- College of Biotechnology and Bioengineering, Zhejiang University of Technology, Hangzhou, China
Horizontal gene transfer (HGT) plays an important role in bacterial evolution. It is well accepted that DNA is pulled/pushed into recipient cells by conserved membrane-associated DNA transport systems, which allow the entry of only single-stranded DNA (ssDNA). However, recent studies have uncovered a new type of natural bacterial transformation in which double-stranded DNA (dsDNA) is taken up into the cytoplasm, thus complementing the existing methods of DNA transfer among bacteria. Regulated by the stationary-phase regulators RpoS and cAMP receptor protein (CRP), Escherichia coli establishes competence for natural transformation with dsDNA, which occurs in agar plates. To pass across the outer membrane, a putative channel, which may compete for the substrate with the porin OmpA, may mediate the transfer of exogenous dsDNA into the cell. To pass across the inner membrane, dsDNA may be bound to the periplasmic protein YdcS, which delivers it into the inner membrane channel formed by YdcV. The discovery of cell-to-cell contact-dependent plasmid transformation implies the presence of additional mechanism(s) of transformation. This review will summarize the current knowledge about mechanisms of HGT with an emphasis on recent progresses regarding non-canonical mechanisms of natural transformation. Fully understanding the mechanisms of HGT will provide a foundation for monitoring and controlling multidrug resistance.
Introduction
Horizontal gene transfer (HGT) drives the evolution of bacteria. Transfer of antibiotic resistance genes (ARGs) plays an important role in the development of multidrug resistance (MDR) in bacteria (Forsberg et al., 2012). There are three “classical" methods of DNA transfer in nature: bacterial conjugation, natural transformation, and transduction (von Wintersdorff et al., 2016). Via HGT, exogenous DNA can be transferred from one bacterium to another even if they are only distantly related (Chen et al., 2005; Burton and Dubnau, 2010). With the accumulation of genes involved in different resistance mechanisms from the exogenous DNA, bacteria are able to acquire MDR rapidly. For example, Acinetobacter and Enterobacter strains carrying the NDM-1 plasmid or mcr-1 plasmid, which contain a group of resistance genes, can tolerate even last-resort antibiotics (Yong et al., 2009; Wang and Sun, 2015; Liu et al., 2016; Shen et al., 2016; Zheng et al., 2017). Understanding the mechanisms of DNA transfer in bacteria would provide new strategies to help address the ongoing challenge of multi-drug-resistant bacteria in the future.
Natural bacterial transformation and conjugation have been found in bacteria and archaea. Two different membrane protein complexes consisting of conserved proteins are responsible for pulling in and pushing out DNA during natural bacterial transformation and conjugation, respectively (Chen and Dubnau, 2004; Claverys et al., 2009; Burton and Dubnau, 2010; Johnston et al., 2014; Cabezon et al., 2015; Ilangovan et al., 2015, 2017). These DNA transport systems deliver single-stranded DNA (ssDNA) either from the donor cell (for conjugation) or into the recipient cell (for transformation) (Chen and Dubnau, 2004; Claverys et al., 2009; Burton and Dubnau, 2010; Johnston et al., 2014; Cabezon et al., 2015; Ilangovan et al., 2015, 2017). Recent studies have revealed two new types of DNA transfer in Escherichia coli. One of these methods has been shown to be independent of the conserved proteins for the transport of ssDNA during natural transformation or bacterial conjugation. Instead, double-stranded DNA (dsDNA) is taken up into the cytoplasm and internalized by E. coli cells on solid agar plates (Sun et al., 2006, 2009, 2013; Sun, 2011, 2016; Zhang et al., 2012). The other method of DNA transfer is dependent on cell-to-cell contact and DNA transfer occurs within a colony on agar plates (Maeda et al., 2004, 2006; Etchuuya et al., 2011; Sobue et al., 2011; Kurono et al., 2012; Matsuda et al., 2012; Matsumoto et al., 2016). DNA transfer via this method is sensitive to DNase I, indicating that DNA that is transported into the recipient cell is naked (rather than protein-protected). Although DNA transfer occurs on agar plates via both of the above two transformation methods, no evidence shows similarities between DNA transfer mechanisms of the two methods. In the former transformation method, DNA transfer occurs in the absence of donor cells. Whereas, in the latter transformation method, donor cells are required for DNA transfer in the colony. During bacterial conjugation, physical contact between the donor and the recipient cells is required. Nonetheless, the cell-to-cell contact-dependent plasmid transformation is different from conjugation in that DNA transfer is not mediated by mobile elements (Maeda et al., 2004; Kurono et al., 2012; Matsuda et al., 2012). In this review, we will first discuss the mechanisms of classical natural bacterial transformation and conjugation. Then, the non-canonical DNA transfer on agar plates will be described in detail, with an emphasis on how DNA is pulled into cells.
Pull Dna in During Natural Transformation
Natural transformation was discovered in Streptococcus pneumoniae in 1928 (Griffith, 1928). Induced by a heptadecapeptide pheromone, naturally transformable bacteria show a special physiological state termed “competence”, during which they are capable of pulling in exogenous DNA (Berka et al., 2002; Ogura et al., 2002). The mechanism of DNA transfer during natural transformation is well conserved among Gram-positive (G+) (e.g., Bacillus subtilis and S. pneumoniae) and Gram-negative (G−) bacterial species (e.g., Neisseria gonorrhoeae, Haemophilus influenzae, and Vibrio cholerae), as well as archaea (Chen and Dubnau, 2004; Claverys et al., 2009; Burton and Dubnau, 2010; Johnston et al., 2014; Cabezon et al., 2015; Ilangovan et al., 2015, 2017; Veening and Blokesch, 2017). Although conditions for competence induction vary widely among bacterial species (Aas et al., 2002; Berka et al., 2002; Claverys et al., 2006; Veening and Blokesch, 2017), proteins involved in DNA uptake are highly conserved even among distantly related bacteria (Johnston et al., 2014), except for Helicobacter pylori, which uses a conjugation-like system for DNA uptake during natural transformation (Smeets and Kusters, 2002). Here, the conserved DNA uptake system in bacteria is described.
G− bacteria have an outer membrane (OM), whereas G+ bacteria have not. During natural transformation, G− bacteria need to pull DNA across both the OM and the inner membrane (IM), whereas G+ bacteria need to overcome the barrier of the peptidoglycan layer, which is much thicker and denser, and needs to be weakened before translocation of DNA across the IM. In this process, dsDNA is pulled across the OM in G− bacteria and ssDNA is pulled across the IM in both G+ and G− bacteria. The general mechanism underlying DNA transfer during natural transformation is summarized in Figure 1.
