
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 July 2018
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01670
 Francesco Iannelli1*
Francesco Iannelli1* Francesco Santoro1
Francesco Santoro1 Maria Santagati2
Maria Santagati2 Jean-Denis Docquier1
Jean-Denis Docquier1 Elisa Lazzeri1
Elisa Lazzeri1 Gabiria Pastore1
Gabiria Pastore1 Marco Cassone1†
Marco Cassone1† Marco R. Oggioni1†
Marco R. Oggioni1† Gian M. Rossolini1†
Gian M. Rossolini1† Stefania Stefani2
Stefania Stefani2 Gianni Pozzi1*
Gianni Pozzi1*The mef(A) gene was originally identified as the resistance determinant responsible for type M resistance to macrolides, a phenotype frequently found in clinical isolates of Streptococcus pneumoniae and Streptococcus pyogenes. MefA was defined as a secondary transporter of the major facilitator superfamily driven by proton-motive force. However, when characterizing the mef(A)-carrying elements Tn1207.1 and Φ1207.3, another macrolide resistance gene, msr(D), was found adjacent to mef(A). To define the respective contribution of mef(A) and msr(D) to macrolide resistance, three isogenic deletion mutants were constructed by transformation of a S. pneumoniae strain carrying Φ1207.3: (i) Δmef(A)–Δmsr(D); (ii) Δmef(A)–msr(D); and (iii) mef(A)–Δmsr(D). Susceptibility testing of mutants clearly showed that msr(D) is required for macrolide resistance, while deletion of mef(A) produced only a twofold reduction in the minimal inhibitory concentration (MIC) for erythromycin. The contribution of msr(D) to macrolide resistance was also studied in S. pyogenes, which is the original host of Φ1207.3. Two isogenic strains of S. pyogenes were constructed: (i) FR156, carrying Φ1207.3, and (ii) FR155, carrying Φ1207.3/Δmsr(D). FR155 was susceptible to erythromycin, whereas FR156 was resistant, with an MIC value of 8 μg/ml. Complementation experiments showed that reintroduction of the msr(D) gene could restore macrolide resistance in Δmsr(D) mutants. Radiolabeled erythromycin was retained by strains lacking msr(D), while msr(D)-carrying strains showed erythromycin efflux. Deletion of mef(A) did not affect erythromycin efflux. This data suggest that type M resistance to macrolides in streptococci is due to an efflux transport system of the ATP-binding cassette (ABC) superfamily, in which mef(A) encodes the transmembrane channel, and msr(D) the two ATP-binding domains.
Macrolides are antibiotic compounds composed of 14 (erythromycin and clarithromycin), 15 (azithromycin), or 16 (josamycin, spiramycin, tylosin)-membered lactones to which amino and/or neutral sugars are linked (Roberts et al., 1999; Roberts, 2008). Resistance to macrolides in streptococci is usually associated with two major mechanisms: (i) target site modification, arising from the presence of erm(B) or erm(A) subclass erm(TR) belonging to the class of erm (erythromycin ribosome methylation) methylase genes (Leclercq and Courvalin, 1991; Seppala et al., 1998) and (ii) drug efflux, associated to the presence of mef(A) (macrolide efflux) genes (Clancy et al., 1996; Sutcliffe et al., 1996; Tait-Kamradt et al., 1997). Methylation of 23S rRNA causes a reduced binding to macrolide, lincosamide, and streptogramin B antibiotics (MLSB phenotype), whereas active efflux of macrolides confers a low level of resistance to resistance only to 14- and 15-membered macrolides, but not to 16-membered macrolides, lincosamides, and streptogramin B antibiotics (M phenotype; Weisblum, 1995; Roberts et al., 1999).
The macrolide efflux mef(A) gene was originally described in Streptococcus pyogenes (Clancy et al., 1996) while the allelic variant mef(E) was first described in Streptococcus pneumoniae (Tait-Kamradt et al., 1997). These variants are highly homologous (about 90% nucleotide identity) and are grouped in the same mef(A) class of macrolide resistance genes (Roberts et al., 1999). The mef(A) gene was not found only in S. pyogenes and S. pneumoniae, but also in a wide variety of other streptococcal species such as Streptococcus agalactiae, Streptococcus mitis, Streptococcus oralis, and Streptococcus salivarius, in other Gram-positive genera including Corynebacterium, Enterococcus, Micrococcus, and Staphylococcus, and in Gram-negative genera such as Acinetobacter, Bacteroides, and Neisseria (Roberts et al., 1999; Klaassen and Mouton, 2005; Santagati et al., 2009). In some countries, the mef(A) gene has become more common than erm(B) in macrolide-resistant S. pneumoniae and S. pyogenes (Green et al., 2006; Reinert et al., 2006; Rudolph et al., 2013) and it has been used as a marker for molecular epidemiology studies (Daly et al., 2004; Klaassen and Mouton, 2005; Amezaga and McKenzie, 2006). The mef(A) allelic variants are carried by different genetic elements. In S. pneumoniae, we described a 7244-bp non-conjugative element named Tn1207.1 carrying mef(A), whereas mef(E) was found to be carried by the 5532-bp pneumococcal genetic element (mega; Santagati et al., 2000; Gay and Stephens, 2001; Del Grosso et al., 2002, 2006). In S. pyogenes, mef(A) is carried by Φ1207.3, a 52,491-bp prophage which we found in the erythromycin-resistant clinical strain 2812A, transferable to a variety of streptococcal species and whose left 7244-bp sequence is 100% identical to pneumococcal Tn1207.1 (Santagati et al., 2003; Pozzi et al., 2004; Iannelli et al., 2014a). Other mef(A)-carrying prophages in clinical isolates of S. pyogenes include Φ10394.4 (Banks et al., 2003), Φm46.1, and its variant VP_00501.1 (Brenciani et al., 2004, 2010; Vitali et al., 2016).
At first, Mef(A) was defined as a secondary transporter of the major facilitator superfamily (MFS)1 and considered to be the only gene product responsible for type M macrolide resistance, as shown by cloning and functional characterization in Escherichia coli (Clancy et al., 1996). However, in mef(A)-carrying genetic elements such as Tn1207.1 and Φ1207.3, another macrolide resistance gene, msr(D), was always found adjacent to mef(A), and msr(D) was shown to contribute to the macrolide efflux resistance of streptococcal strains carrying the mef(A)–msr(D) tandem pair (Iannelli et al., 2004; Ambrose et al., 2005; Vitali et al., 2016; Zhang et al., 2016; Tatsuno et al., 2018).
