
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 23 August 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01962
 Nathan Horn1†
Nathan Horn1† Arun K. Bhunia2,3*
Arun K. Bhunia2,3*The incidence of foodborne outbreaks and product recalls is on the rise. The ability of the pathogen to adapt and survive under stressful environments of food processing and the host gastrointestinal tract may contribute to increasing foodborne illnesses. In the host, multiple factors such as bacteriolytic enzymes, acidic pH, bile, resident microflora, antimicrobial peptides, and innate and adaptive immune responses are essential in eliminating pathogens. Likewise, food processing and preservation techniques are employed to eliminate or reduce human pathogens load in food. However, sub-lethal processing or preservation treatments may evoke bacterial coping mechanisms that alter gene expression, specifically and broadly, resulting in resistance to the bactericidal insults. Furthermore, environmentally cued changes in gene expression can lead to changes in bacterial adhesion, colonization, invasion, and toxin production that contribute to pathogen virulence. The shared microenvironment between the food preservation techniques and the host gastrointestinal tract drives microbes to adapt to the stressful environment, resulting in enhanced virulence and infectivity during a foodborne illness episode.
Foodborne illnesses cause considerable morbidity, mortality, and economic losses globally. The World Health Organization (WHO) estimates approximately 2 billion illnesses resulting in over 1 million deaths caused by 22 major foodborne pathogens (Kirk et al., 2015). The European Food Safety Authority (EFSA) reported an upward trend of foodborne outbreaks (5,196) in 2013 in 28 member states and 4 non-member states (EFSA, 2015). In the United States of America, the incidence of food-related disease outbreaks and product recalls are on the rise (Murphree et al., 2012; Gould et al., 2013). The foodborne illness is blamed for approximately 48 million cases, 128,000 hospitalizations, 3,000 deaths annually (Scallan et al., 2011), and about 72 billion dollars in economic losses (Scharff, 2012). In 2014, the Foodborne Diseases Active Surveillance Network (FoodNet) from 10 U.S. geographic areas reported 19,542 infections, 4,445 hospitalizations, and 71 deaths (Crim et al., 2015). Increased incidence of foodborne outbreaks and product recalls can be attributed to increased surveillance and reporting, modernization of food processing and agricultural practices, food consumption habits such as the desire for more natural preservative-free foods, increased at-risk populations, more accurate detection methods, antimicrobial resistance, and pathogens with improved adaptation and survivability upon exposure to stressors (Alvarez-Ordóñez et al., 2015, 2017; Begley and Hill, 2015; Bhunia, 2018).
Fascinating similarities exist between the food processing/preservation techniques and the host innate defense strategies. Therefore, the modern food processing practices (Van Boekel et al., 2010; Zhou et al., 2010; Davidson et al., 2013; Alvarez-Ordóñez et al., 2017), could “prime” microbes to be more invasive in the gut due to the ability of pathogens to withstand sub-lethal processing treatments and altered gene expression. Advanced food processing and preservation techniques are designed to reduce pathogen or toxin load or eliminate them from food; however, studies have suggested that food preservation techniques could create a sub-lethal environment (Capozzi et al., 2009). It is widely accepted that the sub-lethal treatments may trigger a bacterial stress response that results in changes in gene expression, leading to not only enhanced bacterial resistance to antimicrobials or preservation conditions but also enhanced pathogen survivability and virulence (Wesche et al., 2009; Spector and Kenyon, 2012; Verraes et al., 2013; Sun, 2014; NicAogáin and O’Byrne, 2016; Dawoud et al., 2017; Esbelin et al., 2018). Similarly, to overcome pathogenic microbial assault, a series of host defenses is strategically placed throughout the orogastric and intestinal tract (Fang et al., 2016). The host defense system includes enzymes, acidic pH, bile, resident microflora, antimicrobial peptides, mucus, and innate and adaptive immune responses, which help prevent or minimize pathogen colonization, invasion, and overall pathogenesis in a host (Sleator et al., 2009; Swaggerty et al., 2009; Garrett et al., 2010; Kamada et al., 2013; Bhunia, 2018).
Another intriguing observation is that bacterial exposure to various food preservation, minimal processing, or sub-lethal treatment can change the nature and scale of antibiotic resistance in microbes (McMahon et al., 2007; Verraes et al., 2013) thus creating a situation where clinical management of these pathogens would be difficult. The current review explores the similarities that exist in the microenvironment of food preservation techniques and the host gastrointestinal tract aiding bacterial adaptation and readiness for increased infectivity (Figure 1).
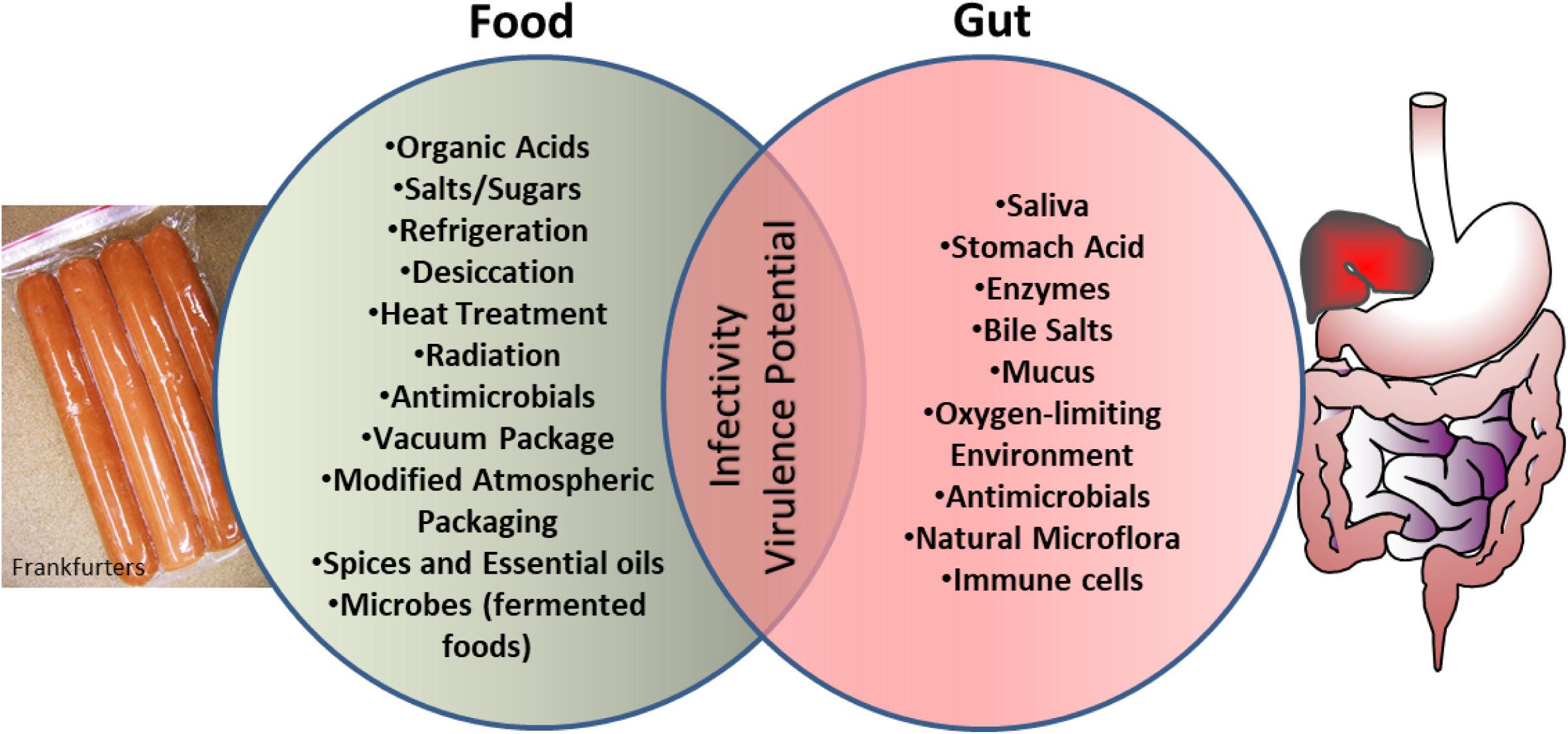
FIGURE 1. Schematics showing microbial exposure to shared stress environment between food processing conditions and human gut, which prime microbes for enhanced virulence and infectivity.
The objective of both host defense mechanisms and food preservation techniques is to eliminate or reduce microbial pathogens load; however, in many instances, the food processing treatments are sub-lethal and may trigger genes that are responsible for stress response. The stress response proteins help microbes to repair from injury and to cope with sub-lethal treatments. In addition, certain virulence genes are also upregulated under the stressful environment (Wesche et al., 2009; Dong and Schellhorn, 2010; Alvarez-Ordóñez et al., 2017). The food associated stressors are originated from a series of physical and chemical processing treatments (Table 1) that are employed to minimize or eliminate bacterial growth in food including drying (desiccation), heating, irradiation, ultrasound, sonication, electric pulse, hydrostatic pressure, acids, ethanol, salts and sugars, natural plant, or microbe-derived antimicrobial preservatives, modified atmospheric packaging, and oxidative treatments (Capozzi et al., 2009; Zhou et al., 2010; Davidson et al., 2013; Wang et al., 2016; Esbelin et al., 2018).
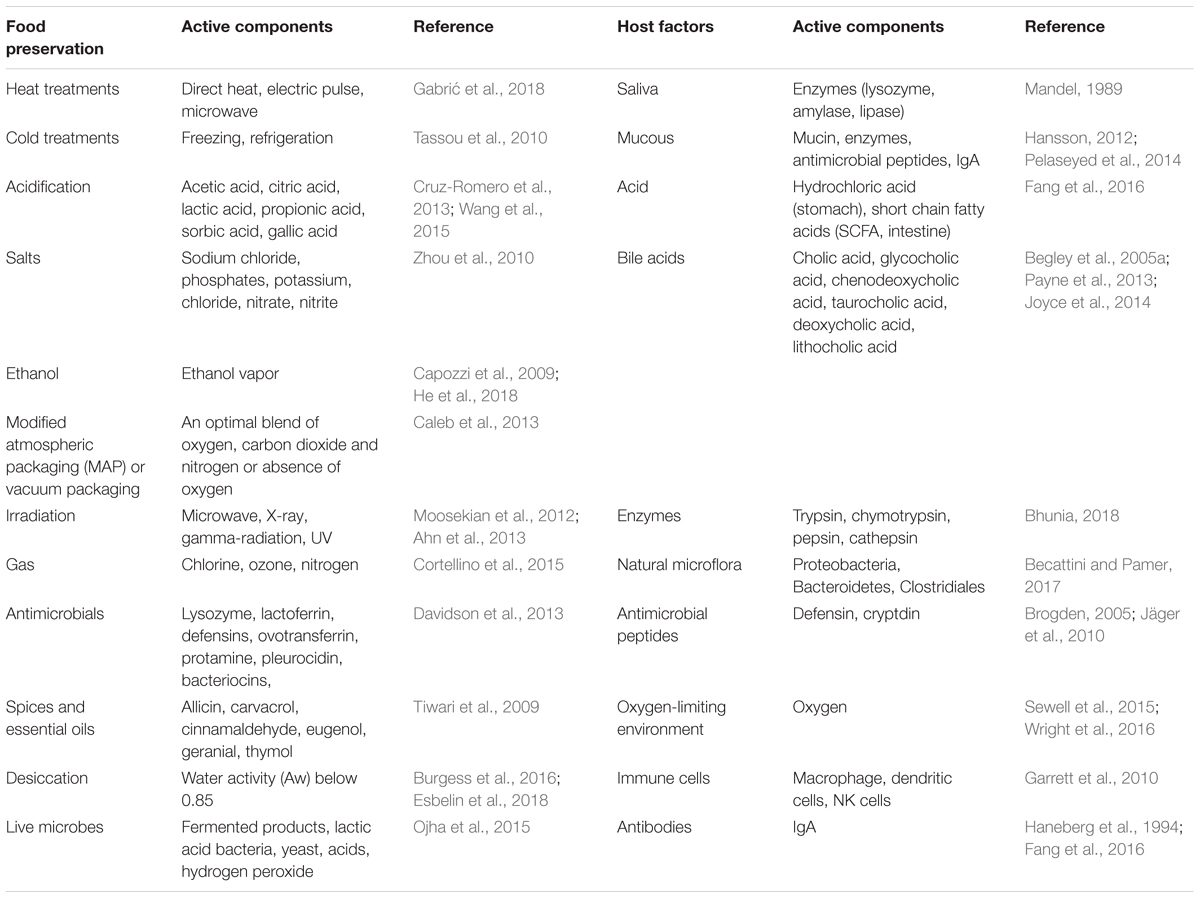
TABLE 1. Food processing/preservation conditions and host factors that may induce stress in pathogens.
