
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 July 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01508
This article is part of the Research Topic Antimicrobial Resistance and the Food Chain: A One Health Perspective View all 14 articles
The objective of this study was to test the prevalence of virulence-associated markers and antimicrobial resistance in 624 C. jejuni isolated from poultry food chain, i. e., chicken feces (n = 160), poultry carcasses (n = 157), poultry meat (n = 152) and from humans (n = 155). All human strains were positive for 9 out of 13 putative virulence genes responsible for expression of pathogenic factors involved in different stages of the infection. The presence of all markers was also high in strains from chicken feces, carcasses and meat although not all of them were identified in 100% of the isolates. On the other hand, the virB11, wlaN, and iam putative pathogenic genes were detected in only 1.9, 15.2, and 20.5% of strains, respectively. C. jejuni isolates, irrespective of the origin, were highly resistant to ciprofloxacin (92.5% isolates), followed by nalidixic acid (88.9%) and tetracycline (68.4%). In case of ciprofloxacin, significantly more isolates from poultry feces, carcasses and meat were resistant than those obtained from humans and the same relationship was observed for tetracycline where the isolates from chicken feces were more often resistant than C. jejuni of carcasses and meat origin. A low number of strains was resistant to streptomycin (18.4% isolates) and only 5 strains (0.8%) displayed resistance to erythromycin. A relationship between resistance to fluoroquinolones and presence of selected pathogenic markers was observed, e.g., from 83.3% strains with the virB11 to 93.4% with the docA genes were resistant to ciprofloxacin. The isolates that did not possess any of the pathogenic traits were also mainly resistant to this antimicrobial, although the number of such strains was usually low, except virB11 (612 isolates), wlaN (529 strains), and iam (496 isolates). Furthermore, resistance to tetracycline was somehow associated with the presence of the virulence associated genes wlaN and virB11 (56.8 and 75.0% isolates, respectively). The present study shows a high antimicrobial resistance to quinolones and tetracycline of C. jejuni isolated along poultry food chain and from patients with diarrhea, which was closely correlated with the presence of several virulence genes playing a role in the pathogenesis of Campylobacter infection.
Campylobacter, mainly Campylobacter jejuni, is one of the most common causes of foodborne bacterial infections worldwide (Allos, 2001; Bolton, 2015; Kaakoush et al., 2015; Tresse et al., 2017). Campylobacteriosis is also the most commonly reported zoonosis in the European Union (EU) with 246,307 confirmed cases and the notification rate 66.3 per 100,000 population in 2016 [EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2017]. The majority of the infections (83.6%) was caused by C. jejuni [EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2017]. The main transmission route of Campylobacter to humans is handling, preparation and consumption of contaminated food, especially of poultry origin (Allos, 2001; Park, 2002; Humphrey et al., 2007; Tresse et al., 2017). C. jejuni does not cause clinical diseases in poultry, but poultry carcasses have been frequently contaminated in the slaughterhouse due to a high prevalence of these bacteria in the intestinal tract of chickens; therefore, poultry carcasses can serve as the source of these microorganisms to humans (Humphrey et al., 2007; van Gerwe et al., 2010). Although mortality in humans is low, morbidity due to post-infectious sequelae such as Guillain-Barré syndrome, reactive arthritis and irritable bowel syndrome is significant (Allos, 2001; Humphrey et al., 2007; Iovine, 2013; Tresse et al., 2017).
The pathogenesis of Campylobacter infection is complex and still poorly understood. However, it is believed that the expression of genes involved in motility, colonization, epithelial cell invasion, and toxin production play an important role in the disease development (Humphrey et al., 2007; Dasti et al., 2010). Mobility of the bacterial cells, involving the coordination of several genes (i.e., flaA and flhA), is essential for passage through the stomach and gut environment (Park, 2002) where Campylobacter produces several cell-surface proteins (encoded by the cadF, docA, racR, virB11, ciaB, and iam genes) that promote to adhere to and invade intestinal epithelial cells (Konkel et al., 1997; Carvalho et al., 2001; Dasti et al., 2010). The bacteria can also excrete several cytotoxins (encoded by the cdtA, cdtB, cdtC, wlaN genes) that contribute to the development of the disease (Hickey et al., 2000; Tresse et al., 2017). Furthermore, C. jejuni is able to produce superoxide dismutase enzyme (encoded by the sodB marker), which catalyzes the breakdown of superoxide radicals and it is one of the bacterial major defense mechanisms against oxidative damage (Pesci et al., 1994).
There has been an increasing trend of antimicrobial resistance in Campylobacter isolated within the food chain and humans in recent years (Melero et al., 2012; Piccirillo et al., 2013; Wieczorek et al., 2013b; Ma et al., 2014; Abdollahpour et al., 2015). Macrolides (i.e., erythromycin) and fluoroquinolones (i.e., ciprofloxacin) are considered as the first- and second-choice of antimicrobials, respectively for the treatment of human Campylobacter infections (Ge et al., 2013; Iovine, 2013). Most campylobacteriosis cases are usually self-limiting and do not require hospitalization and antimicrobial treatment. However, the therapy is required in children with fever, increasing bloody diarrhea, and in eldery or immunocompromised patients with severe and prolonged systemic disorders (Allos, 2001; Ge et al., 2013; Tresse et al., 2017). In these cases, macrolides (mostly erythromycin and azithromycin) are usually the first-choice antibiotics whereas fluoroquinolones and, to a less extent, tetracycline are alternative options (Allos, 2001; Iovine, 2013). A significant use of antimicrobials in animals and in humans has leaded to an increase in antibiotic-resistant Campylobacter population (Humphrey et al., 2007; Ge et al., 2013). Thus, monitoring of C. jejuni resistance is highly relevant to public health.
The objective of this study was to investigate the prevalence of virulence-associated genes and antimicrobial resistance in C. jejuni isolated from poultry food chain and from humans.
Sampling and Campylobacter isolation from chickens was performed in years 2014 and 2016 using the procedure as described earlier (Wieczorek et al., 2013b). Briefly, intact ceca from 10 birds were taken after evisceration, the content was pooled and one loop-full (10 μl) of the material was streaked directly on Karmali agar (Campylobacter Agar Base + Campylobacter Supplement; Oxoid, UK) and Campylobacter blood-free agar (Oxoid) with CCDA selective supplement (Oxoid) and incubated at 41.5°C ± 1°C for at least 48 h ± 2 h in a microaerobic atmosphere generated using CampyGen kit (Oxoid). Campylobacter from poultry feces was isolated by direct plating on two agar plates (Karmali and Campylobacter blood-free) as described above. After incubation, the plates were examined for morphologically typical Campylobacter colonies (grayish, often with a metallic sheen, flat and moist with a tendency to spread) and colony identity was confirmed by microscopic examination of morphology and motility, microaerobic growth at 25°C, and the presence of oxidase. From each fecal sample one presumptive Campylobacter isolate was then confirmed by PCR as described previously (Wieczorek et al., 2013b). A total of 160 confirmed C. jejuni isolates were used for the present analyzis.
