dermatol. 20, 456–470. doi: 10.1111/j.1365-3164.2009.00825.x
Foulston, L., Elsholz, A. K., De Francesco, A. S., and Losick, R. (2014). The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. mBio 5:e01667-14. doi: 10.1128/mBio.01667-14
Frees, D., Chastanet, A., Qazi, S., Sørensen, K., Hill, P., Msadek, T., et al. (2004). Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54, 1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x
Gaupp, R., Schlag, S., Liebeke, M., Lalk, M., and Götz, F. (2010). Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms. J. Bacteriol. 12010, 2385–2394. doi: 10.1128/JB.01472-09
Graham, J. P., Evans, S. L., Price, L. B., and Silbergeld, E. K. (2009). Fate of antimicrobial-resistant enterococci and staphylococci and resistance determinants in stored poultry litter. Environ. Res. 109, 682–689. doi: 10.1016/j.envres.2009.05.005
Guo, X., Mroz, T. A., Popkin, B. M., and Zhai, F. (2000). Structural change in the impact of income on food consumption in China, 1989-1993. Econ. Dev. Cult. Change 48, 737–760. doi: 10.1086/452475
Hamed, R. B., Gomez-Castellanos, J. R., Henry, L., Ducho, C., McDonough, M. A., and Schofield, C. J. (2013). The enzymes of beta-lactam biosynthesis. Nat. Prod. Rep. 30, 21–107. doi: 10.1039/C2NP20065A
Huttner, A., Harbarth, S., Carlet, J., Cosgrove, S., Goossens, H., Holmes, A., et al. (2013). Antimicrobial resistance: a global view from the 2013 World Healthcare-Associated Infections Forum. Antimicrob. Resist. Infect. Control 2:31. doi: 10.1186/2047-2994-2-31
Josefsson, E., Hartford, O., O’Brien, L., Patti, J. M., and Foster, T. (2001). Protection against experimental Staphylococcus aureus arthritis by vaccination with clumping factor A, a novel virulence determinant. J. Infect. Dis. 184, 1572–1580. doi: 10.1086/324430
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. doi: 10.1093/nar/gkw1092
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kaplan, J. B., Izano, E. A., Gopal, P., Karwacki, M. T., Kim, S., Bose, J. L., et al. (2012). Low levels of β-lactam antibiotics nduce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 3:e00198-12. doi: 10.1128/mBio.00198-12
Kotrba, P., Inui, M., and Yukawa, H. (2001). Bacterial phosphotransferase system (PTS) in carbohydrate uptake and control of carbon metabolism. J. Biosci. Bioeng. 92, 502–517. doi: 10.1016/S1389-1723(01)80308-X
Lin, J., Chen, X., Chen, C., Hu, J., Zhou, C., Cai, X., et al. (2018). Durably antibacterial and bacterially antiadhesive cotton fabrics coated by cationic fluorinated polymers. ACS Appl. Mater. Interfaces 10, 6124–6136. doi: 10.1021/acsami.7b16235
Liu, J., Deng, Y., Peters, B. M., Li, L., Li, B., Chen, L., et al. (2016a). Transcriptomic analysis on the formation of the viable putative non-culturable state of beer-spoilage Lactobacillus acetotolerans. Sci. Rep. 6:36753. doi: 10.1038/srep36753
Liu, J., Zhou, R., Peters, B. M., Li, B., Lin, C., Chuang, T., et al. (2016b). Viable but non-culturable State and toxin gene expression of enterohemorrhagic Escherichia coli O157 under cryopreservation. Res. Microbiol. 101, 56–67. doi: 10.1016/j.resmic.2016.11.002
Miao, J., Liang, Y., Chen, L., Wang, W., Wang, J., Li, B., et al. (2017). Formation and development of Staphylococcus biofilm: with focus on food safety. J. Food Saf. 7:e12358. doi: 10.1128/mBio.01157-16
Moormeier, D. E., Bose, J. L., Horswill, A. R., and Bayles, K. W. (2014). Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio 5:e01341-14. doi: 10.1128/mBio.01341-14
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNASeq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Neopane, P., Nepal, H. P., Shrestha, R., Uehara, O., and Abiko, Y. (2018). In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int. J. Gen. Med. 11, 25–32. doi: 10.2147/IJGM.S153268
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
Pereira, S. F., Henriques, A. O., Pinho, M. G., de Lencastre, H., and Tomasz, A. (2007). Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 189, 3525–3531. doi: 10.1128/JB.00044-07
Poupel, O., Proux, C., Jagla, B., Msadek, T., and Dubrac, S. (2018). SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog. 14:e1006917. doi: 10.1371/journal.ppat.1006917
Price, L. B., Johnson, E., Vailes, R., and Silbergeld, E. (2005). Fluoroquinolone-resistant campylobacter isolates from conventional and antibiotic-free chicken products. Environ. Health Perspect. 113, 557–560. doi: 10.1289/ehp.7647
Ranieri, M. R. M., Whitchurch, C. B., and Burrows, L. L. (2018). Mechanisms of biofilm stimulation by subinhibitory concentrations of antimicrobials. Curr. Opin. Microbiol. 45, 164–169. doi: 10.1016/j.mib.2018.07.006
Rees, W. D., and Hay, S. M. (1995). The biosynthesis of threonine by mammalian cells: expression of a complete bacterial biosynthetic pathway in an animal cell. Biochem. J. 309, 999–1007. doi: 10.1042/bj3090999
Rivals, I., Personnaz, L., Taing, L., and Potier, M. C. (2007). Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23, 401–407.
