- 1Department of Stomatology, Nanfang Hospital, Southern Medical University, Guangzhou, China
- 2Department of Neurobiology, School of Basic Medical Science, Southern Medical University, Guangzhou, China
- 3Department of Stomatology, The Affiliated Shenzhen Maternity and Child Healthcare Hospital of the South Medical University, Shenzhen, China
The bacterial second messenger cyclic diguanylate monophosphate (c-di-GMP) regulates a series of cellular functions, including biofilm formation, motility, virulence, and other processes. In this study, we confirmed the presence of several c-di-GMP related genes and evaluated their activities and functions in Lactobacillus species. Bioinformatic and biochemical analyses revealed that Lactobacillus acidophilus La-14 have an active c-di-GMP phosphodiesterase (PdeA) that may act in the metabolic cycle of c-di-GMP. A GGDEF protein (DgcA) induced two c-di-GMP-dependent phenotypes (low motility and high production of curli fimbriae) in Escherichia coli by heterologously expressed in vivo but showed no diguanylate cyclases activity in vitro while in the expression without the N-terminal transmembrane domain. The degenerated EAL-domain protein (PdeB), encoded by the last gene in the gts operon, serve as a c-di-GMP receptor which may be associated with exopolysaccharide (EPS) synthesis in L. acidophilus. Heterologously expressed GtsA and GtsB, encoded by the gts operon, stimulated EPS and biofilm formation in E. coli BL21. Constitutive expression in L. acidophilus revealed that a high concentration of intracellular DgcA levels increased EPS production in L. acidophilus and enhanced the co-aggregation ability with E. coli MG1655, which may be beneficial to the probiotic properties of Lactobacillus species. Our study imply that the c-di-GMP metabolism-related genes, in L. acidophilus, work jointly to regulate its functions in EPS formation and co-aggregation.
Introduction
Cyclic diguanylate monophosphate (c-di-GMP), formed by the condensation of two GTP molecules, is a secondary messenger that is widely distributed in bacteria and is involved in the regulation of multiple bacterial physiological functions (Hengge, 2009). Opposing activities of diguanylate cyclases (DGCs) containing the GGDEF domain and phosphodiesterases (PDEs) containing EAL or HD-GYP domains control cellular c-di-GMP homeostasis (Römling et al., 2013). Genes encoding GGDEF and EAL protein families are distributed unevenly among the genomes of various species. For example, Staphylococcus aureus possesses only one GGDEF protein, GdpS, without DGC activity in vitro and is involved in virulence regulation through an RNA-dependent pathway (Holland et al., 2008; Römling et al., 2013). By comparison, more than 90 genes potentially encoding c-di-GMP-metabolizing enzymes were predicted in Vibrio vulnificus (Römling et al., 2013). These GGDEF or EAL domains, in tandem with other signaling domains and located in the cytoplasm or cytomembranes, precisely regulate local intracellular c-di-GMP concentrations by responding to diverse upstream activating signals. C-di-GMP regulates a variety of physiological processes, including cell-cell interactions (Matsuyama et al., 2016; Lin C. S. et al., 2017), biofilm formation and dispersal (Ha and O'Toole, 2015; Skariyachan et al., 2018), cell motility (Orr and Lee, 2016), and the responses to a variety of external stimulation, such as oxygen (Burns et al., 2016), nitric oxide (Rinaldo et al., 2018), and light (Blain-Hartung et al., 2017). The c-di-GMP signaling pathway is present in many Gram-negative bacteria but is less reported in Gram-positive bacteria (Purcell and Tamayo, 2016). In recent years, however, the existence of a c-di-GMP signaling pathway has also been confirmed in many Gram-positive bacteria, such as Streptomyces coelicolor (den Hengst et al., 2010), Clostridium difficile (Purcell et al., 2012), Bacillus subtilis (Gao et al., 2013), and Listeria monocytogenes (Chen et al., 2014). In these species, c-di-GMP signaling primarily regulates flagellum synthesis, production of adhesion factor in response to surface contact, and production of extracellular polymeric substances (Purcell and Tamayo, 2016; Bedrunka and Graumann, 2017a).
The recent discovery of c-di-GMP signaling in Firmicutes prompted us to focus on the species of Lactobacillus, especially Lactobacillus acidophilus. So far, the c-di-GMP-metabolizing enzymes in Lactobacillus have been poorly characterized except for a degenerated EAL-domain protein (Lp_2714) in Lactobacillus plantarum, surmised as a transmembrane protein involved in regulating polysaccharide synthesis (Brown et al., 2011; Purcell and Tamayo, 2016). The well-known probiotic strain L. acidophilus is one of the major species generally recognized as safe (GRAS; Martínez et al., 2012). L. acidophilus is Gram-positive, produces acid through fermenting sugars into lactic acid, grows readily at rather low pH values (below 5.0), and is a probiotic microorganism that mainly inhabits the human intestines, oral cavities, and vagina (Bâati et al., 2000). As a typical probiotic, L. acidophilus can alleviate lactose intolerance (Kim and Gilliland, 1983), abdominal pain, and irritable bowel syndrome (Rousseaux et al., 2007) as well as modulate dendritic and T cell function (Konstantinov et al., 2008). Among the intestinal microbiota, L. acidophilus shows a strong autoaggregation phenotype and has been demonstrated to efficiently coaggregate with some pathogenic strains in vitro (Collado et al., 2008). The exopolysaccharide (EPS) produced by L. acidophilus possesses bioactive components with various health benefits, such as antioxidative properties and inducing cytotoxicity in two colon cancer cell lines (Deepak et al., 2016). Meanwhile, EPS also plays an important role in protecting microbes from adverse conditions, such as lysozyme osmosis as well the presence of bacteriophages, copper ions, or nisin (Looijesteijn et al., 2001).
In this study, we evaluated the possible role of c-di-GMP in regulating the probiotic properties of L. acidophilus for the first time. We identified the genes and operons related to the c-di-GMP signaling pathway by bioinformatic and transcriptional analyses of L. acidophilus. Soluble proteins were expressed and purified for subsequent evaluation. In vivo and in vitro activity assays were performed for assessing the function of c-di-GMP-related enzymes. We also confirmed a c-di-GMP-specific receptor by an in vitro binding test. The proteins (LA14_RS07015 and LA14_RS07020) were overexpressed in vivo to monitor relevant phenotypes that may be associated with c-di-GMP modulation. The regulatory function of c-di-GMP related genes in EPS formation was also evaluated in L. acidophilus.
Materials and Methods
Bioinformatics
Gene identities for annotated c-di-GMP-related proteins of L. acidophilus La-14 were obtained from the NCBI genome (RefSeq: NC_021181.2). Conserved domain analysis was derived from the SMART (http://smart.embl-heidelberg.de/) and Pfam (Finn et al., 2016) databases. Signal peptide and transmembrane helices were predicted using SignalP 4.0 (Petersen et al., 2011) and TMHMM 2.0 (Möller et al., 2001), respectively. Soft Berry BPROM (Solovyev and Salamov, 2011) and ProOpDB (Taboada et al., 2012) were employed to predict bacterial promoters and operons, respectively. Comparative alignment and homologous proteins searching were performed using NCBI COBLAT and BLASTP, respectively (Papadopoulos and Agarwala, 2007).
