- 1Department of Chemical and Biological Engineering, University at Buffalo, The State University of New York, Buffalo, NY, United States
- 2Key Laboratory of Marine Drugs, Chinese Ministry of Education, School of Medicine and Pharmacy, Ocean University of China, Qingdao, China
- 3Laboratory for Marine Drugs and Bioproducts, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Salicylate 2-O-β-D-glucoside (SAG) is a plant-derived natural product with potential utility as both an anti-inflammatory and as a plant protectant compound. Heterologous biosynthesis of SAG has been established in Escherichia coli through metabolic engineering of the shikimate pathways and introduction of a heterologous biosynthetic step to allow a more directed route to the salicylate precursor. The final SAG compound resulted from the separate introduction of an Arabidopsis thaliana glucosyltransferase enzyme. In this study, a range of heterologous engineering parameters were varied (including biosynthetic pathway construction, expression plasmid, and E. coli strain) for the improvement of SAG specific production in conjunction with a system demonstrating improved plasmid stability. In addition, the glucoside moiety of SAG was systematically varied through the introduction of the heterologous oliose and olivose deoxysugar pathways. Production of analogs was observed for each newly constructed pathway, demonstrating biosynthetic diversification potential; however, production titers were reduced relative to the original SAG compound.
Introduction
Plants have dedicated metabolism for the production of salicylate and a glycosylated version, salicylate 2-O-β-D-glucoside (SAG), which is often stored intracellularly until external stress is encountered (Vlot et al., 2009; Rivas-San Vicente and Plasencia, 2011). At which point, the reversion of SAG to salicylate allows the bioactivity of the latter compound to combat various biological threats to the plant system. Salicylate is also a central component of aspirin and, as such, SAG has the potential to possess similar anti-inflammatory properties.
These various bioactivities of SAG prompted us to explore its production through a heterologous bacterial host. Leveraging the knowledge and prior studies associated with engineering the shikimate pathway of Escherichia coli (Lin et al., 2014), we generated a production host supportive of high titer levels of salicylate (>1 g/L) (Ahmadi et al., 2016). This work included the introduction of an Irp9 salicylate synthase gene from Yersinia enterocolitica, which streamlined metabolism toward this precursor (Figure 1; Pelludat et al., 2003; Kerbarh et al., 2005). The introduction of a glucosyltransferase gene (ugt74f1) from Arabidopsis thaliana enabled conversion to the final SAG compound (Ahmadi et al., 2016).
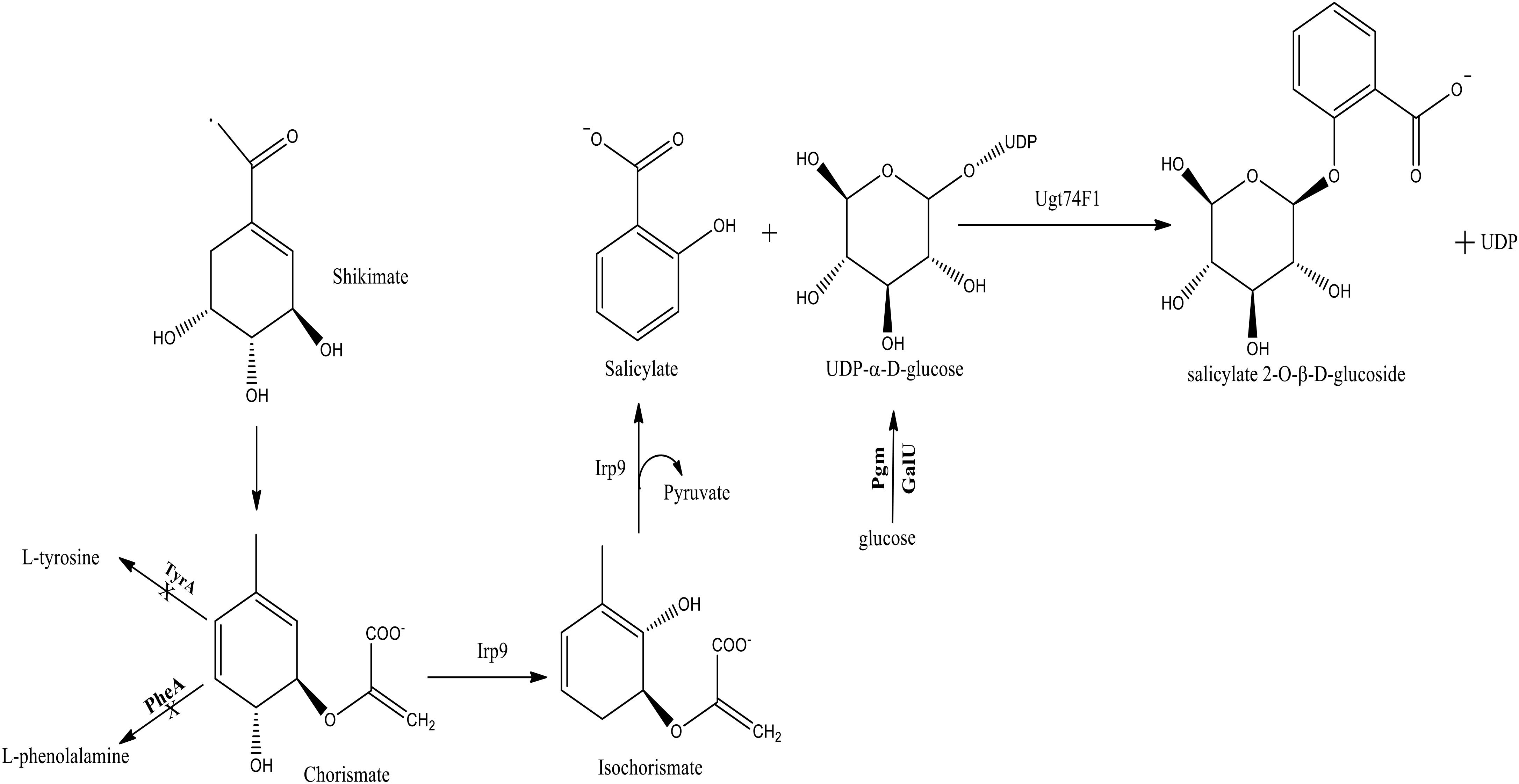
FIGURE 1. The native and heterologous metabolic pathway for salicylate 2-O-β-D-glucoside (SAG) biosynthesis established within E. coli. The native TyrA, PheA, Pgm, and GalU steps were either deleted or over-expressed (bold); whereas the heterologous Irp9 and Ugt74F1 steps were introduced from Yersinia enterocolitica and Arabidopsis thaliana, respectively.
