
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 August 2018
Sec. Microbiotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01925
A correction has been applied to this article in:
Corrigendum: Characterization of ML-005, a Novel Metaproteomics-Derived Esterase
A novel gene encoding for a lipolytic enzyme, designated ML-005, was recently identified using a functional metaproteomics approach. We heterologously expressed this protein in Escherichia coli and biochemically characterized it. ML-005 exhibited lipolytic activity toward short-chained substrates with the preferred substrate being p-nitrophenyl-butyrate, suggesting that ML-005 is an esterase. According to homology analysis and site-directed mutagenesis, the catalytic triad of the enzyme was identified as Ser-99, Asp-164, and His-191. Its optimal pH was determined to be at pH 8. Optimal activity was observed at 45°C. It also exhibited temperature, pH and salt tolerance. Residual relative activity after incubating at 50–60°C for 360 min was above 80% of its initial activity. It showed tolerance over a broad range of pH (5–12) and retained most of its initial activity. Furthermore, incubating ML-005 in 1 – 5M NaCl solution had negligible effect on its activity. DTT, EDTA, and ß-mercaptoethanol had no significant effect on ML-005’s activity. However, addition of PMSF led to almost complete inactivation consistent with ML-005 being a serine hydrolase. ML-005 remains stable in the presence of a range of metal ions, but addition of Cu2+ significantly reduces its relative activity. Organic solvents have an inhibitory effect on ML-005, but it retained 21% of activity in 10% methanol. SDS had the most pronounced inhibitory effect on ML-005 among all detergents tested and completely inactivated it. Furthermore, the Vmax of ML-005 was determined to be 59.8 μM/min along with a Km of 137.9 μM. The kcat of ML-005 is 26 s-1 and kcat/Km is 1.88 × 105 M-1 s-1.
Lipolytic enzymes have the ability to catalyze the cleavage and synthesis of carboxyl esters (Arpigny and Jaeger, 1999). They are generally classified into two major families according to their substrate specificity: carboxylesterases (3.1.1.1) or “esterases” are active against smaller triglycerides with shorter fatty-acid side chains (<C10), while triacylglycerol hydrolases or “lipases” prefer hydrolyzing water-insoluble triglycerides with longer fatty acid chains (>C10) (Bornscheuer, 2002; Chahiniana and Sarda, 2009; Lopes et al., 2011). Both lipases and esterases show the characteristic α/β hydrolase fold (Ollis et al., 1992). The active site of lipolytic enzymes include a nucleophilic serine residue, a histidine residue and an aspartate or glutamate residue. The serine residue is part of a highly conserved pentapeptide motif called the catalytic elbow, with a consensus motif Gly – X – Ser – X – Gly. In some cases the first glycine residue can be replaced by an alanine resulting in an Ala – X – Ser – X – Gly motif (Dartois et al., 1994; Mala and Takeuchi, 2008). The serine residue is essential for enzyme functionality and lipolytic enzymes are thus part of the “serine-hydrolase” family (Simon and Cravatt, 2010).
Lipolytic enzymes are one of the most important classes of biocatalysts currently in use: they belong to the few enzymes that are produced at an industrial scale exceeding 104 tons per year (Kourist and Gaßmeyer, 2015). According to recent market research, the global enzyme market revenue is expected to reach $ 10.7 billion by 2024 with lipolytic enzymes gaining in importance (Global Market Insights, 2017). They are widely used as industrial catalysts, in the detergent, food and leather industries. Increasing demand in recent years to move away from petrochemical-based industrial processes to more environmentally friendly, bio-based industrial processes is a contributing factor in the increasing interest to utilize lipolytic enzymes in various other industries like pulp and paper, textile, cosmetics industries and biodiesel production (Awaji et al., 1998; Schmid and Verger, 1998; Barron et al., 1999; Mangla et al., 2013; Sharma and Kanwar, 2014). The industrial relevance of lipases is part due to their unique set of versatile characteristics. They are generally resilient against harsh conditions. They can have a broad substrate specificity, but also display chain-length selectivity and enantioselectivity. Lipases and esterases generally do not require any cofactors, they possess a broad pH activity profile, and their relative stability at room temperature in various organic solvents makes them highly attractive for various industries (Gupta et al., 2004; Lotti and Alberghina, 2007). All these factors have led to a renewed research interest in establishing novel enzymatic screening methods, characterizing new enzymes and engineering enzymes with industrially relevant properties.
The search of novel lipolytic enzymes is classically done through functional screening of a multitude of microorganisms for lipolytic activity. This remains a time-proven approach and has led to the discovery of numerous enzymes (Ogawa, 1999). However, this approach can be time-consuming and, more importantly, a majority of the environmental microbes is “non-screenable,” as it is difficult to culture them under laboratory conditions. Rappé and Giovannoni (2003) showed that typically less than 1% of the microbes of an environmental sample grow under “laboratory conditions.” To counter the problem of the uncultivable microbial diversity, total DNA isolated from a microbial community, the so-called “metagenome,” provides an attractive solution (Schloss and Handelsman, 2003). Fragments of this total DNA can be cloned and transformed into desired host systems and the resulting library functionally screened for activity. Due to its unbiased nature, library-based metagenomic analysis led to the discovery of numerous enzymes and continues to be widely used (Henne et al., 2000; Faoro et al., 2012). However, such a screening library typically needs to be massive and optimizing the protein expression-system requires substantial effort, making the screening potentially time-consuming and expensive. Increase in computing power in the past decade has opened up “in silico screening” methods. DNA from metagenomic sources is sequenced using “Next Gen” high-sensitivity and high-throughput methods and the resulting sequence database can then be searched for structural motifs of known enzymes by automated bioinformatical analysis (Widmann et al., 2010; Yin et al., 2012). As enzymes are pre-screened in silico, only a substantially reduced number of clones need to be biochemically tested for activity, resulting in drastically reduced cost and increased efficiency (Kusnezowa and Leichert, 2017). Rapid advancements in sequencing technologies and drastic reduction in sequencing costs (Goodwin et al., 2016) has led to its widespread adoption in the scientific community. A number of lipolytic enzymes have been identified and biochemically characterized through sequence-based metagenomics (Kwoun Kim et al., 2004; Sharma et al., 2009; Masuch et al., 2015).
Progressing from the above-mentioned DNA-only approaches, we recently described a functional metaproteomics approach, which combines the specificity of activity-based screening with an unbiased meta-omics approach. Through this approach we successfully identified 14 lipolytically active protein spots on a 2D gel derived from the enriched metaproteome of a soil sample harvested from a restaurant’s used cooking oil disposal site. Among those, we identified ML-005, an esterase from a hitherto uncharacterized family, which we could heterologously express in E. coli (Sukul et al., 2017).
In the present study, we determine, in detail, the biochemical and enzymatic properties of ML-005, the first lipolytic enzyme found through this functional metaproteomics approach. ML-005 is an esterase with a preference for short-chained substrates. It showed a temperature optimum at 45°C and high stability up to 60°C. It is moderately resistant to organic solvents and detergents, active in neutral to alkaline pH and exhibits high pH- and halotolerance.
A detailed description of how ML-005 was identified has been published previously (Figure 1; Sukul et al., 2017). In short, oil-contaminated soil samples were collected from a restaurant grease disposal site and used for enriching microorganisms with lipolytic activity. Subsequently, the proteins and the DNA contained in the samples were isolated. The proteins were then separated on a 2D gel. In-gel activity assays based on the fluorogenic substrate methylumbelliferyl butyrate enabled us to identify 14 lipolytically active proteins. These proteins were then identified using mass spectrometry by searching against a protein database built from the corresponding metagenomic data. One of the proteins we identified was ML-005, a lipid hydrolase that was hitherto unknown. All metagenomic sequences obtained in the previous study including ML-005’s sequence have been deposited to the European Nucleotide Archive1 under project number PRJEB16064 and sample accession number ERP017906. The protein sequence of ML-005 can be found in Supplementary Data Sheet S2.FASTA.
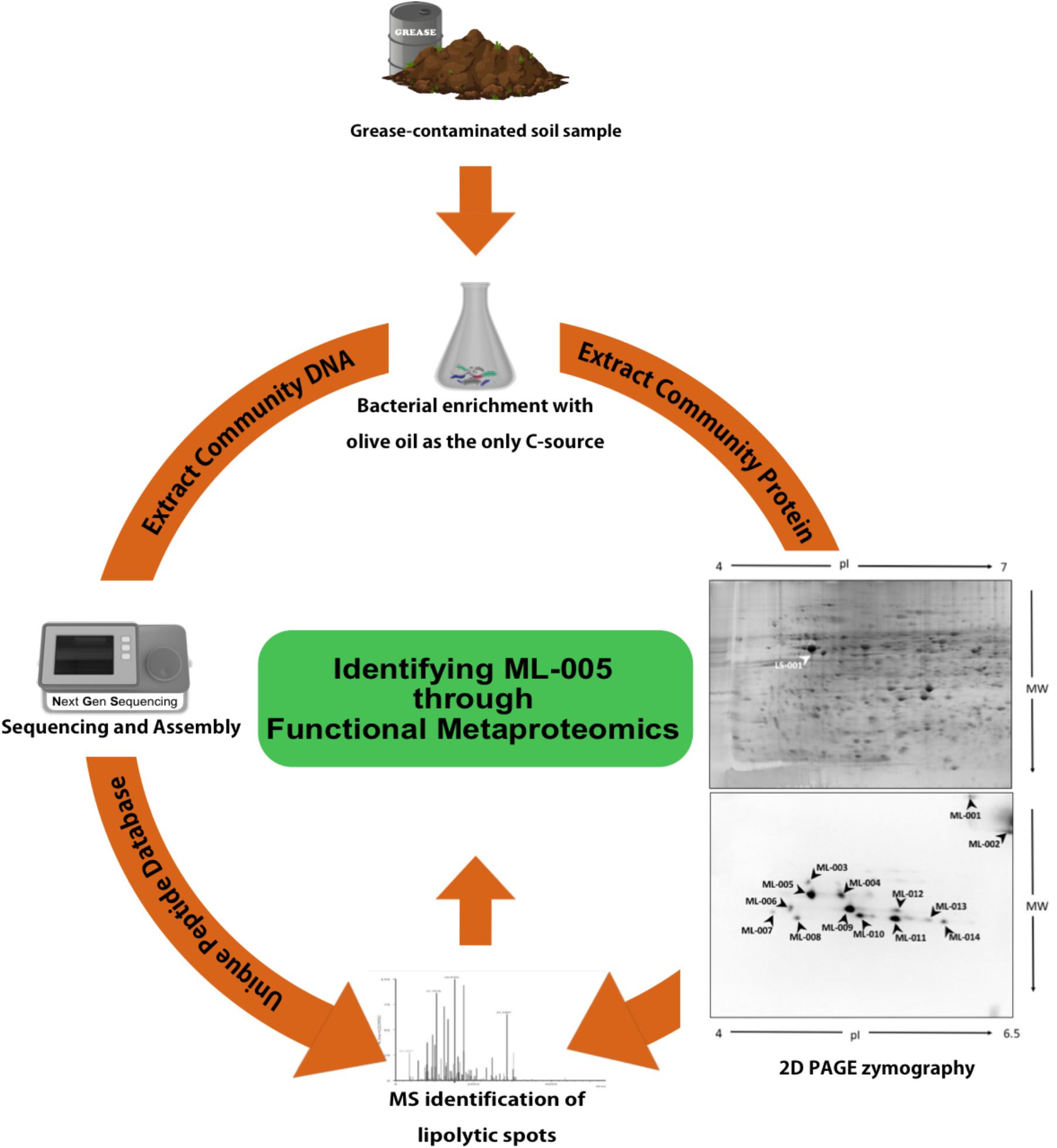
FIGURE 1. Functional Metaproteomics workflow used to discover ML-005. Grease-contaminated soil sample was used for bacterial enrichment with olive oil as the sole carbon source. Community DNA was extracted, sequenced and annotated. Community protein was separated in a 2D gel and 2D PAGE zymography was carried out to visualize protein spots harboring lipolytic activity. Protein spots were identified using mass-spectrometry, searching against a protein database derived from the metagenomic DNA sequences. ML-005 was selected for this study for further characterization. Functional Metaproteomics has the immediacy of an activity-based approach, while still retaining the comprehensive information of the metagenome (Sukul et al., 2017).
