- Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
Previously, several genes, including LRA1–LRA4 and LRAR, involved in rhamnose utilization pathway, were discovered in Pichia pastoris GS115; among them, LRA3 and LRA4 were considered as key rate-determining step enzymes. A P. pastoris expression platform based on the strong rhamnose-inducible promoter PLRA3 did not meet the demands of industrial application due to poor production of recombinant proteins. To enhance recombinant protein production of this expression platform, a genetically engineered strain, P. pastoris GS115m, with decreased rhamnose metabolic flux was developed from P. pastoris GS115 by replacement of the rhamnose-inducible promoter PLRA4 with another much weaker rhamnose-inducible promoter, PLRA2. Grown in MRH and YPR media using rhamnose as the main carbon source, the engineered strain showed decreased growth rate and maximal biomass compared with the parental strain. More importantly, grown in rhamnose-containing MRH and YPR media, the recombinant engineered strain harboring a β-galactosidase gene lacB, whose expression was regulated by rhamnose-inducible PLRA3, yielded substantial increases, of 2.5- and 1.5-fold, respectively, in target protein production over the parental strain. Additionally, grown in MRH and YPR media, the engineered strain had remarkable cell flocculation and rapid sedimentation with the increasing of cell density, providing an effective and convenient separation of the fermentation supernatant from strain cells. The engineered strain is a promising expression host for industrial production of target proteins due to its advantages over the parental strain as follows: (i) improved production of recombinant proteins, and (ii) remarkable cell flocculation and rapid sedimentation.
Introduction
A previous study preliminarily investigated several rhamnose-inducible promoters in Pichia pastoris. Among them, the transcription activities of two promoters, PLRA3 and PLRA4, were higher than those of other promoters, including PAS_chr4_0338 promoter (PLRA1), PAS_chr4_0339 promoter (PLRA2), PAS_chr4_0340 promoter (PLRAR) (Liu et al., 2016), which indicated that LRA3 and LRA4 were rate-determining step enzymes related to rhamnose metabolism. Recombinant proteins under the control of the two promoters, especially PLRA3, could be efficiently produced using rhamnose as the inducer (Liu et al., 2016). Thereby, a rhamnose-inducible P. pastoris expression platform was established based on PLRA3. In this platform, rhamnose acted as the inducer for the following: (i) the transcription activation of genes involved in the rhamnose metabolic pathway to produce enzymes to metabolize rhamnose as a carbon source for P. pastoris growth, and (ii) the transcription activation of PLRA3 to produce target recombinant proteins. It was also demonstrated that both high rhamnose concentrations and long induction durations were helpful for producing high yields of target proteins. Additionally, from the perspective of industrial safety, using edible rhamnose instead of flammable and hazardous methanol as an inducer made this expression system a promising platform for the production of some valuable recombinant proteins, such as food-grade proteins and biopharmaceutical products (Vogl et al., 2013). However, the industrial application of this system is severely limited due to the relatively low yields of target proteins. It is, therefore, necessary to optimize the system to greatly enhance production of target proteins.
To optimize an expression system, several key factors, such as expression host strains, expression vectors, fermentation conditions, fermentation processes, and so on, need to be considered. Expression host strains play significant roles in the massive production of target products, and many expression host strains have been modified for use in industry (Choi et al., 2003; Demain and Vaishnav, 2009; Lee and Park, 2010). Strain engineering, mostly by means of genetic modification, provides an effective approach to improve production of target products (Idiris et al., 2010; Gottardi et al., 2017; Hossain et al., 2017). Recently, metabolic engineering strategies – for example, promoter exchange – have been widely used to rebalance metabolic flux related to synthesis of target products in rational designs of expression host strains (Tai and Stephanopoulos, 2013; Qiu et al., 2014; Willenbacher et al., 2016; Vidya et al., 2017; Wang et al., 2017; Wen et al., 2017). Strain engineering using a metabolic engineering strategy aims to improve production of secondary metabolites of interest, and heterologous recombinant proteins as terminal products were seldom reported or analyzed.
As mentioned above, LRA4 was one of the enzymes determining the rate of rhamnose metabolism in P. pastoris, and down-regulation of LRA4 expression could decrease the metabolic flux of rhamnose metabolism, leading to changes in the profile of P. pastoris. One of the expected profiles would be a decreased rhamnose metabolic rate, which would lead to the following two features: (i) a slower growth rate of the engineered strain grown in medium using rhamnose as the main carbon source; and (ii) a longer duration of high concentrations of residual rhamnose in the medium, leading to improved production of recombinant proteins controlled under the rhamnose-inducible PLRA3. Thereby, the engineered strain would differ from the parental strain in growth profile and production of the recombinant protein. Based on the above assumption, the genetically engineered strain P. pastoris GS115m, with altered rhamnose metabolic flux, was generated from P. pastoris GS115 via promoter replacement, substituting the rhamonose-inducible promoter PLRA4 with the much weaker rhamnose-inducible promoter PLRA2. Grown under specific conditions, the engineered strain significantly differed from the parental strain with regard to several factors, such as growth rate, maximal biomass, and cell flocculation and sedimentation. The engineered strain exhibited an obvious comparative advantage over the parental strain in yielding recombinant proteins.
Materials and Methods
Strains and Media
Escherichia coli Trans1-T1 (TransGen, Beijing, China) was used for gene cloning; P. pastoris GS115 (American Type Culture Collection: 20864) was the parental strain for the engineered strain and was also the host for gene expression. Plasmid pGHLRA3αLacB, strains P. pastoris GS115/LacB and P. pastoris GS115/gfp, in which lacB and gfp expression is regulated under PLRA3, was described in a previous study (Liu et al., 2016). P. pastoris GS115, P. pastoris GS115/LacB, or P. pastoris GS115/gfp were used as the control strain for P. pastoris GS115m, P. pastoris GS115m/LacB, or P. pastoris GS115m/gfp, respectively, in this study. All strains used in this study are listed in Table 1.
MD and MR media contained 1.34% yeast nitrogen base, 4 × 10−5% biotin, and 2% dextrose for MD or 2% rhamnose for MR. MRH and MDH media contained all the components in the MD and MR media, respectively, as well as 0.004% histidine. YPR and YPD media contained 1% yeast extract, 2% peptone, and 2% rhamnose for YPR or 2% dextrose for YPD. To prepare solid medium, agar was supplemented into the above media to a final concentration of 2% (w/v).
The primers used for PCR are listed in Table 2.
Construction of Different LRA4-Complemented Strains With LRA4 Disruption
It was previously confirmed that the LRA4-disrupted strain could not survive on rhamnose as the sole carbon source. Plasmids harboring different lengths of PLRA3 (210, 140, 120, and 100 bp) were constructed based on the pGHLRA3 plasmid, and LRA4 was then ligated into these vectors via the SnaBI and NotI restriction sites. Also, 2 μg DNA of each resultant plasmid digested by SwaI were transformed into the LRA4-disrupted strain, and positive transformants were screened on MR medium and further verified by PCR.
Construction of the Engineered Strain P. pastoris GS115M With Altered Rhamnose Metabolic Flux
A 220-bp DNA fragment, designated as PLRA2, comprising putative PLRA2, and a 1130-bp DNA fragment, designated as C0341, containing the whole open reading frame and putative transcription terminator of LRA4, were amplified from genomic DNA of P. pastoris GS115 using primer pairs PLRA2-F and PLRA2-R and C0341-F and C0341-R, respectively. Then, the two DNA fragments, PLRA2 and C0341, were fused into the expression cassette E0341, in which PLRA2 was located upstream of the start codon of LRA4. The zeocin-resistant gene (zeocin), a selective marker for screening positive P. pastoris transformants, was obtained from the pPICzα plasmid via PCR using primers zeoncin-F and zeoncin-R. C0341up, a 570-bp DNA fragment upstream of the start codon of LRA4, was amplified by PCR using C0341up-F and C0341up-R as primers. Finally, the three fragments, C0341up, zeocin, and E0341, were ligated into a DNA fragment by overlap-extension PCR. The resultant DNA fragment was introduced into P. pastoris GS115 via electroporation. Positive P. pastoris GS115m transformants were screened on YPD medium containing zeocin (100 μg/ml) and verified by PCR and DNA sequencing.
