
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 July 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01644
This article is part of the Research Topic Metabolic Adjustments and Gene Expression Reprogramming for Symbiotic Nitrogen Fixation in Legume Nodules View all 12 articles
The Bradyrhizobium sp. DOA9 strain displays the unusual properties to have a symbiotic plasmid and to fix nitrogen during both free-living and symbiotic growth. Sequence genome analysis shows that this strain contains the structural genes of dinitrogenase (nifDK) and the nifA regulatory gene on both the plasmid and chromosome. It was previously shown that both nifDK clusters are differentially expressed depending on growth conditions, suggesting different mechanisms of regulation. In this study, we examined the functional regulatory role of the two nifA genes found on the plasmid (nifAp) and chromosome (nifAc) that encode proteins with a moderate level of identity (55%) and different structural architectures. Using gusA (β-glucuronidase) reporter strains, we showed that both nifA genes were expressed during both the free-living and symbiotic growth stages. During symbiosis with Aeschynomene americana, mutants in only one nifA gene were not altered in their symbiotic properties, while a double nifA mutant was drastically impaired in nitrogen fixation, indicating that the two NifA proteins are functionally redundant during this culture condition. In contrast, under in vitro conditions, the nifAc mutant was unable to fix nitrogen, and no effect of the nifAp mutation was detected, indicating that NifAc is essential to activate nif genes during free-living growth. In accordance, the nitrogenase fixation deficiency of this mutant could be restored by the introduction of nifAc but not by nifAp or by two chimeric nifA genes encoding hybrid proteins with the N-terminus part of NifAc and the C-terminus of NifAp. Furthermore, transcriptional analysis by RT-qPCR of the WT and two nifA mutant backgrounds showed that NifAc and NifAp activated the expression of both chromosome and plasmid structural nifDK genes during symbiosis, while only NifAc activated the expression of nifDKc during free-living conditions. In summary, this study provides a better overview of the complex mechanisms of regulation of the nitrogenase genes in the DOA9 strain that involve two distinct NifA proteins, which are exchangeable during symbiosis for the activation of nif genes but not during free-living growth where NifAc is essential for the activation of nifDKc.
Rhizobium-legume symbiosis is considered as the major contributor of biologically fixed nitrogen to terrestrial ecosystems. The reduction of atmospheric N2 is catalyzed by the nitrogenase enzyme complex, which requires high-energy input in the form of ATP and electrons to break the triple bond. In addition, this enzymatic complex is highly sensitive to molecular oxygen, which irreversibly inactivates the enzyme. Diazotrophic bacteria have evolved sophisticated regulatory circuits of their nitrogen fixation (nif) genes in response to oxygen and nitrogen availability to prevent unnecessary energy consumption and permit the synthesis of the nitrogenase complex only during the proper environmental conditions (Burris and Roberts, 1993; Fischer, 1994).
A master regulator of nitrogen fixation is the NifA protein, which acts in association with the RNA polymerase sigma factor RpoN (σ54) to activate the expression of nif genes by binding to an upstream activating sequence (UAS; 5′-TGT-N10-ACA-3′). The NifA proteins show a typical three-domain structure. The N-terminal GAF domain is a ubiquitous signaling motif found in signaling and sensory proteins from all three kingdoms of life (Ho et al., 2000). The central domain interacts with the σ54-RNA polymerase and possesses ATPase activity, while the C-terminal domain shows a helix-turn-helix (HTH) motif involved in DNA-binding. The activity of NifA is directly sensitive to molecular oxygen, and in some cases, is also directly affected by combined nitrogen (Kullik et al., 1989; Souza et al., 1999; Steenhoudt and Vanderleyden, 2000). In addition, the nifA gene is subjected to transcriptional regulation, although the mechanisms vary depending on the rhizobial strain. For example, in Sinorhizobium meliloti, nifA expression is activated by the FixLJ two-component regulatory system in response to low oxygen tension, while in Bradyrhizobium japonicum, the fixR-nifA operon is controlled by the redox-responsive two-component system RegSR (Bauer et al., 1998).
The rhizobia generally display only one nifA copy, but one exception has been described for Mesorhizobium loti, which contains two nifA genes, nifA1 and nifA2, both located on the symbiotic island (Nukui et al., 2006). The nifA1 gene is most similar to the nifA of Rhizobium etli, R. leguminosarum, and S. meliloti, and it is found in an identical genomic context associated with other nif genes, while nifA2 is most similar to nifA from B. japonicum and is not found in the vicinity of known nif genes (Sullivan et al., 2013). Interestingly, the two nifA genes are not functionally redundant, since the M. loti nifA2 mutant form nodules that do not fix nitrogen, while the nifA1 mutant is not affected symbiotically (Nukui et al., 2006).
Another example of the presence of two nifA genes has recently emerged with the analysis of the genome sequence of the non-photosynthetic Bradyrhizobium sp. DOA9 strain (Okazaki et al., 2015). This bacterium, isolated from rice paddy fields using Aeschynomene americana as a trap legume (Noisangiam et al., 2012), displays several unusual properties. First, unlike all described bradyrhizobia, this strain contains a symbiotic megaplasmid (pDOA9) that harbors nod and nif genes (Okazaki et al., 2015). Second, on both the chromosome and the plasmid, it can be distinguished the nitrogenase nifHDK genes that encode the subunits of the nitrogenase complex. In both cases, the nifHDK genes are split into two clusters, nifH and nifDK (Okazaki et al., 2015). Third, as described for photosynthetic bradyrhizobia, the bacteria that do contain a chromosomal nifV gene can fix nitrogen in both the free living and symbiotic states (Wongdee et al., 2016). Data from previous research indicated that both nifDK clusters contribute to nitrogenase activity during symbiosis with A. americana, while the nifDK cluster found on the chromosome is the major contributor to the nitrogenase activity of the bacteria under free-living conditions (Wongdee et al., 2016). These data indicate that the two nifDK clusters identified in the DOA9 strain should be differentially regulated. This is supported by the fact that the DOA9 display two nifA genes but also two rpoN homologous genes, both located on each replicon. The simple explanation is that the nifA found on the chromosome (nifAc) and the nifA found on the plasmid (nifAp) specifically regulated the nifDK cluster found on the replicon where nifA is present. However, crosstalk between these two regulatory circuits would also be expected, given that NifDKp proteins require the expression of the accessory nif genes to form a functional nitrogenase. In particular, the nifENX genes whose products are needed for synthesis of the iron-molybdenum cofactor of nitrogenase, exist as a unique copy and are found downstream of the nifDKc cluster.
Thus, in the present work, we aimed to investigate in more detail the regulatory functions of the two nifA genes identified for the Bradyrhizobium sp. DOA9. In a first approach, we analyzed the expression levels of both nifA genes under different culture conditions using translational fusions to gusA (β-glucuronidase). We then analyzed the contribution of each regulatory protein to the control of bacterial nitrogenase activity under free-living and symbiotic states by constructing single and double nifA mutants. Finally, the expression level of several nif genes in three different backgrounds, the DOA9 wild-type (WT), ΔnifAc, and ΔnifAp mutant strains, were analyzed to identify which genes are activated by NifAc and NifAp.
The Bradyrhizobium sp. DOA9 WT was obtained from the School of Biotechnology, Suranaree University of Technology, Thailand, while all mutants were constructed in the Laboratoire des Symbioses Tropicales et Méditerranéennes (LSTM), France. These bacterial strains were grown at 28°C for 4 days in Yeast extract-mannitol (YM) medium (Vincent, 1970) or a BNM-B minimal medium (Renier et al., 2011). Escherichia coli strains were grown in LB medium at 37°C. When required, the media were supplemented with the appropriate antibiotics at the following concentrations: 100 μg/ml kanamycin, 200 μg/ml streptomycin, 20 μg/ml nalidixic acid, and 20 μg/ml cefotaxime.