To assess the relative contribution of mef(A) and msr(D) to macrolide efflux, we generated in-frame isogenic mutants of S. pneumoniae and S. pyogenes carrying mef(A)-msr(D) to be used in functional studies. Mutant strains were tested by (i) determining the minimal inhibitory concentration (MIC) of erythromycin and (ii) assaying the actual intracellular accumulation of radiolabeled erythromycin. Results of functional studies were in accordance with bioinformatics analysis predicting that the tandem mef(A)-msr(D) gene pair encodes an efflux transport system of the ATP-binding cassette (ABC) superfamily.
Protein sequence analysis was performed with the softwares Phyre2 (Kelley et al., 2015) and TMpred. Conserved aminoacids were identified with the PSI-BLAST multiple sequence alignment. Terminator sequence was predicted with RNAstructure ver. 5.02 (Mathews Lab). Nucleotide sequence analysis was performed using Microbial Nucleotide BLAST with the Megablast algorithm2.
Streptococcal strains used in this work and their relevant properties are reported in Table 1. Bacteria were routinely grown in Tryptic Soy Broth (TSB) or Tryptic Soy Agar (Difco) supplemented with 3% horse blood at 37°C. When required, 500 μg/ml streptomycin, 10 μg/ml novobiocin, 0.5 μg/ml erythromycin, 100 μg/ml spectinomycin, and 3 μg/ml chloramphenicol were added to both liquid and solid media.
To allow construction of mutants deleted for the macrolide resistance genes object of this study, and to facilitate further analysis, it was essential to work in a S. pneumoniae strain which is readily transformable and whose genomic sequence is available (Hoskins et al., 2001). For this reason, we transferred Φ1207.3 to the chromosome of unencapsulated laboratory strains of S. pneumoniae which are derived from type 2 strain D39 (Iannelli et al., 1999; Pearce et al., 2002).
Integration of Tn1207.1 or Φ1207.3 into the chromosome of S. pneumoniae occurs within the cds of celB, a gene involved in DNA uptake during transformation, and disruption of celB leads to impairment of competence for genetic transformation in S. pneumoniae strains carrying Tn1207.1 or Φ1207.3 (Santagati et al., 2000, 2003). For this reason, strain FR125 was constructed in which celB was deleted and integration of Φ1207.3 after mating occurred at a different chromosomal location (Iannelli and Pozzi, unpublished results). Subsequently, strain FR183, a transformable S. pneumoniae DP1004 derivative strain carrying Φ1207.3 integrated elsewhere than celB, was obtained by transformation with the chromosomal DNA purified from FR125 (Table 1).
Mating experiments were performed as already described (Iannelli and Pozzi, 2007). Briefly, cells were grown separately in presence of appropriate antibiotics until the end of exponential phase (OD590nm = 0.8). Cells were mixed at 1:10 ratio (Donor/Recipient), harvested by centrifugation and plated. Following 4-h incubation, cells were harvested by scraping the plates. Selection of recombinants was carried out with a multilayer plating procedure. Transfer frequencies were determined by plating alone each parent strain that was also checked for spontaneous antibiotic resistance acquisition.
Oligonucleotide primers used for mutagenesis, sequencing, and PCR selection of recombinant strains are listed in Supplementary Table S1.
Gene splicing by overlap extension (gene SOEing) was used to generate the mutagenic constructs for gene deletions as already described (Pearce et al., 2002; Iannelli and Pozzi, 2004). The deletion of mef(A) and msr(D) genes was obtained with a mutagenic construct containing the ami/cat cassette flanked at the left by a 195-bp DNA fragment corresponding to nucleotides 3112–3307 of Φ1207.3 and at the right by a fragment corresponding to the 532 nucleotides located downstream of the msr(D) stop codon. The ami/cat chloramphenicol-resistance cassette was obtained with the primer pair IF38/IF39 using the E. coli pEVP3 plasmid as template (Claverys et al., 1995). The flanking regions were amplified, respectively, with the primer pairs IF105/F35 and IF183/IF175 from chromosomal DNA of FR183. To minimize possible polar effects, the mef(A) coding sequence (cds) was deleted in-frame by allelic replacement with the cds of the cat chloramphenicol-resistance gene. The cat cds was amplified with primer pair IF184/IF39 from a Ωcat(pC194)-carrying strain (Iannelli et al., 2014b) and flanked by a 1116-bp DNA region located upstream mef(A) start codon and a DNA fragment spanning the 449 nucleotides downstream of the mef(A) stop codon, obtained, respectively, with the primer pairs IF176/IF182 and IF185/F20 from FR183 chromosome. A genetic construct containing the ami/spc cassette flanked by the 281-bp and 532-bp DNA fragments located respectively upstream and downstream of msr(D) cds was produced to inactivate msr(D). The primer couple IF100/IF101 was used to amplify the spectinomycin-resistance cassette from the pR412 plasmid (Martin et al., 2000). This cassette was flanked by the msr(D) flanking segments amplified with the primer pair IF171/IF174 from FR183 chromosome. Linear PCR constructs were used directly as donor DNA in transformation experiments. Mutant strains were selected for acquisition of chloramphenicol or spectinomycin resistance and the desired mutations were confirmed by sequencing. PCR-based selection of S. pneumoniae complemented strains was carried out with the primers pair IF169/IF170 according to the protocol already described (Iannelli and Pozzi, 2004).
The MIC was assessed by microdilution techniques as suggested by the (Clinical and Laboratory Standards Institute [CLSI], 2017). Briefly, MICs were determined as follows: bacterial cells were grown until exponential growth phase (OD590nm = 0.25) in TSB medium and stored at -70°C in 10% glycerol, then frozen cultures were thawed and diluted at 5 × 104 CFU/ml in TSB broth containing serial twofold dilutions of antibiotic, and incubated at 37°C for 18 h. Bacterial growth was determined turbimetrically using the microplate ELISA reader VERSAmax (Molecular Devices). Results are presented as the geometric mean and are derived from at least three experiments. S. pneumoniae ATCC49619 was used as reference control strain as recommended (Clinical and Laboratory Standards Institute [CLSI], 2017).