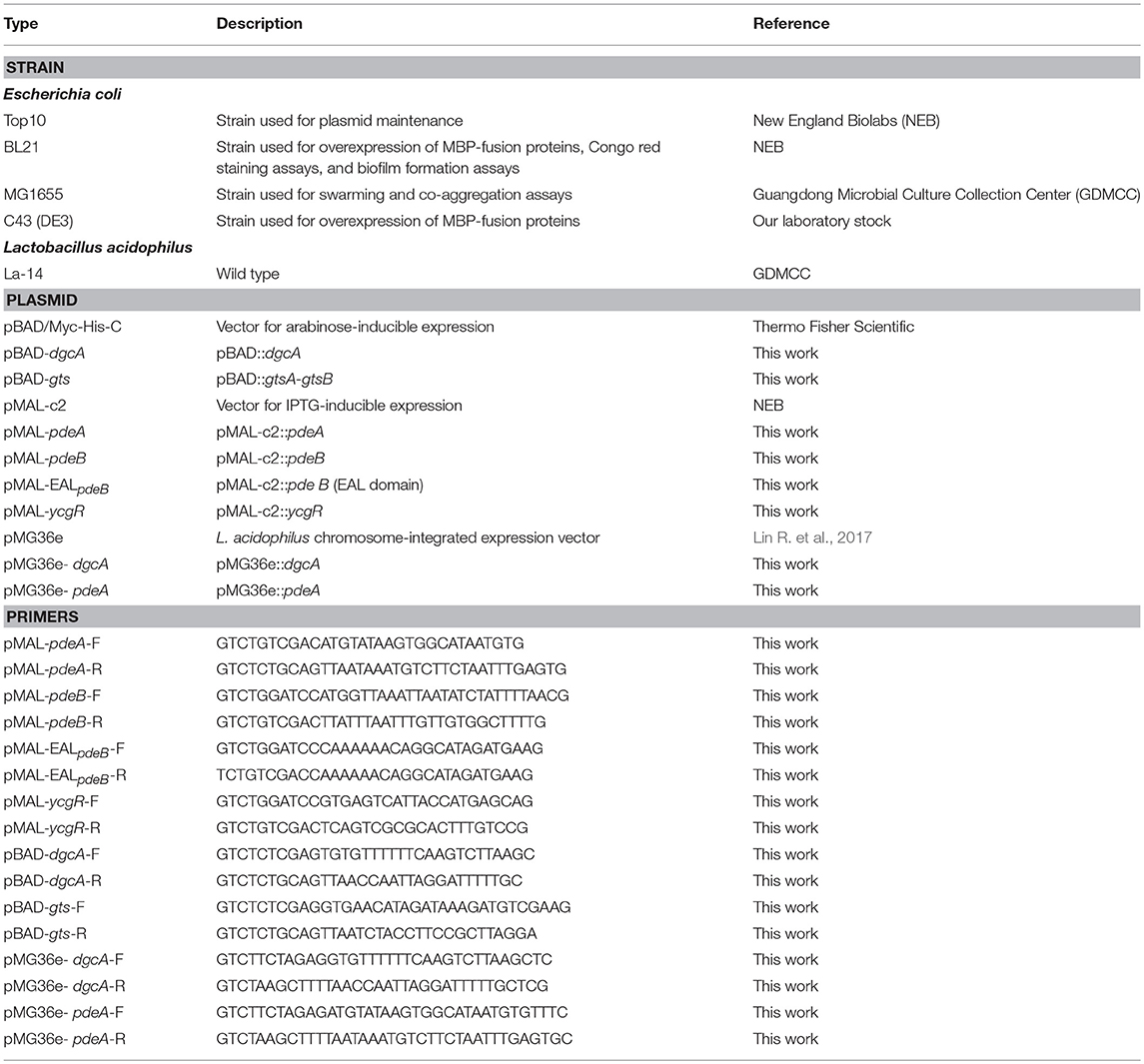
Strain Construction
Putative DGC and glycosyltransferase (gts) genes were cloned into the pBAD-Myc-His vector carrying an ampicillin resistance gene and an L-arabinose-inducible promoter (Table 1). For measuring enzymatic activity and binding assays in vitro, the genes of interest were cloned into pMAL-c2, which contains a maltose-binding protein (MBP) for purification. The constitutively expressing plasmid pMG36e was used to express DGC and PDE proteins in L. acidophilus La-14. Escherichia coli was routinely grown in LB medium containing relevant antibiotics and under appropriate temperatures. L. acidophilus was grown in MRS medium containing relevant antibiotics at 37°C and was transformed via electroporation as described previously (Lin R. et al., 2017). Briefly, cells were cultured in MRS broth medium with 0.05% cysteine-HCl at 37°C for 48 h until optical density at 600 nm (OD600) reached 0.6. The culture was then diluted 1:25 in 100 mL of MRS broth with 0.5 M sucrose and 0.05% cysteine-HCl and left to grow for ~24 h until OD600 reached 0.8. The culture was cooled for 10 min and then cell pellets were harvested and washed twice with 0.5 M sucrose buffer, followed by an additional wash with transformation buffer (10 mM ammonium and 0.5 M sucrose; pH 6.0) and re-suspension in 400 μL transformation buffer. The recombinant plasmid was transformed into L. acidophilus cells by electroporation using a MicroPulser™ Electroporator (Bio-Rad, Hercules, CA, USA) at 1.5 Kv/cm. Transformed bacteria were re-suspended in MRS broth and cultured at 37°C for 1 h, followed by plating on MRS agar (1.5% w/v) containing 0.5 μg/mL erythromycin and incubation at 37°C for 48 h. Positive colonies of transformed bacteria were identified by PCR and target gene sequencing.
Transcriptional Analysis
To characterize operon regulation of the dgcA, pdeA, pdeB, gtsA, and gtsB genes, total RNA was extracted and purified. Briefly, an overnight culture of L. acidophilus La-14 was added into MRS medium and incubated until the late exponential phase. The cells were collected and treated with lysozyme and RNA was extracted using RNAiso reagent (Takara, Shiga, Japan). After treatment with DNA Eraser, the RNA was reverse transcribed into cDNA according to the PrimeScript RT Master Mix Kit (Takara) protocol.
Swarming and Congo Red Dye Binding Assays
Congo red binding assays were used to determine bacterial EPS production. LB (E. coli) or MRS (L. acidophilus) agar plates containing 50–80 μg/mL Congo red was treated at 30°C for 48 or 72 h. For swarming assays, LB plates were made with 0.5% agar supplemented with 0.5% L-arabinose (Harshey and Matsuyama, 1994; Paul et al., 2010). Overnight cultured cells were used to inoculate the plates and then incubated at 37°C for 24 h.
Protein Overexpression and Purification
During MBP-PdeA, MBP-EALpdeB and MBP-YcgR fusion protein expression, IPTG (final concentration, 0.6 mM) was added to exponentially growing E. coli BL21 for a 3-h induction at 37°C. For MBP-PdeB fusion protein expression, IPTG (final concentration, 0.3 mM) was added to exponentially growing E. coli C43 for a 12-h induction at 30°C. After induction, cell pellets were harvested by centrifugation at 6,000 × g for 10 min. Cell pellets were resuspended in lysis buffer containing 150 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 20 mM Tris-HCl (pH 7.4), 17 μg/mL PMSF, and protease inhibitor cocktail (Roche, Basel, Switzerland). After sonication and centrifugation, the clarified lysates were loaded onto a pre-equilibrated amylose column (NEB, Ipswich, MA, USA) which was subsequently washed with 12 column volumes of column buffer (150 mM NaCl, 1 mM EDTA, and 20 mM Tris-HCl, pH 7.4). MBP-fusion proteins were eluted with column buffer containing 10 mM maltose that was subsequently exchanged with PDE activity assay buffer or c-di-GMP binding assay buffer using Amicon Ultra-15 mL Centrifugal Filter Units (Merck Millipore, Burlington, MA, USA). Purified proteins were detected by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and protein concentration was measured by the BCA protein assay (Pierce, Rockford, IL, USA).
PDE Assays
PDE assays were performed as previously described (Schmidt et al., 2005). Briefly, the PDE assay components were incubated with 10 μM enzyme (MBP-PdeA or MBP-EALpdeB) for 1 h at 37°C in buffer containing 50 mM Tris-HCl (pH 9.35), 5 mM MgCl2, 50 mM NaCl, 0.5 mM EDTA, and 100 μM c-di-GMP (Biolog, Bremen, Germany). To monitor the hydrolysis rates of c-di-GMP, the reactions were stopped by adding CaCl2 (final concentration, 10 mM) at various time points and then samples were boiled for 5 min and centrifuged. The supernatant was filtered through a 0.22 μm filter and analyzed by reversed-phase high performance liquid chromatography (HPLC; Waters, Milford, MA, USA). Reactants (15 μL) were injected into a TC-C18 column (15 × 4.6 cm; Agilent, Santa Clara, CA, USA) and separated by reversed-phase HPLC with a buffer system based on the gradient program described previously (Ryjenkov et al., 2005).
c-di-GMP Binding Assays
Differential radial capillary action of ligand assay (DRaCALA) was performed as described previously (Fang et al., 2014) with some modifications. Briefly, MBP-fusion protein in binding buffer (300 mM NaCl, 1 mM EDTA, 10% glycerol, and 50 mM Tris-HCl, pH 7.5) was mixed with 0.5 μM 2′-fluo-aminohexylcarbamoyl-c-di-GMP (Fluo-c-di-GMP; Biolog) and incubated for 20 min at room temperature. Fluo-c-di-GMP was competed away with cold nucleotides in different concentrations. Then, 2 μL of the mixture was spotted on nitrocellulose membranes (Merck Millipore) in triplicate. The Typhoon FLA 9000 scanner (excitation wavelength, 473 nm; GE Healthcare, Chicago, IL, USA) was used to detect membrane fluorescence. The dissociation constant of specific protein-ligand interactions was measured by altering the protein concentration.
Equilibrium dialysis experiments were performed as previously described (Ryjenkov et al., 2006). MBP-EALpdeB (16 μM) was placed into one chamber of the Dispo Equilibrium DIALYZER (10 kDa cut off; Harvard Apparatus, Holliston, MA, USA) with binding buffer. C-di-GMP (1–50 μM) in an equivalent volume (70 μL) was placed in the other chamber. The dialyzers were slowly agitated for 24 h at room temperature to reach equilibrium. Samples from each chamber were boiled for 5 min and centrifuged. The supernatants were then filtered through a 0.22 μm micro filter. For quantification, 50 μM of GDP (final concentration) was added to each sample. Reactants (15 μL) were injected into a TC-C18 column (15 × 4.6 cm; Agilent) and separated by reversed-phase HPLC with a buffer system based on the gradient program described previously (Ryjenkov et al., 2005).
Biofilm and EPS Formation Assays
The ability of bacteria in forming stable biofilms was assessed using cells growing in 96-well plates according to a previous method (O'Toole and Kolter, 1998) with some modifications. For E. coli, different concentrations of L-arabinose were added to the exponentially growing cultures (OD600 = 0.6–0.7) and then 200-μL aliquots of each culture were used to inoculate each of four wells. Plates were incubated at 30°C for 24 h. For biofilm quantification, the media were discarded from microtiter plates to remove unbound cells and then the plates were gently washed twice by TBS. After air-drying, the adherent bacteria were stained with 100 μL 0.1% crystal violet for 15 min at room temperature and then the plates were gently washed twice. The bound dye was extracted from the stained cells by adding 200 μL of an ethanol/acetone (8:2) mixture. Biofilm formation was then quantified by measuring OD600.
EPS formation was evaluated with Congo red dye binding assays and confocal laser scanning fluorescence microscopy as described previously (Wu et al., 2016). L. acidophilus La-14 and its derivatives were grown in MRS broth with 0.5 μg/mL erythromycin for 24 h and the cultures were harvested and diluted 1:100 with MRS medium, after which 5 mL of diluted culture was added to 6-well plates with coverslips placed at the bottom of each well. After incubation for 120 h in 5% CO2 at 37°C, the coverslips were gently washed twice with sterile Tris-buffered saline (TBS) to remove unbound bacteria and then stained with calcofluor-white (Sigma-Aldrich, St. Louis, MI, USA) for 15 min at room temperature in the dark to stain the EPS. The coverslips were then gently washed two times with sterile TBS and observed with a Nikon A1 confocal laser microscope (Nikon, Tokyo, Japan) using the 351-nm line. The stained EPS then appeared blue during confocal fluorescence microscopy analysis. At least five independent fields were collected at 60 × magnification per experiment and three independent experiments were performed. Image J software (version 1.43; NIH) was used to calculate the area covered by the germs.