The innate defense in the gastrointestinal tract is robust and generally effective in preventing foodborne pathogen interaction with the host. The gastrointestinal tract is over 23 ft long in an adult human. Multiple antimicrobial factors are present from mouth to rectum, that is, the entire length of the gastrointestinal tract (Table 1). Saliva in the mouth contains bacteriolytic enzymes (lysozyme), and gastric juice in the stomach contains hydrochloric acid and digestive enzymes, while the small and large intestine contain antimicrobial peptides (defensins, cathelicidin, cryptdin, elafin, etc.), bile, natural microflora, mucus, secretory IgA, oxygen-limiting environment, epithelial barrier, and submucosal immune cells (Brogden, 2005; Wesche et al., 2009; Garrett et al., 2010; Jäger et al., 2010; Sleator and Hill, 2010; Becattini and Pamer, 2017; Bhunia, 2018). Pathogens encounter multiple host-induced stresses in the intestine from acidic pH, nutrient limitation, low iron, oxidative and nitrosative stress, bile salts, free fatty acids, DNA damage, oxygen-limitation, and temperature in the gut (Louis and O’Byrne, 2010; Fang et al., 2016). If these factors fail to completely inactivate the microbes, they may be a source of stressors. Sub-lethal stressors, causing bacterial damage, elicit a bacterial stress response to initiate repair, or protect cells from stressors and increase the likelihood of survival.
In the food system, microbial growth can be retarded or inhibited through acid shock from fermentation, preservatives (organic acids), and acid washes (Davidson et al., 2013). In addition, microbes may be exposed to the alkaline stress originating from the processing equipment, which is often treated with detergents or sanitizers (Capozzi et al., 2009; Wesche et al., 2009).
For microbial inactivation, organic acids such as lactic acid, acetic acid, citric acid, propionic acid, sorbic acid, and benzoic acids at a pH range of 4 to 6 are used. The antimicrobial action is mediated by both dissociated and undissociated ions depending on the final pH of the food matrix, affecting transmembrane proton motive force, inactivation of enzymes, cell injury, and cell death (Theron and Lues, 2007; Cruz-Romero et al., 2013; Wang et al., 2015). Furthermore, acidic pH can also cause damage to RNA and/or DNA (and subsequently alter protein synthesis), damage to membranes, spore coats, or through sequestration of cations regulating bacterial metabolic processes (Begley and Hill, 2015).
In the host gastric environment, an acidic pH 2 or lower is generally effective as a microbial barrier. There are, however, instances where bacteria can pass through the acidic barrier of the gut due to inconsistent acid secretions in the stomach, neutralization of acid by food or beverages or bacterial coping mechanisms or acid tolerance (Louis and O’Byrne, 2010). Acid-adapted bacteria such as Salmonella (Tsai and Ingham, 1997), Escherichia coli O157:H7 (Hsin-Yi and Chou, 2001; Foster, 2004), and Listeria monocytogenes (Gahan et al., 1996) can survive in the highly acidic environment (as low as pH 2.5). E. coli, Shigella, and L. monocytogenes use a glutamate decarboxylase (GAD) system to mitigate acidic pH (Cotter et al., 2005b). Besides, in L. monocytogenes, F1F0-ATPase and arginine deaminase, and in L. innocua a new type of ATP binding universal stress response protein (USP) also help in acid adaptation (Tremonte et al., 2016). In general, foodborne acid-adapted pathogens have a greater chance of survival during the orogastric passage and thus are more invasive than the non-acid adapted microbes.
Expression of acid shock proteins aids in bacterial coping mechanisms to survive acid conditions below a pH of two (Wesche et al., 2009; Dong and Schellhorn, 2010; Fang et al., 2016). Previous studies showed, for example, that during acid-shock, Salmonella enterica serovar Typhimurium (S. Typhimurium) induced expression of 60 genes related to stress resistance (Audia et al., 2001). In the course of acid exposure, sigma E (σE) is highly activated and it increases bacterial survival in the acidified phagosomal vacuole in macrophages or dendritic cells (Muller et al., 2009). This allows Salmonella to evade the host immune system by avoiding oxidative stress in the vacuole (Crouch et al., 2005). Further studies show S. Typhimurium contains several two-component systems that are involved in virulence. Specifically, the EnvZ-OmpR is activated in response to acid stress, which enhances the type three-secretion system (TTSS) genes enhancing cellular replication (Fass and Groisman, 2009). CpxR-CpxA contributes to gut colonization and Salmonella-induced colitis (Fujimoto et al., 2018). In the presence of antimicrobial peptides, CpxR-CpxA-regulated genes are upregulated and contribute to the gut inflammation.
During exposure to gastric acid, enterohemorrhagic E. coli (EHEC) such as E. coli O157:H7 develop acid resistance through activation of an alternative sigma factor, RpoS (Barnett Foster, 2013). Research shows that acid-resistance can be acquired in the ruminant gut, leading to potential food contamination by more virulent acid- and cold-tolerant EHEC (Lin et al., 1996; Callaway et al., 2009). Acid-resistant EHEC also can tolerate the acidic environment (pH 1–3) in the human stomach. The acid resistance is governed by three genetic regulatory elements, RpoS; arginine decarboxylase (adiA) and its regulator, CysB; GAD (gadA or gadB); and γ-amino butyric acid antiporter (gadC) (Lim et al., 2010). Additionally, acid-resistant EHEC has been shown to alter gene expression patterns for adhesion and flagellar proteins, enhancing their ability to colonize the gut, although acid-resistance does not appear to induce Shiga-toxin mediated virulence (Barnett Foster, 2013). Furthermore, in EHEC, sensing of acyl-homoserine lactone appears to activate the transcription regulator SdiA which in turn upregulates locus of enterocyte effacement (LEE) pathogenicity island that encodes gene products required for attachment and effacement lesion and GAD promoting acid resistance (Hughes et al., 2010). Therefore, exposure to acids during food processing or storage enhances acid tolerance, thus ensuring bacterial safe transit through the stomach during foodborne infection. Likewise, contamination of food products with acid tolerant bacteria from meat animals helps bacterial resistance to acids used during food processing or preservation.
In food preservation, freeze-drying and storage in salt solution serve to eliminate bacteria or mitigate growth. Salts of sodium (NaCl), potassium (KCl), nitrate (NaNO3), or nitrite (NaNO2) are common food preservatives due to their exertion of osmotic stress on microbes. Osmotic stress, both a natural host defense and common in food preservation, mitigates bacterial growth and survival. Salt inhibits bacteria by disrupting the osmotic balance between the intracellular and cytoplasmic membrane (Wesche et al., 2009). Osmotic stress induces the filamentous appearance of bacterial pathogens such has been seen in Salmonella, E. coli, Listeria, and Cronobacter (Geng et al., 2003; Burgess et al., 2016). The endopeptidase that is required for cell division is downregulated during bacterial growth in the osmotic environment; hence, the cells are elongated (Burgess et al., 2016).
In the gut, bacteria are exposed to a hyperosmotic challenge, especially the bile salts, which is equivalent to 0.3 M NaCl and other ionic species (Chowdhury et al., 1996; Sleator and Hill, 2002). Therefore, osmoadaptation helps bacterial survival and increased virulence in a host. Various osmoregulatory systems become active in osmoadapted organisms, which include the production of osmoprotective compounds such as ProU in Enterobacter, ProP in E. coli, PutP in Staphylococcus aureus, and OpuC in L. monocytogenes (Sleator and Hill, 2002). Furthermore, many pathogenic bacteria commonly carry virulence genes and antibiotic resistance associated with ion transporters (Ganz and Nemeth, 2015; White et al., 2017). Harris et al. (2012) showed that E. coli O157:H7 exposed to 2% salt solution exhibited increased production of the Shiga toxin, which in part was due to activation of recA gene expression, indicating that osmotic stressors, similar to those that occur during meat processing, contribute to pathogen virulence.
Ethanol, starvation, and osmotic stress also increase microbial resistance to various antimicrobials (antibiotics) (Capozzi et al., 2009), induce biofilm formation, and persister traits (Poole, 2012). In persister cells, the gene loci, toxin–antitoxin (TA) is activated, thus antitoxin is degraded allowing the toxin to inhibit cellular processes such as DNA replication and protein translation, maintaining a non-replicative lifestyle (Helaine and Kugelberg, 2014; Page and Peti, 2016; Fisher et al., 2017). Persister phenotype helps bacteria to survive in an unfavorable condition such as nutrient limitation, extreme pH, and DNA damage by expressing high levels of intracellular guanosine tetraphosphate and guanosine pentaphosphate (p)ppGpp (Harms et al., 2016; Fisher et al., 2017). Many foodborne pathogens exhibit such trait which helps their persistence in food processing environment and in the host (Abee et al., 2016; Buchanan et al., 2017; Fisher et al., 2017; Wu et al., 2017). Osmotic stress increases microbial resistance to antibiotics and helps develop persister state, thus present a challenge for inactivation by sanitizers in the food system or by therapeutic antibiotics in humans.
Multiple antimicrobial proteins of prokaryotic or eukaryotic origin are being used or under investigation for potential use as food preservatives (Table 1; Garcia et al., 2010; Juneja et al., 2012). Bacteriocins such as nisin, pediocin, and reuterine are produced by lactic acid bacteria and are used or being considered for use in food preservation (Perez et al., 2014; Singh, 2018). Bacteriophages and endolysins are also considered for food preservation (Garcia et al., 2010; Schmelcher and Loessner, 2016; Goodridge et al., 2018). In addition, antimicrobial proteins from molds such as natamycin, tylosin, and polylysine are used in certain food products. Animal origin antimicrobials include chitosan, lysozyme, lactoferrin, lactoperoxidase, ovotransferrin, protamine, pleuricidin, and defensins have been considered for food preservation (Tiwari et al., 2009; Juneja et al., 2012; Davidson et al., 2013; Ray and Bhunia, 2014). Antimicrobial treatment may induce persister traits such as seen in L. monocytogenes after exposure to the antibiotics norfloxacin (Knudsen et al., 2013) or nisin (Wu et al., 2017). Likewise, Salmonella, E. coli, Staphylococcus, and others also exhibit persister phenotype after exposure to antimicrobials, which helps bacterial survival (Helaine and Kugelberg, 2014; Fisher et al., 2017). Pathogens in persister state in food can bloom in a host after consumption and can cause disease (Lewis, 2007).
The plant-derived phenolic compounds as secondary metabolites are originated from the metabolism of phenolic acids, flavonoids, stilbenes, lignans, and tannins in the gut. These phenolic compounds also exert an antimicrobial effect on pathogens (Selma et al., 2009) and may induce stress in pathogens.
The host-derived metabolic compounds, such as enzymes, proteins, and immunoglobulins can also exert an antimicrobial effect. Host enzymes such as lysozyme or phospholipase disrupt microbial cell membranes (Mandel, 1989). Host immune proteins, such as IgA or innate immune proteins lipocalin-2, inactivate microbes. Lipocalin-2 production is stimulated by the host inflammatory response and binds siderophores, thus limiting iron uptake and preventing microbial growth (Flo et al., 2004; Raffatellu et al., 2009). Commensal microbes can protect the host from pathogenic microbes through competitive exclusion and production of antimicrobial peptides (bacteriocins) although some pathogenic microbes can evade such a barrier (Cotter et al., 2005a; Hibbing et al., 2010; Becattini and Pamer, 2017).
Modified atmospheric packaging (MAP) is a minimal processing practice to prevent pathogen growth. In MAP, an optimal blend of oxygen, carbon dioxide, and nitrogen is present within a high barrier or permeable package (Caleb et al., 2013). During MAP and vacuum packaging of food products, oxygen level is minimal or absent, which in turn limits oxygen availability for aerobic biochemical processes and in some cases this induces a bacterial stress response in aerobic bacteria (Wesche et al., 2009; Poole, 2012). In the host gastrointestinal tract, the oxygen level gradually decreases from proximal small intestine to distal large intestine and provides a favorable growth environment for anaerobic pathogens. Since many foodborne pathogens are aerobic or facultative anaerobe thus they remain in close proximity to the mucosal epithelial cells where they gain access to oxygen from host cells during colonization of the gut. For example, Salmonella expresses type 1 fimbriae that facilitate the bacterial invasion of oxygen-containing host cells and the fimbriae expression is high during aerobic growth but not in an anaerobic environment (Ernst et al., 1990; Hakalehto et al., 2007). Furthermore, for survival under anaerobic environment, pathogens also employ different strategies. In the course of Salmonella infection, epithelial cells generate reactive oxygen species (ROS), which react with thiosulfate produced by the gut microbiota and convert it into tetrathionate, a terminal electron acceptor, which is used by the bacterium for growth (Winter et al., 2010; Behnsen et al., 2015).
Listeria monocytogenes was shown to up-regulate 28 genes under anaerobic condition (Mueller-Herbst et al., 2014), of these, lmo0355 encoding fumarate reductase expression was high (Figure 2). In addition, generation of a proton motive force via F1F0-ATPase was essential for growth under the anaerobic environment (Mueller-Herbst et al., 2014). While, in the aerobic environment, a redox-responsive transcription factor, spxA1, is necessary for the growth of L. monocytogenes (Whiteley et al., 2017). In addition, L. monocytogenes uses a cytochrome bd-type (CydAB) terminal oxidase for respiration under aerobic environment while a cytochrome aa3-type menaquinol oxidase (QoxAB) for respiration under reduced oxygen levels possibly during host infection (Corbett et al., 2017). Under the anaerobic condition in the presence of central carbon metabolism intermediates, such as acetate, citrate, fumarate, pyruvate, lactate, and succinate, L. monocytogenes expresses reduced listeriolysin O (LLO) but the higher invasion of cultured cell lines (Wallace et al., 2017). Anaerobic growth also promotes enhanced L. monocytogenes adhesion and invasion (Andersen et al., 2007; Burkholder and Bhunia, 2009). Increased cell invasion could be attributed to increased expression of internalin B (InlB) during anaerobic growth, involved in adhesion and invasion of mucosal epithelial cells and hepatic cells (Lindén et al., 2008).