Frozen starter culture was diluted 100-fold in 100 ml warm TSB medium and grown at 37°C until an optical density at 590 nm of 0.25. Radiolabeled erythromycin [N-methyl-14C] (50 μCi/mmol, purchased from Bio Trend, Germany) was added at a final concentration of 0.2 μg/ml (Sutcliffe et al., 1996). Erythromycin uptake was assessed by taking 5-ml samples from each culture every 10 min for 40 min after the addition of [14C]erythromycin, filtering samples with prewet GF/C glass microfiber filters (Whatman), and washing the filters twice with 5 ml of a 0.9% NaCl and 1 μg/ml erythromycin solution (Sutcliffe et al., 1996). Filters were dried O.N., dissolved in 10 ml of Insta-Gel Plus liquid (Packard) and the [14C] erythromycin cell-associated was counted with a TRI-CARB 2000 CA Liquid Scintillation Analyzer (Packard).
In clinical isolates of macrolide resistant streptococci, the macrolide efflux gene, mef(A), is found in tandem with msr(D), a gene which encodes for an ATP-binding transporter (Santagati et al., 2000, 2003; Gay and Stephens, 2001; Banks et al., 2003; Brenciani et al., 2004). Bioinformatic analysis of the prototypic tandem mef(A)-msr(D) from Tn1207.1 (Santagati et al., 2000) and their flanking regions revealed that they constitute a two-gene operon which is 3290 bp long, with only one putative promoter sequence -35 TTGCTT, extended -10 TGTGTTAAAAT (nucleotides 2890–2895 and 2908–2918, respectively, Genbank AF227520) located upstream of the mef(A) start codon and a single predicted terminator sequence (nucleotides 6143–6179, Genbank AF227520) located downstream of the msr(D) stop codon. As previously demonstrated (Ambrose et al., 2005; Chancey et al., 2015), RT-qPCR analysis confirmed that mef(A) and msr(D) genes were part of a single transcriptional unit (data not shown). Sequence analysis of the Mef(A) protein with TMpred showed the presence of 12 predicted transmembrane domains, which have the potential to form six transmembrane helices (Figure 1). When the protein sequence of Msr(D) was analyzed by the PSI-BLAST multiple sequence alignment, it was possible to show the presence of two nucleotide binding domains (NBDs) typical of ATP-binding transporters (spanning amino acids 12–154, and 304–463, Genbank AAG12999.1), and a long predicted alpha helical structure (spanning amino acids 181–227) between the two NBDs (Figure 1). Altogether, mef(A) and msr(D) appear to constitute a two-gene efflux transport system of the ABC superfamily, mef(A) encodes the pore (transmembrane channel), and msr(D) the two ATP-binding domains.
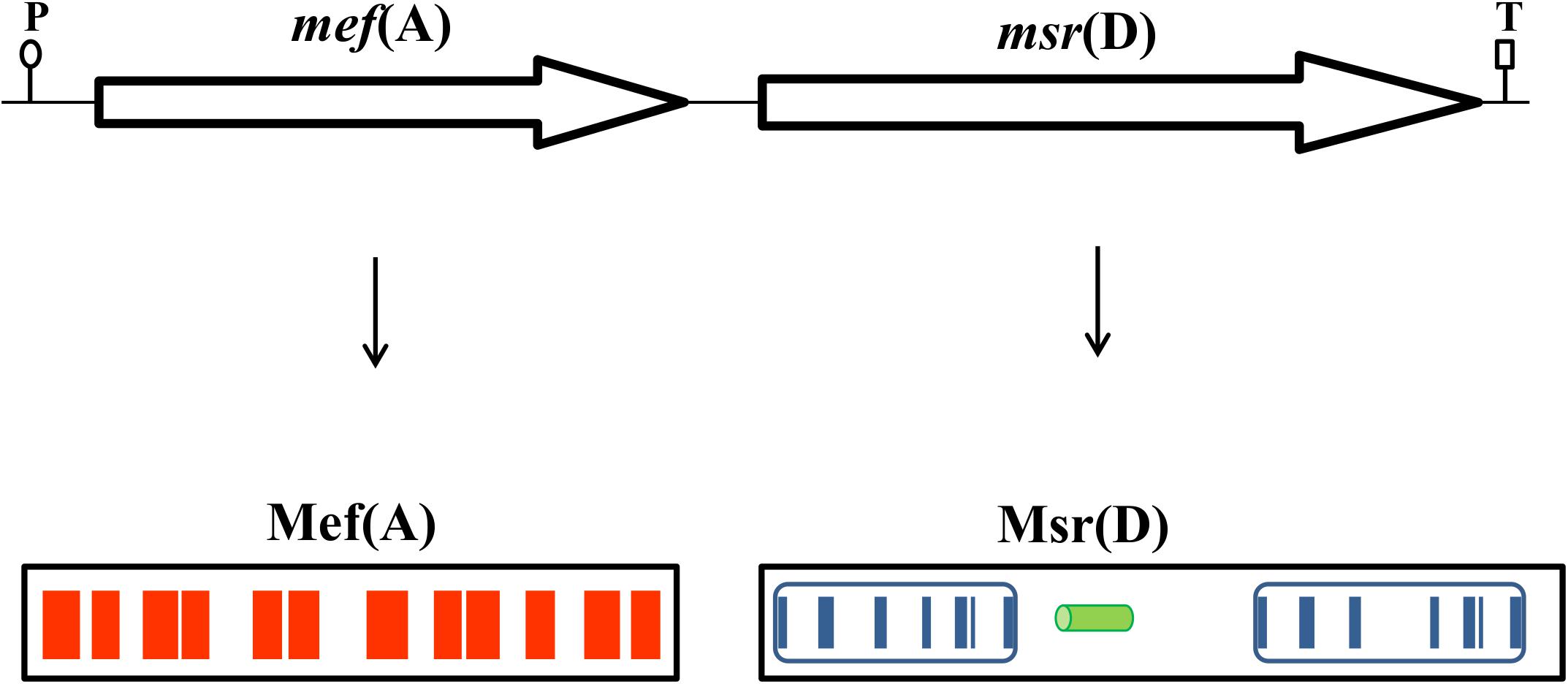
FIGURE 1. The mef(A)–msr(D) operon is 3290-bp-long and includes a single promoter sequence (P) and a terminator sequence (T). Mef(A) contains 12 transmembrane domains spanning the whole protein (red bars). Msr(D) contains two nucleotide binding domains (NBDs, blue boxes) connected by an alpha helical structure (green cylinder). Each NBD has seven motifs (blue bars): (1) the “A-loop” that provides an aromatic side chain residue that interacts with the adenine ring of bound nucleotide (phenylalanine in both NBDs); (2) the “Q-loop” involved in the interaction with the transmembrane domain and with the γ-phosphate through a water bond; (3) the “P-loop” (or “Walker-A” motif) that binds the nucleotide; (4) the “LSGGQ motif” (also called “C-loop” or “ABC signature motif”) which contacts the nucleotide in the ATP-bound state; (5) the “Walker-B” motif that has a conserved glutamate residue that orchestrates the nucleophilic attack on ATP via a water molecule; (6) the “D-loop” involved in the contact between the two NBDs through an interaction with the Walker-A asparagine residue; and (7) the “switch motif” that contains a histidine side chain thought to contribute to the catalytic reaction. Both C-loop motifs of Msr(D), MSGGE and LSGGE, are not identical to the consensus (LSGGQ).