Co-Aggregation Assays
Co-aggregation assays were performed as previously described (Collado et al., 2008; Johnson and Klaenhammer, 2016) with some modifications. Bacterial suspensions for co-aggregation were prepared following the autoaggregation assay protocol. Then, the same volumes of cell suspensions (1 mL) of different probiotic and pathogenic strains were mixed together in pairs and vortexed for 10 s and incubated at room temperature without agitation. OD600 of the suspensions were measured during a 5-h incubation period. The percentage of co-aggregation was calculated using , where Apat and Aprobio represent the OD600 of pathogenic and probiotic bacterial suspensions, respectively, and Amix represents the mixture OD600 at different time points.
Results
Analysis of Genes Related to the c-di-GMP Signaling Pathway in L. acidophilus
C-di-GMP is synthesized by DGC from two GTP molecules and is hydrolyzed by PDE to pGpG. DGC family proteins contain a conserved Gly-Gly-Asp-Glu-Phe (GGDEF) sequence motif, whereas PDE family proteins contain a conserved EAL or HD-GYP motif. The L. acidophilus La-14 genome (NCBI reference sequence: NC_021181.2) contains a gene (LA14_RS07000, dgcA) encoding the GGDEF domain and two genes (LA14_RS07005, pdeA; LA14_RS07010, pdeB) encoding the EAL domain (Figure 1A); these genes may be involved in the metabolic cycle of c-di-GMP. The EAL-only proteins (PdeA and PdeB) can serve as either active PDEs (class I) or inactive enzymes (class III; El Mouali et al., 2017).
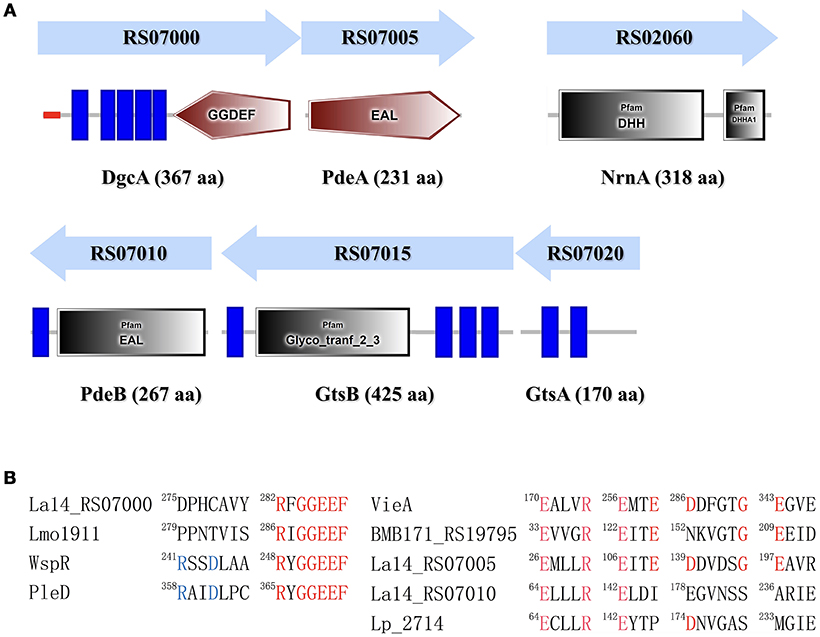
Figure 1. Identification of proteins involved in the c-di-GMP signaling pathway in Lactobacillus acidophilus La-14. (A) Domain symbols are derived from the SMART and Pfam database. In the upper panel, the domain structures are shown under the predicted operon arrangements with gene numbers. The putative name and number of amino acids in each protein are listed at the bottom. Predicted signal peptide and transmembrane regions are shown in red stripes and blue rectangles, respectively. (B) Amino acid sequence alignment of the conserved active site residues for the GGDEF domain (left) and the EAL domain (right). Residues known to be conserved and catalytically important are shown in red. The RXXD motif of the I-site for c-di-GMP binding is shown in blue. Experimentally characterized GGDEF domains are from L. monocytogenes (Lmo1911) (Chen et al., 2014), Pseudomonas aeruginosa (WspR, De et al., 2009), and Caulobacter crescentus (PleD, Paul et al., 2004). Experimentally proven EAL domain sequences are from Vibrio cholerae (VieA) (Tamayo et al., 2005), Bacillus thuringiensis (BMB171_RS19795, Fu et al., 2018), and L. plantarum (LP_2714, Brown et al., 2011).
L. acidophilus La-14 has only one GGDEF domain-containing protein (DgcA; NCBI reference sequence, WP_011254455.1) associated with DGC activity. The N-terminal domain of the predicted DGC protein contains one signal peptide and five transmembrane helices that may sense external signals to regulate c-di-GMP synthesis (Figure 1A). Amino acid sequence alignment (Figure 1B) showed that La14_RS07000 possesses a conserved active site (RxGGDEF) but lacks an inhibitory site (RxxD), similar to L. monocytogenes Lmo1911 (Chen et al., 2014).
The EAL domain protein (Figure 1), La14_RS07005 (PdeA; WP_011254456.1), contains only one EAL domain with conserved residues for c-di-GMP hydrolysis (Tchigvintsev et al., 2010). Bioinformatic analysis predicted that it also lacks the conserved loop 6 [DFG(A/S/T)(G/A)(Y/F)(S/A/T)(S/A/G/V/T)] and adjacent domain that can potentially promote dimerization for enhancing enzymatic activity (Rao et al., 2009). La-14 shared extensive similarity with the NCFM strain during alignment of L. acidophilus genomes (Stahl and Barrangou, 2013). According to ProOpDB, dgcA and pdeA were predicted to belong to the same operon in strain NFCM, whereas we found the opposite prediction in strain La-14. Subsequent biochemical analyses were needed to clarify this contradiction (see section Operon Transcriptional Analysis). The amino acid sequence of another EAL domain protein, La14_RS07010 (PdeB; WP_003548090.1; Figure 1), contains two fractions, a membrane targeting signal sequence and an EAL domain without the residues required for catalysis. Although PdeB appears to lack hydrolysis ability, it retains the c-di-GMP binding site and the conserved EXLXR motif, suggesting that it acts as a receptor protein as previously described (Minasov et al., 2009; Chou and Galperin, 2016). From the c-di-GMP census [http://ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html], there is no other predictable c-di-GMP receptor except for PdeB from the sequence analysis of L. acidophilus NCFM. Thus, the neighboring genes of pdeB—La14_RS07015 and La14_RS07020—emerged as the main genes of interest in our study.
The GtsB protein (WP_011254457.1), encoded by the La14_RS07015 gene nearby pdeB, was described as a glycosyltransferase that functions in the synthesis of cellulose, which is similar to BcsA and PgaC function in Rhodobacter sphaeroides and E.coli, respectively (Steiner et al., 2013; Morgan et al., 2014). Overall, BcsA and GtsB shared 25% amino acid identity and 37% sequence similarity and both belong to glycosyltransferase family 2. GtsB contains an N-terminal and three C-terminal transmembrane domains as well as a predicted cytoplasmic glyco_tranf_2_3 domain (Figure 1A). GtsA (WP_003548094.1), encoded by La14_RS07020 upstream of the pdeB and gtsB genes, was predicted to be a transmembrane protein without any conserved domains. Similar to PgaC-PgaD complex, the membrane-anchored GtsA subunit, together with the GtsB, may form a glycosyltransferase complex (Steiner et al., 2013).
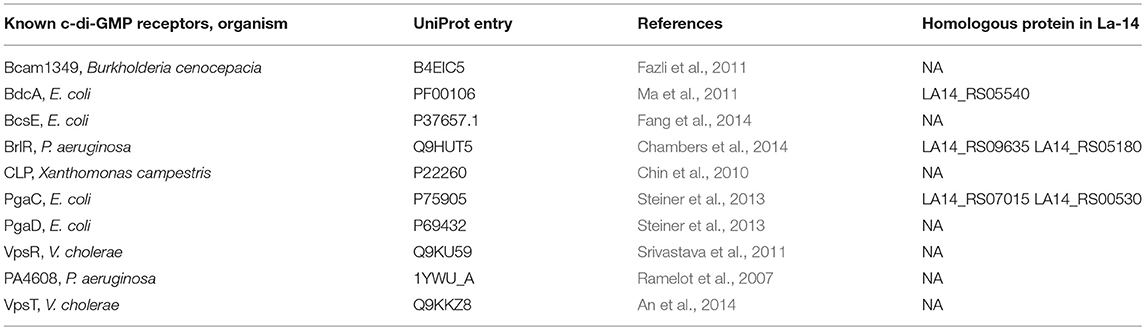
In Pseudomonas aeruginosa, the intermediate molecule pGpG, produced by EAL domains were confirmed to be eventually hydrolyzed to GMP by oligoribonuclease (Cohen et al., 2015). Firmicutes lacks oligoribonuclease but have its homologs protein family nanoRNases (Nrn; clusters of orthologous group: COG0618) instead (Orr et al., 2015). In NCBI protein database, we found an oligoribonuclease functional homologs NrnA (La14_RS02060, WP_011254146.1) which may be responsible for degradation of pGpG in L. acidophilus La-14 (Figure 1A). Besides, according to known c-di-GMP receptors, BlastP was used to search the homologous proteins in La-14. Several putative c-di-GMP receptors were listed in Table 2, but their binding capacity should be confirmed by the biochemical analyses.
