- 1College of Biological and Pharmaceutical Engineering, Wuhan Polytechnic University, Wuhan, China
- 2Wuhan Institute of Virology, Chinese Academy of Science, Wuhan, China
The whole-genome transcriptional response of Pseudomonas putida KT2440 to stress-inducing concentrations of zinc was analyzed in this study by RNA sequencing to thoroughly investigate the bacterial cell response to zinc toxicity. The data revealed that different levels of zinc stress strongly affected the transcription of genes from the following categories: metal transport genes, genes involved in membrane homeostasis, oxidative-stress-responding genes, and genes associated with basic cellular metabolism. At the lowest zinc dose, only several genes associated with metal transport and membrane homeostasis were strongly influenced. At the intermediate zinc dose, transcriptional changes of genes belonging to these two categories were highly pronounced. In addition, the intermediate zinc stress produced high levels of oxidative stress, and influenced amino acid metabolism and respiratory chains of P. putida. At the highest zinc dose, the induction of genes responsible for Fe–S cluster biogenesis was the most remarkable feature. Moreover, upregulation of glyoxylate cycle was observed. In summary, the adaptation of the cell envelope, the maintenance of metal homeostasis and intracellular redox status, and the transcriptional control of metabolism are the main elements of stress response, which facilitates the survival of P. putida KT2440 in zinc-polluted environments.
Introduction
Zn2+ is an essential transition metal ion that plays important roles in enzyme catalysis, protein structure, and transcriptional regulation in organisms from all three kingdoms (Blencowe and Morby, 2003). However, excess zinc is toxic to cells in a variety of ways, such as replacing other metal ions from enzymes, modifying stability of biomolecules, and damaging cells' antioxidant defense systems (Hobman et al., 2007; Nejdl et al., 2014; Krezel and Maret, 2016). This dual behavior has made developing resistance systems necessary for organisms to survive in zinc-polluted environments.
Metal resistance systems in bacteria are abundant and widespread (Silver and Phung, 2005; Barnett et al., 2012). Maintaining low levels of cytosolic metal ions is a key strategy to withstand metal toxicity (Aguilar-Barajas et al., 2010). Bacteria are devoid of subcellular compartments, and the main mechanisms that control cellular zinc concentration are limited to the precisely regulated processes of zinc influx, efflux, and sequestration (Blencowe and Morby, 2003; Gadd, 2010). Zinc can be transported into bacterial cells via ZnuACB, a high-affinity Zn2+ uptake system (Ammendola et al., 2007; Bhubhanil et al., 2014; Tanaka et al., 2018), and some other broad-spectrum metal ion transporters or channels, such as HmtA (Gonzalez et al., 2018), MntABC (Tanaka et al., 2018) and Pit (Beard et al., 2008). Once the intracellular zinc is present above a certain threshold concentration, excess zinc is effluxed from the cytoplasm or periplasmic space by members of cation diffusion family, resistance–nodulation–cell division family, and P-type ATPase (Nies, 2007). Extracellular sequestration is an important mechanism in immobilizing effluxed metal ions to prevent their re-entry (Taghavi et al., 2009). Numerous bacteria produce metabolites that result in precipitation of metals, such as phosphate, sulfur, and siderophores (Etesami, 2018). Furthermore, intracellular sequestration may be involved in zinc homeostasis in some bacteria, since several metallothionein-like proteins were identified and purified, which bind multiple zinc ions with high stability toward protons (Blindauer et al., 2002).
In recent years, genome-wide transcriptional analyses of Escherichia coli and Bacillus subtilis in metal ion stress have strongly implied that it is not only the metal-transport genes that confer resistance to the metal, but also the activation of several different stress response systems. According to microarray analysis, zinc does not significantly induce oxidative stress responses in E. coli; however, three important membrane stress-related regulons, cpxRA, rpoE, and basRS, are activated (Lee et al., 2005; Yamamoto and Ishihama, 2005; Hobman et al., 2007). In non-pathogenic E. coli, activated CpxR increases the production of membrane chaperons and protease, which alleviates periplasmic stress (Dbeibo et al., 2018), whereas basRS controls the biogenesis of capsular- and lipo-polysaccharides (Hobman et al., 2007). RpoE-mediated signaling pathway is potently activated by outer membrane protein assembly defects (Barchinger and Ades, 2013). The major role of rpoE regulon is considered to be restoring outer membrane protein folding by inducing chaperon production to maintain nascent outer membrane proteins in folding-competent states and by increasing the expression of β-barrel assembly machinery (Grabowicz and Silhavy, 2017). Furthermore, the amino acid metabolism pathways are also fine-tuned during zinc exposure. For example, both E. coli and B. subtilis upregulate the syntheses of cysteine and histidine after zinc treatment (Hobman et al., 2007).
Pseudomonas putida is a metabolically versatile saprophytic bacterium with great adaptability to diverse environments (Cánovas et al., 2003). Although the genomic sequence data deposited in Genbank allow for the listing of putative gene involved in zinc homeostasis, they do not provide functional evidence. Recently, Mumm et al. (2016) detected the proteomic responses of P. putida PaW85 to zinc excess by inoculating it into LB medium containing 0.6 mmol L−1 ZnSO4. However, cellular responses of P. putida to different levels of zinc stress remain poorly understood. To obtain insights into the dose-dependent transcriptional responses of P. putida to external zinc, the transcriptomes of P. putida KT2440 were sequenced in this study. Our results revealed that different sets of genes were activated or repressed in response to elevated levels of zinc ions. These results provide in-depth understanding of the adaptation mechanisms used by P. putida to survive in zinc-contaminated environments.
Materials and Methods
Bacterial Strains and Culture Conditions
P. putida KT2440 were routinely grown in LB medium at 30°C. Before being used as inoculates, they were cultured for 30 h to reach the stationary phase. A semi-synthetic cation-defined medium (CDM) (Pederick et al., 2015) was used to study the effect of zinc exposure. The exponential phased cells (about 1.0 × 107 CFU mL−1) were treated with different zinc sulfate concentrations and incubated at 30°C. At intervals, aliquots of control and treated cultures were diluted and plated on CDM agar plates. The plates were incubated at 30°C for 40 h, and the viable cells were counted. The growth inhibition of P. putida KT2440 was calculated according to the following equation: growth inhibition = (cell concentration of control samples – cell concentration of zinc treated samples)/cell concentration of control samples × 100%.
Cultivating P. putida KT2440 for studying the gene transcription under zinc stress was done by first inoculating 100 mL CDM medium with 1 mL stationary P. putida culture. This culture was also allowed to grow to a cell density of 1.0 × 107 CFU mL−1. Afterward, the culture was divided into equal portions in glass tubes. One tube was used as control, whereas the others were challenged with 0.2, 1.5, and 2.5 mmol L−1 zinc sulfate, respectively. After zinc treatment for 1 h (about half a generation time), each subculture was immediately collected, and the cells were harvested by centrifugation (8,000 g, 1 min). The pellets were stabilized by using RNAprotect bacteria reagent (Qiagen, Valencia, CA, USA) and then stored at −80°C before RNA extraction. Three biological replicates were performed for each treatment.
RNA-seq Analysis
Total RNAs were extracted from each sample using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and treated by DNAase I. Ribosomal RNA was removed from the total RNA samples using the MICROBExpress bacterial mRNA enrichment kit (Ambion, Austin, Texas, USA) according to manufacturer's protocol. Total and messenger RNA quantities and quality were assessed by Nanodrop spectrophotometer and electrophoresis on a 1% denaturing agarose gel. cDNA library preparation and RNA sequencing was performed by Beijing Genomics Institute (Beijing, China). Libraries were constructed using the TruSeq stranded mRNA-seq sample preparation kit (Illumina, San Diego, CA, USA), and sequencing was performed with an Illumina Hiseq 2500 platform (Illumina, San Diego, CA, USA) in rapid mode with 150 nt read length.