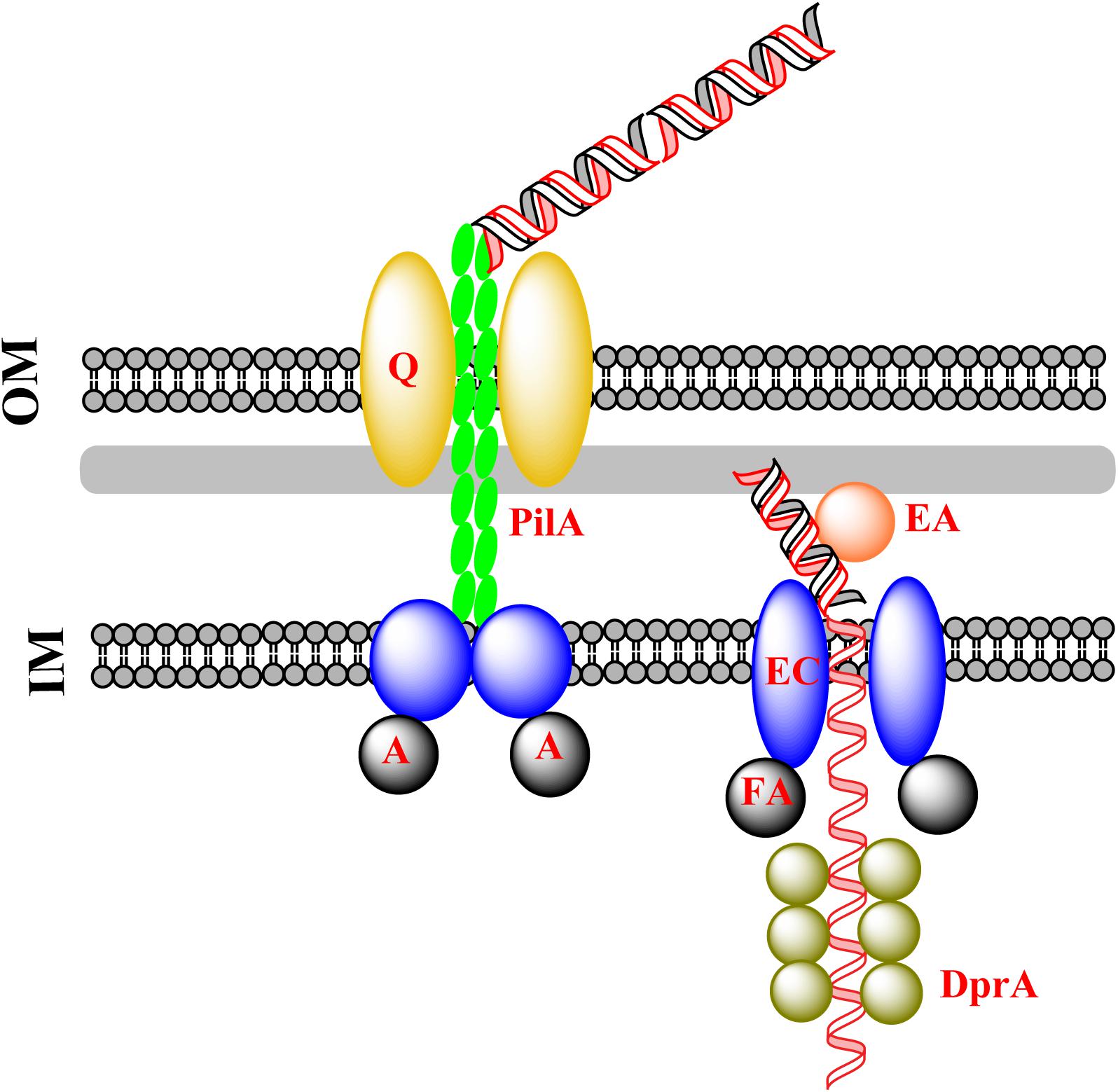
FIGURE 1. Classical DNA uptake during natural transformation. Exogenous DNA is pulled into the cytoplasm by the extension and retraction of pseudopili, as a consequence of the assembly and disassembly of pseudopilin multimers (PilA). This is followed by the transfer of dsDNA across the OM protein PilQ/HofQ (for G− bacteria only). The DNA receptor (ComEA) mediates the transfer of one strand of DNA across the IM channel formed by ComEC with the assistance of the ATPase ComFA, accompanied by degradation of the other strand of DNA. The incoming ssDNA is protected by DprA which conveys it to RecA for homologous recombination in the cytoplasm.
Crossing the OM
For G− bacteria, a sophisticated protein complex is assembled in the OM, where the complex binds exogenous DNA and drags it into the periplasm (Figure 1). The assembly and disassembly of a type IV pilus causes a fiber-like pseudopilus to be extruded out of and hauled back into the pore-forming OM proteins (Chen and Dubnau, 2004). The pore that accommodates exogenous DNA is 6–6.5 nm in diameter, and is formed by PilQ, a secretin that is 15 nm wide and 34 nm long with five rings and an extraordinary stable “cone” and “cup” structures (Chen and Dubnau, 2004). The pore cavity is large enough to accommodate dsDNA (∼2.4 nm) (Collins et al., 2001; Assalkhou et al., 2007; Burkhardt et al., 2011). Accompanied by the extension and retraction of type IV pili, DNA is transported across the OM through the pore (Laurenceau et al., 2013; Salzer et al., 2014, 2016; Leong et al., 2017). Between the OM and IM (the periplasm), the incoming DNA is bound by the substrate-binding protein ComEA, which prevents DNA from slipping by means of a “Brownian Ratcheting” mechanism (Inamine and Dubnau, 1995; Provvedi and Dubnau, 1999; Berge et al., 2002; Takeno et al., 2012; Seitz et al., 2014; Salzer et al., 2016).
Crossing the IM
Bound by ComEA in the periplasm, exogenous DNA is translocated across the IM via a pore formed by ComEC, also named Rec2 in some G− bacteria (e.g., H. influenzae), a widely conserved IM protein for translocation of DNA across the IM (Barouki and Smith, 1985; Berge et al., 2002; Draskovic and Dubnau, 2005; Sinha et al., 2012; Baker et al., 2016; Salzer et al., 2016). It has been proposed that ComEC acts as both a translocase and nuclease during DNA translocation (Chen and Dubnau, 2004; Claverys et al., 2009; Burton and Dubnau, 2010; Johnston et al., 2014; Veening and Blokesch, 2017). In this process, one strand of the dsDNA is translocated into the cytoplasm, simultaneously the degradation of the other strand occurs (Barouki and Smith, 1985; Berge et al., 2002; Baker et al., 2016). In silico analysis of ComEC indicates that the β-lactamase-like domain at the C-terminus may function as a nuclease and Domain of Unknown function 4131 at the N-terminus has a DNA binding domain (Baker et al., 2016). It is possible that the dsDNA that is bound to the N-terminus of ComEC is divided into two strands of ssDNA and one of them is degraded by the nuclease at the C-terminus.