All DNA fragments were amplified using the primers listed in Supplementary Table S1. To construct the reporter strains, DOA9-Pm-fixRnifAc and DOA9-Pm-nifAp, the 500-bp upstream region of fixR and the nifAp operon were amplified by PCR and cloned into the plasmid pVO155-npt2-cefo-npt2-gfp. This plasmid, which is a derivative of the pVO155 plasmid (Oke and Long, 1999), could not replicate in the Bradyrhizobium strains. The plasmid carries the promoterless gusA, gfp, kanamycin, and cefotaxime genes under the constitutive promoter npt2 (Okazaki et al., 2016). To construct the two DOA9ΩnifA (insertion) mutants, 300 to 400 base pairs (bp) of the internal sequence of each nifA gene were amplified by PCR and cloned into the plasmid pVO155-npt2-cefo-npt2-gfp. To construct the DOA9ΔnifA deletion mutants, the upstream and downstream regions (between 700 and 1000-bp) of each nifA gene were amplified and merged using overlap extension PCR. Then, the fragment was cloned into the plasmid pK18 mob-cefo-sacB. This plasmid carries the sacB gene, which induces bacterial death in the presence of sucrose and the kanamycin-resistance gene (Tsai and Alley, 2000) as well as the cefotaxime gene under the npt2 promoter that was added to the KpnI site. The various constructed plasmids were transferred into the DOA9 strain by mating, followed by the insertion or deletion of the selected mutants as previously described (Wongdee et al., 2016).
For the DOA9ΔnifAc mutant, the complete nifAc, nifAp, or hybrid of nifAc and nifAp genes were amplified and cloned downstream of the npt2 promoter into the pMG103-npt2-cefo plasmid that harbored a cefotaxime resistance gene (Wongdee et al., 2016). This plasmid is stable and replicative in the DOA9 strain. To construct the hybrid nifA genes, the 5′-region of nifAc and the 3′-region of nifAp were amplified and merged using overlap extension PCR. The constructed plasmids were introduced into the competent cells of the DOA9ΔnifAc mutant by electroporation. The complemented strains were selected on YM plates supplemented with 20 μg/ml cefotaxime and 20 μg/ml nalidixic acid.
The symbiosis efficiency of the Bradyrhizobium DOA9 strain and its derivatives were tested with A. americana No. 281 collected from the LSTM greenhouse. The seeds were surface sterilized by immersion in sulfuric acid under shaking for 45 min. Seeds were thoroughly washed with sterile distilled water and incubated overnight in sterile water. Seeds were transferred for 1 day at 37°C in the darkness on 0.8% agar plates for germination. Plantlets were transferred onto the top of the test tubes and covered by aluminum paper for hydroponic culture in buffered nodulation medium (BNM) (Ehrhardt et al., 1992). Plants were grown in a 28°C growth chamber with a 16-h light and 8-h dark regime and 70% humidity. Seven days after transfer, each seedling was inoculated with 1 ml of cell suspension resulting from a 5-day-old bacterial culture washed in BNM and adjusted to an optical density of one at 600 nm. For nodulation and the nitrogen fixation assay, 10 to 20 plants per condition were taken at 20 days post-inoculation (dpi) and analyzed for the number of nodules and nitrogenase activity as previously described (Bonaldi et al., 2010). The experiments were performed in duplicate.
To follow the GUS activity in the nodules elicited by the reporter strains, 30- to 40-μm-thick sections from fresh nodule samples were prepared using a vibratome (VT1000S; Leica, Nanterre, France) and incubated at 37°C in the dark in GUS assay buffer for 1 h, as described in Bonaldi et al. (2010). After staining, the sections were mounted and observed under bright-field illumination with a macroscope (Nikon AZ100; Champigny-sur-Marne, France).
To determine the nitrogenase enzyme activity under free-living conditions, the Bradyrhizobium sp. strain DOA9 and derivatives were grown in 10-ml test tubes hermetically closed (BD Vacutainer, Franklin Lakes, NJ, United States) containing 2 ml of semisolid BNM-B medium (agar 0.8% w/v). The BNM-B medium is a synthetic plant growth medium (Ehrhardt et al., 1992) supplemented with a carbon (10 mM succinate) and a cocktail of vitamins (riboflavin at 0.2 μg/ml, biotin at 0.12 μg/ml, thiamine-HCl at 0.8 μg/ml, myo-inositol at 0.5 μg/ml, p-aminobenzoic acid at 0.1 μg/ml, nicotinic acid at 0.5 μg/ml, calcium pantothenate at 0.8 μg/ml, and cyanocobalamin at 1 ng/ml) to support growth of Bradyrhizobium strains (Renier et al., 2011). It is to note that the BNM-B medium was not supplemented with a nitrogenous source but the bacteria growth is possible thanks to the dinitrogen and oxygen present in the air constituting the initial headspace of the test tube. Just after closing hermitically the tubes, acetylene gas (1 ml) was injected to a final concentration of 10%. The cultures were then incubated at 28°C without shaking, and the gas samples were analyzed at 7 dpi for ethylene production by gas chromatography, as previously described (Renier et al., 2011).
The two DOA9 reporter strains were grown for 4 days in YM medium, collected, and washed with BNM-B medium as described above. Bottles of 150-ml sealed with rubber stoppers and containing 55-ml of BNM-B medium and 95-ml of air were then inoculated with DOA9 reporter strains to an initial OD600 of 0.05. The cultures were then incubated at 28°C without shaking. After 7 days, the bacterial culture was removed from the bottle, and GUS activity was measured using the substrate p-nitrophenyl glucuronide (PNPG) as described by Jefferson (1987). β-glucuronidase units were calculated according to Miller (1972).
The expression of genes involved in nitrogen fixation of strain DOA9 was determined from cells grown under free-living conditions and bacteroids obtained from nodules of A. americana under symbiotic conditions. For free-living conditions, the bacterial cells were grown in 150-ml bottles as described just above. For harvesting, cultures were added to a 1:10 volume of “stop solution” [10% Tris-HCl-buffered phenol (pH 8) in ethanol], and cells removed from the liquid medium by centrifugation for 10 min (10,000 rpm, 4°C). The cell pellets were frozen in liquid nitrogen and stored at -80°C. Analysis under symbiotic conditions and RNA isolation from bacteroids were processed from approximately 1 g of frozen nodules by homogenization with a tungsten carbide bead (3 mm; Qiagen, Hilden, Germany) in 2-ml microcentrifuge tubes. Total RNA was isolated from the free-living bacterial cells, and the nodules were disrupted with a hot (65°C) phenol-extraction procedure that was previously described (Babst et al., 1996). RNA was purified and treated with DNase using mini-prep kits (Qiagen, Valencia, CA, United States). Then, the cDNA was synthesized with iScript TM Reverse transcription Supermix for RT-qPCR (Bio-Rad, Hercules, CA, United States). Then, 10–50 ng of each cDNA sample was added to PowerUPTM SYBRTM Green master mixed buffer (Applied Biosystems, United States, Canada), and the appropriate amount of specific primers (listed in Supplementary Table S2) were used in the qRT-PCR analyses using an annealing temperature at 55°C for all reactions. The expression of target genes was relatively compared with the expression of the housekeeping gene, dnaK, using QuantStudio Design & Analysis Software from Applied Biosystems.
Two nifA homologous genes can be identified in the Bradyrhizobium DOA9 strain. One copy found on the chromosome, termed nifAc, is located approximately 6 kb from the nifDKENX operon and found just downstream of the fixR gene (Figure 1A). In B. japonicum, nifA is also found downstream of fixR. It has been shown that the two genes are part of the same transcript (Thöny et al., 1987), suggesting that fixR nifAc also forms an operon in the DOA9 strain. Downstream of this operon, a gene (fer) encoding a 4Fe-4S ferredoxin and a suf operon composed of four genes (sufB, sufC, sufD, and sufS) were identified and have been shown to function in the assembly of iron-sulfur clusters (Takahashi and Tokumoto, 2002). The other copy found on the plasmid, known as nifAp, is surrounded by genes of unknown function, and no known nif or fix genes are found in the vicinity. The two corresponding NifA proteins are clearly distinct and are of different lengths; NifAc (579 aa) and NifAp (503 aa) display only 52% identity. A Pfam analysis to identify functional domains showed that NifAc displays a classical NifA architecture with a N-terminal GAFdomain, a central sigma 54 interaction domain and a C-terminal HTHdomain. NifAp shows a less classical structure with only the presence of the central and HTH domains (Figure 1B). The divergence of NifAp is not limited to the absence of the N-terminal GAF domain, since phylogenetic analysis showed that this protein formed an outgroup that was well separated from the NifA proteins identified in Bradyrhizobium strains (Figure 1D).