Preparation of a Recombinant Engineered Strain Expressing LACB or GFP
Pichia pastoris GS115m cells were cultured in YPD medium to an OD600 of 1.3∼1.5 to prepare electrocompetent cells using the Pichia Expression Kit (catalog no. K1710-01). We introduced 5 μg of the pGHLRA3αLacB or pGHLRA3gfp plasmids, linearized by restriction enzyme Swa I, into P. pastoris GS115m cells by electroporation. The cells were then spread on MD medium and grown at 28°C to form colonies.
To screen the LacB-expressing recombinant engineered strain, transformants were carefully picked and transferred to MR medium with 40 μl of 40 g/L X-gal at 28°C for 48 h. Colonies with blue plaques were the β-galactosidase-producing strain P. pastoris GS115m/LacB. To screen for the gfp-expressing recombinant engineered strain, transformants were grown in YPD medium; chromosomal DNA was then prepared as templates for amplification of gfp by PCR. The transformants harboring gfp in their chromosomal DNA were considered positive transformants P. pastoris GS115m/gfp.
Total RNA Preparation and Real-Time PCR
Three strains, P. pastoris GS115, P. pastoris GS115/LacB, and P. pastoris GS115m/LacB, were grown in YPR to an OD600 of about 12.0. The cells were harvested by centrifugation (12,000 g × 5 min) at 4°C and immediately stored at −80°C. Preparation of total RNA from different P. pastoris cells, removal of trace DNA, cDNA synthesis, and real-time PCR assays were performed using previously described methods (Liu et al., 2016).
Analysis of Growth of Different P. pastoris Strains
A single colony from each strain (P. pastoris GS115, P. pastoris GS115m, and P. pastoris GS115/ΔLRA4) was inoculated into YPD medium and grown for 48 h at 28°C with shaking. The cultures were centrifuged, washed twice with sterile water, suspended in the same volume of sterile water, inoculated into 200 ml of fresh medium (MRH, MDH, YPR, or YPD) at 1% (v/v), and then grown at 28°C with shaking. Every 12 h, the wet cell weight (WCW) of the cultures was measured and recorded to analyze growth rate and cell biomass. WCW was estimated as follows: 10 ml of fermentation cultures was sampled into preweighed 15 ml tube and was centrifuged at 10,000 g for 2 min. The supernatants were discarded, and the tube was reweighed. WCW was then calculated.
Residual rhamnose concentration in medium was measured at A540 by 3,5-dinitrosalicylic acid (DNS) assay (Miller, 1959).
LACB Expression and Enzyme Activity Assay
A single colony of different lacB-harboring Pichia cells was inoculated into YPD medium and cultured for 36 h at 28°C with shaking at 200 rpm. Cultures were inoculated into MR or YPR medium at 1% (v/v) and then grown at 28°C with shaking (200 rpm). The activity and production of β-galactosidase in the culture supernatant were determined at intervals.
β-Galactosidase activity was assayed as follows: 800 μl of 0.25% (w/v) ortho-nitrophenyl-β-galactoside (oNPG) dissolved in phosphate-citrate buffer (50 mM, pH 5.2) were pre-incubated at 60°C for 5 min; then 200 μl of enzyme solution were added, and incubated at 60°C for another 15 min; 1 ml of 10% trichloroacetic acid was then used to terminate the reaction, followed by the addition of 2 ml of 1 M Na2CO3. The absorbance of the solution was measured at 420 nm. Under standard conditions (pH 5.2, 60°C, 15 min), the amount of enzyme releasing 1 μmol of o-nitrophenol per minute was defined as one unit of β-galactosidase.
GFP Expression and Determination of Green Fluorescence Intensity
Recombinant Pichia cells harboring gfp were inoculated into YPD medium and cultured overnight at 28°C with shaking at 200 rpm. Cultures were then transferred into BMRY medium at 1% (v/v), followed by incubation for 48 h at 28°C with shaking. Cells were collected by centrifugation at 8,000 g for 2 min, washed once with sterile water, and then resuspended in 0.9% NaCl solution. Green fluorescence intensity in recombinant cells was monitored with a laser scanning confocal microscope (Nikon, Japan) at 2 s per image to collect a 1024 × 1024-pixel image.
Observation of Cell Flocculation and Sedimentation of P. pastoris Strains
A single colony of each strain (P. pastoris GS115, P. pastoris GS115m, P. pastoris GS115/LacB, and P. pastoris GS115m/LacB) was inoculated into YPD medium and then grown at 28°C with shaking at 200 rpm. Cultures from each strain were inoculated into 200 ml of fresh YPR or YPD at 1% (v/v) and then grown for 36 h at 28°C with shaking. We transferred 6 ml of culture from each strain into a bottle, and cell flocculation and sedimentation were observed when standing for different durations.
Results
Growth Characteristics of Strains Complemented With LRA4 Under Control of PLRA3
It was confirmed that PLRA3 length is positively correlated with transcription level and production of target proteins when the length is less than 210 bp (Liu et al., 2016). Additionally, the production of LRA4, which is a rate-limiting enzyme in rhamnose metabolism, would be expected to determine the growth rate of Pichia cells. To verify the relationship between the growth rate of Pichia cells and LRA4 production, the growth rates of Pichia cells in which the expression of LRA4 was under the control of different 5′-deleted PLRA3 were investigated in LRA4-complemented strains.
Colonies of transformants harboring long DNA fragments, 140- and 210-bp, containing PLRA3 controlling the high-level expression of LRA4 were clearly visible on MR medium after a 48-h incubation, and the number of transformants was steady from 48 to 240 h (Figure 1A). Meanwhile, transformants harboring a short DNA fragment, 120-bp, containing PLRA3 directing the low expression of LRA4 grew until 144 h, and the number of transformants increased afterward (Figure 1A). In addition, an LRA4-disrupted strain and an LRA4-disrupted strain containing a 100-bp DNA fragment of PLRA3 to regulate LRA4 expression did not form colonies during the test (Figure 1A). The above results showed that growth rates of LRA4-complemented strains were indeed dependent on the length of PLRA3. As the length of PLRA3 determined its transcription activity, in other words, the growth rates of LRA4-complemented strains relied on the transcription activities of PLRA3 with different length. Therefore, exchange of promoters of key genes to change expression levels of key enzymes would to some extent affect physical and physiological profiles of the engineered strains.