The channel for DNA translocation is assumed to be formed by two ComEC monomers with seven transmembrane segments (Draskovic and Dubnau, 2005). Given that over-expression of ComEC is toxic to the cell, the structure of ComEC remains unresolved (Draskovic and Dubnau, 2005). The driving force for translocation of ssDNA across the IM may be provided by an ATPase (ComFA), which is also widely conserved in bacteria (Londono-Vallejo and Dubnau, 1994; Takeno et al., 2011; Chilton et al., 2017; Diallo et al., 2017). After translocation of ssDNA, DprA, and RecA bind ssDNA and catalyze the formation of joint DNA molecules for homologous recombination (Mortier-Barriere et al., 2007; Dwivedi et al., 2013; Yadav et al., 2013, 2014; Duffin and Barber, 2016; Diallo et al., 2017; Hovland et al., 2017; Le et al., 2017). In this way, the incoming foreign ssDNA displaces one strand of the chromosomal dsDNA (Mortier-Barriere et al., 2007), followed by being converted to homogeneous dsDNA through DNA replication.
The Transfer of Dna During Bacterial Conjugation
Bacterial conjugation was first discovered in E. coli (Lederberg and Tatum, 1946). Relying on cell-to-cell contact, DNA can be pushed out of a donor cell and transported into a recipient cell during bacterial conjugation. A group of modular mobile genetic elements, known as integrative and conjugative elements (ICEs) or conjugative transposons (Franke and Clewell, 1981), has been found in many bacterial genomes (Wozniak and Waldor, 2010; Bi et al., 2012; Cury et al., 2017). ICEs can transfer from one bacterium to another, facilitating the spread of ARGs in environment (Wozniak and Waldor, 2010; Bi et al., 2012; Cury et al., 2017). The transfer of conjugative DNA across the membrane of the donor bacterium relies on a large membrane-associated protein complex, that belongs to the type IV secretion system (T4SS) (Goessweiner-Mohr et al., 2013; Cabezon et al., 2015; Ilangovan et al., 2015; Figure 2). Components of the T4SS for conjugation are encoded by genes of either self-replicable conjugative plasmids or ICEs in the chromosomal DNA of the donor bacterium (Cabezon et al., 2015; Ilangovan et al., 2015; Johnson and Grossman, 2015). The mechanism of DNA transfer is summarized in Figure 2.
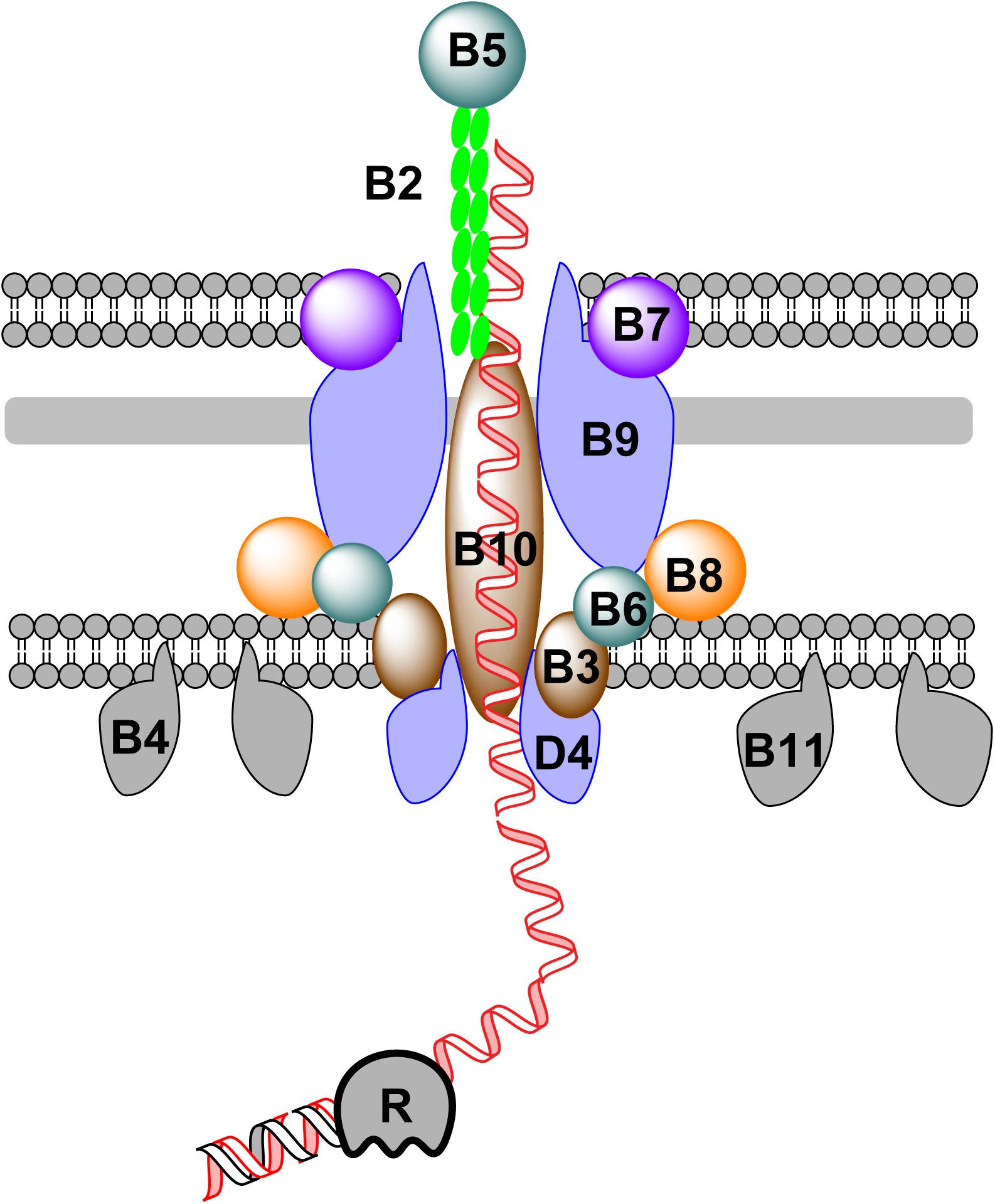
FIGURE 2. DNA transfer during bacterial conjugation. Conjugative DNA is processed into ssDNA by a relaxase (R) in the cytoplasm of the donor bacterium. To further transport ssDNA, a group of membrane and periplasmic proteins are assembled together to form a large complex, which can be subdivided into four distinct parts: (1) the pilus is formed by the assembly of pilins (VirB2) with adhesins (VirB5) at the distal end; (2) the OM component, which consists of VirB7, VirB9, and the C-terminus of VirB10; (3) the periplasmic component which consists of VirB8, VirB10, and VirB6; and (4) the IM component, formed of VirB3, VirB6, VirB8, and VirB10. Additionally, three hexameric ATPases (VirB4, VirB11, and VirD4) are attached to the IM to provide energy during DNA transfer.
