
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 September 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02119
This article is part of the Research Topic Harnessing Useful Rhizosphere Microorganisms for Pathogen and Pest Biocontrol, Volume II View all 33 articles
Tomato bacterial canker disease, caused by Clavibacter michiganensis subsp. michiganensis (Cmm) is a destructive disease and has been a serious concern for tomato industries worldwide. Previously, a rhizosphere isolated strain of Pseudomonas sp. 23S showed antagonistic activity toward Cmm in vitro. This Pseudomonas sp. 23S was characterized to explore the potential of this bacterium for its use in agriculture. Pseudomonas sp. 23S possesses ability to solubilize inorganic phosphorus, and to produce siderophores, indole acetic acid, and hydrogen cyanide. The strain also showed antagonistic activity against Pseudomonas syringae pv. tomato DC 3000. A plant assay indicated that Pseudomonas sp. 23S could promote growth of tomato seedlings. The potential of treating tomato plants with Pseudomonas sp. 23S to reduce the severity of tomato bacterial canker by inducing systemic resistance (ISR) was investigated using well characterized marker genes such as PR1a [salicylic acid (SA)], PI2 [jasmonic acid (JA)], and ACO [ethylene (ET)]. Two-week-old tomato plants were treated with Pseudomonas sp. 23S by soil drench, and Cmm was inoculated into the stem by needle injection on 3, 5, or 7 days post drench. The results indicated that plants treated with Pseudomonas sp. 23S, 5 days prior to Cmm inoculation significantly delayed the progression of the disease. These plants, after 3 weeks from the date of Cmm inoculation, had significantly higher dry shoot and root weight, higher levels of carbon, nitrogen, phosphorus, and potassium in the leaf tissue, and the number of Cmm population in the stem was significantly lower for the plants treated with Pseudomonas sp. 23S. From the real-time quantitative PCR (qRT-PCR) analysis, the treatment with Pseudomonas sp. 23S alone was found to trigger a significant increase in the level of PR1a transcripts in tomato plants. When the plants were treated with Pseudomonas sp. 23S and inoculated with Cmm, the level of PR1a and ACO transcripts were increased, and this response was faster and greater as compared to plants inoculated with Cmm but not treated with Pseudomonas sp. 23S. Overall, the results suggested the involvement of SA signaling pathways for ISR induced by Pseudomonas sp. 23S.
Bacterial canker disease, caused by Clavibacter michiganensis subsp. michiganensis (Cmm) is one of the most destructive diseases in tomato (Gleason et al., 1993; de León et al., 2011). It has been reported in both greenhouse and field tomato production worldwide, and has caused substantial crop losses (Chang et al., 1992a; Hausbeck et al., 2000; de León et al., 2011). Once plants are infected by Cmm, initial marginal leaf necrosis symptoms widen and lead to wilting of all leaves while canker develops on the stem, and the whole plants can be stunted and severely wilted leading to death (de León et al., 2011; Sen et al., 2015). Cmm inoculum can originate from infected soils, seeds, transplants, tomato debris in soil, and operating tools and equipment. The bacteria can enter the plants through wounds and natural openings such as stomata and hydathodes after which they move to the xylem and multiply rapidly (Carlton et al., 1998; Gartemann et al., 2003; Sharabani et al., 2013). Farming practices such as tying, pruning, clipping, spraying and harvesting can cause a high level of secondary infection spread to nearby healthy plants via workers’ fingers and tools (Ark, 1944; Gleason et al., 1993). Despite the seriousness of this disease, no control methods have been found to be completely effective. As no Cmm-resistant seeds are commercially available, current control primarily relies on the use of pathogen-free certified seeds and transplants, good hygiene, disinfection of all tools, and crop rotations (Xu et al., 2015). Hence an effective control method for bacterial canker is urgently needed.
Use of plant growth-promoting rhizobacteria (PGPR) as biocontrol agents offers an ecological means to manage disease problems in agriculture. PGPR are rhizosphere free-living bacteria that colonize plant roots and have beneficial effects on plant growth (Kloepper and Schroth, 1978; Kloepper et al., 1989; Bouizgarne, 2013). The biocontrol ability of PGPR can be attributed to two general mechanisms: direct antagonism against pathogens or induction of systemic resistance throughout the plant. Production of antimicrobial compounds, such as antibiotic metabolites, and bacteriocin has been observed from many PGPR, and their inhibitory actions against pathogens contribute to reduction of plant diseases (Subramanian and Smith, 2015). In addition to direct suppressive effects on the pathogens, PGPR can trigger systemic resistance throughout the plant. PGPR-mediated induced systemic resistance (ISR) is often achieved by priming (Pieterse et al., 2014). Priming is characterized as potentiated activation of defense responses, which are subsequently induced upon pathogen attack, resulting in enhanced plant defense capacity (Conrath et al., 2006).
Although many ISR-inducing PGPR have been discovered, signaling and activation mechanisms of the ISR are not completely understood. The involvement of three plant hormones, salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) have been well documented. Conventionally, SA is believed to be involved in systemic acquired resistance (SAR), which is induced by pathogens attack and follows induction of PR proteins. PGPR-mediated ISR is known to be dependent on JA and ET signaling; it is a SA-independent process and does not lead to induction of PR proteins (Van Loon and Bakker, 2005; Van Wees et al., 2008; Pieterse et al., 2014). As more research on ISR has been conducted, however, evidence of SA-dependent ISR has been observed for some PGPR (De Meyer et al., 1999; Tjamos et al., 2005; Schuhegger et al., 2006; Conn et al., 2008; Rudrappa et al., 2008; Niu et al., 2012).
Regarding tomato bacterial canker, several PGPR having antagonistic activities toward Cmm have been isolated and studied (Amkraz et al., 2010; Lanteigne et al., 2012; Deng et al., 2015; Aksoy et al., 2017). Among them, the induction of ISR was only reported for Pseudomonas putida (CKPp9; Aksoy et al., 2017). This ISR was accompanied by induction of significant amounts of phenolic compounds, which contributed to the disease reduction.
Our laboratory has been working to develop PGPR-based technologies for agriculture. As a part of our work on identifying new PGPR (Jung et al., 2014), we isolated a rhizobacterium that inhibited the growth of Cmm in vitro (Supplementary Figure S1). According to the 16S rRNA sequencing, this bacterium was identified as a strain of Pseudomonas, and is thus referred to here as Pseudomonas sp. 23S.
This study was conducted to achieve three objectives. The first objective was to characterize the newly isolated Pseudomonas sp. 23S for important PGPR traits. Our second objective was then to determine whether Pseudomonas sp. 23S induces ISR in tomato plants and reduces the disease severity specifically, bacterial canker, caused by Cmm. Not all PGPR possess ISR-inducing ability, and the question as to whether Pseudomonas sp. 23S, which has direct antagonistic activity against Cmm, is also able to induce ISR is meaningful to answer, because this could greatly enhance the use of this bacterium as a biocontol agent. Given that initial work reported in this paper did show that the bacterium was an ISR inducer, our final objective was then to determine whether treatment with Pseudomonas sp. 23S causes changes in the transcript levels of defense-related genes, specifically PR1a, PI2 and ACO. Investigating the transcript levels of these three genes could help to determine the possible involvement of SA, JA and/or ET in the ISR response, and to understand the ISR signaling pathway used by Pseudomonas sp. 23S specific to this biotic stress.
Pseudomonas sp. 23S was grown in Nutrient Broth (NB, Difco; 8 g L−1) media at 28°C, at 100 rpm. C. michiganensis subsp. michiganensis strain 930 (Cmm) was provided by Agriculture, Pecheries et Alimentation, Quebec. Cmm was grown in NB media at 28°C, at 150 rpm. Both bacteria were maintained as a glycerol stock in −80°C.
Pseudomonas sp. 23S was assessed for important PGPR traits: (i) phosphorous solubilization; (ii) siderophore production; (iii) hydrogen cyanide production; (iv) indole acetic acid production, and; (v) antagonistic activity against an important phytopathogenic bacterium. The abilities to solubilize phosphorous and to produce indole acetic acid were investigated since these traits improve plant growth. Production of siderophores and hydrogen cyanide, both of which can suppress phytopathogenic bacteria, was studied for the biocontrol traits. Phosphorous solubilization was studied using Pikovskaya medium (PVK; Pikovskaya, 1948) and the National Botanical Research Institute’s phosphate growth medium (NBRIP; Nautiyal, 1999). The two types of plates were used to corroborate the results since the PVK plate could sometime give variable results (Nautiyal, 1999). The Fiske and Subbarow method (Fiske and Subbarow, 1925) was applied for quantitative evaluation. Siderophore production was studied using the chrome azurol S (CAS) assay developed by Alexander and Zuberer (1991). For quantitative assessment, percent siderophore production was calculated by using the following formula:
Where, Ar represents the absorbance of reference (CAS assay solution plus growth medium) at 630 nm and As represents the sample (CAS assay solution plus bacterial supernatant) at 630 nm (Schwyn and Neilands, 1987; Alexander and Zuberer, 1991; Ghosh et al., 2015).
For the hydrogen cyanide, Pseudomonas sp. 23S was grown in Kings B medium (per L of distilled H2O: proteose peptone No.3 20 g, glycerol 10 mL, K2HPO4 1.5 g, MgSO4 1.5 g), in which glycerin serves as a precursor molecule (Knowles, 1976; Askeland and Morrison, 1983; Schippers et al., 1990). Indole acetic acid production was evaluated as described by Deaker et al. (2011). In the NB medium where Pseudomonas sp. 23S was grown, DL-tryptophan (TM 7425 Sigma) was added to serve as a precursor of IAA, at two concentrations 0.5 g L−1 or 1.0 g L−1. Antagonistic activity of Pseudomonas sp. 23S was assessed against Pseudomonas syringae pv. tomato DC3000 (provided by Dr. Diane Cuppels, AAFC, London). Pseudomonas syringe pv. tomato DC3000 was grown in Kings B media and 100 μL of 2-day-old culture was spread on Kings B agar plates. A sterile filter-paper disk, with 10 μL of Pseudomonas sp. 23S culture was placed on each pathogen inoculated plate, and the plates was sealed with parafilm and incubated for at least 2 days at 28°C, to observe development of inhibition zones.
Tomato seeds (Bush Beefsteak 351; Stroke Seeds Inc., Thorold, ON, Canada) were surface-sterilized by soaking in 3% (v/v) hypochlorite solution for 3 min, washing thoroughly with water, and drying overnight. The seeds were sown in pots (7.5 mm diameter; 2 seeds pot−1) filled with a mix of sand and turface (5:5). The pots were washed with bleach, and a mix of sand and turface was autoclaved prior to use. The seedlings were thinned to leave 1 plant pot−1 after emergence. After 7 days from the day of seeding, 50 mL suspension of Pseudomonas sp. 23S (108 cfu mL−1 in 10 mM MgSO4) was applied to each pot as a soil drench. For the control treatment, 50 mL of 10 mM MgSO4 was applied to each pot. The seedlings were grown in a growth chamber with a 14/10 h photoperiod and a 25/23°C day/night temperature. Sterilized water was applied as needed. After 3 and 7 days from Pseudomonas sp. 23S treatment (10 and 14-day-old seedlings), the population of Pseudomonas sp. 23S around the roots was enumerated. For this, the seedling was removed from the soil, shaken to dislodge the soil as much as possible, and the root was cut from the plant. The root was then ground with a mortar and pestle in 500 μL of 10 mM MgSO4. The solution was centrifuged (10,000 rpm, 1 min) to remove the debris, and 100 μL of its serial dilution was plated on Pseudomonas Isolation Agar plates (PIA; Difco). The plates were incubated overnight at 28°C, after which the number of colonies formed was counted. Eleven days after Pseudomonas sp. 23S treatment (18-day-old seedlings), the whole plants were harvested. The shoots were dried in an incubator for 2 days at 60°C and shoot dry weight was determined. The roots were first scanned (Modified Epson Expression 10000XL, Regent Instruments Inc., Quebec, QC, Canada) at 400 dots per inch (dpi) resolution and then, images were analyzed by using WinRHIZO software (Reagent Instruments Inc.) to study morphological features and later used for determination of root dry weight. There were five biological replicates per treatment at each time point for the population study, and there were seven biological replicates for the dry weight study. The experiment was conducted twice. A student’s t-test was applied to determine significant differences between control and bacterial treatments.