The swab samples were collected directly after immersion chilling (0–4°C) but before further processing from the neck skin and the skin surface under the wings of the broiler carcasses and immediately transported to the laboratory in Amies transport medium with charcoal (Medlab, Poland). Campylobacter bacteria were isolated as described (Wieczorek et al., 2013a). Briefly, the swabs were placed in 5 ml of Bolton enrichment broth (Oxoid) supplemented with vancomycin, cefoperazone, trimethoprim, and amphotericin B and incubated as above. The cultures were then plated onto Kamali agar (Oxoid) and Campylobacter blood-free agar with CCDA selective supplement (Oxoid) and incubated at 41.5°C ± 1°C for 48 h ± 2 h under microaerobic conditions. From each sample one presumptive Campylobacter isolate was confirmed using PCR as described previously (Wieczorek et al., 2013b). During 5 years (2012–2016) a total of 157 confirmed C. jejuni were collected for the current investigation.
The Campylobacter strains from chicken meat (n = 152) were recovered in years 2010–2012 and 2015–2016 using the ISO 10272-1 standard and C. jejuni isolates were confirmed with the PCR method as described for the broiler carcasses.
A total of 155 C. jejuni isolates were obtained during the period of 2011–2016 from patients with diarrhea using standard culturing techniques. Rectal swabs were directly streaked onto mCCDA agar (Oxoid) and incubated at 41.5°C ± 1°C for 48 h ± 2 h under microaerobic conditions to grow only thermophilic campylobacters. Then, typical Campylobacter colonies were selected for further investigation using standard biochemical tests C. jejuni was identified with PCR as described previously (Vandamme et al., 1997).
Altogether, 624 C. jejuni were isolated and stored at −80°C until further analyzes.
Campylobacter isolates were tested for the presence of the following virulence genes: flaA and flhA (involved in motility), cadF, docA, racR, virB11 (responsible for adhesion and colonization) cdtA, cdtB, cdtC, wlaN (cytotoxin production). Additionally, the gene markers such as ciaB and iam responsible for the invasiveness of Campylobacter, and sodB (stress response) were also amplified. The PCR primer sequences and annealing temperatures are shown in Table S1.
A microbroth dilution method was used to establish the minimum inhibitory concentrations (MICs) of C. jejuni isolates to 6 antimicrobials (gentamicin, streptomycin, erythromycin, ciprofloxacin, nalidixic acid, and tetracycline) using the Sensititre® custom susceptibility plates, EUCAMP (Trek Diagnostics, UK). The strains were sub-cultured twice on Columbia agar (Oxoid) at 41.5°C for 48 h under microaerobic conditions. The minimum inhibitory concentration of the antimicrobial agents was determined using Mueller-Hinton Broth (Oxoid) supplemented with 2–2.5% horse blood (Trek). The plates were incubated at 37°C for 48 h under microaerophilic conditions and read using the Vision® system (Trek). The obtained results were determined according to the established breakpoints (Table S2). The antimicrobials and cut off values used for the interpretation of the MIC results were in accordance with EUCAST (Sifr et al., 2015) and the European Union Reference Laboratory for Antimicrobial Resistance.
The chi-square test with Yates' correction was used to examine differences in prevalence of virulence marker genes and antimicrobial resistance of C. jejuni isolated from different sources as well as to identify associations between antimicrobial resistance and presence of virulence marker genes. P < 0.05 was considered as significant. Confidence intervals (CIs) with 95% of confidence level in resistant strains were also calculated.
Overall, the presence of virulence marker genes among analyzed C. jejuni isolates was high, especially for the genes associated with motility, adhesion and colonization, (except the virB11 marker), cytotoxin production (except wlaN), and invasiveness (except iam) of the bacteria (Table 1). In case of human isolates, all of them were positive for 9 out of 13 putative virulence gene markers tested, i.e., flaA, cadF, docA, racR, cdtA, cdtB, cdtC, ciaB, and sodB. The vast majority of such strains (99.4%) were also positive for the flhA gene. The presence of all these markers was also high in strains from chicken feces, carcasses and meat although not all of them were identified in 100% of the isolates (Table 1). On the other hand, a low prevalence of the putative pathogenic marker genes was associated with the virB11 which was detected in only 12 (1.9%) of the total number of strains. This gene was not identified among C. jejuni of poultry meat origin and in only one isolate recovered from poultry feces. Furthermore, the wlaN and iam genes were found in 15.2 and 20.5% of all strains tested, respectively (Table 1).

Table 1. Distribution of virulence associated and environmental resistance genes in C. jejuni tested.
The results of antimicrobial resistance of the C. jejuni isolates are shown in Table 2. Overall, most of the strains were resistant to ciprofloxacin (total 577; 92.5% isolates), nalidixic acid (555; 88.9%) and, in a less extent, to tetracycline (427; 68.4%). There were statistical differences in the resistance rates between strains recovered from poultry chain and humans. In case of CIP significantly more isolates from poultry feces, carcasses and meat were resistant than those obtained from human patients (P < 0.005, P < 0.05, and P < 0.005, respectively). The same relationship was observed for tetracycline where the isolates from chicken feces were more often resistant than C. jejuni of carcasses and meat origin (P < 0.05 and P < 0.0001, respectively). A low number of isolates, irrespective of the origin, was resistant to streptomycin (115; 18.4% isolates) and higher resistance rates was observed among strains from chicken feces compared to the isolates from meat (P < 0.001) and humans (P < 0.0001). It was also found that only 5 of 624 strains (0.8%) displayed resistance to erythromycin (Table 2).
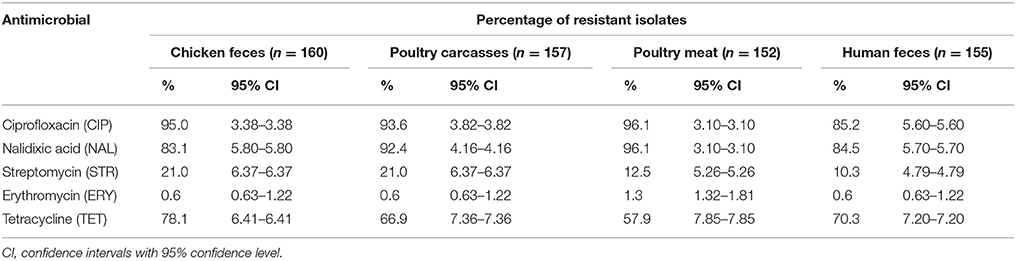
Table 2. Antimicrobial resistance of C. jejuni isolated from different sources. Values are in % ± 95% CI.