The mef(A) gene sequence (GenBank AF227520) was used as a query to interrogate the database of 20,187 complete Microbial genomes (accessed in May 2018). The mef(A) gene was found in 37 genomes, of those: 29 were streptococcal genomes and eight belonged to other genera (namely Turicibacter, Clostridium, Enterococcus, Clostridioides, Gardnerella, and Bacillus). In 33 out of 37 cases, the mef(A) gene was in tandem with msr(D), while in the remaining four cases, msr(D) was substituted by a gene coding for a putative nucleotidyltransferase. No information on macrolide resistance was available for these four genome strains.
To define the respective contribution of mef(A) and msr(D) gene products to macrolide efflux resistance, isogenic deletion mutants were constructed in S. pneumoniae for the mef(A)–msr(D) tandem pair carried by two different genetic elements (Figure 2). Three isogenic deletion mutants were constructed in the Φ1207.3-carrying strain FR183: (i) FP39, Δmef(A)–Δmsr(D); (ii) FP40, Δmef(A); and (iii) FP59, Δmsr(D). In FP39, a 2748-bp DNA fragment (position 3308–6055, GenBank AY657002) containing mef(A) and msr(D) was deleted and replaced with the 850-bp ami/cat cassette. In FP40, a 1218-bp DNA fragment (position 3255–4472, GenBank AY657002) containing the cds of mef(A) was deleted and replaced in-frame with the 651-bp cds of the cat gene. In FP59, a 1464-bp DNA fragment (position 4592–6055, GenBank AY657002) containing the msr(D) cds was deleted and replaced with the 894-bp ami/spc cassette (Figure 2 and Table 1).
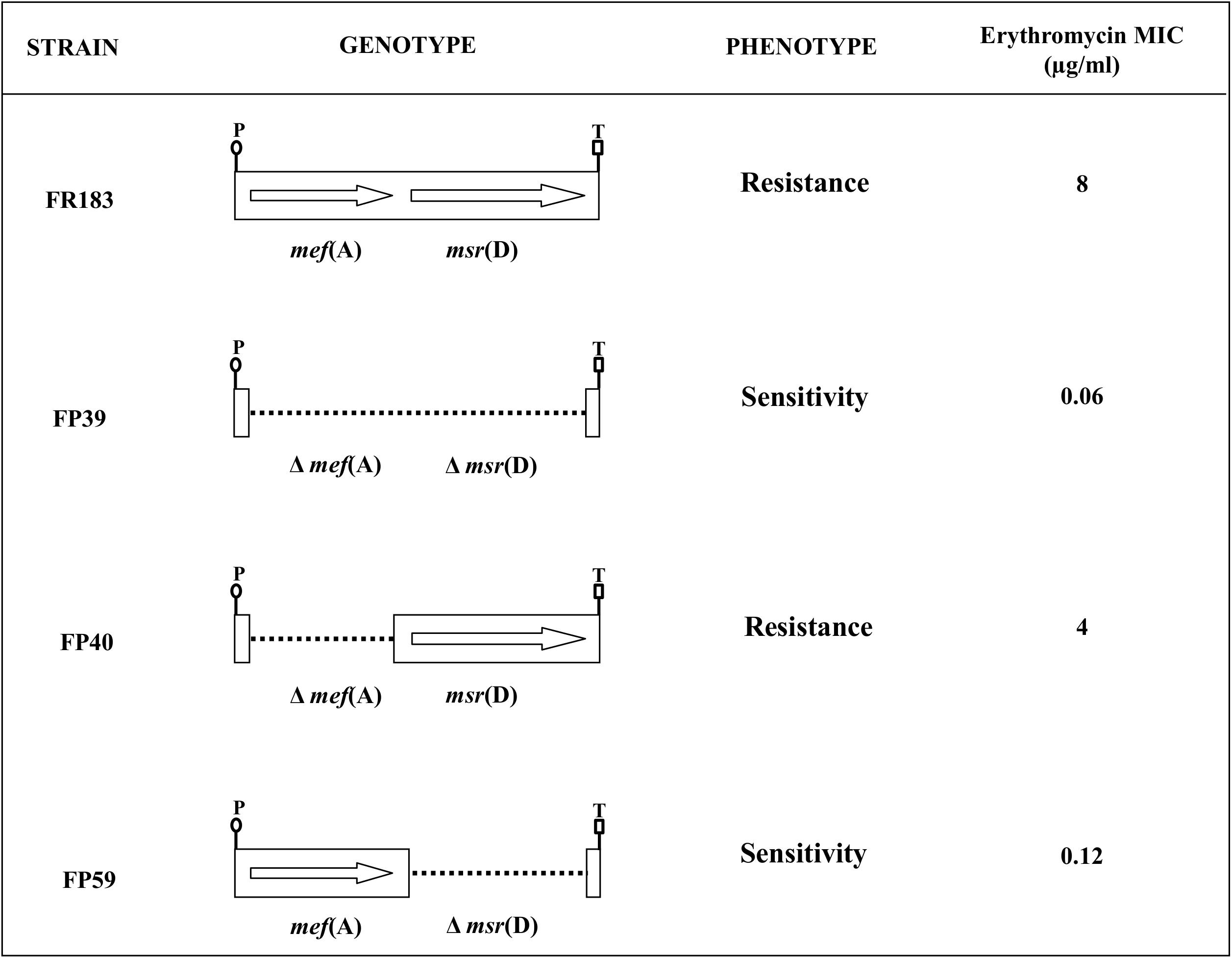
FIGURE 2. The mef(A)-msr(D) operon, carried by Φ1207.3, confers type M resistance to macrolides. Isogenic mutants were produced in a Φ1207.3-carrying S. pneumoniae strain. CDSs were deleted by allelic replacement with mutagenic antibiotic resistance cassettes. In order to minimize polar effect, mef(A) CDS was in frame deleted with the chloramphenicol resistance cat CDS. The genotype and relative erythromycin resistance phenotype of the pneumococcal isogenic strains are schematized. The genes are reported as arrows while dotted lines indicate gene deletions; the transcriptional promoter (P) and the putative transcriptional terminator (T) of the operon are reported.