Tomato seeds were surface-sterilized as described above. The seeds were sown into pots (13 mm diameter) filled with agromix (G10). The plants were grown in a plant growth chamber under the following conditions: 16/8 h of photoperiod, 25/20°C of day/night temperature, and 65% of relative humidity; they were watered daily. Once true leaves emerged, half-strength Hoagland solution was provided once a week (Hoagland and Arnon, 1950; PlantMedia #30630037-5). The experiment was organized following a factorial design with two levels of Pseudomonas sp. 23S treatments (+ and −), and two levels of Cmm inoculation (+ and −). Treatments consisted of: (1) Cont, without Pseudomonas sp. 23S treatment, and Cmm inoculation (negative control); (2) Pse, treated with Pseudomonas sp. 23S; (3) Cmm, without Pseudomonas sp. 23S treatment, and inoculated with Cmm; and, (4) P+C, treated with Pseudomonas sp. 23S, and inoculated with Cmm. After 2 weeks from sowing, the plants were treated with Pseudomonas sp. 23S. Each plant received 100 mL of the Pseudomonas sp. 23S cells suspended in 10 mM MgSO4 (approximately 108 cfu mL−1) for Pse and P+C treatments and 100 mL of 10 mM MgSO4 for Cont and Cmm treatments. The Cmm inoculation was conducted on one of the 4 days, after 1, 3, 5, and 7 days from the date of Pseudomonas sp. 23S treatment (corresponding to 15, 17, 19, and 21 day-old plants); 10 μL of Cmm cells suspended in 10 mM MgSO4 (approximately 108 cfu mL−1) for Cmm and P+C, or 10 μL of 10 mM MgSO4 (approximately 108 cfu mL−1) was inoculated by injecting into the main stem, where the cotyledon emerged, in each plant using a syringe (31 gauge needle, Thermo Scientific #3170513). The number of wilted leaves was counted for monitoring the disease progression. Area under disease progress curve (AUDPC) was calculated based on the formula Σ[(di+1 + di)/2](ti+1 - ti), where di+1 and di are the percentage of wilted leaves and ti+1 and ti are the days after Cmm inoculation (Xu et al., 2012). After 3 weeks from the date of Cmm inoculation, plants were harvested, and shoots (leaves and stems) and root dry weight, plant height, and leaf areas were measured. For each plant, a stem piece (approximately 1 cm) was sampled from 2-cm above the Cmm inoculation site in order to evaluate the Cmm population (number of cells per gram of tissue). The stem piece was weighed and ground with 1 mL of 10 mM MgSO4 using mortar and pestle. The extract was centrifuged at 10,000 rpm for 1 min, and 10-fold serial dilutions of the supernatant were plated on Cmm-selective agar plates (Ftayeh et al., 2011). The number of Cmm colonies were enumerated after 3–4 days after incubation at 28°C. Dried shoot tissues were ground to fine powder with mortar and pestle and used to analyze carbon (C), nitrogen (N), phosphorus (P), and potassium (K) contents. An elemental analyzer (Model NC2500; CE Instruments, Milan, Italy) was used for the C and N analysis. For the P and K analysis, a flow injection analyzer and atomic absorption spectrophotometer were used, respectively (Parkinson and Allen, 1975). First, the tissues were digested in sulfuric acid and peroxide with the addition of catalysts (lithium and selenium) at 340°C for approximately 3 h. The content was diluted to 100 mL and used for the flow injection analyzer. Phosphorous was measured colorimetrically at 880 nm following a complexation with ammonium molybdate (Lachat Instruments QuikChem Method 13-115-01-1-B, 6645 West Mill Road, Milwaukee, WI, United States). Potassium was read on a 10-fold diluted subsample (from the same diluted sample used for the P analysis) by emission on a Varian 220FS (now part of Agilent) atomic absorption spectrophotometer. Seven plants were sampled for each inoculation time (1, 3, 5, and 7 days) per treatment, and the experiment was repeated twice. Since the results from these two experiments were comparable, they were combined and presented in this paper.
Two analyses were performed to study: (1) the effects of Pseudomonas sp. 23S on defense-related genes of tomato plants, and (2) the effects of Pseudomonas sp. 23S on defense-related genes of tomato plants that are infected with bacterial canker by Cmm. For the first analysis, the plants were treated with Pseudomonas sp. 23S with cells suspended in 10 mM MgSO4 (approximately 108 cfu mL−1), and applied as a soil drench, 2 weeks after sowing. For control plants, 100 mL of 10 mM MgSO4 was applied to each plant in the same manner. At 1, 3, 5, and 7 days after the date of Pseudomonas sp. 23S application, the shoot was harvested (biomass pooled for each four replicate plants), flash-frozen with liquid nitrogen, and stored in −80°C for subsequent real-time quantitative PCR (qRT-PCR) analysis. For the second analysis, tomato plants were treated in the same manner as described for the first analysis, with a 5 day-time interval between Pseudomonas sp. 23S application and Cmm inoculation. At 1, 3, 5, and 7 days after the date of Cmm inoculation, the shoot was harvested (pooled for the four replicate plants), flash-frozen with liquid nitrogen, and stored at −80°C for subsequent qRT-PCR analysis. For both situations, the plants were grown under growth chamber condition as described above, and the experiment was conducted three times with independent biological replicates. For the qRT-PCR-analysis, total RNA from tomato leaves was extracted using TRIzol Reagent (Thermo Fisher Scientific catalog#: 15596026), and the RNAs were treated with DNase I (AmbionTM DNaseI, Thermo Fisher Scientific catalog#: AM2222) according to the manufacturer’s instructions. The integrity of the extracted RNA was checked on agarose gel electrophoresis, and its purity and concentration were assessed by a ND-1000 spectrophotometer (NanoDrop). Complementary DNA (cDNA) was synthesized using an iScript Advanced cDNA Synthesis Kit (Bio-Rad, catalog#: 1725037), following the manufacturer’s instructions. The cDNA was diluted to 400 ng uL−1 and stored in −20°C for qPCR. Primer sequences, linear equations, correlation coefficients (R2), and reaction efficiencies for each gene used in this study are provided in Table 1. The qPCR was conducted on a CFX Connect Real Time System (Bio-Rad) with Green-2-Go qPCR Mastermix (Biobasic, catalog#: QPCR004-S), using the cycling program of: 95°C for 10 min for the enzyme activation step, 95°C for 15 s for the initial denaturation step, 60°C for 60 s for annealing and extension, repeated for 40 cycles (PR1: 52°C; PI2: 56°C; ACO: 52°C: GAPDH: 56.4°C). Each plate consisted of three technical replicates from the three independent biological replicates. The Ct value obtained was normalized against the housekeeping gene GAPDH, and the relative gene expression (fold change) was calculated using 2−ΔΔCT method (Livak and Schmittgen, 2001).
To explore its potential, Pseudomonas sp. 23S was studied for general PGPR traits: phosphorous (P) solubilization, production of siderophores, of hydrogen cyanide, of indole acetic acid, and antagonistic activity against phytopathogens. Pikovskaya (PVK) and National Botanical Research Institute’s phosphate growth medium (NBRIP) were used in P solubilization assay. In both PVK and NBRIP plates, the Pseudomonas sp. 23S inoculation resulted in a halo around the disk (Supplementary Figure S2.1; approximately 1 mm for each plate). The quantitative assay showed that Pseudomonas sp. 23S solubilized 5.84 μg mL−1 (±0.85) of inorganic phosphorus. The chrome azurol S (CAS) assay was performed to assess siderophore production. The Pseudomonas sp. 23S inoculation changed the blue color of CAS plates to orange (Supplementary Figure S2.2; size of halo was 4 mm). The quantitative assay showed that its production is 50.7% (±3.38). Hydrogen cyanide (HCN) production was examined by change in color soaked in picric acid. The negative control plate, where media had been applied, was bright yellow (Supplementary Figure S2.3a) whereas the positive control plate, where HCN-positive bacterium had been applied, was bright orange (Supplementary Figure S2.3c). Pseudomonas sp. 23S containing plate was neither this bright yellow nor bright orange, but rather a light orange color (Supplementary Figure S2.3b). To determine whether Pseudomonas sp. 23S could produce indole acetic acid (IAA), two concentrations of tryptophan, 0.5 g mL−1 and 1.0 g L−1, were used as a precursor for production of IAA. The quantitative assay indicated that Pseudomonas sp. 23S produced 1.96 μg IAA mL−1 (±0.09) at 0.5 g L−1 tryptophan and 2.72 μg IAA mL−1 (±0.07) at 1.0 g L−1 tryptophan. Antagonistic activity of Pseudomonas sp. 23S against Pseudomonas syringe pv. tomato DC 3000 was assessed. Pseudomonas sp. 23S inhibited the growth of the Pseudomonas syringe pv. tomato DC 3000, as indicated by inhibition zones around the disk (Supplementary Figure S2.4; the size of the inhibition zone was 4 mm).
Pseudomonas sp. 23S was applied as a soil drench and its effect on growth was examined for tomato seedlings. As Figure 1 shows, Pseudomonas sp. 23S treated seedlings were visually bigger, and the dry weights of their shoots and roots were significantly higher (approximately 47% increase; Figures 1d–f) than those of control seedlings (Figures 1a–c). Roots of Pseudomonas sp. 23S treated seedlings appeared finer and longer (Figures 1j–l) than the roots of control seedlings (Figures 1g–i). Based on the root scanning analysis, root length, volume, and surface area of the Pseudomonas sp. 23S treated seedlings were significantly greater than those of control seedlings (Table 2). When the viable number of Pseudomonas sp. 23S cells around the root was enumerated, it was 105.08 (±0.12) and 105.49 (±0.14) colony forming units per seedling after 3 and 7 days, respectively.