Raw data from Illumina sequencing platform were trimmed using Skewer version 0.2.2 (Jiang et al., 2014). Quality control was performed using Fast QC version 0.11.5 (Babraham Institute, Babraham, Cambridgeshire, UK). The (i) short reads (<20 nucleotides), (ii) adapter-dimer reads, (iii) reads with an N ratio (the number of unknown nucleotides/the number of total nucleotides) of greater than 5%, and (iv) reads with more than 20% low-quality nucleotides (Phred quality score < 10) were removed. Rockhopper 2 (McClure et al., 2013; Tjaden, 2015) was then used to align the remaining reads to the P. putida KT2440 genome (Winsor et al., 2016) and calculated the level of gene expression. Adjusted P-values (q-values < 0.01) were used for controlling the false discovery rate. Global gene transcription similarities/dissimilarities among samples were examined by non-metric multidimensional scaling (nMDS) (SPSS 20.0, Chicago, IL, USA).
Validation of RNA-seq Results by RT-qPCR
Reverse transcription quantitative PCR (RT-qPCR) was used to validate the RNA-seq results. A total of 15 genes were used for validation. The gene-specific primers used in this study are shown in Table S1, and their specificity was confirmed by melting curve analysis. rsd/algQ (the gene coding the regulator of sigma factor RpoD) was used as the internal reference because it has high transcriptional stability under polymetallic stressed conditions (Gómez-Sagasti et al., 2015). RT-qPCR analyses were performed with the same RNA samples used for RNAseq analysis. For cDNA synthesis, 1 μg of RNA was reverse transcribed using PrimeScript RT reagent kit (Takara, Dalian, China). The cDNAs were quantitatively analyzed with a Bio-Rad iCycler machine (Bio-Rad, Berkeley, CA, USA) using Sybr Green. The following program was used: 95°C for 1 min, followed by 40 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 15 s. Melting temperature-determining dissociation steps were performed at 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. The relative expression ratio was calculated as the relative quantity of the target gene transcript under zinc treatment conditions divided by the relative quantity of the target gene transcript under control conditions. Normality tests (Shapiro–Wilk) were conducted for the fold-change data of RNA-seq and RT-qPCR. As all groups followed the normal distribution, Pearson's correlation coefficient was calculated to determine the level of association among variables (SPSS 20.0, Chicago, IL, USA).
Results and Discussion
Effect of Sublethal Concentrations of Zinc on P. putida KT2440 Growth
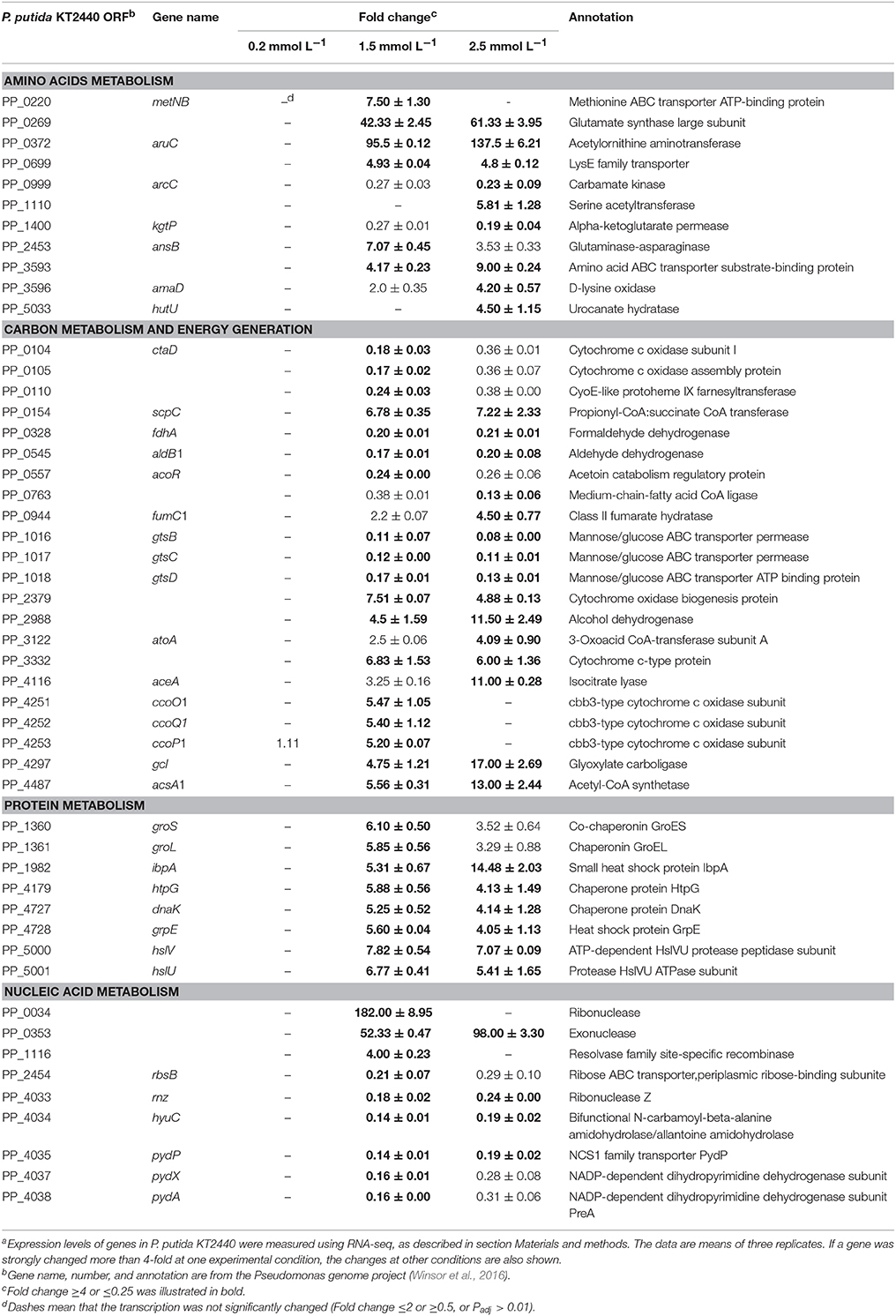
Cell growth in CDM medium was monitored for 7 h, and changes in cell concentration following treatment with increasing concentrations of ZnSO4 were measured. Figure 1 shows that zinc sulfate concentration of 0.1 mmol L−1 did not affect the specific growth rate. Conversely, 3.0 mmol L−1 zinc sulfate almost completely inhibited the growth of P. putida KT2440. When 0.2, 1.5, and 2.5 mmol L−1 zinc were present, about 5%, 40%, and 80% inhibition rates of cell growth were observed in the 6-h-cultured samples. In the following RNAseq analysis, 0.2, 1.5, and 2.5 mmol L−1 of zinc sulfate were used to determine the transcriptional responses of P. putida KT2440 to different levels of zinc stress.
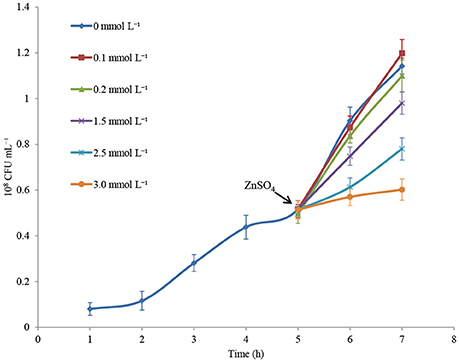
Figure 1. Growth curves of P. putida KT2440 cultured in CDM medium or in CDM medium supplemented with zinc. Addition of zinc is indicated by arrow. The values are means for triplicate cultures. The error bars indicate standard deviations.