Database similarity searches and alignment was performed using BLAST (Marchler-Bauer et al., 2005). The amino acid sequence was analyzed for molecular weight and extinction coefficient using ProtParam on the ExPASy Server (Gasteiger et al., 2005). The three-dimensional structure of ML-005 was predicted using the Phyre2 server2 (Kelley et al., 2015). The pdb file generated by Phyre was visualized and edited with PyMOL (Schrödinger, LLC, 2015).
The sequences of ML-005, Esterases EstA from Pseudomonas aeruginosa, Esterase B from Burkholderia gladioli, Esterase EstC from Streptomyces coelicolor, Esterase EstD from Thermotoga maritima, and Carboxylesterase NP from Bacillus subtilis were queried against the uniref50 database (release 2018_05, 23-May-2018, Suzek et al., 2015) using the blastp command of the blast command line applications version 2.7.1 (Altschul et al., 1990) with the parameters “-outfmt 10 -evalue 0.0001 -max_target_seqs 100000.” This essentially returned all homologous sequence clusters of these proteins contained in the uniref50 database that form local alignments with an e-value below 10-4 in a comma separated table file format. The resulting homologs were then ordered in Excel for Mac version 16.14.1 (Microsoft Corporation, Redmond, WA, United States) by “alignment length” and all results with an alignment length below 80% of the length of the original query protein were dismissed. Next the resulting, shortened table was ordered by “% identity” and only results with an identity greater or equal to 35% were considered for further analysis. A batch file was created from these tables containing the uniref50 identifier and the start and end position of the matching alignment in the subject sequence. This batch file was used to retrieve the aligning parts of the subject sequences as a fasta file from the uniref50 database using the blast command line applications’ blastdbcmd command with the “-entry_batch” parameter.
The acquired sequences were imported in Geneious v11.1.4.3 (Kearse et al., 2012) and aligned using MUSCLE v3.8.425 (Edgar, 2004) using default settings. The multiple alignment was used to build a phylogenetic tree using FastTree v2.1.54 (Price et al., 2010) using default settings. FastTree infers approximately maximum-likelihood phylogenetic trees with Jones-Taylor-Thornton (JTT) model of amino acid evolution.
A detailed description of the heterologous expression und protein purification procedure has been described previously (Sukul et al., 2017). In short, the codon-optimized and artificially synthesized ML-005 gene sequence (645 bp) was cloned into the pET22b expression vector (Merck Millipore, Billerica, MA, United States) containing a T7-promotor. pET22b additionally codes for a C-terminal His6-tag that was thus fused to ML-005 (For the vector sequence including insert see Supplementary Data Sheet S3.FASTA). ML-005 expression from pET22b in E. coli BL21 was induced by the addition of IPTG (1 mM), and was allowed to proceed overnight at 20°C under constant shaking. The cells were harvested by centrifugation (45 min at 6,500 ×g and 4°C), were resuspended in in approximately 50 ml of Tris–HCl-buffer (50 mM, pH 7.1), and disrupted by passing the cell suspension through the Constant Cell Disruption System (Constant Systems, Low March, United Kingdom) at 1.9 kbar three times. The resulting lysate was then centrifuged (50 min at 6,500 ×g and 4°C) to remove the insoluble fraction from the soluble proteins. The supernatant was then filtered using 250 ml Filtropur V50 vacuum filters (Sarstedt, Nümbrecht, Germany). ML-005 was purified from this soluble protein fraction using an ÄKTApurifier (GE Healthcare, Uppsala, Sweden) with a 5 ml HisTrap HP Ni-NTA-column (GE Healthcare, Uppsala, Sweden). The column was equilibrated using Buffer A (50 mM sodium phosphate, 300 mM NaCl, pH 8.0). The soluble protein fraction was loaded onto the column and ML-005 was eluted with 10–12% Buffer B (50 mM sodium phosphate, 300 mM NaCl, 500 mM imidazole, pH 8.0). The eluate was dialyzed overnight at 4°C against 50 mM sodium phosphate buffer (pH 8) using a Spektra/Por Dialysis Membrane 12–14 kDa (Spectrumlabs, Rancho Dominguez, United States).
Lipolytic activity of purified ML-005 was determined spectrophotometrically using p-nitrophenyl (pNP) esters as substrates. pNP-esters are hydrolyzed through lipolytic activity and the released p-nitrophenol is yellow and can be detected spectrophotometrically at 405 nm. Unless otherwise described, 38.3 nM (3.92 mg/ml) of purified ML-005 in 50 mM sodium phosphate buffer (pH 8.0) were used to carry out the pNP-assays in triplicates. Temperature was varied for different reactions. The hydrolysis reaction was initialized by adding 50 μM of substrate. Reaction volume was 2000 μl. The resulting reaction was followed through a time-course-measurement at 405 nm. One unit (U) of activity was defined as the amount of enzyme releasing 1 μmol of p-nitrophenol per minute under the mentioned assay conditions. The extinction coefficient was calculated using a calibration curve. The absorption of different concentrations of p-nitrophenol in 50 mM sodium phosphate buffer (pH 8.0) at 25°C was measured at 405 nm.
Substrate specificity of ML-005 toward pNP-esters with variable acyl chain lengths was measured in 50 mM sodium phosphate buffer (pH 8.0) at 25°C using p-nitrophenyl acetate (C2), p-nitrophenyl butyrate (C4), p-nitrophenyl caprylate (8), p-nitrophenyl decanoate (C10), p-nitrophenyl laurate (C12), p-nitrophenyl myristate (C14), and p-nitrophenyl palmitate (C16).
Temperature optimum was determined in the range between 20 and 60°C, in 5°C degree steps, with p-nitrophenyl butyrate as the substrate. ML-005 was incubated for 30 min at the necessary temperature and subsequently the activity assay was performed at the same temperature. Temperature-stability of ML-005 was determined for the same temperature points. To determine stability, ML-005 was incubated in the appropriate temperature for 60 and 360 min followed by its residual activity measurement at 25°C and pH 8.
pH optimum was determined in the range between 5 and 9.5, in steps of 0.5, with p-nitrophenyl butyrate as the substrate. Stability of ML-005 was determined by incubating at pH 4–13 for 360 min followed by the measurement of its residual activity at 25°C and pH 8.
Stability toward high NaCl concentrations was measured by incubating ML-005 at NaCl concentrations of 1 to 5 M. An initial measurement was taken after 3 h and compared with measurements taken after 7 days.
The influence of metal ions was determined using a range of metal salts (CaCl2, CuCl2, FeCl2, KCl, LiCl, MgCl2, MnCl2, NaCl, and NiCl2) at a final concentration of 1 mM.
The effect of inhibitors was determined by using ethylenediaminetetraacetic acid (EDTA), phenylmethylsulfonyl fluoride (PMSF), β-mercaptoethanol (β-ME), and dithiothreitol (DTT) at final concentrations of 1 mM.
Effect of detergents on ML-005 was determined by using sodium dodecyl sulfate (SDS), 3-(3-cholamidopropyl) dimethylammonio-1-propanesulfonate (CHAPS), Tween 20, Tween 80, and Triton X-100 at a final concentration of 1% (v/v) or (w/v) for CHAPS and SDS.
The effect of organic solvents were determined using dimethyl sulfoxide (DMSO), dimethylformamide (DMF), methanol, isopropanol, and acetone at final concentrations of 1 and 10% (v/v).
Control was defined as the reaction mixture in 50 mM sodium phosphate buffer (pH 8.0) without added metal ions, inhibitors, detergents or organic solvents.
Reaction temperature was 25°C and pH was 8.
The Michaelis Menten kinetics were investigated with different concentrations (10–600 μM) of pNP-butyrate as substrate. The reaction was carried out in 50 mM sodium phosphate buffer (pH 8.0) at the optimum temperature of 45°C. Km, kcat, and the turnover number was determined, as well as the catalytic efficiency (kcat/Km). Vmax and Km were determined through Graphpad Prism using its “Enzyme kinetics – Michaelis-Menten” function. kcat and kcat/Km were calculated based on these results.
Mutations were generated for the catalytic triad residues using the QuikChange method (Stratagene, La Jolla, CA, United States) using the primer pairs listed in Table 1. The QuikChange PCR was carried out in a Biometra TProfessional thermal cycler (Whatman Biometra, Goettingen, Germany). 150 ng of plasmid DNA were added to 125 ng each of forward and reverse primer (Table 1), 2 mM dNTPs (Thermo Scientific), 2.5 U of Pfu DNA Polymerase (Thermo Scientific), and 5 μl 10 × Pfu DNA Polymerase buffer in a volume of 50 μl. The mutated plasmids were then synthesized by 16 cycles of 30 s 95°C (denaturation), 30 s 58°C (annealing), and 5 min 68°C (elongation), followed by cooling at 4°C until the sample was further processed. The QuikChange PCR product was then digested using DpnI (10 U) for 1 h at 37°C to remove the parental DNA template. Finally the sample was transformed into E. coli XL1-Blue cells and the mutagenesis confirmed by sequencing.