Sensitivity to erythromycin of the S. pneumoniae strains carrying mef(A)–msr(D) and of their isogenic mutants was tested in liquid medium. It was clearly shown that msr(D) is required for macrolide resistance (Figure 1 and Table 2). Deletion of mef(A) produced only a twofold reduction of the MIC, while deletion of msr(D) produced a 64-fold MIC decrease (8 to 0.12 μg/ml), and when both genes were deleted, the MIC decreased 128-folds (Table 2). Identical results were also obtained using the allelic variants of mef(A) and msr(D) originally found in a macrolide-resistant clinical strain of S. pneumoniae formerly described as a “mef(E)-positive isolate” (GenBank AF376746) (Del Grosso et al., 2002, 2004). Deletion mutants were constructed and tested for erythromycin resistance. Results on MIC reduction were identical to those observed for the mef(A)–msr(D) alleles described above (data not shown).
To confirm the role of msr(D) in determining type M resistance to macrolides, deletion of msr(D) was complemented in S. pneumoniae FP59 carrying Φ1207.3/Δmsr(D). Competent cells of FP59 were transformed with a 2206-bp PCR fragment containing msr(D) obtained from wild type Φ1207.3, using primer pair IF171/IF175. Transformants were selected by PCR for the presence of the msr(D) gene, as previously described (Iannelli and Pozzi, 2004). A total of four transformants were selected out of 186 tested. All four showed acquisition of erythromycin resistance, with an MIC value of 8 μg/ml. A representative transformant was named FP92, checked by DNA sequencing and used as a control in further experiments.
The contribution of msr(D) to macrolide resistance was also studied in S. pyogenes, which is the original host of Φ1207.3 (Santagati et al., 2003). Two isogenic strains of S. pyogenes were constructed: (i) FR156, carrying Φ1207.3, and (ii) FR155, carrying Φ1207.3/Δmsr(D) (Table 1). Φ1207.3 transfer from S. pneumoniae donors to S. pyogenes recipients occurred at frequencies of 10-7 recombinants per donor. FR155 lacking msr(D) was susceptible to erythromycin (MIC of 0.12 μg/ml), whereas FR156 was resistant, with an MIC of 8 μg/ml (Table 2). These data show that in S. pyogenes the presence of the mef(A) gene alone determines a fourfold increase of the MIC for erythromycin, while when both mef(A) and msr(D) are present, the MIC is increased by 256-folds (Table 2).
The Φ1207.3/Δmsr(D) S. pyogenes FR155 was complemented in trans transferring a wild type Φ1207.3 element from FR122. Recombinants were selected for erythromycin resistance acquisition and in a representative recombinant, FR160, the presence of both the wild type and mutated form of Φ1207.3 was confirmed by DNA sequencing. FR160 showed an erythromycin MIC value of 8 μg/ml.
The function of mef(A) and msr(D) was studied by testing incorporation of radiolabeled erythromycin in the deletion mutants (Figure 3). Radiolabeled erythromycin was retained/incorporated by strains lacking msr(D), while msr(D)-carrying strains showed erythromycin efflux. This was observed both in S. pneumoniae (Figure 3A) and in S. pyogenes (Figure 3B). Accumulation of radiolabeled erythromycin was maximum in the Δmef(A)–Δmsr(D) S. pneumoniae mutant. The deletion of mef(A) did not affect erythromycin efflux: Δmef(A) mutant showed an erythromycin accumulation comparable to that of the wild type strain, even if the kinetics of accumulation were different (Figure 3A). Both the S. pneumoniae and the S. pyogenesΔmsr(D) mutants showed an intracellular accumulation of erythromycin which peaked already 10 min after erythromycin addition (Figures 3A,B). These results were confirmed in a series of control efflux assays which included (i) testing deletion mutants derived a “mef(E)-positive strain” carrying mef(A)–msr(D) allelic variants and (ii) testing complemented strain FP92 (data not shown). Taken together, these results suggest that type M resistance to macrolides in streptococci is due to an efflux transport system of the ABC superfamily, in which mef(A) encodes the transmembrane channel, and msr(D) the two ATP-binding domains.
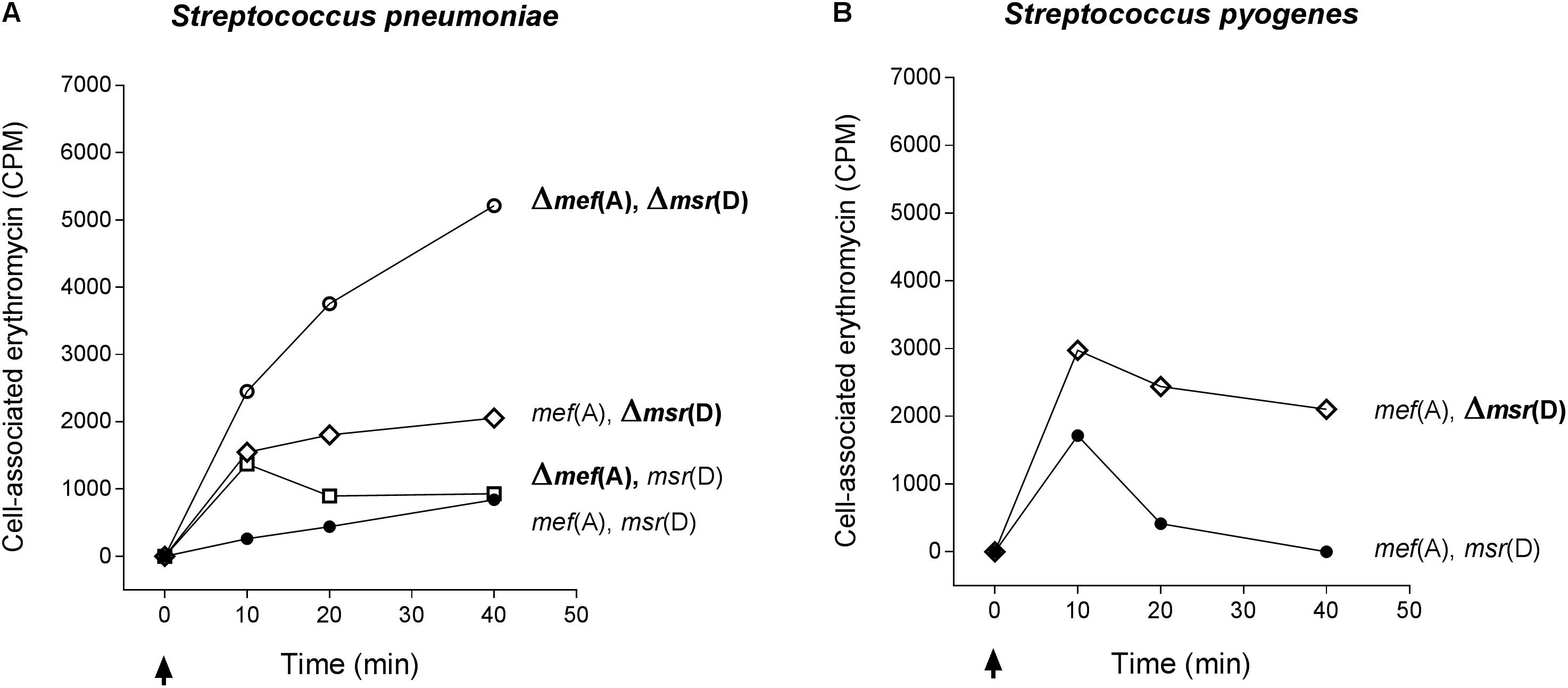
