- Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (Ministry of Education), Wuhan University School of Pharmaceutical Sciences, Wuhan, China
Microbial natural products (NPs) especially of the Streptomyces genus have been regarded as an unparalleled resource for pharmaceutical drugs discovery. Moreover, recent progress in sequencing technologies and computational resources further reinforces to identify numerous NP biosynthetic gene clusters (BGCs) from the genomes of Streptomyces. However, the majority of these BGCs are silent or poorly expressed in native strains and remain to be activated and investigated, which relies heavily on efficient genome editing approaches. Accordingly, numerous strategies are developed, especially, the most recently developed, namely, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) system reveals remarkable higher accuracy and efficiency for genome editing in various model organisms including the Streptomyces. In this mini review, we highlight the application of CRISPR/Cas9-based approaches in Streptomyces, focus on the editing of BGCs either in vivo or in vitro, as well as target cloning of large-sized BGCs and heterologous expression in a genetically manipulatable host, for discovery, characterization, reengineering, and production of potential pharmaceutical drugs.
Introduction
Streptomyces species are known for the most prolific antibiotic producers and have provided a large number of clinical drugs during past decades. However, discovery of natural product (NP) drugs from these talented bacteria has suffered a blow after the Golden Age of NP discovery in 1950s–1960s, that is severely influenced by high-throughput screening of synthetic libraries and the low efficiency of traditional top-down screening strategies (Li and Vederas, 2009). Recently, great advances in next-generation sequencing technologies and computational resources reacquaint microbial genomes and are regarded as a huge reservoir of untapped NP biosynthetic gene clusters (BGCs; Rutledge and Challis, 2015; Weber and Kim, 2016; Kim et al., 2017); moreover, a vast majority of uncultured microorganisms in environments provide limitless possibilities for NP drugs discovery (Banik and Brady, 2010; Katz et al., 2016). For Streptomyces, the most gifted bacteria are supposed to possess 20–50 BGCs in a single genome, that greatly exceed the identified compounds (Challis, 2014; Baltz, 2017). Nevertheless, most of BGCs are silent or poorly expressed in native hosts under conventional laboratory culture conditions. To activate these cryptic BGCs, high-efficient approaches for genome editing and BGC engineering garner widespread attention and become a rapidly advancing field for NP drugs discovery (Hsu et al., 2014; Rutledge and Challis, 2015; Choi and Lee, 2016; Jakociunas et al., 2016; Li et al., 2017a; Ren et al., 2017; Zou et al., 2018).
Compared with other model organisms, like Escherichia coli and Saccharomyces cerevisiae, Streptomyces strains show poverty in genetic manipulation and most are recalcitrant for genome editing. In Streptomyces, recombinase-mediated homologous recombination has been commonly used for genome editing; however, the related protocols are often laborious and time-consuming (Gust et al., 2003; Komatsu et al., 2010; Fernandez-Martinez and Bibb, 2014; Li et al., 2017a). Until recently, application of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) system, especially the CRISPR/Cas9 system, has greatly facilitated high-efficiency genome editing (Jinek et al., 2012; Choi and Lee, 2016). Likewise, CRISPR/Cas9-based genome editing approaches have greatly accelerated insights into Streptomyces derived NP drugs. In this mini review, we summarize the recent developments and challenges of CRISPR/Cas9-based approaches for editing BGCs of Streptomyces; moreover, cloning and assembly of intact BGCs for heterologous expression are also emphasized.
Crispr/Cas9 Advances the Genome Editing
CRISPR/Cas system functions as adaptive immune system in numerous bacteria and archaea, of which RNAs harboring “spacer” sequence from previously exposed bacteriophages help Cas proteins recognize and cleave the specific exogenous DNA (Barrangou et al., 2007; Grissa et al., 2007; Horvath and Barrangou, 2010). Since CRISPR/Cas system exhibits higher specificity and accuracy on sequence targeting, it has become excellent choice for precision genome editing (Jinek et al., 2012). CRISPR/Cas9, a type II CRISPR/Cas system, originally employs CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) to form crRNA-tracrRNA duplex and then assists Cas9 nuclease to recognize and cleave target DNA harboring trinucleotide protospacer adjacent motif (PAM) and a 5’ end of 20 nucleotides complementary to the spacers (Deltcheva et al., 2011; Jinek et al., 2012; Hsu et al., 2014; Nishimasu et al., 2014). System reprogramming that fuses crRNA and tracrRNA into a synthetic single guide RNA (sgRNA) greatly facilitates preparation of transcripts and significantly promotes the application of CRISPR/Cas9 system (Jinek et al., 2012; Hsu et al., 2014). Reprogrammed CRISPR/Cas9 system has since been successfully used in a variety of organisms, including S. cerevisiae (DiCarlo et al., 2013), Drosophila melanogaster (Gratz et al., 2013), Caenorhabditis elegans (Friedland et al., 2013), plants (Jiang et al., 2013), and human embryos (Baltimore et al., 2015).