Protein-coated conjugative ssDNA is transported across the IM, periplasm and OM through a membrane channel formed by a group of proteins encoded by the conjugative DNA molecule (Goessweiner-Mohr et al., 2013; Cabezon et al., 2015; Ilangovan et al., 2015). Inside the channel, the conjugative pilus (formed by pilins) is responsible for pushing ssDNA out of membrane. The mechanism of DNA transfer via conjugation has been best exemplified by the Vir system. To export DNA out of the donor cell, a conjugative plasmid encodes a complicated membrane protein complex. During conjugation, a plasmid- or ICE-encoded relaxase creates a nick in one strand of the conjugative DNA at the oriT site, followed by ssDNA translocation across the channel formed by components of the T4SS and replication of the remaining strand, either independently from or in concert with conjugation (Ilangovan et al., 2017). During translocation across cell membranes, three ATPases (VirD4, VirB4, and VirB11) provide the energy for DNA transport (Chen et al., 2005; Cabezon et al., 2015; Ilangovan et al., 2015). In natural transformation and conjugation, different types of pili participate in the movement of DNA. Competence pili or pseudopili mediate the transfer of dsDNA across the membrane during natural transformation, whereas conjugative pili mediates the transfer of ssDNA across the membrane during conjugation (Cabezon et al., 2015; Ilangovan et al., 2015). In both cases, the assembly/disassembly of pili drives the movement of transferring DNA. It remains unclear how conjugative DNA is further transported in the recipient cell.
The Uptake of dsDna in E. coli
E. coli has long been thought not to be naturally transformable. In this century, the natural transformation of E. coli has been observed initially on nutrient-deficient agar plates and later on nutrient-rich agar plates (Tsen et al., 2002; Sun et al., 2006). Although natural plasmid transformation of E. coli shows single-hit kinetics, implying that dsDNA may enter the cell (Sun et al., 2009), there are basic differences between natural and chemical transformation. First, natural transformation is promoted by an increased concentration of agar, whereas chemical transformation relies on high concentrations of divalent ions (i.e., Ca2+, Mg2+, or Mn2+) (Sun et al., 2009). However, the stimulating effect of agar on transformation is not due to an increase in Ca2+, Mg2+, or Mn2+ concentration (Sun et al., 2009). It remains unclear whether osmotic pressure and/or any other biological/physical factor(s) contribute to the increase of transformation on plates with a high concentration of agar. Second, an OM protein, OmpA, plays opposite roles in natural and chemical transformation of E. coli: it promotes chemical transformation but suppresses natural transformation (Sun et al., 2013). Third, exponentially growing E. coli cells are often employed for preparing chemically competent cells with the highest efficiency, and chemical transformation occurs in a liquid, whereas the natural transformation of stationary-phase E. coli cells is regulated by the transcriptional regulator RpoS and the cyclic AMP (cAMP) – cAMP receptor protein (CRP), and these cells can acquire exogenous DNA exclusively on agar plates (Zhang et al., 2012; Guo et al., 2015). The functions of RpoS or the cAMP-CRP complex in the chemical transformation of E. coli have not been found.
Natural transformation of E. coli also differs from that of other naturally transformable bacteria in that the conserved DNA uptake machinery is not required for the uptake of exogenous dsDNA in E. coli (Sun et al., 2009). Some components of the conserved DNA uptake machinery are believed to function in using DNA as a nutrient in E. coli (Finkel and Kolter, 2001; Palchevskiy and Finkel, 2006). Nevertheless, attempts to confirm these observations in three independent laboratories have not succeeded (Sun, 2011; Johnston et al., 2014). DNA, which has been thought to serve as the sole carbon source could not account for cell growth, implying that other nutrient sources should be present in the culture (Sun, 2011; Johnston et al., 2014). It is possible that degraded DNA in the minimal culture serves as a source of building blocks for the synthesis of new DNA in bacteria. During the natural transformation of E. coli, new ABC transporter proteins have been shown to participate in DNA transfer (Sun et al., 2009; Sun, 2016). These transporters are different from the known classical DNA uptake proteins that mediate natural bacterial transformation. The mechanism of this new type of DNA transfer is proposed in Figure 3.
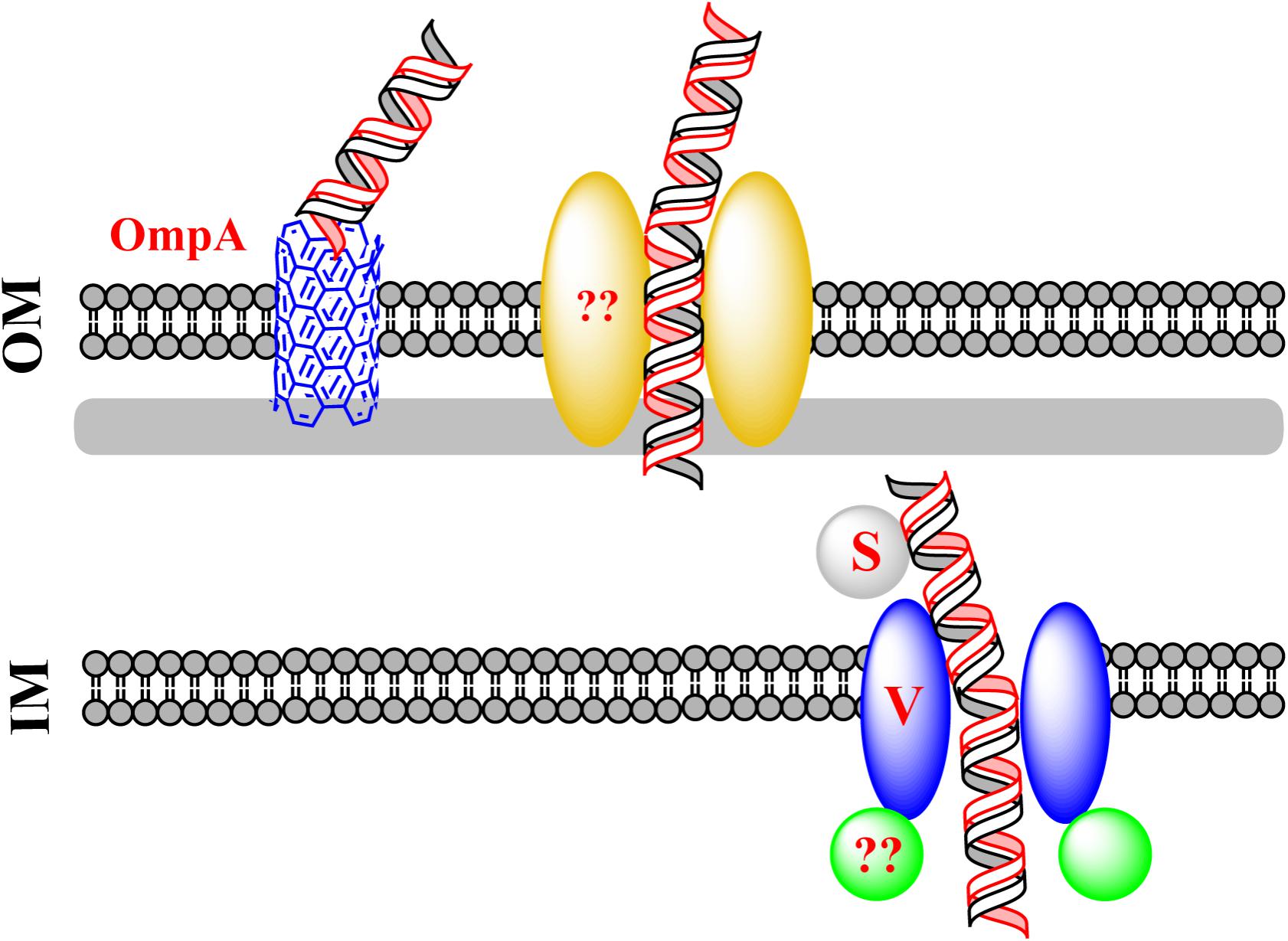
FIGURE 3. A new route for dsDNA transfer in Escherichia coli. Via an unidentified channel, exogenous DNA transfers across the OM. The pore-forming protein OmpA can compete for DNA with the unidentified channel. To pass across the IM, the incoming DNA binds the substrate-binding protein YdcS and is translocated from the periplasm to the cytoplasm via an IM channel formed by YdcV. It remains unclear whether a plasmid enters E. coli as intact circular or linear dsDNA.