In the work presented herein, we were interested in improving SAG production through the application of various heterologous production parameters that spanned the E. coli production strain and several expression plasmids and associated components. In addition, based upon previous work by us and others toward glycodiversification of heterologous natural products, we tested the expanded analog potential of the original SAG compound through the systematic incorporation of two isomeric deoxysugar pathways.
Materials and Methods
Plasmids and Strains
All cloning procedures were completed in E. coli TOP10 through which all recombinant plasmids were transferred and propagated. The irp9 gene from Y. enterocolitica genomic DNA, the pgm and galU genes from E. coli K-12 MG1655, and a codon-optimized ugt74F1 (salicylate glucosyltransferase gene) from A. thaliana were amplified by PCR with primers listed in Supplementary Table S1. Plasmids pMKA-41 and pGEX-UDP bearing these genes were constructed as described previously (Ahmadi et al., 2016) and used as the templates for the PCR reactions conducted in the current work. The PCR products were gel-purified and then digested with restriction enzyme pairs NheI/SalI (for irp9), XbaI/SalI (for galU), and NdeI/SalI (for pgm and ugt74F1). Digested irp9, pgm, galU, and ugt74F1 were separately ligated into similarly digested pET28a to yield pET28-irp9, pET28-pgm, pET28-galU, and pET28a-ugt74F1. Plasmid pET28-pgm was digested with XbaI/SalI and the insert transferred to an SpeI/SalI digested pET28-galU to yield pET28-galU-pgm. In the same way, the XbaI/SalI digested ugt74F1 fragment from pET28-ugt74F1 was inserted into SpeI/SalI digested pET28-irp9 to construct pET28-irp9-ugt74F1. Plasmid pET28-irp9-ugt74F1 was digested with XbaI/SalI and the insert ligated to SpeI/SalI digested pET28-galU-pgm to generate pRQS1. The pRQS1 galU-pgm-irp9-ugt74F1 cassette featured in Figure 2 was digested with NheI/BsiWI and ligated into pETcoco-1 for subsequent digestion and transfer of the same cassette (using XbaI/SalI) to pBAD33, yielding pRQS2 and pRQS3, respectively. A full list of plasmids and strains are presented in Supplementary Table S2 and detailed plasmid maps are provided in Supplementary Figures S1, S2.
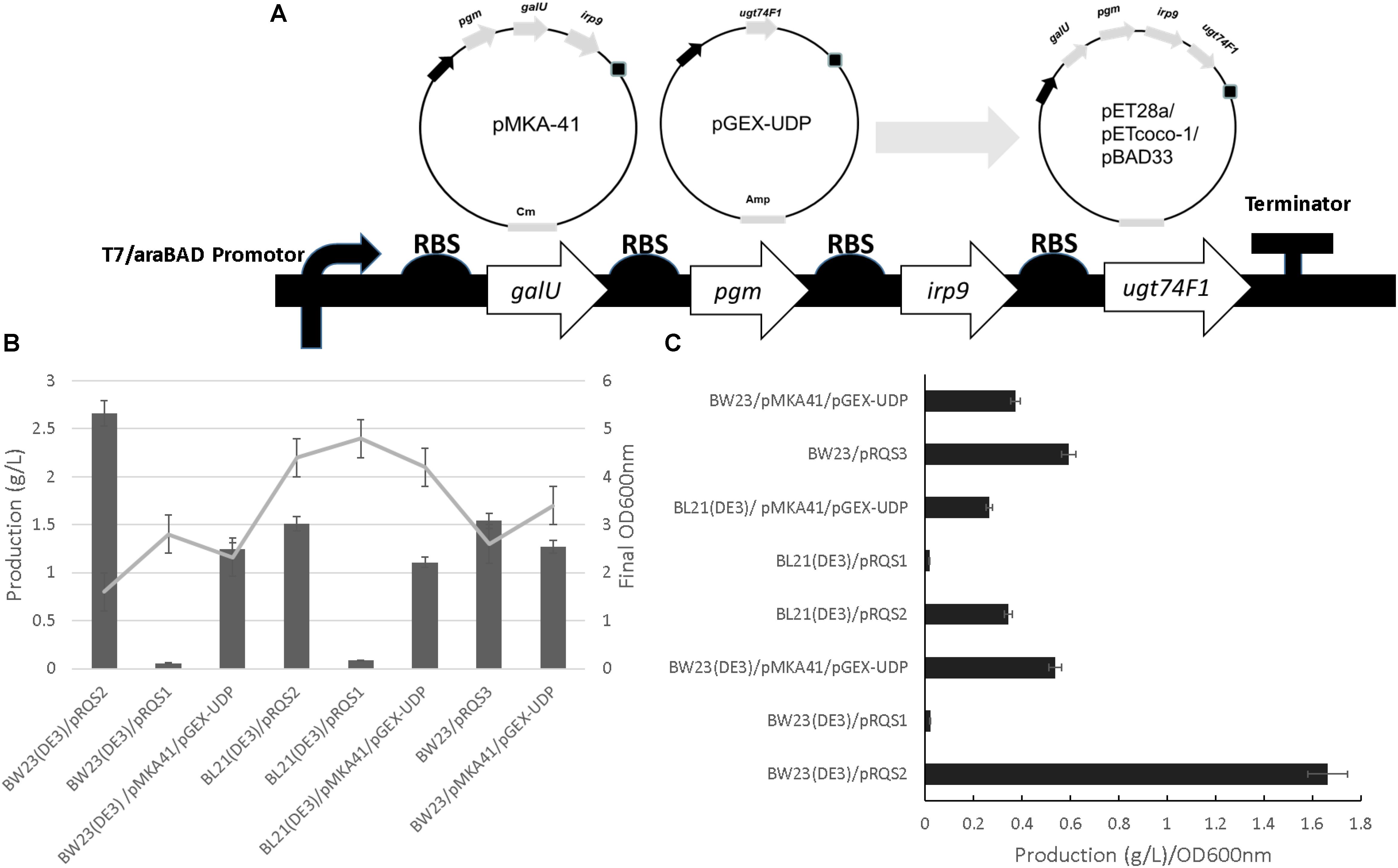
FIGURE 2. Salicylate 2-O-β-D-glucoside plasmid construction and heterologous production. (A) The integration of irp9, galU, pgm, and ugt74F1 into pET28a, pETcoco-1, and pBAD33 expression plasmid backgrounds, yielding pRSQ1, 2, and 3, respectively. Final OD600nm and production levels (B) and normalized production per cellular density (C) for SAG heterologous production strains.