In Vivo Strategies for Genome Editing in Streptomyces
Streptomyces are of utmost importance for novel NP drugs discovery, of which the investigating process relies heavily on high-efficiency genome editing. In Streptomyces, classic genome editing commonly achieves through homologous recombination with a suicide or temperature-sensitive or self-replicative plasmid, and requires intensive and time-consuming screening process. The application of CRISPR/Cas9 system for genome editing in Streptomyces started in 2015, and since then related approaches have been tremendously developed. As shown in Table 1, diversified approaches are widely used to edit or refactor BGCs for NP drugs discovery and characterization.
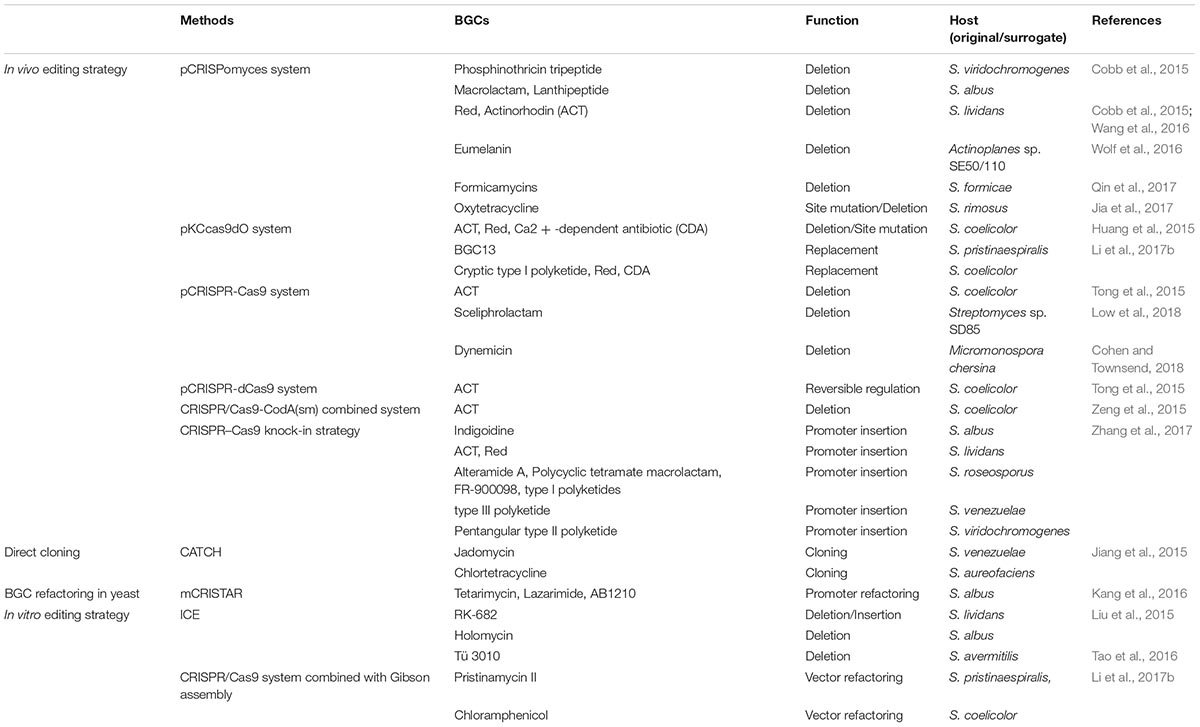
TABLE 1. Application of CRISPR/Cas9 strategies for genome/BGC editing in Streptomyces and some rare actinomycetes.
Cobb et al. (2015) first introduced CRISPR/Cas9 system for genome editing in Streptomyces. The pCRISPomyces-2 system equips a codon-modified Cas9 nuclease driven by a strong promoter, a sgRNA expression cassette, and a 2 kb homology repair template (HRT). It first specifically generates a double-strand break (DSB) at target site by Cas9 nuclease under the guidance of sgRNA harboring a custom-designed spacer, and then repairs the resulting chromosome break by homology-dependent repair (HDR) system in the presence of HRT and introduces chromosomal deletions ranging from 20 bp to 31 kb with an efficiency ranging from 70 to 100% (Figure 1A). Multiplex genome editing may be achieved by equipping multiplex sgRNA cassettes and corresponding repairing templates in a pCRISPomyces system, and excision of 31 kb BGC of undecylprodigiosin (Red) in Streptomyces lividans 66 has thus successfully obtained.