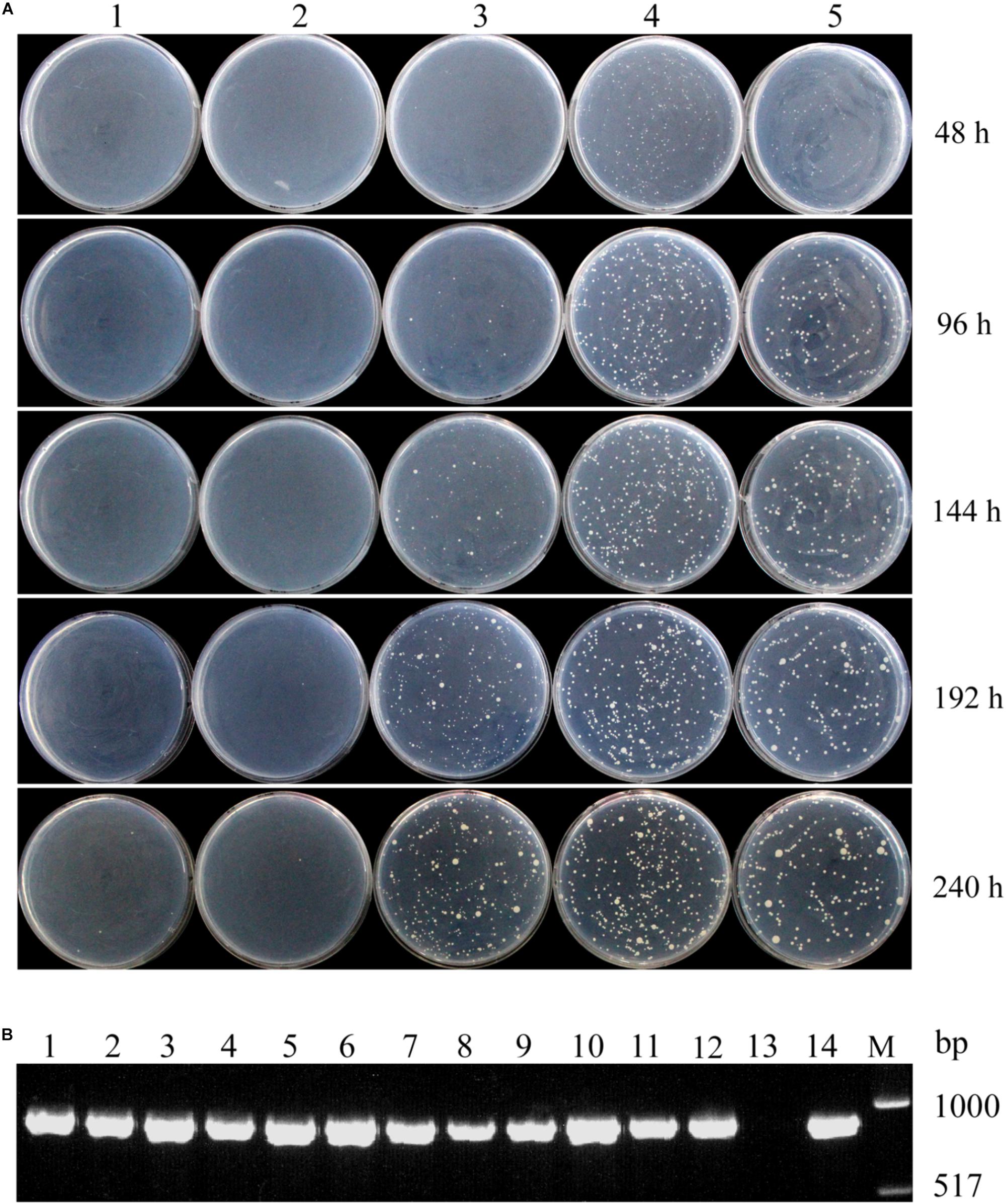
FIGURE 1. Growth profiles of different strains complemented with LRA4 under various PLRA3 lengths. (A) Colonies formed by LRA4-disrupted strains transformed by different plasmids harboring various lengths of PLRA3 controlling the expression of LRA4. 1, LRA4-disrupted strain; 2, 3, 4, and 5, colonies formed by LRA4-disrupted strains complemented with LRA4 under control of various lengths of PLRA3: 100, 120, 140, and 210 bp, respectively. (B) Verification of positive transformants via PCR. An 890-bp DNA fragment of LRA4 was amplified from chromosomal DNA of different strains. Lanes 1–4, 5–8, and 9–12, strains complemented with LRA4 under control of various lengths of PLRA3 (210, 140, and 120 bp, respectively); lane 13, LRA4-disrupted strain; lane 14, wild-type Pichia pastoris strain; lane M, DNA marker.
To further confirm that these transformants were LRA4-complemented strains, chromosomal DNA from 12 transformants harboring different lengths of PLRA3 (210, 140, and 120 bp) were extracted and used as templates for PCR to amplify LRA4. Expected PCR products with a molecular weight of about 900 bp were successfully amplified (Figure 1B), indicating that these transformants harbored LRA4.
Decreased Expression of LRA4 in the Engineered Strain With Perturbed Rhamnose Metabolic Flux
To further confirm the relationship between LRA4 expression level and the growth rate of Pichia cells and to investigate the effects of rhamnose metabolic flux rebalance on P. pastoris, the engineered strain P. pastoris GS115m was developed via a promoter exchange in which LRA4 expression was controlled by the much weaker (compared with its native promoter PLRA4) promoter PLRA2.
The transcription profiles of genes related to rhamnose metabolism, such as LRA1, LRA2, LRA3, LRAR, and LRA4, in several related strains were examined using real-time PCR. In P. pastoris GS115 and the recombinant strain P. pastoris GS115/LacB grown in medium containing rhamnose as the carbon source, LRA4 transcription levels were about fourfold higher than those of LRA2; however, in the engineered P. pastoris GS115m/LacB strain, it was almost equal to that of LRA2; as a control, LRA3 in each strain remained steady with higher transcription levels than LRA2 (Figure 2A). Simultaneously, transcription profiles of genes LRA1, LRA2, LRA3, LRAR, and LRA4 in P. pastoris GS115/LacB and P. pastoris GS115m/LacB were also investigated. The transcription level of LRA4 in P. pastoris GS115m/LacB greatly decreased while transcription profiles of other genes keep similar in the two strains (Figure 2B).
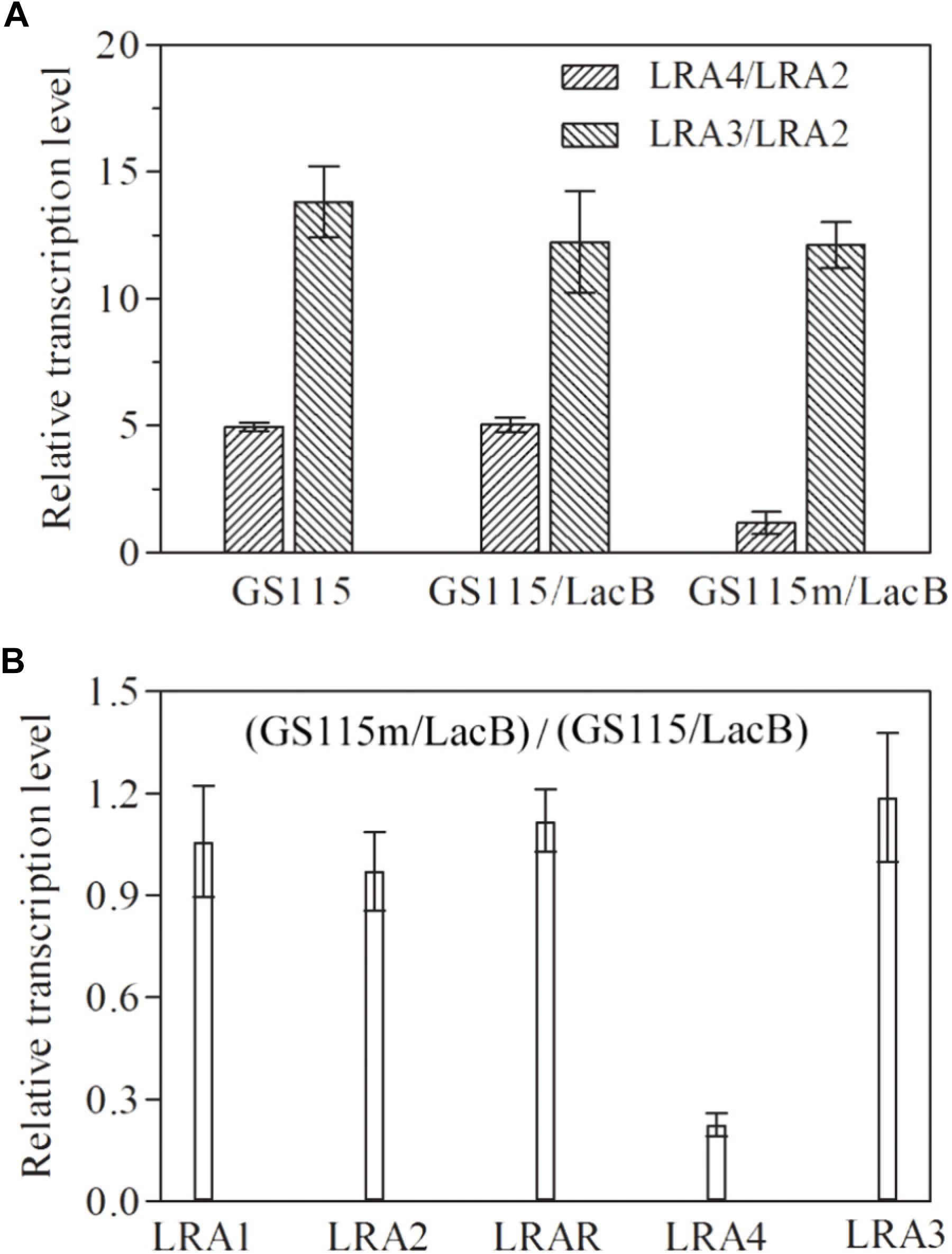
FIGURE 2. Relative transcription levels of rhamnose utilization related genes (LRA1, LRA2, LRAR, LRA3, and LRA4) in various strains grown in YPR. (A) The ratios of LRA4/LRA2 and LRA3/LRA2 in GS115, GS115/LacB, and GS115m/LacB are shown on the vertical axis. (B) Relative transcription levels of LRA1, LRA2, LRAR, LRA3, and LRA4 in GS115m/LacB, using these in GS115/LacB as controls. The GAPDH gene was used as a reference, and the relative expression value of LRA2 in each strain was designated as 1. Each test was performed in triplicate, and the results are presented as means ± SD of three replicates. GS115, P. pastoris GS115; GS115/LacB, P. pastoris GS115 harboring lacB; GS115m/LacB, P. pastoris GS115m harboring lacB.