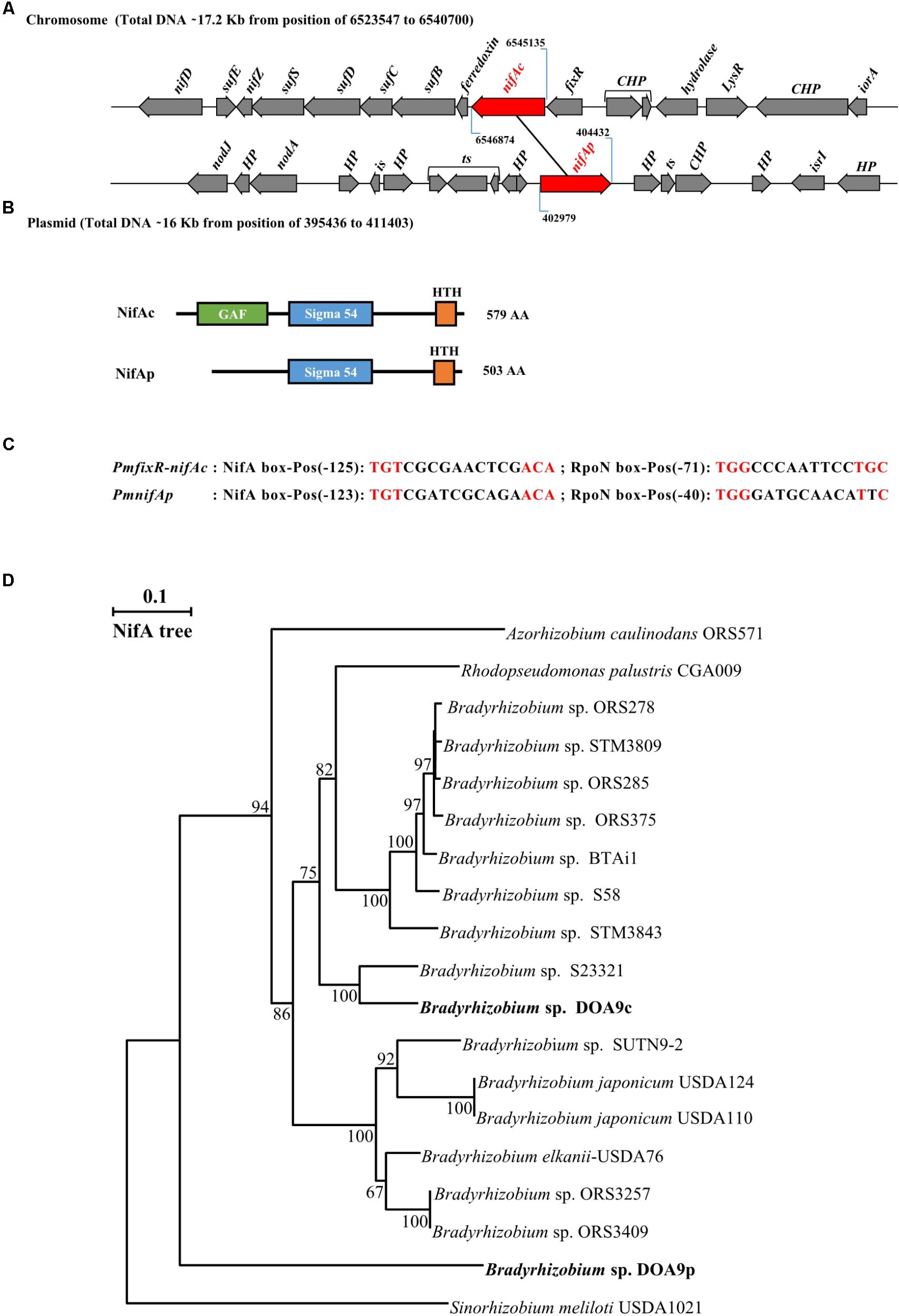
FIGURE 1. Bradyrhizobium sp. strain DOA9 strain displays two distinct nifA genes. (A) Genetic organization of the two nifA genes (in red) located on both the chromosome and plasmid of the DOA9 strain. nifAc, nifA located on the chromosome; nifAp, nifA located on the plasmid; CHP: Conserved Hypothetical protein, HP: Hypothetical protein, is: intregrase and ts: transposase. (B) Predicted domain structure of both NifA proteins. (C) Putative NifA and RpoN boxes identified in promoter region of fixR-nifAc operon and nifAp. Pos: position of 5′ end nucleotide of motif relative to annotated start codon. (D) NifA phylogenetic tree showing relationship between NifA of bradyrhizobia. Sequences were aligned by CLUSTALX, and the tree was generated using the neighbor-joining method (Saitou and Nei, 1987) and displayed using NJPLOT (Perrière and Gouy, 1996). Bootstrap values, expressed as percentages of 1000 replications, are shown at branching points.
Interestingly, in both cases, a close examination of the promoter regions of the fixRnifAc operon and nifAp permitted the identification by manual analysis of a putative NifA and a RpoN binding sites, suggesting that both NifA proteins could autoregulate their own expression level and that of their homolog (Figure 1C).
To analyze the expression of the two nifA genes identified in the Bradyrhizobium DOA9 strain, we constructed two reported strains (DOA9-Pm-fixRnifAc and DOA9-Pm-nifAp) by integrating the nonreplicative plasmid pVO155-npt2-cefo-npt2-gfp, which carries a promoterless gusA gene (Okazaki et al., 2016) downstream of the promoter region of the fixRnifAc operon and nifAp gene. Since the DOA9 strain was isolated using A. americana as a trap, we analyzed these two reporter strains in this host plant. Observations at 14 dpi showed that both reporter strains were able to nodulate and fix nitrogen similar to the WT-strain, indicating that the integration of the pVO155 plasmid in these two promoter regions did not alter the symbiotic performance of the strain (Figures 2A–G). Cytological analysis revealed a β-glucuronidase activity in the nodules, which was elicited by the two reporter strains, in contrast to the WT-nodules for which no activity could be detected (Figures 2E–G). Although X-gluc (5-Bromo-4-chloro-3-indolyl-β-D-glucuronide cyclohexylamine salt) staining is a qualitative measurement of gene expression, the naked eye could observe that the nodules elicited by DOA9-Pm-nifAp displayed a more intense blue color than those elicited by the DOA9-Pm-fixRnifAc reporter strain (Figures 2F,G), which indicates slight differences in the expression of the two nifA genes.