Transcriptome Features Under Zinc Treatment
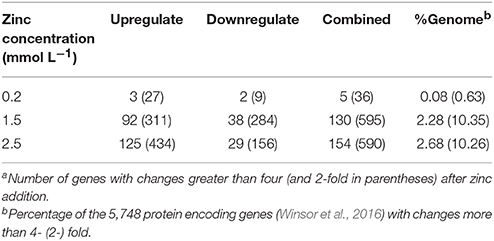
Illumina Hiseq 2500 platform produced an average read length of about 170 bp. The number of reads obtained for each sample ranged from 11.3 to 15.8 million (Table S2), with about 83.4% mapping the P. putida KT2440 genome. To obtain an overview of changes in gene transcriptions elicited by zinc stress, nMDS analysis was performed to visualize the total similarity of the different transcription profiles. In the nMDS plot, distance indicated the unique association of different samples, and the samples that appeared close were those with close proximity (Liu et al., 2017). As shown in Figure 2, there was greater variability between the different experimental conditions than within each biological replicate group. Smaller differences in overall gene transcription profiles between the 0.2 mmol L−1 zinc treated and the control samples were observed. Zinc treatment at two higher concentrations led to transcription profiles that deviated significantly from that of the untreated cells. Overall, 849 genes (14.77% of the genome) were influenced by more than two-fold in at least one condition, and 213 genes were strongly up- or downregulated by at least 4-fold. As expected, 0.2 mmol L−1 zinc affected a relatively small set of genes under these treatment conditions. 2.5 mmol L−1 zinc had the greatest effect on the cell, with a 4-fold or greater upregulation of 125 (2.17% of the genome) and downregulation of 29 genes (0.50% of the genome) (Table 1). Analysis of the strongly influenced genes with known functions revealed that all three levels of zinc stress mainly affected the transcription of genes from the following four categories (Figure 3): metal homeostasis genes (7.51%), genes involved in cell envelope structure (6.57%), antioxidant enzyme encoding genes (6.57%), and genes associated with basic cellular metabolism (24.41%). The functions of hypothetical proteins and newly identified noncoding RNAs (Table S3) are poorly understood, their roles in zinc homeostasis are not discussed in this study.
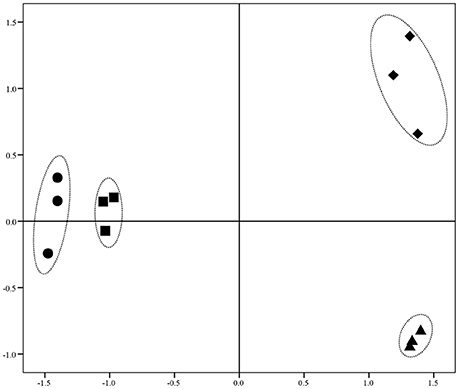
Figure 2. nMDS analysis of zinc treated samples via their transcriptional profiles. Increasing distance between points on the nMDS plot equates to the increasing dissimilarity in transcriptional profiles. The symbols •, ■, ▴, and ♦ represent transcriptional profiles obtained from the samples treated with 0, 0.2, 1.5, and 2.5 mmol L−1 zinc, respectively.
Figure 3. Distribution of genes whose transcription levels were strongly affected by zinc treatment. The top 11 functional categories affected by zinc are illustrated. Genes involved in polyamine transport, p-hydroxybenzoic acid efflux, biotin synthesis, and sulfonate assimilation are sorted into an artificial group designed as “others”.
Genes Involved in Metal Transport
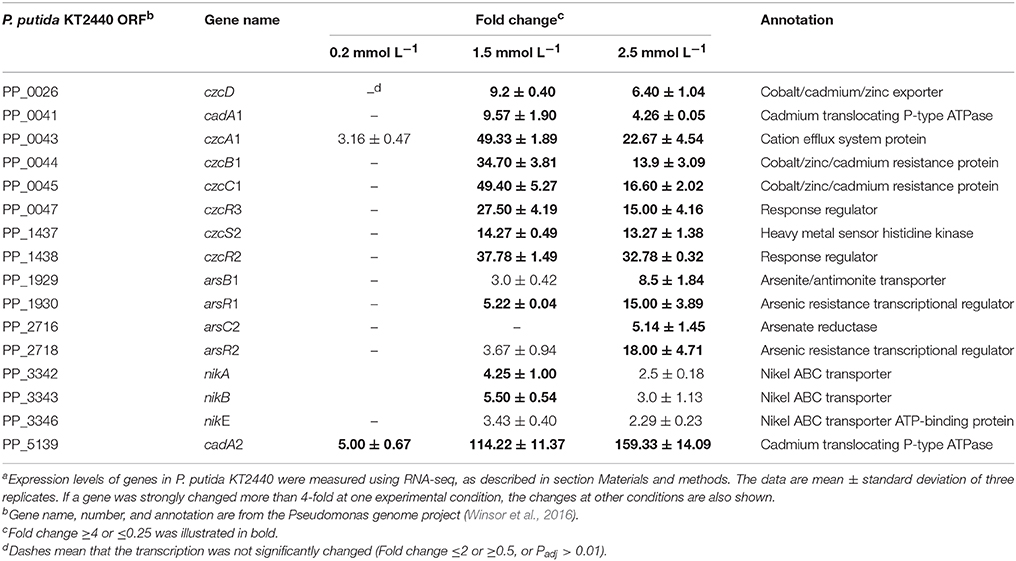
Controlling zinc ions transport is the most effective detoxification mechanism employed by bacteria to cope with zinc stress. Genome analysis has revealed that P. putida KT2440 evolved an unexpectedly large variety of genes involved in metal homeostasis, among which about 40% of zinc-transport related genes appeared to be clustered into an 8 kb region around the chromosome replication origin, including cadRA1, czcDRS1, czcCBA1, and znuABC1 (Cánovas et al., 2003; Wu et al., 2011). For the zinc-transport related genes located far from the ori region, most of them seem to be generated by duplication, such as cadRA2, czcDRS2, czcCBA2, and znuABC2. Gene duplication is a major mechanism through which genes with new functions are generated during evolution (Serres et al., 2009; Kondrashov, 2012; Katju and Bergthorsson, 2013). The benefits of gene duplication in metal resistance can be clearly seen in Cupriavidus metallidurans (von Rozycki and Nies, 2009). The transcription of cadA1, cadA2, czcCBA1, and czcCBA2 in P. putida KT2440 has been analyzed by Leedjärv et al. (2008) after cloning their promoter regions into reporter plasmids. In accordance with their report, only cadA1, cadA2, and czcCBA1 responded to increased zinc ions in this study (Table 2). Moreover, cadA2 was induced in all the three zinc treatment conditions. Under the intermediate level of zinc stress, cadA1 and czcA1 were upregulated by 9.57-fold and 49.33-fold, respectively; in addition, the transcription of czcD was significantly upregulated (fold change = 9.20). Different inducibilities were also observed among some of the other duplicated genes, such as czcRS, the two component system that has been proven to be involved in the induction of czcCBA in C. metallidutans and P. aeruginosa (Große et al., 2004; Perron et al., 2004) with the presence of high levels of several divalent metal ions. The two complete duplications of czcRS in P. putida KT2440 appeared to be sub-functionalized. czcR1 was constitutely transcripted in the control samples (Expression value = 234, calculated by Rockhopper), which may act as a readily available metal-sensing system to cope with sudden increased metal stress. czcR2 and czcR3 positively responded to 1.5 mmol L−1 zinc, which increased their transcription by 37.78-fold and 27.50-fold, respectively. As the zinc concentrations increased to 2.5 mmol L−1, a slight decrease was observed in the transcription of these zinc-related efflux systems compared with that in the 1.5 mmol L−1 zinc treatment. However, nik operon (nikA, nikB, and nikE), which encodes a nickel-import system, and four genes encoding arsenate resistance systems were significantly upregulated. Zinc can replace Ni ions from their enzymes (Aguilar-Barajas et al., 2010); therefore, nickel uptake ATPase was reasonably upregulated to counter the Ni defect induced by zinc toxicity. P. putida KT2440 evolved two asr operons, arsRBCH1 and arsRBCH2, both of them appeared to be upregulated, which was also confirmed by RT-PCR. Arsenic resistance is mainly mediated by the arsRBC genes, which is usually found in most bacteria (Cánovas et al., 2003). arsC encodes an arsenic reductase that transform As(V) into As(III); ArsB is a secondary transporter responsible for the extrusion of As(III), Sb(III), or Bi(III) using the proton motive force; arsR encodes a transcription regulator that de-represses arsBC expression (Cánovas et al., 2003; Moore and Gaballa, 2005). As arsenic and zinc share little similarity, it hard to believe that arsC also effluxing zinc out of the cells. We inferred that the upregulation of these two operons was caused by the unspecific induction of the arsR. In B. subtilis the transcription of arsR was not only induced by As(III) and Bi(III), but also strongly by Cd(II) and Ag(I) (Moore and Gaballa, 2005).