The double-knockout BW mutant (BW23) was constructed as described in previous work (Ahmadi et al., 2016) through deletion of pheA and tyrA, with these deletions improving both SA and SAG production. The λDE3 prophage was integrated into the BW23 chromosome through co-infection using a λDE3 Lysogenization Kit (Novagen), yielding BW23(DE3). By doing so, the new host is equipped with the λDE3 recombinant phage DNA encoding for the T7 RNA polymerase, therefore, allowing the expression of the galU-pgm-irp9-ugt74F1 cassette though the T7 promotor within the pRQS1 and pRQS2 constructs. The pRSQ plasmids were transformed into corresponding strains through the standard heat-shock transformation protocol (Supplementary Table S3), and the resulting strains were stored as 20% glycerol stocks at -80°C.
The gene fragment for irp9 was liberated from pET28-irp9 via XbaI/SalI digestion and then ligated into SpeI/SalI digested pET28-galU-pgm to construct pET28-galU-pgm-irp9, which was then XbaI/SalI digested and ligated into pET21c to generate pRQS4. Plasmids pGJZ1, 2, 3, and 4 (Zhang et al., 2015), containing genes from the oleandomycin, chromomycin, and urdamycin A polyketide biosynthetic pathways, were used to produce two pairs of deoxysugar pathways for oliose and olivose. These four plasmids were integrated with codon-modified urdGT to construct pGJZ1-GT, pGJZ2-GT, pGJZ3-GT, and pGJZ4-GT (Supplementary Figure S3) which were, respectively, co-transferred with pRQS4 into BL21(DE3) (Supplementary Figure S4 and Supplementary Table S4) to provide the complete biosynthetic pathways for SAG analogs.
Culture Conditions and Medium Components
The bacterial culture medium and associated chemical and analytical components were obtained from Sigma-Aldrich (St. Louis, MO, United States) or Thermo Fisher Scientific (Waltham, MA, United States). The DNA-manipulating agents, including restriction enzymes, T4 DNA ligase, Phusion High-Fidelity PCR Master Mix, and associated reagents were purchased from New England Biolabs (Ipswich, MA, United States). PCR primers (Supplementary Table S1) were obtained from Eurofins Genomics (Huntsville, AL, United States).
Respective glycerol stocks of producing strains from Supplementary Table S3 were used to initiate overnight 3 mL cultures at 37°C with shaking in lysogeny broth (LB) medium prior to inoculating (1% v/v) 25 mL of M9Y medium which is formulated with (per liter): Na2HPO4⋅7H2O (12.8 g); KH2PO4 (3 g); NaCl (0.5 g); NH4Cl (1 g); yeast extract (1 g), glycerol (10 g), glucose (2.5 g), MgSO4⋅7H2O (246.5 mg), and CaCl2⋅2H2O (14.7 mg) supplemented with micronutrients including (per liter) vitamin B1 (2.0 mg), H3BO3 (1.25 mg), NaMoO4⋅2H2O (0.15 mg), CoCl2⋅6H2O (0.7 mg), CuSO4⋅5H2O (0.25 mg), MnCl2⋅4H2O (1.6 mg), and ZnSO4⋅7H2O (0.3 mg). After inoculation, cultures were incubated at 30°C with shaking for 2 days with induction initiated using 200 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) and/or 3 mg/mL arabinose when the culture OD600nm reached 0.4–0.6. As needed, plasmid selection in both liquid and solid medium was maintained with 100 mg/L ampicillin, 50 mg/L kanamycin, and 20 mg/L chloramphenicol.