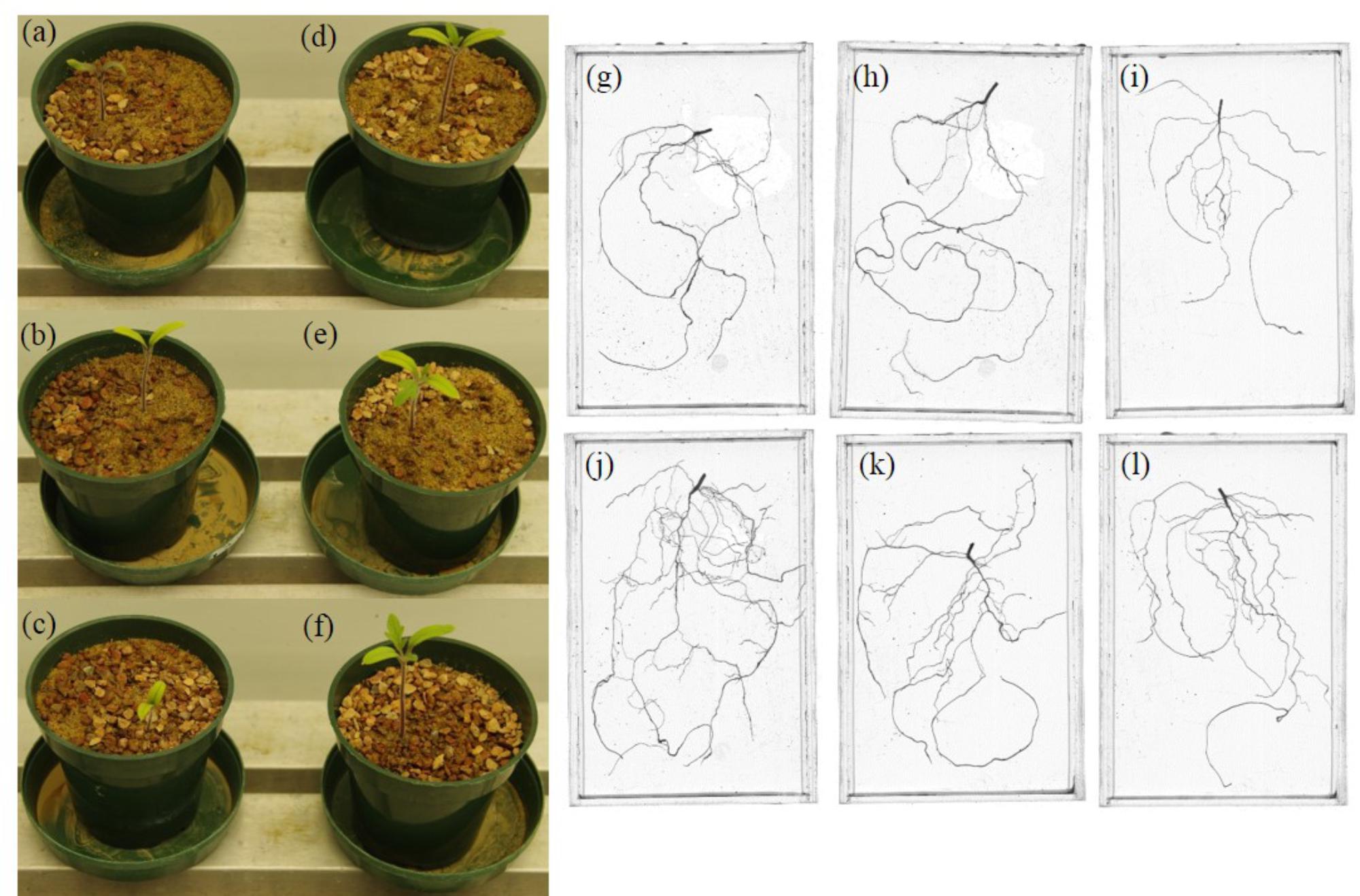
FIGURE 1. Pseudomonas sp. 23S promoted the growth of tomato seedlings. Seedlings from control (a–c) and from Pseudomonas sp. 23S treatment (d–f), and roots from control (g–i) and from Pseudomonas sp. 23S treatment (j–l).
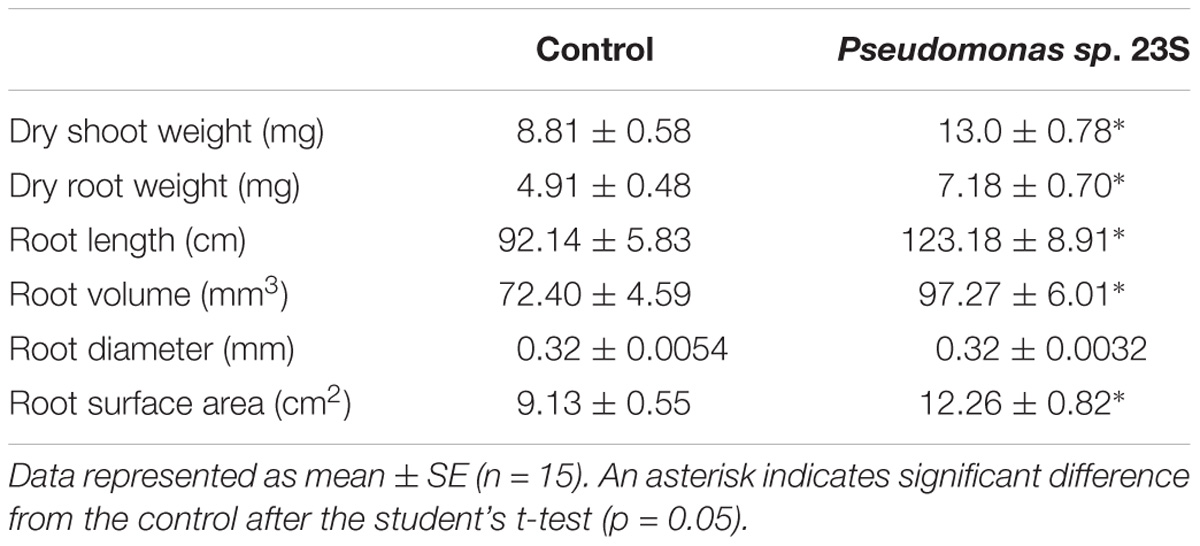
TABLE 2. Pseudomonas sp. 23S treatment increased dry weight of shoots and roots, and improved root length, volume and surface area.
The effects of Pseudomonas sp. 23S on the disease progression were studied for tomato plants infected with Cmm when the time interval between Pseudomonas sp. 23S application and Cmm inoculation dates were 3, 5, and 7 days (Figures 2A–C). The percentage of wilted leaves increased over time under 3-day-interval and reached more than 80% at 21 days post-Cmm inoculation (Figure 2A). Most of these plants were dead; the main stems were broken at the site where the Cmm was inoculated. When the interval was 5 days, the Cmm treatment resulted in a disease progression similar to the Cmm treatment for the 3-day-interval. However, the disease progression for the P+C treatment was significantly slower, and the percentage of wilted leaves was about 60% at 21 days post-Cmm inoculation (Figure 2B). For these plants, the main stems were not broken, the symptom observed in most severely infected plants following the Cmm treatment (Supplementary Figure S3). Under the 7-day-interval, the disease progression was slower than the disease progression at 3- and 5-day intervals. The percentage of wilted leaves was also smaller at 21 days post-Cmm inoculation, less than 60% for the Cmm treatment (Figure 2C). Under the 5-day interval, the AUDPC from the P+C treatment was significantly lower than that from the Cmm treatment (239 for P+C treatment and 576 for Cmm treatment; Supplementary Figure S4). There were no disease symptoms observed for the Cont and Pse treatments at any time.
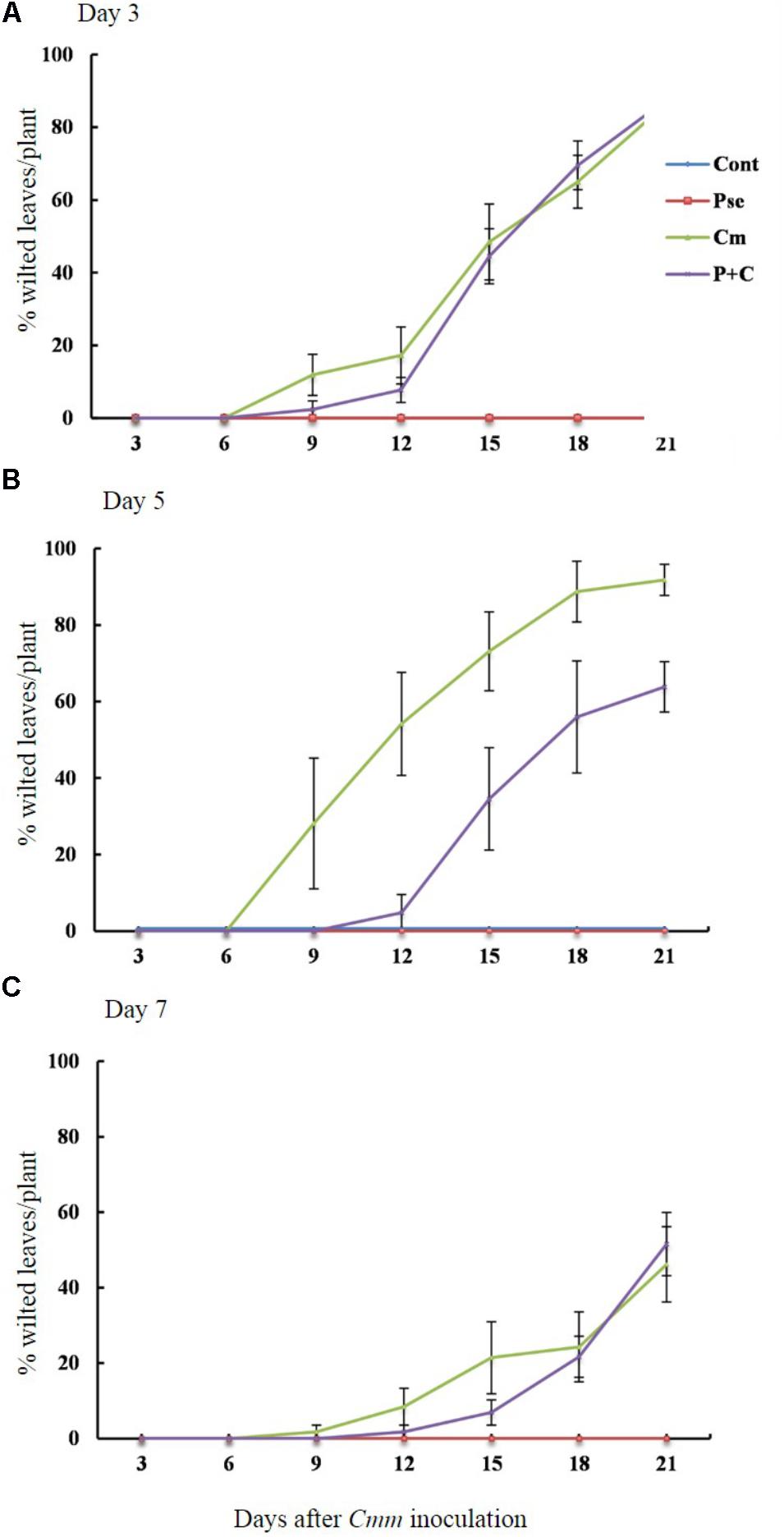
FIGURE 2. Pseudomonas sp. 23S treatment, 5 days prior to Cmm inoculation, delayed progression of bacterial canker. Two-week-old tomato plants were treated with Pseudomonas sp. 23S by soil drench, and after 3 days (A), 5 days (B), or 7 days (C), Cmm was inoculated in the main stem by needle injection. The number of wilted leaves was counted every 3 days for 3 weeks. Treatments are: Cont, control; Pse, treated with Pseudomonas sp. 23S; Cmm, inoculated with Cmm; and, P+C, treated with Pseudomonas sp. 23S, and inoculated with Cmm. Cont and Pse treatments showed no disease symptom throughout the experiment (n = 14, p = 0.05).
To determine whether Pseudomonas sp. 23S treatment results in improvement of plant biomass, the dry weights of shoots and roots for the tomato plants grown under our experimental conditions were measured (Figures 3A,B). The shoot dry weights of the Cont and Pse treatments were not significantly different for any of the intervals (Figure 3A). With an interval of 3 and 5 days, the shoot dry weights of the Cont and Pse treatments were significantly different from those of the Cmm and P+C treatments while they were not significantly different with an interval of 7 days. Under the 3 and 7-days intervals, the shoot dry weights of the Cmm and P+C treatments were not significantly different. On the other hand, the shoot dry weight was greater for the P+C treatments than Cmm treatments under the 5-day-interval. Similar trends were found for the root dry weight, plant height, and leaf areas (Figure 3B and Supplementary Figure S5).