The MICs distribution of all 624 C. jejuni isolates tested is shown in Table 3. Among strains resistant to ciprofloxacin (total 577 isolates), several demonstrated a high resistance rates showing the MIC values ≥16 mg/L (229 strains; 39.7%). The majority of such isolates was recovered from chicken feces (84 out of 152; 55.3%) whereas only 38 of 132 (28.8%) highly resistant strains were isolated from humans (Table 3). A very high resistance rate was observed for tetracycline (total 427 isolates) where 395 (92.5%) displayed the MIC values ≥64 mg/L. These highly resistant strains were recovered from all sources, i.e., poultry ceca (94.4% isolates), carcasses (98.1%), meat (89.8%), and humans (96.3%). Almost all isolates resistant to streptomycin (n = 115 strains) showed a high resistance pattern manifested with MICs ≥16 mg/l (total 114; 99.1% strains). The majority of these C. jejuni (93; 81.6% isolates) had MIC above 16 mg/L and they were recovered from all but poultry meat sources (Table 3). A very few strains were resistant to erythromycin (a total of 5 isolates); however, 4 of them demonstrated a high MIC values (≥128 mg/L). These strains were only isolated along poultry meat chain (Table 3).
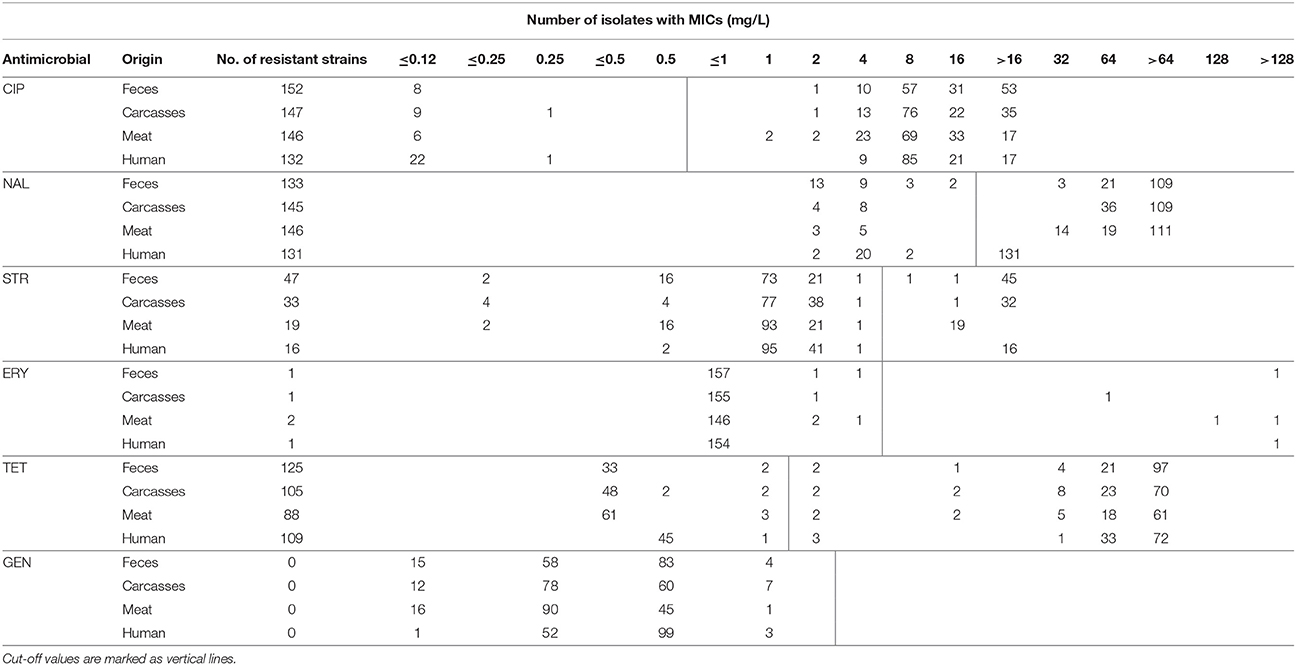
Table 3. Antimicrobial resistance and distribution of MICs among C. jejuni isolated from different origins.
Table 4 shows the prevalence of each virulence gene among all 624 C. jejuni isolates that were either resistant or sensitive to ciprofloxacin, nalidixic acid, streptomycin or tetracycline. The vast majority of strains resistant to CIP or NAL were positive for virulence markers tested, between 83.3% with the virB11 gene (only 12 such isolates identified) to 93.4% with the docA marker. On the other hand, the isolates that did not possess any of the pathogenic genes were also mainly resistant to these two antimicrobials, although the number of such strains was usually low, except virB11 (612 isolates), wlaN (529 strains), and iam (496 isolates). No statistical differences were identified among isolates resistant to ciprofloxacin and in C. jejuni resistant to NAL such differences were detected in strains positive and negative for the cdt toxin genes.
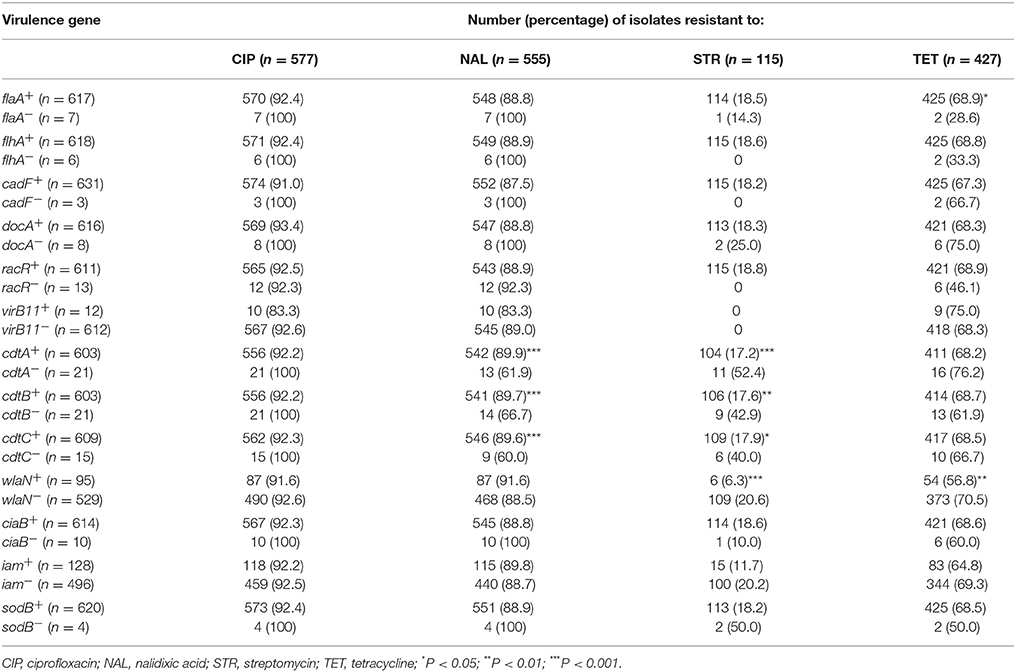
Table 4. Relationship between virulence genes and antimicrobial resistance patterns in all C. jejuni tested.