Plasmid Stability Assay
Salicylate 2-O-β-D-glucoside producing strains were cultured in 25 mL production medium containing appropriate antibiotics at 37°C and 250 rpm until the OD600nm reading reached 0.4–0.6. At this point, cultures were cooled to 22°C, induced for gene expression as described above, and incubated an additional 5 days. Plasmid stability analysis was completed on the first and fifth days after cultures were induced. At these time points, dilutions from each culture were spread evenly on LB agar plates for incubation overnight at 37°C. Thirty colonies from each plate were selected and transferred to LB agar plates containing (1) combined antibiotics according to associated gene expression plasmids, (2) 1 mM IPTG, and (3) 1 mM IPTG + combined antibiotics as indicated in Table 1. Resulting colony development was then recorded, presented as a percentage of transferred colony growth on LB agar containing no antibiotics, and compared between strains.
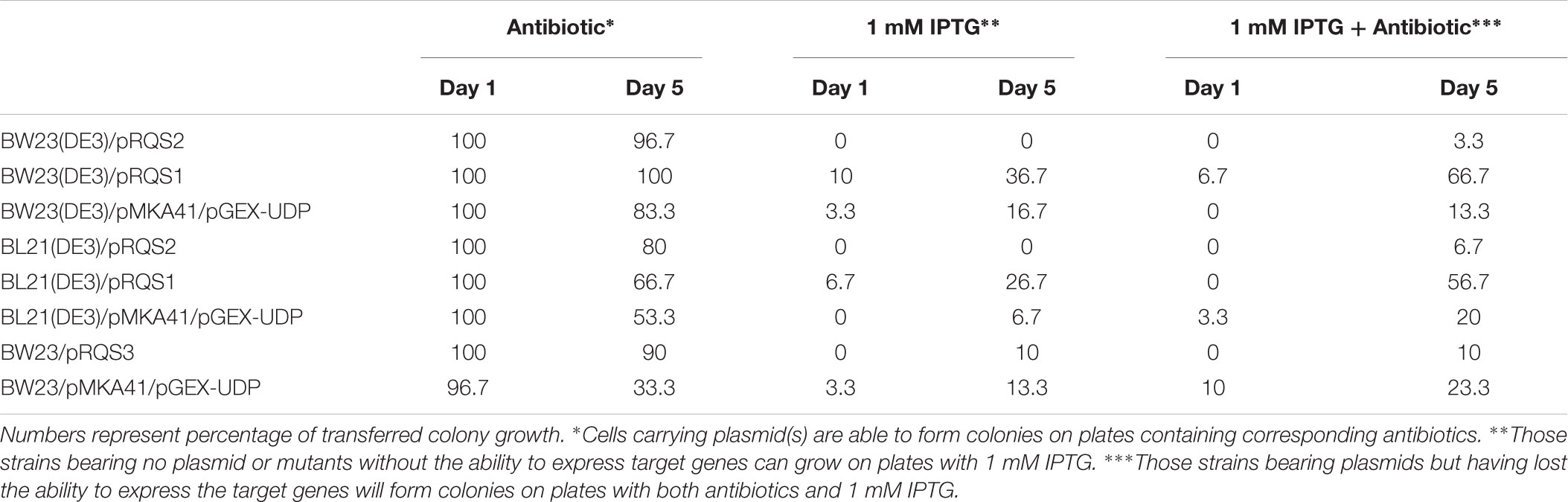
TABLE 1. Plasmid stability comparison with antibiotic selection, IPTG induction, and combined IPTG induction and antibiotic selection.
Salicylate 2-O-β-D-glucoside (SAG) Production Quantification
Post-culture, 1 mL of acetone was added per 50 mL culture and a 1 mL sample was centrifuged. Supernatant (50 μL) was analyzed by HPLC as described previously (Dean et al., 2005). Briefly, SAG and associated analogs were quantified using a ZORBAX Eclipse XDB-C18 column connected to an Agilent 1100 system equipped with a diode array detector. Solvent A was 0.1% acetic acid in water, solvent B was methanol, and a flow rate of 1 mL/min was used across the following gradient: 5–20% solvent B over 10 min; 20–80% solvent B over 5 min; 80% solvent B maintained for 5 min; reset to 5% solvent B. An absorbance wavelength of 274 nm was used for both SAG and associated analog quantification (Ahmadi et al., 2016). Peak area quantification was conducted compared to a standard calibration curve of pure SAG (Toronto Research Chemicals, Toronto, ON, Canada).
LC-MS Analysis for SAG Analog Assessment
A 1 mL SAG analog culture sample was centrifuged and 50 μL of supernatant was used for analysis. LC-MS was performed using an API 3000 Triple Quad LC-MS with a Turbo Ion Spray source (PE Sciex) coupled with a Shimadzu Prominence LC system. Chromatography was performed through a Waters XTerra C18 column (5 mm, 2.1 mm × 250 mm) and MS analysis was conducted in positive ion mode. Following a 3 μL injection from the 50 μL sample, a linear gradient of 5–95% acetonitrile (balance water; both solutions containing 0.1% formic acid) was used for 20 min at a flow rate of 0.2 mL/min.