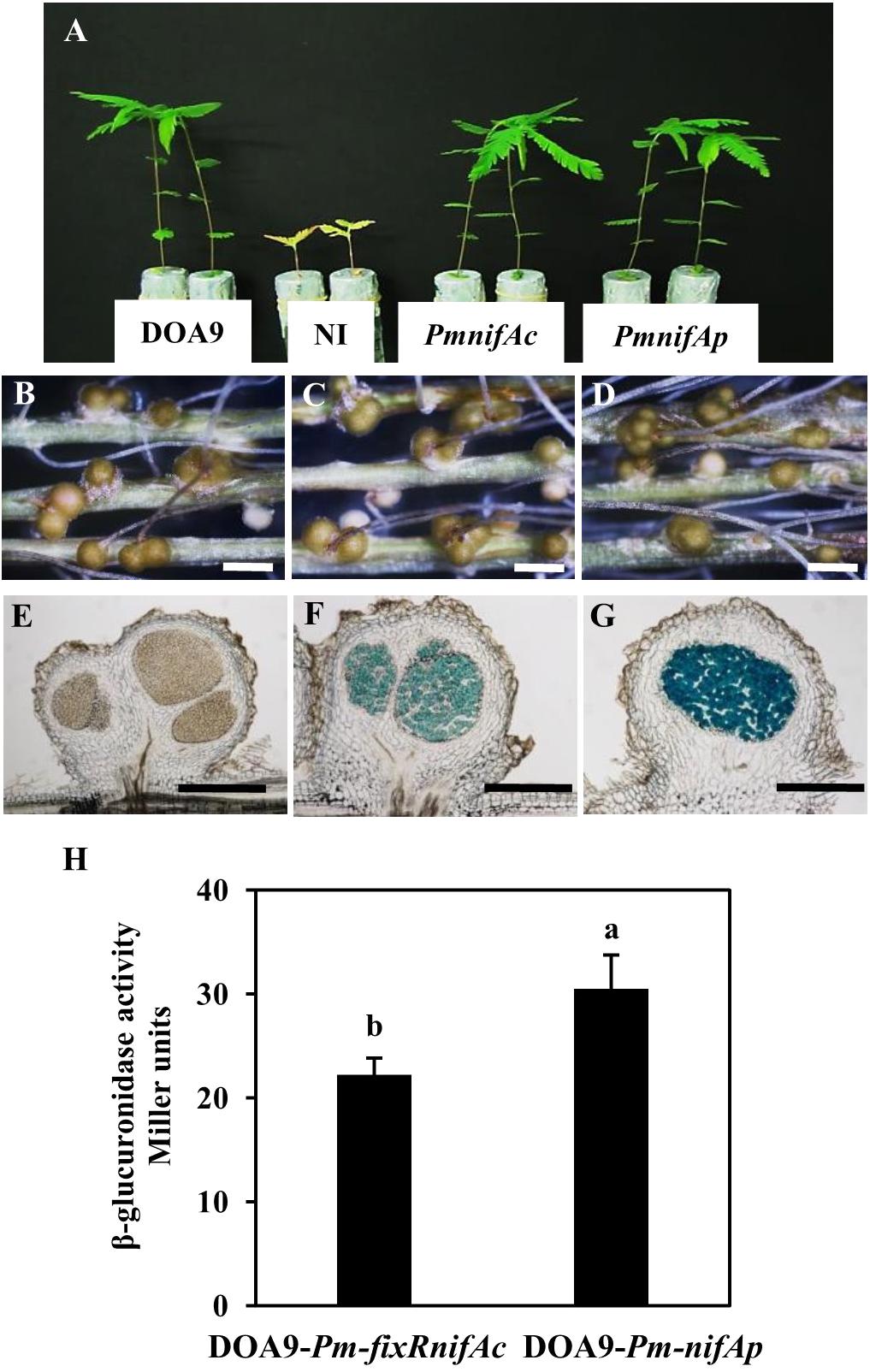
FIGURE 2. The two nifA genes of Bradyrhizobium sp. DOA9 strain are expressed during symbiosis with Aeschynomene americana. (A) Comparison of growth of plants (aerial part) non-inoculated (NI) or inoculated with WT and reporter strains DOA9-Pm-fixRnifAc and DOA9-Pm-nifAp (at 14 dpi). (B–D) Root nodules observed with a stereomicroscope stereomicroscope. (B) Nodules elicited by WT; (C) Nodules elicited by DOA9-Pm-fixRnifAc; (D) Nodules elicited by DOA9-Pm-nifAp. (E–G) Expression of gusA reporter gene revealed on 40 μm nodule sections stained with X-Gluc. Nodules elicited by WT (E), DOA9-Pm-fixRnifAc (F) and DOA9-Pm-nifAp (G). Scale bars are 1 mm for (B–D) and 250 μm (E–G). (H), β-glucuronidase activity of DOA9-Pm-fixRnifAc and DOA9-Pm-nifAp reporter strains grown after 7 days of culture under free-living conditions (microaerobiosis and not combined nitrogen source). Different letters above error bars indicate significant differences at P < 0.05 (Tukey’s HSD test).
Similar observations were also made during free-living growth under microaerobic conditions and the absence of a combined nitrogen source. Indeed, after 7 days of culture in these conditions, the β-glucuronidase activity measured for DOA9-Pm-nifAp (30 Miller unit) was higher than that detected for the DOA9-Pm-fixRnifAc reporter strain (22 Miller Unit) (Figure 2H). Taken together, these data indicate that the two nifA genes identified in the DOA9 strain are expressed during symbiotic and free-living conditions and that in both conditions, the level of expression of nifAp is slightly higher than that of nifAc.
The NifA protein has been shown to be essential for symbiotic nitrogen fixation in several rhizobia (Szeto et al., 1984; Schetgens et al., 1985; Fischer et al., 1986; Iismaa and Watson, 1989). To appreciate the relative importance of each nifA gene identified in DOA9 during symbiosis, we constructed various nifA mutants, single nifA mutants, either by insertion (DOA9ΩnifAc and DOA9ΩnifAp) or deletion (DOA9ΔnifAc and DOA9ΔnifAp), and a double nifA mutant (DOA9ΔnifAp::ΩnifAc). As shown in Figures 3A–C and Supplementary Figure S1, the plants inoculated with the different single mutants displayed no significant difference from those inoculated with the WT strain in terms of their growth, the number of nodules formed or the measured nitrogenase activity indicating that the single mutation of the nifAc or nifAp gene had no impact on the symbiotic performance of the strain. In contrast, the plants inoculated with the double nifA mutant (ΔnifAp::ΩnifAc) displayed a strict fix minus phenotype (Figure 3B), and the growth of the plants was similar to that of the non-inoculated plants. Notably, the double nifA mutant induced nodules that were smaller and displayed symptoms of senescence (they were white instead of pink, indicating the absence of leghemoglobin and the central tissue was digested) (Figures 3D–G). Taken together these data suggest that the two NifA proteins identified in the DOA9 strain are functionally redundant during symbiosis and that at least one functional NifA protein is absolutely required for symbiotic nitrogenase activity, as observed in other rhizobia.
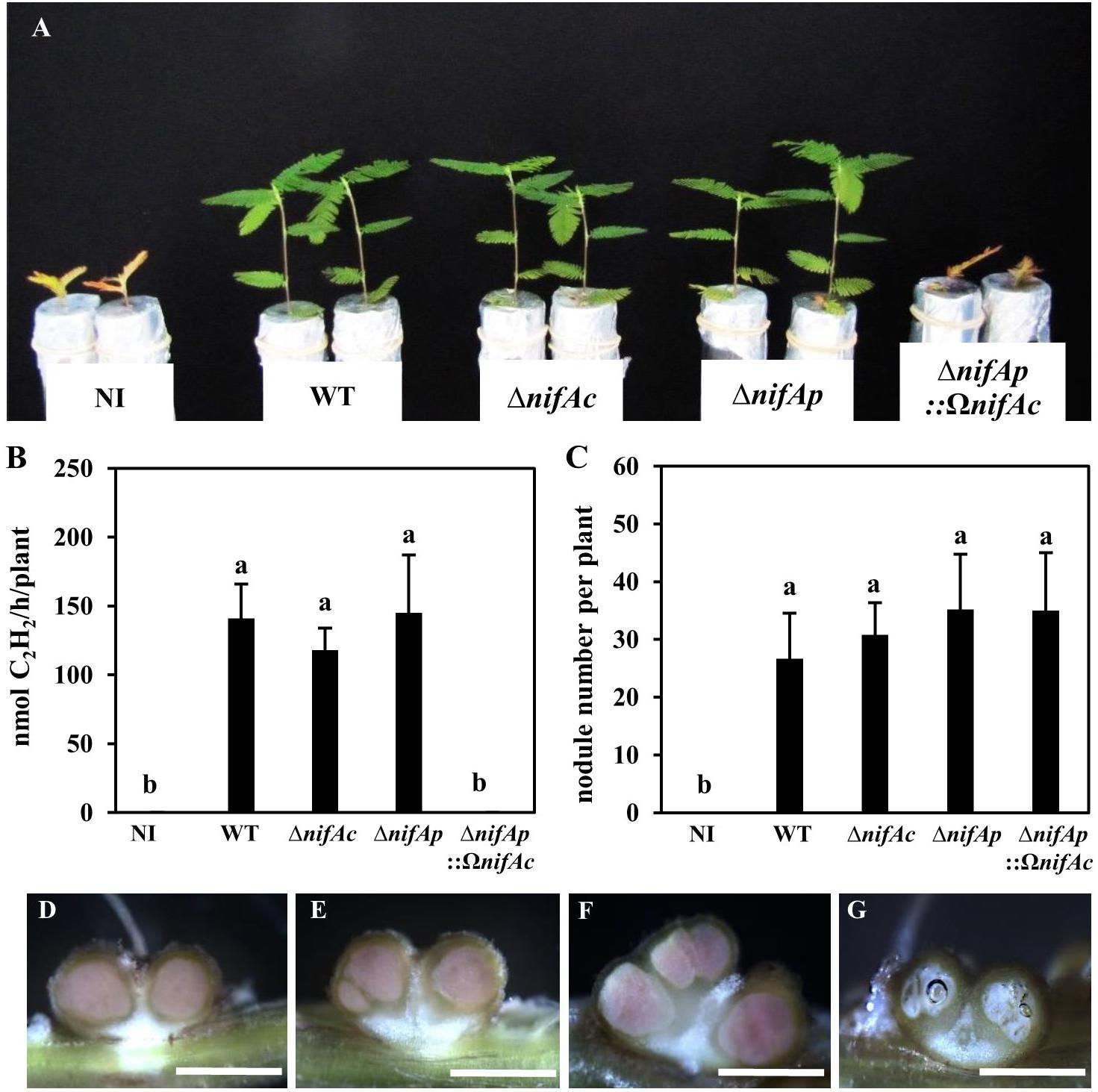
FIGURE 3. The two nifA genes in Bradyrhizobium sp. DOA9 strain are functionally redundant during symbiosis with Aeschynomene americana. (A) Comparison of plant growth (aerial part) non-inoculated (NI) or inoculated with WT and mutant strains DOA9ΔnifAc, DOA9ΔnifAp and DOA9ΔnifAp::ΩnifAcDOA9 (at 20 dpi). (B) Amount of acetylene-reducing activity (ARA) in A. americana plants inoculated with WT and mutant strains. (C) Number of nodules per plant inoculated by WT and nifA mutant strains (D–F) Transversal sections of nodules elicited by WT (D), DOA9ΔnifAc (E), DOA9ΔnifAp (F), and DOA9ΔnifAp::ΩnifAc (G) mutants. Scale bars are 1 mm for (D–G). In (B,C), error bars represent standard error (n = 10). Different letters above error bars indicate significant differences at P < 0.05 (Tukey’s HSD test).
