
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 28 September 2018
Sec. Microbiotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02308
 Gaofeng Ni1*
Gaofeng Ni1* Sebastian Canizales1†
Sebastian Canizales1† Elias Broman1
Elias Broman1 Domenico Simone1
Domenico Simone1 Viraja R. Palwai1
Viraja R. Palwai1 Daniel Lundin1
Daniel Lundin1 Margarita Lopez-Fernandez1†
Margarita Lopez-Fernandez1† Tom Sleutels2
Tom Sleutels2 Mark Dopson1
Mark Dopson1Thiocyanate is a toxic compound produced by the mining and metallurgy industries that needs to be remediated prior to its release into the environment. If the industry is situated at high altitudes or near the poles, economic factors require a low temperature treatment process. Microbial fuel cells are a developing technology that have the benefits of both removing such toxic compounds while recovering electrical energy. In this study, simultaneous thiocyanate degradation and electrical current generation was demonstrated and it was suggested that extracellular electron transfer to the anode occurred. Investigation of the microbial community by 16S rRNA metatranscriptome reads supported that the anode attached and planktonic anolyte consortia were dominated by a Thiobacillus-like population. Metatranscriptomic sequencing also suggested thiocyanate degradation primarily occurred via the ‘cyanate’ degradation pathway. The generated sulfide was metabolized via sulfite and ultimately to sulfate mediated by reverse dissimilatory sulfite reductase, APS reductase, and sulfate adenylyltransferase and the released electrons were potentially transferred to the anode via soluble electron shuttles. Finally, the ammonium from thiocyanate degradation was assimilated to glutamate as nitrogen source and carbon dioxide was fixed as carbon source. This study is one of the first to demonstrate a low temperature inorganic sulfur utilizing microbial fuel cell and the first to provide evidence for pathways of thiocyanate degradation coupled to electron transfer.
The toxic compound thiocyanate (SCN−) is generated by the mining and metallurgy industries during gold recovery [up to 4000 mg/L (Kantor et al., 2017)] with cyanide and it should be removed from wastewaters before being released to recipient water bodies (Zagury et al., 2004; Van Zyl et al., 2011). Chemical removal of SCN− is both inefficient and costly (Van Zyl et al., 2011). In contrast, biodegradation is relatively inexpensive and can completely remove SCN− and other contaminants such as cyanide (Kantor et al., 2015). However, a complicating factor in treating mining wastewaters in cold climates, such as in northern Sweden, is that the water temperature rarely exceeds approximately 12°C and approaches 0°C in winters (Liljeqvist et al., 2011). As a result, economic constraints require the development of (bio)remediation processes for contaminant removal that operate at low temperatures.
A variety of chemolithotrophic and heterotrophic bacterial genera metabolize SCN−, allowing them to be candidate organisms for SCN− removal, including Thiobacillus (Happold et al., 1958), Paracoccus (Ghosh and Roy, 2007), Pseudomonas (Chapatwala et al., 1998), and Arthrobacter (Betts et al., 1979). There are two major SCN− degradation pathways and the overall reaction is shown in Eq. 1. In the ‘COS pathway’ (Eqs. 2, 3), SCN− is initially hydrolyzed into ammonia and carbonyl sulfide (COS), and the carbonyl sulfide is subsequently oxidized into carbon dioxide and sulfate in an energy yielding reaction with a key enzyme in this pathway being the cobalt-coordinating thiocyanate hydrolase (Scn) (Katayama et al., 1992, 1993; Kim and Katayama, 2000).
The second ‘cyanate pathway’ (Eqs. 4, 5) proposes that SCN− is used as the sole nitrogen and/or sulfur source for growth (Stratford et al., 1994) by initially hydrolyzing it into sulfide and cyanate (OCN−). The intermediate cyanate is subsequently converted to ammonium by cyanase (Cyn) and the sulfide is oxidized into sulfate or tetrathionate (Youatt, 1954; Sung and Fuchs, 1988; Stratford et al., 1994).
In a recent metagenomic study, two laboratory-scale SCN− degrading bioreactors showed predominance of Thiobacillus spp. with a chemolithotrophic lifestyle (Kantor et al., 2015). Genes encoding the major SCN− degrading enzymes including Scn and Cyn were present (Kantor et al., 2015).
Bioelectrochemical Systems (BESs) are versatile technologies that utilize the interaction of microbial catalysts at an electrode interface. BESs can either recover electrical energy or produce a product, such as methane (Geppert et al., 2016), acetate (Jourdin et al., 2015), copper (Rodenas Motos et al., 2015), or ammonia (Kuntke et al., 2018). Typical electron donors tested in BESs are organic substrates such as acetate, glucose, and synthetic or real wastewaters [reviewed in Pant et al. (2010)]. With few exceptions, the use of inorganic sulfur compounds as substrates are rarely studied. Exceptions include sulfide and tetrathionate from synthetic or industrial wastewaters (Rabaey et al., 2006; Sulonen et al., 2015; Ni et al., 2016). In general, lowering the operational temperature of these systems will negatively impact reaction rates, alter enzyme-substrate interactions, and ultimately lead to protein cold-denaturation (Makhatadze and Privalov, 1994; Georlette et al., 2004). Lower temperatures also reduce the available energy from the desired processes and is why most of these studies are performed in mesophilic conditions (Dopson et al., 2015). Nonetheless, being able to function in cold temperature is a prerequisite should the biotechnology be implemented in areas where winter climates decrease the temperature of wastewaters (Liljeqvist et al., 2011), and low temperature BESs can be a potential remedy for this situation. Apart from saving heating costs, they can harvest a huge potential energy reserve when applied to marine sediments (Bond et al., 2002; Reimers et al., 2006), inactivate competing processes for electrons such as methanogenesis and acetogenesis when organic substrates are utilized (Lu et al., 2011), and improve Coulombic efficiencies (CE) as demonstrated in a study in which hydrogen production at 4°C was achieved with a CE above 92% (Lu et al., 2011). However, low temperature BES studies using inorganic sulfur compounds have not been published and rarely have studies utilized metatranscriptomics to investigate the active microbial species and their metabolic processes.
Metatranscriptomics is the direct extraction and characterization of the total RNA from a community of interest (Zhi et al., 2014), and provides information regarding the active metabolic processes occurring at the time of RNA extraction. It is powerful in profiling the microbial community composition and the metabolic processes of interest. In this study, we report the first endeavor of a such approach to investigate a novel low temperature SCN− degrading microbial consortium able to facilitate electrical current generation in microbial fuel cells (MFCs, a type of BES). The metatranscriptomic data were used to characterize the active microbial community under the engineered conditions, with the focus on the mechanisms for SCN− degradation coupled to extracellular electron transfer (EET) to the electrode.
Duplicate MFCs (designated as MFC A and B) were constructed from Plexiglas and the volumes of the anode and cathode chambers were 66 and 33 cm3, respectively. The anode had two flow chambers with the inner chamber open on both sides that was filled with a graphite felt material (FMI Composite Ltd., Galashiels, Scotland) while the outer compartment was open on the inner side. These two compartments provided space for the graphite felt and for the electrolyte to flow. The cathode compartment consisted of a single flow chamber sealed with a flat graphite plate. A cation exchange membrane (CMI-7000, Membranes International INC., United States) separated the electrolytes between the anode and cathode compartments. An anaerobic environment was maintained using silicon rubber layers placed on both sides of the flow chambers, being pressed tightly together. The graphite felt was connected to an external cable via a gold wire woven through the felt. Reference electrodes (Ag/AgCl, Sigma-Aldrich Co., LLC; 203 mV vs standard hydrogen electrode) were submerged in 3M KCl solution connected to the anode or cathode electrolyte via glass capillaries (ProSense, Netherlands). The external resistance was initially kept at 1000 Ω and switched to 560 Ω from day 150 to 180 on for MFC A and B, respectively. The cathode, anode, and membrane potentials plus the cell voltage were measured with a digital multimeter (BS3604W, Clas Ohlson, Sweden) on a daily basis. The electrical current production was calculated from the cell voltage and external resistance. The Coulombic efficiency was estimated based on the complete oxidation from thiocyanate to sulfate, according to Eq. 6, where Δ[SCN−] was the decrease of thiocyanate in mol; F is the faraday constant (96485 C/mol); and electrical current (I) was expressed in A. The experiment was carried out in a temperature-controlled room maintained at 8°C.