Crossing the OM
Escherichia coli has a complete set of genes that potentially encode components of the classical DNA uptake machinery. These genes are homologous to the conserved DNA uptake genes in other naturally transformable bacteria. Comparative genomic analysis predicts that, in E. coli, putative DNA uptake genes hofQ and gspD may encode proteins forming a channel for transporting DNA across the OM, and ppdD may encode pilins for the assembly of the competence pili or pseudopili which pull exogenous DNA in (Finkel and Kolter, 2001; Claverys and Martin, 2003; Sun et al., 2009). However, inactivation of hofQ, gspD, or ppdD does not affect natural transformation with dsDNA, suggesting that the conserved DNA uptake machinery for translocation of ssDNA does not mediate dsDNA transfer across the OM in E. coli (Sun et al., 2009). To identify the OM pore used for DNA transport during natural transformation of E. coli, the pore-forming protein OmpA was evaluated, considering that OmpA performs functions in bacteriophage infection and bacterial conjugation. Inactivation of ompA increases natural transformation by 7- to 60-fold while decreasing chemical transformation by ∼10-fold, suggesting that OmpA blocks DNA transfer during natural transformation but promotes DNA transfer in artificial transformation (Sun et al., 2013). OmpA is unlikely to form an open gate under natural conditions, but can be switched to the open state with the molecular force of electrostatic interaction (i.e., salt-bridge), that drives structural transition of a protein under different conditions (Hong et al., 2006). The closed and open states of the gate are dependent on the formation of salt bridges of Arg138-Glu52 and Lys82-Glu128, respectively (Hong et al., 2006). During natural transformation of E. coli, DNA may pass across an unidentified channel that competes with OmpA for transforming DNA. The putative channel may be consisted of OM components (i.e., OM protein and pili/psedopili) of a DNA transport system and pulls DNA into the cell on the LB-agar plate. In the default “gate-closed” state, OmpA traps the transforming DNA, making it unable to reach the right channel for completing transformation. By contrast, during chemical transformation or electroporation, a high concentration of Ca2+ or an electric current helps open the gate, allowing DNA to pass across the channel formed by OmpA (Sun et al., 2013).
Crossing the IM
Based on a membrane topology study, the conserved IM protein called ComEC is predicted to mediate the translocation of ssDNA during classical natural transformation (Chen and Dubnau, 2004; Claverys et al., 2009; Johnston et al., 2014). However, inactivation of the ComEC homolog YcaI in E. coli does not affect natural plasmid transformation of E. coli, suggesting that DNA is translocated into the cytoplasm via a different route (Sun et al., 2009). The single-hit kinetics in natural plasmid transformation of E. coli suggest that the establishment of a plasmid in the cytoplasm is mediated not by the annealing of partially overlapping opposite ssDNA derived from two independent plasmid monomers, but by a new route, i.e., the transfer of dsDNA across the IM of bacteria (Sun et al., 2009). Screening of RpoS-targeted transformation-related genes has identified ydcS and ydcV, which are located in the same ABC-transporter operon (Sun, 2016). Inactivation of ydcS and ydcV reduces natural transformation 6.7- and 9.5-fold, respectively (Sun, 2016). Chemical transformation is also reduced by Inactivation of ydcS, whereas the chemical transformation in a ydcV mutant is not reduced as compared to its wild-type counterpart (Sun, 2016). According to the Transporter Classification Database (TCDB1), ydcS and ydcV are predicted to encode proteins for binding a substrate in the periplasm and for translocation of the substrate across IM (Saier et al., 2014). YdcS has been shown as a PHB synthase in the periplasm (Dai and Reusch, 2008). The mutant lacking ydcT (a putative ATPase encoding gene) in the same operon as ydcS and ydcV, is naturally transformed with slightly but obviously reduced frequency (less than 50%) (Sun, 2016), indicating that this gene is also involved in natural plasmid transformation of E. coli. Nonetheless, with respect to significantly reduced transformation frequency in the ydcV and the ydcS mutants, ydcT seems to have only a minor effect on DNA transport. It is likely that additional energy source is required for efficient transport of dsDNA across the IM.
Cell-To-Cell Transfer of Non-Conjugative Plasmids
Cell-to-cell contact is often required for bacterial conjugation. Of note, plasmid transformation of E. coli in colonies proceeds on the surface of agar plates and cell-to-cell contact is required for the transfer of plasmids that do not carry conjugative functions (Maeda et al., 2004, 2006). Cell-to-cell contact-dependent plasmid transformation occurs not only within the same genus but also across genera (Wang et al., 2007). In some naturally transformable G− bacteria (e.g., H. influenzae and N. gonorrhoeae), a short DNA sequence (named DUS) can increase DNA uptake (Claverys and Martin, 2003; Wang and Sun, 2015; Zheng et al., 2017). During cell-to-cell contact-dependent plasmid transformation of E. coli, an 88 bp DNA sequence promotes DNA transfer (Sobue et al., 2011). Screening of the Keio collection (Baba et al., 2006), a comprehensive library of E. coli knock-out mutants defective in non-essential genes, has identified rodZ, whose product regulates the rod-shape of the cell, as an essential gene for cell-to-cell contact-dependent transformation (Kurono et al., 2012). Because the homologs of DNA uptake genes (e.g., ycaI) have not been revealed to be essential genes, conventional DNA uptake machinery is not likely to be involved in cell-to-cell contact-dependent plasmid transformation of E. coli. Recently, cell-to-cell contact-dependent transformation is discovered in B. subtilis (Zhang et al., 2018). However, inactivation of the competence regulator ComK, that controls the expression of conserved DNA uptake genes, abolishes cell-to-cell DNA transfer (Zhang et al., 2018), indicating that mechanisms of the cell-to-cell DNA transfer in E. coli and B. subtilis are basically different.