Operon Transcriptional Analysis
Through bioinformatics prediction, a promoter region at position −264 or −653 upstream of dgcA was found. Amplified product A (dgcA to position −831) contained both predicted promoter regions, while amplified product B (dgcA to position −638) only contained the promoter region at −264. Based on the principle that promoter sequences can't be transcribed, the corresponding size of B appeared while the A fragment did not (Figure 2), suggesting that the promoter sequence of dgcA is at position −653. Amplified products in the C (dgcA to pdeA), E (gtsA to gtsB), and F (gtsB to pdeB) regions indicate that dgcA and pdeA form an operon, while pdeB, gtsB, and gtsA form another operon named gts on the L. acidophilus chromosome. Therefore, the results suggest that dgcA and pdeA are under the control of a single promoter in an operon and are involved in c-di-GMP cycling.
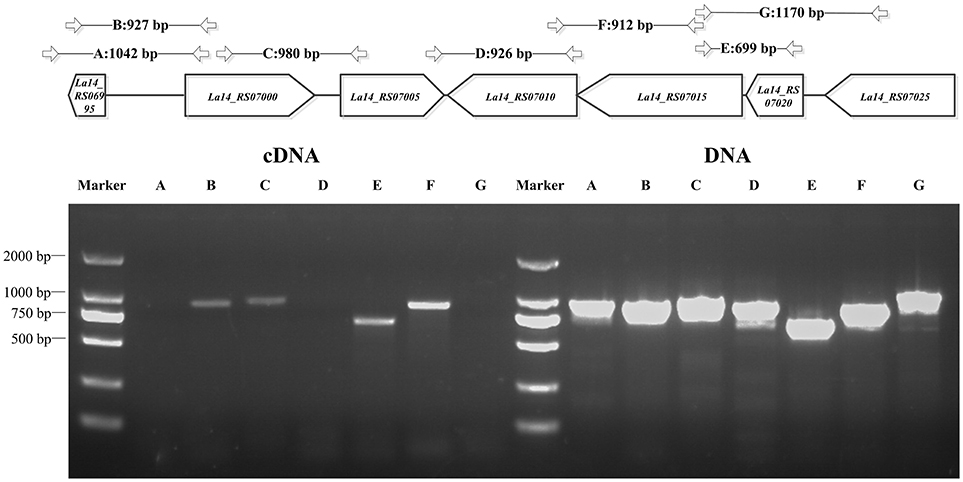
Figure 2. PCR for L. acidophilus transcriptional analysis. Upper panel: genes of interest (in the box) on the chromosome and the length of amplified PCR products (in bp) with specific primer pairs (Table S1) that span the sequences, dgcA to its −831 or −638 bp positions, dgcA to pdeA, pdeA to pdeB, gtsA to gtsB, gtsB to pdeB, and La14_7025 to gtsA. Lower panel: 1% agarose gel PCR analysis of dgcA, pdeA, pdeB, gts, and adjacent genes with specific primer pairs that were used to amplify both gDNA (right half) and cDNA (left half) from L. acidophilus. The letters A–G correspond to the amplified products of gene sequences indicated in the upper panel. The results indicate that the amplified A, D, and G PCR products are non-consecutive, while B, C, E, and F are consecutive in cDNA. Lanes: M, 2,000 bp DNA Ladder; cDNA, La-14 complementary DNA; DNA, La-14 genomic DNA.
DGC Activity Assays in vivo
The DGC activity of L. acidophilus DgcA was analyzed by Congo red staining and swarming motility assays on Congo red plates and 0.6% agar plates, respectively. The binding of Congo red was associated with the production of EPS or curli fimbriae (Olsén et al., 1989). When concentrations of the inducer L-arabinose increased, colonies expressing DgcA were red-stained, dry, and rough compared with the empty vector-containing negative control (Figure 3A). In swarming motility assays for assessing another c-di-GMP-dependent phenotype, the motility of E. coli MG1655 containing the pBAD-dgcA plasmid was highly inhibited compared with the control group (Figure 3B). Both assays suggested that colonies expressing DgcA contained a higher content of c-di-GMP.
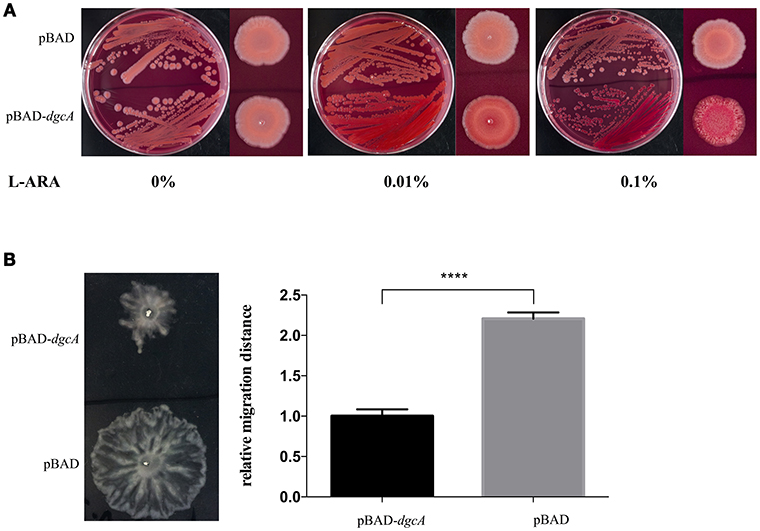
Figure 3. Congo red staining and swarming motility assays. (A) Congo red staining of the EPS-producing strain BL21 caused by L. acidophilus DgcA, indicative of its DGC activity. DgcA was expressed with the plasmid pBAD-Myc-His. LB agar contained 0–0.1% L-arabinose. (B) Inhibition of motility in swarming plates (0.5%) of strain MG1655 by DgcA supports its DGC activity. LB agar contained 0.5% arabinose. Average results from three independent tests of the swarm zones. ****significant difference (p < 0.0001). GraphPad Prism 6.1 was used to perform Student's t-tests.
PDE Activity Assays in vitro
To directly measure c-di-GMP PDE activity in vitro, PdeA and PdeB were expressed and purified to determine whether they can hydrolyze c-di-GMP. PdeA and PdeB were purified as MBP-fusion proteins (Figure 4A). The PdeA enzymatic reaction product corresponded to the retention time of the pGpG [5′-phosphoguanylyl-(3′ → 5′)-guanosine] standard, indicating that PdeA was able to hydrolyze c-di-GMP to pGpG. However, the PdeB enzymatic reaction product corresponded to the retention time of the c-di-GMP standard, indicating that PdeB does not possess PDE activity in vitro (Figures 4B,C).
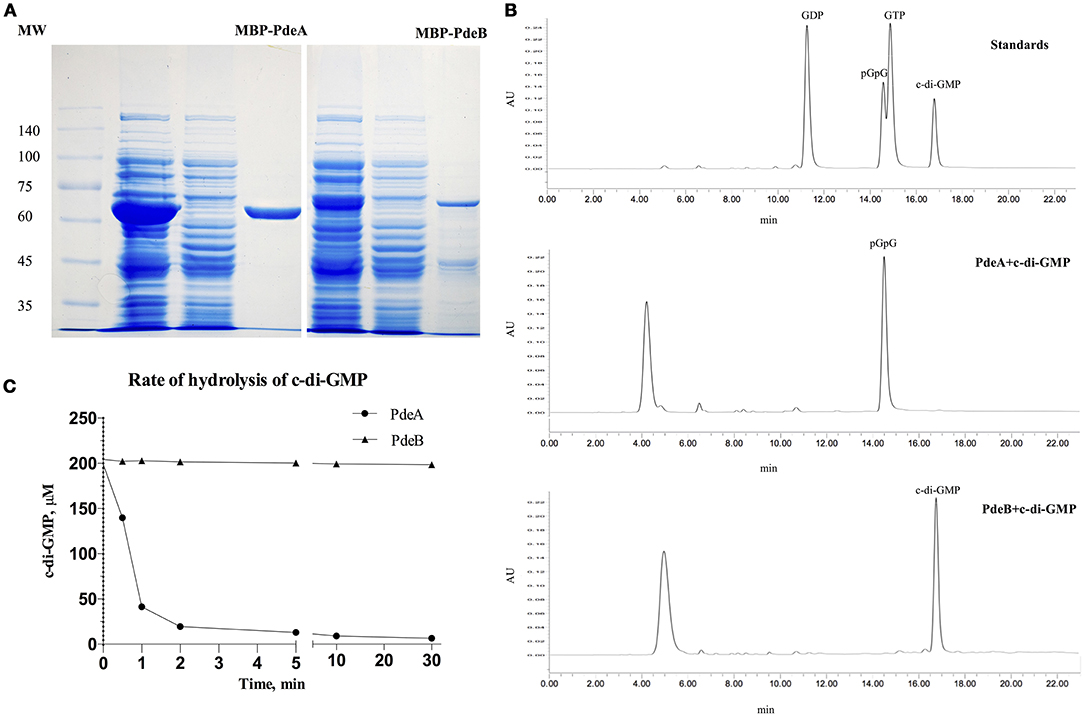
Figure 4. Phosphodiesterase (PDE) activity and HPLC assays. (A) 12% SDS-PAGE of L. acidophilus PdeA (MBP-PdeA) and PdeB (MBP-PdeB) affinity purification to be used in PDE assays. (B) HPLC chromatograms showing the standards and products of PDE assays. (C) PDE activities of MBP-PdeA and MBP-PdeB were monitored based on the hydrolysis rate of c-di-GMP measured by HPLC.
PdeB Protein is a c-di-GMP Receptor
To demonstrate that the PdeB protein acts as a c-di-GMP-specific receptor, we overexpressed its EAL domain (MBP-EALPdeB) containing the ELLLR substrate binding site as an MPB-fusion protein (Figure 5A) and tested its ability to bind c-di-GMP through DRaCALA and equilibrium dialysis. According to the results of the competitive binding assay, excessive unlabeled c-di-GMP competed for Fluo-c-di-GMP and MBP-EALPdeB binding effectively (P < 0.001; Figure 5B), whereas GTP and pGpG did not, indicating that PdeB can bind c-di-GMP specifically. EALPdeB bound c-di-GMP with a Kd of 4.871 ± 0.89 μM and a Bmax of 1.158 ± 0.07 μM c-di-GMP (μM protein)−1 (Figure 5C). Kd value was in the range of 0.1–13 μM, which was consistent with the Kd ranges of other EAL domain (EXLXR motif)-based proteins (FimX or LapD; Chou and Galperin, 2016).