FIGURE 3. Efflux assay in S. pneumoniae (A) and S. pyogenes (B) isogenic mutant strains. The Δmef(A)–Δmsr(D) S. pneumoniae mutant showed the maximum accumulation of radiolabeled erythromycin. Erythromycin efflux was not affected by the deletion of mef(A). S. pneumoniae and S. pyogenesΔmsr(D) mutants showed an intracellular accumulation of erythromycin peaking 10 min after erythromycin addition. Genotype is indicated on the right of each graph. Cell-associated erythromycin is reported as counts per minute (CPM). Arrows indicate the time point at which radiolabeled erythromycin was added. Full circles indicate the wild type strain carrying mef(A) and msr(D), open circles the mef(A) and msr(D) deletion mutant, open diamonds the msr(D) deletion mutant, and open squares the mef(A) deletion mutant. In all curves, the CPM baseline value at time 0 was subtracted from the CPM values of each time point.
In this work, protein sequence analysis showed that Mef(A) contains 12 transmembrane segments and Msr(D) contains two ATP-binding domains (ABC domains). Analysis of bacterial genomes showed that mef(A) and msr(D) are found in tandem in 33 out of 37 cases. We constructed isogenic deletion mutants in S. pneumoniae and S. pyogenes to define the respective contribution of mef(A) and msr(D) to macrolide resistance. Physiological studies by MIC determinations and efflux assays, carried out in the original streptococcal hosts, showed that msr(D) is essential for erythromycin resistance and drug efflux, whereas mef(A) deletion produces a twofold reduction of MIC and does not affect efflux. Thus, it appears that mef(A) and msr(D) encode an ABC transporter involved in macrolide efflux with Mef(A) as the transmembrane channel, and Msr(D) as the cytoplasmatic ATP-binding protein. ABC transporters are multidomain membrane proteins minimally composed of two transmembrane domains and two ATP-binding domains. Transmembrane domains are responsible for binding and transport, while ATP-binding domains for coupling the energy of ATP hydrolysis to conformational changes in the transmembrane domains. These four domains may belong to a single protein or to different proteins.
In E. coli, the majority of transporters are secondary transporters and rely on proton-motive force as a source of energy3. Streptococci are fermentative organisms that use ATP as primary source of energy; accordingly, the majority of transporters are ATP-dependent. Because of these metabolic differences, it is likely that Mef(A) works as a major facilitator when expressed in E. coli (Clancy et al., 1996), while in the original streptococcal host, Mef(A) is coupled to the cognate ATP-binding protein Msr(D) to function as an efflux system.
In this work, we show that the msr(D) gene (i) is always in tandem with mef(A) (Figure 1), constituting a single transcriptional unit, and (ii) confers type M resistance to macrolides by an ATP-dependent efflux mechanism. The msr(D) gene product, Msr(D), is phylogenetically classified in the ABC-F family of ABC transporters together with other ATP-binding proteins, such as Vga and Lsa, which confer resistance also to other ribosome targeting antibiotics such as lincosamides, streptogramins, pleuromutilins, and ketolides. Proteins in this family share a general architecture with two NBDs and a long, mostly alpha helical, linker sequence connecting them. The spectrum of antibacterial resistances conferred is variable among members of this family, with some proteins giving resistance to multiple classes of antibiotics (Lenart et al., 2015; Sharkey et al., 2016).
Since deletion of mef(A) did not affect erythromycin efflux and produced only a twofold reduction on erythromycin MIC, it is possible that in the absence of Mef(A), Msr(D) uses an alternative transmembrane protein to pump the antibiotic out of the cell. A BlastP search in the genomes of S. pneumoniae TIGR4 and S. pyogenes M1 revealed that there are, respectively, three and one hypothetical transmembrane proteins homologous to Mef(A). Site-specific mutagenesis of the mef(A) homologous genes will be a first step for the identification of alternative Msr(D) cognate transmembrane domains which could complement the Mef(A) function.
FI and GPo designed the experiments. FI, FS, MS, J-DD, EL, GPa, and MC performed the experimental work. FI, FS, MO, GR, SS, and GPo analyzed and interpreted the data. FI, FS, and GPo wrote the paper.