C. jejuni resistant to tetracycline were also associated with many of the virulence genes identified although such correlation was not as strongly expressed as for CIP and NAL. The percentage of strains resistant to TET and positive for the pathogenic markers was from 75.0% (virB11 gene; only 12 positive isolates) and 68.9% (flaA and racR genes) to 56.8% (wlaN marker). Statistically significant differences were only observed between the isolates with/without the flaA and wlaN markers (Table 4).
The present study provides the results on analysis of the prevalence putative gene markers and antimicrobial resistance among C. jejuni isolates along poultry food chain and humans with diarrhea. The genes associated with bacterial motility (flaA and flhA) and adhesion to epithelial cells (cadF, docA, and racR), which are the key mechanisms in the development of Campylobacter infection, were identified in most or even all isolates from the analyzed sources, especially from persons suffering from campylobacteriosis (Allos, 2001; Humphrey et al., 2007; Tresse et al., 2017). These findings provide further evidence that flagellar and adhesion genes are highly conserved among C. jejuni as previously suggested by several authors (Datta et al., 2003; Müller et al., 2006; Thakur et al., 2010; Koolman et al., 2015; Lapierre et al., 2016). Only few isolates (1.9% in total) were positive for the virB11 gene encoding a putative type IV secretion system involved in adherence of campylobacters to the gut epithelial cells (Bacon et al., 2000). Most of the virB11-positive isolates were identified in the current investigation among human C. jejuni which may suggest the role of this marker in pathogenesis of the diarrhea. There are also information that this gene is more often absent in human isolates and therefore may not be involved in virulence and pathogenesis of campylobacteriosis (Datta et al., 2003; Müller et al., 2006; Talukder et al., 2008).
Several strains were negative for the wlaN gene responsible for the production of β-1,3 galactosyltransferase involved in cell wall synthesis (only 15.2% positive isolates) but the absence of this marker has been previously observed (Datta et al., 2003; Talukder et al., 2008; Koolman et al., 2015). On the other hand, Kim et al. (2016) identified the wlaN gene among 100% of 63 human and in 78.6% of 42 animal C. jejuni isolated tested in Korea. The product of the wlaN gene shows ganglioside mimicking structures and thus may be involved in developing of Guillain–Barre' syndrome after C. jejuni infection (Thakur et al., 2010; Kim et al., 2016; Lapierre et al., 2016).
Other often prevalent virulence marker determinants included cdtA, cdtB, and cdtC cytotoxin genes which cause an important role in diarrhea by interfering with the division and differentiation of the intestinal crypt cells (mean prevalence of 96.6–97.6% positive strains). As it has been shown in previous investigations all three subunits are required for full toxin activity (Park, 2002; Lapierre et al., 2016). Interestingly, all 155 human C. jejuni isolates tested were positive for three cytotoxin subunit genes. However, some strains of poultry origin were negative for one or two subunit determinants which may suggest that they were not able to express the entire product or the toxin genes were not identified with the primers used in the study due to e.g., point mutations in the coding region (Bang et al., 2004).
Other genes involved in stress response and invasiveness, which are important for Campylobacter survival in the intestinal tract (sodB and ciaB), were in a high prevalence among all strains analyzed in the current study (99.4 and 98.4% of the isolates; including all positive C. jejuni of human origin). The product of the ciaB marker, which play a role both in the invasiveness and in colonization of the epithelial cells, was identified in campylobacters by other authors either in a lower percentage (Ziprin et al., 2001; Hanning et al., 2010) or in similar one to the present study (Raeisi et al., 2017). Since the ciaB and sodB genes are important in the initial stages of colonization, the high prevalence of these markers in C. jejuni currently tested, especially among strains isolated from humans with diarrhea, may suggest that these bacteria were able to overcome the stress conditions during passage through the intestinal tract and then induce the disease.
C. jejuni isolates tested, regardless the origin, were most frequently resistant to quinolones (ciprofloxacin and nalidixic acid; 92.5 and 88.9% of total isolates, respectively). A total of 39.7 and 76.0% of ciprofloxacin- and nalidixic acid-resistant C. jejuni investigated had the MIC values ≥16 mg/l and ≥32 mg/l, respectively. The cause of such high resistance to quinolones of strains isolated along chicken-production chain could be related to a broad use of enrofloxacin in veterinary medicine, especially in poultry production, and are thought to play a role in the spread of resistance to human isolates (Griggs et al., 2005; Iovine, 2013). A high rate of resistance to quinolones has also been reported previously in Poland, both among isolates of poultry and human origins (Wardak et al., 2007; Wieczorek et al., 2013b; Andrzejewska et al., 2015; Szczepanska et al., 2017; Wozniak-Biel et al., 2018). Data described in the recent EFSA/ECDC antimicrobial resistance report for 2016 have shown that C. jejuni isolated from humans were in average in 54.6% resistant to ciprofloxacin (information from 17 countries; no data from Poland) [EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2018]. Even higher resistance rate to ciprofloxacin was noted for C. jejuni isolated from broiler meat (mean value 64.9% strains; data from only 6 EU Member States) but Poland has not provided such information. On the other hand, the vast majority of the isolates from broilers displayed resistance to ciprofloxacin (93.2%) which was much higher than the EU mean rate (66.9%) [EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2018]. Such high percentage of C. jejuni resistant to quinolones in Poland may be due to a broad use of these antimicrobials in animal husbandry. According to the recent European Medicines Agency report, in Poland from 2011 to 2015 an increase was observed in sales (in mg for population correction unit, PCU) of fluoroquinolones used in veterinary medicine (EMA, 2017). In 2011, the proportion of total sales for fluoroquinolones was 5.7% whereas this figure in 2015 was 6.2%. In 2015, the sales of fluoroquinolones were 8.56 mg/PCU, while average value for 25 European countries described in the report in that year were 2.75 mg/PCU. Fluoroquinolones are considered by the World Health Organization as critical drugs for the treatment of humans, therefore investigation of Campylobacter resistance to these antimicrobials in food-producing animals is important for the public health. A high resistance to ciprofloxacin has been reported among human C. jejuni isolates in Korea (96.8%), China (93.1%), Qatar (63.2%), Estonia (67.9%), international travelers (from 50.8% in Africa to 75.0% in Asia), and the United Arab Emirates (85.4%) (Sonnevend et al., 2006; Unicomb et al., 2006; Kim et al., 2016; Zhou et al., 2016; Post et al., 2017). Strains from other countries have shown lower rates of resistance, e.g., 30.5% in Canada, 8.4% in Finland, 2% in Australia, and between 0 and 9% in Sweden (Osterlund et al., 2003; Ghunaim et al., 2015; Riley et al., 2015; Olkkola et al., 2016). It has been considered that the absence or low prevalence of C. jejuni ciprofloxacin-resistant isolates in some countries has been attributed previously to restricting the use of fluoroquinolones in food-producing animals (Osterlund et al., 2003; Griggs et al., 2005; Unicomb et al., 2006).