These results suggested that the expression of LRA4 in the engineered strain was greatly down-regulated after promoter exchange, and the engineered strain developed as expected.
Growth Profiles of the Engineered Strain
In the rhamnose-inducible promoter-based P. pastoris expression platform, rhamnose plays two roles: it served as a carbon source for P. pastoris growth and acted as an inducer for the transcription activation of PLRA3 to produce recombinant proteins. It was proposed that rhamnose metabolism affected these two processes. Due to down-regulation of LRA4 expression, we expected the rhamnose metabolic rate of P. pastoris GS115m to decrease compared with that of the parental strain P. pastoris GS115, leading to the slower growth of P. pastoris GS115m when rhamnose was used as the sole carbon source.
To verify the above hypothesis, the growth profiles of three strains, P. pastoris GS115, P. pastoris GS115m, and P. pastoris GS115/ΔLRA4, which cannot utilize rhamnose as a carbon source, grown in different media were analyzed. The specific growth rate of P. pastoris GS115m grown in MRH medium, which contained 2% (w/v) rhamnose as the sole carbon source, was low compared with that of the parental strain, 0.07 vs. 0.124 h−1, although the two strains could survive on L-rhamnose (Figure 3A). It was simultaneously observed that the maximal biomass (25 mg/ml) of P. pastoris GS115m was much lower than that of the parental strain (33 mg/ml), and the maximal biomass of P. pastoris GS115m was 75.8% of that of the parental strain (Figure 3A). Grown in YPR medium using rhamnose as the main carbon source, the specific growth rates of P. pastoris GS115m and the parental strain were also different, 0.131 vs. 0.149 h−1, and the maximal biomass of P. pastoris GS115m was 80.3% of that of the parental strain (Figure 3C). However, no obvious differences in the growth rate and maximal biomass of the two strains were observed when they were grown in the following media: MDH using glucose as the sole carbon source and YPD using glucose as the main carbon source (Figures 3B,D). Based on these results, when rhamnose was the main carbon source, the perturbation in rhamnose metabolic flux affected P. pastoris; its effects included, but were not limited to, the growth rate and the maximal cell biomass.
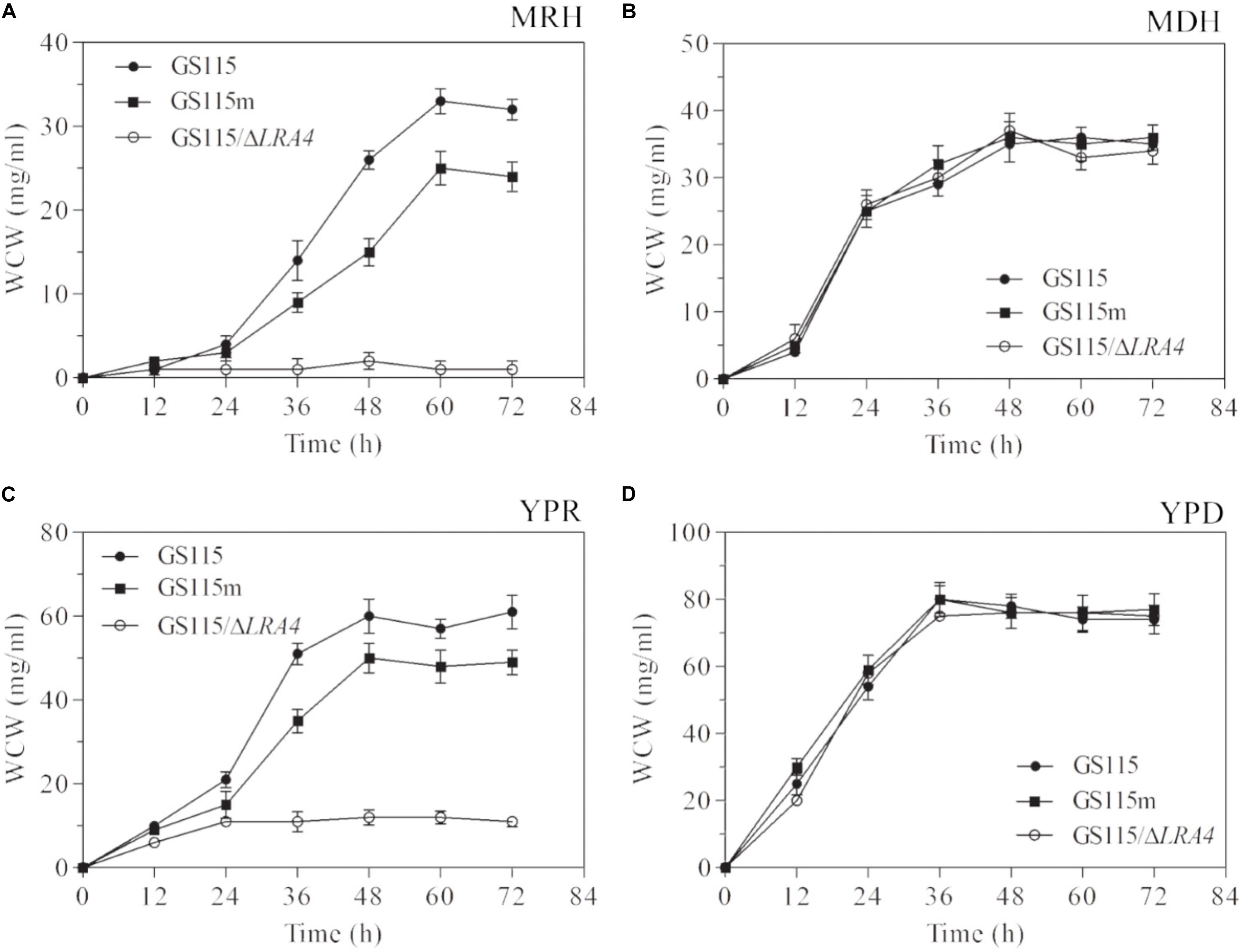
FIGURE 3. Growth characteristics of strains when grown in media MRH (A), MDH (B), YPR (C), or YPD (D). WCW, wet cell weight. GS115, P. pastoris GS115; GS115m, P. pastoris GS115m; GS115/ΔLRA4, P. pastoris GS115 with LRA4 disruption. Each test was performed in triplicate, and the results are presented as means ± SD of three replicates.
The concentrations of residual rhamnose in the cultures of the engineered strains presented higher than these in two non-genetically engineered strains, P. pastoris GS115 and P. pastoris GS115/LacB (Figure 4), which suggested that down-regulated expression of LRA4 indeed decreased rhamnose metabolism rate in the engineered strains.