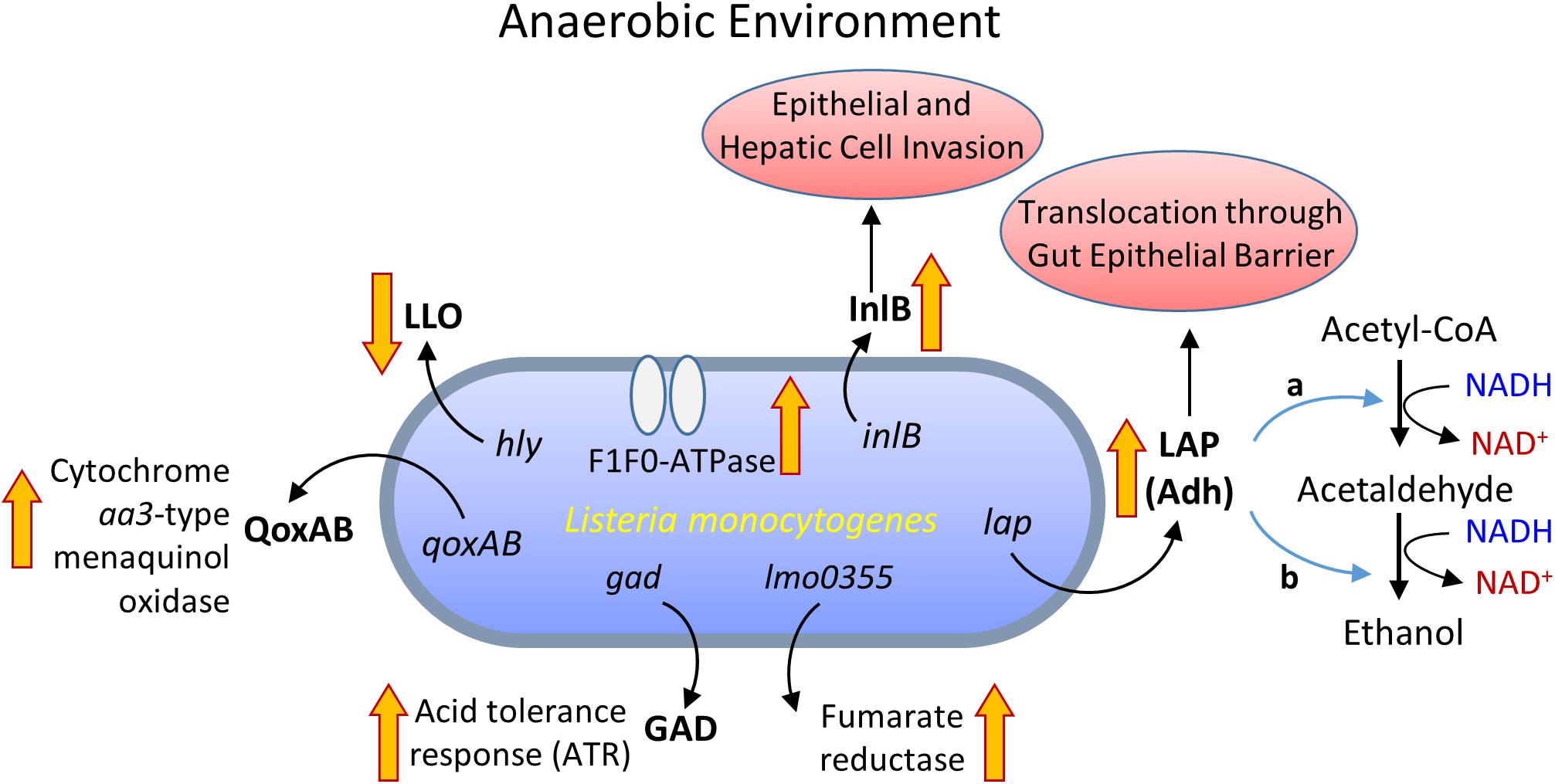
FIGURE 2. Metabolic activity and virulence gene expression in Listeria monocytogenes under anaerobic growth environment. LLO, listeriolysin O; InlB, internalin B; QoxAB, cytochrome aa3-type menaquinol oxidase; GAD, glutamate decarboxylase; LAP, Listeria adhesion protein or alcohol acetaldehyde dehydrogenase (Adh) consists of bifunctional acetaldehyde dehydrogenase (a) and alcohol dehydrogenase (b).
Listeria monocytogenes also expressed high levels of Listeria adhesion protein (LAP) also known as alcohol acetaldehyde dehydrogenase (Adh), which catalyzes the conversion of acetyl-CoA to acetaldehyde and from acetaldehyde to ethanol during growth under the anaerobic environment (Figure 2; Jagadeesan et al., 2010; Mueller-Herbst et al., 2014). The LAP is responsible for L. monocytogenes adhesion to host intestinal epithelial cells (Pandiripally et al., 1999; Jaradat et al., 2003; Wampler et al., 2004) and bacterial paracellular translocation across the gut intestinal epithelial barrier (Burkholder and Bhunia, 2010, 2013; Kim and Bhunia, 2013; Drolia et al., 2018). Under anaerobic condition, in vitro, L. monocytogenes expresses a high level of LAP and induces increased adhesion to Caco-2 cells and increased extra-intestinal dissemination in mice (Burkholder et al., 2009). This implies that oxygen-limiting vacuum-packaged food likely helps the bacterium to adapt and invade host upon entering the host gastrointestinal tract through contaminated food (Andersen et al., 2007; Burkholder et al., 2009; Figure 2). The oxygen-limiting condition also increases acid tolerance and aids L. monocytogenes transit from the stomach to the intestine (Sewell et al., 2015), possibly due to upregulation of GAD (Gahan and Hill, 2014).
In EHEC, low oxygen can stimulate expression of virulence factors such as Sfp fimbriae which enhances bacterial colonization in the gut (Müsken et al., 2008; Barnett Foster, 2013) through upregulation of TTSS and EspA (Schüller and Phillips, 2010). Likewise, S. flexneri, under anaerobic environment, showed differential upregulation of 528 genes, of which 228 genes were influenced by fumarate and nitrate reduction regulator (FNR) (Vergara-Irigaray et al., 2014). Furthermore, genes encoding TTSS, required for bacterial invasion of host cells and pathogenesis were also upregulated under anaerobiosis (Vergara-Irigaray et al., 2014). Vibrio cholerae under oxygen-limiting condition also induces cholera toxin (CT), toxin-coregulated pili (TCP), and AphB, a transcriptional activator of TcpP expression in the host (Krishnan et al., 2004; Liu et al., 2011). This bacterium may use a thiol-based switch system to sense intestinal environment for virulence protein expression (Liu et al., 2011).
Campylobacter is a microaerophilic pathogen with a strict requirement for oxygen (O2), hydrogen (H2), and carbon dioxide (CO2); however, under anaerobic condition, Campylobacter expresses several putative virulence factors (Lee et al., 2014) for increased motility and epithelial cell invasion (Mills et al., 2012).
In case of anaerobic pathogens such as Clostridium botulinum, C. perfringens, and C. difficile growth is supported by the oxygen-deficient environment of food. Upon entry into the host through contaminated food, they can find a niche for colonization in the host intestine (Rossetto et al., 2014; Lessa et al., 2015; Abt et al., 2016; Freedman et al., 2016). Innate host defense may include the release of the stress hormone, epinephrine, and norepinephrine, which severely affect virulence gene expression and iron acquisition and quorum sensing abilities of anaerobic pathogens (Boyanova, 2017). Overall, the microbes that are adapted to the oxygen-limiting environment of the food are well equipped to not only survive in the human intestine but also show enhanced colonization, invasion, and pathogenesis.
Reactive oxygen species include hydrogen peroxide (H2O2) and superoxide (), which exert oxidative stress in microbes that grow aerobically. ROS damages DNA, membranes, and proteins (Imlay, 2003). Bacteria encounter oxidative stress in both food/food processing environment, such as H2O2-based disinfectants used for sanitization of processing equipment or food contact/non-contact surfaces. ROS are also present in immune cells, especially in neutrophils and macrophages in the host. L. monocytogenes express 2-cys peroxiredoxin (prx) to cope with increasing concentration of H2O2 in vitro but not in a mouse model where a prx mutant strain did not show growth defect in mouse liver or spleen (Kim et al., 2007). However, this contrasts with another study (Dons et al., 2014) where authors observed reduced virulence of a prx mutant in mice. In addition, H2O2 induced stress also results in increased transcription of σB and kat at 37°C but not at 20°C and L. monocytogenes exhibits higher resistance to H2O2 at 20°C and petite colony phenotype (Ochiai et al., 2017).
Campylobacter jejuni also regulates oxidative stress defense for survival (Kim et al., 2015). To neutralize superoxide mediated oxidative stress or cell damage, many pathogens including E. coli (Carlioz and Touati, 1986), C. jejuni (Pesci et al., 1994; Purdy and Park, 1994), S. Typhimurium (Fang et al., 1999), S. flexneri (Franzon et al., 1990), S. aureus (Kanafani and Martin, 1985), and L. monocytogenes (Vasconcelos and Deneer, 1994) express superoxide dismutase (SOD) (Chiang and Schellhorn, 2012). Furthermore, the multidrug efflux system is over-expressed in microbes during oxidative stress; thus they exhibit resistance to multiple antimicrobials including antibiotics (Poole, 2012). In addition, oxidative stress can induce biofilm formation and promote persister trait (Zhang, 2014).
Another significant host challenge to microbes is exposure to bile in the intestine. Bile contains acids (deoxycholic acid and lithocholic acid), salts, and enzymes that, at least in part, are responsible for the cellular breakdown of microbes through membrane and DNA damage (Begley et al., 2005a; Merritt and Donaldson, 2009; Sistrunk et al., 2016). As expected, there is a degree of microbial resistance to bile components (Gunn, 2000; Louis and O’Byrne, 2010). Specifically, the lipopolysaccharide (LPS) structure in the outer membrane in Gram-negative bacteria and the presence of specific porins protect cell membrane from bile. Studies have shown that Gram-negative pathogens expressing the OmpF porin contain a greater degree of tolerance to bile components. Furthermore, expression of the AcrAB efflux pump on some pathogens such as in E. coli and S. flexneri may contribute to bile resistance (Thanassi et al., 1997; Nickerson et al., 2017). In the presence of bile, S. flexneri also forms a biofilm and induces genes responsible for multidrug resistance and virulence (Nickerson et al., 2017). An additional coping mechanism to bile, common in gut microflora, is the presence of bile salt hydrolase (BSH) (Jones et al., 2008; Joyce et al., 2014).
Studies have shown that EHEC modulate virulence factor expression in the presence of bile during passage through gut (Barnett Foster, 2013). With bile as an environmental cue, EHEC and Shigella can up-regulate AcrAB efflux pump and lipid A modification pathway, allowing for improved membrane integrity (Rosenberg et al., 2003; Kus et al., 2011; Nickerson et al., 2017). L. monocytogenes also expresses BSH and BilE (bile salt exclusion protein), regulated by sigma B (σB) to neutralize or exclude the effect of bile thus survive in the intestine (Begley et al., 2005b; Sleator et al., 2005; Sleator and Hill, 2010). In the presence of bile acid deoxycholate, C. jejuni expresses virulence genes including Campylobacter invasion antigen (ciaB) and other virulence genes for the enhanced invasion of epithelial cells (Malik-Kale et al., 2008; Novik et al., 2010). In general, enteric pathogens have developed elegant strategies to survive in the presence of bile in the intestine, and capable of causing serious illness. However, if food were contaminated with bile resistant microbes, control would be much more difficult due to their cross protection against other inhibitors.
Microbial byproducts, such as butyrate and the quorum-signaling molecule autoinducer-3 (AI-3) may alter bacterial virulence genes (Barnett Foster, 2013). Previous research suggests that the butyrate concentration may affect adhesion properties and upregulation of siderophores (Barnett Foster, 2013). Secretion of AI-3 has been shown to increase EHEC motility through flagellar biosynthesis (Clarke et al., 2006). The two-component PhoQ/PhoP system in Salmonella is activated in the presence of antimicrobial peptides, rendering protection and further activation of the PmrB/PmrA that influences membrane structure during exposure to a stressor (Gunn, 2000).