Many isolates displayed resistance to tetracycline (68.4%), especially C. jejuni recovered from chicken feces (78.1%) but also from humans with diarrhea (70.3%). It may suggest that poultry can serve as important reservoir of such strains for humans. Recent studies also indicated that C. jejuni of poultry sources were often resistant to tetracycline at the rates from 32.3% of the isolates in Chile (Lapierre et al., 2016), 75.5% in Iran (Raeisi et al., 2017), 79.4% in China (Han et al., 2016) up to 83.5% in the USA (Ladely et al., 2017). Investigations performed earlier in Poland showed different resistance levels to this antimicrobial, ranging from 9.0% (Rozynek et al., 2008), 31.1% (Wieczorek et al., 2015), 42.3% (Andrzejewska et al., 2015), 46.5% (Wieczorek and Osek, 2015), 51.1% (Szczepanska et al., 2017) up to even 100% of C. jejuni tested (Wozniak-Biel et al., 2018). Human C. jejuni isolates resistant to tetracycline were also identified during studies in several countries where the percentage of positive strains was from 2.1% (Olkkola et al., 2016), 24.3% (Lapierre et al., 2016), 42.9% (Mäesaar et al., 2016), 48.3% (Post et al., 2017), 64.4% (Riley et al., 2015) to 74.6% (Kim et al., 2016). Furthermore, such resistant bacteria were also previously detected in Poland among isolates from patients with diarrhea at the constantly increasing level still lower that identified in the current investigation, i.e., 13.7% (Wardak et al., 2007), 17.5% (Rozynek et al., 2009), 39.1% (Szczepanska et al., 2017) and up to 40% (Wardak and Szych, 2010). Recent results from EFSA/ECDC report show that 42.8% of C. jejuni from humans isolated in the European Union were resistant to tetracycline (lack information from Poland) [EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2018]. Similar resistance levels were observed for the isolates from broiler meat (48.6%) and broilers (50.7%). However, the last value was much higher in Poland (71.6% of resistant strains). This can be the result of a broad use of tetracyclines in veterinary medicine at the level of 42.9 mg/PCU (EMA, 2017).
Erythromycin is the drug of choice for the treatment of C. jejuni infections and resistance levels observed in all isolates regardless the origin were very low (0.8% resistant strains in total). The percentage of macrolide-resistant isolates recovered previously from poultry chain was usually also low but higher than obtained in the present investigation, and ranged from 2.2 to 26.0% (Han et al., 2016; Lapierre et al., 2016; Mäesaar et al., 2016; Ladely et al., 2017; Raeisi et al., 2017). Analyzes of such isolates in Poland demonstrated that erythromycin resistant levels among C. jejuni were rather low, from 0% (Wieczorek et al., 2013b; Wieczorek and Osek, 2015; Wozniak-Biel et al., 2018), 2.4% (Wieczorek et al., 2015), 3.0% (Andrzejewska et al., 2015) to 3.3% (Szczepanska et al., 2017). At the EU level, the percentage of such resistant strains in 2016 was 2.2% for broiler meat and 1.3% (0% in Poland) for broilers [EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2018].
In the present study only one isolate (0.6%) of human origin displayed resistance to erythromycin which was a lower rate than identified in similar strains by other authors: 1.5% (Lapierre et al., 2016; Cha et al., 2017), 3.9% (Riley et al., 2015), 4.8% (Kim et al., 2016) and 8.6% (Ghunaim et al., 2015). Recent EFSA/ECDC report demonstrated that mean European level of C. jejuni resistance to erythromycin was 2.1% out of 21,993 isolates tested (no data from Poland) [EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2018]. There are not many results concerning resistance to macrolides of C. jejuni from human patients in Poland; however, the previous investigations demonstrated that either all isolates tested were sensitive to azithromycin and/or erythromycin (Wardak et al., 2007; Szczepanska et al., 2017) or only a low percentage displayed resistance to erythromycin, i.e., 0.4 and 1.7% strains (Rozynek et al., 2008, 2009).
Analysis of relationship between the presence of putative virulence genes and antimicrobial resistance of C. jejuni isolates did not show clear correlations. The percentages of strains with pathogenic factor that were either resistant or sensitive to antimicrobials tested were similar, although some statistical differences were identified, especially among isolates resistant to streptomycin with and without the cdt and wlaN genes responsible for cytotoxin production. Furthermore, single strains negative for the flhA, cadF, and racR markers were sensitive for streptomycin but the number of such isolates were very low to draw any conclusions. We have demonstrated that C. jejuni with the virulence markers tested were mostly resistant to ciprofloxacin, which is used for treatment of humans with campylobacteriosis or in patients with presumed Campylobacter infections not confirmed by laboratory analyzes. Positive and negative associations between virulence genes and antimicrobial resistance have been previously identified in other bacterial pathogens (McGowan-Spicer et al., 2008; Adib et al., 2014). It seems that the presence of antimicrobial resistance and potential virulence factors are both important for development of the disease. Therefore, further investigation on the interactions between virulence markers and antimicrobial resistance as well as on molecular relationship of positive and negative isolates are needed to better known the nature of Campylobacter pathogenesis. We have made a preliminary study concerning genetic similarity of antimicrobial sensitive and resistant C. jejuni using the multilocus sequence typing method (MLST) as described previously (Wieczorek et al., 2017). The results, based on sequence types (STs), demonstrated a high diversity of the isolates in both groups. Strains resistant to ciprofloxacin (n = 570) were mainly associated with ST464 (58; 10.2% isolates), ST257 (53; 9.3%), and ST6461 (37; 6.5%) whereas C. jejuni sensitive to this antimicrobial (n = 47) were classified to other genotypes, e.g., ST583 (6; 12.8% strains), ST122 (4; 8.5%) and ST51 (4; 8.5%). Different STs were also found among tetracycline resistant (ST464; ST257; ST6461) and sensitive isolates (ST50; ST137; ST2036) as well as streptomycin resistant (ST6411; ST6461; ST5397) and sensitive (ST464; ST257; ST50) bacteria tested. A correlation between resistance to tetracycline and quinolones and MLST sequence type 464 among C. jejuni isolated from poultry meat was also identified in France (Guyard-Nicodème et al., 2015). Furthermore, it was previously shown that ST464 is more generally associated with quinolone resistance (Wirz et al., 2010). Further broader analyzes are needed for molecular comparison and assessment of association between sequence types, antimicrobial resistance and presence of virulence marker genes among C. jejuni isolated from poultry food chain and humans with diarrhea.