Concluding Remarks
Classical mechanisms of HGT (i.e., natural transformation and conjugation) share common features in that ssDNA is pushed out or pulled into the cell with the assistance of membrane-associated protein complexes. Recent studies have uncovered new types of DNA transfer, which are independent of the classical DNA uptake or conjugation machineries, suggesting that other therapeutic targets should be considered in the fight against ARGs. The conserved proteins involved in transporting or protecting DNA may serve as targets for limiting the transfer of ARGs and in turn for reducing MDR in bacteria (Goessweiner-Mohr et al., 2013). On the other hand, the discovery of non-classical HGT mechanisms suggests that controlling the spread of ARGs is more complicated than previously thought.
Author Contributions
DS conceived the idea and wrote the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (Grant Nos. 31670084, 31100071) and the Zhejiang Provincial Natural Science Foundation of China (Grant Nos. LY16C010003, Y3110237).
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Aas, F. E., Wolfgang, M., Frye, S., Dunham, S., Lovold, C., and Koomey, M. (2002). Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46, 749–760. doi: 10.1046/j.1365-2958.2002.03193.x
Assalkhou, R., Balasingham, S., Collins, R. F., Frye, S. A., Davidsen, T., Benam, A. V., et al. (2007). The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology 153, 1593–1603. doi: 10.1099/mic.0.2006/004200-0
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:msb4100050. doi: 10.1038/msb4100050
Baker, J. A., Simkovic, F., Taylor, H. M., and Rigden, D. J. (2016). Potential DNA binding and nuclease functions of ComEC domains characterized in silico. Proteins 84, 1431–1442. doi: 10.1002/prot.25088
Barouki, R., and Smith, H. O. (1985). Reexamination of phenotypic defects in rec-1 and rec-2 mutants of Haemophilus influenzae Rd. J. Bacteriol. 163, 629–634.
Berge, M., Moscoso, M., Prudhomme, M., Martin, B., and Claverys, J. P. (2002). Uptake of transforming DNA in Gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45, 411–421. doi: 10.1046/j.1365-2958.2002.03013.x
Berka, R. M., Hahn, J., Albano, M., Draskovic, I., Persuh, M., Cui, X., et al. (2002). Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43, 1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x
Bi, D., Xu, Z., Harrison, E. M., Tai, C., Wei, Y., He, X., et al. (2012). ICEberg: a web-based resource for integrative and conjugative elements found in bacteria. Nucleic Acids Res. 40, D621–D626. doi: 10.1093/nar/gkr846
Burkhardt, J., Vonck, J., and Averhoff, B. (2011). Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27. J. Biol. Chem. 286, 9977–9984. doi: 10.1074/jbc.M110.212688
Burton, B., and Dubnau, D. (2010). Membrane-associated DNA transport machines. Cold Spring Harb. Perspect. Biol. 2, a000406. doi: 10.1101/cshperspect.a000406
Cabezon, E., Ripoll-Rozada, J., Pena, A., De La Cruz, F., and Arechaga, I. (2015). Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 39, 81–95. doi: 10.1111/1574-6976.12085
Chen, I., Christie, P. J., and Dubnau, D. (2005). The ins and outs of DNA transfer in bacteria. Science 310, 1456–1460. doi: 10.1126/science.1114021
Chen, I., and Dubnau, D. (2004). DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249. doi: 10.1038/nrmicro844
Chilton, S. S., Falbel, T. G., Hromada, S., and Burton, B. M. (2017). A conserved metal binding motif in the Bacillus subtilis vompetence protein ComFA enhances transformation. J. Bacteriol. 199:e00272-17. doi: 10.1128/JB.00272-17
Claverys, J. P., and Martin, B. (2003). Bacterial “competence” genes: signatures of active transformation, or only remnants? Trends. Microbiol. 11, 161–165. doi: 10.1016/S0966-842X(03)00064-7
Claverys, J. P., Martin, B., and Polard, P. (2009). The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33, 643–656. doi: 10.1111/j.1574-6976.2009.00164.x
Claverys, J. P., Prudhomme, M., and Martin, B. (2006). Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60, 451–475. doi: 10.1146/annurev.micro.60.080805.142139
Collins, R. F., Davidsen, L., Derrick, J. P., Ford, R. C., and Tonjum, T. (2001). Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183, 3825–3832. doi: 10.1128/JB.183.13.3825-3832.2001
Cury, J., Touchon, M., and Rocha, E. P. C. (2017). Integrative and conjugative elements and their hosts: composition, distribution and organization. Nucleic Acids Res. 45, 8943–8956. doi: 10.1093/nar/gkx607
Dai, D., and Reusch, R. N. (2008). Poly-3-hydroxybutyrate synthase from the periplasm of Escherichia coli. Biochem. Biophys. Res. Commun. 374, 485–489. doi: 10.1016/j.bbrc.2008.07.043
Diallo, A., Foster, H. R., Gromek, K. A., Perry, T. N., Dujeancourt, A., Krasteva, P. V., et al. (2017). Bacterial transformation: ComFA is a DNA-dependent ATPase that forms complexes with ComFC and DprA. Mol. Microbiol. 105, 741–754. doi: 10.1111/mmi.13732
Draskovic, I., and Dubnau, D. (2005). Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol. Microbiol. 55, 881–896. doi: 10.1111/j.1365-2958.2004.04430.x
Duffin, P. M., and Barber, D. A. (2016). DprA is required for natural transformation and affects pilin variation in Neisseria gonorrhoeae. Microbiology 162, 1620–1628. doi: 10.1099/mic.0.000343
Dwivedi, G. R., Sharma, E., and Rao, D. N. (2013). Helicobacter pylori DprA alleviates restriction barrier for incoming DNA. Nucleic. Acids. Res. 41, 3274–3288. doi: 10.1093/nar/gkt024
Etchuuya, R., Ito, M., Kitano, S., Shigi, F., Sobue, R., and Maeda, S. (2011). Cell-to-cell transformation in Escherichia coli: a novel type of natural transformation involving cell-derived DNA and a putative promoting pheromone. PLoS One 6:e16355. doi: 10.1371/journal.pone.0016355
Finkel, S. E., and Kolter, R. (2001). DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183, 6288–6293. doi: 10.1128/JB.183.21.6288-6293.2001
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Franke, A. E., and Clewell, D. B. (1981). Evidence for conjugal transfer of a Streptococcus faecalis transposon (Tn916) from a chromosomal site in the absence of plasmid DNA. Cold Spring Harb. Symp. Quant. Biol. 45(Pt 1), 77–80. doi: 10.1101/SQB.1981.045.01.014
Goessweiner-Mohr, N., Arends, K., Keller, W., and Grohmann, E. (2013). Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid 70, 289–302. doi: 10.1016/j.plasmid.2013.09.005
Griffith, F. (1928). The significance of Pneumococcal types. J. Hyg. 27, 113–159. doi: 10.1017/S0022172400031879