To determine whether the two nifA genes were also exchangeable during free-living conditions, we analyzed the nitrogenase activity of the different constructed nifA mutants after 7 days of culture in semisolid BNM medium. The nifAc mutants including DOA9ΩnifAc and DOA9ΔnifAc were obviously unable to fix nitrogen in their free-living state (Table 1). In contrast, DOA9ΩnifAp and DOA9ΔnifAp mutants displayed nitrogenase activity similar to the WT strain. These data indicate that NifAc is essential for nitrogenase activity during the free-living condition, while NifAp does not play a significant role in this condition. As expected, it was found that the double nifA mutant was not able to fix nitrogen in the free-living state (Table 1).
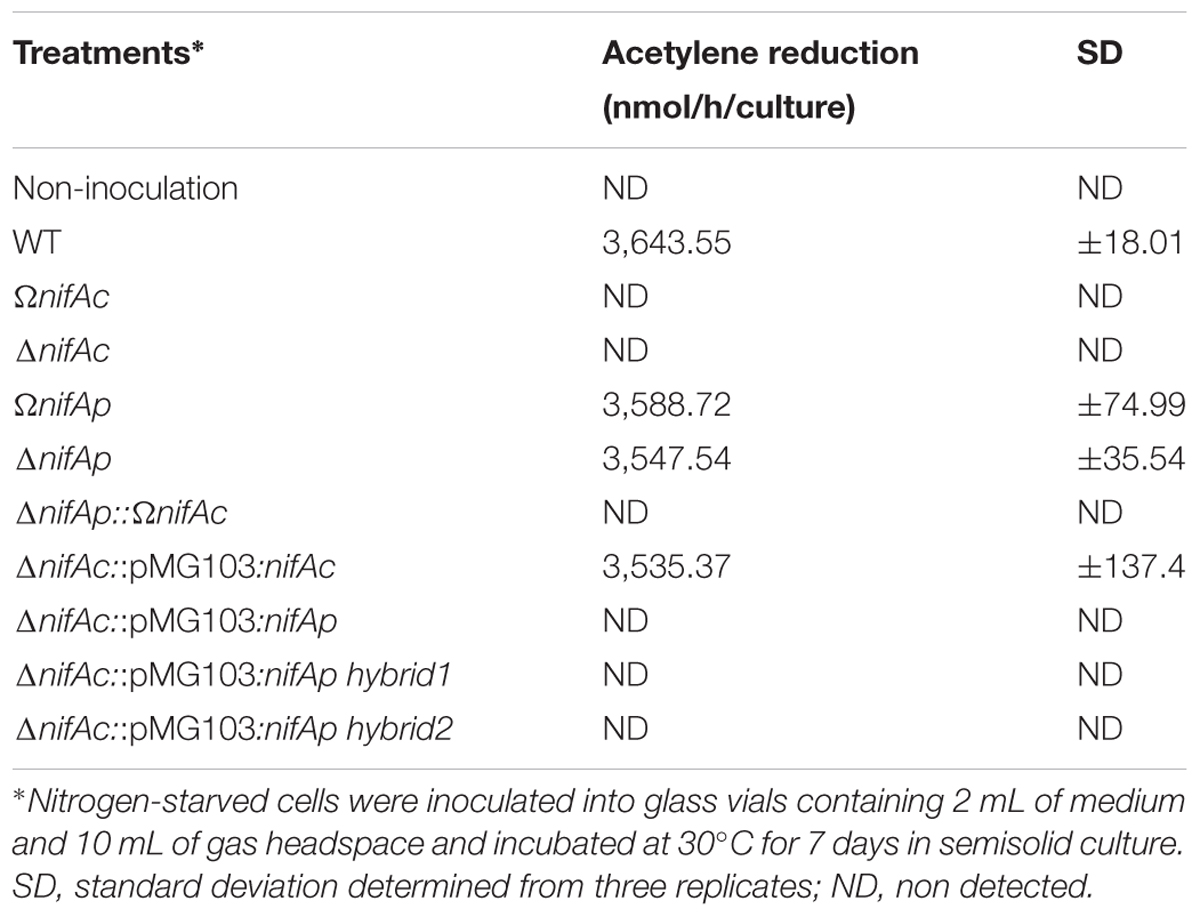
TABLE 1. Nitrogenase activity in Bradyrhizobium sp. DOA9 (WT) and nifA mutant strains grown under free-living conditions as described in Material and Methods.
Because NifAp lacks the N-terminal GAF domain, a simple hypothesis would be to postulate that NifAp protein is not active under free-living conditions, due to the absence of this functional domain. To check this hypothesis, we constructed two chimeric NifA hybrid proteins, one corresponding to the almost complete NifAp, to which has been added the first 70 AA of NifAc, the second corresponding to the sigma 54 interaction domain and HTH domain of NifAp (from the AA 150 to 503), to which was added the complete GAF domain of NifAc (the first 220 AA). These constructs were cloned into the pMG103 plasmid under the constitutive npt2 promoter and reintroduced into the DOA9ΔnifAc mutant (Supplementary Figure S2). As controls, we also reintroduced the complete nifAc or nifAp gene using the same plasmid and npt2 promoter. As shown in Table 1, only the reintroduction of the complete nifAc gene completely restored the nitrogenase activity of the DOA9ΔnifAc mutant. No gain of function was observed for all other constructs. This suggests that simple addition of the GAF domain is not sufficient to rebuild functional activity in NifAp protein under free-living conditions.
The analysis of nifA mutants on plants suggested that the two NifA proteins are functionally redundant, but considering that a functional redundancy of the nifHDK genes found on the chromosome and the plasmid was also reported (Wongdee et al., 2016), we cannot completely exclude the possibility that the absence of phenotype observed for the single nifA mutants results in fact to this last redundancy. In other words, we can ask whether each NifA protein activates only one specific set of nifHDK genes, or both sets found on the chromosome and the plasmid.
To answer this question, we used quantitative reverse transcription PCR (qRT-PCR) to analyze the level of expression of the structural nif genes (nifDKc, nifHc, nifDKp, nifHp) as well as controls, namely, the two regulatory nifA genes (nifAc, nifAp) in both the WT and the two ΔnifAc and ΔnifAp mutants. As shown in Figure 4A, during symbiosis, the expression profiles of all structural nif genes is well conserved in the WT and the two nifA mutants. This clearly demonstrates that in this condition, the two NifA proteins can activate the expression of the two nifHDK sets, definitively confirming their functional redundancy in this condition. In contrast, during free-living growth, we observed that the ΔnifAc mutant differed drastically from the WT and the ΔnifAp mutant (Figure 4B). Indeed, in the ΔnifAc mutant, no expression of nifDKc and p was detected and the expression levels of nifHc and p were also extremely low. These data, which were in concordance with the absence of nitrogenase activity detected in this mutant, confirms the essential role of NifAc protein in the activation of nif genes under free-living conditions. In addition, for this mutant, no expression of nifAp could be detected, indicating that NifAc also activates the expression of nifAp which is in accordance with the presence of putative NifA and RpoN binding sites found in the nifAp promoter region.