Genes Involved in Cell Envelope Homeostasis
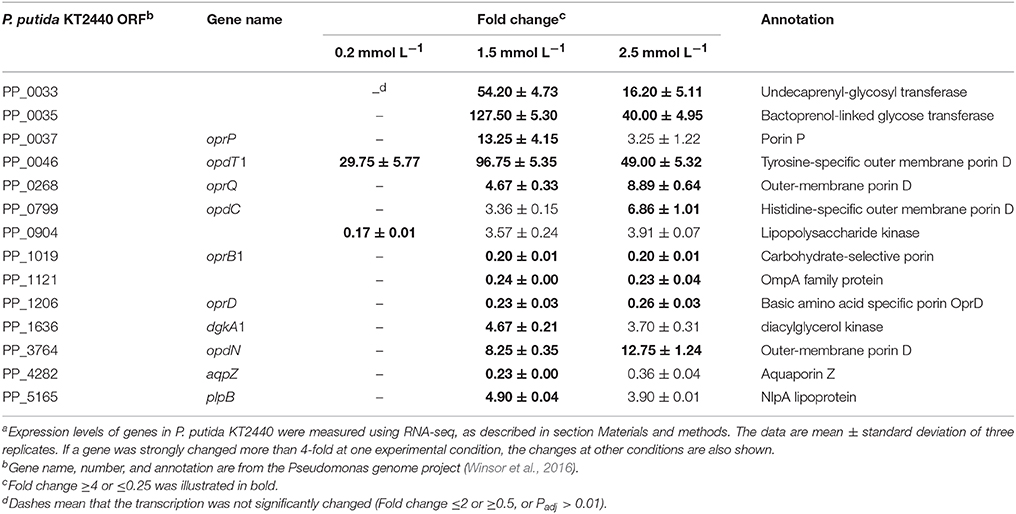
The bacterial cell envelope comprises the inner cell membrane and/or the cell wall, which provides structural integrity to the cell, and acts as a sensory interface and molecular sieve mediating both the controlled solute transportation and information flow (Jordan et al., 2008). Maintaining the cellular envelope balance in the presence of stress conditions is crucial for the bacteria's survival. Previous studies have demonstrated that zinc ions act as a strong envelope-perturbing agent in E. coli (Hobman et al., 2007). Moreover, zinc ions induce membrane stress responses in P. aeruginosa PA14 and P. putida PaW85 as the ColRS two-component signal transduction system is activated (Nowicki et al., 2015; Mumm et al., 2016). In this study, 1.5 mmol L−1 zinc induced the transcription of dgkA1 (fold change = 4.67), which is responsible for the glycerophospholipid biosynthesis, and plpB (fold change = 4.90), whose product is NlpA lipoprotein (Table 3). In E. coli, NlpA lipoprotein is an inner membrane protein contributing to the biogenesis of outer membrane vesicles (Bodero et al., 2007). The induction of NlpA in P. putida suggests that the formation of outer membrane vesicles was enhanced. Accelerating the formation of outer membrane vesicles was proven to be an adaptive response of P. putida to several kinds of environmental stress, such as heat shock, high NaCl or EDTA concentration (Baumgarten et al., 2012). The released outer membrane vesicles lead to an increased cell surface hydrophobicity as well as to a higher tendency to form biofilms (Baumgarten et al., 2012).
Unlike the plasma membrane, the outer membrane of gram-negative bacteria is relatively permeable to small molecules. A group of proteins, known as porins, form fluid-filled channels in the outer membrane that allow hydrophilic solute to diffuse across in to the periplasmic space (Blencowe and Morby, 2003). The exposure of P. putida KT2440 to zinc greatly altered the transcription of several porin encoding genes. For example, the Pseudomonads phosphate-specific porin, OprP (Pongprayoon et al., 2009), was upregulated by 13.25-fold under intermediate zinc stress. The zinc-specific induction of oprP may explain the need to enhance the phospholipid biosynthesis and maintain the membrane integrity. Two porins that facilitated the diffusion of tyrosine (OpdT1) and histidine (OpdC, only induced by 2.5 mmol L−1 zinc) were also upregulated. oprD was commonly downregulated by 1.5 mmol L−1 (fold change = 0.23) and 2.5 mmol L−1 (fold change = 0.26) zinc treatment in this study. In P. aeruginosa, the transcription of oprD was reduced by copper and zinc (Caille et al., 2007). Nevertheless, OprD was not directly involved in heavy metal resistance because an oprD knock-out mutant was as susceptible as the wild-type to heavy metals (Perron et al., 2004). CzcRS is a regulatory system that connects czcCBA and oprD expression; unphosphorylated CzcR repress the transcription oprD, whereas the amount of active phosphorylated CzcR is critical for inducing czcCBA (Perron et al., 2004). Therefore, oprD downregulation is a secondary effect linked to czcCBA overtranscription. Another porin commonly downregulated under the intermedium and high zinc stress is a carbohydrate-specific outer membrane porin, OprB1. The repression of oprB1 appeared to contract with the requirement of more energy because much cellular damage must be restored. Other channels were possibly used to compensate for its downregulation.