Although not fully understood, the host gut signaling hormones epinephrine and norepinephrine play a role in the induction of EHEC virulence genes responsible for chemotaxis, biofilm formation and bacterial adhesion to enterocytes (Bansal et al., 2007; Moreira and Sperandio, 2010; Barnett Foster, 2013). EHEC receptor kinase sensor QseE is, for example, sensitive to epinephrine, and its activation enhances EHEC colonization (Moreira and Sperandio, 2010; Sperandio and Nguyen, 2012). QseC histidine kinase sensor helps Salmonella to sense AI-3, epinephrine or norepinephrine for increased motility, invasion of epithelial cells and survival inside macrophages (Moreira et al., 2010). In Campylobacter, norepinephrine also increases bacterial growth rate, motility, cell invasion, and disruption of the epithelial tight junction (Cogan et al., 2007). These indicate that stress and consequent infection may dictate the severity of foodborne diseases.
Bacteria have evolved to have stressor coping mechanisms, namely, the ability to sense the environment in the gut (or preserved food) and elicit changes in gene expression to cope with specific stressors (NicAogáin and O’Byrne, 2016). Two mechanisms are used by microbes to alter gene and protein expression during exposure to stressors, the signal transduction system and alternative sigma factors such as σB, σS, σE, σF, σN, RpoE, RpoH, and RpoS (Kazmierczak et al., 2005). The signal transduction system is a coping mechanism in which a membrane-associated sensor is phosphorylated due to an external signal. A response regulator is subsequently activated, which plays a major role in the secretion of cationic peptides by the innate immune system (Louis and O’Byrne, 2010). Generally, the response of stress stimuli in damaged bacteria leads to expression of factors to guide RNA polymerase for inducing repair or to express proteins required for survival (Wesche et al., 2009). Alternative sigma factors play an important role in promoter recognition and production of cellular proteins to support virulence mechanisms (Kazmierczak et al., 2005). σB plays a major role in exposure to acids, salts, and bile (Kazmierczak et al., 2005; Louis and O’Byrne, 2010). In response to a stress, such as low pH, the stressosome is activated and the phosphatase RsbU, which subsequently allows RsbV anti-anti-sigma factor to bind to anti-sigma factor RsbW rendering the σB free to guide the RNA polymerase.
Another alternative sigma factor, RpoS, plays a role in Salmonella and E. coli acid tolerance and virulence (Hengge-Aronis and Storz, 2000; Foster, 2004; Dong and Schellhorn, 2010). Stress-induced activation of RpoS activates the esp genes in pathogenic E. coli essential for attachment and effacement lesion during pathogenesis (Laaberki et al., 2006). Furthermore, activation of RpoS helps V. cholera to evade host immune system allowing the pathogen to be localized in the intestinal lumen and consequent shedding into the environment (Conner et al., 2016). The molecular basis of the RpoS regulator includes stress-cue activation of anti-adaptor proteins, which release the protein RssB that forms a RssB-RpoS complex. This complex is further degraded by the ClpXP protease rendering a free RpoS that guides the RNA polymerase (Jaishankar and Srivastava, 2017).
σE is another alternative sigma factor activated by periplasmic stress leading to the activation of genes involved in degradation and refolding of damaged proteins (Rhodius and Mutalik, 2010). Heat, cold, and ethanol stressors have been shown to activate σE (Rowley et al., 2006). Activation of σE in Gram-negative bacteria is initiated by stress perception. Specifically, unfolded proteins (generated from oxidative stress for example) interact with membrane-associated proteases (DegS and RseP) and degrade RseA releasing σE. The free RseA- σE complex is then tagged by SspB and subsequently degraded by protease ClpXP allowing for σE to interact with the RNA polymerase, guiding expression of proteins to enhance protein synthesis (Louis and O’Byrne, 2010; Jaishankar and Srivastava, 2017).
Another significant factor to consider with environmental induction of pathogen virulence is cross-protection, that is, resistance to multiple environmental stressors derived from bacterial exposure to a specific environmental stressor (Capozzi et al., 2009; Alvarez-Ordóñez et al., 2015, 2017). For example, several studies have shown that starvation stress induces heat tolerance and resistance to oxidative stress in pathogenic E. coli and Serratia marcescens (Jenkins et al., 1988; Leenanon and Drake, 2001; Pittman et al., 2015).
Adaptation of S. Typhimurium to acid stress is linked to cross-protection against salt and oxidative stress (Leyer and Johnson, 1993). Likewise, acid-adapted Salmonella, E. coli O157:H7 and L. monocytogenes showed increased resistance to heat (Mazzotta, 2001; Haberbeck et al., 2017). Conversely, bacterial growth at higher temperatures can also evoke cross-protection against other lethal treatments. For example, S. Enteritidis, grown at 37°C showed increased cell membrane fluidity, acid resistance, and RpoS expression; while at 42°C, bacteria showed increased heat resistance and RpoH expression, and decreased RpoS expression (Yang et al., 2014). At 42°C, S. Enteritidis also showed induction of virulence-related genes, spvR, hilA, and avrA (Yang et al., 2014). In response to heat shock, the presence of damaged or denatured proteins is thought to be stimuli for activation of alternative sigma factors and subsequent activation of heat shock proteins that are involved in protein repair (Wesche et al., 2009). Kobayashi et al. (2005) showed that heat treatment (55°C for 15 min) resulted in up-regulation of 19 heat-inducible and 12 oxidative-stress and DNA damage-inducible genes in S. Enteritidis. In a separate study, Sirsat et al. (2011) showed that sublethal heat treatment (42°C for 10 or 30 min) in S. Typhimurium induced genes encoded in SPI-2 and SPI-5 especially transcription of genes encoding fimbriae and Rpo regulons. Sub-lethal heat stressed S. Typhimurium also showed increased adhesion and invasion of intestinal enterocyte-like Caco-2 cells and virulence (Burkholder and Bhunia, 2009; Sirsat et al., 2011; Dawoud et al., 2017).
Escherichia coli O157:H7 exposed to progressively intensifying milder heat (54–60°C) treatment displayed higher resistance to high hydrostatic pressure possibly by activating RpoS and RpoH (Gayán et al., 2016). Interestingly, in C. botulinum, exposure to prolonged heat stress (45°C), heat shock genes and members of the SOS regulons were activated while genes encoding neurotoxin (botA) synthesis was downregulated (Selby et al., 2017). In L. monocytogenes, growth at 37°C induced its adaptation to acid (Shen et al., 2014), salts (6% NaCl), and hydrogen peroxide (Bergholz et al., 2012).
In the presence of low temperatures, cold stress proteins have been shown to play important roles in microbial viability (example, L. monocytogenes, E. coli, and Bacillus cereus) (Wesche et al., 2009; Saldivar et al., 2018). Subsequent to cellular sensing of abnormally cold temperatures, alternative sigma factors are induced, leading to expression of cold-shock proteins that aid in membrane fluidity, protein folding, and nutrient uptake (Wouters et al., 2000). Interestingly, pre-exposure to cold stress also increases bacterial cross protection against osmotic stress. L. monocytogenes exposed to refrigerated temperature showed increased survival in 3% NaCl, and showed higher expression of proteins required for maintenance of cell wall and cellular processes such as osmolyte transporters, amino acid metabolism, and lipid biosynthesis (Pittman et al., 2014).
Pre-exposure to organic acid salts (potassium lactate), L. monocytogenes exhibited resistance against food antimicrobials such as nisin, lauric arginate, and 𝜖-polylysine. This cross-protection involves activation of two-component response-regulator, VirRS (Kang et al., 2015). Similarly, in the presence of NaCl (6%), L. monocytogenes exhibited resistance against nisin (Bergholz et al., 2013). In a food product such as in soft cheese environment (containing acid and salts), L. monocytogenes displayed cross-protection against antimicrobials thus may help the bacterium to overcome antimicrobials encountered in the host intestine (Melo et al., 2015).
Growth under oxygen-limiting environment also increases bacterial acid tolerance (Sewell et al., 2015) and resistant to bile salts (Payne et al., 2013; Wright et al., 2016), and survival in the gastrointestinal tract such was seen in L. monocytogenes. Conversely, sub-lethal exposure to antibiotics renders L. monocytogenes to shift to anaerobic metabolism and the bacterium made reduced levels of virulence proteins including LLO, InlB, and LAP; however, exhibited increased bacterial tolerance to multiple other antibiotics (Knudsen et al., 2016; Zhu et al., 2018).
Adaptation to ethanol caused S. Enteritidis to be more resistant to acid and showed upregulation of acid tolerant regulator, RpoS (He et al., 2018). Similarly, prolonged exposure (24 h) of acidic stress protected S. aureus against non-thermal plasma treatment exhibiting reduced cell membrane damage, membrane potential, and intracellular enzyme activity (Liao et al., 2018). Altogether, cross-protection must be considered a significant bacterial coping mechanism that may have broad implications in regards to pathogen survival in the gut, resistance to antimicrobials and increased virulence.
Studies have shown that food environment significantly affects microbial pathogenesis (Ponder, 2017) and antimicrobial resistance depending on the specific stressors (discussed above) microbes encounter in a food (Table 2). L. monocytogenes cultured in ready-to-eat (RTE) meat matrices appeared to have increased invasiveness in a cell culture J774A.1 (Larsen et al., 2010; Lin et al., 2010) or in a mouse (Mahoney and Henriksson, 2003) model. In fresh cut melon inoculated with L. monocytogenes, and stored at 10°C for 2 days, the bacterium was highly invasive to Caco-2 cells (Colás-Medà et al., 2017). Similarly, L. monocytogenes showed increased invasiveness to Caco-2 cells when stored in pasteurized milk than raw milk at 4°C (Pricope-Ciolacu et al., 2013). L. monocytogenes obtained from a fermented sausage and cured cooked ham stored under 10°C for up to 4 weeks were also highly invasive to Caco-2 cells (Larsen et al., 2010). Furthermore, L. monocytogenes grown in meat juice differentially expressed high levels of virulence genes such as gad, σB, sod, and inlA (Rantsiou et al., 2012). Likewise, in liver pâté, prfA, σB, and inlA were upregulated (Olesen et al., 2010) and in salami, acidic (σB and lmo0669) and osmotic (gbuA and lmo1421) stress-related genes were upregulated (Mataragas et al., 2015).
Bacillus species grown in reconstituted infant formula containing glucose exhibited increased expression of enterotoxin production and cytotoxicity when tested on Caco-2 and HEp-2 cell lines (Rowan et al., 2001). S. aureus also showed increased production of enterotoxin in various food products including cheese, ham, and sausage containing high levels of salts (Schelin et al., 2011). In cheese under the acidified environment (containing Lactococcus lactis), genes for enterotoxin sea, sel, and seh, and the stress response genes, dnaK, sodA, and others were upregulated while the stress regulator, σB, and agr were downregulated (Cretenet et al., 2011). S. Typhimurium grown sequentially in soil, lettuce and cut lettuce and stored under modified atmosphere (MAP) conditions showed increased survival in simulated gastric and intestinal juice but exhibited reduced adhesion and invasion of Caco-2 cells in vitro suggesting the food matrix or environmental factors may have differential effect on Salmonella virulence when analyzed in vitro (Oliveira et al., 2011).
Natural inhibitors exist in the host gastrointestinal tract to eliminate pathogenic bacteria including acidic pH, low oxygen levels, changes in osmolarity, antimicrobials, and ROS and oxidative stress. Similarly, food preservation and sanitation techniques are employed to eliminate foodborne pathogens. Bacterial stress or injury happens when chemical or physical treatment results in a sub-lethal damage to the microorganisms. The general bacterial response to sub-lethal stress triggers gene expression through activation of signal transduction systems and alternative sigma factors. Through environmental cues, alternative sigma factors are activated that guides RNA polymerase, gene reprogramming, and production of proteins to support bacterial coping with a stress event. Therefore, the stressed bacteria have improved survival strategy such as biofilm formation, resistance to antimicrobials, and persister state, which under favorable condition can bloom and show increased virulence. Furthermore, exposure to a primary stressor may lead to the onset of cross-resistance (or adaptation) to multiple secondary stressors, antibiotic resistance, and increased virulence. Due to the similarity of stressors between the food preservation treatments and the gut innate defense, and their impact on pathogen physiology and behavior, it is a grim reality that the food processing treatments could prime the microbial pathogens for enhanced survival and infectivity in a host (Lin et al., 1996; Lim et al., 2010; Alvarez-Ordóñez et al., 2015, 2017; Buchanan et al., 2017; Fisher et al., 2017). Therefore, the modern food processing and production practices employed today, ironically are a curse in disguise, and possibly a major contributing factor for the emergence of increased incidence of foodborne illnesses, outbreaks, and fatality around the globe.
The current trend is to use minimal processing or hurdle approach to reduce pathogen load in foods; therefore, we need to have a greater understanding of the sub-lethal effect of such processing treatments on microorganisms, especially their physiology, behavior or pathogenesis. Furthermore, new and emerging technologies such as ultra violet (UV) rays (Kim et al., 2017), X-ray (Mahmoud et al., 2016), cold plasma (Niemira, 2012; Lu et al., 2014), and bacteriophages (Schmelcher and Loessner, 2016; Goodridge et al., 2018; Shahin and Bouzari, 2018) are attractive and are being used in industrial settings. These processing methods directly affect genetic elements of microbes; however, it is unknown, the impact of sub-lethal treatment or prolonged exposure to microbial physiology, virulence, and infectivity. UV, X-rays, and bacteriophages are known to cause gene deletion or insertion (mutation) in microbes, and survival and spread of such microbes could pose a grave danger to our food safety and public health management practices.