Ser-99 was mutated to Ala-99, Asp-164 was mutated to Asn-164, and His-191 was mutated to Asn-191. Following successful mutations, mutated strains (S99A, D164N, and H191N) were cloned into pET22b expression vector and heterologously expressed in E. coli BL21. 2 ml of the overexpressed cells were harvested, disrupted through sonication (amplitude: 80%, Cycle: 0.5 s, 3 × 1 min) using a Vial Tweeter (Hielscher, Teltow, Germany). The resulting crude extract was centrifuged (13,000 ×g, 20 min, 4°C) to remove cell debris. The supernatant was used for spectrophotometric lipolytic activity determination using pNP-butyrate. Reaction was carried out in 50 mM sodium phosphate buffer (pH 8.0) at 25°C.
A BLASTp search of ML-005 showed it to be a serine hydrolase family protein similar to B. subtilis YdeN (UniProtKB – P96671), a predicted esterase with a canonical alpha/beta hydrolase fold (Accession No. COG3545 at NCBI) (Marchler-Bauer et al., 2005; The UniProt Consortium, 2017). The sequence comparison of ML-005 with P96671 showed a sequence identity of 28.44% (Figure 2A). Furthermore, a nucleophilic serine residue at position 99 was embedded in a pentapeptide motif composed of Ala-His-Ser-Leu-Gly (97-101). The serine together with aspartic acid residue at position 164 and the histidine residue at position 191 were in silico predicted to form the catalytic triad. Three dimensional structure of ML-005 was modeled using the Phyre2 server. The catalytic triad residues were found to be in spatial proximity to each other (Figure 2B).
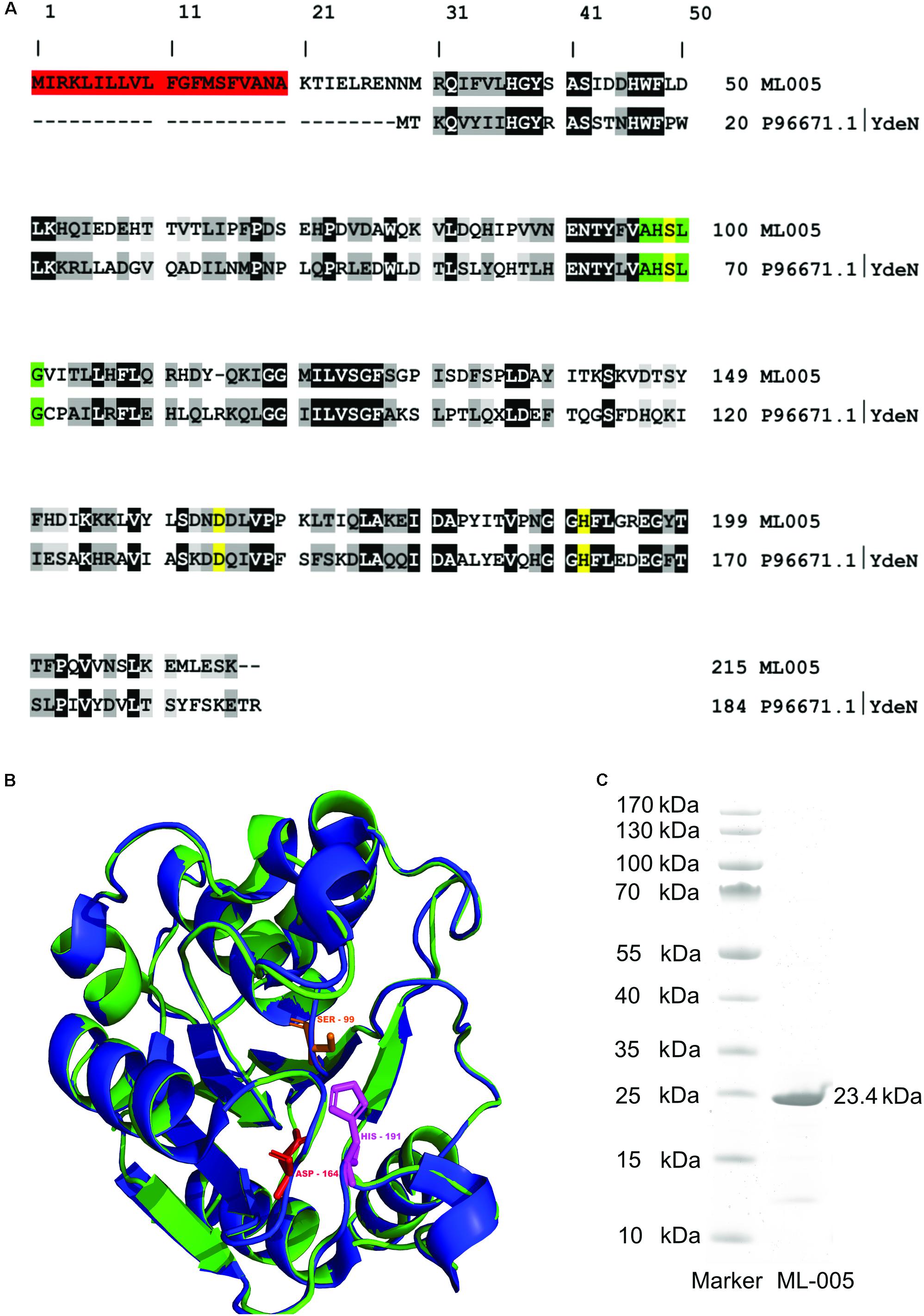
FIGURE 2. (A) Sequence alignment of ML-005 with the uncharacterized esterase YdeN from Bacillus subtilis (UniProt accession number P96671.1) showed a sequence identity of 28.44% (highlighted in black, similar amino acids in shades of gray). Residues forming the catalytic triad i.e., serine, (99), aspartic acid (164), and histidine (191) are highlighted in yellow. These residues form an integral part of the active site. The pentapeptide Ala – His – Ser – Leu – Gly motif is highlighted in green. This represents the nucleophilic elbow and is a conserved structure found within lipolytic enzymes. Residues 1–20 form a signal peptide (red) which was processed in E. coli (see panel C). (B) Three dimensional structure of ML-005 was modeled using the Phyre2 server (blue ribbon; http://www.sbg.bio.ic.ac.uk/phyre/). Potential catalytic triad residues (His-191, Asp-164, and Ser-99) were predicted to be in close proximity to each other. The structural alignment with YdeN from B. subtilis (green ribbon) shows substantial similarity (C) ML-005 was cloned into a pET22b vector with a T7 promoter system and a C-terminal His6-tag und purified to homogeneity. The purified ML-005 band showed a size that approximates 23.4 kDa, consistent with the calculated mass of His6-tagged ML-005 with a removed signal peptide.
The molecular weight of ML-005 was determined to be 24.6 kDa using ProtParam on the ExPASy Server (Gasteiger et al., 2005). With the additional C-terminal His6-tag added to the recombinant protein, its theoretical mass was determined to be 25.6 kDa. SignalP (Ferrari et al., 2014) predicted that the initial 20 amino acids are part of a signal peptide sequence (Supplementary Figure S1). This hypothesis was confirmed after purification. Purified ML-005 appeared at an apparent size below 25 kDa on an SDS–PAGE, significantly smaller than the predicted 25.6 kDa, indicating the cleavage of the signal-peptide from the newly synthesized protein (see also Supplementary Figure S2).
ML-005 was purified to homogeneity (>85%) via a C-terminal His6-tag from a pET22b expression vector using E. coli BL21 (DE3) as a heterologous host (Figure 2C).
To determine the substrate specificity of ML-005, various chain-lengths of p-nitrophenyl esters were tested (C2-C16) (Figure 3A). The maximum hydrolysis activity was observed with p-nitrophenyl butyrate (C4). An increase in chain-length resulted in lower of activity. pNP-octonoate (C8) showed 66.1% of activity when compared to ML-005’s preferred substrate, pNP-butyrate (14.1 U mg-1), while pNP-decanoate (C10), pNP-dodecanoate (C12), and pNP-myristate (C14) retained 11%, 2%, and less than 1%, respectively, while ML-005 showed no detectable enzymatic activity against pNP-palmitate (C16) (Figure 3A). This affinity toward short-chained, water-soluble esters classifies ML-005 as a carboxylesterase (EC 3.1.1.1). This classification is consistent with the predicted structure (Figure 2B), which lacks a hydrophobic lid, a feature normally present in true lipases and absent from esterases (Brady et al., 1990; Winkler et al., 1990). In contrast, triacylglycerol hydrolase or lipases are generally active against water insoluble long-chained esters (Bornscheuer, 2002). In ML-005 serine (99) along with histidine (191) and glutamate (164) were predicted to be part of the catalytic triad in silico. To experimentally confirm the structure-based prediction of the catalytic triad, we mutagenized these three residues. As expected, all three mutants S99A, D164N and H191N showed negligible activity (<1%) compared to wild type, thus experimentally confirming ML-005 to be a classic serine hydrolase (Figure 3B).