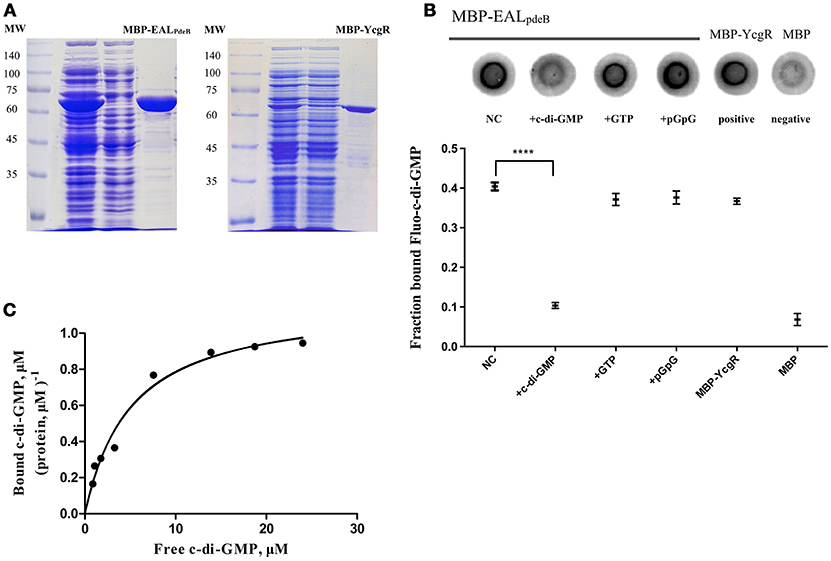
Figure 5. DRaCALA and equilibrium dialysis experiments for the c-di-GMP-specific receptor. (A) SDS-PAGE of MBP-EALpdeB and MBP-YcgR overexpressed in BL21 and purified with amylose resin. The EAL domain of PdeB (residues 32–267) with an ELLLR motif was fused with MBP and used in c-di-GMP binding assays. (B) Binding of Fluo-c-di-GMP to PdeB (MBP-EALpdeB) in DRaCALA. Upper panel: Fluo-c-di-GMP in the presence of increasing concentrations of unlabeled c-di-GMP as a competitor. Lower panel: graph of FB for each sample with averages indicated by a horizontal bar of three independent experiments. ****Significant difference (p < 0.0001). GraphPad Prism 6.1 was used to perform Student's t-tests. (C) Equilibrium binding between c-di-GMP and MBP-EALpdeB. The chart represents the ratio of bound c-di-GMP per protein unit in the dialysis chamber vs. the concentration of free c-di-GMP in another chamber.
Overexpression of GtsA and GtsB Increases EPS Synthesis in E. coli
Cellulose and poly-N-acetylglucosamine increase biofilm formation of E. coli on abiotic surfaces (Wang et al., 2004). To analyze the function of the gts operon, we cloned its coding sequence of gtsA and gtsB into the pBAD-Myc-His vector, followed by E. coli (BL21) transformation. Utilizing heterologous expression allowed us to assess the effects of the protein of interest without additionally impacting protein-protein interactions. On Congo red plates, compared with control, the expression of pBAD-gts resulted in red color colonies but with a lesser extent than pBAD-dgcA (Figures 6A,B). Moreover, we performed a crystal violet staining assay to examine biofilm formation. Under induction, an increase in biofilm formation was observed in the colonies expressing pBAD-dgcA and pBAD-gts (Figure 6C). The pBAD-gts group exhibited a somewhat similar phenotype to the pBAD-dgcA group, which may be due to the lack of PdeB c-di-GMP receptor activation. These results suggest that gts is associated with the formation of bacterial EPS.
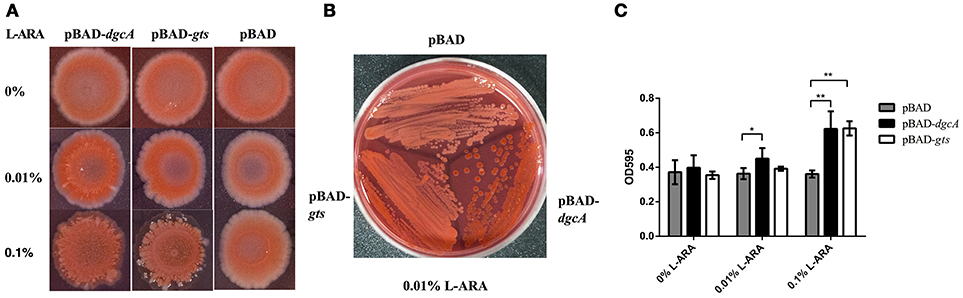
Figure 6. Congo red staining and biofilm formation assays. (A,B) Congo red staining of the EPS-producing strain BL21 was caused by L. acidophilus DgcA or Gts. Proteins were expressed by cloning the corresponding genes into the pBAD-Myc-His vector. LB agar contained 0–0.1% L-arabinose. (C) Biofilm formation of E. coli BL21 transformed with different recombinant plasmids in 96-well plates with 0–0.1% L-arabinose. Bars denote mean values of the data from three biological replicates. *significant difference (p < 0.05); **significant difference (p < 0.01). GraphPad Prism 6.1 was used to perform Student's t-tests.
Intracellular DgcA and PdeA Levels Regulate Bacterial Form and EPS Formation in L. acidophilus
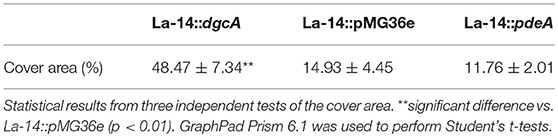
After analyzing the functional components of c-di-GMP signaling in L. acidophilus, we determined the phenotypes associated with increased intracellular DgcA or PdeA levels. We overexpressed DgcA or PdeA in L. acidophilus La-14 and analyzed their roles in EPS formation. The covered area on the coverslip surface by La-14 and its recombinant strains was shown in Table 3. Compared with the vector control, the strain expressing DgcA adhered more biomass to the coverslip, formed a smaller size of cell and more compact structures (Figure 7Aa), whereas the strain expressing PdeA grew in short rod-shaped chains, and stained in lighter blue (Figure 7Ac). Among these photos, the difference of EPS production level was not obvious. Then we performed a Congo red assay to detect EPS and found that the strain expressing DgcA exhibited redder colonies compared with other strains (Figure 7B). This result suggest that DgcA may promote the formation of EPS in L. acidophilus.
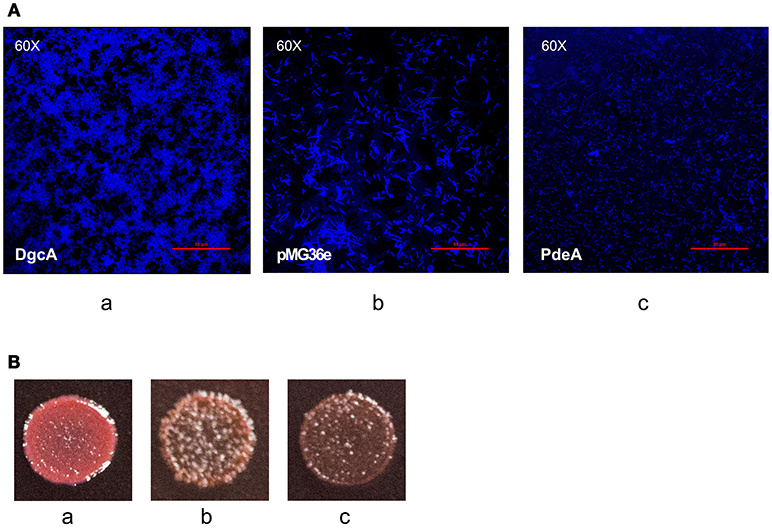
Figure 7. EPS formation analysis with confocal microscopy and Congo red staining. (A) Confocal microscopy for examining (a) La-14::dgcA; (b) La-14::pMG36e; (c) La-14::pdeA. The blue color indicates EPS formation. (B) Congo red staining of the La-14 strain is indicative of EPS production. (a) La-14::dgcA; (b) La-14::pMG36e; (c) La-14::pdeA.
DgcA/PdeA-Induced L. acidophilus EPS Promotes Co-Aggregation With E. coli
In the presence of other bacteria or fungi, lactobacilli strains usually exhibit strong co-aggregation phenotypes, which is a characteristic of probiotics (Collado et al., 2008; Chew et al., 2015). To test the role of DgcA/PdeA-induced EPS formation in L. acidophilus, the co-aggregation of this strain compared with E. coli was evaluated. The settling rate was determined within 5 h. During the first hour, the three strains showed similar autogenesis rates. From 1 to 5 h, the co-aggregation rate of La-14::dgcA was significantly faster (P < 0.01) than those of La-14::pMG36e and La-14::pdeA (Figure 8). However, there was no significant difference between La-14::pMG36e and La-14::pdeA throughout the assay.