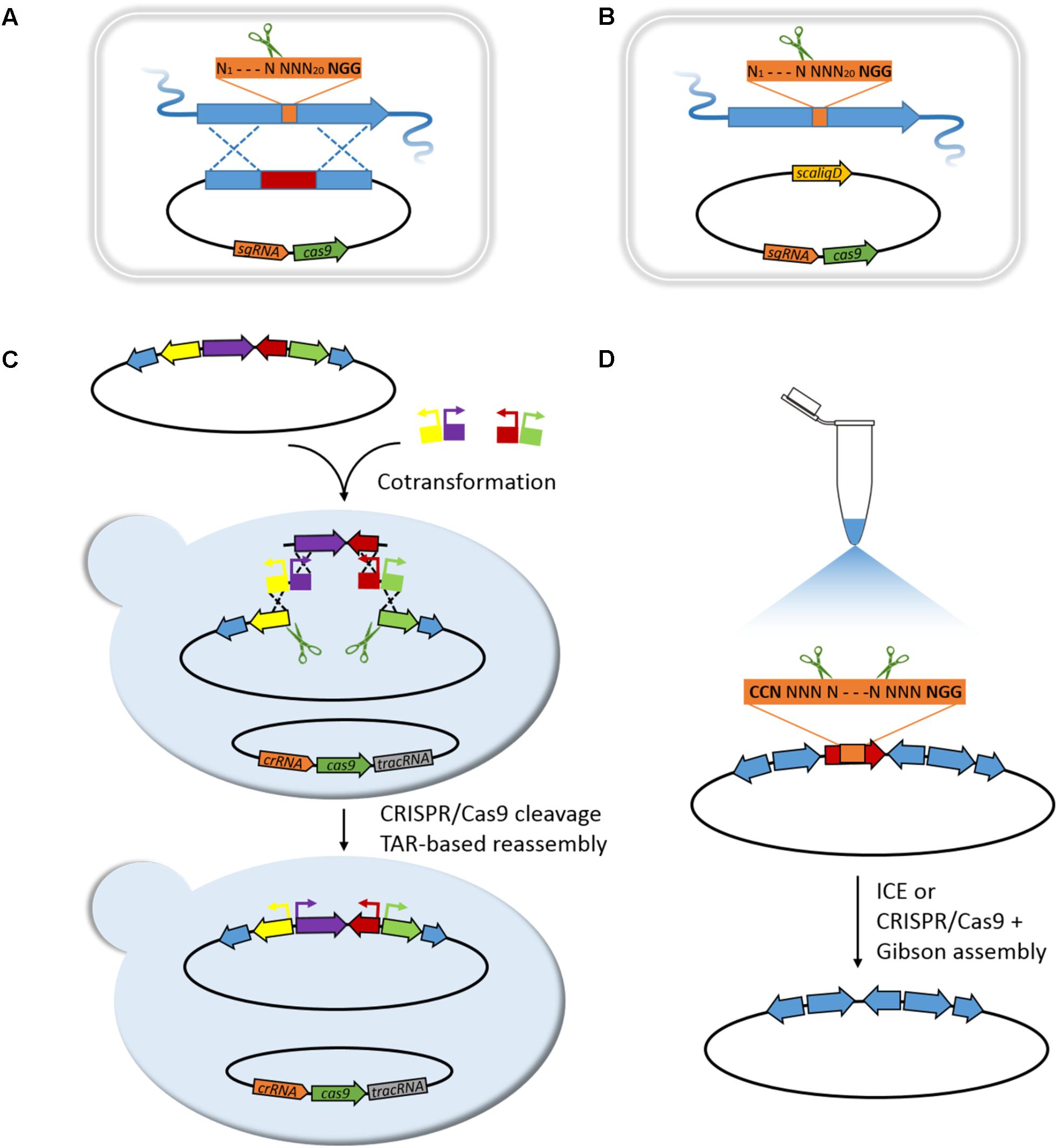
FIGURE 1. Editing of BGCs based on CRISPR/Cas9 strategies. (A) HDR-mediated editing in Streptomyces. Gene deletion, point mutation, or promoter substitution can be performed, respectively, when the repairing template carries corresponding deletion, point mutation, or promoter replacement. Multiplex loci editing can be achieved by equipping multiplex sgRNA cassettes and corresponding HRTs. (B) NHEJ-mediated editing in Streptomyces. Small sized deletion, insertion, or substitution (mostly 1–3 bp) close to the target site can be achieved by using a reconstituted NHEJ system that co-expressing the scaligD in S. coelicolor. (C) mCRISTAR for BGC refactoring in yeast. (D) In vitro editing of BGC by ICE or SRISPR/Cas9 coupling Gibson assembly.