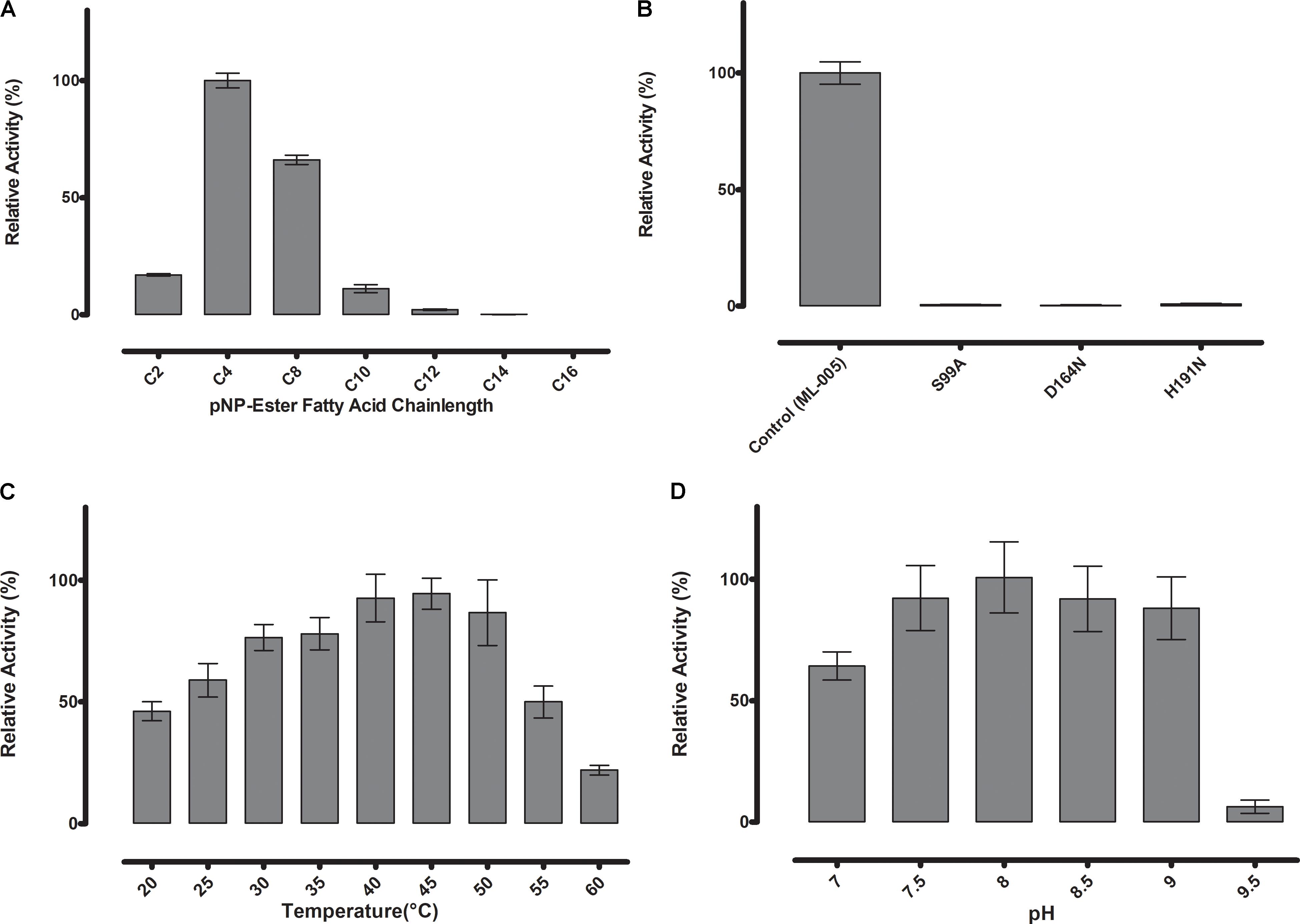
FIGURE 3. (A) Substrate specificity of ML-005 showed clear preference for p-nitrophenyl butyrate (C4). Substrate with a chain length of C8 retained approximately 66% of the maximum activity, whereas activity dropped significantly with longer chains. (B) Serine (99), aspartic acid (164), and histidine (191) constituting the catalytic triad were mutated and negligible activity was observed for any of the mutants. (C) Temperature optimum was observed to be at 45°C. (D) pH optimum was observed to be at pH 8, however, activity of ML-005 was largely stable between pH 7.5 and 9. At pH 7, still approximately 64% of maximum activity was retained. pH 9.5 resulted in a drastic loss of activity.
In order to justify an in-depth characterization of ML-005’s biochemical properties, we compared ML-005 to 5 other microbial esterases that have been well characterized. These included Esterases EstA from P. aeruginosa (Wilhelm et al., 1999), Esterase EstB from B. gladioli (Wagner et al., 2002), Esterase EstC from S. coelicolor (Brault et al., 2012), Esterase EstD from T. maritima (Levisson et al., 2007), and Carboxylesterase NP from B. subtilis (Quax and Broekhuizen, 1994). A phylogenetic tree revealed that ML-005 formed a distinct branch with 70 clusters representing 825 proteins found in the uniref50 database (Figure 4). To our knowledge, none of these proteins have been characterized so far. We therefore decided to biochemically characterize this metagenomic enzyme as the first representative of this novel family in more detail.
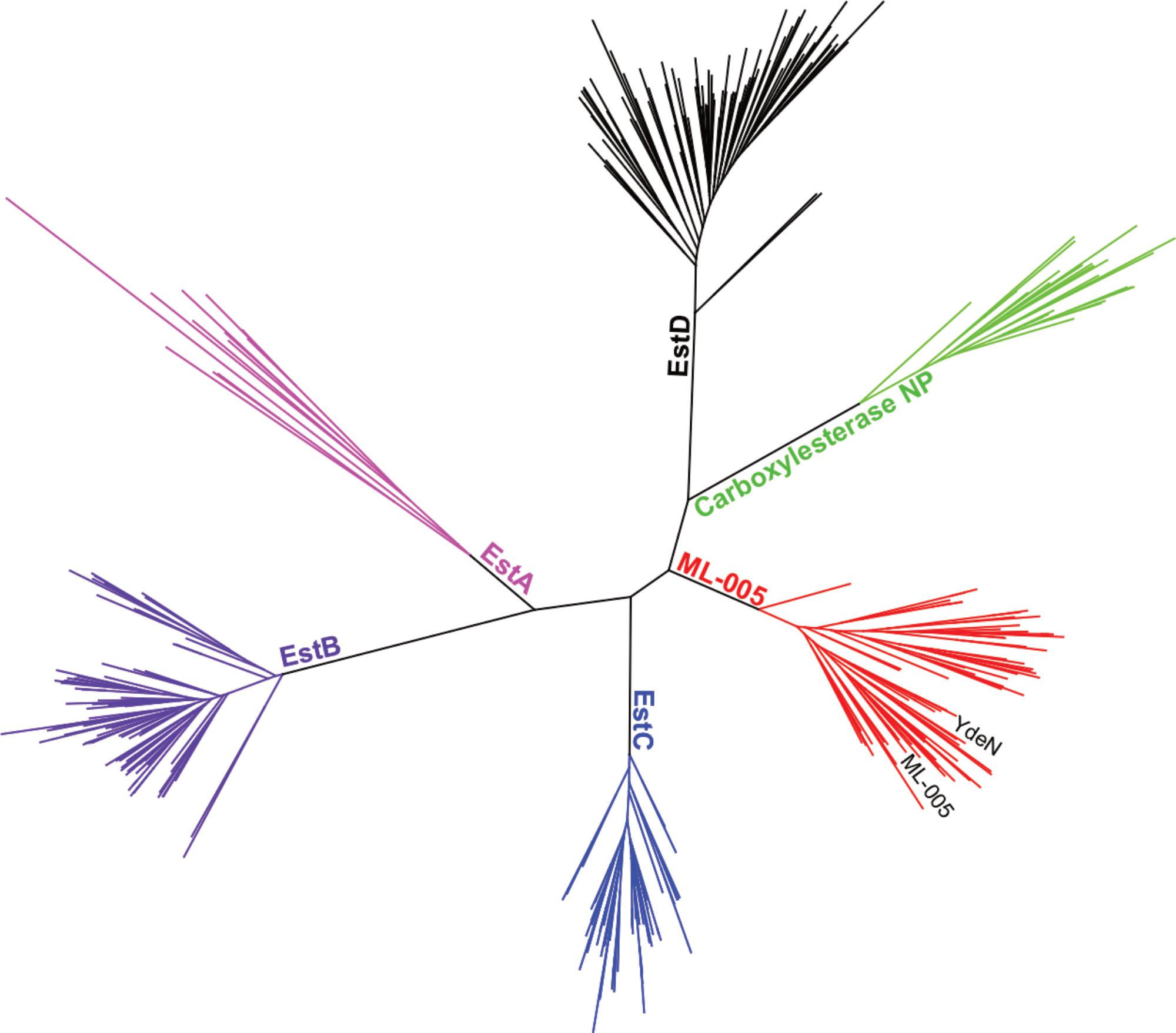
FIGURE 4. Phylogenetic comparison of ML-005 and 5 other well-characterized bacterial esterases. Tree of lipase sequences with 35% identity to ML-005 and to esterases EstA from P. aeruginosa, EstB from B. gladioli, EstC from S. coelicolor, EstD from T. maritima and Carboxylesterase NP from B. subtilis. ML-005 is member of an as of yet uncharacterized family of esterases, which also contains YdeN from B. subtilis.
Thermostability and pH-optimum are critical parameters when choosing biocatalysts. We therefore measured the hydrolytic activity of ML-005 at temperatures ranging from 20 to 60°C. The optimum temperature for ML-005 was determined to be 45°C (Figure 3C). At 55°C, it still retained 50% of its activity and approximately 22% activity was retained at 60°C. This suggests a slightly thermophilic activity profile. Purified ML-005 exhibited high activity between pH 7.5 and pH 9 with pH 8 being the optimum pH with the highest activity (Figure 3D). At pH 7, ML-005 had 64% of maximum activity while pH 9.5 resulted in a drastic breakdown in activity with only 6% of maximum activity remaining.
The stability of ML-005 was determined by measuring the residual activity with p-nitrophenyl-butyrate (C4) at optimum conditions after incubation at a wide range of temperature, pH and NaCl concentration. After incubation at 60°C for 360 min ML-005 still retained an activity of 80% when compared to optimum conditions (Figure 5B). ML-005 retained activity close to its optimum activity when incubated between pH 5 to pH 12 for 360 min. Incubating ML-005 at pH 4 led to a decrease in activity to approximately 46%, while on the basic side, incubating ML-005 at pH 13 resulted in the loss of most of its activity (2%) (Figure 5C). Incubating ML-005 at 1 to 5 M of NaCl concentration, even for 7 days, showed only negligible effect on its activity (Figure 5D), indicating that ML-005 is a halotolerant enzyme.