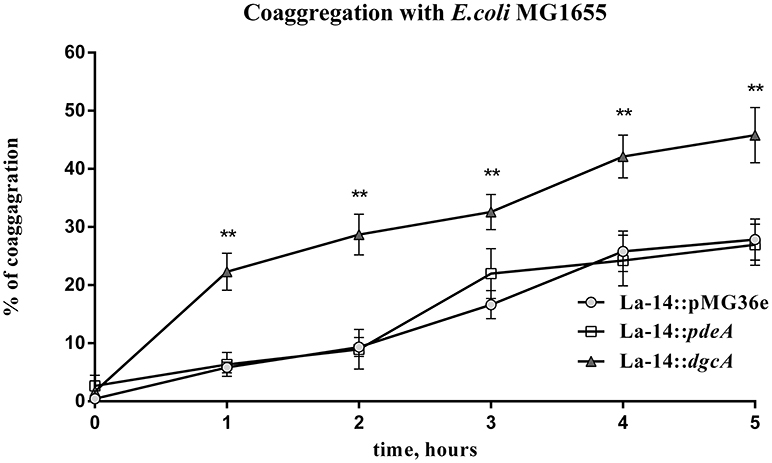
Figure 8. L. acidophilus co-aggregation assay with E. coli MG1655. Co-aggregation abilities of recombinant La-14 with potential pathogen E. coli MG1655 were measured in 96-well polystyrene plates. The triangle represents La-14::dgcA; circle, La-14::pMG36e; square, La-14::pdeA. Data represent the average results from two independent experiments of each strain grown in three wells. **significant difference (p < 0.01) between La-14::dgcA and control group (La-14::pMG36e). GraphPad Prism 6.1 was used to perform Student's t-tests.
Discussion
Bacteria adapt to environmental stresses through changes in EPS and proteinaceous appendages. Such adaptations are regulated in various bacteria by proteins with GGDEF and EAL domains, which involve the second messenger c-di-GMP. The GGDEF domain acts as a nucleotide cyclase for c-di-GMP synthesis, whereas the EAL domain acts as a phosphodiesterase for c-di-GMP degradation. As L. acidophilus is a probiotic and a significant bacterium present in the digestive tract, it is important to investigate the probiotic properties of probiotics. In the present study, we aimed to address whether there are c-di-GMP regulator-related genes in the L. acidophilus genome, how the genes are organized into a cluster to form the operons, and the roles these genes play in conducting their biological functions.
First, we collected La-14 genomic information through the NCBI (GenBank) website. Subsequently, bioinformatic analysis for c-di-GMP related genes and proteins were performed. In combination with our experimental data, we confirmed the presence of one Dgc gene (dgcA), two copies of Pde (pdeA and pdeB), one copy of nanoRNases gene (nrnA), and one copy of each Gts subunit gene, gtsA (subunit A) and gtsB (subunit B), on the L. acidophilus chromosome (Figure 1A). Then.we confirmed that dgcA and pdeA belong to one operon while pdeB, gtsA, and gtsB belong to another.
The conformation of GGDEF and EAL domains affects the catalytic function of DGC and PDE enzymes, respectively (Chan et al., 2004). In our experiment, PdeA exhibited PDE activities in vitro and DgcA induced two c-di-GMP-dependent phenotypes (low motility and high production of curli fimbriae) in E. coli by heterologously expressed in vivo. However, even though PdeB has no enzymatic activity, it can bind to c-di-GMP as a receptor. Moreover, we found that the GGDEF and EAL motifs of DgcA and PdeA, respectively, were homologous to the conserved motifs of other bacterial strains in which the two enzymes retain their catalytic activities (Figure 1B). Similarly, the amino acid sequences of DgcA and PdeA in La-14 also showed higher sequence homology with other members of the Lactobacillus family such as L. amylovorus, L. kalixensis, and L. reuteri (Figures S1A,B). This suggests that the c-di-GMP signaling pathway seem to be involved in Lactobacillus strain behavior regulation throughout evolution. However, the EAL motif of PdeB showed less homology with other known PDEs (Figure 1B). We believe that the structural differences in PdeB resulted in its lack of c-di-GMP hydrolase activity. In the EAL domain, the conserved motif DDFG(T/A)GYSS plays an important role in positioning Mg2+ for catalytic activity (Römling, 2009); however, there is no such motif in PdeB and instead, the EGVNSSARIE motif is present (Figure 1B). In fact, several EAL domain-containing PDEs with variations in some of these conserved residues lack PDE activity but retain a regulatory role. Furthermore, the GGDEF domain in DgcA lacks the conserved RxxD motif that is used as an inhibitory site for receiving feedback regulation by c-di-GMP in other bacterial strains (Figure 1B). DgcA expressed without the N-terminal hydrophobic sequence exhibited no activity in vitro (data not shown) after we observed no DgcA activity when the full-length dcgA gene was transformed in bacterial cells. This may be due to the truncation of its N-terminal transmembrane domain, as this domain is disadvantageous for expression of soluble proteins. In addition, lack of the transmembrane domain, which may be essential for receiving external signals, will prevent the GGDEF domains from forming active homodimers (Paul et al., 2007). Many DGCs and PDEs contain responsive regulator (REC) domains that can receive input signals for responding to environmental stimulation. A well-characterized P. aeruginosa strain contains several DGCs and PDEs that regulate cellular c-di-GMP levels and sense input signals, such as chemoattractants (WspR) and oxygen-deprived conditions (SadC), to alter intracellular c-di-GMP levels (O'Connor et al., 2012; Schmidt et al., 2016). For Gram-negative bacteria, the transmembrane GGDEF protein is generally located on the inner membrane and can form components of the response pathway with sensory proteins located in the periplasm or outer membrane (Kim and Harshey, 2016; Schmidt et al., 2016). However, the study of transmembrane GGDEF proteins is relatively poor in Gram-positive bacteria with only a single layer of membrane structure. For example, DgcK, a typical transmembrane DGC protein, has a synergistic effect with the degenerated GGDEF-transmembrane protein Ydak to regulate the production of an unknown EPS in Gram-positive B. subtilis (Bedrunka and Graumann, 2017b). There are five transmembrane helices in DgcA with a similar structure to DgcK, which belongs to the 5TMR-LYT family (5 transmembrane receptors of the LytS-YhcK type; PF07694), although they lack similarity in amino acid sequence alignment. However, the type of input signals that activate DgcK via 5TMR-LYT remain unknown.
We showed that DgcA is responsible for several phenotypes involved in biofilm formation, EPS synthesis, and co-aggregation in L. acidophilus. EPSs of probiotics are important in alleviating lactose intolerance, enhancing immunity against pathogens, and reducing mutagenic enzymes, such as β-glucuronidase, nitroreductase, and choloylglycine hydrolase (de Roos and Katan, 2000). In our experiments, DgcA expressed without the N-terminal hydrophobic sequence exhibited no activity in vitro, so we tried to prove the DGC activity by in vivo assay referred to a previous study (Chen et al., 2014; Purcell and Tamayo, 2016). The related results can be compared from the intracellular expression of pBAD vector and pBAD-dgcA (Figures 3, 6). Especially, when we in vivo expressed empty pMG36e vector and pMG36e-dgcA in La-14 respectively the EPS formation and co-aggregation are significantly higher (Figures 7, 8). All these functional tests proved that the DgcA has its activity in vivo. The functions of DgcA protein could be achieved in vivo by both through a c-di-GMP dependent (Chen et al., 2014) and independent (Holland et al., 2008) mechanism. On the other hand, the PDE activity of PdeA and the c-di-GMP receptor (PdeB) have been confirmed in assays in vitro. Combined with the evidence of in vivo assay, DgcA may be involved in c-di-GMP metabolism in L. acidophilus. Because of the concentration of c-di-GMP is hardly measured in the DgcA expressed bacterial lysate (containing complex components, data not shown) with HPLC used in our experiments, so the functions of DgcA could be achieved in vivo by both of the mechanisms which we will identify in the next step. Overexpression of PdeA resulted in changes in structure, but with no phenotypic changes observed in Congo red or co-aggregation assays. We hypothesized that this phenomenon was due to the low background concentration of intracellular c-di-GMP levels in L. acidophilus La-14 (only one copy of diguanylate cyclase gene in its genome).
It has been shown that L. acidophilus EPS is responsible for cell co-aggregation, which is an important characteristic of Lactobacillus that plays a critical role in its vitality (Goh and Klaenhammer, 2010). The operon gts encoded a BcsA-like glycosyltransferase (GtsB) and a hypothetical protein (GtsA) with double transmembrane loops, whose function appears to be involved in bacterial capsule biosynthesis, like cellulose or polymeric N-acetyl-glucosamine synthases and is associated with bacterial biofilm formation (Itoh et al., 2005; O'Gara, 2007; Morgan et al., 2013). Considering the above information, our data indicate that the genes La14_RS07015 to La14_RS07020 may be involved in L. acidophilus EPS formation through an unknown synthesis pathway (Figure 6). GtsB may serve as a poly-beta-1,6-N-acetyl-D-glucosamine or a catalytic subunit of poly-beta-1,4-D-glucopyranose synthase, while GtsA functions as synthase regulatory subunit. PdeB may bind to c-di-GMP to allosterically modulate enzymatic functions of GtsA/B through protein-protein interactions. The function of gts operon-encoded proteins in L. acidophilus may be similar to the Pss EPS synthase in L. monocytogenes or the cellulose synthase in R. sphaeroides (Omadjela et al., 2013; Chen et al., 2014; Köseoglu et al., 2015). Based on the references and our experiments, we speculate that c-di-GMP can bind with the PdeB and induce conformation changes through allosteric regulation of PdeB. This allosteric effect will remove the inhibitory interactions on the PdeB-GtsA/B complex or activate the idle state of GtsA/B to enhance the catalytic activity of GtsA/B (subunits A and B forming a glycosyltransferase) in EPS synthesis. The resulting production of EPS may increase the intercellular adhesion capacity of L. acidophilus and promote it to a higher aggregative state, both of which are characteristics of L. acidophilus as a probiotic. This allows L. acidophilus to colonize the host (oral cavity, gastrointestinal tract, and vagina) more easily and provides an advantage during bacterial competition in biofilms. Although the composition of L. acidophilus EPS remains unclear, we uncovered a potential regulatory pathway where input signals regulate L. acidophilus EPS production via intracellular DgcA and PdeA, allowing for physiological changes in the bacteria to cope with changes in the external environment.