Statistical Evaluation
Data presented were generated from three independent experiments, and error bars represent standard deviation values.
Results
As outlined in Figure 2, cellular parameters were engineered to improve specific SAG production. First, the production system designed previously for SAG formation [represented by plasmids pMKA-41 and pGEX-UDP (Ahmadi et al., 2016)] was organized into one operon introduced to three different expression plasmids (Figure 2A). Plasmids pETcoco-1 and pET28a allowed operon expression from the T7 promoter system coupled to a bacterial strain containing the T7 RNA polymerase (encoded within DE3 cellular variants). The pBAD33 plasmid featured expression driven by an arabinose inducible promoter system (Guzman et al., 1995). Plasmids were then introduced to strains BW23(DE3) and BL21(DE3) to accommodate the T7-based plasmids or BW23 for the pBAD33 plasmid system (also used for pMKA-41 and pGEX-UDP).
A production comparison revealed that the BW23(DE3)/pRSQ2 strain generated the best relative SAG levels based upon volumetric and specific titer comparisons (Figures 2B,C and Supplementary Figure S5). When comparing performance by either SAG titer or production per cell density, the BW23(DE3)/pRSQ2 strain demonstrated a twofold to fivefold improvement relative to the original BW23/pMKA-41/pGEX-UDP system. Of the new expression plasmids tested, pRSQ1 showed the lowest levels of SAG production. Of the production strains tested, the BW23(DE3) background, engineered to support SAG metabolic channeling and to accommodate the strong T7 promoter, demonstrated the best overall titers.
The plasmids used in this study included a low-copy option (pRSQ2; pETcoco-1 [OriV/S, 1–2 copies per cell]) and two medium copy options: pRSQ1 (pET28a [pBR322, ∼40 copies per cell]) and pRSQ3 (pBAD33 [pACYC184/p15A]). Table 1 presents plasmid stability data for the associated SAG production strains. From this analysis, the RSQ2 plasmid shows the best overall stability when tested for plasmid maintenance over time. The consolidated SAG biosynthetic pathways across plasmids pRSQ1-3 showed improved stability relative to the original dual expression plasmid system reliant upon pMKA-41 and pGEX-UDP. The same set of strains were also tested for stability when exposed to either IPTG induction or IPTG induction with antibiotic selection Novagen (1999). The plasmid stability data under these conditions indicate that, with the exception of pRSQ2, the newly constructed systems suffer from a combination of plasmid loss and mutant formation that results in lack of gene expression. From this perspective, the order of plasmid stability would be pRSQ2, pRSQ3, and pRSQ1.