The microbial inoculum was a previously described low temperature, autotrophic denitrifying culture using SCN− as the electron donor (Broman et al., 2017). The anolyte consisted of mineral salts medium (Broman et al., 2017) except that the electron acceptor (nitrate) was excluded, the catholyte consisted of 50 mM potassium ferricyanide. The initial SCN− concentration was approximately 6 mM, a value chosen based on the SCN− concentration in a Boliden AB (Sweden) process water from a sulfide mineral processing plant (Broman et al., 2017). SCN− concentration was analyzed by cyanolysis (Kelly et al., 1969) as described in Dopson and Lindström (1999) without the incubation step with tetrathionate. Additional SCN− was periodically added to the anolyte when the concentration decreased close to zero, carbonate was supplied as the carbon source.
RNA extraction from planktonic cells from the two MFCs was performed at the end of the experiment (i.e., day 272) by sampling the anolyte, immediately mixing (1:1 ratio) with RNA fix solution (Feike et al., 2012), and cell capture on a 0.22 μm filter. RNA extraction from cells attached to the graphite felt was carried out at the same time by immediately soaking the felt in the RNA fixative followed by vigorous shaking of the felt-containing solution and then cell capture with a 0.22 μm filter. Community RNA was extracted from both planktonic and attached cells according to the manufacturer’s instruction using the RNeasy midi kit for isolation of total RNA from Bacteria (Qiagen, Germany). The extracted RNA was used to generate cDNA with the Ovation® RNA-Seq System V2 (NuGEN, United States) according to manufacturer’s instructions. The generated cDNA was purified using MinElute Reaction Cleanup Kit (Qiagen, Germany). Metatranscriptome library preparation was carried out using the Illumina Hiseq Truseq Nano DNA Library Prep Kit for NeoPrep at SciLifeLab. Clustering was done by ‘cBot’ and samples were sequenced on HiSeq2500 (HiSeq Control Software 2.2.58/RTA 1.18.64) with a 2 × 126 bp setup using ‘HiSeq SBS Kit v4’ chemistry. The Bcl to FastQ conversion was performed using bcl2fastq-1.8.4 from the CASAVA software suite. The quality scale used was Sanger / phred33 / Illumina 1.8+.
Details of metatranscriptomic analysis is reported in Supplementary File 1. Briefly, the active members of MFC communities were identified through the phylogenetic placement of 16S rRNA reads, which were a subset from the total datasets extracted with SortMeRNA (version 2.1b), using default parameters (Kopylova et al., 2012) and 16S rRNA reference databases provided by the authors on the tool repository1. The phylogenetic placement was performed as follows: 16S rRNA reads were aligned to a reference multiple alignment (RMA) with PaPaRa v2.5 using default parameters (Berger and Stamatakis, 2011), then inserted into a related reference phylogenetic tree (RPT) by re-optimization of RPT edge lengths through the Evolutionary Placement Algorithm (EPA) implemented in RAxML (version 8.2.10) (Stamatakis, 2014). Two separate RPTs/RMAs were used for Archaea and Bacteria, respectively, including SSU sequences from Anantharaman et al. (2016) and Hug et al. (2016). Only the most reliable phylogenetic placements (likelihood weight ratio of ≥ 0.90) were considered for downstream analyses. Abundances of reads at each tree node were determined with the guppy version 1.1 utility of the pplacer version 1.1.alpha17 package (Matsen et al., 2010), summarized at the genus level and reported in Figure 2.
The mRNA reads were aligned against the NCBI NR database with an e-value < 0.001 using Diamond 0.9.10 (Buchfink et al., 2015) in conjunction with BLASTX (Altschul et al., 1990). The read alignments were imported into MEGAN 6 with default LCA-settings (Huson and Mitra, 2012) and classified into taxonomy and proteins based on the MEGAN available protein accession to taxonomy (March, 2018) and InterPro2GO (November, 2016 version) databases. The name of each individual read was used to link taxonomic affiliation with InterPro proteins. Sample counts were normalized among samples as CPM values (counts per million sequences; i.e., (x/sum of sample) × 1 000 000). Statistical testing of the assigned protein classification between the planktonic and electrode-attached communities was conducted using the function exactTest from the edgeR package in R with false discovery threshold of 0.05 (Robinson et al., 2010).
The raw and metatranscriptomic sequences were submitted to the NCBI BioProject database with accession number PRJNA473625.
Duplicate MFCs were inoculated with a SCN− degrading culture and operated at 8°C for 272 days. Since no electrical current was produced in the first 118 days, potentially due to the insufficient activity from the microbiome, re-inoculation took place on days 93 (MFC B), 94 (MFC A), 99 (both MFCs), and 111 (both MFCs; Supplementary Files 2, 3). An increase in electrical current production occurred on day 119 and 129 for MFCs A and B, respectively (Figure 1). Before the increase of current was observed, the concentration of SCN− remained stable in both MFCs (6.37 ± 0.06 mM, n = 3 for MFC A; 6.51 ± 0.14 mM, n = 8 for MFC B). On experimental day 176, the maximum current density normalized to anode volume was 675.0 and 592.6 mA/m3 for MFCs A and B, respectively, that gave 20.2 and 17.8 mA/m2 as normalized to the projected anode/membrane surface area. At that time, the SCN− degradation rates for MFCs A and B were 0.123 mM/day and 0.068 mM/day, respectively. In conjunction with the electrical current production, the concentration of SCN− in both systems decreased, indicating the produced electrical current was dependent on the oxidation of SCN−. In general, a low temperature negatively impacts on the microbial enzyme machinery and reduces electrochemical reaction rates (Dopson et al., 2015); furthermore, psychrophiles generally have longer doubling times compared to mesophiles (Ferroni et al., 1986). These factors may explain the long start-up time of the electrical current production, which has also been observed in other low temperature BES studies (Bond et al., 2002; Reimers et al., 2006).