This study was supported by the European Commission grants ANTIRESDEV HEALTH-F3-2009-241446 and Italian Ministry of University and Research, project PRIN 2012 (2012WJSX8K).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01670/full#supplementary-material
Ambrose, K. D., Nisbet, A. D., and Stephens, D. S. (2005). Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible. Antimicrob. Agents Chemother. 49, 4203–4209. doi: 10.1128/AAC.49.10.4203-4209.2005
Amezaga, M. R., and McKenzie, H. (2006). Molecular epidemiology of macrolide resistance in beta-haemolytic streptococci of lancefield groups A, B, C and G and evidence for a new mef element in group G streptococci that carries allelic variants of mef and msr(D). J. Antimicrob. Chemother. 57, 443–449. doi: 10.1093/jac/dki490
Banks, D. J., Porcella, S. F., Barbian, K. D., Martin, J. M., and Musser, J. M. (2003). Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phenotypically diverse clones of group A Streptococcus. J. Infect. Dis. 188, 1898–1908. doi: 10.1086/379897
Brenciani, A., Bacciaglia, A., Vignaroli, C., Pugnaloni, A., Varaldo, P. E., and Giovanetti, E. (2010). Fm46.1, the main Streptococcus pyogenes element carrying mef(A) and tet(O) genes. Antimicrob. Agents Chemother. 54, 221–229. doi: 10.1128/AAC.00499-09
Brenciani, A., Ojo, K. K., Monachetti, A., Menzo, S., Roberts, M. C., Varaldo, P. E., et al. (2004). Distribution and molecular analysis of mef(A)-containing elements in tetracycline-susceptible and -resistant Streptococcus pyogenes clinical isolates with efflux-mediated erythromycin resistance. J. Antimicrob. Chemother. 54, 991–998. doi: 10.1093/jac/dkh481
Chancey, S. T., Bai, X., Kumar, N., Drabek, E. F., Daugherty, S. C., Colon, T., et al. (2015). Transcriptional attenuation controls macrolide inducible efflux and resistance in Streptococcus pneumoniae and in other gram-positive bacteria containing mef/mel(msr(D)) elements. PLoS One 10:e0116254. doi: 10.1371/journal.pone.0116254
Clancy, J., Petitpas, J., Dib-Hajj, F., Yuan, W., Cronan, M., and Kamath, A. V., et al. (1996). Molecular cloning and fuctional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22, 867–879. doi: 10.1046/j.1365-2958.1996.01521.x
Claverys, J. P., Dintilhac, A., Pestova, E. V., Martin, B., and Morrison, D. A. (1995). Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164, 123–128. doi: 10.1016/0378-1119(95)00485-O
Clinical and Laboratory Standards Institute [CLSI] (2017). Performance Standards for Antimicrobial Susceptibility Testing, 27th Edn, Wayne, PA: CLSI supplement Clinical and Laboratory Standards Institute.
Daly, M. M., Doktor, S., Flamm, R., and Shortridge, D. (2004). Characterization and prevalence of mefA, mefE, and the associated mrs(D) gene in Streptococcus pneumoniae clinical isolates. J. Clin. Microbiol. 42, 3570–3574. doi: 10.1128/JCM.42.8.3570-3574.2004
Del Grosso, M., Camilli, R., Iannelli, F., Pozzi, G., and Pantosti, A. (2006). The mef(E)-carrying genetic element (mega) of Streptococcus pneumoniae: insertion sites and association with other genetic elements. Antimicrob. Agents Chemother. 50, 3361–3366. doi: 10.1128/AAC.00277-06
Del Grosso, M., Iannelli, F., Messina, C., Santagati, M., Petrosillo, N., and Stefani, S. et al. (2002). Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 50, 1230–1240. doi: 10.1128/JCM.40.3.774-778.2002
Del Grosso, M., Scotto d’Abusco, A., Iannelli, F., Pozzi, G., and Pantosti, A. (2004). Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48, 2037–2042. doi: 10.1128/AAC.48.6.2037-2042.2004
Gay, K., and Stephens, D. S. (2001). Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184, 56–65. doi: 10.1086/321001
Green, M. D., Beall, B., Marcon, M. J., Allen, C. H., Bradley, J. S., Dashefsky, B., et al. (2006). Multicentre surveillance of the prevalence and molecular epidemiology of macrolide resistance among pharyngeal isolates of group A streptococci in the USA. J. Antimicrob. Chemother. 57, 1240–1243. doi: 10.1093/jac/dkl101
Hoskins, J., Alborn, W. E., Arnold, J., Blaszczak, L. C., Burgett, S., Dehoff, B. S., et al. (2001). Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183, 5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001
Iannelli, F., Pearce, B. J., and Pozzi, G. (1999). The type 2 capsule locus of Streptococcus pneumoniae. J. Bacteriol. 81, 2652–2654.
Iannelli, F., Santagati, M., Docquier, J. D., Cassone, M., Oggioni, M. R., Rossolini, G. M., et al. (2004). “Type M resistance to macrolides in streptococci is not due to the mef(A) Gene, but to mat(A) encoding an ATP-dependent efflux pump,” in Proceedings of the Forty fourth Interscience Conference on Antimicrobial Agents and Chemoterapy, Washington, DC.
Iannelli, F., and Pozzi, G. (2004). Method for introducing specific and unmarked mutations into the chromosome of Streptococcus pneumoniae. Mol. Biotechnol. 26, 81–86. doi: 10.1385/MB:26:1:81
Iannelli, F., and Pozzi, G. (2007). “Protocol for conjugal transfer of genetic elements in Streptococcus pneumoniae,” in Molecular Biology of Streptococci, eds R. Hakenbeck and G. S. Chhatwal (Norfolk: Horizon Scientific Press), 525–529.
Iannelli, F., Santagati, M., Santoro, F., Oggioni, M. R., Stefani, S., and Pozzi, G. (2014a). Nucleotide sequence of conjugative prophage Phi1207.3 (formerly Tn1207.3) carrying the mef(A)/msr(D) genes for efflux resistance to macrolides in Streptococcus pyogenes. Front. Microbiol. 5:687. doi: 10.3389/fmicb.2014.00687
Iannelli, F., Santoro, F., Oggioni, M. R., and Pozzi, G. (2014b). Nucleotide sequence analysis of integrative conjugative element Tn5253 of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58, 1235–1239. doi: 10.1128/AAC.01764-13
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Klaassen, C. H., and Mouton, J. W. (2005). Molecular detection of the macrolide efflux gene: to discriminate or not to discriminate between mef(A) and mef(E). Antimicrob. Agents Chemother. 49, 1271–1278. doi: 10.1128/AAC.49.4.1271-1278.2005
Lanie, J. A., Ng, W. L., Kazmierczak, K. M., Andrzejewski, T. M., Davidsen, T. M., Wayne, K. J., et al. (2007). Genome sequence of avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189, 38–51. doi: 10.1128/JB.01148-06
Leclercq, R., and Courvalin, P. (1991). Bacterial resistance to macrolides, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35, 1267–1272. doi: 10.1128/AAC.35.7.1267
Lenart, J., Vimberg, V., Vesela, L., Janata, J., and Balikova Novotna, G. (2015). Detailed mutational analysis of vga(A) interdomain linker: implication for antibiotic resistance specificity and mechanism. Antimicrob. Agents Chemother. 59, 1360–1364. doi: 10.1128/AAC.04468-14
Martin, B., Prudhomme, M., Alloing, G., Granadel, C., and Claverys, J. P. (2000). Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38, 867–878. doi: 10.1046/j.1365-2958.2000.02187.x
Pearce, B. J., Iannelli, F., and Pozzi, G. (2002). Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res. Microbiol. 153, 243–247. doi: 10.1016/S0923-2508(02)01312-8
Pozzi, G., Iannelli, F., Oggioni, M. R., Santagati, M., and Stefani, S. (2004). Genetic elements carrying macrolide efflux genes in streptococci. Curr. Drug Targets Infect. Disord. 4, 203–206. doi: 10.2174/1568005043340641
Reinert, R. R., Ringelstein, A., van der Linden, M., Cil, M. Y., Al-Lahham, A., and Schmitz, F. J. (2006). Molecular epidemiology of macrolide-resistant Streptococcus pneumoniae isolates in Europe. J. Clin. Microbiol. 43, 1294–1300. doi: 10.1128/JCM.43.3.1294-1300.2005
Roberts, M. C., Sutcliffe, J., Courvalin, P., Jensen, L. B., Rood, J., and Seppala, H. (1999). Nomenclature for macrolide and macrolide -lincosamde-streptogramine B resistance determinants. Antimicrob. Agents Chemother. 43, 2823–2830.