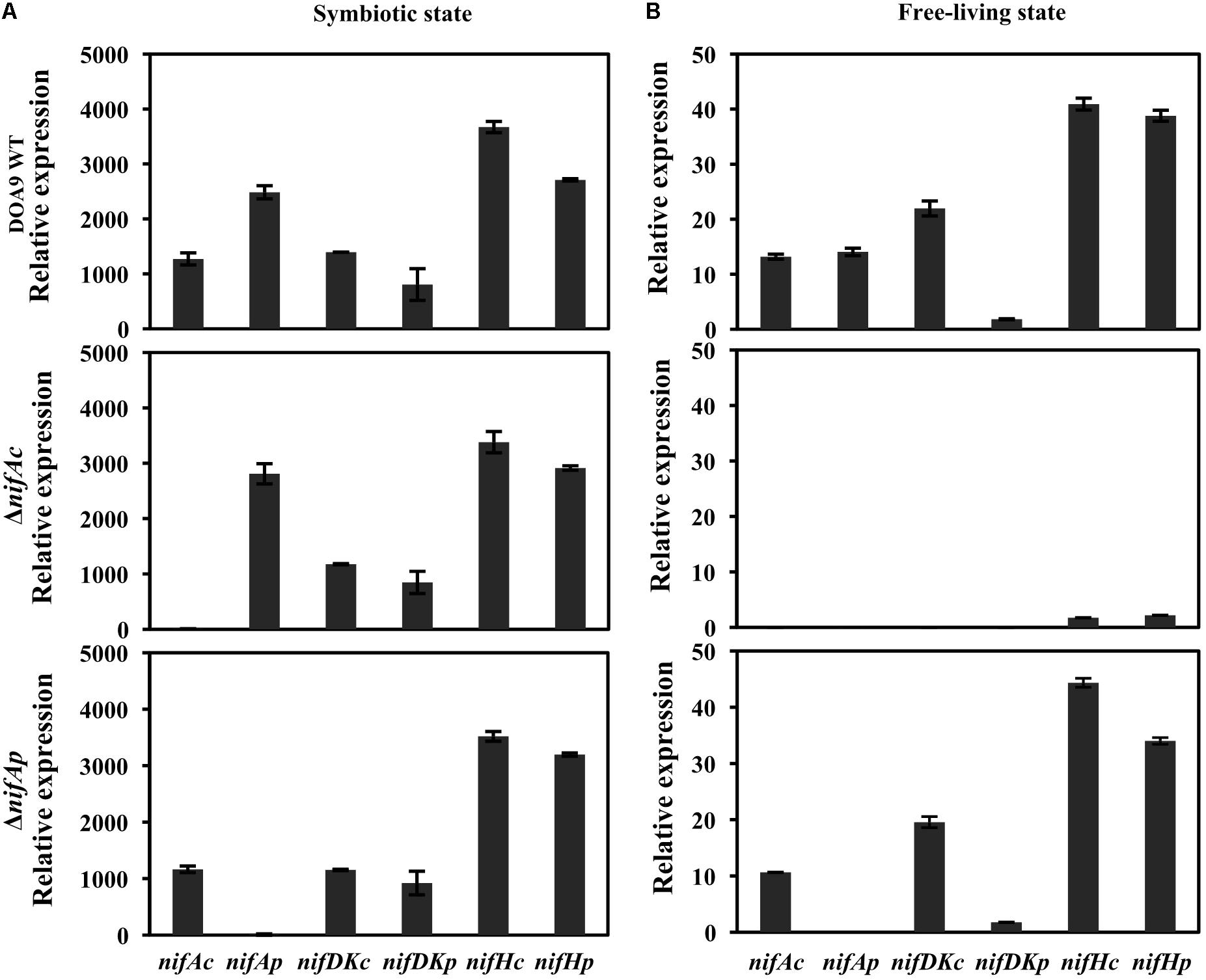
FIGURE 4. Expression of nitrogen-fixing genes (nifDKc, nifDKp, nifHc, and nifHp) and nifAc, nifAp genes in Bradyrhizobium sp. DOA9 (WT) and nifA mutant strains grown under symbiosis (A) or free-living conditions (B). Bacteroid cells were obtained from A. americana nodules at 20 days post-inoculation (A), and bacterial cells grown under free-living conditions were obtained after 7 days of culture in BNM-B medium without glutamate (B). Total RNA was extracted and subjected to quantitative reverse transcription-PCR (qRT-PCR) with an internal standard of dnaK. All data were from one representative experiment that was repeated three times. Error bars indicate standard deviation.
In contrast, the pattern of expression of the nif genes remains very similar between the WT strain and the ΔnifAp mutant during free-living conditions, confirming that NifAp is dispensable during this condition. Interestingly, in these culture conditions, nifDKc was expressed at a far higher level than nifDKp. These data are in agreement with our previous study, which showed that NifDKc was the major contributor to the bacterial nitrogenase activity under free-living conditions (Wongdee et al., 2016).
Taken together, these data confirm that the two NifA proteins are exchangeable in the activation of nif genes during symbiosis, but not during free-living conditions, where NifAc is essential.
In this study, we showed that the two nifA genes present in the Bradyrhizobium sp. DOA9 strain are expressed during both symbiotic and free-living growth and encoded functional proteins. In particular, we observed that these two NifA are perfectly exchangeable for the regulation of nitrogen fixation during symbiosis. These data were unexpected, given the moderate level of identity between these two proteins and the difference observed in their architectural organization (see Discussion below). Furthermore, in the only other example rhizobial strain reported to contain two nifA genes, Mesorhizobium loti, there was no functional redundancy observed between these two NifA proteins since the ΔnifA2 mutant gave a Fix- phenotype, while no symbiotic defect was observed for the ΔnifA1 mutant (Sullivan et al., 2001; Nukui et al., 2006). In fact, the regulatory role of NifA1 in this last bacterium is unclear, since the expression of NifA-regulated genes, i.e., those containing NifA and RpoN binding sites boxes in their promoter, were drastically impacted in the ΔnifA2 mutant but not in the ΔnifA1 background (Sullivan et al., 2013).
The origin of these two nifA genes in the DOA9 strain is puzzling. Their localization on different replicons and their moderate level of identity suggest that they were separately acquired, rather than via duplication of a single gene. In both cases, a Blast search using the amino acid sequence of NifAp or NifAc returned as best hits Bradyrhizobium NifA homologs, suggesting that both nifA genes derived from a common ancestor. However, while the percentage of identity of NifAc with the other bradyrhizobial NifA ranges between 91 to 76 %, this percentage drops to 55-50% for NifAp. In accordance, a phylogenetic analysis (Figure 1D) clearly showed that NifAp forms an outgroup from the bradyrhizobial NifA proteins. Furthermore, nifAp is found in an unusual genomic context, as no known nif or fix genes are found in the vicinity, in contrast to the other rhizobial nifA genes that were always found associated with genes involved in nitrogen fixation (Fischer, 1994). The presence of insertion sequence elements belonging to the IS3 family surrounding nifAp suggests the possibility that nifAp could have been separated from nif genes by a transposition event (Supplementary Text S1).
In all of the rhizobia in which nifA has been studied, it has been shown that NifA is absolutely required to activate nitrogen fixation during symbiosis (Szeto et al., 1984; Schetgens et al., 1985; Fischer et al., 1986; Iismaa and Watson, 1989). If the plant perceives that the nodules are ineffective, a sanctioning program is rapidly triggered (Westhoek et al., 2017), such as observed in this study for the double nifA mutant for which the nodules were senescing. Therefore, we can assume that NifA is essential for the symbiotic life of rhizobia and that there is a high selective pressure to maintain its functionality. DOA9 contains two nifA genes, and this likely relaxed this selective pressure and permitted a higher evolution rate of one homologous gene, i.e., nifAp. On the other hand, although nifAp has strongly diverged from the other nifA, NifAp remains functional, as it can activate both nifHDKc and p during symbiosis in the absence of NifAc. This suggests that maintaining two functional NifA proteins in DOA9 strain may be selectively advantageous even if there is an overlap in their regulatory function.