Furthermore, there seems to be a scarcity of information linking pathogen virulence in humans to sub-lethal stress exposure in the meat or companion animals. Several interesting aspects that need future investigations such as pathogenesis and virulence gene expression in pathogens in a meat animal model (for example swine) where the animal has experienced multiple stressors (for example changes in nutrition, transportation, or management procedures). A better understanding of the relationship between food production and preservation techniques and induction of pathogen virulence may lead to techniques and procedures to reduce foodborne illnesses.
NH and AB designed the study, reviewed literature, and relevant articles and wrote the manuscript. AB made the figures and tables. All authors read and approved the manuscript.
The research in author’s laboratory was supported by Agricultural Research Service of the United States Department of Agriculture (USDA-ARS) project number 8072-42000-072-02G, and the USDA National Institute of Food and Agriculture (NIFA), the National Academy of Science (US-AID) Award No. AID-263-A-15-00002, and the Center for Food Safety Engineering at Purdue University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Chad W. Coakley is acknowledged for critical reading of the manuscript. Our sincerest apology for the unintentional omission of any relevant research articles of colleagues working in this field.
Abee, T., Koomen, J., Metselaar, K., Zwietering, M., and Den Besten, H. (2016). Impact of pathogen population heterogeneity and stress-resistant variants on food safety. Annu. Rev. Food Sci. Technol. 7, 439–456. doi: 10.1146/annurev-food-041715-033128
Abt, M. C., Mckenney, P. T., and Pamer, E. G. (2016). Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol. 14, 609–620. doi: 10.1038/nrmicro.2016.108
Ahn, D. U., Kim, I. S., and Lee, E. J. (2013). Irradiation and additive combinations on the pathogen reduction and quality of poultry meat. Poult. Sci. 92, 534–545. doi: 10.3382/ps.2012-02722
Alvarez-Ordóñez, A., Broussolle, V., Colin, P., Nguyen-The, C., and Prieto, M. (2015). The adaptive response of bacterial food-borne pathogens in the environment, host and food: implications for food safety. Int. J. Food Microbiol. 213, 99–109. doi: 10.1016/j.ijfoodmicro.2015.06.004
Alvarez-Ordóñez, A., López, M., and Prieto, M. (2017). “Role of stress response on microbial ecology of foods and its impact on the fate of food’borne microorganisms,” in Quantitative Microbiology in Food Processing: Modeling the Microbial Ecology ed. A. de Souza Sant’Ana (Hoboken, NJ: Wiley), 631–648.
Andersen, J. B., Roldgaard, B. B., Christensen, B. B., and Licht, T. R. (2007). Oxygen restriction increases the infective potential of Listeria monocytogenes in vitro in Caco-2 cells and in vivo in guinea pigs. BMC Microbiol. 7:55. doi: 10.1186/1471-2180-7-55
EFSA (2015). The european union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 13:4329.
Audia, J. P., Webb, C. C., and Foster, J. W. (2001). Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291, 97–106. doi: 10.1078/1438-4221-00106
Bansal, T., Englert, D., Lee, J., Hegde, M., Wood, T. K., and Jayaraman, A. (2007). Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157: H7 chemotaxis, colonization, and gene expression. Infect. Immun. 75, 4597–4607. doi: 10.1128/IAI.00630-07
Barnett Foster, D. (2013). Modulation of the enterohemorrhagic E. coli virulence program through the human gastrointestinal tract. Virulence 4, 315–323. doi: 10.4161/viru.24318
Becattini, S., and Pamer, E. G. (2017). Multifaceted defense against Listeria monocytogenes in the gastro-intestinal lumen. Pathogens 7:E1. doi: 10.3390/pathogens7010001
Begley, M., Gahan, C. G., and Hill, C. (2005a). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Begley, M., and Hill, C. (2015). Stress adaptation in foodborne pathogens. Annu. Rev. Food Sci. Technol. 6, 191–210. doi: 10.1146/annurev-food-030713-092350
Begley, M., Sleator, R. D., Gahan, C. G., and Hill, C. (2005b). Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73, 894–904. doi: 10.1128/IAI.73.2.894-904.2005
Behnsen, J., Perez-Lopez, A., Nuccio, S.-P., and Raffatellu, M. (2015). Exploiting host immunity: the Salmonella paradigm. Trends Immunol. 36, 112–120. doi: 10.1016/j.it.2014.12.003
Bergholz, T. M., Bowen, B., Wiedmann, M., and Boor, K. J. (2012). Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl. Environ. Microbiol. 78, 2602–2612. doi: 10.1128/AEM.07658-11
Bergholz, T. M., Tang, S., Wiedmann, M., and Boor, K. J. (2013). Nisin resistance of Listeria monocytogenes is increased by exposure to salt stress and is mediated via LiaR. Appl. Environ. Microbiol. 79, 5682–5688. doi: 10.1128/AEM.01797-13
Bhunia, A. (2018). Foodborne Microbial Pathogens: Mechanisms and Pathogenesis. Berlin: Springer. doi: 10.1007/978-1-4939-7349-1
Boyanova, L. (2017). Stress hormone epinephrine (adrenaline) and norepinephrine (noradrenaline) effects on the anaerobic bacteria. Anaerobe 44, 13–19. doi: 10.1016/j.anaerobe.2017.01.003
Brogden, K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250. doi: 10.1038/nrmicro1098
Buchanan, R. L., Gorris, L. G., Hayman, M. M., Jackson, T. C., and Whiting, R. C. (2017). A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 75, 1–13. doi: 10.1016/j.foodcont.2016.12.016
Burgess, C. M., Gianotti, A., Gruzdev, N., Holah, J., Knøchel, S., Lehner, A., et al. (2016). The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int. J. Food Microbiol. 221, 37–53. doi: 10.1016/j.ijfoodmicro.2015.12.014
Burkholder, K., and Bhunia, A. (2009). Salmonella enterica serovar Typhimurium adhesion and cytotoxicity during epithelial cell stress is reduced by Lactobacillus rhamnosus GG. Gut Pathog. 1:14. doi: 10.1186/1757-4749-1-14
Burkholder, K. M., and Bhunia, A. K. (2010). Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation, and induces expression of LAP receptor Hsp60. Infect. Immun. 78, 5062–5073. doi: 10.1128/IAI.00516-10
Burkholder, K. M., Kim, K.-P., Mishra, K., Medina, S., Hahm, B.-K., Kim, H., et al. (2009). Expression of LAP, a SecA2-dependent secretory protein, is induced under anaerobic environment. Microbes Infect. 11, 859–867. doi: 10.1016/j.micinf.2009.05.006
Burkholder, K. M., and Bhunia, A. K. (2013). “Listeria monocytogenes and Host Hsp60 – An invasive pairing,” in Moonlighting Cell Stress Proteins in Microbial Infections, Heat Shok Proteins, ed. B. Henderson (Dordrecht: Springer),267–282. doi: 10.1007/978-94-007-6787-4_17
Caleb, O. J., Mahajan, P. V., Al-Said, F. A.-J., and Opara, U. L. (2013). Modified atmosphere packaging technology of fresh and fresh-cut produce and the microbial consequences—a review. Food Bioproc. Technol. 6, 303–329. doi: 10.1007/s11947-012-0932-4
Callaway, T. R., Carr, M., Edrington, T., Anderson, R. C., and Nisbet, D. J. (2009). Diet, Escherichia coli O157: H7, and cattle: a review after 10 years. Curr. Issues Mol. Biol. 11, 67–79.
Capozzi, V., Fiocco, D., Amodio, M. L., Gallone, A., and Spano, G. (2009). Bacterial stressors in minimally processed food. Int. J. Mol. Sci. 10, 3076–3105. doi: 10.3390/ijms10073076
Carlioz, A., and Touati, D. (1986). Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5, 623–630.
Chiang, S. M., and Schellhorn, H. E. (2012). Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525, 161–169. doi: 10.1016/j.abb.2012.02.007
Chowdhury, R., Sahu, G. K., and Das, J. (1996). Stress response in pathogenic bacteria. J. Biosci. 21, 149–160. doi: 10.1007/BF02703105
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Nat. Acad. Sci. U.S.A. 103, 10420–10425. doi: 10.1073/pnas.0604343103
Cogan, T. A., Thomas, A. O., Rees, L. E., Taylor, A. H., Jepson, M. A., Williams, P. H., et al. (2007). Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut 56, 1060–1065. doi: 10.1136/gut.2006.114926
Colás-Medà, P., Viñas, I., Oliveira, M., Anguera, M., Serrano, J. C., and Abadias, M. (2017). Exposure to minimally processed pear and melon during shelf life could modify the pathogenic potential of Listeria monocytogenes. Food Microbiol. 62, 275–281. doi: 10.1016/j.fm.2016.10.016
Conner, J. G., Teschler, J. K., Jones, C. J., and Yildiz, F. H. (2016). Staying alive: Vibrio cholerae’s cycle of environmental survival, transmission, and dissemination. Microbiol. Spectr. 4:10.1128/microbiolspec.VMBF-0015-2015 doi: 10.1128/microbiolspec.VMBF-0015-2015
Corbett, D., Goldrick, M., Fernandes, V. E., Davidge, K., Poole, R. K., Andrew, P. W., et al. (2017). Listeria monocytogenes has both a bd-type and an aa3-type terminal oxidase which allow growth in different oxygen levels and both are important in infection. Infect. Immun. doi: 10.1128/IAI.00354-17 [Epub ahead of print].
Cortellino, G., Gobbi, S., Bianchi, G., and Rizzolo, A. (2015). Modified atmosphere packaging for shelf life extension of fresh-cut apples. Trends Food Sci. Technol. 46, 320–330. doi: 10.1016/j.tifs.2015.06.002
Cotter, P. D., Hill, C., and Ross, R. P. (2005a). Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788. doi: 10.1038/nrmicro1273
Cotter, P. D., Ryan, S., Gahan, C. G. M., and Hill, C. (2005b). Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71, 2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005
Cretenet, M., Nouaille, S., Thouin, J., Rault, L., Stenz, L., François, P., et al. (2011). Staphylococcus aureus virulence and metabolism are dramatically affected by Lactococcus lactis in cheese matrix. Environ. Microbiol. Rep. 3, 340–351. doi: 10.1111/j.1758-2229.2010.00230.x
Crim, S. M., Griffin, P. M., Tauxe, R., Marder, E. P., Gilliss, D., Cronquist, A. B., et al. (2015). Preliminary incidence and trends of infection with pathogens transmitted commonly through food-foodborne diseases active surveillance network, 10 US sites, 2006-2014. MMWR Morb. Mortal. Wkly. Rep. 64, 495–499.
Crouch, M. L., Becker, L. A., Bang, I. S., Tanabe, H., Ouellette, A. J., and Fang, F. C. (2005). The alternative sigma factor σE is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol. Microbiol. 56, 789–799. doi: 10.1111/j.1365-2958.2005.04578.x
Cruz-Romero, M., Murphy, T., Morris, M., Cummins, E., and Kerry, J. (2013). Antimicrobial activity of chitosan, organic acids and nano-sized solubilisates for potential use in smart antimicrobially-active packaging for potential food applications. Food Control 34, 393–397. doi: 10.1016/j.foodcont.2013.04.042
Davidson, P. M., Critzer, F. J., and Taylor, T. M. (2013). Naturally occurring antimicrobials for minimally processed foods. Annu. Rev. Food Sci. Technol. 4, 163–190. doi: 10.1146/annurev-food-030212-182535
Dawoud, T. M., Davis, M. L., Park, S. H., Kim, S., Kwon, Y. M., Jarvis, N., et al. (2017). The potential link between thermal resistance and virulence in Salmonella: a review. Front. Vet. Sci. 4:93. doi: 10.3389/fvets.2017.00093
Dong, T., and Schellhorn, H. E. (2010). Role of RpoS in virulence of pathogens. Infect. Immun. 78, 887–897. doi: 10.1128/IAI.00882-09
Dons, L. E., Mosa, A., Rottenberg, M. E., Rosenkrantz, J. T., Kristensson, K., and Olsen, J. E. (2014). Role of the Listeria monocytogenes 2-Cys peroxiredoxin homologue in protection against oxidative and nitrosative stress and in virulence. Pathog. Dis. 70, 70–74. doi: 10.1111/2049-632X.12081
Drolia, R., Tenguria, S., Durkes, A. C., Turner, J. R., and Bhunia, A. K. (2018). Listeria adhesion protein induces intestinal epithelial barrier dysfunction for bacterial translocation. Cell Host Microbe 23, 470–484. doi: 10.1016/j.chom.2018.03.004
Ernst, R., Dombroski, D., and Merrick, J. (1990). Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella Typhimurium. Infect. Immun. 58, 2014–2016.