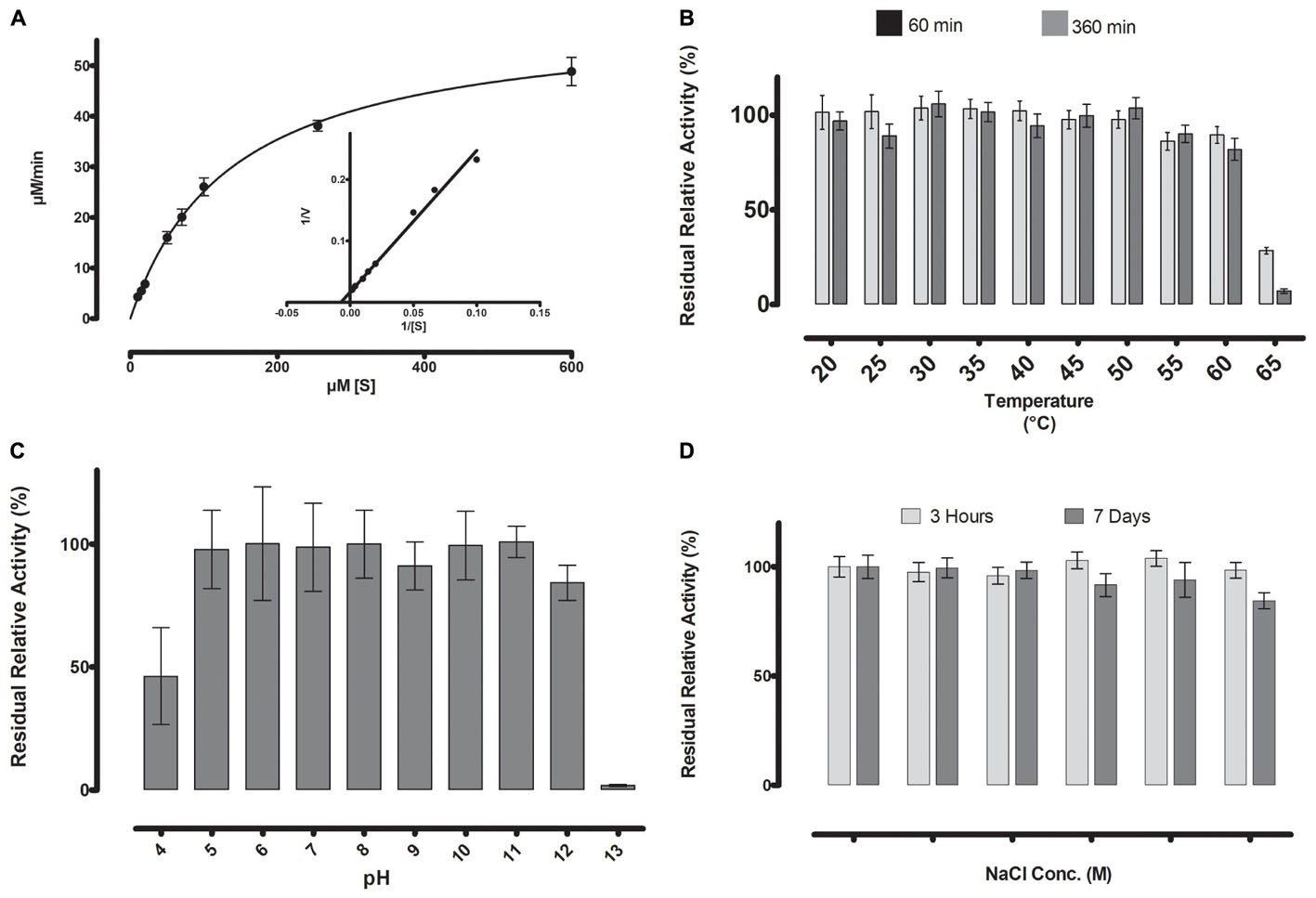
FIGURE 5. (A) Michaelis-Menten kinetics were observed for ML-005 with pNP-butyrate. Vmax of ML-005 was determined to be 59.8 μM/min along with a Km of 137.9 μM. The kcat of ML-005 is 26 s-1 and kcat/Km is 1.88 × 105 M-1 s-1. (B) ML-005 showed temperature tolerance from 20 to 60°C. (C) ML-005 showed tolerance over a broad range of pH (5–12) and retained most of its activity. At pH 4 it retained approximately 50% of its activity while pH 13 almost completely deactivated ML-005. (D) ML-005 showed halotolerance when incubated in increasing NaCl concentrations. After 7 days of incubation at close to saturated NaCl solution (5M), ML-005 still retained most of its activity.
With the optimal conditions at hand, we determined ML-005’s Michaelis-Menten kinetics by measuring its activity at various substrate concentrations at pH 8 and 45°C (Figure 5A). The Vmax of ML-005 was determined to be 59.8 μM/min and the Km was determined to be 137.9 μM. The kcat of ML-005 is 26 s-1 and its catalytic efficiency kcat/Km is 1.88 × 105 M-1 s-1.
To further determine ML-005’s resilience, the modulating or inhibiting effects of various compounds on its activity were studied (Figure 6).
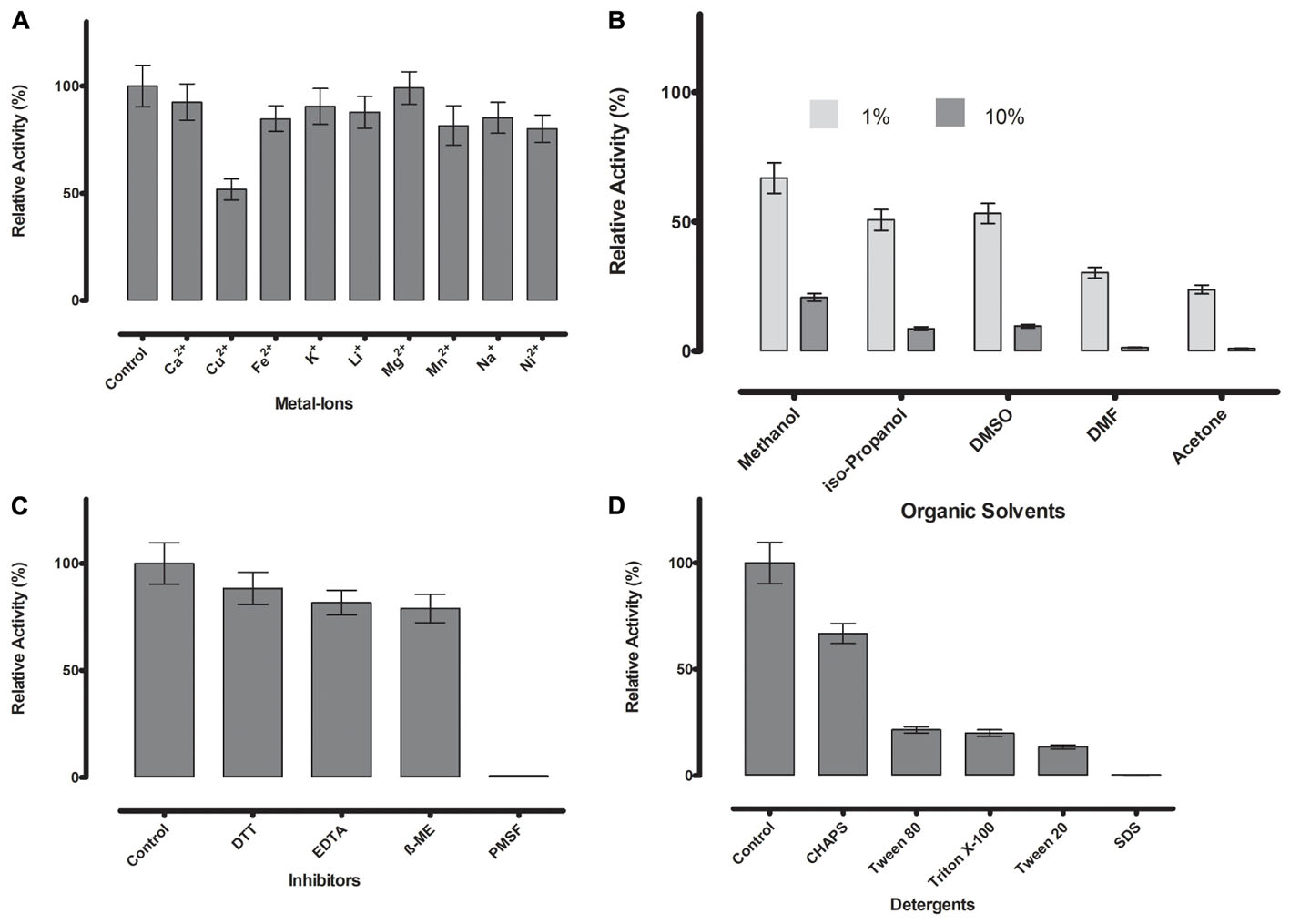
FIGURE 6. (A) Metal ions showed negligible effect on ML-005 at a concentration of 1 mM, with copper showing the most drastic effect by inhibiting ML-005 by approximately 50%. (B) Organic solvents had an overall inhibiting effect on ML-005 without exception, but ML-005 was moderately stable in the presence of even 10% Methanol. (C) All inhibitors showed moderate inhibiting effect at a concentration of 1 mM, with relative activity staying at around 80%. Only PMSF showed almost complete inhibition of ML-005, consistent with an active-site serine. (D) Detergents at 1% were also found to have an overall inhibiting effect. CHAPS showed the least effect with 66% relative activity and SDS inactivating ML-005 completely with negligible remaining activity.
Divalent cations are known to have inhibitory or stimulating effect on lipase and esterase activity (Gupta et al., 2004). Most are considered to have an inhibitory effect, but the majority of metal ions we tested (Ca2+, Fe2+, K+, Li+, Mg2+, Mn2+, Na+, and Ni2+) showed no or negligible effects at 1 mM concentration in our assay (Figure 6A). Only Cu2+ reduced the activity substantially to 52%, a known phenomenon with lipolytic enzymes (Hiol et al., 2000).
Resilience against organic solvents is of particular interest for industrial applications, as esterases are widely used in processes like the synthesis of pharmaceutical and fine chemicals where these solvents are present (Fuciños et al., 2012). All tested organic solvents showed an inhibiting effect on ML-005. 1% of acetone, DMF, DMSO, isopropanol and methanol diminished the activity to approximately 24, 30, 53, 51, and 67% of the original activity, respectively. 10% organic solvent diminished ML-005 activity even more, nevertheless ML-005 still retained 21% of its activity in 10% Methanol (Figure 6B), making it potentially usable in applications such as methyl-esterification of organic substrates.
Presence of DTT, EDTA and ß-mercaptoethanol at a concentration of 1 mM had a slight inhibiting effect on ML-005 with 88, 81, and 79% of remaining activity compared to control, respectively. However, based on these results we conclude that neither structural disulfides or coordinated metals play a role in ML-005’s activity (Figure 6C). PMSF inactivated ML-005 completely with only 0.7% residual relative activity. PMSF is most likely acting as a specific inhibitor and irreversibly deactivates ML-005 by binding to the nucleophilic serine of the active site (Narayanan and Jones, 2015). This further confirmed to us that, unlike some lipases, ML-005 cannot resist the specific inhibition of PMSF by physically hindering access to the active site with the help of a hydrophobic lid (Côté and Shareck, 2008).