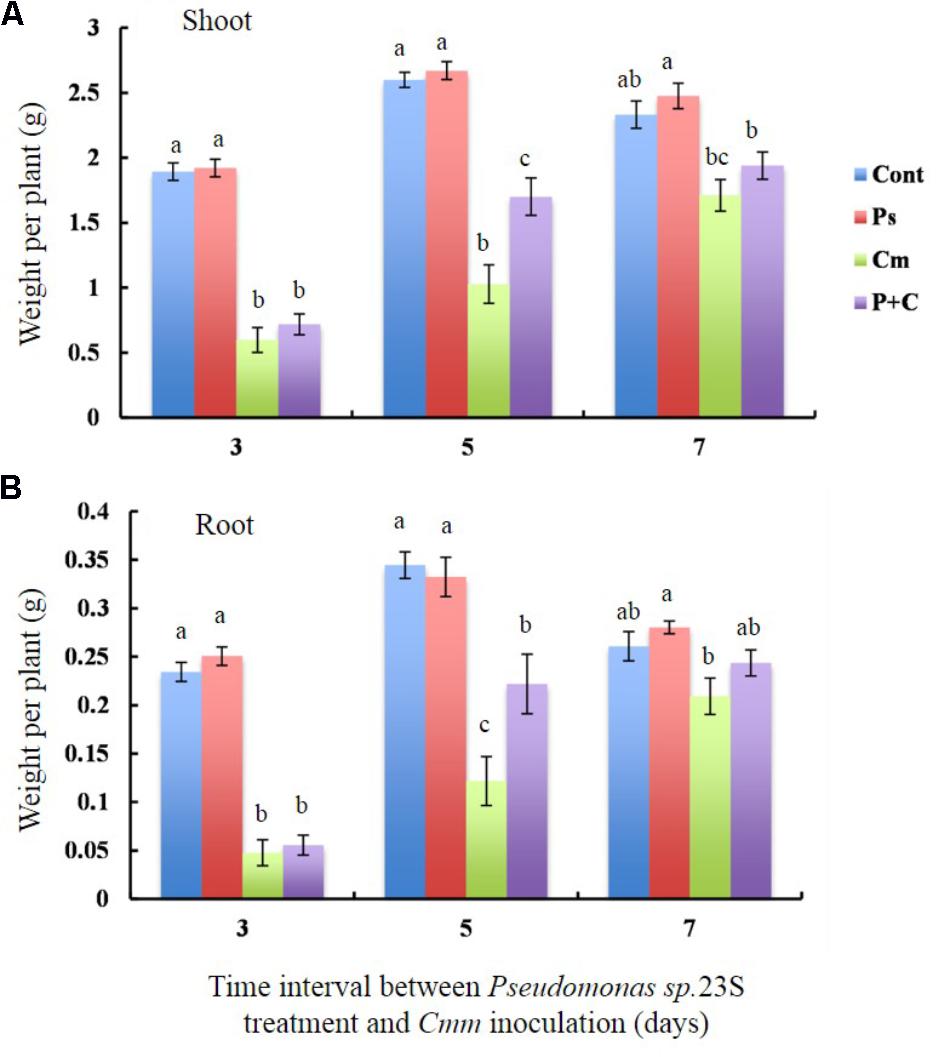
FIGURE 3. Pseudomonas sp. 23S treatment, 5 days prior to Cmm inoculation, increased the weights of shoots and roots. Two-week-old tomato plants were treated with Pseudomonas sp. 23S (or 10 mM MgSO4) by soil drench, and after 3, 5, or 7 days, Cmm (or 10 mM MgSO4) was inoculated into the main stem by needle injection. The plants were harvested after 3 weeks and sampled for: (A) weight of dry shoots and (B) weight of dry roots. Error bars indicate standard error of the mean. Association with different letters indicates statistical significance based on ANOVA followed by Tukey’s multiple comparison test. Treatments are: Cont, control; Pse, treated with Pseudomonas sp. 23S; Cmm, inoculated with Cmm; and, P+C, treated with Pseudomonas sp. 23S, and inoculated with Cmm (n = 14, p = 0.05).
At the harvest (21 days post-Cmm inoculation), a 1-cm-length stem piece above the inoculation site was taken and used for counting the colony forming units (cfu) of Cmm presence (Table 3). The number of cfu for the P+C treatment was significantly lower than that for the Cmm treatment when the interval was 5 days, while no difference was detected when the intervals were 3 and 7 days.
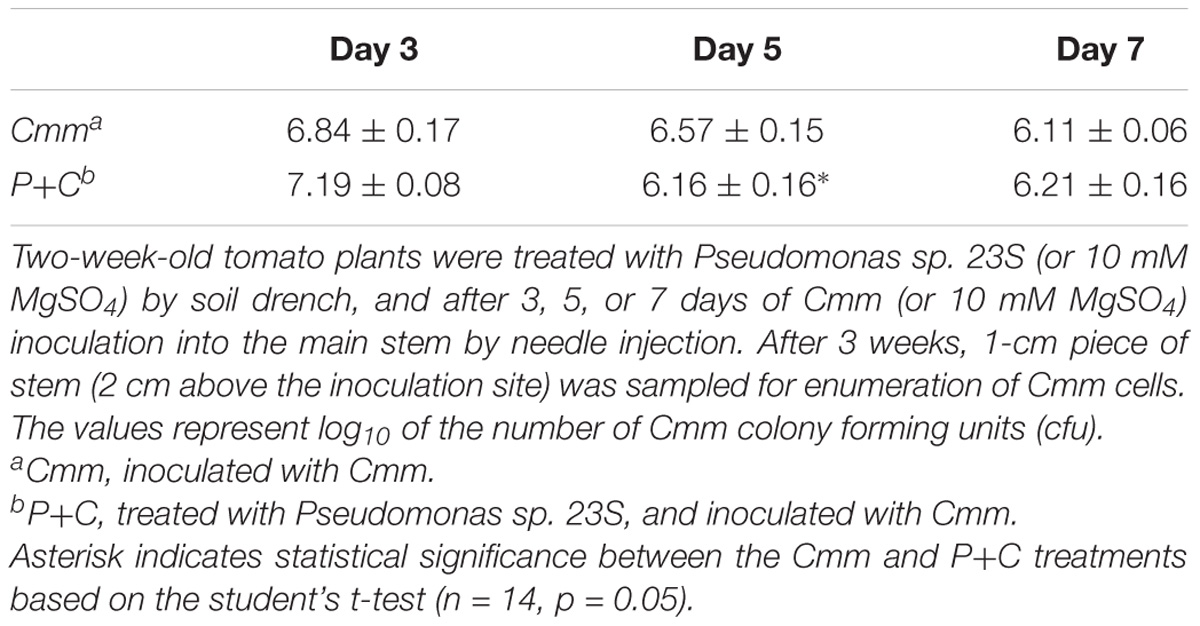
TABLE 3. Pseudomonas sp. 23S treatment, 5-day prior to Cmm inoculation, reduced the Cmm population in the stem.
Nutrient levels of shoots, specifically nitrogen (N), phosphorus (P), potassium (K), and carbon (C) were measured to study the effects of the Pseudomonas sp. 23S treatment (Figures 4A–D). For all the nutrients that were measured, the levels were not significantly different between the Cont and Pse treatments under any of the interval times. On the other hand, the levels of the nutrients from the Cmm and P+C treatments were significantly lower than those of the Cont and Pse treatments. With an interval of 5 days, the levels of N, P, K, and C for the P+C treatment were significantly higher than those of the Cmm treatment. In addition, the nutrient levels of P and K for the P+C treatment were not different from those of the Cont treatment (Figures 4B,C). Under the 7-day-interval, the levels of the Cmm treatment tended to be lower but overall, the levels for all the nutrients were not very different among the treatments (Figures 4A–D).
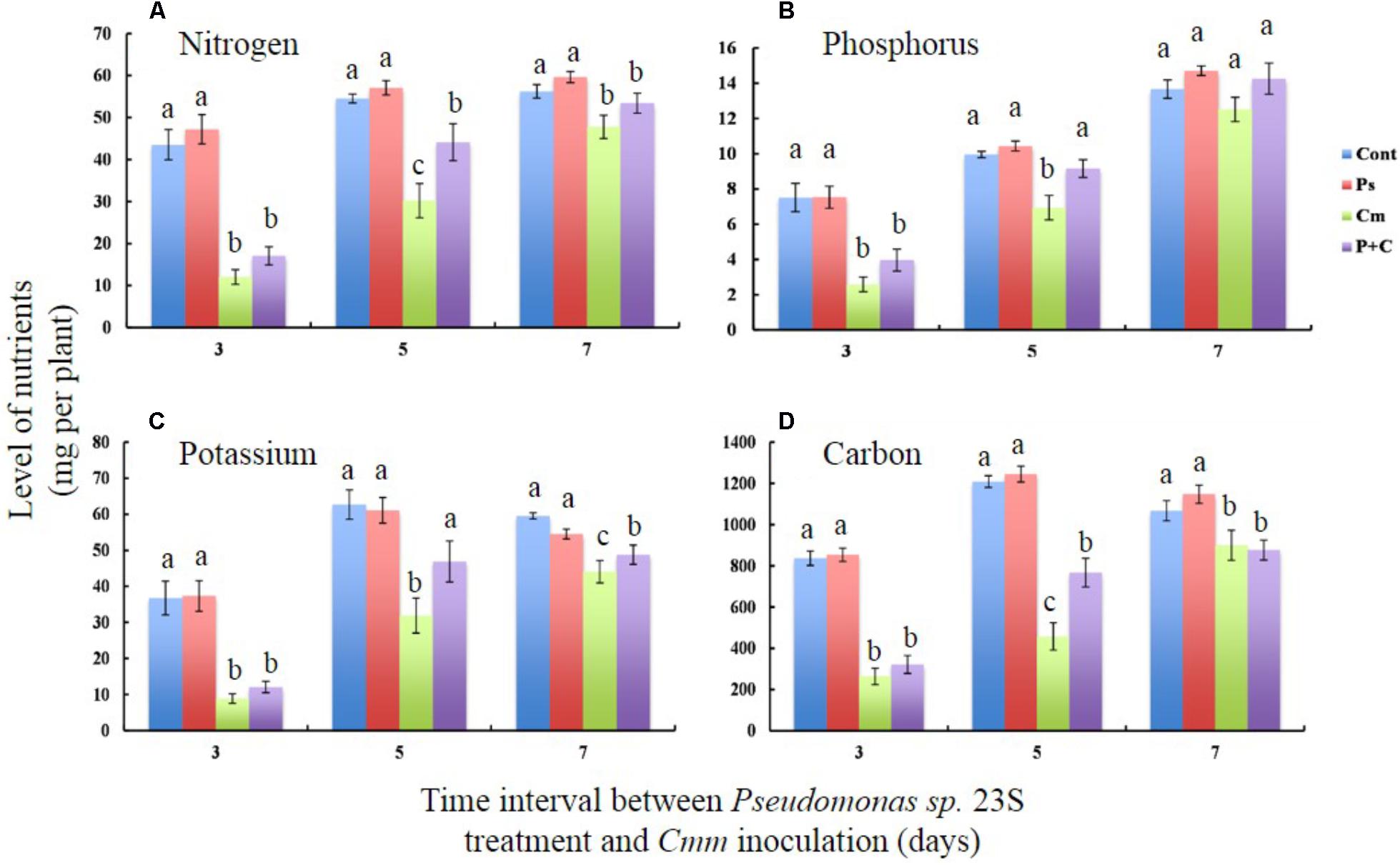
FIGURE 4. Pseudomonas sp. 23S treatment, 5 days prior to Cmm inoculation, improved nutrient levels of leaf tissue. Two-week-old tomato plants were treated with Pseudomonas sp. 23S (or 10 mM MgSO4) by soil drench, and after 3, 5, or 7 days, Cmm (or 10 mM MgSO4) was inoculated into the main stem by needle injection. The plants were harvested after 3 weeks. Nutrients analyzed are (A) Nitrogen, (B) Phosphorus, (C) Potassium, and (D) Carbon. Association with different letters indicates statistical significance based on ANOVA followed by Tukey’s multiple comparison test. Treatments are: Cont, control; Pse, treated with Pseudomonas sp. 23S; Cmm, inoculated with Cmm; and, P+C, treated with Pseudomonas sp. 23S, and inoculated with Cmm (n = 14, p = 0.05).