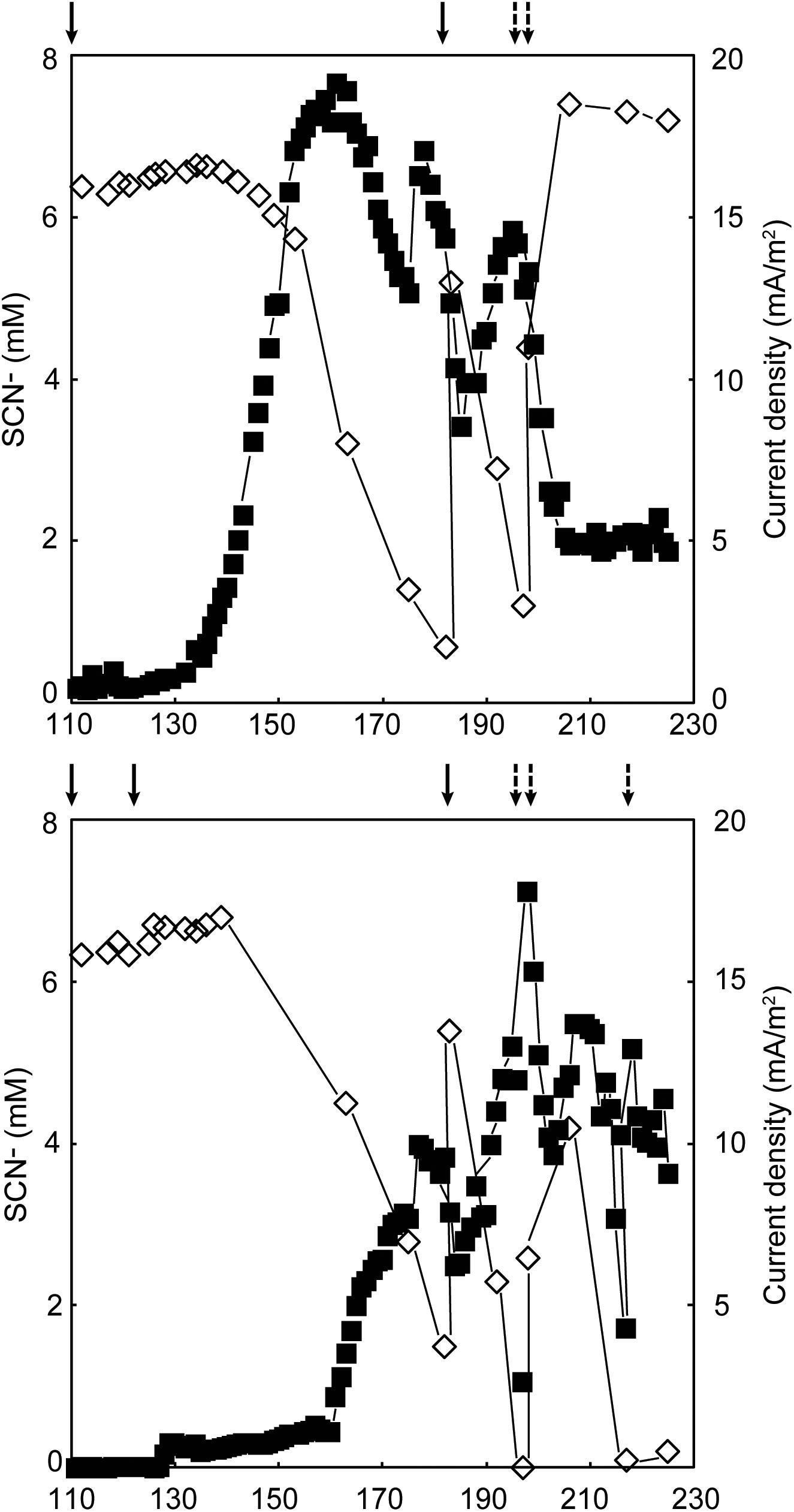
FIGURE 1. Electrical current production  corresponding to the right axis and thiocyanate degradation
corresponding to the right axis and thiocyanate degradation  corresponding to the left axis in MFCs (A) and (B) (top and bottom, respectively). Dashed arrows indicate addition of thiocyanate and the solid arrows designate the MFCs were re-started with fresh anolyte and inoculum. No thiocyanate degradation or electrical current production occurred prior to day 100 and one outlier electrical current value was removed from both MFCs.
corresponding to the left axis in MFCs (A) and (B) (top and bottom, respectively). Dashed arrows indicate addition of thiocyanate and the solid arrows designate the MFCs were re-started with fresh anolyte and inoculum. No thiocyanate degradation or electrical current production occurred prior to day 100 and one outlier electrical current value was removed from both MFCs.
Based on Eq. 1, the theoretical anode potential at pH 7 and 8°C was calculated as −476 mV (against Ag/AgCl). For both MFC A and B, the measured anode potential values were more positive than the theoretical (Supplementary Files 2, 3). As electrochemically active microorganism(s) transfer electrons to the electrode, the anode potential would develop toward the more negative theoretical value when the electrical circuit was open (measured as anode ‘open cell potential’). However, since the graphite-felt material had a relatively high electrical capacity and a slow electron transfer to the electrode was caused by low temperature; plus, no biofilm formation was observed which would facilitate electron transfer; it would take time for the anode potential to reach the theoretical value. This was confirmed when MFC A was operated in open cell and the anode potential decreased from 296 to 2 mV after 25.5 h (experimental days 261 – 262) and was still decreasing.
The Coulombic efficiencies between days 111 – 272 were 2.5 and 1.0% for MFCs A and B, respectively, (Supplementary File 4). Compared to other psychrophilic BES studies (reviewed in Dopson et al., 2015), both the produced electrical current and Coulombic efficiency were relatively low and could be for several reasons. Firstly, carbon dioxide fixation for biomass growth by some members of the anodic microbial consortium (described below) restricted the available electrons that could be harvested as electrical current. Secondly, although N2 gas was continuously flushed in the anolyte circulation bottle, trace levels of oxygen (a competing electron acceptor) can diffuse into the MFC via rubber tubing and graphite plates. Thirdly, competing energy conservation reactions induced by cold stress may have occurred (discussed below).
The original inoculum was obtained from an anaerobic SCN− degrading stirred tank bio-reactor (Broman et al., 2017). However, since the terminal electron acceptor (nitrate) was removed in this study, electrons were suggested to pass to the solid electrode surface. Community RNA was extracted from both the planktonic cells and those attached to the graphite felt (electrode) from the replicate MFCs. A total of 170 million read pairs were obtained across the four samples (MFC anode/anolyte plus MFC B anode/anolyte), of which 78% remained after quality trimming. 8% of the trimmed reads were identified as 16S rRNA; 4% of the trimmed reads were identified as mRNA, of which 88% was assigned to a known protein in the InterPro database (Supplementary File 5). Although using 16S rRNA data as a proxy for microbial activity has been questioned (Blazewicz et al., 2013), here we use the 16S rRNA read counts to infer a ‘protein synthesis potential’ (Thureborn et al., 2017). The dominant 16S rRNA reads (between 63 and 76%) were aligned to the Thiobacillus genus and primarily to T. denitrificans across the four samples (Figure 2). In addition, 16S rRNA reads also aligned with species including Methyloversatilis universalis, Salinibacterium amurskyense, Azorhizobium caulinodans, and Mucilaginibacter paludis. The type species of the genus Thiobacillus, T. thioparus can grow anaerobically with thiosulfate and nitrate (Katayama and Hiroshi, 1978). The presence of Thiobacillus spp. was in accordance with previous studies in which Thiobacillus populations were the dominating strains for SCN− degradation (Kantor et al., 2015, 2017), and also suggested a novel electrochemically active trait for populations within this genus.