Figure 3 outlines a schematic to test the flexibility of the SAG pathway to generate analogs resulting from alternative glycosylation patterning. In particular, isomeric variants of the oliose and olivose deoxysugars were tested for glycodiversification of the incoming salicylate precursor (Aguirrezabalaga et al., 2000). Initial analog production efforts with the Ugt74F1 salicylate glucosyltransferase resulted in minimal product formation (data not shown). As an alternative, the urdamycin system glycotransferase (UrdGT) was used due to previously observed flexibility in glycosylation patterning (Hoffmeister et al., 2000, 2003). Using HPLC and LC-MS analysis, data supporting analog formation were generated for each deoxysugar variant (Figures 4A,B and Supplementary Figures S6, S7). However, production levels were significantly reduced compared to those from the original SAG production systems (Figure 2).
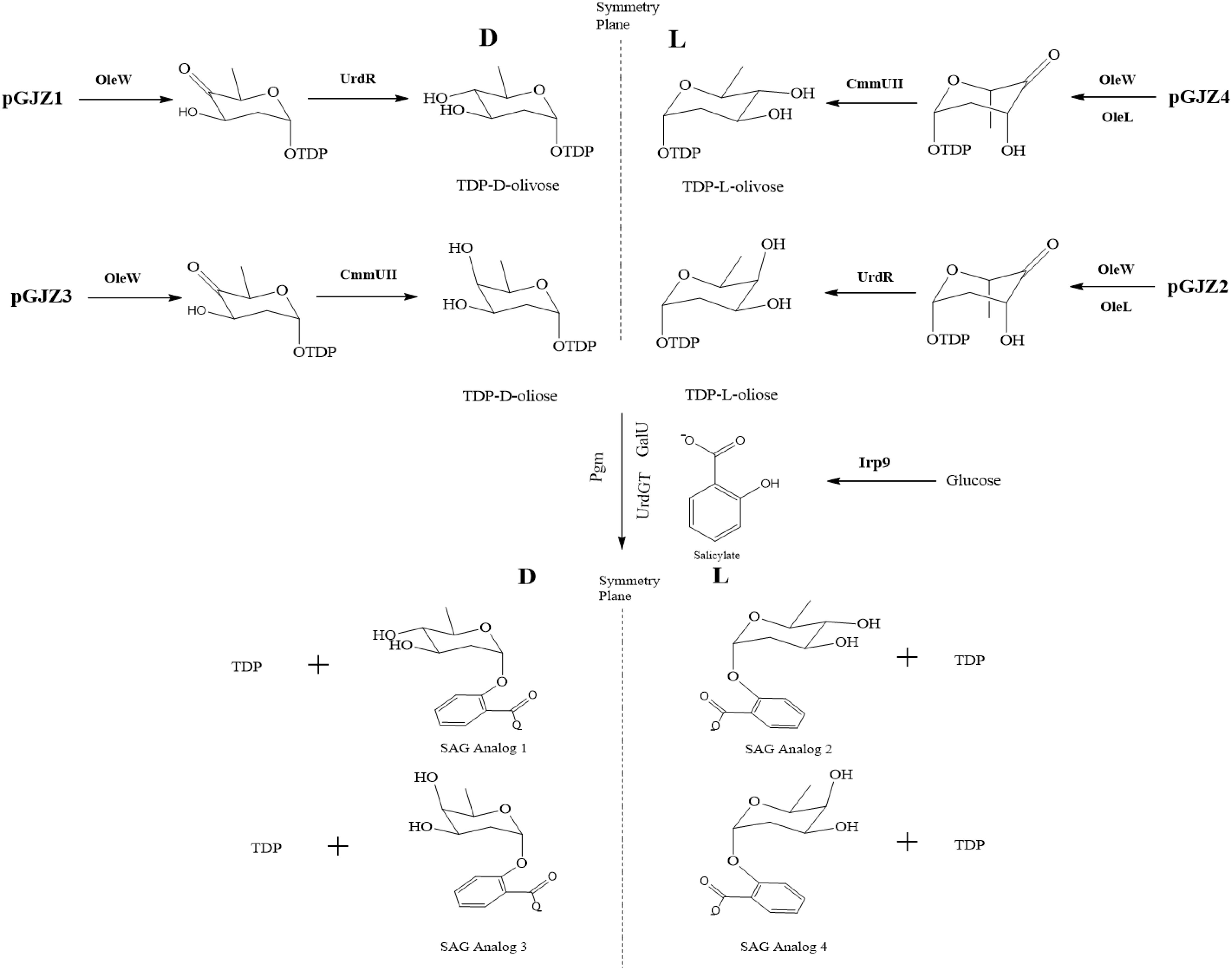
FIGURE 3. Biosynthetic pathways for the production of SAG analogs. pGJZ plasmids 1–4 encode pairs of chiral-symmetric olivose and oliose deoxysugars to glycodiversify salicylate.
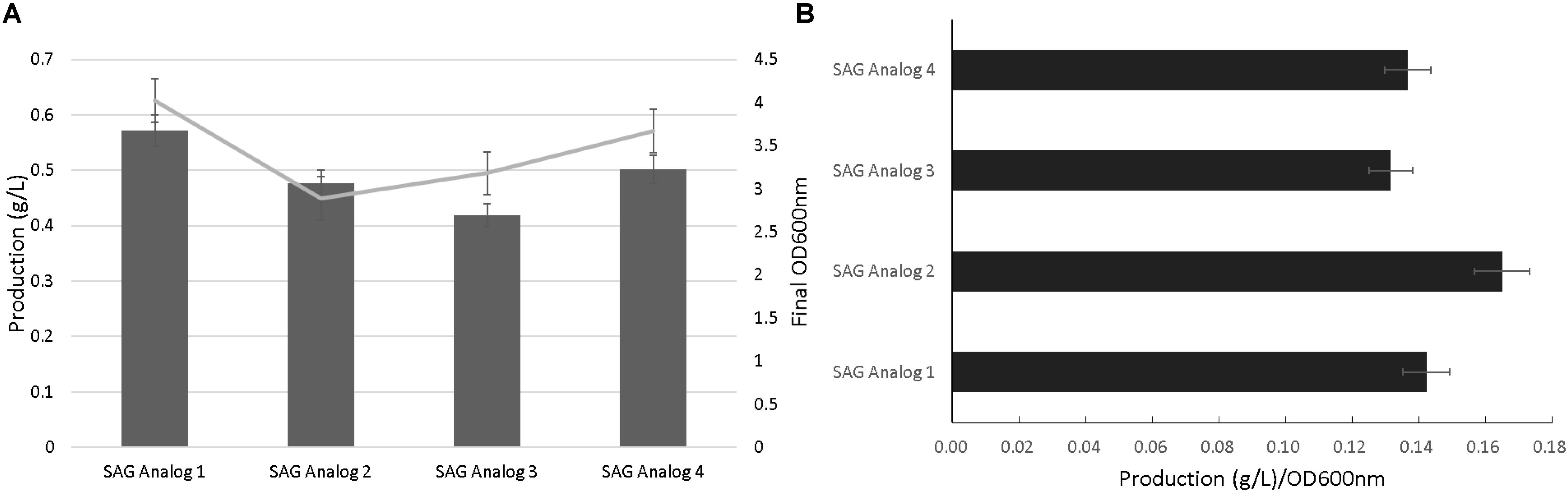
FIGURE 4. Salicylate 2-O-β-D-glucoside analog production. Final OD600nm and production levels (A) and normalized production per cellular density (B).
Discussion
A combination of metabolic engineering and gene expression design resulted in plasmids pRSQ1, 2, and 3. Each plasmid design consolidated genes needed for SAG biosynthesis. Furthermore, the plasmids featured different copy numbers and promoter strengths and, when combined with E. coli strains engineered to support gene expression and SAG production, allowed for a systematic evaluation of final product values. Production levels were best for strain BW23(DE3)/pRSQ2, which featured a metabolically engineered strain to streamline carbon flow to SAG production and support the strong T7 promoter driving expression from pRSQ2. Of note, this particular expression plasmid was the lowest copy version of those tested. pRSQ1 featured T7-based gene expression but from a higher copy plasmid; pRSQ3 utilized an arabinose-inducible system within pBAD33 (similar to use previously with the control system BW23/pMDA-41/pGEX-UDP). This particular study focused only on SAG production as a function of plasmid biosynthetic pathway design (Figure 2); however, future engineering approaches, such as tuning biosynthetic steps via expression variation, will likely be needed to maximally drive complete salicylate to SAG formation (Ahmadi et al., 2016).