Slightly afterward, three different groups successively applied CRISPR/Cas9 system in Streptomyces for diverse applications. The pKCcas9dO system by Lu group similarly revealed high editing efficiency of CRISPR/Cas9 system in Streptomyces coelicolor M145 for single gene/BGC deletion, as well as multiplex genes/BGCs deletions (Huang et al., 2015). Besides, a point mutation editing strategy that CRISPR/Cas9 cleaves chromosome DNA at specific site guided by synthetic sgRNA, and then the HDR in S. coelicolor helps repair DSB in assistance of HRT with designed point mutation (AAG of 262–264 nucleotides in rpsL was changed to GAA), has performed to convert Lys88 to Glu in rpsL (Figure 1A). Tong et al. (2015) have thoroughly investigated editing efficiency when repairing Cas9-generated site-specific DSBs by non-homologous end joining (NHEJ) system in S. coelicolor A3(2). It revealed an incomplete NHEJ system in S. coelicolor that lacking a core component LigD, and led to randomly sized deletions around target site. Reconstitution of this defective NHEJ system by complementing Streptomyces carneus derived ScaligD has increased editing efficiency up to 77% and qualified the mutations to 1–3 bp deletion/insertion/substitution in most cases (Figure 1B). Moreover, high precision genome editing efficiency near 100% achieved when supplying the HRT. In the study, CRISPRi using a catalytically inactive Cas9 nuclease (dCas9) has also been developed, to target promoter region or open reading frame of actIORF1 for reversible regulation of actinorhodin production in S. colicolor. In the same year, Sun group developed an extraordinary CRISPR/Cas9-CodA(sm) combined system, using CodA(sm), the D314A mutant of cytosine deaminase to convert 5-fluorocytosine to toxic 5-fluorouracil, as an efficient counter-selection approach to select for progenies lost recombinant plasmid, which greatly accelerates screening process (Zeng et al., 2015). Besides, a most significant feature that differs from above three systems is application of a segregationally unstable sti- pIJ101-derived shuttle vector. The behavior of self-replication with high copy number of about 50 per chromosome of this deliver vector can produce large amounts of single-strand plasmid DNA and plenty of template DNAs, resulting in dramatically high efficiency of double cross-over recombination and frequency of target mutant (Zeng et al., 2015). All the above reports facilitate rapid progress for genome editing in Streptomyces, since CRISPR/Cas9 helps select against wild-type sequence in the presence of HRTs. CRISPR/Cas9 system also enables activation of cryptic BGCs in Streptomyces. Zhao group utilized CRISPR/Cas9-mediated knock-in strategy for efficient and precise insertion of constitutive promoters upstream of main biosynthetic operons or pathway-specific activators, and triggered production of novel NPs of different classes in multiple Streptomyces species (Figure 1A; Zhang et al., 2017).
To date, CRISPR/Cas9 system has been applied for genome editing in Streptomyces for 3 years, the high specificity and efficiency made it the most attractive technology in that field. Its application has now extended to many non-model Streptomyces strains, like Streptomyces formicae from the African fungus-growing plant-ant Tetraponera penzigi (Qin et al., 2017), Streptomyces rimosus with distinctive chromosome terminal and core regions (Jia et al., 2017), Streptomyces sp. SD85 from tropical mangrove sediments (Low et al., 2018), and some rare actinomycetes like Actinoplanes sp.SE50/110 (Wolf et al., 2016) and Micromonospora chersina (Cohen and Townsend, 2018). However, in vivo application of this fascinating technology in Streptomyces is confined to the strains are genetically tractable, missing out on a vast amount of precious BGC resources from genetically intractable strains or yet uncultured strains. In that case, acquiring and refactoring of intact BGCs for heterologous expression in a genetically tractable surrogate host could be alternatively considered.
Crispr/Cas9-Mediated Bgcs Cloning and Refactoring for Heterologous Expression
Cloning of Large-Sized BGCs
A variety of cryptic BGC awakening approaches, like pathway-specific/global regulator manipulation, promoter refactoring, and ribosome engineering, have been used for NPs discovery in Streptomyces. However, most require genetic manipulation of native strains thus are constrained in genetically intractable strains or BGCs from environmental DNA (eDNA; Rutledge and Challis, 2015; Weber et al., 2015; Zhang et al., 2016). Strategies for heterologous expression of BGCs in a genetically manipulatable host can perfectly circumvent above bottleneck, but cloning and editing of large-sized BGCs (sometimes over 100 kb) remain challenging. For cloning large-sized DNAs, classic strategies generally utilize randomly digested genomic libraries; however, the screening process is always laborious and it is arduous for packaging intact BGCs over 100 kb in a single vector. Previous precision cloning strategies often utilize restriction enzymes (REs) to release target BGCs that subsequently acquired by coupling diverse DNA capturing strategies. Linear–linear homologous recombination (LLHR) uses RecE/T mediated homologous recombination for direct capture of REs generated genome segments and is widely used for direct cloning of NP BGCs from Streptomyces (Fu et al., 2012; Nah et al., 2017). Gibson assembly coupling REs cleavage is also used for capturing BGCs, and accordingly, the conglobatin cluster has target cloned by Leadlay group (Zhou et al., 2015). Transformation-associated recombination (TAR) cloning uses homologous recombination in S. cerevisiae to capture REs generated BGC segments, and has employed for cloning BGCs of taromycin A (Yamanaka et al., 2014), alterochromide (Ross et al., 2015), and thiotetronates (Tang et al., 2015). However, these REs-dependent approaches are severely constrained for broader application since appropriate RE cutting sites do not regularly exist close to BGC terminals. CRISPR/Cas9 system perfectly addresses the limitation, that target cleaves the DNA guided by a synthetic sgRNA, allowing target cloning of large-sized BGCs. Wang et al. (2015) tentatively applied CRISPR/Cas9 system as REs in vitro to linearize a large vector (22 kb) and subsequently seamlessly assembled with a small DNA using Gibson assembly (Wang et al., 2015). For precision acquiring large-sized DNAs harboring NP BGCs, Jiang et al. (2015) developed Cas9-assisted targeting of chromosome segments (CATCH), which allows target cloning of intact BGCs up to 100 kb that cleaved by CRISPR/Cas9 at specific sites guided by custom-designed sgRNAs and subsequent target captured by Gibson assembly (Jiang et al., 2015). Simultaneously, Lee et al. (2015) combined CRISPR/Cas9 with TAR cloning that employs homologous recombination in yeast to target capture CRISPR/Cas9 released chromosomal segments and dramatically accelerated capture efficiency of TAR cloning up to 32% (Lee et al., 2015). Soon after, CRISPR/Cas9 system coupling TAR cloning was further applied to construct even megabase-sized DNA segments. Zhou et al. (2016) developed Cas9-facilitated homologous recombination assembly (CasHRA), which co-introduces large circular DNAs into S. cerevisiae and release the target DNA segments by CRISPR/Cas9 for subsequent assembly by homologous recombination. It provides an alternative for assembly of large-sized BGCs over 100 kb, using DNAs obtained from cosmid libraries of Streptomyces or eDNA. However, it involves assembly steps and tends to be time-consuming.