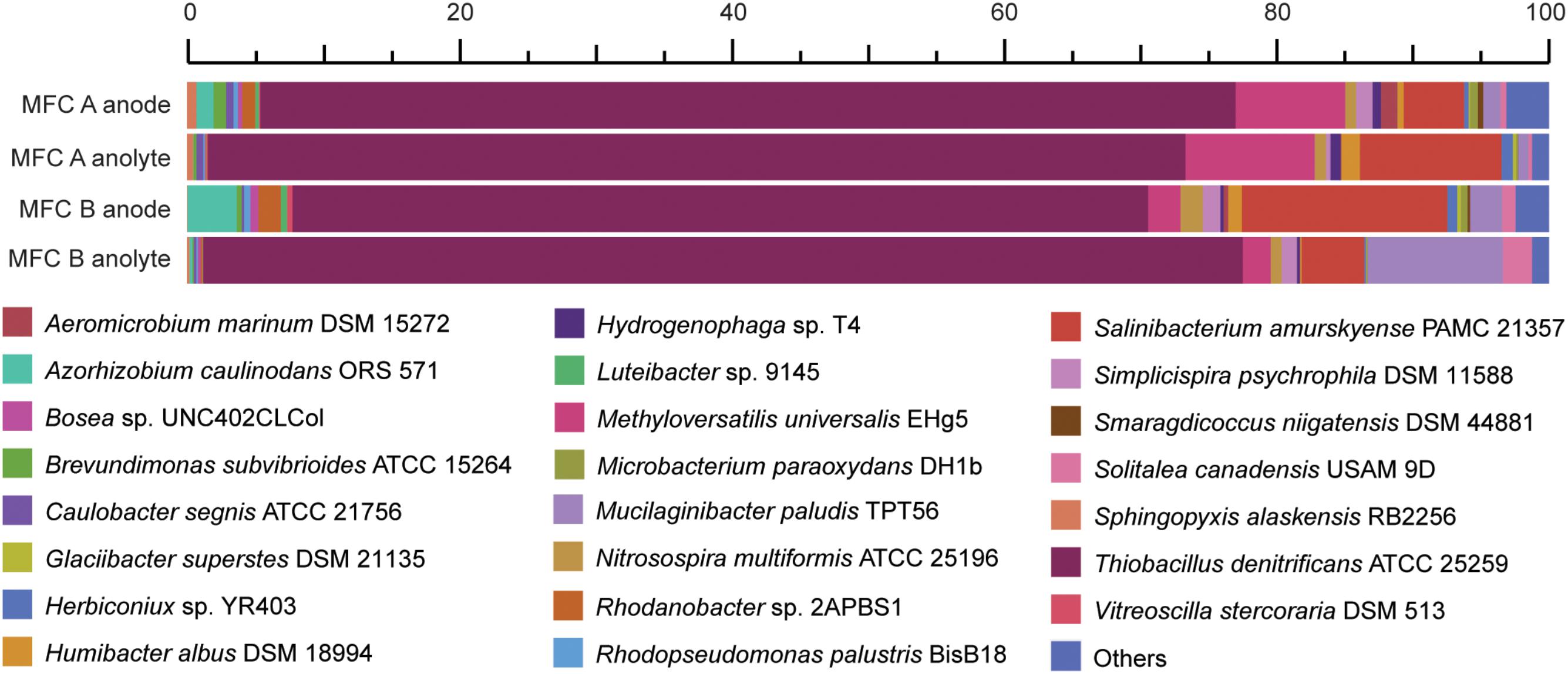
FIGURE 2. Relative abundance of the two MFCs anode attached and anolyte microbiomes based on high throughput 16S rRNA data. The duplicate MFCs (termed MFC A and MFC B) were producing an electrical current when the cells were harvested and fixed for metatranscriptomic sequencing.
An abundance of mRNA reads were not taxonomically assigned with better precision than Bacteria (7%) or Proteobacteria (49%) (Supplementary File 6). Based on the high activity of T. denitrificans according to 16S rRNA analysis (Figure 2), these Bacteria and Proteobacteria populations suggested by mRNA analysis were potentially from the Thiobacillus genus. Finally, default edgeR analysis gave no statistically significant differences regarding the discussed genes coding for sulfur, carbon, nitrogen metabolism as well as electron transfer and adaptation to low temperature between the electrode-attached and planktonic communities based on mRNA analysis (Supplementary File 6).
Genes coding for both the ‘COS’ and ‘cyanate’ pathways for thiocyanate degradation to sulfide were identified in the duplicate MFCs (Figure 3 and Supplementary File 6). The key gene in the ‘COS pathway’ is thiocyanate hydrolase (scn; Kantor et al., 2015) that was attributed to the unclassified Bacteria (0 – 67 CPM) and unclassified Proteobacteria (0 – 53 CPM) across the four samples. However, mRNA reads for the second gene in the ‘COS’ pathway (carbonyl sulfide hydrolase) were not identified in either of the MFCs. Although mRNA reads for thiocyanate dehydrogenase that catalyzes the first stage in the ‘cyanate’ pathway was not identified, the key cyanase gene (cyn) in this pathway (Kantor et al., 2015) was present. The majority of mRNA reads for cyn were attributed to unclassified Proteobacteria (9 – 284 CPM) and Betaproteobacteriales (3 – 151 CPM) and with lower numbers of mRNA reads assigned to e.g., a Thiobacillus population. Of the key genes for the two pathways, the greater number of mRNA reads for cyanase (Cyn) suggested a higher activity of the ‘cyanate’ pathway.
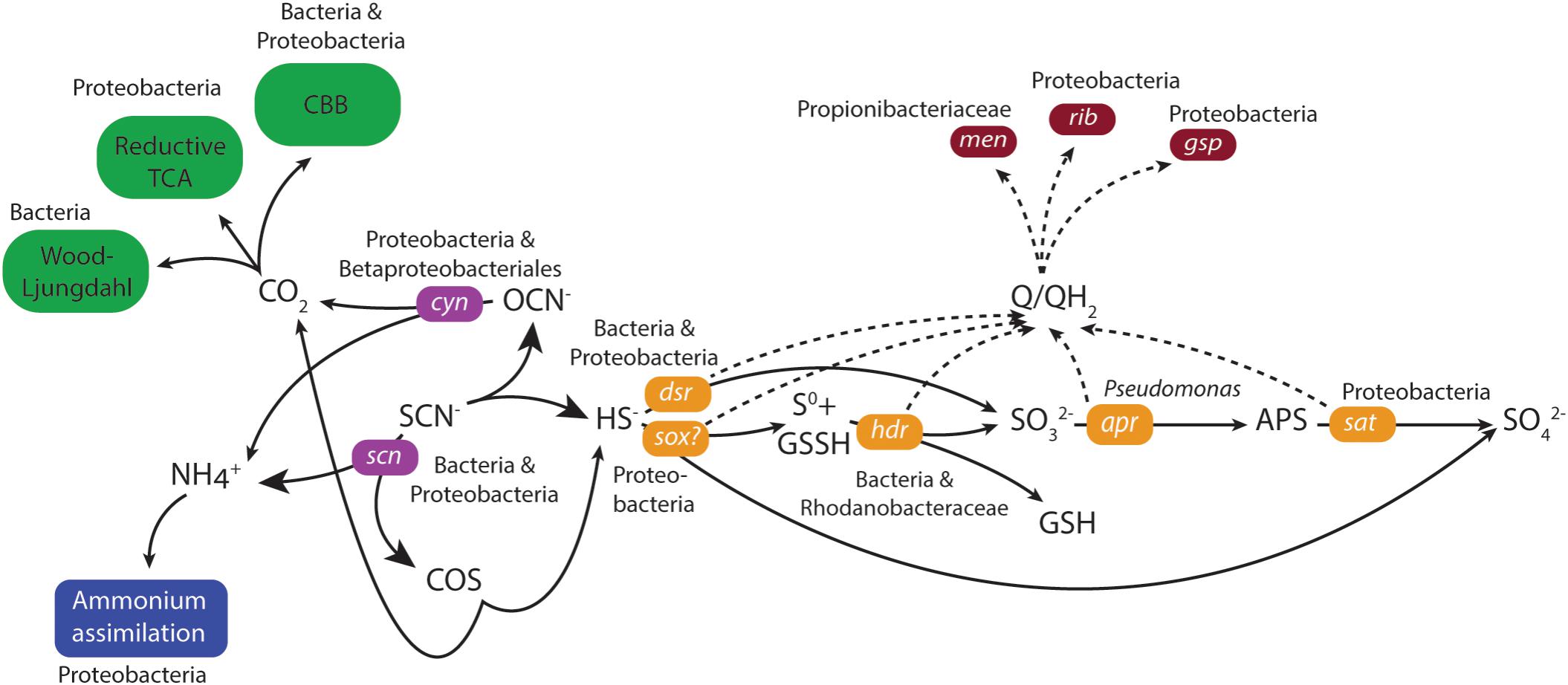
FIGURE 3. Model of thiocyanate degradation together with sulfur, carbon and nitrogen metabolism coupled to EET based upon mRNA data. Solid lines indicate the flow of metabolites and dashed lines the flow of electrons. The primarily assigned populations based upon mRNA data were indicated. Green indicates CO2 fixation, blue indicates ammonium assimilation, purple indicates thiocyanate degradation, orange indicates sulfur metabolism, and dark red indicates electron transfer.