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor panormalage for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Scherr, T. D., Heim, C. E., Morrison, J. M., and Kielian, T. (2014). Hiding in plain sight: interplay between staphylococcal biofilms and host immunity. Front. Immunol. 5:37. doi: 10.3389/fimmu.2014.00037
Silva, W. J., Seneviratne, J., Parahitiyawa, N., Rosa, E. A., Samaranayake, L. P., and Del Bel Cury, A. A. (2008). Improvement of XTT assay performance for studies involving Candida albicans biofilms. Braz. Dent. J. 19, 364–369. doi: 10.1590/S0103-64402008000400014
Smith, T. C., Gebreyes, W. A., Abley, M. J., Harper, A. L., Forshey, B. M., Male, M. J., et al. (2013). Methicillin-resistant Staphylococcus aureus in pigs and farm workers on conventional and antibiotic-free swine farms in the USA. PLoS One 8:e63704. doi: 10.1371/journal.pone.0063704
Soutourina, O., Poupel, O., Coppée, J. Y., Danchin, A., Msadek, T., and Martin-Verstraete, I. (2009). CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol. Microbiol. 73, 194–211. doi: 10.1111/j.1365-2958.2009.06760.x
Stewart, P. S., and Costerton, J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/S0140-6736(01)05321-1
Storey, J. D. (2002). A direct approach to false discovery rates. J. R. Stat. Soc. B 64, 479–498. doi: 10.1111/1467-9868.00346
Swarupa, V., Chaudhury, A., and Sarma, P. V. G. K. (2018). Iron enhances the peptidyl deformylase activity and biofilm formation in Staphylococcus aureus. 3Biotech 8:32. doi: 10.1007/s13205-017-1050-9
Tan, X., Qin, N., Wu, C., Sheng, J., Yang, R., Zheng, B., et al. (2015). Transcriptome analysis of the biofilm formed by methicillin-susceptible Staphylococcus aureus. Sci. Rep. 5:11997. doi: 10.1038/srep11997
Tchieu, J. H., Norris, V., Edwards, J. S., and Saier, M. H. Jr. (2001). The complete phosphotranferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3, 329–346.
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U.S.A. 112, 5649–5654. doi: 10.1073/pnas.1503141112
Velasco, A. M., Leguina, J. I., and Lazcano, A. (2002). Molecular evolution of the lysine biosynthetic pathways. J. Mol. Evol. 55, 445–459. doi: 10.1007/s00239-002-2340-2
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. doi: 10.1093/nar/gkq603
Wang, X., and Wood, T. K. (2011). Toxin/antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microb. 77, 5577–5583. doi: 10.1128/AEM.05068-11
Waxman, D. J., and Strominger, J. L. (1983). Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu. Rev. Biochem. 52, 825–869. doi: 10.1146/annurev.bi.52.070183.004141
Xu, Z., Hou, Y., and Shirtliff, M. E. (2018). “Transcriptomic study on Staphylococcus biofilm under low concentration of antibiotics,” in Proceedings of the 28th European Congress of Clinical Microbiology and Infectious Diseases, Madrid.
Xu, Z., Xie, J., Peters, B. M., Li, B., Li, L., Yu, G., et al. (2017). Longitudinal surveillance on antibiogram of important gram-positive pathogens in Southern China, 2001 to 2015. Microb. Pathog. 103, 80–86. doi: 10.1016/j.micpath.2016.11.013
Yan, C., Dinh, Q. T., Chevreuil, M., Garnier, J., Roose-Amsaleg, C., Labadie, P., et al. (2013). The effect of environmental and therapeutic concentrations of antibiotics on nitrate reduction rates in river sediment. Water Res. 47, 3654–3662. doi: 10.1016/j.watres.2013.04.025
Yarwood, J. M., Bartels, D. J., Volper, E. M., and Greenberg, E. P. (2004). Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004
You, Y., and Silbergeld, E. K. (2014). Learning from agriculture: understanding low-dose antimicrobials as drivers of resistome expansion. Front. Microbiol. 5:284. doi: 10.3389/fmicb.2014.00284
Zapun, A., Contreras-Martel, C., and Vernet, T. (2008). Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32, 361–385. doi: 10.1111/j.1574-6976.2007.00095.x
Keywords: transcriptomics, RNA-sequencing, Staphylococcus aureus, biofilm formation, ampicillin
Citation: Liu J, Yang L, Hou Y, Soteyome T, Zeng B, Su J, Li L, Li B, Chen D, Li Y, Wu A, Shirtliff ME, Harro JM, Xu Z and Peters BM (2018) Transcriptomics Study on Staphylococcus aureus Biofilm Under Low Concentration of Ampicillin. Front. Microbiol. 9:2413. doi: 10.3389/fmicb.2018.02413
Received: 30 May 2018; Accepted: 20 September 2018;
Published: 30 October 2018.
Edited by:
Liang Xue, Guangdong Institute of Microbiology (CAS), ChinaReviewed by:
Chunlei Shi, Shanghai Jiao Tong University, ChinaCopyright © 2018 Liu, Yang, Hou, Soteyome, Zeng, Su, Li, Li, Chen, Li, Wu, Shirtliff, Harro, Xu and Peters. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenbo Xu, emhlbmJvLnh1QGhvdG1haWwuY29t Aiwu Wu, YWl3d3U2NkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.