CRISPR/Cas9-Mediated Editing of BGCs
Acquiring of intact BGCs of interest is the first step to heterologously investigate the novel NP drugs. Editing of acquired BGCs is generally required for successful heterologous expression. Routine strategies for BGCs editing are always constrained by difficulty of handling large-sized DNAs, and are always laborious. λ-Red recombination mediated PCR-targeting has often used for editing BGC by creating gene replacements/deletions; however, residues like antibiotic selection markers or FRT sequence remain at editing sites, and unintended recombination may raise from repetitive sequences of such modular PKS or NRPS genes (Gust et al., 2003; Yamanaka et al., 2014). λ-Red recombination also enables promoter refactoring or domains/modules exchange for characterization of NPs biosynthesis (Nguyen et al., 2006; Du et al., 2013). Recently, a more facile promoter refactoring approach based on homologous recombination in S. cerevisiae has been developed by Brady group, that enables multiplex promoter refactoring in a single TAR reaction (Montiel et al., 2015). Based on this, production of eDNA-derived indolotryptoline antiproliferative agents, lazarimides A and B, was activated. Nevertheless, the refactoring rate is relatively low. Homologous recombination in yeast could be greatly improved if specific DSBs are introduced at recombination sites (Storici et al., 2003; Storici and Resnick, 2006; Lee et al., 2015). Accordingly, Brady group developed multiplexed-CRISPR-TAR (mCRISTAR) approach, which introduces CRISPR/Cas9 system to specifically create DSBs across target recombination sites (Kang et al., 2016). With mCRISTAR, multiplex CRISPR/Cas9 generated operon fragments can be reassembled with synthetic promoter cassettes by homologous recombination, and are capable of achieving four promoters exchange simultaneously in a single round using one auxotrophic marker selection, with efficiency up to 80% (Figure 1C). General applicability of mCRISTAR has been validated by applying to activate three different cryptic BGCs coding for tetarimycin, lazarimide, and AB1210, indicating a powerful and promising technology for discovery of novel NP drugs from cryptic BGCs resource.
In contrast to the above in vivo strategies for BGC editing based on homologous recombination in E. coli or S. cerevisiae, Sun group developed a new in vitro CRISPR/Cas9-mediated editing (ICE) system for high-efficient BGCs editing (Liu et al., 2015). ICE system allows a complete in vitro operating process with normal molecular operations, which cleaves BGCs at specific sites guided by synthetic sgRNAs and ligates the blunt ends that are repaired by T4 polymerase, to create gene in-frame deletion/replacement/insertion mutations (Figure 1D). With ICE system, BGCs of tetronate RK-682 and dithiolopyrrolone holomycin were readily edited (Liu et al., 2015), especially for Tü 3010, a particular thiotetronate antibiotic, various gene in-frame deletions were rapidly constructed and accordingly deciphered biosynthesis of this exceptional thiotetronate structure (Tao et al., 2016). Soon afterward Lu group utilized a similar in vitro approach that coupling CRISPR/Cas9 system with Gibson assembly to refactor the bacterial artificial chromosome vector harboring BGC of pristinamycin II for following multiplexed site-specific genome engineering in Streptomyces (Figure 1D; Li et al., 2017b). The above two examples indicate that in vitro application of CRISPR/Cas9 could be of wide applicability for BGCs editing, especially coupling the subsequent heterologous expression of BGCs for NP drugs discovery, characterization, and engineering. Nevertheless, optimization of in vitro strategies for multiplex loci refactoring of BGCs is of great necessity, and coupling of CRISPR/Cas9 system with Gibson assembly may preliminarily address the problem.