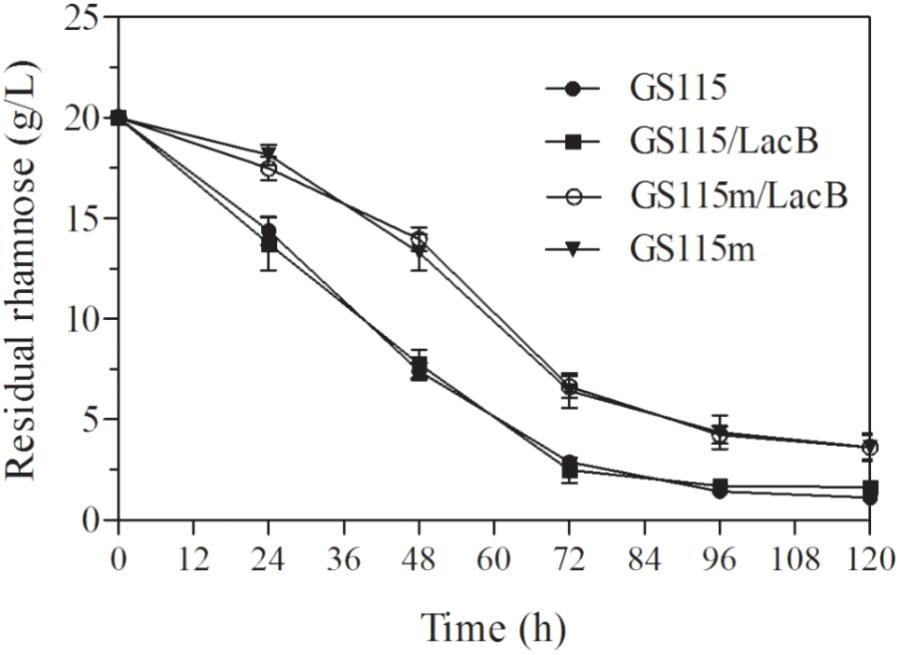
FIGURE 4. Concentrations of residual rhamnose in cultures of strains grown in YPR at different duration. GS115, P. pastoris GS115; GS115/LacB, P. pastoris GS115/LacB; GS115m, P. pastoris GS115m; GS115m/LacB, P. pastoris GS115m/LacB. Each test was performed in triplicate, and the results are presented as means ± SD of three replicates.
Enhanced Production of Target Protein in the Engineered Strain
The engineered strain differed from the parental strain with regard to growth rate and maximal cell biomass. However, we focused on whether the engineered strain could enhance the production of target proteins. Two lacB-harboring strains, P. pastoris GS115/LacB and P. pastoris GS115m/LacB, were used to examine this feature. The two strains plus P. pastoris GS115 were grown in 200 ml of YPR and MRH media, respectively, and the β-galactosidase activities in the supernatant were determined. The β-galactosidase activities in the supernatants from P. pastoris GS115/LacB and P. pastoris GS115m/LacB cultures increased with incubation time from 0 to 72 h, reaching a maximum at 96 h. The maximal β-galactosidase activities of P. pastoris GS115m/LacB in YPR and MRH media yielded 1.5- and 2.5-fold improvements over those of P. pastoris GS115/LacB, respectively (Figures 5A,B). Estimated according to specific activity of β-galactosidase (575 U/mg), β-galactosidase amounts in the supernatants reached to 84.1 μg/ml for P. pastoris GS115m/LacB and 31.6 μg/ml for P. pastoris GS115/LacB when grown in YPR medium, and 58.3 μg/ml for P. pastoris GS115m/LacB and 19.1 μg/ml for P. pastoris GS115/LacB when grown in MRH medium.
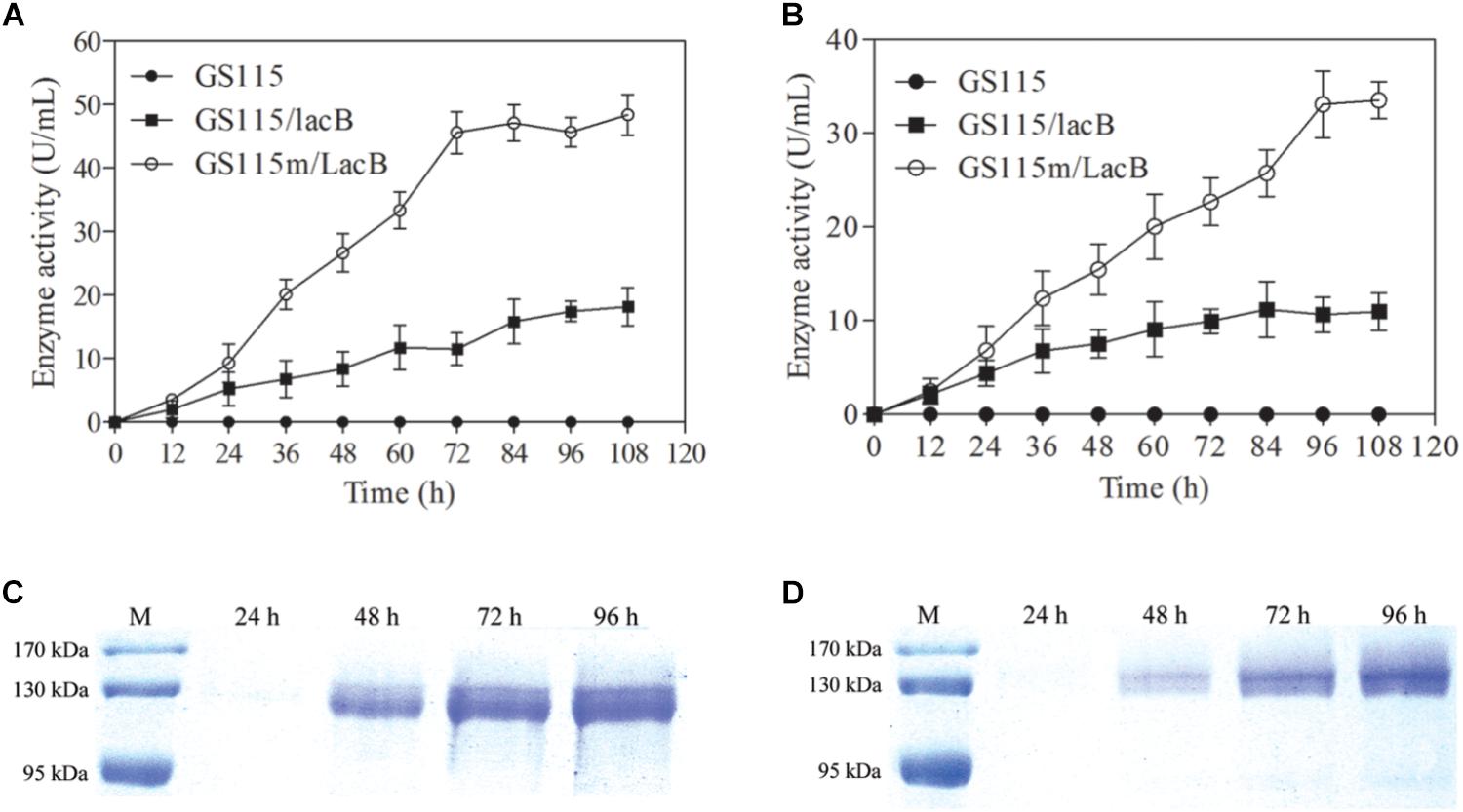
FIGURE 5. β-Galactosidase production in culture supernatants of different strains grown in different media. β-Galactosidase activity in culture supernatants of strains grown in YPR (A) or MRH (B). β-Galactosidase production in culture supernatants of GS115m/LacB (C) and GS115/LacB (D) in YPR. GS115, P. pastoris GS115; GS115/LacB, P. pastoris GS115/LacB; GS115m/LacB, P. pastoris GS115m/LacB. Each test was performed in triplicate, and the results are presented as means ± SD of three replicates.
Twenty microliter of culture supernatants from P. pastoris GS115/LacB and P. pastoris GS115m/LacB grown in YPR were loaded for SDS-PAGE, and β-galactosidase amounts were further examined using SDS-PAGE (Figures 5C,D). The results were consistent with the β-galactosidase activity analysis. P. pastoris GS115m/LacB had a higher yield of recombinant proteins compared to P. pastoris GS115/LacB.
To further investigate the expression of recombinant proteins in the engineered strain, the intracellular expression of gfp was monitored in different strains grown in YPR (Figure 6). Mean green fluorescence intensity in P. pastoris GS115/gfp and P. pastoris GS115m/gfp was 573 and 1223, respectively. The results also indicated that the expression of the target gene in the engineered strain was higher than that in the wild strain.