Guo, M., Wang, H., Xie, N., and Xie, Z. (2015). Positive effect of carbon sources on natural transformation in Escherichia coli: role of low-level cyclic AMP (cAMP)-cAMP receptor protein in the derepression of rpoS. J. Bacteriol. 197, 3317–3328. doi: 10.1128/JB.00291-15
Hong, H., Szabo, G., and Tamm, L. K. (2006). Electrostatic couplings in OmpA ion-channel gating suggest a mechanism for pore opening. Nat. Chem. Biol. 2, 627–635. doi: 10.1038/nchembio827
Hovland, E., Beyene, G. T., Frye, S. A., Homberset, H., Balasingham, S. V., Gomez-Munoz, M., et al. (2017). DprA from Neisseria meningitidis: properties and role in natural competence for transformation. Microbiology 163, 1016–1029. doi: 10.1099/mic.0.000489
Ilangovan, A., Connery, S., and Waksman, G. (2015). Structural biology of the gram-negative bacterial conjugation systems. Trends. Microbiol. 23, 301–310. doi: 10.1016/j.tim.2015.02.012
Ilangovan, A., Kay, C. W. M., Roier, S., El Mkami, H., Salvadori, E., Zechner, E. L., et al. (2017). Cryo-EM structure of a relaxase reveals the molecular basis of DNA unwinding during bacterial conjugation. Cell 169, 708.e12–721.e12. doi: 10.1016/j.cell.2017.04.010
Inamine, G. S., and Dubnau, D. (1995). ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J. Bacteriol. 177, 3045–3051. doi: 10.1128/jb.177.11.3045-3051.1995
Johnson, C. M., and Grossman, A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49, 577–601. doi: 10.1146/annurev-genet-112414-055018
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J. P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196. doi: 10.1038/nrmicro3199
Kurono, N., Matsuda, A., Etchuya, R., Sobue, R., Sasaki, Y., Ito, M., et al. (2012). Genome-wide screening of Escherichia coli genes involved in execution and promotion of cell-to-cell transfer of non-conjugative plasmids: rodZ (yfgA) is essential for plasmid acceptance in recipient cells. Biochem. Biophys. Res. Commun. 421, 119–123. doi: 10.1016/j.bbrc.2012.03.127
Laurenceau, R., Pehau-Arnaudet, G., Baconnais, S., Gault, J., Malosse, C., Dujeancourt, A., et al. (2013). A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog. 9:e1003473. doi: 10.1371/journal.ppat.1003473
Le, S., Serrano, E., Kawamura, R., Carrasco, B., Yan, J., and Alonso, J. C. (2017). Bacillus subtilis RecA with DprA-SsbA antagonizes RecX function during natural transformation. Nucleic. Acids. Res. 45, 8873–8885. doi: 10.1093/nar/gkx583
Lederberg, J., and Tatum, E. L. (1946). Gene recombination in Escherichia coli. Nature 158, 558. doi: 10.1038/158558a0
Leong, C. G., Bloomfield, R. A., Boyd, C. A., Dornbusch, A. J., Lieber, L., Liu, F., et al. (2017). The role of core and accessory type IV pilus genes in natural transformation and twitching motility in the bacterium Acinetobacter baylyi. PLoS One 12:e0182139. doi: 10.1371/journal.pone.0182139
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Londono-Vallejo, J. A., and Dubnau, D. (1994). Mutation of the putative nucleotide binding site of the Bacillus subtilis membrane protein ComFA abolishes the uptake of DNA during transformation. J. Bacteriol. 176, 4642–4645. doi: 10.1128/jb.176.15.4642-4645.1994
Maeda, S., Ito, M., Ando, T., Ishimoto, Y., Fujisawa, Y., Takahashi, H., et al. (2006). Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli. FEMS Microbiol. Lett. 255, 115–120. doi: 10.1111/j.1574-6968.2005.00072.x
Maeda, S., Sawamura, A., and Matsuda, A. (2004). Transformation of colonial Escherichia coli on solid media. FEMS Microbiol. Lett. 236, 61–64. doi: 10.1111/j.1574-6968.2004.tb09627.x
Matsuda, A., Kurono, N., Kawano, C., Shirota, K., Hirabayashi, A., Horino, M., et al. (2012). Genome-wide screen for Escherichia coli genes involved in repressing cell-to-cell transfer of non-conjugative plasmids. Biochem. Biophys. Res. Commun. 428, 445–450. doi: 10.1016/j.bbrc.2012.10.098
Matsumoto, A., Sekoguchi, A., Imai, J., Kondo, K., Shibata, Y., and Maeda, S. (2016). Natural Escherichia coli strains undergo cell-to-cell plasmid transformation. Biochem. Biophys. Res. Commun. 481, 59–62. doi: 10.1016/j.bbrc.2016.11.018
Mortier-Barriere, I., Velten, M., Dupaigne, P., Mirouze, N., Pietrement, O., Mcgovern, S., et al. (2007). A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130, 824–836. doi: 10.1016/j.cell.2007.07.038
Ogura, M., Yamaguchi, H., Kobayashi, K., Ogasawara, N., Fujita, Y., and Tanaka, T. (2002). Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 184, 2344–2351. doi: 10.1128/JB.184.9.2344-2351.2002
Palchevskiy, V., and Finkel, S. E. (2006). Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J. Bacteriol. 188, 3902–3910. doi: 10.1128/JB.01974-05
Provvedi, R., and Dubnau, D. (1999). ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol. Microbiol. 31, 271–280. doi: 10.1046/j.1365-2958.1999.01170.x
Saier, M. H. Jr., Reddy, V. S., Tamang, D. G., and Vastermark, A. (2014). The transporter classification database. Nucleic Acids Res. 42, D251–D258. doi: 10.1093/nar/gkt1097
Salzer, R., Joos, F., and Averhoff, B. (2014). Type IV pilus biogenesis, twitching motility, and DNA uptake in Thermus thermophilus: discrete roles of antagonistic ATPases PilF, PilT1, and PilT2. Appl. Environ. Microbiol. 80, 644–652. doi: 10.1128/AEM.03218-13
Salzer, R., Kern, T., Joos, F., and Averhoff, B. (2016). The Thermus thermophilus comEA/comEC operon is associated with DNA binding and regulation of the DNA translocator and type IV pili. Environ. Microbiol. 18, 65–74. doi: 10.1111/1462-2920.12820
Seitz, P., Pezeshgi Modarres, H., Borgeaud, S., Bulushev, R. D., Steinbock, L. J., Radenovic, A., et al. (2014). ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae cells. PLoS Genet. 10:e1004066. doi: 10.1371/journal.pgen.1004066
Shen, Z., Wang, Y., Shen, Y., Shen, J., and Wu, C. (2016). Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 16:293. doi: 10.1016/S1473-3099(16)00061-X
Sinha, S., Mell, J. C., and Redfield, R. J. (2012). Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J. Bacteriol. 194, 5245–5254. doi: 10.1128/JB.00671-12
Smeets, L. C., and Kusters, J. G. (2002). Natural transformation in Helicobacter pylori: DNA transport in an unexpected way. Trends. Microbiol 10, 159–162; discussion 162. doi: 10.1016/S0966-842X(02)02314-4
Sobue, R., Kurono, N., Etchuya, R., and Maeda, S. (2011). Identification of a novel DNA element that promotes cell-to-cell transformation in Escherichia coli. FEBS Lett. 585, 2223–2228. doi: 10.1016/j.febslet.2011.05.040
Sun, D. (2011). Etude de la Transformation Plasmidique Naturelle D’escherichia Coli Et De Ses Relations Eventuelles la compétence Programmée Pour la Transformation Génétique et la Compétence Dite Nutritionnelle. Ph.D. thesis, avec, Toulouse.