Conclusion
In conclusion, CRISPR/Cas9 system has proved to be a powerful technology for genome editing or BGC refactoring due to the outstanding features, like higher sequence specificity, artificial guided targeting, and high editing efficiency. Its applications of genome editing specialized for Streptomyces are still relatively narrower in range, especially for the strains little studied. Thus, more efficient and convenient CRISP/Cas tools are of urgent requirement. For instance, diversified CRISPR/Cas systems like Cpf1 (Zetsche et al., 2015; Fonfara et al., 2016; Li et al., 2016), the newly identified class 2 type V CRISPR/Cas protein, xCRISPR/Cas9 (Hu et al., 2018), the most recently evolved CRISPR/Cas9 system with broad PAM compatibility, and even the CRISPR/Cas systems from Streptomyces (Choi and Lee, 2016) could be introduced for diverse applications in Streptomyces, to advance the researches on NP drugs and open a new era for NP drugs discovery.
Author Contributions
WT and YS wrote the manuscript. All authors revised and approved the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant Nos. 31770069 and 31300061).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SC and handling Editor declared their shared affiliation.
References
Baltimore, D., Berg, P., Botchan, M., Carroll, D., Charo, R. A., Church, G., et al. (2015). Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science 348, 36–38. doi: 10.1126/science.aab1028
Baltz, R. H. (2017). Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 44, 573–588. doi: 10.1007/s10295-016-1815-x
Banik, J. J., and Brady, S. F. (2010). Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. Curr. Opin. Microbiol. 13, 603–609. doi: 10.1016/j.mib.2010.08.012
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Challis, G. L. (2014). Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 41, 219–232. doi: 10.1007/s10295-013-1383-2
Choi, K. R., and Lee, S. Y. (2016). CRISPR technologies for bacterial systems: current achievements and future directions. Biotechnol. Adv. 34, 1180–1209. doi: 10.1016/j.biotechadv.2016.08.002
Cobb, R. E., Wang, Y., and Zhao, H. (2015). High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth. Biol. 4, 723–728. doi: 10.1021/sb500351f
Cohen, D. R., and Townsend, C. A. (2018). A dual role for a polyketide synthase in dynemicin enediyne and anthraquinone biosynthesis. Nat. Chem. 10, 231–236. doi: 10.1038/nchem.2876
Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. doi: 10.1038/nature09886
DiCarlo, J. E., Norville, J. E., Mali, P., Rios, X., Aach, J., and Church, G. M. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic. Acids. Res. 41, 4336–4343. doi: 10.1093/nar/gkt135
Du, D., Zhu, Y., Wei, J., Tian, Y., Niu, G., and Tan, H. (2013). Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene clusters. Appl. Microbiol. Biotechnol. 97, 6383–6396. doi: 10.1007/s00253-013-4836-7
Fernandez-Martinez, L. T., and Bibb, M. J. (2014). Use of the meganuclease I-SceI of Saccharomyces cerevisiae to select for gene deletions in actinomycetes. Sci. Rep. 4:7100. doi: 10.1038/srep07100
Fonfara, I., Richter, H., Bratovic, M., Le Rhun, A., and Charpentier, E. (2016). The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521. doi: 10.1038/nature17945
Friedland, A. E., Tzur, Y. B., Esvelt, K. M., Colaiacovo, M. P., Church, G. M., and Calarco, J. A. (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10, 741–743. doi: 10.1038/nmeth.2532
Fu, J., Bian, X., Hu, S., Wang, H., Huang, F., Seibert, P. M., et al. (2012). Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol. 30, 440–446. doi: 10.1038/nbt.2183
Gratz, S. J., Cummings, A. M., Nguyen, J. N., Hamm, D. C., Donohue, L. K., Harrison, M. M., et al. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. doi: 10.1534/genetics.113.152710
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172
Gust, B., Challis, G. L., Fowler, K., Kieser, T., and Chater, K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 100, 1541–1546. doi: 10.1073/pnas.0337542100
Horvath, P., and Barrangou, R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170. doi: 10.1126/science.1179555
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Hu, J. H., Miller, S. M., Geurts, M. H., Tang, W., Chen, L., Sun, N., et al. (2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. doi: 10.1038/nature26155