Lastly we tested the effect of detergents on ML-005 (Figure 6D). All detergents showed inhibition with CHAPS, SDS, Tween 20, Tween 80, and Triton X-100 showing a relative activity of 66, 0.2, 13, 21, and 19%, respectively, at a 1% concentration. ML-005 showed the most resilience against CHAPS and was most susceptible to the effects of SDS.
ML-005 was identified in a previous study through a functional metaproteomics approach. In this preceding study, enrichment cultures from grease-contaminated soil samples were used to grow oil-metabolizing bacteria. The metaproteome acquired from the enrichment cultures was then screened for lipolytic activity. The metagenome was simultaneously extracted from the same biological samples followed by its sequencing and annotation. This enabled us to excise ML-005 based on its lipolytic activity and identify it through MS by searching against a protein database derived from the metagenome (Sukul et al., 2017). In the present study we carry out a comprehensive characterization of this novel esterase ML-005.
Sequence analysis revealed that ML-005 is a distant homolog of YdeN from B. subtilis. YdeN is a canonical alpha/beta hydrolase, which is part of a family of esterases that to our knowledge has not been biochemically characterized yet. To our knowledge, characterization of ML-005 is the first characterization of a member of this novel family and could help provide information about the characteristics of this family of lipolytic enzymes. The crystal structure of B. subtilis YdeN has been resolved previously (pdb – 1UXO) and provided insights over its putative functionality. The presence of a complete catalytic triad indicated hydrolytic activity but the absence of a hydrophobic lid suggested that it is possibly active against water-soluble esters or various thioesters (Janda et al., 2004). In the case of ML-005, a nucleophilic serine residue embedded in the characteristic pentapeptide motif composed of Ala-His-Ser-Leu-Gly strongly suggested hydrolytic activity. While non-catalytic alpha-beta hydrolases have been previously identified, these typically lacked the nucleophilic serine (Wilson et al., 2004). The pentapeptide in ML-005 is modified from the more common Gly-X-Ser-X-Gly, a highly conserved sequence feature of lipolytic enzymes (Mala and Takeuchi, 2008). Such a modified pentapeptide is typically found in thermostable lipases from Bacillus spp. and Geobacillus spp. (Cho et al., 2000; Masomian et al., 2016). ML-005 has a mesophilic to thermophilic temperature preference. It showed a relatively high optimum temperature at 45°C while still retaining 50% of its activity at 55°C. This is in contrast to thermostable lipases from Bacillus spp. and Geobacillus spp. where the optimum temperature range is between 55 and 70°C (Masomian et al., 2016). ID-1 from Bacillus thermoleovorans showed even higher optimal temperature at 75°C. On the other hand ML-005 showed higher temperature stability. ID-1 retained 50% of its activity after exposure at 60°C for 60 min, whereas ML-005 retained approximately 90% of its activity after 60 min and approximately 80% of its activity after 360 min (Cho et al., 2000). In addition to its elevated temperature profile, ML-005 is active in neutral to alkaline pH. This is similar to esterases EstOF4 and Carboest; both from Bacillus spec (Karpushova et al., 2004; Rao et al., 2013). At pH 9.5 ML-005 is reversibly inactivated, but it did show a substantially broad pH-tolerance and regained close to its full activity in optimal buffer conditions even after long incubation at pH 5 on the acidic side and pH 12 on the basic side. This is in contrast to multiple esterases characterized, where the tolerances were confined to a narrower pH range (Alex et al., 2014; Kang et al., 2017).
For lipolytic enzymes, substrate specificity may be used to distinguish between carboxylesterases and lipases (Lopes et al., 2011). Numerous carboxylesterases, active against water-soluble short-chained esters, have been previously characterized. Esterases like Ly-2 from Brevundimonas or B2T from Pelagibacterium showed their preference for C2/C4 sidechains while Rv0045c from Mycobacterium showed preference for a C6 substrate and even others like EstMY isolated from metagenomic origin showed preference for a C8 substrate (Guo et al., 2010; JunGang et al., 2010; Jiang et al., 2012; Zhang et al., 2017). This is in contrast to lipases which are active against long-chained esters. For example, Acinetobacter LipA or SRT-9 from Pseudomonas aeruginosa showed optimal activity for longer sidechains ranging from C10-C18 (Kok et al., 1995; Borkar et al., 2009; Kumar et al., 2016). ML-005 is an esterase with a preference for short-chained substrates. It shows its maximum activity toward pNP-C4 with its activity decreasing with increasing chain length. This bears similarity to other esterases found previously like est_p1 (Peng et al., 2011).
We also determined the kinetic properties of ML-005 against pNP-butyrate. With a kcat of 26 s-1, and a KM of 137.9 μM and a catalytic efficiency of kcat/Km of 1.88 × 105 M-1 s-1, ML-005 has enzymatic parameters fairly typical for esterases already characterized over the years. FNE, lipG, HydS14, and AT4 showed lower catalytic efficiency than ML-005, with HydS14 showing a catalytic efficiency as low as 1.17 × 104 M-1 s-1 (Lee et al., 2006; Park et al., 2006; Yu et al., 2010; Sriyapai et al., 2015). EstI, EStEP16, and PDF1Est showed better catalytic properties than ML-005 with EStEP16 showing an efficiency of 1.58 × 106 M-1 s-1 (Choi et al., 2004; Ay et al., 2011; Zhu et al., 2013).
ML-005 is a serine hydrolase with the nucleophilic serine integrally embedded in the active site. 3D-structure of ML-005 was modeled using Phyre2 and Ser99, Asp164, and His191 were found to be in close proximity to each other. These residues coordinate together to form a charge-relay-network at the active site and are key to the functionality of esterases and lipases (Brady et al., 1990; Dodson and Wlodawer, 1998). Consequently, our mutagenesis of these three amino acid residues resulted in a virtually complete loss of activity. Furthermore, PMSF, a serine inhibitor, deactivates ML-005. While there are known lipases and esterases that are resistant to PMSF (Makhzoum et al., 1996; Abramić et al., 1999; De Santi et al., 2016), a majority of esterases do not sterically hinder the PMSF to access the active site and are as such sensitive against it (Xin and Ying, 2013; Zhu et al., 2015; Bakir and Metin, 2016; Samoylova et al., 2018).
Mg2+, Ca2+, and Na+ have been shown to have a stimulating effect on lipolytic enzymes (Rathi et al., 2001; Sharma et al., 2002), however, we did not observe a significant effect with these metals. Cu2+ at 1 mM reduced the activity substantially to 51.71%, this phenomenon has been previously observed (Hiol et al., 2000).
In general terms, all tested organic solvents showed substantial inhibitory effect on ML-005. Only with 1% methanol, ML-005 showed 67% remaining activity and even with 10% methanol, 21% of activity remained. While there are a number of known solvent-resistant lipolytic enzymes, there are many that are unstable in organic solvents (Doukyu and Ogino, 2010). Protein engineering provides an interesting solution to the problem. For Geobacillus stearothermophilus lipase T6, two protein engineering approaches, random mutagenesis and structure-guided consensus provided mutants that showed stability improvement of 23-fold and 66-fold, respectively, with elevated half-life in 70% methanol making it viable for biodiesel production (Dror et al., 2014). Directed evolution was also used for producing the methanol-tolerant Dieselzyme 4, which outperforms the industrially used lipase from Burkholderia cepacia for biodiesel production (Korman et al., 2013). Protein engineering approaches may even be used to change substrate specificity in specific organic solvents. In the case of the well-known industrial lipase Candida antarctica Lipase B (CALB), a point mutation in the stereospecificity pocket resulted in 270-fold increase of the specificity constant for the acylation of bulky, non-native substrates in cyclohexane, when compared to wildtype (Hudson et al., 2005).
All detergents showed inhibitory effect while SDS resulted in a drastic loss of activity. Detergents have not only consequences for enzyme-detergent interaction but also for enzyme-substrate interaction. Activity can be promoted in some lipases where surfactant binding results in an increase of activity due to greater availability of substrate and increased access to the active site (Delorme et al., 2011) Exposure to SDS, an anionic detergent, may lead to tertiary conformational changes and thus may result in loss of activity (Rao et al., 2013).
Over the last decade, significant steps are being taken to transition from a petrochemical-based economy toward a sustainable bio-based economy. Biocatalysis is a necessary piece of the puzzle and searching for novel enzymes with novel characteristics continue to be worthwhile. Intrinsically stable enzymes provide a promising starting point for protein engineering endeavors to evolve enzymes that can be used for highly specialized industrial applications (Bornscheuer et al., 2012; Bornscheuer and Kourist, 2017). ML-005 is a robust esterase that is tolerant toward elevated temperature and a broad range of pH and may provide an interesting starting point for directed evolution.
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher. All strains used in this study are available upon request from the authors.
PS and LL designed the experiments. NL and PS purified ML-005. PS performed the enzymatic characterization of ML-005. PS and LL wrote the manuscript.
Principal funding for this work was provided by the European Research Council under the European Union’s Seventh Framework Program (FP7/2007-2013)/ERC Grant agreement n°281384–FuMe to LL. Part of this research was funded by the DFG Research Training Group GRK 2341 “Microbial Substrate Conversion (MiCon)”. This manuscript was written in part during a Writing Retreat funded by the Ruhr-Universität Bochum Research School RURSplus.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01925/full#supplementary-material
FIGURE S1 | SignalP prediction server predicted the presence of a signal peptide with a cleavage site between position 20 and 21 (Ferrari et al., 2014).
FIGURE S2 | Alignment of ML-005 with and without signal sequence and comparison with Bacillus subtilis YdeN (P96671).
DATA SHEET S2.FASTA | Supplementary File 1: Protein sequence of ML-005.
DATA SHEET S3.FASTA | Supplementary File 2: DNA sequence of pML-005.
Abramić, M., Leščić, I., Korica, T., Vitale, L., Saenger, W., and Pigac, J. (1999). Purification and properties of extracellular lipase from Streptomyces rimosus. Enzyme Microb. Technol. 25, 522–529. doi: 10.1016/S0141-0229(99)00077-0
Alex, D., Shainu, A., Pandey, A., and Sukumaran, R. K. (2014). Esterase active in polar organic solvents from the yeast Pseudozyma sp. NII 08165. Enzyme Res. 2014:494682. doi: 10.1155/2014/494682
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arpigny, J. L., and Jaeger, K.-E. (1999). Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343(Pt 1), 177–183. doi: 10.1042/bj3430177
Awaji, H., Fujita, Y., Heldt-Hansen, H. P., Sharyou, M., Shimoto, H., As, N. N., et al. (1998). Lipase-catalyzed ester hydrolysis.