An important step in the prevention and control of campylobacteriosis in humans is identification and characterization of C. jejuni that pose the greatest risk to human health, i.e., the isolates which have virulence traits and are resistant to antimicrobials used in treatment of the infection. The goal of this study was to assess the prevalence of markers in C. jejuni associated with pathogenesis of the disease and to identify such virulence genes among isolates recovered along poultry food chain. It was shown that strains with crucial pathogenic factors responsible for C. jejuni motility (flaA, flhA), adherence and colonization (docA, racR), toxin production (cdt), invasiveness (ciaB), and stress response (sodB) were highly conserved among isolates of different origin. In contrast, the virB11, wlaN, and iam were relatively rare and therefore, their role in the pathogenesis of the disease should be further evaluated. It was also found that the majority of C. jejuni tested was resistant to ciprofloxacin, nalidixic acid, and tetracycline but they were mostly sensitive to erythromycin and streptomycin. Isolates resistant to quinolones were mostly classified to ST464 subtype as tested by MLST. We have also provided a broad data on the correlation between the presence of key virulence factors and identified interactions between these genes and antimicrobial resistance, especially to macrolides and quinolones. The results of this study show a high prevalence of several pathogenic markers, but it is difficult to predict how virulent or less virulent a particular C. jejuni isolate may be in vivo during human infection. Therefore, further studies must be performed on the presence or absence of putative pathogenic factors, antimicrobial resistance and molecular relationship among C. jejuni food and clinical isolates to provide more information on the pathogenesis of Campylobacter infection. Although the exact nature and effects of these two markers for pathogenicity of C. jejuni are not yet clear, the results of the present investigations provide a basis for future research important for a public health risk.
KW and JO conceived the study and contributed material from the poultry chain; TW provided the human C. jejuni isolates; KW and JO planned the study; KW and TW performed the experiments; KW and JO analyzed the data and drafted the paper; all authors critically read and approved the final version.
This study was financially supported by National Science Centre, Poland, on the basis of Decision UMO-2014/15/B/NZ7/00874.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Edyta Denis and Katarzyna Półtorak for their technical assistance in specimen collection and processing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01508/full#supplementary-material
Abdollahpour, N., Zendehbad, B., Alipour, A., and Khayatzadeh, J. (2015). Wild-bird feces as a source of Campylobacter jejuni infection in children's playgrounds in Iran. Food Control 50, 378–381. doi: 10.1016/j.foodcont.2014.09.007
Adib, N., Ghanbarpour, R., Solatzadeh, H., and Alizade, H. (2014). Antibiotic resistance profile and virulence genes of uropathogenic Escherichia coli isolates in relation to phylogeny. Trop. Biomed. 31, 17–25.
Allos, B. M. (2001). Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32, 1201–1206. doi: 10.1086/319760
Andrzejewska, M., Szczepanska, B., Spica, D., and Klawe, J. J. (2015). Trends in the occurrence and characteristics of Campylobacter jejuni and Campylobacter coli isolates from poultry meat in Northern Poland. Food Control 51, 190–194. doi: 10.1016/j.foodcont.2014.11.014
Bacon, D. J., Alm, R. A., Burr, D. H., Hu, L., Kopecko, D. J., Ewing, C. P., et al. (2000). Involvement of a plasmid in virulence of Campylobacter 81-176. Infect. Immun. 68, 4384–4390. doi: 10.1128/IAI.68.8.4384-4390.2000
Bang, D. D., Borck, B., Nielsen, E. M., Scheutz, F., Pedersen, K., and Madsen, M. (2004). Detection of seven virulence and toxin genes of Campylobacter jejuni isolates from Danish turkeys by PCR and cytolethal distending toxin production of the isolates. J. Food Prot. 67, 2171–2177. doi: 10.4315/0362-028X-67.10.2171
Bolton, D. J. (2015). Campylobacter virulence and survival factors. Food Microbiol. 48, 99–108. doi: 10.1016/j.fm.2014.11.017
Carvalho, A. C. T., Ruiz-Palacios, G. M., Ramos-Cervantes, P., Cervantes, L. E., Jiang, X., and Pickering, L. K. (2001). Molecular characterization of invasive and noninvasive Campylobacter jejuni and Campylobacter coli isolates. J. Clin. Microbiol. 39, 1353–1359. doi: 10.1128/JCM.39.4.1353-1359.2001
Cha, W., Mosci, R. E., Wengert, S. L., Venegas Vargas, C., Rust, S. R., Bartlett, P. C., et al. (2017). Comparing the genetic diversity and antimicrobial resistance profiles of Campylobacter jejuni recovered from cattle and humans. Front. Microbiol. 8:818. doi: 10.3389/fmicb.2017.00818
Dasti, J. I., Tareen, A. M., Lugert, R., Zautner, A. E., and Gross, U. (2010). Campylobacter jejuni: a brief overview on pathogenicity associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. 300, 205–211. doi: 10.1016/j.ijmm.2009.07.002
Datta, S., Niwa, H., and Itoh, K. (2003). Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 52, 345–348. doi: 10.1099/jmm.0.05056-0
EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control) (2017). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 15:5077. doi: 10.2903/j.efsa.2017.5077
EMA (2017). European Medicines Agency, European Surveillance of Veterinary Antimicrobial Consumption. Sales of Veterinary Antimicrobial Agents in 30 European countries in 2015 (EMA/184855/2017).
EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control) (2018). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 16:5182. doi: 10.2903/j.efsa.2018.5182
Ge, B., Wang, F., Sjolund-Karlsson, M., and McDermott, P. F. (2013). Antimicrobial resistance in Campylobacter: susceptibility testing methods and resistance trends. J. Microbiol. Methods 95, 57–67. doi: 10.1016/j.mimet.2013.06.021
Ghunaim, H., Behnke, J. M., Aigha, I., Sharma, A., Doiphode, S. H., Deshmukh, A., et al. (2015). Analysis of resistance to antimicrobials and presence of virulence/stress response genes in Campylobacter isolates from patients with severe diarrhoea. PLoS ONE 10:e0119268. doi: 10.1371/journal.pone.0119268
Griggs, D. J., Johnson, M. M., Frost, J. A., Humphrey, T., Jørgensen, F., and Piddock, L. J. V. (2005). Incidence and mechanism of ciprofloxacin resistance in Campylobacter spp. isolated from commercial chicken flocks in the United Kingdom before, during, and after fluoroquinolone treatment. Antimicrob. Agents Chemother. 49, 699–707. doi: 10.1128/AAC.49.2.699-707.2005
Guyard-Nicodème, M., Rivoal, K., Houard, E., Rose, V., Quesne, S., Mourand, G., et al. (2015). Prevalence and characterization of Campylobacter jejuni from chicken meat sold in French retail outlets. Int. J. Food Microbiol. 203, 8–14. doi: 10.1016/j.ijfoodmicro.2015.02.013
Han, X., Zhu, D., Lai, H., Zeng, H., Zhou, K., Zou, L., et al. (2016). Prevalence, antimicrobial resistance profiling and genetic diversity of Campylobacter jejuni and Campylobacter coli isolated from broilers at slaughter in China. Food Control 69, 160–170. doi: 10.1016/j.foodcont.2016.04.051
Hanning, I., Biswas, D., Herrera, P., Roesler, M., and Ricke, S. C. (2010). Prevalence and characterization of Campylobacter jejuni isolated from pasture flock poultry. J. Food Sci. 75, 496–502. doi: 10.1111/j.1750-3841.2010.01747.x
Hickey, T. E., McVeigh, A. L., Scott, D. A., Michielutti, R. E., Bixby, A., Carroll, S. A., et al. (2000). Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect. Immun. 68, 6535–6541. doi: 10.1128/IAI.68.12.6535-6541.2000
Humphrey, T., O'Brien, S., and Madsen, M. (2007). Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117, 237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006
Iovine, N. M. (2013). Resistance mechanisms in Campylobacter jejuni. Virulence 4, 230–240. doi: 10.4161/viru.23753
Kaakoush, N. O., Castaño-Rodríguez, N., Mitchell, H. M., and Man, S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28, 687–719. doi: 10.1128/CMR.00006-15
Kim, J. S., Lee, M. Y., Kim, S. J., Jeon, S.-E., Cha, I., Hong, S., et al. (2016). High-level ciprofloxacin-resistant Campylobacter jejuni isolates circulating in humans and animals in Incheon, Republic of Korea. Zoonozes Public Health 63, 545–554. doi: 10.1111/zph.12262
Konkel, M. E., Garvis, S. G., Tipton, S. L., Anderson, D. E. Jr., and Cieplak, W. Jr. (1997). Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol. Microbiol. 24, 953–963. doi: 10.1046/j.1365-2958.1997.4031771.x
Koolman, L., Whyte, P., Burgess, C., and Bolton, D. (2015). Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 12, 424–432. doi: 10.1089/fpd.2014.1883
Ladely, S. R., Berrang, M. E., Meinersmann, R. J., and Cox, N. A. (2017). Campylobacter multi-locus sequence types and antimicrobial susceptibility of broiler cecal isolates: a two year study of 143 commercial flocks. J. Food Saf. 37:e12366. doi: 10.1111/jfs.12366
Lapierre, L., Gatica, M. A., Riquelme, V., Vergara, C., Yañez, J. M., San Martin, B., et al. (2016). Characterization of antimicrobial susceptibility and its association with virulence genes related to adherence, invasion, and cytotoxicity in Campylobacter jejuni and Campylobacter coli isolates from animals, meat, and humans. Microbial Drug Resist. 22, 432–444. doi: 10.1089/mdr.2015.0055
Mäesaar, M., Kramarenko, T., Meremäe, K., Sögel, J., Lillenberg, M., Häkkinen, L., et al. (2016). Antimicrobial resistance profiles of Campylobacter spp. isolated from broiler chicken meat of Estonian, Latvian and Lithuanian origin at Estonian retail level and from patients with severe enteric infections in Estonia. Zoonozes Public Health 63, 89–96. doi: 10.1111/zph.12208
Ma, L., Wang, Y., Shen, J., Zhang, Q., and Wu, C. (2014). Tracking Campylobacter contamination along a broiler chicken production chain from the farm level to retail in China. Int. J. Food Microbiol. 181, 77–84. doi: 10.1016/j.ijfoodmicro.2014.04.023
McGowan-Spicer, L. L., Fedorka-Cray, P. J., Frye, J. G., Meinersmann, R. J., Barrett, J. B., and Jackson, C. R. (2008). Antimicrobial resistance and virulence of Enterococcus faecalis isolated from retail food. J. Food Protoc. 71, 760–769. doi: 10.4315/0362-028X-71.4.760
Melero, B., Juntunen, P., Hänninen, M.-L., Jaime, I., and Rovira, J. (2012). Tracing Campylobacter jejuni strains along the poultry meat production chain from farm to retail by pulsed-field gel electrophoresis, and the antimicrobial resistance of isolates. Food Microbiol. 32, 124–128. doi: 10.1016/j.fm.2012.04.020
Müller, J., Schulze, F., Müller, W., and Hänel, I. (2006). PCR detection of virulence-associated genes in Campylobacter jejuni strains with differential ability to invade Caco-2 cells and to colonize the chick gut. Vet. Microbiol. 113, 123–129. doi: 10.1016/j.vetmic.2005.10.029
Olkkola, S., Nykäsenoja, S., Raulo, S., Llarena, A.-K., Kovanen, S., Kivistö, R., et al. (2016). Antimicrobial resistance and multilocus sequence types of Finnish Campylobacter jejuni isolates from multiple sources. Zoonoses Public Health 63, 10–19. doi: 10.1111/zph.12198
Osterlund, A., Hermann, M., and Kahlmeter, G. (2003). Antibiotic resistance among Campylobacter jejuni/coli strains acquired in Sweden and abroad: a longitudinal study. Scand. J. Infect. Dis. 35, 478–481. doi: 10.1080/00365540310010949
Park, S. F. (2002). The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int. J. Food Microbiol. 74, 177–188. doi: 10.1016/S0168-1605(01)00678-X
Pesci, E. C., Cottle, D. E., and Pickett, C. L. (1994). Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62, 2687–2694.
Piccirillo, A., Dotto, G., Salata, C., and Giacomelli, M. (2013). Absence of class 1 and class 2 integrons among Campylobacter jejuni and Campylobacter coli isolated from poultry in Italy. J. Antimicrob. Chemother. 68, 2683–2685. doi: 10.1093/jac/dkt242
Post, A., Martiny, D., van Waterschoot, N., Hallin, M., Maniewski, U., Bottieau, E., et al. (2017). Antibiotic susceptibility profiles among Campylobacter isolates obtained from international travelers between 2007 and 2014. Eur. J. Clin. Microbiol. Infect. Dis. 36, 2101–2107. doi: 10.1007/s10096-017-3032-6
Raeisi, M., Khoshbakht, R., Ghaemi, E. A., Bayani, M., Hashemi, M., Seyedghasemi, N. S., et al. (2017). Antimicrobial resistance and virulence-associated genes of Campylobacter spp. isolated from raw milk, fish, poultry, and red meat. Microb. Drug Resist. 23, 925–933. doi: 10.1089/mdr.2016.0183
Riley, A., Eshaghi, A., Olsha, R., Allen, V. G., and Patel, S. N. (2015). Antibiotic susceptibility of clinical isolates of Campylobacter jejuni and Campylobacter coli in Ontario, Canada during 2011-2013. Diagn. Microbiol. Infect. Dis. 83, 292–294. doi: 10.1016/j.diagmicrobio.2015.07.020
Rozynek, E., Dzierzanowska-Fangrat, K., Korsak, D., Konieczny, P., Wardak, S., Szych, J., et al. (2008). Comparison of antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from humans and chicken carcasses in Poland. J. Food Prot. 71, 602–607. doi: 10.4315/0362-028X-71.3.602
Rozynek, E., Dzierzanowska-Fangrat, K., Szczepanska, B., Wardak, S., Szych, J., Konieczny, P., et al. (2009). Trends in antimicrobial susceptibility of Campylobacter isolates in Poland (2000-2007). Pol. J. Microbiol. 58, 111–115.