Huang, H., Zheng, G., Jiang, W., Hu, H., and Lu, Y. (2015). One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim. Biophys. Sin. (Shanghai) 47, 231–243. doi: 10.1093/abbs/gmv007
Jakociunas, T., Jensen, M. K., and Keasling, J. D. (2016). CRISPR/Cas9 advances engineering of microbial cell factories. Metab. Eng. 34, 44–59. doi: 10.1016/j.ymben.2015.12.003
Jia, H., Zhang, L., Wang, T., Han, J., Tang, H., and Zhang, L. (2017). Development of a CRISPR/Cas9-mediated gene-editing tool in Streptomyces rimosus. Microbiology 163, 1148–1155. doi: 10.1099/mic.0.000501
Jiang, W., Zhao, X., Gabrieli, T., Lou, C., Ebenstein, Y., and Zhu, T. F. (2015). Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat. Commun. 6:8101. doi: 10.1038/ncomms9101
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41:e188. doi: 10.1093/nar/gkt780
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Kang, H. S., Charlop-Powers, Z., and Brady, S. F. (2016). Multiplexed CRISPR/Cas9- and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast. ACS Synth. Biol. 5, 1002–1010. doi: 10.1021/acssynbio.6b00080
Katz, M., Hover, B. M., and Brady, S. F. (2016). Culture-independent discovery of natural products from soil metagenomes. J. Ind. Microbiol. Biotechnol. 43, 129–141. doi: 10.1007/s10295-015-1706-6
Kim, H. U., Blin, K., Lee, S. Y., and Weber, T. (2017). Recent development of computational resources for new antibiotics discovery. Curr. Opin. Microbiol. 39, 113–120. doi: 10.1016/j.mib.2017.10.027
Komatsu, M., Uchiyama, T., Omura, S., Cane, D. E., and Ikeda, H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 2646–2651. doi: 10.1073/pnas.0914833107
Lee, N. C., Larionov, V., and Kouprina, N. (2015). Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 43:e55. doi: 10.1093/nar/gkv112
Li, J. W., and Vederas, J. C. (2009). Drug discovery and natural products: end of an era or an endless frontier? Science 325, 161–165. doi: 10.1126/science.1168243
Li, L., Jiang, W., and Lu, Y. (2017a). New strategies and approaches for engineering biosynthetic gene clusters of microbial natural products. Biotechnol. Adv. 35, 936–949. doi: 10.1016/j.biotechadv.2017.03.007
Li, L., Zheng, G., Chen, J., Ge, M., Jiang, W., and Lu, Y. (2017b). Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab. Eng. 40, 80–92. doi: 10.1016/j.ymben.2017.01.004
Li, S. Y., Zhao, G. P., and Wang, J. (2016). C-Brick: A new standard for assembly of biological parts using Cpf1. ACS Synth. Biol. 5, 1383–1388. doi: 10.1021/acssynbio.6b00114
Liu, Y., Tao, W., Wen, S., Li, Z., Yang, A., Deng, Z., et al. (2015). In vitro CRISPR/Cas9 system for efficient targeted DNA editing. mBio 6, e01714–15. doi: 10.1128/mBio.01714-15
Low, Z. J., Pang, L. M., Ding, Y., Cheang, Q. W., Le Mai Hoang, K., Thi Tran, H., et al. (2018). Identification of a biosynthetic gene cluster for the polyene macrolactam sceliphrolactam in a Streptomyces strain isolated from mangrove sediment. Sci. Rep. 8:1594. doi: 10.1038/s41598-018-20018-8
Montiel, D., Kang, H. S., Chang, F. Y., Charlop-Powers, Z., and Brady, S. F. (2015). Yeast homologous recombination-based promoter engineering for the activation of silent natural product biosynthetic gene clusters. Proc. Natl. Acad. Sci. U.S.A. 112, 8953–8958. doi: 10.1073/pnas.1507606112
Nah, H. J., Pyeon, H. R., Kang, S. H., Choi, S. S., and Kim, E. S. (2017). Cloning and heterologous expression of a large-sized natural product biosynthetic gene cluster in Streptomyces species. Front. Microbiol. 8:394. doi: 10.3389/fmicb.2017.00394
Nguyen, K. T., Ritz, D., Gu, J. Q., Alexander, D., Chu, M., Miao, V., et al. (2006). Combinatorial biosynthesis of novel antibiotics related to daptomycin. Proc. Natl. Acad. Sci. U.S.A. 103, 17462–17467. doi: 10.1073/pnas.0608589103
Nishimasu, H., Ran, F. A., Hsu, P. D., Konermann, S., Shehata, S. I., Dohmae, N., et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. doi: 10.1016/j.cell.2014.02.001
Qin, Z., Munnoch, J. T., Devine, R., Holmes, N. A., Seipke, R. F., Wilkinson, K. A., et al. (2017). Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chem. Sci. 8, 3218–3227. doi: 10.1039/c6sc04265a
Ren, H., Wang, B., and Zhao, H. (2017). Breaking the silence: new strategies for discovering novel natural products. Curr. Opin. Biotechnol. 48, 21–27. doi: 10.1016/j.copbio.2017.02.008