Roberts, M. C. (2008). Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282, 147–159. doi: 10.1111/j.1574-6968.2008.01145.x
Rudolph, K., Bulkow, L., Bruce, M., Zulz, T., Reasonover, A., and Harker-Jones, M., et al. (2013). Molecular resistance mechanisms of macrolide-resistant invasive Streptococcus pneumoniae isolates from Alaska, 1986 to 2010. Antimicrob. Agents Chemother. 57, 5415–5422. doi: 10.1128/AAC.00319-13
Salles, C., Creancier, L., Claverys, J. P., and Mejean, V. (1992). The high level streptomycin resistance gene from Streptococcus pneumoniae is a homologue of the ribosomal protein S12 gene from Escherichia coli. Nucleic Acids Res. 20:6103. doi: 10.1093/nar/20.22.6103
Santagati, M., Iannelli, F., Cascone, C., Campanile, F., Oggioni, M. R., Stefani, S., et al. (2003). The novel conjugative transposon Tn1207.3 carries the macrolide efflux gene mef(A) in Streptococcus pyogenes. Microb. Drug Resist. 9, 243–247. doi: 10.1089/107662903322286445
Santagati, M., Iannelli, F., Oggioni, M. R., Stefani, S., and Pozzi, G. (2000). Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44, 2585–2587. doi: 10.1128/AAC.44.9.2585-2587.2000
Santagati, M., Lupo, A., Scillato, M., Di Martino, A., and Stefani, S. (2009). Conjugal mobilization of the mega element carrying mef(E) from Streptococcus salivarius to Streptococcus pneumoniae. FEMS Microbiol. Lett. 290, 79–84. doi: 10.1111/j.1574-6968.2008.01408.x
Santoro, F., Oggioni, M. R., Pozzi, G., and Iannelli, F. (2010). Nucleotide sequence and functional analysis of the tet(M)-carrying conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 308, 150–158. doi: 10.1111/j.1574-6968.2010.02002.x
Scott, J. R., and Fischetti, V. A. (1983). Expression of Streptococcal M protein in Escherichia coli. Science 221, 758–760. doi: 10.1126/science.6192499
Seppala, H., Skurnik, M., Soini, H., Roberts, M. C., and Huovinen, P. (1998). A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42, 257–262.
Sharkey, L. K., Edwards, T. A., and O’Neill, A. J. (2016). ABC-F Proteins mediate antibiotic resistance through ribosomal protection. mBio 7:e01975. doi: 10.1128/mBio.01975-15
Smith, M. D., and Guild, W. R. (1979). A plasmid in Streptococcus pneumoniae. J. Bacteriol. 137, 735–739.
Sutcliffe, J., Tait-Kamradt, A., and Wondrack, L. (1996). Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40, 1817–1824.
Tait-Kamradt, A., Clancy, J., Cronan, M., Dib-Hajj, F., Wondrack, L., Yuan, W., et al. (1997). mefE is necessary for erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41, 2251–2255.
Tatsuno, I., Isaka, M., Masuno, K., Hata, N., Matsumoto, M., and Hasegawa, T. (2018). Functional predominance of msr(D), which is more effective as mef(A)-associated than mef(E)-associated, over mef(A)/mef(E) in macrolide resistance in Streptococcus pyogenes. Microb. Drug Resist. doi: 10.1089/mdr.2017.0277 [Epub ahead of print].
Vitali, L. A., Di Luca, M. C., Prenna, M., and Petrelli, D. (2016). Correlation between genetic features of the mef(A)-msr(D) locus and erythromycin resistance in Streptococcus pyogenes. Diagn. Microbiol. Infect. Dis. 84, 57–62. doi: 10.1016/j.diagmicrobio.2015.08.007
Weisblum, B. (1995). Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39, 577–585. doi: 10.1128/AAC.39.3.577
Keywords: ABC transporter, macrolide efflux, Φ1207.3, prophage, mef(A), msr(D), Streptococcus pneumoniae, Streptococcus pyogenes
Citation: Iannelli F, Santoro F, Santagati M, Docquier J-D, Lazzeri E, Pastore G, Cassone M, Oggioni MR, Rossolini GM, Stefani S and Pozzi G (2018) Type M Resistance to Macrolides Is Due to a Two-Gene Efflux Transport System of the ATP-Binding Cassette (ABC) Superfamily. Front. Microbiol. 9:1670. doi: 10.3389/fmicb.2018.01670
Received: 22 February 2018; Accepted: 04 July 2018;
Published: 31 July 2018.
Edited by:
Fiona Walsh, Maynooth University, IrelandCopyright © 2018 Iannelli, Santoro, Santagati, Docquier, Lazzeri, Pastore, Cassone, Oggioni, Rossolini, Stefani and Pozzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Iannelli, ZnJhbmNlc2NvLmlhbm5lbGxpQHVuaXNpLml0 Gianni Pozzi, Z2lhbm5pLnBvenppQHVuaXNpLml0
†Present address: Marco Cassone, Division of Geriatric and Palliative Care Medicine, University of Michigan, Ann Arbor, MI, United States
Marco R. Oggioni, Department of Genetics, University of Leicester, Leicester, United Kingdom
Gian Maria Rossolini, Department of Experimental and Clinical Medicine, University of Florence, Florence, Italy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.