Esbelin, J., Santos, T., and Hébraud, M. (2018). Desiccation: an environmental and food industry stress that bacteria commonly face. Food Microiol. 69, 82–88. doi: 10.1016/j.fm.2017.07.017
Fang, F. C., Degroote, M. A., Foster, J. W., Bäumler, A. J., Ochsner, U., Testerman, T., et al. (1999). Virulent Salmonella Typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc. Natl. Acad. Sci. U.S.A. 96, 7502–7507. doi: 10.1073/pnas.96.13.7502
Fang, F. C., Frawley, E. R., Tapscott, T., and Vázquez-Torres, A. (2016). Bacterial stress responses during host infection. Cell Host Microbe 20, 133–143. doi: 10.1016/j.chom.2016.07.009
Fass, E., and Groisman, E. A. (2009). Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 12, 199–204. doi: 10.1016/j.mib.2009.01.004
Fisher, R. A., Gollan, B., and Helaine, S. (2017). Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464. doi: 10.1038/nrmicro.2017.42
Flo, T. H., Smith, K. D., Sato, S., Rodriguez, D. J., Holmes, M. A., Strong, R. K., et al. (2004). Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921. doi: 10.1038/nature03104
Foster, J. W. (2004). Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2, 898–907. doi: 10.1038/nrmicro1021
Franzon, V., Arondel, J., and Sansonetti, P. (1990). Contribution of superoxide dismutase and catalase activities to Shigella flexneri pathogenesis. Infect. Immun. 58, 529–535.
Freedman, J., Shrestha, A., and Mcclane, B. (2016). Clostridium perfringens enterotoxin: action, genetics, and translational applications. Toxins 8:E73. doi: 10.3390/toxins8030073
Fujimoto, M., Goto, R., Haneda, T., Okada, N., and Miki, T. (2018). Salmonella Typhimurium CpxRA two-component system contributes to gut colonization in Salmonella-induced colitis. Infect. Immun. doi: 10.1128/IAI.00280-18 [Epub ahead of print].
Gabrić, D., Barba, F., Roohinejad, S., Gharibzahedi, S. M. T., Radojčin, M., Putnik, P., et al. (2018). Pulsed electric fields as an alternative to thermal processing for preservation of nutritive and physicochemical properties of beverages: a review. J. Food Proc. Eng. 41:e12638. doi: 10.1111/jfpe.12638
Gahan, C., and Hill, C. (2014). Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front. Cell. Infect. Microbiol. 4:9. doi: 10.3389/fcimb.2014.00009
Gahan, C., O’driscoll, B., and Hill, C. (1996). Acid adaptation of Listeria monocytogenes can enhance survival in acidic foods and during milk fermentation. Appl. Environ. Microbiol. 62, 3128–3132.
Ganz, T., and Nemeth, E. (2015). Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 15, 500–510. doi: 10.1038/nri3863
Garcia, P., Rodriguez, L., Rodriguez, A., and Martinez, B. (2010). Food biopreservation: promising strategies using bacteriocins, bacteriophages and endolysins. Trends Food Sci. Technol. 21, 373–382. doi: 10.1016/j.tifs.2010.04.010
Garrett, W. S., Gordon, J. I., and Glimcher, L. H. (2010). Homeostasis and inflammation in the intestine. Cell 140, 859–870. doi: 10.1016/j.cell.2010.01.023
Gayán, E., Cambré, A., Michiels, C. W., and Aertsen, A. (2016). Stress-induced evolution of heat resistance and resuscitation speed in Escherichia coli O157:H7 ATCC 43888. Appl. Environ. Microbiol. 82, 6656–6663. doi: 10.1128/AEM.02027-16
Geng, T., Kim, K. P., Gomez, R., Sherman, D. M., Bashir, R., Ladisch, M. R., et al. (2003). Expression of cellular antigens of Listeria monocytogenes that react with monoclonal antibodies C11E9 and EM-7G1 under acid-, salt- or temperature-induced stress environments. J. Appl. Microbiol. 95, 762–772. doi: 10.1046/j.1365-2672.2003.02035.x
Goodridge, L., Fong, K., Wang, S., and Delaquis, P. (2018). Bacteriophage-based weapons for the war against foodborne pathogens. Curr. Opin. Food Sci. 20, 69–75. doi: 10.1016/j.cofs.2018.03.017
Gould, L. H., Mungai, E. A., Johnson, S. D., Richardson, L. C., Williams, I. T., Griffin, P. M., et al. (2013). Surveillance for foodborne disease outbreaks - United States, 2009-2010. Morb. Mortal. Wkly. Rep. 62, 41–47.
Gunn, J. S. (2000). Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2, 907–913. doi: 10.1016/S1286-4579(00)00392-0
Haberbeck, L. U., Wang, X., Michiels, C., Devlieghere, F., Uyttendaele, M., and Geeraerd, A. H. (2017). Cross-protection between controlled acid-adaptation and thermal inactivation for 48 Escherichia coli strains. Int. J. Food Microbiol. 241, 206–214. doi: 10.1016/j.ijfoodmicro.2016.10.006
Hakalehto, E., Pesola, J., Heitto, L., Närvänen, A., and Heitto, A. (2007). Aerobic and anaerobic growth modes and expression of type 1 fimbriae in Salmonella. Pathophysiology 14, 61–69. doi: 10.1016/j.pathophys.2007.01.003
Haneberg, B., Kendall, D., Amerongen, H. M., Apter, F. M., Kraehenbuhl, J. P., and Neutra, M. R. (1994). Induction of specific immunoglobulin A in the small intestine, colon-rectum, and vagina measured by a new method for collection of secretions from local mucosal surfaces. Infect. Immun. 62, 15–23.
Hansson, G. C. (2012). Role of mucus layers in gut infection and inflammation. Curr. Opin. Microbiol. 15, 57–62. doi: 10.1016/j.mib.2011.11.002
Harms, A., Maisonneuve, E., and Gerdes, K. (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268
Harris, S. M., Yue, W.-F., Olsen, S. A., Hu, J., Means, W. J., Mccormick, R. J., et al. (2012). Salt at concentrations relevant to meat processing enhances Shiga toxin 2 production in Escherichia coli O157: H7. Int. J. Food Microbiol. 159, 186–192. doi: 10.1016/j.ijfoodmicro.2012.09.007
He, S., Cui, Y., Qin, X., Zhang, F., Shi, C., Paoli, G. C., et al. (2018). Influence of ethanol adaptation on Salmonella enterica serovar Enteritidis survival in acidic environments and expression of acid tolerance-related genes. Food Microbiol. 72, 193–198. doi: 10.1016/j.fm.2017.12.005
Helaine, S., and Kugelberg, E. (2014). Bacterial persisters: formation, eradication, and experimental systems. Trends Microbiol. 22, 417–424. doi: 10.1016/j.tim.2014.03.008
Hibbing, M. E., Fuqua, C., Parsek, M. R., and Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25. doi: 10.1038/nrmicro2259
Hsin-Yi, C., and Chou, C.-C. (2001). Acid adaptation and temperature effect on the survival of E. coli O157: H7 in acidic fruit juice and lactic fermented milk product. Int. J. Food Microbiol. 70, 189–195. doi: 10.1016/S0168-1605(01)00538-4
Hughes, D. T., Terekhova, D. A., Liou, L., Hovde, C. J., Sahl, J. W., Patankar, A. V., et al. (2010). Chemical sensing in mammalian host–bacterial commensal associations. Proc. Natl. Acad. Sci. U.S.A. 107, 9831–9836. doi: 10.1073/pnas.1002551107
Imlay, J. A. (2003). Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418. doi: 10.1146/annurev.micro.57.030502.090938
Jagadeesan, B., Koo, O. K., Kim, K. P., Burkholder, K. M., Mishra, K. K., Aroonnual, A., et al. (2010). LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology 156, 2782–2795. doi: 10.1099/mic.0.036509-0
Jäger, S., Stange, E. F., and Wehkamp, J. (2010). Antimicrobial peptides in gastrointestinal inflammation. Int. J. Inflam. 2010:910283. doi: 10.4061/2010/910283
Jaishankar, J., and Srivastava, P. (2017). Molecular basis of stationary phase survival and applications. Front. Microbiol. 8:2000. doi: 10.3389/fmicb.2017.02000
Jaradat, Z. W., Wampler, J. W., and Bhunia, A. K. (2003). A Listeria adhesion protein-deficient Listeria monocytogenes strain shows reduced adhesion primarily to intestinal cell lines. Med. Microbiol. Immunol. 192, 85–91.
Jenkins, D., Schultz, J., and Matin, A. (1988). Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J. Bacteriol. 170, 3910–3914. doi: 10.1128/jb.170.9.3910-3914.1988
Jones, B. V., Begley, M., Hill, C., Gahan, C. G., and Marchesi, J. R. (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Nat. Acad. Sci. U.S.A. 105, 13580–13585. doi: 10.1073/pnas.0804437105
Joyce, S. A., Shanahan, F., Hill, C., and Gahan, C. G. (2014). Bacterial bile salt hydrolase in host metabolism: potential for influencing gastrointestinal microbe-host crosstalk. Gut Microbes 5, 669–674. doi: 10.4161/19490976.2014.969986
Juneja, V. K., Dwivedi, H. P., and Yan, X. (2012). Novel natural food antimicrobials. Annu. Rev. Food Sci. Technol. 3, 381–403. doi: 10.1146/annurev-food-022811-101241
Kamada, N., Chen, G. Y., Inohara, N., and Núñez, G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Kanafani, H., and Martin, S. E. (1985). Catalase and superoxide dismutase activities in virulent and nonvirulent Staphylococcus aureus isolates. J. Clin. Microbiol. 21, 607–610.
Kang, J., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2015). VirR-mediated resistance of Listeria monocytogenes against food antimicrobials and cross-protection induced by exposure to organic acid salts. Appl. Environ. Microbiol. 81, 4553–4562. doi: 10.1128/AEM.00648-15
Kazmierczak, M. J., Wiedmann, M., and Boor, K. J. (2005). Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69, 527–543. doi: 10.1128/MMBR.69.4.527-543.2005
Kim, H., and Bhunia, A. K. (2013). Secreted Listeria adhesion protein (Lap) influences Lap-mediated Listeria monocytogenes paracellular translocation through epithelial barrier. Gut Pathog. 5:16. doi: 10.1186/1757-4749-5-16
Kim, J.-C., Oh, E., Kim, J., and Jeon, B. (2015). Regulation of oxidative stress resistance in Campylobacter jejuni, a microaerophilic foodborne pathogen. Front. Microbiol. 6:751. doi: 10.3389/fmicb.2015.00751
Kim, K.-P., Hahm, B.-K., and Bhunia, A. (2007). The 2-Cys peroxiredoxin-deficient Listeria monocytogenes displays impaired growth and survival in the presence of hydrogen peroxide in vitro but not in mouse organs. Curr. Microbiol. 54, 382–387. doi: 10.1007/s00284-006-0487-6
Kim, M.-J., Tang, C. H., Bang, W. S., and Yuk, H.-G. (2017). Antibacterial effect of 405 ± 5 nm light emitting diode illumination against Escherichia coli O157: H7, Listeria monocytogenes, and Salmonella on the surface of fresh-cut mango and its influence on fruit quality. Int. J. Food Microbiol. 244, 82–89. doi: 10.1016/j.ijfoodmicro.2016.12.023
Kirk, M. D., Pires, S. M., Black, R. E., Caipo, M., Crump, J. A., Devleesschauwer, B., et al. (2015). World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 12:e1001921. doi: 10.1371/journal.pmed.1001921
Knudsen, G. M., Fromberg, A., Ng, Y., and Gram, L. (2016). Sublethal concentrations of antibiotics cause shift to anaerobic metabolism in Listeria monocytogenes and induce phenotypes linked to antibiotic tolerance. Front. Microbiol. 7:1091. doi: 10.3389/fmicb.2016.01091
Knudsen, G. M., Ng, Y., and Gram, L. (2013). Survival of bactericidal antibiotic treatment by a persister subpopulation of Listeria monocytogenes. Appl. Environ. Microbiol. 79, 7390–7397. doi: 10.1128/AEM.02184-13
Kobayashi, H., Miyamoto, T., Hashimoto, Y., Kiriki, M., Motomatsu, A., Honjoh, K.-I., et al. (2005). Identification of factors involved in recovery of heat-injured Salmonella Enteritidis. J. Food Prot. 68, 932–941. doi: 10.4315/0362-028X-68.5.932
Krishnan, H., Ghosh, A., Paul, K., and Chowdhury, R. (2004). Effect of anaerobiosis on expression of virulence factors in Vibrio cholerae. Infect. Immun. 72, 3961–3967. doi: 10.1128/IAI.72.7.3961-3967.2004
Kus, J. V., Gebremedhin, A., Dang, V., Tran, S.-L., Serbanescu, A., and Foster, D. B. (2011). Bile salts induce resistance to polymyxin in enterohemorrhagic Escherichia coli O157: H7. J. Bacteriol. 193, 4509–4515. doi: 10.1128/JB.00200-11
Laaberki, M.-H., Janabi, N., Oswald, E., and Repoila, F. (2006). Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157: H7: interplay between Ler, GrlA, HNS and RpoS. Int. J. Food Microbiol. 296, 197–210. doi: 10.1016/j.ijmm.2006.02.017
Lapidot, A., and Yaron, S. (2009). Transfer of Salmonella enterica serovar Typhimurium from contaminated irrigation water to parsley is dependent on curli and cellulose, the biofilm matrix components. J. Food Prot. 72, 618–623. doi: 10.4315/0362-028X-72.3.618
Larsen, M. H., Koch, A. G., and Ingmer, H. (2010). Listeria monocytogenes efficiently invades Caco-2 cells after low-temperature storage in broth and on deli meat. Foodborne Pathog. Dis. 7, 1013–1018. doi: 10.1089/fpd.2009.0470
Lee, H., Ma, R., Grimm, M. C., Riordan, S. M., Lan, R., Zhong, L., et al. (2014). Examination of the anaerobic growth of Campylobacter concisus strains. Int. J. Microbiol. 2014:476047. doi: 10.1155/2014/476047
Leenanon, B., and Drake, M. (2001). Acid stress, starvation, and cold stress affect poststress behavior of Escherichia coli O157: H7 and nonpathogenic Escherichia coli. J. Food Prot. 64, 970–974. doi: 10.4315/0362-028X-64.7.970
Lessa, F. C., Mu, Y., Bamberg, W. M., Beldavs, Z. G., Dumyati, G. K., Dunn, J. R., et al. (2015). Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 825–834. doi: 10.1056/NEJMoa1408913
Lewis, K. (2007). Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56. doi: 10.1038/nrmicro1557
Leyer, G., and Johnson, E. (1993). Acid adaptation induces cross-protection against environmental stresses in Salmonella Typhimurium. Appl. Environ. Microbiol. 59, 1842–1847.