Sun, D. (2016). Two different routes for double-stranded DNA transfer in natural and artificial transformation of Escherichia coli. Biochem. Biophys. Res. Commun. 471, 213–218. doi: 10.1016/j.bbrc.2016.01.137
Sun, D., Wang, B., Zhu, L., Chen, M., and Zhan, L. (2013). Block and boost DNA transfer: opposite roles of OmpA in natural and artificial transformation of Escherichia coli. PLoS One 8:e59019. doi: 10.1371/journal.pone.0059019
Sun, D., Zhang, X., Wang, L., Prudhomme, M., Xie, Z., Martin, B., et al. (2009). Transforming DNA uptake gene orthologs do not mediate spontaneous plasmid transformation in Escherichia coli. J. Bacteriol. 191, 713–719. doi: 10.1128/JB.01130-08
Sun, D., Zhang, Y., Mei, Y., Jiang, H., Xie, Z., Liu, H., et al. (2006). Escherichia coli is naturally transformable in a novel transformation system. FEMS Microbiol. Lett. 265, 249–255. doi: 10.1111/j.1574-6968.2006.00503.x
Takeno, M., Taguchi, H., and Akamatsu, T. (2011). Role of ComFA in controlling the DNA uptake rate during transformation of competent Bacillus subtilis. J. Biosci. Bioeng. 111, 618–623. doi: 10.1016/j.jbiosc.2011.02.006
Takeno, M., Taguchi, H., and Akamatsu, T. (2012). Role of ComEA in DNA uptake during transformation of competent Bacillus subtilis. J. Biosci. Bioeng. 113, 689–693. doi: 10.1016/j.jbiosc.2012.02.004
Tsen, S. D., Fang, S. S., Chen, M. J., Chien, J. Y., Lee, C. C., and Tsen, D. H. (2002). Natural plasmid transformation in Escherichia coli. J. Biomed. Sci. 9, 246–252. doi: 10.1159/000059425
Veening, J. W., and Blokesch, M. (2017). Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat. Rev. Microbiol. 15, 621–629. doi: 10.1038/nrmicro.2017.66
von Wintersdorff, C. J., Penders, J., Van Niekerk, J. M., Mills, N. D., Majumder, S., Van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Wang, B., and Sun, D. (2015). Detection of NDM-1 carbapenemase-producing Acinetobacter calcoaceticus and Acinetobacter junii in environmental samples from livestock farms. J. Antimicrob. Chemother. 70, 611–613. doi: 10.1093/jac/dku405
Wang, X., Li, M., Yan, Q., Chen, X., Geng, J., Xie, Z., et al. (2007). Across genus plasmid transformation between Bacillus subtilis and Escherichia coli and the effect of Escherichia coli on the transforming ability of free plasmid DNA. Curr. Microbiol. 54, 450–456. doi: 10.1007/s00284-006-0617-1
Wozniak, R. A., and Waldor, M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563. doi: 10.1038/nrmicro2382
Yadav, T., Carrasco, B., Hejna, J., Suzuki, Y., Takeyasu, K., and Alonso, J. C. (2013). Bacillus subtilis DprA recruits RecA onto single-stranded DNA and mediates annealing of complementary strands coated by SsbB and SsbA. J. Biol. Chem. 288, 22437–22450. doi: 10.1074/jbc.M113.478347
Yadav, T., Carrasco, B., Serrano, E., and Alonso, J. C. (2014). Roles of Bacillus subtilis DprA and SsbA in RecA-mediated genetic recombination. J. Biol. Chem. 289, 27640–27652. doi: 10.1074/jbc.M114.577924
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Zhang, X., Jin, T., Deng, L., Wang, C., Zhang, Y., and Chen, X. (2018). Stress-induced, highly efficient, donor cell-dependent cell-to-cell natural transformation in Bacillus subtilis. J. Bacteriol. 200:e00267-18. doi: 10.1128/JB.00267-18
Zhang, Y., Shi, C., Yu, J., Ren, J., and Sun, D. (2012). RpoS regulates a novel type of plasmid DNA transfer in Escherichia coli. PLoS One 7:e33514. doi: 10.1371/journal.pone.0033514
Keywords: antibiotic resistance gene, multidrug resistance, DNA transfer, natural transformation, conjugation, Escherichia coli
Citation: Sun D (2018) Pull in and Push Out: Mechanisms of Horizontal Gene Transfer in Bacteria. Front. Microbiol. 9:2154. doi: 10.3389/fmicb.2018.02154
Received: 23 March 2018; Accepted: 22 August 2018;
Published: 06 September 2018.
Edited by:
Peter Mullany, University College London, United KingdomReviewed by:
Baltasar Mayo, Consejo Superior de Investigaciones Científicas (CSIC), SpainVictor Krylov, I. I. Mechnikov Research Institute of Vaccines and Sera (RAS), Russia
J. Tony Pembroke, University of Limerick, Ireland
Copyright © 2018 Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongchang Sun, c3VuZGNoQHpqdXQuZWR1LmNu
 Dongchang Sun
Dongchang Sun