Sifr,é, E., Salha, B. A., Ducournaua, A., Floch, P., Chardon, H., Mégraud, F., et al. (2015). EUCAST recommendations for antimicrobial susceptibility testing applied to the three main Campylobacter species isolated in humans. J. Microbiol. Methods 119, 206–213. doi: 10.1016/j.mimet.2015.10.018
Sonnevend, A., Rotimi, V. O., Kolodziejek, J., Usmani, A., Nowotny, N., and Pál, T. (2006). High level of ciprofloxacin resistance and its molecular background among Campylobacter jejuni strains isolated in the United Arab Emirates. J. Med. Microbiol. 55, 1533–1538. doi: 10.1099/jmm.0.46744-0
Szczepanska, B., Andrzejewska, M., Spica, D., and Klawe, J. J. (2017). Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. 17:80. doi: 10.1186/s12866-017-0991-9
Talukder, K., Aslam, M., Islam, Z., Azmin, I., and Duttad, D. (2008). Prevalence of virulence genes and cytolethal distending toxin production in Campylobacter jejuni isolates from diarrheal patients in Bangladesh. J. Clin. Microbiol. 46, 1485–1488. doi: 10.1128/JCM.01912-07
Thakur, S., Zhao, S., McDermontt, P., Harbottle, H., Abbott, J., English, L., et al. (2010). Antimicrobial resistance, virulence, and genotypic profile comparison of Campylobacter jejuni and Campylobacter coli isolated from humans and retail meats. Foodborne Pathog. Dis. 7, 835–844. doi: 10.1089/fpd.2009.0487
Tresse, O., Alvarez-Ordóñez, A., and Connerton, I. F. (2017). Editorial: about the foodborne pathogen Campylobacter. Front. Microbiol. 8:1908. doi: 10.3389/fmicb.2017.01908
Unicomb, L. E., Ferguson Stafford, R. J., Ashbolt, R., Kirk, M. D., Becker, N. G., Patel, M. S., et al. (2006). Low-level fluoroquinolone resistance among Campylobacter jejuni isolates in Australia. Clin. Infect. Dis. 42, 1368–1374. doi: 10.1086/503426
Vandamme, P., Van Doorn, L. J., al Rashid, S. T., Quint, W. G. V., van der Plas, J., Chan, V. L., et al. (1997). Campylobacter hyoilei Alderton et al. 1995 and Campylobacter coli Wron and Chatelain 1973 are subjective synonyms. Int. J. Syst. Bacteriol. 47, 1055–1060. doi: 10.1099/00207713-47-4-1055
van Gerwe, T., Bouma, A., Wagenaar, J. A., Jacobs-Reitsma, W. F., and Stegeman, A. (2010). Comparison of Campylobacter levels in crops and ceca of broilers at slaughter. Avian Dis. 54, 1072–1074. doi: 10.1637/9113-101809-ResNote.1
Wardak, S., and Szych, J. (2010). Tigecicline susceptibilities of tetracycline-resistant Campylobacter jejuni clinical strains isolated in Poland. Med. Dośw. Mikrobiol. 62, 345–350.
Wardak, S., Szych, J., Zasada, A. A., and Gierczynski, R. (2007). Antibiotic resistance of Campylobacter jejuni and Campylobacter coli clinical isolates from Poland. Antimicrob. Agents Chemother. 51, 1123–1125. doi: 10.1128/AAC.01187-06
Wieczorek, K., Denis, E., Lachtara, B., and Osek, J. (2017). Distribution of Campylobacter jejuni multilocus sequence types isolated from chickens in Poland. Poult. Sci. 96, 703–709. doi: 10.3382/ps/pew343
Wieczorek, K., Denis, E., Lynch, O., and Osek, J. (2013a). Molecular characterization and antibiotic resistance profiling of Campylobacter isolated from cattle in polish slaughterhouses. Food Microbiol. 34, 130–136. doi: 10.1016/j.fm.2012.12.003
Wieczorek, K., Denis, E., and Osek, J. (2015). Comparative analysis of antimicrobial resistance and genetic diversity of Campylobacter from broilers slaughtered in Poland. Int. J. Food Microbiol. 210, 24–32. doi: 10.1016/j.ijfoodmicro.2015.06.006
Wieczorek, K., Kania, I., and Osek, J. (2013b). Prevalence and antimicrobial resistance of Campylobacter spp. isolated from poultry carcasses in Poland. J. Food Prot. 76, 1451–1455. doi: 10.4315/0362-028X.JFP-13-035
Wieczorek, K., and Osek, J. (2015). A five-year study on prevalence and antimicrobial resistance of Campylobacter from poultry carcasses in Poland. Food Microbiol. 49, 161–165. doi: 10.1016/j.fm.2015.02.006
Wirz, S. E., Overesch, G., Kuhnert, P., and Korczak, B. M. (2010). Genotype and antibiotic resistance analyses of Campylobacter isolates from ceca and carcasses of slaughtered broiler flocks. Appl. Environ. Microbiol. 76, 6377–6386. doi: 10.1128/AEM.00813-10
Wozniak-Biel, A., Bugla-Płoskonska, G., Kielsznia, A., Korzekwa, A., Tobiasz, A., Korzeniowska-Kowal, A., et al. (2018). High prevalence of resistance to fluoroquinolones and tetracycline Campylobacter spp. isolated from poultry in Poland. Microb. Drug Resist. 24, 314–322. doi: 10.1089/mdr.2016.0249
Zhou, J., Zhang, M., Yang, W., Fang, Y., Wang, G., and Hou, F. (2016). A seventeen-year observation of the antimicrobial susceptibility of clinical Campylobacter jejuni and the molecular mechanisms of erythromycin-resistant isolates in Beijing, China. Int. J. Infect. Dis. 42, 28–33. doi: 10.1016/j.ijid.2015.11.005
Keywords: Campylobacter jejuni, poultry food chain, humans, virulence genes, antimicrobial resistance, zoonotic pathogen
Citation: Wieczorek K, Wołkowicz T and Osek J (2018) Antimicrobial Resistance and Virulence-Associated Traits of Campylobacter jejuni Isolated From Poultry Food Chain and Humans With Diarrhea. Front. Microbiol. 9:1508. doi: 10.3389/fmicb.2018.01508
Received: 26 January 2018; Accepted: 18 June 2018;
Published: 04 July 2018.
Edited by:
David Rodriguez-Lazaro, University of Burgos, SpainReviewed by:
Beatrix Stessl, Veterinärmedizinische Universität Wien, AustriaCopyright © 2018 Wieczorek, Wołkowicz and Osek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacek Osek, am9zZWtAcGl3ZXQucHVsYXd5LnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.