Oxidative Stress Responding Genes
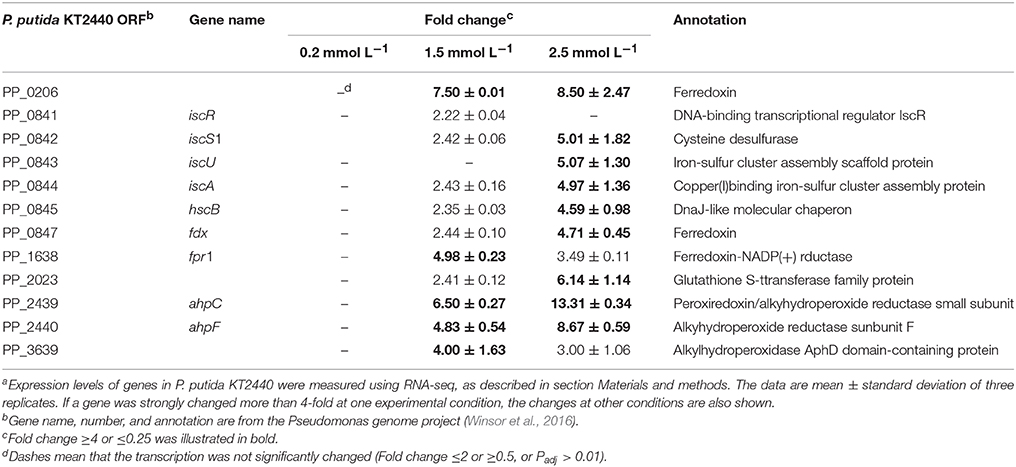
A number of studies performed on biological systems have shown that redox-active metals can undergo redox cycling reactions and produce reactive oxygen species (ROS) (Jomova et al., 2012). Although many transition metal ions have no redox activity under physiological conditions, they can cause oxidative responses by damaging cell's antioxidant defense systems (Hobman et al., 2007). Alhasawi et al. (2014) confirmed that zinc toxicity led to oxidative stress in P. fluorescence; a twofold increase in oxidized protein was observed in the zinc-challenged cells compared with the control. Our transcriptomic data showed that low level of zinc stress did not enhance the transcription of antioxidant enzyme encoding genes. However, alkylhydroperoxide reductase was upregulated with increasing zinc concentration (Table 4). This thilol-specific peroxidase catalyzes the reduction of hydrogen peroxide and organic hydroperoxides to their respective alcohols (Harris, 2003). The induction of ferredoxin-NADP reductase was observed under the 1.5 mmol L−1 zinc treatment, which is critical for the maintenance of appropriate levels of NADPH (Ray et al., 2013). Compared with the bacteria exposed to the intermediate zinc dose, the upregulation of isu operon responsible for Fe–S cluster biogenesis was the most remarkable feature of the P. putida cells under high zinc stress. The isu operon is under the transcriptional control of the IscR repressor, which contains three cysteine residues and is shown to be an Fe–S protein (Schwartz et al., 2001). Holo-IscR is able to repress transcription of the isu operon. When Fe–S cluster assembly is disturbed by ROS or iron limitation, apo-IscR predominates, and isc expression increases to meet the demand (Ayala-Castro et al., 2008). Directly replacing Fe from the Fe–S clusters is another way for zinc to perturb the function of Fe–S clusters (Xu and Imlay, 2012), which also increases the amount of free Fe ions to participate in Fenton reaction. Therefore, zinc-induced oxidative stress is further enhanced.
Genes Involved in Basic Cellular Metabolism
Among all the genes whose transcription was strongly influenced, about 24.41% of them were involved in basic cellular metabolism. Of these, 11 amino acid metabolism genes were up or downregulated by more than 4-fold after zinc treatment (Table 5). The synthesis of glutamate was enhanced, as revealed by the great upregulation of glutamate synthase (PP_0269). The transcription of hutU and PP_1110 were strongly increased under the high zinc stress. HutU is involved in the second step of the subway that synthesizes L-glutamate from L-histidine, whereas PP_1110 encodes serine acetyltransferase which catalyzes the first step that convert L-serine to L-cysteine. Glutamate and cysteine are two substrates for glutathione synthesis, and their alteration could reflect increased synthesis of glutathione. Moreover, glutamate was also shown to play an important role in the bacteria adaption to noxious conditions caused by Ni (Ray et al., 2013). Among all the up-regulated genes associated with amino acid metabolism, the most up-regulated gene was aruC which encodes an acetylornithine aminotransferase. In P. aeruginosa, this enzyme palys dual roles in arginine metabolism, which catalyze the transamination of L-glutamate with N-2-acetyl-L-glutamate 5-semialdehyde when functioning in arginine biosynthesis and transaminates L-ornithine with 2-oxoglutarate when participating in L-arginine degradation (Voellmy and Leisinger, 1975; Schomburg et al., 2008). Therefore, AruC could maintain the balance between L-arginine and L-glutamate and fine-tune the glutamate pool. The upregulation of lysine and methionine transporters indicated that zinc-stressed P. putida exhibited more demand for some other amino acids. Simply obtaining amino acids from the environment provided a more energy-efficient way than provoking their de novo synthesis.
In addition to amino acid metabolism genes, transcriptional changes in the genes involved in carbon source metabolism and energy generation were also observed. The only carbon source presented in the CDM medium was glucose. Metabolic flux analysis revealed that glucose is mainly assimilated through Enter–Doudoroff pathway in P. putida, generating most of the pyruvate; then the flow of carbon is directed toward the citric acid cycle (TCA cycle) under aerobic conditions (Sudarsan et al., 2014). After zinc treatment, most of the genes involved in the Enter–Doudoroff pathway and TCA cycle appeared to be stable, only the fumarate hydratase encoding gene (fumC1) was upregulated under the high zinc stress. aceA was another gene that only upregulated by 2.5 mmol L−1 zinc, which encodes an enzyme in the glyoxylate shunt (GS) that catalyzes the cleavage of isocitrate to succinate and glyoxylate. The GS in bacterial physiology has traditionally been associated with the demand for gluconeogenesis, as induced by carbon-source limitation. However, growing evidence suggests that oxidative stress activate the GS (Ahn et al., 2016). Under usual conditions, cells produce ROS as by-products of their aerobic respiration or nutrient oxidation (Hobman et al., 2007). GS bypasses two NADH-generation steps in TCA cycle, which diminishes the electrons flux funneled into respiration. Therefore, activation of GS limits the further exacerbation of oxidative stress induced by zinc. According to Alhasawi et al. (2014), P. fluorescence tends to use another way to reduced NADH generation under zinc stress. The activities of TCA cycle enzymes, such as isocitrate dehydrogenase, α-ketoglutarate dehydrogenase, and malate dehydrogenase, were markedly diminished. To survive in changing environmental conditions, P. putida KT2440 evolved branched respiratory chains containing five different terminal oxidases with different affinities for oxygen and capabilty to pump protons (Morales et al., 2006). Regulation of these oxidases is quite complex and has not been completely unraveled (Follonier et al., 2013). Although the cytochrome Cbb3-1 oxidase (PP_4251, PP_4252, and PP_4253) was remarkably upregulated and the Aa3-type (PP_0104, and PP_0105) was downregulated under the intermediate zinc stress, the underlying mechanism remains unknown.
Another group of genes whose transcription was altered in zinc-treated samples was that of genes involved in protein folding and degradation. Six chaperone encoding genes (groES, groEL, ibpA, dnaK, grpE, and htpG) were upregulated after 1.5 and 2.5 mmol L−1 zinc treatment, suggesting a greater protein folding efficiency. An ATP-dependent protease belonging to Clp family (HslUV) was also induced. Clp proteases comprise proteolytic and regulatory components (Manara et al., 2012); some of them play a decisive role in determining the density of proteins under both normal and stressed conditions (Gómez-Sagasti et al., 2015). The upregulation of hslUV might be required in response to zinc stress to recycle unnecessary proteins rapidly or to remove proteins denatured by zinc ions. Pertaining to nucleic acid metabolism, the recycling of nucleotide or base was also enhanced because an exonuclease (PP_0353) was greatly upregulated and a nucleobase cation symporter (PP_4035) as well as the genes (PP_4037, and PP_4038) responsible for pyrimidine degradation were strongly downregulated.