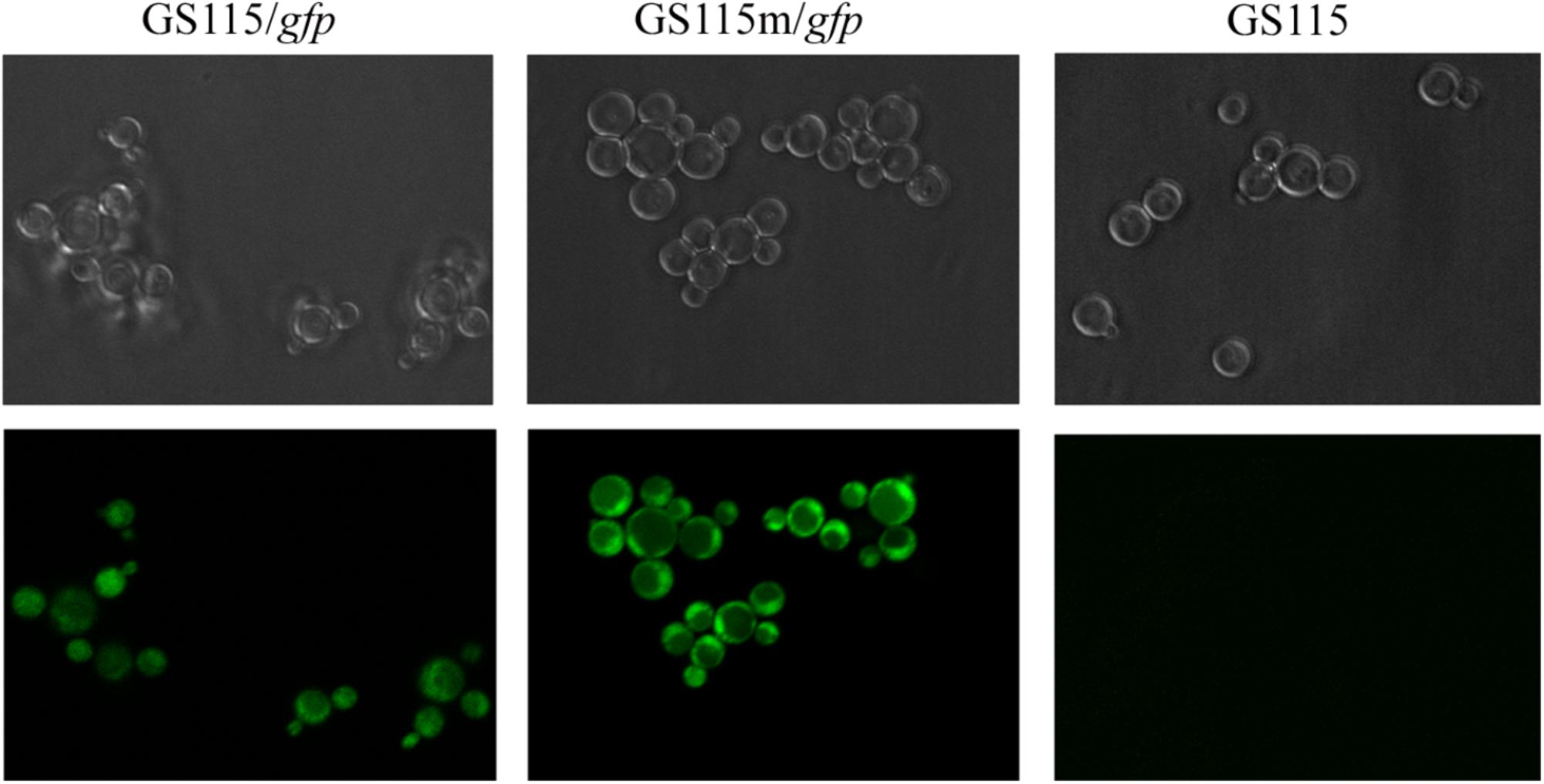
FIGURE 6. Green fluorescence in different strains grown in YPR. GS115/gfp, P. pastoris GS115 harboring gfp; GS115m/gfp, P. pastoris GS115 harboring gfp; GS115, P. pastoris GS115.
Cell Flocculation and Sedimentation in the Engineered Strains P. pastoris GS115M and P. pastoris GS115M/LACB
Rhamnose metabolism was important for the survival of Pichia cells when used as the sole source of carbon and energy. Changes in the rhamnose metabolic flux in Pichia cells may lead to numerous physiological alterations in addition to those affecting the growth rate and production of recombinant proteins.
Pichia pastoris GS115/LacB and P. pastoris GS115m/LacB were grown in rhamnose-containing medium YPR. The two strains did not present cell flocculation at low cell density (OD600 below 3.0) (Figures 7A,B); at high cell density (OD600 beyond 6.0), the engineered strain presented cell flocculation whereas P. pastoris GS115 did not (Figure 7C).
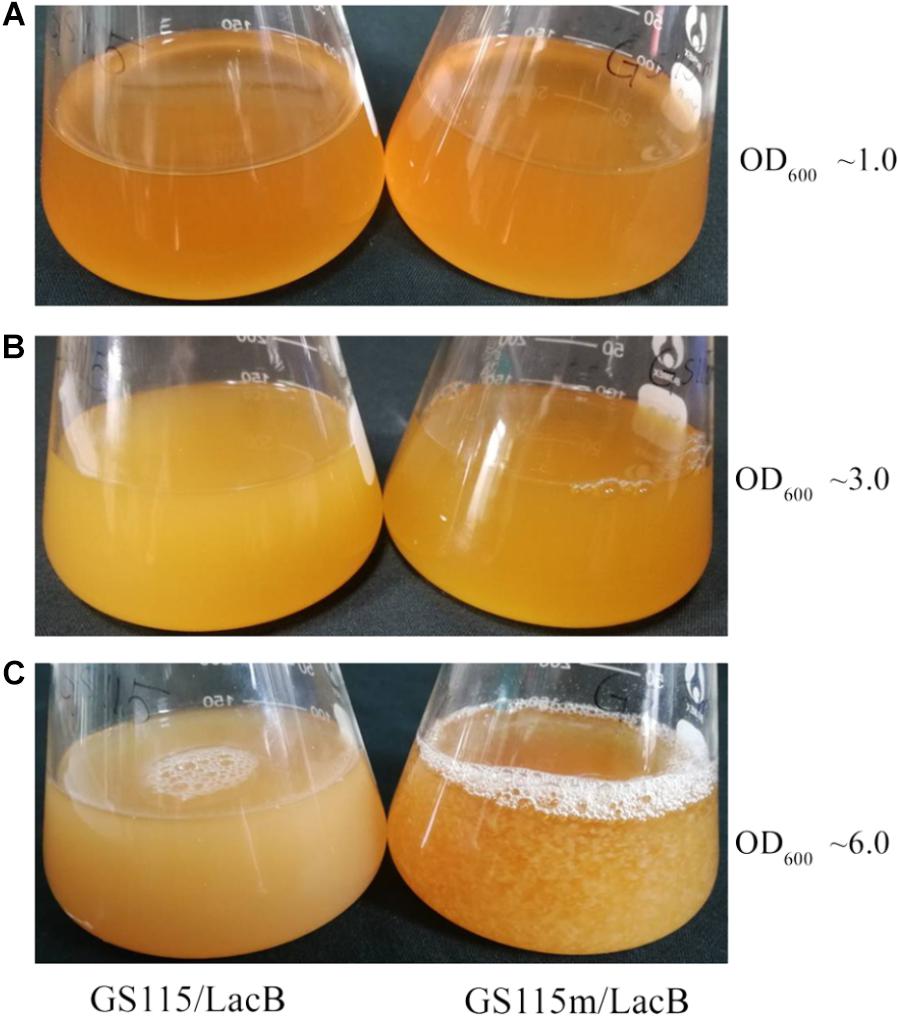
FIGURE 7. Cell flocculation profiles of GS115/LacB and GS115m/LacB grown in YPR at different cell density. (A) OD600 ∼1.0; (B) OD600 ∼3.0; (C) OD600 ∼6.0. GS115/LacB, P. pastoris GS115/LacB; GS115m/LacB, P. pastoris GS115m/LacB.