The generated sulfide from thiocyanate degradation was suggested to be oxidized by the highly expressed dissimilatory sulfite reductase (Dsr) that functions in reverse to produce sulfite (Schedel and Trüper, 1979; Kantor et al., 2015). mRNA reads for dsr were predominantly attributed to either the unclassified Bacteria (4387 – 7294 CPM) or unclassified Proteobacteria (2275 – 5914 CPM) in the four metatranscriptomes (Figure 3 and Supplementary File 6). The resultant sulfite can then be metabolized to form adenosine 5′-phosphosulfate catalyzed by adenylylsulphate reductase (Apr) (Kappler and Dahl, 2001) and subsequently to sulfate via the membrane-bound dissimilatory adenylyl-sulfate reductase (Sat) (Peck, 1962). A higher number of mRNA reads were identified for sat (543 – 4255 CPM) than for apr (< 27 CPM in both MFCs) attributed to unclassified Proteobacteria. The gene for sulfur carrier accessory protein (tus) is commonly found together with dsr (Stockdreher et al., 2014) and its mRNA reads were primarily attributed to unclassified Proteobacteria (375 – 754 CPM). An alternative sulfide oxidation pathway was also present, involving a truncated sox complex (soxABCXY) primarily attributed to unclassified Proteobacteria and unclassified Bacteria that could oxidize sulfide generating either sulfur or sulfate (Kantor et al., 2017). Finally, a very low level of mRNA reads were identified for the anaerobic elemental sulfur oxidizing hdr in MFC A (Osorio et al., 2013).
The generation of an electrical current alongside thiocyanate degradation after a lag phase of more than 100 days strongly suggested electrons from thiocyanate and intermediate inorganic sulfur compound oxidation were transported to the anode facilitated by the anodic microbiome. No genetic pathways for carrying out extracellular transfer (EET) in the Thiobacillus genus are reported and therefore, the potential presence in our MFCs of known genes and pathways for EET in the extensively studied genera Geobacter and Shewanella were investigated. Outer membrane multi-heme c-type cytochromes encoded by genes such as mtrAC, cymA, and omcAESBZ in Shewanella and Geobacter species are critical in carrying out electron transfer from the microorganisms to electrodes in MFCs (Gorby et al., 2006; Holmes et al., 2006; Bretschger et al., 2007; Shi et al., 2007; Richter et al., 2009). In addition, the capacity of Shewanella and Geobacter to produce an electrical current is severely impaired when the type II secretion and type IV pilin biosynthesis pathways that involves the genes gspDG and pilAD are deficient (Reguera et al., 2005; Gorby et al., 2006; Bretschger et al., 2007; Richter et al., 2009; Vargas et al., 2013). Finally, although they have other functions within the cell (Wissenbach et al., 1990; Vitreschak et al., 2002), redox active electron shuttles such as menaquinone and quinone intermediates encoded by the menC gene, as well as riboflavin have been shown to be crucial in the ability to perform EET in Shewanella species (Newman and Kolter, 2000; Myers and Myers, 2004; Marsili et al., 2008). The mRNA data did not support EET involving known c-type cytochromes or conductive pili. mRNA reads for the type II secretion pathway genes gspDEGHM were present, primarily attributed to unclassified Proteobacteria (180 – 900 CPM; Figure 3 and Supplementary File 6) or Thiobacillus (3 – 359 CPM). mRNA reads for the type IV pilin genes pilAD were absent, but mRNA reads for a pilin fimbrial protein were mainly attributed to a Rhodanobacter sp. (15 – 154 CPM) and unclassified Proteobacteria (15 – 110 CPM). mRNA reads for menaquinone biosynthesis genes menBC were primarily assigned to the family Propionibacteriaceae (0 – 183 CPM) while mRNA reads for riboflavin biosynthesis proteins RibABD were mostly assigned to unclassified Proteobacteria (111 – 291 CPM). The presence of mRNA reads for type II secretion as well as menaquinone plus riboflavin biosynthesis suggested the anodic microbial community could have carried out EET using soluble electron shuttles. This claim was supported by the decrease and subsequent recovery in current density upon the removal and addition of new medium during operation of the duplicate MFCs (Figure 1 and Supplementary Files 2, 3). However, it cannot be ruled out that other, presently unknown mechanisms were responsible for EET in the selected community.
The proposed thiocyanate conversion to ammonium by both degradation pathways raises the possibility of energy conservation by ammonium oxidation coupled to sulfate reduction (Fdz-Polanco et al., 2001). However, the suggested predominance of the ‘cyanate pathway’ whereby SCN− is used as the sole nitrogen source for growth (Stratford et al., 1994); the lack of mRNA reads for known ammonium oxidation genes; and no observed mRNA reads for the dsrK gene that codes for energy conservation during sulfate reduction all suggested that this did not occur. Instead, the ammonium was likely used as a nitrogen source and assimilated primarily by Type I glutamine synthetase for which mRNA reads were mostly assigned to unclassified Proteobacteria (539 – 985 CPM; Figure 3 and Supplementary File 6) followed by Microbacteriaceae (Actinobacteria, 53 – 169 CPM).
All described species in the Thiobacillus genus fix carbon dioxide for cellular growth (Boden et al., 2017) and reconstruction of the thiocyanate degrading Thiobacillus-like species genome contained genes coding for the Calvin-Benson-Bassham (CCB) cycle (Kantor et al., 2015). However, mRNA reads encoding the key CBB cycle enzyme ribulose biphosphate carboxylase (RuBisCO) were missing although seven other genes in the cycle (that also have other functions) encoding phosphoglycerate kinase, erythrose phosphate dehydrogenase, and triosephosphate isomerase had CPMs mostly attributed to unclassified Proteobacteria (Figure 3 and Supplementary File 6). In addition, the key enzyme ATP-citrate lyase in the reductive TCA cycle was present; carbon monoxide dehydrogenase from the Wood-Ljungdahl pathway (0 – 248 CPM) were identified in unclassified Bacteria. This suggested that several carbon fixation pathways were present and despite all described Thiobacillus species using the CBB pathway, it was not clearly demonstrated for the MFC communities.
Along with reduced abiotic reaction and diffusion rates, low temperatures affect microbes by hindering folding of proteins into their three-dimensional structure, coiling and uncoiling DNA, and over-stabilization of mRNA (D’Amico et al., 2006; Casanueva et al., 2010; De Maayer et al., 2014). In response, psychrophiles use a number of strategies to combat low temperature although in many cases, these systems also have other functions within the cell. Cold shock proteins (CSPs) and cold-inducible proteins (CIPs) overlap with each other and are expressed during both short and long-term exposure to cold (Phadtare and Inouye, 2004; Horn et al., 2007; Barria et al., 2013). One example of CSPs and CIPs are chaperones that remove low temperature related mRNA and DNA secondary structures and thus, aid ribosomes and RNA polymerases to function. In addition to CSPs and CIPs, psychrophiles respond to low temperature by producing compatible solutes that protect against freezing (Casanueva et al., 2010) and by altering their membrane structure to increase flexibility (Chintalapati et al., 2004).