Our study demonstrated that L. acidophilus might have a complete signaling system, regulating intracellular c-di-GMP levels, or a c-di-GMP-independent mechanism (depending on the direct evidence whether the DgcA could synthesize c-di-GMP to be got), both of which in turn could regulate EPS synthesis and coaggregation. However, some questions remain regarding c-di-GMP signaling in Lactobacillus, including whether the transmembrane protein DgcA actually synthesize c-di-GMP in L. acidophilus and how DgcA is involved in upstream signaling to control c-di-GMP synthesis, the composition of Gts EPS, and whether Gts EPS contributes to other phenotypes in L. acidophilus. Further studies should be conducted to better understand this process.
Author Contributions
JH designed and did the experiments with gene construction, culture experiments, biochemical tests, analyzed data, and wrote the manuscript. WR and JS did the experiments with biochemical tests. WY and FW provided overall directions and contributed to revising the manuscript.
Funding
Supported by National Natural Science Foundation of China (81100747) and the Guangdong Science and Technology Planning Project, China (2014A020211016).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01935/full#supplementary-material
Figure S1. Multiple alignment results using a column-based method. The red color indicates highly conserved columns and the blue indicates less conserved ones. (A) Amino acid sequence alignment of conserved DgcA residues with other homologous proteins from L. amylovorus, L. kalixensis, L. crispatus, L. frumenti, L. vaginalis, and L. reuteri. (B) Amino acid sequence alignment of conserved PdeA residues with other homologous proteins from L. amylovorus, L. crispatus, L. kalixensis, L. vaginalis, L. frumenti, and L. reuteri.
Table S1. Primers used in the analysis of operon transcription.
References
An, S. Q., Caly, D. L., McCarthy, Y., Murdoch, S. L., Ward, J., Febrer, M., et al. (2014). Novel cyclic di-GMP effectors of the YajQ protein family control bacterial virulence. PLoS Pathog. 10:e1004429. doi: 10.1371/journal.ppat.1004429
Bâati, L., Fabre-Gea, C., Auriol, D., and Blanc, P. J. (2000). Study of the cryotolerance of Lactobacillus acidophilus: effect of culture and freezing conditions on the viability and cellular protein levels. Int. J. Food Microbiol. 59, 241–247. doi: 10.1016/S0168-1605(00)00361-5
Bedrunka, P., and Graumann, P. L. (2017a). Subcellular clustering of a putative c-di-GMP-dependent exopolysaccharide machinery affecting macro colony architecture in Bacillus subtilis. Environ. Microbiol. Rep. 9, 211–222. doi: 10.1111/1758-2229.12496
Bedrunka, P., and Graumann, P. L. (2017b). New functions and subcellular localization patterns of c-di-GMP components (GGDEF Domain Proteins) in B. subtilis. Front. Microbiol. 8:794. doi: 10.3389/fmicb.2017.00794
Blain-Hartung, M., Rockwell, N. C., and Lagarias, J. C. (2017). Light-regulated synthesis of cyclic-di-GMP by a bidomain construct of the cyanobacteriochrome Tlr0924 (SesA) without stable dimerization. Biochemistry 56, 6145–6154. doi: 10.1021/acs.biochem.7b00734
Brown, R., Marchesi, J. R., and Morby, A. P. (2011). Functional characterisation of Lp_2714, an EAL-domain protein from Lactobacillus plantarum. Biochem. Biophys. Res. Commun. 411, 132–136. doi: 10.1016/j.bbrc.2011.06.112
Burns, J. L., Rivera, S., Deer, D. D., Joynt, S. C., Dvorak, D., and Weinert, E. E. (2016). Oxygen and bis(3′,5′)-cyclic dimeric guanosine monophosphate binding control oligomerization state equilibria of diguanylate cyclase-containing globin coupled sensors. Biochemistry 55, 6642–6651. doi: 10.1021/acs.biochem.6b00526
Chambers, J. R., Liao, J., Schurr, M. J., and Sauer, K. (2014). BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol. Microbiol. 92, 471–487. doi: 10.1111/mmi.12562
Chan, C., Paul, R., Samoray, D., Amiot, N. C., Giese, B., Jenal, U., et al. (2004). Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 101, 17084–17089. doi: 10.1073/pnas.0406134101
Chen, L. H., Köseoglu, V. K., Güvener, Z. T., Myers-Morales, T., Reed, J. M., D'Orazio, S. E., et al. (2014). Cyclic di-GMP-dependent signaling pathways in the pathogenic Firmicute Listeria monocytogenes. PLoS Pathog. 10:e1004301. doi: 10.1371/journal.ppat.1004301
Chew, S. Y., Cheah, Y. K., Seow, H. F., Sandai, D., and Than, L. T. (2015). Probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 exhibit strong antifungal effects against vulvovaginal candidiasis-causing Candida glabrata isolates. J. Appl. Microbiol. 118, 1180–1190. doi: 10.1111/jam.12772
Chin, K. H., Lee, Y. C., Tu, Z. L., Chen, C. H., Tseng, Y. H., Yang, J. M., et al. (2010). The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J. Mol. Biol. 396, 646–662. doi: 10.1016/j.jmb.2009.11.076
Chou, S. H., and Galperin, M. Y. (2016). Diversity of cyclic Di-GMP-binding proteins and mechanisms. J. Bacteriol. 198, 32–46. doi: 10.1128/JB.00333-15
Cohen, D., Mechold, U., Nevenzal, H., Yarmiyhu, Y., Randall, T. E., Bay, D. C., et al. (2015). Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 11359–11364. doi: 10.1073/pnas.1421450112
Collado, M. C., Meriluoto, J., and Salminen, S. (2008). Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 226, 1065–1073. doi: 10.1007/s00217-007-0632-x
de Roos, N. M., and Katan, M. B. (2000). Effects of probiotic bacteria on diarrhea, lipid metabolism, and carcinogenesis: a review of papers published between 1988 and 1998. Am. J. Clin. Nutr. 71, 405–411. doi: 10.1093/ajcn/71.2.405
De, N., Navarro, M. V., Raghavan, R. V., and Sondermann, H. (2009). Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 393, 619–633. doi: 10.1016/j.jmb.2009.08.030
Deepak, V., Ramachandran, S., Balahmar, R. M., Pandian, S. R., Sivasubramaniam, S. D., Nellaiah, H., et al. (2016). In vitro evaluation of anticancer properties of exopolysaccharides from Lactobacillus acidophilus in colon cancer cell lines. In vitro Cell. Dev. Biol. Anim. 52, 163–173. doi: 10.1007/s11626-015-9970-3
den Hengst, C. D., Tran, N. T., Bibb, M. J., Chandra, G., Leskiw, B. K., and Buttner, M. J. (2010). Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78, 361–379. doi: 10.1111/j.1365-2958.2010.07338.x
El Mouali, Y., Kim, H., Ahmad, I., Brauner, A., Liu, Y., Skurnik, M., et al. (2017). Stand-alone EAL domain proteins form a distinct subclass of EAL proteins involved in regulation of cell motility and biofilm formation in enterobacteria. J. Bacteriol. 199:e00179-17. doi: 10.1128/JB.00179-17
Fang, X., Ahmad, I., Blanka, A., Schottkowski, M., Cimdins, A., Galperin, M. Y., et al. (2014). GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol. Microbiol. 93, 439–452. doi: 10.1111/mmi.12672
Fazli, M., O'Connell, A., Nilsson, M., Niehaus, K., Dow, J. M., Givskov, M., et al. (2011). The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 82, 327–341. doi: 10.1111/j.1365-2958.2011.07814.x
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Fu, Y., Yu, Z., Liu, S., Chen, B., Zhu, L., Li, Z., et al. (2018). c-di-GMP regulates various phenotypes and insecticidal activity of gram-positive Bacillus thuringiensis. Front. Microbiol. 9:45. doi: 10.3389/fmicb.2018.00045
Gao, X., Mukherjee, S., Matthews, P. M., Hammad, L. A., Kearns, D. B., and Dann, C. R. (2013). Functional characterization of core components of the Bacillus subtilis cyclic-di-GMP signaling pathway. J. Bacteriol. 195, 4782–4792. doi: 10.1128/JB.00373-13
Goh, Y. J., and Klaenhammer, T. R. (2010). Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 76, 5005–5012. doi: 10.1128/AEM.00030-10
Ha, D. G., and O'Toole, G. A. (2015). c-di-GMP and its Effects on Biofilm Formation and Dispersion: a Pseudomonas aeruginosa review. Microbiol. Spectr. 3:MB-0003–2014. doi: 10.1128/microbiolspec.MB-0003-2014
Harshey, R. M., and Matsuyama, T. (1994). Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. U.S.A. 91, 8631–8635. doi: 10.1073/pnas.91.18.8631
Hengge, R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. doi: 10.1038/nrmicro2109
Holland, L. M., O'Donnell, S. T., Ryjenkov, D. A., Gomelsky, L., Slater, S. R., Fey, P. D., et al. (2008). A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J. Bacteriol. 190, 5178–5189. doi: 10.1128/JB.00375-08
Itoh, Y., Wang, X., Hinnebusch, B. J., Preston, J. R., and Romeo, T. (2005). Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187, 382–387. doi: 10.1128/JB.187.1.382-387.2005
Johnson, B. R., and Klaenhammer, T. R. (2016). AcmB is an S-Layer-associated beta-N-acetylglucosaminidase and functional autolysin in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 82, 5687–5697. doi: 10.1128/AEM.02025-16
Kim, H. K., and Harshey, R. M. (2016). A diguanylate cyclase acts as a cell division inhibitor in a two-step response to reductive and envelope stresses. mBio 7:e00822-16. doi: 10.1128/mBio.00822-16
Kim, H. S., and Gilliland, S. E. (1983). Lactobacillus acidophilus as a dietary adjunct for milk to aid lactose digestion in humans. J. Dairy Sci. 66, 959–966. doi: 10.3168/jds.S0022-0302(83)81887-6