Cell flocculation accelerated the sedimentation of strain cells. Four strains, P. pastoris GS115, P. pastoris GS115m, P. pastoris GS115/LacB, and P. pastoris GS115m/LacB, were grown in YPD or YPR medium for about 36 h at 28°C; then, 6 ml of each culture were transferred into a bottle for flocculation and sedimentation analysis. In YPD medium, the cell flocculation and sedimentation of each strain were similar, and sedimentation was present during all test durations: 30 min (Figure 8A), 60 min (Figure 8B), 120 min (Figure 8C), and 240 min (Figure 8D). When cultured in YPR medium, cell flocculation occurred in the engineered strains (P. pastoris GS115m and P. pastoris GS115m/LacB), and cell sedimentation rates improved. The engineered strains almost fully settled within 30 min (Figure 8E), whereas the sedimentation of the parental strains (P. pastoris GS115, P. pastoris GS115/LacB) was present throughout the test at durations of 30 min (Figure 8E), 60 min (Figure 8F), 120 min (Figure 8G), and 240 min (Figure 8H). Rapid cell flocculation and sedimentation also happened to the engineered strains (P. pastoris GS115m and P. pastoris GS115m/LacB) when grown in MRH medium (Figures 8I–P), although rates of cell flocculation and sedimentation were low compared these when grown in YPR medium. These results suggested that more pronounced cell flocculation and more rapid sedimentation occurred in engineered strains grown in rhamnose-containing media (MRH and YPR) than in those grown in non-rhamnose containing media (MDH and YPD).
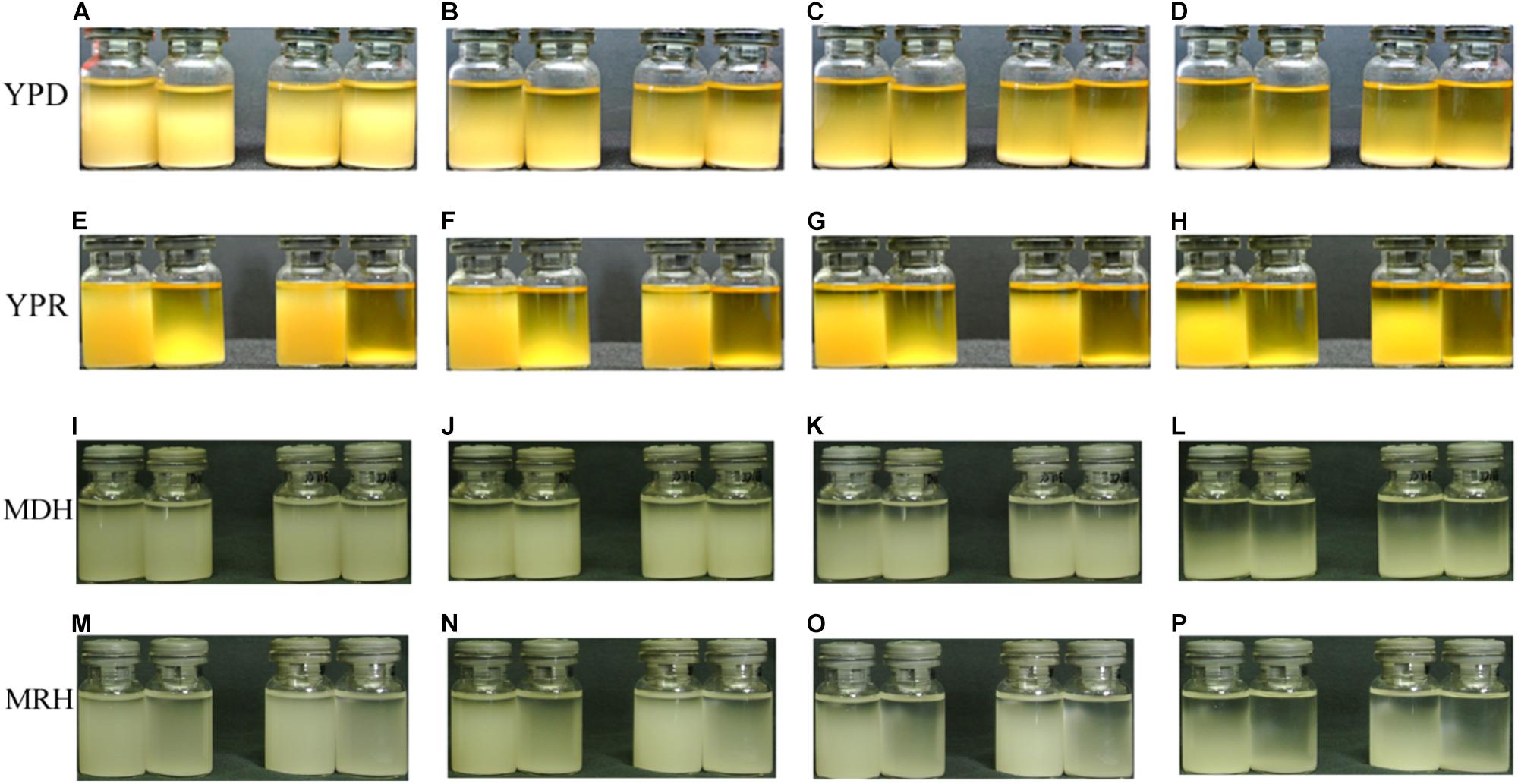
FIGURE 8. Cell flocculation and sedimentation of different strains in different media. (A,E,I,M) 30 min; (B,F,J,N) 60 min; (C,G,K,O) 120 min; (D,H,L,P) 240 min. Cultures from left to right in (A–P) are P. pastoris GS115, P. pastoris GS115m, P. pastoris GS115/LacB, and P. pastoris GS115m/LacB, respectively.
Discussion
Great progress in metabolic engineering facilitated the engineering of microbes to improve production of target products of interest (Kruyer and Peralta-Yahya, 2017). Most optimize biochemical pathways to redirect metabolic flux; for example, increasing the production of bottleneck enzymes or/and decreasing the byproduct-related gene expression yields high-level production of desired secondary metabolites, including pharmaceuticals, biofuels, and material precursors (Zhang, 2015). This study sought to improve the production of recombinant proteins instead of secondary metabolites via metabolic engineering, and the results provide insight into the engineering of other strains designed to improve the production of recombinant proteins.
To enhance the production of recombinant proteins in a rhamnose-inducible promoter PLRA3-based P. pastoris expression platform, the engineered strain with down-regulated expression of one key rhamnose metabolism-related gene, LRA4, was acquired. Decreases in both the growth rate and the maximal cell biomass of the engineered strain occurred in medium MRH using rhamnose as the sole carbon source, which was consistent with the fact that LRA4 is a step-limiting enzyme involved in rhamnose metabolism. Moreover, down-regulated expression of LRA4 led to slower growth of the engineered strain due to a lower rate of consumption of rhamnose. However, the increases in cell flocculation and in the sedimentation rate of the engineered strains grown in rhamnose-containing media (MRH and YPR) were unexpected. One possible explanation for these findings may be as follows. Cell flocculation, a mechanism for responding to environmental changes, is usually correlated with environmental stress, including limited carbon source availability (Tanneberger et al., 2007). Decreases in the rhamnose metabolic rate, which led to a lower energy supply for cell growth, may have delivered an inaccurate message to the engineered strain to the effect that the carbon source available in the medium was poor, leading cells to flocculate despite the enough availability of rhamnose in the growth medium. Cell flocculation presents numerous advantages in industrial fermentation processes, e.g., providing an effective and convenient separation of the fermentation supernatant from strain cells; and these were comprehensively described by Soares (Soares, 2011). To present, at least four aspects of the engineered strain (the decrease in growth rate, low maximal biomass, remarkable cell flocculation, and rapid sedimentation) resulted from a rebalance of the rhamnose metabolic flux, although the mechanisms underpinning the cell flocculation in rhamnose-containing media, especially YPR, are unknown.
As mentioned in Section “Introduction,” in a rhamnose-inducible expression system, rhamnose acts not only as the carbon source for growth but also as the inducer of the transcription activation of PLRA3 to direct the expression of the gene of interest. It was shown that recombinant protein production closely depends on rhamnose concentration and induction duration, with high rhamnose concentrations and long induction durations helpful for producing high-level yields of target proteins in this expression platform. In the engineered strain, a lower rhamnose metabolic rate resulted in a higher concentration of residual rhamnose. When combined with longer induction duration, this would lead the rhamnose-inducible promoter PLRA3 to produce higher levels of recombinant proteins. This explained the fact that recombinant protein yields by the engineered strain were higher than those in the parental strain.