Liao, X., Li, J., Suo, Y., Ahn, J., Liu, D., Chen, S., et al. (2018). Effect of preliminary stresses on the resistance of Escherichia coli and Staphylococcus aureus toward non-thermal plasma (NTP) challenge. Food Res. Int. 105, 178–183. doi: 10.1016/j.foodres.2017.11.010
Lim, J. Y., Yoon, J. W., and Hovde, C. J. (2010). A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 20, 5–14.
Lin, C.-S., Wang, C., Tsai, H.-J., and Chou, C.-H. (2010). Growth of Listeria monocytogenes on a RTE-meat matrix enhances cell invasiveness to mouse J774A.1 macrophages. Int. J. Food Microbiol. 144, 199–201. doi: 10.1016/j.ijfoodmicro.2010.09.021
Lin, J., Smith, M. P., Chapin, K. C., Baik, H. S., Bennett, G. N., and Foster, J. W. (1996). Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62, 3094–3100.
Lindén, S., Bierne, H., Sabet, C., Png, C., Florin, T., Mcguckin, M., et al. (2008). Listeria monocytogenes internalins bind to the human intestinal mucin MUC2. Arch. Microbiol. 190, 101–104. doi: 10.1007/s00203-008-0358-6
Liu, Z., Yang, M., Peterfreund, G. L., Tsou, A. M., Selamoglu, N., Daldal, F., et al. (2011). Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc. Nat. Acad. Sci. U.S.A. 108, 810–815. doi: 10.1073/pnas.1014640108
Louis, P., and O’Byrne, C. P. (2010). Life in the gut: microbial responses to stress in the gastrointestinal tract. Sci. Prog. 93, 7–36. doi: 10.3184/003685009X12605525292307
Lu, H., Patil, S., Keener, K. M., Cullen, P., and Bourke, P. (2014). Bacterial inactivation by high-voltage atmospheric cold plasma: influence of process parameters and effects on cell leakage and DNA. J. Appl. Microbiol. 116, 784–794. doi: 10.1111/jam.12426
Mahmoud, B., Nannapaneni, R., Chang, S., and Coker, R. (2016). Effect of X-ray treatments on Escherichia coli O157: H7, Listeria monocytogenes, Shigella flexneri, Salmonella enterica and inherent microbiota on whole mangoes. Lett. Appl. Microbiol. 62, 138–144. doi: 10.1111/lam.12518
Mahoney, M., and Henriksson, A. (2003). The effect of processed meat and meat starter cultures on gastrointestinal colonization and virulence of Listeria monocytogenes in mice. Int. J. Food Microbiol. 84, 255–261. doi: 10.1016/S0168-1605(02)00400-2
Malik-Kale, P., Parker, C. T., and Konkel, M. E. (2008). Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J. Bacteriol. 190, 2286–2297. doi: 10.1128/JB.01736-07
Mandel, I. D. (1989). The role of saliva in maintaining oral homeostasis. J. Am. Dent. Assoc. 119, 298–304. doi: 10.14219/jada.archive.1989.0211
Mataragas, M., Rovetto, F., Bellio, A., Alessandria, V., Rantsiou, K., Decastelli, L., et al. (2015). Differential gene expression profiling of Listeria monocytogenes in Cacciatore and Felino salami to reveal potential stress resistance biomarkers. Food Microbiol. 46, 408–417. doi: 10.1016/j.fm.2014.09.003
Mazzotta, A. S. (2001). Thermal inactivation of stationary-phase and acid-adapted Escherichia coli O157: H7, Salmonella, and Listeria monocytogenes in fruit juices. J. Food Prot. 64, 315–320. doi: 10.4315/0362-028X-64.3.315
McMahon, M. A. S., Xu, J., Moore, J. E., Blair, I. S., and Mcdowell, D. A. (2007). Environmental stress and antibiotic resistance in food-related pathogens. Appl. Environ. Microbiol. 73, 211–217. doi: 10.1128/AEM.00578-06
Melo, J., Andrew, P., and Faleiro, M. (2015). Listeria monocytogenes in cheese and the dairy environment remains a food safety challenge: the role of stress responses. Food Res. Int. 67, 75–90. doi: 10.1016/j.foodres.2014.10.031
Merritt, M. E., and Donaldson, J. R. (2009). Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J. Med. Microbiol. 58, 1533–1541. doi: 10.1099/jmm.0.014092-0
Mills, D. C., Gundogdu, O., Elmi, A., Bajaj-Elliott, M., Taylor, P. W., Wren, B. W., et al. (2012). Increase in Campylobacter jejuni invasion of intestinal epithelial cells under low-oxygen coculture conditions that reflect the in vivo environment. Infect. Immun. 80, 1690–1698. doi: 10.1128/IAI.06176-11
Moosekian, S. R., Jeong, S., Marks, B. P., and Ryser, E. T. (2012). X-ray irradiation as a microbial intervention strategy for food. Annu. Rev. Food Sci. Technol. 3, 493–510. doi: 10.1146/annurev-food-022811-101306
Moreira, C. G., and Sperandio, V. (2010). The epinephrine/norepinephrine/autoinducer-3 interkingdom signaling system in Escherichia coli O157:H7. Microb. Endocrinol. 874, 247–261. doi: 10.1007/978-1-4419-5576-0_12
Moreira, C. G., Weinshenker, D., and Sperandio, V. (2010). QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect. Immun. 78, 914–926. doi: 10.1128/IAI.01038-09
Mueller-Herbst, S., Wustner, S., Muhlig, A., Eder, D., Fuchs, T. M., Held, C., et al. (2014). Identification of genes essential for anaerobic growth of Listeria monocytogenes. Microbiology 160, 752–765. doi: 10.1099/mic.0.075242-0
Muller, C., Bang, I. S., Velayudhan, J., Karlinsey, J., Papenfort, K., Vogel, J., et al. (2009). Acid stress activation of the σE stress response in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 71, 1228–1238. doi: 10.1111/j.1365-2958.2009.06597.x
Murphree, R., Garman, K., Phan, Q., Everstine, K., Gould, L. H., and Jones, T. F. (2012). Characteristics of foodborne disease outbreak investigations conducted by foodborne diseases active surveillance network (FoodNet) Sites, 2003–2008. Clin. Infect. Dis. 54, S498–S503. doi: 10.1093/cid/cis232
Müsken, A., Bielaszewska, M., Greune, L., Schweppe, C. H., Müthing, J., Schmidt, H., et al. (2008). Anaerobic conditions promote expression of Sfp fimbriae and adherence of sorbitol-fermenting enterohemorrhagic Escherichia coli O157: NM to human intestinal epithelial cells. Appl. Environ. Microbiol. 74, 1087–1093. doi: 10.1128/AEM.02496-07
NicAogáin, K., and O’Byrne, C. P. (2016). The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front. Microbiol. 7:1865. doi: 10.3389/fmicb.2016.01865
Nickerson, K. P., Chanin, R. B., Sistrunk, J. R., Rasko, D. A., Fink, P. J., Barry, E. M., et al. (2017). Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect. Immun. 85:e01067-16 doi: 10.1128/IAI.01067-16
Niemira, B. A. (2012). Cold plasma decontamination of foods. Annu. Rev. Food Sci. Technol. 3, 125–142. doi: 10.1146/annurev-food-022811-101132
Novik, V., Hofreuter, D., and Galán, J. E. (2010). Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect. Immun. 78, 3540–3553. doi: 10.1128/IAI.00109-10
Ochiai, Y., Yamada, F., Yoshikawa, Y., Mochizuki, M., Takano, T., Hondo, R., et al. (2017). Sequential transition of the injury phenotype, temperature-dependent survival and transcriptional response in Listeria monocytogenes following lethal H2O2 exposure. Int. J. Food Microbiol. 259, 52–58. doi: 10.1016/j.ijfoodmicro.2017.08.001
Ojha, K. S., Kerry, J. P., Duffy, G., Beresford, T., and Tiwari, B. K. (2015). Technological advances for enhancing quality and safety of fermented meat products. Trends Food Sci. Technol. 44, 105–116. doi: 10.1016/j.tifs.2015.03.010
Olesen, I., Thorsen, L., and Jespersen, L. (2010). Relative transcription of Listeria monocytogenes virulence genes in liver pâtés with varying NaCl content. Int. J. Food Microbiol. 141, S60–S68. doi: 10.1016/j.ijfoodmicro.2010.01.042
Oliveira, M., Wijnands, L., Abadias, M., Aarts, H., and Franz, E. (2011). Pathogenic potential of Salmonella Typhimurium DT104 following sequential passage through soil, packaged fresh-cut lettuce and a model gastrointestinal tract. Int. J. Food Microbiol. 148, 149–155. doi: 10.1016/j.ijfoodmicro.2011.05.013
Page, R., and Peti, W. (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214. doi: 10.1038/nchembio.2044
Pandiripally, V. K., Westbrook, D. G., Sunki, G. R., and Bhunia, A. K. (1999). Surface protein p104 is involved in adhesion of Listeria monocytogenes to human intestinal cell line, Caco-2. J. Med. Microbiol. 48, 117–124. doi: 10.1099/00222615-48-2-117
Payne, A., Schmidt, T. B., Nanduri, B., Pendarvis, K., Pittman, J. R., Thornton, J. A., et al. (2013). Proteomic analysis of the response of Listeria monocytogenes to bile salts under anaerobic conditions. J. Med. Microbiol. 62, 25–35. doi: 10.1099/jmm.0.049742-0
Pelaseyed, T., Bergstrom, J. H., Gustafsson, J. K., Ermund, A., Birchenough, G. M. H., Schutte, A., et al. (2014). The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 260, 8–20. doi: 10.1111/imr.12182
Perez, R. H., Zendo, T., and Sonomoto, K. (2014). Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb. Cell Fact. 13:S3. doi: 10.1186/1475-2859-13-S1-S3
Pesci, E. C., Cottle, D. L., and Pickett, C. L. (1994). Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62, 2687–2694.
Pittman, J. R., Buntyn, J. O., Posadas, G., Nanduri, B., Pendarvis, K., and Donaldson, J. R. (2014). Proteomic analysis of cross protection provided between cold and osmotic stress in Listeria monocytogenes. J. Proteome Res. 13, 1896–1904. doi: 10.1021/pr401004a
Pittman, J. R., Kline, L. K. C., and Kenyon, W. J. (2015). Carbon-starvation induces cross-resistance to thermal, acid, and oxidative stress in Serratia marcescens. Microorganisms 3, 746–758. doi: 10.3390/microorganisms3040746
Ponder, M. A. (2017). “The effects of food composition on foodborne illness infectious dose and host susceptibility,” in Foodborne Pathogens, eds J. B. Gurtler, M. P. Doyle and J. L. Kornacki (New York, NY: Springer), 469-494.
Poole, K. (2012). Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 67, 2069–2089. doi: 10.1093/jac/dks196
Pricope-Ciolacu, L., Nicolau, A. I., Wagner, M., and Rychli, K. (2013). The effect of milk components and storage conditions on the virulence of Listeria monocytogenes as determined by a Caco-2 cell assay. Int. J. Food Microbiol. 166, 59–64. doi: 10.1016/j.ijfoodmicro.2013.05.027
Purdy, D., and Park, S. F. (1994). Cloning, nucleotide sequence and characterization of a gene encoding superoxide dismutase from Campylobacter jejuni and Campylobacter coli. Microbiology 140, 1203–1208. doi: 10.1099/13500872-140-5-1203
Raffatellu, M., George, M. D., Akiyama, Y., Hornsby, M. J., Nuccio, S. -P., Paixao, T. A., et al. (2009). Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486. doi: 10.1016/j.chom.2009.03.011
Rantsiou, K., Greppi, A., Garosi, M., Acquadro, A., Mataragas, M., and Cocolin, L. (2012). Strain dependent expression of stress response and virulence genes of Listeria monocytogenes in meat juices as determined by microarray. Int. J. Food Microbiol. 152, 116–122. doi: 10.1016/j.ijfoodmicro.2011.08.009
Ray, B., and Bhunia, A. (2014). “Control by antimicrobial preservatives and bacteriophages,” in Fundamental Food Microbiology, 5th Edn. (Baca Raton, FL: CRC Press), 489–503.