RT-qPCR Validation
To confirm RNA-seq data, the expression levels of 15 genes were examined via RT-qPCR in triplicate. For all 15 genes, the same expression trend was detected under RT-qPCR and RNAseq analyses (Figure 4). Additionally, the observed fold changes for each gene were moderately correlated (r2 = 0.83, Figure 5). Therefore, the RT-qPCR results confirm the accuracy and consistency of the RNA-seq data. Moreover, several genes previously known to be induced or repressed by zinc further validate the capability of RNA-seq experiments to identify candidate genes responding to toxic concentrations of external zinc.
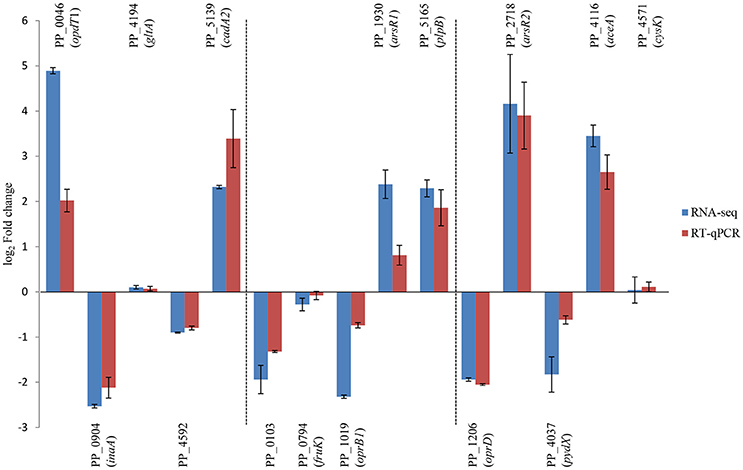
Figure 4. Validation of RNA-seq results by RT-qPCR. Rectangles represent transcription changes of selected genes as measured by RNA-seq and RT-qPCR. Bars illustrate standard deviations of three replicates.
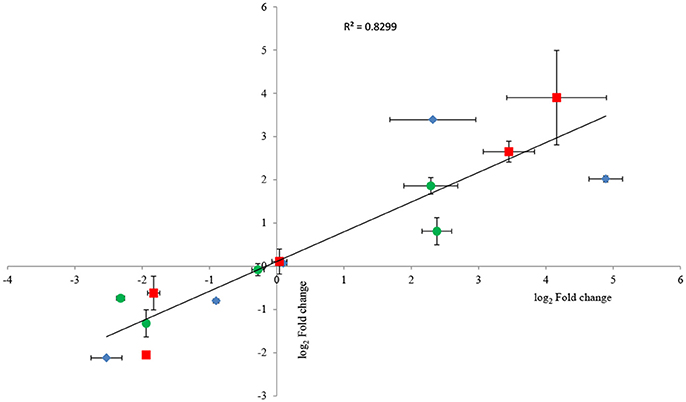
Figure 5. Correlation plot between fold changes of selected genes observed in RNA-seq and their corresponding fold-change values in RT-qPCR analysis. Horizontal bars represent standard deviations for RT-qPCR data, and vertical bars illustrate standard deviations of RNA-seq results. Three replicates were carried out for both RNA-seq and RT-qPCR analysis. As all groups followed the normal distribution, Pearson's correlation coefficient was calculated to determine the level of correlation. The symbols ♦, •, and ■ represent the genes selected from the samples under the low, intermediate, and high zinc stress, respectively.
Comparative Analysis of Gene Expression Patterns
In 2016, Mumm et al. (2016) analyzed the response of P. putida and its colR-deficient strain to zinc excess at the whole-proteome level. Although different culture conditions could cause different cellular responses (Kim et al., 2013), we still found quite a few similarities in gene expression patterns. Both proteomic and transcriptomic analysis revealed that the most upregulated genes under zinc stress were the genes responsible for metal or multidrug efflux systems, whereas the most downregulated genes were some porin-encoding genes. Besides, cell envelop was fine-tuned with the regulation of lipoprotein synthesis. The induction of oxidative response genes and genes involved in Fe-S cluster synthesis were not observed at the proteome level, which indicated that 0.6 mmol L−1 zinc ions in LB medium only generated a relatively low level of zinc stress.
E. coli is another bacterial species whose response to zinc has been intensively analyzed. Genome analysis revealed that P. putida evolved a greater number of genes responsible for metal resistance than E. coli. Our results and the data reported by Mumm et al. (2016) indicated that some other differences were observed at transcriptomic and proteomic levels. E. coli and P. putida both enhanced the synthesis of lipopolysaccharides or lipoproteins that constitute outer membrane, however, only P. putida greatly altered the expression of porins. In addition, these two strains may use different ways to chelate cellular free zinc ions. In E. coli, positive regulation of cysteine synthesis operon was a remarkable feature in both nutrient-rich and nutrient-limited medium containing low concentrations of zinc (Hobman et al., 2007). Large amounts of cysteine residues were needed to play a role in transient trapping of excess free zinc ions prior to export (Yamamoto and Ishihama, 2005; Etesami, 2018). In P. putida, upregulation of cysteine synthesis was not observed under low or intermediate zinc stress; only serine acetyltransferase which catalyzes the first step that convert serine to cysteine was moderately upregulated at a really high zinc concentration.
Conclusion
The transcriptional response of P. putida KT2440 to elevated concentrations of zinc was analyzed in this study. After comparing the different RNA-seq datasets obtained, 213 genes were found to be transcriptionally changed by more than 4-fold. Zinc ions have no redox activity under normal conditions. Many previous researches also confirmed that zinc excess did not cause cellular oxidative stress. However, this is true only for the bacteria under low zinc concentrations. As the concentration increased, zinc ions clearly induced the generation of ROS in P. putida cells, as evidenced by the upregulation of alkylhydroperoxide reductase and ferredoxin-NADPH reductase. At the highest zinc dose, the central carbon source metabolism pathways (glycolysis and TCA cycle) of P. putida were very stable, whereas the synthesis of glutamate and the recycle use of protein and nucleotide were activated. GS shunt was also upregulated at the highest zinc concentration, which limited the further exacerbation of oxidative stress by decreasing the electron flux funneled into respiration chain. Although zinc-stressed P. putida cells tended to use different terminal enzymes in their branched respiration chain, the underlying mechanism is poorly understood and merits further investigation.
Data Availability Statement
The RNA-seq datasets for this study can be found in the NCBI short read archive database under the Bioproject accession number PRJNA450701.
Author Contributions
PL and XC conceived the experiment. JP and LM performed the experiment. PL wrote the manuscript.
Funding
This research was financially supported by the National Natural Science Foundation of China (Grant no. 31700100) and the Hubei Provincial Natural Science Foundation of China (Grant no. 2017CFB207).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01669/full#supplementary-material
Abbreviations
CDM, cation-defined medium used in this study; RT-qPCR, reverse transcription quantitative PCR; nMDS, non-metric multidimensional scaling; ROS, reactive oxygen species; GS, glyoxylate shunt; TCA cycle, citric acid cycle.
References
Aguilar-Barajas, E., Ramirez-Diaz, M. I., Riveros-Rosas, H., and Cervantes, C. (2010). “Heavy metal resistance in Pseudomonads,” in Pseudomonas, eds J. L. Ramos and A. Filloux (Berlin; Heidelberg: Springer), 255–282.