Previously, it was verified that β-galactosidase expression level under PLRA3 was 40% of that under the most widely used promoter PAOX1 in P. pastoris (Liu et al., 2016). In this study, the β-galactosidase production of P. pastoris GS115m/LacB in YPR and MRH media presented 2.5- and 3.5-fold of those of P. pastoris GS115/LacB, respectively. In other words, production of target proteins under PLRA3 in the engineered strain was comparable with that under PAOX1 in wild strain, and recombinant gene expression level in this expression system had potential for industry.
Rhamnose is much expensive compared with dextrose. It is no doubt that recombinant protein production is cost-competitive if dextrose can be used as the carbon source. To present, dextrose is usually combined with constitutive promoters to produce recombinant proteins in P. pastoris. However, some recombinant proteins, which are lethal or toxic to P. pastoris growth, cannot be expressed using dextrose combined with constitutive promoters. When lethal or toxic recombinant proteins would be produced, dextrose can be first used as carbon source for P. pastoris growth, and then rhamnose is used as the inducer and carbon source to produce recombinant proteins once cell biomass is up to a certain value. So, dextrose and rhamnose are cooperative for producing lethal or toxic recombinant proteins in P. pastoris. Additionally, the strict and strong methanol-inducible promoter PAOX1 was usually used for heterologous protein production in P. pastoris. However, PAOX1 is not proper for recombinant protein production because of disadvantages of the inducer, methanol: highly flammable and hazardous to health. Using edible rhamnose as the inducer made rhamnose-inducible promoters be excellent in food-grade and therapeutically important recombinant proteins in P. pastoris.
In summary, down-regulated expression of LRA4 encoding a step-limiting enzyme resulted in a decrease in the rhamnose metabolic rate in the engineered strain, leading to several advantageous features for industrial applications, including remarkable cell floccution, rapid sedimentaion, and the high-level productivity of recombinant proteins. Besides, based on a transcriptional activity analysis of PLRA3 and PLRA4, LRA3 was a more important step-limiting enzyme than LRA4. Strain engineering based on LRA3, or/and LRA4, would be performed in a future study.
Author Contributions
BL and WZ designed the study. XZ, CY, XX, and YwZ performed the experiments. YZ, XX, and ZZ analyzed the data. XZ, CY, and XX wrote the manuscript. All authors participated in the discussion of the research and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31671802).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Choi, B. K., Bobrowicz, P., Davidson, R. C., Hamilton, S. R., Kung, D. H., Li, H. J., et al. (2003). Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc. Natl. Acad. Sci. U.S.A. 100, 5022–5027.
Demain, A. L., and Vaishnav, P. (2009). Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 27, 297–306. doi: 10.1016/j.biotechadv.2009.01.008
Gottardi, M., Reifenrath, M., Boles, E., and Tripp, J. (2017). Pathway engineering for the production of heterologous aromatic chemicals and their derivatives in Saccharomyces cerevisiae: bioconversion from glucose. FEMS Yeast Res. 17:fox035. doi: 10.1093/femsyr/fox035
Hossain, G. S., Yuan, H. B., and Li, J. H. (2017). Metabolic engineering of Raoultella ornithinolytica BF60 for production of 2,5-Furandicarboxylic acid from 5-Hydroxymethylfurfural. Appl. Environ. Microbiol. 83, e2312–e2316.
Idiris, A., Tohda, H., Kumagai, H., and Takegawa, K. (2010). Engineering of protein secretion in yeast: strategies and impact on protein production. Appl. Microbiol. Biotechnol. 86, 403–417. doi: 10.1007/s00253-010-2447-0
Kruyer, N. S., and Peralta-Yahya, P. (2017). Metabolic engineering strategies to bio-adipic acid production. Curr. Opin. Biotechnol. 45, 136–143. doi: 10.1016/j.copbio.2017.03.006
Lee, S. Y., and Park, J. H. (2010). Integration of systems biology with bioprocess engineering: L-threonine production by systems metabolic engineering of Escherichia coli. Adv. Biochem. Eng. Biotechnol. 120, 1–19. doi: 10.1007/10_2009_57
Liu, B., Zhang, Y. W., Zhang, X., Yan, C. L., Zhang, Y. H., Xu, X. X., et al. (2016). Discovery of a rhamnose utilization pathway and rhamnose-inducible promoters in Pichia pastoris. Sci. Rep. 6:2735. doi: 10.1038/srep27352
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428.
Qiu, Y. M., Xiao, F., Wei, X. T., Wen, Z. Y., and Chen, S. W. (2014). Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl. Microbiol. Biotechnol. 98, 8895–8903. doi: 10.1007/s00253-014-5978-y
Soares, E. V. (2011). Flocculation in Saccharomyces cerevisiae: a review. J. Appl. Microbiol. 110, 1–18. doi: 10.1111/j.1365-2672.2010.04897.x
Tai, M., and Stephanopoulos, G. (2013). Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 15, 1–9. doi: 10.1016/j.ymben.2012.08.007
Tanneberger, K., Kirchberger, J., Bar, J., Schellenberger, W., Rothemund, S., Kamprad, M., et al. (2007). A novel form of 6-phosphofructokinase - identification and functional relevance of a third type of subunit in Pichia pastoris. J. Biol. Chem. 282, 23687–23697.
Vidya, J., Sajitha, S., Ushasree, M. V., Sindhu, R., Binod, P., Madhavan, A., et al. (2017). Genetic and metabolic engineering approaches for the production and delivery of L-asparaginases: an overview. Bioresour. Technol. 245, 1775–1781. doi: 10.1016/j.biortech.2017.05.057
Vogl, T., Hartner, F. S., and Glieder, A. (2013). New opportunities by synthetic biology for biopharmaceutical production in Pichia pastoris. Curr. Opin. Biotechnol. 24, 1094–1101. doi: 10.1016/j.copbio.2013.02.024
Wang, C. H., Zhang, C., and Xing, X. H. (2017). Metabolic engineering of Escherichia coli cell factory for highly active xanthine dehydrogenase production. Bioresour. Technol. 245, 1782–1789. doi: 10.1016/j.biortech.2017.05.144
Wen, Z. Q., Minton, N. P., Zhang, Y., Li, Q., Liu, J. L., Jiang, Y., et al. (2017). Enhanced solvent production by metabolic engineering of a twin-clostridial consortium. Metab. Eng. 39, 38–48. doi: 10.1016/j.ymben.2016.10.013
Willenbacher, J., Mohr, T., Henkel, M., Gebhard, S., Mascher, T., Syldatk, C., et al. (2016). Substitution of the native srfA promoter by constitutive Pveg in two B. subtilis strains and evaluation of the effect on surfactin production. J. Biotechnol. 224, 14–17. doi: 10.1016/j.jbiotec.2016.03.002
Keywords: Pichia pastoris, rhamnose-inducible promoter, promoter exchange, rhamnose metabolic, cell flocculation
Citation: Yan C, Xu X, Zhang X, Zhang Y, Zhang Y, Zhang Z, Zhang W and Liu B (2018) Decreased Rhamnose Metabolic Flux Improved Production of Target Proteins and Cell Flocculation in Pichia pastoris. Front. Microbiol. 9:1771. doi: 10.3389/fmicb.2018.01771
Received: 16 May 2018; Accepted: 16 July 2018;
Published: 02 August 2018.
Edited by:
Christopher Bagwell, Pacific Northwest National Laboratory (DOE), United StatesReviewed by:
Huansheng Cao, Arizona State University, United StatesWeerawat Runguphan, National Center for Genetic Engineering and Biotechnology (BIOTEC), Thailand
Copyright © 2018 Yan, Xu, Zhang, Zhang, Zhang, Zhang, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhang, emhhbmd3ZWkwMkBjYWFzLmNu; endfYmlvQGNhYXMuY24=; Bo Liu, bGl1Ym8wMUBjYWFzLmNu
†These authors have contributed equally to this work.
 Chengliang Yan†
Chengliang Yan† Wei Zhang
Wei Zhang