The identified cold adaptation systems included mRNA reads for the CspA (Phadtare and Inouye, 2008) and Clp protease (Skinner and Trempy, 2001) that were mostly attributed to unclassified Proteobacteria (479 – 1113 and 1410 – 3435 CPM, respectively; Figure 3 and Supplementary File 6). In addition, mRNA reads were identified for several molecular chaperones including Trigger factor (Tig; 797 – 5124 CPM) that functions together with GroEL (Kandror and Goldberg, 1997) and the SecB protein export chaperone (47 – 158 CPM), both were mostly identified from unclassified Proteobacteria. mRNA reads for the compatible solute synthesis enzyme betaine aldehyde dehydrogenase (BetB; 142 – 998 CPM) were identified from unclassified Proteobacteria and a glycine betaine ABC transporter were primarily identified from Actinobacteria (ProVWX, 0 – 179 CPM). mRNA reads coding for proteins involved in transcription and translation include DeaD RNA helicases that were identified from several populations with the Actinobacteria. NusA (293 – 1365 CPM) that involves in termination and anti-termination of transcription (Jones and Inouye, 1994), and DNA gyrase (516 – 2006 CPM) that relieves the strain of double stranded DNA being unwound (Morais Cabral et al., 1997) were primarily attributed to unclassified Proteobacteria. Finally, mRNA reads from several membrane alteration genes that were mainly attributed to unclassified Proteobacteria and in the case of N-acetylglucosaminyl transferase, also to the Thiobacillus genus, suggested that cold induced alteration to the cell membrane occurred. The identification of mRNA reads for cold adaptation suggested the cells were under stress in the low temperature MFCs and this may have contributed to the low Coulombic efficiency.
In this study, the extraction and analysis of 16S rRNA reads from the metatranscriptomes suggested that Thiobacillus was the most abundant and active genus. Metatranscriptomic analysis suggested that the anodic microbial consortium could degrade thiocyanate while the resultant sulfide was oxidized for energy conservation; ammonium was assimilated; and carbon dioxide was fixed via various pathways. It was also revealed that the consortium potentially utilized multiple mechanisms to acclimate to the low temperature including CSPs, cold inducible proteins, and molecular chaperones. Based on mRNA analysis from the metatranscriptomes, these processes were primarily associated with Bacterial and Proteobacteria populations, potentially attributed to Thiobacillus. Furthermore, mRNA analysis revealed no significant difference between the planktonic and electrode-attaching communities. Finally, our findings demonstrated for the first time, that an autotrophic psychrophilic microbial consortium facilitated electrical current generation from thiocyanate degradation in a microbial fuel cell. The Thiobacillus population potentially capable of carrying out EET enriched the inventory of electrochemically active microorganisms, these insights could benefit future industrial-scale low temperature remediation of thiocyanate, as well as a more diverse application of BESs.
GN and MD designed the study. SC and GN defined materials and configuration of the MFCs. SC and GN maintained and analyzed the long-term operation of the MFCs. GN and ML-F extracted nucleic acids. GN, DS, EB, VP, and DL designed and carried out bioinformatic analyses. GN, MD, and TS interpreted the data. GN, MD, DS, and EB drafted the manuscript. All authors read and approved the manuscript for submission.
The research leading to these results has received funding from the European Union Seventh Framework Program (FP7/2012-2016) under grant agreement number 282970 (BioElectroMET).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank our consortium members for comments on the manuscript National Genomics Infrastructure (NGI) and Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) for providing assistance in massive parallel sequencing and computational infrastructure. The computations were performed on resources provided by SNIC through UPPMAX under Project b2013127.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02308/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anantharaman, K., Brown, C. T., Hug, L. A., Sharon, I., Castelle, C. J., Probst, A. J., et al. (2016). Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7:13219. doi: 10.1038/ncomms13219
Barria, C., Malecki, M., and Arraiano, C. M. (2013). Bacterial adaptation to cold. Microbiology 159, 2437–2443. doi: 10.1099/mic.0.052209-0
Berger, S. A., and Stamatakis, A. (2011). Aligning short reads to reference alignments and trees. Bioinformatics 27, 2068–2075. doi: 10.1093/bioinformatics/btr320
Betts, P., Rinder, D., and Fleeker, J. (1979). Thiocyanate utilization by an Arthrobacter. Can. J. Microbiol. 25, 1277–1282. doi: 10.1139/m79-201
Blazewicz, S. J., Barnard, R. L., Daly, R. A., and Firestone, M. K. (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 7, 2061–2068. doi: 10.1038/ismej.2013.102
Boden, R., Hutt, L. P., and Rae, A. W. (2017). Reclassification of Thiobacillus aquaesulis (Wood & Kelly, 1995) as Annwoodia aquaesulis gen. nov., comb. nov., transfer of Thiobacillus (Beijerinck, 1904) from the Hydrogenophilales to the Nitrosomonadales, proposal of Hydrogenophilalia class. nov. within the ’Proteobacteria’, and four new families within the orders Nitrosomonadales and Rhodocyclales. Int. J. Syst. Evol. Microbiol. 67, 1191–1205. doi: 10.1099/ijsem.0.001927
Bond, D. R., Holmes, D. E., Tender, L. M., and Lovley, D. R. (2002). Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295, 483–485. doi: 10.1126/science.1066771
Bretschger, O., Obraztsova, A., Sturm, C. A., Chang, I. S., Gorby, Y. A., Reed, S. B., et al. (2007). Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl. Environ. Microbiol. 73, 7003–7012. doi: 10.1128/AEM.01087-07
Broman, E., Jawad, A., Wu, X., Christel, S., Ni, G., Lopez-Fernandez, M., et al. (2017). Low temperature, autotrophic microbial denitrification using thiosulfate or thiocyanate as electron donor. Biodegradation 28, 287–301. doi: 10.1007/s10532-017-9796-7
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Casanueva, A., Tuffin, M., Cary, C., and Cowan, D. A. (2010). Molecular adaptations to psychrophily: the impact of ‘omic’ technologies. Trends Microbiol. 18, 374–381. doi: 10.1016/j.tim.2010.05.002
Chapatwala, K. D., Babu, G. R. V., Vijaya, O. K., Kumar, K. P., and Wolfram, J. H. (1998). Biodegradation of cyanides, cyanates and thiocyanates to ammonia and carbon dioxide by immobilized cells of Pseudomonas putida. J. Ind. Microbiol. Biotechnol. 20, 28–33. doi: 10.1038/sj.jim.2900469
Chintalapati, S., Kiran, M. D., and Shivaji, S. (2004). Role of membrane lipid fatty acids in cold adaptation. Cell. Mol. Biol. 50, 631–642.
D’Amico, S., Collins, T., Marx, J. C., Feller, G., and Gerday, C. (2006). Psychrophilic microorganisms: challenges for life. EMBO Rep. 7, 385–389. doi: 10.1038/sj.embor.7400662
De Maayer, P., Anderson, D., Cary, C., and Cowan, D. A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. doi: 10.1002/embr.201338170
Dopson, M., and Lindström, E. B. (1999). Potential role of Thiobacillus caldus in arsenopyrite bioleaching. Appl. Environ. Microbiol. 65, 36–40.