Ahn, S., Jung, J., Jang, I. A., Madsen, E. L., and Park, W. (2016). Role of glyoxylate shunt in oxidative stress response. J. Biol. Chem. 291, 11928–11938. doi: 10.1074/jbc.M115.708149
Alhasawi, A., Auger, C., Appanna, V. P., Chahma, M., and Appanna, V. D. (2014). Zinc toxicity and ATP production in Pseudomonas fluorescens. J. Appl. Microbiol. 117, 65–73. doi: 10.1111/jam.12497
Ammendola, S., Pasquali, P., Pistoia, C., Petrucci, P., Petrarca, P., Rotilio, G., et al. (2007). High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect. Immun. 75, 5867–5876. doi: 10.1128/IAI.00559-07
Ayala-Castro, C., Saini, A., and Outten, F. W. (2008). Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 72, 110–125. doi: 10.1128/MMBR.00034-07
Barchinger, S. E., and Ades, S. E. (2013). Regulated proteolysis: control of the Escherichia coli σE-dependent cell envelope stress response. Subcell. Biochem. 66, 129–160. doi: 10.1007/978-94-007-5940-4_6
Barnett, J. P., Millard, A., Ksibe, A. Z., Scanlan, D. J., Schmid, R., and Blindauer, C. A. (2012). Mining genomes of marine cyanobacteria for elements of zinc homeostasis. Front. Microbiol. 3:142. doi: 10.3389/fmicb.2012.00142
Baumgarten, T., Vazquez, J., Bastisch, C., Veron, W., Feuilloley, M. G. J., Nietzsche, S., et al. (2012). Alkanols and chlorophenols cause different physiological adaptive response on the level of cell surface properties and membrane vesicle formation in Pseudomonas putida DOT-T1E. Appl. Microbiol. Biotechnol. 93, 837–845. doi: 10.1007/s00253-011-3442-9
Beard, S. J., Hashim, R., Wu, G., Binet, M. R., Hughes, M. N., and Poole, R. K. (2008). Evidence for the transport of zinc ions via the pit inorganic phosphate transport system in Escherichia coli. FEMS Microbiol. Lett. 2184, 231–235. doi: 10.1111/j.1574-6968.2000.tb09019.x
Bhubhanil, S., Sittipo, P., Chaoprasid, P., Nookabkaew, S., Sukchawalit, R., and Mongkolsuk, S. (2014). Control of zinc homeostasis in Agrobacterium tumefaciens via zur and the zinc uptake genes znuABC and zinT. Microbiology 160, 2452–2463. doi: 10.1099/mic.0.082446-0
Blencowe, D. K., and Morby, A. P. (2003). Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 27, 291–311. doi: 10.1016/S0168-6445(03)00041-X
Blindauer, C. A., Harrison, M. D., Robinson, A. K., Parkinson, J. A., Bowness, P. W., Sadler, P. J., et al. (2002). Multiple bacteria encode metallothioneins and SmtA-like zinc fingers. Mol. Microbiol. 45, 1421–1432. doi: 10.1046/j.1365-2958.2002.03109.x
Bodero, M. D., Pilonieta, M. C., and Munson, G. P. (2007). Repression of the inner membrane lipoprotein NlpA by RNA in enterotoxigenic Escherichia coli. J. Bacteriol. 189, 1627–1632. doi: 10.1128/JB.01714-06
Caille, O., Rossier, C., and Perron, K. (2007). A copper-activated two-component system interacts with zinc and impenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 189, 4561–4568. doi: 10.1128/JB.00095-07
Cánovas, D., Cases, I., and de Lorenzo, V. (2003). Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 5, 1242–1256. doi: 10.1111/j.1462-2920.2003.00463.x
Dbeibo, L., van Rensburg, J. J., Smith, S. N., Fortney, K. R., Gangaiah, D., Gao, H., et al. (2018). Evaluation of CpxRA as a therapeutic target for uropathogenic Escherichia coli infections. Infect. Immun. 86, e00798–e00817. doi: 10.1128/IAI.00798-17
Etesami, H. (2018). Bacterial mediated alleviation of heavy metal stress and decreased accumulation of metals in plant tissues: mechanisms and future prospects. Ecotoxicol. Environ. Saf. 147, 175–191. doi: 10.1016/j.ecoenv.2017.08.032
Follonier, S., Escapa, I. F., Fonseca, P. M., Henes, B., Panke, S., Zinn, M., et al. (2013). New insights on the reorganization of gene transcription in Pseudomonas putida KT2440 at elevated pressure. Microb. Cell. Fact. 12, 30. doi: 10.1186/1475-2859-12-30
Gadd, G. M. (2010). Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156, 609–643. doi: 10.1099/mic.0.037143-0
Gómez-Sagasti, M. T., Becerril, J. M., Epelde, L., Alkorta, I., and Garbisu, C. (2015). Early gene expression in Pseudomonas fluorescence exposed to a polymetallis solution. Cell Biol. Toxicol. 31, 39–81. doi: 10.1007/s10565-015-9294-9
Gonzalez, M. R., Ducret, V., Leoni, S., and Perron, K. (2018). Pseudomonas aeruginosa zinc homeostasis: key issues for an opportunistic pathogen. Biochim. Biophys. Acta. doi: 10.1016/J.bbagrm.2018.01.018. [Epub ahead of print].
Grabowicz, M., and Silhavy, T. J. (2017). Encelope stress responses: an interconnected safety net. Trends Biochem. Sci. 42, 232–242. doi: 10.1016/j.tibs.2016.10.002
Große, C., Anton, A., Hoffmann, T., Franke, S., Schleuder, G., and Nies, D. H. (2004). Identification of a regulatory pathway that controls the heavy-metal resistance system Czc via promoter czcNp in Ralstonia metallidurans. Arch. Microbiol. 182, 109–118. doi: 10.1007/s00203-004-0670-8
Harris, J. R. (2003). Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40. doi: 10.1016/S0968-0004(02)00003-8
Hobman, J. L., Yamamoto, K., and Oshima, T. (2007). “Transcriptiomic responses of bacterial cells to sublethal metal ion stress,” in Molecular Microbiology of Heavy Metals, eds D. H. Nies and S. Silver (Berlin; Heidelberg: Springer), 73–115.