The transcript levels of PR1a, PI2, and ACO were studied in tomato plants 1, 3, 5, and 7 days after soil drench treatment with Pseudomonas sp. 23S (Figures 5A–C). The transcript levels of PR1a, a marker gene of salicylic acid activity were higher for the Pseudomonas sp. 23S treatment as compared with those of the Control treatment at all the time points (Figure 5A). Its transcript abundance reached highest at day 3 (10-fold), then diminished at days 5 (fourfold) and 7 (fivefold). The transcript levels of PI2 and ACO were not different between Control and Pseudomonas sp. 23S treatments at any of the time points (Figures 5B,C).
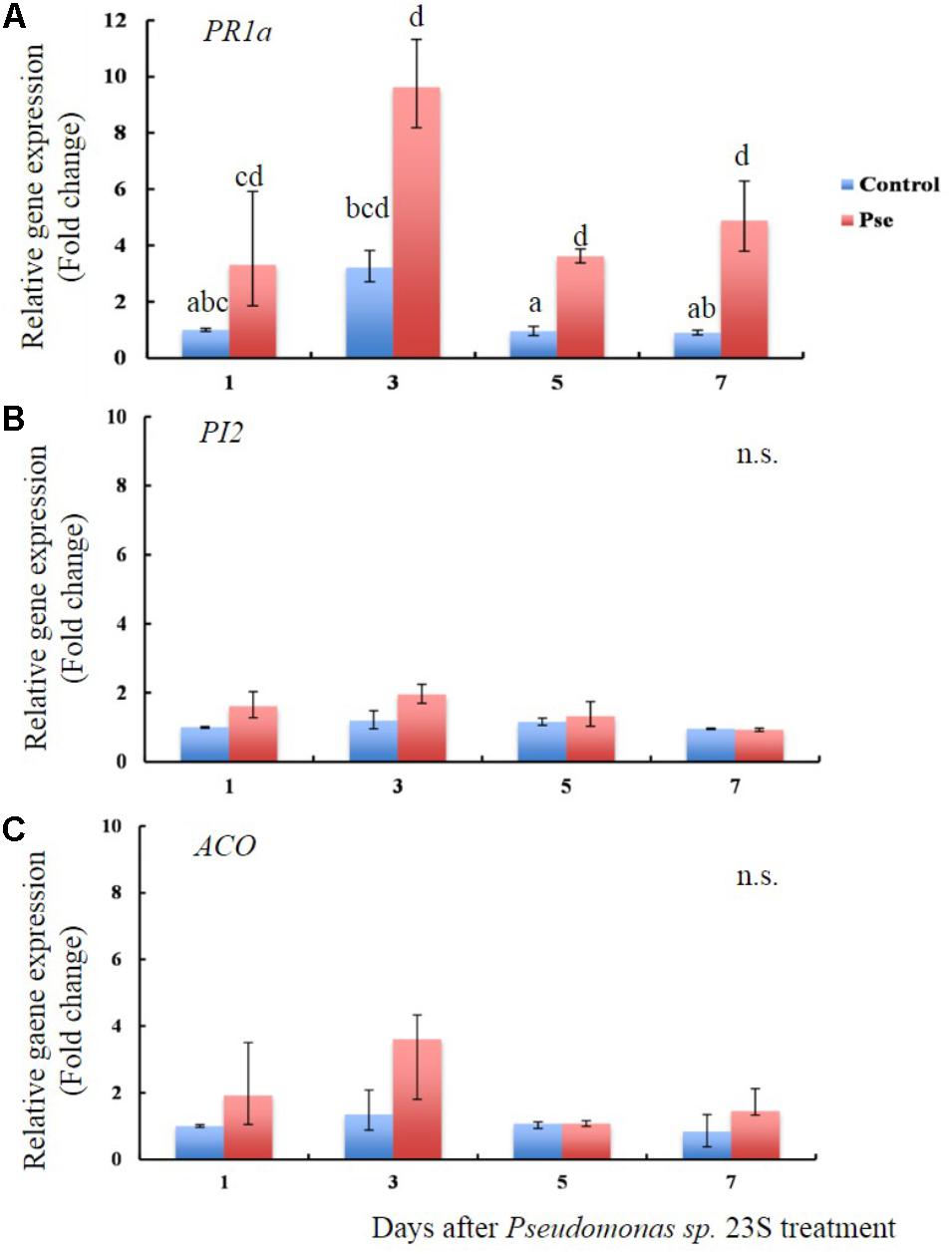
FIGURE 5. Pseudomonas sp. 23S treatment induced an increase in the transcript level of PR1a. Two-week old tomato plants were treated with Pseudomonas sp. 23S (Pse) or 10 mM MgSO4 (Control) and after 1, 3, 5, and 7 days, the shoots were harvested and used for RNA extraction and real-time qPCR. Genes analyzed are (A) PR1a, (B) PI2, and (C) ACO. Error bars indicate standard error of the mean. Association with different letters indicates statistical significance based on ANOVA followed by Tukey’s multiple comparison test (p = 0.05).
Transcript levels of PR1a, PI2, and ACO were also examined when tomato plants treated with Pseudomonas sp. 23S by soil drench and inoculation of Cmm into the main stem by needle injection after 5 days (Figures 6A–C). Day 5 was chosen because the previous physiological experiment, described above, indicated that the disease severity was smallest when Cmm was inoculated after 5 days, rather than 3 or 7 days. The transcript level of PR1a was not different among treatments at day 1; however, at days 3 and 5, the P+C treatment resulted in significantly higher transcript levels than other treatments (54 fold at day 3 and 58 fold at day 5; Figure 6A). At day 7, its transcript level was still higher (55 fold), and the transcript levels for the Pse and Cmm treatments were also as high as that of the P+C treatment (34 fold for the Pse and 75 fold for Cmm treatment; Figure 6A). The transcript levels of PI2 were relatively high at day 1 for all of the treatments (87, 25, 48, and 68 fold for the Cont, Pse, Cmm and P+C treatments, respectively) as compared with those at day 3, 5, 7 (Figure 6B). In addition, they were variable among biological replicates, resulting in large standard errors. No differences were detected among treatments at days 1, 3 and 5. At day 7, the transcript level of the Cmm treatment (ninefold greater than the control) was significantly higher than other treatments. The transcript level of ACO gradually increased going from days 1 to 7 (Figure 6C). For every time point: the transcript level of the Cont treatment was the lowest; the level of the Pse and Cmm treatments were similar to or slightly higher than those of the Cont treatment; and, no difference was detected between the Pse and Cmm treatments. The P+C treatment was always the highest among the treatments (1.5, 1.75, 2.5, and 4.5 fold relative to the Cont, Pse, Cmm and P+C treatments, respectively; Figure 6C).
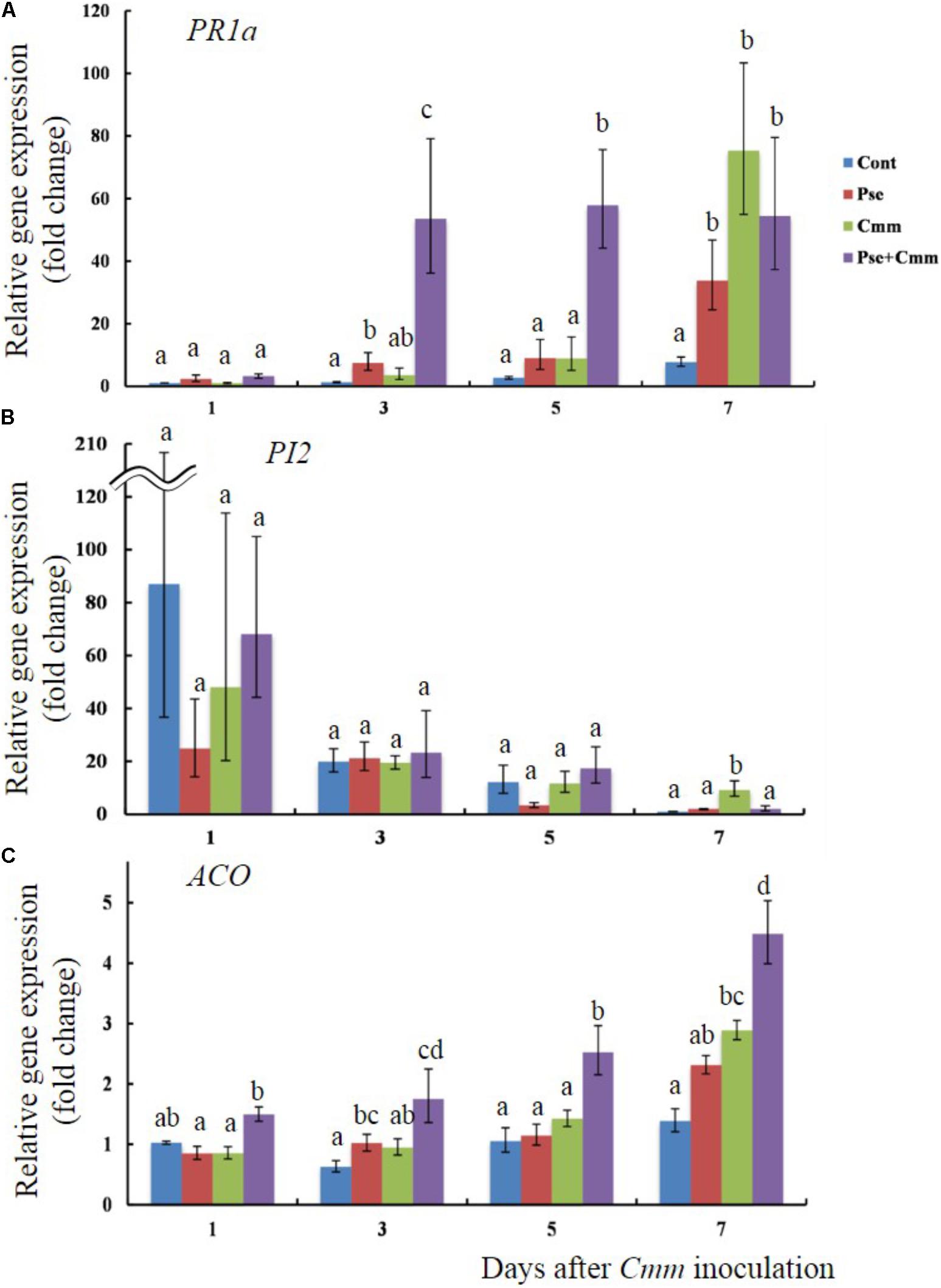
FIGURE 6. Pseudomonas sp. 23S treatment, 5 days prior to Cmm inoculation, induced faster and greater response in transcript levels of PR1a and ACO. Two-week old tomato plants were treated with Pseudomonas sp. 23S (or 10 mM MgSO4) and after 5 days, Cmm (or 10 mM MgSO4) was inoculated into the stem by needle injection: Cont, control; Pse, treated with Pseudomonas sp. 23S; Cmm, inoculated with Cmm; and, P+C, treated with Pseudomonas sp. 23S, and inoculated with Cmm. After 1, 3, 5, and 7 days, the shoots were harvested and used for RNA extraction, and real-time qPCR. Genes analyzed are (A) PR1a, (B) PI2, and (C) ACO. Error bars indicate standard error of the mean. Association with different letters indicates statistical significance based on ANOVA followed by Bonferroni multiple comparison (p = 0.05).