The major striking difference between NifAp and NifAc or the other bradyrhizobial NifA is the lack of an N-terminal GAF domain. There are several reports in various diazotrophic bacteria indicating that the GAF domain plays a key role in the modulation of NifA activity. For example, in Azotobacter vinelandii, the GAF domain binds 2-oxoglutarate, a key metabolic signal of carbon status, the presence of which influences the interaction with the antiactivator protein NifL (Martinez-Argudo et al., 2004). In the same vein, in Herbaspirillum seropedicae, NifA regulation by ammonium involves its N-terminal GAF domain and the signal transduction protein GlnK (Aquino et al., 2015). In contrast, to our knowledge, no functional role has been attributed to the N-terminal GAF domain of the rhizobial NifA proteins. It does not play an obvious role, since its deletion in the NifA of S. meliloti or B. diazoefficiens does not impair the ability of the protein to activate nif genes (Beynon et al., 1988; Fischer et al., 1988). Furthermore, it exists among rhizobia, one example (R. leguminosarum), for which NifA naturally lacks this GAF domain and which, despite this, maintains its essential role in the activation of nif genes during symbiosis indicating that this domain is dispensable, at least under this culture condition (Iismaa and Watson, 1989). Therefore, it is not so surprising that NifAp maintains a regulatory role during symbiosis, despite the lack of a N-terminal GAF domain. It is more surprising that NifAp is not functional during free-living growth. Our RT-qPCR analysis clearly showed that in free-living conditions, nifAp was expressed, which excludes the possibility that the lack of complementation of the ΔnifAc mutant is due to the absence of NifAp synthesis. It is possible that the GAF domain, which is dispensable during symbiosis, plays a more prominent role in NifA activity under in vitro conditions. We tested this hypothesis by constructing two chimeric NifA hybrid proteins containing the N-terminal part of NifAc and the C-terminal part of NifAp, but these constructs did not restore the free-living nitrogen fixing deficiency of the ΔnifAc mutant. Nevertheless, we cannot completely reject this hypothesis because it is possible that these hybrid NifA proteins did not have the correct conformation. Further studies at the protein level remain necessary to better understand the function and mode of action of both NifA homologs under free-living conditions.
The mechanism of regulations involving NifA in the DOA9 strain are certainly more complex than expected, considering that this strain also contains two rpoN homologous genes found on both the plasmid and chromosome. NifA activates nif gene expression and other genes by forming a complex with RpoN (Gong et al., 2007; Hauser et al., 2007). We can ask whether each NifA protein can form a complex with both RpoN proteins and depending on the NifA and RpoN composition, whether the activity of this complex and its affinity for a DNA binding motif differs. Intriguingly, while the promoter regions of nifDKc and nifHc contain a perfectly conserved NifA binding site (5′-TGT-N10-ACA-3′), only a nonconventional site differing by one nucleotide has been identified in the upstream regions of nifDKp, nifHp (Wongdee et al., 2016). Analysis of the expression of these nif genes suggests that these slight variations do not impact the ability of the two NifA proteins to activate the nifDK and nifH genes on both the chromosome and plasmid during symbiosis, since a similar level of expression of these different nif genes was observed for the 3 bacterial backgrounds tested (WT and the two nifA mutants). However, at the same time, under free-living growth, nifDKc was more highly expressed than nifDKp. Therefore, we cannot exclude the hypothesis that these slight variations in the upstream activating sequence differentially impact the affinity of NifA (at least for NifAc) according to the environmental conditions. Behind the control of nif genes, the NifA protein influences various cellular processes in rhizobia (Fischer et al., 1986; Gong et al., 2007; Hauser et al., 2007). Thus, it is attractive to speculate that the presence of multiple NifA and RpoN proteins in the DOA9 strain facilitates the switch from a free-living to a symbiotic lifestyle and vice-versa, allowing better control of the expression of various sets of genes.
JW, NT, NB, PT, and EG conceived the experiments. JW, NT, PT, and EG conducted the experiments. JW, NT, PT, and EG analyzed the results and wrote the paper. All authors reviewed the manuscript.
This work was supported by the ARTS and BEST programs from IRD-France (Fellowship respectively to JW and PT) and the Franco-Thai PHC Siam program (project SIAM N°29589XA). This work was also supported by Suranaree University of Technology and the Office of the Higher Education Commission under the NRU project of Thailand.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01644/full#supplementary-material
FIGURE S1 | The two nifA genes in Bradyrhizobium sp. DOA9 strain are functionally redundant during symbiosis with Aeschynomene americana. (A) Comparison of plant growth (aerial part) non-inoculated (NI) or inoculated with WT and insertion mutant strains DOA9ΩnifAc and DOA9ΩnifAp (at 20 dpi). (B–D) Root nodules observed with a fluorescent stereomicroscope equipped with a green fluorescent protein (GFP) filter. (B) Nodules elicited by WT; (C) Nodules elicited by DOA9ΩnifAc; (D) Nodules elicited by DOA9ΩnifAp. (E) Acetylene-reducing activity (ARA) in A. americana plants inoculated with WT and insertion mutant strains DOA9ΩnifAc and DOA9ΩnifAp. (F) Number of nodules per plant elicited by WT and DOA9ΩnifAc and DOA9ΩnifAp. (G-I) Cross section of nodule elicited by WT (G) and mutants DOA9ΩnifAc (H) and DOA9ΩnifAp (I). Scale bars are 250 μm for (D–G). In (E,F), error bars represent standard error (n = 10). Different letters above error bars indicate significant differences at P < 0.05 (Tukey’s HSD test).
FIGURE S2 | Bradyrhizobium sp. strain DOA9 strain displays two distinct nifA genes. (A) Sequence alignment of NifAc and NifAp. Arrows and boxes indicate different portions of NifAp and NifAc used to form chimeric NifA proteins (B). The color boxes indicate NifAc protein domains including GAF (blue), sigma factor 54 (δ54) interaction (purple), and HTH (green) domains. (B) Schematic representation of different versions of nifA introduced into plasmid pMG103-npt2-cefo under control of the constitutive nptll promoter. Each constructed plasmid was transferred into DOA9ΔnifAc cells for complementation experiments (see Table 1).
TABLE S1 | Primers used in this study.
TABLE S2 | Primers used in the qRT-PCR experiment.
TEXT S1 | Analysis of Insertion sequences in the nifAp surrounding region.
Aquino, B., Stefanello, A. A., Oliveira, M. A., Pedrosa, F. O., Souza, E. M., Monteiro, R. A., et al. (2015). Effect of point mutations on Herbaspirillum seropedicae NifA activity. Braz. J. Med. Biol. Res. 48, 683–690. doi: 10.1590/1414-431X20154522
Babst, M., Hennecke, H., and Fischer, H. M. (1996). Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol. Microbiol. 19, 827–839. doi: 10.1046/j.1365-2958.1996.438968.x
Bauer, E., Kaspar, T., Fischer, H. M., and Hennecke, H. (1998). Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR. J. Bacteriol. 180, 3853–3863.