Ay, F., Karaoglu, H., Inan, K., Canakci, S., and Belduz, A. O. (2011). Cloning, purification and characterization of a thermostable carboxylesterase from Anoxybacillus sp. PDF1. Protein Expr. Purif. 80, 74–79. doi: 10.1016/j.pep.2011.06.019
Bakir, Z. B., and Metin, K. (2016). Purification and characterization of an alkali-thermostable lipase from thermophilic Anoxybacillus flavithermus HBB 134. J. Microbiol. Biotechnol. 26, 1087–1097. doi: 10.4014/jmb.1512.12056
Barron, L. J., Hernndez, I., Bilbao, A., Flanagan, C. E., Njera, A. I., Virto, M., et al. (1999). Changes in lipid fractions and sensory properties of Idiazabal cheese induced by lipase addition. J. Dairy Res. 71, 372–379. doi: 10.1017/S0022029904000135
Borkar, P. S., Bodade, R. G., Rao, S. R., and Khobragade, C. N. (2009). Purification and characterization of extracellular lipase from a new strain: Pseudomonas aeruginosa SRT 9. Braz. J. Microbiol. 40, 358–366. doi: 10.1590/S1517-838220090002000028
Bornscheuer, U. T. (2002). Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 26, 73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x
Bornscheuer, U. T., Huisman, G. W., Kazlauskas, R. J., Lutz, S., Moore, J. C., and Robins, K. (2012). Engineering the third wave of biocatalysis. Nature 485, 185–194. doi: 10.1038/nature11117
Bornscheuer, U. T., and Kourist, R. (2017). “Evolving enzymes for biocatalysis,” in Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids: Production of Fuels and Chemicals, ed. S. Y. Lee (Cham: Springer), 1–17. doi: 10.1007/978-3-319-31421-1_217-1
Brady, L., Brzozowski, A. M., Derewenda, Z. S., Dodson, E., Dodson, G., Tolley, S., et al. (1990). A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature 343, 767–770. doi: 10.1038/343767a0
Brault, G., Shareck, F., Hurtubise, Y., Lépine, F., and Doucet, N. (2012). Isolation and characterization of EstC, a new cold-active esterase from Streptomyces coelicolor A3(2). PLoS One 7:e32041. doi: 10.1371/journal.pone.0032041
Chahiniana, H., and Sarda, L. (2009). Distinction between esterases and lipases: comparative biochemical properties of sequence-related carboxylesterases. Protein Pept. Lett. 16, 1149–1161. doi: 10.2174/092986609789071333
Cho, A. R., Yoo, S. K., and Kim, E. J. (2000). Cloning, sequencing and expression in Escherichia coli of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol. Lett. 186, 235–238. doi: 10.1111/j.1574-6968.2000.tb09110.x
Choi, Y. J., Miguez, C. B., and Lee, B. H. (2004). Characterization and heterologous gene expression of a novel esterase from Lactobacillus casei CL96. Appl. Environ. Microbiol. 70, 3213–3221. doi: 10.1128/AEM.70.6.3213-3221.2004
Côté, A., and Shareck, F. (2008). Cloning, purification and characterization of two lipases from Streptomyces coelicolor A3 (2). Enzyme Microb. Technol. 42, 381–388. doi: 10.1016/j.enzmictec.2008.01.009
Dartois, V., Coppée, J. Y., Colson, C., and Baulard, A. (1994). Genetic analysis and overexpression of lipolytic activity in Bacillus subtilis. Appl. Environ. Microbiol. 60, 1670–1673.
De Santi, C., Altermark, B., Pierechod, M. M., Ambrosino, L., de Pascale, D., and Willassen, N.-P. (2016). Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries. BMC Biochem. 17:1. doi: 10.1186/s12858-016-0057-x
Delorme, V., Dhouib, R., Canaan, S., Fotiadu, F., Carrière, F., and Cavalier, J.-F. (2011). Effects of surfactants on lipase structure, activity, and inhibition. Pharm. Res. 28, 1831–1842. doi: 10.1007/s11095-010-0362-9
Dodson, G., and Wlodawer, A. (1998). Catalytic triads and their relatives. Trends Biochem. Sci. 23, 347–352. doi: 10.1016/S0968-0004(98)01254-7
Doukyu, N., and Ogino, H. (2010). Organic solvent-tolerant enzymes. Biochem. Eng. J. 48, 270–282. doi: 10.1016/j.bej.2009.09.009
Dror, A., Shemesh, E., Dayan, N., and Fishman, A. (2014). Protein engineering by random mutagenesis and structure-guided consensus of Geobacillus stearothermophilus lipase T6 for enhanced stability in methanol. Appl. Environ. Microbiol. 80, 1515–1527. doi: 10.1128/AEM.03371-13
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Faoro, H., Glogauer, A., Couto, G. H., de Souza, E. M., Rigo, L. U., Cruz, L. M., et al. (2012). Characterization of a new Acidobacteria-derived moderately thermostable lipase from a Brazilian Atlantic Forest soil metagenome. FEMS Microbiol. Ecol. 81, 386–394. doi: 10.1111/j.1574-6941.2012.01361.x
Ferrari, A. R., Gaber, Y., and Fraaije, M. W. (2014). A fast, sensitive and easy colorimetric assay for chitinase and cellulase activity detection. Biotechnol. Biofuels 7:37. doi: 10.1186/1754-6834-7-37
Fuciños, P., González, R., Atanes, E., Sestelo, A. B. F., Pérez-Guerra, N., Pastrana, L., et al. (2012). Lipases and esterases from extremophiles: overview and case example of the production and purification of an esterase from Thermus thermophilus HB27. Methods Mol. Biol. 861, 239–266. doi: 10.1007/978-1-61779-600-5_15
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook, ed. J. M. Walker (Totowa, NJ: Humana Press), 571–607. doi: 10.1385/1-59259-890-0:571
Global Market Insights (2017). Enzymes Market Size. Available at: https://www.gminsights.com/industry-analysis/enzymes-market [accessed February 26, 2018]
Goodwin, S., McPherson, J. D., and McCombie, W. R. (2016). Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351. doi: 10.1038/nrg.2016.49
Guo, J., Zheng, X., Xu, L., Liu, Z., Xu, K., Li, S., et al. (2010). Characterization of a Novel Esterase Rv0045c from Mycobacterium tuberculosis. PLoS One 5:e13143. doi: 10.1371/journal.pone.0013143
Gupta, R., Gupta, N., and Rathi, P. (2004). Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 64, 763–781. doi: 10.1007/s00253-004-1568-8
Henne, A., Schmitz, R. A., Bömeke, M., Gottschalk, G., and Daniel, R. (2000). Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 66, 3113–3116. doi: 10.1128/AEM.66.7.3113-3116.2000
Hiol, A., Jonzo, M. D., Rugani, N., Druet, D., Sarda, L., and Comeau, L. C. (2000). Purification and characterization of an extracellular lipase from a thermophilic Rhizopus oryzae strain isolated from palm fruit. Enzyme Microb. Technol. 26, 421–430. doi: 10.1016/S0141-0229(99)00173-8
Hudson, E., Eppler, R., and Clark, D. (2005). Biocatalysis in semi-aqueous and nearly anhydrous conditions. Curr. Opin. Biotechnol. 16, 637–643. doi: 10.1016/j.copbio.2005.10.004
Janda, I., Devedjiev, Y., Cooper, D., Chruszcz, M., Derewenda, U., Gabrys, A., et al. (2004). Harvesting the high-hanging fruit: the structure of the YdeN gene product from Bacillus subtilis at 1.8 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 1101–1107. doi: 10.1107/S0907444904007188
Jiang, X., Huo, Y., Cheng, H., Zhang, X., Zhu, X., and Wu, M. (2012). Cloning, expression and characterization of a halotolerant esterase from a marine bacterium Pelagibacterium halotolerans B2T. Extremophiles 16, 427–435. doi: 10.1007/s00792-012-0442-3
JunGang, L., KeGui, Z., and WenJun, H. (2010). Cloning and biochemical characterization of a novel lipolytic gene from activated sludge metagenome, and its gene product. Microb. Cell Fact. 9:83. doi: 10.1186/1475-2859-9-83
Kang, L.-J., Meng, Z.-T., Hu, C., Zhang, Y., Guo, H.-L., Li, Q., et al. (2017). Screening, purification, and characterization of a novel organic solvent-tolerant esterase, Lip2, from Monascus purpureus strain M7. Extremophiles 21, 345–355. doi: 10.1007/s00792-016-0907-x
Karpushova, A., Brümmer, F., Barth, S., Lange, S., and Schmid, R. D. (2004). Cloning, recombinant expression and biochemical characterisation of novel esterases from Bacillus sp. associated with the marine sponge Aplysina aerophoba. Appl. Microbiol. Biotechnol. 67, 59–69. doi: 10.1007/s00253-004-1780-6
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kok, R. G., Thor, J. J., Nugteren Roodzant, I. M., Brouwer, M. B. W., Egmond, M. R., Nudel, C. B., et al. (1995). Characterization of the extracellular lipase, LipA, of Acinetobacter calcoaceticus BD413 and sequence analysis of the cloned structural gene. Mol. Microbiol. 15, 803–818. doi: 10.1111/j.1365-2958.1995.tb02351.x
Korman, T. P., Sahachartsiri, B., Charbonneau, D. M., Huang, G. L., Beauregard, M., and Bowie, J. U. (2013). Dieselzymes: development of a stable and methanol tolerant lipase for biodiesel production by directed evolution. Biotechnol. Biofuels 6:70. doi: 10.1186/1754-6834-6-70
Kourist, R., and Gaßmeyer, S. (2015). “Biocatalysis and enzyme engineering,” in Biotechnology, eds U. Kück and N. Frankenberg-Dinkel (Berlin: DE GRUYTER), 25–45. doi: 10.1515/9783110342635-004
Kumar, A., Dhar, K., Kanwar, S. S., and Arora, P. K. (2016). Lipase catalysis in organic solvents: advantages and applications. Biol. Proced. Online 18:2. doi: 10.1186/s12575-016-0033-2
Kusnezowa, A., and Leichert, L. I. (2017). In silico approach to designing rational metagenomic libraries for functional studies. BMC Bioinformatics 18:267. doi: 10.1186/s12859-017-1668-y