Pseudomonas sp. 23S was shown to possess key PGPR traits, but its degree differed among traits. Specifically, production of siderophores and indole acetic acids were clearly demonstrated. Since siderophores facilitate iron acquisition, especially under iron-limited conditions, siderophore-production represents a biocontrol mechanism for suppression of root diseases by rhizobacteria (Schippers et al., 1987; O’Sullivan and O’Gara, 1992; Shanmugaiah et al., 2015). Similar to past findings (Idris et al., 2007), the amount of IAA produced by Pseudomonas sp. 23S was dependent on the concentration of the IAA precursor tryptophan. Bacteria-produced IAA is known to alter root architecture and support plant development (Dodd et al., 2010; Bhattacharyya and Jha, 2012). In addition, Pseudomonas sp. 23S inhibited the growth of Pseudomonas syringae pv. tomato DC 3000 that causes bacterial speck disease of tomato (Solanum lycopersicum) and Arabidopsis thaliana (Whalen et al., 1991), suggesting potential for Pseudomonas sp. 23S as a biocontrol agent for this disease. On the other hand, Pseudomonas sp. 23S may not be a very good phosphorous (P) solubilizer, as the amount of P solubilized was relatively low as compared to other phosphorus-solubilizing PGPR (Rahi et al., 2010). Pseudomonas sp. 23S was shown to be a moderate producer of hydrogen cyanide (HCN) a volatile, antibiotic, secondary metabolite, which can contribute to disease suppression by some biocontrol bacteria (Voisard et al., 1989; Defago and Haas, 1990; Haas and Keel, 2003). Since HCN is known to inhibit Cmm growth (Lanteigne et al., 2012), production of HCN may partly explain the antagonistic activity that Pseudomonas sp. 23S has against Cmm.
While in vitro assay revealed the potential of Pseudomonas sp. 23S as a PGPR, the plant assay demonstrated that Pseudomonas sp. 23S has plant-growth promoting effects on tomato seedlings. In this experiment, the substrate was probably not the source of nutrients for seedlings, or for Pseudomonas sp. 23S, because it was composed of a mix of sand and turface, and the seedlings were supplied with water only. Pseudomonas sp. 23S might have utilized the nutrients from root exudates and synthesized chemicals that might have growth-promoting effects on the tomato seedlings. Many rhizobacteria are known to convert the root exudate tryptophan to IAA (Frankenberger and Muhammad, 1995), and as the in vitro assay suggested that Pseudomonas sp. 23S was an IAA producer, this mechanism could explain the enhanced root development of the plants treated with Pseudomonas sp. 23S. Pseudomonas sp. 23S was also shown to be a good colonizer of tomato roots, as it colonized the roots and established in good numbers within 3 days, similar to previous reports (Lugtenberg et al., 1999; Yan et al., 2003). We also tried to isolate Pseudomonas sp. 23S from inside the root tissue by sterilizing the root surface, but we were not able to do so (data not shown). Hence, Pseudomonas sp. 23S seems to reside only on the root surface (rhizoplane) and in the soil around the root (rhizosphere), not the inside the root, and causes plant growth promotion.
Pseudomonas sp. 23S triggered ISR, which probably contributed to the reduction of bacterial canker severity by Cmm in tomato plants when Cmm was inoculated 5 days after the Pseudomonas sp. 23S application. In this study, Pseudomonas sp. 23S was applied before Cmm inoculation, and the two bacteria were spatially separated since the Pseudomonas sp. 23S was applied as soil drench and Cmm was injected to the main stem by syringe needle. We tested for the presence of Pseudomonas sp. 23S in the stem samples by using a selective plate assay, but the bacteria were never detected (data not shown). Thus, direct contact between Cmm of the Pseudomonas sp. 23S was not likely to occur, and the bacterial canker reduction that was observed must have been a result of ISR effects. Cmm can survive as an endophyte in tomato plants, but induction of disease symptoms requires a certain minimum population level, generally 108 cfu g−1 of plant tissue (Gartemann et al., 2003). Compared to this number, as well as to the reports from past studies (Balaji et al., 2008; Sharabani et al., 2013), the Cmm population in the stem samples in our study was relatively small, although leaf wilting was clearly observed. This could be because the tomato plants used in our study were younger than those used in other studies (17–21 vs. 28 days). Since we observed the disease progression over 3 weeks, we cannot elaborate on the effects of Pseudomonas sp. 23S application on older plants or at later growth stages. However, most plants inoculated with Cmm, but without the Pseudomonas sp. 23S treatment, were severely infected and would not be able to recover as the main stems were completely broken down. Treatment with Pseudomonas sp. 23S reduced the Cmm population and limited the disease severity as indicated by the fact that these plants stood straight and their leaves were unwilted or less wilted. This effect is certainly important considering that the protection of plants at this stage is more critical than that of older plants that are more resistant to the disease.
In our experimental system, the age and/or size of the plants at the time of the Cmm-inoculation might have influenced the disease progression within plants. The date of visual symptom appearance, and the severity of bacterial canker are affected by temperature, plant age, inoculum concentration, and cultivar (Chang et al., 1992b). In young tomato plants, the disease symptoms caused by Cmm are known to appear earlier and they are more susceptible to infection than mature plants (Chang et al., 1992b; Sharabani et al., 2013). This may explain our observation that disease progression was relatively slow for the plants at the 7-day-interval. Nevertheless, at the plants at the 5-day interval the plants are younger than those at the 7-day interval and thus, the slower disease progression observed at this interval is most probably due to ISR, and not to an age-related resistance.
In our study, 5 days was the optimal interval between Pseudomonas sp. 23S application and Cmm inoculation, in terms of alleviating bacterial canker. Past studies showed that several days are required for ISR to develop and to deliver resistance against various phytopathogens (Babu et al., 2015). The different time interval between PGPR treatment and pathogen inoculation could be related to the population size of the PGPR. The protection by PGPR-mediated ISR is said to be apparent only when the roots were colonized by PGPR at a specific threshold population size (Raaijmakers et al., 1995). Also, Zhang et al. (2004) indicate “quorum sensing effects,” where a certain bacterial population density is essential to produce a signal molecule that is involved in provoking ISR.
The results showed that the Pseudomonas sp. 23S increased the transcript level of the PR1a, but not of PI2 and ACO, suggesting that the ISR induced by the Pseudomonas sp. 23S may involve the salicylic acid (SA) pathway, rather than jasmonic acid (JA) or ethylene (ET). The PR1a gene codes for a pathogenesis-related protein, and has been used as a marker gene for salicylic acid resistance induction (Park et al., 2001; Block et al., 2005; Niu et al., 2012; Martínez-Medina et al., 2013). PI2 gene codes for a proteinase inhibitor and is induced by wounding and jasmonic acid (JA; Peña-Cortés et al., 1995; Peiffer et al., 2009; Niu et al., 2012; Martínez-Medina et al., 2013). ACO is a gene coding for 1-aminocyclopropane-1-carboxylix acid (ACC) oxidase, the level of which is related to ethylene (ET) production since ACO is an enzyme that converts ACC into ethylene (Stearns and Glick, 2003; Yim et al., 2014). Although JA/ET are generally considered to be key hormone(s) for ISR response, which is mediated by non-pathogenic plant growth promoting bacteria (PGPR), different results have been reported from more recent studies (De Vleesschauwer and Höfte, 2009), and the ISR by the Pseudomonas sp. 23S seems to fall into this new trend. The signaling pathway for ISR seems species specific, that is specific to the rhizobacterium, and pathogen involved (Ryu et al., 2003; Djavaheri, 2007; Conn et al., 2008). Researchers agree that hormone crosstalk plays an important role in regulating ISR. Regarding SA-and JA/ET pathways, antagonistic interaction has been documented from many studies (Koornneef and Pieterse, 2008), and this might apply to our case, where Pseudomonas sp. 23S induced SA response but not JA and ET. The antagonistic interaction between SA and JA may be the outcome of resource allocation, costs of induction, or a means for the plant to adaptively tailor its responses to different enemies and a target for manipulation by enemies (Thaler et al., 2012). Generally, SA-dependent defense response is said to be effective against biotrophic pathogens, while JA/ET-dependent defense response is effective against necrotrophic pathogens (Sorokan et al., 2013). In this respect, Cmm would be a suitable target for the SA-dependent ISR because Cmm is considered as a biotrophic pathogen (Eichenlaub and Gartemann, 2011). We cannot exclude the possibility that JA and ET are not involved in the ISR provoked by the Pseudomonas sp. 23S. Mutant plants impaired in SA, JA, and ET pathways could be utilized to confirm whether these hormones are required for ISR.
Furthermore, the results demonstrated priming effect of the Pseudomonas sp. 23S treatment. Faster and/or greater response of defense-genes - priming has been observed for many ISR-inducing PGPR (Pieterse et al., 2014). Accumulation response of PR1a after Cmm inoculation was faster and quantitatively greater with the plant pre-treated with the Pseudomonas sp. 23S than the plants without the pre-treatment. Since the Pseudomonas sp. 23S treatment alone also induces accumulation response of PR1a, the prior- Pseudomonas sp. 23S treatment probably prepares the plants for the pathogen attack, by making this response faster and greater and enhancing the defense capacity of the plants. Priming may explain the disease reduction observed under the 5-day interval in our study. To understand the effects of the Pseudomonas sp. 23S treatment for disease reduction, studying the responses of other defense-related genes (e.g., other PR proteins, defense-related enzymes) and whether they do also show priming effects would be helpful.
Faster and greater accumulation response was also observed for the ACO, but the situation may be different from the PR1, because the ACO transcript abundance was not affected by Pseudomonas sp. 23S treatment alone. From past studies, ethylene is known to play a critical role in bacterial canker of tomato. Plants with reduced ethylene production or impairment of ethylene perception results in decrease in the disease severity (Balaji et al., 2008; Savidor et al., 2011), and thus host-derived ethylene is suggested to be a requirement for the disease development by Cmm. Our results that the Pseudomonas sp. 23S treatment alone did not significantly affect the ACO abundance but the same treatment did after Cmm inoculation supports the past studies in that ethylene is involved in the disease infection by Cmm. At the same time, the fact that Pseudomonas sp. 23S treatment can make this response faster and greater in quantitatively and alleviate the disease may suggest that the Pseudomonas sp. 23S might have modulated the role of ethylene. This consequently could influence the disease development by Cmm and thus might have contributed to the disease reduction as observed in our study.
The transcript level of PI2 showed different trend from that of PR1a or ACO, elevated on day 1, especially for the control treatment. This may be due to the damaging nature of applying the mock (needle injection) as the transcript level decreased at later time points. For Cmm inoculated plants, however, its level remained high at day 7. Treatment with Pseudomonas sp. 23S might have explained this: while the Cmm inoculated plants must combat Cmm invasion, the same plants were less affected by the prior Pseudomonas sp. 23S treatment due to ISR effects.
In this study, we investigated Pseudomonas sp. 23S, which was previously isolated based on the in vitro antagonistic activity against Cmm. The characterization study of Pseudomonas sp. 23S revealed great potential of this strain in agriculture, both for plant growth promotion and as a biocontrol agent. Future study could investigate its effectiveness in field condition. This study also demonstrated that Pseudomonas sp. 23S could induce ISR in tomato plants and reduce the severity of tomato bacterial canker disease that is caused by Cmm. The best time interval between the Pseudomonas sp. 23S treatment and Cmm inoculation for reducing the severity of bacterial canker was 5-days in our experimental system, which used drench application of Pseudomonas sp., stem inoculation of Cmm, and young tomato plants; this interval, as well as the effectiveness of ISR, could change with different methods, timing of bacterial application and of pathogen inoculation, and plant ages. Such information would be useful, especially for the commercial use of this bacterium in the future. Our study also suggested that the ISR by Pseudomonas sp. 23S may involve SA in its signaling pathway. However, the possibility of JA and/or ET involvement should not be ignored. Mutant plants with impaired hormonal pathways could be studied in the context of a Cmm infection to confirm their involvement. In addition, our results provided new insights on the role of ethylene in disease development of Cmm. Further studies to elucidate the signaling pathways of Pseudomonas sp. 23S ISR would certainly add knowledge for understanding molecular mechanism of ISR induced by PGPR but it would provide useful information regarding the disease strategies taken by Cmm.