Rhodius, V. A., and Mutalik, V. K. (2010). Predicting strength and function for promoters of the Escherichia coli alternative sigma factor, σE. Proc. Nat. Acad. Sci. U.S.A. 107, 2854–2859. doi: 10.1073/pnas.0915066107
Rosenberg, E. Y., Bertenthal, D., Nilles, M. L., Bertrand, K. P., and Nikaido, H. (2003). Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48, 1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x
Rossetto, O., Pirazzini, M., and Montecucco, C. (2014). Botulinum neurotoxins: genetic, structural and mechanistic insights. Nat. Rev. Microbiol. 12, 535–549. doi: 10.1038/nrmicro3295
Rowan, N. J., Deans, K., Anderson, J. G., Gemmell, C. G., Hunter, I. S., and Chaithong, T. (2001). Putative virulence factor expression by clinical and food isolates of Bacillus spp. after growth in reconstituted infant milk formulae. Appl. Environ. Microbiol. 67, 3873–3881. doi: 10.1128/AEM.67.9.3873-3881.2001
Rowley, G., Spector, M., Kormanec, J., and Roberts, M. (2006). Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4, 383–394. doi: 10.1038/nrmicro1394
Saldivar, J. C., Davis, M. L., Johnson, M. G., and Ricke, S. C. (2018). “Listeria monocytogenes adaptation and growth at low temperatures: mechanisms and implications for foodborne disease,” in Food and Feed Safety Systems and Analysis, eds S. C. Ricke, G. G. Atungulu, C. E. Rainwater, S. H. Park (Cambridge, MA: Academic Press), 227–248. doi: 10.1016/B978-0-12-811835-1.00013-0
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Scharff, R. (2012). Economic burden from health losses due to foodborne illness in the United States. J. Food Prot. 75, 123–131. doi: 10.4315/0362-028X.JFP-11-058
Schelin, J., Wallin-Carlquist, N., Thorup Cohn, M., Lindqvist, R., and Barker, G. C. (2011). The formation of Staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence 2, 580–592. doi: 10.4161/viru.2.6.18122
Schmelcher, M., and Loessner, M. J. (2016). Bacteriophage endolysins: applications for food safety. Curr. Opin. Biotechnol. 37, 76–87. doi: 10.1016/j.copbio.2015.10.005
Schüller, S., and Phillips, A. D. (2010). Microaerobic conditions enhance type III secretion and adherence of enterohaemorrhagic Escherichia coli to polarized human intestinal epithelial cells. Environ. Microbiol. 12, 2426–2435. doi: 10.1111/j.1462-2920.2010.02216.x
Selby, K., Mascher, G., Somervuo, P., Lindström, M., and Korkeala, H. (2017). Heat shock and prolonged heat stress attenuate neurotoxin and sporulation gene expression in group I Clostridium botulinum strain ATCC 3502. PLoS One 12:e0176944. doi: 10.1371/journal.pone.0176944
Selma, M. V., Espin, J. C., and Tomas-Barberan, F. A. (2009). Interaction between phenolics and gut microbiota: role in human health. J. Agric. Food Chem. 57, 6485–6501. doi: 10.1021/jf902107d
Sewell, D., Allen, S. C., and Phillips, C. A. (2015). Oxygen limitation induces acid tolerance and impacts simulated gastro-intestinal transit in Listeria monocytogenes J0161. Gut Pathog. 7:11. doi: 10.1186/s13099-015-0058-0
Shahin, K., and Bouzari, M. (2018). Bacteriophage application for biocontrolling Shigella flexneri in contaminated foods. J. Food Sci. Technol. 55, 550–559. doi: 10.1007/s13197-017-2964-2
Shen, Q., Soni, K. A., and Nannapaneni, R. (2014). Influence of temperature on acid-stress adaptation in Listeria monocytogenes. Foodborne Pathog. Dis. 11, 43–49. doi: 10.1089/fpd.2013.1611
Singh, V. P. (2018). Recent approaches in food bio-preservation-a review. Open Vet. J. 8, 104–111. doi: 10.4314/ovj.v8i1.16
Sirsat, S. A., Burkholder, K. M., Muthaiyan, A., Dowd, S. E., Bhunia, A. K., and Ricke, S. C. (2011). Effect of sublethal heat stress on Salmonella Typhimurium virulence. J. Appl. Microbiol. 110, 813–822. doi: 10.1111/j.1365-2672.2011.04941.x
Sistrunk, J. R., Nickerson, K. P., Chanin, R. B., Rasko, D. A., and Faherty, C. S. (2016). Survival of the fittest: how bacterial pathogens utilize bile to enhance infection. Clin. Microbiol. Rev. 29, 819–836. doi: 10.1128/CMR.00031-16
Sleator, R. D., and Hill, C. (2002). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26, 49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x
Sleator, R. D., and Hill, C. (2010). Compatible solutes: the key to Listeria’s success as a versatile gastrointestinal pathogen? Gut Pathog. 2:20. doi: 10.1186/1757-4749-2-20
Sleator, R. D., Watson, D., Hill, C., and Gahan, C. G. M. (2009). The interaction between Listeria monocytogenes and the host gastrointestinal tract. Microbiology 155, 2463–2475. doi: 10.1099/mic.0.030205-0
Sleator, R. D., Wemekamp-Kamphuis, H. H., Gahan, C. G. M., Abee, T., and Hill, C. (2005). A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 55, 1183–1195. doi: 10.1111/j.1365-2958.2004.04454.x
Spector, M. P., and Kenyon, W. J. (2012). Resistance and survival strategies of Salmonella enterica to environmental stresses. Food Res. Int. 45, 455–481. doi: 10.1016/j.foodres.2011.06.056
Sperandio, V., and Nguyen, Y. (2012). Enterohemorrhagic E. coli (EHEC) pathogenesis. Front. Cell. Infect. Microbiol. 2:90. doi: 10.3389/fcimb.2012.00090
Swaggerty, C. L., Pevzner, I. Y., He, H., Genovese, K. J., Nisbet, D. J., Kaiser, P., et al. (2009). Selection of broilers with improved innate immune responsiveness to reduce on-farm infection by foodborne pathogens. Foodborne Pathog. Dis. 6, 777–783. doi: 10.1089/fpd.2009.0307
Tassou, S. A., Lewis, J. S., Ge, Y. T., Hadawey, A., and Chaer, I. (2010). A review of emerging technologies for food refrigeration applications. Appl. Therm. Eng. 30, 263–276. doi: 10.1016/j.applthermaleng.2009.09.001
Thanassi, D. G., Cheng, L. W., and Nikaido, H. (1997). Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179, 2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997
Theron, M. M., and Lues, J. F. R. (2007). Organic acids and meat preservation: a review. Food Rev. Int. 23, 141–158. doi: 10.1080/87559120701224964
Tiwari, B. K., Valdramidis, V. P., O’Donnell, C. P., Muthukumarappan, K., Bourke, P., and Cullen, P. J. (2009). Application of natural antimicrobials for food preservation. J. Agric. Food Chem. 57, 5987–6000. doi: 10.1021/jf900668n
Tremonte, P., Succi, M., Coppola, R., Sorrentino, E., Tipaldi, L., Picariello, G., et al. (2016). Homology-based modeling of Universal Stress Protein from Listeria innocua up-regulated under acid stress conditions. Front. Microbiol. 7:1998. doi: 10.3389/fmicb.2016.01998
Tsai, Y.-W., and Ingham, S. C. (1997). Survival of Escherichia coli O157: H7 and Salmonella spp. in acidic condiments. J. Food Prot. 60, 751–755. doi: 10.4315/0362-028X-60.7.751
Van Boekel, M., Fogliano, V., Pellegrini, N., Stanton, C., Scholz, G., Lalljie, S., et al. (2010). A review on the beneficial aspects of food processing. Mol. Nutr. Food Res. 54, 1215–1247. doi: 10.1002/mnfr.200900608
Vasconcelos, J., and Deneer, H. G. (1994). Expression of superoxide dismutase in Listeria monocytogenes. Appl. Environ. Microbiol. 60, 2360–2366.
Vergara-Irigaray, M., Fookes, M. C., Thomson, N. R., and Tang, C. M. (2014). RNA-seq analysis of the influence of anaerobiosis and FNR on Shigella flexneri. BMC Genomics 15:438. doi: 10.1186/1471-2164-15-438
Verraes, C., Van Boxstael, S., Van Meervenne, E., Van Coillie, E., Butaye, P., Catry, B., et al. (2013). Antimicrobial resistance in the food chain: a review. Int. J. Environ. Res. Pub. Health 10, 2643–2669. doi: 10.3390/ijerph10072643
Wallace, N., Newton, E., Abrams, E., Zani, A., and Sun, Y. (2017). Metabolic determinants in Listeria monocytogenes anaerobic listeriolysin O production. Arch. Microbiol. 199, 827–837. doi: 10.1007/s00203-017-1355-4
Wampler, J. L., Kim, K. P., Jaradat, Z., and Bhunia, A. K. (2004). Heat shock protein 60 acts as a receptor for the Listeria adhesion protein in Caco-2 cells. Infect. Immun. 72, 931–936. doi: 10.1128/IAI.72.2.931-936.2004
Wang, C., Chang, T., Yang, H., and Cui, M. (2015). Antibacterial mechanism of lactic acid on physiological and morphological properties of Salmonella Enteritidis, Escherichia coli and Listeria monocytogenes. Food Control 47, 231–236. doi: 10.1016/j.foodcont.2014.06.034
Wang, C.-Y., Huang, H.-W., Hsu, C.-P., and Yang, B. B. (2016). Recent advances in food processing using high hydrostatic pressure technology. Crit. Rev. Food Sci. Nutr. 56, 527–540. doi: 10.1080/10408398.2012.745479
Wesche, A. M., Gurtler, J. B., Marks, B. P., and Ryser, E. T. (2009). Stress, sublethal injury, resuscitation, and virulence of bacterial foodborne pathogens. J. Food Prot. 72, 1121–1138. doi: 10.4315/0362-028X-72.5.1121
White, P., Joshi, A., Rassam, P., Housden, N. G., Kaminska, R., Goult, J. D., et al. (2017). Exploitation of an iron transporter for bacterial protein antibiotic import. Proc. Natl. Acad. Sci. U.S.A. 114, 12051–12056. doi: 10.1073/pnas.1713741114
Whiteley, A. T., Ruhland, B. R., Edrozo, M. B., and Reniere, M. L. (2017). A redox-responsive transcription factor is critical for pathogenesis and aerobic growth of Listeria monocytogenes. Infect. Immun. 85:e00978-16. doi: 10.1128/IAI.00978-16
Winter, S., Thiennimitr, P., Winter, M., Butler, B., and Huseby, D. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Wouters, J. A., Rombouts, F. M., Kuipers, O. P., De Vos, W. M., and Abee, T. (2000). The role of cold-shock proteins in low-temperature adaptation of food-related bacteria. Syst. Appl. Microbiol. 23, 165–173. doi: 10.1016/S0723-2020(00)80001-6
Wright, M. L., Pendarvis, K., Nanduri, B., Edelmann, M. J., Jenkins, H. N., Reddy, J. S., et al. (2016). The effect of oxygen on bile resistance in Listeria monocytogenes. J. Proteomics Bioinformatics 9, 107–119. doi: 10.4172/jpb.1000396
Wu, S., Yu, P.-L., and Flint, S. (2017). Persister cell formation of Listeria monocytogenes in response to natural antimicrobial agent nisin. Food Control 77, 243–250. doi: 10.1016/j.foodcont.2017.02.011
Yang, Y., Khoo, W. J., Zheng, Q., Chung, H.-J., and Yuk, H.-G. (2014). Growth temperature alters Salmonella Enteritidis heat/acid resistance, membrane lipid composition and stress/virulence related gene expression. Int. J. Food Microbiol. 172, 102–109. doi: 10.1016/j.ijfoodmicro.2013.12.006
Zhang, Y. (2014). Persisters, persistent infections and the Yin–Yang model. Emerg. Microbes Infect. 3:e3. doi: 10.1038/emi.2014.3
Zhou, G., Xu, X., and Liu, Y. (2010). Preservation technologies for fresh meat–A review. Meat Sci. 86, 119–128. doi: 10.1016/j.meatsci.2010.04.033
Keywords: human pathogen, food processing, pathogenesis, infection, stress response and adaptation, pathogen survival, gut, immunity
Citation: Horn N and Bhunia AK (2018) Food-Associated Stress Primes Foodborne Pathogens for the Gastrointestinal Phase of Infection. Front. Microbiol. 9:1962. doi: 10.3389/fmicb.2018.01962
Received: 04 April 2018; Accepted: 02 August 2018;
Published: 23 August 2018.
Edited by:
Vittorio Capozzi, University of Foggia, ItalyReviewed by:
Christina S. Faherty, Harvard Medical School, United StatesCopyright © 2018 Horn and Bhunia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arun K. Bhunia, Ymh1bmlhQHB1cmR1ZS5lZHU=
†Present address: Nathan Horn, BioMatrix International, Princeton, MN, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.