Dopson, M., Ni, G., and Sleutels, T. H. (2015). Possibilities for extremophilic microorganisms in microbial electrochemical systems. FEMS Microbiol. Rev. 40, 164–181. doi: 10.1093/femsre/fuv044
Fdz-Polanco, F., Fdz-Polanco, M., Fernandez, N., Uruena, Garcia, P. A., and Villaverde, S. (2001). Combining the biological nitrogen and sulfur cycles in anaerobic conditions. Water Sci. Technol. 44, 77–84. doi: 10.2166/wst.2001.0469
Feike, J., Jürgens, K., Hollibaugh, J. T., Krüger, S., Jost, G., and Labrenz, M. (2012). Measuring unbiased metatranscriptomics in suboxic waters of the central Baltic Sea using a new in situ fixation system. ISME J. 6, 461–470. doi: 10.1038/ismej.2011.94
Ferroni, G., Leduc, L., and Todd, M. (1986). Isolation and temperature characterization of psychrotrophic strains of Thiobacillus ferrooxidans from the environment of a uranium mine. J. Gen. Appl. Microbiol. 32, 169–175. doi: 10.2323/jgam.32.169
Georlette, D., Blaise, V., Collins, T., D’amico, S., Gratia, E., Hoyoux, A., et al. (2004). Some like it cold: biocatalysis at low temperatures. FEMS Microbiol. Rev. 28, 25–42. doi: 10.1016/j.femsre.2003.07.003
Geppert, F., Liu, D., Van Eerten-Jansen, M., Weidner, E., Buisman, C., and Ter Heijne, A. (2016). Bioelectrochemical power-to-gas: state of the art and future perspectives. Trends Biotechnol. 34, 879–894. doi: 10.1016/j.tibtech.2016.08.010
Ghosh, W., and Roy, P. (2007). Chemolithoautotrophic oxidation of thiosulfate, tetrathionate and thiocyanate by a novel rhizobacterium belonging to the genus Paracoccus. FEMS Microbiol. Lett. 270, 124–131. doi: 10.1111/j.1574-6968.2007.00670.x
Gorby, Y. A., Yanina, S., Mclean, J. S., Rosso, K. M., Moyles, D., Dohnalkova, A., et al. (2006). Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. U.S.A. 103, 11358–11363. doi: 10.1073/pnas.0604517103
Happold, F., Jones, G., and Pratt, D. (1958). Utilization of thiocyanate by Thiobacillus thioparus and T. thiocyanoxidans. Nature 182, 266–267. doi: 10.1038/182266a0
Holmes, D. E., Chaudhuri, S. K., Nevin, K. P., Mehta, T., Methe, B. A., Liu, A., et al. (2006). Microarray and genetic analysis of electron transfer to electrodes in Geobacter sulfurreducens. Environ. Microbiol. 8, 1805–1815. doi: 10.1111/j.1462-2920.2006.01065.x
Horn, G., Hofweber, R., Kremer, W., and Kalbitzer, H. R. (2007). Structure and function of bacterial cold shock proteins. Cell. Mol. Life Sci. 64, 1457–1470. doi: 10.1007/s00018-007-6388-4
Hug, L. A., Baker, B. J., Anantharaman, K., Brown, C. T., Probst, A. J., Castelle, C. J., et al. (2016). A new view of the tree of life. Nat. Microbiol. 1:16048. doi: 10.1038/nmicrobiol.2016.48
Huson, D. H., and Mitra, S. (2012). Introduction to the analysis of environmental sequences: metagenomics with MEGAN. Meth. Mol. Biol. 856, 415–429. doi: 10.1007/978-1-61779-585-5_17
Jones, P. G., and Inouye, M. (1994). The cold-shock response–a hot topic. Mol. Microbiol. 11, 811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x
Jourdin, L., Grieger, T., Monetti, J., Flexer, V., Freguia, S., Lu, Y., et al. (2015). High acetic acid production rate obtained by microbial electrosynthesis from carbon dioxide. Environ. Sci. Technol. 49, 13566–13574. doi: 10.1021/acs.est.5b03821
Kandror, O., and Goldberg, A. L. (1997). Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proc. Natl. Acad. Sci. U.S.A. 94, 4978–4981. doi: 10.1073/pnas.94.10.4978
Kantor, R. S., Huddy, R. J., Iyer, R., Thomas, B. C., Brown, C. T., Anantharaman, K., et al. (2017). Genome-resolved meta-omics ties microbial dynamics to process performance in biotechnology for thiocyanate degradation. Environ. Sci. Technol. 51, 2944–2953. doi: 10.1021/acs.est.6b04477
Kantor, R. S., Van Zyl, A. W., Van Hille, R. P., Thomas, B. C., Harrison, S. T. L., and Banfield, J. F. (2015). Bioreactor microbial ecosystems for thiocyanate and cyanide degradation unravelled with genome-resolved metagenomics. Environ. Microbiol. 17, 4929–4941. doi: 10.1111/1462-2920.12936
Kappler, U., and Dahl, C. (2001). Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 203, 1–9. doi: 10.1111/j.1574-6968.2001.tb10813.x
Katayama, Y., and Hiroshi, K. (1978). Characteristics of Thiobacillus thioparus and its thiocyanate assimilation. Can. J. Microbiol. 24, 804–810. doi: 10.1139/m78-135
Katayama, Y., Kanagawa, T., and Kuraishi, H. (1993). Emission of carbonyl sulfide by Thiobacillus thioparus grown with thiocyanate in pure and mixed cultures. FEMS Microbiol. Lett. 114, 223–227. doi: 10.1111/j.1574-6968.1993.tb06577.x
Katayama, Y., Narahara, Y., Inoue, Y., Amano, F., Kanagawa, T., and Kuraishi, H. (1992). A thiocyanate hydrolase of Thiobacillus thioparus. A novel enzyme catalyzing the formation of carbonyl sulfide from thiocyanate. J. Biol. Chem. 267, 9170–9175.
Kelly, D. P., Chambers, L., and Trudinger, P. (1969). Cyanolysis and spectrophotometric estimation of trithionate in mixture with thiosulfate and tetrathionate. Anal. Chem. 41, 898–901. doi: 10.1021/ac60276a029
Kim, S.-J., and Katayama, Y. (2000). Effect of growth conditions on thiocyanate degradation and emission of carbonyl sulfide by Thiobacillus thioparus THI115. Water Res. 34, 2887–2894. doi: 10.1016/S0043-1354(00)00046-4
Kopylova, E., Noé, L., and Touzet, H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. doi: 10.1093/bioinformatics/bts611
Kuntke, P., Sleutels, T., Arredondo, M. R., Georg, S., Barbosa, S., Ter Heijne, A., et al. (2018). (Bio)electrochemical ammonia recovery: progress and perspectives. Appl. Microbiol. Biotechnol. 102, 3865–3878. doi: 10.1007/s00253-018-8888-6
Liljeqvist, M., Sundkvist, J. E., Saleh, A., and Dopson, M. (2011). Low temperature removal of inorganic sulfur compounds from mining process waters. Biotechnol. Bioeng. 108, 1251–1259. doi: 10.1002/bit.23057
Lu, L., Ren, N., Zhao, X., Wang, H., Wu, D., and Xing, D. (2011). Hydrogen production, methanogen inhibition and microbial community structures in psychrophilic single-chamber microbial electrolysis cells. Energy Environ. Sci. 4, 1329–1336. doi: 10.1039/c0ee00588f
Makhatadze, G. I., and Privalov, P. L. (1994). Hydration effects in protein unfolding. Biophys. Chem. 51, 291–309. doi: 10.1016/0301-4622(94)00050-6
Marsili, E., Baron, D. B., Shikhare, I. D., Coursolle, D., Gralnick, J. A., and Bond, D. R. (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105, 3968–3973. doi: 10.1073/pnas.0710525105
Matsen, F. A., Kodner, R. B., and Armbrust, E. V. (2010). pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11:538. doi: 10.1186/1471-2105-11-538
Morais Cabral, J. H., Jackson, A. P., Smith, C. V., Shikotra, N., Maxwell, A., and Liddington, R. C. (1997). Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388, 903–906. doi: 10.1038/42294
Myers, C. R., and Myers, J. M. (2004). Shewanella oneidensis MR-1 restores menaquinone synthesis to a menaquinone-negative mutant. Appl. Environ. Microbiol. 70, 5415–5425. doi: 10.1128/AEM.70.9.5415-5425.2004
Newman, D. K., and Kolter, R. (2000). A role for excreted quinones in extracellular electron transfer. Nature 405, 94–97. doi: 10.1038/35011098
Ni, G., Christel, S., Roman, P., Wong, Z. L., Bijmans, M. F., and Dopson, M. (2016). Electricity generation from an inorganic sulfur compound containing mining wastewater by acidophilic microorganisms. Res. Microbiol. 167, 568–575. doi: 10.1016/j.resmic.2016.04.010
Osorio, H., Mangold, S., Denis, Y., Nancucheo, I., Johnson, D. B., Bonnefoy, V., et al. (2013). Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 79, 2172–2181. doi: 10.1128/AEM.03057-12
Pant, D., Van Bogaert, G., Diels, L., and Vanbroekhoven, K. (2010). A review of the substrates used in microbial fuel cells (MFCs) for sustainable energy production. Bioresour. Technol. 101, 1533–1543. doi: 10.1016/j.biortech.2009.10.017
Peck, H. Jr. (1962). V. Comparative metabolism of inorganic sulfur compounds in microorganisms. Bacteriol. Rev. 26:67.