Konstantinov, S. R., Smidt, H., de Vos, W. M., Bruijns, S. C., Singh, S. K., Valence, F., et al. (2008). S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U.S.A. 105, 19474–19479. doi: 10.1073/pnas.0810305105
Köseoglu, V. K., Heiss, C., Azadi, P., Topchiy, E., Güvener, Z. T., Lehmann, T. E., et al. (2015). Listeria monocytogenes exopolysaccharide: origin, structure, biosynthetic machinery and c-di-GMP-dependent regulation. Mol. Microbiol. 96, 728–743. doi: 10.1111/mmi.12966
Lin, C. S., Liu, Y., Li, Y., Jun, Ting, H., Kohli, G. S., Cai, Z., et al. (2017). Reduced intracellular c-di-GMP content increases expression of quorum sensing-regulated genes in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 7:451. doi: 10.3389/fcimb.2017.00451
Lin, R., Zhang, Y., Long, B., Li, Y., Wu, Y., Duan, S., et al. (2017). oral immunization with recombinant Lactobacillus acidophilus expressing espA-Tir-M confers protection against enterohemorrhagic Escherichia coli O157:H7 challenge in mice. Front. Microbiol. 8:417. doi: 10.3389/fmicb.2017.00417
Looijesteijn, P. J., Trapet, L., de Vries, E., Abee, T., and Hugenholtz, J. (2001). Physiological function of exopolysaccharides produced by Lactococcus lactis. Int. J. Food Microbiol. 64, 71–80. doi: 10.1016/S0168-1605(00)00437-2
Ma, Q., Yang, Z., Pu, M., Peti, W., and Wood, T. K. (2011). Engineering a novel c-di-GMP-binding protein for biofilm dispersal. Environ. Microbiol. 13, 631–642. doi: 10.1111/j.1462-2920.2010.02368.x
Martínez, M. G., Prado, Acosta, M., Candurra, N. A., and Ruzal, S. M. (2012). S-layer proteins of Lactobacillus acidophilus inhibits JUNV infection. Biochem. Biophys. Res. Commun. 422, 590–595. doi: 10.1016/j.bbrc.2012.05.031
Matsuyama, B. Y., Krasteva, P. V., Baraquet, C., Harwood, C. S., Sondermann, H., and Navarro, M. V. (2016). Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 113, E209–E218. doi: 10.1073/pnas.1523148113
Minasov, G., Padavattan, S., Shuvalova, L., Brunzelle, J. S., Miller, D. J., Baslé, A., et al. (2009). Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J. Biol. Chem. 284, 13174–13184. doi: 10.1074/jbc.M808221200
Möller, S., Croning, M. D., and Apweiler, R. (2001). Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17, 646–653. doi: 10.1093/bioinformatics/17.7.646
Morgan, J. L., McNamara, J. T., and Zimmer, J. (2014). Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol. 21, 489–496. doi: 10.1038/nsmb.2803
Morgan, J. L., Strumillo, J., and Zimmer, J. (2013). Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493, 181–186. doi: 10.1038/nature11744
O'Connor J. R Kuwada N. J Huangyutitham V Wiggins P. A Harwood C. S. (2012). Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol. Microbiol. 86, 720–729. doi: 10.1111/mmi.12013
O'Gara, J. P. (2007). ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188. doi: 10.1111/j.1574-6968.2007.00688.x
Olsén, A., Jonsson, A., and Normark, S. (1989). Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338, 652–655. doi: 10.1038/338652a0
Omadjela, O., Narahari, A., Strumillo, J., Mélida, H., Mazur, O., Bulone, V., et al. (2013). BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 110, 17856–17861. doi: 10.1073/pnas.1314063110
Orr, M. W., Donaldson, G. P., Severin, G. B., Wang, J., Sintim, H. O., Waters, C. M., et al. (2015). Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc. Natl. Acad. Sci. U.S.A. 112, E5048–E5057. doi: 10.1073/pnas.1507245112
Orr, M. W., and Lee, V. T. (2016). A PilZ domain protein for chemotaxis adds another layer to c-di-GMP-mediated regulation of flagellar motility. Sci. Signal. 9:fs16. doi: 10.1126/scisignal.aai8859
O'Toole, G. A., and Kolter, R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461. doi: 10.1046/j.1365-2958.1998.00797.x
Papadopoulos, J. S., and Agarwala, R. (2007). COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. doi: 10.1093/bioinformatics/btm076
Paul, K., Nieto, V., Carlquist, W. C., Blair, D. F., and Harshey, R. M. (2010). The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38, 128–139. doi: 10.1016/j.molcel.2010.03.001
Paul, R., Abel, S., Wassmann, P., Beck, A., Heerklotz, H., and Jenal, U. (2007). Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J. Biol. Chem. 282, 29170–29177. doi: 10.1074/jbc.M704702200
Paul, R., Weiser, S., Amiot, N. C., Chan, C., Schirmer, T., Giese, B., et al. (2004). Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18, 715–727. doi: 10.1101/gad.289504
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Purcell, E. B., McKee, R. W., McBride, S. M., Waters, C. M., and Tamayo, R. (2012). Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 194, 3307–3316. doi: 10.1128/JB.00100-12
Purcell, E. B., and Tamayo, R. (2016). Cyclic diguanylate signaling in Gram-positive bacteria. FEMS Microbiol. Rev. 40, 753–773. doi: 10.1093/femsre/fuw013
Ramelot, T. A., Yee, A., Cort, J. R., Semesi, A., Arrowsmith, C. H., and Kennedy, M. A. (2007). NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins 66, 266–271. doi: 10.1002/prot.21199
Rao, F., Qi, Y., Chong, H. S., Kotaka, M., Li, B., Li, J., et al. (2009). The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J. Bacteriol. 191, 4722–4731. doi: 10.1128/JB.00327-09
Rinaldo, S., Giardina, G., Mantoni, F., Paone, A., and Cutruzzolà, F. (2018). Beyond nitrogen metabolism: nitric oxide, cyclic-di-GMP and bacterial biofilms. FEMS Microbiol. Lett. 365:fny029. doi: 10.1093/femsle/fny029
Römling, U. (2009). Rationalizing the evolution of EAL domain-based cyclic di-GMP-specific phosphodiesterases. J. Bacteriol. 191, 4697–4700. doi: 10.1128/JB.00651-09
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Rousseaux, C., Thuru, X., Gelot, A., Barnich, N., Neut, C., Dubuquoy, L., et al. (2007). Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat. Med. 13, 35–37. doi: 10.1038/nm1521
Ryjenkov, D. A., Simm, R., Römling, U., and Gomelsky, M. (2006). The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281, 30310–30314. doi: 10.1074/jbc.C600179200
Ryjenkov, D. A., Tarutina, M., Moskvin, O. V., and Gomelsky, M. (2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187, 1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005
Schmidt A., Hammerbacher A. S., Bastian M., Nieken K. J., Klockgether J., Merighi M., et al. (2016). Oxygen-dependent regulation of c-di-GMP synthesis by SadC controls alginate production in Pseudomonas aeruginosa. Environ. Microbiol. 18, 3390–3402. doi: 10.1111/1462-2920.13208
Schmidt, A. J., Ryjenkov, D. A., and Gomelsky, M. (2005). The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187, 4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005
Skariyachan, S., Sridhar, V. S., Packirisamy, S., Kumargowda, S. T., and Challapilli, S. B. (2018). Recent perspectives on the molecular basis of biofilm formation by Pseudomonas aeruginosa and approaches for treatment and biofilm dispersal. Folia Microbiol. 63, 413–432. doi: 10.1007/s12223-018-0585-4
Solovyev, V., and Salamov, A. (2011). “Automatic Annotation of Microbial Genomes and Metagenomic Sequences,” in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, ed R. W. Li (Nova Science Publishers), 61–78.
Srivastava, D., Harris, R. C., and Waters, C. M. (2011). Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J. Bacteriol. 193, 6331–6341. doi: 10.1128/JB.05167-11
Stahl, B., and Barrangou, R. (2013). Complete genome sequence of probiotic strain Lactobacillus acidophilus La-14. Genome Announc. 1:e00376-13. doi: 10.1128/genomeA.00376-13
Steiner, S., Lori, C., Boehm, A., and Jenal, U. (2013). Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 32, 354–368. doi: 10.1038/emboj.2012.315
Taboada, B., Ciria, R., Martinez-Guerrero, C. E., and Merino, E. (2012). ProOpDB: prokaryotic operon database. Nucleic Acids Res. 40, D627–D631. doi: 10.1093/nar/gkr1020
Tamayo, R., Tischler, A. D., and Camilli, A. (2005). The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280, 33324–33330. doi: 10.1074/jbc.M506500200
Tchigvintsev, A., Xu, X., Singer, A., Chang, C., Brown, G., Proudfoot, M., et al. (2010). Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 402, 524–538. doi: 10.1016/j.jmb.2010.07.050
Wang, X., Preston, J. F., and Romeo, T. (2004). The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004
Keywords: c-di-GMP signaling, Lactobacillus acidophilus, GGDEF domain, EAL domain, c-di-GMP receptor, exopolysaccharide
Citation: He J, Ruan W, Sun J, Wang F and Yan W (2018) Functional Characterization of c-di-GMP Signaling-Related Genes in the Probiotic Lactobacillus acidophilus. Front. Microbiol. 9:1935. doi: 10.3389/fmicb.2018.01935
Received: 11 February 2018; Accepted: 30 July 2018;
Published: 29 August 2018.
Edited by:
Satoshi Tsuneda, Waseda University, JapanReviewed by:
Aindrila Mukhopadhyay, Lawrence Berkeley National Laboratory (LBNL), United StatesMatias Castro, Fundación Ciencia and Vida, Chile
Copyright © 2018 He, Ruan, Sun, Wang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjuan Yan, NjQ1NjEzMDUzQHFxLmNvbQ==
Fang Wang, ZmFuZ3dhbmc1NUB5YWhvby5jb20=
 Jiahui He
Jiahui He Wenhao Ruan
Wenhao Ruan Jieli Sun1
Jieli Sun1 Fang Wang
Fang Wang Wenjuan Yan
Wenjuan Yan