Beynon, J. L., Williams, M. K., and Cannon, F. C. (1988). Expression and functional analysis of the Rhizobium meliloti nifA gene. EMBO J. 7, 7–14. doi: 10.1002/j.1460-2075.1988.tb02777.x
Bonaldi, K., Gherbi, H., Franche, C., Bastien, G., Fardoux, J., Barker, D., et al. (2010). The Nod factor-independent symbiotic signaling pathway: development of Agrobacterium rhizogenes-mediated transformation for the legume Aeschynomene indica. Mol. Plant Microbe Interact. 23, 1537–1544. doi: 10.1094/MPMI-06-10-0137
Burris, R. H., and Roberts, G. P. (1993). Biological nitrogen fixation. Annu. Rev. Nutr. 1993, 17–35. doi: 10.1146/annurev.nu.13.070193.001533
Ehrhardt, D. W., Atkinson, E. M., and Long, S. R. (1992). Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256, 998–1000. doi: 10.1126/science.10744524
Fischer, H. M. (1994). Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58, 352–386. s
Fischer, H. M., Alvarez-Morales, A., and Hennecke, H. (1986). The pleiotropic nature of symbiotic regulatory mutants: Bradyrhizobium japonicum nifA gene is involved in control of nif gene expression and formation of determinate symbiosis. EMBO J. 5, 1165–1173. doi: 10.1002/j.1460-2075.1986.tb04342.x
Fischer, H. M., Bruderer, T., and Hennecke, H. (1988). Essential and non-essential domains in the Bradyrhizobium japonicum NifA protein: identification of indispensible cysteine residues potentially involved in redox reactivity and/or metal binding. Nucleic Acids Res. 16, 2207–2224. doi: 10.1093/nar/16.5.2207
Gong, Z., Zhu, J., Yu, G., and Zou, H. (2007). Disruption of nifA gene influences multiple cellular processes in Sinorhizobium meliloti. J. Genet. Genomics 34, 783–789. doi: 10.1016/S1673-8527(07)60089-7
Hauser, F., Pessi, G., Friberg, M., Weber, C., Rusca, N., Lindemann, A., et al. (2007). Dissection of the Bradyrhizobium japonicum NifA-RpoN regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol. Genet. Genomics 278, 255–271. doi: 10.1007/s00438-007-0246-9
Ho, Y. S., Burden, L. M., and Hurley, J. H. (2000). Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19, 5288–5299. doi: 10.1093/emboj/20.6.1483
Iismaa, S. E., and Watson, J. M. (1989). The nifA gene product from Rhizobium leguminosarum biovar trifolii lacks the N-terminal domain found in other NifA proteins. Mol. Microbiol. 3, 943–955. doi: 10.1111/j.1365-2958.1989.tb00244.x
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS fusion system. Plant Mol. Biol. Rep. 5, 387–405. doi: 10.1007/BF02667740
Kullik, I., Hennecke, H., and Fischer, H. M. (1989). Inhibition of Bradyrhizobium japonicum nifA-dependent nif gene activation by oxygen occurs at the NifA protein level and is irreversible. Arch. Microbiol. 151, 191–197. doi: 10.1007/BF00413129
Martinez-Argudo, I., Little, R., and Dixon, R. (2004). Role of the amino-terminal GAF domain of the NifA activator in controlling the response to the antiactivator protein NifL. Mol. Microbiol. 52, 1731–1744. doi: 10.1111/j.1365-2958.2004.04089.x
Miller, J. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory
Noisangiam, R., Teamtisong, K., Tittabutr, P., Boonkerd, N., Toshiki, U., Minamisawa, K., et al. (2012). Genetic diversity, symbiotic evolution, and proposed infection process of Bradyrhizobium strains isolated from root nodules of Aeschynomene americana L. in Thailand. Appl. Environ. Microbiol. 78, 6236–6250. doi: 10.1128/AEM.00897-12
Nukui, N., Minamisawa, K., Ayabe, S., and Aoki, T. (2006). Expression of the 1-aminocyclopropane- 1-carboxylic acid deaminase gene requires symbiotic nitrogen-fixing regulator gene nifA2 in Mesorhizobium loti MAFF303099. Appl. Environ. Microbiol. 72, 4964–4969. doi: 10.1128/AEM.02745-05
Okazaki, S., Noisangiam, R., Okubo, T., Kaneko, T., Oshima, K., Hattori, M., et al. (2015). Genome analysis of a novel Bradyrhizobium sp. DOA9 carrying a symbiotic plasmid. PLoS One. 10:e0117392. doi: 10.1371/journal.pone.0117392
Okazaki, S., Tittabutr, P., Teulet, A., Thouin, J., Fardoux, J., Chaintreuil, C., et al. (2016). Rhizobium-legume symbiosis in the absence of Nod factors: two possible scenarios with or without the T3SS. ISME J. 10, 64–74. doi: 10.1038/ismej.2015.103
Oke, V., and Long, S. R. (1999). Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32, 837–849. doi: 10.1046/j.1365-2958.1999.01402.x
Perrière, G., and Gouy, M. (1996). WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78, 364–369. doi: 10.1016/0300-9084(96)84768-7
Renier, A., Maillet, F., Fardoux, J., Poinsot, V., Giraud, E., and Nouwen, N. (2011). Photosynthetic Bradyrhizobium sp. strain ORS285 synthesizes 2-O-methyl-fucosylated lipochitooligo-saccharides for nod gene-dependent interaction with Aeschynomene plants. Mol. Plant Microbe Interact. 24, 1440–1447. doi: 10.1094/MPMI-05-11-0104
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Schetgens, R. M. P., Hontely, J. G. J., van den Bos, R. C., and van Kammen, A. (1985). Identification and phenotypical characterization of a cluster of fix genes, including a nif regulatory gene, from Rhizobium leguminosarum PRE. Mol. Gen. Genet. 200, 368–374. doi: 10.1007/BF00425719
Souza, E. M., Pedrosa, F. O., Drummond, M., Rigo, L. U., and Yates, M. G. (1999). Control of Herbaspirillum seropedicae NifA activity by ammonium ions and oxygen. J. Bacteriol. 181, 681–684.
Steenhoudt, O., and Vanderleyden, J. (2000). Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 24, 487–506. doi: 10.1111/j.1574-6976.2000.tb00552.x
Sullivan, J. T., Brown, S. D., and Ronson, C. W. (2013). The NifA-RpoN regulon of Mesorhizobium loti strain R7A and its symbiotic activation by a novel LacI/GalR-family regulator. PLoS One 8:e53762. doi: 10.1371/journal.pone.0053762
Sullivan, J. T., Brown, S. D., Yocum, R. R., and Ronson, C. W. (2001). The bio operon on the acquired symbiosis island of Mesorhizobium sp. strain R7A includes a novel gene involved in pimeloyl-CoA synthesis. Microbiol. 147, 1315–1322. doi: 10.1099/00221287-147-5-1315
Szeto, W. W., Zimmerman, J. L., Sundaresan, V., and Ausubel, F. M. (1984). A Rhizobium meliloti symbiotic regulatory gene. Cell 36, 1035–1043. doi: 10.1016/0092-8674(84)90053-9
Takahashi, Y., and Tokumoto, U. (2002). A third bacterial system for the assembly of iron-sulfurclusters with homologs in archaea and plastids. J. Biol. Chem. 277, 28380–28383. doi: 10.1074/jbc.C200365200
Thöny, B., Fischer, H. M., Anthamatten, D., Bruderer, T., and Hennecke, H. (1987). The symbiotic nitrogen fixation regulatory operon (fixRnifA) of Bradyrhizobium japonicum is expressed aerobically and is subject to a novel, nifA-independent type of activation. Nucleic Acids Res. 15, 8479–8499. doi: 10.1093/nar/15.20.8479
Tsai, J. W., and Alley, M. R. (2000). Proteolysis of the McpA chemoreceptor does not require the Caulobacter major chemotaxis operon. J. Bacteriol. 182, 504–507. doi: 10.1128/JB.182.2.504-507.2000
Vincent, J. M. (1970). A manual for the practical study of root-nodule bacteria. Handbook No. 15. Hoboken, NJ: Blackwell Scientific Publications.
Westhoek, A., Field, E., Rehling, F., Mulley, G., Webb, I., Poole, P. S., et al. (2017). Policing the legume-Rhizobium symbiosis: a critical test of partner choice. Sci. Rep. 7:1419. doi: 10.1038/s41598-017-01634-2
Wongdee, J., Songwattana, P., Nouwen, N., Noisangiam, R., Fardoux, J., Chaintreuil, C., et al. (2016). nifDK clusters located on the chromosome and megaplasmid of Bradyrhizobium sp. strain DOA9 contribute differently to nitrogenase activity during symbiosis and free-living growth. Mol. Plant Microbe Interact. 29, 767–773. doi: 10.1094/MPMI-07-16-0140-R
Keywords: NifA, Bradyrhizobium, symbiosis, legume, nitrogen, nitrogenase, Rhizobium
Citation: Wongdee J, Boonkerd N, Teaumroong N, Tittabutr P and Giraud E (2018) Regulation of Nitrogen Fixation in Bradyrhizobium sp. Strain DOA9 Involves Two Distinct NifA Regulatory Proteins That Are Functionally Redundant During Symbiosis but Not During Free-Living Growth. Front. Microbiol. 9:1644. doi: 10.3389/fmicb.2018.01644
Received: 18 May 2018; Accepted: 02 July 2018;
Published: 24 July 2018.
Edited by:
Kiwamu Minamisawa, Tohoku University, JapanReviewed by:
Christian Staehelin, Sun Yat-sen University, ChinaCopyright © 2018 Wongdee, Boonkerd, Teaumroong, Tittabutr and Giraud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Panlada Tittabutr, cGFubGFkYUBzdXQuYWMudGg= Eric Giraud, ZXJpYy5naXJhdWRAaXJkLmZy
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.