Ross, A. C., Gulland, L. E., Dorrestein, P. C., and Moore, B. S. (2015). Targeted capture and heterologous expression of the Pseudoalteromonas alterochromide gene cluster in Escherichia coli represents a promising natural product exploratory platform. ACS Synth. Biol. 4, 414–420. doi: 10.1021/sb500280q
Rutledge, P. J., and Challis, G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523. doi: 10.1038/nrmicro3496
Storici, F., Durham, C. L., Gordenin, D. A., and Resnick, M. A. (2003). Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl. Acad. Sci. U.S.A. 100, 14994–14999. doi: 10.1073/pnas.2036296100
Storici, F., and Resnick, M. A. (2006). The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 409, 329–345. doi: 10.1016/S0076-6879(05)09019-1
Tang, X., Li, J., Millan-Aguinaga, N., Zhang, J. J., O’Neill, E. C., Ugalde, J. A., et al. (2015). Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol. 10, 2841–2849. doi: 10.1021/acschembio.5b00658
Tao, W., Yurkovich, M. E., Wen, S., Lebe, K. E., Samborskyy, M., Liu, Y., et al. (2016). A genomics-led approach to deciphering the mechanism of thiotetronate antibiotic biosynthesis. Chem. Sci. 7, 376–385. doi: 10.1039/c5sc03059e
Tong, Y., Charusanti, P., Zhang, L., Weber, T., and Lee, S. Y. (2015). CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth. Biol. 4, 1020–1029. doi: 10.1021/acssynbio.5b00038
Wang, J. W., Wang, A., Li, K., Wang, B., Jin, S., Reiser, M., et al. (2015). CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning. Biotechniques 58, 161–170. doi: 10.2144/000114261
Wang, Y., Cobb, R. E., and Zhao, H. (2016). High-efficiency genome editing of Streptomyces species by an engineered CRISPR/Cas system. Methods Enzymol. 575, 271–284. doi: 10.1016/bs.mie.2016.03.014
Weber, T., Charusanti, P., Musiol-Kroll, E. M., Jiang, X., Tong, Y., Kim, H. U., et al. (2015). Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol. 33, 15–26. doi: 10.1016/j.tibtech.2014.10.009
Weber, T., and Kim, H. U. (2016). The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth. Syst. Biotechnol. 1, 69–79. doi: 10.1016/j.synbio.2015.12.002
Wolf, T., Gren, T., Thieme, E., Wibberg, D., Zemke, T., Puhler, A., et al. (2016). Targeted genome editing in the rare actinomycete Actinoplanes sp. SE50/110 by using the CRISPR/Cas9 System. J. Biotechnol. 231, 122–128. doi: 10.1016/j.jbiotec.2016.05.039
Yamanaka, K., Reynolds, K. A., Kersten, R. D., Ryan, K. S., Gonzalez, D. J., Nizet, V., et al. (2014). Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. U.S.A. 111, 1957–1962. doi: 10.1073/pnas.1319584111
Zeng, H., Wen, S., Xu, W., He, Z., Zhai, G., Liu, Y., et al. (2015). Highly efficient editing of the actinorhodin polyketide chain length factor gene in Streptomyces coelicolor M145 using CRISPR/Cas9-CodA(sm) combined system. Appl. Microbiol. Biotechnol. 99, 10575–10585. doi: 10.1007/s00253-015-6931-4
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. doi: 10.1016/j.cell.2015.09.038
Zhang, M. M., Wang, Y., Ang, E. L., and Zhao, H. (2016). Engineering microbial hosts for production of bacterial natural products. Nat. Prod. Rep. 33, 963–987. doi: 10.1039/c6np00017g
Zhang, M. M., Wong, F. T., Wang, Y., Luo, S., Lim, Y. H., Heng, E., et al. (2017). CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. doi: 10.1038/nchembio.2341 [Epub ahead of print].
Zhou, J., Wu, R., Xue, X., and Qin, Z. (2016). CasHRA (Cas9-facilitated Homologous Recombination Assembly) method of constructing megabase-sized DNA. Nucleic Acids Res. 44:e124. doi: 10.1093/nar/gkw475
Zhou, Y., Murphy, A. C., Samborskyy, M., Prediger, P., Dias, L. C., and Leadlay, P. F. (2015). Iterative mechanism of macrodiolide formation in the anticancer compound conglobatin. Chem. Biol. 22, 745–754. doi: 10.1016/j.chembiol.2015.05.010
Keywords: natural product, Streptomyces, biosynthetic gene cluster, genome editing, CRISPR/Cas9
Citation: Tao W, Yang A, Deng Z and Sun Y (2018) CRISPR/Cas9-Based Editing of Streptomyces for Discovery, Characterization, and Production of Natural Products. Front. Microbiol. 9:1660. doi: 10.3389/fmicb.2018.01660
Received: 30 March 2018; Accepted: 04 July 2018;
Published: 24 July 2018.
Edited by:
Eung-Soo Kim, Inha University, South KoreaReviewed by:
Yinhua Lu, Shanghai Institutes for Biological Sciences (CAS), ChinaSisun Choi, Inha University, South Korea
Copyright © 2018 Tao, Yang, Deng and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuhui Sun, eWhzdW5Ad2h1LmVkdS5jbg==
 Weixin Tao
Weixin Tao Anna Yang
Anna Yang Zixin Deng
Zixin Deng Yuhui Sun
Yuhui Sun