YT wrote the manuscript, conducted the growth chamber experiments, gene expression studies, and analysis of the data. J-BC and DS guided and supervised the overall study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge Ministère de l’Agriculture, des Pêcheries et de l’ Alimentation du Québec (MAPAQ), Engineering Research Council (NSERC) of Canada, and the Canadian Networks of Centres of Excellence (BioFuelNet, Canada) for supporting the project. We are grateful for Hélène Lalande (McGill University) for phosphorus and potassium analyses, and for Werda Saeed (McGill University) for carbon and nitrogen analyses of plant tissues. We thank Alex Martel (McGill University) for helping real-time qPCR. We greatly appreciate Dr. Sowmyalakshmi Subramanian (McGill University) for advice during the experiments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02119/full#supplementary-material
FIGURE S1 | Anti-Cmm activity of Pseudomonas sp. 23S in vitro. One hundred micro liter of Cmm culture was spread on Nutrient Broth Yeast Extract (NBYE) agar. A sterile filter-paper-disk (6 mm diameter) was placed on the agar surface. Five micro liter of the newly isolated bacterium culture (on the right), and Nutrient Broth (on the left) were applied on the respective disks. The plate was sealed with parafilm and incubated at 28°C for 2 days. Zone of inhibition = 5 mm.
FIGURE S2 | Characterization of Pseudomonas sp. 23S for PGPR traits. 1 (a) Phosphorus solubilization on a PVK, NB media (top), Pseudomonas sp. 23S (left and right), and positive control bacterium (bottom). 1 (b) Phosphorus solubilization on a PBRIP agar plate, NB media (left), and of Pseudomonas sp. 23S (right). 2 Siderophore production, NB media (left), and Pseudomonas sp. 23S culture (right). 3 Hydrogen cyanide production, (a) Kings B media, (b) Pseudomonas sp. 23S, and (c) a positive control bacterium. 4 Antagonistic activities against Pseudomonas syringae pv. tomato DC3000.
FIGURE S3 | Representive plant photos showing the effects of Pseudomonas sp. 23S treatment 5-day prior to Cmm inoculation. Two-week-old tomato plants were treated with Pseudomonas sp. 23S by soil drench, and after 5 days, Cmm was inoculated in the main stem by needle injection: (A) Cont, control; (B) Pse, treated with Pseudomonas sp. 23S; (C) Cmm, inoculated with Cmm; and (D) P+C, treated with Pseudomonas sp. 23S, and inoculated with Cmm.
FIGURE S4 | Area under disease progression curve (AUDPC). Two-week-old tomato plants were treated with Pseudomonas sp. 23S by soil drench, and after 5 days, Cmm was inoculated in the main stem by needle injection. The AUDPC was based on the percentage of wilted leaves during 3 weeks after the date of Cmm inoculation (presented by Figure 2 in the main text): Cmm, inoculated with Cmm; and P+C, treated with Pseudomonas sp. 23S, and inoculated with Cmm. Error bars indicate standard error of the mean. An asterisk indicates significant difference from the Cmm treatment after ANOVA followed by Tukey’s multiple comparison test (n = 14, p = 0.05).
FIGURE S5 | Effects of Pseudomonas sp. 23S treatment 5 days prior to Cmm inoculation on (A) plant height, and (B) leaf area. Two-week-old tomato plants were treated with Pseudomonas sp. 23S (or 10 mM MgSO4) by soil drench, and after 3, 5, or 7 days, Cmm (or 10 mM MgSO4) was inoculated into the main stem by needle injection. The plant height and leaf areas were measured. Error bars indicate standard error of the mean. Association with different letters indicate statistical significance based on ANOVA followed by Tukey’s multiple comparison test. Treatments are: Cont, control; Pse, treated with Pseudomonas sp. 23S; Cmm, inoculated with Cmm; and, P+C, treated with Pseudomonas sp. 23S, and inoculated with Cmm (n = 14, p = 0.05).
Aksoy, H. M., Kaya, Y., Ozturk, M., Secgin, Z., Onder, H., and Okumus, A. (2017). Pseudomonas putida–induced response in phenolic profile of tomato seedlings (Solanum lycopersicum L.) infected by Clavibacter michiganensis subsp. michiganensis. Biol. Control 105, 6–12. doi: 10.1016/j.biocontrol.2016.11.001
Alexander, D. B., and Zuberer, D. A. (1991). Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12, 39–45. doi: 10.1007/BF00369386
Amkraz, N., Boudyach, E. H., Boubaker, H., Bouizgarne, B., and Aoumar, A. A. B. (2010). Screening for fluorescent pseudomonades, isolated from the rhizosphere of tomato, for antagonistic activity toward Clavibacter michiganensis subsp. michiganensis. World J. Microbiol. Biotechnol. 26, 1059–1065. doi: 10.1007/s11274-009-0270-5
Askeland, R. A., and Morrison, S. M. (1983). Cyanide production by Pseudomonas fluorescens and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 45, 1802–1807.
Babu, A. N., Jogaiah, S., Ito, S. I., Nagaraj, A. K., and Tran, L. S. P. (2015). Improvement of growth, fruit weight and early blight disease protection of tomato plants by rhizosphere bacteria is correlated with their beneficial traits and induced biosynthesis of antioxidant peroxidase and polyphenol oxidase. Plant Sci. 231, 62–73. doi: 10.1016/j.plantsci.2014.11.006
Balaji, V., Mayrose, M., Sherf, O., Jacob-Hirsch, J., Eichenlaub, R., Iraki, N., et al. (2008). Tomato transcriptional changes in response to Clavibacter michiganensis subsp. michiganensis reveal a role for ethylene in disease development. Plant Physiol. 146, 1797–1809. doi: 10.1104/pp.107.115188
Bhattacharyya, P. N., and Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Block, A., Schmelz, E., O’Donnell, P. J., Jones, J. B., and Klee, H. J. (2005). Systemic acquired tolerance to virulent bacterial pathogens in tomato. Plant Physiol. 138, 1481–1490. doi: 10.1104/pp.105.059246
Bouizgarne, B. (2013). “Bacteria for plant growth promotion and disease management,’,” in Bacteria in Agrobiology: Disease Management, ed. D. Maheshwari (Berlin: Springer), 15–47. doi: 10.1007/978-3-642-33639-3_2
Carlton, W. M., Braun, E. J., and Gleason, M. L. (1998). Ingress of Clavibacter michiganensis subsp. michiganensis into tomato leaves through hydathodes. Phytopathology 88, 525–529. doi: 10.1094/PHYTO.1998.88.6.525
Chalupowicz, L., Cohen-Kandli, M., Dror, O., Eichenlaub, R., Gartemann, K. H., Sessa, G., et al. (2010). Sequential expression of bacterial virulence and plant defense genes during infection of tomato with Clavibacter michiganensis subsp. michiganensis. Phytopathology 100, 252–261. doi: 10.1094/PHYTO-100-3-0252
Chang, R. J., Ries, S. M., and Pataky, J. K. (1992a). Local sources of Clavibacter michiganensis subsp. michiganensis in the development of bacterial canker on tomatoes. Phytopathology 82, 553–560. doi: 10.1094/Phyto-82-553
Chang, R. J., Ries, S. M., and Pataky, J. K. (1992b). Effects of temperature, plant age, inoculum concentration, and cultivar on the incubation period and severity of bacterial canker of tomato. Plant Dis. 76, 1150–1155. doi: 10.1094/PD-76-1150
Conn, V. M., Walker, A. R., and Franco, C. M. M. (2008). Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Mol. Plant Microbe Interact. 21, 208–218. doi: 10.1094/MPMI-21-2-0208
Conrath, U., Beckers, G. J., Flors, V., García-Agustín, P., Jakab, G., Mauch, F., et al. (2006). Priming: getting ready for battle. Mol. Plant Microbe Interact. 19, 1062–1071. doi: 10.1094/MPMI-19-1062
de León, L., Siverio, F., López, M. M., and Rodríguez, A. (2011). Clavibacter michiganesis subsp. michiganensis, a seedborne tomato pathogen: healthy seeds are still the goal. Plant Dis. 95, 1328–1338. doi: 10.1094/PDIS-02-11-0091
De Meyer, G., Capieau, K., Audenaert, K., Buchala, A., Métraux, J. P., and Höfte, M. (1999). Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol. Plant Microbe. Interact. 12, 450–458. doi: 10.1094/MPMI.1999.12.5.450
De Vleesschauwer, D., and Höfte, M. (2009). Rhizobacteria-induced systemic resistance. Adv. Bot. Res. 51, 223–281. doi: 10.1016/S0065-2296(09)51006-3
Deaker, R., Kecskés, M. L., Rose, M. T., Amprayn, K., Ganisan, K., Tran, T. K. C., et al. (2011). Practical Methods for the Quality Control of Inoculant Biofertilisers. Canberra: Australian Centre for International Agricultural Research (ACIAR).
Defago, G., and Haas, D. (1990). Pseudomonas as antagonists of soilborne plant pathogens: modes of action and genetic analysis. Soil Biochem.6:291.
Deng, P., Wang, X., Baird, S. M., and Lu, S. E. (2015). Complete genome of Pseudomonas chlororaphis strain UFB2, a soil bacterium with antibacterial activity against bacterial canker pathogen of tomato. Stand. Genomic Sci. 10:117. doi: 10.1186/s40793-015-0106-x
Djavaheri, M. (2007). Iron-Regulated Metabolites of Plant Growth-Promoting Pseudomonas fluorescens WCS374: their Role in Induced Systemic Resistance. Doctoral dissertation, Utrecht University, Utrecht.
Dodd, I. C., Zinovkina, N. Y., Safronova, V. I., and Belimov, A. A. (2010). Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 157, 361–379. doi: 10.1111/j.1744-7348.2010.00439.x
Eichenlaub, R., and Gartemann, K. H. (2011). The Clavibacter michiganensis subspecies: molecular investigation of gram-positive bacterial plant pathogens. Ann. Rev. Phytopathol. 49, 445–464. doi: 10.1146/annurev-phyto-072910-095258
Fiske, C. H., and Subbarow, Y. (1925). A colorimetric determination of phosphorus. J. Biol. Chem. 66, 375–400.
Frankenberger, J. W. T., and Muhammad, A. (1995). Phytohormones in Soils: Microbial Production and Function. New York, NY: Marcel Dekker Inc.
Ftayeh, R. M., von Tiedemann, A., and Rudolph, K. W. (2011). A new selective medium for isolation of Clavibacter michiganensis subsp. michiganensis from tomato plants and seed. Phytopathology 101, 1355–1364. doi: 10.1094/PHYTO-02-11-0045
Gartemann, K. H., Kirchner, O., Engemann, J., Gräfen, I., Eichenlaub, R., and Burger, A. (2003). Clavibacter michiganensis subsp. michiganensis: first steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium. J. Biotechnol. 106, 179–191. doi: 10.1016/j.jbiotec.2003.07.011
Ghosh, S. K., Pal, S., and Chakraborty, N. (2015). The qualitative and quantitative assay of siderophore production by some microorganisms and effect of different media on its production. Int. J. Chem. 13, 1621–1629.
Gleason, M. L., Gitaitis, R. D., and Ricker, M. D. (1993). Recent progress in understanding and controlling bacterial canker of tomato in eastern North America. Plant Dis. 77, 1069–1076. doi: 10.1094/PD-77-1069
Haas, D., and Keel, C. (2003). Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41, 117–153. doi: 10.1146/annurev.phyto.41.052002.095656
Hausbeck, M. K., Bell, J., Medina-Mora, C., Podolsky, R., and Fulbright, D. W. (2000). Effect of bactericides on population sizes and spread of Clavibacter michiganensis subsp. michiganensis on tomatoes in the greenhouse and on disease development and crop yield in the field. Phytopathology 90, 38–44. doi: 10.1094/PHYTO.2000.90.1.38
Hoagland, D. R., and Arnon, D. I. (1950). The Water-Culture Method for Growing Plants Without Soil. Berkeley, CA: Circular, California Agricultural Experiment Station.