One likely contributor to the heightened production observed for pRSQ2 was incorporation of the highly stable OriV/S replication system (Shizuya et al., 1992; Monaco and Larin, 1994; Tao and Zhang, 1998). As such, even though the copy number for this plasmid was reduced, stability was improved as indicated within the results presented in Table 1. The strong T7 expression system likely compensated for the reduced copy number level (Golomb and Chamberlin, 1974; Studier and Moffatt, 1986); whereas, the higher copy number T7 system represented by pRSQ1 showed lower relative SAG production and higher plasmid loss. The pRSQ2 system has the added advantage (not tested in this study) of plasmid copy-up capability (Wild et al., 1996, 2001, 2002; Wild and Szybalski, 2004). Thus, there is the potential for further increased production through the stable maintenance of the pRSQ2 plasmid during the growth phase of the host system followed by induction of both SAG biosynthesis and plasmid amplification to spur subsequent product generation.
Our group and others have studied natural product glycodiversification, which offers a directed means of natural product structural variation (Thibodeaux et al., 2007, 2008; Williams et al., 2008; Jiang et al., 2013; Zhang et al., 2015; Fang et al., 2018). Given the glycosylated nature of SAG, we were interested if this product could also accommodate alternative sugar moieties. In conducting this work, we relied on a series of deoxysugar pathways previously incorporated into polyketide biosynthesis to generate erythromycin analogs (Zhang et al., 2015). However, contingent upon this strategy working is the flexibility of a glycosylation enzymatic step capable of accepting new substrate groups. Use of the original glucosyltransferas (Ugt74F1) resulted in minimal analog production. As a result, we turned to an alternative glycotransferase from the urdamycin biosynthetic pathway [recognized for substrate flexibility (Hoffmeister et al., 2002; Luzhetskyy et al., 2005) and the same source as some of the deoxysugar pathways genes], which resulted in the production level of the analogs presented in Figure 4. The ability for the urdamycin glycotransferase to accommodate novel SAG analogs supports the general theme of glycodiversification applied to this compound. As in the case of several previous examples of analogs produced as a result of biosynthetic pathway modification (Jiang and Pfeifer, 2013; Zhang et al., 2015), titer levels of the SAG analogs were significantly reduced compared to the original product, likely to do the new substrates limiting the catalytic activity of the glycosyltransferase. We also note that additional analytical work is needed to fully chemically characterize these new analogs. However, indication of novel analogs provides a basis for future studies to test potential variation in bioactivity in applications that range from inflammatory relief [as we have tested previously with the original SAG compound (Ahmadi et al., 2016)] or plant stress protection.
Author Contributions
GZ and BP designed and supervised the study. RQ executed the experimental plan.
Funding
The authors recognize funding provided by the NYS Pollution Prevention Institute through a grant from the NYS Department of Environmental Conservation. Any opinions, findings, conclusions, or recommendations expressed are those of the author(s) and do not necessarily reflect the views of the Department of Environmental Conservation. The authors also acknowledge funding support from the following agencies: SUNY 4E and NSF (1550378).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02241/full#supplementary-material
References
Aguirrezabalaga, I., Olano, C., Allende, N., Rodriguez, L., Brana, A. F., Mendez, C., et al. (2000). Identification and expression of genes involved in biosynthesis of L-oleandrose and its intermediate L-olivose in the oleandomycin producer Streptomyces antibioticus. Antimicrob. Agents Chemother. 44, 1266–1275. doi: 10.1128/AAC.44.5.1266-1275.2000
Ahmadi, M. K., Fang, L., Moscatello, N., and Pfeifer, B. A. (2016). E. coli metabolic engineering for gram scale production of a plant-based anti-inflammatory agent. Metab. Eng. 38, 382–388. doi: 10.1016/j.ymben.2016.10.001
Dean, J. V., Mohammed, L. A., and Fitzpatrick, T. (2005). The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 221, 287–296. doi: 10.1007/s00425-004-1430-3
Fang, L., Zhang, G., El-Halfawy, O., Simon, M., Brown, E. D., and Pfeifer, B. A. (2018). Broadened glycosylation patterning of heterologously produced erythromycin. Biotechnol. Bioeng. doi: 10.1002/bit.26735 [Epub ahead of print].
Golomb, M., and Chamberlin, M. (1974). Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J. Biol. Chem. 249, 2858–2863.