Phadtare, S., and Inouye, M. (2004). Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J. Bacteriol. 186, 7007–7014. doi: 10.1128/JB.186.20.7007-7014.2004
Phadtare, S., and Inouye, M. (2008). “Cold-shock protein,” in Psychrophiles: from Biodiversity to Biotechnology, eds R. Margesin, F. Schinner, J.-C. Marx, and C. Gerday (Berlin: Springer-Verlag), 191–209. doi: 10.1007/978-3-540-74335-4_12
Rabaey, K., Van De Sompel, K., Maignien, L., Boon, N., Aelterman, P., Clauwaert, P., et al. (2006). Microbial fuel cells for sulfide removal. Environ. Sci. Technol. 40, 5218–5224. doi: 10.1021/es060382u
Reguera, G., Mccarthy, K. D., Mehta, T., Nicoll, J. S., Tuominen, M. T., and Lovley, D. R. (2005). Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101. doi: 10.1038/nature03661
Reimers, C., Girguis, P., Stecher, H., Tender, L., Ryckelynck, N., and Whaling, P. (2006). Microbial fuel cell energy from an ocean cold seep. Geobiology 4, 123–136. doi: 10.1111/j.1472-4669.2006.00071.x
Richter, H., Nevin, K. P., Jia, H., Lowy, D. A., Lovley, D. R., and Tender, L. M. (2009). Cyclic voltammetry of biofilms of wild type and mutant Geobacter sulfurreducens on fuel cell anodes indicates possible roles of OmcB, OmcZ, type IV pili, and protons in extracellular electron transfer. Energy Environ. Sci. 2, 506–516. doi: 10.1039/b816647a
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodenas Motos, P., Ter Heijne, A., Van Der Weijden, R., Saakes, M., Buisman, C. J., and Sleutels, T. H. (2015). High rate copper and energy recovery in microbial fuel cells. Front. Microbiol. 6:527. doi: 10.3389/fmicb.2015.00527
Schedel, M., and Trüper, H. (1979). Purification of Thiobacillus denitrificans siroheme sulfite reductase and investigation of some molecular and catalytic properties. Biochim. Biophys. Acta 568, 454–466. doi: 10.1016/0005-2744(79)90314-0
Shi, L., Squier, T. C., Zachara, J. M., and Fredrickson, J. K. (2007). Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 65, 12–20. doi: 10.1111/j.1365-2958.2007.05783.x
Skinner, M. M., and Trempy, J. E. (2001). Expression of clpX, an ATPase subunit of the Clp protease, is heat and cold shock inducible in Lactococcus lactis. J. Dairy Sci. 84, 1783–1785. doi: 10.3168/jds.S0022-0302(01)74615-2
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stockdreher, Y., Sturm, M., Josten, M., Sahl, H.-G., Dobler, N., Zigann, R., et al. (2014). New proteins involved in sulfur trafficking in the cytoplasm of Allochromatium vinosum. J. Biol. Chem. 289, 12390–12403. doi: 10.1074/jbc.M113.536425
Stratford, J., Dias, A. E. O., and Knowles, C. J. (1994). The utilization of thiocyanate as a nitrogen source by a heterotrophic bacterium: the degradative pathway involves formation of ammonia and tetrathionate. Microbiology 140, 2657–2662. doi: 10.1099/00221287-140-10-2657
Sulonen, M. L., Kokko, M. E., Lakaniemi, A.-M., and Puhakka, J. A. (2015). Electricity generation from tetrathionate in microbial fuel cells by acidophiles. J. Hazard. Mater. 284, 182–189. doi: 10.1016/j.jhazmat.2014.10.045
Sung, Y.-C., and Fuchs, J. A. (1988). Characterization of the cyn operon in Escherichia coli K12. J. Biol. Chem. 263, 14769–14775.
Thureborn, P., Hu, Y. O., Franzetti, A., Sjöling, S., and Lundin, D. (2017). Accumulation of DNA in an anoxic sediment-rDNA and rRNA presence of members of the microbial community from Landsort Deep, the Baltic Sea. PeerJ Prepr. 5:e2051v2.
Van Zyl, A. W., Harrison, S. T., and Van Hille, R. P. (2011). Biodegradation of Thiocyanate by a Mixed Microbial Population. Aachen: Mine Water-Managing the Challenges.
Vargas, M., Malvankar, N. S., Tremblay, P.-L., Leang, C., Smith, J. A., Patel, P., et al. (2013). Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. mBio 4:e00105-13. doi: 10.1128/mBio.00105-13
Vitreschak, A. G., Rodionov, D. A., Mironov, A. A., and Gelfand, M. S. (2002). Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30, 3141–3151. doi: 10.1093/nar/gkf433
Wissenbach, U., Kroger, A., and Unden, G. (1990). The specific functions of menaquinone and demethylmenaquinone in anaerobic respiration with fumarate, dimethylsulfoxide, trimethylamine N-oxide and nitrate by Escherichia coli. Arch. Microbiol. 154, 60–66. doi: 10.1007/BF00249179
Youatt, J. B. (1954). Studies on the metabolism of Thiobacillus thiocyanoxidans. Microbiology 11, 139–149.
Zagury, G. J., Oudjehani, K., and Deschênes, L. (2004). Characterization and availability of cyanide in solid mine tailings from gold extraction plants. Sci. Total Environ. 320, 211–224. doi: 10.1016/j.scitotenv.2003.08.012
Keywords: MFC, thiocyanate degradation, extracellular electron transfer, low temperature, metatranscriptomics
Citation: Ni G, Canizales S, Broman E, Simone D, Palwai VR, Lundin D, Lopez-Fernandez M, Sleutels T and Dopson M (2018) Microbial Community and Metabolic Activity in Thiocyanate Degrading Low Temperature Microbial Fuel Cells. Front. Microbiol. 9:2308. doi: 10.3389/fmicb.2018.02308
Received: 14 June 2018; Accepted: 10 September 2018;
Published: 28 September 2018.
Edited by:
Pascal E. Saikaly, King Abdullah University of Science and Technology, Saudi ArabiaReviewed by:
Krishnaveni Venkidusamy, University of South Australia, AustraliaCopyright © 2018 Ni, Canizales, Broman, Simone, Palwai, Lundin, Lopez-Fernandez, Sleutels and Dopson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaofeng Ni, Z2FvZmVuZy5uaUBsbnUuc2U=
†Present address: Sebastian Canizales, Wetsus, European Centre of Excellence for Sustainable Water Technology, Leeuwarden, Netherlands; Margarita Lopez-Fernandez, Helmholtz-Zentrum Dresden-Rossendorf, Dresden, Germany
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.