Idris, E. E., Iglesias, D. J., Talon, M., and Borriss, R. (2007). Tryptophan-dependent production of indole-3- acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol. Plant Microbe Interact. 20, 619–626. doi: 10.1094/MPMI-20-6-0619
Jung, W. J., Mabood, F., Souleimanov, A., Whyte, L. G., Niederberger, T. D., and Smith, D. L. (2014). Antibacterial activity of antagonistic bacterium Bacillus subtilis DJM-51 against phytopathogenic Clavibacter michiganense subsp. michiganense ATCC 7429 in vitro. Microb. Pathog. 77, 13–16. doi: 10.1016/j.micpath.2014.10.008
Kloepper, J. W., Lifshitz, R., and Zablotowicz, R. M. (1989). Free-living bacterial inocula for enhancing crop productivity. Trends Biotechnol. 7, 39–44. doi: 10.1016/0167-7799(89)90057-7
Kloepper, J. W., and Schroth, M. N. (1978). “Plant growth-promoting rhizobacteria on radishes,” in Proceedings of the 4th International Conference on Plant Pathogenic Bacteria, Poole, 879–882.
Koornneef, A., and Pieterse, C. M. (2008). Cross talk in defense signaling. Plant Physiol. 146, 839–844. doi: 10.1104/pp.107.112029
Lanteigne, C., Gadkar, V. J., Wallon, T., Novinscak, A., and Filion, M. (2012). Production of DAPG and HCN by Pseudomonas sp. LBUM300 contributes to the biological control of bacterial canker of tomato. Phytopathology 102, 967–973. doi: 10.1094/PHYTO-11-11-0312
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Ráez, J. A., Verhage, A., Fernández, I., García, J. M., Azcón-Aguilar, C., Flors, V., et al. (2010). Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. J. Exp. Bot. 61, 2589–2601. doi: 10.1093/jxb/erq089
Lugtenberg, B. J., Kravchenko, L. V., and Simons, M. (1999). Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1, 439–446. doi: 10.1046/j.1462-2920.1999.00054.x
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Martínez-Medina, A., Fernández, I., Sánchez-Guzmán, M. J., Jung, S. C., Pascual, J. A., and Pozo, M. J. (2013). Deciphering the hormonal signalling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front. Plant Sci. 4:206. doi: 10.3389/fpls.2013.00206
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
Niu, D. D., Wang, C. J., Guo, Y. H., Jiang, C. H., Zhang, W. Z., Wang, Y. P., et al. (2012). The plant growth-promoting Rhizobacterium Bacillus cereus AR156 induces resistance in tomato with induction and priming of defence response. Biocontrol Sci. Technol. 22, 991–1004. doi: 10.1080/09583157.2012.706595
O’Sullivan, D. G., and O’Gara, F. (1992). Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol. Rev. 56, 662–676.
Park, K., Ahn, I. P., and Kim, C. H. (2001). Systemic resistance and expression of the pathogenesis-related genes mediated by the plant growth-promoting rhizobacterium Bacillus amyloliquefaciens EXTN-1 against anthracnose disease in cucumber. Mycobiology 29, 48–53. doi: 10.1080/12298093.2001.12015759
Parkinson, J. A., and Allen, S. E. (1975). A wet oxidation procedure suitable for the determination of nitrogen and mineral nutrients in biological material. Commun. Soil Sci. Plant Anal. 6, 1–11. doi: 10.1080/00103627509366539
Peiffer, M., Tooker, J. F., Luthe, D. S., and Felton, G. W. (2009). Plants on early alert: glandular trichomes as sensors for insect herbivores. New Phytol. 184, 644–656. doi: 10.1111/j.1469-8137.2009.03002.x
Peña-Cortés, H., Fisahn, J., and Willmitzer, L. (1995). Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc. Natl. Acad. Sci. U.S.A. 92, 4106–4113. doi: 10.1073/pnas.92.10.4106
Pieterse, C. M., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C., and Bakker, P. A. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Microbiology 17, 362–370.
Raaijmakers, J. M., Leeman, M., Van Oorschot, M. M., Van der Sluis, I., Schippers, B., and Bakker, P. A. H. M. (1995). Dose-response relationships in biological control of fusarium wilt of radish by Pseudomonas spp. Phytopathology 85, 1075–1080. doi: 10.1094/Phyto-85-1075
Rahi, P., Pathania, V., Gulati, A., Singh, B., Bhanwra, R. K., and Tewari, R. (2010). Stimulatory effect of phosphate-solubilizing bacteria on plant growth, stevioside and rebaudioside-A contents of Stevia rebaudiana Bertoni. Appl. Soil Ecol. 46, 222–229. doi: 10.1016/j.apsoil.2010.08.008
Raj, S. N., Shetty, N. P., and Shetty, H. S. (2004). Seed bio-priming with Pseudomonas fluorescens isolates enhances growth of pearl millet plants and induces resistance against downy mildew. Int. J. Pest Manage. 50, 41–48. doi: 10.1080/09670870310001626365
Rudrappa, T., Czymmek, K. J., Paré, P. W., and Bais, H. P. (2008). Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. doi: 10.1104/pp.108.127613
Ryu, C. M., Hu, C. H., Reddy, M. S., and Kloepper, J. W. (2003). Different signaling pathways of induced resistance by rhizobacteria in Arabidopsis thaliana against two pathovars of Pseudomonas syringae. New Phytol. 160, 413–420. doi: 10.1046/j.1469-8137.2003.00883.x
Savidor, A., Teper, D., Gartemann, K. H., Eichenlaub, R., Chalupowicz, L., Manulis-Sasson, S., et al. (2011). The Clavibacter michiganensis subsp. michiganensis–tomato interactome reveals the perception of pathogen by the host and suggests mechanisms of infection. J. Proteome Res. 11, 736–750. doi: 10.1021/pr200646a
Schippers, B., Bakker, A. W., and Bakker, P. A. H. M. (1987). Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practises. Annu. Rev. Phytopathol. 25, 339–358. doi: 10.1146/annurev.py.25.090187.002011
Schippers, B., Bakker, A. W., Bakker, P. A. H. M., and Van Peer, R. (1990). Beneficial and deleterious effects of HCN-producing Pseudomonas on rhizosphere interactions. Plant Soil 129, 75–83. doi: 10.1007/BF00011693
Schuhegger, R., Ihring, A., Gantner, S., Bahnweg, G., Knappe, C., Vogg, G., et al. (2006). Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 29, 909–918. doi: 10.1111/j.1365-3040.2005.01471.x
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Sen, Y., van der Wolf, J., Visser, R. G., and van Heusden, S. (2015). Bacterial canker of tomato: current knowledge of detection, management, resistance, and interactions. Plant Dis. 99, 4–13. doi: 10.1094/PDIS-05-14-0499-FE
Shanmugaiah, V., Nithya, K., Harikrishnan, H., Jayaprakashvel, M., and Balasubramanian, N. (2015). “Biocontrol mechanisms of siderophores against bacterial plant pathogens,” in Sustainable Approaches to Controlling Plant Pathogenic Bacteria, ed. K. K. Bastas (Boca Raton, FL: CRC Press), 167–190. doi: 10.1201/b18892-9
Sharabani, G., Shtienberg, D., Borenstein, M., Shulhani, R., Lofthouse, M., Sofer, M., et al. (2013). Effects of plant age on disease development and virulence of Clavibacter michiganensis subsp. michiganensis on tomato. Plant Pathol. 62, 1114–1122. doi: 10.1111/ppa.12013
Song, Y., Chen, D., Lu, K., Sun, Z., and Zeng, R. (2015). Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant Sci. 6:786. doi: 10.3389/fpls.2015.00786
Sorokan, A. V., Burkhanova, G. F., and Maksimov, I. V. (2013). ““The interplay between salicylic and jasmonic acid during phytopathogenesis,”,” in Salicylic Acid, ed. M. Alymeni (Berlin: Springer), 277–297. doi: 10.1007/978-94-007-6428-6_12
Stearns, J. C., and Glick, B. R. (2003). Transgenic plants with altered ethylene biosynthesis or perception. Biotechnol. Adv. 21, 193–210. doi: 10.1016/S0734-9750(03)00024-7
Subramanian, S., and Smith, D. L. (2015). Bacteriocins from the rhizosphere microbiome – from an agriculture perspective. Front. Plant Sci. 6:909. doi: 10.3389/fpls.2015.00909
Thaler, J. S., Humphrey, P. T., and Whiteman, N. K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. doi: 10.1016/j.tplants.2012.02.010
Tjamos, S. E., Flemetakis, E., Paplomatas, E. J., and Katinakis, P. (2005). Induction of resistance to Verticillium dahliae in Arabidopsis thaliana by the biocontrol agent K-165 and pathogenesis-related proteins gene expression. Mol. Plant Microbe Interact. 18, 555–561. doi: 10.1094/MPMI-18-0555
Van Loon, L. C., and Bakker, P. A. H. M. (2005). “Induced systemic resistance as a mechanism of disease suppression by rhizobacteria,” in PGPR: Biocontrol and biofertilization, ed. Z. A. Siddiqui (Dordrecht: Springer),39–66.
Van Wees, S. C., Van der Ent, S., and Pieterse, C. M. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448. doi: 10.1016/j.pbi.2008.05.005
Voisard, C., Keel, C., Haas, D., and Dèfago, G. (1989). Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8, 351–358.
Whalen, M. C., Innes, R. W., Bent, A. F., and Staskawicz, B. J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. doi: 10.1105/tpc.3.1.49
Xu, X., Kumar, A., Deblais, L., Pina-Mimbela, R., Nislow, C., Fuchs, J. R., et al. (2015). Discovery of novel small molecule modulators of Clavibacter michiganensis subsp. michiganensis. Front. Microbiol. 6:1127. doi: 10.3389/fmicb.2015.01127
Xu, X., Rajashekara, G., Paul, P. A., and Miller, S. A. (2012). Colonization of tomato seedlings by bioluminescent Clavibacter michiganensis subsp. michiganensis under different humidity regimes. Phytopathology 102, 177–184. doi: 10.1094/PHYTO-03-11-0090
Yan, Z., Reddy, M. S., and Kloepper, J. W. (2003). Survival and colonization of rhizobacteria in a tomato transplant system. Can. J. Microbiol. 49, 383–389. doi: 10.1139/w03-051
Yim, W. J., Kim, K. Y., Lee, Y. W., Sundaram, S. P., Lee, Y., and Sa, T. M. (2014). Real time expression of ACC oxidase and PR-protein genes mediated by Methylobacterium spp. in tomato plants challenged with Xanthomonas campestris pv. vesicatoria. J. Plant Physiol. 171, 1064–1075. doi: 10.1016/j.jplph.2014.03.009
Keywords: tomato, Pseudomonas, Clavibacter michiganensis subsp. michiganensis, PGPR, biocontrol, induced systemic resistance
Citation: Takishita Y, Charron J-B and Smith DL (2018) Biocontrol Rhizobacterium Pseudomonas sp. 23S Induces Systemic Resistance in Tomato (Solanum lycopersicum L.) Against Bacterial Canker Clavibacter michiganensis subsp. michiganensis. Front. Microbiol. 9:2119. doi: 10.3389/fmicb.2018.02119
Received: 30 March 2018; Accepted: 20 August 2018;
Published: 11 September 2018.
Edited by:
Jesús Mercado-Blanco, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Alex Williams, University of Sheffield, United KingdomCopyright © 2018 Takishita, Charron and Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donald L. Smith, ZG9uYWxkLnNtaXRoQG1jZ2lsbC5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.