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Hoffmeister, D., Drager, G., Ichinose, K., Rohr, J., and Bechthold, A. (2003). The C-Glycosyltransferase UrdGT2 is unselective toward D- and L-configured nucleotide-bound rhodinoses. J. Am. Chem. Soc. 125, 4678–4679. doi: 10.1021/ja029645k
Hoffmeister, D., Ichinose, K., Domann, S., Faust, B., Trefzer, A., Drager, G., et al. (2000). The NDP-sugar co-substrate concentration and the enzyme expression level influence the substrate specificity of glycosyltransferases: cloning and characterization of deoxysugar biosynthetic genes of the urdamycin biosynthetic gene cluster. Chem. Biol. 7, 821–831. doi: 10.1016/S1074-5521(00)00029-6
Hoffmeister, D., Wilkinson, B., Foster, G., Sidebottom, P. J., Ichinose, K., and Bechthold, A. (2002). Engineered urdamycin glycosyltransferases are broadened and altered in substrate specificity. Chem. Biol. 9, 287–295. doi: 10.1016/S1074-5521(02)00114-X
Jiang, M., and Pfeifer, B. A. (2013). Metabolic and pathway engineering to influence native and altered erythromycin production through E. coli. Metab. Eng. 19C, 42–49. doi: 10.1016/j.ymben.2013.05.005
Jiang, M., Zhang, H., Park, S. H., Li, Y., and Pfeifer, B. A. (2013). Deoxysugar pathway interchange for erythromycin analogues heterologously produced through Escherichia coli. Metab. Eng. 20, 92–100. doi: 10.1016/j.ymben.2013.09.005
Kerbarh, O., Ciulli, A., Howard, N. I., and Abell, C. (2005). Salicylate biosynthesis: overexpression, purification, and characterization of Irp9, a bifunctional salicylate synthase from Yersinia enterocolitica. J. Bacteriol. 187, 5061–5066. doi: 10.1128/JB.187.15.5061-5066.2005
Lin, Y., Sun, X., Yuan, Q., and Yan, Y. (2014). Extending shikimate pathway for the production of muconic acid and its precursor salicylic acid in Escherichia coli. Metab. Eng. 23, 62–69. doi: 10.1016/j.ymben.2014.02.009
Luzhetskyy, A., Vente, A., and Bechthold, A. (2005). Glycosyltransferases involved in the biosynthesis of biologically active natural products that contain oligosaccharides. Mol. Biosyst. 1, 117–126. doi: 10.1039/b503215f
Monaco, A. P., and Larin, Z. (1994). YACs, BACs, PACs and MACs: artificial chromosomes as research tools. Trends Biotechnol. 12, 280–286. doi: 10.1016/0167-7799(94)90140-6
Pelludat, C., Brem, D., and Heesemann, J. (2003). Irp9, encoded by the high-pathogenicity island of Yersinia enterocolitica, is able to convert chorismate into salicylate, the precursor of the siderophore yersiniabactin. J. Bacteriol. 185, 5648–5653. doi: 10.1128/JB.185.18.5648-5653.2003
Rivas-San Vicente, M., and Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. doi: 10.1093/jxb/err031
Shizuya, H., Birren, B., Kim, U. J., Mancino, V., Slepak, T., Tachiiri, Y., et al. (1992). Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. U.S.A. 89, 8794–8797. doi: 10.1073/pnas.89.18.8794
Studier, F. W., and Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130. doi: 10.1016/0022-2836(86)90385-2
Tao, Q., and Zhang, H. B. (1998). Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors. Nucleic Acids Res. 26, 4901–4909. doi: 10.1093/nar/26.21.4901
Thibodeaux, C. J., Melancon, C. E. III, and Liu, H. W. (2008). Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. Int. Ed. Engl. 47, 9814–9859. doi: 10.1002/anie.200801204
Thibodeaux, C. J., Melancon, C. E., and Liu, H. W. (2007). Unusual sugar biosynthesis and natural product glycodiversification. Nature 446, 1008–1016. doi: 10.1038/nature05814
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wild, J., Hradecna, Z., Posfai, G., and Szybalski, W. (1996). A broad-host-range in vivo pop-out and amplification system for generating large quantities of 50- to 100-kb genomic fragments for direct DNA sequencing. Gene 179, 181–188. doi: 10.1016/S0378-1119(96)00487-8
Wild, J., Hradecna, Z., and Szybalski, W. (2001). Single-copy/high-copy (SC/HC) pBAC/oriV novel vectors for genomics and gene expression. Plasmid 45, 142–143.
Wild, J., Hradecna, Z., and Szybalski, W. (2002). Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res. 12, 1434–1444. doi: 10.1101/gr.130502
Wild, J., and Szybalski, W. (2004). Copy-control pBAC/oriV vectors for genomic cloning. Methods Mol. Biol. 267, 145–154. doi: 10.1385/1-59259-774-2:145
Williams, G. J., Gantt, R. W., and Thorson, J. S. (2008). The impact of enzyme engineering upon natural product glycodiversification. Curr. Opin. Chem. Biol. 12, 556–564. doi: 10.1016/j.cbpa.2008.07.013
Keywords: salicylate, salicylate 2-O-β-D-glucoside, metabolic engineering, E. coli, analog
Citation: Qi R, Pfeifer BA and Zhang G (2018) Engineering Heterologous Production of Salicylate Glucoside and Glycosylated Variants. Front. Microbiol. 9:2241. doi: 10.3389/fmicb.2018.02241
Received: 21 July 2018; Accepted: 03 September 2018;
Published: 20 September 2018.
Edited by:
Dipesh Dhakal, Sun Moon University, South KoreaReviewed by:
Joong-Hoon Ahn, Konkuk University, South KoreaPrakash Parajuli, University of Maryland, Baltimore, United States
Copyright © 2018 Qi, Pfeifer and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Blaine A. Pfeifer, YmxhaW5lcGZAYnVmZmFsby5lZHU= Guojian Zhang, Z3VvamlhbnpAYnVmZmFsby5lZHU=; emhhbmdndW9qaWFuQG91Yy5lZHUuY24=
 Ruiquan Qi
Ruiquan Qi Blaine A. Pfeifer
Blaine A. Pfeifer Guojian Zhang1,2,3*
Guojian Zhang1,2,3*