Kwoun Kim, H., Jung, Y. J., Choi, W. C., Ryu, H. S., Oh, T. K., and Lee, J. K. (2004). Sequence-based approach to finding functional lipases from microbial genome databases. FEMS Microbiol. Lett. 235, 349–355. doi: 10.1111/j.1574-6968.2004.tb09609.x
Lee, M.-H., Lee, C.-H., Oh, T. K., Song, J. K., and Yoon, J.-H. (2006). Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl. Environ. Microbiol. 72, 7406–7409. doi: 10.1128/AEM.01157-06
Levisson, M., van der Oost, J., and Kengen, S. W. (2007). Characterization and structural modeling of a new type of thermostable esterase from Thermotoga maritima. FEBS J. 274, 2832–2842. doi: 10.1111/j.1742-4658.2007.05817.x
Lopes, D. B., Fraga, L. P., Fleuri, L. F., and Macedo, G. A. (2011). Lipase and esterase: to what extent can this classification be applied accurately? Food Sci. Technol. 31, 603–613. doi: 10.1590/S0101-20612011000300009
Lotti, M., and Alberghina, L. (2007). “Lipases: molecular structure and function,” in Industrial Enzymes, eds J. Polaina, and A. P. McCabe (Dordrecht: Springer), 263–281. doi: 10.1007/1-4020-5377-0_16
Makhzoum, A., Owusu-Apenten, R. K., and Knapp, J. S. (1996). Purification and properties of lipase from Pseudomonas fluorescens strain 2D. Int. Dairy J. 6, 459–472. doi: 10.1016/0958-6946(95)00064-X
Mala, J. G. S., and Takeuchi, S. (2008). Understanding structural features of microbial lipases–an overview. Anal. Chem. Insights 3, 9–19. doi: 10.4137/ACI.S551
Mangla, V., Ravi, B., Mehra, S., Mehrotra, S., and Mehrotra, R. (2013). Biofuels: an overview with special emphasis on biodiesel and the role of lipase enzyme in its synthesis. Curr. Chem. Biol. 7, 96–103. doi: 10.2174/2212796811206020104
Marchler-Bauer, A., Anderson, J. B., Cherukuri, P. F., DeWeese-Scott, C., Geer, L. Y., Gwadz, M., et al. (2005). CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33, D192–D196. doi: 10.1093/nar/gki069
Masomian, M., Rahman, R. N., Salleh, A. B., and Basri, M. (2016). Analysis of comparative sequence and genomic data to verify phylogenetic relationship and explore a new subfamily of bacterial lipases. PLoS One 11:e0149851. doi: 10.1371/journal.pone.0149851
Masuch, T., Kusnezowa, A., Nilewski, S., Bautista, J. T., Kourist, R., and Leichert, L. I. (2015). A combined bioinformatics and functional metagenomics approach to discovering lipolytic biocatalysts. Front. Microbiol. 6:1110. doi: 10.3389/fmicb.2015.01110
Narayanan, A., and Jones, L. H. (2015). Sulfonyl fluorides as privileged warheads in chemical biology. Chem. Sci. 6, 2650–2659. doi: 10.1039/C5SC00408J
Ogawa, J. (1999). Microbial enzymes: new industrial applications from traditional screening methods. Trends Biotechnol. 17, 13–20. doi: 10.1016/S0167-7799(98)01227-X
Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., et al. (1992). The alpha/beta hydrolase fold. Protein Eng. 5, 197–211. doi: 10.1093/protein/5.3.197
Park, H. J., Kim, Y. J., and Kim, H. K. (2006). Expression and characterization of a new esterase cloned directly from Agrobacterium tumefaciens genome. J. Microbiol. Biotechnol. 16, 145–148.
Peng, Q., Zhang, X., Shang, M., Wang, X., Wang, G., Li, B., et al. (2011). A novel esterase gene cloned from a metagenomic library from neritic sediments of the South China Sea. Microb. Cell Fact. 10:95. doi: 10.1186/1475-2859-10-95
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Quax, W. J., and Broekhuizen, C. P. (1994). Development of a new Bacillus carboxyl esterase for use in the resolution of chiral drugs. Appl. Microbiol. Biotechnol. 41, 425–431. doi: 10.1007/BF00939031
Rao, L., Xue, Y., Zheng, Y., Lu, J. R., and Ma, Y. (2013). A novel alkaliphilic bacillus esterase belongs to the 13th bacterial lipolytic enzyme family. PLoS One 8:e60645. doi: 10.1371/journal.pone.0060645
Rappé, M. S., and Giovannoni, S. J. (2003). The uncultured microbial majority. Annu. Rev. 57, 369–394. doi: 10.1146/annurev.micro.57.030502.090759
Rathi, P., Saxena, R. K., and Gupta, R. (2001). A novel alkaline lipase from Burkholderia cepacia for detergent formulation. Process Biochem. 37, 187–192. doi: 10.1016/S0032-9592(01)00200-X
Samoylova, Y. V., Sorokina, K. N., Romanenko, M. V., and Parmon, V. N. (2018). Cloning, expression and characterization of the esterase estUT1 from Ureibacillus thermosphaericus which belongs to a new lipase family XVIII. Extremophiles 22, 271–285. doi: 10.1007/s00792-018-0996-9
Schloss, P. D., and Handelsman, J. (2003). Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14, 303–310. doi: 10.1016/S0958-1669(03)00067-3
Schmid, R. D., and Verger, R. (1998). Lipases: interfacial enzymes with attractive applications. Angew. Chem. Int. Ed. 37, 1608–1633. doi: 10.1002/(SICI)1521-3773(19980703)37:12<1608::AID-ANIE1608>3.0.CO;2-V
Schrödinger, LLC (2015). The PyMOL Molecular Graphics System, Version∼1.8. New York, NY: Schrödinger LLC.
Sharma, R., Soni, S. K., Vohra, R. M., Gupta, L. K., and Gupta, J. K. (2002). Purification and characterisation of a thermostable alkaline lipase from a new thermophilic Bacillus sp. RSJ-1. Process Biochem. 37, 1075–1084. doi: 10.1016/S0032-9592(01)00316-8
Sharma, S., and Kanwar, S. S. (2014). Organic solvent tolerant lipases and applications. Sci. World J. 2014:625258. doi: 10.1155/2014/625258
Sharma, V. K., Kumar, N., Prakash, T., and Taylor, T. D. (2009). MetaBioME: a database to explore commercially useful enzymes in metagenomic datasets. Nucleic Acids Res. 38, D468–D472. doi: 10.1093/nar/gkp1001
Simon, G. M., and Cravatt, B. F. (2010). Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J. Biol. Chem. 285, 11051–11055. doi: 10.1074/jbc.R109.097600
Sriyapai, P., Kawai, F., Siripoke, S., Chansiri, K., and Sriyapai, T. (2015). Cloning, expression and characterization of a thermostable esterase HydS14 from Actinomadura sp. Strain S14 in Pichia pastoris. IJMS 16, 13579–13594. doi: 10.3390/ijms160613579
Sukul, P., Schäkermann, S., Bandow, J. E., Kusnezowa, A., Nowrousian, M., and Leichert, L. I. (2017). Simple discovery of bacterial biocatalysts from environmental samples through functional metaproteomics. Microbiome 5:28. doi: 10.1186/s40168-017-0247-9
Suzek, B. E., Wang, Y., Huang, H., McGarvey, P. B., and Wu, C. H. (2015). UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932. doi: 10.1093/bioinformatics/btu739
The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. doi: 10.1093/nar/gkw1099
Wagner, U. G., Petersen, E. I., Schwab, H., and Kratky, C. (2002). EstB from Burkholderia gladioli: a novel esterase with a β-lactamase fold reveals steric factors to discriminate between esterolytic and β-lactam cleaving activity. Protein Sci. 11, 467–478. doi: 10.1110/ps.33002
Widmann, M., Juhl, P. B., and Pleiss, J. (2010). Structural classification by the lipase engineering database: a case study of Candida antarctica lipase A. BMC Genomics 11:123. doi: 10.1186/1471-2164-11-123
Wilhelm, S., Tommassen, J., and Jaeger, K.-E. (1999). A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 181, 6977–6986.
Wilson, R. A., Maughan, W. N., Kremer, L., Besra, G. S., and Fütterer, K. (2004). The structure of Mycobacterium tuberculosis MPT51 (FbpC1) defines a new family of non-catalytic α/β hydrolases. J. Mol. Biol. 335, 519–530. doi: 10.1016/j.jmb.2003.11.001
Winkler, F. K., D’Arcy, A., and Hunziker, W. (1990). Structure of human pancreatic lipase. Nature 343, 771–774. doi: 10.1038/343771a0
Xin, L., and Ying, Y. H. (2013). Purification and characterization of an extracellular esterase with organic solvent tolerance from a halotolerant isolate, Salimicrobium sp. LY19. BMC Biotechnol. 13:108. doi: 10.1186/1472-6750-13-108
Yin, Y., Mao, X., Yang, J., Chen, X., Mao, F., and Xu, Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451. doi: 10.1093/nar/gks479
Yu, S., Zheng, B., Zhao, X., and Feng, Y. (2010). Gene cloning and characterization of a novel thermophilic esterase from Fervidobacterium nodosum Rt17-B1. Acta Biochim. Biophys. Sin. 42, 288–295. doi: 10.1093/abbs/gmq020
Zhang, Y., Hao, J., Zhang, Y.-Q., Chen, X.-L., Xie, B.-B., Shi, M., et al. (2017). Identification and characterization of a novel salt-tolerant esterase from the deep-sea sediment of the South China Sea. Front. Microbiol. 8:441. doi: 10.3389/fmicb.2017.00441
Zhu, Y., Li, J., Cai, H., Ni, H., Xiao, A., and Hou, L. (2013). Characterization of a new and thermostable esterase from a metagenomic library. Microbiol. Res. 168, 589–597. doi: 10.1016/j.micres.2013.04.004
Keywords: esterase, lipase, metagenomics, metaproteomics, biocatalysis
Citation: Sukul P, Lupilov N and Leichert LI (2018) Characterization of ML-005, a Novel Metaproteomics-Derived Esterase. Front. Microbiol. 9:1925. doi: 10.3389/fmicb.2018.01925
Received: 24 April 2018; Accepted: 30 July 2018;
Published: 22 August 2018.
Edited by:
Pankaj Kumar Arora, Babasaheb Bhimrao Ambedkar University, IndiaReviewed by:
Ramón Alberto Batista-García, Universidad Autónoma del Estado de Morelos, MexicoCopyright © 2018 Sukul, Lupilov and Leichert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lars I. Leichert, bGFycy5sZWljaGVydEBydWhyLXVuaS1ib2NodW0uZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.