Jiang, H., Lei, R., Ding, S. W., and Zhu, S. (2014). Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15:182. doi: 10.1186/1471-2015-15-182
Jomova, K., Baros, S., and Valko, M. (2012). Redox active metal-induced oxidative stress in biological systems. Transit. Metal Chem. 37, 127–134. doi: 10.1007/s11243-012-9583-6
Jordan, S., Hutchings, M. I., and Mascher, T. (2008). Cell envelope stress response in gram-positive bacteria. FEMS Microbiol. Rev. 32, 107–146. doi: 10.1111/j.1574-6976.2007.00091.x
Katju, V., and Bergthorsson, U. (2013). Copy-number changes in evolution: rates, fitness effects and adaptive significance. Front. Genet. 4:273. doi: 10.3389/fgene.2013.00273
Kim, J., Oliveros, J. C., Nikel, P. I., de Lorenzo, V., and Silva-Rocha, R. (2013). Transcriptomic fingerprinting of Pseudomonas putida under alternative physiological regimes. Environ. Microbiol. Rep. 5, 883–891. doi: 10.1111/1758-2229.12090
Kondrashov, F. A. (2012). Gene duplication as a mechanism of genomic adaption to a changing environment. Proc. Biol. Sci. 279, 5048–5057. doi: 10.1098/rspb.2012.1108
Krezel, A., and Maret, W. (2016). The biological inorganic chemistry of zinc ions. Arch. Biochem. Biophys. 611, 3–19. doi: 10.1016/j.abb.2016.04.010
Lee, L. J., Barrett, J. A., and Poole, R. K. (2005). Genome-wide transcriptional response of chemostat-cultured Escherichia coli to zinc. J. Bacteriol. 187, 1124–1134. doi: 10.1128/JB.187.3.1124-1134.2005
Leedjärv, A., Ivask, A., and Virta, M. (2008). Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J. Bacteriol. 190, 2680–2689. doi: 10.1128/JB.01494-07
Liu, P., Xiong, X., Wang, S., and Miao, L. (2017). Population dynamics and metabolite analysis of yeasts involved in a Chinese miscellaneous-flavor liquor fermentation. Ann. Microbiol. 67, 553–565. doi: 10.1007/s13213-017-1286-y
Manara, A., DalCorso, G., Baliardini, C., Farinati, S., Cecconi, D., and Furini, A. (2012). Pseudomonas putida response to cadmium: changes in membrane and cytosolic proteomes. J. Proteome Res. 11, 4169–4176. doi: 10.1021/pr300281f
McClure, R., Balasubramanian, D., Sun, Y., Bobrovskyy, M., Sumby, P., and Genco, C. A. (2013). Computional analysis of bacterial RNA-seq data. Nucleic. Acids Res. 41:e140. doi: 10.1093/nar/gkt444
Moore, C. M., and Gaballa, A. (2005). Genetic and physiological responses of Bacillus subtilis to metal ions stress. Mol. Microbiol. 57, 27–40. doi: 10.1111/j.1365-2958.2005.04642.x
Morales, G., Ugidos, A., and Rojo, F. (2006). Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidase. Environ. Microbiol. 8, 1764–1774. doi: 10.1111/j.1462-2920.2006.01061.x
Mumm, K., Ainsaar, K., Kasvandik, S., Tenson, T., and Hõrak, R. (2016). Response of Pseudomonas putida to zinc excess determined at the proteome level: pathways dependent and independent of ColRS. J. Proteome Res. 15, 4349–4368. doi: 10.1021/acs.jproteome.6b00420
Nejdl, L., Ruttkay-Nedecky, B., Kudr, J., Krizkova, S., Smerkova, K., Dostalova, S., et al. (2014). DNA interaction with zinc ions. Int. J. Biol. Macromol. 64, 281–287. doi: 10.1016/j.ijbiomac.2013.12.013
Nies, D. H. (2007). “Bacterial transition metal homeostasis,” in Molecular Microbiology of Heavy Metals, eds D. H. Nies and S. Silver (Berlin; Heidelberg: Springer), 117–142.
Nowicki, E. M., O'Brien, J. P., Brodbelt, J. S., and Trent, M. S. (2015). Extracellular zinc induces phosphoethanolamine addition to Pseudomonas aeruginosa lipid A via the ColRS two-component system. Mol. Microbiol. 97, 166–178. doi: 10.1111/mmi.13018
Pederick, V. G., Eijkelkamp, B. A., Begg, S. L., Ween, M. P., McAllister, L. J., Paton, J. C., et al. (2015). ZnuA and zinc homeostasis in Pseudomonas aeruginosa. Sci. Rep. 5, 13139. doi: 10.1038/srep13139
Perron, K., Caille, O., Rossier, C., Van Delden, C., Dumas, J. L., and Köhler, T. (2004). CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 279, 8761–8768. doi: 10.1074/jbc.M312080200
Pongprayoon, P., Beckstein, O., Wee, C. L., and Sansom, M. S. (2009). Simulation of anion transport through OprP reveal the molecular basis for high affinity and selectivity for phosphate. Proc. Natl. Acad. Sci. U. S. A. 106, 21614–21618. doi: 10.1073/pnas.0907315106
Ray, P., Girard, V., Gault, M., Job, C., Bonneu, M., Mandrand-Berthelot, M. A., et al. (2013). Pseudomonas putida KT2440 response to nickel or cobalt induced stress by quantitative proteomics. Metallomics 5, 68–79. doi: 10.1039/c2mt20147j
Schomburg, D., Schomburg, I., and Chang, A. (2008). “Class 2. Transferase ?,” in Springer Handbook of Enzymes, eds D. Schomburg and I. Schomburg (Berlin; Heidelberg: Springer), 342–346.
Schwartz, C. J., Giel, J. L., Patschkowski, T., Luther, C., Ruzicka, F. J., Beinert, H., et al. (2001). IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. U. S. A. 98, 14895–14900. doi: 10.1073/pnas.251550898
Serres, M. H., Kerr, A. R., McCormack, T. J., and Riley, M. (2009). Evolution by leaps: gene duplication in bacteria. Biol. Direct. 4, 46. doi: 10.1186/1745-6150-4-46
Silver, S., and Phung, L. T. (2005). A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 32, 587–605. doi: 10.1007/s10295-005-0019-6
Sudarsan, S., Dethlefsen, S., Blank, L. M., Siemann-Herzberg, M., and Schmid, A. (2014). The functional structure of central carbon metabolism in Pseudomonas putida KT2440. Appl. Environ. Microbiol. 80, 5292–5303. doi: 10.1128/AEM.01643-14
Taghavi, S., Lesaulnier, C., Monchy, S., Wattiez, R., Mergeay, M., and van der Lelie, D. (2009). Lead resistance in Cupriavidus metallidurans CH34: interplay between plasmid and chromosomally-located functions. Antonie van Leeuwenhoek 96, 171–182. doi: 10.1007/s10482-008-9289-0
Tanaka, K. J., Song, S., Mason, K., and Pinkett, H. W. (2018). Selective substrate uptake: the role of ATP-binding cassette (ABC) importers in pathogenesis. Biochim. Biophys. Acta. 1860, 868–877. doi: 10.1016/j.bbamem.2017.08.011
Tjaden, B. (2015). De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol. 16, 1. doi: 10.1186/s13059-014-0572-2
Voellmy, R., and Leisinger, T. (1975). Dual role for N-2-acetylornithine 5-aminotransferase from Pseudomonas aeruginosa in arginine biosynthesis and arginine catabolism. J. Bacteriol. 122, 799–809.
von Rozycki, T., and Nies, D. H. (2009). Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie van Leeuwenhoek 96, 115–139. doi: 10.1007/s10482-008-9284-5
Winsor, G. L., Griffiths, E. J., Lo, R., Dhillon, B. K., Shay, J. A., and Brinkman, F. S. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653. doi: 10.1093/nar/gkv1227
Wu, X., Monchy, S., Taghavi, S., Zhu, W., Ramos, J., and van der Lelie, D. (2011). Comparative genomics and functional analysis of niche-specific adaption in Pseudomonas putida. FEMS Microbiol. Rev. 35, 299–323. doi: 10.1111/j.1574-6976.2010.00249.x
Xu, F. F., and Imlay, J. A. (2012). Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl. Environ. Microbiol. 78, 3614–3621. doi: 10.1128/AEM.07368-11
Keywords: Pseudomonas putida, zinc, transcriptome analysis, stress response, metal transport, membrane homeostasis, oxidative stress, basic metabolism
Citation: Peng J, Miao L, Chen X and Liu P (2018) Comparative Transcriptome Analysis of Pseudomonas putida KT2440 Revealed Its Response Mechanisms to Elevated Levels of Zinc Stress. Front. Microbiol. 9:1669. doi: 10.3389/fmicb.2018.01669
Received: 09 May 2018; Accepted: 04 July 2018;
Published: 24 July 2018.
Edited by:
Daniela De Biase, Sapienza Università di Roma, ItalyReviewed by:
David Richardson, University of East Anglia, United KingdomUlisses Padua Pereira, Universidade Estadual de Londrina, Brazil
Copyright © 2018 Peng, Miao, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pulin Liu, plliu@whpu.edu.cn
 Jun Peng
Jun Peng Lihong Miao1
Lihong Miao1 Xi Chen
Xi Chen Pulin Liu
Pulin Liu